FUS-dependent liquid–liquid phase separation is a critical process in the early activation of the DNA damage response and in the recruitment of key proteins, which facilitates the proper assembly of double-strand break repair complexes.
Abstract
RNA-binding proteins (RBPs) are emerging as important effectors of the cellular DNA damage response (DDR). The RBP FUS is implicated in RNA metabolism and DNA repair, and it undergoes reversible liquid–liquid phase separation (LLPS) in vitro. Here, we demonstrate that FUS-dependent LLPS is necessary for the initiation of the DDR. Using laser microirradiation in FUS-knockout cells, we show that FUS is required for the recruitment to DNA damage sites of the DDR factors KU80, NBS1, and 53BP1 and of SFPQ, another RBP implicated in the DDR. The relocation of KU80, NBS1, and SFPQ is similarly impaired by LLPS inhibitors, or LLPS-deficient FUS variants. We also show that LLPS is necessary for efficient γH2AX foci formation. Finally, using superresolution structured illumination microscopy, we demonstrate that the absence of FUS impairs the proper arrangement of γH2AX nanofoci into higher-order clusters. These findings demonstrate the early requirement for FUS-dependent LLPS in the activation of the DDR and the proper assembly of DSB repair complexes.
Introduction
Unrepaired DNA damage can lead to genome instability, a hallmark of cancer cells. To counteract DNA damage, cells have evolved a complex cellular response, commonly referred to as DNA damage response (DDR). The early DDR events have been best elucidated at sites of DNA double-strand breaks (DSBs), which are the most dangerous type of DNA lesions. After the occurrence of a DSB, the sensor protein kinases DNA-dependent protein kinase (DNA-PK), ataxia telangiectasia mutated (ATM), and ATM and Rad3-related (ATR) are rapidly activated, and the KU70/KU80 (XRCC6/XRCC5) heterodimer is recruited to the broken DNA ends. Phosphorylation of the histone variant H2AX at S139 (known as γH2AX) by ATM serves as an early mark of DNA damage and as a platform for the recruitment of early DDR factors such as MDC1 and the MRN complex (consisting of MRE11, RAD50, and NBS1; Stucki and Jackson, 2006). In mammalian cells, the accumulation of DDR factors at sites of DNA damage gives rise to subnuclear foci that can be readily visualized microscopically. Using superresolution microscopy, we and others have recently shown that these foci correspond to clusters of nanodomains, and this clustering is required for an efficient DNA damage repair (Lopez Perez et al., 2016; Natale et al., 2017). In mammals, DSBs are eventually repaired via two main pathways, depending on the cell cycle phase: homologous recombination (HR), which repairs DSBs in late S and G2 phases, and nonhomologous end joining (NHEJ), which is active throughout the entire cell cycle.
In addition to canonical DDR factors, large-scale proteomic and genomic studies have identified several RNA-binding proteins (RBPs) as potential novel DDR factors, either as targets of the apical DDR kinases (Matsuoka et al., 2007) or as proteins that, when absent, lead to the activation of the DDR (Paulsen et al., 2009). RBPs have been shown to contribute both directly and indirectly to genome stability. For example, the loss of pre-mRNA splicing or mRNA export factors can favor the accumulation of RNA:DNA hybrids (R-loops) that can be processed to DSBs (Aguirre et al., 2005; Chuang et al., 2019). In addition, an increasing number of studies have shown that several RBPs are recruited to sites of DNA damage and participate in DSB repair (Mikolaskova et al., 2018).
The multifunctional DNA/RNA-binding protein fused in sarcoma (FUS) is involved in splicing, translation, and mRNA transport (Dormann and Haass, 2013). Both in vivo and in vitro observations point toward a role for FUS in maintaining genome stability. Mice lacking FUS are hypersensitive to ionizing radiation and show defects in spermatogenesis and chromosomal instability (Hicks et al., 2000; Kuroda et al., 2000). In vitro, FUS stimulates the formation of DNA loops between complementary DNA molecules, structures that correspond to one of the first steps in HR (Baechtold et al., 1999). In cells, FUS is recruited very early to sites of DNA damage (Aleksandrov et al., 2018; Mastrocola et al., 2013), and its silencing leads to an impairment of DSB repair by both HR and NHEJ (Mastrocola et al., 2013; Michelini et al., 2017). In addition, FUS is an ATM and DNA-PK substrate (Deng et al., 2014b; Gardiner et al., 2008).
The N-terminal region of FUS (residues 1–165) is a highly conserved low-complexity domain (LCD) composed primarily of glutamine, glycine, serine, and tyrosine (QGSY-rich). This domain mediates protein–protein interactions and drives the aggregation of FUS into protein inclusions (Sun et al., 2011). Several studies have shown that the LCD of FUS undergoes a reversible dynamic phase transition between a disperse state, liquid droplets, and hydrogels (Kato et al., 2012; Murakami et al., 2015; Patel et al., 2015). FUS liquid–liquid phase separation (LLPS) occurs both in vivo and in vitro at physiological concentrations (Burke et al., 2015; Murakami et al., 2015).
It is increasingly recognized that LLPS provides a molecular basis for the formation of subcellular membraneless organelles such as Cajal bodies, paraspeckles, and stress granules (SGs; Boeynaems et al., 2018). Paraspeckles are subnuclear compartments that assemble on the long noncoding RNA (lncRNA) NEAT1, which induces phase separation of four core RBPs: splicing factor proline- and glutamine-rich (SFPQ), non-POU domain-containing octamer-binding (NONO), FUS, and RBM14 (Hirose et al., 2019), all of which contain LCDs of variable length (Harrison and Shorter, 2017). The LCDs of FUS and RBM14 are required for in vitro phase separation and in vivo paraspeckle formation (Hennig et al., 2015). SFPQ, its paralog NONO, and RBM14 are also RBPs implicated in DNA repair (West et al., 2016). SFPQ silencing was reported to sensitize cells to DNA cross-linking and alkylating agents and to reduce DSB repair by HR (Rajesh et al., 2011). SFPQ has DNA reannealing and strand-invasion activity that may lead to the formation of DNA loop structures (Akhmedov and Lopez, 2000). In addition, SFPQ, NONO, and RBM14 promote NHEJ (Bladen et al., 2005; Jaafar et al., 2017; Marchesini et al., 2017).
Despite all the observations implicating FUS and SFPQ in DNA damage repair, their precise molecular function remains to be fully elucidated. Here we show that FUS is required for the recruitment of SFPQ and the retention of KU80 on DSBs. Consistent with recent results indicating a role for LLPS in DNA damage repair (Kilic et al., 2019; Pessina et al., 2019), we show that LLPS inhibitors impair the formation of γH2AX and 53BP1 foci and the proper recruitment of FUS, SFPQ, KU80, and NBS1. Moreover, LLPS-deficient variants of FUS affect accumulation of SFPQ and KU80 at sites of laser-induced DNA damage. Finally, we demonstrate that FUS is needed for the higher-order clustering of γH2AX chromatin nanodomains, which is required for efficient DNA damage repair (Natale et al., 2017).
Results
FUS-KO sensitizes human cells to DNA damage
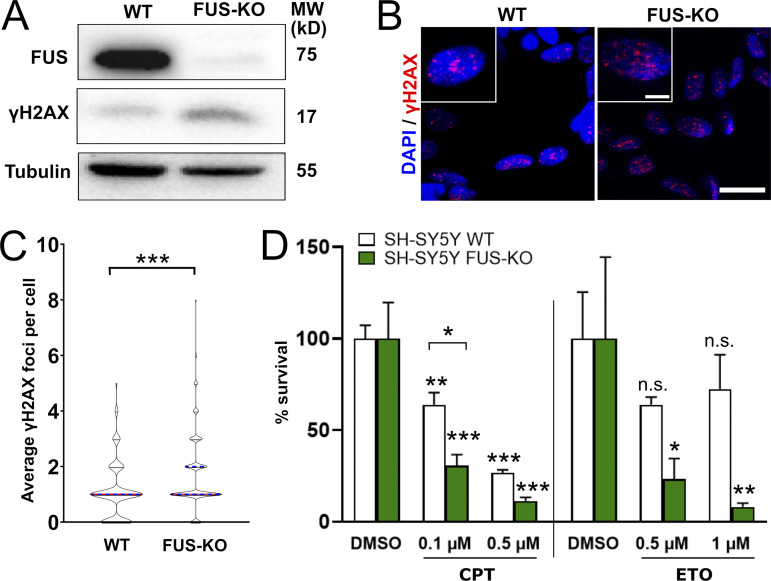
To shed light on the role of FUS in DDR and genome stability, we generated human HeLa and SH-SY5Y cells in which the FUS gene was knocked out by CRISPR-trap genome editing technique (Reber et al., 2018). Western blot analysis revealed that HeLa and SH-SY5Y FUS-KO cells display an 8.1-fold and 2.6-fold increase in the phosphorylation of H2AX (γH2AX), respectively (Fig. 1 A, and Fig. S1 A, respectively). Consistently, immunofluorescence analyses showed increased formation of γH2AX foci in FUS-KO cells (Fig. 1, B and C; and Fig. S1, B and C). These observations suggest that, in the absence of FUS, cells generate more DNA damage, repair DNA damage less efficiently, or both. Indeed, transient transfection of HeLa FUS-KO cells with exogenous Flag-tagged FUS reduced the number of γH2AX foci by 37% (Fig. 1 D).
Figure 1.
Loss of FUS results in accumulation of DNA damage and sensitization to genotoxic insult in HeLa cells. (A) Total extracts of WT and FUS-KO HeLa cells were analyzed by Western blotting with anti-FUS and anti-γH2AX antibodies (loading control: Tubulin). FUS-KO cells display an 8.1-fold increase in the level of endogenous γH2AX in comparison to WT cells. (B) Representative confocal micrographs of γH2AX foci in WT and FUS-KO HeLa cells. Scale bar: 20 µm. Cropped single cells are enlarged 2× (scale bar: 5 µm). (C) Quantification of γH2AX foci. The number of foci per nucleus was counted using ImageJ. WT cells have an average of 1.02 foci per cell, compared with 1.82 in FUS-KO cells. Data are from two biological replicates (170 cells each). Statistics: Student’s t test. (D) HeLa FUS-KO cells were transiently transfected with a plasmid expressing FUS-Flag and stained with anti-Flag and anti-γH2AX. Foci were quantified by ImageJ. Data are from two biological replicates (65 cells each; only transfected cells were included). Statistics: Student’s t test. (E) HeLa WT and FUS-KO cell viability assessed by Trypan blue staining upon treatment with CPT (0.1 or 0.5 µM) or ETO (0.5 or 1 µM). Percentage survival was calculated by normalizing the number of surviving cells by their respective DMSO group. Statistics: two-way ANOVA, Bonferroni post hoc test. In all panels: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S1.
Loss of FUS results in accumulation of DNA damage and sensitization to genotoxic insult in SH-SY5Y cells. (A) Total extracts of WT and FUS-KO SH-SY5Y cells were analyzed by Western blotting with anti-FUS and anti-γH2AX antibodies. FUS-KO cells display a 2.6-fold increase in the level of endogenous γH2AX in comparison to WT cells. Tubulin was used as loading control. MW, molecular weight. (B) Representative confocal micrographs of γH2AX foci in WT and FUS-KO SH-SY5Y cells. Scale bar: 20 µm. Cropped single cells are enlarged 2× (scale bar: 5 µm). (C) Quantification of B. The number of foci per nucleus was counted using ImageJ and plotted as a violin plot. Data from two biological replicates, with 170 cells per replicate. The average foci number in WT cells is 1.01, compared with 1.71 in FUS-KO cells. Statistics: Student’s t test (***, P < 0.001). (D) SH.SY5Y WT and FUS-KO cell viability assessed by Trypan blue staining upon treatment with increasing concentrations of CPT (0.1 or 0.5 µM) or ETO (0.5 or 1 µM). Statistics: two-way ANOVA followed by Bonferroni post hoc test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
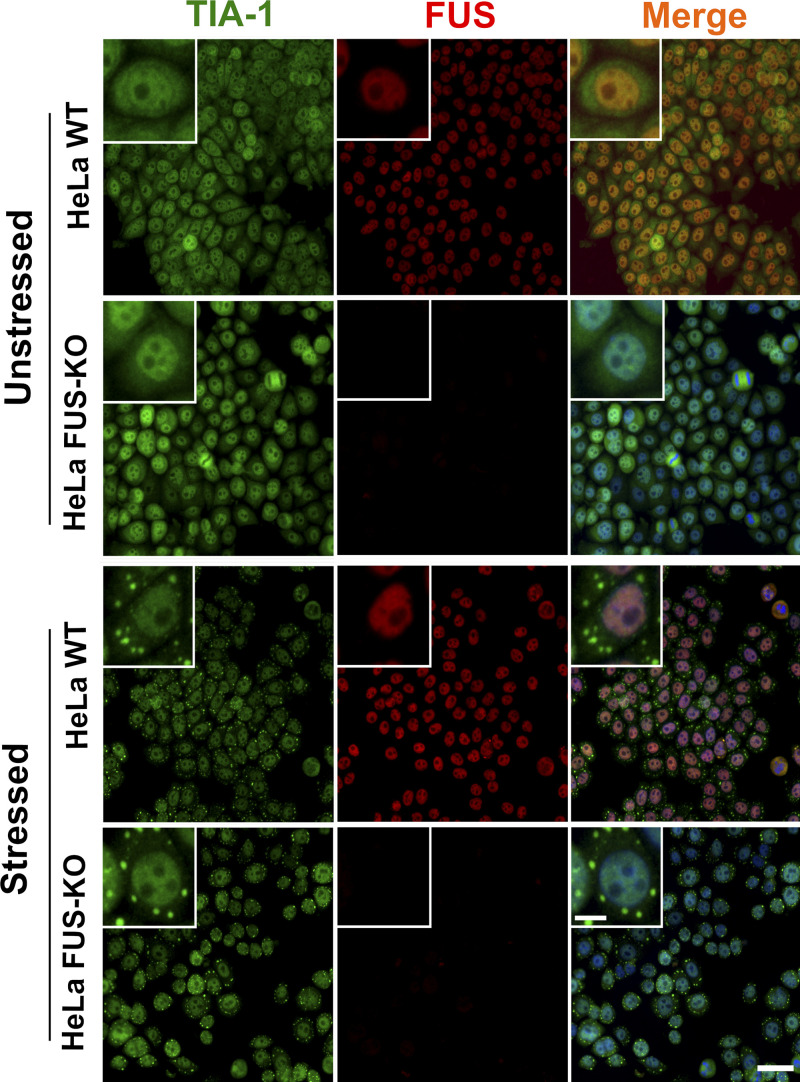
To exclude that increased DDR activation in FUS-KO cells could result from alternative splicing alterations in DNA damage genes, we reanalyzed the RNA sequencing dataset of SH-SY5Y FUS-KO cells (Reber et al., 2016) using gene set enrichment analysis (Mootha et al., 2003; Subramanian et al., 2005) and concluded that the KO of FUS did not affect alternative splicing of DNA damage genes (data not shown). We also verified that the KO of FUS did not affect the assembly of cytoplasmic membraneless organelles by assessing SG formation upon treatment with an oxidative stress inducer, sodium arsenite (Fig. S2).
