Abstract
The conversion of the glycerophospholipid phosphatidic acid (PA) into diacylglycerol (DAG) is essential for the biosynthesis of membrane phospholipids and storage fats. Importantly, both PA and DAG can also serve signaling functions in the cell. The dephosphorylation of PA that yields DAG can be executed by two different classes of enzymes, Mg2+-dependent lipins and Mg2+-independent lipid phosphate phosphatases. Here, I will discuss the current status of research directed at understanding the roles of these enzymes in insect development and metabolism. Special emphasis will be given to studies in the model organism Drosophila melanogaster.
Keywords: Phosphatidate phosphatases, lipins, nutrient signaling, lipid phosphate phosphatases, Wunen, Lazaro
Graphical Abstract
1. Introduction
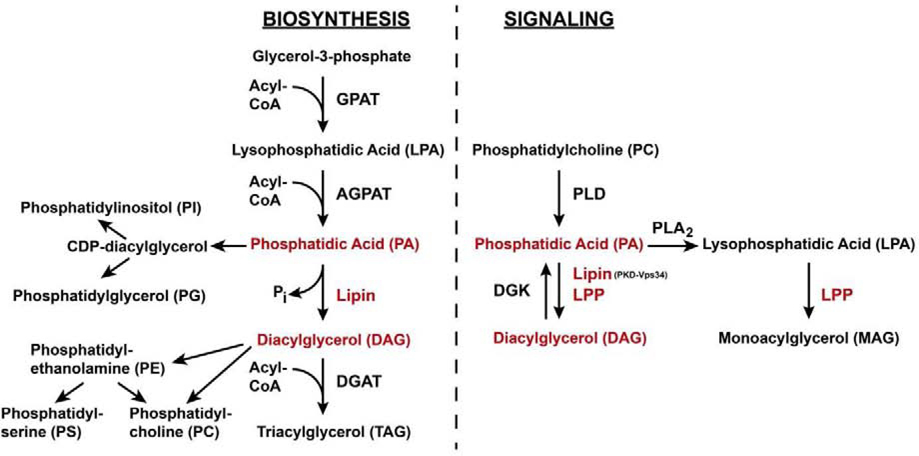
Lipids are essential structural and functional components of cells. A central intermediate in the synthesis of both membrane phospholipids and neutral fats is the glycerophospholipid phosphatidic acid (PA). PA can be produced in three different ways, (1) acylation of lysophosphatidic acid (LPA), (2) hydrolysis of phosphatidylcholine (PC), or (3) phosphorylation of diacylglycerol (DAG) (Fig. 1). The de novo synthesis of PA in the glycerol-3-phosphate pathway by acylation of LPA is an essential intermediate step in the synthesis of all glycerophospholipids. It is also the penultimate step in the synthesis of triacylglycerols (TAG) that are used as energy stores. Enzymes of the glycerol-3-phosphate pathway are located at the endoplasmatic reticulum. Some of them, including 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), the enzyme that produces PA by acylation of LPA, are also found at the surface of fat droplets (Jacquemyn et al., 2017; Wilfling et al., 2013).
Figure 1.
Integration of phosphatidic acid and phosphatidic acid phosphatases (Lipin and LPP) into biosynthetic and signaling pathways. Substrates and products of PA phosphatases and the enzymes themselves are highlighted in red.
Apart from its role as a biosynthetic precursor, PA can have structural and signaling functions. Compared to other phospholipids, PA is underrepresented in the biological membranes of eukaryotic cells, constituting only 1–4% of total cellular lipid in mammalian and yeast cells (Selvy et al., 2011; van Meer et al., 2008). PA content is equally low in membranes of insect cells (Carvalho et al., 2012). Local enrichment, however, can have profound effects. It can bring about changes in membrane properties that may, for instance, lead to increased membrane curvature. Such a structural role could be the mechanistic underpinning of PA’s established function in intracellular vesicle trafficking in both mammals and Drosophila (Kooijman et al., 2003; LaLonde et al., 2006; Roth et al., 1999; Siddhanta and Shields, 1998). Local production of PA can also serve signaling purposes. PA derived from hydrolysis of PC by phospholipase D (PLD) is involved in a number of different signaling processes including nutrient signaling through the kinase target of rapamycin (TOR) and growth factor signaling mediated by receptor tyrosine kinases and Ras (Foster, 2013; Selvy et al., 2011; Zhao et al., 2007). Phosphorylation of DAG by diacylglycerol kinase (DGK) produces PA that plays a regulatory role in the vertebrate immune system and in photoreceptor function in Drosophila (Mérida et al., 2008) (Fig. 1). Conversion of DAG into PA by DGK may also serve structural functions as exemplified by the role of yeast DGK1 in nuclear envelope biogenesis (G.-S. Han et al., 2008).
