Abstract
Densitometric analysis is often used to quantify NaV1.1 protein on immunoblots, although the sensitivity and dilution linearity of the method are usually poor. Here we present a protocol for quantification of NaV1.1 in mouse brain tissues using a Meso Scale Discovery-Electrochemiluminescence (MSD-ECL) method. MSD-ECL is based on ELISA (enzyme-linked immunosorbent assay) and uses electrochemiluminescence to produce measurable signals. Two different antibodies are used in this assay to capture and detect NaV1.1 respectively in brain tissue lysate. The specificity of the antibodies is confirmed by Scn1a gene knock-out tissue. The calibration curve standards used in this assay were generated with mouse liver lysate spiked with mouse brain lysate, instead of using a recombinant protein. We showed that this method was qualified and used for quantification of NaV1.1 in mouse brain tissues with specificity, accuracy and precision.
Keywords: NaV1.1 , Scn1a, Dravet syndrome, Quantification, Meso Scale Discovery, MSD, Electrochemiluminescence, ECL, Immunoassay, Mouse, Brain
Background
NaV1.1, also known as the alpha subunit of voltage gated sodium channel, type I, is a transmembrane protein encoded by the Scn1a gene ( Meisler et al., 2010 ). Decreased expression of functional NaV1.1 leads to Dravet syndrome (DS), a severe early-onset epileptic encephalopathy ( Dravet et al., 2005 ). Expression of NaV1.1 in biological samples has been used as a non-clinical pharmacological biomarker for DS and can be measured using densitometric analysis of immunoblots. Densitometry methods are often not as accurate and sensitive as standard immunoassays. In addition, some NaV1.1 antibodies may cross react with other voltage gated sodium channels (VGSCs), including NaV1.2, NaV1.3 and NaV1.8, due to homology of the protein sequences. We developed a specific and sensitive method to quantify NaV1.1 protein in mouse brain tissue using a Meso Scale Discovery-Electrochemiluminescence (MSD-ECL) method. MSD-ECL method is ELISA (enzyme-linked immunosorbent assay) based and uses electrochemiluminescence to produce measurable signals based on labeled antibody binding to the target ( Kuhle et al., 2016 ). Scheme of antibody binding in MSD-ECL assay described in this protocol is shown in Figure 1. The MSD-ECL method has advantages of low background and high sensitivity. The specificity of this assay was confirmed by using capture and detection antibodies that were validated using Scn1a knock-out (Scn1a-/-) mouse brain tissue (Figure 2). Since NaV1.1 is a transmembrane protein, reproducible production of a high-quality reference standard in cells could be challenging. Therefore, in this method, reference standards composed of mouse liver lysate spiked with mouse brain lysate was utilized. Mouse liver lysate was selected as a diluent to maintain equivalent protein load in each standard since it did not show detectable levels of NaV1.1 protein expression (Figure 3). This “fit for purpose” MSD-ECL method was qualified and used for quantification of NaV1.1 in mouse brain tissues with specificity, accuracy and precision. This method is limited, however, to have a relative narrow dynamic range of detection: The upper limit of quantification (ULOQ) is set to 100% of the NaV1.1 expression in adult mouse brain. Further dilution of some sample lysate may be necessary if the NaV1.1 expression in those samples exceed the ULOQ in the initial run.
Figure 1. Scheme of antibody binding in MSD-ECL assay described in this protocol.
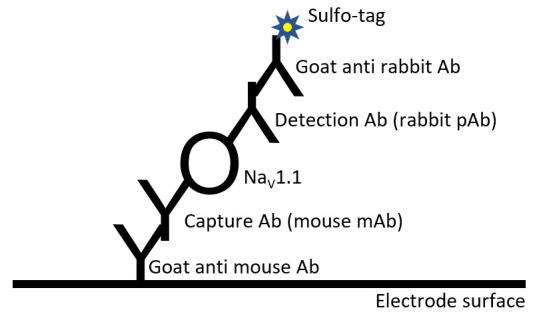
The electrode surface of each well is pre-coated with goat anti-mouse antibody (Ab). A mouse monoclonal antibody (mAb) is used as capture Ab for NaV1.1. A rabbit polyclonal antibody (pAb) is used as detection Ab. The capture and detection antibodies bind to different epitopes within NaV1.1. A Sulfo-tag labeled goat anti-rabbit Ab produces electrochemiluminescence signal.
Figure 2. Examples of NaV1.1 antibody validation. Immunoblotting was performed with brain lysate from a Scn1a-/- mouse (mid lane) and two wild type (WT) littermates (flanking lanes).