Figure S2.
Loss of FUS does not affect SG assembly. HeLa WT and FUS-KO cells were stressed by treatment with 0.5 mM sodium arsenite for 1 h and immunostained for FUS and TIA-1 (a eukaryotic SG marker). Consistent with previous observations (Sama et al., 2013; Hock et al., 2018), FUS remained nuclear in WT cells upon arsenite incubation, and SGs could be detected in the cytoplasm of both WT and FUS-KO cells. Scale bar: 50 µm (10 µm for cropped image).
Next, we investigated whether the KO of FUS results in DNA damage sensitization. WT and FUS-KO HeLa and SH-SY5Y cells were compared for their sensitivity to DNA damage induced by camptothecin (CPT) and by etoposide (ETO), inhibitors of topoisomerase I and II, respectively. The viability was assessed by Trypan blue staining. FUS-KO cells showed a reduction in cell viability, similar to what had been reported in FUS knockdown lines (Ward et al., 2014). Upon exposure to either CPT or ETO, both HeLa FUS-KO and SH-SY5Y FUS-KO cells showed a significantly stronger reduction in viability relative to their respective WT cells (Fig. 1 E, and Fig. S1 D, respectively). Overall, these observations indicate that FUS is required for the maintenance of genome integrity.
FUS KO affects ATM-dependent signaling and recruitment of DDR factors
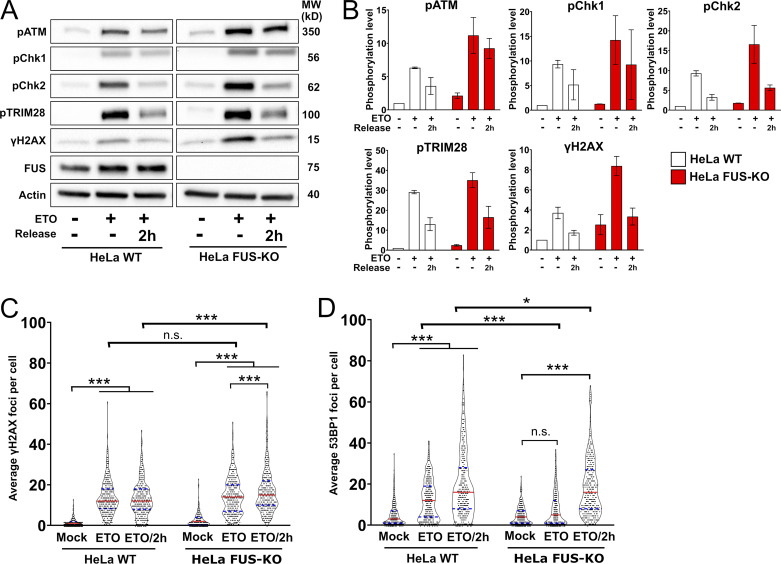
Next, we asked whether FUS could be directly involved in DDR signaling. HeLa WT and FUS-KO cells were treated with ETO for 1 h and allowed to recover in ETO-free medium for 2 h (ETO release). Western blot analysis showed that DMSO-treated FUS-KO cells display a basal activation of the ATM-dependent DDR signaling, in addition to increased phosphorylation of H2AX (Fig. 2, A and B). This observation strengthens the idea that, in the absence of FUS, the DDR is chronically activated, and is consistent with previous reports that showed increased γH2AX immunoreactivity in neurons expressing amyotrophic lateral sclerosis–linked FUS mutants (Wang et al., 2013). Upon incubation of cells with ETO, we observed increased phosphorylation of ATM, CHK1, CHK2, and TRIM28, in addition to increased levels of γH2AX in both cell lines (Fig. 2, A and B). We also assessed ATR and BRCA1, but we failed to detect consistent changes in their phosphorylation level (Fig. S3 A). In FUS-KO cells, the phosphorylation of ATM, CHK1, CHK2, and TRIM28 was slightly higher compared with WT cells, possibly reflecting preexisting DDR activation. Interestingly, 2 h after ETO release, while phosphorylation of most proteins started to reduce in WT cells, it remained higher in FUS-KO cells (Fig. 2, A and B). This indicates that the activation of the DDR, and in particular of the ATM pathway, occurred normally in the absence of FUS, but that DNA damage signaling persisted longer after release from the genotoxic treatment. These data reinforce the concept that cells lacking FUS generate more DNA damage than WT cells upon ETO treatment, repair the DNA damage less efficiently, or both.
Figure 2.
Loss of FUS perturbs DDR signaling and foci formation upon genotoxic insult. (A) Representative Western blot of DDR proteins in HeLa WT and FUS-KO cells upon ETO treatment. Cells were treated with 10 µM ETO for 1 h and were allowed to recover in ETO-free medium for 2 h (ETO release). Cells were collected at the indicated time points, lysed in the presence of phosphatase inhibitors, separated on a gradient SDS-PAGE, and processed for Western blotting (loading control: Actin). MW, molecular weight. (B) Quantification of the blots from two independent experiments as in A (n = 2). (C) Quantification of ETO-induced γH2AX foci. Data are from two biological replicates (170 cells each). Statistics: two-way ANOVA, Bonferroni post hoc test. (D) Quantification of ETO-induced 53BP1 foci analyzed as in C. *, P < 0.05; ***, P < 0.001.
Figure S3.
FUS is required for efficient DSB repair and DNA damage foci formation upon genotoxic insult. (A) DDR activation in HeLa WT and FUS-KO cells upon ETO treatment. Cells were treated with 10 µM ETO for 1 h and were allowed to recover in ETO-free medium for 2 h (ETO release). Cells were collected at the indicated time points, lysed in the presence of phosphatase inhibitors, separated on a gradient SDS-PAGE, and processed for Western blotting (loading control: Actin). ATR and BRCA1 were probed on the same blot as the proteins shown in Fig. 2 B. MW, molecular weight. (B) DSB repair efficiency was quantified in U2OS cells containing a stably integrated NHEJ reporter system. Cells were silenced for FUS, KU80, or both. Right: Western blot demonstrating the silencing of the respective proteins. Data are presented as the mean ± SEM (experiments done in triplicate, with at least 10,000 cells analyzed per experiment). Statistics: one-way ANOVA, followed by Bonferroni post hoc test. *, P < 0.05. (C) DSB repair efficiency was quantified in U2OS cells containing a stably integrated HR reporter system. Cells were silenced for FUS, TOPBP1, or both. Right: Western blot demonstrating the silencing of the respective proteins. Statistical analysis as in A. ***, P < 0.001. (D) HeLa WT and FUS-KO cells were stained with γH2AX and counterstained with DAPI. Cells were treated with DMSO, ETO for 1 h, or ETO plus 2-h recovery from ETO treatment (ETO/2h). These are representative figures for graph shown in Fig. 2 C. Scale bar: 20 µm. (E) HeLa WT and FUS-KO cells were stained with 53BP1 and counterstained with DAPI. Cells were treated with DMSO, ETO for 1 h, or ETO plus 2-h recovery from ETO treatment (ETO/2h). These are representative figures for graph shown in Fig. 2 D. Scale bar: 20 µm. (F) 53BP1 expression is affected by neither KO of FUS nor by the ETO exposure. Loading control: Tubulin.
To evaluate the impact of FUS depletion on DSB repair, we used U2OS cells stably transfected with GFP reporter constructs that allow the measurement of HR- and NHEJ-mediated repair (Gunn and Stark, 2012). Consistent with previous reports (Mastrocola et al., 2013; Wang et al., 2013), we observed that depletion of FUS affected both DSB repair pathways (Fig. S3, B and C).
Next, we determined whether the absence of FUS affected the formation of DNA damage foci. As shown in Fig. 2 C, after 1-h ETO exposure, we observed a similar number of γH2AX foci in HeLa WT and FUS-KO cells, indicating that the absence of FUS does not prevent the initial sensing of DSBs. However, after 2-h recovery from ETO (ETO/2h), FUS-KO cells had significantly more γH2AX foci than WT cells (Fig. 2 C, and Fig. S3 D). In addition, FUS-KO cells showed significantly less 53BP1 foci than WT cells after 1-h ETO (Fig. 2 D, and Fig. S3 E), which was not due to differential expression or degradation of 53BP1 (Fig. S3 F). 53BP1 foci formation was restored 2 h after recovery (Fig. 2 D), indicating that the absence of FUS results in a delay in the assembly of DNA repair complexes.
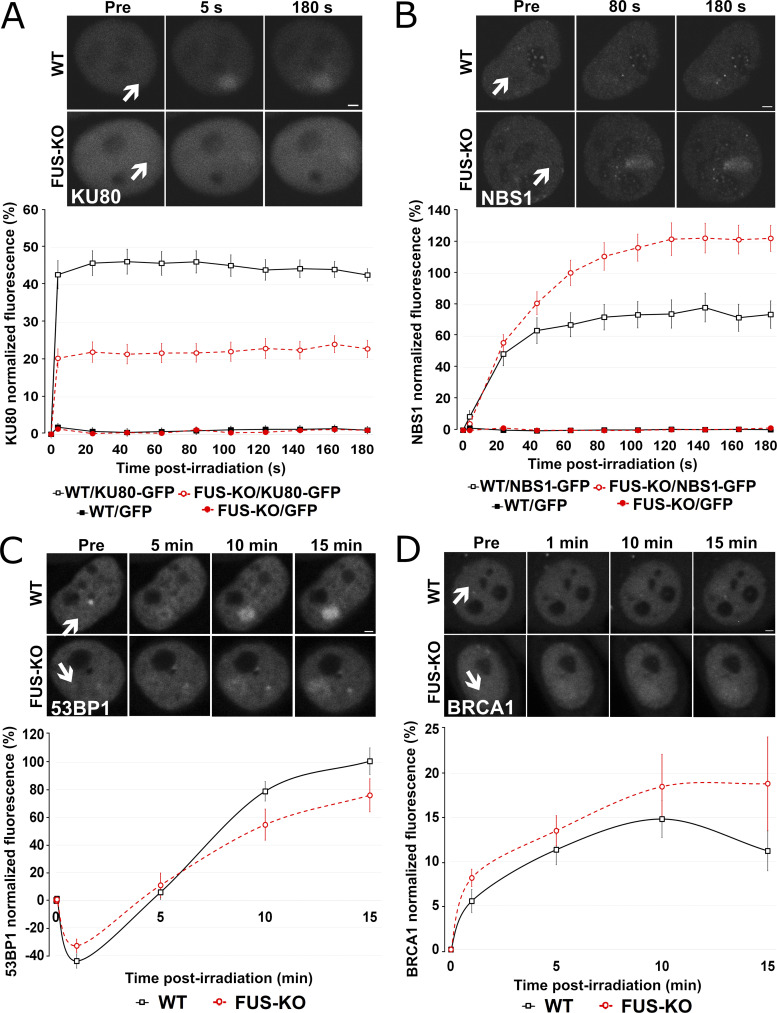
To gain insight into the molecular function of FUS during early DDR events, we analyzed the recruitment kinetics to DNA damage sites of two apical DDR factors, the 86-kD subunit of the KU heterodimer and NBS1, one of the subunits of the MRN complex. HeLa WT and FUS-KO cells transiently expressing KU80-GFP or NBS1-GFP were subjected to microirradiation with a 405-nm laser to induce time-specific and localized DNA damage. Real-time recording revealed that, in WT cells, KU80-GFP was promptly recruited to γH2AX-positive irradiation spots (Fig. S4 A), reaching a peak of recruitment within 5 s after microirradiation and remaining until the end of the assessment (180 s; Fig. 3 A). In FUS-KO cells, KU80 showed similar recruitment kinetics, but its accumulation was severely impaired. In contrast to KU80, NBS1 showed slower recruitment kinetics, reaching a peak ∼130 s after microirradiation (Fig. 3 B). However, in the absence of FUS, NBS1 was more efficiently recruited compared with WT cells. Transfection levels of GFP-tagged proteins in WT and FUS-KO cells were comparable (Fig. S4, B and C). Overall, these observations demonstrate that FUS plays an apical role in DDR activation, in particular in the retention of KU80 at the DNA broken ends. In addition, our results suggest that when stable KU80 binding is impaired, the MRN complex can more effectively gain access to the DNA ends.
Figure S4.
Microirradiation experiments are not influenced by differential expression of KU80-GFP or NBS1-GFP in WT versus FUS-KO cell lines. (A) KU80-GFP is recruited to the microirradiated area and colocalizes with γH2AX staining. Scale bar: 2 µm. (B) The raw fluorescence intensity of HeLa WT and FUS-KO cells transiently transfected with KU80-GFP was assessed to rule out the possibility that differential expression levels could affect the observed effect in Fig. 3 A. (C) The raw fluorescence intensity of HeLa WT and FUS-KO cells transiently transfected with NBS1-GFP was assessed to rule out the possibility that differential expression levels could affect the observed effect in Fig. 3 B.
