Abstract
To fully decipher the immunogenicity of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike protein, it is essential to assess which part is highly immunogenic in a systematic way. We generate a linear epitope landscape of the Spike protein by analyzing the serum immunoglobulin G (IgG) response of 1,051 coronavirus disease 2019 (COVID-19) patients with a peptide microarray. We reveal two regions rich in linear epitopes, i.e., C-terminal domain (CTD) and a region close to the S2′ cleavage site and fusion peptide. Unexpectedly, we find that the receptor binding domain (RBD) lacks linear epitope. We reveal that the number of responsive peptides is highly variable among patients and correlates with disease severity. Some peptides are moderately associated with severity and clinical outcome. By immunizing mice, we obtain linear-epitope-specific antibodies; however, no significant neutralizing activity against the authentic virus is observed for these antibodies. This landscape will facilitate our understanding of SARS-CoV-2-specific humoral responses and might be useful for vaccine refinement.
Keywords: COVID-19, SARS-CoV-2, Spike protein, linear epitope landscape, neutralizing antibodies
Graphical abstract
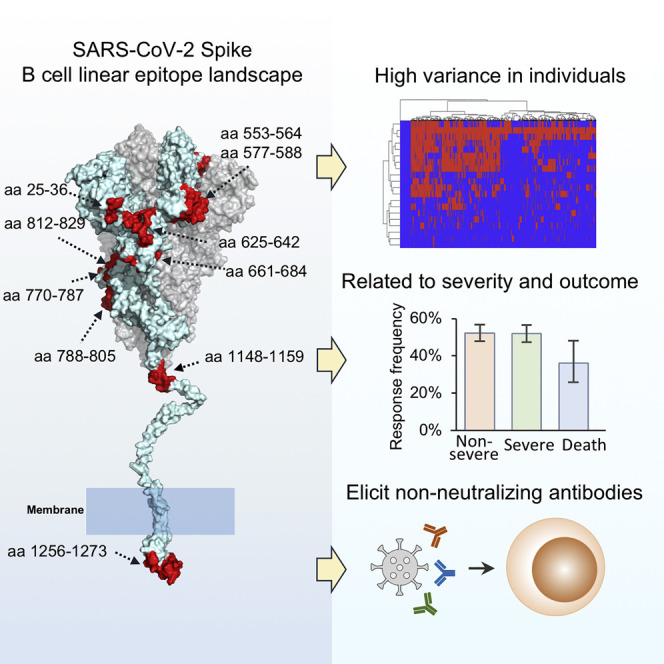
Li et al. construct a B cell linear epitope landscape of SARS-CoV-2 Spike protein, based on a large cohort of COVID-19 patients. The epitope responses were related to disease severity and outcome but mainly elicit non-neutralizing antibodies.
Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wu et al., 2020b; Zhou et al., 2020b), which is still causing an unfolding global pandemic. By February 3, 2021, 103,972,191 cases had been diagnosed and 2,255,496 lives had been claimed (https://coronavirus.jhu.edu/map.html; Dong et al., 2020a). Recently, several COVID-19 vaccines have been successfully developed and approved by the FDA (US Food and Drug Administration) (Baden et al., 2021; Polack et al., 2020), and more candidates are on the road (Dong et al., 2020b; Krammer, 2020). They will be undoubtedly helpful to combat this pandemic. However, there are still several unelucidated immunological questions related with SARS-CoV-2 infection that need much more effort to answer to benefit disease therapy as well as refinement of vaccine design (Jeyanathan et al., 2020).
The genome of SARS-CoV-2 encodes 27 proteins, and among them, the Spike protein plays a central role in the binding and entry of the virus to the host cell. The Spike protein is cleaved into S1 and S2 at furin and S2′ sites by specific proteases (Andersen et al., 2020). The Spike protein is highly glycosylated with 21 N-glycosylation sites (Watanabe et al., 2020). The Spike protein, and more specifically the receptor biding domain (RBD), is currently the target most focused on for the development of COVID-19 neutralizing antibodies and vaccines (Baum et al., 2020; Cao et al., 2020; Hansen et al., 2020; Wec et al., 2020; Wu et al., 2020d; Yuan et al., 2020a). Actually, RBD is immunodominant to elicit SARS-CoV-2-specific antibodies, RBD immunoglobulin G (IgG) response highly correlates with the S1 subunit IgG level (Jiang et al., 2020a; Premkumar et al., 2020), and importantly, RBD antibody level also highly correlates neutralizing activities of sera from patients (Iyer et al., 2020; Premkumar et al., 2020).