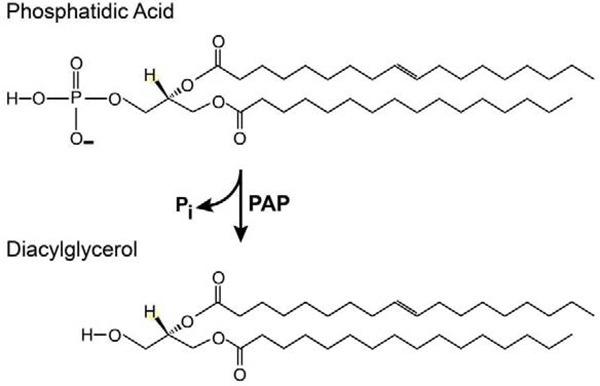
The focus of this review will be the conversion of PA into DAG by phosphatidate phosphatases (PAP; also referred to as phosphatidate phosphate hydrolases) (Fig. 2). Like PA, DAG can have signaling functions. Particularly well characterized are these functions for DAG produced by phospholipase C that binds to and activates protein kinases C and D (Mochly-Rosen et al., 2012; Roy et al., 2017). Similar to the production of PA, PA’s conversion to DAG is compartmentalized and carried out by separate groups of PAP enzymes that serve specific roles. Early biochemical characterization of PA phosphatases in both insects and mammals indicated that enzymatic activity could be stimulated by Mg2+ ions (Hirano and Gilbert, 1967; Jamdar and Fallon, 1973a). In 1973, Jamdar and Fallon reported the presence of at least two different types of PA phosphatases in rat adipose tissue that differed by their dependence on Mg2+, one being Mg2+-independent (Jamdar and Fallon, 1973b). Today, these groups are distinguished as lipins (formerly PAP1 enzymes; Mg2+-dependent) and lipid phosphate phosphatases (LPPs; formerly PAP2 enzymes; Mg2+-independent) (Brindley and Pilquil, 2009). Here, I will review studies that have identified roles of these types of PA phosphatases in insect development and metabolism.
Figure 2.
Reaction catalyzed by PAP enzymes. The general structure of phosphatidic acids and diacylglycerols is shown. Different species of these glycerolipids exist in cells that are specified by their fatty acid side chains.
2. Lipins
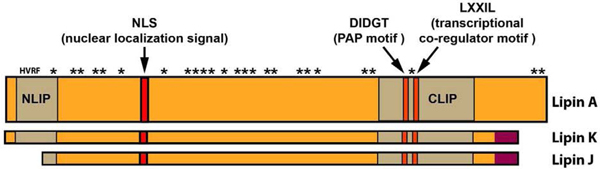
The first gene encoding an animal PAP1 enzyme was identified in 2001 in the mouse. However, it took six more years until the product was shown to catalyze the conversion of PA into DAG (Donkor et al., 2007; Peterfy et al., 2001). Mutations in the gene, originally referred to as the fatty liver dystrophy (fld) gene (Langner et al., 1989), cause fatty liver and a severe lipodystrophy in mice (Peterfy et al., 2001). Upon cloning of the gene, it was renamed as lipin-1 (Lpin1) (Peterfy et al., 2001). The mouse and human genomes encode three lipin paralogs, whereas Drosophila and other insects have a single lipin gene (Alves-Bezerra and Gondim, 2012; Csaki and Reue, 2010; Kanost et al., 2016; Ugrankar et al., 2011; Valente et al., 2010). The basic structure of lipins, as outlined in figure 3 for Drosophila Lipin, as well as the basic functions of these enzymes, are conserved in all eukaryotes, from yeasts and plants to protozoans and vertebrates (Peterfy et al., 2001; Pillai et al., 2017).
Figure 3.
Structure of the Drosophila Lipin protein. Lipin A is the most widely expressed isoform, whereas Lipin K dominates in the central nervous system and Lipin J is specific to the male gonads (Valente et al., 2010). The N-terminal and C-terminal NLIP and CLIP domains are highly conserved between lipins from yeasts, plants, and animals (Peterfy et al., 2001). CLIP harbors two conserved sequence motifs characteristic of PA phosphatases and transcriptional co-regulators. The asterisks indicate serine and threonine phosphorylation sites identified by mass spectrometry (Bodenmiller et al., 2008; Bridon et al., 2012). The HVRF motif is a recognition site of protein phosphatase PP-1c (Kok et al., 2014).
Characterization of insect lipins has so far been limited to Drosophila melanogaster (Schmitt et al., 2015; Ugrankar et al., 2011; Valente et al., 2010). Similar to the phenotype caused by mutations in the mouse Lpin1 gene, fruit fly larvae lacking Lipin display a severe lipodystrophy and increased hemolymph sugar levels indicating insulin resistance (Schmitt et al., 2015; Ugrankar et al., 2011). The Lipin protein is broadly expressed in Drosophila tissues including the fat body, the gut, the Malpighian tubules, the nervous system, the gonads, and the endocrine ring gland (Ugrankar et al., 2011; Valente et al., 2010). The Lipin gene encodes at least three protein isoforms that have been characterized to some extent, and the presence of additional isoforms is predicted (FlyBase version FB2020_03, released June 16, 2020). Some of the characterized isoforms show stage and tissue specificity in their expression (Valente et al., 2010). They differ in their C and N termini, but all contain intact catalytic and transcriptional co-regulator motifs. The conserved sequence motifs are part of the CLIP domain that is highly conserved between lipins from different species. A second highly conserved domain, the NLIP domain is located near the N terminus of the protein (Fig. 3). This domain contains an HVRF motif which constitutes a binding site for the catalytic subunit of protein phosphatase 1 (PP-1c). Interestingly, deletion of this motif completely abrogates PAP activity and nuclear translocation of lipin-1, indicating that the NLIP domain is required for both enzymatic and nuclear functions of lipin-1 (Kok et al., 2014). The Drosophila Lipin J isoform, which is specifically expressed in testis, lacks this conserved motif (Valente et al., 2010) (Fig. 3). Consistent with the predicted enzymatic inactivity of this isoform, TAG levels are low in testis, raising the question what the function of Lipin J is in this tissue. In general, it will be an important goal for future studies to identify functional differences between the Lipin isoforms. With today’s availability of CRISPR/Cas9 mutagenesis, this should be an achievable task.
