Abstract
A novel poxvirus was discovered in Crocodilurus amazonicus (Teiidae) presenting with a debilitating skin disease. The generated first genome sequence of a reptilian poxvirus revealed the closest phylogenetic relationship to avipoxviruses, highlighting potential virus exchanges between avian and reptilian species.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-021-04975-6.
Poxviruses are large enveloped viruses containing a double-stranded DNA genome of 130-360 kb. They are important pathogens of high public health and economic impact and are able to infect a wide range of host species, ranging from insects to mammals. Within the family Poxviridae, two subfamilies (Chordopoxvirinae and Entomopoxvirinae) have been defined based on their hosts, which are either vertebrates or insects [1, 2]. The subfamily Chordopoxvirinae is divided into 18 genera, which include a total of 52 species [3].
Poxviruses infecting non-mammalian species belong to the genera Avipoxvirus and Crocodylidpoxvirus. As also observed in mammalian hosts, poxviruses of avian and crocodilian species are primarily associated with skin lesions but can also affect the upper respiratory and gastrointestinal tract [4, 5]. Poxvirus infections of reptiles other than crocodilians have been described in the literature, but no sequence information or further characterization of these viruses is available [6–11].
The species affected in this study, Crocodilurus amazonicus (“crocodile tegu”), is part of the family Teiidae. These lizards are native to the Amazon and Orinoco basins in South America [12] and belong to one of only two genera of living semi-aquatic teiids, and as such, they inhabit seasonally flooded forested areas near riverbanks or other watercourses. Their mean body temperature, 31.2°C, is higher than the environmental temperature but relatively low compared to that of other teiids, which could be due to their association with water habitats. Their variable diet consists of insects, other small reptiles, and frogs [13, 14].
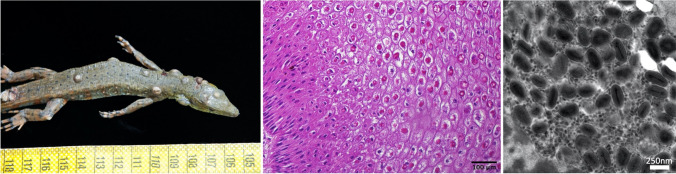
In 2019, five C. amazonicus, owned by a private collector in Austria, were presented with skin lesions and weight loss. Due to the severity of the clinical signs, one animal had to be euthanized and was sent for further diagnostic evaluation to the pathologists of the Research Institute of Wildlife Ecology, University of Veterinary Medicine, Vienna, Austria. The animal was underweight and had multiple elevated, partially ulcerated skin lesions with a diameter of up to 4 mm on the back and the dorsal areas of head, neck, and tail that did not extend into the underlying muscle (Fig. 1A). Lesions were subsequently analyzed histologically, revealing multifocally and focally extensive epidermal hyperplasia of up to 10-15 times the normal thickness with a severely thickened stratum spinosum forming rete ridges. Keratinocytes showed ballooning degeneration and contained large eosinophilic, intracytoplasmic viral inclusion bodies (Bollinger bodies) up to 20 µm in size (Fig. 1B). Large numbers of poxvirus-like particles were detected within these lesions by transmission electron microscopy (Fig. 1C).
Fig. 1.
Pathologic and histologic examination. (A) Macroscopic presentation of skin lesions, represented by multiple, up to lentil-sized, elevated, partially ulcerated foci. This scale bar is in centimeters. (B) Hematoxylin-eosin-stained tissue section of an affected area of skin, displaying severe ballooning of the cells and prominent, eosinophilic intracytoplasmatic inclusion bodies. (C) Multiple poxvirus-like particles in skin lesions detected by transmission electron microscopy of uranyl-acetate-stained tissue sections.
A sample taken from a skin lesion tested positive for the presence of poxvirus DNA when a pan-poxvirus PCR for high-GC-content poxviruses developed by Li et al. was used [15]. Comparison of this 630-bp amplicon, which corresponded to part of the gene encoding the DNA-dependent RNA polymerase subunit rpo147 ortholog, to sequences available in public databases revealed the closest relationship, with 80% sequence identity, to members of the genus Avipoxvirus. This isolate was tentatively named "teiidaepox virus 1" (TePV-1). Cultivation in freshly isolated chicken embryo fibroblasts was attempted at different temperatures (22, 28 and 37°C), but this did not result in detectable virus replication, either by PCR testing of the supernatant or the development of a cytopathic effect. In order to characterize the virus further, we determined the sequence of the virus genome, employing a combination of Illumina sequencing technology (150-bp paired-end reads) and nanopore sequencing (MinION, Oxford Nanopore Technologies [ONT]) using DNA extracted from skin. Briefly, an aseptically dissected piece of skin containing lesions of approximately 10 mg was mechanically homogenized in 60 µl of PBS in a TissueLyser II at 30 Hz for 3 min (QIAGEN). For nanopore sequencing, 120 µl of lysis buffer was added, and the rest of the DNA preparation was performed following the manufacturer’s instructions for preparation of DNA from tissues using a QIAamp DNA Mini Kit. For Illumina sequencing, DNA was extracted using an NEB genomic DNA extraction kit according to the manufacturer’s instructions. The quality of the DNA preparation was checked using Genomic DNA ScreenTape (Agilent) on a 4200 TapeStation (Agilent) at the VetCore genomics facility. The library for Illumina sequencing was prepared using an NEBNext Ultra II DNA Library Prep Kit (New England Biolabs) according to the manufacturer’s protocol and quality controlled using a fragment analyzer at the Vienna BioCenter Core Facilities before being sequenced using Illumina MiSeq chemistry. The raw reads were quality controlled, and adapter sequences were removed before commencing data analysis. For nanopore sequencing, the DNA was processed according to the protocol for sequencing of genomic DNA by ligation (SQK-LSK109) provided by ONT, and the library was loaded onto a Nanpore Flongle Flowcell (ONT).
