Abstract
Background
The coronavirus disease-19 (COVID-19) emerged in Wuhan, China and rapidly spread worldwide. Researchers are trying to find a way to treat this disease as soon as possible. The present study aimed to identify the genes involved in COVID-19 and find a new drug target therapy. Currently, there are no effective drugs targeting SARS-CoV-2, and meanwhile, drug discovery approaches are time-consuming and costly. To address this challenge, this study utilized a network-based drug repurposing strategy to rapidly identify potential drugs targeting SARS-CoV-2. To this end, seven potential drugs were proposed for COVID-19 treatment using protein-protein interaction (PPI) network analysis. First, 524 proteins in humans that have interaction with the SARS-CoV-2 virus were collected, and then the PPI network was reconstructed for these collected proteins. Next, the target miRNAs of the mentioned module genes were separately obtained from the miRWalk 2.0 database because of the important role of miRNAs in biological processes and were reported as an important clue for future analysis. Finally, the list of the drugs targeting module genes was obtained from the DGIDb database, and the drug-gene network was separately reconstructed for the obtained protein modules.
Results
Based on the network analysis of the PPI network, seven clusters of proteins were specified as the complexes of proteins which are more associated with the SARS-CoV-2 virus. Moreover, seven therapeutic candidate drugs were identified to control gene regulation in COVID-19. PACLITAXEL, as the most potent therapeutic candidate drug and previously mentioned as a therapy for COVID-19, had four gene targets in two different modules. The other six candidate drugs, namely, BORTEZOMIB, CARBOPLATIN, CRIZOTINIB, CYTARABINE, DAUNORUBICIN, and VORINOSTAT, some of which were previously discovered to be efficient against COVID-19, had three gene targets in different modules. Eventually, CARBOPLATIN, CRIZOTINIB, and CYTARABINE drugs were found as novel potential drugs to be investigated as a therapy for COVID-19.
Conclusions
Our computational strategy for predicting repurposable candidate drugs against COVID-19 provides efficacious and rapid results for therapeutic purposes. However, further experimental analysis and testing such as clinical applicability, toxicity, and experimental validations are required to reach a more accurate and improved treatment. Our proposed complexes of proteins and associated miRNAs, along with discovered candidate drugs might be a starting point for further analysis by other researchers in this urgency of the COVID-19 pandemic.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12896-021-00680-z.
Introduction
A novel coronavirus (i.e., COVID-19) has led to the emergence of a major outbreak in the world. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the main cause of coronavirus disease which has turned to become an international concern worldwide [1–4]. According to the latest reports of the COVID-19 situation in the world, as of 15 November 2020, 53.7 million confirmed cases and 1.3 million deaths occurred worldwide [5]. SARS-CoV-2 is a positive-sense single-stranded RNA genome. Based on the latest genome assemblies of this virus, it contains 12 proteins, 11 genes, GC 38%, and the size of 29.9 KB (https://www.ncbi.nlm.nih.gov/genome), with a 5′-cap structure and a 3′-poly-A tail [6].
miRNAs are small molecules of non-coding RNA that inhibit the translation of mRNAs in prokaryotes and eukaryotes [7, 8]. Recent evidence has confirmed the role of pathologic processes in miRNAs, including inflammatory responses and viral infection. It has been demonstrated that miR-9, miR-98, miR-223, and miR-214 expressions in COVID-19-infected host cells should be changed, and consequently, leading to amendments in cytokines manufacturing [9].
A distinguishing proof of host factors, especially genes for contamination is basic to educate systems regarding COVID-19 pathogenesis, uncover varieties in having powerlessness, and recognize novel host-coordinated treatments, which may have viability against current and future pandemic coronaviruses [10]. Recent research has represented and validated anti-viral genes (e.g., CABIN1, HIRA, and ASF1A). These genes potentially provide protection from SARS-CoV-2 [11, 12]. A number of studies have focused on the immune system [12–15]. Melo et al. [16] detailed a moderate interferon (IFN) reaction to SARS-CoV-2 disease in essential cells and indicated that IFN can decrease SARS-CoV-2 replication in vitro. Moreover, Zhou et al. [17] and Xiong et al. [18] analyzed the inborn invulnerable reaction to SARS-CoV-2 contamination in the bronchoalveolar lavage liquid (BALF). These examinations detailed the noteworthy upregulation of a subset of interferon-invigorated qualities (ISGs) which are legitimately identified with an antiviral action (e.g., ISG15, IFIH1, MX1, OAS1–3, and IFITMs). Prasad et al. reported that several genes of the host (e.g., OAS1–3, IRF7, IRF9, STAT1, and IFIH1) are exceptionally communicated and profoundly related to reactions to viral contaminations [19]. Likewise, Cava et al. found that nine genes (i.e., LRRK2, ACSL5, HSD17B4, EPHX1, MCCC2, GSTA4, ACACA, HGD, and ROS1) were positively correlated with angiotensin-converting enzyme 2 while one gene (CRIP2) was negatively correlated with this enzyme [20].
miRNA-based therapy could be proposed for SARS-CoV-2 treatment through viral genome suppression [21]. Antibodies or anther antiviral medications are not yet accessible for COVID-1 9 contamination medicines [22]. In a short time after this pandemic infection, many scientists seek to identify the involved host genes and proteins in diseases to find a new therapy [17, 23].
Concerning the current speed of SARS-CoV-2 spread in the world, the drug repurposing strategy is a more immediate and effective method of drug discovery [24, 25]. Recently, multiple studies have utilized network-based and computational approaches for repurposing candidate drugs for COVID-19 [26, 27]. Network-based approaches have demonstrated their effectiveness in the identification of repurposable drugs for various human diseases during the last decade [28, 29].
