Abstract
RNA viruses rapidly mutate, which can result in increased virulence, increased escape from vaccine protection, and false-negative detection results. Targeted detection methods have a limited ability to detect unknown viruses and often provide insufficient data to detect coinfections or identify antigenic variants. Random, deep sequencing is a method that can more fully detect and characterize RNA viruses and is often coupled with molecular techniques or culture methods for viral enrichment. We tested viral culture coupled with third-generation sequencing for the ability to detect and characterize RNA viruses. Cultures of bovine viral diarrhea virus, canine distemper virus (CDV), epizootic hemorrhagic disease virus, infectious bronchitis virus, 2 influenza A viruses, and porcine respiratory and reproductive syndrome virus were sequenced on the MinION platform using a random, reverse primer in a strand-switching reaction, coupled with PCR-based barcoding. Reads were taxonomically classified and used for reference-based sequence building using a stock personal computer. This method accurately detected and identified complete coding sequence genomes with a minimum of 20× coverage depth for all 7 viruses, including a sample containing 2 viruses. Each lineage-typing region had at least 26× coverage depth for all viruses. Furthermore, analyzing the CDV sample through a pipeline devoid of CDV reference sequences modeled the ability of this protocol to detect unknown viruses. Our results show the ability of this technique to detect and characterize dsRNA, negative- and positive-sense ssRNA, and nonsegmented and segmented RNA viruses.
Keywords: metagenomic, MinION, RNA viruses, sequencing, strand-switching
Introduction
RNA viruses are common etiologic agents of animal diseases. Many of these diseases, such as influenza,43 Newcastle disease (ND),6 epizootic hemorrhagic disease (EHD),10 infectious bronchitis (IB),8 and porcine respiratory and reproductive syndrome (PRRS)56 are global economic and/or health burdens for domestic and wild animal populations. Moreover, RNA viruses account for most emerging diseases given the swift production of genetic variants, enabling rapid evolution for adapting to environments and hosts.52 The low fidelity of viral-encoded, RNA-dependent RNA polymerases significantly contributes to genetic diversity, causing viral mutations up to a million times higher compared to cells.41 Additionally, genetically related viral species with segmented genomes (e.g., reoviruses and orthomyxoviruses) can reassort genomic segments, resulting in increased genetic variation. Enhanced virulence, resistance to vaccines, and ability for novel tissue tropism can also occur as a result of nucleotide mutations and reassortment events.9,12,40 Therefore, there is a need for rapid detection of variants through whole-genome sequencing (WGS) for proper diagnosis, treatment, control, and prevention.13
The molecular detection of RNA viruses is traditionally done using methods such as PCR, real-time PCR (rtPCR), PCR cloning for Sanger sequencing, and in situ hybridization.11,16 However, these methods use targeted approaches that require prior knowledge of the viral genome for detection and are inefficient for the discovery of novel viruses, mixed infections, and identifying whole genomes. The advent of next-generation sequencing (NGS) has permitted new techniques that circumvent some issues with targeted RNA sequencing. Platforms such as Illumina allow untargeted deep sequencing for novel virus detection, but can be expensive and labor intensive, particularly for WGS.39 Major drawbacks of these platforms are the generation of large volumes of raw data and short reads that require high-performance computers and extensive computational analysis.22 Another limitation of untargeted RNA sequencing across all platforms is the low relative abundance of viral RNA compared to host cellular RNA, which may require depletion of ribosomal RNA (rRNA) and/or enrichment of viral RNA to obtain an ample number of reads for viral strain detection.18,27 For these reasons, using NGS for accurate, viral WGS remains challenging.
The long-read sequencing technology provided by MinION sequencing (Oxford Nanopore Technologies [ONT]) has enabled rapid, inexpensive, high-throughput WGS of viruses.38 MinION-based viral metagenomic studies have accurately sequenced partial genomic reads for foot-and-mouth disease virus typing17 and performed transcriptomic analysis of several herpesviruses (varicella zoster virus37; herpes simplex virus 13; suid herpesvirus 130) from cell culture by using protocols developed by ONT. These studies targeted the 3’-poly(A)–tailed RNA by using an adapter-tailed oligo(dT) VN primer for reverse transcription with strand-switching (or template-switching). Strand-switching (similar to 5’ rapid amplification of cDNA ends [RACE]) only needs one primer-binding site for the synthesis of full-length cDNA, which is advantageous for sequencing methods by reducing the chances of primer-binding mismatches and bioinformatic complexity to reassemble the sequences.57 However, not all viruses have poly-adenylated RNA,12 and the previous studies were not focused on WGS (or whole CDS [coding sequences]). Thus, our aim was to test if replacing the adapter-tailed oligo(dT) VN primer with an adapter-tailed random hexamer-based primer would simultaneously detect and provide whole CDS coverage for characterization of various RNA viruses with different genome compositions from culture. We selected viruses to include a double-stranded RNA virus, positive-stranded viruses, negative-stranded viruses, and segmented viruses (epizootic hemorrhagic disease virus 2 [EHDV-2; Reoviridae, Orbivirus]; infectious bronchitis virus [IBV; Coronaviridae, Gammacoronavirus, Avian coronavirus]; porcine reproductive and respiratory syndrome virus [PRRSV; Arteriviridae, Betaarterivirus, Betaarterivirus suid 2]; bovine viral diarrhea virus [BVDV; Flaviviridae, Pestivirus, Pestivirus B]; canine distemper virus [CDV; Paramyxoviridae, Morbillivirus, Canine morbillivirus]; and 2 influenza A viruses [IAV; Orthomyxoviridae, Alphainfluenzavirus, Influenza A virus] isolated from a dog and a pig). This approach provides rapid, complete CDS for unknown RNA viruses in culture fluids and demonstrates the utility of the random hexamer-based, strand-switching primer for MinION library synthesis.
Materials and methods
Samples
The EHDV-2 isolate was propagated on cattle pulmonary artery endothelial cells (CPAE) from spleen and lung homogenate from a white-tailed deer (Odocoileus virginianus) collected in Georgia in 2016 at the Southeastern Cooperative Wildlife Disease Study at the University of Georgia (UGA; Athens, GA); CPAE cells are also persistently infected with BVDV. The CDV sample was isolated from the brain of an infant, female raccoon (Procyon lotor) from Kentucky in 2018 using African green monkey kidney cells expressing canine-signaling lymphocytic activation molecule (Vero-Dog SLAM cell line) in the Athens Veterinary Diagnostic Laboratory (AVDL; UGA). The canine IAV sample was collected from a nasal swab of a 7-y-old, male, Boxer dog in 2015 from Georgia and was cultured in embryonated chicken eggs at the Poultry Diagnostic and Research Center (PDRC; UGA). The Center for Vaccines and Immunology, UGA, provided the swine IAV sample, which was isolated in 2019 from a 6-mo-old, female, Hampshire-cross pig from Georgia after testing positive for IAV by immunohistochemistry and PCR. Swine IAV from lung homogenate was propagated on Madin–Darby canine kidney (MDCK) cells. The IBV sample (Mass vaccine) was cultured in embryonated chicken eggs and supplied by the PDRC. Isolate VR2385 of PRRSV was cultured on MARC-145 cells at the Veterinary Diagnostic Laboratory, Iowa State University (Ames, IA). Reverse-transcription real-time PCR (RT-rtPCR) was conducted for BVDV, EHDV-2,10 CDV, canine IAV, and IBV (Table 1). The CDV (AVDL), canine IAV (PDRC), and IBV (PDRC) RT-rtPCR assays were conducted using in-house methods on the isolates provided. For BVDV, RT-rtPCR was performed using a modified protocol26 (SuperScript III first-strand synthesis SuperMix for qRT-PCR; Invitrogen) and amplified in a thermocycler (CFX96 touch real-time PCR detection system; Bio-Rad).
Table 1.
Sample background information and reverse-transcription real-time PCR (RT-rtPCR) results.
| Virus | Original host | Propagation system | RT-rtPCR (Ct) | Collection | |
|---|---|---|---|---|---|
| State (USA) | Year | ||||
| BVDV | BVDV-positive CPAE cells | CPAE | 24.9 | NA | NA |
| CDV | North American raccoon | Vero-Dog SLAM | 20.4 | Kentucky | 2018 |
| EHDV | White-tailed deer | CPAE | 15.2 | Georgia | 2016 |
| IBV | NA | ECE | 12.5 | NA | NA |
| Canine IAV | Dog | ECE | 16.9 | Georgia | 2015 |
| Swine IAV | Hampshire-cross pig | MDCK | ND | Georgia | 2019 |
| PRRSV | NA | MARC-145 | ND | NA | NA |
BVDV= bovine viral diarrhea virus; CDV = canine distemper virus; Ct = cycle threshold; CPAE = cattle pulmonary artery endothelial; ECE = embryonated chicken eggs; EHDV = epizootic hemorrhagic disease virus; IAV = influenza A virus; IBV = infectious bronchitis virus; MARC = cloned African green monkey kidney cell line; MDCK = Madin–Darby canine kidney; NA = not applicable; ND = not determined; PRRSV = porcine reproductive and respiratory syndrome virus.
