Abstract
Bone strength is controlled by both bone mass, and the organization and quality of the bone material. The current standard method for measuring bone mass in mouse and rat studies is micro-computed tomography. This method typically uses a single threshold to identify bone material in the cortical and trabecular regions. However, this single threshold method obscures information about the mineral content of the bone material and depends on normal morphology to separately analyze cortical and trabecular structures. To extend this method to identify bone mass at multiple density levels, we have established a protocol for unbiased selection and application of multiple thresholds using a standard laboratory-based micro-computed tomography instrument. This non-invasive method can be applied to longitudinal studies and archived samples and provides additional information about bone structure and strength.
Keywords: Cortical bone, Micro-computed tomography, Bone structure, Bone development, Bone mass, Mineralization
Background
Micro-computed tomography is a well-established standardized method for measuring bone mass and structure in experimental mice and rats. Typically, the inner network of bone (trabecular) is analyzed separately from the thick outer shell of bone (cortical). Identifying trabecular and cortical regions is usually either manual or uses algorithm-based segmentation on the basis of morphology ( Buie et al., 2007 ; Bouxsein et al., 2010 ). A single threshold within each region is used to define bone: cortical bone is detected using a high threshold, and trabecular bone at a lower threshold ( Ansari et al., 2018 ; McGregor et al., 2019 ). Recently however, while analyzing a genetically altered mouse model with severely disrupted bone structure ( Dmp1Cre:Socs3f/f), it was not possible to delineate cortical and trabecular structures ( Cho et al., 2017 ; Walker et al., 2020 ), and standard segregation of bone into trabecular and cortical was not meaningful. Such segregation would also be challenging in other mouse models where cortical bone structure is abnormal, such as during ageing or in chronic kidney disease when it is porous ( Piemontese et al., 2017 ; Metzger et al., 2020 ), or in conditions with poorly formed cortex such as osteopetrosis ( Abe et al., 2000 ). Using a single threshold also reduces the amount of information that can be gained from micro-computed tomography, in which raw images show multiple levels of gray that are reduced to a single level for quantitation.
To overcome this problem and move away from the dichotomy of “trabecular” and “cortical” bone, we made use of multi-level Otsu thresholding to measure bone mass at multiple density levels. The Otsu method makes use of an algorithm that segments pixels from the raw images into different classes based on the gray level intensities within the image (Otsu, 1979). Initially, we applied this method to the metaphyseal region ( Walker et al., 2020 ) because it contains both immature and mature cortical bone, with the most mature bone at greatest distance from the growth plate (Rauch, 2012). Using this new method applied to a slice-by-slice analysis of the metaphyseal region enabled quantification of the process by which normal cortical bone matures over time, specifically, the replacement of a low density structure with a more robust higher-density structure ( Walker et al., 2020 ). It also revealed a higher proportion of low density cortical bone in the mature skeleton of Dmp1Cre:Socs3f/f mice ( Walker et al., 2020 ), which we previously reported to have impaired strength, even though the structure was normal when measured by a single-threshold method ( Cho et al., 2017 ). We have also used this method to identify deposition of new bone resulting in a change in bone shape due to pharmacologically-induced muscle hypertrophy (Chan et al., Submitted).
This unbiased method of measuring bone at multiple densities is non-destructive and can be used to measure changes in bone over time, since it can be used in micro-computed tomography scans of live animals ( Walker et al., 2020 ). The method may also be applied to the study of bone development ( Bortel et al., 2015 ), and to the analysis of human bone, particularly during ageing ( Zebaze et al., 2010 ). The method may also be helpful for providing new methods for age estimation in bioarchaeology and forensic anthropology ( Maggiano et al., 2015 ), and would provide valuable new information in study of other species, such as measuring bone mass at different density levels that have been observed with egg-laying ( Squire et al., 2017 ) and hibernation (McGee- Lawrence et al., 2009 ).
We describe here the use of this method as it is applied to the metaphyseal region of fixed murine femora which contains both immature and mature bone, since this is likely to be the most common application. The same approach can be used for smaller or different regions, and for scans of bones taken from mice, or other species, including longitudinal studies under anaesthetic. This method can also be applied to archived scans, in which case the protocol need only be followed from Procedure C onwards. This protocol assumes a basic familiarity with micro-computed tomography.
