Abstract
Background
Propofol is a commonly used anesthetic for cancer surgery. Previous studies have shown that propofol has an anticancer role in various cancers, including lung cancer. This study aimed to investigate the role of propofol in lung cancer and its underlying mechanism.
Methods
Cell proliferation was determined by cell counting kit‐8 (CCK‐8) and colony formation assays. Flow cytometry and transwell assays were used to detect cell apoptosis and invasion, respectively. Glycolysis was evaluated by detecting glucose consumption, lactate production and ATP/ADP ratios. The levels of circular RNA erb‐b2 receptor tyrosine kinase 2 (circ‐ERBB2), microRNA‐7‐5p (miR‐7‐5p) and forkhead box M1 (FOXM1) were tested by quantitative real‐time PCR and Western blot. The binding relationship between miR‐7‐5p and circ‐ERBB2/FOXM1 was verified by dual‐luciferase reporter assay. Moreover, in vivo experiments were performed by establishing a mouse xenograft model.
Results
Propofol suppressed cell proliferation, invasion and glycolysis and expedited apoptosis in lung cancer cells. Circ‐ERBB2 and FOXM1 were upregulated, while miR‐7‐5p was decreased in lung cancer tissues and cells. Propofol suppressed lung cancer cell progression by regulating circ‐ERBB2. Additionally, miR‐7‐5p directly interacted with circ‐ERBB2 and FOXM1. Also, propofol played an antitumor role in lung cancer via modulating miR‐7‐5p or FOXM1. Moreover, circ‐ERBB2 knockdown enhanced the suppressive effect of propofol on tumor growth in vivo.
Conclusions
Propofol inhibited lung cancer progression via mediating circ‐ERBB2/miR‐7‐5p/FOXM1 axis, which might provide an effective therapeutic target for lung cancer therapy.
Keywords: circ‐ERBB2, FOXM1, lung cancer, miR‐7‐5p, propofol
Propofol inhibited lung cancer progression via modulating the circ‐ERBB2/miR‐7‐5p/FOXM1 network. These findings might provide an effective therapeutic target for lung cancer.
INTRODUCTION
Lung cancer ranks first in cancer‐related deaths globally, accounting for about 18.4%. 1 The high incidence and mortality of lung cancer poses a huge threat to public health problems, and its five‐year survival rate is only about 4%–17%. 2 In recent years, molecular diagnosis has been widely used in the treatment of lung cancer patients, 3 but the prognosis of lung cancer patients is still unsatisfactory. Hence, in‐depth exploration of the molecular mechanism of lung cancer progression is essential for improving poor prognosis.
In addition, studies have corroborated that the administration of anesthetics during cancer resection exerts a suppressive effect on tumor recurrence or metastasis. 4 Propofol is a short‐acting intravenous anesthetic commonly used in surgery and is related to the pathophysiological process of many diseases. 5 , 6 Existing studies have identified that propofol plays an anticancer role in various cancers by regulating non‐coding RNAs and multiple signaling pathways. 7 Nevertheless, the underlying mechanism of propofol‐induced antitumor activity remains largely unknown.
In recent years, long noncoding RNA (lncRNA) has been extensively studied in cancer. For example, cytoplasmic lncRNAs can play an anticancer role via activating the p53 pathway. 8 Wang et al. found that LINC00336 hindered ferroptosis via acting as a competing endogenous RNA (ceRNA) in lung cancer. 9 In comparison, the research on circular RNAs (circRNAs) is relatively rare and needs further exploration. CircRNAs are endogenous RNA molecules with no 5′ to 3′ polarity obtained by back‐splicing, which results in their high resistance to exonuclease. 10 Mounting reports have validated that circRNAs participate in tumor development and progression by regulating a variety of biological functions, including cell growth, invasion, metabolism and angiogenesis. 11 In addition, several studies have demonstrated that circRNAs act as critical regulators in lung cancer. 12 , 13 High‐throughput sequencing results showed that hsa_circRNA_102063 (hsa_circ_0007766) derived from erb‐b2 receptor tyrosine kinase 2 (ERBB2) gene was highly expressed in lung cancer. Xu et al. found that hsa‐circ‐0007766 (circ‐ERBB2) accelerated the malignant growth of gastric carcinoma via directly targeting microRNA‐1233–3p. 14 However, the expression pattern and role of circ‐ERBB2 in propofol‐exposed lung cancer have not been studied.
Moreover, increasing evidence has corroborated that circRNAs disrupt the suppressive effect of microRNAs (miRNAs) on target genes via serving as miRNA sponges. 15 Also, miRNAs aberrantly expressed in lung cancer have become promising diagnostic and prognostic biomarkers through their ability to regulate biological networks. 16 For example, interference of miR‐324‐3p impeded lung cancer progression via regulating ALX4 to inactivate the MAPK pathway. 17 Furthermore, upregulation of miR‐550a‐5p contributed to the growth of lung adenocarcinoma by suppressing LIMD1. 18 Additionally, Xiao et al. suggested that miR‐7‐5p was conspicuously decreased in NSCLC, and its overexpression hindered NSCLC tumorigenesis through negatively regulating NOVA2. 19 Nonetheless, the role of miR‐7‐5p in propofol‐treated lung cancer remains unknown.
Hence, we first verified the antitumor effect of propofol. Furthermore, we investigated the possible association between propofol and circ‐ERBB2, and explored the potential mechanism of propofol in regulating lung cancer progression.
METHODS
Clinical samples
Lung cancer tissues (n = 31) and adjacent normal tissues (n = 31) were collected from lung cancer patients undergoing surgical resection at Henan Provincial People's Hospital. This research was ratified by the Ethics Committee of Henan Provincial People's Hospital. All patients carefully read and signed their written informed consent. All tissues were immediately stored at −80°C for follow‐up studies.
Cell culture and propofol treatment
Human bronchial epithelial cells (HBE) were purchased from Procell Life Science & Technology Co., Ltd. Four lung cancer cell lines (SK‐MES‐1, H1650, Calu‐3, H460, H1299 and A549) were commercially acquired from American Type Culture Collection (ATCC). All cells were cultured in RPMI‐1640 medium (Youkang Biotech) containing 10% fetal bovine serum (FBS; Youkang Biotech) with 5% CO2 at 37°C.