Total RNA extraction
All viruses, other than PRRSV, were fully processed at the University of Georgia. To test the protocol in 2 different laboratories, library preparation and sequencing of PRRSV and a CDV replicate were conducted at Virginia Tech University (Blacksburg, VA). Total RNA for CDV, EHDV-2, BVDV, swine IAV, and canine IAV was extracted using 1 mL of culture supernatant (Trizol LS reagent; Thermo Fisher) following the manufacturer’s protocol. Total RNA was eluted in 88.5 µL of nuclease-free water (Qiagen). DNase treatment was performed (RNase-free DNase set; Qiagen) and then purified (RNeasy MinElute cleanup kit; Qiagen) per the manufacturer’s instructions. Total RNA for IBV was extracted (QIAamp viral RNA mini kit; Qiagen) following the manufacturer’s protocol. At Virginia Tech, RNA was extracted from PRRSV and one replicate of CDV (QIAamp viral RNA mini kit; Qiagen) according to the manufacturer’s instructions. Concentrations were then measured (Qubit RNA HS assay kit, Qubit 3.0 fluorometer; Thermo Fisher).
Strand-switching cDNA synthesis
Strand-switching cDNA synthesis for MinION sequencing was completed by modifying the 1D PCR barcoding cDNA (SQK-LSK108) protocol from ONT. Reverse transcription was performed by combining 8 µL of total RNA, 2 µL of 1 µM PCR-RH-RT primer (5’-/5Phos/ACTTGCCTGTCGCTCTATCTTCNNNNNN-3’; synthesized by Integrated DNA Technologies [IDT], with standard desalting; ONT adapter sequence is underlined), and 1 µL of 10 mM dNTPs. The reaction mixture was incubated at 65°C for 5 min, then snap-cooled in an ice-water slurry for 1 min. Then 4 µL of 5× RT buffer, 1 µL of 100 mM DTT, 1 µL of 40 U/µL RNaseOUT (Invitrogen), and 2 µL of 10 µM strand-switching oligo (PCR_Sw_mod_3G: 5’-TTTCTGTTGGTGCTGATATTGCTGCCATTACGGCCmGmGmG-3’; sequence provided by ONT; synthesized by IDT with HPLC purification) were added and incubated at 42°C for 2 min. SuperScript IV reverse transcriptase (1 µL; Invitrogen) was added, and the reaction was incubated under the following conditions: 30 min at 50°C, 10 min at 42°C, and 10 min at 80°C. The cDNA was bead-purified (KAPA pure beads, Kapa Biosystems; or AMPure XP beads, Beckman Coulter) at 0.7× beads:solution ratio.
Barcoding PCR
The reverse-transcribed cDNA was amplified following ONT’s 1D PCR barcoding cDNA (SQK-LSK108) protocol (PCR barcoding expansion 1-12 [EXP-PBC001] kit, ONT; LongAmp Taq 2× master mix, New England Biolabs) with the following thermocycling conditions: 95°C for 3 min; 18 cycles of 95°C for 15 s, 62°C for 15 s, 65°C for 22 min; 65°C for 23 min. The barcoded, amplified DNA was bead-purified at a 0.8× ratio.
MinION library preparation and sequencing
After the barcoding PCR, 2 or 3 samples were pooled by equal volume to make a total volume of 47 µL. Each viral sample in our study was pooled with samples from other experiments that are not directly relevant to our study. Seven MinION libraries were prepared from the pooled barcoded amplicons (Ligation sequencing kit SQK-LSK109; ONT) following the 1D amplicon/cDNA by ligation per ONT’s instructions. Briefly, the pooled samples were end-prepped (NEBNext FFPE repair mix; NEBNext end repair/dA-tailing module; New England Biolabs) and bead-purified at 1.0× bead volume. Then, sequencing adapters (ONT) were ligated onto the end-prepped library (NEBNext quick T4 DNA ligase; New England Biolabs) and bead-purified (0.4× beads ratio) with the long fragment buffer (ONT). The final libraries were combined with sequencing buffer and loading beads per ONT’s instructions, and were sequenced on used or new FLO-MIN106 R9.4 flowcells (ONT), except for the PRRSV and replicated CDV library, which were sequenced on a FLO-MIN107 R9.5, with the MinION Mk1b sequencer. Per the pre-run quality control check for previously used flowcells, a minimum of 1,000 active pores were required for sequencing. To maximize data generation from single flowcells, used flowcells were nuclease-flushed, before or after sequencing, by adding a mixture of 10 µL of DNase I and 190 µL of DNase I reaction buffer (New England Biolabs) to the priming port and incubated for 30 min at room temperature. Sequencing was initiated by MinKNOW v.18.12.4–19.05.0 (ONT) using the 48-h sequencing protocol with the live basecalling option turned off. During the run, available FAST5 files were processed as below to estimate the number and length of viral reads to estimate sequencing time required to obtain sufficient genome coverage.
Pre-processing of raw MinION sequence data
The FAST5 files produced from sequencing for IBV, CDV, swine and canine IAV, and EHDV-2 and BVDV libraries were processed using an in-house script that sequentially basecalled, demultiplexed, and trimmed adapters on a MacBook Pro (3.1 GHz Intel Core i7, 8GB) using OS X El Capitan v.10.11.6. In summary, reads were basecalled using the CPU version of Guppy v.2.1.3 (ONT) by defining the appropriate configurations for the flowcell (FLO-MIN106) and kit (SQK-LSK109) used for sequencing and library preparation. Calibration strand detection and filtering was enabled with the –calib_detect parameter. The basecalled fastq files, with a qscore ≥7, were sorted into a “pass” folder using the –qscore-filtering parameter and used for further analysis. Then, reads were demultiplexed using Porechop v.0.2.4 (https://github.com/rrwick/Porechop) based on barcodes (-b output_dir) with –require_two_barcodes setting enabled; 1,000,000 reads were aligned to all known adapter sets (–check_reads 1000000). Adapters with 99% identity were trimmed from read ends (–adapter_threshold 99), and chimeric reads with middle adapters were removed.
The raw FAST5 files produced for PRRSV and the CDV replicate from Virginia Tech were basecalled using the GPU version of Guppy v.3.1.5 on a Dell Precision 3060 Tower (Intel Core i7, 31.3GB, NVIDIA GeForce GTX 1080) using Ubuntu 16.04 LTS. Guppy was initiated using the same parameters as above with the flowcell configuration defined as “FLO-MIN107”. The basecalled reads were then trimmed and demultiplexed using Porechop with the same parameters described previously.
Virus classification and lineage typing
After basecalling, demultiplexing, and adapter trimming, reads were classified with Centrifuge v.1.0.423 by the lowest taxonomical rank with default parameters, with the addition of allowing 50 assignments (or hits) for each read (-k 50) using a custom index built for each library based on the propagation system. Custom indices were constructed using an exhaustive search for complete genome sequences of all possible viruses infecting vertebrates downloaded from the NCBI nucleotide database, as of 15 March 2019, that included 80,550 viral reference genomes (Suppl. Table 1). Low-complexity regions in sequences were masked using dustmasker (NCBI C++ Toolkit, https://ncbi.github.io/cxx-toolkit). Dustmasked whole genomes of the species of cell line (see below), and/or the bovine genome (GCF_002263795.1_ARS-UCD1.2) to account for fetal bovine serum used in cell culture, were obtained using centrifuge-download and concatenated with the dustmasked vertebrate virus sequences to classify reads from host(s) cellular RNA and prevent misalignment of host reads to viral sequences. Using the concatenated files, indices were built by centrifuge-build with default parameters. The indices were then used to cluster reads based on short alignments to respective host and viral sequences to determine the presence and abundance of viral species in each sample using the standard out file. Reads to unexpected viral species were clustered and compared to GenBank accessions using web-based nucleotide BLASTn (https://blast.ncbi.nlm.nih.gov) with default settings. The percentage of viral reads and host reads were determined by removing duplicate reads and then dividing the number of reads clustered by the number of total reads after demultiplexing.
