Abstract
Abstract
Olive leaf as an agricultural waste contains valuable bioactive compounds that are mainly used for pharmaceutical and cosmetic industries. Lately the major component, oleuropein, has gained extra attention due to the anti-viral activity against SARS-CoV-2 that causes Coronavirus disease (Covid-19). In this study, extraction of the bioactive compounds from olive leaves was conducted using a non-conventional and green method. New generation green solvents, natural deep eutectic solvents (NADES) were used in combination with ultrasound assisted extraction. Screening of NADES type, temperature, and particle size were investigated using one-pot-at-a-time method while, NADES amount and liquid-to-solid ratio were optimized using experimental design. The results were evaluated in terms of total polyphenol yield (YTP), total flavonoid yield (YTF) and antiradical activity (AAR). At the optimized conditions, the highest total polyphenol yield and the highest total flavonoid yield were achieved with choline chloride–fructose–water (CFW) (5:2:5) as 187.31 ± 10.3 mg gallic acid equivalent g−1 dw and 12.75 ± 0.6 mg apigenin equivalent g−1 dw, respectively. The extracts were also analyzed for oleuropein, caffeic acid and luteolin contents. The highest amount of oleuropein and caffeic acid were extracted by glucose–fructose–water (GFW) (1:1:11) as 1630.80 mg kg−1 dw and 112.77 mg kg−1 dw, respectively.
Graphic Abstract
Supplementary Information
The online version of this article (10.1007/s12649-021-01411-3) contains supplementary material, which is available to authorized users.
Keywords: Olive leaf; Natural deep eutectic solvents; Ultrasound assisted extraction; Green extraction, experimental design, response surface methodology
Statement of Novelty
This study presents the first time use of novel natural deep eutectic solvents for the extraction from olive leaves using ultrasound assisted extraction and contributes for improving and broadening the use of natural deep eutectic solvents for the extraction of bioactive compounds from different sources within ‘green extraction’ domain.
Introduction
Olive leaf (Olea europaea) extracts have been regarded as valuable items since ancient times. It is known that pharaohs were mummified using olive leaf extracts by Egyptians [1]. In the next years, olive leaf extracts were used for health purposes such as healing fevers, and afterwards healing tropical diseases such as malaria [1]. In 1854, the treatment of these diseases could be executed formally by the olive leaf extracts [2]. Later on, research activities on olive leaves revealed promising significant effects such as antioxidant capacity [3–5], antifungal activity [6], antibacterial activity [7], anti-HIV property [8], vasodilator effect [9], and hypoglycemic effect [10] both in vivo and in vitro. Regarding these properties, olive leaf components have been under research for their potential anti-viral effect against SARS-CoV-2 that causes Coronavirus disease (Covid-19). Lately, remarkable results were presented on the blockage of the SARS-CoV-2 spike protein-ACE-2 interface by oleuropein dimer and dihydro oleuropein [11]. Additionally, demethyloleuropein was reported to block SARS-CoV-2 main protease [11]. On the other hand, oleuropein, quercetin, luteolin-7-glucoside, apigenin-7-glucoside, catechin and epicatechin-gallate were reported to be investigated as the potential inhibitor of Covid-19 main protease [12].
The valuable content of olive leaves is comprised of mainly phenolics and several flavonoids. Principally five groups of phenolic compounds present in the olive leaves, as oleuropeosides (oleuropein and verbascoside), flavones (luteolin-7-glucoside, apigenin-7-glucoside, diosmetin-7-glucoside, luteolin, and diosmetin); flavonols (rutin); flavan-3-ols (catechin), and substituted phenols (tyrosol, hydroxytyrosol, vanillin, vanillic acid, and caffeic acid). One of the major compounds in olive leaves is oleuropein, which is followed by hydroxytyrosol [13].
There are approximately 890 million olive trees in the world and 172 million of them are in Turkey, covering around 1.8 million ha of area [14]. Olive cultivation is a significant issue for Turkey both in economic and cultural aspects. During the harvesting of olives together with pruning stages, considerable amount of by-product mainly consisting of olive leaves are accumulated, that is almost 25 kg per tree [13]. This biomass is generally used to feed animals or burned out to remove. Considering the curing, healing and nutritional properties of the leaves, valorization of these by-products have a great importance.
Researchers working on this subject reported successful conventional extraction methodologies from olive leaves such as the use of dimethyl sulfoxide [4], hexane [15], ethanol [16] and methanol [17] as solvents. However, the requirement of long extraction times was the bottleneck of the conventional methods. It was later reported that the extraction rate could be enhanced by the change of the type of the solvent or by increasing the agitation rate or using high temperatures. However, considering the reduction of both the phenolic content and antioxidant capacity at high temperatures, researchers studied on alternative procedures named as non-conventional extraction processes. Non-conventional procedures aim to enhance extraction yield, decrease the cost and enhance the selectivity of the extraction. These procedures include the use of ultrasound, microwave, supercritical fluid extraction, pressurized liquid extraction, pulsed electric fields and high voltage electrical discharges [18–23]. Among these non-conventional methods, ultrasound assisted extraction is considered to be one of the most interesting techniques because it is simple, efficient and also cheap [24]. Ultrasound is reported to enhance mass transfer mainly by inducing cavitation. Gas bubbles lead to high localized pressures and micro-streaming that disrupt the plant tissue; therefore, enhancing the intracellular substances into the solvent [25]. Beyond these, ultrasound creates interfacial instabilities and efficient compressions and expansions influencing external and internal mass transfers. Additionally, this principal non-conventional method is regarded as a green extraction method due to the reduction in energy consumption and ensuring safety as well as sustainability [23, 26, 27]. Ultrasound-assisted extraction is known to improve the efficiency of the green solvents, enhance both the extraction yield and rate, additionally known to be safe for the heat sensitive components [27]. Successful green extractions using ultrasound from olive leaves were previously reported in the literature [28–35].
In terms of the nature of the extraction solvent, the use of non-toxic, natural and renewable substances has gained great importance in the last two decades due to ecological aspects. To promote sustainable extraction processes and the utilization of green chemistry, non-petroleum derived solvents have been encouraged in many fields of research [36]. From this point of view, deep eutectic solvents (DESs) are good candidates as extraction solvents. They are new generation green solvents, non-toxic, recyclable, non-flammable and they have low vapor pressure [37–39]. They are mostly composed of natural substances and numerous types of DESs can be formed easily in the laboratory. DESs that are prepared using natural substances are called natural deep eutectic solvents, NADESs [40]. NADESs are reported to probably occur in living cells and involve in many processes in the cell such as biosynthesis, solubilization and also storage of different poorly water-soluble metabolites and unstable compounds in cells [40]. Therefore, they are very attractive solvents to be used in a broad research areas such as drug delivery systems, bone-therapy scaffolds, and other food, pharmaceutical and cosmetics related applications such as extractions [41].
In the literature, many studies have been performed using NADESs such as; extraction from olive oil [42], grape and olive pomaces [43, 44], Firus carica L. [45], Greek medicinal plants [46], almond, sesame, cinnamon and olive oil [47] and agro-food waste [48]. There is also an increasing interest of the use of DESs on the extraction from olive leaves especially in the last 2 years [49–52].
However, the combination of the use of NADES and ultrasound assisted extraction was only reported by Dedousi et al. [53] and Mouratoglou et al. [48], who investigated sodium potassium tartarate–glycerol–water (7:1:2), and choline chloride–glycerol (1:3) together with sodium acetate–glycerol (1:3), respectively.
In this study, the aim is to (i) screen novel NADESs for the ultrasound assisted extraction of bioactive compounds from olive leaves for the first time, (ii) optimize the extraction process, (iii) present an advantageous green procedure that encourage the scale-up for industrial applications.
Materials and Methods
Chemicals and Reagents
Olive leaves were harvested from Burhaniye, Balıkesir/Turkey in 2018. Olive leaves were washed with distilled water and dried overnight at 45 °C. Dried leaves were grounded using a domestic blender (Profilo Mambo, 500 W) and separated to three different sizes using molecular sieves as < 106 µm, 106–425 µm and 425–1400 µm (Endecotts, Octagon 200, England). Dried and grounded olive leaves were stored at − 20 °C until further use. Choline chloride (C1879), lactic acid (27714), glycerol (G5516), apigenin (10,798), gallic acid (G7384), 1,1-diphenyl-2-picrylhydrazyl (D9132) (DPPH), Folin–Ciocalteu reagent (9252) were purchased from Sigma. Ethylene glycol (1.009.49), d-fructose (104.007), malonic acid (800,387) were obtained from Merck whereas d-glucose (A3666) and d-sucrose (A2211) were from Applichem. All other chemicals were of reagent grade.
Preparation of NADESs
NADESs were prepared by mixing required amount of the components in a screw-capped bottle and heating till a clear liquid was formed. Choline chloride was dried under vacuum over silica gel in a desiccator prior to use. The NADESs prepared and used in this study are as follows: glucose–fructose–sucrose–water (1:1:1:11) (GFSW), glucose–fructose–water (1:1:11) (GFW), glucose–sucrose–water (1:1:11) (GSW), fructose–sucrose–water (1:1:11) (FSW) [54], choline chloride (ChCl)–glucose–water (5:2:5) (CGW), ChCl–fructose–water (5:2:5) (CFW), ChCl–sucrose–water (4:1:4) (CSW) [55], ChCl–lactic acid (1:2) (CLa) [44], ChCl–malonic acid (1:1) (CMa) [55], ChCl–ethylene glycol (1:2) (CEG) [38] and ChCl–glycerol (1:2) (CGly) [56].
Ultrasound Assisted Extraction
Certain amount of grounded leaves (< 106 µm, 106–425 µm and 425–1400 µm) were placed in a screw cap-tube with a certain amount of (8.61–90%) NADES. After mixing thoroughly, the tubes were placed in a temperature controlled sonication bath (Elma S30H, Singer, Germany) at a certain temperature (55–75 °C) for 60 min, at a sonication power of 140 W, a frequency of 37 kHz, and an acoustic energy density (AED) of 35 W L−1. The extract was filtered and the clear supernatant was used for the analyses. 50% (v/v) aqueous methanol was also used for comparison (30 mL g−1, 75 °C).
The effects of different NADES type, temperature (55–75 °C) and particle size were investigated using one-pot-at-a-time method on the total YTP, YTF and AAR. On the other hand, optimization was performed to investigate the amount of selected NADES (%) and liquid-to-solid ratio (RL/S) on the total YTP and YTF.