Out of RBD, other regions of Spike or S1 subunit can also elicit strong antibodies (Shrock et al., 2020; Wang et al., 2020a), and some epitopes-elicited antibodies have been demonstrated to exhibit neutralizing activities (Chi et al., 2020; Li et al., 2020a; Poh et al., 2020). However, full investigation of the Spike epitopes that can elicit antibodies, in particular neutralizing antibodies, has been not reported. Furthermore, if the epitopes elicit non-neutralizing antibodies, the beneficial or detrimental functions in disease development or vaccination are also poorly understood. Despite beneficial function of any virus-binding antibody due to ADCC (antibody-dependent cellular cytotoxicity) effect to help to eliminate pathogens, ADE (antibody-dependent enhancement) and pro-inflammation caused by antibodies are unneglectable concerns (Liu et al., 2019; Vabret et al., 2020). Although whether ADE occurs in the context of SARS-CoV-2 infection remains unclear, the facts that higher antibody titers and neutralizing plasma activities in patients are associated with more-severe conditions (Jiang et al., 2020a; Long et al., 2020a) and cytokine profiles resemble those in macrophage activation syndrome (Jeyanathan et al., 2020; Mehta et al., 2020) warrant further investigation of ADE as well as VAED (vaccine-associated enhanced disease) (Haynes et al., 2020). In addition, non-neutralizing antibodies may compete with and suppress the neutralizing antibody production through a widely studied mechanism called immunodominance (Abbott and Crotty, 2020; Cirelli et al., 2019). So non-neutralizing antibodies should be avoided to maximum the production of neutralizing antibody as well as minimize the likelihood of disease enhancement for vaccine design (Abbott and Crotty, 2020; Jeyanathan et al., 2020). Collectively, elucidation of high antigenic epitopes and investigation of the neutralization property of these epitopes are quite essential to refine vaccine design.
In the present study, through a large cohort of COVID-19 patients and controls, we build a full linear epitope landscape of B cells against SARS-CoV-2 Spike protein, identifying that C-terminal domain (CTD) on S1 unit and fusion peptide (FP) region on S2 unit are two antigenic hot areas, although RBD lacks linear epitopes. Combined with tensive clinical data, we found responsive epitope numbers and some specific linear epitopes are associated with disease severity. No neutralizing activities were detected for the antibodies elicited by the linear peptides immunized in mice.
Results
The IgG linear epitope landscape of the SARS-CoV-2 Spike protein
To reveal the immunogenic linear epitopes of the Spike protein, a peptide microarray with full coverage of the Spike protein was updated from an original version (Li et al., 2020a). Because B cell linear epitopes for antibody recognition typically span 3–8 continuous amino acids (aas) (Larman et al., 2011; Shi et al., 2019) and peptides of 10–12 aas are commonly used for immunization to develop antibodies (Wang et al., 2020c), to largely cover possible linear epitopes, we synthesized peptides of 12 aas with 6-aa overlap for every two adjacent peptides. A total of 211 peptides across the S protein (Spike protein) were selected (Figure S1; Table S1). Sera were collected from two groups, 1,051 COVID-19 patients and 528 controls (Table S2), and individually analyzed on the peptide microarray. By plotting the signal intensities of all the samples against each peptide, a linear epitope landscape was constructed; for a better overview, the landscape was aligned to the sequence of the Spike protein (Figure 1 ). To assure specificity, all the control samples (Table S2) were also analyzed on the peptide microarray. Almost all the peptides were negative for all the control samples, although significant binding was observed for many of the peptides when probed with COVID-19 sera. This indicates that the positive bindings are SARS-CoV-2 specific.