Before contig assembly, the nanopore reads were aligned to the genome sequence of penguinpox virus using bowtie2-2.2.8 [16]. Nanopore reads aligning to this sequence were subsequently used for contig assembly of the Illumina reads with SPAdes 3.14.0 [17]. This straightforward approach, combining short reads with reads of up to 30 kb length, resulted in the ab initio assembly of two contigs with a length of 116,666 and 46,221 bp, respectively. These contigs had overlapping ends and could therefore easily be merged into one genome assembly with a final length of 166,425 bp and a GC content of 35.5%. The overlap between the two contigs was confirmed by site-specific PCR and Sanger sequencing. The coverage of the Illumina reads was 124 (138,058 aligned reads), and that of the nanopore sequencing reads was 125 (11,248 aligned reads out of 94,061; average length of aligned reads = 1848). The length distribution of the aligned reads and a histogram of the coverage distribution are shown Supplementary File 1.
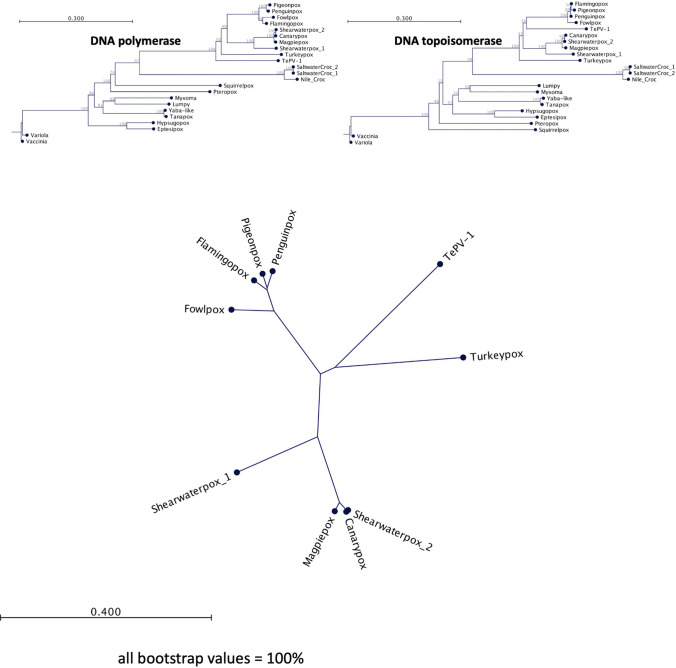
Phylogenetic analysis of the genome sequence of TePV-1 and the amino acid sequences of the putative DNA polymerase (highest amino acid sequence identity, 74.3%, to flamingopox virus, MF678796.1) and DNA topoisomerase (highest amino acid sequence identity, 76.0%, to fowlpox virus, NC_002188.1) revealed that TePV-1 is most closely related to members of the genus Avipoxvirus (Fig. 2).
Fig. 2.
Phylogenetic relationship of TePV-1 to other members of the subfamily Chordopoxvirinae. (A) Neighbor-joining tree based on the DNA polymerase protein sequence. (B) Neighbor-joining tree based on the DNA topoisomerase protein sequence. (C) Neighbor-joining tree of the full genome sequences based on avipoxviruses. All branches had a bootstrap value of 100. All trees were generated after multiple sequence alignment in CLC Workbench with 1,000 replicates. The GenBank accession numbers for sequences employed in this analysis are as follows: canarypox virus, NC_005309.1; eptesipox virus, NC_035460.1; flamingopox virus, MF678796.1; fowlpox virus, NC_002188.1; hypsugopox virus, MK860688.1; lumpy skin disease virus, NC_003027.1; magpiepox virus, MK903864.1; myxoma virus, NC_001132.2; Nile crocodilepox virus, DQ356948.1; penguinpox virus, KJ859677.1; pigeonpox virus: KJ801920.1; pteropox virus, NC_030656.1; saltwater crocodile poxvirus 1, MG450915.1; saltwater crocodile poxvirus 2, MG450916.1; shearwaterpox virus 1, KX857216.1; shearwaterpox virus 2, KX857215.1; squirrelpox virus, NC_022563.1; tanapox virus, EF420157.1; turkeypox virus, KP728110.2; vaccinia virus, NC_006998.1; variola virus, NC_001611.1; Yaba-like disease virus, NC_002642.1.
Using CLC Workbench, 154 open reading frames coding for ≥100 amino acids were detected, and these sequences were compared to the proteome of other avipoxviruses, using BLASTp. The results of this analysis are presented in Table 1. Apart from the presence of ankyrin repeat proteins at the very beginning and the end of the coding region of the TePV-1 genome, the ORF arrangement was the same as in other avipoxviruses, even though the TePV-1 genome is significantly shorter. Interestingly, nine reading frames coding for proteins of 101-206 aa could not be related to any other sequences based on amino acid sequence identity or the presence of conserved domains. Seven ORFs were found to encode proteins that are not related to other poxvirus proteins but are related to proteins found in eukaryotes (see Supplementary Table 1) and could have been acquired by horizontal gene transfer [18]. These sequences, as well as the assembly site of the two contigs and the transitions to the inverted terminal repeats were confirmed by specific PCR and Sanger sequencing.
Table 1.