Design of Experiments and Response Surface Methodology
To consider the influence of the two of the critical parameters, an experimental design was performed. The design included the amount of NADES (%) and liquid-to-solid ratio (RL/S) as independent variables and YTP and YTF, as responses. A circumscribed central composite design (CCD) was used to determine and optimize the parameters to accomplish maximum extraction efficiency of phenolic substances and flavonoids. The range of the factors were obtained from preliminary experiments.
Design-Expert® 9.0 (Stat-Ease, Inc., USA) was used for the experimental design and statistical analysis. Circumscribed central composite design included axial points beyond the factorial points. Fourteen runs were conducted which included the replication of five runs at the central point. The replications were used to estimate the experimental uncertainty variance. The runs were performed randomly to prevent systematic bias. In order to derive an equation expressing the relation between the independent variables and response, stepwise regression analysis was performed for the data collected from experimental runs. To confirm the results, the experiments were run again at optimum level of independent variables. Adequacy of the model was evaluated through analysis of variance (ANOVA). ‘Backward elimination’ was performed to remove the insignificant terms (p > 0.05) which leaded the improvement of the significance of the model. The visualization of the model was performed by using Response Surface plots. For statistical calculations, the relation between the coded values and actual values are described by the following equation:
| 1 |
Here , describes the dimensionless value of an independent variable; , real value of an independent variable; , real value of an independent variable at the center point; and , step change of real value of the variable i corresponding to a variation of a unit for the dimensionless value of the variable i.
Most of the relationship of the independent variables and the responses were calculated by the second order polynomial. The quadratic model is expressed as;
| 2 |
where is the predicted response, and represent the variables or parameters, is the offset term, is the linear effect, is the first order interaction effect and is the squared effect. The coded and actual factors for the experimental design are listed in Table 1.
Table 1.
Independent variables and their coded and actual values used for optimization
| Independent variable | Unit | Symbol | Code levels | ||||
|---|---|---|---|---|---|---|---|
| − α | −1 | 0 | 1 | + α | |||
| NADES | % | A | 8.61 | 20 | 47.5 | 75 | 86.39 |
| RL/S | mL g−1 | B | 9.64 | 20 | 45.0 | 70 | 80.36 |
Determination of Total Polyphenol Yield
Total polyphenol yield was determined as reported by Blidi et al. [57] Olive leaf extract (0.02 mL) was mixed with water (0.78 mL) and Folin–Ciocalteu reagent (0.05 mL) was added to the mixture and left for 1 min at room temperature. Then, sodium carbonate [20% (v/v)] was added and the mixture was incubated at room temperature in the dark for 1 h. The absorbance was read at 750 nm and the total polyphenol concentration (CTP) was calculated from the calibration curve prepared using gallic acid. Total polyphenol yield was expressed in mg gallic acid equivalents (GAE) g−1 of dry weight (dw) from Eq. (3)
| 3 |
Here, V is the volume of the extraction medium (L) and m is the dry weight of the material (g).
Determination of Total Flavonoid Yield
Total flavonoid yield was determined as reported by Lee et al. [58]. Diethylene glycol (10 mL), olive leaf extract (1 mL) and 1 N NaOH solution (1 mL) were mixed in a test tube and incubated at room temperature for 30 min. The absorbance was measured at 420 nm. The total flavonoid content was calculated as mg apigenin equivalents (ApE) per g of dry weight and was calculated using Eq. (4).
| 4 |
Determination of the Antiradical Activity
Antiradical activity assay was performed using the method reported by Shehata et al. [59]. An aliquot of extract (0.025 mL) sample was mixed with 100 µM DPPH solution in methanol (0.975 mL). The absorbance at 515 nm was read immediately after mixing (A515(i)) and after exactly 30 min (A515(f)). AAR was calculated using Eq. (5).
| 5 |
Here CDPPH is the initial concentration of DPPH, in mol L−1; CTP is the total polyphenol concentration of the extract, in mg GAE L−1.
HPLC–ESI–QTOF-MS Analysis
Analyses were performed using an Agilent 1200 Liquid Chromatography system (Agilent Technologies, Palo Alto, CA, USA) equipped with a standard autosampler. The HPLC column was Inertsil Diol C18 (3 µm, 4.6 × 100 mm), with a flow rate of 0.4 mL min−1 at 25 °C. The mobile phase consisted of 20 mM ammonium format in water (A) (60%) and acetonitrile (B) (40%), and analyses was performed using isocratic mode. The injection volume was 5 µL. The HPLC system was coupled to a Q-TOF mass spectrometer equipped with a Jet Stream ionization source (Agilent 6530, Agilent Technologies, Palo Alto, CA, USA) operating in negative ion mode. JSI-QTOF-MS parameters were; drying gas temperature, 300 °C; drying gas flow, 8 L min−1 and nebulizing gas pressure, 40 psi. Detection was carried out within a mass range of 60–1000 m/z. The MS/MS analyses were acquired by automatic fragmentation where the three most intense mass peaks where fragmented. Nitrogen was used as drying, nebulizing and collision gas.
Statistics
All extractions were carried out in duplicate. All determinations were carried out at least in triplicate and values were averaged. Statistics was performed with ANOVA with Design-Expert® 9.0 (Stat-Ease, Inc., USA).
Results and Discussion
Effect of NADES Type
Eleven green solvents were utilized to investigate the effect of different types of NADESs on the total polyphenol yield, total flavonoid yield and antiradical activity, using one-pot-at-a-time method. The solvents used were grouped in four classes, as sugar based NADESs, choline chloride-sugar based NADESs, acid based NADESs and polyalcohol based NADESs and the properties are presented in Table 2.
Table 2.
The properties of natural deep eutectic solvents
| Group | NADES type | Density (g cm−3) | Viscosity (mPa s) | pH | Conductivity (µS cm−1) | Water activity, aw |
|---|---|---|---|---|---|---|
| Sugar based NADESs | GFSW (1:1:1:11) | 1.3657 [56] (40 °C) | 983.304 [56] (40 °C) | 4.76* | 0.09* | 0.689* |
| GFW (1:1:11) | 1.3006 [54] | N/A | 4.48* | 0.83* | 0.798* | |
| GSW (1:1:11) | 1.3511 [54] | N/A | 5.83* | 0.24* | 0.754* | |
| FSW (1:1:11) | 1.3597 [54] | N/A | 5.24* | 0.26* | 0.776* | |
| ChCl-sugar based NADESs | CFW (5:2:5) | 1.2095 [60] (30 °C) | 598 [60] (30 °C) | 1.96 [55] | 1399 [60] (30 °C) | 0.16* |
| CGW (5:2:5) | 1.2094 [60] (30 °C) | 584 [60] (30 °C) | 2.32 [55] | 2820 [60] (30 °C) | 0.159* | |
| CSW (4:1:4) | 1.2164* | 853.3 [61] (30 °C) |
4.03 [55] 5.29* |
471* | 0.158* | |
| Acid based NADESs | CLa (1:2) | 1.134 [44] (40 °C) | 29.5 [44] (40 °C) | 0.46* | 1764* | 0.208* |
| CMa (1:1) | 1.2112 [60] (30 °C) | 616 [60] (30 °C) | 0 [55] | 732 [60] (30 °C) | 0.089* | |
| Polyalcohol based NADESs | CEG (1:2) | 1.120 [62] | 37 [62] | 3.96* | 7960* (24 °C), 7610 (20 °C) [62] | < 0.03* |
| CGly (1:2) | 1.181*, 1.180 [63] | 351*, 259 [64] | 3.91* | 1189*, 1300 [39] | < 0.03* |
*Measured in our laboratory
**The properties given in the table are at 25 °C unless otherwise stated
N/A not available
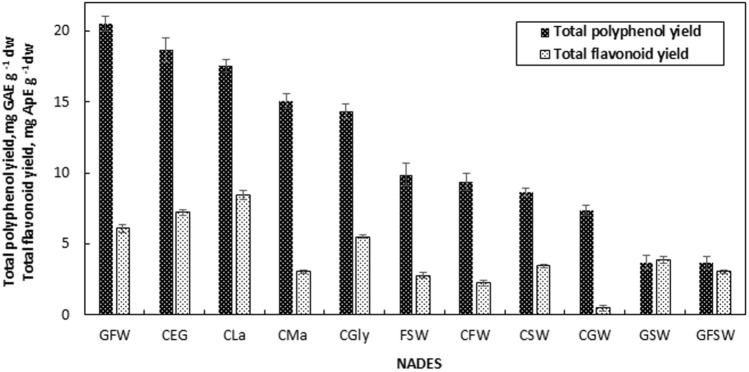
Figure 1 shows the effect of NADES type on the total polyphenol yield and total flavonoid yield of the extracts. Among NADESs used, GFW provided the highest polyphenol yield as 20.49 ± 0.50 mg GAE g−1 dw, followed by CEG and CLa as 18.65 ± 0.85 and 17.53 ± 0.43 mg GAE g−1 dw, respectively. On the other hand, the highest YTF was obtained with CLa as 8.44 ± 0.30 mg ApE g−1 dw, followed by CEG and GFW as 7.23 ± 0.20 and 6.10 ± 0.30 mg GAE g−1 dw, respectively.
Fig. 1.
The effect of NADES type on total polyphenol yield and total flavonoid yield of the extracts [liquid-to-solid ratio 50 mL g−1, 65 °C, 90% (v/v) NADES, particle size 425–1400 µm]
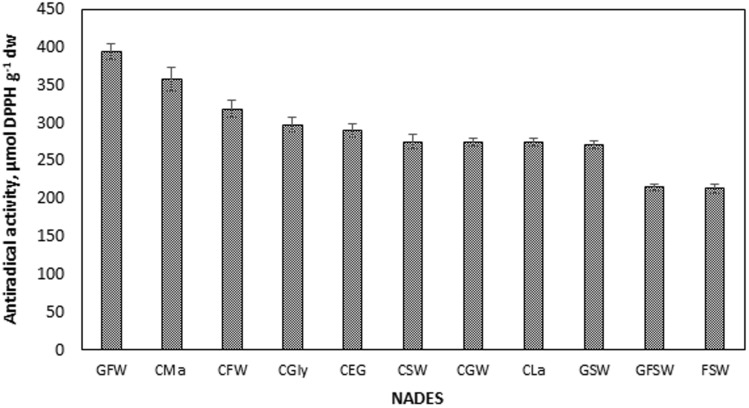
Antiradical activities of the extracts are presented in Fig. 2. The highest value was obtained using GFW as 394.49 ± 10.58 µmol DPPH g−1 dw, followed by CMa and CFW as, 357.94 ± 15.37 µmol DPPH g−1 dw and 318.70 ± 12.05 µmol DPPH g−1 dw, respectively. Consequently, GFW, CEG and CLa were the NADESs that showed up for the polyphenol and flavonoid yields. These three pioneering NADESs belong to the subclasses of sugar based, polyalcohol based and acid based NADESs. On the other hand, despite providing lower polyphenol and flavonoid yield, CFW was the best among ChCl-sugar based NADESs. Considering the properties of NADESs given in Table 1, GFW has the lowest pH among sugar based NADESs as 4.48 and the handling was easy with GFW due to its low viscosity, especially when compared to GFSW. Similarly, CFW has the lowest pH among ChCl–sugar based NADESs as 1.96. Despite the close pH values of CEG and CGly, CEG was found to provide higher polyphenol and flavanoid yields. Another advantage was the lower viscosity of CEG than CG (Table 1).
