Abstract
Objectives
The worldwide emergence of antibiotic resistance calls for effective exploitation of existing antibiotics. Antibiotic combinations with different modes of action can synergize for successful treatment. In the present study, we used microcalorimetry screening to identify synergistic combination treatments against clinical MDR isolates. The synergistic effects were validated in a murine infection model.
Methods
The synergy of meropenem combined with colistin, rifampicin or amikacin was tested on 12 isolates (1 Escherichia coli, 5 Klebsiella pneumoniae, 3 Pseudomonas aeruginosa and 3 Acinetobacter baumannii) in an isothermal microcalorimeter measuring metabolic activity. One A. baumannii strain was tested with two individual pairings of antibiotic combinations. The microcalorimetric data were used to predict in vivo efficacy in a murine peritonitis/sepsis model. NMRI mice were inoculated intraperitoneally and after 1 h treated with saline, drug X, drug Y or X+Y. Bacterial load was determined by cfu in peritoneal fluid and blood after 4 h.
Results
In vitro, of the 13 combinations tested on the 12 strains, 3 of them exhibited a synergistic reduction in MIC (23% n = 3/13), 5 showed an additive effect (38.5% n = 5/13) and 5 had indifferent or antagonistic effects (38.5% n = 5/13). There was a significant correlation (P = 0.024) between microcalorimetry-screening FIC index values and the log reduction in peritoneal fluid from mice that underwent combination treatment compared with the most effective mono treatment. No such correlation could be found between chequerboard and in vivo results (P = 0.16).
Conclusions
These data support microcalorimetic metabolic readout to predict additive or synergistic effects of combination treatment of MDR infections within hours.
Introduction
The ability to treat and eradicate bacterial infection with antibiotics is at the very core of modern medicine. Worldwide, the number of pathogenic bacterial strains resistant to multiple types of antibiotics is increasing and the WHO has called for action on a global scale to combat bacterial MDR.1,2
The increase in novel types of resistance combined with the limited development of new types of antibiotics leaves few other options than optimized treatment with currently available drugs.
One possible approach to optimize the treatment of resistant bacteria with current antibiotics is treatments combining two or more types of antibiotics. Combination therapy can have several benefits, such as giving initial broad-spectrum coverage therapy of severely infected patients before diagnosis or being effective against polymicrobial infections.3,4 When treating MDR strains, combination treatment may provide synergistic effects where otherwise resistant strains become susceptible when two or more drugs are administered.5 The synergistic effects can be a result of one drug directly targeting the resistance mechanisms of the other drug, such as the inhibition of β-lactamase enzymes, or can be caused by drugs indirectly targeting resistance by blocking signalling pathways.6
The standard method of detecting additive or synergistic effects of multidrug treatment as part of routine diagnostics is cumbersome, uncertain and slow. Biomass-based disc cross-diffusion assays, microtitre synergy assays and gradient tests are all associated with an extended incubation period before being evaluated, which may result in a late start of appropriate antibiotic treatment and aggravation of the infection.
Microcalorimetry is an emerging technology in microbiology for metabolic phenotyping that has recently been shown to provide a fast and reliable analysis of MICs for clinical isolates.7 Antibiotic treatment of bacterial strains with a specific metabolic phenotype can provide an assessment of a specific treatment efficacy within hours of inoculation. Microcalorimetry allows MIC determination for a metabolic phenotype and provides a very sensitive measurement with the possibility to assess single or combination drug efficacy with minimal assay development.
Our aims with this study were to: (i) investigate the correlation between antibiotic combination effectiveness in vitro and the clearance rate of MDR bacteria in a murine peritonitis/sepsis infection model; and (ii) compare the predictive value of traditional biomass-based and novel metabolic phenotype-based synergy screening assays. To the best of our knowledge, this is the largest in vivo investigation of the correlation between the in vitro synergy/additive/indifferent activity of combinations of antibiotics and the outcome of in vivo reduction of infectious load (cfu).
Materials and methods
Clinical isolates and study design
The isolates studied (n = 12) were obtained by four clinical microbiological laboratories (Hospital Universitario Ramón y Cajal, Spain; Erasmus Medical Center, The Netherlands; Rigshospitalet, Copenhagen, Denmark; and Careggi University Hospital, Florence, Italy) within the European Project H2020-SME-Inst-2–2016-2017 (grant agreement 784514) (Table 1). These isolates expressing MDR phenotypes, including ESBLs and carbapenemases, were used for in vitro synergy tests, time–kill assays and chequerboard assays. Findings from in vitro assays were evaluated in vivo in a murine peritonitis/sepsis model. One Acinetobacter baumannii strain (Ab_27) was tested with two different drug combinations, meropenem/rifampicin and meropenem/colistin, as these combinations showed indifferent and synergistic effects, respectively, on the same strain. This was done to test strain/drug combination specificity in both screening assays and the in vivo model.
