Abstract
Abstract
A most discussed topic of the new decade, COVID-19 is an infectious disease caused by the recently discovered SARS-CoV-2. With an exceedingly high transmission rate, COVID-19 has affected almost all the countries in the world. Absent any vaccine or specific treatment, the humanity is left with nothing but the legacy method of quarantine. However, quarantine can only be effective when combined with early diagnosis of suspected cases. With their high sensitivity and unmatched specificity, biosensors have become an area of interest for development of novel diagnostic methods. Compared to the more traditional diagnostics, nanobiotechnology introduces biosensors as different diagnostics with greater versatility in application. Today, a growing number of analytes are being accurately identified by these nanoscopic sensing machines. Several reports of validated application with real samples further strengthen this idea. As of recent, there has been a rise in the number of studies on portable biosensors. Despite the slow progression, certain devices with embedded biosensors have managed to be of diagnostic value in several countries. The perceptible increase in development of mobile platforms has revolutionized the healthcare delivery system in the new millennium. The present article reviews the most recent advancements in development of diagnostic nanobiosensors and their application in the clinical fields.
Key points
• There is no specific treatment for highly transmissible SARS-CoV-2.
• Early diagnosis is critical for control of pandemic.
• Highly sensitive/specific nanobiosensors are emerging assets against COVID-19.
Keywords: COVID-19, SARS-CoV-2, 2019-nCoV, Nanobiotechnology, Biosensor, Early diagnosis, Pandemic
Introduction
A global concern in the new decade, COVID-19 is a viral disease associated with severe acute respiratory syndrome (Lai et al. 2020; Sheervalilou et al. 2020). In December 2019, a cluster of peculiar cases with pneumonia was reported in Wuhan, China (WHO 2020c). In the beginning, the causative virus was called 2019-nCoV. However, it was officially recognized as “SARS-CoV-2” due to its great similarity to SARS-CoV, the pathogen behind the original SARS outbreak (Cascella et al. 2020). With an astonishingly rapid person-to-person transmission, COVID-19 was declared to be a pandemic on March 12, 2020 (WHO 2020a). As of February 9, 2021, nearly 106,321,987 confirmed cases of COVID-19 have been reported globally, resulting in 2,325,282 deaths (https://covid19.who.int/). Over the last two decades, coronaviruses (CoVs) have been involved in several important outbreaks of respiratory diseases. The severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) epidemics are two such incidents (Cascella et al. 2020) that were instigated by SARS-CoV and MERS-CoV, respectively (de Wit et al. 2016). COVID-19 is the third large-scale epidemic, and the first pandemic to have been caused by a coronavirus (Ksiazek et al. 2003). With a total of 10,000 cases, the previous two outbreaks concluded with a mortality rate of 10% for SARS and a strikingly high 37% for MERS. However, in the case of COVID-19, an overwhelmingly high number of patients have been reported, which is nearly 90 times greater than the cumulative cases of SARS and MERS (Mahase 2020). With due attention to the expeditious transmission of COVID-19, there is no doubt that secondary prevention measures by means of early diagnosis can immensely contribute to the management of the situation (Qiu et al. 2020).
Being zoonotic viruses, CoVs are major culprits for mild to moderate upper respiratory tract (URT) infection in both humans and animals (Li et al. 2020a). According to its severity, the disease is classified into three categories: mild (81%), severe (14%), and critical (5%). Mild CoV disease is commonly manifested as mild pneumonia. Severe disease, on the other hand, is characterized by shortness of breath with a respiratory rate of 30 or more, which often results in oxygen saturations below 93%. Few patients deteriorate and develop critical conditions associated with life-threatening systemic complications such as acute respiratory distress syndrome, arrhythmias, and shock and eventually, die of multiple organ failure (Wang et al. 2020; Wu and McGoogan 2020).
A lack of specific manifestations in the early phase of COVID-19 has made its diagnosis an arduous challenge for clinicians (Huang et al. 2020). Despite their high accuracy, genome sequencing methods have not received favorable regards in terms of rapid diagnosis (Wu et al. 2020). As of today, reverse transcription-polymerase chain reaction (RT-PCR) is the widely accepted technique for diagnosis of COVID-19. Nevertheless, erroneous results like false-positive and false-negative reports have made this method a fairly questionable tool for early diagnosis of COVID-19 (Qiu et al. 2020). Apart from imperfect results, the technique is neither cost-effective nor time-friendly, as it requires skilled personnel and takes several hours to generate results (Chu et al. 2020).
Presently, COVID-19 is propagating around the world with an unprecedentedly high transmission rate for a disease caused by a coronavirus. Furthermore, cases of COVID-19 transmission from asymptomatic patients have also been reported (Bai et al. 2020). It is estimated that each patient infected with SARS-CoV-2 can transmit the pathogen to approximately two individuals (Li et al. 2020b). Since there are no vaccines or specific medicines for prevention and treatment of COVID-19, successful containment of this sensitive situation can only be achieved with early detection of the infection in asymptomatic patients (Zhu et al. 2020).