Fig. 2.
The effect of NADES type on total antiradical activities (AAR) of the extracts [liquid-to-solid ratio 50 mL g−1, 65 °C, 90% (v/v) NADES, particle size 425–1400 µm]
Mourtzinos et al. [65] reported that CEG provided the highest extraction efficiency among the polyalcohol based DESs used in the extraction of olive leaves. This result is compatible with the findings of this study. Due to the polar nature of EG, it is likely to show dipole-type and H-bond interactions with phenolic compounds. On the other hand, the lower extraction efficiency with glycerol based DESs compared to EG may arise from the branched structure of glycerol creating a steric hindrance [47].
When acid based NADESs were compared within, the use of CLa resulted in higher YTP, YTF and AAR than CMa. This may be due to the higher viscosity of CMa (Table 1) that led to lower mass transfer during the extraction process. CLa was also reported to provide higher extraction yield than other organic acid based DESs tested by Alañón et al. [50]. However, they reported higher extraction ability for polyalcohol based DESs than acid base DESs [50]. On the other hand, Şahin et al. [52] declared that carboxylic acid based DESs resulted in more efficient extracts in terms of oleuropein when compared to other DESs they tested.
Consequently, considering the effect of NADES type on total polyphenol yield, total flavonoid yield and total antiradical activities of the extracts; GFW, CFW, CLa and CEG were the four NADESs selected to be used in further experiments.
Effect of the Amount of NADES
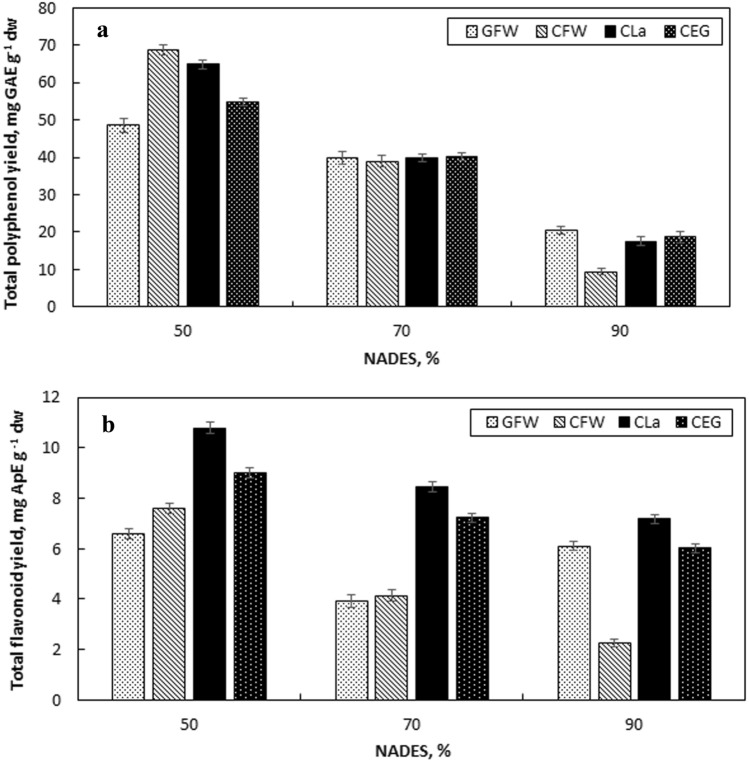
The effect of NADES type was tested at a high amount, as 90% (v/v) in this study. The aim was to use as much NADES amount as possible to take the advantage of the solubility capacity of the green solvents to extract bioactive compounds. Successful extractions with 90% were also reported in the literature [66]. However, the viscosity of the green solvents change significantly with the addition of water, affecting the mass transfer rate and therefore, the extraction capacity [56]. Additionally, water as a polar solvent, increases the polarity of the NADESs, facilitating the extraction of polar compounds [41]. Also, many studies reported the enhancement of the extraction performance with the addition of water to DESs at a specific range [49, 50, 53, 56]. Therefore, lower NADES amounts such as 50, 70 and 90% (v/v) were tested and the results are presented in Fig. 3 and Table 3. For all four types of NADESs used, YTP and YTF were found to be at their highest values with 50% (v/v) amount (Fig. 3a, b). When NADES content was decreased to 50%, approximately 3.2-fold increase was detected at YTP, while 1.4-fold increase was detected at YTF in comparison with 90% (v/v). The highest YTP was obtained using 50% CFW as 68.66 ± 1.4 mg GAE g−1 dw and the highest YTF was obtained using 50% CLa as 10.78 ± 0.22 mg GAE g−1 dw.
Fig. 3.
The effect of the amount of NADES on total polyphenol yield (a) and total flavonoid yield (b) of the extracts (liquid-to-solid ratio 50 mL g−1, 65 °C, particle size 425–1400 µm)
Table 3.
Effects of NADES amount, temperature, liquid-to-solid ratio and particle size on the antiradical activity of the extracts
| Antiradical activity of the extracts | ||||
|---|---|---|---|---|
| GFW | CFW | CLa | CEG | |
| Amount of NADES [% (v/v), liquid-to-solid ratio = 50 mL g−1, T = 65 °C, particle size = 425–1400 µm] | ||||
| 50 | 463.99 ± 13.1 | 463.77 ± 9.8 | 374.77 ± 7.6 | 458.63 ± 12.0 |
| 70 | 455.94 ± 11.6 | 453.71 ± 10.5 | 373.43 ± 9.2 | 457.51 ± 8.7 |
| 90 | 394.50 ± 9.5 | 318.70 ± 11.2 | 274.38 ± 8.8 | 290.37 ± 13.2 |
| Temperature, T (°C, liquid-to-solid ratio = 50 mL g−1, NADES = 50%, particle size = 425–1400 µm) | ||||
| 55 | 461.76 ± 11.9 | 468.69 ± 10.7 | 424.42 ± 9.9 | 463.33 ± 12.5 |
| 65 | 463.99 ± 13.1 | 463.77 ± 9.8 | 374.77 ± 7.6 | 458.63 ± 12.0 |
| 75 | 461.98 ± 10.6 | 461.98 ± 9.8 | 416.14 ± 12.4 | 460.86 ± 11.7 |
| Effect of particle size [µm, liquid-to-solid ratio = 50 mL g−1, NADES = 50% (v/v), T = 75 °C] | ||||
| < 160 | 433.50 ± 13.3 | 437.50 ± 12.6 | 342.50 ± 12.1 | 457.50 ± 8.4 |
| 160–425 | 457.50 ± 11.1 | 462.50 ± 10.7 | 394.00 ± 12.4 | 461.00 ± 9.8 |
| 425–1400 | 461.98 ± 10.6 | 461.98 ± 9.8 | 416.14 ± 12.4 | 460.86 ± 11.7 |
Similarly, the highest antiradical activities were found using 50% of NADESs as 463.99 ± 13.1 µmol DPPH g−1 dw with GFW, 463.77 ± 9.8 µmol DPPH g−1 dw with CFW, 374.77 ± 7.6 µmol DPPH g−1 dw with CLa and 458.63 ± 12.0 µmol DPPH g−1 dw with CEG (Table 3). In the literature, a similar optimum water content as 43.3% (v/v) was reported for CEG (1:2) [50] while much lower optimum water content 20% (w/v) was reported for glycerol–glycine–water (7:1:3) [49]. Therefore, the optimum water content varies depending on the nature and viscosity of the DES.
According to the results, NADES amount was determined to be very significant parameter on the extraction yields. Therefore, it was selected as a factor to be optimized using experimental design.
Effect of Temperature
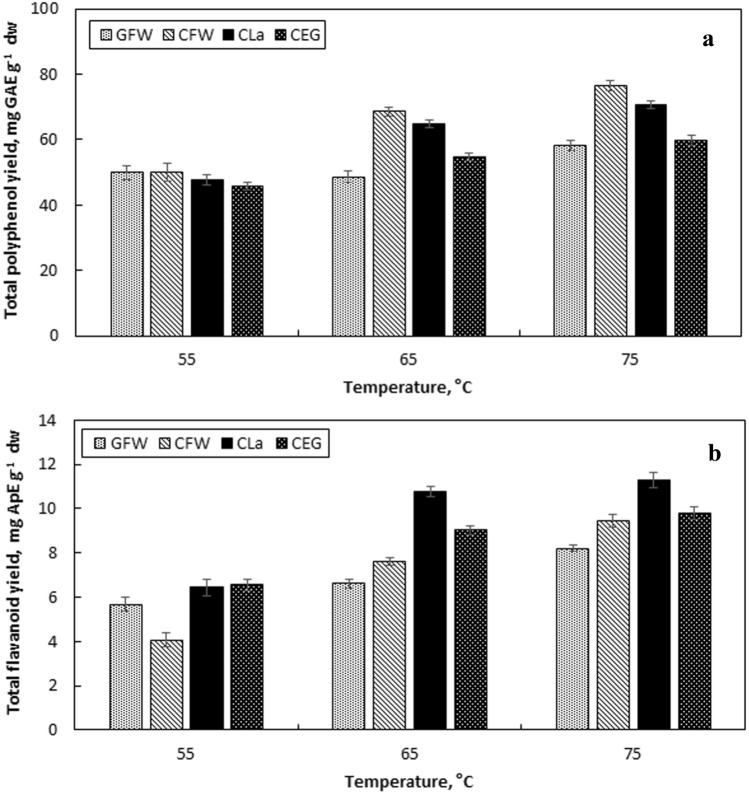
The effect of temperature was investigated at 55, 65 and 75 °C. Higher values were not tested due to possible negative effect of high temperature on the phenolic content and antioxidant capacity [65]. According to the results, 75 °C was found to provide the highest YTP and YTF (Fig. 4a, b). CFW was found to provide the highest YTP as 76.62 ± 0.99 mg GAE g−1 dw while CLa provided the highest YTF as 11.29 ± 0.35. In terms of AAR, similar values were obtained at tested temperature range (Table 2). The viscosity of NADES decrease with the increase in the temperature and therefore facilitates the penetration of the solvent to the plant. This destructs the intermolecular interaction in the plant and leads to increased extraction at high temperatures [67].