Materials and Reagents
-
Fixed or frozen murine bone samples (e.g., tibiae or femora) for analysis
Note: We use samples collected and immediately fixed overnight in 4% paraformaldehyde (PFA) in a phosphate buffered saline solution that have been stored in 70% ethanol. Others recommend using samples that have been freshly frozen and slowly defrosted and scanned in saline.
Calibration rod pairs, comprising epoxy resin with embedded fine calcium hydroxyapatite (CaHA) powder at concentrations of 0.25 and 0.75 g/cm3, diameters of 2mm (Bruker Skyscan, SKY016946)
Sterile screw tubes with standard caps (Scientific Specialities Inc., Sure seal 2240-00)
Non-sterile plain non-woven swabs (Multisorb, catalog number: 2902305-01)
70% ethanol
Equipment
Micro-computed tomography scanner (Bruker Skyscan 1276, or similar)
Software
NRecon (version 1.7.1.0), Bruker https://www.bruker.com
DataViewer (version1.5.4), Bruker https://www.bruker.com
CT Analyzer (version 1.17.7.2), Bruker https://www.bruker.com
Microsoft Excel or similar
GraphPad Prism (version 8.3.1), https://www.graphpad.com
Procedure
-
Scanning bone samples and calibration standards
Preparation of samples: We wrap fixed bone samples in cotton gauze soaked in 70% ethanol, sufficient gauze is used so that the samples sit firmly within the screw-cap tubes and do not move if inverted gently.
Scan bones according to your standard method. We scan murine bones at the following settings: 9 μm voxel resolution (or smaller), 0.25 mm aluminium filter, 55 kV voltage, 200 μA current, rotation 0.4°, frame averaging = 2.
At the same time, and using the same scanning settings, scan both calibration rods.
-
Reconstruct images using your standard protocol. You may find it helpful to refer to method notes from your manufacturer (e.g., For Bruker instruments, “An overview of NRecon: reconstructing the best images from your microCT scan Method note MCT-062”, available when you register at https://www.brukersupport.com/). We reconstruct with SkyScan software NRecon using the following settings:
Smoothing – 1
Object larger than field of view – deselected
Ring artefact reduction – 6
Beam hardening compensation – 35%
CS rotation – automatic
The attenuation coefficient (density) – 0-0.14
-
Define the region of interest for measuring bone structure
Note: These instructions describe the process for defining the femoral metaphyseal region.
Open sample files in DataViewer.
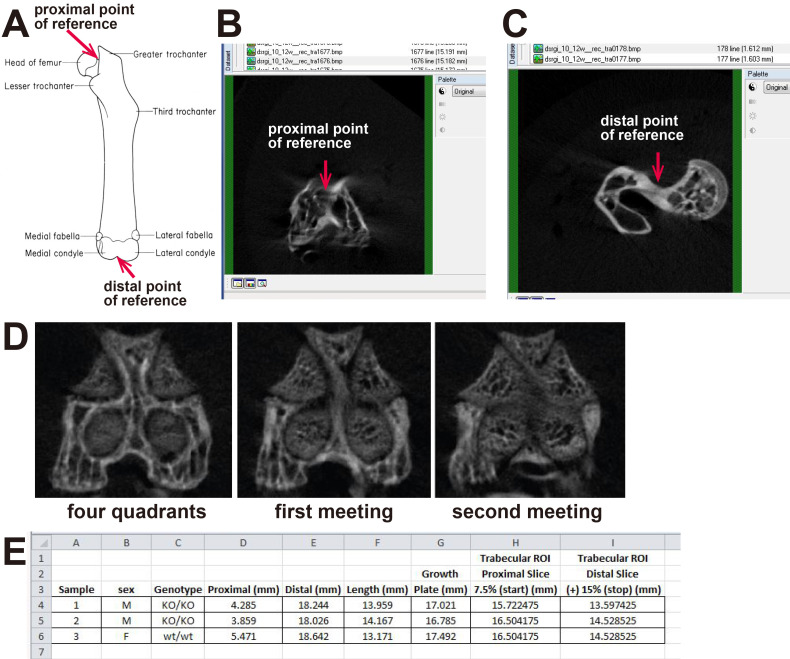
Orientate the sample in all three planes so that it is vertical and symmetrically aligned (Figure 1). Using the coronal plane, manipulate the bone orientation with the mouse while holding ‘Ctrl’. By directing the mouse to the center of the growth plate, the second transverse view can be manipulated so that the bone is orientated with the patella at the top of the image (see Figure 1). Use the sagittal view to align the bone horizontally.