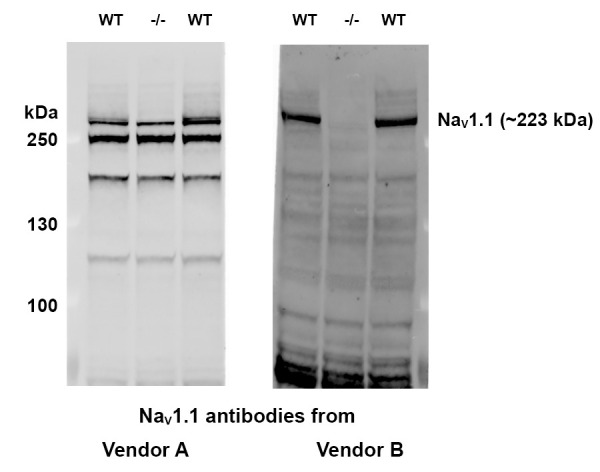
The NaV1.1 antibody from vendor A (left panel) is not specific and may cross react with other VGSCs homologs. The NaV1.1 antibody from vendor B (right panel) is specific since NaV1.1 protein band is not seen in the lane of Scn1a-/- brain lysate. Therefore, NaV1.1 antibody from Vendor B is found to be suitable for MSD-ECL assay development. (Protein ladder: PageRuller Plus)
Figure 3. Evaluation of mouse liver lysate as diluent for brain lysate.
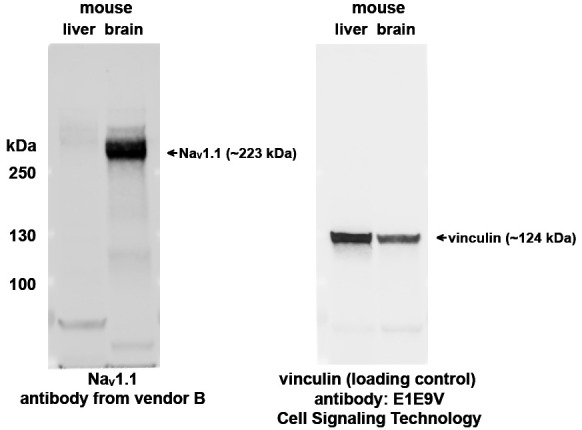
Immunoblotting was performed with mouse liver lysate and brain lysate with a validated NaV1.1 antibody (from vendor B, the same antibody as in Figure 2). Vinculin immunoblotting was shown for loading control. (Protein ladder: PageRuller Plus)
Materials and Reagents
2-ml Eppendorf microcentrifuge tubes, LoBind for protein (Fisher Scientific, catalog number: 13-698-795)
1.5-ml Eppendorf microcentrifuge tubes, LoBind for protein (Fisher Scientific, catalog number: 13-698-794)
0.2 µm nitrocellulose membrane (GE Healthcare Life Sciences, catalog number: 10600004)
MULTI-ARRAY 96 Small Spot GAM Plate (Meso Scale Discovery, catalog number: L45MA-2)
Scn1atm1Kea 129S6.Scn1a+/− mice (The Jackson Laboratory, catalog number: 37107-Jax)
Liquid nitrogen
PageRuller Plus prestained protein ladder (Thermo Fisher Scientific, catalog number: 26620)
Halt protease inhibitor cocktail (100×) (Thermo Fisher Scientific, catalog number: 78429)
Capture antibody, mouse anti NaV1.1 monoclonal antibody (NeuroMab, catalog number: 75-023)
Detection antibody, rabbit anti NaV1.1 polyclonal antibody (Alomone, catalog number: ASC-001)
Anti-Rabbit antibody, Goat, Sulfo-Tag labeled (Meso Scale Discovery, catalog number: R32AB-1)
Anti-vinculin antibody (E1E9V), rabbit monoclonal antibody (Cell Signaling Technology, catalog number: 13901)
Phosphate-buffered saline (PBS), pH 7.4 (Thermo Fisher Scientific, catalog number: 10010023)
TGX acrylamide gradient gel (Bio-Rad, catalog number: 5671124)
ECL Prime blocking reagent (GE Healthcare Life Sciences, catalog number: RPN418V)
ECL Plus Western Blotting Substrate (Thermo Fisher Scientific, catalog number: 32132)
MSD Blocker B, 2 grams (Meso Scale Discovery, catalog number: R93BB-2)
MSD read buffer T (4×) (Meso Scale Discovery, catalog number: R92TC-2)
TBS, 10× (Bio-Rad, catalog number: 170-6435)
10% Tween-20 (Bio-Rad, catalog number: 1610781)
TX-100 (Sigma-Aldrich, catalog number: T8787-100ML)
Nonidet P-40, IGEPAL CA-630 (Sigma-Aldrich, catalog number: 56741-250ML-F)
Sodium deoxycholate (Sigma-Aldrich, catalog number: D6750-25G)
0.5 M EDTA pH 8.0 (Ambion, catalog number: AM9260G)
BCA protein assay kit (Pierce, catalog number: 23227)
MSD lysis buffer (see Recipes and Note 1)
Equipment
Meso Quickplex SQ 120 (Meso Scale Discovery, model: SQ120)
Microplate shaker (Corning, model: S2020-P4-COR)
Analytical balance (Mettler Toledo, model: AB54-S)
Cryo-cup grinder (Biospec, model: 206)
Teflon coated pestle and mortar tissue grinder Size A. Chamber volume 10 ml (Thomas Scientific, model: 3431D76)