Considering the above-mentioned explanations, the present study aimed to identify the genes involved in COVID-19 and find a new drug target therapy. Currently, there are no effective drugs targeting SARS-CoV-2 and drug discovery approaches are considered costly and time-consuming. In this regard, this study applied a network-based drug repurposing strategy in order to rapidly identify potential drugs targeting SARS-CoV-2. For this purpose, we used protein-protein interaction (PPI) network and computational tools with resources for genomics and proteomics. Appropriate drugs might be useful for understanding viral disease mechanisms and designing and developing anti-viral agents.
Material and methods
Dataset and preprocessing
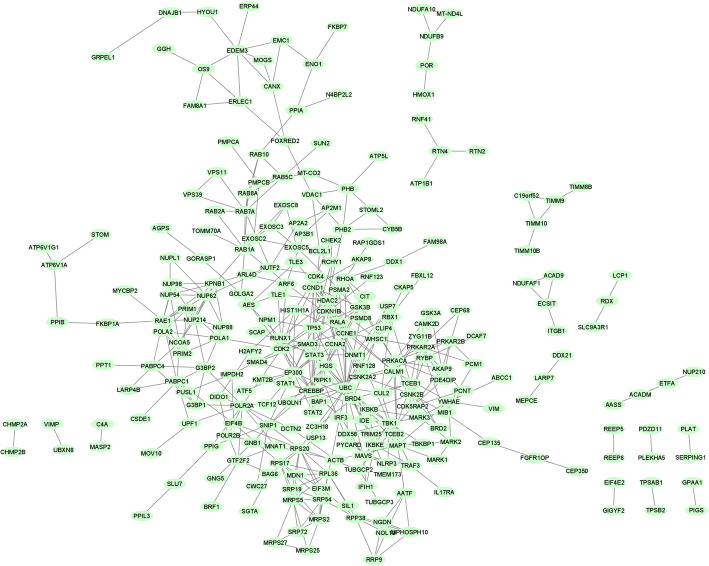
In this work, coronavirus interaction data release was downloaded from the BioGRID database [30] containing the most up-to-date version of genes interacted in the COVID-19. This dataset contained 928 entries (https://downloads.thebiogrid.org/BioGRID/Latest-Release), and 524 genes remained after preprocessing. For preprocessing, all data were checked out and the duplicate gene symbols were eliminated, and then the missing gene symbols were extracted from the NCBI database using the Entrez ID and imported into the data. Next, the PPI network was extracted for our gene dataset (Fig. 1) by applying the STRING database, version 11.0 [31]. To this end, our search was limited to those experimentally validated interactors which had direct interaction with our gene list. Afterward, the PPI network was plotted using Cytoscape 3.6.0 [32].
Fig. 1.
Global PPI network based on Gathered 524 Proteins. Note. PPI: Protein-protein interaction. Every node corresponds to a protein, and edges show experimentally validated physical interactions
Clustering and network analysis
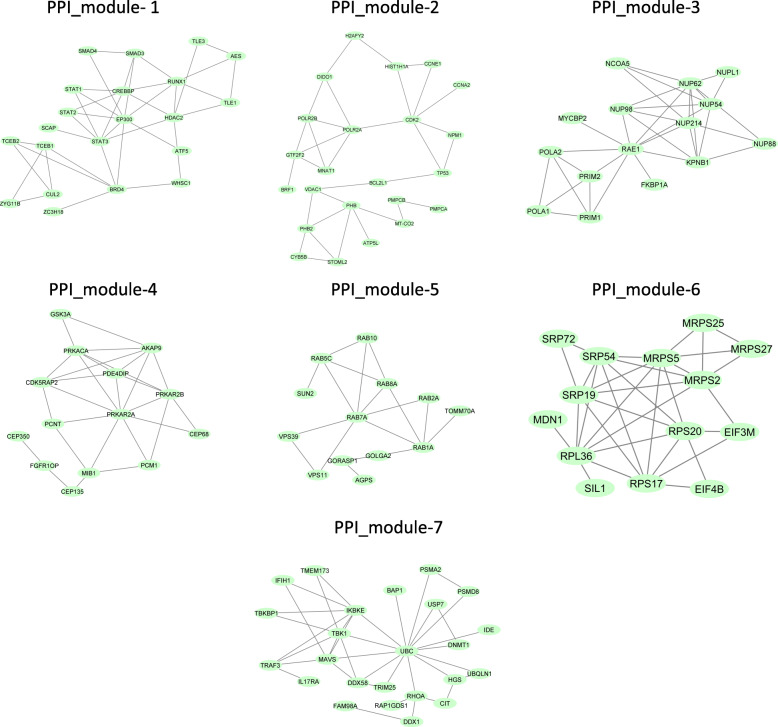
For the next analysis of this biological network, ClusterViz [33] was utilized for clustering the highly physically interconnected modules of proteins (protein complexes). The cluster analysis of biological networks (e.g., PPI or gene networks) is one of the most common strategies for detecting protein complexes or functional modules. Clustervize is a user-friendly and platform-independent plugin for cytoscape which facilitates the operation of the user [33]. ClusterViz uses different algorithms to perform clustering analysis. The fast agglomerate edge clustering (FAG-EC) algorithm was selected based on the aim of this study. FAG-EC is a fast agglomerative hierarchical clustering algorithm that functions based on edge clustering coefficients. First, this algorithm calculates the edge clustering coefficient for each edge in the network. Then, edges are sorted in a non-increasing order according to clustering coefficients. The complexes of proteins are detected according to the bottom-up condensing the hierarchical clustering algorithm. Due to the low complexity and fast computational power, the FAG-EC algorithm is effective for analyzing large protein networks [34]. The selected parameters of the algorithm included DefinitionWay: Weak, In/OutThreshold: 1.0, Overlapped: true, CliqueSizeThreshold: 3, and OutputThreshold: 10. Totally, seven different PPI clusters (i.e., PPI complexes) were discovered based on the analysis. Figure 2 shows these seven PPI modules, and the list of genes for every cluster (module) is provided in supplementary file S2.