Save a new (transaxial) dataset with the corrected orientation.
Open the new correctly oriented dataset in CTan.
Measure femur length by identifying the distal and proximal ends of the bone, and the slice containing the growth plate in CTan (Figure 2A). We define the distal end of the bone as the optical slice at which the medial and lateral condyle meet (Figure 2B). We define the proximal end of the bone as the slice where the femoral head and the trochanter first meet (Figure 2C). In the distal femur, we identify the growth plate by identifying the site where the four quadrants of the femoral growth plate begin to merge. As you scroll from the distal end towards the center of the bone, you will notice the presence of four circular quadrants of the growth plate, that enlarge and meet (Figure 2D). The medial and lateral growth plate regions meet first. We call this the first meeting. As you continue to scroll, the anterior and posterior growth plate regions meet. We call this convergence of the growth plate “the second meeting” and define it as the “growth plate” slice. Use the measurement values for each landmark to calculate bone length by subtracting the distal measurement from the proximal measurement (Figure 2E).
Calculate the length of the metaphyseal region of interest (ROI) for each sample based on the bone length. We do this so that the same anatomical region is measured in each sample, regardless of whether bone growth is modified. In the original publication using this method, we defined the distal end of the region of interest as the slice at a distance equal to 7.5% of the femur length from the growth plate (see Figure 2A ( Walker et al., 2020 ) for a representation of this region of interest); this normally encompasses the secondary spongiosa. We define the proximal end of the metaphyseal region as the slice located an additional 15% of the femur length from the newly defined top measurement. Calculate these values in Microsoft Excel using the landmarks identified in Step 6, above (Figure 2E).
Select the ROI icon to enable a “Region of Interest” drop down menu. From here, select an “elliptical” region that encompasses both trabecular and cortical bone within the top slice. Repeat this for the bottom slice of the ROI. The CTan software will interpolate the ROI for the slices in-between. Manually check the ROI generated on each slice by scrolling through the dataset to ensure that the entirety of the bone is contained within the whole ROI. Save this as a new metaphyseal dataset.
-
Define four bone density thresholds using the Otsu algorithm in control samples
Note: This should be performed separately for each study, unless you aim to compare volumes of bone at defined densities between studies. In the latter case, ensure that all scanning and reconstruction settings are the same.
-
Using the “Custom Processing” function, create a task list with the following steps:
Thresholding: Multilevel (Otsu method), With a 3D space, Number of threshold levels: 4, Select “Inside VOI”, Output color of class thresholding: Mean intensity in class.
Save Bitmaps: Select the following options: Image inside ROI, BMP format, Copy shadow projection, copy dataset log file. This will save bitmaps that you can use to check the Otsu-based thresholds if any unexpected results are obtained.
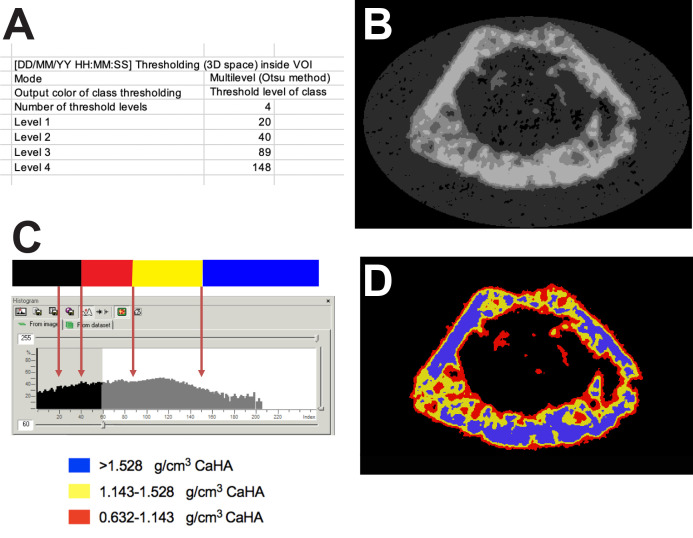
Load all samples from your control group, and run the above custom processing using CTAn’s batch manager (BATman). This will generate an output file for each sample that reports 4 Otsu-based thresholds (Figure 3A).
