Abstract
Giant cell tumor of bone (GCTB) is a locally aggressive lesion of intermediate malignancy. Malignant transformation of GCTB is a rare event. In 2013, the humanized monoclonal antibody against receptor activator of nuclear factor-κb-Ligand (RANKL) denosumab was approved for treatment of advanced GCTB. Since then, several reports have questioned the role of denosumab during occasional malignant transformation of GCTB. We report on three patients with H3F3A-mutated GCTBs, treated with denosumab. The tissue samples were analysed by histomorphology, immunohistochemistry, and in two instances by next generation panel sequencing of samples before and after treatment. One patient had a mutation of ARID2 in the recurrence of the GCTB under treatment with denosumab. One patient developed a pleomorphic sarcoma and one an osteoblastic osteosarcoma during treatment. Sequencing revealed a persisting H3F3A mutation in the osteosarcoma while the pleomorphic sarcoma lost the H3F3A mutation; however, a FGFR1 mutation, both in the recurrence and in the pleomorphic sarcoma persisted. In addition, the pleomorphic sarcoma showed an AKT2 and a NRAS mutation. These data are inconclusive concerning the role denosumab plays in the event of malignant progression/transformation of GCTB and point to diverging pathways of tumor progression of GCTB associated with this treatment.
Subject terms: Cancer, Bone cancer, Cancer genetics, Cancer, Genetics research, Cancer, Bone cancer
Introduction
Giant cell tumor of bone (GCTB) was first described by Jaffe et al.1 in 1940 and makes up about five percent of all primary bone lesions2,3; GCTB is typically located in the epiphyseal region of the long tubular bones such as the distal femur and the proximal tibia3,4. Based on the WHO-Classification for tumors of soft tissue and bone, GCTB is considered to be an aggressive and rarely metastasizing tumor and therefore is regarded as tumor of intermediate malignancy (ICD-O: 9250/1)5. Metastatic lesions most commonly occur in the lungs4,6. In rare cases, a high-grade malignant neoplasia is identified arising in GCTB (primary malignancy in GCTB or after previous radiotherapy (in both instances classified as ICD-O: 9250/3)).
Histologically, the GCTB mainly consists of three different cell types. These are the neoplastic spindle-shaped stromal cells that show enhanced synthesis of receptor activator of nuclear factor-κb ligand (RANKL) and the large multinucleated osteoclast-like giant cells and their monocytic precursors expressing the corresponding receptor, i.e., receptor activator of nuclear factor-κb (RANK)7,8. The stroma cells are the neoplastic component of GCTB and harbour a characteristic point mutation at the histone gene H3F3A leading to a substitution of glycine by tryptophan at position 34 (G34W). The increased synthesis of the RANKL by the neoplastic cells leads to enhanced bone resorption by the osteoclast-like giant cells through the RANK/RANKL signalling pathway, which in turn are responsible for the locally aggressive growth.
Intralesional curettage is the primary treatment option for GCTB6. The rate of local recurrence after intralesional curettage ranges from 30 to 40%3,9. Denosumab was first approved for the treatment of osteoporosis under the brand name PROLIA in 2010 and was later approved for the treatment of GCTB in 2013 under the brand name XGEVA. Denosumab is a humanized monoclonal IgG2-anti-RANK-Ligand-antibody. This monoclonal antibody binds to RANKL and inhibits the interaction between the spindle-like stromal cells and the osteoclast-like giant cells, and thereby prevents local bone resorption, thus mimicking the effect of osteoprotegerin, a physiological RANKL-antagonist6,7.
Clinical case presentation
Patient one is a female, first diagnosed with GCTB in 2015 at the age of 33. The tumor measured 12 cm and was located in the pelvis; she was treated by a complete (R0) resection. In 08/2016 a denosumab treatment was started (120 mg subcutaneously every 4th week) until 06/2017 due to recurrence in the pelvis confirmed by a biopsy. The patient is well and shows no further signs of progression.
Patient two is a male that was diagnosed with a tumor of the sacrum measuring 13 cm in 03/2014 at the age of 20. A biopsy was performed and the diagnosis of GCTB was confirmed by detection of the H3F3A G34W mutation. In 04/2014 an incomplete resection of the tumor with instillation of alcohol 90% was performed. A fistula, which developed shortly after the first surgery, was resected.
In 12/2014, nine months after resection of the GCTB, a recurrence was diagnosed and denosumab treatment was started with 120 mg every 4 weeks until 05/2016. In 2017 the patient underwent palliative surgery. A CT-Scan showed a large local recurrence and pulmonary as well as liver lesions highly suspicious of metastases. The tumor mass was resected in 07/2017 and a high-grade osteosarcoma with angioinvasion harbouring the H3F3A G34W mutation was diagnosed.