For propofol treatment, cells were stimulated with different concentrations (5, 10, and 15 μg/ml) of propofol (Solarbio) dissolved with dimethyl sulfoxide (DMSO; Solarbio) for 48 h. In addition, cells exposed to the equal amount of DMSO (Solarbio) served as blank control. In subsequent experiments, the cells were incubated with propofol for 48 h.
Cell transfection
Circ‐ERBB2 overexpression vector (circ‐ERBB2) and the control (pCD5‐ciR), miR‐7‐5p mimics (miR‐7‐5p) and the control (miR‐NC), circ‐ERBB2 small interfering RNA (si‐circ‐ERBB2) and the control (si‐NC), miR‐7‐5p inhibitor (anti‐miR‐7‐5p) and the control (anti‐miR‐NC), forkhead box M1 (FOXM1) overexpression vector (FOXM1) and negative control (pcDNA) were synthesized from Genechem. Lipofectamine 3000 (Invitrogen) was applied for cell transfection when cell confluence reached ~80%.
Cell viability assay
The treated H1299 and A549 cells were plated into 96‐well plates and incubated for 24 h. Afterwards, 10 μl of cell counting kit‐8 solution (CCK‐8; Boster) was injected into each well and cultivated for 4 h. Finally, the optical density at 450 nm was measured using a microplate reader (Bio‐Rad).
Colony formation assay
After different treatments, H1299 and A549 cells (100 cells/well) were injected into six‐well plates. After 14 days of cultivation, the colonies were fixed with formaldehyde (Solarbio) and stained with 0.1% crystal violet (Solarbio). Finally, the colonies were calculated under a microscope (Olympus).
Flow cytometry
The cell apoptosis rate was monitored via Annexin V‐FITC Apoptosis Detection Kit (Vazyme). Briefly, the treated H1299 and A549 cells were harvested and washed with PBS (Solarbio). Afterwards, the cells were stained with Annexin V‐FITC and propidium iodide (PI), and the apoptotic cells were determined using flow cytometer (Beckman Coulter).
Transwell assay
Cell invasion capacity was evaluated via 24‐well transwell chambers (Corning) equipped with Matrigel (Corning). The treated H1299 and A549 cells were seeded into the upper chamber. Meanwhile, the bottom chamber was filled with 10% FBS. Following 24 h of cultivation, the invaded cells were stained with 0.1% crystal violet (Solarbio) and counted under a microscope (Olympus) at 100× magnification.
Detection of glucose consumption, lactate production and ATP/ADP ratios
Glycolysis was assessed by measuring glucose consumption, lactate production and ATP/ADP ratios. Glucose consumption and lactate production were detected in lung cancer cell culture medium by using Glucose Assay Kit and Lactic Acid Kit (Abcam) according to the manufacturer's instructions. Relative ATP/ADP ratios were measured using ApoSENSOR ADP/ATP Ratio Assay Kit (BioVision) following the manufacturer's instructions.
Quantitative real‐time PCR (qRT‐PCR)
Total RNA was extracted using TRIzol reagent (Leagene). For RNase R assay, 2 μg of RNA was reacted with RNase R (3 U/μg; Epicenter). Subsequently, cDNA was obtained using the specific cDNA synthesis kit (Takara). Next, qRT‐PCR reaction was carried out using SYBR Green Master Mix (Takara). Additionally, GAPDH and U6 (for miR‐7‐5p) were regarded as endogenous controls. The primers are shown below: circ‐ERBB2‐F: 5′‐GCCCTGGTCACCTACAACAC‐3′, circ‐ERBB2‐R: 5′‐TGGATATCAGGGACAGGCAGT‐3′; ERBB2‐F: 5′‐TGCAGGGAAACCTGGAACTC‐3′, ERBB2‐R: 5′‐ACAGGGGTGGTATTGTTCAGC‐3′; miR‐7‐5p‐F: 5′‐GCGCTGGAAGACTAGTGATTTTG‐3′, miR‐7‐5p‐R: 5′‐GTGCAGGGTCCGAGGT3‐3′; FOXM1‐F: 5′‐CGTCGGCCACTGATTCTCAAA‐3′, FOXM1‐R: 5′‐GGCAGGGGATCTCTTAGGTTC‐3′; GAPDH‐F: 5′‐GCACCGTCAAGGCTGAGAAC‐3′, GAPDH‐R: 5′‐ATGGTGGTGAAGACGCCAGT‐3′; U6‐F: 5′‐CTCGCTTCGGCAGCACATA‐3′, U6‐R: 5′‐AACGCTTCACGAATTTGCGT‐3′.
Dual‐luciferase reporter assay
The sequences of circ‐ERBB2 or FOXM1 3′UTR harboring the putative miR‐7‐5p binding site were inserted into pmirGLO vector (Promega) to form WT‐circ‐ERBB2 or WT‐FOXM1 3′UTR reporter. Simultaneously, a mutant reporter (MUT‐circ‐ERBB2 or MUT‐FOXM1 3′UTR) was constructed by changing the predicted miR‐7‐5p binding site. Finally, the corresponding vector and miR‐NC or miR‐7‐5p were cotransfected into H1299 and A549 cells, and luciferase activity was measured via Dual‐Lucy Assay Kit (Solarbio).
Western blot assay
After extraction with RIPA lysis buffer (Beyotime, Shanghai, China), the protein was quantified via BCA Protein Assay Kit (Beyotime). Afterwards, the protein was separated by 10% SDS‐PAGE and transferred onto PVDF membranes (Millipore). Following blocking with 5% non‐fat milk, the membranes were probed with primary antibodies against FOXM1 (1:1500, ab180710, Abcam) or β‐actin (1:2500, ab8227, Abcam). Afterwards, the membranes were incubated with secondary antibody (1:20000, ab205718, Abcam). Finally, the protein signal was measured via ECL system (Beyotime).
Xenograft assay
Animal experiments were conducted following the Guide for the Care and Use of Laboratory Animals and were approved by the Animal Research Ethics Committee of Henan Provincial People's Hospital. BALB/c nude mice aged five weeks were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. and randomly divided into four groups (n = 8 for each group). Lentivirus harboring circ‐ERBB2 short hairpin RNA (sh‐circ‐ERBB2) or the control (sh‐NC) were bought from Genechem and transfected into A549 cells. Subsequently, A549 cells (4 × 106) were subcutaneously injected into the backs of nude mice. After eight days, the mice were injected with DMSO or propofol (45 mg/kg) every three days, and tumor volume was monitored. The mice were sacrificed 23 days later, the xenograft tumors were weighed and harvested for RNA level analysis.