Once the viral species was identified, custom lineage-typing Centrifuge indices were also built to classify beyond the viral species and assess for the possibility of mixed infections of the same virus species, as described previously for IBV.7 Briefly, for each main lineage per virus, one complete sequence of the lineage-typing region was selected and then combined with the respective genome of the cell line. One sequence per lineage is required because some lineages are overrepresented in GenBank and, as a result of the scoring system in Centrifuge, these overrepresented lineages would be unequally weighted.7 Indices were constructed using the following: 20 N-terminal protease fragment (Npro) sequences for BVDV55 with the bovine genome (GCF_002263795.1_ARS-UCD1.2), 13 hemagglutinin (H) gene sequences for CDV36 with the African green monkey genome (GCF_000409795_Chlorocebus_sabeus_1.1), 9 VP2 sequences for EHDV5 with the bovine genome (GCF_002263795.1_ARS-UCD1.2), 32 spike 1 (S1) sequences for IBV50 with the chicken genome (GCF_000002315.4_Gallus_gallus-5.0), 18 hemagglutinin (HA) sequences and 11 neuraminidase (NA) sequences for IAV44 with the canine genome (GCF_000002285.3_CanFam3.1) or chicken genome (GCF_000002315.6_GRCg6a), and 18 ORF5 sequences for PRRSV21 with the swine genome (GCF_000003025.6_Sscrofa11.1; Suppl. Table 2). All sequences were dustmasked, assigned a unique taxonomy identification number, with the exception of the IAV sequences, and indices were built with default settings using centrifuge-build. Viral lineage typing was evaluated by aligning the individual trimmed and barcoded FASTQ files produced from Porechop to the respective custom lineage-typing indexes with Centrifuge, and reads were clustered by taxonomy identifications that represent potential lineage types in the sample. Each cluster of reads, representing potentially different lineages, were then used for reference-based consensus building using Geneious v.11.1.3 (Biomatters) to build a consensus for each lineage using the “Map to Reference” tool with medium sensitivity, re-iterating up to 5 times, and using the same reference sequences chosen for generating the custom Centrifuge lineage-typing databases. Consensus sequences derived for each potential lineage were analyzed using BLASTn with default settings. Results from BLASTn were sorted by query coverage and subject coverage. Subjects with the highest query and subject coverages and identical bit-scores were considered the “top hit(s)” to identify the virus and lineage in each cluster. A coinfection was defined as different clusters of reads creating consensus sequences that matched to different lineages; reads could be parsed accordingly for final consensus building. If no coinfection was detected, all viral reads were used for consensus building.
Viral genome consensus sequence generation
After viruses were lineage typed for each sample, all reads that aligned to respective viral species from Centrifuge output were imported into Geneious for whole-genome, reference-based consensus building with the “Map to Reference” tool, after removing duplicate reads, using the same methods as above. Consensus sequence building was performed with the requirement of at least 20× coverage at each base. References used for mapping were selected from using the “top hit” result in BLASTn from the lineage-typing analysis. Given the known errors within long homopolymer regions during MinION sequencing,32 consensus sequences were manually inspected, and homopolymer areas that resulted in frameshifts were manually edited. The CDS regions were extracted from each edited consensus sequence and analyzed using BLASTn. For viruses with gapped genomes (e.g., CDV, IBV, and PRRSV), complete CDS and noncoding intergenic regions were extracted and analyzed with BLASTn. The “top hit” for each consensus sequence was determined using the same criteria as above. The CDS regions for lineage-typing regions for each virus were also extracted and analyzed with BLASTn. The best “top hit”, percent pairwise identity, mean, minimum, maximum, and standard deviation of base coverage, and percentage of genome coverage were evaluated for each viral genome and lineage-typing region consensus sequences with the statistics tool in Geneious.
Simulating novel virus identification
In addition to the above method, sequencing data from the CDV sample were analyzed in a manner that simulated novel virus identification by constructing an all vertebrate virus, custom Centrifuge index with the omission of all canine morbillivirus sequences downloaded from NCBI as of 12 April 2019 (Suppl. Table 1). The index was assembled as described previously with African green monkey genome. Trimmed and demultiplexed reads from the CDV sample were classified with Centrifuge using parameters described above. Reads were grouped by taxonomy ID and exported to Geneious to build consensus sequences as described previously. The consensus was analyzed with BLASTn by excluding canine morbillivirus (taxid: 11232) in the search set. For comparison, Kraken-style reports were created using centrifuge-kreport from the Centrifuge output files from the CDV-absent pipeline and the CDV-present pipeline from above. Output files were visualized using Pavian v.1.0.4 To discern the phylogeny of the “novel” virus among other pathogens in the Morbillivirus genus, the final genome CDS and phosphoprotein (P) gene CDS consensus sequences were aligned with 18 complete genome CDS and P gene CDS acquired from GenBank using the neighbor-joining algorithm in ClustalW48 with default settings. Phylogenetic trees for complete genome CDS and P gene CDS were inferred using the maximum-likelihood statistical method based on the Tamura 3-parameter substitution model47 with bootstrap values calculated at 1,000 replicates in MEGA X.24
Alternative sequencing for comparison
Sanger sequencing was performed for the 5′-UTR region of BVDV, H gene of CDV, and VP2 segment of EHDV-2 to confirm the identity of isolates and to compare MinION consensus sequences for the lineage-typing regions. Briefly, using 8 µL of total RNA from each sample, cDNA was synthesized (SuperScript III first-strand synthesis system for RT-PCR; Invitrogen) with random hexamers (50 ng/µL) following the manufacturer’s instructions. The cDNA from each sample was amplified (DreamTaq Green PCR master mix, 2×; Thermo Fisher) following the manufacturer’s protocol, with 10 µM of each respective primer28,46,51 (Table 2) targeting partial sequences of the lineage-typing regions, and 1 µL of cDNA. Thermocycling conditions for PCR amplification of the 5’-UTR region for BVDV were as follows: 95°C for 5 min; 34 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 30 s; 72°C for 5 min. Thermocycling conditions for PCR amplification of the H gene for CDV were as follows: 95°C for 5 min; 40 cycles of 95°C for 30 s, 50°C for 30 s, 72°C for 1.5 min; 72°C for 10 min. Thermocycling conditions for PCR amplification of the VP2 segment for EHDV were as follows: 95°C for 3 min; 40 cycles of 95°C for 30 s, 57°C for 30 s, 72°C for 45 s; 72°C for 5 min. Electrophoresis with 1.0% agarose gel was performed to confirm PCR products. Amplicons were then purified (QIAquick PCR purification kit; Qiagen) following the manufacturer’s protocol and eluted in 30 µL of nuclease-free water (Qiagen). Final concentration and purity were measured (NanoDrop 2000 spectrophotometer; Thermo Fisher). The purified PCR products and 5 µM of each primer (Table 2) were submitted to GENEWIZ (South Plainfield, NJ) for bidirectional Sanger sequencing. A sample of the swine IAV was sent to St. Jude Children’s Research Hospital (Memphis, TN) for Illumina sequencing for other research. Consensus sequences for the HA and NA segments were made available for comparison to the MinION sequences.
Table 2.
Reverse-transcription PCR primer information for Sanger sequencing.
| Virus | Target region | Primer name | Primer sequence (5′→3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| BVDV | 5’-UTR | 324 | ATGCCCT/ATAGTAGGACTAGCA | 288 | Vilček, et al.51 |
| 326 | TCAACTCCATGTGCCATGTAC | ||||
| CDV | H | 204 (+) | GAATTCGACTTCCGCGATCTCC | 1,160 | Martella et al.28 |
| 232 (–) | TAGGCAACACCACTAATTTRGACTC | ||||
| EHDV | VP2 | EHDV-2F | TGGTGAAAATACGGTATATAACC | 246 | Sun et al.46 |
| EHDV-2R | GTTCAAATTCATCTGGGCTCATACT |
BVDV = bovine viral diarrhea virus; CDV = canine distemper virus; EHDV = epizootic hemorrhagic disease virus; H = hemagglutinin; UTR = untranslated region.
Partial lineage-typing sequences from Sanger sequencing for BVDV, CDV, and EHDV, and full-length lineage-typing sequences from Illumina for swine IAV, were compared with the full-length lineage-typing consensus sequences from MinION using Geneious Alignment with default settings. Pairwise identity of the alignment was calculated with Geneious.