The patient died in 08/2017 due to tumor progression and sepsis.
Patient three, a woman, is a follow-up initially published by Aponte-Tinao et al.10 in 2015. These authors reported the case of a 20-year-old female who was first diagnosed with GCTB in 2009. The GCTB was located at the right proximal tibia and was treated by intralesional curettage. She was diagnosed with a recurrence of GCTB about one year later in 2010 and was treated by en-bloc resection. The following two years were uneventful until a follow-up CT-scan showed a second recurrence in 2013 and denosumab therapy was started. The first application was a subcutaneous dose of 360 mg followed by 120 mg subcutaneously every 4 weeks. About one year after beginning of treatment with denosumab, the patient noticed a palpable, painful mass in the popliteal fossa. A CT-Scan showed that the mass included two sections of different density. An open biopsy was performed and histologic workup showed a high-grade undifferentiated pleomorphic sarcoma besides the GCTB. Subsequently an above-knee amputation was performed. Several tissue blocks were available for further histological analysis of the resection specimen. The patient is well and shows no signs of progression.
Methods
The samples were analysed by conventional histology using haematoxylin–eosin (HE) staining of sections of paraffin-embedded tissue. Immunohistochemistry was performed as described using a mutation specific monoclonal antibody for detection of the H3F3A G34W mutation (Anti H3.3 G34W clone 31-1145-00; RevMab Biosciences, San Francisco, CA, USA; dilution 1:400); for detection of proliferation indices the Ki-67 antibody (M7240, Dako, Glostrup, Denmark; dilution 1:200) was applied11.
The alkaline phosphatase/RED detection system (Dako) was used for immunohistochemistry on formalin-fixed and paraffin-embedded tissue via the avidin–biotin-complex-method. The samples were pseudonymized according to the German law for correct usage of archival tissue for clinical research. The research was approved by the local ethics committee of the University of Ulm (reference 369/17) and was in compliance with the ethical principles of the World Medical Association Declaration of Helsinki. Informed consent was obtained from all patients.
Isolation of tumor DNA from FFPE tissue
For isolation of genomic DNA from the formalin fixed paraffin embedded (FFPE) tissue samples, 5 μm tissue slices were transferred to glass slides. To estimate the area containing the tumor, HE stained FFPE tissue slices (2 µm) were validated by an expert pathologist. The tumor-harbouring areas of the FFPE tissue were subjected to a DNA extraction procedure using the QIAamp DNA FFPE tissue kit (QIAGEN, Hilden, Germany) according to manufacturer’s instruction. DNA purity and concentration were determined fluorometrically (Qubit 2.0; Invitrogen, Carlsbad, CA, USA).
We further performed Sanger-Sequencing for H3F3A G34W for all tissue samples of patient 1 and patient 3. Gene sequencing (Sanger) was performed according to a diagnostic standard protocol. The cell line A498 was used as negative control. The graphs were generated with FinchTV 1.4.0 (Geospiza Inc., Seattle Washington, USA)12.
Next generation sequencing
For molecular characterization of both tumor tissue and ctDNA, we employed a targeted re-sequencing methodology using the GeneRead V2 chemistry (QIAGEN, Hilden, Germany) and a custom-made re-sequencing panel including primers for all exons of a panel of 37 genes (primer sequences and locations of target areas are available upon request). Target enrichment, amplicon processing, and library generation were performed according to the manufacturer’s instructions. For target enrichment, we included 10–40 ng (DNA from FFPE tumor and non-neoplastic tissue). Successful target enrichment and library generation was checked using the High Sensitivity DNA kit on a bioanalyzer device (Agilent, Santa Clara, CA, USA). Libraries were diluted to 10 pM solutions and the sequencing was performed on a MiSeq platform (Illumina, San Diego, CA, USA) using the V2 chemistry. Mean read depth on target region was 2000–8000-fold and 99% of bases were covered at 96–100% on average. The resulting fastq files were subjected to further analysis using the GeneRead web based analysis tool (http://ngsdataanalysis.sabiosciences.com/NGS2/), the Biomedical Workbench software package (QIAGEN, Hilden, Germany), and the Variant Studio software (Illumina, San Diego, CA, USA)13. All identified mutations were manually re-analyzed using the Integrated Genome Viewer Software (Broad Institute, MA, USA).
Single nucleotide polymorphisms (SNP) detected in non-tumor-tissue were excluded from further analysis.