Annotation of ORFs in the genome of TePV-1 encoding proteins larger than 100 aa. The start and end of ORFs located on the positive strand are shown in bold. The length of each ORF is given in base pairs. "Protein" indicates the highest-rated BLAST hit for this ORF, which is further specified by its accession number (GenBank no.), location in the genome (ref loc), the % amino acid sequence identity (% identity), the alignment length (ali), the e-value and the species. AC, Anolis carolinensis; CG, Cricetulus griseus; CNPV, canarypox virus; EA, Equus asinus; FePV2, pigeonpox virus; FGPV, flamingopox virus; FWPV, fowlpox virus; LA, Lingula anatina; MPPV, magpiepox virus; N., ORF number; PEPV, penguinpox virus; PM, Protobothrops mucrosquamatus; SWPV, shearwaterpox virus; TKPV, turkeypox virus
| N | Start | End | Length | Protein | GenBank no. | ref loc | % identity | ali | e-value | Species |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 843 | 1463 | 621 | SWPV1-308, conserved hypothetical protein | ARF02867.1 | 323041..323703 | 59.545 | 220 | 2.43E-87 | SWPV-1 |
| 2 | 1841 | 3643 | 1803 | Ankyrin repeat family protein | AUD40350.1 | 271364..273295 | 33.667 | 600 | 4.68E-93 | FGPV |
| 3 | 3583 | 4626 | 1044 | Serpin family protein | AUD40336.1 | 256018..257064 | 34.582 | 347 | 2.93E-74 | FGPV |
| 4 | 4670 | 6163 | 1494 | Ankyrin repeat protein | YP_009046459.1 | 264529..266415 | 41.04 | 519 | 2.61E-122 | FeP2 |
| 5 | 6183 | 7934 | 1752 | Ankyrin repeat protein | YP_009046459.1 | 264529..266415 | 42.484 | 612 | 1.26E-157 | FeP2 |
| 6 | 7991 | 8368 | 378 | EFc-like protein | ALA62542.1 | 185548..185937 | 34.711 | 121 | 2.27E-25 | TKPV |
| 7 | 8412 | 8969 | 558 | Ankyrin repeat protein | YP_009046231.1 | 281304..281861 | 39.572 | 187 | 8.05E-45 | PEPV |
| 8 | 9035 | 9832 | 798 | Ankyrin repeat protein | AYO89833.1 | 274191..275423 | 42.146 | 261 | 9.34E-66 | FWPV |
| 9 | 9321 | 9668 | 348 | No similarity found | ||||||
| 10 | 9853 | 11604 | 1752 | Ankyrin repeat protein | YP_009046027.1 | 33882..35669 | 46.259 | 588 | 6.97E-175 | PEPV |
| 11 | 11632 | 12234 | 603 | Hypothetical protein | YP_009046028.1 | 35800..36411 | 55.051 | 198 | 5.22E-85 | PEPV |
| 12 | 12608 | 13561 | 954 | G-protein-coupled receptor family protein | AUD40130.1 | 29544..30545 | 36.824 | 296 | 3.84E-67 | FGPV |
| 13 | 13541 | 15733 | 2193 | alkaline phosphodiesterase | AUD40134.1 | 33068..35515 | 42.665 | 743 | 0 | FGPV |
| 14 | 15749 | 16861 | 1113 | SWPV1-041, DNase II-like protein | ARF02656.1 | 47782..48978 | 41.842 | 380 | 2.09E-97 | SWPV-1 |
| 15 | 16866 | 17279 | 414 | Hypothetical protein | AUD40139.1 | 40555..40962 | 52.239 | 134 | 1.03E-48 | FGPV |
| 16 | 17303 | 17776 | 474 | Hypothetical protein | ALA62391.1 | 22080..22559 | 43.312 | 157 | 4.06E-44 | TKPV |
| 17 | 17779 | 18213 | 435 | dUTP pyrophosphatase | AUD40142.1 | 41945..42382 | 68.056 | 144 | 4.29E-72 | FGPV |
| 18 | 18219 | 18749 | 531 | B-cell lymphoma 2 | AUD40143.1 | 42434..42961 | 34.078 | 179 | 2.11E-28 | FGPV |
| 19 | 18785 | 19798 | 1014 | Serpin family protein | YP_009046279.1 | 49969..50982 | 46.154 | 338 | 1.91E-111 | FeP2 |
| 20 | 20154 | 21833 | 1680 | DNA ligase | AYP74252.1 | 247565..249259 | 62.186 | 558 | 0 | FWPV |
| 21 | 21865 | 22923 | 1059 | Putative serpin | AYP74251.1 | 246462..247523 | 50.992 | 353 | 1.06E-133 | FWPV |
| 22 | 22961 | 24649 | 1689 | Semaphorin | YP_009046047.1 | 57639..59369 | 48.772 | 570 | 0 | PEPV |
| 23 | 24724 | 25494 | 771 | GNS1/SUR4 | AUD40150.1 | 51121..51906 | 78.516 | 256 | 1.57E-157 | FGPV |
| 24 | 25547 | 26011 | 465 | Late transcription factor VLTF-2 | AXY04490.1 | 52429..52893 | 76.623 | 154 | 5.41E-89 | FWPV |
| 25 | 25648 | 25971 | 324 | Rifampicin resistance protein | AXY04491.1 | 52647..52856 | 76.562 | 64 | 1.14E-27 | FWPV |
| 26 | 26032 | 27693 | 1662 | Rifampicin resistance protein | ALA62402.1 | 33712..35373 | 75.769 | 553 | 0 | TKPV |
| 27 | 27710 | 28579 | 870 | mRNA capping enzyme | YP_009046288.1 | 61089..61958 | 79.585 | 289 | 1.20E-174 | FeP2 |
| 28 | 28576 | 29709 | 1134 | Class I histocompatibility antigen, F10 alpha chain-like | XP_015684078.2 | 46 | 164 | 5.00E-20 | PM | |
