Abstract
The in vitro cell adhesion assay is a quantitative method for measuring selective cell adhesion to specific proteins. Traditionally, cell adhesion assays employ purified protein immobilized on a solid glass or plastic surface. Here, we describe a transient 293T cell transfection-based cell adhesion assay to study selective cell adhesion of a specific cell type to a protein of interest. In this protocol, 293T cells are transfected with a mammalian expression plasmid containing mSiglec1 cDNA or an empty plasmid as a mock control and are then cultured to form a monolayer. Subsequently, these Siglec1-expressing and mock-transfected 293T cell monolayers are used for cell adhesion assays with GFP-expressing B16F10 cells. The number of GFP+ cancer cells adhering to each 293T monolayer is a quantitative mean to compare the selective adhesiveness of cancer cells to Siglec1. This method eliminates the need to express and purify the protein of interest to perform in vitro cell adhesion assays and can easily be performed with difficult-to-purify proteins while maintaining their native in situ structure.
Keywords: Cell-cell adhesion assay, Cell adhesion, Cancer cell adhesion assay, 293T, Transient transfection, Siglec1, Fluorescence microscopy
Background
Cell-to-cell interactions are critical for biological processes such as tissue development, regeneration, and morphogenesis, as well as immune responses and cancer metastasis (Gumbiner, 1996). The cell adhesion molecules that meditate these cell-to-cell interactions can be lectins, integrins, cadherins, selectins, lymphocyte homing receptors, or immunoglobulin super family receptors ( Janiszewska et al., 2020 ). Interactions between a cell surface receptor and counter-receptor on cell pairs initiate the stable cell-to-cell interactions and activation of tissue-specific signal transduction. Studying these interactions by using methods such as adhesion assays, co-immunoprecipitations, or pull-down assays is of utmost importance to understand the consequences of an individual protein-protein interaction in complex cellular processes. In cancer biology, cell-to-cell interactions are of particular interest for evaluating the role of host organ stroma in providing initial anchorage and a growth-promoting environment to metastatic cells ( Chen et al., 2011 ; Singh and Choi, 2019).
The in vitro cell adhesion assay is a simple low-cost method for investigating interactions between specific cell types and a protein of interest (Kucik and Wu, 2005). Conventionally, adhesion assays employ purified protein immobilized on a solid surface, such as the wells of an enzyme-linked immunosorbent assay (ELISA) or cell culture plate (Chen, 2012). However, the molecular interactions that mediate the cell-to-cell contacts are often weak and low in affinity (Jiang and Barclay, 2009). This renders conventional cell adhesion assays inefficient in such situations. Additionally, it is cumbersome to perform cell adhesion assays with difficult-to-purify proteins. Recently, our group developed a 293T cell transient transfection-based cell adhesion assay to demonstrate interactions between highly sialylated B16F10 murine melanoma cells and sialic acid binding protein Siglec1 by using fluorescence microscopy (Singh and Choi, 2019). Here, we systematically describe the experimental procedures of 293T cell transient transfection-based cell adhesion assay adapted from our previously published work (Singh and Choi, 2019). This cell adhesion assay can be broadly applied to evaluate interactions between other receptors and cell types in addition to those mentioned above. The key advantage of this protocol is that, unlike the conventional adhesion assays, this method does not require purified proteins. Additionally, this method can be useful for studying interactions where the weak binding affinity with the protein of interest is the major restriction of the cell adhesion assay. The high expression of the protein of interest on the 293T cell surface may provide more interaction sites to bind to cells in the adhesion assay. Consequently, this improves cell adhesion by increasing the avidity (accumulated strength of low affinity interactions) of the interaction by forming more ligation bonds. This protocol can also be adapted to visualize protein-specific complex signaling events such as focal adhesion by using antibody staining and immunofluorescence microscopy.