Results
Patient 1
Morphology
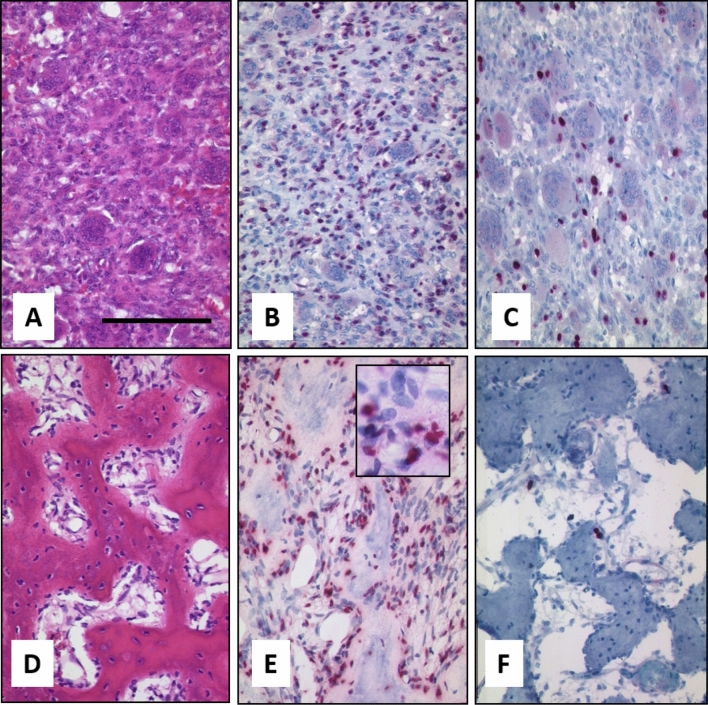
Tissue blocks of the primary biopsy, the resection specimen, and the recurrence under denosumab therapy were available. Histological analysis of the biopsy and resection specimen showed a giant cell tumor with typical morphology, consisting of osteoclastic giant cells and a mononuclear spindle cell compartment. The primary tumor showed a strong positivity for the H3.3 G34W detecting antibody in the nuclei of the stromal compartment. Nuclei of osteoclastic giant cells were negative. A second mononuclear cell population was detected being negative for H3.3 G34W which most probably presents osteoclastic precursors. Ki-67 rate was about 5% in the mononuclear cell population. After treatment the tumor revealed a dramatic change in morphology. The number of H3.3 G34W-positive osteoblastic cells was greatly reduced; positive mononuclear cells were still detectable along strands of neoformated osteoid. We detected an intermingled spindle cell compartment negative for H3.3 G34W. The Ki-67 index dropped to less than 1% (Fig. 1). The presence of the H3F3A G34W mutation was further proven by Sanger sequencing.
Figure 1.
Morphology of Case 1. (A) Typical GCTB with osteoclastic giant cells and intermingled mononuclear cells before therapy (bar = 100 µm). Neoplastic mononuclear cells are positive in a H3F3A G34W staining in the nuclei (B) and about 5% of cells are Ki-67 positive (C). After denosumab therapy GCTB shows induction of neoformated osteoid and complete reduction of giant cells (D), while H3F3A G34W-positive cells are still present (E); insert shows H3F3A-negative spindle cells. Proliferation has dropped to 1% in a Ki67 staining (F).
Panel sequencing of microdissected tissue of the resected tumor and the biopsy of the recurrence revealed a mutation of ARID2 (chromosome 12; position (GRCh37) 46205208, c.292G > A; p.E98K) with an allele frequency of 48% in the tumor recurrence under denosumab treatment. Neither the non-tumorous tissue nor the GCTB before treatment with denosumab harboured this mutation.
Patient 2
Morphology
The biopsy and resection specimen revealed a GCTB with typical morphology of osteoclastic giant cells and mononuclear spindle cells. Ki-67 rate was about 5%. Immunochemistry and sequencing confirmed the diagnosis of GCTB with positive staining for the mutation H3F3A G34W. This diagnosis was verified by Sanger-Sequencing.