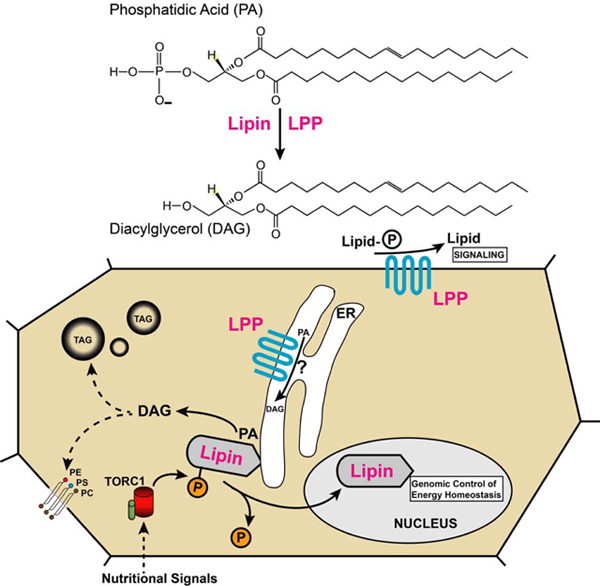
Lipins are highly phosphorylated proteins and the phosphorylation status controls intracellular localization of the protein. When nutrients are scarce, Drosophila Lipin and other lipins migrate into the cell nucleus. Nuclear translocation of mouse lipin-1 and Drosophila Lipin is under control of the nutrient-sensitive target of rapamycin (TOR) C1 pathway (Peterson et al., 2011; Schmitt et al., 2015). TOR is a serine/threonine kinase that has been shown to directly phosphorylate lipin-1, leading to retention of the protein in the cytoplasm (Peterson et al., 2011). Upon dephosphorylation, when TORC1 activity is low, dephosphorylated forms of lipins translocate to the ER and into the cell nucleus. Phosphatases involved in dephosphorylation of lipin-1 are CTDNEP1 and NEP1-R1, and potentially the above-mentioned pP-1c (S. Han et al., 2012; Kim et al., 2007; Wu et al., 2011). The Drosophila homolog of CTDNEP1, Dullard, genetically interacts with Lipin in wing vein formation (Liu et al., 2011). However, it yet has to be shown that Dullard directly contributes to the dephosphorylation of Lipin.
Flies expressing reduced amounts of Lipin exhibit increased sensitivity to starvation (Ugrankar et al., 2011). A similar increase is observed in flies generated by CRISPR/Cas9 mutagenesis that entirely rely on Lipin lacking the nuclear translocation signal (NLS) located between the conserved NLIP and CLIP domains (Fig. 3). The LipinΔNLS flies have altered gene expression profiles in both the starved and fed state, which correlate with decreased survival during starvation and, notably, increased survival in the fed state (S.E. Hood, X.V. Kofler, Q. Chen, J. Scott, J. Ortega and M.L., manuscript in preparation). Genes showing changed expression in LipinΔNLS flies are involved in energy metabolism, feeding behavior, and the immune response. Specifically, in the starved state, genes involved in lipolysis and carbohydrate breakdown are upregulated, suggesting that the flies exhaust energy reserves more rapidly than control flies. In the fed state, on the other hand, the changes in gene expression suggest a shift from fatty acid β-oxidation to lipogenesis, which would predict reduced energy production in LipinΔNLS flies. Indeed, in support of this hypothesis, LipinΔNLS flies exhibit a lowered metabolic rate. In the mouse, the genomic response to lipin-1 overexpression in the liver points to a similar involvement of the protein in the control of lipid and energy metabolism (Finck et al., 2006). Thus, the genomic responses to lipin are at least partially conserved between mammals and insects.
The CLIP domain harbors a transcriptional co-regulator motif and studies in mice and yeasts suggest that nuclear lipins indeed function as transcriptional co-regulators (Finck et al., 2006; Santos-Rosa et al., 2005). However, the corresponding mechanism of action has yet to be confirmed for Drosophila Lipin or the Lipin protein of another insect. Interestingly, mouse lipin-1 can alter nuclear localization of SREBP1, a key transcription factor in the activation of lipogenic genes (Kunte et al., 2006; Porstmann et al., 2008). This implies that a subset of lipin-1-responsive genes is controlled through this indirect mechanism (Peterson et al., 2011). However, it is unknown if Drosophila Lipin has a similar effect on SREBP1 localization.
In both mice and Drosophila, tissues that lack lipin exhibit insulin resistance, which leads to an increase in hemolymph sugar levels in fruit fly larvae and makes Lpin1 mutant mice more susceptible to atherosclerosis (Reue et al., 2000; Schmitt et al., 2015). Genetic mosaic studies in Drosophila have shown that insulin pathway activity is cell autonomously downregulated in fat body cells that lack the PAP activity provided by Lipin. A nuclear function of Lipin does not seem to be required for generating insulin sensitivity, suggesting that it is the change in the intracellular concentration of a metabolite or metabolites in the wake of PAP reduction that causes insulin resistance (Schmitt et al., 2015). Candidates for this function are ceramides, which are elevated in cultured myotubes after Lpin1 knockdown. The increase in insulin resistance that is observed in these cells after Lpin1 knockdown is ameliorated by blocking ceramide synthesis (Huang et al., 2017). It will be important to determine in more detail in future studies the mechanism by which Drosophila Lipin influences insulin sensitivity and how it exerts its effects on gene expression.