Figure 3.
Loss of FUS changes the pattern of recruitment of HR- and NHEJ-related proteins to DSBs. (A) HeLa WT and FUS-KO cells were transiently transfected with a KU80-GFP expressing plasmid. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of the normalized fluorescence intensity of KU80-GFP recruitment at the microirradiated sites. All microirradiation experiments were performed in two biological replicates (with 10 cells each, except for BRCA1 recruitment, which was done in four biological replicates with 10 cells each). (B) HeLa WT and FUS-KO cells were transiently transfected with a NBS1-GFP–expressing plasmid. Upper panel: Representative micrographs of selected time points. Lower panel: Time course for NBS1-GFP recruitment. (C) HeLa WT and FUS-KO cells were transiently transfected with a 53BP1-GFP–expressing plasmid. Upper panel: Representative micrographs of selected time points. Lower panel: Time course for 53BP1-GFP recruitment. (D) HeLa WT and FUS-KO cells were transiently transfected with BRCA1-GFP–expressing plasmids. Upper panel: Representative micrographs of selected time points. Lower panel: Time course for BRCA1-GFP recruitment. In all graphs, data are plotted as normalized average ± SEM. Scale bars: 2 µm. Arrows indicate microirradiated area.
We next studied the recruitment of the effector proteins 53BP1 and BRCA1. In HeLa WT cells, 53BP1-GFP displayed a biphasic behavior: after microirradiation, fluorescence decreased in the first 5 min and then progressively increased, reaching a peak after 15 min (Fig. 3 C). The initial reduction in 53BP1-GFP fluorescence may possibly be due to chromatin reorganization (Izhar et al., 2015) that leads to a transient 53BP1 eviction, allowing the recruitment of DDR sensor proteins. In FUS-KO cells, the initial decrease in fluorescence was less pronounced than in WT cells. In addition, consistent with the delayed appearance of 53BP1 foci observed upon ETO incubation of FUS-KO cells, we observed a reduction in the maximal recruitment of 53BP1-GFP (Fig. 3 C). To determine if FUS could contribute to the choice of DSB repair pathway, we also tested the recruitment of BRCA1-GFP. As shown in Fig. 3 D, BRCA1 was recruited slightly more efficiently and appeared to be retained longer at DNA damage sites in FUS-KO compared with WT cells. Since 53BP1 was shown to inhibit BRCA1 accumulation at DSB sites (Escribano-Díaz et al., 2013), the increased BRCA1 recruitment observed in FUS-KO cells could be a consequence of the impaired accumulation of 53BP1 rather than the result of the absence of FUS. This hypothesis is supported by the results of the GFP repair reporter assay (Fig. S3, B and C; Mastrocola et al., 2013; Wang et al., 2013), which suggest that FUS is important for both HR- and NHEJ-mediated DSB repair.
FUS promotes the recruitment of SFPQ to sites of DNA damage
We reported earlier (Reber et al., 2016) the interactome analysis of FUS. Gene ontology analysis revealed a substantial number of proteins involved in the DDR, including PARP1/2, XRCC6/KU70, and XRCC5/KU80 (Table 1). In addition, we found a significant enrichment for RBPs with LCDs (Table 2), some of which have been linked to DNA damage repair. Among potential FUS interactors, we further explored the functional interaction with SFPQ, because of its involvement in the DDR (Jaafar et al., 2017; Salton et al., 2010) and because, together with FUS, it is an essential structural component of paraspeckles (Hennig et al., 2015).
Table 1. DDR-related FUS interactors.
| Uniprot ID | Protein name |
|---|---|
| Q92499 | ATP-dependent RNA helicase DDX |
| P54132 | Bloom syndrome protein |
| P49674 | Casein kinase I isoform ε |
| P24941 | Cyclin-dependent kinase 2 |
| Q03468 | DNA excision repair protein ERCC-6 |
| P11388 | DNA topoisomerase 2-α |
| Q92547 | DNA topoisomerase 2-binding protein 1 |
| O60870 | DNA/RNA-binding protein KIN17 |
| P29372 | DNA-3-methyladenine glycosylase |
| P78527 | DNA-dependent protein kinase catalytic subunit |
| P24928 | DNA-directed RNA polymerase II subunit RPB1 |
| Q9Y5B9 | FACT complex subunit SPT16 |
| Q08945 | FACT complex subunit SSRP1 |
| Q9UBU8 | Mortality factor 4-like protein 1 |
| Q15014 | Mortality factor 4-like protein 2 |
| Q15233 | Non-POU domain-containing octamer-binding protein |
| P06748 | Nucleophosmin |
| P09874 | Poly[ADP-ribose] polymerase 1 |
| Q9UGN5 | Poly[ADP-ribose] polymerase 2 |
| Q9C0J8 | Pre-mRNA 3′ end processing protein WDR33 |
| Q9UMS4 | Pre-mRNA-processing factor 19 |
| Q9HCS7 | Pre-mRNA-splicing factor SYF1 |
| P25789 | Proteasome subunit α type-4 |
| P35251 | Replication factor C subunit 1 |
| P35250 | Replication factor C subunit 2 |
| P40938 | Replication factor C subunit 3 |
| P35249 | Replication factor C subunit 4 |
| P40937 | Replication factor C subunit 5 |
| Q96PK6 | RNA-binding protein 14 |
| Q9Y230 | RuvB-like 2 |
| P23246 | Splicing factor, proline- and glutamine-rich |
| Q9NTJ3 | Structural maintenance of chromosomes protein 4 |
| Q13263 | Transcription intermediary factor 1-β |
| P07948 | Tyrosine-protein kinase Lyn |
| Q14694 | Ubiquitin carboxyl-terminal hydrolase 10 |
| P13010 | X-ray repair cross-complementing protein 5 |
| P12956 | X-ray repair cross-complementing protein 6 |
Compiled from Reber et al. (2016).
Table 2. LCD-containing FUS interactors.
| Gene name | Uniprot ID | Repeats in LCD | References for DDR involvement |
|---|---|---|---|
| HNRNPA1 | P09651 | 2 × [G/S]Y[G/S] | |
| HNRNPA2/B1 | P22626 | 7 × [G/S]Y[G/S] | |
| HNRNPF | P52597 | 2 × YXXQ, 5 × [S/G]Y[S/G} | |
| HNRNPH1 | P31943 | 7 × [G/S]Y[G/S] | |
| HNRNPH3 | P31942 | 6 × [G/S]Y[G/S] | |
| HNRNPK | P61978 | 2 × [G/S]Y[G/S] | Moumen et al., 2013 |
| HNRNPR | O43390 | 1 × YNQ, 1 × YGQQ | Sui et al., 2015 |
| HNRNPUL1 | Q9BUJ2 | 8 × YXQ, 8 × [G/S]Y[G/S] | Harrison and Shorter, 2017; Polo et al., 2012 |
| MATR3 | P43243 | Salton et al., 2010 | |
| RBM14 | Q96PK6 | 19 × Y[G/N/A/S]AQ, 2 × [S/G]YG | Simon et al., 2017 |
| SFPQ | P23246 | 1 × SYQ | Rajesh et al., 2011; Salton et al., 2010 |
| TAF15 | Q86X94 | 12 × [G/S]Y[G/S], 9 × Y[G/S]Q |
Compiled from Hennig et al. (2015) and Couthouis et al. (2011).
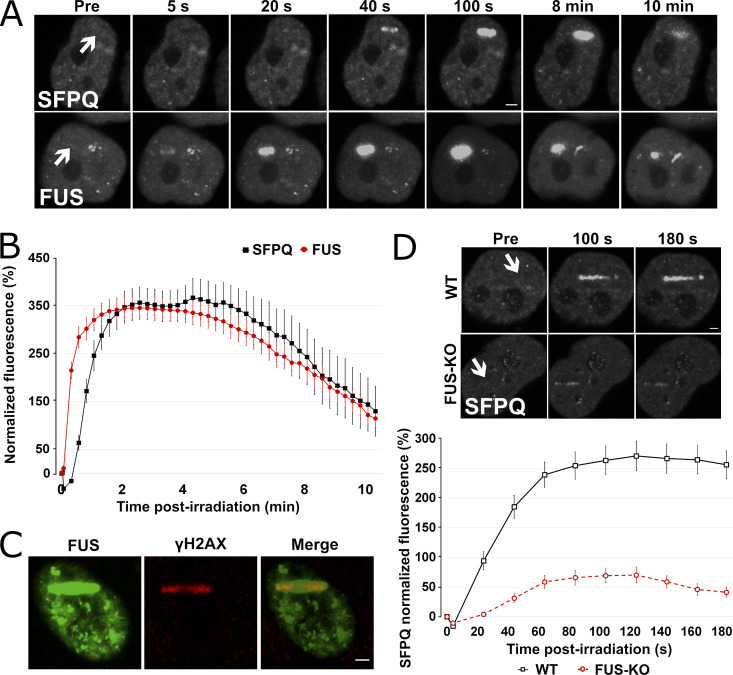
To determine whether SFPQ functions in DNA repair depended on FUS, we first compared the recruitment kinetics of the two proteins (Fig. 4, A and B). HeLa WT cells expressing either GFP-FUS or GFP-SFPQ were subjected to laser microirradiation to induce localized DNA damage, as confirmed by anti-γH2AX staining (Fig. 4 C). While FUS was promptly recruited and reached a maximum after 40 s, the redistribution of SFPQ to laser-induced damage sites was slower, reaching a peak after ∼100 s (Fig. 4, A and B). Then, we examined whether the recruitment of SFPQ is altered in the absence of FUS. As shown in Fig. 4 D, SFPQ accumulation was severely delayed and impaired in FUS-KO cells, indicating that the recruitment of SFPQ to DSB sites is FUS dependent (Video 1). Since laser microirradiation also generates single-strand breaks (SSBs), we tested whether the lack of FUS also affected the recruitment of XRCC1, a protein involved in base excision repair and the SSB repair pathway, by cotransfecting FUS-KO cells with both SFPQ-GFP and XRCC1-RFP. XRCC1 was efficiently recruited to DNA damage sites after laser microirradiation both in WT and in FUS-KO cells, indicating that FUS is specifically required for the recruitment of SFPQ (Fig. S5, A–C).
Figure 4.
FUS recruitment to DSBs precedes SFPQ, and its absence strongly reduces SFPQ accumulation. (A) WT HeLa cells were transiently transfected either with SFPQ-GFP or FUS-GFP plasmid and then laser microirradiated. (B) Comparison of FUS and SFPQ recruitment kinetics. (C) HeLa cells were transiently transfected with a GFP-tagged FUS expression plasmid and then laser microirradiated. Cells were then immunostained for γH2AX. H2AX is phosphorylated at laser microirradiation sites and colocalized with FUS-GFP. (D) HeLa WT and FUS-KO cells were transiently transfected with a SFPQ-GFP–expressing plasmid. Upper panel: Representative micrographs of selected time points (see Video 1). Lower panel: Time course for SFPQ-GFP recruitment. In all graphs, data are plotted as normalized average ± SEM. Scale bars: 2 µm. Arrows indicate microirradiated area.
Video 1.
SFPQ recruitment in HeLa WT versus FUS-KO cells. HeLa WT and FUS-KO cells were transiently transfected with SFPQ-GFP plasmid and laser microirradiated in the area indicated. Video was made by 183 sequential micrographs (3 pre- and 180 post-irradiation) and runs at 10× speed. It is possible to observe that the absence of FUS both delays and impairs the recruitment of SFPQ. Scale bar: 5 µm. Playback: 30 frames/s.
Figure S5.
Loss of FUS specifically affects SFPQ but not XRCC1 recruitment to DSB. (A) WT HeLa cells were transiently cotransfected with SFPQ-GFP and XRCC1-RFP plasmids and submitted to laser microirradiation as described in Materials and methods. Recruitment of these proteins was assessed for a 3-min period, and images were taken every 20 s. Scale bar: 2 µm. (B) Recruitment and accumulation of SFPQ, as shown in Fig. 4 D, is severely impaired in FUS-KO cells. (C) Recruitment of XRCC1 is very high (saturated fluorescence signal) and the same for WT and FUS-KO cells. Error bars represent SE.