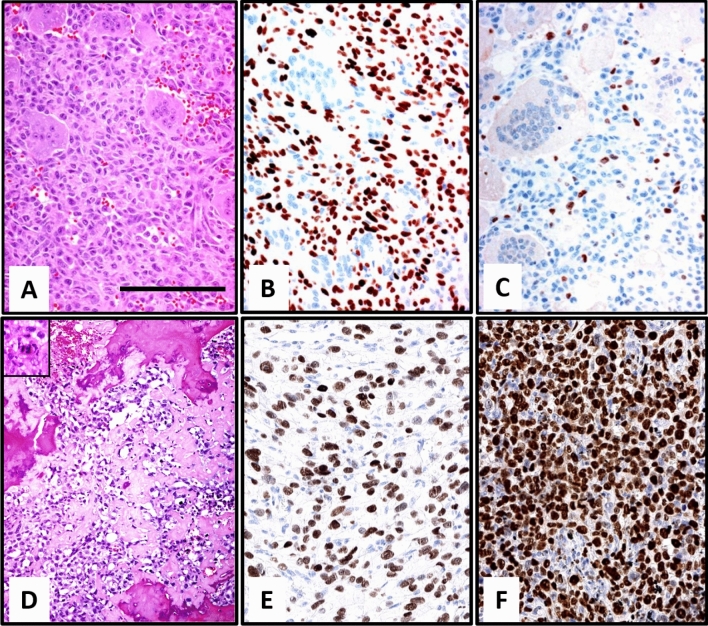
Morphology of the resected recurrence revealed a pleomorphic high-grade sarcoma with necrosis and presence of atypical mitotic figures. The sarcoma showed intermingled lace-like neoplastic osteoid and was classified as a high-grade osteosarcoma; an angioinvasion was present. The sarcoma harboured the H3F3A G34W mutation as shown by immunohistochemistry in all tumor cells and a Ki-67 index of 90% (Fig. 2). This presence of the mutation was further confirmed by Sanger sequencing for H3F3A G34W mutation.
Figure 2.
Morphology of Case 2. Typical morphology of GCTB with osteoclastic giant cells and mononuclear spindle cells (A) neoplastic mononuclear cells with positive H3F3A G34W staining (B) (bar = 100 µm). Staining for Ki-67 shows about 5% positive cells (C). The sarcoma showed presents with polymorphic tumor cells andatypical mitotic figures with necrosis intermingled lace-like neoplastic osteoid intermingled with necrotic bone. An atypical mitotic figure is shown in the insert (D). The sarcoma harboured the H3F3A G34W mutation (E) and had a high rate of Ki-67 (F).
Patient 3
Morphology
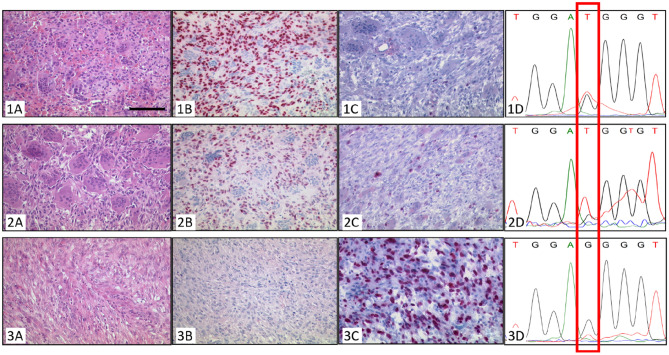
The morphology of this tumor has been described in details by Aponte et al.10. In brief, the initial GCTB revealed a typical morphology with osteoclastic giant cells and a mononuclear H3F3A G34W-positive neoplastic tumor population as shown by immunohistological staining and sequencing both of the primary tumor and the recurrence. The Ki-67 index was about 5%. The sarcoma after denosumab treatment showed a high-grade sarcoma (not otherwise specified) with spindle-like to pleomorphic tumor cells and extensive necrosis and a Ki-67 index of 50%.
The sarcoma was negative in an immunohistochemical staining with the monoclonal antibody for detection of H3F3A G34W.
Sequencing of DNA from microdissected tissue of the H3F3A G34W-negative tumor showed a very weak peak for a mutation-specific thymine that was below the detection threshold as shown in Fig. 3.
Figure 3.
Morphology of Case 3. (1) Biopsy. (2) Resection specimen. (3) Sarcoma. Typical GCTB with osteoclastic giant cells and intermingled mononuclear cells in the biopsy (1A) and the resection specimen (2A) as well as neoplastic mononuclear cells with positive H3F3A G34W staining in the nuclei (1B,2B) (bar = 100 µm). Staining for Ki-67 shows about 1% of positive cells (1C,2C). Sanger-Sequencing for H3F3A G34W shows the mutation specific thymine in tissue of the biopsy (1D) as well as the resection specimen (2D). The sarcoma showed spindle to pleomorphic tumor cells and extensive necrosis (3A) and a high Ki-67 index (3C), however it did not stain positive for H3F3A G34W (3B). Sanger-sequencing of DNA from microdissected tissue of the H3F3A G34W-negative tumor showed a very weak peak for a mutation-specific thymine (3D).