Lipin is a broadly expressed gene in Drosophila, suggesting that it functions not only in fat body development and fat storage. A function in adult development is suggested by the occasional occurrence of notched wings in the few adult flies that the hypomorphic Lipine00680 mutant produces (Ugrankar, 2011). Indeed, recent studies have shown that expression of Lipin in the wing imaginal disc is required for normal wing development, apparently through a requirement for disc patterning by morphogenetic BMP signaling (Duy Binh et al., 2019; Liu et al., 2011; Raftery and Umulis, 2012). It will be interesting to further dissect the mechanism through which Lipin interacts with signaling pathways during wing development. More generally, it will be important to address in future studies whether lipins, similar to the PLDs, play roles in signaling processes and, if so, how. In a first step in this direction, it has been shown that mouse lipin-1 is required for activation of the protein kinase D (PKD)-Vps34 phosphatidylinositol 3-kinase signaling pathway that controls autolysosome formation during autophagy in mouse skeletal muscle (P. Zhang et al., 2014). This is the first example showing that a lipin can produce DAG that acts as a signaling molecule. While the source of the PA used in this pathway remains unknown, one obvious candidate is the breakdown of PC by PLD.
3. Lipid phosphate phosphatases (LPPs)
The cloning of the first cDNA encoding a vertebrate LPP from mouse kidney was reported in 1996. This was shortly followed by a report describing the Drosophila Wunen protein and its homology to the vertebrate LPP (Kai et al., 1996; N. Zhang et al., 1997). In addition to wunen (wun), the Drosophila genome harbors six genes that encode proteins with homology to vertebrate LPPs. All contain six putative transmembrane domains and three other domains that are characteristic of LPPs (Garcia-Murillas et al., 2006). While four of the LPP homologs are as yet uncharacterized, three of them, Wun, Wunen 2 (Wun2), and Lazaro (Laza), have undergone some functional characterization (Burnett and Howard, 2003; N. Zhang et al., 1997; 1996). In contrast, the mammalian genome encodes only three LPPs, LPP1, LPP2, and LPP3. Therefore, it appears that LPPs have diversified to a larger extent in the invertebrate lineage compared to vertebrates.
Like lipins, LPPs can dephosphorylate PA to generate DAG. However, LPPs have a broader substrate specificity that includes other lipid phosphates such as LPA, sphingosine 1-phosphate (S1P), ceramide 1-phosphate, and diacylglycerol pyrophosphate (Brindley and Pilquil, 2009). Therefore, identification of the specific substrate(s) and product(s) involved is a crucial, but challenging task when studying the biological roles of LPPs. In contrast to lipins, which are soluble proteins, LPPs are integral membrane proteins that are anchored in membranes through six transmembrane domains. When located in the plasma membrane of the cell, they exhibit extracellular catalytic activity that contributes to cell-cell signaling. LPPs are also found in the membranes of the ER and Golgi apparatus where catalytic activity is presumably directed toward the lumen (Tang et al., 2015). While preferred extracellular substrates of mammalian LPPs are LPA and S1P, PA is being discussed as a major substrate of intracellular activities of LPPs (Tang et al., 2015). For instance, LPP3 present in the Golgi apparatus of HeLa cells dephosphorylates de novo synthesized PA to generate DAG that is required for retrograde vesicle trafficking from the Golgi to the ER. Reduction of LPP3 disrupts this process (Gutiérrez-Martínez et al., 2013).
The Wun and Wun2 proteins of Drosophila are most similar to mammalian LPP3. That LPP3 represents the Wun/Wun2 ortholog in mammals is supported by the observation that LPP3, but not LPP1, shows the same biological activity in Drosophila as Wun (Burnett and Howard, 2003). Both Wun and Wun2 play an essential role in germ cell migration during Drosophila embryogenesis. Expression in the cells of somatic tissues repels migrating primordial germ cells that are homing in on the developing gonads during embryogenesis. Loss of wun and wun2 results in a scattered distribution of the germ cells, which fail to reach their gonadal target tissue (Renault and Lehmann, 2006). It is believed that the catalytic activity of Wun/Wun2 degrades an extracellular phospholipid and, thus, contributes to formation of a chemotactic gradient that guides germ cell migration (LeBlanc and Lehmann, 2017). Biochemical assays show that, in vitro, a preferred substrate of Wun is LPA, whereas activity toward PA is negligible. However, although human LPP3 exhibits the same biological activity in germ cell repulsion as Wun when expressed in Drosophila, it metabolizes LPA only very poorly. Moreover, mouse LPP1, which dephosphorylates LPA very efficiently, is ineffective in Drosophila. These data strongly suggest that LPA may not be the in vivo target of Wun, but another, yet to be identified substrate (Burnett and Howard, 2003). This conclusion is further supported by the apparent absence of homologs of the vertebrate LPA and S1P receptors in Drosophila (Ile et al., 2012).