Fig. 2.
Obtained PPI modules from PPI networks. Note. Protein-protein interaction. The green circles represent each module genes
mRNA-miRNA bipartite sub-network reconstruction
In the next step, the experimentally validated miRNA targets of genes were searched in the miRWalk 2.0 database in order to reconstruct the bipartite of mRNA-miRNA sub-network for each PPI module [35]. Eventually, seven bipartite mRNA-miRNA sub-networks were obtained after removing duplicate connections. In these sub-networks, each node demonstrates the genes or their miRNA targets, and each edge shows the connection between the nodes. Moreover, the sub-networks were plotted and analyzed using Cytoscape 3.6.0 [32]. The degree of each node represents the number of connections of that node with other nodes of the network. A node with a higher degree has a more important and remarkable role in the network. In general, 10 miRNAs with the highest degree for each sub-network were determined in this phase of our analysis. The details of the reconstructed bipartite mRNA-miRNA sub-networks, along with network information are reported in supplementary file S2.
Functional annotation and pathway enrichment analysis of the identified clusters of genes in SARS-CoV-2 infection
In the next phase, the functional enrichment analysis was performed for each sub-network in order to identify the biological mechanisms of important genes and miRNAs. The applied tools and databases were the gene ontology (GO) tool of DAVID1 [36] for functional enrichment analysis and the KEGG2 [37] database for the pathway enrichment analysis of genes.
Gene set enrichment analysis (GSEA)
We performed the gene set enrichment analysis as a validation method to test whether the predicted anti-COVID-19 repurposed drugs can counteract the gene expression perturbations caused by the virus. To this end we utilized the Enrichr [38] database to perform the Connectivity Map (CMAP) analysis [39]. The main concept of CMAP analysis is to compare a disease-specific gene signature with the drug-specific gene expression profiles using a comprehensive perturbation database like Connectivity Map [39, 40] that elucidate the connections between diseases, genes and drugs. We used our data set containing 524 genes as COVID-19-host signatures to evaluate the therapeutic effects of predicted drugs. To perform the CMAP analysis we submitted our genes in Enrichr database to retrieve the genes expressed up or down in cells treated with different drugs. Two data sets named CMAP-up and CMAP-down containing genes up-regulated or down-regulated respectively by various drugs were extracted. We searched for our identified repurposed drugs in CMAP data sets.
Results
PPI network and clustering analysis
The PPI network of 524 genes was extracted from the String database (Fig. 1). Details of the PPI network are provided in supplementary file S6. Then, clustering analysis was performed to find highly interacted modules of proteins. Seven modules of proteins were found, which are shown in Fig. 2 (More information related to these modules is provided in supplementary file S7).
mRNA-miRNA bipartite sub-networks
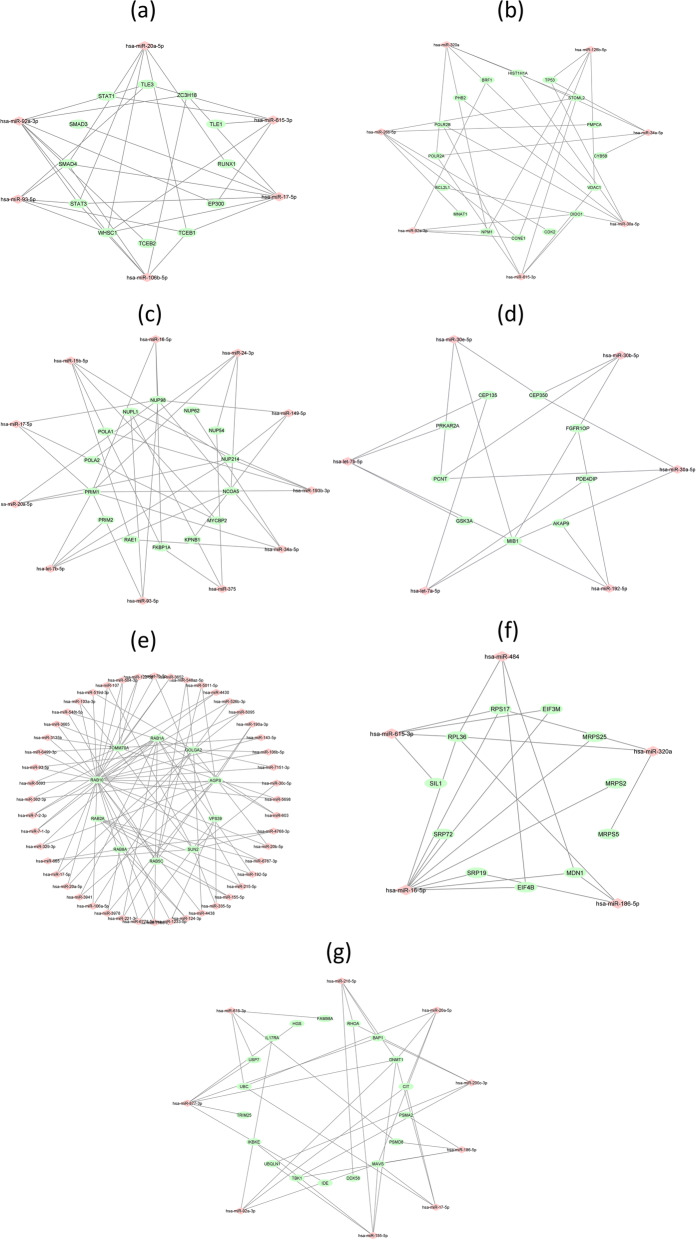
Seven mRNA-miRNA bipartite sub-networks were reconstructed from the determined PPI modules and their targeting miRNAs. Table 1 presents the detailed properties of these networks (These networks are available as supplementary files S1 in detail). miRNAs with a higher degree in the network are more effective with a more important role in post-transcriptional gene regulation processes. Eventually, high-degree miRNAs (i.e., hub miRNAs) and the associated target genes were selected as a sub-network of the original network for further investigations so that to reduce the analysis complexity. Figure 3 depicts these seven mRNA-miRNA bipartite sub-networks. The details of these seven mRNA-miRNA bipartite sub-networks are available in supplementary file S2.