| 29 | 29667 | 30548 | 882 | Major histocompatibility complex class I-related gene protein | XP_008122411.1 | 35 | 172 | 2.00E-16 | CNPV | |
| 30 | 30526 | 32436 | 1911 | NPH-1 transcription termination factor | AUD40154.1 | 55108..57021 | 78.74 | 635 | 0 | FGPV |
| 31 | 32409 | 33104 | 696 | muT motif expression regulator | YP_009046053.1 | 65721..66398 | 62.667 | 225 | 3.39E-102 | PEPV |
| 32 | 33088 | 33723 | 636 | mutT motif containing protein | AXY04496.1 | 58126..58821 | 71.226 | 212 | 3.25E-109 | FWPV |
| 33 | 34048 | 34533 | 486 | RNA polymerase subunit RPO18 | YP_009046293.1 | 66549..67034 | 83.23 | 161 | 8.20E-98 | FeP2 |
| 34 | 34520 | 36421 | 1902 | Early transcription factor small subunit | AUD40159.1 | 60145..62046 | 90.047 | 633 | 0 | FGPV |
| 35 | 36402 | 38759 | 2358 | NTPase | AUD40160.1 | 62027..64402 | 79.267 | 791 | 0 | FGPV |
| 36 | 38788 | 39444 | 657 | Uracil DNA glycosylase | AXY04504.1 | 67861..68517 | 76.389 | 216 | 6.82E-127 | FWPV |
| 37 | 39461 | 40600 | 1140 | SWPV1-075, conserved hypothetical protein | ARF02682.1 | 81697..82893 | 41.071 | 392 | 1.85E-88 | SWPV-1 |
| 38 | 40859 | 41215 | 357 | Putative glutathione peroxidase | QGM48717.1 | 80344..80754 | 68.908 | 119 | 1.91E-59 | MPPV |
| 39 | 41169 | 41540 | 372 | Hypothetical protein | NP_955112.1 | 100368..100847 | 44.898 | 98 | 4.02E-29 | CNPV |
| 40 | 41859 | 42653 | 795 | Virion protein | YP_009046067.1 | 80684..81493 | 47.212 | 269 | 3.94E-82 | PEPV |
| 41 | 42841 | 43218 | 378 | Glutaredoxin | ALA62425.1 | 53825..54202 | 68 | 125 | 9.15E-62 | TKPV |
| 42 | 43161 | 43859 | 699 | Putative transcriptional elongation factor | AUD40182.1 | 80826..81503 | 64 | 225 | 8.93E-102 | FGPV |
| 43 | 43853 | 44164 | 312 | Hypothetical protein | AXY04521.1 | 79590..79901 | 66.019 | 103 | 3.22E-48 | FWPV |
| 44 | 44188 | 46041 | 1854 | SWPV1-097, putative metalloprotease | ARF02699.1 | 95995..97896 | 65.561 | 633 | 0 | SWPV-1 |
| 45 | 46028 | 48064 | 2037 | RNA helicase NPH-II | AYP74219.1 | 203411..205456 | 72.485 | 676 | 0 | FWPV |
| 46 | 48079 | 49338 | 1260 | Virion core proteinase | AYP74218.1 | 202111..203376 | 75.059 | 421 | 0 | FWPV |
| 47 | 49339 | 50508 | 1170 | DNA-binding protein | YP_009046080.1 | 94650..95822 | 63.333 | 390 | 0 | PEPV |
| 48 | 50376 | 50708 | 333 | No similarity found | ||||||
| 49 | 51076 | 51828 | 753 | DNA-binding phosphoprotein | ALA62437.1 | 65235..66116 | 50.883 | 283 | 3.47E-101 | TKPV |
| 50 | 52033 | 52968 | 936 | DNA-binding virion protein | QGM48748.1 | 99500..100432 | 81.935 | 310 | 0 | MPPV |
| 51 | 53112 | 55055 | 1944 | Hypothetical protein | AUD40195.1 | 92674..94644 | 48.558 | 659 | 0 | FGPV |
| 52 | 54985 | 55380 | 396 | Virion core protein | QGM48751.1 | 102503..102898 | 51.145 | 131 | 2.17E-47 | MPPV |
| 53 | 55688 | 58654 | 2967 | DNA polymerase | AUD40198.1 | 95291..98260 | 74.291 | 988 | 0 | FGPV |
| 54 | 58646 | 59452 | 807 | Hypothetical protein | AUD40199.1 | 98252..99100 | 71.269 | 268 | 3.94E-141 | FGPV |
| 55 | 59454 | 61169 | 1716 | Hypothetical protein | AUD40200.1 | 99093..100808 | 71.979 | 571 | 0 | FGPV |
| 56 | 61190 | 66739 | 5550 | B22R family protein | YP_009046096.1 | 118822..124590 | 47.199 | 1928 | 0 | PEPV |
| 57 | 64481 | 64939 | 459 | No similarity found | ||||||
| 58 | 66827 | 67372 | 546 | RNA polymerase subunit | AYP74202.1 | 171000..171548 | 79.006 | 181 | 1.84E-105 | FWPV |
| 59 | 67375 | 69501 | 2127 | Hypothetical protein | AUD40205.1 | 118803..120956 | 55.23 | 717 | 0 | FGPV |
| 60 | 69473 | 70891 | 1419 | PolyA polymerase large subunit PAP-L | YP_009046336.1 | 126407..127825 | 79.025 | 472 | 0 | FeP2 |
| 61 | 70885 | 71202 | 318 | DNA binding virion core phosphoprotein | AUD40207.1 | 122355..122699 | 70.175 | 114 | 9.14E-54 | FGPV |
| 62 | 71281 | 71880 | 600 | Hypothetical protein | AYO89698.1 | 121669..122301 | 38.756 | 209 | 3.96E-46 | FWPV |
| 63 | 71929 | 72375 | 447 | Hypothetical protein | YP_009046339.1 | 128991..129440 | 81.081 | 148 | 1.22E-88 | FeP2 |
| 64 | 72732 | 73865 | 1134 | SWPV1-124, putative palmitylated EEV envelope lipase | ARE67652.1 | 142768..143904 | 75.067 | 373 | 0 | SWPV-1 |