Materials and Reagents
T25 flask (Corning, catalog number: 430639)
40 μm nylon cells strainer (Falcon, catalog number: 352340)
Chamber slides, non-coated (Cell culture slides 8 well, SPL Life Sciences, catalog number: 30108) or pre-coated (Lab-Tek II CC2 chamber slide, Nunc, catalog number: 154941)
15 ml conical tube (SPL Life Science, catalog number: 50015)
Human embryonic kidneys (HEK) 293T cells (ATCC, catalog number: CRL-3216)
Green fluorescent protein (GFP) expressing mouse melanoma cell line B16F10 (B16-GFP for brevity; derivative of B16F10 cells, ATCC, catalog number: CRL-6475)
Dulbecco's Modified Eagle Medium (DMEM; Welgene, catalog number: LM 001-05)
Ca2+/Mg2+ free Dulbecco's phosphate-buffered saline (DPBS; Welgene, catalog number: LB 001-02)
Trypsin-EDTA 0.05% (Gibco, catalog number: 25300054)
Trypsin-EDTA 0.25% (Gibco, catalog number: 25200056)
Enzyme-free PBS based cell dissociation buffer (Gibco, catalog number: 13151014)
Gelatin (Sigma-Aldrich, Catalog number: G1890-100G)
Full length mouse Siglec1 cDNA (Transomic Technologies, catalog number: BC141335) cloned into mammalian expression vector pd18, a derivative of pcDNA 3 ( Hayden et al., 1996 )
Lipofectamine 3000 (Invitrogen, catalog number: L3000015)
Bovine serum albumin (BSA; RMBIO, catalog number: BSA-BSH)
4% Paraformaldehyde (PFA) solution (Biosesang, catalog number: PC2031-100-00)
Alexa fluor 647-conjugated anti-mouse Siglec1 antibody (BioLegend, clone 3D6.112, catalog number: 142408)
DAPI containing mounting medium (Vector Laboratories, catalog number: H-1200)
Triton X-100 (Biosesang, Korea, catalog number: T1020)
Fetal bovine serum (FBS; RMBIO, catalog number: FBS-BBT-5XM)
Trypan Blue solution (Sigma-Aldrich, Catalog number: T8154-100ML)
Opti-MEM reduced serum medium, GlutaMAX supplemented (Gibco, catalog number: 51985034)
Fingernail polish to seal the edges of the coverslip mounted slides (Missha, Able C&C Co., Ltd, South Korea)
Blocking and staining buffer (see Recipes)
2% PFA solution in complete DMEM (see Recipes)
Equipment
Water bath (Lab Companion, model: BW-20G)
CO2 incubator (Thermo Scientific, model: 3131)
P1000 pipette (Eppendorf, catalog number: 3123000063)
Zeiss Laser Scanning Microscope 780 (Carl Zeiss, model: LSM 780)
Inverted microscope (Olympus, model: CKX31)
Centrifuge (Eppendorf, model: 5810 R)
Bio-Hazard Safety Cabinet, Class II (Vision Scientific Co., LTD, Korea, model: VS-1400LSN)
Software
Imaris (Oxford instruments, version 8.1, https://imaris.oxinst.com), or ImageJ (https://imagej.nih.gov/ij/)
Procedure
-
Maintenance of 293T and B16-GFP cell cultures
Warm DMEM supplemented with 10% FBS (hereafter referred to as complete DMEM) and DPBS in a 37 °C water bath for at least 15 min.
Wash ~90% confluent cells in a T25 flask with 3 ml of DPBS, aspirate the DPBS, and add 0.5 ml of Trypsin-EDTA. Use 0.05% Trypsin-EDTA for 293T cells and 0.25% Trypsin-EDTA for B16-GFP cells.
Incubate the cells at 37 °C in a 5% CO2 incubator for 2-5 min, add 3 ml of prewarmed complete DMEM, and transfer the cell suspensions to a 15 ml sterile conical tube.
Spin down the cells at 300 × g for 3 min at room temperature, discard the supernatant, and resuspend the cell pellet in 15 ml of complete DMEM.
Dispense 4 ml of the cell suspension into each T25 flask, and then incubate it for 3 days in a 37 °C 5% CO2 incubator before subculturing again.