Check the BMP files to ensure they correctly represent the amount of bone visualized. You will notice that the images for the lowest threshold will include much non-bone material. This lowest threshold will not be used for analysis of your samples (Figure 3B).
Collate the threshold values derived for all the samples in a excel worksheet.
Calculate the average threshold values for each level. These newly obtained threshold values will be used to complete the 3-level thresholding for all samples.
-
-
Use the results from scanning the calibration standards to calculate the equivalent calcium hydroxyapatite levels (g/cm3) represented by your thresholds (Figures 3C, 3D)
Note: These are required for reporting, so your levels can be reproduced and understood by other researchers who may be using different instruments (reporting the 0-255 gray levels will not achieve this).
Follow the protocol in “MN009 BMD calibration in CTan ‘HU and BMD calibration in Bruker-MicroCT CT-analyser’ September 2005 page 5-19” (found at https://www.brukersupport.com/) as follows.
In CTAn, under the Binary Selection tab, select the “From dataset” tab. Type in the gray level threshold for Level 2 (in our example is 40). Select the “Bone mineral density” tab in this window and scroll down to find the first BMD value that is not greyed out. In our example, the threshold level 40 corresponds to a mineral density of 0.632 g/cm3 CaHA.
Repeat this process for threshold Levels 3 and 4.
Note that these values are not universal and will differ if the scanning parameters are changed. Therefore, it is important to scan the phantoms under the same settings when you scan your samples and calibrate CTAn for each analysis.
-
Measure bone mass in all samples at all three thresholds
In CTAn’s Batch manager (BATman) function, load all sample BMP files and ROIs from the datasets defined in Procedure B; for each sample, this is a single dataset including both trabecular and cortical bone.
-
Make a new Custom Processing task list to define the bone area, and the volume of bone at each threshold as follows:
Thresholding: Select the following: Global, and add the Level 2 threshold value calculated in Procedure C. In our example it is 40. Leave the maximum at 255.
ROI Shrink Wrap: Select the following: 2D space, stretch over holes 20 pixels; this creates an ROI that includes only the region within the periosteum, including cortical and trabecular bone, that is to be measured.
Reload image.
Thresholding for low density bone: Select the following: Global, and add the Level 2 threshold value calculated in Procedure C. In our example it is 40. Set the maximum (minus 1) at the Level 3 threshold. In our example it is 88.
2D Analysis: Select “All results”, deselect “Append summary results to file”. This will generate Bone Area measurements for each slice included in the dataset, within the low density threshold (Level 2 to Level 3) and will automatically place them into a file called ‘[sample name].batman.csv’. Note that the value for Total Area will be identical in all analysis files generated.
3D Analysis: Create a .csv file for the results of the low density threshold (Level 2 to Level 3) bone analysis. Save results as both text table and single text line and link the Excel file you just created. This will list the volume of bone (Bone Volume) within the Level 2 and Level 3 thresholds (low density), for all samples. Note that the value for Total Area will be identical in all analysis files generated.
Save Bitmaps: Apply ROI, Select the following: BMP file format, Copy Shadow Projection and Copy Dataset log file. You can customize the name of the subfolder where the low-density BMP files are to be saved.
Reload image.
Thresholding for mid density bone: Global, and add the Level 3 threshold value calculated in Procedure C. In our example it is 89. Set the maximum (minus 1) at the Level 4 threshold. In our example it is 147.
2D Analysis: Select “All results”, deselect “Append summary results to file”. This will generate Bone Area measurements for each slice included in the dataset, within the mid density threshold (Level 3 to Level 4) and will automatically place them into the file called ‘[sample name].batman.csv’.