Statistical analysis
All data are presented as mean ± standard deviation by using Graphpad Prism 7.0 software (GraphPad). The differences were analyzed by Student's t‐test and one‐way analysis of variance. The association between miR‐7‐5p and circ‐ERBB2/FOXM1 was tested by Spearman's correlation analysis. p < 0.05 was considered statistically significant.
RESULTS
Propofol inhibited the proliferation, invasion and glycolysis of lung cancer cells and induced apoptosis
To determine the effect of propofol on the malignancy of lung cancer cells, HBE, H1299 and A549 cells were exposed to various doses of propofol (5, 10, and 15 μg/ml). CCK‐8 assay revealed that propofol significantly reduced the viability of H1299 and A549 cells in a dose‐dependent manner, but had no effect on the viability of HBE cells (Figure 1(a)–(c)). In addition, colony formation, flow cytometry and transwell assays showed that propofol restrained cell proliferation (Figure 1(d)) and invasion (Figure 1(f)) and promoted cell apoptosis (Figure 1(e)) in a concentration‐dependent manner. In addition, propofol remarkably reduced glucose consumption (Figure 1(g)), lactate production (Figure 1(h)) and ATP/ADP ratios (Figure 1(i)) in a dose‐dependent manner, indicating that propofol treatment hindered the glycolysis of lung cancer cells. Additionally, 10 μg/ml propofol, which reduced cell viability to ~50%, was selected for subsequent experiments.
FIGURE 1.
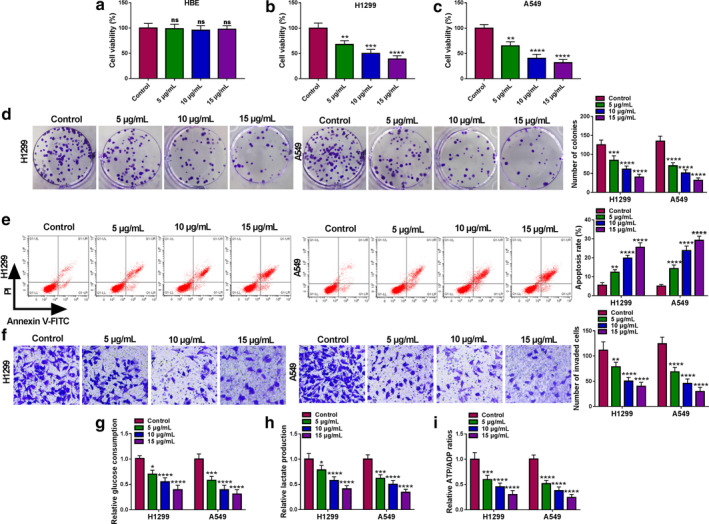
Propofol inhibited the proliferation, invasion and glycolysis of lung cancer cells and induced apoptosis. HBE, H1299 and A549 cells were treated with different doses of propofol (5, 10, and 15 μg/ml) for 48 h. (a–c) Cell viability was determined by CCK‐8 assay. (d) Cell proliferation was assessed by colony formation assay. (e) The apoptosis rate of H1299 and A549 cells was measured by flow cytometry. (f) Cell invasion was evaluated by transwell assay. (g–i) Relative glucose consumption, lactate production and ATP/ADP ratios were detected using the appropriate kit. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Circ‐ERBB2 was upregulated in lung cancer tissues and cells
To explore the potential function of circ‐ERBB2 in lung cancer, the expression pattern of circ‐ERBB2 in lung cancer tissues and cells was first determined. According to GSE158695 expression dataset, hsa_circRNA_102063 (hsa_circ_0007766; circ‐ERBB2) was significantly upregulated in lung cancer tissues relative to normal controls (Figure 2(a)). Consistently, qRT‐PCR analysis showed that circ‐ERBB2 level was markedly increased in lung cancer tissues compared to normal tissues (Figure 2(b)). Compared with human bronchial epithelial cells (HBE), circ‐ERBB2 level was remarkably elevated in lung cancer cells (SK‐MES‐1, H1650, Calu‐3, H460, H1299 and A549) (Figure 2(c)). Additionally, RNase R digestion assay showed that circ‐ERBB2 was more resistant to RNase R than linear ERBB2, indicating that circ‐ERBB2 was a stable circRNA (Figure 2(d) and (e)).
FIGURE 2.
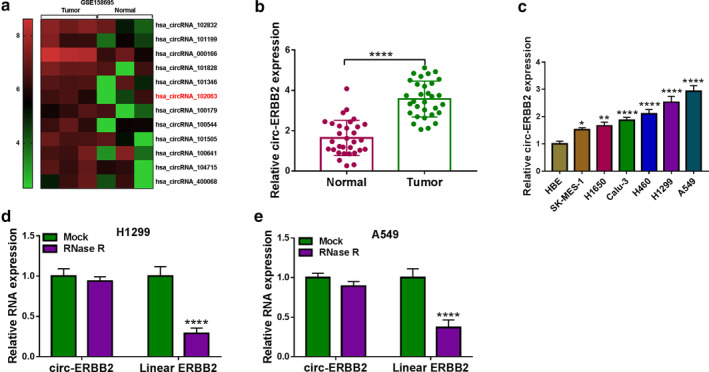
Circ‐ERBB2 was upregulated in lung cancer tissues and cells. (a) The heat map showed abnormally expressed circRNAs in lung cancer tissues and normal tissues. (b) Circ‐ERBB2 level in lung cancer tissues (n = 31) and adjacent normal tissues (n = 31) was measured by qRT‐PCR. (c) The expression of circ‐ERBB2 in HBE cells and lung cancer cells (SK‐MES‐1, H1650, Calu‐3, H460, H1299 and A549) was examined by qRT‐PCR. (d and e) After RNase R treatment, the levels of circ‐ERBB2 and linear ERBB2 were tested using qRT‐PCR. **p < 0.01, ***p < 0.001, ****p < 0.0001
Circ‐ERBB2 reversed the suppressive effect of propofol on lung cancer cell progression
Next, we examined the expression of circ‐ERBB2 in H1299 and A549 cells exposed to different concentrations of propofol. The results suggested that propofol dose‐dependently decreased the level of circ‐ERBB2 (Figure 3(a)). To investigate whether circ‐ERBB2 played a role in the regulation of propofol in lung cancer, H1299 and A549 cells transfected with pCD5‐ciR or circ‐ERBB2 were treated with 10 μg/ml propofol. First, qRT‐PCR showed that the overexpression efficiency of circ‐ERBB2 was significant (Figure 3(b)). In addition, introduction of circ‐ERBB2 restored the reduction in circ‐ERBB2 level caused by propofol treatment (Figure 3(c)). CCK‐8 and colony formation assays revealed that upregulation of circ‐ERBB2 abolished the inhibitory effect of propofol exposure on lung cancer cell proliferation (Figure 3(d) and (e)). Flow cytometry and transwell assays showed that overexpression of circ‐ERBB2 reversed the promotion of propofol on lung cancer cell apoptosis and the inhibition of propofol on cell invasion (Figure 3(f) and (g)). Additionally, propofol exposure reduced glucose consumption, lactate production and ATP/ADP ratios, while these changes were reversed by upregulating circ‐ERBB2 (Figure 3(h)–(j)). Collectively, these data indicated that propofol alleviated lung cancer cell progression via regulating circ‐ERBB2.