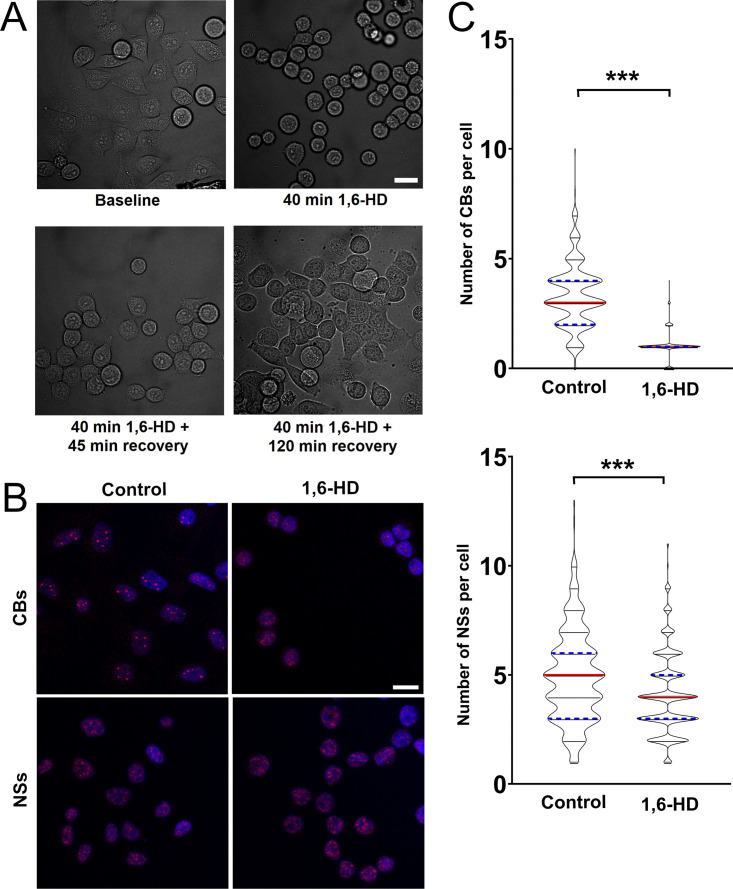
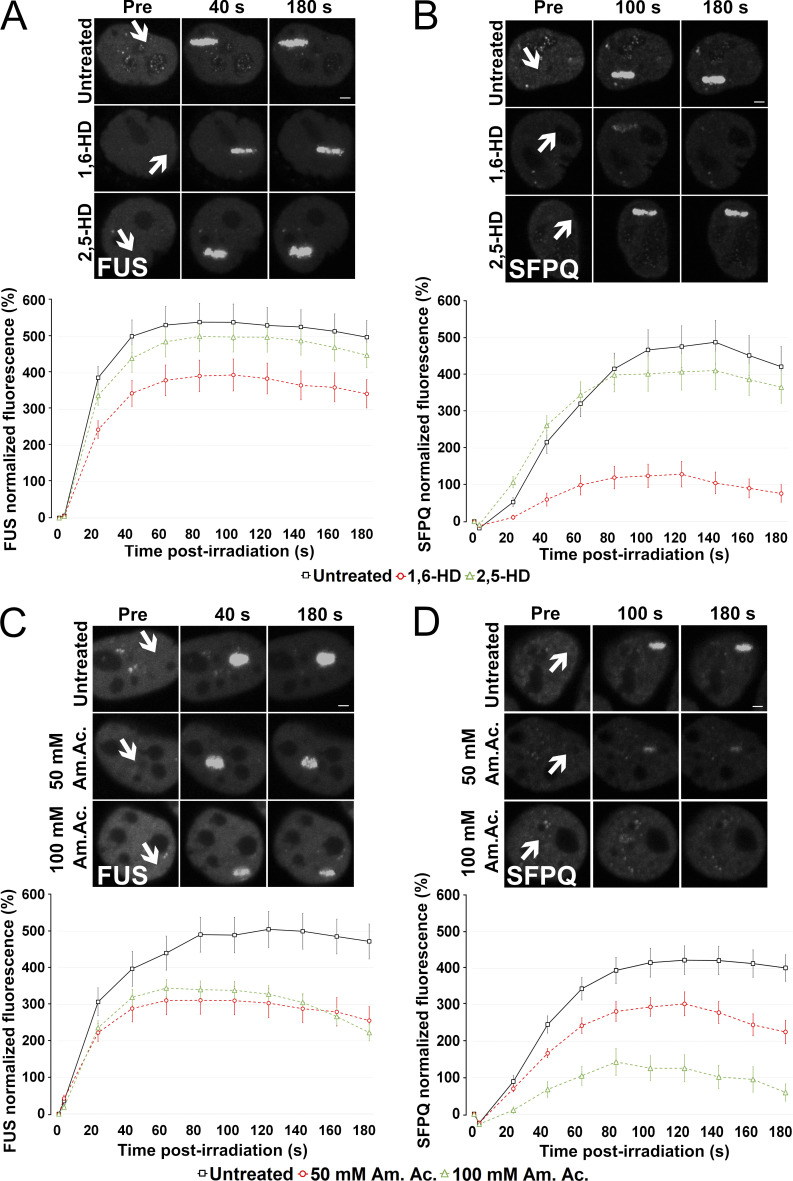
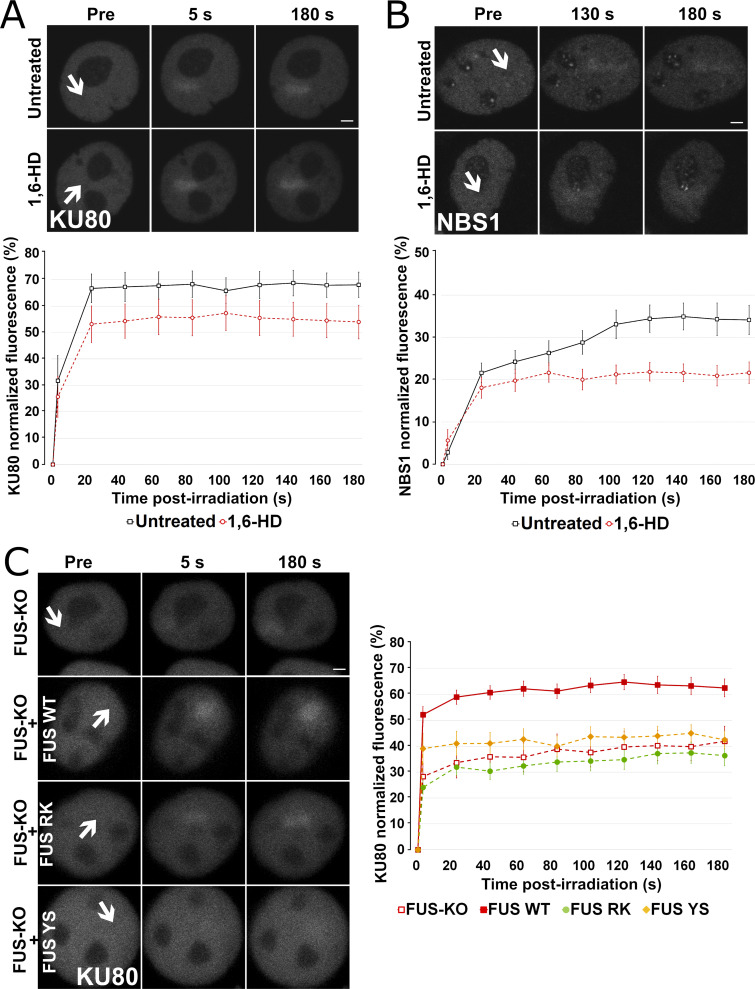
Both FUS and SFPQ LCDs were shown to liquid–liquid phase separate and to form hydrogels in vitro (Lee et al., 2015; Yamazaki et al., 2018). FUS is able to drive LLPS at low protein concentrations and physiological salt concentrations (Wang et al., 2018). Thus, we next tested whether the recruitment of FUS and SFPQ at DNA damage sites is dependent on LLPS and, more specifically, whether the recruitment of SFPQ is dependent on FUS-induced LLPS. We performed microirradiation experiments in the presence of chemicals that were previously shown to disrupt phase separation in vivo. First, we exposed HeLa cells to 2% 1,6-hexanediol (1,6-HD, 30 min), an aliphatic alcohol that is known to dissolve various cytoplasmic and nuclear membraneless compartments in vivo by disrupting their multivalent hydrophobic interactions (Allodi et al., 2016; Kroschwald et al., 2015; Updike et al., 2011; Yamazaki et al., 2018), and which was shown to partially dissolve FUS polymers in vitro (Allodi et al., 2016; Kato and McKnight, 2018). At this concentration, we observed that cells were able to recover their normal morphology 2 h after withdrawal from the alcohol (Fig. S6 A). Cajal bodies were partially disrupted, while nuclear speckles were affected to a lesser extent (Fig. S6, B and C). These observations indicate that this 1,6-HD treatment condition was mild enough not to disrupt all subcellular structures. Incubation of transiently transfected HeLa cells with 1,6-HD reduced FUS relocation to DNA damage sites (Fig. 5 A), an effect that was even more dramatic on the recruitment of SFPQ (Fig. 5 B). As control, we performed experiments in the presence of 2% 2,5-HD, a less hydrophobic isomer of 1,6-HD that does not affect LLPS (Allodi et al., 2016; Kato and McKnight, 2018). The presence of 2,5-HD did not affect FUS or SFPQ relocation (Fig. 5, A and B).
Figure S6.
2% 1,6-HD treatment does not irreversibly affect the morphology and vitality of cells. (A) Bright-field micrograph of HeLa cells treated with 1,6-HD as described in Materials and methods. Cells were allowed to recover in alcohol-free medium. Cells returned to a normal morphology within 120 min after 1,6-HD withdrawal. Scale bar: 20 µm. (B) Representative images of HeLa WT cells treated or not with 1,6-HD for 30 min and stained for Cajal bodies (CBs, α-coilin antibody) or nuclear speckles (NSs, α-SC-35 antibody). Scale bar: 20 µm. (C) Quantification of Cajal bodies and nuclear speckles in untreated and 1,6-HD–treated cells. HeLa cells were treated with 2% 1,6-HD for 30 min and then stained with DAPI and either anti-coilin or anti-SC35 antibodies. Quantification was performed as in Materials and methods. Experiments were done in duplicate, and 200 cells were analyzed per experiment. Statistics: Student’s t test (***, P < 0.001).
Figure 5.
LLPS is required for FUS and SFPQ recruitment to DNA damage sites. (A) HeLa WT cells were transiently transfected with a FUS-GFP–expressing plasmid and incubated with either 2% 1,6-HD or 2% 2,5-HD for 30 min before laser microirradiation. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of FUS recruitment. (B) HeLa WT cells were transiently transfected with a SFPQ-GFP–expressing plasmid and treated as in A. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of SFPQ recruitment. (C) HeLa WT cells were transiently transfected with a FUS-GFP–expressing plasmid and incubated with 50 or 100 mM Am. Ac. for 30 min before laser microirradiation. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of FUS recruitment. (D) HeLa WT cells were transiently transfected with a SFPQ-GFP–expressing plasmid and treated as in C. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of SFPQ recruitment. In all graphs, data are plotted as normalized average ± SEM. Scale bars: 2 µm. Arrows indicate microirradiated area.
To further substantiate the role of LLPS in FUS and SFPQ accumulation at laser-induced DNA damage sites, we assessed the effect of ammonium acetate (Am. Ac.), which promptly permeates cells and inhibits RNA-protein gelation without perturbing intracellular pH (Hamaguchi et al., 1997; Jain and Vale, 2017). At 100-mM concentration, Am. Ac. can effectively lead to the disappearance of nuclear CAG-repeat RNA foci and of nuclear speckles, ribonucleoprotein bodies that depend on ionic interactions (Jain and Vale, 2017). Recently, it was shown that Am. Ac. can rapidly dissolve 53BP1 foci (Pessina et al., 2019). Therefore, we incubated cells for 30 min before laser microirradiation with either 50 mM or with 100 mM Am. Ac. As shown in Fig. 5, C and D, similar to what we had observed in the presence of 1,6-HD, Am. Ac. severely affected the relocation of both FUS and SFPQ to DNA damage sites. Overall, these results support the idea that LLPS occurs at sites of DNA damage and is required for the efficient recruitment of FUS and SFPQ.
To determine if the impaired recruitment of SFPQ in FUS-KO cells could be due to the absence of FUS-induced LLPS, we took advantage of LLPS-deficient FUS constructs previously characterized by Wang et al. (2018). We tested the FUS LCD Y→S (YS) and RNA-binding domain R→K (RK) variants, which strongly affect phase separation, and as a control, the FUS LCD Q→G (QG) variant, which instead affects the hardening of droplets (Wang et al., 2018). As shown in Fig. 6, A and B, while WT FUS and the Q/G FUS variant were similarly recruited to DNA damage sites, the accumulation of Y/S and R/K FUS variants was significantly reduced. To test whether these FUS variants influence the recruitment of SFPQ, HeLa FUS-KO cells were cotransfected with plasmids expressing SFPQ-GFP and one of the FUS-mCherry constructs (WT, YS, QG, or RK). While complementation of FUS-KO cells with the WT FUS or the QG FUS variant increased SFPQ recruitment, expression of the YS FUS variant did not improve its relocation (Fig. 6, A and C). In the presence of the RK FUS variant, SFPQ was initially recruited but prematurely released from the DSB sites. Overall, these observations implicate a requirement for FUS-driven LLPS for the efficient recruitment and retention of SFPQ at DNA damage sites.
Figure 6.
LLPS-deficient FUS variants do not rescue SFPQ recruitment. HeLa FUS-KO cells were transiently cotransfected with one mCherry FUS construct (WT or the mutants RK, YS, or QG) and SFPQ-GFP before laser microirradiation. (A) Representative confocal micrographs of the recruitment of SFPQ and FUS in HeLa FUS-KO cells transiently transfected with both SFPQ and a FUS construct (WT or RK, YS, or QG mutants). (B) Time course of recruitment of WT and mutant FUS-mCherry. (C) Time course of SFPQ-GFP in FUS-KO cells either not transfected with FUS or transfected with FUS WT, FUS RK, FUS YS, or FUS QG. In all graphs, data are plotted as normalized average ± SEM. Scale bars: 2 µm. Arrows indicate microirradiated area.