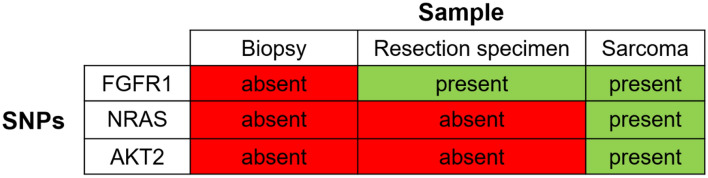
In the panel-sequencing we found a mutation of FGFR1 (chromosome 8; position (GRCh37) 38287303; c354G > A; p.E118) in the resection specimen of the GCTB and in the sarcoma with an allele frequency of 10%, which was not found in the biopsy. We found two additional mutations in the sarcoma tissue for AKT2 (chromosome 19; position (GRCh37) 40742052) and NRAS (chromosome 1; position (GRCh37) 115256669) with a frequency of 26% and 48%, respectively; these two mutations were not present in the initial biopsy or in the resection specimen (Fig. 4) and present intronic mutations with no effect on the protein structure. Supplementary Figs. 1 and 2 summarize the clinical and molecular findings.
Figure 4.
Summary of SNPs found in the different tissue samples (Case 3). SNPs in the resection: FGFR1 (chromosome 8; position (GRCh37) 38287303; c354G > A; p.E118). SNPs in the pleomorphic sarcoma: FGFR1 (chromosome 8; position (GRCh37) 38287303; c354G > A; p.E118); AKT2 (chromosome 19; position (GRCh37) 40742052); NRAS (chromosome 1; position (GRCh37) 115256669).
Discussion
GCTB is characterized by an enhanced paraneoplastic secretion of RANKL, leading to a shift of normal bone homeostasis to enforced bone resorption due to an increased number of induced non-neoplastic osteoclasts3,14. Denosumab was first approved for treatment of GCTB in 2013 in advanced stage and is a humanized monoclonal IgG2 antibody. Through specific binding to RANKL denosumab blocks the RANK/RANKL signalling. This treatment leads to striking changes in morphology of GCTB characterized by reduction of both, the neoplastic cell compartment and the osteoclasts14. Due to blockage of the RANK/RANKL axis enhanced osteoid neoformation of bone is observed by H3F3A-non-mutated osteoblasts and fibroblastoid spindle cells11.. Mak et al. showed that denosumab reduced the number of osteoclasts and RANKL secretion in vitro, however the neoplastic stromal cells continued to grow after reduction of denosumab, even though at a slower proliferation rate15. The authors conclude that after denosumab treatment the H3F3A-mutated tumor cell pool may reside in the tissue and be reactivated during recurrence15. Besides the role of the RANK axis in the control of bone remodelling and as a crucial factor in GCTB growth, RANKL has an important role for the differentiation of B and T cells as well as for the survival of dendritic cells16,17; by this mechanism, enhanced RANKL synthesis in GCTB may lead to immunosuppression and therefore promote neoplastic transformation18.
On the other hand, nuclear factor κb upregulation by enhanced RANK signalling is discussed to interfere with regulation of oncogenes18. It has been shown that RANKL upregulates the expression of semaphorin 3A20, and may thereby affect growth of cartilage and bone21. Furthermore, in a comparative proteome study of GCTB before and after denosumab therapy several differentially expressed proteins were identified including metalloproteinase 9 being downregulated after denosumab treatment19. Interestingly, we noted a reduction of proliferation of the GCTB after treatment in the recurrences of patient one, however we further detected a H3F3A-negative mononuclear population most probably corresponding to re-populating osteoblasts. The suppression of RANK signalling may therefore generate a micro milieu favourable for malignant transformation.