FIGURE 3.
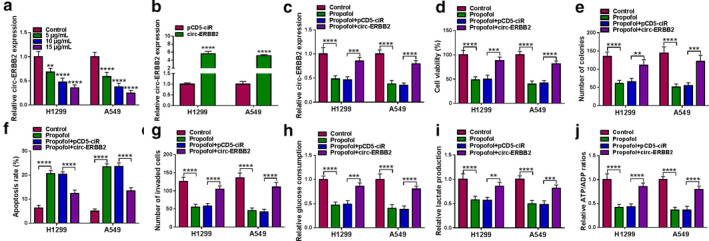
Circ‐ERBB2 reversed the suppressive effect of propofol on lung cancer cell progression. (a) circ‐ERBB2 expression was measured by qRT‐PCR in H1299 and A549 cells exposed to different doses of propofol (5, 10, and 15 μg/ml) for 48 h. (b) The overexpression efficiency of circ‐ERBB2 was determined by qRT‐PCR. (c–j) H1299 and A549 cells were introduced with pCD5‐ciR or circ‐ERBB2, followed by treatment with propofol (10 μg/ml) for 48 h. Circ‐ERBB2 expression (c), cell viability (d), colony number (e), apoptosis rate (f) and cell invasion (g) were detected by qRT‐PCR, CCK‐8, colony formation, flow cytometry and transwell assays. (h–j) Relative glucose consumption, lactate production and ATP/ADP ratios were examined via the appropriate kits. **p < 0.01, ***p < 0.001, ****p < 0.0001
Circ‐ERBB2 directly interacted with miR‐7‐5p
In addition, we predicted miRNAs that might bind to circ‐ERBB2 and screened seven miRNAs (miR‐1205, miR‐187, miR‐377, miR‐503, miR‐767‐3p, miR‐940 and miR‐7‐5p) that were downregulated in lung cancer. Then, the expression of seven miRNAs was detected by qRT‐PCR in A549 cells transfected with pCD5‐ciR or circ‐ERBB2. The results showed that the downregulation of miR‐7‐5p was the most significant after circ‐ERBB2 overexpression, so miR‐7‐5p was selected for further research (Figure S1). Moreover, we predicted that miR‐7‐5p might be combined with circ‐ERBB2 through the online database circular RNA interactome (Circinteractome) (Figure 4(a)). As shown in Figure 4(b), miR‐7‐5p level was strikingly increased in the miR‐7‐5p mimics group compared with the miR‐NC group. Afterwards, the binding relationship between miR‐7‐5p and circ‐ERBB2 was verified by dual‐luciferase reporter assay, and the results showed that miR‐7‐5p mimics strikingly decreased the luciferase activity of WT‐circ‐ERBB2 in lung cancer cells (Figure 4(c) and (d)). Compared with the control group, miR‐7‐5p expression was prominently decreased in lung cancer tissues and cells (Figure 4(e) and (g)). In addition, Spearman's correlation analysis showed that circ‐ERBB2 level was negatively correlated with miR‐7‐5p level in lung cancer tissues (Figure 4(f)). Additionally, qRT‐PCR analysis showed that the knockdown efficiency of circ‐ERBB2 was significant after transfection with si‐circ‐ERBB2 in H1299 and A549 cells (Figure 4(h)). In addition, circ‐ERBB2 silence significantly increased miR‐7‐5p level, and circ‐ERBB2 overexpression strikingly reduced miR‐7‐5p level (Figure 4(i)). These data demonstrated that circ‐ERBB2 acted as a sponge for miR‐7‐5p.
FIGURE 4.