LLPS is required to maintain the integrity of DNA damage foci
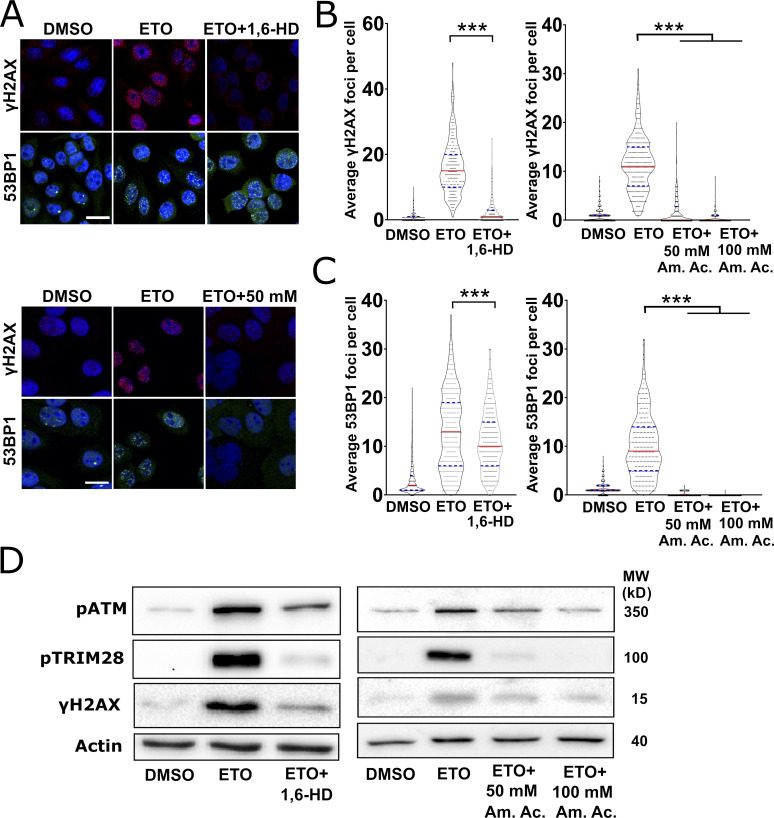
We then speculated that the actual formation of DDR foci might involve some form of phase separation. We first tested this hypothesis by exposing HeLa cells to either 2% 1,6-HD or to 50 mM or 100 mM Am. Ac., together with ETO, for 30 min (Fig. 7, A and B). While 1,6-HD treatment severely impaired the formation of γH2AX foci, 53BP1 foci were only partially dissolved. In the presence of Am. Ac., γH2AX foci formation was severely impaired, similar to what we had observed with the aliphatic alcohol. Am. Ac. also affected 53BP1 foci formation, as previously reported (Pessina et al., 2019). In addition to compromising foci integrity, 1,6-HD and Am. Ac. treatments compromised the phosphorylation of ATM, H2AX, and TRIM28 (Fig. 7 C). To test whether the recruitment of key DDR factors to sites of DNA damage is also dependent on LLPS, we assessed the effect of the aliphatic alcohol on the recruitment of KU80-GFP (Fig. 8 A) and NBS1-GFP (Fig. 8 B). We observed a reduction in the recruitment of both DDR factors in the presence of 1,6-HD, but not in the presence of the control alcohol 2,5-HD. Overall, these data indicate that LLPS is required for the proper formation of DNA damage foci and activation of the DDR signaling cascade.
Figure 7.
LLPS is required for DDR activation and foci formation. (A) Representative confocal micrographs of HeLa cells that were treated with ETO alone, ETO plus 1,6-HD (upper micrographs), or ETO plus 50 or 100 mM Am. Ac. (lower micrographs) before immunostaining for γH2AX or 53BP1. Scale bars: 20 µm. (B) Quantification of γH2AX 1 foci in the experiments in A. In the HD experiment, 200 cells were analyzed per condition, and in the Am. Ac. experiment, 150 cells per condition, and experiments were performed in duplicate. Graphs represent the number of foci per cell and are shown as violin plots with all samples. Statistics: one-way ANOVA and Bonferroni post hoc test. (C) Quantification of 53BP1 foci in the experiments in A. Experiments, quantifications, and statistics were performed as described in B. (D) Western blot analysis of total extracts prepared from HeLa cells treated with ETO alone, ETO and 2% 1,6-HD, or ETO and Am. Ac. (50 or 100 µM). Phosphorylation of ATM, TRIM28, and H2AX was assessed (loading control: Actin). MW, molecular weight. ***, P < 0.001.
Figure 8.
LLPS is required for the proper recruitment of HR- and NHEJ-related proteins. (A) HeLa WT cells were transiently transfected with KU80-GFP–expressing plasmids and then incubated with 2% 1,6-HD for 30 min before laser microirradiation. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of KU80 recruitment. (B) HeLa WT cells were transiently transfected with NBS1-GFP–expressing plasmids and then incubated with 2% 1,6-HD for 30 min before laser microirradiation. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of NBS1 recruitment. (C) Recruitment of KU80-GFP in FUS-KO cells either untransfected or transfected with FUS WT, FUS RK, or FUS YS. Upper panel: Representative micrographs of selected time points. Lower panel: Time course of KU80 recruitment. In all graphs, data are plotted as normalized average ± SEM. Scale bars: 2 µm. Arrows indicate microirradiated area.
To strengthen the link between LLPS of FUS and the DDR, we tested the effect of the reconstitution of FUS-KO cells with the two FUS variants that are the most affected in LLPS, the YS and RK mutations, on KU80 recruitment (Bogaert et al., 2018; Maharana et al., 2018). Both FUS variants were unable to rescue KU80 accumulation at laser-induced DNA damage sites (Fig. 8 C), providing further evidence for the requirement of FUS LLPS for the efficient assembly of the DDR machinery.
FUS is required for γH2AX cluster formation
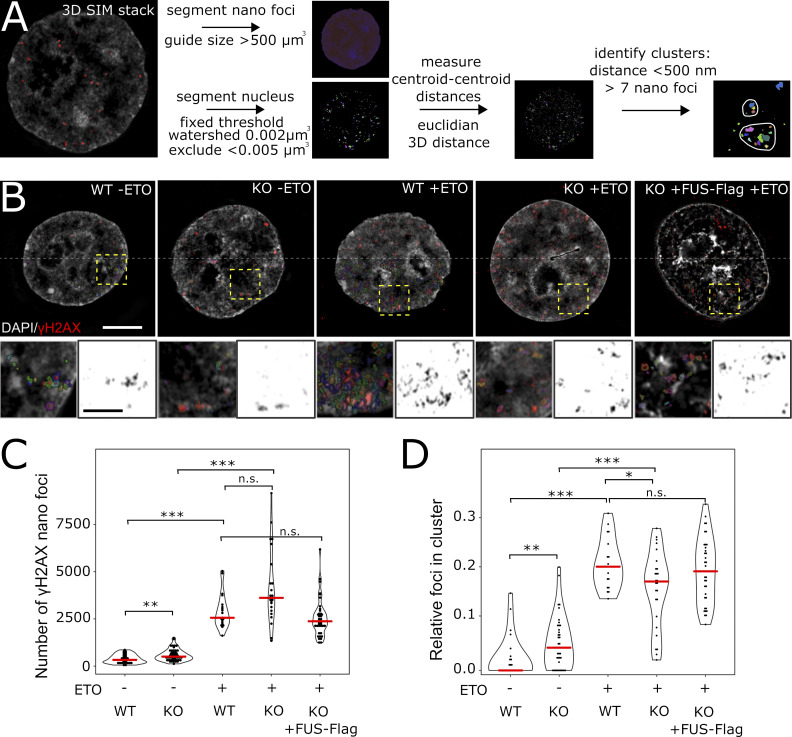
We have recently shown using 3D structured illumination microscopy (SIM) that γH2AX foci consist of spatially clustered γH2AX nanofoci (Natale et al., 2017). Given that FUS-KO cells have defective recruitment of DNA damage factors and formation of DNA damage foci, we next assessed whether FUS may be required for the proper organization of the 3D arrangement of γH2AX-decorated chromatin. We thus quantified γH2AX nanofoci in WT and FUS-KO cells treated with 10 µM ETO for 1 h using 3D-SIM (Fig. 9, A–C).
Figure 9.
FUS is required for γH2AX nanofoci clustering. (A) Image analysis workflow for 3D-SIM γH2AX cluster analysis. Nuclei and γH2AX nanofoci are segmented; centroid-centroid distances for the nanofoci are computed; and nanofoci with a centroid-centroid distance shorter than 500 nm are assigned to the same cluster. Clusters are quantified as structures that contain more than seven nanofoci. (B) Images of HeLa WT and FUS-KO cells treated for 1 h with ETO or DMSO, respectively. Additionally, HeLa-FUS-KO cells transiently transfected with FUS-Flag plasmid were analyzed after ETO treatment. The main images show the merged DAPI and γH2AX nanofoci in pseudo-color (scale: 5 µm). The lower half of the image shows the outline of the segmented nanofoci. Crop areas are highlighted with yellow boxes, and the magnified regions are shown below (scale bar: 1 µm). (C) Quantification of nanofoci per cell in the different conditions indicated. Red lines indicate the median. Statistics: Wilcoxon’s rank sum test. (D) Frequency of nanofoci found in clusters, as compared with the total number of nanofoci in the cell. Red lines indicate the median. Statistics as in C. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Fig. 9 A displays the workflow for foci segmentation and cluster identification. Consistent with the data obtained by confocal microscopy (Fig. 1, B and C), DMSO-treated WT control cells exhibited a low number of γH2AX nanofoci, while foci were significantly increased in control FUS-KO cells (Fig. 9 C). Upon ETO treatment, we did not observe a significant difference between the total number of nanofoci in WT versus FUS-KO cells (Fig. 9 C), similar to the results we obtained by confocal microscopy.
We then studied the spatial distribution of γH2AX nanofoci in terms of cluster formation. We assessed the relative number of nanofoci contained in clusters compared with the total number of nanofoci per cell. Unsurprisingly, considering the extremely low number of γH2AX nanofoci in DMSO-treated WT cells, we found the fraction of γH2AX nanofoci clusters in these cells to be extremely low. In contrast, the fraction of clusters was higher in DMSO-treated FUS-KO cells, probably due to their increased total number of γH2AX nanofoci. Upon ETO treatment, while WT cells showed a significantly increased fraction of γH2AX nanofoci organized in typical clusters, these were significantly reduced in FUS-KO cells (Fig. 9, B and D), indicating that FUS is required for the proper organization of the 3D arrangement of γH2AX-decorated chromatin. To support this conclusion, we transiently transfected HeLa FUS-KO cells with an FUS-Flag construct and quantified the number of γH2AX nanofoci and clusters upon ETO treatment (Fig. 9 B, right panel). Unlike untransfected FUS-KO cells, FUS-Flag transfected FUS-KO cells showed a similar total number of total clustered nanofoci as WT cells (Fig. 9, C and D). These findings demonstrate that FUS is an additional factor, besides CTCF (CCCTC-binding factor) and direct chromatin contacts (Collins et al., 2020; Hwang et al., 2019; Natale et al., 2017), that is required for the orchestrated formation of γH2AX-decorated chromatin domains. These domains are the essential basis for the subsequent steps of DSB repair, and our results indicate that the defective DNA damage repair in FUS-KO cells is associated with inefficient clustering of γH2AX nanofoci.
Discussion
The results reported here provide the first evidence that FUS plays an important early role in the DDR by promoting LLPS at DNA damage sites, thereby physically contributing to the efficient recruitment of key DDR factors. Previous reports have already implicated FUS in DNA damage repair (Mastrocola et al., 2013; Singatulina et al., 2019; Wang et al., 2013), and more recently, review articles have proposed that FUS, and more generally RBPs, contribute to the DDR and to DNA repair by promoting phase separation at damage sites (D’Alessandro and d’Adda di Fagagna, 2017; Kai, 2016; Patel et al., 2015). However, in vivo experimental evidence of their molecular function in this process remained to be established.
We screened DDR proteins for their recruitment to laser-induced damage sites in FUS-KO cell lines. We found that FUS is required at a very early step for the retention at DSBs of the 80-kD subunit of the DSB sensor KU70/80. Interestingly, whereas in FUS-KO cells the retention of KU80 is impaired, the recruitment of NBS1, one of the three subunits of the MRN complex, is increased by >50%. On the one hand, these observations may explain at least in part the higher phosphorylation of ATM, and of downstream DDR factors, that we observed in FUS-KO cells, since NBS1 directly interacts with ATM (Falck et al., 2005), and the MRN complex is required for ATM activation (Lee and Paull, 2005). On the other hand, they are consistent with the idea that, due to its high abundance and strong affinity for DNA, KU heterodimer is the first DDR factor that binds to the DNA broken ends irrespective of the cell cycle phase (Shibata et al., 2018). KU70/80 is then removed by the endonucleolytic activity of the MRN complex (Chanut et al., 2016; Myler et al., 2017) or by phosphorylation of KU70 (Lee et al., 2016). Therefore, KU70/80 is recruited but not efficiently retained in the absence of FUS, allowing the MRN complex to successfully compete for binding to the DNA broken ends. However, this is not sufficient for efficient repair, since we and others have demonstrated that the silencing of FUS affects both HR- and NHEJ-mediated repair (Mastrocola et al., 2013; Wang et al., 2013). This justified the continuing quest for FUS-dependent events during the DDR.
Indeed, FUS is also required for the relocation of 53BP1 to DNA damage sites, and consistently, the formation of 53BP1 foci is delayed in FUS-KO cells. In line with the inhibitory role that 53BP1 exerts on BRCA1 at DSB sites (Escribano-Díaz et al., 2013), we observed that in FUS-KO cells, in which 53BP1 recruitment at laser-induced DNA damage sites is impaired, the association of BRCA1 is more efficient. We also discovered that FUS is necessary for the recruitment to damage sites of SFPQ, an RBP that had already been implicated in DNA damage repair (Bladen et al., 2005; Jaafar et al., 2017; Rajesh et al., 2011; Simon et al., 2017). Intriguingly, FUS and SFPQ are both core components of paraspeckles, phase-separated membraneless organelles that assemble on the NEAT1 lncRNA, which induces LLPS via interaction with SFPQ/NONO (Yamazaki et al., 2018). The purified LCD of FUS forms a hydrogel at high concentrations in vitro, and this region is essential for paraspeckle formation in vivo (Hennig et al., 2015). Based on these observations, it was tempting to hypothesize that FUS and SFPQ play a similar role in the DDR, inducing LLPS at DSBs to promote the efficient assembly of DNA repair complexes. Indeed, we show that efficient relocation of SFPQ requires LLPS-competent FUS.
Consistent with a physical role of FUS in triggering the formation of DDR protein assemblies around DSBs through LLPS, we found that γH2AX foci are sensitive to treatment with two different LLPS inhibitors, indicating that LLPS occurs very early during the DDR. 53BP1 foci were also recently shown to be phase-separated droplet-like compartments (Kilic et al., 2019; Pessina et al., 2019). Interestingly, we observed that γH2AX foci are more sensitive to the LLPS inhibitor 1,6-HD compared with 53BP1 foci: while γH2AX foci were completely dissolved by 2% 1,6-HD, disassembly of 53BP1 foci could only be observed with a concentration of ≥4% (data not shown and Kilic et al., 2019). Since 1,6-HD can disrupt structures that depend on weak interactions between sticky LCDs but is ineffective in disrupting solid amyloid-like aggregates (Kroschwald et al., 2015), our observations suggest that 53BP1 foci are more solid-like in comparison with γH2AX foci, possibly because of their different protein composition. Remarkably, however, the number of both γH2AX and 53BP1 foci decreased upon addition of Am. Ac., which can prevent RNA gelation and can disrupt nuclear speckles, ribonucleoprotein bodies that depend on ionic interactions (Jain and Vale, 2017; Kroschwald et al., 2015). The effect of Am. Ac. on γH2AX and 53BP1 foci is consistent with a role for RNA in the earliest DDR events (Bonath et al., 2018; Francia et al., 2012; Michelini et al., 2017).