Table 1.
The details of the mRNA-miRNA sub-networks
| Cluster sub-networks | mRNA no. | miRNA no. | Interactions no. |
|---|---|---|---|
| Sub-network-1 | 21 | 532 | 678 |
| Sub-network-2 | 23 | 431 | 557 |
| Sub-network-3 | 15 | 409 | 493 |
| Sub-network-4 | 14 | 300 | 364 |
| Sub-network-5 | 13 | 385 | 433 |
| Sub-network-6 | 14 | 200 | 234 |
| Sub-network-7 | 24 | 360 | 463 |
mRNA Messenger RNA, miRNA microRNA
Fig. 3.
mRNA-miRNA Bipartite Sub-networks. Note. Green circles and red diamonds represent the genes and the miRNAs, respectively. a sub-network 1, b sub-network 2, c sub-network 3, d sub-network 4, e sub-network 5, f sub-network 6, and g sub-network 7.
According to our previously explained hub-node selection criteria, some hub miRNAs and their target genes were considered as a sub-network for each cluster network. Sub-network 1 contains 18 nodes (6 miRNAs and 12 genes) and 34 interactions. In addition, sub-networks 2–7 include 23, 25, 15, 58, 16, and 27 nodes, as well as 32, 38, 19, 96, 22, and 39 interactions, respectively (Fig. 3 and Supplementary file S2). The list of miRNAs for sub-networks is reported in Table 2.
Table 2.
The details of the mRNA-miRNA Sub-networks
| Sub-network no. | miRNA no. | List of miRNAs |
|---|---|---|
| 1 | 6 | miR-106b-5p, miR-17-5p, miR-20a-5p, miR-615-3p. miR-92a-3p, miR-93-5p |
| 2 | 7 | miR-106b-5p, miR-17-5p, miR-20a-5p, miR-615-3p, miR-92a-3p, miR-93-5p |
| 3 | 11 | let-7b-5p, miR-149-5p, miR-15b-5p, miR-16-5p, miR-17-5p, miR-193b-3p, miR-20a-5p, miR-24-3p,miR-34a-5p, miR-375, miR-93-5p |
| 4 | 6 | let-7a-5p, let-7b-5p, miR-192-5p, miR-30a-5p, miR-30b-5p, miR-30e-5p |
| 5 | 48 | miR-93-5p, miR-7156-3p, miR-7151–3p, miR-7-2-3p, miR-7-1-3p, miR-6787-3p, miR-6778-5p, miR-665, miR-6499-3p, miR-603, miR-5698, miR-5693, miR-548 t-5p, miR-548az-5p, miR-526b-3p, miR-519d-3p, miR-5095, miR-504-3p, miR-5011-5p, miR-4768-3p, miR-4438, miR-4430, miR-3978, miR-3941, miR-3665, miR-3652, miR-362-3p, miR-335-5p, miR-329-3p, miR-3135b, miR-30c-5p, miR-221–3p, miR-215-5p, miR-20b-5p, miR-20a-5p, miR-192-5p, miR-190a-3p, miR-17-5p, miR-155-5p, miR-143-5p, miR-124-3p, miR-1233-5p, miR-122-5p, miR-107, miR-106b-5p, miR-106a-5p, miR-103a-3p, let-7b-5p |
| 6 | 5 | miR-16-5p, miR-186-5p, miR-320a, miR-484, miR-615-3p |
| 7 | 9 | miR-155-5p, miR-17-5p, miR-186-5p, miR-200c-3p, miR-20a-5p, miR-218-5p, miR-615-3p, miR-877-3p, miR-92a-3p |
mRNA Messenger RNA, miRNA microRNA
Totally, 92 hub miRNAs were identified in all seven sub-networks of which 69 cases were unique miRNAs.
Functional annotation and pathway enrichment analysis of PPI modules in SARS-CoV-2 infection
To study the underlying biological functions of genes in gene clusters, the DAVID [36, 41] was used to apply GO enrichment and pathway analysis. It should be noted that only those significant terms with P < 0.01 were considered in this regard. The findings of the GO enrichment analysis for each cluster of genes are provided as supplementary file S4. As reported in this file, the most significant terms for PPI-modules 1–7 were transcription from RNA polymerase II promoter, cell cycle G1/S phase transition, mitotic cell cycle phase transition, single-organism organelle organization, endomembrane system organization, translation, and negative regulation of type I interferon production, respectively.
The KEGG database was used for the pathway enrichment analysis of the genes in each PPI-module and the most significant pathway for every PPI-module, except for PPI-module_4 is reported in Table 3. There was no significant pathway for PPI-module_4. Other significant pathways for all seven PPI-modules are listed in supplementary file S5.
Table 3.