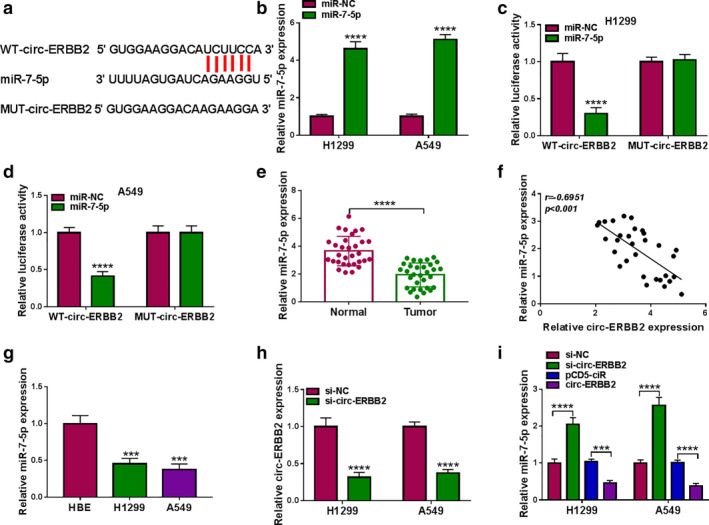
Circ‐ERBB2 directly interacted with miR‐7‐5p. (a) The potential binding sequence between circ‐ERBB2 and miR‐7‐5p was speculated through circular RNA interactome (Circinteractome). (b) The overexpression efficiency of miR‐7‐5p was confirmed by qRT‐PCR after transfection with miR‐7‐5p mimics. (c and d) The targeting relationship was validated by dual‐luciferase reporter assay. (e) The expression of miR‐7‐5p in lung cancer tissues (n = 31) and matched normal tissues (n = 31) was detected by qRT‐PCR. (f) Spearman's correlation coefficient was utilized to assess the relationship between circ‐ERBB2 and miR‐7‐5p. (g) The level of miR‐7‐5p in HBE and lung cancer cells (H1299 and A549) was detected by qRT‐PCR. (h) The transfection efficiency of circ‐ERBB2 siRNA was examined by qRT‐PCR. (i) miR‐7‐5p level was measured by qRT‐PCR in H1299 and A549 cells transfected with si‐NC, si‐circ‐ERBB2, pCD5‐ciR or circ‐ERBB2. ***p < 0.001, ****p < 0.0001
Propofol suppressed lung cancer cell progression via modulating miR‐7‐5p
As shown in Figure 5(a), propofol remarkably increased miR‐7‐5p expression in a dose‐dependent manner. Thus, we speculated that miR‐7‐5p might be involved in the regulation of propofol in lung cancer development. First, H1299 and A549 cells were transfected with anti‐miR‐NC or anti‐miR‐7‐5p, and qRT‐PCR assay showed that miR‐7‐5p level was markedly reduced after the introduction of anti‐miR‐7‐5p (Figure 5(b)). Subsequently, the transfected cells were exposed to 10 μg/ml propofol, and the results suggested that transfection of anti‐miR‐7‐5p abated the increase in miR‐7‐5p level induced by propofol exposure (Figure 5(c)). Rescue experiments revealed that knockdown of miR‐7‐5p overturned the suppressive effect of propofol on proliferation (Figure 5(d) and (e)) and invasion (Figure 5(g)) and the promotion of propofol on apoptosis (Figure 5(f)) in H1299 and A549 cells. Additionally, downregulation of miR‐7‐5p rescued the reduction in glucose consumption, lactate production and ATP/ADP ratios caused by propofol treatment (Figure 5(h) and (j)). These data confirmed that propofol inhibited lung cancer cell development by modulating miR‐7‐5p. In addition, a series of functional experiments showed that miR‐7‐5p overexpression enhanced the suppressive effect of propofol on lung cancer cell progression (Figure S2).
FIGURE 5.
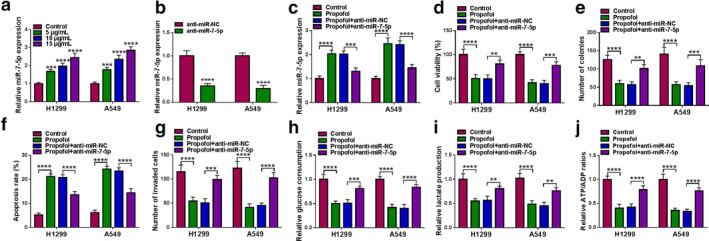
Propofol suppressed lung cancer cell progression via modulating miR‐7‐5p. (a) miR‐7‐5p level was detected by qRT‐PCR in H1299 and A549 cells stimulated with different doses of propofol (5, 10, and 15 μg/ml) for 48 h. (b) The transfection efficiency of miR‐7‐5p inhibitor was verified by qRT‐PCR. (c–j) H1299 and A549 cells were transduced with anti‐miR‐NC or anti‐miR‐7‐5p, and then exposed to propofol (10 μg/ml) for 48 h. (c) The level of miR‐7‐5p; (d) cell viability; (e) number of colonies; (f) apoptosis rate; and (g) cell invasion capacity were assessed by corresponding methods. (h–j) Relative glucose consumption, lactate production and ATP/ADP ratios were detected via the appropriate kit. **p < 0.01, ***p < 0.001, ****p < 0.0001
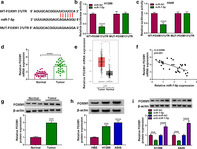
miR‐7‐5p directly targeted FOXM1
According to the StarBase2.0 online database, the putative binding site of miR‐7‐5p was located in FOXM1 3′UTR (Figure 6(a)). Next, dual‐luciferase reporter assay exhibited that miR‐7‐5p mimics prominently reduced the luciferase activity of WT‐FOXM1 3′UTR in lung cancer cells (Figure 6(b) and (c)). Moreover, qRT‐PCR analysis showed that FOXM1 level was significantly increased in lung cancer tissues relative to normal tissues (Figure 6(d)), which was consistent with the previous data (Figure 6(e)). Furthermore, Spearman's correlation coefficient showed that miR‐7‐5p level was negatively correlated with FOXM1 level in lung cancer tissues (Figure 6(f)). As expected, FOXM1 protein level was remarkably elevated in lung cancer tissues and cells compared with the control group (Figure 6(g) and (h)). In addition, miR‐7‐5p upregulation impeded the expression of FOXM1, and miR‐7‐5p knockdown induced the expression of FOXM1 (Figure 6(i)). Thus, these data indicated that miR‐7‐5p negatively regulated FOXM1 expression.
FIGURE 6.