Interestingly, wun2 is not only required in somatic tissues but also expressed in the germ cells themselves and required for their survival. Since overexpression of either wun or wun2 in somatic tissues causes germ cell death, it is believed that germ cell survival requires uptake by germ cells of a lipid that Wun2 generates by breakdown of the phospholipid that serves as the chemotactic agent (Hanyu-Nakamura et al., 2004; Renault et al., 2004; Slaidina and Lehmann, 2017). It will be interesting to see if it is possible to visualize gradient formation and to elucidate the molecular nature of the gradient-forming phospholipid. In this context, it is interesting to note that, in a Drosophila model of muscular dystrophy, increased levels of S1P can suppress the dystrophic muscle phenotype. Reducing expression of wun has the same effect, suggesting that S1P is the biologically active target of Wun, at least in this system (Pantoja et al., 2013). During normal muscle development, Wun is required for cell migration that leads to proper formation of longitudinal muscles that ensheathe the gut (Stepanik et al., 2016). Wunens are not only required for muscle development and germ cell migration, but also for normal development of the tracheae and the heart. Analysis of tracheal defects in wunen mutants indicated a requirement of wun, but not wun2, for proper functioning of septate junctions between tracheal cells leading to defects in the integrity of the tracheal lumen (Ile et al., 2012). The mechanistic basis for heart defects in wun and wun2 mutants is less well understood. Wun/wun2 mutant embryos exhibit a ‘broken-hearted’ phenotype in which cardioblasts fail to properly associate with pericardial cells and bilaterally positioned cardioblasts fail to move medially leading to breaks in the cardiac lumen. Wunens are expressed in both pericardial cells and the overlying ectoderm and mutant phenotypes can only be fully rescued if wun function is restored in both tissues (Haack et al., 2014). The implication of wunens with not only germ cell migration, but also tracheal, muscle, and heart development indicates that these lipid phosphatases have broad functions in Drosophila. This conclusion is also supported by the broad expression patterns of wunens in larval and adult tissues (Flybase, version FB2020_03, released June 16, 2020). It will be interesting to further dissect these functions and to explore whether wunens also function intracellularly and in PA dephosphorylation as their mammalian LPP3 counterpart.
A third Drosophila LPP that has been functionally characterized is Laza (Garcia-Murillas et al., 2006; Kwon and Montell, 2006; Kwon et al., 2008). Laza is required for phototransduction in the Drosophila complex eye and the regulation of thermotactic behavior. In the eye, overexpression of laza enhances the retinal degeneration phenotype of mutations in the DGK-encoding rdgA gene and loss of laza suppresses the phenotype (Garcia-Murillas et al., 2006). Before degeneration sets in, mutations in laza also attenuate and shorten the light response (Kwon and Montell, 2006). Retinal PA levels are reduced in rdgA3 flies and this reduction is strongly enhanced when LPP activity is increased. Reduction of laza also suppresses retinal degeneration induced by overexpression of PLD, supporting a model in which PLD-produced PA feeds into DAG production that causes degeneration (Kwon and Montell, 2006). Together, the data support a model in which Laza specifically metabolizes PA and cooperates with the rdgA-encoded DGK and PLD to adjust PA levels during phototransduction (Garcia-Murillas et al., 2006). It is currently unclear how PA influences photoreceptor function mechanistically. Phototransduction is accomplished by the opening of TRP Ca2+ channels that is regulated by a phospholipase C (PLC)-controlled pathway (Hardie and Juusola, 2015). PA does not seem to directly alter activity of the TRP Ca2+ channels. It has been proposed that, instead, altered PA levels act through an effect on the production of phosphatidylinositol, which is needed for feeding substrates into the PLC-controlled pathway that controls the TRP channels (Garcia-Murillas et al., 2006). TRP Ca2+channels acting downstream of a PLC pathway also control thermotactic behavior that responds to mutations in laza. Thus, Laza seems to have a more general role in controlling TRP channel activity (Kwon et al., 2008).
4. Conclusion and outlook
PA phosphatases have been primarily studied in mammalian systems and much remains to be learned about the functions of these enzymes in insects, in particular in non-drosophilids. There are still many questions that can be addressed using Drosophila by taking advantage of the genetic toolbox available for this model organism. To what extent are Lipin and the wunens integrated into signaling pathways in which PA or DAG function as ligands of downstream effectors? What are the tissue-specific functions of these PA phosphatases in the larval and adult stages? Lipin is, for instance, strongly expressed in the endocrine ring gland and in Malpighian tubules. Are there specific requirements for the presence of Lipin in hormone production or in excretion? Similarly, wun is strongly expressed in adult salivary glands and the larval and adult digestive tract. Wun2 shows relatively high expression in the ovary compared to other tissues. What are the roles that these two LPPs have in these tissues? A long-standing open question is the molecular nature of the phospholipid or -lipids that are the substrates of the exo-enzymatic activity of Wun and Wun2. Experimental evidence is needed to support the model in which uptake of a Wun2 product by primordial germ cells supports germ cell survival. What is this product and how does it prevent germ cell death? There are major open questions regarding the mechanism by which nuclear Lipin exerts its effects on gene expression. So far, it has not been shown that Lipin can bind to specific loci on fat body polytene chromosomes or that it can be located on chromosomal loci by ChIP assays. Which role, if any, does the PAP activity play in nuclear functions of Lipin? Finally, what are the functions of the four LPPs encoded in the Drosophila genome that have so far remained uncharacterized? It will be exciting to see these, and other questions answered in the coming months and years.
Highlights.
Phosphatidic acid phosphatases convert phosphatidic acid into diacylglycerol
Two classes: Mg2+-dependent lipins and Mg2+-independent lipid phosphate phosphatases (LPPs)
Lipins have essential cytoplasmic and nuclear functions in energy homeostasis
LPPs are transmembrane proteins with broader substrate specificity
Insect LPPs function in development, germ cell migration, and phototransduction
Acknowledgements
Work in the author’s laboratory is supported by the National Institutes of Health [grant number 1R15DK114748-01] and the Arkansas Biosciences Institute.