Fig. 4.
The effect of temperature on total polyphenol yield (a) and total flavonoid yield (b) of the extracts [liquid-to-solid ratio 50 mL g−1, 50% (v/v) NADES, particle size 425–1400 µm]
Our results were found to be compatible with, Alañón et al. [50] who reported 79.6 °C as the optimum temperature for the extraction of phenolics from olive leaf. On the other hand Athanadiadis et al. reported an enhancement in the extraction kinetics from olive leaf at 80 °C [49].
Effect of Particle Size
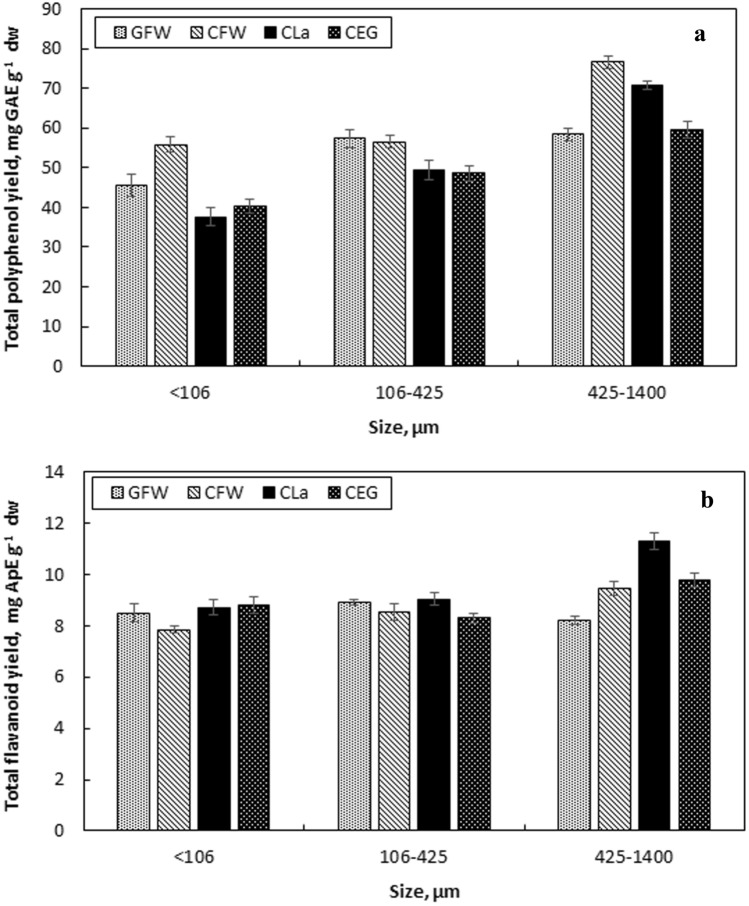
To investigate the effect of the particle size, grounded olive leaves were fractionated into three different particle sizes. The leaf particles that were < 106 µm and between 106 and 425 µm provided similar values, whereas 425–1400 µm provided higher YTP and YTF for all NADESs used (Fig. 5). CFW and CLa were the NADESs that let the highest YTP and YTF as 76.62 ± 1.5 mg GAE g−1 dw and 11.29 ± 0.35, respectively. In terms of AAR, the particles between 160 and 425 µm and 425–1400 µm provided very close results except for CLa. CLa provided the highest AAR for 425–1400 µm size. Therefore, the optimum particle size was detected as 425–1400 µm. This results is consistent with the recommended average particle size that is reported to be 0.4–0.8 mm [41].
Fig. 5.
The effect of particle size on total polyphenol yield (a) and total flavonoid yield (b) of the extracts [liquid-to-solid ratio 50 mL g−1, 50% (v/v) NADES, 75 °C]
Process Optimization by Response Surface Methodology
The effects of the temperature, particle size and the type and the amount of NADESs were investigated using one-pot-at-a-time method. The optimum values were identified as; 75 °C, 425–1400 µm and 50% (v/v) NADES (GFW, CFW, CLa and CEG). The results showed that a fine tuning of the amount of NADES (%) would provide higher extractions efficiency. Beyond these, another significant parameter on the extraction is the solid-to-liquid ratio (RL/S). Therefore, the effect of solid-to-liquid ratio (RL/S) and the amount of NADES (%) on YTP and YTF were investigated in detail, using an experimental design.
Experimental design was performed by using CCD and the responses were revealed by RSM. This let to find out the joint effects of the two factors; NADES amount and RL/S on the responses; total polyphenol yield and total flavonoid yield. Four pioneering NADESs from each group, as GFW, CFW, CLa, and CEG, were used in the experimental design and each of them were optimized separately. The experimental design, levels of the two independent variables and responses are tabulated in Table 4.
Table 4.
Central composite design matrix for NADESs (experimental variables and responses)
| Run | Variables | Responses | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NADES (%) | RL/S (mL g−1) | GFW | CFW | CLa | CEG | |||||||
| Level | Actual value | Level | Actual value | Total polyphenol yield (mg GAE g−1 dw) | Total flavonoid yield (mg ApE g−1 dw) | Total polyphenol yield (mg GAE g−1 dw) | Total flavonoid yield (mg ApE g−1 dw) | Total polyphenol yield (mg GAE g−1 dw) | Total flavonoid yield (mg ApE g−1 dw) | Total polyphenol yield (mg GAE g−1 dw) | Total flavonoid yield (mg ApE g−1 dw) | |
| A | B | |||||||||||
| 1 | 0 | 47.50 | 0 | 45.00 | 101.79 | 7.78 | 166.95 | 12.43 | 133.36 | 10.16 | 92.24 | 9.11 |
| 2 | 0 | 47.50 | 0 | 45.00 | 89.79 | 7.50 | 195.00 | 11.52 | 111.58 | 9.78 | 98.24 | 8.65 |
| 3 | − 1 | 20.00 | − 1 | 20.00 | 82.38 | 6.84 | 90.36 | 9.76 | 74.98 | 7.53 | 81.94 | 7.56 |
| 4 | + 1 | 75.00 | + 1 | 70.00 | 83.11 | 6.14 | 3.87 | 8.39 | 3.32 | 1.26 | 2.14 | 6.89 |
| 5 | + 1 | 75.00 | − 1 | 20.00 | 72.01 | 5.19 | 120.03 | 7.66 | 81.05 | 9.32 | 73.35 | 3.90 |
| 6 | 0 | 47.50 | 0 | 45.00 | 110.50 | 7.69 | 157.59 | 12.60 | 114.00 | 9.00 | 95.00 | 8.11 |
| 7 | − 1 | 20.00 | + 1 | 70.00 | 77.79 | 6.57 | 144.39 | 13.27 | 85.00 | 8.47 | 83.5 | 7.20 |
| 8 | 0 | 47.50 | 0 | 45.00 | 98.68 | 7.15 | 180.69 | 11.83 | 108.55 | 8.3 | 87.8 | 7.53 |
| 9 | 0 | 47.50 | 0 | 45.00 | 116.24 | 8.33 | 154.02 | 11.20 | 118.25 | 9.58 | 76.68 | 8.30 |
| 10 | 0 | 47.50 | − α | 9.64 | 11.81 | 3.8 | 112.05 | 9.48 | 91.18 | 9.03 | 80.32 | 4.20 |
| 11 | 0 | 47.50 | 0 | 45.00 | 93.44 | 7.33 | 175.11 | 11.90 | 105.00 | 10.00 | 80.00 | 7.82 |
| 12 | 0 | 47.50 | + α | 80.36 | 84.61 | 6.46 | 7.50 | 10.91 | 9.37 | 1.52 | 3.77 | 6.90 |
| 13 | + α | 86.39 | 0 | 45.00 | 75.57 | 5.46 | 96.03 | 7.27 | 80.90 | 3.54 | 51.79 | 5.90 |
| 14 | − α | 8.61 | 0 | 45.00 | 83.12 | 7.20 | 119.37 | 9.89 | 94.66 | 7.75 | 72.24 | 7.30 |
Table 5 shows the ANOVA results for both of the responses; total polyphenol yield (mg GAE g−1 dw) and total flavonoid yield (mg ApE g−1 dw) obtained with four different NADESs. On the other hand, the mathematical models representing the responses in the experimental region and also the R2 values are presented in Table 6. ANOVA results, p values and R2 in accordance implied the reliability of the models to predict the responses.
Table 5.