Table 1.
Characteristics, MICs and resistance profiles for the isolates
| Isolate (code) | Source | Year of isolation | Clone | Mechanism of resistance | Origin | Patient’s ward | MEM MIC (mg/L) | AMK MIC (mg/L) | CST MIC (mg/L) | RIF MIC (mg/L) | Other resistance profile |
|---|---|---|---|---|---|---|---|---|---|---|---|
| K. pneumoniae (RYC-Kp12) | Madrid, Spain | 2018 | ST-147 | plasmid AmpC+ porin deficiency+ SHV-11 | blood culture | Gastroenterology | 1 | 32 | 0.5 | 64 | TZP, CTX, CAZ, FEP, ATM, IPM, GEN, TOB, CIP, SXT |
| K. pneumoniae (RYC-Kp17) | Madrid, Spain | 2018 | ST-307 | SHV-28+TEM-1 +CTX-M-15 | blood culture | Surgical ICU | 8 | 8 | 0.5 | 32 | TZP, CTX, CAZ, FEP, ATM, GEN, CIP, |
| K. pneumoniae (RYC-Kp30) | Madrid, Spain | 2018 | ST-307 | KPC-3+CTX-M-15 | respiratory sample | Nephrology | 16 | <8 | 4 | 16 | TZP, CTX, CAZ, FEP, ATM, IPM, GEN, TOB, CIP, SXT |
| K. pneumoniae (RYC-Kp47) | Madrid, Spain | 2018 | ST-11 | OXA-48+CTX-M- 15+SHV-11 | urine | Emergency | 16 | 32 | 0.5 | – | TZP, CTX, CAZ, FEP, ATM, IPM, CIP, FOS |
| K. pneumoniae (RYC-Kp46) | Madrid, Spain | 2018 | ST-307 | KPC-3+TEM- 1+SHV-28 | abscess | Vascular Surgery | 4 | <8 | 0.5 | >64 | TZP, CTX, CAZ, FEP, ATM, IPM, GEN, CIP, SXT |
| P. aeruginosa (RYC-Pa20) | Madrid, Spain | 2018 | ST-244 | unknown | rectal swab | Preventive Medicine | 2 | 64 | 0.5 | 32 | TZP, CAZ, FEP, ATM, IPM, LVX, SXT |
| A. baumannii (EMC-Ab43) | Rotterdam, The Netherlands | 2015 | ST-448 | TEM-1+OXA-23 | wound fluid | Dermatology | 32 | >64 | 4 | 2 | TZP, IPM, CAZ, AMK, CIP, LVX |
| A. baumannii (EMC-Ab55) | Rotterdam, The Netherlands | 2016 | ST-391 | OXA-23 | sputum | ICU | 32 | >64 | 4 | 64 | TZP, CAZ, GN, TOB, SXT, CIP, CST |
| Escherichia coli (EMC-Ec-NFM-104) | Copenhagen, Denmark | 2010 | ST-131 | SHV-26+IMP-1 | blood | not available | 32 | >64 | 4 | 64 | TZP, CTX, CAZ, CIP |
| A. baumannii (KI-Ab27) | Iran | 2011 | ST-947 | OXA-23+OXA-51 | unknown | not available | 32 | 32 | 8 | 32 | IPM, CIP |
| P. aeruginosa (KI-Pa15) | Italy | 2003 | ST-235 | VIM-1 | unknown | not available | 32 | 32 | 4 | 64 | TZP, CAZ, IPM, CIP |
| P. aeruginosa (KI-Pa12) | Italy | 2004 | ST-111 | VIM-2 | unknown | not available | 16 | 16 | 0.5 | – | TZP, CAZ, IPM, CIP |
MEM, meropenem; AMK, amikacin; CST, colistin; RIF, rifampicin; TZP, piperacillin/tazobactam; CTX, cefotaxime (not tested against P. aeruginosa and A. baumannii); CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; IPM, imipenem; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole (not tested against P. aeruginosa and A. baumannii); FOS, fosfomycin; LVX, levofloxacin.