Teflon coated pestle and mortar tissue grinder Size B. Chamber volume 30 ml (Thomas Scientific, model: 3431D88)
5-speed drill press (Wen, catalog number: 4208)
Refrigerated Centrifuge (Eppendorf, model: 5430R)
Dry Blotting System (Thermo Fisher Scientific, model: iBlot 2)
Laser scanner (GE Healthcare Life Sciences, model: Typhoon FLA 9500)
Software
MSD Discovery Workbench 4.0 (Meso Scale Diagnostics, LLC., www.mesoscale.com)
GraphPad Prism 8.0 (GraphPad Software, www.graphpad.com)
Procedure
-
Preparation of tissue lysate
-
Prepare mouse liver lysate as diluent for MSD calibration curve standards
Perfuse an adult mouse liver with PBS, pH 7.4 (see Note 2) through the portal vein until the color of liver turns yellow or white ( Cabral et al., 2018 ). Cut off the liver tissue and weigh on an analytical balance (Mettler Toledo).
Freeze the liver tissue in liquid nitrogen and pulverize it using a liquid nitrogen cooled mortar and pestle Cryo-cup grinder (Biospec).
Add ice-chilled MSD lysis buffer to the pulverized liver tissue in the amount of 2 ml of buffer per 100 mg of tissue. Homogenized the tissue with a motor-driven Teflon coated mortar and pestle tissue grinder, size B (Thomas Scientific) on ice, at a speed of 740 rpm for 30 strokes.
Transfer the homogenate to 2-ml Eppendorf tubes and centrifuge at 16K rcf, 4 °C, for 15 min with a refrigerated centrifuge (Eppendorf).
Transfer the supernatant (liver lysate) to a new tube without disturbing the precipitation (containing nuclei and cellular debris) and avoid taking any fat or oil droplet floating on the surface of the lysate. Keep the supernatant on ice.
Measure the protein concentration of liver lysate following a protocol using the Bicinchoninic acid (BCA) method (Pierce, see Note 3). The protein concentration typically ranges from 5 to 7 mg/ml for liver lysate obtained from this process. Adjust protein concentration to 4 mg/ml with the MSD lysis buffer.
-
Prepare mouse brain lysate for MSD calibration curve standards and for test samples
Prepare brain lysate for standards using adult mouse brains, as NaV1.1 expression is developmentally regulated and stays at a high level in adulthood ( Han et al., 2020 ). Prepare brain lysate in similar way as liver lysate, except that brain tissue is not pulverized in liquid nitrogen before homogenization, and 1.5-ml of MSD lysis buffer is added to every 100 mg of brain tissue. Smaller sized Teflon coated mortar and pestle tissue grinder, size A (Thomas Scientific) could be used for homogenization of test samples. The protein concentration typically ranges from 5 to 7 mg/ml for brain lysate obtained from this process. At the end of preparation, adjust the concentration of brain lysate to 4 mg/ml with MSD lysis buffer.
-
-
Qualification and characterization of MSD-ECL assay
-
Validate the specificity of antibodies with Scn1a-/- mouse brain tissue (see Note 4).
Prepare brain lysate from Scn1a-/- mice and wild type (WT) littermates. We bred the Scn1atm1Kea 129S6.Scn1a+/− mice (The Jackson Laboratory) to create Scn1a-/- mice. Alternatively, NaV1.1 null tissue could be obtained from CRISPR engineered Scn1a knock-out cell culture.