Results
MinION sequencing, viral classification, and lineage typing
Seven libraries were prepared for 7 cultured samples for 7 different viruses by using random hexamer-primed, strand-switching for RT-PCR–based barcoding, and pooled sequencing using MinION. Sequencing occurred for a minimum of ~2 h and 45 min and a maximum of ~47 h and 37 min to obtain 567,780–6,984,000 raw reads. For reference, total sequencing time varied between runs based on the time of day the run was started, the number of viral reads detected early in the sequencing run, and the ability to reuse a flowcell (i.e., if the flowcell was not to be reused, then it was often sequenced to near exhaustion) but total time was not compared between libraries. After the raw reads were basecalled, demultiplexed, and trimmed, 7,523–1,173,058 reads were assigned to barcodes of interest and used for viral classification (Table 3).
Table 3.
Sequencing data from Porechop and Centrifuge.
| Virus | Approximate total run time (h:min) | Porechop | Viral reads (%) | Host reads (%) | |
|---|---|---|---|---|---|
| Sample total reads | Sample total bases | ||||
| BVDV & EHDV | 8:20 | 756,373 | 299,505,703 | 12.7 | 75.2 |
| CDV | 3:20 | 183,206 | 63,116,488 | 2.10 | 80.4 |
| CDV* | 47:37 | 75,492 | 33,681,975 | 0.97 | 45.8 |
| IBV | 7:40 | 7,523 | 11,361,873 | 63.3 | 2.50 |
| Canine IAV | 2:45 | 745,382 | 341,878,915 | 36.6 | 53.2 |
| Swine IAV | 5:20 | 1,173,058 | 450,544,038 | 31.1 | 63.4 |
| PRRSV* | 47:37 | 28,418 | 18,889,591 | 57.1 | 14.3 |
BVDV = bovine viral diarrhea virus; CDV = canine distemper virus; EHDV = epizootic hemorrhagic disease virus; IAV = influenza A virus; IBV = infectious bronchitis virus; PRRSV = porcine reproductive and respiratory syndrome virus.
Libraries were prepared and sequenced at Virginia Tech University.
Barcoded reads for each of the 7 libraries were aligned to custom-built indices with Centrifuge that detected 7 different viruses belonging to 3 main types of RNA viral genomes: BVDV (positive-sense ssRNA), CDV (negative-sense ssRNA), EHDV-2 (dsRNA), IBV (positive-sense ssRNA), canine IAV (negative-sense ssRNA), swine IAV (negative-sense ssRNA), and PRRSV (positive-sense ssRNA). Additionally, 3 of the viruses (EHDV-2, canine IAV, swine IAV) are comprised of segmented genomes. The randomly primed strand-switching method was also able to classify a sample containing EHDV-2 and BVDV. All viruses detected were categorized with viral reads representing 0.97–63.3% of all reads sequenced for each sample with percentage of host reads of 2.5–80.4% (Table 3). Reads to unexpected viral species were analyzed with BLASTn and were determined to be short alignments to various host, often ribosomal, sequences.
Viral sequences were parsed and further classified into lineage types using custom-built lineage-typing indices for Centrifuge. Reads were clustered based on the lineage-typed alignment, and consensus sequences were built using Geneious. Although the Centrifuge alignments suggested the possibility of 2–19 lineages per virus isolate, only 1 lineage per species was detected in each isolate after consensus building of each potential lineage (Table 4).
Table 4.
Consensus results for coding sequences (CDS) of lineage-typing regions for each virus.
| Virus | Lineage-typing region | Consensus length (bp) | BLASTn top hit | Lineage | Pairwise identity (%) | Depth of coverage | |||
|---|---|---|---|---|---|---|---|---|---|
| Description | Accession | Mean | Min. | Max. | |||||
| BVDV | 5’-UTR | 381 | BVDV-2 95-1501 | MH231130 | ND | 97.9 | 12,395.6 | 10,115 | 13,980 |
| N pro | 504 | BVDV-2 McCart_C | MH806438 | 2a | 96.2 | 14,783.5 | 13,286 | 15,908 | |
| CDV | H | 1,824 | CDV A75/17† | AF164967 | America II | 96.5 | 817.3 | 151 | 1,739 |
| CDV* | H | 1,824 | CDV A75/17† | AF164967 | America II | 96.5 | 105.1 | 33 | 176 |
| EHDV | VP2 | 2,949 | EHDV-2 OV617 | MK958997 | 2w | 99.9 | 523.7 | 43 | 896 |
| IBV | S1 | 3,489 | Avian coronavirus strain Ma5 | KY626045 | GI-L4 | 100.0 | 287.5 | 219 | 350 |
| Canine IAV | HA | 1,701 | IAV (A/canine/Georgia/101875/2015 (H3N2))† | MF173286 | H3g1 | 100.0 | 13,981.0 | 960 | 21,463 |
| NA | 1,410 | IAV (A/canine/Georgia/95391/2015 (H3N2))† | KX570998 | N2g2 | 99.8 | 22,968.0 | 674 | 37,816 | |
| Swine IAV | HA | 1,698 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198216 | H1g4 | 99.6 | 2,360.7 | 424 | 3,148 |
| NA | 1,410 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198218 | N2g2 | 99.8 | 4,752.8 | 517 | 6,928 | |
| PRRSV* | ORF5 | 603 | PRRSV VR2385 | JX044140 | Type 2, Lineage 5 | 99.7 | 546.2 | 470 | 588 |
BVDV = bovine viral diarrhea virus; CDV = canine distemper virus; EHDV = epizootic hemorrhagic disease virus; Gx-Lx = genotype–lineage; H = hemagglutinin; HA = hemagglutinin; IAV = influenza A virus; IBV = infectious bronchitis virus; NA = neuraminidase; ND = not determined; Npro = N-terminal protease; ORF = open reading frame; PRRSV = porcine reproductive and respiratory syndrome virus; S1 = spike 1.
Libraries were prepared and sequenced at Virginia Tech University.
BLASTn results showed 2–29 sequence alignments with identical bit-scores, query coverage, and pairwise identities to other isolates. The provided “top hit” was included in those alignments.
The complete CDS for each virus’s lineage-typing region had at least 33× depth of coverage with a mean range of 105.1–22,968.0 (Table 4). The BLASTn comparison for the genotyping regions resulted in 96.5–100.0% pairwise identity with each “top hit” (Table 4). Using the Npro region, the lineage of BVDV was determined as genotype BVDV-2a, with 96.2% identity with the McCart_C strain. Sequences from both libraries of CDV were genotyped as America II, with 96.5% identity of the H gene to A75/17. The genotype for EHDV-2 was determined as EHDV-2w, with 99.9% identity to the OV215 strain using the VP2 segment. IBV was determined to be genotype 1, lineage 4, with 100.0% identity of the S1 gene to the Ma5 strain. The HA and NA segments of the canine IAV showed 100.0% and 99.8% identity, respectively, to an H3N2 subtype circulating in dogs in North America. The porcine IAV was determined to be an H1N2 subtype similar to other North American porcine IAVs, with 99.6% identity to the HA segment and 99.8% identity to the NA segment. The ORF5 for PRRSV had 99.7% identity to VR2385 and was determined as a genotype 2, lineage 5.
Viral genome consensus sequence evaluation
For all 7 viruses, complete genome CDS were acquired with a minimum of 26× depth of each base, with the exception of the replicated CDV with 90.2% genome coverage at 20× depth (Table 5). Additionally, complete CDS for all segments of 3 segmented viruses (EHDV-2 [dsRNA], swine IAV and canine IAV [negative-sense ssRNA]), were obtained with a minimum of 43× depth of coverage and an average of 99.9% identity across all segments. The mean depth for each virus was 72.1–28,014.8. The complete genome CDS consensus sequences had a high pairwise identity of 97.4–100.0% to their respective “top hit” when using BLASTn to compare sequences with GenBank (Table 5). The complete CDS consensus sequences for BVDV, EHDV, and canine IAV were deposited in GenBank under the following accessions: BVDV = MN824468; EHDV = MN824457–MN824466; canine IAV = MN812282–MN812289. The complete CDS with intergenic regions for CDV was also deposited in GenBank as accession MN824467.
Table 5.