-
Preparation of gelatin-coated chamber slides
Gelatin coating of the glass slides helps the cells to adhere firmly to the slides and stops them from detaching during culture, as well as during subsequent adhesion assay and staining procedures.
Add 300 μl of 0.1% Gelatin Solution to each well of the chamber slide aseptically in a biosafety cabinet.
Rock the slide gently to spread the solution evenly in the whole well and place the slide with the lid on in a 37 °C CO2 incubator for 1 h.
Aspirate the gelatin solution from the wells and wash two times with DPBS, then briefly let it dry in the biosafety cabinet.
-
Immediately use the slide to culture the 293T cells, or store it at room temperature for up to one week.
Alternatively, pre-coated chambers slides can also be used.
-
Transfection of 293T cells
The day before transfection (Day 1), prepare the 293T cell suspension as described in Procedure A.
Count the cells using standard trypan blue exclusion (https://www.abcam.com/protocols/counting-cells-using-a-haemocytometer) and seed 1 × 105 cells in 500 μl complete DMEM in each well of the gelatin-coated or pre-coated chamber slide. Seed cells in the required number of wells on the basis of your experimental design.
-
The next day (Day 2), perform the following steps to prepare transfection mixture:
Prepare two tubes, Tube A and Tube B, separately. The following quantities (Table 1) are for transfection of one well; multiply the volumes by the number of replicates to scale up. Prepare transfection mix with 25% additional volume to account for pipetting and dispensing errors. Prepare the transfection mix in a similar manner for the mock transfection as well.
Mix tube A and tube B and incubate for 5 min at room temperature.
Replace the complete DMEM in the wells of the chamber slide with 500 μl Opti-MEM.
Add 50 μl of the transfection mix from Step C4 to each well dropwise. Rock the slide gently a few times and incubate at 37 °C in a 5% CO2 incubator.
Six hours after the transfection, gently remove the lipofectamine-containing medium and replace the medium with DMEM containing 5% FBS and incubate the slide at 37 °C in a 5% CO2 incubator until Day 3.
On Day 3, the transfected slides can be used to perform cell adhesion assay.
-
Preparation of B16-GFP single-cell suspension
To prepare the B16-GFP cells for the cell adhesion assay, dissociate cells that are 60%-70% confluent in a T25 flask using an enzyme-free cell dissociation buffer. This non-enzymatic cell dissociation is suitable for cell adhesion assays that require intact cell surface proteins to facilitate binding interactions.
Discard the growth medium from the flask and wash the cells twice with 3 ml Ca2+- and Mg2+-free DPBS.
Add 2 ml of prewarmed (37 °C in a water bath) enzyme-free cell dissociation buffer in a T25 flask and place the flask in a 37 °C 5% CO2 incubator.
Observe the cells every two minutes under the microscope. When the cells roundup (4-5 min after adding enzyme-free cell dissociation buffer), firmly tap the flask against the palm of your gloved hand to promote cell detachment.
Add 5 ml of prewarmed complete DMEM to the flask and resuspend the cells by pipetting up and down. Transfer the cell suspension to a 15-ml conical tube. Add 5 ml of complete DMEM again to the flask to collect the remaining cells and add this to the same conical tube.
Centrifuge the cells at 300 × g for 3 min at room temperature and resuspend the cell pellet in 3 ml of complete DMEM. Pass the cell suspension through a 40-µm nylon cell strainer to remove cell clumps and to prepare single-cell suspensions.
Count the cells using standard trypan blue exclusion and adjust the cell number to 1 × 105 cells/500 μl by diluting the cells using complete DMEM.
-
Cell adhesion assay
Remove the culture medium from the transfected 293T cells (from Procedure C) using a P1000 pipette and add 1 × 105 B16-GFP cells in 500 µl of complete DMEM per well (for both mock and Siglec1-transfected wells).
Incubate the slide at 37 °C in a 5 % CO2 incubator for 5 min. The incubation time should be decided empirically for different protein-cell pairs to minimize the non-specific adherence of cells to the monolayer.