MIC testing, chequerboard assay and synergy testing based on metabolic phenotype screening
MICs were determined by standard broth microdilution following ISO recommendations (ISO 20776–1)8 and interpreted following EUCAST 2019 criteria.9 In the absence of clinical breakpoints, the epidemiological cut-off (‘ECOFF’) values were used to define resistance phenotypes (https://mic.eucast.org/Eucast2/). The chequerboard assay was performed in Mueller–Hinton broth (MH-II) with an inoculum of 106 cfu/mL, as previously described.10 Combinations tested were meropenem/amikacin, meropenem/colistin and meropenem/rifampicin at different concentrations (Table S1, available as Supplementary data at JAC Online). The concentrations used in the chequerboard assay and metabolic phenotype screening, as well as in time–kill assays, were chosen as standard concentrations around the MICs for the strains. The synergy study using metabolic phenotype screening was based on the same principle for the determination of the MIC values using calScreener® (Symcel, Sweden) running calView software (Symcel, Sweden).7 In both assays, the results were given in terms of FIC index (FICI) (MICX+Y/MICX+MICX+Y/MICY; e.g. X = meropenem and Y = amikacin); <0.5 being synergistic, 0.5–1.0 being additive and >1.0 being indifferent.
Time–kill curves
Time–kill assays were performed in MH-II with meropenem, amikacin, colistin and rifampicin alone, as well as in combinations, using an inoculum of 105–106 cfu/mL. Colony counts were assessed at 1, 3, 5, 7, 24 and 48 h. A ≥3 log10 cfu/mL reduction of the original inoculum, when tested with a single antibiotic, was considered bactericidal. A reduction of ≥2 log10 cfu/mL by the antibiotic combination compared with that of the most active compound was defined as synergy. Indifference was defined as a <2 log10 reduction with the combination compared with that obtained with the most active single antibiotic. Antagonism was defined as a ≥2 log10 increase in viable count when using the combination compared with that obtained with the most active drug alone.
In vivo murine peritonitis/sepsis model
All in vivo experiments were conducted under the supervision of the Danish Animal Ethical Council under license 2017–15-0201–01274.
Each experiment included 36 outbred female NMRI mice (7 weeks old, weight 26–30 g) (Taconic, Germany) divided into five groups: 4 mice in a control group for determining the baseline infection load at the start of treatment, 8 mice treated at t = 1 (1 h) with antibiotic X, 8 mice treated at t = 1 (1 h) with antibiotic Y, 8 mice treated at t = 1 (1 h) with both X and Y, and 8 mice treated at t = 1 (1 h) with saline (0.9% NaCl) as a control.
At t = 0, mice were inoculated intraperitoneally with 500 μL of bacterial suspension containing 10−7 cfu/mL and 5% (w/v) porcine mucin (Sigma, USA). One hour later, blood and peritoneal fluid were sampled from four mice for determining the level of infection before treatment. After euthanizing the mice by cervical dislocation, a peritoneal wash was performed by injecting 2 mL of sterile saline (intraperitoneally), performing gentle massage of the abdomen for 1 min and then opening the abdomen aseptically for sampling of the peritoneal fluid with a pipette. During the study, at t = 1, t = 3 and t = 5, the mice were observed and their clinical scores were noted according to the grades that can be found in the Supplementary data available at JAC Online.
The remaining mice in treatment groups were injected (subcutaneously in the thigh) with 200 μL of antibiotic X, antibiotic Y, X and Y or saline (e.g. amikacin alone, meropenem alone, amikacin and meropenem together or saline). Mice treated with two antibiotics were given each in separate thighs.
At t = 3, after 2 h of treatment, four mice from each group were removed and blood and peritoneal fluid samples were taken as described above. For tests involving types of antibiotics with a low half-life (<1 h) in mice, these antibiotics were boosted with a second injection at t = 3 (Table 2). At t = 5, the last four mice in each group were removed and samples were taken as described above.
Table 2.
Specific doses given during in vivo experiments, half-lives and human doses
| Antibiotic | Dose at t = 1 h (mg/mouse) | Dose at t = 3 h (mg/mouse) | Total dose (mg/mouse) | Half-life reported in mice (h) | Normal human dose (mg/kg/day) |
|---|---|---|---|---|---|
| Amikacin | 2.4 | 2.4 | 4.8 | 0.3-0.5422 | 15 |
| Colistin | 0.6 | 0.6 | 1.2 | 0.5323 | 2.5–5 |
| Meropenem | 4.8 | 4.8 | 9.6 | 0.5124 | 30–40 |
| Rifampicin | 2.4 | 2.4 | 1225 | 15 |
Antibiotic treatment regimens for the murine peritonitis/sepsis model for amikacin, colistin, meropenem and rifampicin. Milligrams per mouse injected 1 or 3 h post-infection, total dose (mg/mouse) and the half-life reported in mice (h) with references for these, as well as the normal human dose (mg/kg/day) of the particular antibiotic.