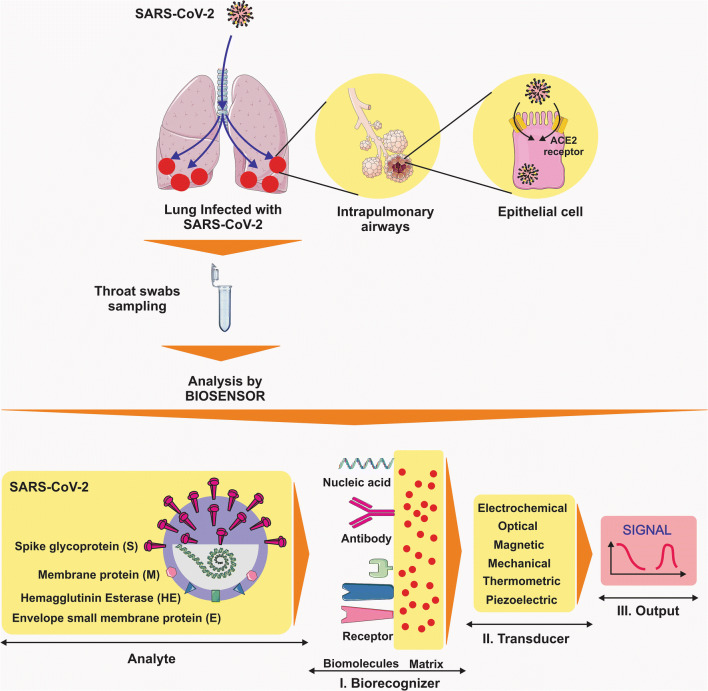
With their high sensitivity, immunological diagnostic tests are valuable assets in precise detection of viral antigens, providing a seamless pathway for rapid diagnosis of COVID-19 (Seo et al. 2020). Narrowing down the scope, the lesser-known “nanobiosensors” have also been clinically tested amid major viral outbreaks such as influenza (Saylan and Denizli 2020). A growing body of evidence is now suggesting biosensors as a reliable alternative solution to the time-consuming process of diagnosis (Soler et al. 2019) (Fig. 1). The present review paper seeks to convey a brief yet illustrative account of biosensors and their potential application in the detection of SARS-CoV-2 infection.
Fig. 1.
The COVID-19 diagnosis by biosensors. (Abbreviations: SARS-CoV-2, severe respiratory syndrome coronavirus 2)
SARS-CoV-2 biology
Commonly isolated from different animal species, CoVs belong to a family of positive-sense betacoronavirus with a single-stranded RNA at their core (Perlman and Netland 2009). The densely packed genetic material is responsible for the expression of four major proteins in betacoronavirus (β-CoV), especially SARS-CoV-2. These structural proteins are known as spike (S), small envelope (E), matrix (M), and nucleocapsid (N) (de Wit et al. 2016). Based on recent investigations, SARS-CoV-2 preferably targets angiotensin-converting enzyme II (ACE2) as a receptor for entry to the cell. ACE2 is also a receptor of interest for the much older SARS-CoV (Li et al. 2003). SARS-CoV-2 is internalized into its host cell once a high-affinity binding is formed between the viral spike protein and ACE2 (Wrapp et al. 2020). In this regard, SARS-CoV-2 shares greater similarity with SARS-CoV than MERS-CoV (Lu et al. 2020).
For unexplained reasons, CoVs can infiltrate from one species to another, causing illness in humans that might range from a benign common cold to severe malevolent disease. It is speculated that CoVs might have originated from bat species and then moved to other mammalian hosts before afflicting humans, i.e., the Himalayan palm civet in the case of SARS-CoV. Unknown newly emerging strain with dynamics yet to be elucidated, SARS-CoV-2 is also suspected of having originated in animals (Cascella et al. 2020).
The prominence of early diagnosis
Following identification of the first cases, unprotected exposure to certain animal species was assumed to be the primary route of transmission. The reason was that the disease was strongly linked to a particular seafood market in China, as the majority of patients had a history of visiting the place. However, after a monumental burst in the number of cases, it was clarified that the major mechanism at large was human-to-human transmission. Later, it was revealed that patients with no symptoms at all could also transmit the infection to other people. Transmission of COVID-19 is mediated through tiny droplets expelled into the air by coughing and sneezing. According to analyses on the propagation pattern of SARS-CoV-2 in China, close contact between individuals is suggested to be a prerequisite for transmission of the virus. Hence, the majority of patients either come from the same community or work in the public healthcare system (Cascella et al. 2020).
The incubation period of COVID-19 can span from 3 to 7 days. Occasionally, this period can last as long as 2 weeks (Li et al. 2020b). The doubling time, through which the number of patients doubles, is estimated to be 7 days for COVID-19. This is in contrast to the much shorter 3 days reported for the SARS-CoV epidemic. On average, each COVID-19 patient is capable of spreading the virus to two other individuals (Bauch et al. 2005). These data unanimously indicate that absent any treatment, the only promissory strategy for slowing down the rate of transmission is most probably early diagnosis of the infection.
Routine molecular diagnostics
Being a top priority, accurate laboratory diagnosis is of pre-eminent value in prevention and control of potential outbreaks (Qiu et al. 2020). In their latest interim guidance laboratory testing for the 2019 novel coronavirus disease, the WHO asserts that several molecular assays have been developed for this cause (Organization 2020). Among various diagnostics, the more traditionally practiced modalities include hemagglutination assay (HA) inhibition, complement fixation, immunofluorescence, viral culture, and PCR (Saeed and Wu 2020).