| 65 | 73897 | 75696 | 1800 | EEV maturation protein | AXY04551.1 | 129455..131347 | 46.825 | 630 | 0 | FWPV |
| 66 | 75727 | 77052 | 1326 | Ser/Thr kinase | AYP74192.1 | 153725..155065 | 75.688 | 436 | 0 | FWPV |
| 67 | 77030 | 77671 | 642 | Hypothetical protein | AXY04554.1 | 134129..134770 | 69.484 | 213 | 6.53E-106 | FWPV |
| 68 | 78116 | 78655 | 540 | HAL3 domain containing protein | AXY04556.1 | 135430..135981 | 68.306 | 183 | 6.98E-96 | FWPV |
| 69 | 79127 | 79822 | 696 | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial | XP_003216736.1 | 38 | 121 | 1.00E-07 | AC | |
| 70 | 79860 | 80702 | 843 | Glutamate-rich protein 3 isoform X3 | XP_013402139.1 | 37 | 101 | 6.00E-06 | LA | |
| 71 | 80169 | 80498 | 330 | No similarity found | ||||||
| 72 | 80787 | 82091 | 1305 | Hypothetical protein | NP_955168.1 | 170735..172057 | 60.277 | 433 | 0 | CNPV |
| 73 | 82286 | 82864 | 579 | SWPV1-137, conserved hypothetical protein | ARE67661.1 | 158056..158622 | 71.123 | 187 | 1.15E-98 | SWPV-1 |
| 74 | 82817 | 83839 | 1023 | Putative virion core protein | NP_955171.1 | 172777..173823 | 77.011 | 348 | 0 | CNPV |
| 75 | 83894 | 84676 | 783 | Late transcription factor VLTF-1 | AYP74173.1 | 129528..130310 | 87.308 | 260 | 3.61E-177 | FWPV |
| 76 | 84681 | 85688 | 1008 | Poxvirus myristoylprotein | AXY04569.1 | 160397..161407 | 60.417 | 336 | 1.45E-159 | FWPV |
| 77 | 85689 | 86420 | 732 | SWPV2-ORF161, putative myristylated IMV envelope protein | ARE67397.1 | 201657..202388 | 86.364 | 242 | 4.06E-165 | SWPV-2 |
| 78 | 86699 | 87607 | 909 | Hypothetical protein | YP_009046124.1 | 171203..172108 | 75.908 | 303 | 0 | PEPV |
| 79 | 87633 | 88391 | 759 | DNA-binding virion core VP8 | YP_009046362.1 | 156784..157545 | 67.984 | 253 | 1.64E-132 | FeP2 |
| 80 | 88392 | 88772 | 381 | Hypothetical protein | AYO89725.1 | 164799..165188 | 62.016 | 129 | 1.32E-54 | FWPV |
| 81 | 88729 | 89169 | 441 | Putative IMV membrane protein | QGM48809.1 | 169617..170063 | 76.224 | 143 | 4.18E-80 | MPPV |
| 82 | 89186 | 90079 | 894 | PolyA polymerase small subunit | AUD40238.1 | 158947..159873 | 70.27 | 296 | 4.29E-158 | FGPV |
| 83 | 90076 | 90636 | 561 | RNA polymerase subunit 22 | AUD40239.1 | 159870..160430 | 75.275 | 182 | 3.22E-102 | FGPV |
| 84 | 90629 | 91039 | 411 | Membrane protein | AXY04578.1 | 166439..166852 | 77.686 | 121 | 4.38E-70 | FWPV |
| 85 | 91075 | 94941 | 3867 | RNA polymerase subunit RPO147 | YP_009046131.1 | 175641..179504 | 88.276 | 1288 | 0 | PEPV |
| 86 | 94945 | 95445 | 501 | Protein tyrosine phosphatase | YP_009046369.1 | 164161..164661 | 79.518 | 166 | 1.86E-100 | FeP2 |
| 87 | 95459 | 96028 | 570 | Putative viral membrane protein | AXY04581.1 | 171281..171853 | 79.894 | 189 | 1.78E-117 | FWPV |
| 88 | 96025 | 96891 | 867 | Virion envelope protein (p35) | YP_009046371.1 | 165428..166429 | 50.617 | 324 | 1.64E-121 | FeP2 |
| 89 | 96892 | 99279 | 2388 | RNA polymerase-associated protein 94 | AUD40245.1 | 166990..169389 | 79.099 | 799 | 0 | FGPV |
| 90 | 99382 | 99870 | 489 | VLTF-4 | YP_009046373.1 | 168979..169503 | 46.286 | 175 | 3.66E-34 | FeP2 |
| 91 | 99871 | 100821 | 951 | SWPV2-ORF177, DNA topoisomerase | ARE67414.1 | 217385..218335 | 75.633 | 316 | 0 | SWPV-2 |
| 92 | 100826 | 101287 | 462 | SWPV1-166, conserved hypothetical protein | ARE67699.1 | 196050..196511 | 58.17 | 153 | 5.47E-61 | SWPV-1 |
| 93 | 101533 | 104082 | 2550 | mRNA capping enzyme large subunit | AXY04588.1 | 177778..180333 | 75.972 | 849 | 0 | FWPV |
| 94 | 104052 | 104474 | 423 | CNPV194 virion protein | NP_955217.1 | 228919..229341 | 56.835 | 139 | 4.89E-50 | CNPV |
| 95 | 104492 | 105904 | 1413 | Sodium-dependent lysophosphatidylcholine symporter 1 isoform X2 | XP_027254970.1 | 47 | 473 | 2.00E-135 | CG | |
| 96 | 106631 | 107491 | 861 | p28-like protein | AYP74141.1 | 99505..100464 | 51.736 | 288 | 5.16E-114 | FWPV |
| 97 | 107493 | 108848 | 1356 | CNPV206 putative photolyase | NP_955229.1 | 237537..238955 | 65.778 | 450 | 0 | CNPV |
| 98 | 108875 | 109471 | 597 | N1R/p28 family protein | YP_009046151.1 | 198797..199537 | 33.166 | 199 | 2.99E-29 | PEPV |
| 99 | 109519 | 110163 | 645 | CNPV209 N1R/p28-like protein | NP_955232.1 | 240422..241354 | 33.005 | 203 | 9.90E-36 | CNPV |