ANOVA results for the responses obtained with NADESs
| NADES | Model | Source | Sum of squares | df | Mean square | F | P | Inference |
|---|---|---|---|---|---|---|---|---|
| Total polyphenol yield (mg GAE g−1 dw) | ||||||||
| GFW | Reduced cubic model | Model | 6899.26 | 6 | 1149.88 | 8.34 | 0.0066 | S |
| A-NADES % | 30.92 | 1 | 30.92 | 0.22 | 0.6502 | |||
| B-R | 2649.92 | 1 | 2649.92 | 19.23 | 0.0032 | |||
| AB | 61.54 | 1 | 61.54 | 0.45 | 0.5254 | |||
| A2 | 408.35 | 1 | 408.35 | 2.96 | 0.1289 | |||
| B2 | 3907.74 | 1 | 3907.74 | 28.35 | 0.0011 | |||
| A2B | 1162.70 | 1 | 1162.70 | 8.44 | 0.0228 | |||
| Residual | 964.71 | 7 | 137.82 | |||||
| Lack of fit | 456.66 | 2 | 228.33 | 2.25 | 0.2013 | NS | ||
| Pure error | 508.05 | 5 | 101.61 | |||||
| Cor total | 7863.97 | 13 | ||||||
| CFW | Quadratic model | Model | 42,326.51 | 5 | 8465.30 | 23.23 | 0.0001 | S |
| A-NADES % | 2586.88 | 1 | 2586.88 | 7.10 | 0.0286 | |||
| B-R | 5511.77 | 1 | 5511.77 | 15.12 | 0.0046 | |||
| AB | 7241.16 | 1 | 7241.16 | 19.87 | 0.0021 | |||
| A2 | 6846.47 | 1 | 6846.47 | 18.79 | 0.0025 | |||
| B2 | 21,862.78 | 1 | 21,862.78 | 59.99 | < 0.0001 | |||
| Residual | 2915.71 | 8 | 364.46 | |||||
| Lack of fit | 1746.26 | 3 | 582.09 | 2.49 | 0.1750 | NS | ||
| Pure error | 1169.46 | 5 | 233.89 | |||||
| Cor total | 45,242.22 | 13 | ||||||
| CLa | Quadratic model | Model | 17,260.19 | 5 | 3452.04 | 21.08 | 0.0002 | S |
| A-NADES % | 1129.78 | 1 | 1129.78 | 6.90 | 0.0303 | |||
| B-R | 4204.76 | 1 | 4204.76 | 25.68 | 0.0010 | |||
| AB | 1925.02 | 1 | 1925.02 | 11.76 | 0.0090 | |||
| A2 | 1810.20 | 1 | 1810.20 | 11.06 | 0.0105 | |||
| B2 | 8743.32 | 1 | 8743.32 | 53.40 | < 0.0001 | |||
| Residual | 1309.90 | 8 | 163.74 | |||||
| Lack of fit | 808.04 | 3 | 269.35 | 2.68 | 0.1575 | NS | ||
| Pure error | 501.86 | 5 | 100.37 | |||||
| Cor total | 18,570.09 | 13 | ||||||
| CEG | Quadratic model | Model | 10,999.23 | 5 | 2199.85 | 15.31 | 0.0006 | S |
| A-NADES % | 1766.28 | 1 | 1766.28 | 12.29 | 0.0080 | |||
| B-R | 3956.41 | 1 | 3956.41 | 27.53 | 0.0008 | |||
| AB | 1323.87 | 1 | 1323.87 | 9.21 | 0.0162 | |||
| A2 | 910.71 | 1 | 910.71 | 6.34 | 0.0360 | |||
| B2 | 3284.65 | 1 | 3284.65 | 22.85 | 0.0014 | |||
| Residual | 1149.83 | 8 | 143.73 | |||||
| Lack of fit | 786.46 | 3 | 262.15 | 3.61 | 0.1006 | NS | ||
| Pure error | 363.38 | 5 | 72.68 | |||||
| Cor total | 12,149.06 | 13 | ||||||
| Total flavonoid yield (mg ApE g−1 dw) | ||||||||
| GFW | Reduced cubic model | Model | 17.63 | 6 | 2.94 | 16.00 | 0.0009 | S |
| A-NADES % | 2.58 | 1 | 2.58 | 14.03 | 0.0072 | |||
| B-R | 3.54 | 1 | 3.54 | 19.26 | 0.0032 | |||
| AB | 0.37 | 1 | 0.37 | 2.03 | 0.1976 | |||
| A2 | 2.12 | 1 | 2.12 | 11.56 | 0.0114 | |||
| B2 | 9.53 | 1 | 9.53 | 51.91 | 0.0002 | |||
| A2B | 1.19 | 1 | 1.19 | 6.46 | 0.0385 | |||
| Residual | 1.29 | 7 | 0.18 | |||||
| Lack of fit | 0.43 | 2 | 0.22 | 1.27 | 0.3590 | NS | ||
| Pure error | 0.85 | 5 | 0.17 | |||||
| Cor total | 18.92 | 13 | ||||||
| CFW | Quadratic model | Model | 42.48 | 5 | 8.50 | 18.62 | 0.0003 | S |
| A-NADES % | 14.33 | 1 | 14.33 | 31.42 | 0.0005 | |||
| B-R | 4.90 | 1 | 4.90 | 10.74 | 0.0112 | |||
| AB | 1.94 | 1 | 1.94 | 4.25 | 0.0731 | |||
| A2 | 18.22 | 1 | 18.22 | 39.93 | 0.0002 | |||
| B2 | 4.33 | 1 | 4.33 | 9.49 | 0.0151 | |||
| Residual | 3.65 | 8 | 0.46 | |||||
| Lack of fit | 2.24 | 3 | 0.75 | 2.65 | 0.1605 | NS | ||
| Pure error | 1.41 | 5 | 0.28 | |||||
| Cor total | 46.13 | 13 | ||||||
| CLa | Quadratic model | Model | 115.94 | 5 | 23.19 | 27.16 | < 0.0001 | S |
| A-NADES % | 16.17 | 1 | 16.17 | 18.94 | 0.0024 | |||
| B-R | 39.34 | 1 | 39.34 | 46.07 | 0.0001 | |||
| AB | 20.25 | 1 | 20.25 | 23.72 | 0.0012 | |||
| A2 | 19.29 | 1 | 19.29 | 22.59 | 0.0014 | |||
| B2 | 23.96 | 1 | 23.96 | 28.06 | 0.0007 | |||
| Residual | 6.83 | 8 | 0.85 | |||||
| Lack of fit | 4.38 | 3 | 1.46 | 2.97 | 0.1358 | NS | ||
| Pure error | 2.45 | 5 | 0.49 | |||||
| Cor total | 122.77 | 13 | ||||||
| CEG | Quadratic model | Model | 27.55 | 5 | 5.51 | 17.69 | 0.0004 | S |
| A-NADES % | 4.43 | 1 | 4.43 | 14.21 | 0.0055 | |||
| B-R | 5.20 | 1 | 5.20 | 16.69 | 0.0035 | |||
| AB | 2.81 | 1 | 2.81 | 9.01 | 0.0170 | |||
| A2 | 4.14 | 1 | 4.14 | 13.28 | 0.0065 | |||
| B2 | 11.98 | 1 | 11.98 | 38.45 | 0.0003 | |||
| Residual | 2.49 | 8 | 0.31 | |||||
| Lack of fit | 0.87 | 3 | 0.29 | 0.89 | 0.5072 | NS | ||
| Pure error | 1.62 | 5 | 0.32 | |||||
| Cor total | 30.04 | 13 | ||||||
S significant, NS not significant
Table 6.
Mathematical equations expressing the responses in coded factors
| NADES | Models (Coded Factors) | R2 | Adj R2 | CV (%) |
|---|---|---|---|---|
| Total polyphenol yield (mg GAE g−1 dw) | ||||
| GFW | 101.74 – 1.9 *A + 25.74 * B + 3.92 * A * B − 7.44 * A2 − 23.00 * B2 − 24.11 * A2 * B | 0.877 | 0.772 | 13.92 |
| CFW | 171.56 – 17.98 * A − 26.25 * B − 42.55 * A * B − 30.45 * A2 − 54.41 * B2 | 0.936 | 0.895 | 15.51 |
| CLa | 115.12 – 11.88 * A − 22.93 * B − 21.94 *A * B − 15.66 * A2 − 34.41* B2 | 0.930 | 0.885 | 14.79 |
| CEG | 88.33 – 14.86 * A − 22.24 * B − 18.19 * A * B − 11.11 * A2 − 21.09 * B2 | 0.905 | 0.846 | 17.14 |
| Total flavonoid yield (mg ApE g−1 dw) | ||||
| GFW | 7.63 − 0.57 * A + 0.94 * B + 0.31 * A * B − 0.54 * A2 − 1.14 * B2 – 0.77 * A2 * B | 0.932 | 0.874 | 6.42 |
| CFW | 11.91 – 1.34 * A + 0.78 * B − 0.70 *A * B -1.57 * A2 − 0.77 * B2 | 0.921 | 0.871 | 6.38 |
| CLa | 9.47 – 1.42 * A − 2.22 * B − 2.25 * A* B − 1.62 * A2 − 1.80 * B2 | 0.944 | 0.909 | 12.29 |
| CEG | 8.25 − 0.74 * A + 0.81 * B + 0.84 * A * B -0.75 * A2 − 1.27 * B2 | 0.917 | 0.865 | 7.86 |
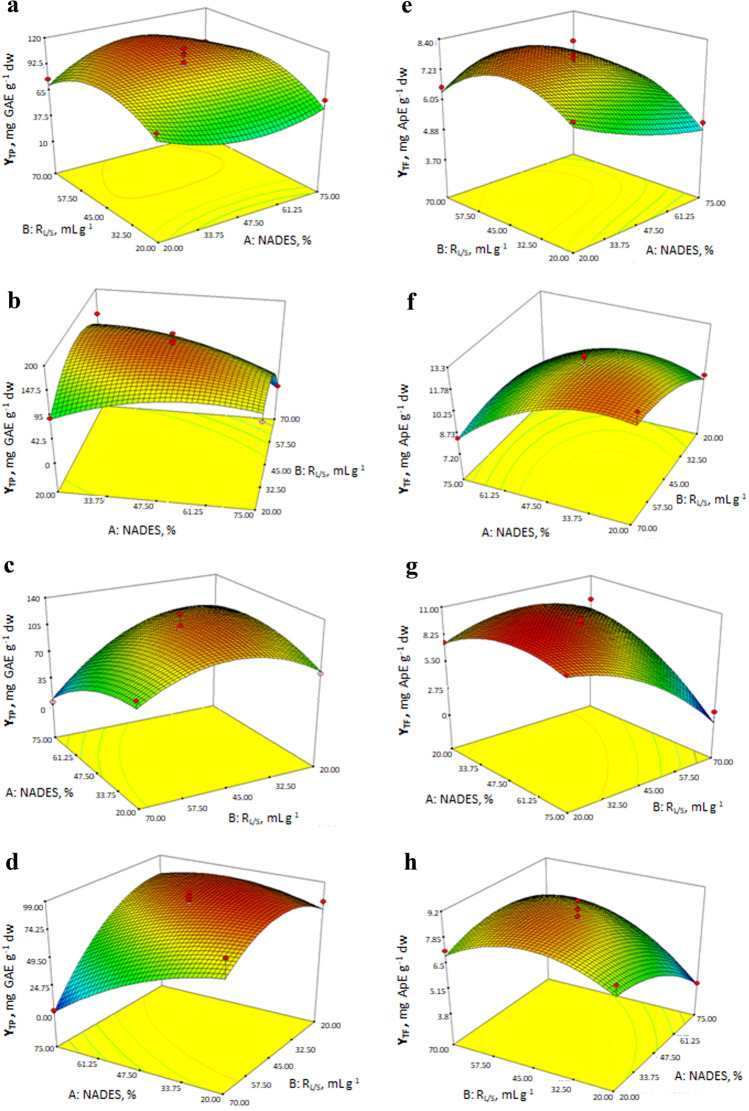
First of all, total polyphenol yield values obtained with four different NADESs are discussed. The model F-value of 8.34 for GFW indicated a statistically significant reduced cubic model. The significant model terms of this equation, that was modified using ‘backward elimination’, were identified as B, B2 and A2B (Table 6). YTP values obtained with GFW were in the range of 116.24–11.81 mg GAE g−1 dw. When response surface is analysed (Fig. 6a), a unique 3D plot was observed representing the reduced cubic model. High values of YTP was achieved around 45–70 mL g−1 of RL/S and 33.75–61.25% of GFW. At low values of RL/S, especially around 20–32.5 mL g−1, YTP was low regardless of the amount of GFW, as indicated in green color.
