Sir,
We read with great interest the report by Peterson et al.1 detailing 10 molecular assays for prediction of susceptibilities in nucleic acid amplification test specimens of Neisseria gonorrhoeae in Canada. Antimicrobial resistance in N. gonorrhoeae is an urgent global health threat, as the organism has developed resistance to every class of antibiotic currently available.2 Dual therapy with ceftriaxone and azithromycin is recommended in most settings, although some countries use ceftriaxone monotherapy.3,4
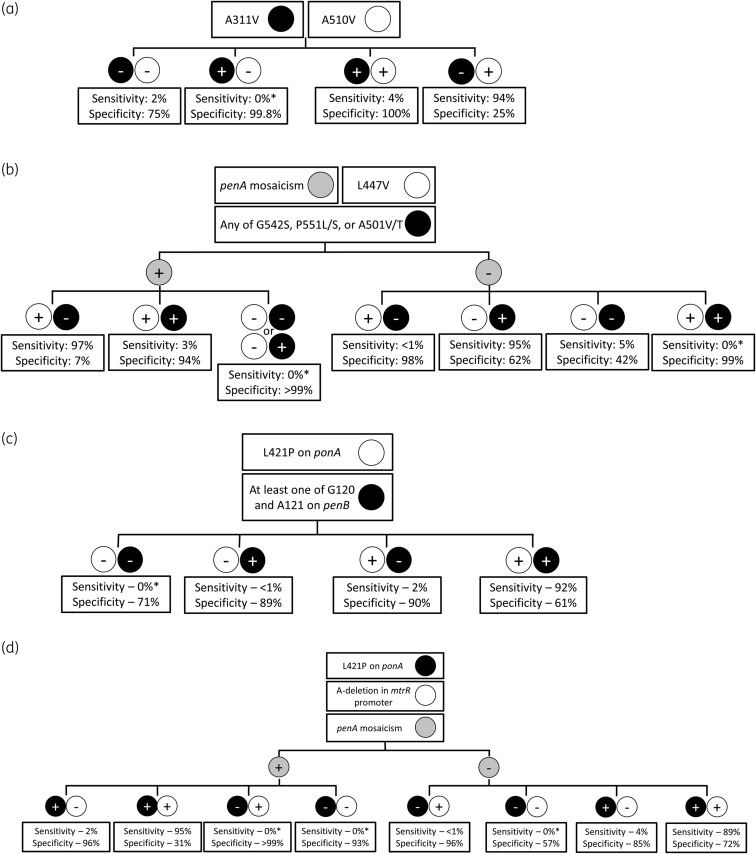
We compiled a global set of publicly available genetic data of N. gonorrhoeae isolates from 73 published reports through 15 October 2019 to propose four molecular algorithms to predict decreased susceptibility to ceftriaxone (defined as an MIC >0.064 mg/L).5 That breakpoint was based upon prior work using 0.06 mg/L to define decreased susceptibility to ceftriaxone, as a midpoint between susceptibility and resistance.6–8 The dataset included N. gonorrhoeae strains isolated from 1996 to 2019 and are from 23 countries, with the 5 largest countries represented by the USA (30%), Russia (14%), Canada (12%), New Zealand (11%) and China (8%). Sensitivities were calculated by dividing the number of isolates with decreased susceptibility to ceftriaxone and the genetic mutation by the total number of isolates with decreased susceptibility to ceftriaxone. Specificities were calculated by dividing the number of isolates with susceptibility to ceftriaxone without the genetic mutation by the total number of isolates with susceptibility to ceftriaxone. Our proposed algorithms differ in whether they (i) utilized penA or non-penA genes (penB, ponA, mtrR) and (ii) utilized penA mosaicism. These four algorithms are not intended to be used simultaneously. Rather, they show the flexibility in using different genetic targets that can be used in different settings. Each resulted in either high sensitivity or specificity with moderate complementary specificity and sensitivity (Figure 1), with our highest sensitivity and specificity values being 95% and 62%, respectively.
Figure 1.