| 100 | 110429 | 110941 | 513 | Hypothetical protein | ALA62492.1 | 122400..122924 | 34.969 | 163 | 6.26E-24 | TKPV |
| 101 | 110916 | 112124 | 1209 | SWPV1-208, C4L/C10L-like protein | ARF02782.1 | 232534..233796 | 43.796 | 411 | 1.05E-115 | SWPV-1 |
| 102 | 110927 | 111544 | 618 | No similarity found | ||||||
| 103 | 112126 | 112803 | 678 | Late transcription factor VLTF-3 | AXY04607.1 | 198622..199299 | 88.889 | 225 | 2.66E-149 | FWPV |
| 104 | 113020 | 114966 | 1947 | Virion core protein P4b | AXY04609.1 | 199529..201502 | 80.864 | 648 | 0 | FWPV |
| 105 | 115027 | 115521 | 495 | Immunodominant virion protein | ALA62497.1 | 130871..131458 | 34.01 | 197 | 6.65E-30 | TKPV |
| 106 | 115560 | 116054 | 495 | RNA polymerase subunit RPO19 | YP_009046400.1 | 196511..197020 | 76.923 | 169 | 1.69E-89 | FeP2 |
| 107 | 116049 | 117170 | 1122 | SWPV2-ORF229, conserved hypothetical protein | ARE67480.1 | 268134..269255 | 65.952 | 373 | 2.06E-179 | SWPV-2 |
| 108 | 116797 | 117099 | 303 | No similarity found | ||||||
| 109 | 117173 | 119299 | 2127 | Early transcription factor large subunit VETF-L | AYO90282.1 | 203339..205468 | 83.216 | 709 | 0 | FWPV |
| 110 | 118121 | 118438 | 318 | No similarity found | ||||||
| 111 | 119354 | 120256 | 903 | Intermediate transcription factor 3 | AUD40279.1 | 203735..204640 | 79.07 | 301 | 0 | FGPV |
| 112 | 120449 | 123115 | 2667 | Virion core protein P4a | AXY04616.1 | 207389..210064 | 73.46 | 893 | 0 | FWPV |
| 113 | 123132 | 123908 | 777 | Hypothetical protein | AXY04617.1 | 210082..210906 | 74.088 | 274 | 3.08E-142 | FWPV |
| 114 | 123903 | 124352 | 450 | Virion protein | QGM48882.1 | 229297..229803 | 56.875 | 160 | 7.46E-54 | MPPV |
| 115 | 125401 | 126492 | 1092 | Putative myristoylated membrane protein | AUD40289.1 | 210114..211220 | 69.919 | 369 | 0 | FGPV |
| 116 | 126500 | 127078 | 579 | Phosphorylated virion membrane protein | AXY04624.1 | 213785..214381 | 72.727 | 198 | 2.15E-100 | FWPV |
| 117 | 127096 | 128472 | 1377 | CNPV257 DNA helicase, transcriptional elongation | NP_955280.1 | 286822..288210 | 74.891 | 458 | 0 | CNPV |
| 118 | 128696 | 129028 | 333 | Conserved hypothetical protein | QGM48893.1 | 234410..234748 | 82.143 | 112 | 1.09E-62 | MPPV |
| 119 | 129027 | 130310 | 1284 | Processivity factor | YP_009046183.1 | 227993..229291 | 59.767 | 430 | 0 | PEPV |
| 120 | 130313 | 130765 | 453 | Holliday junction resolvase | AUD40295.1 | 215103..215588 | 67.105 | 152 | 1.83E-75 | FGPV |
| 121 | 130785 | 131936 | 1152 | Intermediate transcription factor VITF-3 | QGM48896.1 | 236523..237674 | 74.413 | 383 | 0 | MPPV |
| 122 | 131963 | 135436 | 3474 | RNA polymerase subunit RPO132 | YP_009046420.1 | 213357..216830 | 90.147 | 1157 | 0 | FeP2 |
| 123 | 135425 | 136993 | 1569 | A-type inclusion like protein | NP_955287.1 | 295207..297018 | 55.017 | 598 | 0 | CNPV |
| 124 | 135968 | 136303 | 336 | No similarity found | ||||||
| 125 | 137029 | 138375 | 1347 | A-type inclusion protein | YP_009046188.1 | 236280..237698 | 49.895 | 477 | 4.20E-170 | PEPV |
| 126 | 138376 | 138798 | 423 | Hypothetical protein | AYP74103.1 | 57016..57438 | 72.857 | 140 | 2.63E-78 | FWPV |
| 127 | 138802 | 139713 | 912 | RNA polymerase subunit RP035 | AXY04635.1 | 226511..227419 | 60.726 | 303 | 2.60E-138 | FWPV |
| 128 | 140010 | 140351 | 342 | Hypothetical protein | YP_009046427.1 | 221822..222163 | 61.947 | 113 | 6.59E-48 | FeP2 |
| 129 | 140352 | 140714 | 363 | Hypothetical protein | AYP74099.1 | 55034..55393 | 45.763 | 118 | 1.49E-35 | FWPV |
| 130 | 140703 | 141446 | 744 | Virion assembly protein | YP_009046195.1 | 240113..241027 | 84.836 | 244 | 6.67E-161 | PEPV |
| 131 | 141490 | 142044 | 555 | SWPV1-245, C-type lectin-like EEV protein | ARF02806.1 | 267315..267863 | 61.932 | 176 | 4.63E-81 | SWPV-1 |
| 132 | 142089 | 142520 | 432 | Hypothetical protein | NP_955296.1 | 302592..303416 | 38.095 | 147 | 4.07E-24 | CNPV |
| 133 | 142505 | 143281 | 777 | Hypothetical protein | AXY04907.1 | 232786..233643 | 48 | 250 | 1.63E-75 | FWPV |
| 134 | 143273 | 143875 | 603 | Hypothetical protein | AYP74093.1 | 49333..50022 | 37.864 | 206 | 1.42E-35 | FWPV |