The Significant Biological KEGG Pathways for All Seven Clusters
| Cluster no. | KEGG_PATHWAY | p-Value |
|---|---|---|
| PPI_module_1 | Pathways in cancer | 1.436E-8 |
| PPI_module_2 | Small cell lung cancer | 6.149E-5 |
| PPI_module_3 | RNA transport | 1.072E-6 |
| PPI_module_4 | There is no significant pathway | – |
| PPI_module_5 | AMPK signaling pathway | 0.003 |
| PPI-module_6 | Ribosome | 3.936E-4 |
| PPI-module_7 | RIG-I-like receptor signaling pathway | 9.115E-6 |
KEGG Kyoto encyclopedia gene and genomes
Identification of candidate drugs as a gene regulator
To identify some candidate drugs for repurposing against SARS-CoV-2 as potential therapies, the DGIdb3 [42] was used to consolidate the information of drug-gene interactions from multiple databases.
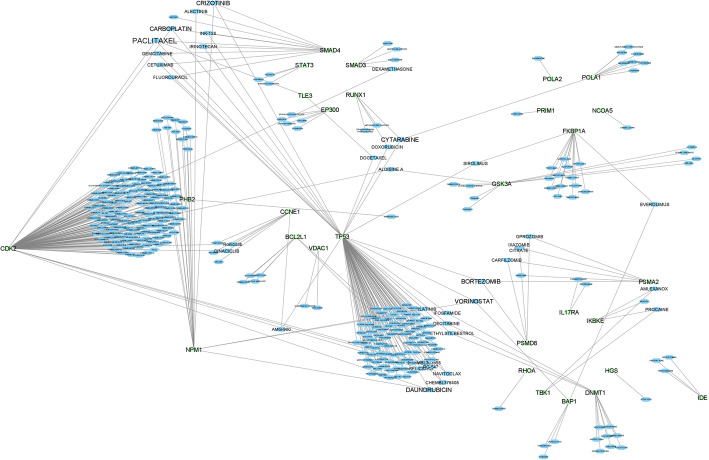
The DGIdb was applied to identify drugs that target module genes. In this regard, first, module genes were separately imported into DGIdb, and then drug-gene interactions were obtained for module genes by limiting drugs to approved drugs. After obtaining drug-gene interactions for all modules, the entire drug-gene interactions were gartered and reconstructed as a single drug-gene network (Fig. 4).
Fig. 4.
Drug-Gene Interaction Network. Note. The blue hexagon and green circle nodes are drugs and PPI module genes, respectively. The higher degree of the drug node represents a larger related font size. PACLITAXEL has four and BORTEZOMIB, CARBOPLATIN, CRIZOTINIB, CYTARABINE, DAUNORUBICIN, and VORINOSTAT have three target genes
Using this database, 28, 267, 26, 9, and 31 drugs were found for PPI_module_1, PPI_module_2, PPI_module_3, PPI_module_4, and PPI_module_7, respectively. However, no drug was found for clusters 5 and 6 PPI modules. Then, the cytoscape (Version 3.6) was utilized to visualize these data through a drug-gene network (Fig. 4). As shown in this network, some drugs have more than degree one, implying that these drugs regulate more than one gene and thus are more important in the COVID-19 gene network. According to our findings, the most remarkably identified candidate drugs for COVID-19 were PACLITAXEL with four interactions and BORTEZOMIB, CARBOPLATIN, CRIZOTINIB, CYTARABINE, DAUNORUBICIN, and VORINOSTAT with three interactions with the genes associated with the coronavirus infection. The above-mentioned drugs can be repurposed for treating COVID-19. More related drugs and their obtained interacted genes are available in detail in supplementary file S3.
Gene set enrichment analysis and candidate drugs validation
In the last step to further evaluate the validation of repurposable drugs against SARS-CoV-2, we performed GSEA by querying the Enrichr CMAP database. Two data sets named CMAP-up and CMAP-down respectively containing 5667 and 5164 drugs and associated up-regulated or down-regulated genes obtained. We considered our network-based identified drugs which had contractions with 2 genes at least and searched for them in CMAP data sets. As it is shown in Table 4, a number of 11 repurposable drugs which identified in previous analysis are validated by CMAP analysis. Each row in Table 4 demonstrates the drug with related degree in obtained gene-drug network along with affected up-regulated and down-regulated genes. According to the results, PACLITAXEL, DAUNORUBICIN and VORINOSTAT that were the most potent therapeutic candidate drugs are specially validated by GSEA.
Table 4.