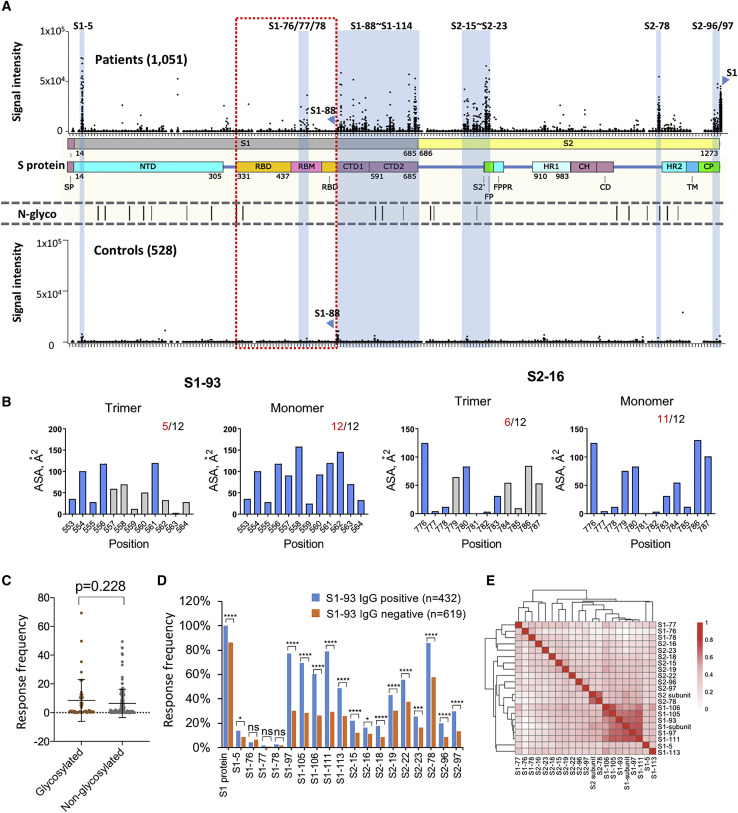
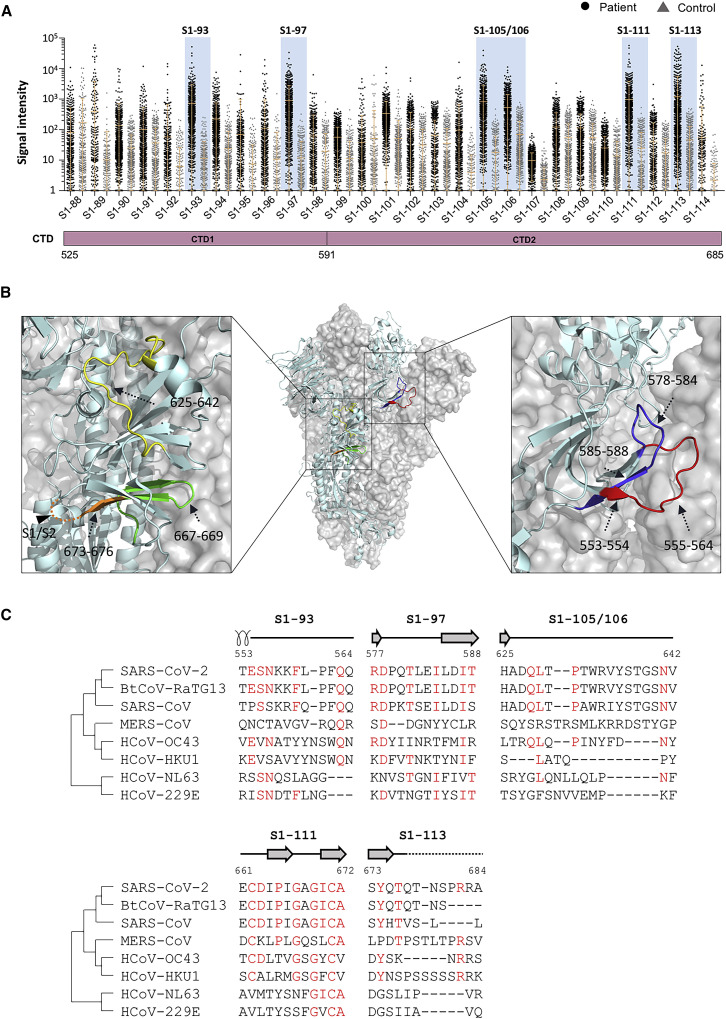
Figure 1.