All samples were stored at 4°C and were processed within 1 h of removal from animals. Samples were 10-fold serially diluted in saline before being plated onto solid lactose agar plates (‘blue plates’ based on a modified Conradi–Drigalski medium containing 10g/L detergent, 1g/L Na2S2O3·H2O, 0.1g/L bromothymol blue, 9g/L lactose and 0.4g/L glucose, pH8.0; SSI, Copenhagen, Denmark) for cfu counts. Beside cfu, data are presented as log reduction as per the untreated saline group. Additional information about animal experiments can be found in the Supplementary data available at JAC Online. A synergistic effect of combination treatment in the model was defined as significantly increased eradication by the combination compared with the eradication of the best mono treatment at the experimental endpoint after 4 h of treatment.
WGS
Genomic DNA from freshly subcultured bacterial colonies was extracted using an automated extraction system with the EZ1 DNA Tissue Kit (Qiagen, Hilden, Germany). DNA concentrations were measured using a Qubit™ 3.0 fluorometer (Thermo Scientific, MA, USA). Sequencing libraries were prepared using the Nextera XT Kit (Illumina, San Diego, CA, USA). Paired-end, short-read sequencing (150 bp) was performed on a HiSeq 2500 system (Illumina) at the Science for Life Laboratory, Stockholm, Sweden. Quality control of the sequencing data was performed using FastQC v0.11.8 (Babraham Bioinformatics, UK). Trimmed reads were assembled using SPAdes (version 3.13.1).11 MLST of the study isolates was performed using microSALT (https://github.com/Clinical-Genomics/microSALT) and detection of antimicrobial resistance genes was performed online using CARD (https://card.mcmaster.ca/analyze).
Statistics
Statistical analysis was performed in Prism 8.0 (GraphPad, USA). cfu counts rely on two technical replicates for each sample. Kruskal–Wallis one-way analysis was used to test non-parametric cfu data with Bonferroni multicomparison. Non-parametric data were correlated using Spearman correlation. Data were fitted with a non-linear curve fit with a P < 0.05 correlation coefficient according to a t-test. Data were tested for normality using the Shapiro–Wilk test. P values below 0.05 were considered significant.
Results
Antimicrobial susceptibility characteristics of isolates
The MIC values of meropenem, colistin, amikacin and rifampicin and their combinations, as well as resistance profiles for other antimicrobials, are presented in Table 1.
FICI of combination treatment found using microcalorimetry
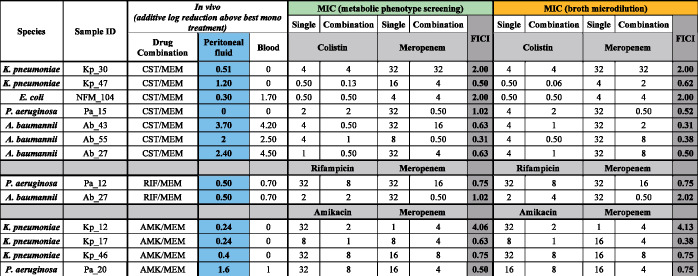
Regarding the meropenem and colistin combination, four isolates (Pa_15, Ab_43, Ab_55 and Ab_27) showed a ∑FIC ≤0.5 using the chequerboard assay, indicating synergy of these drugs. However, using the metabolic phenotype screening, only Ab_55 showed a ∑FIC ≤0.5 with this combination. In contrast, strain Kp_47 showed a comparable synergy result with the microcalorimetry assay, but not when using the chequerboard assay. The meropenem/rifampicin combination was not synergistic at any of the concentrations tested with either method. The meropenem and amikacin combination showed a synergistic effect using the chequerboard assay for isolate Kp_17, but not with the microcalorimetry assay. In contrast, a different isolate (Pa_20) showed a synergistic effect with the metabolic phenotype screening, but not with the chequerboard assay. Interestingly, with this combination, one isolate showed a high FICI with both assays (Kp_12). The overall comparison between the chequerboard assay and the calorimetry assay revealed that 8 out of 13 samples belonged to the same category (synergy/additive/indifferent) and that 5 out of 13 samples had identical FICI values (see Figure 1).
Figure 1.