Genome sequencing
Genome sequencing is regarded as a universal language since many countries employ target genes that, if not identical, are highly similar to each other. RNA-dependent RNA polymerase (RdRp) and open reading frame 1ab (ORF1ab) stand among the most used sequences around the world (Qiu et al. 2020). In spite of its high accuracy, genome sequencing is not a readily applicable method for rapid detection of COVID-19. This is primarily due to the time-consuming process of sequencing and the expensive equipment demanded by this prestigious technique (Wu et al. 2020).
Reverse transcriptase polymerase chain reaction
RT-PCR is a reliable method for detection of emerging pathogens. To this date, real-time RT-PCR has successfully been applied for identification of SARS-CoV-2 in specimens of respiratory secretions from suspected cases (WHO 2020b). With its unrivaled sensitivity, RT-PCR is the method of choice for uncovering viral RNA through the instantaneous generation of multiple copies from a specific template sequence. Several studies on RT-PCR amid the recent epidemic have reported an astonishing sensitivity of 3.7 RNA copies for accurate detection of SARS-CoV-2 (Corman et al. 2020).
A mostly triumphant diagnostic method, RT-PCR, can fail on various occasions when confounding factors are present, e.g., spurious nucleic acid sequences (Qiu et al. 2020). Else, a single RT-PCR test takes a minimum of 3 h to be completed, including the time allocated for preparation of viral RNA, which is an important step due to its direct effect on the accuracy of the test (Seo et al. 2020). Time and time again, the method also struggles to correctly report an ongoing infection in asymptomatic patients (Zhang et al. 2020). There are accounts of certain false-negative reports by RT-PCR for cases that had already been confirmed to have SARS-CoV-2 infection (Xie et al. 2020). Accordingly, in clinical experience, a single negative RT-PCR result is not considered sufficient evidence to exclude the possibility of COVID-19, since the positive rate for this test can be as low as 30% (Zhang and Zhao 2020), especially if the sample is taken from the upper respiratory tract (Qiu et al. 2020). Regrettably, the COVID-19 outbreak is most obstreperous in regions where allocation of enough resources for PCR testing is not plausible (Chu et al. 2020).
Based on past clinical experience with swine flu (Moscona 2005), late diagnosis of COVID-19 might result in high mortality rates as well (Sakai-Tagawa et al. 2010). On that account, it would be a sensible strategy to screen suspected cases by another equally sensitive yet superiorly reliable diagnostic method (Qiu et al. 2020).
New approaches: nanobiotechnology/biosensors
Error-free detection of pathogens is perhaps the cornerstone of point-of-care testing (Saylan and Denizli 2020). With their distinctive characteristics, biosensors have recently become a sought-after area of interest, due mostly to the unrivaled sensitivity and specificity they provide in real-time (Liu and Guo 2012). As of today, an expansive array of different substances has been appraised for designing biosensors; however, gold-, magnetic-, and carbon-based nanomaterials appear to be the most promising options in developing clinically effective nanobiosensors (Akbarzadeh et al. 2006; Norouzi et al. 2019; Saeed and Wu 2020; Shakeri-Zadeh et al. 2020).
Every functional biosensor consists of recognition elements and transducers that identify the target and translate potential responses to recognizable signals, respectively. Biosensors are commonly classified into five categories with distinctive chemical properties: electrochemical, magnetic, thermal, optic, and piezoelectric. This novel family of sensing small contraptions has evolved to be a sophisticated facility in identification of infinitesimal analytes, e.g., peptides, nucleic acids, toxins, and pathogens (Saylan and Denizli 2020).
Virus detection
Infectious diseases are notoriously renowned for their pervasive nature (Saylan and Denizli 2020). They have always been an obstinate source of trepidation due to the paucity of diagnostic tests and treatment modalities (Demirci and Inci 2019). To this date, several promising attempts have been made at application of nanobiosensors in sensitive fields such as diagnosis (Chang et al. 2018) and laboratory medicine (Choi et al. 2018). Through the last few years, new recognition molecules have been examined to enhance their detection capacity and bring new properties to the game (Justino et al. 2013; Saylan et al. 2019). Time and time again, biosensors have been sought for diagnosis of serious maladies, e.g., influenza, hepatitis B, Ebola virus disease, Zika fever, and HIV infection (Saylan and Denizli 2020) (Table 1 (Bai et al. 2012; Diltemiz et al. 2013; Kim et al. 2019; Tam et al. 2009; Vollmer et al. 2008)). New evidence indicates that biosensors can be of clinical value in early detection of SARS-CoV-2.
Table 1.