Gently remove the media containing non-adherent cells using a P1000 pipette.
Wash the wells by filling them with 1 ml of complete DMEM and gently pipetting up and down several times so as not to damage the 293T cell monolayer. Repeat this washing step three times.
After the last wash, remove the DMEM medium and add 500 μl of prewarmed 2% PFA diluted in complete DMEM and incubate the slide for 10 min at 37 °C in a 5% CO2 incubator.
Remove the PFA solution and gently wash each well with 1 ml of DPBS. Repeat this washing step three times and proceed to antibody staining.
-
Antibody staining
Block the formaldehyde-fixed slides in blocking and staining buffer (1× DPBS/1% BSA/0.3% Triton X-100) for 30 min at room temperature.
Prepare the Alexa Fluor 647-conjugated anti-mSiglec1 antibody by diluting it as indicated on the manufacturer datasheet (5 μg/ml) in blocking and staining buffer (see Recipes) (https://www.biolegend.com/en-us/products/alexa-fluor-647-anti-mouse-cd169-siglec-1-antibody-9835).
Remove the blocking solution and add 150 μl of the diluted antibody to each well of the chamber slide. Incubate for 1 h at room temperature.
Wash each slide well three times in 1 ml of DPBS for 5 min. Remove the chambers carefully and coverslip the slides with Vector lab mounting Reagent with DAPI. Use fingernail polish to seal the edges of the coverslip and keep the slide at room temperature for 5 min to let it dry completely.
-
Image acquisition
Acquire the images on a LSM 780 or any other confocal microscope using standard microscopic techniques.
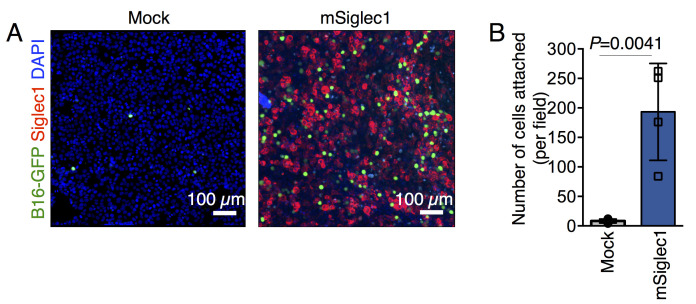
Acquire Z-stack images using 10× magnification and with 1 μm Z-stack interval (Figure 1A).
Table 1. Transfection mixture preparation.
| Component | Amount per well | |
| Tube A | Opti-MEM medium | 25 μl |
| Lipofectamine 3000 Reagent | 1.5 μl | |
| Tube B | Opti-MEM medium | 25 μl |
| Purified pd18-mSiglec1 plasmid or empty plasmid DNA for mock transfection (1-5 μg/μl) | 500 ng | |
| P3000 reagent (2 μl/μg plasmid) | 1 μl |
Figure 1. Cell adhesion assays.
A. Representative images of adherent GFP-expressing B16F10 cells on monolayers of mSiglec-1-expressing and mock-transfected 293T cells. B. Quantification of the number of adherent B16-GFP cells on indicated monolayers. Data are mean ± S.D.; n = 4 biologically independent experiments. The P-value was calculated with a two-tailed, unpaired t-test (Singh and Choi, 2019).
Data analysis
The Imaris Spots tool provides a procedure to automatically detect and count the point-like structures. The following link provides a video tutorial for using the Imaris Spots tool: https://youtu.be/8ldNUmWNbB0?list=PLkGKyuSIzfzqB24QUcj65Xhvesj0XhRJD&t=1016.
In the ‘Objects’ toolbar of the ‘Surpass view,’ click the ‘Spots’ option. This opens the Spots creation wizard. The creation wizard guides the user through the steps to obtain optimal parameters.
Select the source channel which shows GFP+ cells. The Spots wizard automatically detects the threshold to insert the Spots. Alternatively, to modify the threshold setting, change the automatic setting of the wizard to manual setting by clicking the manual box and set the lower threshold to include all of the GFP+ cells. Each selected GFP+ cell is represented by a gray spot.