Fig. 6.
3D plots of the response surfaces of the response YTP for GFW (a), CFW (b), CLa (c), CEG (d) and YTF for GFW (e), CFW (f), CLa (g), and CEG (h)
In the case of CFW, quadratic model was found to express the responses thoroughly for the working space (Table 6). The maximum and minimum values were obtained as, 195.00 and 3.87 mg GAE g−1 dw. All of the model terms were found to be significant. High values of YTP could be obtained at average values of the working space. At the high levels of the both of the independent variables, YTP reached its lowest values as indicated with blue colour (Fig. 6b). Similar to the effect of the high levels of the independent variables, low levels also let to a decrease in YTP (green area), but not as dramatic as the blue area.
YTP values were in the range of 133.36 and 3.32 mg GAE g−1 dw when CLa was used for the extractions (Fig. 6c). The ANOVA results indicated that the response could be expressed by a quadratic model (Table 6). Additionally, all of the model terms were identified to be statistically significant. The quadratic surface showed that high values of the response could be obtained at medium values of RL/S together with relatively low amount of CLa. On the other hand, low values were obtained at highest levels of the independent variables. YTP was found to increase with decreasing amount of NADES at constant RL/S, while it showed an increasing and decreasing trend with increasing RL/S at constant amount of NADES.
The design of the experiments conducted using CEG resulted in a quadratic model for YTP, indicating A, B, AB, A2 and B2 as significant terms (Table 6). The quantity changed between 98.24 and 2.14 mg GAE g−1 dw. High values of YTP could be achieved at both lower RL/S and NADES values (Fig. 6d) as indicated in red. YTP decreased dramatically with the decrease in the CEG amount at RL/S=70 mL g−1. A similar decrease was also observed with increasing RL/S at constant NADES amount. The lowest level of the response surface showed up for high levels of both of the variables.
When the four selected NADESs are compared in terms of the YTP values obtained, CFW was found to provide the highest values.
The response surface plots of YTF are presented in Fig. 6e–h. In the case of GFW, the response values fitted best in a reduced cubic model (Table 6). The reduction was performed in order to eliminate the insignificant factors by ‘backward elimination’. The significant model terms were identified as A, B, A2, B2 and A2B. The highest and lowest values obtained were 8.33 and 3.80 mg ApE g−1 dw, respectively. The 3D plot showing the response surface indicated that the highest values of YTF were achieved at lower values of NADES amount and at medium values of RL/S. On the other hand, the lowest YTF values were obtained at low RL/S (20–30 mL g−1) and high values of NADES amount (47.5–75%) (Fig. 6e). However, decreasing amount of GFW had a positive effect on YTF around 20–30 mL g−1 of RL/S. Additionally, YTF showed an increasing and decreasing trend at constant amount of NADES.
Quadratic model was obtained as the equation to describe the total flavonoid yield for CFW extractions. The values were in the range of 13.27–7.23 mg ApE g−1 dw. A, B, A2 and B2 were the significant model terms (Table 6). The 3D plot indicated a clear increase of YTF at low amount of CFW together with high level of RL/S (Fig. 6f). On the other hand, a slight shift of the working space to higher values of RL/S would provide a better view of the entire reddish area.
The ANOVA results of the design for CLa showed that predicted R2 (0.717) was in reasonable agreement with adjusted R2 (0.909). All of the model terms were found to be significant (Table 6). Total flavonoid yield values were in the range of 10.16–1.26 mg ApE g−1 dw. Response surface plot indicated that highest YTF values were achieved at medium to low values of RL/S but medium to high lower values of CLa (Fig. 6g). NADES amount did not have a dramatic effect on YTF at constant low values of RL/S.
CEG also resulted in a quadratic model to describe the response surface for YTF, with the significant terms as A, B, AB, A2 and B2 (Table 6). The quantities changed between 9.11 and 3.90 mg ApE g−1 dw. All of the terms of the equation representing the responses were shown to be significant. On the other hand, predicted R2 (0.717) was found to be in reasonable agreement with adjusted R2 (0.865). It was observed that high amount of NADES together with low amount RL/S resulted in very low values of YTF. On the other hand, when NADES amount decreased YTF increased at constant RL/S (Fig. 6h). The high extraction yield for flavonoid were obtained at lower NADES amount and medium values of RL/S.
When the four selected NADESs are compared in terms of the YTF values CFW was found to provide the highest responses while GFW provided the lowest responses.
The optimization of the responses was also performed using Design Expert. The experimental conditions providing the highest values of the responses YTP and YTF were predicted separately using Design Expert. The predictions of the program for YTF indicated that flavonoid yield would not change significantly between its own optimum conditions or the other response’s (YTP) optimum conditions. Moreover, the desirability function that was used to optimize both of the responses predicted lower YTP than a single optimization. Considering the choices, the predicted optimized conditions for one of the responses, YTP were used in the experiments and the results are given at Table 7. Additionally, antiradical activity assays were also performed and presented at the optimum conditions. According the results, the difference between predicted and experimental results were found to be lower than 6%, which showed the convenience of the experimental design. The highest YTP ,YTF and AAR were obtained with CFW as 187.31 ± 10.3 mg GAE g−1 dw, 12.75 ± 0.6 mg ApE g−1 dw and 480 ± 26 µmol DPPH g−1 dw, respectively.
Table 7.
Optimized conditions for the ultrasound assisted extraction with NADESs
| NADES | Optimum conditions | YTP, mg GAE g−1 dw Predicted | YTP, mg GAE g−1 dw Experimental | YTF, mg ApE g−1 dw Predicted | YTF, mg ApE g−1 dw Experimental | AARµmol DPPH g−1 dw |
|---|---|---|---|---|---|---|
| GFW | 47.09%, 63.27 mL g−1 | 116.64 ± 4.6 | 122.47 ± 4.7 | 8.06 ± 0.4 | 8.46 ± 0.5 | 470 ± 27.2 |
| CFW | 42.69%, 40.66 mL g−1 | 175.38 ± 8.2 | 187.31 ± 10.3 | 11.91 ± 0.7 | 12.75 ± 0.6 | 480 ± 26 |
| CLa | 40.00%, 38.84 mL g−1 | 119.28 ± 6.2 | 124.05 ± 4.7 | 10.02 ± 0.5 | 10.42 ± 0.5 | 435 ± 28.2 |
| CEG | 50.00%, 30.85 mL g−1 | 93.65 ± 6.1 | 99.45 ± 6.6 | 7.88 ± 0.48 | 8.12 ± 0.4 | 472 ± 22.1 |
*Extraction conditions: T = 75 ℃, t = 1 h
The Principle Bioactive Compounds Detected in the Olive Leaf Extracts
The extracts obtained at the optimum conditions were subjected to LC–MS analysis for the detection of oleuropein, luteolin and caffeic acid and compared with MeOH extract which was obtained at non-optimized conditions. According to the results (Table 8), NADESs were found to extract comparable amounts with MeOH. The highest oleuropein content was achieved with GFW as 1630.80 mg kg−1 dw, followed by CEG as 1031.57 mg kg−1 dw; whereas MeOH provided 1221.17 mg kg−1 dw. GFW was also found to provide highest caffeic acid as 112.77 mg kg−1 dw, followed by MeOH as 41.54 mg kg−1 dw. GFW could extract higher amount of oleuropein and caffeic acid than MeOH, showing the comparable extraction performance of the NADES with the organic solvent. On the other hand, MeOH extract provided the highest amount of luteolin as 2.59 mg kg−1 dw followed by GFW and CFW extracts, as 1.34 mg kg−1 dw and 0.49 mg kg−1 dw, respectively.
Table 8.
Bioactive compounds extracted from olive leaves
| NADES/Solvent | Oleuropein (mg kg−1 dw) | Caffeic acid (mg kg−1 dw) | Luteolin (mg kg−1 dw) |
|---|---|---|---|
| GFW | 1630.80 | 112.77 | 1.34 |
| CFW | 853.46 | 0.00 | 0.49 |
| CLa | 290.07 | 0.09 | 0.01 |
| CEG | 1031.57 | 0.08 | 0.25 |
| MeOH | 1221.17 | 41.54 | 2.59 |
According to the results, oleuropein was found to be the most abundant compound among the three phenolics analyzed. This was inevitable since it is also the most abundant phenolic compound in the olive leaf [13]. Despite the highest YTP and YTF values were obtained with CFW, higher amount of oleuropein, caffeic acid and luteolin could be detected in GFW extracts. This may be due to the modification of the extracted substances into their derivatives that could not be analyzed. Moreover, other phenolic substances may be present in CFW extract. The results showed that NADESs can be good candidates to be used as an alternative of conventional solvents.