Perform SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) with 50 µg of brain lysate on each lane of a precast TGX acrylamide gradient gel (Bio-Rad). Run SDS-PAGE at 120 V for ~90 min.
Transfer total protein to a 0.2 µm nitrocellulose membrane (GE Healthcare Life Sciences) with a dry blotting system (Thermo Fisher Scientific). Use the following transfer program for protein transfer: step 1: 20 V for 1 min; step 2: 23 V for 4 min; step 3: 25 V for 5 min.
Perform immunoblotting with the test antibodies. Block the membrane with 2% ECL Prime blocking reagent (GE Healthcare Life Sciences) for 30 min. Use Tris-buffered saline containing 0.1% Tween-20 (TBS-T) to prepare the blocking buffer. Use the blocking buffer as diluent for primary and secondary antibodies incubation. Wash the nitrocellulose membrane with TBS-T between antibody incubation. Develop immunoblotting signals with the ECL Plus Western Blotting Substrate (Thermo Fisher Scientific) and capture the immunoblot images with a laser scanner (GE Healthcare Life Sciences). Antibodies specific to NaV1.1 should detect a protein band at about 223 kDa with WT mouse brain lysate but not with Scn1a-/- mouse brain lysate. An example of NaV1.1 specific antibody validation is given in Figure 2.
-
Evaluation of mouse liver lysate as diluent for MSD calibration curve standards
Perform SDS-PAGE with 50 µg of mouse liver lysate or brain lysate on each lane. Transfer total protein to a 0.2 µm nitrocellulose membrane and perform immunoblotting with a validated NaV1.1 antibody. Antibodies specific to NaV1.1 should detect a protein band about 223 kDa with brain lysate but not with the diluent (liver lysate in this case). Results confirming mouse liver lysate as a suitable diluent for brain lysate are shown in Figure 3.
-
Preparation of MSD calibration curve standards
Prepare MSD calibration curve standards by mixing WT mouse brain lysate (4 mg/ml) and WT mouse liver lysate (4 mg/ml) at various ratios (Table 1). Store MSD calibration curve standards in small aliquots at -80 °C to avoid multiple freeze-thaw cycles (see Note 5).
-
Characterization and acceptance criteria for the MSD-ECL assay
-
Accuracy and precision
Perform MSD-ECL assay with standards and quality controls (QCs) in duplicate wells. We typically prepare high concentration quality control (HQC) at 70-75% level and medium concentration quality control (MQC) at 45-55% level of the largest non-zero calibrator (STD12 in this case). We prepare low concentration quality control (LQC) approximately 3 times the smallest non-zero calibrator (STD2 in this case). Prepare the QCs using mouse liver lysate as diluent (see Note 5).
As a measure of assay accuracy (A), the mean back-calculated concentrations for all standards should be ± 15% of their nominal values. 75% of the non-zero standards should meet the above criteria. The mean back-calculated concentrations for QCs should be ± 20% of their nominal values.
As a measure of assay precision (P), the % CV between duplicates should be ≤ 20%.
-
Lower limit of quantification (LLOQ) and upper limit of quantification (ULOQ)
LLOQ is defined by the smallest non-zero calibrator with MSD signal at least 3 times greater than the blank (STD1) or greater than blank plus 10 times the standard deviation (SD) of the blank. LLOQ should meet the accuracy and precision criteria.
ULOQ is defined by the largest non-zero calibrator (STD12 in this case) that meets the accuracy and precision criteria.
-
Specificity
Perform MSD-ECL assay with brain lysate from Scn1a-/- mouse following steps in Procedure C. The MSD signal from Scn1a-/- brain lysate should be below the lower limit of quantification (BLQ).
-
Dilution linearity
The three QC samples (HQC, MQC and LQC) serve the purpose of evaluating the dilution linearity of the assay. If more stringent test is desired, MSD-ECL assay could be performed with 4 or more levels of dilution for the QC samples. The mean back-calculated concentrations for QCs should be ± 20% of their nominal values.
Establishment of dilution linearity helps to extend the dynamic range of the standard curve beyond 100% of NaV1.1 expression in WT mouse brain. Test samples for which anticipated NaV1.1 expression is higher than WT mouse brain tissues or showing levels above ULOQ can be diluted using the established dilution factors.