3D Analysis: Create a .csv file for the results of the mid density threshold (Level 2 to Level 3) bone analysis. Save results as both text table and single text line and link the Excel file you just created. This will list the volume of bone (Bone Volume) within the Level 3 and Level 4 thresholds (mid density), for all samples.
Save Bitmaps: Apply ROI, Select the following: BMP file format, Copy Shadow Projection and Copy Dataset log file. You can customize the name of the subfolder where the mid density BMP files are to be saved.
Reload image.
Thresholding for high density bone: Select the following: Global, and add the Level 4 threshold value calculated in Procedure C. In our example it is 148. Set the maximum at 255.
2D Analysis: Select “All results”, deselect “Append summary results to file”. This will generate Bone Area measurements for each slice included in the dataset, within the high density threshold (Level 4 to maximum) and will automatically place them into the file called ‘[sample name].batman.csv’.
3D Analysis: Create a .csv file for the results of the high-density threshold (Level 4 to maximum) bone analysis. Save results as both text table and single text line and link the Excel file you just created. This will list the volume of bone (Bone Volume) from Level 4 to the maximum threshold (high density) for all samples.
Save Bitmaps: Apply ROI, Select BMP file format, Copy Shadow Projection and Copy Dataset log file. You can customize the name of the subfolder where the high BMP files are to be saved.
Run the above custom processing using BATman batch processing. This will take some hours, depending on the number of samples being assessed.
Figure 1. DataViewer screen shot showing a femur oriented correctly in all three planes.
Note that each axis (blue and green) lies central in each orientation, and the growth plate is balanced evenly on the red axis.
Figure 2. Measuring femur length and defining a metaphyseal region of interest in CTan.
A. Diagram showing the anatomical locations we use to measure femoral bone length. B. Typical image showing the top slice of a femoral metaphyseal region of interest. C. Typical image showing bottom slice of a femoral metaphyseal region of interest. D. Typical images showing how the regions of the growth plate change as you scroll from the distal end (four quadrants) to the first and second meeting points of the growth plate. E. Example of a spreadsheet used to calculate femur length and the region of interest from micro-CT scans. Image in panel A is taken from http://www.informatics.jax.org/cookbook/figures/figure42.shtml.
Figure 3. An example of the four thresholds applied to a femoral region of interest.
A. Screenshot of the 4 levels derived from the automated Otsu thresholding method from a single sample. B. BMP file showing the four levels applied to a slice through the femur. C. A histogram of pixel intensities for a metaphyseal region of interest with arrows to show the Otsu levels defined in panel A, showing which level is discarded, and the calibrated density values that correspond to those levels. D. A pseudo-color image of panel B showing the three bone density levels measured.
Data analysis
Procedure E above will generate a summary of the 3D measurements for each sample, at each density threshold. The values for low, mid and high-density Bone Volume will be in their respective separate comma-separated value (CSV) files that you created. Another CSV file will be automatically generated (named ‘[sample name].batman.csv’ when BATman was used to analyze a batch of samples, or ‘[sample name].ctan.csv’ if you have analyzed a single sample). This can be found within the sample folder. This file contains a summary of the 2D measurements, followed by individual measurements for each image slice at each threshold. They appear in the order they are listed in the BATman task list (i.e., low density, then mid-density, then high density). You will see them as you scroll down.
-
Analyzing average Bone Volume/Total Volume (BV/TV) at three levels of bone density for the entire region of interest.
In Excel, open the three CSV files that you created for each density level. These files contain a summary of 3D values for each sample, in a common file based on each density threshold (low, mid or high).
Copy and paste the values for Bone Volume/Total Volume (BV/TV) at each density level. These values describe individual volumes of low, mid and high-density bone for each sample, as a percentage of the total bone volume (i.e., sum of low, mid and high density bone) (see Walker et al., 2020 Figure 2B for an example). You can also extract the bone volume data (BV), and present absolute bone volumes at each density.
-
Analyzing and graphing Bone Area/Total Area (B.Ar/T.Ar) at three levels of bone density for each slice within the region of interest.