In addition, we observed that LLPS inhibition impairs the recruitment of FUS, SFPQ, KU80, and NBS1 and affects DDR signaling. In fact, FUS forms LLPS-specific interactions with additional DNA damage repair proteins (Reber et al., 2019 Preprint). Our findings are consistent with two recent studies showing that LLPS is necessary for the subsequent recruitment of the downstream effector 53BP1 to DNA damage foci (Kilic et al., 2019; Pessina et al., 2019).
To support the idea that FUS-dependent LLPS at DSBs is required for the efficient assembly of DNA repair complexes, we investigated the structure of H2AX foci formed in the absence of FUS using superresolution microscopy. We reported previously that γH2AX foci formed following ionizing radiation exposure consist of spatially clustered nanofoci of ∼200-nm diameter (Natale et al., 2017). Here we characterized γH2AX foci in WT and FUS-KO cells treated with ETO. We found that FUS contributes to the spatial clustering of γH2AX foci. The impairment of the structural organizations of γH2AX nanofoci in FUS-KO cells upon genotoxic damage is reminiscent of what we observed upon depletion of the chromatin architectural protein CTCF (Natale et al., 2017). Thus, we propose that FUS exerts an early role in DDR by promoting LLPS. This, and the presence of CTCF, contributes to the 3D organization of chromatin around DSBs, enabling the activation of an efficient DDR.
The question remains open as to what recruits FUS to broken DNA ends. In this regard, the formation of a complex between KU and FUS is supported by several recent reports (Abbasi and Schild-Poulter, 2019; Morchikh et al., 2017). Based on our data and on the study by Aleksandrov et al. (2018), who measured, clustered, and modeled the kinetics of recruitment and dissociation of 70 DNA repair proteins to laser-induced DNA damage sites, we propose that FUS relocates to DSBs, where it promotes LLPS together with SFPQ. This stabilizes KU70/80 on the broken DNA ends and leads to the further recruitment of multiple proteins required for DSB repair. Intriguingly, several of these proteins are RBPs, which supports the idea that RNAs are involved in DSB repair (Michelini et al., 2017; Pessina et al., 2019). How FUS and other RBPs are recruited to broken ends remains to be established. Most likely, this occurs through binding to poly-ADP ribose chains that are deposited at sites of DNA damage by PARP1, which is among the first proteins that are recruited (have half-times between 1.8 and 3.7 s; Aleksandrov et al., 2018). Both FUS and the SFPQ/NONO heterodimer have been shown to bind to poly (ADP-ribose) (Krietsch et al., 2012; Mastrocola et al., 2013). While further work is required to understand the role of all the different RBPs that have been involved in DNA repair, and also of RNA itself, our data will facilitate mechanistic investigations to uncover their precise activities.
The elucidation of FUS’s role in the activation of the DDR upon DNA damage will also be relevant for the design of novel therapeutic approaches for neurodegenerative diseases and cancers in which FUS plays a pathological role. FUS was initially identified ∼20 yr ago as a fusion oncogene in human myxoid liposarcomas with the C/EBP homologous protein transcription factor. More recently, FUS fusions have been found in Ewing’s sarcoma and acute myeloid leukemia (Shing et al., 2003; Zerkalenkova et al., 2018). These translocations involve the fusion of FUS N-terminal LCD with different transcription factors. In addition, FUS is associated with a variety of neurodegenerative diseases, including amyotrophic lateral sclerosis, frontotemporal lobar degeneration, and polyglutamine diseases (Deng et al., 2014a), where it is found in cytoplasmic inclusions. To explain the pathological role of FUS in neurons, two hypotheses have been proposed: the gain-of-function model, in which FUS gains a toxic function in the cytoplasm through aggregate formation and sequestration of important regulators; alternatively, the relocalization of FUS to the cytoplasm depletes the nuclear protein pool affecting transcription, alternative splicing, and DNA repair. Indeed, H2AX phosphorylation has been observed in motor neurons carrying FUS nuclear localization signal mutations (Naumann et al., 2018; Wang et al., 2013), suggesting that a chronic DDR activation may exacerbate neuroinflammation, contributing to neuronal death. Therefore, elucidation of the molecular function of FUS in the DDR could provide new insight into its role in the pathogenesis of neurodegenerative diseases and in cancer, allowing the design of novel therapeutic strategies.
Materials and methods
Cell lines, cell culture, and treatments
HEK293T cells stably expressing Flag-tagged FUS and HeLa FUS-KO were described in Reber et al. (2016). U2OS cells stably expressing HR and NHEJ repair reporters were a kind gift from J.M. Stark (Beckman Research Institute of City of Hope, Duarte, CA) and were described in Gunn and Stark (2012). HeLa and SH-SY-5Y FUS-KO cells were generated by CRISPR-trap (Reber et al., 2018). All cell lines were tested for mycoplasma contamination and were found to be negative. Cells were cultured in DMEM (high glucose) supplemented with 10% FBS, 2 mM l-glutamine, 100 IU/ml penicillin, and 0.1 mg/ml streptomycin (all EuroClone). Cells were grown at 37°C and 5% CO2 in a humidified incubator.
ETO treatment, where not specified otherwise, was performed for 1 h with 10 µM diluted in DMSO (Enzo Lifesciences). Where specified, cells were treated with 2% 1,6-HD (diluted in growth medium; Sigma-Aldrich), 2% 2,5-HD (diluted in growth medium; Sigma Aldrich), or 50 or 100 mM Am. Ac. (diluted in water; Sigma Aldrich) for 30 min.
Plasmid DNA transfections were performed using Lipofectamine 2000 (Invitrogen), while siRNA transfections were done using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s instructions. The synthetic siRNAs used in this study were purchased from Riboxx Life Sciences.
Trypan blue assay
HeLa WT and FUS-KO cells were seeded at a concentration of 2 × 103 cells/well in 96-well plates. Cells were allowed to attach overnight and were then treated with increasing concentrations of ETO (DMSO, 0.5 or 1 µM) or CPT (DMSO, 0.1 or 0.5 µM) for 18 h. Cells were then detached with trypsin and diluted 1:2 in Trypan blue. Living cells were visualized and counted with a hemocytometer using a bright-field microscope. Data are expressed as the percentage of cells in relation to their respective control (DMSO).
DNA constructs
NBS1-GFP was kindly provided by Dr. A. Nussenzweig (Laboratory of Genome Integrity, National Institutes of Health, Bethesda, MD; Kruhlak et al., 2006). KU80-EGFP was purchased from AddGene (#46958). SFPQ-EGFP was generated by subcloning the SFPQ ORF obtained from the Myc-PSF-WT plasmid (#35183; AddGene). The FUS-EGFP plasmid was generated by inserting the FUS ORF into pcDNA6F-EGFP. The generation of the XRCC1-RFP plasmid (pc1156) is described in detail in Muster et al. (2017). FUS-mCherry plasmids (WT, RK, YS, and QG) were kindly provided by Dr. J. Wang and Dr. S. Alberti (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany; Wang et al., 2018). The sequences of the oligonucleotides/siRNAs used are as follows): siXRCC5, 5′-AAGAGCUAAUCCUCAAGUCUU-3′; siTOPBP1, 5′-CUCACCUUAUUGCAGGAGAdTdT-3′; siFUS1, 5′-AGCCCAUGAUUAAUUUGUATT-3′; siFUS_sh, 5′-GGACAGCAGCAAAGCUAUATT-3′; siRNA pool negative control, catalog no. SKU# K-00100 (iBONI; Riboxx’s design).
Protein collection, quantification, and Western blotting
Adherent cells were washed twice with PBS and then scraped and digested on ice for 30 min with a homemade radioimmunoprecipitation assay medium (50 mM Tris-HCl, pH 7.5, 15 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS, supplemented with protease inhibitors [Roche] and phosphatase inhibitors [Sigma-Aldrich]). Cell extracts were centrifuged at 16,000 g for 20 min at 4°C, and only the soluble fraction (supernatant) was used. Proteins were quantified using a validated bicinchoninic acid Protein Assay Kit (EuroClone) protocol. For Western blotting, the same amount of proteins was loaded, diluted in homemade sample buffer (6× Laemmli sample buffer: 1 M Tris-HCl, pH 6.8, 10% SDS, 40% glycerol, 12% β-mercaptoethanol, and bromophenol blue). Running buffer was prepared with Tris-glycine 1× and SDS 0.1% using a homemade gel (7–12% bis-acrylamide; Bio-Rad), which ran at 100 V for ∼2.5 h. After the run, the proteins were transferred to a nitrocellulose membrane (0.45-µm pore size; Amersham). Transfer was performed at 80 V at 4°C for 1.5–2.5 h using a 20% methanol transfer buffer. Transfer of 53BP1 was done using a modified transfer buffer with 18% methanol and 0.1% SDS. Membranes were stained with Ponceau (Sigma-Aldrich) for 8 min, and images were acquired using a ChemiDoc (Bio-Rad). After removal of the Ponceau by washing once with TBS-T (TBS 1× and Tween 0.05%), membranes were blocked for 1 h with 5% milk or 4% BSA (for phosphorylated proteins), diluted in TBS-T. Membranes were then incubated overnight at 4°C with a primary antibody diluted in 5% milk or 4% BSA. The membrane was then washed three times with TBS-T for 5 min and incubated with a secondary antibody diluted in 5% milk for 1 h at RT. The membrane was finally washed three times with TBS-T for 10 min. Acquisition was made using Cyanagen ECL according to the manufacturer’s protocol, exposing the membrane in the trans-illuminator ChemiDoc (Bio-Rad). The quantification of the signal intensity was performed using Image Lab v6.0.
The antibodies used for Western blot experiments are as follows: rabbit α-γH2AX, #9718; Cell Signaling (1:1,000); rabbit α-pATM, #5883; Cell Signaling (1:1,000); rabbit α-pATR, #2853; Cell Signaling (1:1,000); rabbit α-pCHK1, #2348, Cell Signaling (1:1,000); rabbit α-pCHK2, #2197; Cell Signaling (1:1,000); rabbit α-pTRIM28, A300-767A-T; Bethyl (1:1,000); rabbit α-pBRCA1, #9009; Cell Signaling (1:1,000); rabbit α-53BP1, #4937S; Cell Signaling (1:1,500); rabbit α-FUS, homemade by M.-D. Ruepp (1:3,000; Raczynska et al., 2015); rabbit α-KU80, #2753S; Cell Signaling (1:1,000); mouse α-TOPBP1, sc271043; Santa Cruz Biotechnology (1:1,000); mouse α-Tubulin, sc-5286; Santa Cruz Biotechnology (1:3,000); mouse α-β-Actin, ab8226; Abcam (1:1,000); goat α-rabbit IgG HRP-linked, #7074; Cell Signaling (1:8,000); and horse α-mouse IgG HRP-linked, #7076; Cell Signaling (1:8,000).
Immunofluorescence, confocal imaging, and quantification
Cells were fixed with 4% PFA (diluted in PBS, pH 7.4) at RT for 15 min and, after three washes with PBS, were permeabilized with 0.25% Triton X-100 in PBS for 5 min. Permeabilized cells were blocked in blocking solution (20% FBS and 0.05% Tween in PBS) for 1 h at RT. Primary antibodies were diluted in wash buffer (0.2% BSA in PBS) and incubated for 1 h at RT. After three washes with wash buffer, cells were incubated with the respective secondary antibodies (diluted in wash buffer) for 1 h at RT. After three washes with wash buffer, cells were counterstained with DAPI (D9542; Sigma-Aldrich; 1 µg/ml in PBS) diluted in PBS for 10 min. Coverslips were washed twice with PBS and once with water and mounted onto microscope slides using an antifade mounting medium (FluorSave; Calbiochem). The antibodies used were mouse α-γH2AX, ab26350; Abcam (1:100); rabbit α-53BP1, NB100-305; NovusBio (1:200); rabbit α-Coilin, A.I. Lamond (1:100; University of Dundee, Scotland); mouse α-SC35, S4045; Sigma-Aldrich (1:100); Alexa Fluor 488 goat α-rabbit IgG, A11008; Invitrogen (1:4,000); and Alexa Fluor 647 goat α-mouse IgG, A21235; Invitrogen (1:4,000).
Cell imaging was performed using a confocal microscope ECLIPSE Ti A1 (Nikon) and acquisition software NIS-Elements. Between 6–10 images were taken per coverslip (randomly) using the 60× oil objective. Quantification of DNA damage foci (γH2AX and 53BP1) and subnuclear bodies (Cajal bodies and nuclear speckles) was performed using the software Fiji ImageJ v2.0. To that end, a mask of the nucleus was made using DAPI staining (filter Gaussian blur radius 2 followed by adjusting the threshold and then analyzing particles). Nuclear foci were measured using the function Find Maxima (by setting a noise tolerance and dividing the final image by 255). For baseline experiments (WT vs. FUS-KO), data are shown as the average number of foci per nucleus. For all other experiments, data were normalized by the WT DMSO group (which was considered 100%). At least 100 cells were counted per experiment, which were done at least in duplicate.