-
Storage stability
Store standards and QCs at -80 °C and perform MSD-ECL assay in 1 week, 1 month and 3 months (or longer as appropriate for the study). The mean back-calculated concentrations for all standards and QCs should be ± 15% and ± 20% of their nominal values respectively, while both passing the precision criteria.
-
-
-
Performing MSD-ECL assay
Block the MSD GAM (goat anti-mouse antibody) plates with 5% MSD blocker B (see Note 6) in Tris-Buffered Saline, 0.1% Tween-20 (TBS-T), 100 µl/well. Seal the plates and shake on a microplate shaker (Corning) for 1 h, at room temperature, speed 700 rpm.
Tap blocker B out of the plates over a stack of paper towels. Coat the wells with capture antibody (NeuroMab, 0.95 mg/ml, dilute to 1:200 in 5% Block B/TBS-T), 25 µl/well. Seal the plates and shake for 4 h at room temperature, speed 700 rpm.
Tap the capture antibody out of plates over paper towels. Wash the wells with TBS-T (150 µl/well) for 3 times while shaking the plates, at room temperature, speed 700 rpm. Five minutes for each wash.
Add 25 µl of MSD calibration curve standard (4 mg/ml) or test sample lysate (4 mg/ml) to individual wells. Include high, medium and low concentration quality controls (HQC, MQC and LQC, respectively) on the same plate. Run each standard and test sample as duplicate. An example of MSD plate layout is given in Figure 4. Seal the plates, shake overnight, at 4 °C, speed 700 rpm.
Tap the content out of plates over paper towels. Wash the wells with TBS-T (150 µl/well), for 3 times while shaking the plates, at room temperature, speed 700 rpm. Five minutes for each wash.
Add 25 µl of detection antibody (Alomone, 0.6 mg/ml, dilute to 1:250 in 5% Block B/TBS-T) to each well. Seal the plates and shake for 2 h, at room temperature, speed 700 rpm.
Tap the detection antibody out of plates over paper towels. Wash the wells with TBS-T (150 µl/well), for 3 times while shaking the plates, at room temperature, speed 700 rpm. Five minutes for each wash.
Add 25 µl of Sulfo-tagged anti-rabbit Ab (MSD, 0.5 mg/ml, dilute to 1:250) to each well. Seal the plates and shake for 1 h at room temperature, speed 700 rpm.
Tap the content out of plates over paper towels. Wash the wells with TBS-T (150 µl/well), for 3 times while shaking the plates, at room temperature, speed 700 rpm. Five minutes for each wash.
Add 150 µl of 1× MSD Read Buffer T (diluted in dH2O) to each well. Make sure bubbles are not created when dispensing the read buffer. Read the plates immediately on a Meso Quickplex SQ 120 machine with the MSD Discovery Workbench 4.0 software (Meso Scale Diagnostics, LLC).
Table 1. Preparation of calibration curve standards (STDs).
| STD name |
% NaV1.1 expression (normalized to adult WT mouse brain) |
Brain lysate (ml) | Liver lysate (ml) |
| STD 1 | 0 | 0 | 3 |
| STD 2 | 5 | 0.15 | 2.85 |
| STD 3 | 10 | 0.3 | 2.7 |
| STD 4 | 20 | 0.6 | 2.4 |
| STD 5 | 30 | 0.9 | 2.1 |
| STD 6 | 40 | 1.2 | 1.8 |
| STD 7 | 50 | 1.5 | 1.5 |
| STD 8 | 60 | 1.8 | 1.2 |
| STD 9 | 70 | 2.1 | 0.9 |
| STD 10 | 80 | 2.4 | 0.6 |
| STD 11 | 90 | 2.7 | 0.3 |
| STD 12 | 100 | 3 | 0 |
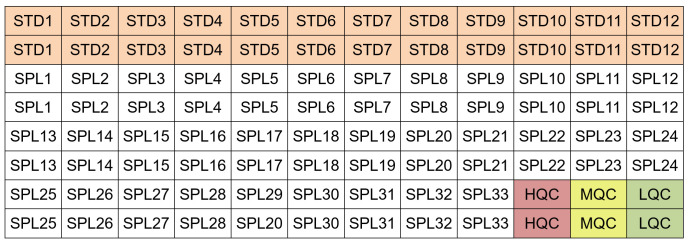
Figure 4. Example of plate layout for a 96-well MSD-ECL assay.
Each standard (STD) and test sample (SPL) was run as duplicate. High, medium and low concentration quality controls (HQC, MQC and LQC, respectively) are included in the plate. One can also run one standard curve and one set of QCs before the test samples and the second set of standard curve and QCs at the end of the test samples.