| 135 | 143925 | 145406 | 1482 | Prostacyclin synthase isoform X1 | XP_008108335.1 | 45 | 486 | 1.00E-128 | AC | |
| 136 | 145447 | 145944 | 498 | SWPV2-ORF270, putative interleukin binding protein | ARE67521.1 | 302929..303510 | 35.897 | 117 | 7.90E-11 | SWPV-2 |
| 137 | 145928 | 146287 | 360 | Epidermal growth factor like protein | YP_009046207.1 | 251214..251585 | 40 | 100 | 3.24E-22 | PEPV |
| 138 | 146287 | 147168 | 882 | Ser/Thr protein kinase | ALA62528.1 | 164710..165612 | 59.932 | 292 | 7.44E-135 | TKPV |
| 139 | 147451 | 148422 | 972 | Hypothetical protein | AXY04659.1 | 243703..244689 | 48.287 | 321 | 1.65E-102 | FWPV |
| 140 | 148454 | 149758 | 1305 | Ankyrin repeat protein | YP_009046446.1 | 240328..241650 | 40.139 | 431 | 2.75E-99 | FeP2 |
| 141 | 149820 | 152030 | 2211 | Ankyrin repeat protein | AXY05185.1 | 248089..250332 | 41.384 | 737 | 0 | FWPV |
| 142 | 152033 | 152608 | 576 | Ankyrin repeat protein | YP_009046450.1 | 247134..248219 | 48.408 | 157 | 7.36E-51 | FeP2 |
| 143 | 152568 | 153041 | 474 | Zinc finger protein 709-like isoform X3 | XP_014701435.1 | 48 | 119 | 9.00E-19 | EA | |
| 144 | 153057 | 154364 | 1308 | SWPV1-279, ankyrin repeat protein | ARF02838.1 | 294643..295941 | 39.25 | 400 | 4.78E-96 | SWPV-1 |
| 145 | 154387 | 155430 | 1044 | Serpin family protein | AUD40336.1 | 256018..257064 | 39.093 | 353 | 2.95E-80 | FGPV |
| 146 | 155482 | 156876 | 1395 | Ankyrin repeat family protein | AUD40338.1 | 257748..259259 | 50 | 462 | 2.60E-153 | FGPV |
| 147 | 156861 | 157838 | 978 | CNPV039 G protein-coupled receptor-like protein | NP_955062.1 | 46721..47704 | 40.122 | 329 | 2.49E-85 | CNPV |
| 148 | 157330 | 157659 | 330 | No similarity found | ||||||
| 149 | 158212 | 159471 | 1260 | Ankyrin repeat family protein | ALA62540.1 | 178533..179828 | 33.412 | 422 | 1.17E-67 | TKPV |
| 150 | 159473 | 159784 | 312 | Late transcription factor VLTF-1 | ALA62543.1 | 186082..186339 | 31.325 | 83 | 4.97E-09 | TKPV |
| 151 | 159825 | 161417 | 1593 | Ankyrin repeat protein | YP_009046233.1 | 282988..284919 | 38.214 | 560 | 9.22E-117 | PEPV |
| 152 | 161398 | 163155 | 1758 | SWPV2-ORF300, ankyrin repeat protein | ARE67552.1 | 332667..334556 | 38.548 | 620 | 1.06E-126 | SWPV-2 |
| 153 | 163142 | 164908 | 1767 | SWPV2-ORF300, ankyrin repeat protein | ARE67552.1 | 332667..334556 | 41.977 | 617 | 2.20E-157 | SWPV-2 |
| 154 | 165195 | 165815 | 621 | SWPV1-308, conserved hypothetical protein | ARF02867.1 | 323041..323703 | 59.545 | 220 | 2.43E-87 | SWPV-1 |
Poxvirus-like lesions and infections have been described in various reptiles, including crocodilians, tortoises, chameleons, and tegus [6–11, 19]. Despite their description in the literature, they have not yet been characterized at the genetic level, except for poxviruses in Nile and saltwater crocodiles (Nile crocodilepox virus [CRV] and saltwater crocodilepox virus subtypes 1 and 2 [SwCRV1/2], respectively) [10, 11]. This first report of the genome sequence of a poxvirus causing disease in a lizard reveals it to be most closely related to avipoxviruses. This is surprising, considering the phylogenetic distance between avian and reptilian species and the differences in homeostasis. The GC content of TePV-1 (35%) is also more similar to that of avipoxviruses than to the known crocodile-infecting poxviruses (62% for CRV, Sw-CRV-1 and -2) [10, 11]. Interestingly, the initial diagnostic PCR only was positive when a primer set for high-GC-content poxviruses was employed. This might indicate that poxviruses of Reptilia are quite variable. Therefore, additional research is warranted to examine the diversity of poxviruses of Reptilia and their species specificity and zoonotic potential.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Coverage of the assembled genome sequence for the short- and long-read next-generation sequencing approaches. The read length distribution for the long-read sequencing approach is shown (DOCX 14 KB)
Supplementary file2 ORFs encoding proteins related to eukaryotic proteins (JPG 485 KB)
Acknowledgments
This research was supported using resources of the VetCore Facility (Transcriptomics Unit) of the University of Veterinary Medicine Vienna.