For SG imaging, HeLa WT and HeLa FUS-KO cells were seeded in 24-well plates fitted with coverslips. 24 h later, the cells were stressed for 1 h by addition of 0.5 mM sodium (meta)arsenite (S7400; Sigma-Aldrich) to the medium. Cells were then fixed in 4% PFA for 30 min at RT. Subsequently, cells were washed 3 × 5 min with PBS and then permeabilized in TBS supplemented with 0.5% Triton X-100 and 6% BSA for 30 min at RT. Primary antibody incubation was conducted in 0.1% Triton X-100 and 6% BSA in TBS overnight at 4°C. Cells were washed 3 × 5 min with PBS, incubated with secondary antibodies in 0.1% Triton X-100 and 6% BSA in TBS for 2 h at RT, and then counterstained with 100 ng/ml DAPI (Sigma-Aldrich) for 10 min at RT. Cells were then washed 2 × 5 min with PBS and subsequently rinsed with H2O before mounting onto microscope slides using Vectashield hardset antifade mounting medium (Vectashield; H-1400). Antibodies used were mouse α-FUS, sc47711; Santa Cruz (1:200); goat α-TIA1, sc1751; Santa Cruz (1:500); Alexa Fluor 488 donkey α-goat IgG, A11055; Invitrogen (1:500); and Alexa Fluor 546 donkey α-mouse IgG, A10036; Invitrogen (1:500).
Laser microirradiation
HeLa WT and/or FUS-KO cells were transiently transfected with the indicated GFP- or mCherry-tagged plasmids 2 d before irradiation. The following day, transfected cells were plated onto 35-mm plates with a glass bottom and allowed to attach overnight. 30 min before irradiation, the cell medium was replaced by phenol red–free medium containing 0.5 µg/ml Hoechst 33342 (B2261; Sigma-Aldrich). Three images were taken as baseline (preirradiation), and then cells were irradiated for 4 s using the 405-nm laser at 25% power. The fluorescence intensity of the irradiated area and of two other nuclear regions of interest (ROIs) and one background ROI were assessed. Protein recruitment to the irradiated area was analyzed by removing the background fluorescence intensity from each of the other ROIs, and then calculating the percentage change at each time point from the average of the three baseline images (preirradiation). Finally, the difference between the fluorescence signal in the irradiated region and the average of the two control regions in the nucleus was calculated. The formula used to calculate the protein recruitment: Recruitment (%) = % from baseline in irradiated ROI − [(% from baseline in control ROI 1 + % from baseline in control ROI 2)/2]. All the microirradiation experiments were performed in a confocal microscope ECLIPSE Ti A1 (Nikon), using a 60× oil objective plus 5× digital zoom. During microirradiation, cell plates were maintained in a humidified chamber at 37°C and 5% CO2. Experiments were done in duplicate, and 10 cells were assessed per experiment (for a total of 20 cells per group), except for the recruitment of BRCA1, in which four independent experiments were performed with 10 cells assessed per experiment (for a total of 40 cells).
HR and NHEJ repair reporter assays
DSB repair assays were performed in Direct Repeats–GFP or EJ5-GFP U2OS cell lines (Gunn and Stark, 2012). The constructs are based on an engineered GFP gene containing recognition sites for the I-SceI endonuclease for enzymatic induction of DSBs. The starting constructs are GFP negative, as the GFP gene is inactivated by an additional exon or by mutations. Successful repair of the I-SceI–induced breaks by NHEJ or HR restores the functional GFP gene. The number of GFP-positive cells, as counted by flow cytometry, provides a quantitative measure of the NHEJ or HR efficiency.
Briefly, cells were transfected with siRNAs. 2 d later, cells were cotransfected with a plasmid expressing I-SceI (pCBA-I-SceI) together with indicated siRNAs using Lipofectamine 2000 (Invitrogen). Cells were harvested 3 d after transfection and subjected to flow cytometric analysis to identify and quantify GFP-positive cells (BD FACS Calibur and CellQuest software). The repair efficiency was scored as the percentage of GFP-positive cells, and data were normalized to a control siRNA treatment in each individual experiment. Experiments were done in triplicate, and ≥10,000 cells were assessed per experiment.
3D SIM imaging and analysis
Cells were plated on high-precision coverslips (18 × 18 mm, #1.5 thickness with low variance). After treatment, cells were fixed with 2% formaldehyde at RT for 10 min and then washed three times (first time by removing only two thirds of fixation solution) with 0.02% Tween 20 in PBS (PBST). Cells were permeabilized with 0.5% Triton X-100 in PBST for 10 min and blocked with 2% BSA in PBST for 1 h at RT. After blocking, primary antibody diluted in blocking solution was added for 1 h at RT, followed by three washes with PBST. Secondary antibody diluted in blocking solution was then added for 1 h at RT, followed by three washes with PBST. Samples were postfixed with 4% formaldehyde for 10 min at RT and then washed three times with PBST. DAPI counterstaining (Sigma Aldrich; 1 µg/ml in PBST) was done for 10 min at RT and then washed once with PBST and once with double distilled H2O before mounting onto a microscope slide using Vectashield antifade mounting medium and sealed with nail polish. The antibodies used were mouse α-Flag M2, F1804; Sigma-Aldrich (1:250); rabbit α-γH2AX, #9718S; Cell Signaling (1:150); Alexa Fluor 488 goat α-rabbit IgG, A11008; Invitrogen (1:4,000); and Alexa Fluor 647 goat α-mouse IgG, A21235; Invitrogen (1:4,000).
A two-step analysis of γH2AX clusters was done with Volocity software v6.1.2 (PerkinElmer) and with the software R (R Foundation). First, aligned 3D-SIM RGB image stacks were used in Volocity, and the respective channels were separated for the segmentation of γH2AX structures and DAPI-stained nuclei. γH2AX segmentation was performed for all cells with the following commands: (1) Find Objects (threshold using intensity, lower: 32, upper: 255); (2) Separate Touching Objects with an object size guide of 0.002 µm3; and (3) Exclude Objects by Size, excluding structures <0.005 µm3. Besides volume measurements, the centroid position of each γH2AX volume was always registered. For the challenging segmentation of nuclei, we used the commands Find Objects, Dilate, Erode, and Fill Holes in Objects with specific settings. To obtain only γH2AX structures within a nucleus, the Intersect and Compartmentalize commands were used. In a second step, the provided information of γH2AX structures (centroid position, etc.) was transferred to R. Here the Euclidean centroid-to-centroid distances were computed for all nanofoci structures within one nucleus, and then nanofoci with a centroid–centroid distance <500 nm were assigned to the same cluster. Obtained clusters were filtered for structures that contained a minimum of seven nanofoci.
Statistical analysis
Bar graphs show average ± SEM, while violin plots display median (red line) and quartiles (blue dotted lines). Statistical analysis was performed using the software IBM SPSS Statistics v26. Data distribution was assumed to be normal, but this was not formally tested. Comparison of two groups was done by two-sided Student’s t test, while more groups were compared by one-way ANOVA. Two variable comparisons were performed using two-way ANOVA. All post hoc analysis, when necessary, was done using Bonferroni post hoc test. Values of P < 0.05 were considered significant. Significant values are shown by *, P < 0.05; **, P < 0.001; and ***, P < 0.001.
Online supplemental material
Fig. S1 shows that loss of FUS results in an accumulation of DNA damage and sensitization to genotoxic insult in SH-SY5Y cells. Fig. S2 shows that loss of FUS does not affect SG assembly. Fig. S3 shows that FUS is required for efficient DSB repair and DNA damage foci formation upon genotoxic insult. Fig. S4 shows that microirradiation experiments are not influenced by differential expression of KU80-GFP or NBS1-GFP in WT versus FUS-KO cell lines. Fig. S5 shows that loss of FUS specifically affects SFPQ but not XRCC1 recruitment to DSBs. Fig. S6 shows that 2% 1,6-HD treatment does not irreversibly affect the morphology and vitality of cells. Video 1 shows SFPQ recruitment in HeLa WT versus FUS-KO cells.
Supplementary Material
Acknowledgments
We are grateful to J. Stark for the U2OS cells (HR/NHEJ reporters), J. Wang and S. Alberti for FUS constructs, and A. Nussenzweig for the NBS1 construct. We thank G. Chenell (Wohl Cellular Imaging Centre, King’s College London, London, UK) for technical support, M. Colombo for bioinformatics support, D. Jutzi for assistance in generating the FUS-KO cells, and M.E. Bianchi for critically reading the manuscript.
This work was partially supported by the Swiss National Fond Sinergia (grant CRSII3_136222 to O. Mühlemann and S.M.L. Barabino), the UK Dementia Research Institute (to M.-D. Ruepp), the NOMIS Foundation (to M.-D. Ruepp), and the Deutsche Forschungsgemeinschaft (grant LE 721/18-1 to H. Leonhardt and CA 198/16-1 to M.C. Cardoso).
The authors declare no competing financial interests.
Author contributions: B.R. Levone conceived and performed the experiments and the data analysis for Figs. 1–8 and associated supplementary data and participated in the preparation of figures and tables. S.C. Lenzken conceived and performed the experiments and the data analysis for Figs. 1 and 2 and associated supplementary data and participated in the preparation of figures and tables. M. Antonaci performed the experiments in Figs. 1 A, 2 A, and 7 C and associated supplementary data. A. Maiser and A. Rapp performed the experiments and data analysis for Fig. 9. F. Conte performed the experiments in Figs. 1 D and 2 A. S. Reber and M.-D. Ruepp generated KO lines and FUS constructs. J. Mechtersheimer performed the experiment in Fig. S2. A.E. Ronchi, O. Mühlemann, and M.-D. Ruepp critically revised the manuscript. H. Leonhardt and M.C. Cardoso supervised the experiments in Fig. 9 and critically revised the manuscript. S.M.L. Barabino conceived the study and wrote the manuscript. All authors provided discussion and data interpretation and contributed to the final version of the manuscript.
References
- Abbasi, S., and Schild-Poulter C.. 2019. Mapping the Ku Interactome Using Proximity-Dependent Biotin Identification in Human Cells. J. Proteome Res. 18:1064–1077. 10.1021/acs.jproteome.8b00771 [DOI] [PubMed] [Google Scholar]
- Aguirre, N., Beal M.F., Matson W.R., and Bogdanov M.B.. 2005. Increased oxidative damage to DNA in an animal model of amyotrophic lateral sclerosis. Free Radic. Res. 39:383–388. 10.1080/10715760400027979 [DOI] [PubMed] [Google Scholar]
- Akhmedov, A.T., and Lopez B.S.. 2000. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 28:3022–3030. 10.1093/nar/28.16.3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrov, R., Dotchev A., Poser I., Krastev D., Georgiev G., Panova G., Babukov Y., Danovski G., Dyankova T., Hubatsch L., et al. 2018. Protein Dynamics in Complex DNA Lesions. Mol. Cell. 69:1046–1061.e5. 10.1016/j.molcel.2018.02.016 [DOI] [PubMed] [Google Scholar]
- Allodi, I., Comley L., Nichterwitz S., Nizzardo M., Simone C., Benitez J.A., Cao M., Corti S., and Hedlund E.. 2016. Differential neuronal vulnerability identifies IGF-2 as a protective factor in ALS. Sci. Rep. 6:25960. 10.1038/srep25960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechtold, H., Kuroda M., Sok J., Ron D., Lopez B.S., and Akhmedov A.T.. 1999. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 274:34337–34342. 10.1074/jbc.274.48.34337 [DOI] [PubMed] [Google Scholar]
- Bladen, C.L., Udayakumar D., Takeda Y., and Dynan W.S.. 2005. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 280:5205–5210. 10.1074/jbc.M412758200 [DOI] [PubMed] [Google Scholar]
- Boeynaems, S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., et al. 2018. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 28:420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert, E., Boeynaems S., Kato M., Guo L., Caulfield T.R., Steyaert J., Scheveneels W., Wilmans N., Haeck W., Hersmus N., et al. 2018. Molecular Dissection of FUS Points at Synergistic Effect of Low-Complexity Domains in Toxicity. Cell Rep. 24:529–537.e4. 10.1016/j.celrep.2018.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonath, F., Domingo-Prim J., Tarbier M., Friedländer M.R., and Visa N.. 2018. Next-generation sequencing reveals two populations of damage-induced small RNAs at endogenous DNA double-strand breaks. Nucleic Acids Res. 46:11869–11882. 10.1093/nar/gky1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, K.A., Janke A.M., Rhine C.L., and Fawzi N.L.. 2015. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell. 60:231–241. 10.1016/j.molcel.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut, P., Britton S., Coates J., Jackson S.P., and Calsou P.. 2016. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat. Commun. 7:12889. 10.1038/ncomms12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, T.W., Lu C.C., Su C.H., Wu P.Y., Easwvaran S., Lee C.C., Kuo H.C., Hung K.Y., Lee K.M., Tsai C.Y., and Tarn W.Y.. 2019. The RNA Processing Factor Y14 Participates in DNA Damage Response and Repair. iScience. 13:402–415. 10.1016/j.isci.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, P.L., Purman C., Porter S.I., Nganga V., Saini A., Hayer K.E., Gurewitz G.L., Sleckman B.P., Bednarski J.J., Bassing C.H., and Oltz E.M.. 2020. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner. Nat. Commun. 11:3158. 10.1038/s41467-020-16926-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis, J., Hart M.P., Shorter J., DeJesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D., et al. 2011. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. USA. 108:20881–20890. 10.1073/pnas.1109434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro, G., and d’Adda di Fagagna F.. 2017. Transcription and DNA Damage: Holding Hands or Crossing Swords? J. Mol. Biol. 429:3215–3229. 10.1016/j.jmb.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Deng, H., Gao K., and Jankovic J.. 2014a. The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 10:337–348. 10.1038/nrneurol.2014.78 [DOI] [PubMed] [Google Scholar]
- Deng, Q., Holler C.J., Taylor G., Hudson K.F., Watkins W., Gearing M., Ito D., Murray M.E., Dickson D.W., Seyfried N.T., and Kukar T.. 2014b. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci. 34:7802–7813. 10.1523/JNEUROSCI.0172-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann, D., and Haass C.. 2013. Fused in sarcoma (FUS): an oncogene goes awry in neurodegeneration. Mol. Cell. Neurosci. 56:475–486. 10.1016/j.mcn.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Escribano-Díaz, C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 49:872–883. 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Falck, J., Coates J., and Jackson S.P.. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 434:605–611. 10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- Francia, S., Michelini F., Saxena A., Tang D., de Hoon M., Anelli V., Mione M., Carninci P., and d’Adda di Fagagna F.. 2012. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 488:231–235. 10.1038/nature11179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, M., Toth R., Vandermoere F., Morrice N.A., and Rouse J.. 2008. Identification and characterization of FUS/TLS as a new target of ATM. Biochem. J. 415:297–307. 10.1042/BJ20081135 [DOI] [PubMed] [Google Scholar]
