Here, we report the coding-complete genome sequences of nine clinical severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants and their mutations. The samples were collected from nine Bangladeshi coronavirus disease 2019 (COVID-19) patients. We have identified the E484K escape mutation and the S359T mutation within the spike protein coding region of the sequenced genomes.
ABSTRACT
Here, we report the coding-complete genome sequences of nine clinical severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants and their mutations. The samples were collected from nine Bangladeshi coronavirus disease 2019 (COVID-19) patients. We have identified the E484K escape mutation and the S359T mutation within the spike protein coding region of the sequenced genomes.
ANNOUNCEMENT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the Betacoronavirus genus in the Coronaviridae family, has been responsible for more than 2 million deaths globally (1). In this study, we performed coding-complete genome sequencing of nine clinical SARS-CoV-2 isolates to observe genomic and inferred proteomic mutational variations (Table 1). The samples were collected at Gonoshasthaya-RNA Molecular Diagnostic and Research Center (Dhaka, Bangladesh) within the period from October to December 2020, with ethical clearance from the National Research Ethics Committee (approval number BMRC/NREC/2019-2022/697). The samples were identified as positive by the novel coronavirus (2019-nCoV) nucleic acid diagnostic kit (Sansure, Inc., China) and had cycle threshold (CT) values ranging from 27 to 30, which, according to the kit’s information, implies high viral load.
TABLE 1.
Genomic features of nine SARS-CoV-2 clinical samples
| Sample no. | BioSample no. | SRA accession no. (raw reads) | GenBank accession no. | Total no. of reads | No. of mapped reads | Avg coverage (×) | Assembly length (bp) | GC content (%) | PANGO lineage | Emerging clade |
|---|---|---|---|---|---|---|---|---|---|---|
| GRBL_S1 | SAMN17359583 | SRR13449688 | MW532093 | 326,252 | 324,957 | 39.98 | 29,847 | 38.01 | B.1.1.103 | 20B |
| GRBL_S2 | SAMN17359584 | SRR13449687 | MW532094 | 7,951,736 | 7,940,015 | 974.5 | 29,858 | 38.01 | B.1.1.103 | 20B |
| GRBL_S3 | SAMN17359585 | SRR13449686 | MW532095 | 208,394 | 203,220 | 25.54 | 29,842 | 38.03 | B.1.1.103 | 20B |
| GRBL_S4 | SAMN17359586 | SRR13449685 | MW532096 | 1,526,918 | 1,473,009 | 187.13 | 29,856 | 38.01 | B.1.1.103 | 20B |
| GRBL_S6 | SAMN17359587 | SRR13449684 | MW532097 | 353,644 | 345,476 | 43.34 | 29,823 | 38.37 | B.1.1.316 | 20B |
| GRBL_S9 | SAMN17359588 | SRR13449683 | MW532098 | 6,758,972 | 6,749,017 | 828.32 | 29,863 | 37.99 | B.1.1.316 | 20B |
| GRBL_S10 | SAMN17359589 | SRR13449682 | MW532099 | 126,716 | 125,886 | 15.53 | 29,850 | 38.00 | B.1.1.25 | 20B |
| GRBL_S11 | SAMN17359590 | SRR13449681 | MW532100 | 11,244,412 | 11,228,723 | 1,378.02 | 29,868 | 37.99 | B.1.1.25 | 20F |
| GRBL_S14 | SAMN17359592 | SRR13449679 | MW532101 | 151,570 | 150,440 | 18.58 | 29,849 | 38.00 | B.1.1.316 | 20B |
The genomic viral RNA was purified from both nasopharyngeal and oropharyngeal swabs from individuals (both male and female patients; age range, 17 to 68 years) suspected of being infected with SARS-CoV-2, using the ReliaPrep viral TNA miniprep system (Promega) according to the manufacturer’s instructions. This was followed by preparation of libraries using Illumina RNA preparation with enrichment in combination with the Illumina respiratory virus oligonucleotide panel v2 according to the manufacturer’s instructions (Illumina, Inc., San Diego, CA). The sequencing was carried out in an Illumina MiniSeq instrument implementing a paired-end protocol (read length, 74 bp). The Fastq sequences were trimmed, quality controlled, and mapped and a consensus sequence was generated using DRAGEN v3.5.1.15 (Illumina) (2). The genome coverages and mutations were initially checked with SAMtools and Snippy in comparison with the Wuhan reference genome (GenBank accession number MN996528) (3–6). The novel mutations were confirmed with the EpiCov tool, integrated in the Global Initiative on Sharing All Influenza Data (GISAID) database (7). All mutations and deletions were confirmed by area-wise coverage using Snippy (5).