Consensus results of whole-genome coding sequences (CDS) for each virus.
| Virus/Segment | Reads per consensus | Consensus length (bp) | BLASTn top hit | Pairwise identity (%) | Depth of coverage | |||
|---|---|---|---|---|---|---|---|---|
| Description | Accession | Mean | Min. | Max. | ||||
| BVDV | 54,561 | 11,689 | BVDV-2 95-1501 | MH231130 | 97.4 | 4,277.3 | 518 | 15,908 |
| CDV | 3,428 | 15,477 | CDV A75/17 | AF164967 | 97.0 | 299.6 | 26 | 1,739 |
| CDV* | 717 | 13,957 | CDV A75/17 | AF164967 | 97.0 | 72.1 | 20 | 176 |
| EHDV | ||||||||
| VP1 | 2,091 | 3,909 | EHDV-2 OV617 | MK958997 | >99.9 | 576.4 | 231 | 1,333 |
| VP2 | 1,423 | 2,949 | EHDV-2 OV617 | MK958997 | 99.9 | 523.7 | 43 | 896 |
| VP3 | 1,807 | 2,700 | EHDV-2 OV215 | MF688818 | 99.9 | 606.3 | 100 | 1,346 |
| VP4 | 1,602 | 1,935 | EHDV-2 OV215 | MF688819 | >99.9 | 625.3 | 218 | 1,205 |
| NS1 | 3,598 | 1,656 | EHDV-2 OV215 | MF688823 | 99.9 | 2,295.5 | 185 | 2,711 |
| VP5 | 3,249 | 1,584 | EHDV-2 OV617 | MK959001 | 100.0 | 1,473.6 | 168 | 2,693 |
| VP7 | 3,415 | 1,050 | EHDV-2 OV215 | MF688822 | 100.0 | 1,751.5 | 117 | 2,804 |
| NS2 | 6,485 | 903 | EHDV-2 OV617† | MK959005 | 99.9 | 2,884.1 | 392 | 3,930 |
| VP6 | 15,516 | 1,080 | EHDV-2 OV617 | MK959002 | 99.9 | 6,335.3 | 196 | 13,629 |
| NS3 | 3,512 | 687 | EHDV-2 OV617 | MK959006 | 100.0 | 2,879.7 | 181 | 3,409 |
| IBV | 4,695 | 26,574 | Avian coronavirus strain Ma5 | KY626045 | 100.0 | 416.3 | 158 | 1,011 |
| Canine IAV | ||||||||
| PB2 | 50,753 | 2,280 | IAV (A/canine/Florida/269770/2015 (H3N2)) | MF173191 | 100.0 | 19,415.2 | 1,136 | 30,101 |
| PB1 | 38,930 | 2,274 | IAV (A/canine/Florida/269770/2015 (H3N2))† | MF173194 | 100.0 | 13,406.6 | 2,339 | 21,726 |
| PA | 28,755 | 2,151 | IAV (A/canine/Florida/269770/2015 (H3N2)) | MF173220 | 100.0 | 1,1662.4 | 923 | 17,815 |
| HA‡ | 33,293 | 1,701 | IAV (A/canine/Georgia/101875/2015 (H3N2))† | MF173286 | 100.0 | 13,981.0 | 960 | 21,463 |
| NP | 32,490 | 1,497 | IAV (A/canine/Georgia/95391/2015 (H3N2))† | KX571004 | 100.0 | 1,3119.1 | 2,544 | 19,511 |
| NA‡ | 41,130 | 1,410 | IAV (A/canine/Georgia/95391/2015 (H3N2))† | KX570998 | 99.8 | 22,968.0 | 674 | 37,816 |
| M1 & M2 | 39,862 | 982 | IAV (A/canine/Texas/2100186/2015 (H3N2))† | MF173280 | 100.0 | 23,887.5 | 962 | 34,210 |
| NEP & NS1 | 3,937 | 838 | IAV (A/canine/Georgia/101875/2015 (H3N2))† | MF173122 | 100.0 | 2,291.5 | 524 | 2,971 |
| Swine IAV | ||||||||
| PB2 | 41,421 | 2,280 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198223 | 99.7 | 9,835.2 | 223 | 33,925 |
| PB1 | 14,908 | 2,274 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198222 | 99.7 | 2,303.0 | 352 | 3,438 |
| PA | 26,431 | 2,151 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198221 | 99.7 | 7,941.8 | 417 | 20,275 |
| HA‡ | 6,673 | 1,698 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198216 | 99.7 | 2,360.7 | 424 | 3,148 |
| NP§ | 64,370 | 1,497 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198219 | 99.4 | 28,014.8 | 305 | 61,449 |
| NA‡ | 8,519 | 1,410 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198218 | 99.8 | 4,752.8 | 517 | 6,928 |
| M1 & M2 | 9,655 | 982 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198217 | 100.0 | 4,855.6 | 178 | 7,536 |
| NEP & NS1 | 3,276 | 838 | IAV (A/swine/Ohio/18TOSU1194/2018 (H1N2))† | MN198220 | 99.9 | 1,637.4 | 283 | 2,076 |
| PRRSV | 16,216 | 14,636 | PRRSV VR2385 | JX044140 | 99.8 | 2,045.4 | 27 | 13,535 |
BVDV = bovine viral diarrhea virus; CDV = canine distemper virus; EHDV = epizootic hemorrhagic disease virus; HA = hemagglutinin; IAV = influenza A virus; IBV = infectious bronchitis virus; M = matrix protein; NA = neuraminidase; NEP = nuclear export protein; NP = nucleocapsid; NS = nonstructural protein; PA = polymerase; PB = polymerase; PRRSV = porcine reproductive and respiratory syndrome virus.
Libraries prepared and sequenced at Virginia Tech University.
BLASTn results showed 2–77 sequence alignments with identical bit-scores, query coverage, and pairwise identities to other isolates. The provided “top hit” was included in those alignments.
Repeated from Table 4 for completeness of genome assessment.
§ Given a large number of reads, only half of the reads classified as the NP gene were used for reference-based consensus building.
Simulating novel virus identification
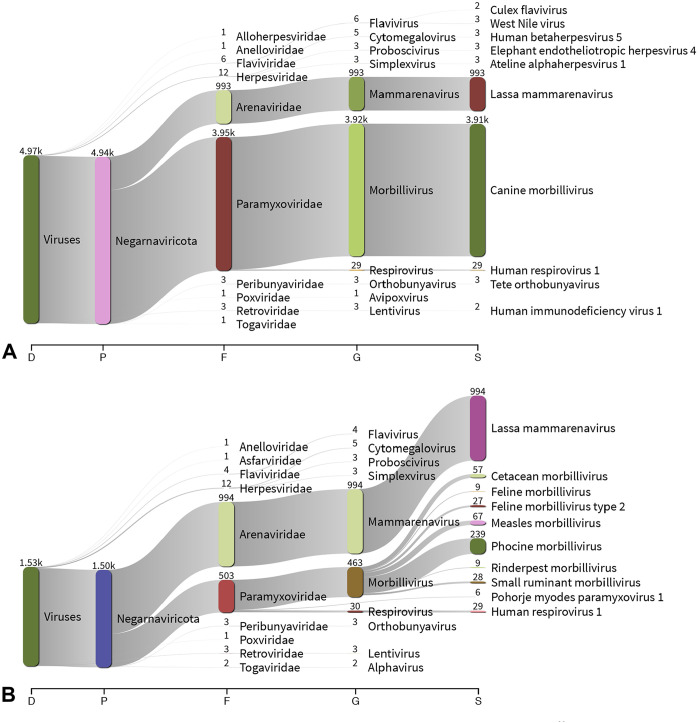
Under simulated conditions in which CDV would be an unknown virus, the Centrifuge standard out file for CDV analyzed showed that ~500 reads aligned to a morbillivirus, with 239 reads aligned to phocine distemper virus (PDV; Paramyxoviridae, Morbillivirus, Phocine morbillivirus) and <70 reads aligning to other more distantly related morbilliviruses: feline morbilliviruses (Feline morbillivirus), rinderpest virus (Rinderpest morbillivirus), peste-des-petits-ruminants virus (Small ruminant morbillivirus), cetacean morbillivirus (Cetacean morbillivirus), and measles virus (Measles morbillivirus; Fig. 1A). In contrast, when CDV was included in the Centrifuge database, ~4,000 reads aligned to CDV, which was the only morbillivirus detected (Fig. 1B). In the CDV-absent analysis, reads scattered across different morbilliviruses, suggesting that the actual species was absent from the database but is most similar to PDV. A total of 251 (12 reads aligned to 3 or more sequences, resulting in these reads not being counted in the Centrifuge standard out file) reads were classified as PDV and were exported to Geneious for reference-based consensus building by aligning to PDV/Wadden_Sea.NLD/1988 (GenBank NC_028249). The consensus sequence was analyzed with BLASTn by excluding canine morbillivirus in the search set, which resulted in 78.7% identity with PDV/Wadden_Sea.NLD/1988 (GenBank KC802221.1) with 78.0% query coverage. Furthermore, phylogenetic analysis of the whole genome CDS and P gene CDS consensus of the dubbed “novel” sequence clustered with PDV but shows sequence divergence suggestive of a species similar to, but different from, PDV (Fig. 2).