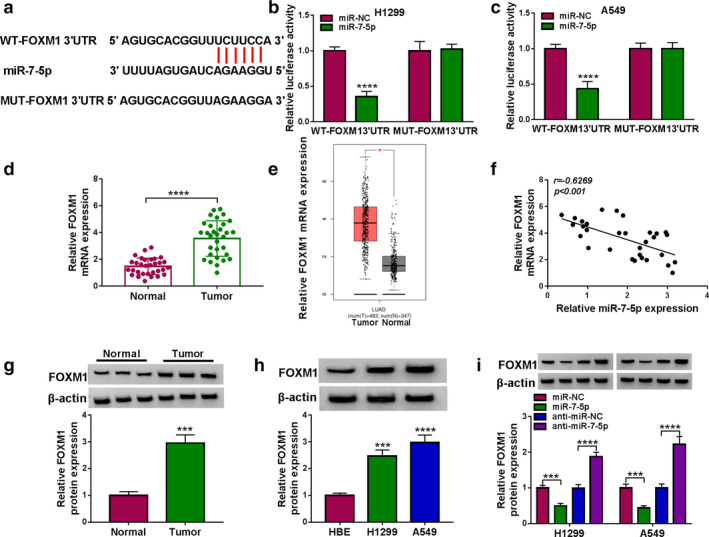
miR‐7‐5p directly targeted FOXM1. (a) The putative binding site between miR‐7‐5p and FOXM1 3′UTR was predicted by StarBase2.0. (b and c) The binding relationship was confirmed by dual‐luciferase reporter assay. (d) FOXM1 mRNA level in lung cancer tissues (n = 31) and matched normal tissues (n = 31) was examined using qRT‐PCR. (e) TCGA data showed that FOXM1 was upregulated in lung cancer tissues. (f) The correlation between miR‐7‐5p and FOXM1 was tested by Spearman's correlation coefficient. (g and h) FOXM1 protein level was detected by Western blot in lung cancer tissues and cells. (i) FOXM1 protein level was measured using Western blot in H1299 and A549 cells transfected with miR‐NC, miR‐7‐5p, anti‐miR‐NC or anti‐miR‐7‐5p. *p < 0.05, ***p < 0.001, ****p < 0.0001
Propofol alleviated lung cancer cell progression by regulating FOXM1
We also examined the expression of FOXM1 in H1299 and A549 cells exposed to different doses of propofol. Western blot analysis showed that propofol dose‐dependently reduced the protein level of FOXM1 (Figure 7(a)). To elucidate the relationship between propofol and FOXM1 in lung cancer progression, pcDNA or FOXM1 was introduced into H1299 and A549 cells before propofol treatment. As shown in Figure 7(b), the overexpression efficiency of FOXM1 was significant. Also, Western blot assay suggested that FOXM1 transfection mitigated the decrease in FOXM1 protein level caused by propofol stimulation (Figure 7(c)). Additionally, propofol exposure suppressed cell proliferation and invasion and accelerated cell apoptosis in H1299 and A549 cells, while these effects were reversed after the introduction of FOXM1 (Figure 7(d)–(g)). Furthermore, upregulation of FOXM1 restored propofol‐mediated inhibition on glucose consumption, lactate production and ATP/ADP ratios (Figure 7(h)–(j)). Altogether, these data indicated that FOXM1 attenuated the effect of propofol on lung cancer cell development.
FIGURE 7.
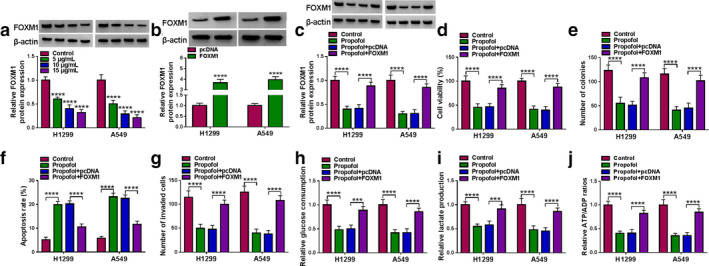
Propofol alleviated lung cancer cell progression by regulating FOXM1. (a) FOXM1 protein level was detected by Western blot in H1299 and A549 cells treated with various doses of propofol (5, 10, and 15 μg/ml) for 48 h. (b) The overexpression efficiency of FOXM1 was validated via Western blot. (c–j) H1299 and A549 cells transfected with pcDNA or FOXM1 were stimulated with propofol (10 μg/ml) for 48 h. (c) FOXM1 protein level; (d) cell viability; (e) number of colonies; (f) apoptosis rate; and (g) cell invasion were evaluated by corresponding methods. (h–j) Relative glucose consumption, lactate production and ATP/ADP ratios were measured using the appropriate kit. ***p < 0.001, ****p < 0.0001
Propofol regulated FOXM1 expression via circ‐ERBB2/miR‐7‐5p/FOXM1 axis
To clarify whether propofol affected lung cancer cell progression through circ‐ERBB2/miR‐7‐5p/FOXM1 axis, we detected FOXM1 protein expression in H1299 and A549 cells after different treatments. As illustrated in Figure 8(a), circ‐ERBB2 transfection recovered the decrease in FOXM1 protein level caused by propofol stimulation. Meanwhile, anti‐miR‐7‐5p transfection restored propofol exposure‐resulted decrease in FOXM1 protein level (Figure 8(b)). Moreover, circ‐ERBB2 silencing remarkably reduced FOXM1 protein expression in H1299 and A549 cells, while this change was abolished by cotransfecting with si‐circ‐ERBB2 and anti‐miR‐7‐5p (Figure 8(c)). These data indicated that propofol regulated lung cancer cell progression via modulating the circ‐ERBB2/miR‐7‐5p/FOXM1 axis.
FIGURE 8.
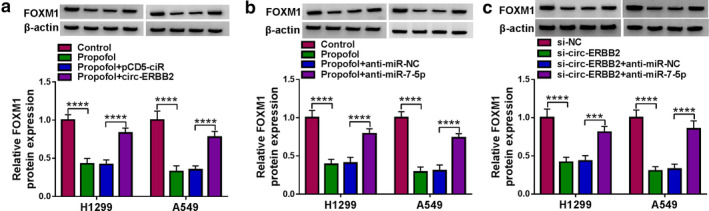
Propofol regulated FOXM1 expression via the circ‐ERBB2/miR‐7‐5p/FOXM1 axis. (a) H1299 and A549 cells transfected with pCD5‐ciR or circ‐ERBB2 were exposed to propofol (10 μg/ml) for 48 h, and FOXM1 protein level was measured by Western blot. (b) H1299 and A549 cells transfected with anti‐miR‐NC or anti‐miR‐7‐5p were treated with propofol (10 μg/ml) for 48 h, and FOXM1 expression was detected by Western blot. (c) FOXM1 protein level was examined by Western blot in H1299 and A549 cells transfected with si‐NC, si‐circ‐ERBB2, si‐circ‐ERBB2 + anti‐miR‐NC or si‐circ‐ERBB2 + anti‐miR‐7‐5p. ***p < 0.001, ****p < 0.0001
Circ‐ERBB2 knockdown enhanced the anticancer effect of propofol in vivo
To investigate the effects of propofol and circ‐ERBB2 on tumorigenesis in vivo, A549 cells harboring sh‐NC or sh‐circ‐ERBB2 were subcutaneously injected into the nude mice. Afterwards, the mice were given DMSO or propofol (45 mg/kg) every three days. As shown in Figure 9(a) and (b), circ‐ERBB2 knockdown markedly decreased tumor volume and weight, and this effect was enhanced by propofol. In addition, transfection of sh‐circ‐ERBB2 reduced circ‐ERBB2 expression and FOXM1 protein level and elevated miR‐7‐5p level, while these trends were reinforced after propofol stimulation (Figure 9(c)–(e)). These data suggested that circ‐ERBB2 silencing strengthened the inhibitory effect of propofol on tumor growth in vivo.
FIGURE 9.