The validated candidate drugs by CMAP analysis
| Drug | Degree | UP | DOWN |
|---|---|---|---|
| PACLITAXEL | 4 | ARL4D, NUP214, LARP7, NEK9, C20ORF27, TOR1AIP1 | POLA1, CCNE1, BSG, FKBP10, GTF2F2, HMOX1 |
| DAUNORUBICIN | 3 | PCM1, NLRX1, C20ORF27, HMOX1 | PLEKHF2, TIMM9, PRIM1, CLIP4 |
| VORINOSTAT | 3 | SLC9A3R1, ATP6V1A, KDELC1, NEU1, ARL4D, ERO1LB, PDE4DIP | CCND1, H2AFY2, ZC3H7A, CDKN1B, USP13, MEPCE |
| DOXORUBICIN | 2 | FKBP10, TPSB2, ARL4D | PLEKHF2, DNAJB1 |
| DEXAMETHASONE | 2 | GSK3A, BRF1, C20ORF27, NUP214, HOOK1, POR | FKBP10, NLRX1, TRIM25, LCP1, HMOX1, RALA, SLC46A3, NPTX1 |
| IRINOTECAN | 2 | BRD2, NGDN | GSK3B, TIAM1 |
| DIETHYLSTILBESTROL | 2 | ERO1LB, CEACAM5, ALB, INHBE, CLIP4 | FOXRED2, PLOD2, POLA1, PLD3, NOL10 |
| DECITABINE | 2 | ERP44, | FKBP10, |
| IFOSFAMIDE | 2 | NEK9, RAB2A, LOX, | FKBP10, RCAN3, |
| SIROLIMUS | 2 | G3BP1, DIDO1, AKAP8L, SUN2, DDAH2, CEACAM5, CEP68, SIGMAR1, NUP88, PDE4DIP, ITGB1, NOL10, POFUT1, NLRX1, FKBP10, F2RL1, ERP44, COL6A1, IFIH1, BRF1, TAPT1, IKBKB, SCARB1, DPY19L1, SMAD3, LOX, SIRT5, CHPF, NEK9, SMAD3, HMOX1, ERMP1, POFUT1, PDE4DIP, PRIM2, | CCNE1, FOXRED2, SUN2, CHEK2, POR, CHPF, RALA, GOLGA3, FKBP10, DDX58, PRKAR2B, TIMM9, CCDC86, SGTA, TRIM25, NMB, FASTKD5, ERO1LB, STC2, VPS11, HMOX1, DDX10, NOL10, RRP9, SIL1, |
| PROCAINE | 2 | G3BP1, NUP88, MASP2 | SLC46A3, SLC9A3R1, PLD3 |
Each row demonstrates the drug with related degree in obtained gene-drug network along with affected up-regulated and down-regulated genes
Discussion
At present, due to the emergency of finding a proper drug for the treatment of COVID-19, many studies are conducted based on drug repurposing strategy to rapidly identify an effective drug from the existing ones based on various host molecular factors [26, 43, 44]. This study analyzed the genes engaged in COVID-19 using a bioinformatics computational approach to identify candidate drugs for repurposing. To this end, the PPI network for coronavirus protein interactions was extracted from the String database. Then, for further analysis, seven clusters of proteins were specified as the complexes of proteins, which are more associated with SARS-CoV-2, followed by retrieving miRNAs associated with the identified clusters of genes. The central idea was to discover novel therapeutic candidate drugs to control gene regulation in COVID-19. Accordingly, the experimentally validated drugs were extracted from DGIDB. Recently, a similar network-based approach has been established to identify dysregulated genes and miRNAs in correlation with breast cancer [45] which demonstrates its feasibility to apply these kinds of studies for other diseases including COVID-19. Fiscon et al. [46] presented a novel network-based algorithm named SAveRUNNER4 regarding drug repurposing for COVID-19. In comparison with our network-based strategy that introduces a gene-drug network, they constructed a drug-disease network in which nodes are both drugs and diseases, and edges are significant associations between drugs and diseases. They applied the algorithm on 14 diseases related to the COVID-19. Focusing on known genes related to SARS with the highest genetic similarity with SARS-CoV-2, they detected 282 candidate repurposable drugs for SARS-CoV-2. In another study, Zhou et al. [26] introduced an integrative drug repurposing strategy to quantify the interactions between the human coronavirus host interactome and drug targets in the human PPI network. For building the COVID host intractome network, they assembled the host proteins of known COVID viruses such as SARS-CoV and MERS-CoV from the literature (119 COVID associated host proteins). Then, they collected drug-target interactions using multiple drug databases and mapped them to the COVID host intractome network. Finally, they identified 16 potential repurposable drugs targeting SARS-CoV-2 by applying network proximity analyses between drug targets and COVID-associated proteins.
A combination of multiple therapeutic agents including hydroxychloroquine, chloroquine, remdesivir, antiviral drugs, and common antibiotics was used for treating patients with COVID-19 [47]. Through long-time clinical uses, there is sufficient evidence of efficacy and safety regarding the application of these treatments for patients infected by the SARS-CoV-2 virus [48, 49]. However, some early studies expressed contradictory ideas in this regard. For instance, Nucsok et al. reported the potential adverse effect of hydroxychloroquine and chloroquine on cardiac function [50]. COVID-19 is a new disease and our related information is limited, and a large proportion of COVID-19 patients have underlying diseases such as cardiac injury. Thus, it is important to investigate the clinical safety of the proposed novel drugs in addition to their efficacy. Our approach in this study focused on the effectiveness of repurposed drugs and clinical safety remains unknown.
The most potent therapeutic candidate drug was PACLITAXEL, which is a clinical anti-cancer agent with antiviral activity [51]. Recently, Al-Motawa et al. [52] established a study applying bioinformatics tools, proteomics data, and a cell model in SARS-CoV-2 to find a suitable drug for COVID-19 based on the enrichment of arginine residues. For this purpose, the reference sequences of the SARS-CoV-2 proteome and the sequences of the human proteome were retrieved, followed by performing receptor binding domain analysis. They revealed that the anti-proliferative activity of PACLITAXEL leads to an increase in the concentration of methylglyoxal in cells, thus they suggested that this drug has the potential for repurposing as a candidate for the treatment of COVID-19.
The other six candidate drugs included BORTEZOMIB, CARBOPLATIN, CRIZOTINIB, CYTARABINE, DAUNORUBICIN, and VORINOSTAT, some of which were previously discovered to be efficient against COVID-19. BORTEZOMIB is an anti-tumor agent whitch induces apoptosis in different kinds of cancers [53]. In a study by Xing et al., BORTEZOMIB was reported to have reversal effects against SARS-CoV-2-induced gene expression [54]. They used 430 samples infected by MERS-CoV or SARS-CoV from different databases to extract differentially expressed genes as disease signatures for the prediction of COVID-19 candidate drugs.
Based on the findings of the current study, VORINOSTAT was the other striking drug, which is an anticancer histone deacetylase (HDAC). A recent study was conducted to identify repurposed drugs in SARS-CoV-2 infection [55] by analyzing dis-regulated genes in response to coronavirus infections including SARS-CoV-2. To this end, a meta-analysis approach was utilized to find common differentially expressed genes in the human hosts infected by various kinds of respiratory viruses. Accordingly, 31 up-regulated genes and 27 drugs for their regulation were found for SARS-CoV-2 cases. Among the reported drugs, VORINOSTAT was observed as an inhibitor of HDAC [55]. Moreover, Sinba et al. [56] reported VORINOSTAT as the up-regulator of angiotensin-converting enzyme 2 (ACE2), which is the human host receptor of SARS-CoV-2 in cell lines.