Molecular algorithms previously reported that utilize (a) penA without mosaicism determination, (b) penA with mosaicism determination, (c) non-penA without mosaicism determination and (d) non-penA with mosaicism determination. Sensitivity and specificity values are for decreased susceptibility to ceftriaxone. Sensitivity was calculated by dividing the number of isolates with decreased susceptibility to ceftriaxone with the targeted genetic modification by the total number of isolates with decreased susceptibility to ceftriaxone. Specificity was calculated by dividing the number of isolates with susceptibility to ceftriaxone without the targeted genetic modification by the total number of isolates with susceptibility to ceftriaxone. Testing for all genetic loci in these algorithms is intended to be done simultaneously and not necessarily in a step-wise fashion. An asterisk indicates that no decreased susceptible strains have been reported with this combination of amino acid alterations.
Peterson et al.1 developed an assay to predict intermediate to decreased susceptibility of N. gonorrhoeae to ceftriaxone using genetic alterations in penA (A311V, A501, N513Y, G545S), ponA (L421P), penB (G120/A121) and mtrR (−35delA). In our analysis, N513Y and G545S were not important predictors of decreased susceptibility to ceftriaxone. Strains with either penA A311V, penA A501 and any two other SNPs with MICs ≥0.125 mg/L (decreased susceptibility) or any three of the aforementioned SNPs with MICs 0.032 to ≥0.125 mg/L (intermediate) were deemed true positive for intermediate/decreased susceptibility, while strains with fewer than three SNPs and MICs <0.125 mg/L were deemed true negative for susceptibility. Applying their assay to a set of Canadian isolates, Peterson et al.1 reported sensitivity and specificity of 99.8% and 89.0%, respectively. Those promising values in a Canadian dataset prompt the question of the generalizability to a global set of N. gonorrhoeae genetic sequences.
Applying all of the genetic targets proposed by Peterson et al.1 to our global collection of isolates, we calculated a comparable sensitivity of 98.4%, but a lower specificity of 67.3%. The decreased specificity is consistent with our observations applying prior assay targets for predicting decreased susceptibility to ceftriaxone in our algorithms, likely due to each of those assays being developed using strains limited to a specific country or region.5 For example, in another report utilizing Canadian strains, Peterson et al.9 found that L421P in ponA resulted in sensitivity and specificity of 100% and 57.4%, respectively, for predicting decreased susceptibility to ceftriaxone. In contrast, we reported 99.6% sensitivity and 45.4% specificity using that mutation in a global set, although both single SNP-based assays suffer from low specificity.5 The genetic diversity of N. gonorrhoeae globally limits the generalizability of those developed assays. This was consistent with our expectations, as one critical limitation of a global SNP-based assay is the geographical and temporal variation due to N. gonorrhoeae’s ability to rapidly evolve resistance mechanisms.
We analysed the geographical distribution of strains producing false positive (falsely predicted to be decreased susceptible) results in our analysis of the Peterson et al.1 targets and found that only 26 (4.2%) of the false positive strains were from Canada. The USA accounted for over half of the strains with false positive results (350/622, 56.3%), followed by Australia (81/622, 13.0%) and New Zealand (81/622, 13.0%). However, this could arise from bias of publicly available whole-genome sequenced strains, as the USA accounted for 30% (1142/3821) of the dataset’s isolates, while Canada accounted for 12% (459/3821), New Zealand accounted for 11% (411/3821) and Australia accounted for 5% (178/3821). Of the US strains, 31% (350/1142) were false positive, while only 6% (26/459) of Canada’s strains were false positive. To study how those molecular assays might perform in another country, we applied the genetic targets reported by Peterson et al.1 to the US subset of sequences and found sensitivity and specificity of 98.1% and 56.0%, respectively. This higher proportion of false positives and decrease in specificity suggest limited generalizability of algorithms created from region-specific strains.