Recent studies on virus monitoring by biosensors
| Sensor | Nanomaterial | Probe | Virus | Sample | LOD, sensitivity/specificity | Efficiency | Ref |
|---|---|---|---|---|---|---|---|
| Optical resonance | PS, silica microsphere | - | Flu A | PBS filled sample cell | High sensitivity | Label-free, real-time optical detection | Vollmer et al. (2008) |
| DNA sensor | MWCNTs | DNA probe | Flu A | The cell filled with 2 mL of buffer solution | 0.5 nM | Direct and label-free detection, response time of DNA sensor: 4 min, highly sensitive, low-cost, potential application in controlling disease | Tam et al. (2009) |
| SPR aptasensor | Streptavidin-biotin, Au surface | DNA-aptamer | AIV H5N1 | Swab samples | R2 = 0.99, AIV range: 0.128–1.28 HAU, negligible signal (<4% of H5N1): from six non-target AIV subtypes | HAU detection: in 1.5 h, rapid and specific detection | Bai et al. (2012) |
| piezoelectric QCM and SPR sensors | Au, quartz crystal | 4-APBA, SA | Flu A | Virus |
LOD: QCM: 4.7×10−2 μM, (0.26 μg ml−1) SPR: 1.28×10−1 μM, (0.72 μg ml−1) in the 95% CI |
Rapid, sensitive, simpler, and less expensive | Diltemiz et al. (2013) |
| LSPR | AuNPs | dsDNA, upE, and ORF 1a | MERS-CoV, TMV | Virus | 1 pmol/μL of 30 bp MERS-CoV | Label-free, rapid and accurate diagnostic, on-site detection especially in resource-limited settings | Kim et al. (2019) |
Abbreviations: MWCNTs carbon multi-walled nanotubes, Flu A influenza A virus, NPs nanoparticles, PBS phosphate-buffered saline, PS polystyrene particles, AIV H1N1 avian influenza virus H5N1, SPR surface plasmon resonance, R2 linearly related, HAU HA unit, QCM quartz crystal microbalance, HA hemagglutinin, Ab antibody, CI confidence interval, 4-APBA 4-aminophenyl boronic acid, SA sialic acid, Au gold, AuNPs gold nanoparticles, dsDNA double-stranded DNA, upE E protein gene, ORF open reading frames, MERS-CoV Middle East respiratory syndrome, TMV tobacco mosaic virus
SARS-CoV-2 assay strategies
Nucleic acid detection
SARS-CoV-2 is an enveloped, nonsegmented single-stranded RNA virus (Guarner 2020). Similar to RT-PCR, the DNA-RNA hybridization technique is extensively employed in various biosensors. The process is predicted by the melting of nucleic acid strands (Zhang et al. 2018), which is the reason behind the utility of nucleic acids in bionanotechnology, as well as clinical diagnosis (Zhang et al. 2012).
Protein detection
Another appropriately relevant approach is to detect the conspicuous S protein on SARS-CoV-2 surface (Seo et al. 2020; Zhang et al. 2020). In a recent investigation, scientists evaluated the biophysical properties of this protein and reported a remarkable binding affinity to ACE2, which was significantly higher than that of SARS-CoV (Wrapp et al. 2020).
Implication of biosensors in SARS-CoV-2 assay
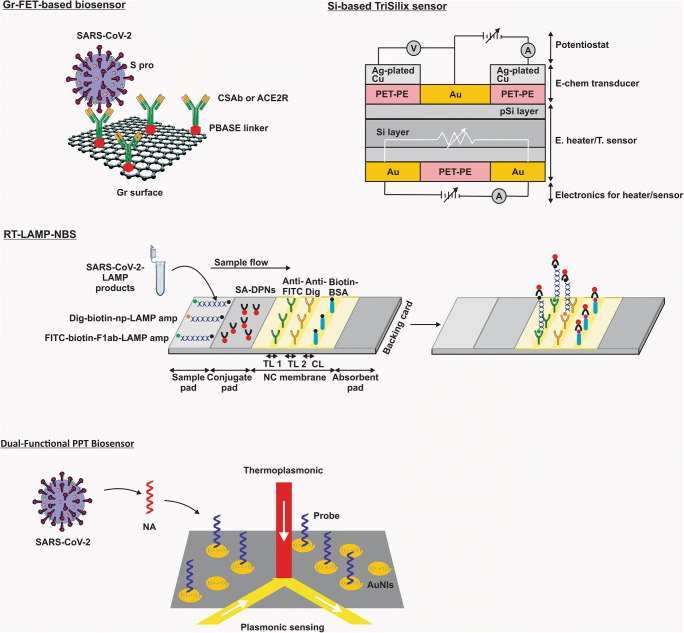
Figure 2 and Table 2 list recent studies on SARS-CoV-2 through biosensors.
Fig. 2.
Biosensors in SARS-CoV-2 detection. (Abbreviations: SARS-CoV-2, severe respiratory syndrome coronavirus 2; FET, field-effect transistor; Gr, graphene; S pro, spike protein; CSAb, SARS-COV spike S1 subunit protein antibody; ACE2R, angiotensin-converting enzyme 2 receptor; PBASE, 1-pyrenebutanoic acid succinimidyl ester; RT-LAMP-NBS, reverse transcription loop–mediated isothermal amplification coupled with nanoparticle-based biosensor assay; TL 1, test line 1; TL 2, test line 2; CL, control line; FITC, fluorescein; Dig, digoxigenin; Biotin-BSA, biotinylated bovine serum albumin; Dig-biotin-np-LAMP amp, Dig-biotin–labeled np-LAMP amplicons; FITC-biotin-F1ab-LAMP amp, FITC-biotin–labeled F1ab-LAMP amplicons; SA-DPNs, dye streptavidin (Crimson Red)–coated polymer nanoparticles; NC membrane, nitrocellulose membrane; pSi, nanoporous silicon; Si, silicon; Ag, Silver; Cu, copper; Au, gold; PET-PE, polyethylene terephthalate-polyethylene; T. sensor, temperature sensor; E. heater, electrical heater; TriSilix, silicon-based integrated point-of-need tri-modal nucleic acid transducer; PPT, plasmonic photothermal; NA, nucleic acid; AuNIs, two-dimensional gold nanoislands)
Table 2.