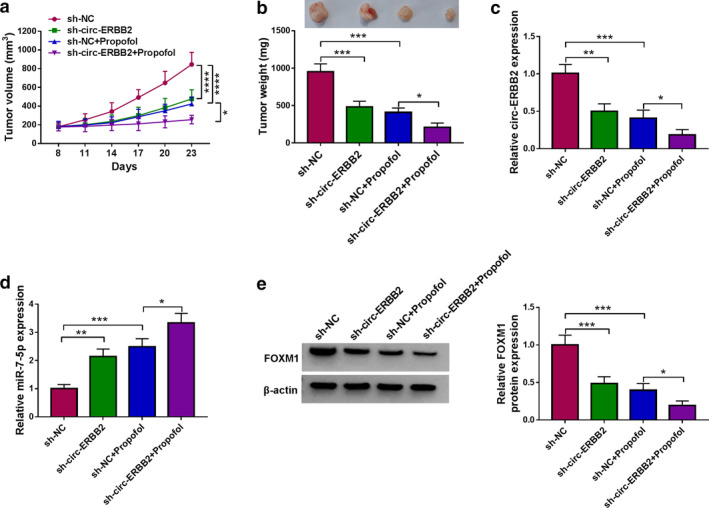
Circ‐ERBB2 knockdown enhanced the anticancer effect of propofol in vivo. A549 cells transfected with sh‐NC or sh‐circ‐ERBB2 were injected into the nude mice. (a) After eight days, the mice were injected with DMSO or propofol (45 mg/kg) every three days, and tumor volume was monitored. (b) After 23 days, the mice were killed and the xenografts were weighed. (c–e) The levels of circ‐ERBB2, miR‐7‐5p and FOXM1 were measured using qRT‐PCR and Western blot. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
DISCUSSION
Recently, several studies have demonstrated that propofol is strongly related to the prognosis of cancer and occupies an important position in the mechanism of cancer progression. 20 Additionally, some references have confirmed that propofol is implicated in lung cancer development. For instance, propofol has been reported to alleviate cell viability and facilitate cell apoptosis in NSCLC by inhibiting miR‐21‐5p and elevating MAPK10. 21 Liu et al. discovered that propofol restrained lung cancer cell growth and metastasis via activating miR‐1284. 22 Also, propofol increased the sensitivity of cisplatin in lung cancer by suppressing Wnt/β‐catenin signaling. 23 In the current research, we unveiled that propofol blocked tumorigenesis and glycolysis in lung cancer, which was in agreement with previous studies. 24 , 25
Propofol regulates tumor progression through various pathways, including mediating the expression of non‐coding RNAs and acting as a regulator of different signaling pathways. 26 For example, propofol attenuated the malignant behaviors of gastric cancer via mediating the circ‐PVT1/miR‐195‐5p/ETS1 network. 27 Also, propofol suppressed tumor development and glycolysis via modulating the circTADA2A/miR‐455‐3p/FOXM1 pathway. 24 Therefore, we speculated that propofol might influence the progression of lung cancer by regulating other circRNAs. In the present research, propofol stimulation reduced the high expression of circ‐ERBB2 in lung cancer. In addition, circ‐ERBB2 overexpression overturned the suppressive effect of propofol on lung cancer malignancies.
In addition, growing evidence has identified that circRNAs regulate a series of biological functions through the ceRNA network. 15 , 28 For example, circ‐ERBB2 triggered gastric cancer development via sponging miR‐503/miR‐637 to upregulate CACUL1/MMP‐19. 29 In the present research, miR‐7‐5p was selected as a potential target for circ‐ERBB2 based on previous references, and this targeting relationship was verified through experiments. In lung cancer, low expression of miR‐7‐5p predicted unfavorable prognosis, and miR‐7‐5p upregulation hindered tumor progression via downregulating PAK2. 30 In this study, propofol exposure remarkably elevated the miR‐7‐5p level. Furthermore, depletion of miR‐7‐5p undermined the suppressive effect of propofol on lung cancer progression.
Previous studies have corroborated that miRNAs contribute to translational repression and mRNA degradation via combining with 3′UTR of mRNAs. 31 Based on bioinformatic and dual‐luciferase reporter analysis, our research validated that FOXM1 directly interacted with miR‐7‐5p. FOXM1 is a transcription factor belonging to the Forkhead box family, which is essential for cell cycle progression. 32 Numerous studies have identified that FOXM1 expression is increased in diverse cancers, including lung carcinoma. 33 In addition, propofol mitigated the malignancy of lung cancer via mediating miR‐1284/FOXM1 axis. 22 In the current study, propofol stimulation prominently decreased FOXM1 level, and upregulation of FOXM1 rescued the inhibition of propofol on lung cancer development. More importantly, propofol reduced the expression of FOXM1 through the circ‐ERBB2/miR‐7‐5p/FOXM1 axis.
In conclusion, propofol inhibited lung cancer progression by modulating the circ‐ERBB2/miR‐7‐5p/FOXM1 network. These findings might provide an effective therapeutic target for lung cancer.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Supporting information
Figure S1 The expression of seven possible target miRNAs (miR‐1205, miR‐187, miR‐377, miR‐503, miR‐767‐3p, miR‐940 and miR‐7‐5p) was measured by qRT‐PCR in A549 cells transfected with pCD5‐ciR or circ‐ERBB2. ***p < 0.001, ****p < 0.0001.
Figure S2 miR‐7‐5p mimics enhanced the repressive effect of propofol on lung cancer cell progression. H1299 and A549 cells were transfected with miR‐NC or miR‐7‐5p, followed by treatment with propofol (10 μg/ml) for 48 h. miR‐7‐5p level (a), cell viability (b), colony number (c), apoptosis rate (d), cell invasion ability (e), glucose consumption (f), lactate production (g) and ATP/ADP ratios (h) were examined via corresponding methods. **p < 0.01, ***p < 0.001, ****p < 0.0001.
Gao J, Ding C, Zhou J, et al. Propofol suppresses lung cancer tumorigenesis by modulating the circ‐ERBB2/miR‐7‐5p/FOXM1 axis. Thorac Cancer. 2021;12:824–834. 10.1111/1759-7714.13856
Contributor Information
Jie Gao, Email: gaoojiee@126.com.