In osteoporosis, denosumab treatment leads to significant increase in bone density; up to now no cases have been described of malignant tumors in patients treated with denosumab for osteoporosis. GCTB and osteoporosis are treated with different doses of denosumab. In osteoporosis 60 mg of denosumab is administered in a subcutaneous injection every six months. For treatment of GCTB a loading dose of 360 mg may be administered, followed by a subcutaneous injection of 120 mg every 4 weeks. The prescribed dose for GCTB is therefore estimated 12-times higher than the dosage used for treatment of osteoporosis. Since the approval for treatment of GCTB there have been at least 15 described cases of malignant transformation of GCTB in association with denosumab treatment10,20–26. Due to these reports the question arises whether the sarcoma is clonally linked to the primary tumor. In patient one we detected an ARID2 mutation limited to the recurrence of the GCTB after denosumab treatment. This finding suggests that mutated ARID2 is a marker during tumor progression under denosumab treatment in this H3F3A-mutated GCTB, although no sarcomatous transformation is present. In line with this finding is that ARID2 was identified to play a critical role in the differentiation of osteoblasts and that a mutation may interfere in this process27. The second patient reported was characterized by a persisting H3F3A mutation in the relapse as well as in the osteosarcoma arising in the sacrum. This finding points to a clonal evolution of the sarcoma under denosumab treatment; the H3F3A mutation can be regarded as a clonal marker in this case. In rare cases, H3F3A mutations have been described in osteosarcoma of the epiphyseal region of mostly older patients as shown by Koelsche et al.28. This finding is in line with a possible transformation of a pre-existing GCTB completely overrun by the sarcoma. In contrast, in patient three, a different mutational profile during progression emerged regarding the H3F3A mutation. The initial GCTB in the biopsy and in the recurrence was H3F3A-mutated, while the sarcoma was negative for this mutation as shown by sequencing and immunohistochemical staining. However, the recurrence of the GCTB and the sarcoma revealed an overlapping mutation for FGFR1, which was not present in the first biopsy. In addition, we detected a mutation in AKT2 and NRAS in the sarcoma. This may point to the evolution of a high-grade sarcoma from a H3F3A-negative, but FGFR1-positive subclone with acquisition of additional mutations in AKT2 and NRAS during tumor progression. One explanation is a transformation of the H3F3A-negative mononuclear cells residing in the tumor after denosumab treatment. Although the detected mutations in NRAS and AKT2 are intronic in the presented GCTB with no effect on the cDNA level these two genes have been shown to play a role in bone physiology and osteosarcomas. While NRAS mutations have been described in dysplastic bone formation29, AKT2 has been shown to play a role in tumor growth by inhibiting cisplatin-induced apoptosis in primary osteosarcomas30,31. Furthermore, Zhu et al. described an enhanced expression of AKT2 in osteosarcoma and that this finding is associated with a more aggressive clinical behaviour and worse outcome32.
Based on our analysis we cannot determine whether or not the transformation is a spontaneous event and occasionally associated with denosumab treatment. Large studies with a total of 2315 patients with GCTB have shown a cumulative incidence of primary malignant giant cell tumor of Bone (PMGCTB) of 1.6% in GCTB compared with 2.4% for secondary malignant giant cell tumor of bone (SMGCTB) following radiotherapy33. PMGCTB usually occurs next to an area of benign GCTB whereas SMGCTB occurs superimposed on a previously treated benign GCTB34. Several studies showed that patients with PMGCTB were of older age compared to patients with SMGCTB34,35. The clinical presentation of PMGCTB and SMGCTB is comparable36. Radiologically PMGCTB is often not distinguishable from benign GCTB whereas SMGCTB often presents findings suspicious of malignancy35.
In a further study on 532 patients receiving denosumab the number of patients with sarcomatous transformation was limited to four patients (1%)26.
Scotto di Carlo et al. reported in a recent study that GCTB shows a more malignant phenotype in a patient with Paget disease of bone which further points to malignant behaviour of GCTB depending on further co-factors37.
Whether the patients reported in literature with sarcomas after denosumab treatment fall in the category of being spontaneously transformed or being associated with denosumab therapy is yet not clear since 12 of them did not receive additional radiotherapy.
In conclusion, our findings point to different molecular profiles of GCTB associated with denosumab treatment and raise the question whether these events are spontaneous or associated with denosumab treatment. To clarify this question, more studies should be performed of GCTB in progression with and without denosumab treatment excluding those patients who received radiotherapy.
Supplementary Information
Acknowledgements
Case 3 was presented at the 89th spring meeting of the Arbeitsgemeinschaft Knochentumoren (AGKT; e.V.) in Heidelberg in 2017.
Author contributions
M.H., K.M., R.M., T.F.E.B. G.M. and P.M. conceived the experiments and analysed the data. M.H. and T.F.E.B. conducted the experiments. A.v.B., M.S., P.R, L.A-T, and B.L. collected material and primary data. M.H., K.M. and T.F.E.B. wrote the manuscript. M.H. and T.F.E.B., designed the figures and tables. All authors reviewed the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85319-x.
References
- 1.JAFFE, H. L. Giant-cell tumour (osteoclastoma) of bone: Its pathologic delimitation and the inherent clinical implications. Ann. R. Coll. Surg. Engl.13, 343–355 (1953). [PMC free article] [PubMed]
- 2.Turcotte RE. Giant cell tumor of bone. Orthop. Clin. N. Am. 2006;37:35–51. doi: 10.1016/j.ocl.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Czerniak, B. & Preceded by (work): Dorfman, H. D. Dorfman and Czerniak’s bone tumors. (Elsevier, 2015).