Recent studies on SARS-CoV-2 diagnosis by biosensors
| Sensor | Nanomaterial | Probe | Subjects | Clinical sample | Primary Ag detection/copy number detection | LOD, sensitivity/specificity | Efficiency | Ref |
|---|---|---|---|---|---|---|---|---|
| FET-based biosensor |
Graphene sheets |
Ab against SARS-CoV-2 S pro | COVID-19 patients, normal subjects | Nasopharyngeal swab | ELISA, real-time RT-PCR |
S pro detection concentration: PBS: 1 fg/ml Clinical transport medium: 100 fg/ml LOD: Culture medium: 1.6 × 101 pfu/ml Clinical samples: 2.42 × 102 copies/ml |
Highly sensitive immunological diagnostic method for COVID-19, sample pretreatment-free Labeling-free Distinguishing the SARS-CoV-2 Ag pro from those of MERS-CoV |
Giwan Seo et al. (2020) |
| RT-LAMP-NBS | Dye streptavidin–coated polymer NPs | LAMP primer sets; F1ab and np | Clinically diagnosed COVID-19 patients, non-COVID-19 patients | Oropharynx swab | - |
Sensitivity: 12 copies/per reaction Specificity: No cross-reactivity |
Sensitivity of SARS-CoV-2 was 100% (33/33), Assay’s specificity was 100% (96/96) The total diagnosis test from sample collection to result interpretation only takes approximately 1 h |
Xiong Zhu et al. (2020) |
| Gr-FET immunosensors | High-quality monolayer single-crystal graphene | CSAb, ACE2 | - | - | - | 0.2-pM concentration | The early screening/diagnosis, fast responding time within 2 min, in a real-time and label-free manner, applied to screen for high-affinity antibodies (CSAb) (with binding constant up to 2×1011 M-1 against the RBD) at concentrations down to 0.1 pM | Xiaoyan Zhang et al. (2020) |
| Silicon-based TriSilix contain electrochemical sensor | Silicon | PCR primers; reverse and forward | - | cDNA fragments produced synthetically | PCR | Detection of 20 fg, equivalent to a single bacterium, at the 30th cycle. Using TriSilix, we also detected the cDNA from SARS-CoV-2 (1 pg), through PCR |
Can chemically amplify and detect pathogen-specific sequences of nucleic acids (NA) quantitatively in real-time Label-free electrochemical sensor |
Estefania Nunez-Bajo et al. (2020) |
|
Dual-functional plasmonic photothermal biosensors |
AuNIs | Nucleic acid hybridization | - |
Actual sample |
- | 0.22 pM | Allows precise detection of the specific target in a multigene mixture, viral disease diagnosis | Guangyu Qiu et al. (2020) |
Abbreviations: COVID-19 coronavirus disease 2019, FET field-effect transistor, Ab antibody, PCR polymerase chain reaction, SARS-CoV-2 sever acute respiratory syndrome coronavirus 2, S pro spike protein, LOD limit of detection, Ag pro antigen protein, MERS-CoV middle east acute respiratory syndrome coronavirus, ELISA enzyme-linked immunosorbent assay, RT-LAMP-NBS reverse transcription loop–mediated isothermal amplification coupled with nanoparticle-based biosensor, NPs nanoparticles, np nucleoprotein, CSAb SARS-COV spike S1 subunit protein antibody, Gr-FET graphene field-effect transistor, Gr graphene, RBD receptor binding domain, ACE2 human angiotensin-converting enzyme 2, TriSilix silicon-based integrated point-of-need tri-modal nucleic acid transducer, AuNIs gold nanoislands
Field-effect transistor–based biosensing devices
In terms of clinical diagnosis, field-effect transistor (FET)–based biosensing devices come with certain benefits, on top of which stands their sharp sensitivity for measuring trace amounts of analytes (Liu et al. 2019). Accordingly, FET-based biosensors could be game-changing assets in point-of-care testing and clinical diagnosis (Seo et al. 2020).
Graphene-based FET biosensors
Graphene is a uniform two-dimensional layer of carbon atoms that are arranged in a hexagonal layout (Cooper et al. 2012). Thanks to its large surface area and high electronic conductivity, this material has been widely used for development of sensing platforms (Geim and Novoselov 2010). Capable of detecting alterations in their surroundings, graphene-based FET (Gr-FET) biosensors excel as a low-noise means of ultrasensitive detection (Zhou et al. 2017). Such unparalleled sensitivity and stability have rendered Gr-FET biosensors a material of choice for immunosensing (Zhang et al. 2020).
Scientists recently devised a special FET-based biosensing device that was able to selectively recognize SARS-CoV-2 S protein in a groundbreaking concentration of 100 fg/ml. The device comprises a FET-containing graphene sheet that was coated with specific antibodies against the S protein on SARS-CoV-2 strains isolated from patients with COVID-19 (Seo et al. 2020). Another investigation reported a successful trial of a unique CoV immunosensor in experimental settings. Similarly, the device was based on Gr-FET biosensors and was capable of detecting SARS-CoV-2 spike protein in concentrations as low as 0.2 pM, a promising achievement that further supports the utility of such biosensor-based devices in clinical diagnosis (Zhang et al. 2020).