MICs (mg/L) of meropenem, colistin, amikacin and rifampicin and their combinations (determined using phenotypic and conventional methods). Additional log reduction achieved by combination treatment above the best mono treatment is also shown. FICI data show the synergistic, indifferent or antagonistic effects of combination treatments. CST, colistin; MEM, meropenem; RIF, rifampicin; AMK, amikacin.
Time–kill assays
Overall, in Enterobacterales, we observed that meropenem exhibited bactericidal activity against five out of six isolates (Kp_12, Kp_17, Kp_46, Kp_47 and Ec_NFM104) when this antimicrobial was used alone after 5 h of incubation. Using meropenem/colistin, meropenem/rifampicin and meropenem/amikacin combinations, we observed the same behaviour. In the remaining isolate (Kp_30), colistin showed bactericidal activity when used alone after 3 h of incubation. Nevertheless, after 12–24 h, regrowth was observed for the Klebsiella pneumoniae isolates. No antagonism was observed with any of these combinations.
In two out of three Pseudomonas aeruginosa isolates (Pa_12 and Pa_20), bactericidal activity was not observed when using the antibiotics alone, but a synergistic bactericidal activity was shown when combining meropenem and rifampicin against Pa_12 and meropenem and amikacin against Pa_20. The synergistic effect was observed after 5 h. In the remaining P. aeruginosa isolate, Pa_15, only colistin showed bactericidal activity. This effect was maintained in combination with meropenem.
Regarding A. baumannii isolates, we observed that colistin exhibited bactericidal activity when this antimicrobial was tested alone against the Ab_27 isolate. Against the other two isolates (Ab_43 and Ab_55), we did not observe bactericidal activity when we used meropenem or colistin alone. Nevertheless, against these two isolates, we observed a synergistic bactericidal effect after 3 h of the meropenem/colistin combination. In addition, against Ab_27, a synergistic effect was observed when we used the meropenem/rifampicin combination. All curves are displayed in Figure S1.
Combination treatment in the in vivo murine peritonitis/sepsis model
At 1, 3 and 5 h after bacterial inoculation, each mouse was evaluated for its degree of symptoms based on the clinical score scheme developed by the Danish Animal Ethical Council (see the Supplementary data available at JAC Online). With all bacterial strains tested, all mice displayed symptoms of the infections after the first hour to a degree corresponding to score 2. There was no change with any treatment or strain combination after 2 or 4 h of treatment.
One hour after inoculation, a control group of four mice was sacrificed to determine the level of infection at the start of treatment. cfu recovered from mice after 1 h of infection varied <1 log both in the peritoneal fluid and in the blood [mean log SD in peritoneal fluid 0.36 (log range 0.03–0.77) and mean log SD in blood 0.49 (log range 0–0.95)].
cfu/mL recovered for each mouse treated or untreated with all strains is shown in Figure S2. In Figure 2(a) representative example of two treatment regimens, tested in vivo against the same A. baumannii strain (Ab_27), is displayed; one treatment where the combination treatment had limited additional eradication (Figure 2a and b) and one treatment where the combination treatment demonstrated significant eradication compared with the best mono treatment (Figure 2c and d). The first in vitro treatment combination with rifampicin and meropenem showed an indifferent effect when screened with microcalorimetry screening (FICI 2.02). In the mouse model, the combination treatment of rifampicin and meropenem improved the eradication compared with the best mono treatment, rifampicin, both after 2 h (P = 0.014) and 4 h (P = 0.014) of treatment in the peritoneal fluid or the blood after 2 h (P = 0.014) and 4 h (P = 0.014). The improved eradication by combination treatment constituted an additional log reduction of 0.5 in the peritoneal fluid and 0.7 in the blood after 4 h of treatment (Figure 2a and b).
Figure 2.
Representative in vivo results of two treatment regimens, one where no synergy could be found (a and b) and one in which the combination treatment had a high degree of additional effect (c and d). Two combination treatments of the same A. baumannii strain, Ab_27. cfu recovered in either the blood or peritoneal fluid at t = 1, t = 3 and t = 5.
The other combination treatment of colistin and meropenem had a synergistic effect using microcalorimetry screening (FICI 0.5). The combination treatment did not give any significant (P = 0.44) additional eradication in the peritoneal fluid after 2 h of treatment, but had a significant (P = 0.029) additional effect after 4 h in peritoneal fluid with an added eradication of 2.4 log (Figure 2c). In addition, the combination treatment did not have any additional effect after 2 h of treatment (P = 0.21) in the blood. However, there was a significantly (P = 0.029) higher eradication by the combination treatment compared with the mono treatment with colistin with an additional eradication of 4.5 log (Figure 2d).