Click the ‘finish’ button on the wizard tool bar to compete the process. The total number of spots (GFP+ cells) can be observed by selecting the statistics tab. An alternative is ImageJ, an open source software that can be used to detect and count total adherent cells by using the analyze particles feature of the program. The detailed procedure can be found in the ImageJ user manual.
Plot the average number of cells attached per field, as indicated in Figure 1B.
Notes
293T cells express a wide range of cell surface molecules, so it is advisable to confirm the expression level of your protein of interest before utilizing 293T cells in your adhesion assay.
An isotype control antibody can be used to ascertain the specificity of the antibody (in this case anti-Siglec1-Alexa 647 antibody) and the transfection efficiency of 293T cells.
A fully intact 293T cells monolayer is important to avoid non-specific attachment of cancer cells to the slide surface. Avoid overgrowth of cells at the time of transfection and keep the cell density at 60%-70% confluency.
A 60%-70% confluency of 293T cells at the time of transfection also ensures consistence and maximum transfection efficiency, thus reducing the variation between experiments.
293T cells are weakly adherent. During the cell adhesion assay and subsequent PFA fixation, antibody staining, and washing steps, care should be taken not to disrupt the 293T monolayer.
Commercially available, ready-to-use, pre-coated chamber slides can also be used in this protocol. They are superior to the in-house gelatin-coated slides as they provide better cell attachment, which helps to maintain the intact cell monolayer during the assay.
Recipes
-
Blocking and staining buffer
1× DPBS
1% BSA
0.3% Triton X-100
To prepare 10 ml, add 30 µl Triton X-100 to 10 ml 1× DPBS and mix well by vortexing. Add 0.1 g BSA and gently vortex to dissolve BSA completely.
-
2% PFA solution in complete DMEM
Mix 5 ml of 4% PFA solution with 5 ml of complete DMEM
Acknowledgments
This work is supported by grants from National Cancer Centre, Korea (NCC-1911261 and NCC-1910050). We acknowledge Mi Ae Kim of National Cancer Centre’s Microscopy Core Facility for technical support. This protocol is adapted from previously published work (Singh and Choi, 2019).
Competing interests
The authors declare no competing financial interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Chen Q., Zhang X. H. and Massague J.(2011). Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell 20(4): 538-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen Y.(2012). Cell Adhesion Assay. Bio-protocol 2(5): e98. [Google Scholar]
- 3. Gumbiner B. M.(1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84(3): 345-357. [DOI] [PubMed] [Google Scholar]
- 4. Hayden M. S., Grosmaire L. S., Norris N. A., Gilliland L. K., Winberg G., Tritschler D., Tsu T. T., Linsley P. S., Mittler R. S., Senter P. D., Fell H. P. and Ledbetter J. A.(1996). Costimulation by CD28 sFv expressed on the tumor cell surface or as a soluble bispecific molecule targeted to the L6 carcinoma antigen. Tissue Antigens 48(4Pt 1): 242-254. [DOI] [PubMed] [Google Scholar]
- 5. Janiszewska M., Primi M. C. and Izard T.(2020). Cell adhesion in cancer: Beyond the migration of single cells. J Biol Chem 295(8): 2495-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang L., and Barclay A. N.(2009). New assay to detect low‐affinity interactions and characterization of leukocyte receptors for collagen including leukocyte‐associated Ig-like receptor-1(LAIR-1). Eur J Immunol 39(4): 1167-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kucik D. F. and Wu C.(2005). Cell-Adhesion Assays. In: Cell Migration. Guan J. L.(Ed.). Methods in Molecular Biology. Humana Press. 43-54. [DOI] [PubMed] [Google Scholar]
- 8. Singh R. and Choi B. K.(2019). Siglec1-expressing subcapsular sinus macrophages provide soil for melanoma lymph node metastasis. Elife 8: e48916. [DOI] [PMC free article] [PubMed] [Google Scholar]