Data analysis
-
Import the signals of MSD calibration curve standards and test samples to the GraphPad Prism 8 software (GraphPad Software). Check if the standards and test samples meet the accuracy and precision criteria. See an example of an MSD-ECL data set in Table 2.
BLQ: Below the limit of quantification
4PL: Four parameter logistic
N/A: Not applicable
Fit standards to a non-linear regression curve (2nd order polynomial or four-parameter logistic). See an example of MSD standard curve fitting in Figure 5.
Interpolate NaV1.1 concentration in each test sample against the standard curve. Any sample with MSD signal above the ULOQ should be diluted with mouse liver lysate and rerun with a new MSD-ECL assay.
Use proper statistic test to compare NaV1.1 expression between different groups. Most used tests are Student’s t-test, Mann Whitney Wilcoxon test and two-way ANOVA. See an example of MSD-ECL data presentation and statistical analysis in Figure 6.
Table 2. Example of MSD calibration curve generation and evaluation of NaV1.1 expression in representative test samples .
| STD name | % NaV1.1 expression (nominal) | MSD signal | % NaV1.1 expression (interpolated) | Recovery (%) | ||||
|---|---|---|---|---|---|---|---|---|
| repeat 1 | repeat 2 | %CV | 2nd order polynomial | 4PL | 2nd order polynomial | 4PL | ||
| STD1 | 0 | 1303 | 1235 | 3.8 | BLQ | BLQ | N/A | N/A |
| STD2 | 5 | 4572 | 4963 | 5.8 | 4.7 | 4.8 | 94.8 | 96.0 |
| STD3 | 10 | 8410 | 9243 | 6.7 | 10.4 | 11.4 | 104.1 | 113.7 |
| STD4 | 20 | 16505 | 17320 | 3.4 | 20.7 | 21.8 | 103.7 | 108.9 |
| STD5 | 30 | 25789 | 25059 | 2.0 | 30.6 | 31.2 | 101.9 | 104.0 |
| STD6 | 40 | 33821 | 33094 | 1.5 | 39.1 | 39.3 | 97.8 | 98.3 |
| STD7 | 50 | 41928 | 47594 | 9.0 | 50.2 | 49.9 | 100.3 | 99.9 |
| STD8 | 60 | 57065 | 54643 | 3.1 | 60.2 | 59.7 | 100.3 | 99.5 |
| STD9 | 70 | 65153 | 62717 | 2.7 | 67.0 | 66.5 | 95.7 | 95.0 |
| STD10 | 80 | 89640 | 77442 | 10.3 | 82.4 | 82.1 | 103.1 | 102.7 |
| STD11 | 90 | 98015 | 92896 | 3.8 | 91.1 | 91.2 | 101.3 | 101.3 |
| STD12 | 100 | 108910 | 104363 | 3.0 | 98.9 | 99.4 | 98.9 | 99.4 |