The log reduction of combination treatments above the best single drug treatment in peritoneal fluid and blood at t = 5 after 4 h of treatment is shown in Figure 1 for all treatments and strains, as well as accompanying FICI values found with microcalorimetry screening and chequerboard screening.
At the termination of the in vivo experiments, after 4 h of treatment, several strains reacted with increased eradication when treated with a combination of the two tested drugs. Of the 13 combinations tested, 5 had an improved eradication of more than 1 log in the peritoneal fluid compared with the best mono treatment against that strain and 5 had more than 1 log improvement in the blood. These improvements in treatment were found in A. baumannii, K. pneumoniae and P. aeruginosa and with combination treatments of colistin and meropenem, and amikacin and meropenem (Figure 1).
FICI value prediction based on metabolic phenotype, as well as optical density assessed in the chequerboard assay, was correlated to improvement in the eradication or lack thereof in the in vivo experiments. There was a significant (P = 0.024) correlation between FICI prediction of a synergistic effect found with metabolic phenotype screening and the additive log reduction of combining two drugs above the reduction of the best mono treatment in the peritoneal fluid (Figure 3a). No significant correlation between cfu reduction in the blood of the mice and FICI could be found (P = 0.2) (Figure 3b). Additionally, it was not possible to confirm any correlation between the FICI values found with in vitro chequerboard assays and the log reduction found in the peritoneal fluid (P = 0.16).
Figure 3.
Correlations and non-linear curve fit between additional eradication (log reduction) from combination treatment above best single drug treatment, as a function of the FICI prediction by the metabolic phenotype screening for peritoneal fluid (triangles) and blood (circles). x-y plot of additional log reduction as a function of FICI from each metabolic phenotype screening. Linear regression, FICI versus log cfu reduction (a and b); non-linear regression (curve fit), log FICI versus normalized response to highest eradication - variable slope (c and d). Panel (a) shows a significant (P = 0.024) correlation between FICI and additional eradication in peritoneal fluid. Panel (b) shows the lack of a significant correlation (P = 0.2) between FICI and additional eradication in blood. P value of Spearman correlation. (c and d) Same data as in panel (a) and panel (b) with normalized response calculated as percentage reduction related to the highest value of reduction, i.e. 4.2 for peritoneal fluid and 4.5 for blood, and fitted with a non-linear regression. The P value of the correlation coefficient for both models was <0.05 based on t-test (GraphPad Prism 8.0).
WGS
Results from the whole-genome sequence analysis of the study isolates with respect to their ST and carriage of genes conferring antimicrobial resistance are depicted in Table 1. Sparing the three K. pneumoniae isolates that belonged to ST-307, all the other isolates included in the study belonged to separate STs. All three A. baumannii isolates (Ab_43, Ab_55 and Ab_27) included in the study were OXA-23 producers; all of these isolates demonstrated a ∑FIC ≤0.5, by chequerboard assay, indicating synergistic activity when testing a combination of meropenem and colistin. Four out of six Enterobacterales isolates harboured blaCTX-M-15 genes, two out of five K. pneumoniae isolates were KPC-3 producers and none of the isolates was identified as an NDM producer. Among the three P. aeruginosa isolates included, two of them were VIM producers; the genetic determinants for carbapenem resistance in Pa_20 could not be determined using CARD. It is very likely that the resistance of isolate Pa_20 is chromosomally mediated; a large number of regulatory and mutational changes often in combination in the same isolate has been shown to be able to confer carbapenem resistance.12 From the present study, we could not assess whether synergy while testing a combination of antibiotics is more common among isolates belonging to a particular ST, carrying a specific resistance mechanism or within a given bacterial species.
Discussion
The initial screening was conducted using both broth microdilution in the chequerboard assay and phenotypic metabolic readout in the calScreener platform. Both assays were able to identify several possible combinations with an additive eradication effect compared with the single-drug treatment. Interestingly, there was a disparity between the identified treatments and strains between the two assays, with only one treatment combination against one K. pneumoniae strain showing a decreased MIC in both assays. A disparity between synergy in different in vitro assays has been described previously, i.e. lack of coherence between the assays.13 The time–kill studies showed a limited number of combinations with additive effects compared with the single-drug treatment. Only four of the combination treatments tested were found to have additive eradication in the time–kill experiments, which again raises the issue of transferability of results between in vitro models for synergistic treatment. Bayer and Morrison14 reported the marked disparity between the time–kill and chequerboard methods to identify synergy for vancomycin/rifampicin treatment regimens against Staphylococcus aureus and called for in vivo validation of these methods. In the cases where combination treatment proved to convey additional eradication in our time–kill assay, the additional eradication all occurred within the first 5 h, suggesting that the time scale of the in vivo experiments is within the limits of an additive effect of the combination found in vitro.