Conclusions
Green, simple and cheap extraction procedure from an agricultural waste, olive leaf, was presented as an alternative to conventional extraction methods, using the potential of green and low cost natural deep eutectic solvents. This extraction method meets many principles of green extraction, such as simple and inexpensive preparation of the solvents, minimum amount of solvent, sustainable production, decreased waste and also the use of safer solvents. The novelty of this study is the first time use of NADESs that are not previously used for the extraction from olive leaves using ultrasound assisted extraction. Moreover glucose–fructose–water—as a firstly presented NADES in the ultrasound assisted extraction from olive leaves, was found to extract higher amount of oleuropein and caffeic acid than MeOH, showing an encouraging possible shift of organic solvents with NADESs for an environmentally-friendly process. The sustainable utilization of the resources, can only be managed by avoiding existing chemical-based methods and by using green and non-conventional methods such as ultrasound assisted-extraction, as presented in this study. In addition to the shift of the extraction method with an environmentally-friendly method, the substitution of the hazardous solvents (especially chlorinated solvents) with the green solvents will result in satisfactory clean processes. Similar changes in the industrial processes—even a partial change will probably mark a new epoch for a clean and healthy earth. The presented procedure is a promising route for the green extraction from olive leaves that will contribute to the elimination of the hazardous processes. On the other hand, the optimum green solvent content that was found to be around 50%, offers a clear away of the disadvantage of the difficulty of pumping and stirring of high viscosity deep eutectic solvents in the industrial scale. Additionally, the extract has the potential to be used without further purification steps, due to the non-toxic and natural structure of natural deep eutectic solvents.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The author would like to thank Dr. Alper Karakaya, Düzen Biological Sciences R&D and Production for the supplement of olive leaves and Zeynep Erdoğan, Institute of Materials Science and Nanotechnology National Nanotechnology Research Center (UNAM), Bilkent University for LC–MS analysis.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wichers HJ, Soler-rivas C, Espı JC. Review Oleuropein and related compounds. J. Sci. Food Agric. 2000;80:1013–1023. doi: 10.1002/(SICI)1097-0010(20000515)80:7<1013::AID-JSFA571>3.0.CO;2-C. [DOI] [Google Scholar]
- 2.Laguerre M, López Giraldo LJ, Piombo G, Figueroa-Espinoza MC, Pina M, Benaissa M, Combe A, Rossignol Castera A, Lecomte J, Villeneuve P. Characterization of olive-leaf phenolics by ESI–MS and evaluation of their antioxidant capacities by the cat assay&. J. Am. Oil Chem. Soc. 2009;86:1215–1225. doi: 10.1007/s11746-009-1452-x. [DOI] [Google Scholar]
- 3.Bouaziz M, Sayadi S. Isolation and evaluation of antioxidants from leaves of a Tunisian cultivar olive tree. Eur. J. Lipid Sci. Technol. 2005;107:497–504. doi: 10.1002/ejlt.200501166. [DOI] [Google Scholar]
- 4.Benavente-García O, Castillo J, Lorente J, Ortuño A, Del Rio JA. Antioxidant activity of phenolics extracted from Olea europaea L. leaves. Food Chem. 2000;68:457–462. doi: 10.1016/S0308-8146(99)00221-6. [DOI] [Google Scholar]
- 5.Andrikopoulos NK, Salta FN, Mylona A, Chiou A, Boskou G. Oxidative stability of edible vegetable oils enriched in polyphenols with olive leaf extract. Food Sci. Technol. Int. 2007;13:413–421. doi: 10.1177/1082013208089563. [DOI] [Google Scholar]
- 6.Korukluoglu M, Sahan Y, Yigit A, Karakas R. Korukluoglu2006_Article_AntifungalActivityOfOliveLeafO. 2006;56:359–362. [Google Scholar]
- 7.Markín D, Duek L, Berdícevsky I. In vitro antimicrobial activity of olive leaves. Mycoses. 2003;46:132–136. doi: 10.1046/j.1439-0507.2003.00859.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee-Huang S, Zhang L, Huang PL, Chang YT, Huang PL. Anti-HIV activity of olive leaf extract (OLE) and modulation of host cell gene expression by HIV-1 infection and OLE treatment. Biochem. Biophys. Res. Commun. 2003;307:1029–1037. doi: 10.1016/s0006-291x(03)01292-0. [DOI] [PubMed] [Google Scholar]
- 9.Zarzuelo A, Duarte J, Jimenez J, Gonzalez M, Utrilla MP. Vasodilator effect of olive leaf. Planta Med. 1991 doi: 10.1055/s-2006-960138. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez M, Zarzuelo A, Gamez MJ, Utrilla MP, Jimenez J, Osuna I. Hypoglycemic activity of olive leaf. Planta Med. 1992;58:513–515. doi: 10.1055/s-2006-961538. [DOI] [PubMed] [Google Scholar]
- 11.Thangavel N, Al Bratty M, Hazmi A, Najmi HA, Ali Alaqi RO. Molecular docking and molecular dynamics aided virtual search of OliveNet™ Directory for Secoiridoids to combat SARS-CoV-2 infection and associated hyperinflammatory responses. Front. Mol. Biosci. 2021 doi: 10.3389/fmolb.2020.627767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adhikari B, Marasini BP, Rayamajhee B, Bhattarai BR, Lamichhane G, Khadayat K, Adhikari A, Khanal S, Parajuli N. Potential roles of medicinal plants for the treatment of viral diseases focusing on COVID-19: a review. Phytother. Res. 2020 doi: 10.1002/ptr.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El SN, Karakaya S. Olive tree (Olea europaea) leaves: potential beneficial effects on human health. Nutr. Rev. 2009;67:632–638. doi: 10.1111/j.1753-4887.2009.00248.x. [DOI] [PubMed] [Google Scholar]
- 14.Ministry of Customs and Trade: 2017 Yılı Zeytin ve Zeytinyağı Raporu. 29 (2018)
- 15.Tabera J, Guinda Á, Ruiz-Rodríguez A, Señoráns FJ, Ibáñez E, Albi T, Reglero G. Countercurrent supercritical fluid extraction and fractionation of high-added-value compounds from a hexane extract of olive leaves. J. Agric. Food Chem. 2004;52:4774–4779. doi: 10.1021/jf049881+. [DOI] [PubMed] [Google Scholar]
- 16.Mylonaki S, Kiassos E, Makris DP, Kefalas P. Optimisation of the extraction of olive (Olea europaea) leaf phenolics using water/ethanol-based solvent systems and response surface methodology. Anal. Bioanal. Chem. 2008;392:977–985. doi: 10.1007/s00216-008-2353-9. [DOI] [PubMed] [Google Scholar]
- 17.Paiva-Martins F, Gordon MH. Isolation and characterization of the antioxidant component 3,4-dihydroxyphenylethyl 4-formyl-3-formylmethyl-4-hexenoate from olive (Olea europaea) leaves. J. Agric. Food Chem. 2001;49:4214–4219. doi: 10.1021/jf010373z. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad-Qasem MH, Cánovas J, Barrajón-Catalán E, Micol V, Cárcel JA, García-Pérez JV. Kinetic and compositional study of phenolic extraction from olive leaves (var. Serrana) by using power ultrasound. Innov. Food Sci. Emerg. Technol. 2013;17:120–129. doi: 10.1016/j.ifset.2012.11.008. [DOI] [Google Scholar]
- 19.Roselló-Soto E, Koubaa M, Moubarik A, Lopes RP, Saraiva JA, Boussetta N, Grimi N, Barba FJ. Emerging opportunities for the effective valorization of wastes and by-products generated during olive oil production process: non-conventional methods for the recovery of high-added value compounds. Trends Food Sci. Technol. 2015;45(2):296–310. doi: 10.1016/j.tifs.2015.07.003. [DOI] [Google Scholar]
- 20.Xynos N, Papaefstathiou G, Psychis M, Argyropoulou A, Aligiannis N, Skaltsounis AL. Development of a green extraction procedure with super/subcritical fluids to produce extracts enriched in oleuropein from olive leaves. J. Supercrit. Fluids. 2012 doi: 10.1016/j.supflu.2012.03.014. [DOI] [Google Scholar]
- 21.Caldas TW, Mazza KEL, Teles ASC, Mattos GN, Brígida AIS, Conte-Junior CA, Borguini RG, Godoy RLO, Cabral LMC, Tonon RV. Phenolic compounds recovery from grape skin using conventional and non-conventional extraction methods. Ind. Crops Prod. 2018;111:86–91. doi: 10.1016/j.indcrop.2017.10.012. [DOI] [Google Scholar]
- 22.Belwal T, Chemat F, Venskutonis PR, Cravotto G, Jaiswal DK, Bhatt ID, Devkota HP, Luo Z. Recent advances in scaling-up of non-conventional extraction techniques: learning from successes and failures. Trends Anal. Chem. 2020;127:115895. doi: 10.1016/j.trac.2020.115895. [DOI] [Google Scholar]
- 23.Anticona M, Blesa J, Frigola A, Esteve MJ. High biological value compounds extraction from citrus waste with non-conventional methods. Foods. 2020;9:2–24. doi: 10.3390/foods9060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W, Xue A, Niu H, Jia Z, Wang J. Optimised ultrasonic-assisted extraction of flavonoids from Folium eucommiae and evaluation of antioxidant activity in multi-test systems in vitro. Food Chem. 2009 doi: 10.1016/j.foodchem.2008.10.079. [DOI] [Google Scholar]
- 25.Knorr D, Ade-Omowaye BIO, Heinz V. Nutritional improvement of plant foods by non-thermal processing. Proc. Nutr. Soc. 2002 doi: 10.1079/pns2002162. [DOI] [PubMed] [Google Scholar]
- 26.Chemat F, Rombaut N, Sicaire AG, Meullemiestre A, Fabiano-Tixier AS, Abert-Vian M. Ultrasound assisted extraction of food and natural products. Mechanisms, techniques, combinations, protocols and applications. A review. Ultrason. Sonochem. 2017;34:540–560. doi: 10.1016/j.ultsonch.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 27.Tiwari BK. Ultrasound: a clean, green extraction technology. Trends Anal. Chem. 2015;71:100–109. doi: 10.1016/j.trac.2015.04.013. [DOI] [Google Scholar]
- 28.Japón-Luján R, Luque-Rodríguez JM, Luque De Castro MD. Dynamic ultrasound-assisted extraction of oleuropein and related biophenols from olive leaves. J. Chromatogr. A. 2006 doi: 10.1016/j.chroma.2005.12.106. [DOI] [PubMed] [Google Scholar]
- 29.Sánchez Ávila N, Capote P, Luque de Castro F. Ultrasound-assisted extraction and silylation prior to gas chromatography–mass spectrometry for the characterization of the triterpenic fraction in olive leaves. J. Chromatogr. A. 2007 doi: 10.1016/j.chroma.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 30.del Contreras MM, Lama-Muñoz A, Espínola F, Moya M, Romero I, Castro E. Valorization of olive mill leaves through ultrasound-assisted extraction. Food Chem. 2020 doi: 10.1016/j.foodchem.2020.126218. [DOI] [PubMed] [Google Scholar]