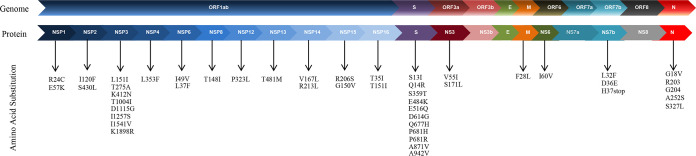
Eight strains (GRBL_S1, GRBL_S2, GRBL_S3, GRBL_S4, GRBL_S6, GRBL_S9, GRBL_S10, and GRBL_S14) belong to the emerging clade 20B, whereas one strain (GRBL_S11) is affiliated with the 20F clade (Table 1). In this announcement, among the multitude of mutations, we are reporting two mutations, one at position 1450 from G (0 evidence) to A (267 evidence) in GRBL_S1, which results in the amino acid substitution of E484K, and one at position 1076 from G (0 evidence) to C (4,559 evidence) in GRBL_S9, which results in S359T in the spike protein region (“evidence” relates to area-wise coverage obtained by Snippy tools for short-read sequences generated by Illumina MiniSeq). The former mutation was reported in the South African variant as an escape mutation (8, 9). The E484K substitution was observed in a cluster containing D614G, P681H, and S13I changes (GRBL_S1), while the S359T amino acid substitution was observed in a cluster containing D614G and A942V changes (GRBL_S9). Additionally, we found cluster substitutions D614G, Q677H, and A871V in the spike protein of GRBL_S10. Within the open reading frame 1ab (ORF1ab) region, we observed amino acid substitutions of I1257S in GRBL_S1 and L151I and T275A in GRBL_S14. The complete list of amino acid substitutions is provided in Fig. 1.
FIG 1.
Complete list of amino acid substitutions in the genomes of nine different clinical SARS-CoV-2 isolates, with reference to isolate WIV04 (GenBank accession number MN996528).
It is also noteworthy that in two samples, GRBL_S2 (2,446 evidence) and GRBL_S3 (68 evidence), the deletion of GATCAT and its subsequent replacement by G at nucleotide position 108 of ORF7b resulted in the introduction of a stop codon, ultimately resulting in a frameshift mutation and D36E substitution, potentially without any loss of function (10). The patients harboring these variants were detected as family members residing within the same household. Substitutions pertaining to the nucleocapsid protein coding region were S327L, G18V, and A252S in samples GRBL_S1, GRBL_S11, and GRBL_S14, respectively. Further investigations are required to identify the effects of the substitutions on the outcomes of coronavirus disease 2019 (COVID-19).
Data availability.
The sequences of nine SARS-CoV-2 genomes were submitted to the GISAID database under the identifiers EPI_ISL_774976, EPI_ISL_775019, EPI_ISL_775020, EPI_ISL_890189, EPI_ISL_775215, EPI_ISL_775218, EPI_ISL_890192, EPI_ISL_890193, and EPI_ISL_890194 and to the NCBI GenBank under the accession numbers MW532093, MW532094, MW532095, MW532096, MW532097, MW532098, MW532099, MW532100, and MW532101 (Table 1). The raw reads were submitted to the NCBI SRA under BioProject accession number PRJNA692653 and SRA accession number SRP302071.
REFERENCES
- 1.World Health Organization. 2020. WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int. Accessed 31 January 2021.
- 2.Goyal A, Kwon H, Lee K, Garg R, Yun S, Hee Kim Y, Lee S, Seob Lee M. 2017. Ultra-fast next generation human genome sequencing data processing using DRAGENTM Bio-IT processor for precision medicine. Open J Genet 7:9–19. doi: 10.4236/ojgen.2017.71002. [DOI] [Google Scholar]
- 3.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etherington GJ, Ramirez-Gonzalez RH, MacLean D. 2015. bio-samtools 2: a package for analysis and visualization of sequence and alignment data with SAMtools in Ruby. Bioinformatics 31:2565–2567. doi: 10.1093/bioinformatics/btv178. [DOI] [PubMed] [Google Scholar]
- 5.Seeman T. 2015. Snippy: rapid haploid variant calling and core SNP phylogeny. https://github.com/tseemann/snippy.
- 6.Koyama T, Platt D, Parida L. 2020. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ 98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall 1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao M, Korsman S, Davies M-A, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Pond SLK, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. 2020. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.12.21.20248640. doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 9.Collier DA, De Marco A, Ferreira IATM, Meng B, Datir R, Walls AC, Kemp SA, Bassi J, Pinto D, Fregni CS, Bianchi S, Tortorici MA, Bowen J, Culap K, Jaconi S, Cameroni E, Snell G, Pizzuto MS, Pellanda AF, Garzoni C, Riva A, Elmer A, Kingston N, Graves B, McCoy LE, Smith KG, Bradley JR, Thaventhiran JJ, Ceron-Gutierrez L, Barcenas-Morales G, Virgin HW, Lanzavecchia A, Piccoli L, Doffinger R, Wills M, Veesler D, Corti D, Gupta RK. 2021. SARS-CoV-2 B.1.1.7 escape from mRNA vaccine-elicited neutralizing antibodies. medRxiv 2021.01.19.21249840. doi: 10.1101/2021.01.19.21249840. [DOI] [Google Scholar]
- 10.Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, Zhuang Y, Kalimuddin S, Low JGH, Tan CW, Chia WN, Mak TM, Octavia S, Chavatte JM, Lee RTC, Pada S, Tan SY, Sun L, Yan GZ, Maurer-Stroh S, Mendenhall IH, Leo YS, Lye DC, Wang LF, Smith GJD. 2020. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio 11:e01610-20. doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences of nine SARS-CoV-2 genomes were submitted to the GISAID database under the identifiers EPI_ISL_774976, EPI_ISL_775019, EPI_ISL_775020, EPI_ISL_890189, EPI_ISL_775215, EPI_ISL_775218, EPI_ISL_890192, EPI_ISL_890193, and EPI_ISL_890194 and to the NCBI GenBank under the accession numbers MW532093, MW532094, MW532095, MW532096, MW532097, MW532098, MW532099, MW532100, and MW532101 (Table 1). The raw reads were submitted to the NCBI SRA under BioProject accession number PRJNA692653 and SRA accession number SRP302071.