Such a disparity is problematic for a screening tool for synergistic treatment combinations in clinical settings. It was, therefore, imperative for us to identify if one or the other assay proved to have clinical validity when it came to predicting the increased success of combination treatment in vivo. We, therefore, applied the well-known murine peritonitis/sepsis model.15 The model has proven a reliable tool for assessing the effectiveness of antibiotics on both resistant and susceptible strains of bacteria, as well as confirming in vitro synergism and antagonism in vivo.16–18 The peritonitis model has, in our view, an advantage over the otherwise well-published mouse thigh model; the latter needs induction of neutropenia to prime the mice for infection, while the mice are left immunocompetent in the peritonitis model, which is more relevant for extrapolation to clinical infections in general.
In an effort to pursue as clinically relevant results as possible, humanized doses were used in all treatments, which demanded two doses of antibiotics at 1 and 3 h after the start of treatment. In most instances, strains found resistant in vitro did, as expected, not exhibit any reduction in bacterial load in vivo, when treated with that drug alone. One such example is meropenem treatment against A. baumannii Ab_27. For the same strain, rifampicin and colistin exhibit a bacteriostatic effect on the bacterial load in vivo. On the other hand, combination treatment with meropenem/colistin reduced bacterial counts with 2.4 and 4.5 log more than colistin alone both in blood and peritoneal fluid, respectively. Especially against A. baumannii, the combination of colistin and meropenem seemed to be highly effective, at least for three out of four strains investigated. Colistin has been recommended for inclusion in combination therapies.19 Several other studies have focused on the colistin/rifampicin combination against A. baumannii as well.20,21 The colistin/meropenem combination regimen may, therefore, be of interest for further investigation.
Our findings support the fact that additional synergistic effects are highly strain and combination specific. What works on one strain does not necessarily have any additional effect on another and, as seen with A. baumannii Ab_27, one combination may have a high additional effect on that strain, whereas another combination may have limited additional effects. The lack of predictable synergistic effects of drug/bug combinations increases the interest in methods with a high predictive outcome, which within 5–6 h could predict a synergistic or additive effect in vivo. The direct correlation between in vitro predictions and the in vivo eradication we found in this study substantiates the possibility of rapid screening for effective combination treatments.
Given that the metabolic phenotype screening platform, in theory, can give an indication of a treatment’s effectiveness within hours, whereas the chequerboard assay requires overnight growth, the metabolic phenotype screening may be advantageous in combination screening. As metabolic phenotype relies on any form of metabolic activity, dormant but still viable cells may be registered as surviving the treatment, whereas biomass-based assays rely on actively dividing cells to register survival through OD or cfu measurements.7 Both systems require more or less the same hands-on time as they both rely on a microplate set-up.
We believe our findings indicate that screening for relevant in vivo combination treatments with the use of microcalorimetry is possible and may allow for personalized medical treatment, with better effect, less complication and a smaller and more specified antibiotic use. However, this needs further study.
Supplementary Material
Funding
The project was performed in collaboration with Symcel, which developed the calScreener technology. Funding was received from the European Union’s Horizon 2020 research and innovation programme (grant 729076).
Transparency declarations
C.G.G. and R.C. are members of the Steering Committee of EUCAST. All other authors: none to declare.
The project was administered by the PIs of the funded project in collaboration with Symcel.
Supplementary data
Supplementary data, including Table S1 and Figures S1 and S2, are available at JAC Online.
References
- 1.WHO. Global Action Plan on Antimicrobial Resistance. 2015. https://www.who.int/antimicrobial-resistance/global-action-plan/en/.
- 2. Sengupta S, Chattopadhyay MK, Grossart HP.. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 2013; 4: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Acar JF. Antibiotic synergy and antagonism. Med Clin North Am 2000; 84: 1391–406. [DOI] [PubMed] [Google Scholar]
- 4. Acar JF, Goldstein F, Chabbert YA.. Synergistic activity of trimethoprim-sulfamethoxazole on Gram-negative bacilli: observations in vitro and in vivo. J Infect Dis 1973; 128 Suppl 3: S470–7. [DOI] [PubMed] [Google Scholar]
- 5. Tamma PD, Cosgrove SE, Maragakis LL.. Combination therapy for treatment of infections with Gram-negative bacteria. Clin Microbiol Rev 2012; 25: 450–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Worthington RJ, Melander C.. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 2013; 31: 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tellapragada C, Hasan B, Antonelli A. et al. Isothermal microcalorimetry minimal inhibitory concentration testing in extensively drug resistant Gram-negative bacilli: a multicentre study. Clin Microbiol Infect 2020; 26: 1413.e1–7. [DOI] [PubMed] [Google Scholar]
- 8.ISO. ISO 20776-1:2019 - Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Broth Micro-Dilution Reference Method for Testing the In Vitro Activity of Antimicrobial Agents Against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. 2019. https://www.iso.org/standard/70464.html.