- 31.Dobrinčić A, Repajić M, Garofulić IE, Tuden L, Dragović-Uzelac V, Levaj B. Comparison of different extraction methods for the recovery of olive leaves polyphenols. Processes. 2020 doi: 10.3390/PR8091008. [DOI] [Google Scholar]
- 32.Lama-Muñoz A, Del Mar Contreras M, Espínola F, Moya M, Romero I, Castro E. Optimization of oleuropein and luteolin-7-o-glucoside extraction from olive leaves by ultrasound-assisted technology. Energies. 2019 doi: 10.3390/en12132486. [DOI] [PubMed] [Google Scholar]
- 33.da Rosa GS, Vanga SK, Gariepy Y, Raghavan V. Comparison of microwave, ultrasonic and conventional techniques for extraction of bioactive compounds from olive leaves (Olea europaea L.) Innov. Food Sci. Emerg. Technol. 2019 doi: 10.1016/j.ifset.2019.102234. [DOI] [Google Scholar]
- 34.Sucharitha P, Satyanarayana S, Reddy K. Pretreatment and optimization of processing conditions for extraction of oleuropein from olive leaves using central composite design. Pharmacogn. Res. 2019 doi: 10.4103/pr.pr_179_18. [DOI] [Google Scholar]
- 35.Yao Q, Shen Y, Bu L, Yang P, Xu Z, Guo X. Ultrasound-assisted aqueous extraction of total flavonoids and hydroxytyrosol from olive leaves optimized by response surface methodology. Prep. Biochem. Biotechnol. 2019 doi: 10.1080/10826068.2019.1630648. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Fabiano-Tixier AS, Tomao V, Cravotto G, Chemat F. Green ultrasound-assisted extraction of carotenoids based on the bio-refinery concept using sunflower oil as an alternative solvent. Ultrason. Sonochem. 2013;20:12–18. doi: 10.1016/j.ultsonch.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Abbott AP, Capper G, Davies DL, Rasheed RK, Tambyrajah V. Novel solvent properties of choline chloride/urea mixtures. Chem. Commun. 2003;1:70–71. doi: 10.1039/b210714g. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, De Oliveira Vigier K, Royer S, Jerome F. Deep eutectic solvents: syntheses, properties and applications. Chem. Soc. Rev. 2012;41:7108–7146. doi: 10.1039/c2cs35178a. [DOI] [PubMed] [Google Scholar]
- 39.García G, Aparicio S, Ullah R, Atilhan M. Deep eutectic solvents: physicochemical properties and gas separation applications. Energy Fuels. 2015;29:2616. doi: 10.1021/ef5028873. [DOI] [Google Scholar]
- 40.Choi YH, van Spronsen J, Dai Y, Verberne M, Hollmann F, Arends IWCE, Witkamp G-J, Verpoorte R. Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology? Plant Physiol. 2011;156:1701–1705. doi: 10.1104/pp.111.178426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cvjetko Bubalo M, Vidović S, Radojčić Redovniković I, Jokić S. New perspective in extraction of plant biologically active compounds by green solvents. Food Bioprod. Process. 2018;109:52–73. doi: 10.1016/j.fbp.2018.03.001. [DOI] [Google Scholar]
- 42.García A, Rodríguez-Juan E, Rodríguez-Gutiérrez G, Rios JJ, Fernández-Bolaños J. Extraction of phenolic compounds from virgin olive oil by deep eutectic solvents (DESs) Food Chem. 2016;197:554–561. doi: 10.1016/j.foodchem.2015.10.131. [DOI] [PubMed] [Google Scholar]
- 43.Panić M, Radić Stojković M, Kraljić K, Škevin D, Radojčić Redovniković I, Gaurina Srček V, Radošević K. Ready-to-use green polyphenolic extracts from food by-products. Food Chem. 2019;283:628–636. doi: 10.1016/j.foodchem.2019.01.061. [DOI] [PubMed] [Google Scholar]
- 44.Chanioti S, Tzia C. Extraction of phenolic compounds from olive pomace by using natural deep eutectic solvents and innovative extraction techniques. Innov. Food Sci. Emerg. Technol. 2018;48:228–239. doi: 10.1016/j.ifset.2018.07.001. [DOI] [Google Scholar]
- 45.Wang T, Jiao J, Gai QY, Wang P, Guo N, Niu LL, Fu YJ. Enhanced and green extraction polyphenols and furanocoumarins from Fig (Ficus carica L.) leaves using deep eutectic solvents. J. Pharm. Biomed. Anal. 2017;145:339–345. doi: 10.1016/j.jpba.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Georgantzi C, Lioliou A-E, Paterakis N, Makris D. Combination of lactic acid-based deep eutectic solvents (DES) with β-cyclodextrin: performance screening using ultrasound-assisted extraction of polyphenols from selected native Greek medicinal plants. Agronomy. 2017;7:54. doi: 10.3390/agronomy7030054. [DOI] [Google Scholar]
- 47.Khezeli T, Daneshfar A, Sahraei R. A green ultrasonic-assisted liquid–liquid microextraction based on deep eutectic solvent for the HPLC–UV determination of ferulic, caffeic and cinnamic acid from olive, almond, sesame and cinnamon oil. Talanta. 2016;150:577–585. doi: 10.1016/j.talanta.2015.12.077. [DOI] [PubMed] [Google Scholar]
- 48.Mouratoglou E, Malliou V, Makris DP. Novel glycerol-based natural eutectic mixtures and their efficiency in the ultrasound-assisted extraction of antioxidant polyphenols from agri-food waste biomass. Waste Biomass Valoriz. 2016;7:1377–1387. doi: 10.1007/s12649-016-9539-8. [DOI] [Google Scholar]
- 49.Athanasiadis V, Grigorakis S, Lalas S, Makris DP. Highly efficient extraction of antioxidant polyphenols from Olea europaea& leaves using an eco-friendly glycerol/glycine deep eutectic solvent. Waste Biomass Valoriz. 2018;9:1985–1992. doi: 10.1007/s12649-017-9997-7. [DOI] [Google Scholar]
- 50.Alañón ME, Ivanović M, Gómez-Caravaca AM, Arráez-Román D, Segura-Carretero A. Choline chloride derivative-based deep eutectic liquids as novel green alternative solvents for extraction of phenolic compounds from olive leaf. Arab. J. Chem. 2018 doi: 10.1016/j.arabjc.2018.01.003. [DOI] [Google Scholar]
- 51.Chakroun D, Grigorakis S, Loupassaki S, Makris DP. Enhanced-performance extraction of olive (Olea europaea) leaf polyphenols using l-lactic acid/ammonium acetate deep eutectic solvent combined with β-cyclodextrin: screening, optimisation, temperature effects and stability. Biomass Convers. Biorefin. 2019 doi: 10.1007/s13399-019-00521-2. [DOI] [Google Scholar]
- 52.Şahin S, Kurtulbaş E, Bilgin M. Special designed deep eutectic solvents for the recovery of high added-value products from olive leaf: a sustainable environment for bioactive materials. Prep. Biochem. Biotechnol. 2020 doi: 10.1080/10826068.2020.1824162. [DOI] [PubMed] [Google Scholar]
- 53.Dedousi M, Mamoudaki V, Grigorakis S, Makris D. Ultrasound-assisted extraction of polyphenolic antioxidants from olive (Olea europaea) leaves using a novel glycerol/sodium-potassium tartrate low-transition temperature mixture (LTTM) Environments. 2017;4:31. doi: 10.3390/environments4020031. [DOI] [Google Scholar]
- 54.Mohammadpour Z, Abdollahi SH, Safavi A. Sugar-based natural deep eutectic mixtures as green intercalating solvents for high-yield preparation of stable MoS2 nanosheets: application to electrocatalysis of hydrogen evolution reaction. ACS Appl. Energy Mater. 2018;1:5896–5906. doi: 10.1021/acsaem.8b00838. [DOI] [Google Scholar]
- 55.Elgharbawy AA. Shedding light on lipase stability in natural deep eutectic solvents. Chem. Biochem. Eng. Q. 2018;32:359–370. doi: 10.15255/cabeq.2018.1335. [DOI] [Google Scholar]
- 56.Dai Y, van Spronsen J, Witkamp GJ, Verpoorte R, Choi YH. Natural deep eutectic solvents as new potential media for green technology. Anal. Chim. Acta. 2013;766:61. doi: 10.1016/j.aca.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 57.Blidi S, Bikaki M, Grigorakis S, Loupassaki S, Makris DP. A comparative evaluation of bio-solvents for the efficient extraction of polyphenolic phytochemicals: apple waste peels as a case study. Waste Biomass Valoriz. 2015;6:1125–1133. doi: 10.1007/s12649-015-9410-3. [DOI] [Google Scholar]
- 58.Lee OH, Lee BY, Lee J, Lee HB, Son JY, Park CS, Shetty K, Kim YC. Assessment of phenolics-enriched extract and fractions of olive leaves and their antioxidant activities. Bioresour. Technol. 2009;100:6107–6113. doi: 10.1016/j.biortech.2009.06.059. [DOI] [PubMed] [Google Scholar]
- 59.Shehata E, Grigorakis S, Loupassaki S, Makris DP. Extraction optimisation using water/glycerol for the efficient recovery of polyphenolic antioxidants from two Artemisia species. Sep. Purif. Technol. 2015;149:462–469. doi: 10.1016/j.seppur.2015.06.017. [DOI] [Google Scholar]
- 60.Zhao BY, Xu P, Yang FX, Wu H, Zong MH, Lou WY. Biocompatible deep eutectic solvents based on choline chloride: characterization and application to the extraction of rutin from Sophora japonica. ACS Sustain. Chem. Eng. 2015;3:2746–2755. doi: 10.1021/acssuschemeng.5b00619. [DOI] [Google Scholar]
- 61.Hayyan M, Mbous YP, Looi CY, Wong WF, Hayyan A, Salleh Z, Mohd-Ali O. Natural deep eutectic solvents: cytotoxic profile. SpringerPlus. 2016;5:913. doi: 10.1186/s40064-016-2575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Longo LS, Jr., Craveiro MV. Deep eutectic solvents as unconventional media for multicomponent reactions. J. Braz. Chem. Soc. 2018;29:1999–2025. doi: 10.21577/0103-5053.20180147. [DOI] [Google Scholar]
- 63.Abbott AP, Harris RC, Ryder KS, D’Agostino C, Gladden LF, Mantle MD. Glycerol eutectics as sustainable solvent systems. Green Chem. 2011;13:82. doi: 10.1039/c0gc00395f. [DOI] [Google Scholar]
- 64.D’Agostino C, Harris RC, Abbott AP, Gladden LF, Mantle MD. Molecular motion and ion diffusion in choline chloride based deep eutectic solvents studied by 1H pulsed field gradient NMR spectroscopy. Phys. Chem. Chem. Phys. 2011;13:21383–21391. doi: 10.1039/c1cp22554e. [DOI] [PubMed] [Google Scholar]
- 65.Mourtzinos I, Anastasopoulou E, Petrou A, Grigorakis S, Makris D, Biliaderis CG. Optimization of a green extraction method for the recovery of polyphenols from olive leaf using cyclodextrins and glycerin as co-solvents. J. Food Sci. Technol. 2016;53:3939–3947. doi: 10.1007/s13197-016-2381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng X, Duan MH, Yao XH, Zhang YH, Zhao CJ, Zu YG, Fu YJ. Green extraction of five target phenolic acids from Lonicera japonica& Flos with deep eutectic solvent. Sep. Purif. Technol. 2016;157:249–257. doi: 10.1016/j.seppur.2015.10.065. [DOI] [Google Scholar]
- 67.Wei ZF, Wang XQ, Peng X, Wang W, Zhao CJ, Zu YG, Fu YJ. Fast and green extraction and separation of main bioactive flavonoids from Radix Scutellariae. Ind. Crops Prod. 2015;63:175–181. doi: 10.1016/j.indcrop.2014.10.013. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.