- Gunn, A., and Stark J.M.. 2012. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920:379–391. 10.1007/978-1-61779-998-3_27 [DOI] [PubMed] [Google Scholar]
- Hamaguchi, M.S., Watanabe K., and Hamaguchi Y.. 1997. Regulation of intracellular pH in sea urchin eggs by medium containing both weak acid and base. Cell Struct. Funct. 22:387–398. 10.1247/csf.22.387 [DOI] [PubMed] [Google Scholar]
- Harrison, A.F., and Shorter J.. 2017. RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 474:1417–1438. 10.1042/BCJ20160499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Iyer K.S., Ho D., Newcombe E.A., et al. 2015. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 210:529–539. 10.1083/jcb.201504117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, G.G., Singh N., Nashabi A., Mai S., Bozek G., Klewes L., Arapovic D., White E.K., Koury M.J., Oltz E.M., et al. 2000. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 24:175–179. 10.1038/72842 [DOI] [PubMed] [Google Scholar]
- Hirose, T., Yamazaki T., and Nakagawa S.. 2019. Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip. Rev. RNA. 10:e1545. 10.1002/wrna.1545 [DOI] [PubMed] [Google Scholar]
- Hock, E.M., Maniecka Z., Hruska-Plochan M., Reber S., Laferrière F., Sonu Sahadevan M.K., Ederle H., Gittings L., Pelkmans L., Dupuis L., et al. 2018. Hypertonic Stress Causes Cytoplasmic Translocation of Neuronal, but Not Astrocytic, FUS due to Impaired Transportin Function. Cell. Rep. 24:987–1000.e7. 10.1016/j.celrep.2018.06.094 [DOI] [PubMed] [Google Scholar]
- Hwang, S.Y., Kang M.A., Baik C.J., Lee Y., Hang N.T., Kim B.G., Han J.S., Jeong J.H., Park D., Myung K., and Lee J.S.. 2019. CTCF cooperates with CtIP to drive homologous recombination repair of double-strand breaks. Nucleic Acids Res. 47:9160–9179. 10.1093/nar/gkz639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhar, L., Adamson B., Ciccia A., Lewis J., Pontano-Vaites L., Leng Y., Liang A.C., Westbrook T.F., Harper J.W., and Elledge S.J.. 2015. A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Rep. 11:1486–1500. 10.1016/j.celrep.2015.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar, L., Li Z., Li S., and Dynan W.S.. 2017. SFPQ•NONO and XLF function separately and together to promote DNA double-strand break repair via canonical nonhomologous end joining. Nucleic Acids Res. 45:1848–1859. 10.1093/nar/gkw1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, A., and Vale R.D.. 2017. RNA phase transitions in repeat expansion disorders. Nature. 546:243–247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, M. 2016. Roles of RNA-Binding Proteins in DNA Damage Response. Int. J. Mol. Sci. 17:310. 10.3390/ijms17030310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M., and McKnight S.L.. 2018. A Solid-State Conceptualization of Information Transfer from Gene to Message to Protein. Annu. Rev. Biochem. 87:351–390. 10.1146/annurev-biochem-061516-044700 [DOI] [PubMed] [Google Scholar]
- Kato, M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic, S., Lezaja A., Gatti M., Bianco E., Michelena J., Imhof R., and Altmeyer M.. 2019. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38:e101379. 10.15252/embj.2018101379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietsch, J., Caron M.C., Gagné J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., and Masson J.Y.. 2012. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 40:10287–10301. 10.1093/nar/gks798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald, S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., and Alberti S.. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 4:e06807. 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak, M.J., Celeste A., Dellaire G., Fernandez-Capetillo O., Müller W.G., McNally J.G., Bazett-Jones D.P., and Nussenzweig A.. 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172:823–834. 10.1083/jcb.200510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T., and Ron D.. 2000. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J. 19:453–462. 10.1093/emboj/19.3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H., and Paull T.T.. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 308:551–554. 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- Lee, K.H., Zhang P., Kim H.J., Mitrea D.M., Sarkar M., Freibaum B.D., Cika J., Coughlin M., Messing J., Molliex A., et al. 2016. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 167:774–788.e17. 10.1016/j.cell.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., Sadowska A., Bekere I., Ho D., Gully B.S., Lu Y., Iyer K.S., Trewhella J., Fox A.H., and Bond C.S.. 2015. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 43:3826–3840. 10.1093/nar/gkv156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Perez, R., Best G., Nicolay N.H., Greubel C., Rossberger S., Reindl J., Dollinger G., Weber K.J., Cremer C., and Huber P.E.. 2016. Superresolution light microscopy shows nanostructure of carbon ion radiation-induced DNA double-strand break repair foci. FASEB J. 30:2767–2776. 10.1096/fj.201500106R [DOI] [PubMed] [Google Scholar]
- Maharana, S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T.M., et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 360:918–921. 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesini, M., Ogoti Y., Fiorini E., Aktas Samur A., Nezi L., D’Anca M., Storti P., Samur M.K., Ganan-Gomez I., Fulciniti M.T., et al. 2017. ILF2 Is a Regulator of RNA Splicing and DNA Damage Response in 1q21-Amplified Multiple Myeloma. Cancer Cell. 32:88–100.e6. 10.1016/j.ccell.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrocola, A.S., Kim S.H., Trinh A.T., Rodenkirch L.A., and Tibbetts R.S.. 2013. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 288:24731–24741. 10.1074/jbc.M113.497974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, S., Ballif B.A., Smogorzewska A., McDonald E.R. III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316:1160–1166. 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- Michelini, F., Pitchiaya S., Vitelli V., Sharma S., Gioia U., Pessina F., Cabrini M., Wang Y., Capozzo I., Iannelli F., et al. 2017. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19:1400–1411. 10.1038/ncb3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolaskova, B., Jurcik M., Cipakova I., Kretova M., Chovanec M., and Cipak L.. 2018. Maintenance of genome stability: the unifying role of interconnections between the DNA damage response and RNA-processing pathways. Curr. Genet. 64:971–983. 10.1007/s00294-018-0819-7 [DOI] [PubMed] [Google Scholar]
- Mootha, V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Morchikh, M., Cribier A., Raffel R., Amraoui S., Cau J., Severac D., Dubois E., Schwartz O., Bennasser Y., and Benkirane M.. 2017. HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol. Cell. 67:387–399.e5. 10.1016/j.molcel.2017.06.020 [DOI] [PubMed] [Google Scholar]
- Moumen, A., Magill C., Dry K.L., and Jackson S.P.. 2013. ATM-dependent phosphorylation of heterogeneous nuclear ribonucleoprotein K promotes p53 transcriptional activation in response to DNA damage. Cell Cycle. 12:698–704. 10.4161/cc.23592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, T., Qamar S., Lin J.Q., Schierle G.S., Rees E., Miyashita A., Costa A.R., Dodd R.B., Chan F.T., Michel C.H., et al. 2015. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 88:678–690. 10.1016/j.neuron.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster, B., Rapp A., and Cardoso M.C.. 2017. Systematic analysis of DNA damage induction and DNA repair pathway activation by continuous wave visible light laser micro-irradiation. AIMS Genet. 4:47–68. 10.3934/genet.2017.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myler, L.R., Gallardo I.F., Soniat M.M., Deshpande R.A., Gonzalez X.B., Kim Y., Paull T.T., and Finkelstein I.J.. 2017. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell. 67:891–898.e4. 10.1016/j.molcel.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale, F., Rapp A., Yu W., Maiser A., Harz H., Scholl A., Grulich S., Anton T., Hörl D., Chen W., et al. 2017. Identification of the elementary structural units of the DNA damage response. Nat. Commun. 8:15760. 10.1038/ncomms15760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann, M., Pal A., Goswami A., Lojewski X., Japtok J., Vehlow A., Naujock M., Günther R., Jin M., Stanslowsky N., et al. 2018. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat. Commun. 9:335. 10.1038/s41467-017-02299-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. 2015. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Paulsen, R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E., et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 35:228–239. 10.1016/j.molcel.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessina, F., Giavazzi F., Yin Y., Gioia U., Vitelli V., Galbiati A., Barozzi S., Garre M., Oldani A., Flaus A., et al. 2019. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 21:1286–1299. 10.1038/s41556-019-0392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S.E., Blackford A.N., Chapman J.R., Baskcomb L., Gravel S., Rusch A., Thomas A., Blundred R., Smith P., Kzhyshkowska J., et al. 2012. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell. 45:505–516. 10.1016/j.molcel.2011.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska, K.D., Ruepp M.D., Brzek A., Reber S., Romeo V., Rindlisbacher B., Heller M., Szweykowska-Kulinska Z., Jarmolowski A., and Schümperli D.. 2015. FUS/TLS contributes to replication-dependent histone gene expression by interaction with U7 snRNPs and histone-specific transcription factors. Nucleic Acids Res. 43:9711–9728. 10.1093/nar/gkv794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajesh, C., Baker D.K., Pierce A.J., and Pittman D.L.. 2011. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 39:132–145. 10.1093/nar/gkq738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber, S., Lindsay H., Devoy A., Jutzi D., Mechtersheimer J., Domanski M., Mühlemann O., Barabino S.M.L., and Ruepp M.-D.. 2019. The phase separation-dependent FUS interactome reveals nuclear and cytoplasmic function of liquid-liquid phase separation. BioRxiv. 10.1101/806158 (Preprint posted October 15, 2019) [DOI] [PMC free article] [PubMed]
- Reber, S., Mechtersheimer J., Nasif S., Benitez J.A., Colombo M., Domanski M., Jutzi D., Hedlund E., and Ruepp M.D.. 2018. CRISPR-Trap: a clean approach for the generation of gene knockouts and gene replacements in human cells. Mol. Biol. Cell. 29:75–83. 10.1091/mbc.E17-05-0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber, S., Stettler J., Filosa G., Colombo M., Jutzi D., Lenzken S.C., Schweingruber C., Bruggmann R., Bachi A., Barabino S.M., et al. 2016. Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 35:1504–1521. 10.15252/embj.201593791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton, M., Lerenthal Y., Wang S.Y., Chen D.J., and Shiloh Y.. 2010. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 9:1568–1576. 10.4161/cc.9.8.11298 [DOI] [PubMed] [Google Scholar]
- Sama, R.R., Ward C.L., Kaushansky L.J., Lemay N., Ishigaki S., Urano F., and Bosco D.A.. 2013. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J. Cell Physiol. 228:2222–2231. 10.1002/jcp.24395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, A., Jeggo P., and Löbrich M.. 2018. The pendulum of the Ku-Ku clock. DNA Repair (Amst.). 71:164–171. 10.1016/j.dnarep.2018.08.020 [DOI] [PubMed] [Google Scholar]
- Shing, D.C., McMullan D.J., Roberts P., Smith K., Chin S.F., Nicholson J., Tillman R.M., Ramani P., Cullinane C., and Coleman N.. 2003. FUS/ERG gene fusions in Ewing’s tumors. Cancer Res. 63:4568–4576. [PubMed] [Google Scholar]
- Simon, N.E., Yuan M., and Kai M.. 2017. RNA-binding protein RBM14 regulates dissociation and association of non-homologous end joining proteins. Cell Cycle. 16:1175–1180. 10.1080/15384101.2017.1317419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singatulina, A.S., Hamon L., Sukhanova M.V., Desforges B., Joshi V., Bouhss A., Lavrik O.I., and Pastré D.. 2019. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep. 27:1809–1821.e5. 10.1016/j.celrep.2019.04.031 [DOI] [PubMed] [Google Scholar]
- Stucki, M., and Jackson S.P.. 2006. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst.). 5:534–543. 10.1016/j.dnarep.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, J., Lin Y.F., Xu K., Lee K.J., Wang D., and Chen B.P.. 2015. DNA-PKcs phosphorylates hnRNP-A1 to facilitate the RPA-to-POT1 switch and telomere capping after replication. Nucleic Acids Res. 43:5971–5983. 10.1093/nar/gkv539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., Diaz Z., Fang X., Hart M.P., Chesi A., Shorter J., and Gitler A.D.. 2011. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 9:e1000614. 10.1371/journal.pbio.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike, D.L., Hachey S.J., Kreher J., and Strome S.. 2011. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192:939–948. 10.1083/jcb.201010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., et al. 2018. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell. 174:688–699.e16. 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.Y., Pan L., Su S.C., Quinn E.J., Sasaki M., Jimenez J.C., Mackenzie I.R., Huang E.J., and Tsai L.H.. 2013. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 16:1383–1391. 10.1038/nn.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, C.L., Boggio K.J., Johnson B.N., Boyd J.B., Douthwright S., Shaffer S.A., Landers J.E., Glicksman M.A., and Bosco D.A.. 2014. A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death Dis. 5:e1572. 10.1038/cddis.2014.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, J.A., Mito M., Kurosaka S., Takumi T., Tanegashima C., Chujo T., Yanaka K., Kingston R.E., Hirose T., Bond C., et al. 2016. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 214:817–830. 10.1083/jcb.201601071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, T., Souquere S., Chujo T., Kobelke S., Chong Y.S., Fox A.H., Bond C.S., Nakagawa S., Pierron G., and Hirose T.. 2018. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell. 70:1038–1053.e7. 10.1016/j.molcel.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Zerkalenkova, E., Panfyorova A., Kazakova A., Baryshev P., Shelihova L., Kalinina I., Novichkova G., Maschan M., Maschan A., and Olshanskaya Y.. 2018. Molecular characteristic of acute leukemias with t(16;21)/FUS-ERG. Ann. Hematol. 97:977–988. 10.1007/s00277-018-3267-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.