- 9.EUCAST. Clinical Breakpoints – Breakpoints and Guidance. http://www.eucast.org/clinical_breakpoints/.
- 10. Garcia LS. Synergism testing: broth microdilution checkerboard and broth macrodilution methods. In: Clinical Microbiology Procedures Handbook. 3rd edn.American Society of Microbiology, 2014; 140–62. [Google Scholar]
- 11. Nurk S, Bankevich A, Antipov D. et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Vol 7821 LNBI. Springer, 2013; 158–70. [Google Scholar]
- 12. De Sá Cavalcanti FL, Mirones CR, Paucar ER. et al. Mutational and acquired carbapenem resistance mechanisms in multidrug resistant Pseudomonas aeruginosa clinical isolates from Recife, Brazil. Mem Inst Oswaldo Cruz 2015; 110: 1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moody JA, Gerding DN, Peterson LR.. Evaluation of ciprofloxacin’s synergism with other agents by multiple in vitro methods. Am J Med 1987; 82: 44–54. [PubMed] [Google Scholar]
- 14. Bayer AS, Morrison JO.. Disparity between timed-kill and checkerboard methods for determination of in vitro bactericidal interactions of vancomycin plus rifampin versus methicillin-susceptible and -resistant Staphylococcus aureus. Antimicrob Agents Chemother 1984; 26: 220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frimodt-Møller N, Knudsen JD, Espersen F.. The mouse peritonitis/sepsis model. In: Handbook of Animal Models of Infection. Academic Press, 1999; 127–36. [Google Scholar]
- 16. Sandberg A, Hessler JHR, Skov RL. et al. Intracellular activity of antibiotics against Staphylococcus aureus in a mouse peritonitis model. Antimicrob Agents Chemother 2009; 53: 1874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frimodt‐Møller N, Thomsen VF.. Interaction between β‐lactam antibiotics and gentamicin against Streptococcus pneumoniae in vitro and in vivo. Acta Pathol Microbiol Scand B 1987; 95: 269–75. [DOI] [PubMed] [Google Scholar]
- 18. Johansen HK, Jensen TG, Dessau RB. et al. Antagonism between penicillin and erythromycin against Streptococcus pneumoniae in vitro and in vivo. J Antimicrob Chemother 2000; 46: 973–80. [DOI] [PubMed] [Google Scholar]
- 19. Petrosillo N, Ioannidou E, Falagas ME.. Colistin monotherapy vs. combination therapy: evidence from microbiological, animal and clinical studies. Clin Microbiol Infect 2008; 14: 816–27. [DOI] [PubMed] [Google Scholar]
- 20. Giamarellos-Bourboulis EJ, Xirouchaki E, Giamarellou H.. Interactions of colistin and rifampin on multidrug-resistant Acinetobacter baumannii. Diagn Microbiol Infect Dis 2001; 40: 117–20. [DOI] [PubMed] [Google Scholar]
- 21. Hogg GM, Barr JG, Webb CH.. In-vitro activity of the combination of colistin and rifampicin against multidrug-resistant strains of Acinetobacter baumannii. J Antimicrob Chemother 1998; 41: 494–5. [DOI] [PubMed] [Google Scholar]
- 22. Craig WA, Redington J, Ebert SC.. Pharmacodynamics of amikacin in vitro and in mouse thigh and lung infections. J Antimicrob Chemother 1991; 27: 29–40. [DOI] [PubMed] [Google Scholar]
- 23. Hengzhuang W, Wu H, Ciofu O. et al. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 2011; 55: 4469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumoto K, Kurihara Y, Kuroda Y. et al. Pharmacokinetics and brain penetration of carbapenems in mice. J Infect Chemother 2016; 22: 346–9. [DOI] [PubMed] [Google Scholar]
- 25. Jayaram R, Gaonkar S, Kaur P. et al. Pharmacokinetics-pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrob Agents Chemother 2003; 47: 2118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.