Abbreviations:
- AGPAT
1-acylglycerol-3-phosphate O-acyltransferase
- DAG
diacylglycerol
- DGAT
diacylglycerol acyltransferase
- DGk
diacylglycerol kinase
- GPAT
glycerol-3-phosphate acyltransferase
- Laza
Lazaro
- LPA
lysophosphatidic acid
- LPP
lipid phosphate phosphatase
- PA
phosphatidic acid
- PAP
phosphatidic acid phosphatase
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PLC
phospholipase C
- PLD
phospholipase D
- PI
phosphatidylinositol
- PS
phosphatidylserine
- S1P
sphingosine 1-phosphate
- TAG
triacylglycerol
- Wun
Wunen
- Wun2
Wunen 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Alves-Bezerra M, Gondim KC, 2012. Triacylglycerol biosynthesis occurs via the glycerol-3-phosphate pathway in the insect Rhodnius prolixus. Biochim. Biophys. Acta 1821, 1462–1471. doi: 10.1016/j.bbalip.2012.08.002 [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R, 2008. PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat. Biotechnol 26, 1339–1340. doi: 10.1038/nbt1208-1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridon G, Bonneil E, Muratore-Schroeder T, Caron-Lizotte O, Thibault P, 2012. Improvement of phosphoproteome analyses using FAIMS and decision tree fragmentation. application to the insulin signaling pathway in Drosophila melanogaster S2 cells. J. Proteome Res 11, 927–940. doi: 10.1021/pr200722s [DOI] [PubMed] [Google Scholar]
- Brindley DN, Pilquil C, 2009. Lipid phosphate phosphatases and signaling. J. Lipid Res 50 Suppl, S225–30. doi: 10.1194/jlr.R800055-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Howard K, 2003. Fly and mammalian lipid phosphate phosphatase isoforms differ in activity both in vitro and in vivo. EMBO Rep. 4, 793–799. doi: 10.1038/sj.embor.embor900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M, Sampaio JL, Palm W, Brankatschk M, Eaton S, Shevchenko A, 2012. Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol 8, 600. doi: 10.1038/msb.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaki LS, Reue K, 2010. Lipins: multifunctional lipid metabolism proteins. Annu. Rev. Nutr 30, 257–272. doi: 10.1146/annurev.nutr.012809.104729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K, 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem 282, 3450–3457. doi: 10.1074/jbc.M610745200 [DOI] [PubMed] [Google Scholar]
- Duy Binh T, L A Pham T, Nishihara T, Thanh Men T, Kamei K, 2019. The Function of Lipin in the Wing Development of Drosophila melanogaster. Int. J. Mol. Sci 20, 3288. doi: 10.3390/ijms20133288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Kelly DP, 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 4, 199–210. doi: 10.1016/j.cmet.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Foster DA, 2013. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol. Metab 24, 272–278. doi: 10.1016/j.tem.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Murillas I, Pettitt T, Macdonald E, Okkenhaug H, Georgiev P, Trivedi D, Hassan B, Wakelam M, Raghu P, 2006. lazaro encodes a lipid phosphate phosphohydrolase that regulates phosphatidylinositol turnover during Drosophila phototransduction. Neuron 49, 533–546. doi: 10.1016/j.neuron.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Martínez E, Fernández-Ulibarri I, Lázaro-Diéguez F, Johannes L, Pyne S, Sarri E, Egea G, 2013. Lipid phosphate phosphatase 3 participates in transport carrier formation and protein trafficking in the early secretory pathway. J. Cell Sci 126, 2641–2655. doi: 10.1242/jcs.117705 [DOI] [PubMed] [Google Scholar]
- Haack T, Schneider M, Schwendele B, Renault AD, 2014. Drosophila heart cell movement to the midline occurs through both cell autonomous migration and dorsal closure. Dev. Biol 396, 169–182. doi: 10.1016/j.ydbio.2014.08.033 [DOI] [PubMed] [Google Scholar]
- Han G-S, O’Hara L, Carman GM, Siniossoglou S, 2008. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem 283, 20433–20442. doi: 10.1074/jbc.M802903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM, 2012. Nuclear envelope phosphatase 1-regulatory subunit 1 (formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J. Biol. Chem 287, 3123–3137. doi: 10.1074/jbc.M111.324350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Kobayashi S, Nakamura A, 2004. Germ cell-autonomous Wunen2 is required for germline development in Drosophila embryos. Development 131, 4545–4553. doi: 10.1242/dev.01321 [DOI] [PubMed] [Google Scholar]
- Hardie RC, Juusola M, 2015. Phototransduction in Drosophila. Curr. Opin. Neurobiol 34, 37–45. doi: 10.1016/j.conb.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Hirano C, Gilbert LI, 1967. Phosphatidate phosphohydrolase in the fat body of Hyalophora cecropia. Journal of Insect Physiology 13, 163–174. [Google Scholar]
- Huang S, Huang S, Wang X, Zhang Q, Liu J, Leng Y, 2017. Downregulation of lipin-1 induces insulin resistance by increasing intracellular ceramide accumulation in C2C12 myotubes. Int. J. Biol. Sci 13, 1–12. doi: 10.7150/ijbs.17149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ile KE, Tripathy R, Goldfinger V, Renault AD, 2012. Wunen, a Drosophila lipid phosphate phosphatase, is required for septate junction-mediated barrier function. Development 139, 2535–2546. doi: 10.1242/dev.077289 [DOI] [PubMed] [Google Scholar]