Biosensor coupled with loop-mediated isothermal amplification
It is possible to run nucleic acid amplification under isothermal conditions without the application of a precision thermal cycler. Called “loop-mediated isothermal amplification” (LAMP), this fairly new method is extremely efficient (Schoepp et al. 2017). LAMP is a tremendously specific technique as it requires validation of 6 or 8 independent regions for successful recognition of target sequence (Chotiwan et al. 2017). RT-LAMP is an upgraded iteration of LAMP that has been integrated with reverse transcription assay for detection of several respiratory viruses, including influenza virus, MERS-CoV, and SARS-CoV (Wong et al. 2018).
Dye streptavidin-coated polymer nanoparticles–based biosensor assay
In a most recent study, scientists ameliorated RT-LAMB and developed a brand-new technique. Duly named “RT-LAMP-NBS,” this original system is a reflective fusion of RT-LAMP coated with a streptavidin-coated nanoparticle-based biosensor (NBS). With a sensitivity of 12 copies per reaction, RT-LAMP-NBS could detect SARS-CoV-2 through a single-step reaction. The analytical sensitivity of this novel assay for detection of SARS-CoV-2 was reported to be 100%, as it accurately identified the virus in 33 oropharyngeal samples collected from 33 patients with COVID-19. Further analyses conducted on 96 individuals without COVID-19 indicated a perfect specificity of 100%. Each diagnostic test in this study only took 1 h to be completed (Zhu et al. 2020).
The point-of-need technology
Point-of-need or PON transducers can amplify select nucleic acid sequences specific to certain pathogens in real time. PON is a facile technology since it does not imperatively require advanced equipment to be manufactured. A wafer-scale iteration of this technology can be produced in a standard laboratory. Hence, this technology appropriately follows the code of sustainability, as it can be assembled anywhere in the world (Nunez-Bajo et al. 2020).
Silicon-based integrated PON transducer
To further lower the costs, scientists developed a silicon-based trimodal transducer with integrated PON technology. Known as “TriSilix,” this customized transducer offered real-time performance in amplification and quantitative detection of nucleic acids specific to pathogens. With their novel TriSilix, scientists were able to maintain PCR at a predetermined temperature, while simply measuring the concentration of amplicons in real time. More importantly, TriSilix managed to discern trace amounts of complementary DNA (cDNA) specific to SARS-CoV-2 from that of SARS-CoV in exceedingly low concentrations (1 pg/ml) (Nunez-Bajo et al. 2020).
Localized surface plasmon resonance biosensing systems
Biosensing systems based on localized surface plasmon resonance (LSPR) are another class of fairly applicable devices for analysis of different molecules (Haes et al. 2005). An optical phenomenon, LSPR, occurs when a surface plasmon is confined within a conductive nanoparticle and excited by a certain wavelength of light that is larger than the size of the nanoparticle (Willets and Van Duyne 2007). LSPR-based devices exhibit great sensitivity to variables such as molecular binding and refractive index, which could be associated with enhanced plasmonic field (Anker et al. 2010). Accordingly, biosensing devices based on LSPR stand among the alternatives that should be appraised for detection of nanoscopic analytes (Qiu et al. 2020).
Localized surface plasmon resonance with gold nanoislands
Combining the plasmonic photothermal (PPT) effect and LSPR, scientists designed a transcendent biosensing device comprising two-dimensional gold nanoislands (AuNIs). They functionalized their system with receptors specific to cDNA based on nucleic acid hybridization. In this regard, the heat resulting from PPT can raise the temperature at which in situ hybridization is running, therefore further facilitating error-free discrimination between two similar nucleic acid sequences. With a lower detection limit than plain LSPR, this AuNI-based system was able to accurately identify selected sequences of cDNA synthesized from SARS-CoV-2 RNA (Qiu et al. 2020).
Conclusion and outlook
With the highly transmissible SARS-CoV-2 at large and no specific treatment at hand, one can sense an urge for development of novel diagnostic methods able to rapidly identify COVID-19 in asymptomatic patients. In doing so, humanity may finally succeed at containing the situation caused by SARS-CoV-2.
Highly sensitive and distinctly specific nanobiosensors are emerging assets that can come to our aid in the ongoing fight against COVID-19. With their excellent biocompatibility and optimal stability, nanobiosensors are soon going to be the new trend in signal detection, promising a finer point-of-care testing in the forthcoming years.
Acknowledgements
This review was conducted under supervision of Zahedan University of Medical Sciences (Grant No: IR.ZAUMS.REC.1399.197).
Author contribution
All authors participated in the investigation, design, interpretation of the studies and analysis of the data, and review of the manuscript.
Code Availability
None.
Declarations
Consent for publication
All the authors agreed.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Habib Ghaznavi, Email: ghaznavih@yahoo.com.
Samideh Khoei, Email: khoei.s@iums.ac.ir, Email: skhoei@gmail.com.