| SPL1 | N/A | 15697 | 15143 | 2.5 | 18.9 | 20.0 | N/A | N/A |
| SPL2 | N/A | 13630 | 14060 | 2.2 | 17.0 | 18.1 | N/A | N/A |
| SPL3 | N/A | 13786 | 13443 | 1.8 | 16.7 | 17.8 | N/A | N/A |
| SPL4 | N/A | 12903 | 12511 | 2.2 | 15.5 | 16.6 | N/A | N/A |
| SPL5 | N/A | 13425 | 16106 | 12.8 | 18.1 | 19.2 | N/A | N/A |
| SPL6 | N/A | 15493 | 14147 | 6.4 | 18.2 | 19.3 | N/A | N/A |
| SPL7 | N/A | 15021 | 16930 | 8.4 | 19.6 | 20.7 | N/A | N/A |
| SPL8 | N/A | 15070 | 15661 | 2.7 | 18.9 | 19.9 | N/A | N/A |
| SPL9 | N/A | 27183 | 30487 | 8.1 | 34.3 | 34.7 | N/A | N/A |
| SPL10 | N/A | 32746 | 29349 | 7.7 | 36.6 | 36.9 | N/A | N/A |
| SPL11 | N/A | 30121 | 29539 | 1.4 | 35.3 | 35.7 | N/A | N/A |
| SPL12 | N/A | 36951 | 34568 | 4.7 | 41.4 | 41.6 | N/A | N/A |
| SPL13 | N/A | 33546 | 36913 | 6.8 | 40.9 | 41.0 | N/A | N/A |
| SPL14 | N/A | 29073 | 31291 | 5.2 | 35.7 | 36.1 | N/A | N/A |
| SPL15 | N/A | 31355 | 26931 | 10.7 | 34.6 | 35.0 | N/A | N/A |
| SPL16 | N/A | 30716 | 34868 | 9.0 | 38.4 | 38.7 | N/A | N/A |
| HQC | 70 | 70022 | 70597 | 0.6 | 72.2 | 71.7 | 103.1 | 102.4 |
| MQC | 50 | 40367 | 43335 | 5.0 | 47.4 | 47.3 | 94.8 | 94.6 |
| LQC | 20 | 15445 | 15775 | 1.5 | 19.2 | 20.2 | 96.0 | 101.0 |
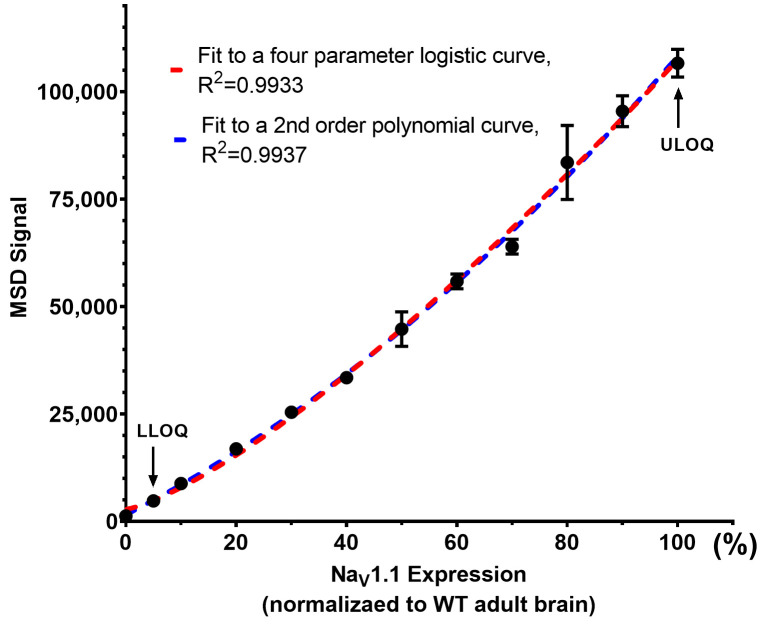
Figure 5. Example of MSD standard curve fitting.
The MSD signals from calibration curve standards (Table 2) were fit to a four-parameter logistic curve (in red) or a 2nd order polynomial curve (in blue). STD12 defines the upper limit of quantification (ULOQ). STD2 defines the lower limit of quantification (LLOQ).
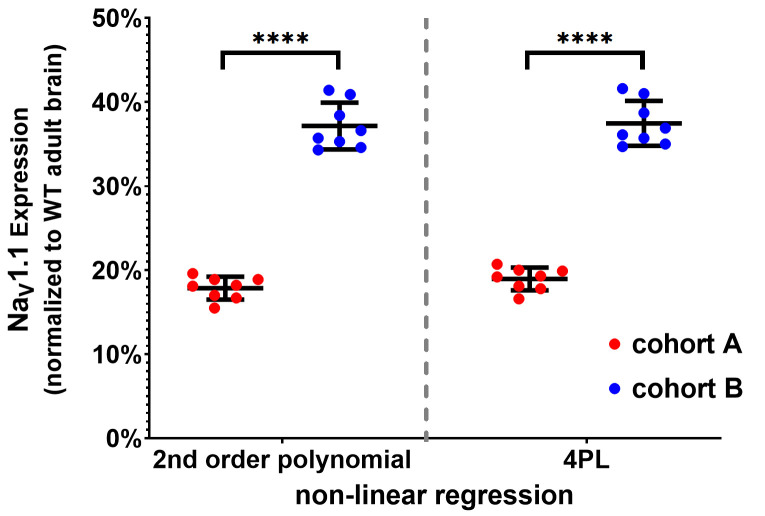
Figure 6. Example of MSD-ECL data presentation and statistical analysis.
Interpolated results of NaV1.1 expression (mean ± SD and individual values, also reported in Table 2) in brain tissues from two cohorts of pre-weaning mice (A and B) were plotted and mean values were compared with unpaired two-tailed Student’s t-test (****, P < 0.0001). Interpolation using 2nd order polynomial (left) and four-parameter logistic (4PL) standard curves (right) yielded similar results.