Author contributions
CR, KS and TR designed the study; MH and CH examined the animals; AKH, AL, AP and GS performed the pathological and histopathological examination; HW and ND provided the electron micrographs; and AA, AK, CR, KS and MM performed the diagnostic PCRs, generated the full genome sequence, analyzed the data and prepared the figures. CR and KS wrote the manuscript and all authors commented on it.
Funding
Open Access funding provided by University of Veterinary Medicine Vienna. It is not applicable for this publication.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Availability of data and material
The sequence of TePV-1 has been submitted to the GenBank database (accession number MT712273). The raw data are available at SRA (BioProject ID: PRJNA681025).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Haller SL, Peng C, McFadden G, Rothenburg S. Poxviruses and the evolution of host range and virulence. Infect Genet Evol. 2014;21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefkowitz EJ, Wang C, Upton C. Poxviruses: Past, present and future. Virus Res. 2006;117:105–118. doi: 10.1016/j.virusres.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Lefkowitz EJ, Dempsey DM, Hendrickson RC, et al. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weli SC, Tryland M. Avipoxviruses: Infection biology and their use as vaccine vectors. Virol J. 2011;8:49. doi: 10.1186/1743-422X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariel E. Viruses in reptiles. Vet Res. 2011;42:1–12. doi: 10.1186/1297-9716-42-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobson ER, Popp JA, Shields RP, Gaskin JM. Poxlike skin lesions in captive caimans. J Am Vet Med Assoc. 1979;175:937–940. [PubMed] [Google Scholar]
- 7.Jacobson ER, Telford SR. Chlamydial and poxvirus infections of circulating monocytes of a flap-necked chameleon (Chamaeleo dilepis) J Wildl Dis. 1990;26:572–577. doi: 10.7589/0090-3558-26.4.572. [DOI] [PubMed] [Google Scholar]
- 8.Stauber E, Gogolewski R. Poxvirus dermatitis in a Tegu lizard (Tupinambis teguixin) J Zoo Wildl Med. 1990;21(2):228–230. [Google Scholar]
- 9.Penrith ML, Nesbit JW, Huchzermeyer FW. Pox virus infection in captive juvenile caimans (Caiman crocodilus fuscus) in South Africa. J S Afr Vet Assoc. 1991;62:137–139. doi: 10.4102/jsava.v62i3.1612. [DOI] [PubMed] [Google Scholar]
- 10.Afonso CL, Tulman ER, Delhon G, et al. Genome of Crocodilepox Virus. J Virol. 2006;80:4978–4991. doi: 10.1128/jvi.80.10.4978-4991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarker S, Isberg SR, Milic NL, et al. Molecular characterization of the first saltwater crocodilepox virus genome sequences from the world’s largest living member of the Crocodylia. Sci Rep. 2018 doi: 10.1038/s41598-018-23955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avila-Pires TCS De (1995) Lizards of Brazilian Amazonia, Nationaal Natuurhistorisch Museum, Leiden.
- 13.Mesquita DO, Colli GR, Costa GC, et al. At the water’s edge: ecology of semiaquatic teiids in Brazilian Amazon. J Herpetol. 2006;40:221–229. doi: 10.1670/123-05a.1. [DOI] [Google Scholar]
- 14.Tattersall GJ, Leite CAC, Sanders CE, et al. Seasonal reproductive endothermy in tegu lizards. Sci Adv. 2016;2:e1500951. doi: 10.1126/sciadv.1500951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–276. doi: 10.1128/JCM.01697-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankevich A, Nurk S, Antipov D, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bratke KA, McLysaght A. Identification of multiple independent horizontal gene transfers into poxviruses using a comparative genomics approach. BMC Evol Biol. 2008;8:67. doi: 10.1186/1471-2148-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marschang RE. Viruses infecting reptiles. Viruses. 2011;3:2087–2126. doi: 10.3390/v3112087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Coverage of the assembled genome sequence for the short- and long-read next-generation sequencing approaches. The read length distribution for the long-read sequencing approach is shown (DOCX 14 KB)
Supplementary file2 ORFs encoding proteins related to eukaryotic proteins (JPG 485 KB)