- Jacquemyn J, Cascalho A, Goodchild RE, 2017. The ins and outs of endoplasmic reticulum-controlled lipid biosynthesis. EMBO Rep. 18, 1905–1921. doi: 10.15252/embr.201643426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamdar SC, Fallon HJ, 1973a. Glycerolipid biosynthesis in rat adipose tissue. I. Properties and distribution of glycerophosphate acyltransferase and effect of divalent cations on neutral lipid formation. J. Lipid Res 14, 509–516. [PubMed] [Google Scholar]
- Jamdar SC, Fallon HJ, 1973b. Glycerolipid synthesis in rat adipose tissue. II. Properties and distribution of phosphatidate phosphatase. J. Lipid Res 14, 517–524. [PubMed] [Google Scholar]
- Kai M, Wada I, Imai S, Sakane F, Kanoh H, 1996. Identification and cDNA cloning of 35-kDa phosphatidic acid phosphatase (type 2) bound to plasma membranes. Polymerase chain reaction amplification of mouse H2O2-inducible hic53 clone yielded the cDNA encoding phosphatidic acid phosphatase. J. Biol. Chem 271, 18931–18938. doi: 10.1074/jbc.271.31.18931 [DOI] [PubMed] [Google Scholar]
- Kanost MR, Arrese EL, Cao X, Chen Y-R, Chellapilla S, Goldsmith MR, Grosse-Wilde E, Heckel DG, Herndon N, Jiang H, Papanicolaou A, Qu J, Soulages JL, Vogel H, Walters J, Waterhouse RM, Ahn S-J, Almeida FC, An C, Aqrawi P, Bretschneider A, Bryant WB, Bucks S, Chao H, Chevignon G, Christen JM, Clarke DF, Dittmer NT, Ferguson LCF, Garavelou S, Gordon KHJ, Gunaratna RT, Han Y, Hauser F, He Y, Heidel-Fischer H, Hirsh A, Hu Y, Jiang H, Kalra D, Klinner C, König C, Kovar C, Kroll AR, Kuwar SS, Lee SL, Lehman R, Li K, Li Z, Liang H, Lovelace S, Lu Z, Mansfield JH, McCulloch KJ, Mathew T, Morton B, Muzny DM, Neunemann D, Ongeri F, Pauchet Y, Pu L-L, Pyrousis I, Rao X-J, Redding A, Roesel C, Sanchez-Gracia A, Schaack S, Shukla A, Tetreau G, Wang Y, Xiong G-H, Traut W, Walsh TK, Worley KC, Wu D, Wu W, Wu Y-Q, Zhang X, Zou Z, Zucker H, Briscoe AD, Burmester T, Clem RJ, Feyereisen R, Grimmelikhuijzen CJP, Hamodrakas SJ, Hansson BS, Huguet E, Jermiin LS, Lan Q, Lehman HK, Lorenzen M, Merzendorfer H, Michalopoulos I, Morton DB, Muthukrishnan S, Oakeshott JG, Palmer W, Park Y, Passarelli AL, Rozas J, Schwartz LM, Smith W, Southgate A, Vilcinskas A, Vogt R, Wang P, Werren J, Yu X-Q, Zhou J-J, Brown SJ, Scherer SE, Richards S, Blissard GW, 2016. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem. Mol. Biol 76, 118–147. doi: 10.1016/j.ibmb.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Dixon JE, 2007. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc. Natl. Acad. Sci. U.S.A 104, 6596–6601. doi: 10.1073/pnas.0702099104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok BPC, Skene-Arnold TD, Ling J, Benesch MGK, Dewald J, Harris TE, Holmes CFB, Brindley DN, 2014. Conserved residues in the N terminus of lipin-1 are required for binding to protein phosphatase-1c, nuclear translocation, and phosphatidate phosphatase activity. J. Biol. Chem 289, 10876–10886. doi: 10.1074/jbc.M114.552612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, Chupin V, de Kruijff B, Burger KNJ, 2003. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 4, 162–174. doi: 10.1034/j.1600-0854.2003.00086.X [DOI] [PubMed] [Google Scholar]
- Kunte AS, Matthews KA, Rawson RB, 2006. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 3, 439–448. doi: 10.1016/j.cmet.2006.04.011 [DOI] [PubMed] [Google Scholar]
- Kwon Y, Montell C, 2006. Dependence on the Lazaro phosphatidic acid phosphatase for the maximum light response. Curr. Biol 16, 723–729. doi: 10.1016/j.cub.2006.02.057 [DOI] [PubMed] [Google Scholar]
- Kwon Y, Shim H-S, Wang X, Montell C, 2008. Control of thermotactic behavior via coupling of a TRP channel to a phospholipase C signaling cascade. Nat. Neurosci 11, 871–873. doi: 10.1038/nn.2170 [DOI] [PubMed] [Google Scholar]
- LaLonde M, Janssens H, Yun S, Crosby J, Redina O, Olive V, Altshuller YM, Choi S-Y, Du G, Gergen JP, Frohman MA, 2006. A role for Phospholipase D in Drosophila embryonic cellularization. BMC Dev. Biol 6, 60–13. doi: 10.1186/1471-213X-6-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner CA, Birkenmeier EH, Ben-Zeev O, Schotz MC, Sweet HO, Davisson MT, Gordon JI, 1989. The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J. Biol. Chem 264, 7994–8003. [PubMed] [Google Scholar]
- LeBlanc MG, Lehmann R, 2017. Domain-specific control of germ cell polarity and migration by multifunction Tre1 GPCR. J. Cell Biol 216, 2945–2958. doi: 10.1083/jcb.201612053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Matsuoka S, Enoki A, Yamamoto T, Furukawa K, Yamasaki Y, Nishida Y, Sugiyama S, 2011. Negative modulation of bone morphogenetic protein signaling by Dullard during wing vein formation in Drosophila. Dev. Growth Differ 53, 822–841. doi: 10.1111/j.1440-169X.2011.01289.x [DOI] [PubMed] [Google Scholar]