Figure 1.
Sankey diagram displaying the top 10 nodes per taxonomic classification level. Centrifuge v.1.0.423 Kraken-style reports from analyzing sequences from canine distemper virus (CDV), with A. pipeline containing CDV sequences and B. novel virus simulation using a pipeline devoid of CDV sequences in custom Centrifuge indices, were visualized with Pavian v.1.0.4 Number of reads assigned to taxonomic classification corresponds with the width of the flow and are listed above each node. Reads hitting to the bovine and African green monkey genomes were excluded. D = domain; F = family; G = genus; P = phylum; S = species.
Figure 2.
Phylogenetic analysis of morbilliviruses of A. whole-genome CDS and B. P gene CDS after a data analysis of CDV sequences using a pipeline devoid of CDV reads to simulate novel virus detection and identification. Each sequence is designated by species abbreviation and GenBank accession. The evolutionary history was inferred by using the maximum-likelihood method and Tamura 3-parameter model.47 These analyses involved 19 nucleotide sequences and 14,578 positions (A) or 1,460 positions (B) in the final dataset. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA X.24 CeMV = cetacean morbillivirus; FeMV = feline morbillivirus; MeV = measles virus; PDV = phocine distemper virus; PPRV = peste-des-petits-ruminants virus; RPV = rinderpest virus. Asterisk (*) indicates sequence generated from the data analysis pipeline with MinION sequencing.
Alternative sequencing and pairwise identity with MinION
Sanger sequencing targeting partial sequences of the lineage-typing regions for BVDV, CDV, and EHDV was used to confirm lineage types in the samples (Table 6). For BVDV, primers targeting a partial sequence of the 5′-UTR were used and resulted in a “top hit” to BVDV-2 isolate 95-1501. Sequencing of the H gene for CDV resulted in a “top hit” to CDV isolate THA/VG. The VP2 segment was targeted for EHDV and resulted in a “top hit” to EHDV-2 isolate OV617. The consensus sequences from Sanger and MinION sequencing had 100.0% pairwise identities for all 3 viruses (Table 6). MinION consensus sequences compared to Sanger for BVDV and EHDV had identical “top hits”. For CDV, MinION sequencing had a “top hit” to CDV A75/15 with 96.5% identity, but the shorter 923-bp fragment from Sanger had a top alignment with CDV THA/VG with 96.5% identity (Table 6). The Sanger sequence is based on smaller fragments of the lineage-typing region and, therefore, the BLASTn-based pairwise identities cannot be directly compared between MinION and Sanger sequences.
Table 6.
Sanger and MinION sequencing consensus pairwise identity results.
| Virus | Target region | MinION | Sanger | Sanger vs. MinION pairwise identity (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Consensus length (bp) | BLASTn top hit description | Pairwise identity (%) | Consensus length (bp) | BLASTn top hit description | Pairwise identity (%) | |||
| BVDV | 5′-UTR | 381 | BVDV-2 95-1501 (MH231130) | 97.9 | 268 | BVDV-2 95-1501 (MH231130) | 99.2 | 100.0 |
| CDV | H | 1,824 | CDV A75/17 (AF164967) | 96.5 | 923 | CDV THA/VG (JX886780)* | 96.5 | 100.0 |
| EHDV | VP2 | 2,949 | EHDV-2 OV617 (MK958998) | 99.9 | 158 | EHDV-2 OV617 (MK958998)* | 100.0 | 100.0 |
BVDV = bovine viral diarrhea virus; CDV = canine distemper virus; EHDV = epizootic hemorrhagic disease virus; H = hemagglutinin; UTR = untranslated region.
BLASTn results showed 5–12 sequence alignments with identical bit-scores, query coverage, and pairwise identities to other isolates. The provided “top hit” was included in those alignments.
Full-length consensus sequences from Illumina sequencing for the HA and NA genes for the swine IAV sample were also used to confirm the lineage. The pairwise identities between MinION and Illumina consensus sequences for the HA gene showed 99.9% with one base pair mismatch in a homopolymer region; the NA gene had 100.0% identity.
Discussion
A large proportion of emerging diseases are caused by RNA viruses53 and, given their mutability, rapid detection is needed; however, this can be hindered because of the limitations of PCR panels used for identification of unknown viruses and can be inefficient for the discovery of coinfections.14 Deep sequencing–based approaches using viral nucleic acid enrichment methods have been described to address this issue, including targeted and untargeted library preparations, such as SISPA.2,34 The methodology in our study demonstrates the application of culture-based viral enrichment followed by using an adapter-modified random hexamer reverse primer with strand-switching and MinION sequencing for accurately detecting and characterizing RNA viruses. RNA viruses with various genome compositions (single-stranded [positive- and negative-sense], double-stranded, and segmented) were used to demonstrate the ability of untargeted strand-switching to obtain complete CDS of genotyping regions and whole genomes with viral culture enrichment methods. Moreover, 2 viruses (EHDV-2 and BVDV) were detected from a single sample, illustrating the utility of the random sequencing approach. Lastly, data analysis for one sample (CDV) was treated as a novel virus, highlighting the feasibility of this method to identify a new or poorly characterized virus.
This random sequencing approach proved to be robust across various RNA viruses in obtaining full-length CDS of the complete genome after using an unbiased, fast aligner to identify the likely virus, followed by reference-based consensus building to identify the lineage type. With at least 26× depth of coverage across the genome for all viruses, whole-genome CDS had 97.0–100.0% identity to the top BLASTn hits. Complete CDS for all segments of 3 segmented viruses (EHDV-2 [dsRNA], swine IAV and canine IAV [negative-sense ssRNA]), were obtained with a minimum of 43× depth of coverage and an average of 99.9% identity across all segments. The best “top hit” for each segment does not match across all segments to the same isolate, which may be because of reassortment events for segmented genomes9,12 and the unavailable sequence data for all segments for some isolates in NCBI.
The alignments of the whole-genome CDS and the lineage-typing regions were similar and consistent with the origin of the samples in our study. The identification of the EHDV as an EHDV-2w, with highest similarity to EHDVs circulating in white-tailed deer in the southeastern United States in 2017, is consistent with the collection of this sample from a white-tailed deer in Georgia in 2016. The sample of canine IAV was determined to be a canine H3N2 strain, matching the typing completed as part of the original diagnostic case workup. The full sequencing provided by this method was able to confirm that the isolate in our study was most similar to canine H3N2 viruses circulating in the southeastern United States in 2015, consistent with the time and geographic location in which this sample was collected. Lineage typing of the swine IAV categorized it as an H1N2 most similar to an H1N2 IAV isolated from swine in 2018 from Ohio. It is possible that the slightly lower percent identity for the swine IAV is a result of relative paucity of sequence data available for 2019 swine IAV at this time. The CDV America II lineage, the classification of CDV in our study, is a common lineage found in North American wildlife,54 and is consistent with the collection of this sample from a raccoon in Kentucky. The sequences for IBV and PRRSV were highly similar to the known sequences of those isolates.
We also tested our protocol in 2 scenarios. The first was the detection of 2 viruses in a single culture system, demonstrating its ability to simultaneously obtain accurate, complete CDS for rapid detection and characterization of viruses in coinfected samples, comparable to other studies using advanced sequencing for analysis of cultured viruses.1 The second was the data analysis under the simulating conditions of CDV being a novel virus. This resulted in read alignments that spanned the Morbillivirus genus. Excluding the background hits, the largest proportion of reads hit to PDV, and consensus building with these reads had a low pairwise identity (78.7%), suggesting it was not a PDV. The phylogenetic divergence gives evidence that the virus in the sample belongs to the Morbillivirus genus and is phylogenetically related to PDV. If CDV was truly an unknown virus, the Centrifuge output would have suggested the identification of a novel or divergent virus that is most similar to PDV sequences, consistent with the known close genetic relationship between CDV and PDV.35 Furthermore, in the event of an unknown viral etiology and given the multiplexing capabilities of MinION sequencing, DNA library approaches can be applied and sequenced concurrently with the random strand-switching library to identify RNA or DNA virus in the sample.19,29 Lastly, although virus isolation is not a novel means for virus enrichment, our study demonstrates that this classic technique, which has recently been underused in deference to PCR assays, remains useful in a veterinary diagnostic laboratory. Not only would viral culture be useful for efficient genetic characterization of the viruses, but viral culture is required for isolation of different strains, which in turn allows for research into the various biological characteristics of any new viruses or viral strains.