The next drug was DAUNORUBICIN, which was previously detected as an inhibitor of SARS-CoV-2 Mpro (Main protease) and reported as a potential therapeutic drug targeting COVID-19 [57]. In addition, DAUNORUBICIN was approved as an anti-cancer drug by the US Food and Drug Administration regulating SARS-CoV-2 interactors (The ABCC1 gene) and having the potential for investigation as a repurposed drug against SARS-CoV-2 infection [58, 59].
Based on the literature review, no study was found to report CARBOPLATIN, CRIZOTINIB, and CYTARABINE, which were introduced in the current study. Therefore, these novel potential drugs should be investigated as a therapy for COVID-19.
This study had some limitations. The study could not investigate the validation of these drugs in vitro and in vivo due to the computational nature of our study method and the restriction of experimental resources. Nevertheless, regarding the imperative need to reach treatment for COVID-19, these identified candidate drugs are required to be validated in experimental studies. Moreover, as mentioned above, our approaches only considered the efficacy aspect of drugs, and thus urgent clinical trials are needed to provide safety data on repurposed drugs for the treatment of COVID-19 patients.
In conclusion, as a strategy for predicting repurposable candidate drugs against COVID-19, the protein-protein interaction network-based method was used to find genes and related miRNAs highly correlated with coronavirus. These computational methods provide efficacious and rapid data for therapeutic purposes. However, further experimental analysis and testing such as clinical applicability, toxicity, and experimental validations are required to reach a more accurate and improved treatment. Our proposed complexes of proteins and associated miRNAs, along with the discovered candidate drugs might be a starting point for further analysis in this urgency of COVID-19 pandemic.
Supplementary Information
Acknowledgements
This work has been performed and supported by the Kerman University of medical sciences and Islamic Azad Univerity-Tabriz branch (unique project identifier: IRKMU.REC.1399.508).
Authors’ contributions
Conceptualization, MA,BS; Methodology, HM,AR; Software, HM; Validation, MA, AAH,HM; Formal analysis, MA, AR,AAH; Investigation, MA, HM; preparation, MA,AR,BS; Writing- review and editing, MA,BS, HM; Visualization, MA, HM; Supervision, HM; Project administration, HM; All authors consent and approved the final version of the manuscript.
Funding
Not applicable.
Availability of data and materials
The data used in this project is available at (https://github.com/habibmoti/COVID-19). All data, raw and processed, is readily available from the corresponding author on request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Database for Annotation Visualization and Integrated Discovery
Kyoto Encyclopedia Gene and Genomes
Drug Gene Interaction database
Searching off-lAbel dRUg aNd NEtwoRk
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li X, et al. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10(2):102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborty C, et al. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci. 2020;24(7):4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty C, et al. The 2019 novel coronavirus disease (COVID-19) pandemic: a zoonotic prospective. Asian Pac J Trop Med. 2020;13(6):242–246. doi: 10.4103/1995-7645.281613. [DOI] [Google Scholar]
- 4.Chakraborty C, et al. Extensive partnership, collaboration, and teamwork is required to stop the COVID-19 outbreak. Arch Med Res. 2020;51(7):728–730. doi: 10.1016/j.arcmed.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Organization, W.H . Coronavirus disease 2019 (COVID-19): situation reports, weekly epidemiological update. 2020. [Google Scholar]
- 6.Long C, Zhong L. Genomics functional analysis and drug screening of SARS-CoV-2. Genes Dis. 2020;7.4:542-50. [DOI] [PMC free article] [PubMed]
- 7.Motieghader H, et al. mRNA–miRNA bipartite network reconstruction to predict prognostic module biomarkers in colorectal cancer stage differentiation. Mol BioSyst. 2017;13(10):2168–2180. doi: 10.1039/C7MB00400A. [DOI] [PubMed] [Google Scholar]
- 8.Habib MG, et al. mRNA and microRNA selection for breast cancer molecular subtype stratification using meta-heuristic based algorithms. Genomics. 2020;112.5:3207-17. [DOI] [PubMed]
- 9.Leon-Icaza SA, Zeng M, Rosas-Taraco AG. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019;1(1):1–1. doi: 10.1186/s41544-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbott TR, et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell. 2020;181(4):865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei J, et al. Genome-wide CRISPR screen reveals host genes that regulate SARS-CoV-2 infection. bioRxiv. 2020:2020.06.16.155101. https://www.biorxiv.org/content/10.1101/2020.06.16.155101v1.abstract.