Finally, our proposed algorithms resulted in slightly lower sensitivity (95%) and specificity (62%) when applying the assay targets of Peterson et al.1 to our global collection of isolates, compared with sensitivity of 98.4% and specificity of 67.3% in their report. Our report used different MIC breakpoints, which might have contributed to the observed differences. The MIC breakpoint of 0.032 mg/L used by Peterson et al.1 was based on treatment failures observed with cefixime (not ceftriaxone), yet most other reports utilize intermediate cut-offs between ceftriaxone susceptibility and resistance in N. gonorrhoeae with MICs of 0.06–0.064 mg/L.6–8 Moreover, strains with fewer than three SNPs are predicted to be susceptible (MICs <0.125 mg/L), while strains with three or more SNPs (excluding penA A311V, penA A501) are predicted to be intermediate/decreased susceptible (MICs ≥0.032 mg/L), leading to overlap in predicted MICs between categories. The complexity of cephalosporin resistance in N. gonorrhoeae is such that there is no consensus between CLSI and EUCAST breakpoints, which has led to varying cut-off points in the literature, making comparisons between reports difficult.
Applying the genetic targets of the Peterson et al.1 assay in a global set of N. gonorrhoeae sequences, we have demonstrated a similar sensitivity, with a decrease in specificity. The lower specificity suggests the assay may not be generalizable to other countries, despite promising results in Canadian isolates. Regardless, the assay developed by Peterson et al.1 shows the potential value of using molecular methods to predict decreased susceptibility to ceftriaxone. Moving forward, we encourage (i) the use of the same MIC cut-offs for assay development to avoid mis-classification and allow for direct comparisons between studies and (ii) the search for a global set of genetic alterations that can be utilized to predict decreased susceptibility to ceftriaxone in N. gonorrhoeae, as a universal molecular assay is much more likely to be commercially viable. To accomplish this goal, continued expansion of genomic surveillance of genetic loci associated with ceftriaxone resistance is critical, and testing algorithms should continuously be evaluated, in conjunction with WGS and phenotypic testing, to ensure accuracy and reliability.
Funding
This work was supported by the National Institutes of Health (R21 AI157817 to J.D.K. and T32MH080634 to P.C.A.).
Transparency declarations
None to declare.
References
- 1. Peterson SW, Martin I, Demczuk W. et al. Multiplex real-time PCR assays for the prediction of cephalosporin, ciprofloxacin and azithromycin antimicrobial susceptibility of positive Neisseria gonorrhoeae nucleic acid amplification test samples. J Antimicrob Chemother 2020; 75: 3485–90. [DOI] [PubMed] [Google Scholar]
- 2. Ohnishi M, Golparian D, Shimuta K. et al. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother 2011; 55: 3538–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. WHO Guidelines for the Treatment of Neisseria gonorrhoeae. https://apps.who.int/iris/handle/10665/246114. 2016. [PubMed]
- 4. Fifer H, Saunders J, Soni S. et al. 2018 UK national guideline for the management of infection with Neisseria gonorrhoeae. Int J STD AIDS 2020; 31: 4–15. [DOI] [PubMed] [Google Scholar]
- 5. Lin EY, Adamson PC, Deng X. et al. Establishing novel molecular algorithms to predict decreased susceptibility to ceftriaxone in Neisseria gonorrhoeae strains. J Infect Dis 2020; doi:10.1093/infdis/jiaa495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allen VG, Seah C, Martin I. et al. Azithromycin resistance is coevolving with reduced susceptibility to cephalosporins in Neisseria gonorrhoeae in Ontario, Canada. Antimicrob Agents Chemother 2014; 58: 2528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gianecini RA, Zittermann S, Oviedo C. et al. Use of whole genome sequencing for the molecular comparison of Neisseria gonorrhoeae isolates with decreased susceptibility to extended spectrum cephalosporins from 2 geographically different regions in America. Sex Transm Dis 2019; 46: 548–55. [DOI] [PubMed] [Google Scholar]
- 8. de Laat MM, Wind CM, Bruisten SM. et al. Ceftriaxone reduced susceptible Neisseria gonorrhoeae in the Netherlands, 2009 to 2017: from penA mosaicism to A501T/V nonmosaicism. Sex Transm Dis 2019; 46: 594–601. [DOI] [PubMed] [Google Scholar]
- 9. Peterson SW, Martin I, Demczuk W. et al. Molecular assay for detection of genetic markers associated with decreased susceptibility to cephalosporins in Neisseria gonorrhoeae. J Clin Microbiol 2015; 53: 2042–8. [DOI] [PMC free article] [PubMed] [Google Scholar]