Sanger sequencing was performed on partial lineage-typing sequences for BVDV, CDV, and EHDV and compared with the full-length lineage-typing regions obtained from MinION sequencing, resulting in 100.0% pairwise identity. Illumina sequencing was completed for full-length sequences of the lineage-typing regions for swine IAV and resulted in ≥99.9% pairwise identity with the full-length CDS from MinION sequencing. The high pairwise identity between the Sanger or Illumina and MinION sequences is consistent with previous results using MinION sequencing,6,7 which demonstrated the ability to obtain accurate MinION sequences by increasing depth of coverage.20,25 Sanger sequencing and BLASTn results for the 5’-UTR for BVDV and VP2 for EHDV matched the MinION lineage-typing region BLASTn results. For the H gene for CDV, Sanger sequencing had a top BLASTn alignment with isolate THA/VG with 96.5%; however, the results did show 96.0% identity with isolate A75/15, comparable to MinION sequencing result of 96.5% identity to isolate A75/15. It is noteworthy that aligning the Sanger and MinION results for the H gene in CDV showed 100.0% pairwise identity. The differences in BLASTn top alignments are attributed to the shorter sequence from Sanger sequencing, causing a slightly lower sequence specificity. Longer fragments, particularly WGS, have shown improved resolution for genotyping analyses.15,33 Additionally, the limitations of partial sequences used in Sanger sequencing are illustrated by the inability of Sanger sequencing to identify a single best hit for EHDV-2 and CDV, which resulted in multiple hits with identical BLASTn scores; whereas, the MinION-based method obtained complete CDS of the VP2 segment and H gene, allowing for a single best hit for these viruses.
In addition to more accurate classification, WGS of viruses is advantageous in providing more data for detection, epidemiology, genotyping, and phylogenetic analysis compared to partial and/or targeted sequences obtained from other classical sequencing methods. The relatively limited data provided by targeted sequencing may require further sequencing and expense to obtain additional biological information. In particular, different genetic sequences are used for phylogenetic analysis and genotyping. Some viruses, such as BVDV, EHDV, and IAV, also have multiple genotyping regions.5,44,55 Furthermore, recombination events are often difficult to identify with partial sequences and are important in investigating host range, virulence, and vaccine evasion.42,45 Thus, routine WGS of viruses gives quick turnaround data for various analyses and could provide more comprehensive databases needed to understand viral evolution.
Random, deep sequencing for many high-throughput sequencing platforms has some caveats such as false hits. As one example (Fig. 1), some reads were classified as lassa mammarenavirus, various herpesviruses, and others. Endogenous retroviruses and some DNA viruses were also frequently found to be present after annotating sequences. After clustering and aligning these reads to GenBank using BLASTn, we determined that the reads were short sequences that matched various host sequences. Reference sequences of the host and propagation systems of the cultured viruses were included in the Centrifuge indices to reduce these misalignments; however, these genomes are not as well described as other commonly studied organisms and may not represent the full sequence diversity of the host genome. Users can adjust alignment settings to help reduce the number of false hits, but as with any test, increasing the specificity of the test will negatively impact sensitivity. Similar to other fields (e.g., background lesions in pathology, growth of nonpathogenic bacteria in bacterial cultures), confirmatory tests may be required, and analysis of deep-sequencing results requires evaluation by a trained user, one experienced with bioinformatic methodology and with knowledge of veterinary infectious diseases.
Although the individual-read error rate with MinION is high compared to other sequencing platforms, this can be mitigated by using reference-based assembly and high depth of coverage at each base for consensus building, as was done in our study.30,31,49 Requiring 20× depth of coverage results in a 0.0344% error rate in the consensus sequence.56 Additionally, the reference-based assembly of reads for consensus building required manual inspection and editing of erroneous bases in homopolymer regions. MinION’s difficulty in accurately sequencing homopolymers is known,32 and one potential site manifested itself in the swine IAV HA CDS, but ONT is continuously improving flowcell design (FLO-MIN R10.3) and data analysis tools (Guppy and nanopolish [https://github.com/jts/nanopolish]) to increase accuracy in the single-read error rate.
Although rapid, cost-effective WGS may be useful in a diagnostic setting, ease of use and robustness across laboratories are key for deployment. For this reason, libraries for CDV and PRRSV were prepared at different laboratories. Both libraries for CDV had similar results; however, the differences can possibly be attributed to the use of a sequencing kit intended for 1D sequencing with a FLO-MIN107 (R9.5) flowcell that is typically used for 1D2 sequencing. The repercussions of the 1D kit with a 1D2 flowcell combination are not well known, but it is of interest to note that trimming the basecalled files resulted in 1,691,792 reads removed because of middle adapters, significantly decreasing the total number of reads assigned to each barcode.
Our study provides promising results for quick identification of unknown cultured RNA viruses by using MinION sequencing with a previously untested random approach. Future studies to compare the various random methods of deep sequencing are required to determine the most efficient methods. As with other deep-sequencing methods,33 MinION-based sequencing will also likely be used to metagenomically detect viruses directly from clinical samples (e.g., serum, swabs, tissues), and work is ongoing to investigate the usage of this random hexamer-based, strand-switching approach in clinical samples. MinION sequencing is a cost-effective way to multiplex samples and achieve long reads for more accurate genome consensus building compared to other short read sequencing technologies. Overall, the addition of full-genome sequencing to more routine diagnostic use will increase the available knowledge regarding sequence diversity and allow for improved tracking of viruses and a better understanding of the genetic determinants of viral pathogenesis.
Supplemental Material
Supplemental material, sj-pdf-1-vdi-10.1177_1040638720981019 for Randomly primed, strand-switching, MinION-based sequencing for the detection and characterization of cultured RNA viruses by Kelsey T. Young, Kevin K. Lahmers, Holly S. Sellers, David E. Stallknecht, Rebecca L. Poulson, Jerry T. Saliki, Stephen Mark Tompkins, Ian Padykula, Chris Siepker, Elizabeth W. Howerth, Michelle Todd and James B. Stanton in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank Vanessa Gauthiersloan, Samantha Day and Erich Linnemann (PDRC, UGA), Clara Kienzle-Dean (Southeastern Cooperative Wildlife Disease Study, UGA), and Pablo Pinyero (Veterinary Diagnostic Laboratory, Iowa State University) for their technical help. We also thank Stacey Schultz-Cherry (St. Jude Children’s Research Hospital) and Daniel Perez (Department of Population Health, UGA) for assistance with Illumina sequencing.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Our project was supported by Agriculture and Food Research Initiative Competitive Grant 2018-67015-28306 from the USDA National Institute of Food and Agriculture. Our project was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Center of Excellence of Influenza Research and Surveillance (CEIRS) under contract HHSN272201400004C (SMT).
ORCID iDs: Kelsey T. Young  https://orcid.org/0000-0003-1163-567X
https://orcid.org/0000-0003-1163-567X
Ian Padykula  https://orcid.org/0000-0003-2889-1864
https://orcid.org/0000-0003-2889-1864
Supplementary material: Supplementary material for this article is available online.
Contributor Information
Kelsey T. Young, Department of Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA
Kevin K. Lahmers, Department of Biomedical Sciences & Pathobiology, VA-MD College of Veterinary Medicine, Virginia Tech University, Blacksburg, VA
Holly S. Sellers, Poultry Diagnostic and Research Center, Department of Population Health, College of Veterinary Medicine, University of Georgia, Athens, GA
David E. Stallknecht, Southeastern Cooperative Wildlife Disease Study Department of Population Health, College of Veterinary Medicine, University of Georgia, Athens, GA
Rebecca L. Poulson, Southeastern Cooperative Wildlife Disease Study Department of Population Health, College of Veterinary Medicine, University of Georgia, Athens, GA
Jerry T. Saliki, Athens Veterinary Diagnostic Laboratory, College of Veterinary Medicine, University of Georgia, Athens, GA
Stephen Mark Tompkins, Center for Vaccines and Immunology, Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, GA.
Ian Padykula, Center for Vaccines and Immunology, Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, GA.
Chris Siepker, Department of Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA.
Elizabeth W. Howerth, Department of Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA
Michelle Todd, Department of Biomedical Sciences & Pathobiology, VA-MD College of Veterinary Medicine, Virginia Tech University, Blacksburg, VA.
James B. Stanton, Department of Pathology, College of Veterinary Medicine, University of Georgia, Athens, GA.