- 12.Gulshan S, Hanania NA, Shim YM. The aging immune system and its relationship to the development of chronic obstructive pulmonary disease. Proc Am Thor Soc. 2009;6.7:573-80. [DOI] [PMC free article] [PubMed]
- 13.Chakraborty C, Sharma AR. Consider TLR5 for new therapeutic development against COVID-19. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha A, et al. Tocilizumab: a therapeutic option for the treatment of cytokine storm syndrome in COVID-19. Arch Med Res. 2020;51(6):595–597. doi: 10.1016/j.arcmed.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya M, Sharma AR. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J Med Virol. 2020;92(6):618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco-Melo D, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181(5):1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Z, et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;181(6):883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong Y, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prasad K, et al. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int J Biol Macromol. 2020;163:1–8. doi: 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cava C, Bertoli G, Castiglioni I. In Silico discovery of candidate Drugs against Covid-19. Viruses. 2020;12(4):404. doi: 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sardar R, et al. Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. bioRxiv. 2020:2020.03.21.001586. https://www.biorxiv.org/content/10.1101/2020.03.21.001586v1.abstract. [DOI] [PMC free article] [PubMed]
- 22.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 23.Murray MF, et al. COVID-19 outcomes and the human genome. Genet Med. 2020;22(7):1175–1177. doi: 10.1038/s41436-020-0832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatia R, Narang RK, Rawal RK. Drug Repurposing - A Promising Tool in Drug Discovery Against CoV-19. Biomed J Sci Tech Res. 2020;28(5):21913–21915. [Google Scholar]
- 25.Saha RP, et al. Repurposing Drugs, Ongoing Vaccine, and New Therapeutic Development Initiatives Against COVID-19. Front Pharmacol. 2020;11(1258). https://www.frontiersin.org/articles/10.3389/fphar.2020.01258/full. [DOI] [PMC free article] [PubMed]
- 26.Zhou Y, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stolfi P, et al. Designing a network proximity-based drug repurposing strategy for COVID-19. Front Cell Dev Biol. 2020;8(1021). https://www.frontiersin.org/articles/10.3389/fcell.2020.545089/full. [DOI] [PMC free article] [PubMed]
- 28.Zeng X, et al. deepDR: a network-based deep learning approach to in silico drug repositioning. Bioinformatics. 2019;35(24):5191–5198. doi: 10.1093/bioinformatics/btz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng F, et al. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat Commun. 2019;10(1):3476. doi: 10.1038/s41467-019-10744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, et al. ClusterViz: a Cytoscape APP for cluster analysis of biological network. IEEE/ACM Trans Comput Biol Bioinform. 2015;12(4):815–822. doi: 10.1109/TCBB.2014.2361348. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Wang J, Chen J. 2008 International Conference on BioMedical Engineering and Informatics. 2008. A Fast Agglomerate Algorithm for Mining Functional Modules in Protein Interaction Networks. [Google Scholar]
- 35.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Kanehisa M, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamb J, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 40.Musa A, et al. A review of connectivity map and computational approaches in pharmacogenomics. Brief Bioinform. 2017;19(3):506–523. doi: 10.1093/bib/bbw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner AH, et al. DGIdb 2.0: mining clinically relevant drug–gene interactions. Nucleic Acids Res. 2016;44.D1:D1036-44. [DOI] [PMC free article] [PubMed]
- 43.Ge Y, et al. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19. bioRxiv. 2020:2020.03.11.986836. https://www.biorxiv.org/content/10.1101/2020.03.11.986836v1.abstract. [DOI] [PMC free article] [PubMed]
- 44.Bhattacharya M, et al. A SARS-CoV-2 vaccine candidate: In-silico cloning and validation. Inform Med Unlocked. 2020;20:100394. doi: 10.1016/j.imu.2020.100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adhami M, et al. Gene co-expression network approach for predicting prognostic microRNA biomarkers in different subtypes of breast cancer. Genomics. 2020;112(1):135–143. doi: 10.1016/j.ygeno.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Fiscon G, et al. SAveRUNNER: A network-based algorithm for drug repurposing and its application to COVID-19. PLoS Comput Biol. 2021;17(2):e1008686. doi: 10.1371/journal.pcbi.1008686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saha A, et al. Probable molecular mechanism of Remdesivir for the treatment of COVID-19: need to know more. Arch Med Res. 2020;51(6):585–586. doi: 10.1016/j.arcmed.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colson P, Rolain JM, Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents. 2020;55(3):105923. doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortegiani A, et al. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naksuk N, Lazar S, Peeraphatdit TB. Cardiac safety of off-label COVID-19 drug therapy: a review and proposed monitoring protocol. Eur Heart J Acute Cardiovasc Care. 2020;9(3):215–221. doi: 10.1177/2048872620922784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277(5698):665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 52.Al-Motawa M, et al. Vulnerabilities of the SARS-CoV-2 virus to proteotoxicity – opportunity for repurposed chemotherapy of COVID-19 infection. bioRxiv. 2020:2020.04.07.029488. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7581855/. [DOI] [PMC free article] [PubMed]
- 53.Landowski TH, et al. Mitochondrial-mediated disregulation of Ca2+ is a critical determinant of Velcade (PS-341/bortezomib) cytotoxicity in myeloma cell lines. Cancer Res. 2005;65(9):3828–3836. doi: 10.1158/0008-5472.CAN-04-3684. [DOI] [PubMed] [Google Scholar]
- 54.Xing J, et al. Reversal of infected host gene expression identifies repurposed drug candidates for COVID-19. bioRxiv. 2020:2020.04.07.030734. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7217282.1/.
- 55.Loganathan T, et al. Host transcriptome-guided drug repurposing for COVID-19 treatment: a meta-analysis based approach. PeerJ. 2020;8:e9357. doi: 10.7717/peerj.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinha S, et al. In vitro and in vivo identification of clinically approved drugs that modify ACE2 expression. Mol Syst Biol. 2020;16(7):e9628. doi: 10.15252/msb.20209628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiménez-Alberto A, et al. Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Comput Biol Chem. 2020;88:107325. doi: 10.1016/j.compbiolchem.2020.107325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gordon DE, et al. A SARS-CoV-2-Human protein-protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv. 2020:2020.03.22.002386. https://www.biorxiv.org/content/10.1101/2020.03.22.002386v3.
- 59.Priebe W, et al. Doxorubicin- and daunorubicin-glutathione conjugates, but not unconjugated drugs, competitively inhibit leukotriene C4 transport mediated by MRP/GS-X pump. Biochem Biophys Res Commun. 1998;247(3):859–863. doi: 10.1006/bbrc.1998.8887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this project is available at (https://github.com/habibmoti/COVID-19). All data, raw and processed, is readily available from the corresponding author on request.