- 4.Alberghini M, et al. Morphological and immunophenotypic features of primary and metastatic giant cell tumour of bone. Virchows Arch. 2010;456:97–103. doi: 10.1007/s00428-009-0863-2. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher, C. D. M., Unni, K. K., Mertens, F., World Health Organization. & International Agency for Research on Cancer. in Pathology and Genetics of Tumours of Soft Tissue and Bone. (IARC Press, 2002).
- 6.Skubitz KM. Giant cell tumor of bone: Current treatment options. Curr. Treat. Options Oncol. 2014;15:507–518. doi: 10.1007/s11864-014-0289-1. [DOI] [PubMed] [Google Scholar]
- 7.Thomas DM. RANKL, denosumab, and giant cell tumor of bone. Curr. Opin. Oncol. 2012;24:397–403. doi: 10.1097/CCO.0b013e328354c129. [DOI] [PubMed] [Google Scholar]
- 8.Raskin KA, Schwab JH, Mankin HJ, Springfield DS, Hornicek FJ. Giant cell tumor of bone. J. Am. Acad. Orthop. Surg. 2013;21:118–126. doi: 10.5435/JAAOS-21-02-118. [DOI] [PubMed] [Google Scholar]
- 9.Campanacci M, Baldini N, Boriani S, Sudanese A. Giant-cell tumor of bone. J. Bone Joint Surg. Am. 1987;69:106–114. doi: 10.2106/00004623-198769010-00018. [DOI] [PubMed] [Google Scholar]
- 10.Aponte-Tinao LA, Piuzzi NS, Roitman P, Farfalli GL. A high-grade sarcoma arising in a patient with recurrent benign giant cell tumor of the proximal tibia while receiving treatment with denosumab. Clin. Orthop. Relat. Res. 2015;473:3050–3055. doi: 10.1007/s11999-015-4249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lüke, J. et al. H3F3A mutation in giant cell tumour of the bone is detected by immunohistochemistry using a monoclonal antibody against the G34W mutated site of the histone H3.3 variant. Histopathology71, 125–133 (2017). [DOI] [PubMed]
- 12.Lübbehüsen, C. et al. Characterization of three novel H3F3A-mutated giant cell tumor cell lines and targeting of their Wee1 pathway. Sci. Rep.9 (2019). [DOI] [PMC free article] [PubMed]
- 13.Berger AW, et al. Targeted deep sequencing of circulating tumor DNA in metastatic pancreatic cancer. Oncotarget. 2018;9:2076–2085. doi: 10.18632/oncotarget.23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tariq, M. U. et al. Spectrum of histological features of Denosumab treated giant cell tumor of bone; potential pitfalls and diagnostic challenges for pathologists. Ann. Diagn. Pathol.45 (2020). [DOI] [PubMed]
- 15.Mak IWY, Evaniew N, Popovic S, Tozer R, Ghert M. A translational study of the neoplastic cells of giant cell tumor of bone following neoadjuvant denosumab. J. Bone Jt. Surg. 2014;96:e127. doi: 10.2106/JBJS.M.01332. [DOI] [PubMed] [Google Scholar]
- 16.Criscitiello C, et al. Crosstalk between bone niche and immune system: Osteoimmunology signaling as a potential target for cancer treatment. Cancer Treat. Rev. 2015;41:61–68. doi: 10.1016/j.ctrv.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Ellis GK, et al. Randomized trial of denosumab in patients receiving adjuvant aromatase inhibitors for nonmetastatic breast cancer. J. Clin. Oncol. 2008;26:4875–4882. doi: 10.1200/JCO.2008.16.3832. [DOI] [PubMed] [Google Scholar]
- 18.Mori K, et al. Receptor activator of nuclear factor-κB ligand (RANKL) directly modulates the gene expression profile of RANK-positive Saos-2 human osteosarcoma cells. Oncol. Rep. 2007;18:1365–1371. [PubMed] [Google Scholar]
- 19.Mukaihara, K. et al. Protein expression profiling of giant cell tumors of bone treated with denosumab. PLoS One11 (2016). [DOI] [PMC free article] [PubMed]
- 20.Thomas D, et al. Denosumab in patients with giant-cell tumour of bone: An open-label, phase 2 study. Lancet Oncol. 2010;11:275–280. doi: 10.1016/S1470-2045(10)70010-3. [DOI] [PubMed] [Google Scholar]