Notes
Protease inhibitors are not included in the MSD lysis buffer used in this protocol. It was reported that dimethylsulfoxide (DMSO), a solvent used for many proteinase inhibitors, can change the properties of proteins in solution, leading to protein denaturation, aggregation, or degradation. DMSO can also change the apparent binding properties of the proteins ( Chan et al., 2017 ). To avoid possible adverse effect of DMSO on this assay, which requires the integrity of the target protein and its binding to antibodies, we explored protocol without proteinase inhibitors and compared it to a similar protocol with protease inhibitors (Thermo Fisher Scientific, Halt Protease Inhibitor Cocktail). We did not find any difference between the two protocols in terms of assay results and storage stability as long as all the standards, QCs and test samples were processed on ice or at 4 °C, and stored at -80 °C in small aliquots.
Perfusion of mouse liver with PBS prior to tissue lysis greatly reduces background for liver lysate.
Prepare the standards for BCA assay with the MSD lysis buffer.
We strongly recommend validating any new lot of polyclonal antibody used in the MSD-ECL assay, since they may come from different immunized animals and the specificity of antibody may be different.
Store calibration curve standards, QCs and test sample lysates in small aliquots at -80 °C to avoid multiple freeze-thaw cycles (although establishment of freeze-thaw stability is recommended).
Blocker A (R93BA-4) for MSD-ECL assay is based on bovine serum albumin (BSA) and causes high background in our hands. Blocker B (R93BB-2) for MSD-ECL is based on milk proteins and results in low background in MSD-ECL assay.
Recipes
-
MSD lysis buffer
Components Amount Final Concentration 10× TBS, pH 7.4 5 ml 1× 10% TX-100 5 ml 1% 10% Nonidet P-40 2.5 ml 0.5% Na deoxycholate 125 mg 0.25% 0.5 M EDTA 100 µl 1 mM Bring up the volume to 50 ml with dH2O
Acknowledgments
This protocol was originally used in the research paper “Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome” ( Han et al., 2020 ).
Competing interests
ZH, AC, M and GL are employees of Stoke Therapeutics, Inc. ZH is one of the inventors on patent/patent application (PCT/US2018/48031; WO/2019/040923) submitted by Stoke Therapeutics, Inc. that covers use of therapeutic agents to promote exclusion of the NMD exon from the NMD exon mRNA encoding NaV1.1.
Ethics
Procedures involving mice were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Stoke Therapeutics, Inc. Approval ID: STK2019-03-CNS. Validity period: 04/11/2019 to 04/11/2022. All in vivo experiment procedures were in accordance with the NIH Guide for the Care and Use of Laboratory Animals. No human subject was involved in this study.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Meisler M. H., O'Brien J. E. and Sharkey L. M.(2010). Sodium channel gene family: epilepsy mutations, gene interactions and modifier effects. J Physiol 11): 1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dravet C., Bureau M., Oguni H., Fukuyama Y. and Cokar O.(2005) Severe myoclonic epilepsy in infancy: Dravet syndrome. Adv Neurol 95: 71-102. [PubMed] [Google Scholar]
- 3. Kuhle J., Barro C., Andreasson U., Derfuss T., Lindberg R., Sandelius Å., Liman V., Norgren N., Blennow K. and Zetterberg H.(2016). Comparison of three analytical platforms for quantification of the neurofilament light chain in blood samples: ELISA, electrochemiluminescence immunoassay and Simoa. Clin Chem Lab Med 54(10): 1655-1661. [DOI] [PubMed] [Google Scholar]
- 4. Cabral F., Miller C. M., Kudrna K. M., Hass B. E., Daubendiek J. G., Kellar B. M., and Harris E. N.(2018). Purification of Hepatocytes and Sinusoidal Endothelial Cells from Mouse Liver Perfusion. Journal of visualized experiments. J Vis Exp(132): 56993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan D. S., Kavanagh M. E., McLean K. J., Munro A. W., Matak-Vinković D., Coyne A. G. and Abell C.(2017). Effect of DMSO on Protein Structure and Interactions Assessed by Collision-Induced Dissociation and Unfolding. Anal Chem 89(18): 9976-9983. [DOI] [PubMed] [Google Scholar]
- 6. Han Z., Chen C., Christiansen A., Ji S., Lin Q., Anumonwo C., Liu C., Leiser S. C., Meena, Aznarez I., Liau G. and Isom L. L., (2020). Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome . Sci Transl Med 12(558): eaaz6100. [DOI] [PubMed] [Google Scholar]