Chengzhi Ding, Email: dingchengzhi3017@163.com.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. [DOI] [PubMed] [Google Scholar]
- 3. Hung YP, Chirieac LR. How should molecular findings be integrated in the classification for lung cancer? Transl Lung Cancer Res. 2020;9:2245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heaney A, Buggy DJ. Can anaesthetic and analgesic techniques affect cancer recurrence or metastasis? Br J Anaesth. 2012;109(Suppl 1):i17i28. [DOI] [PubMed] [Google Scholar]
- 5. Irwin MG, Chung CKE, Ip KY, Wiles MD. Influence of propofol‐based total intravenous anaesthesia on peri‐operative outcome measures: a narrative review. Anaesthesia. 2020;75(Suppl 1):e90–e100. [DOI] [PubMed] [Google Scholar]
- 6. Bateman BT, Kesselheim AS. Propofol as a transformative drug in anesthesia: insights from key early investigators. Drug Discov Today. 2015;20:1012–7. [DOI] [PubMed] [Google Scholar]
- 7. Xu Y, Pan S, Jiang W, Xue F, Zhu X. Effects of propofol on the development of cancer in humans. Cell Prolif. 2020;53:e12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, et al. A G3BP1‐interacting lncRNA promotes Ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26:2329–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90. [DOI] [PubMed] [Google Scholar]
- 11. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–36. [DOI] [PubMed] [Google Scholar]
- 12. Drula R, Braicu C, Harangus A, Nabavi SM, Trif M, Slaby O, et al. Critical function of circular RNAs in lung cancer. WIREs RNA. 2020;11:e1592. [DOI] [PubMed] [Google Scholar]
- 13. Zhang C, Ma L, Niu Y, Wang Z, Xu X, Li Y, et al. Circular RNA in lung cancer research: biogenesis, functions, and roles. Int J Biol Sci. 2020;16:803–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu W, Zhou B, Wu J, Jiang P, Chen H, Yan F. Circular RNA hsa‐circ‐0007766 modulates the progression of gastric carcinoma via miR‐1233‐3p/GDF15 axis. Int J Med Sci. 2020;17:1569–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zagryazhskaya A, Zhivotovsky B. miRNAs in lung cancer: a link to aging. Ageing Res Rev. 2014;17:54–67. [DOI] [PubMed] [Google Scholar]
- 17. Song T, Zhou H, Wei X, Meng Y, Guo Q. Downregulation of microRNA‐324‐3p inhibits lung cancer by blocking the NCAM1‐MAPK axis through ALX4. Cancer Gene Ther. 2020. [DOI] [PubMed] [Google Scholar]
- 18. Guo ZZ, Ma ZJ, He YZ, Jiang W, Xia Y, Pan CF, et al. miR‐550a‐5p functions as a tumor promoter by targeting LIMD1 in lung adenocarcinoma. Front Oncol. 2020;10:570733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao H. MiR‐7‐5p suppresses tumor metastasis of non‐small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao X, Mi Y, Guo N, Luan J, Xu H, Hu Z, et al. The mechanism of propofol in cancer development: an updated review. Asia Pac J Clin Oncol. 2020;16:e3–e11. [DOI] [PubMed] [Google Scholar]
- 21. Wu X, Li X, Xu G. Propofol suppresses the progression of nonsmall cell lung cancer via downregulation of the miR215p/MAPK10 axis. Oncol Rep. 2020;44:487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu WZ, Liu N. Propofol inhibits lung cancer A549 cell growth and epithelial‐mesenchymal transition process by upregulation of microRNA‐1284. Oncol Res. 2018;27:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Huang Y, Lei L, Liu Y. Propofol improves sensitivity of lung cancer cells to Cisplatin and its mechanism. Med Sci Monit. 2020;26:e919786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao H, Wei H, He J, Wang D, Li W, Wang Y, et al. Propofol disrupts cell carcinogenesis and aerobic glycolysis by regulating circTADA2A/miR‐455‐3p/FOXM1 axis in lung cancer. Cell Cycle. 2020;19:2538–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun H, Gao D. Propofol suppresses growth, migration and invasion of A549 cells by down‐regulation of miR‐372. BMC Cancer. 2018;18:1252. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Jiang S, Liu Y, Huang L, Zhang F, Kang R. Effects of propofol on cancer development and chemotherapy: potential mechanisms. Eur J Pharmacol. 2018;831:46–51. [DOI] [PubMed] [Google Scholar]
- 27. Sui H, Zhu C, Li Z, Yang J. Propofol suppresses gastric cancer tumorigenesis by modulating the circular RNAPVT1/miR1955p/E26 oncogene homolog 1 axis. Oncol Rep. 2020;44:1736–46. [DOI] [PubMed] [Google Scholar]
- 28. Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15:995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X, He M, Guo J, Cao T. Upregulation of circular RNA circ‐ERBB2 predicts unfavorable prognosis and facilitates the progression of gastric cancer via miR‐503/CACUL1 and miR‐637/MMP‐19 signaling. Biochem Biophys Res Commun. 2019;511:926–30. [DOI] [PubMed] [Google Scholar]
- 30. Li Q, Wu X, Guo L, Shi J, Li J. MicroRNA‐7‐5p induces cell growth inhibition, cell cycle arrest and apoptosis by targeting PAK2 in non‐small cell lung cancer. FEBS Open Bio. 2019;9:1983–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–65. [DOI] [PubMed] [Google Scholar]
- 32. Halasi M, Gartel AL. FOX(M1) news–it is cancer. Mol Cancer Ther. 2013;12:245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, Zhang J, Cui X, Yang Y, Li M, Qu J, et al. FoxM1: a novel tumor biomarker of lung cancer. Int J Clin Exp Med. 2015;8:3136–40. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The expression of seven possible target miRNAs (miR‐1205, miR‐187, miR‐377, miR‐503, miR‐767‐3p, miR‐940 and miR‐7‐5p) was measured by qRT‐PCR in A549 cells transfected with pCD5‐ciR or circ‐ERBB2. ***p < 0.001, ****p < 0.0001.
Figure S2 miR‐7‐5p mimics enhanced the repressive effect of propofol on lung cancer cell progression. H1299 and A549 cells were transfected with miR‐NC or miR‐7‐5p, followed by treatment with propofol (10 μg/ml) for 48 h. miR‐7‐5p level (a), cell viability (b), colony number (c), apoptosis rate (d), cell invasion ability (e), glucose consumption (f), lactate production (g) and ATP/ADP ratios (h) were examined via corresponding methods. **p < 0.01, ***p < 0.001, ****p < 0.0001.