For each sample, create a new Excel (.xls or .xlsv) file to collate your results. In Excel, open the ‘[sample name].batman.csv’ file that contains the summarized and individual measurements from the 2D analysis. Copy and paste the values for Bone Area Bone Area / Total Area (B.Ar/T.Ar) for each threshold level.
You may want to visualize the way in which B.Ar/T.Ar values change across the entire dataset (i.e., length of the bone). To do this, you first need to associate each 2D slice with its longitudinal position. For example, using Sample 1 from Figure 2E, the first slice corresponds to 7.5% of bone length, which is 1.95 mm from the growth plate, and the last slice ends at 3.14 mm from the growth plate. Type these values in the cell adjacent to the corresponding B.Ar/T.Ar value. Highlight this column, beginning and ending at the first and last values, and use the ‘Fill’ function in Excel to populate the empty cells with incremental values. See source data file for Figure 3 in Walker et al., 2020 for an example. Ensure that these values correctly correspond to the longitudinal positions as calculated above.
-
Use Excel or GraphPad Prism to plot three line graphs visualizing the percentage of B.Ar/T.Ar of low, mid and high density bone along the bone length. Bone length or distance from the growth plate should be placed on the X axis, with B.Ar/T.Ar (%) (or Bone/Cross sectional area (%)) on the Y axis (see Figure 3, supplement 2 of ( Walker et al., 2020 ) and Figure 2C-E (Chan et al., Submitted) for examples). You can use stacked graphs to show all densities on the same axis (see Figure 3B-E ( Walker et al., 2020 ) for an example).
Statistical analysis
Prior to any statistical analysis, it is important to graph individual values for each sample to ensure there has not been any error made along the way. You should see an increase in the proportion of high-density bone with increasing distance from the growth plate, and a decrease in the proportion of low density bone along the same distance. If major outliers are observed, check back on the BMP files generated during the data processing to visualize the threshold regions analyzed and identify any errors.
To determine whether there is an overall difference in the volume of bone at each density level throughout the entire region, we compare overall Bone Volume/Total Volume for the entire region by a one- or two-way ANOVA, depending on the number of factors analyzed.
To determine whether there is a significant difference in the amount of bone at a single density threshold between two genotypes at each slice, we apply a two-way ANOVA using “genotype” and “distance from the growth plate” as categories.
Notes
The most common cause of variability is when samples have been scanned, or reconstructed, under differing conditions. It is vital to ensure this is the same for comparable results.
It is important to check that your scans are of high quality and that your reconstructions are conducted properly – for example, any ring artefact or flaring will interfere with this analysis method.
Acknowledgments
This work was supported by NHMRC Grants 1120978 and 1058625 to NAS. NAS was supported by an NHMRC Senior Research Fellowship and by the SVI Brenda Shanahan Fellowship. St Vincent’s Institute acknowledges the support of the Victorian State Government OIS program.
Competing interests
The authors have no financial or non-financial competing interests to declare.
Ethics
All samples used for developing this method were from studies approved by the St. Vincent’s Health Melbourne Animal Ethics Committee.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Abe S., Watanabe H., Hirayama A., Shibuya E., Hashimoto M. and Ide Y.(2000). Morphological study of the femur in osteopetrotic(op/op) mice using microcomputed tomography. Br J Radiol 73(874): 1078-1082. [DOI] [PubMed] [Google Scholar]
- 2. Ansari N., Ho P. W., Crimeen-Irwin B., Poulton I. J., Brunt A. R., Forwood M. R., Divieti Pajevic P., Gooi J. H., Martin T. J. and Sims N. A.(2018). Autocrine and Paracrine Regulation of the Murine Skeleton by Osteocyte-Derived Parathyroid Hormone-Related Protein. J Bone Miner Res 33(1): 137-153. [DOI] [PubMed] [Google Scholar]