References
- Akbarzadeh A, Fekri Aval S, Sheervalilou R, Fekri L, Zarghami N, Mohammadian MJRCB, Medicine M. Quantum dots for biomedical delivery applications. Rev Cell Biol Mol Med. 2006;2(4):66–78. [Google Scholar]
- Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP (2010) Biosensing with plasmonic nanosensors. Nanoscience and Technology: A Collection of Reviews from Nature Journals. World Sci pp 308-319
- Bai H, Wang R, Hargis B, Lu H, Li Y. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors. 2012;12(9):12506–12518. doi: 10.3390/s120912506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L, Wang M. Presumed asymptomatic carrier transmission of COVID-19. Jama. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauch CT, Lloyd-Smith JO, Coffee MP, Galvani AP. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology. 2005;16:791–801. doi: 10.1097/01.ede.0000181633.80269.4c. [DOI] [PubMed] [Google Scholar]
- Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R (2020) Features, evaluation and treatment coronavirus (COVID-19). StatPearls [Internet]. StatPearls Publishing [PubMed]
- Chang J-E, Lee D-S, Ban S-W, Oh J, Jung MY, Kim S-H, Park S, Persaud K, Jheon S. Analysis of volatile organic compounds in exhaled breath for lung cancer diagnosis using a sensor system. Sens Actuators B Chem. 2018;255:800–807. doi: 10.1016/j.snb.2017.08.057. [DOI] [Google Scholar]
- Choi D-H, Thaxton A, Cheol Jeong I, Kim K, Sosnay PR, Cutting GR, Searson PC. Sweat test for cystic fibrosis: wearable sweat sensor vs. standard laboratory test. J Cyst Fibros. 2018;17(4):e35–e38. doi: 10.1016/j.jcf.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Chotiwan N, Brewster CD, Magalhaes T, Weger-Lucarelli J, Duggal NK, Rückert C, Nguyen C, Luna SMG, Fauver JR, Andre B. Rapid and specific detection of Asian- and African-lineage Zika viruses. Sci Transl Med. 2017;9(388):eaag0538. doi: 10.1126/scitranslmed.aag0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, Liu Y, Ng DY, Wan CK, Yang P, Wang Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DR, D’Anjou B, Ghattamaneni N, Harack B, Hilke M, Horth A, Majlis N, Massicotte M, Vandsburger L, Whiteway E. Experimental review of graphene. ISRN Condens Matter Phys. 2012;2012:1–56. doi: 10.5402/2012/501686. [DOI] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci U, Inci F (2019) Detection, capture and quantification of biological moieties from unprocessed bodily fluids using nanoplasmonic platform. Google Patents
- Diltemiz SE, Ersöz A, Hür D, Keçili R, Say R. 4-Aminophenyl boronic acid modified gold platforms for influenza diagnosis. Mater Sci Eng C. 2013;33(2):824–830. doi: 10.1016/j.msec.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Geim AK, Novoselov KS (2010) The rise of graphene. Nanoscience and technology: a collection of reviews from nature journals. World Sci pp 11-19
- Guarner J (2020) Three emerging coronaviruses in two decades: the story of SARS, MERS, and now COVID-19. Oxford University Press US
- Haes AJ, Chang L, Klein WL, Van Duyne RP (2005) Detection of a biomarker for Alzheimer's disease from synthetic and clinical samples using a nanoscale optical biosensor. J Am Chem Soc 127(7):2264–2271 https://covid19.who.int/ [DOI] [PubMed]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506 [DOI] [PMC free article] [PubMed]
- Justino CI, Rocha-Santos TA, Cardoso S, Duarte AC. Strategies for enhancing the analytical performance of nanomaterial-based sensors. TrAC. 2013;47:27–36. doi: 10.1016/j.trac.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Park M, Hwang J, Kim JH, Chung D-R, Lee K-s, Kang M. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019;4(5):1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W. A novel coronavirus associated with severe acute respiratory syndrome. NEJM. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R (2020) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents 105924 [DOI] [PMC free article] [PubMed]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. NEJM. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Guo X. Carbon nanomaterials field-effect-transistor-based biosensors. NPG Asia Mater. 2012;4(8):e23–e23. doi: 10.1038/am.2012.42. [DOI] [Google Scholar]
- Liu J, Chen X, Wang Q, Xiao M, Zhong D, Sun W, Zhang G, Zhang Z. Ultrasensitive monolayer MoS2 field-effect transistor based DNA sensors for screening of Down syndrome. Nano Lett. 2019;19(3):1437–1444. doi: 10.1021/acs.nanolett.8b03818. [DOI] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Coronavirus: covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ. 2020;368:m641. doi: 10.1136/bmj.m641. [DOI] [PubMed] [Google Scholar]
- Moscona A. Neuraminidase inhibitors for influenza. NEJM. 2005;353(13):1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- Norouzi M, Yasamineh S, Montazeri M, Dadashpour M, Sheervalilou R, Abasi M, Pilehvar-Soltanahmadi Y. Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection. Mater Sci Eng C. 2019;104:110007. doi: 10.1016/j.msec.2019.110007. [DOI] [PubMed] [Google Scholar]
- Nunez-Bajo E, Kasimatis M, Cotur Y, Asfour T, Collins A, Tanriverdi U, Grell M, Kaisti M, Senesi G, Stevenson K (2020) Ultra-low-cost integrated silicon-based transducer for on-site, genetic detection of pathogens. bioRxiv doi. 10.1101/2020.03.23.002931
- Organization WH (2020) Laboratory testing for coronavirus disease 2019 (Covid-19) in suspected human cases: interim guidance, 2 March 2020; WHO/COVID-19/laboratory/2020.4; World Health Organization: Geneva, 2020; https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratoryguidance (Accessed 2 March 2020).
- Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS nano. 2020;14(5):268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Saeed M, Wu A (2020) Biosensors for swine influenza viruses. Nanobiosensors:311–327
- Sakai-Tagawa Y, Ozawa M, Tamura D, Nidom CA, Sugaya N, Kawaoka Y. Sensitivity of influenza rapid diagnostic tests to H5N1 and 2009 pandemic H1N1 viruses. J Clin Microbiol. 2010;48(8):2872–2877. doi: 10.1128/JCM.00439-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saylan Y, Denizli A (2020) Virus detection using nanosensors. Nanosensors for Smart Cities. Elsevier, pp 501-511
- Saylan Y, Erdem Ö, Ünal S, Denizli A. An alternative medical diagnosis method: biosensors for virus detection. Biosensors. 2019;9(2):65. doi: 10.3390/bios9020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp NG, Schlappi TS, Curtis MS, Butkovich SS, Miller S, Humphries RM, Ismagilov RF. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci Transl Med. 2017;9(410):eaal3693. doi: 10.1126/scitranslmed.aal3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shakeri-Zadeh A, Zareyi H, Sheervalilou R, Laurent S, Ghaznavi H, Samadian H. Gold nanoparticle-mediated bubbles in cancer nanotechnology. J Control Release. 2020;330:49–60. doi: 10.1016/j.jconrel.2020.12.022. [DOI] [PubMed] [Google Scholar]
- Sheervalilou R, Shirvaliloo M, Dadashzadeh N, Shirvalilou S, Shahraki O, Pilehvar-Soltanahmadi Y, Ghaznavi H, Khoei S, Nazarlou Z. COVID-19 under spotlight: a close look at the origin, transmission, diagnosis, and treatment of the 2019-nCoV disease. J Cell Physiol. 2020;235(12):8873–8924. doi: 10.1002/jcp.29735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M, Huertas CS, Lechuga LM. Label-free plasmonic biosensors for point-of-care diagnostics: a review. Expert Rev Mol Diagn. 2019;19(1):71–81. doi: 10.1080/14737159.2019.1554435. [DOI] [PubMed] [Google Scholar]
- Tam PD, Van Hieu N, Chien ND, Le A-T, Tuan MA. DNA sensor development based on multi-wall carbon nanotubes for label-free influenza virus (type A) detection. J Immunol Methods. 2009;350(1-2):118–124. doi: 10.1016/j.jim.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Vollmer F, Arnold S, Keng D. Single virus detection from the reactive shift of a whispering-gallery mode. PNAS. 2008;105(52):20701–20704. doi: 10.1073/pnas.0808988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Du Z, Zhu F, Cao Z, An Y, Gao Y, Jiang B. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395(10228):e52. doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO CDC-SR- (2020a) Coronavirus disease 2019 (COVID-19): situation report - 52. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200312-sitrep-52-covid-19.pdf?sfvrsn=e2bfc9c0_4 (Accessed 15 April 2020).
- WHO CDC-TGLTf-niH (2020b) Coronavirus disease (COVID-19) technical guidance: laboratory testing for 2019-nCoV in humans. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance (Accessed 15 April 2020).
- WHO NC-nSR (2020c) Novel coronavirus (2019-nCoV) situation report – 1.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4 (Accessed 15 April 2020).
- Willets KA, Van Duyne RP. Localized surface plasmon resonance spectroscopy and sensing. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- Wong YP, Othman S, Lau YL, Radu S, Chee HY. Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. J Appl Microbiol. 2018;124(3):626–643. doi: 10.1111/jam.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, McGoogan J. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology:200343 [DOI] [PMC free article] [PubMed]
- Zhang Q, Zhao Q (2020) Inactivating porcine coronavirus before nuclei acid isolation with the temperature higher than 56° C damages its genome integrity seriously. BioRxiv. 10.1101/2020.02.20.958785
- Zhang DY, Chen SX, Yin P. Optimizing the specificity of nucleic acid hybridization. Nat Chem. 2012;4(3):208–214. doi: 10.1038/nchem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JX, Fang JZ, Duan W, Wu LR, Zhang AW, Dalchau N, Yordanov B, Petersen R, Phillips A, Zhang DY. Predicting DNA hybridization kinetics from sequence. Nat Chem. 2018;10(1):91–98. doi: 10.1038/nchem.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Qi Q, Jing Q, Ao S, Zhang Z, Ding M, Wu M, Liu K, Wang W, Ling Y (2020) Electrical probing of COVID-19 spike protein receptor binding domain via a graphene field-effect transistor. arXiv preprint arXiv:12529
- Zhou L, Mao H, Wu C, Tang L, Wu Z, Sun H, Zhang H, Zhou H, Jia C, Jin Q. Label-free graphene biosensor targeting cancer molecules based on non-covalent modification. Biosens Bioelectron. 2017;87:701–707. doi: 10.1016/j.bios.2016.09.025. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wang X, Han L, Chen T, Wang L, Li H, Li S, He L, Fu X, Chen S (2020) Reverse transcription loop-mediated isothermal amplification combined with nanoparticles-based biosensor for diagnosis of COVID-19. medRxiv 10.1101/2020.03.17.20037796 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
None.