- Mérida I, Avila-Flores A, Merino E, 2008. Diacylglycerol kinases: at the hub of cell signalling. Biochem. J 409, 1–18. doi: 10.1042/BJ20071040 [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D, Das K, Grimes KV, 2012. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov 11, 937–957. doi: 10.1038/nrd3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja M, Fischer KA, leronimakis N, Reyes M, Ruohola-Baker H, 2013. Genetic elevation of sphingosine 1-phosphate suppresses dystrophic muscle phenotypes in Drosophila. Development 140, 136–146. doi: 10.1242/dev.087791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K, 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet 27, 121–124. doi: 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM, 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420. doi: 10.1016/j.cell.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai AN, Shukla S, Rahaman A, 2017. An evolutionarily conserved phosphatidate phosphatase maintains lipid droplet number and endoplasmic reticulum morphology but not nuclear morphology. Biol Open 6, 1629–1643. doi: 10.1242/bio.028233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung Y-L, Schulze A, 2008. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8, 224–236. doi: 10.1016/j.cmet.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery LA, Umulis DM, 2012. Regulation of BMP activity and range in Drosophila wing development. Current Opinion in Cell Biology 24, 158–165. doi: 10.1016/j.ceb.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault AD, Lehmann R, 2006. Follow the fatty brick road: lipid signaling in cell migration. Curr. Opin. Genet. Dev 16, 348–354. doi: 10.1016/j.gde.2006.06.015 [DOI] [PubMed] [Google Scholar]
- Renault AD, Sigal YJ, Morris AJ, Lehmann R, 2004. Soma-germ line competition for lipid phosphate uptake regulates germ cell migration and survival. Science 305, 1963–1966. doi: 10.1126/science.1102421 [DOI] [PubMed] [Google Scholar]
- Reue K, Xu P, Wang XP, Slavin BG, 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res 41, 1067–1076. [PubMed] [Google Scholar]
- Roth MG, Bi K, Ktistakis NT, Yu S, 1999. Phospholipase D as an effector for ADP-ribosylation factor in the regulation of vesicular traffic. Chem. Phys. Lipids 98, 141–152. doi: 10.1016/s0009-3084(99)00026-2 [DOI] [PubMed] [Google Scholar]
- Roy A, Ye J, Deng F, Wang QJ, 2017. Protein kinase D signaling in cancer: A friend or foe? Biochim Biophys Acta Rev Cancer 1868, 283–294. doi: 10.1016/j.bbcan.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S, 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941. doi : 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Ugrankar R, Greene SE, Prajapati M, Lehmann M, 2015. Drosophila Lipin interacts with insulin and TOR signaling pathways in the control of growth and lipid metabolism. J. Cell Sci 128, 4395–4406. doi: 10.1242/jcs.173740 [DOI] [PubMed] [Google Scholar]
- Selvy PE, Lavieri RR, Lindsley CW, Brown HA, 2011. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev 111, 6064–6119. doi: 10.1021/cr200296t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddhanta A, Shields D, 1998. Secretory vesicle budding from the trans-Golgi network is mediated by phosphatidic acid levels. J. Biol. Chem 273, 17995–17998. doi: 10.1074/jbc.273.29.17995 [DOI] [PubMed] [Google Scholar]
- Slaidina M, Lehmann R, 2017. Quantitative Differences in a Single Maternal Factor Determine Survival Probabilities among Drosophila Germ Cells. Curr. Biol 27, 291–297. doi: 10.1016/j.cub.2016.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanik V, Dunipace L, Bae Y-K, Macabenta F, Sun J, Trisnadi N, Stathopoulos A, 2016. The migrations of Drosophila muscle founders and primordial germ cells are interdependent. Development 143, 3206–3215. doi: 10.1242/dev.134346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Benesch MGK, Brindley DN, 2015. Lipid phosphate phosphatases and their roles in mammalian physiology and pathology. J. Lipid Res 56, 2048–2060. doi: 10.1194/jlr.R058362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrankar R, 2011. Lipin is a central regulator of fat body development and function in Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M, 2011. Lipin is a central regulator of adipose tissue development and function in Drosophila melanogaster. Mol. Cell. Biol 31, 1646–1656. doi: 10.1128/MCB.01335-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente V, Maia RM, Vianna MCB, Paçó-Larson ML, 2010. Drosophila melanogaster lipins are tissue-regulated and developmentally regulated and present specific subcellular distributions. FEBS J. 277, 4775–4788. doi: 10.1111/j.1742-4658.2010.07883.x [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW, 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol 9, 112–124. doi: 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng J-X, Graham M, Christiano R, Fröhlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV, Walther TC, 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24, 384–399. doi: 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Garland M, Dunaway-Mariano D, Allen KN, 2011. Homo sapiens dullard protein phosphatase shows a preference for the insulin-dependent phosphorylation site of lipin1. Biochemistry 50, 3045–3047. doi: 10.1021/bi200336b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhang J, Cheng Y, Howard K, 1996. Identification and genetic analysis of wunen, a gene guiding Drosophila melanogaster germ cell migration. Genetics 143, 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhang J, Purcell KJ, Cheng Y, Howard K, 1997. The Drosophila protein Wunen repels migrating germ cells. Nature 385, 64–67. doi: 10.1038/385064a0 [DOI] [PubMed] [Google Scholar]
- Zhang P, Verity MA, Reue K, 2014. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20, 267–279. doi: 10.1016/j.cmet.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D, 2007. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol 9, 706–712. doi: 10.1038/ncb1594 [DOI] [PubMed] [Google Scholar]