- 21.Chawla S, et al. Safety and efficacy of denosumab for adults and skeletally mature adolescents with giant cell tumour of bone: interim analysis of an open-label, parallel-group, phase 2 study. Lancet. Oncol. 2013;14:901–908. doi: 10.1016/S1470-2045(13)70277-8. [DOI] [PubMed] [Google Scholar]
- 22.Rutkowski P, et al. Surgical downstaging in an open-label phase II trial of denosumab in patients with giant cell tumor of bone. Ann. Surg. Oncol. 2015;22:2860–2868. doi: 10.1245/s10434-015-4634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broehm CJ, Garbrecht EL, Wood J, Bocklage T. Two cases of sarcoma arising in giant cell tumor of bone treated with denosumab. Case Rep. Med. 2015;2015:1–6. doi: 10.1155/2015/767198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park A, Cipriano CA, Hill K, Kyriakos M, McDonald DJ. Malignant transformation of a giant cell tumor of bone treated with denosumab. JBJS Case Connect. 2016;6:e78. doi: 10.2106/JBJS.CC.16.00024. [DOI] [PubMed] [Google Scholar]
- 25.Tsukamoto S, et al. Development of high-grade osteosarcoma in a patient with recurrent giant cell tumor of the ischium while receiving treatment with denosumab. Jpn. J. Clin. Oncol. 2017;47:1090–1096. doi: 10.1093/jjco/hyx112. [DOI] [PubMed] [Google Scholar]
- 26.Chawla S, et al. Denosumab in patients with giant-cell tumour of bone: A multicentre, open-label, phase 2 study. Lancet Oncol. 2019;20:1719–1729. doi: 10.1016/S1470-2045(19)30663-1. [DOI] [PubMed] [Google Scholar]
- 27.Xu F, Flowers S, Moran E. Essential role of ARID2 protein-containing SWI/SNF complex in tissue-specific gene expression. J. Biol. Chem. 2012;287:5033–5041. doi: 10.1074/jbc.M111.279968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koelsche, C. et al. Histone 3. 3 hotspot mutations in conventional osteosarcomas : A comprehensive clinical and molecular characterization of six H3F3A mutated cases. Clin. Sarcoma Res. 1–11, 10.1186/s13569-017-0075-5 (2017). [DOI] [PMC free article] [PubMed]
- 29.Lim YH, et al. Multilineage somatic activating mutations in HRAS and NRAS cause mosaic cutaneous and skeletal lesions, elevated FGF23 and hypophosphatemia. Hum. Mol. Genet. 2014;23:397–407. doi: 10.1093/hmg/ddt429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Y. et al. MiR-200c regulates tumor growth and chemosensitivity to cisplatin in osteosarcoma by targeting AKT2. Sci. Rep.7 (2017). [DOI] [PMC free article] [PubMed]
- 31.Zhang G, Li M, Zhu X, Bai Y, Yang C. Knockdown of Akt sensitizes osteosarcoma cells to apoptosis induced by cisplatin treatment. Int. J. Mol. Sci. 2011;12:2994–3005. doi: 10.3390/ijms12052994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Zhou J, Ji Y, Yu B. Elevated expression of AKT2 correlates with disease severity and poor prognosis in human osteosarcoma. Mol. Med. Rep. 2014;10:737–742. doi: 10.3892/mmr.2014.2314. [DOI] [PubMed] [Google Scholar]
- 33.Palmerini, E., Picci, P., Reichardt, P. & Downey, G. Malignancy in giant cell tumor of bone: A review of the literature. Technol. Cancer Res. Treat.18 (2019). [DOI] [PMC free article] [PubMed]
- 34.Dahlin, D. C. Caldwell lecture. Giant cell tumor of bone: Highlights of 407 cases. AJR. Am. J. Roentgenol.144, 955–960 (1985). [DOI] [PubMed]
- 35.Bertoni F, Bacchini P, Staals EL. Malignancy in giant cell tumor of bone. Cancer. 2003;97:2520–2529. doi: 10.1002/cncr.11359. [DOI] [PubMed] [Google Scholar]
- 36.Dahlin;, M. R. S. U. W. F. S. B. Secondary malignant giant-cell tumor of bone. Clinicopathological assessment of nineteen patients. J. Bone Jt. Surg.68, 1073–1079 (1986). [PubMed]
- 37.Scotto di Carlo, F., Whyte, M. P. & Gianfrancesco, F. The two faces of giant cell tumor of bone. Cancer Lett. 489, 1–8 (2020). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.