References
- 1. Ahasan MS, et al. Complete genome sequence of mobuck virus isolated from a Florida white-tailed deer (Odocoileus virginianus). Microbiol Resour Announc 2019;8:e01324–01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beato MS, et al. Identification and genetic characterization of bovine enterovirus by combination of two next generation sequencing platforms. J Virol Methods 2018;260:21–25. [DOI] [PubMed] [Google Scholar]
- 3. Boldogkői Z, et al. Transcriptomic study of herpes simplex virus type-1 using full-length sequencing techniques. Sci Data 2018;5:180266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breitwieser FP, et al. Pavian: interactive analysis of metagenomics data or microbiome studies and pathogen identification. Bioinformatics 2020;36:1303–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown-Joseph T, et al. Identification and characterization of epizootic hemorrhagic disease virus serotype 6 in cattle co-infected with bluetongue virus in Trinidad, West Indies. Vet Microbiol 2019;229:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butt SL, et al. Rapid virulence prediction and identification of Newcastle disease virus genotypes using third-generation sequencing. Virol J 2018;15:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butt SL, et al. Real-time, MinION-based, amplicon sequencing for lineage typing of infectious bronchitis virus from upper respiratory samples. J Vet Diagn Invest 2020. doi: 10.1177/1040638720910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cavanagh D, Naqi SA. Infectious bronchitis. In: Saif YM, ed. Diseases of Poultry. 11th ed. Iowa State University Press, 2003:101–119. [Google Scholar]
- 9. Chan RWY, et al. Tissue tropism of swine influenza viruses and reassortants in ex vivo cultures of the human respiratory tract and conjunctiva. J Virol 2011;85:11581–11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clavijo A, et al. An improved real-time polymerase chain reaction for the simultaneous detection of all serotypes of epizootic hemorrhagic disease virus. J Vet Diagn Invest 2010;22:588–593. [DOI] [PubMed] [Google Scholar]
- 11. Cobo F. Application of molecular diagnostic techniques for viral testing. Open Virol J 2012;6:104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flint J. Synthesis of RNA from RNA templates. In: Principles of Virology. 4th ed. Vol. 1: Molecular Biology. ASM Press, 2015:157–184. [Google Scholar]
- 13. Gilchrist CA, et al. Whole-genome sequencing in outbreak analysis. Clin Microbiol Rev 2015;28:541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ginocchio CC, et al. Current best practices for respiratory virus testing. J Clin Microbiol 2011;49:S44–S48. [Google Scholar]
- 15. Goldstein EJ, et al. Integrating patient and whole-genome sequencing data to provide insights into the epidemiology of seasonal influenza A (H3N2) viruses. Microb Genom 2018; 4:e000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gullett JC, et al. Quantitative nucleic acid amplification methods for viral infections. Clin Chem 2015;61:72–78. [DOI] [PubMed] [Google Scholar]
- 17. Hanson S, et al. Serotyping of foot-and-mouth disease virus using oxford nanopore sequencing. J Virol Methods 2019;263: 50–53. [DOI] [PubMed] [Google Scholar]
- 18. Head SR, et al. Library construction for next-generation sequencing: overviews and challenges. Biotechniques 2014;56: 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jain M, et al. Improved data analysis for the MinION nanopore sequencer. Nat Methods 2015;12:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain M, et al. MinION analysis and reference consortium: phase 2 data release and analysis of R9.0 chemistry. F1000Res 2017;6:760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang H, et al. Geographic distribution and molecular analysis of porcine reproductive and respiratory syndrome viruses circulating in swine farms in the Republic of Korea between 2013 and 2016. BMC Vet Res 2018;14:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kasibhatla SM, et al. Analysis of next-generation sequencing data in virology—opportunities and challenges. IntechOpen 2016;6:1–33. [Google Scholar]
- 23. Kim D, et al. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 2016;26:1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar S, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Magi A, et al. Characterization of MinION nanopore data for resequencing analyses. Brief Bioinform 2016;18:940–953. [DOI] [PubMed] [Google Scholar]
- 26. Mahlum CE, et al. Detection of bovine viral diarrhea virus by TaqMan reverse transcription polymerase chain reaction. J Vet Diagn Invest 2002;14:120–125. [DOI] [PubMed] [Google Scholar]
- 27. Marston DA, et al. Next generation sequencing of viral RNA genomes. BMC Genomics 2013;14:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martella V, et al. Genotyping canine distemper virus (CDV) by a hemi-nested multiplex PCR provides a rapid approach for investigation of CDV outbreaks. Vet Microbiol 2007;122:32–42. [DOI] [PubMed] [Google Scholar]
- 29. McNaughton AL, et al. Illumina and nanopore methods for whole genome sequencing of hepatitis B virus (HBV). Sci Rep 2019;9:7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moldován N, et al. Multi-platform sequencing approach reveals a novel transcriptome profile of pseudorabies virus. Front Microbiol 2018;8:2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Munnink B, et al. Towards high quality real-time whole genome sequencing during outbreaks using Usutu virus as example. Infect Genet Evol 2019;73:49–54. [DOI] [PubMed] [Google Scholar]
- 32. O’Donnell CR, et al. Error analysis of idealized nanopore sequencing. Electrophoresis 2013;34:2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pallen MJ, et al. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol 2010;13:625–631. [DOI] [PubMed] [Google Scholar]
- 34. Peserico A, et al. Diagnosis and characterization of canine distemper virus through sequencing by MinION nanopore technology. Sci Rep 2019;9:1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfeffermann K, et al. Morbillivirus pathogenesis and virus–host interactions. In: Kielian M, et al., eds. Advances in Virus Research. Vol. 100. Academic Press, 2018:75–98. [DOI] [PubMed] [Google Scholar]
- 36. Piewbang C, et al. Genetic and evolutionary analysis of a new Asia-4 lineage and naturally recombinant canine distemper virus strains from Thailand. Sci Rep 2019;9:3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prazsák I, et al. Long-read sequencing uncovers a complex transcriptome topology in varicella zoster virus. BMC Genomics 2018;19:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016;530:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ranjan R, et al. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun 2016;469:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Read SJ, et al. Molecular techniques for clinical diagnostic virology. J Clin Pathol 2000;53:502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rubio L, et al. Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae. Front Microbiol 2013; 4:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Runckel C, et al. Identification and manipulation of the molecular determinants influencing poliovirus recombination. PLoS Pathog 2013;9:e1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sartore S, et al. The effects of control measures on the economic burden associated with epidemics of avian influenza in Italy. Poult Sci 2010;89:1115–1121. [DOI] [PubMed] [Google Scholar]
- 44. Shi W, et al. A complete analysis of HA and NA genes of influenza A viruses. PLoS One 2010;5:e14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Su S, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016;24:490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun F, et al. Molecular typing of epizootic hemorrhagic disease virus serotypes by one-step multiplex RT-PCR. J Wildl Dis 2014;50:639–644. [DOI] [PubMed] [Google Scholar]
- 47. Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 1992;9:678–687. [DOI] [PubMed] [Google Scholar]
- 48. Thompson JD, et al. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tyler A, et al. Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci Rep 2018;8:10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Valastro V, et al. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect Genet Evol 2016;39:349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vilček Š, et al. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol 1994;136:309–323. [DOI] [PubMed] [Google Scholar]
- 52. Woolhouse MEJ, et al. Assessing the epidemic potential of RNA and DNA viruses. Emerg Infect Dis 2016;22:2037–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Woolhouse MEJ, et al. RNA viruses: a case study of the biology of emerging infectious diseases. In: Atlas RM, Maloy S, eds. One Health: People, Animals, and the Environment. 1st ed. Am Soc Microbiol, 2014:83–97. [Google Scholar]
- 54. Wostenberg DJ, et al. Evidence of two cocirculating canine distemper virus strains in mesocarnivores from northern Colorado, USA. J Wildl Dis 2018;54:534–543. [DOI] [PubMed] [Google Scholar]
- 55. Yeşilbağ K, et al. Variability and global distribution of subgenotypes of bovine viral diarrhea virus. Viruses 2017; 9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang J, et al. High-throughput whole genome sequencing of porcine reproductive and respiratory syndrome virus from cell culture materials and clinical specimens using next-generation sequencing technology. J Vet Diagn Invest 2016;29:41–50. [DOI] [PubMed] [Google Scholar]
- 57. Zhu Y, et al. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 2001;30:892–897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-vdi-10.1177_1040638720981019 for Randomly primed, strand-switching, MinION-based sequencing for the detection and characterization of cultured RNA viruses by Kelsey T. Young, Kevin K. Lahmers, Holly S. Sellers, David E. Stallknecht, Rebecca L. Poulson, Jerry T. Saliki, Stephen Mark Tompkins, Ian Padykula, Chris Siepker, Elizabeth W. Howerth, Michelle Todd and James B. Stanton in Journal of Veterinary Diagnostic Investigation