- 3. Bortel E. L., Duda G. N., Mundlos S., Willie B. M., Fratzl P. and Zaslansky P.(2015). Long bone maturation is driven by pore closing: A quantitative tomography investigation of structural formation in young C57BL/6 mice. Acta Biomater 22: 92-102. [DOI] [PubMed] [Google Scholar]
- 4. Bouxsein M. L., Boyd S. K., Christiansen B. A., Guldberg R. E., Jepsen K. J. and Muller R.(2010). Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 25(7): 1468-1486. [DOI] [PubMed] [Google Scholar]
- 5. Buie H. R., Campbell G. M., Klinck R. J., MacNeil J. A. and Boyd S. K.(2007). Automatic segmentation of cortical and trabecular compartments based on a dual threshold technique for in vivo micro-CT bone analysis. Bone 41(4): 505-515. [DOI] [PubMed] [Google Scholar]
- 6. Chan, A. S. M., McGregor, N. E., Poulton, I. J., Hardee, J. P., Cho, E. H. J., Martin, T. J., Gregorevic, P., Sims, N. A. and Lynch, G. S. Bone geometry is altered by follistatin-induced muscle growth in adult male mice. J Bone Miner Res. Submitted. [DOI] [PMC free article] [PubMed]
- 7. Cho D. C., Brennan H. J., Johnson R. W., Poulton I. J., Gooi J. H., Tonkin B. A., McGregor N. E., Walker E. C., Handelsman D. J., Martin T. J. and Sims N. A.(2017). Bone corticalization requires local SOCS3 activity and is promoted by androgen action via interleukin-6. Nat Commun 8(1): 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maggiano I. S., Maggiano C. M., Tiesler V. G., Chi-Keb J. R. and Stout S. D.(2015). Drifting Diaphyses: Asymmetry in Diametric Growth and Adaptation Along the Humeral and Femoral Length. Anat Rec(Hoboken) 298(10): 1689-1699. [DOI] [PubMed] [Google Scholar]
- 9. McGee-Lawrence M. E., Wojda S. J., Barlow L. N., Drummer T. D., Castillo A. B., Kennedy O., Condon K. W., Auger J., Black H. L., Nelson O. L., Robbins C. T. and Donahue S. W.(2009). Grizzly bears(Ursus arctos horribilis) and black bears(Ursus americanus) prevent trabecular bone loss during disuse(hibernation) . Bone 45(6): 1186-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McGregor N. E., Murat M., Elango J., Poulton I. J., Walker E. C., Crimeen-Irwin B., Ho P. W. M., Gooi J. H., Martin T. J. and Sims N. A.(2019). IL-6 exhibits both cis- and trans-signaling in osteocytes and osteoblasts, but only trans-signaling promotes bone formation and osteoclastogenesis. J Biol Chem 294(19): 7850-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Metzger C. E., Swallow E. A. and Allen M. R.(2020). Elevations in Cortical Porosity Occur Prior to Significant Rise in Serum Parathyroid Hormone in Young Female Mice with Adenine-Induced CKD. Calcif Tissue Int 106(4): 392-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Otsu N.(1979). A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 9: 62-66. [Google Scholar]
- 13. Piemontese M., Almeida M., Robling A. G., Kim H. N., Xiong J., Thostenson J. D., Weinstein R. S., Manolagas S. C., O'Brien C. A. and Jilka R. L.(2017). Old age causes de novo intracortical bone remodeling and porosity in mice. JCI Insight 2(17): e93771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rauch F.(2012). The dynamics of bone structure development during pubertal growth. J Musculoskelet Neuronal Interact 12(1): 1-6. [PubMed] [Google Scholar]
- 15. Squire M. E., Veglia M. K., Drucker K. A., Brazeal K. R., Hahn T. P. and Watts H. E.(2017). Estrogen levels influence medullary bone quantity and density in female house finches and pine siskins. Gen Comp Endocrinol 246: 249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walker E. C., Truong K., McGregor N. E., Poulton I. J., Isojima T., Gooi J. H., Martin T. J. and Sims N. A.(2020). Cortical bone maturation in mice requires SOCS3 suppression of gp130/STAT3 signalling in osteocytes. Elife 9: e56666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zebaze R. M., Ghasem-Zadeh A., Bohte A., Iuliano-Burns S., Mirams M., Price R. I., Mackie E. J. and Seeman E.(2010). Intracortical remodelling and porosity in the distal radius and post-mortem femurs of women: a cross-sectional study. Lancet 375(9727): 1729-1736. [DOI] [PubMed] [Google Scholar]