The IgG linear epitope landscape of the SARS-CoV-2 Spike protein
(A) The signal intensities of 1,051 COVID-19 sera against 197 peptides were obtained by using the peptide microarray. The peptides are listed in the x axis and aligned to the corresponding locations on the Spike protein. As a control, the signal intensities of the S1 protein were also presented. The missing spots are peptides that either could not be synthesized or failed the BSA conjugation (see Table S1 for details). A cohort of 528 control sera were also analyzed on the microarray. In addition, the known N-glycosylation sites (N-glyco) were aligned with the Spike protein. The peptides or regions with significant binding were marked blue. Peptide S1-88 was specifically labeled because significant bindings were also observed for the controls. CD, connector domain; CH, center helix; CP, cytoplasmic; CTD1, C-terminal domain 1; CTD2, C-terminal domain 2; FP, fusion peptide; FPPR, fusion peptide proximal region; HR1, heptad repeat 1; HR2, heptad repeat 2; NTD, N-terminal domain; RBD, receptor biding domain; RBM, receptor binding motif; S2′, protease cleavage site; SP, signaling peptide; TM, trans-membrane.
(B) The area of solvent accessibility (ASA) of each amino acid for S1-93 and S2-16, with regard to the S protein trimer structure (PDB: 6X6P).
(C) The response frequencies of the two groups of peptides with or without the glycosylation site. The p value was calculated with the t test.
(D) The response frequency for each peptide in the two groups as S1-93 IgG positive or negative. The p value was calculated with the χ2 test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
(E) Heatmap of Spearman’s correlation coefficients between the IgG responses of the peptides and related proteins.
To determine which epitopes were highly immunogenic, the criteria were set as an average_signal intensity greater than 3∗Cutoff2 and a response frequency greater than 10% (see STAR Methods for the definitions). A total of 16 peptides were obtained; surprisingly, all of them are outside of the RBD. We hereby define these peptides or epitopes as significant epitopes. Due to the significance of the RBD, we lowered the criteria, i.e., a response frequency greater than 1%, while keeping the average_signal intensity greater than 3∗Cutoff2. Three consecutive epitopes of moderate immunogenicity, S1-76, S1-77, and S1-78, were selected (Table S3). Interestingly, all three epitopes were located within the RBM (receptor binding motif), the binding interface of the Spike protein and ACE2.
Although a few of these immunogenic epitopes are dispersed on the Spike protein, there are two linear epitope “hot” regions that could be immediately recognized: aas 525–685 and aas 770–829, one of which is the CTD and another that covers the S2′ cleavage site and the FP. There are several SARS-CoV-2 epitope-related studies involving small sample sets (Ahmed et al., 2020; Wang et al., 2020b). Our immunogenic epitopes are partially consistent with these studies (Table S3). A relative high consistency was observed between our data, ReScan, and VirScan, a phage display-based strategy (Shrock et al., 2020; Xu et al., 2015; Zamecnik et al., 2020).
To further illustrate the location and distribution of the 19 immunogenic epitopes (Table S3), we mapped them to the 3D structure of the Spike protein (Herrera et al., 2020; Figures S2A and S2B; Table S4). It is clear that most of these epitopes were located on the surface of the Spike protein, which is consistent with the common idea (Emini et al., 1985). Additionally, an accessibility analysis at the amino acid level revealed that most epitopes have at least 5 accessible amino acids on the trimer of the Spike protein and more on the monomer (Figures 1B and S2C). For S1-111, S2-16, S2-18, and S2-19, there are substantially fewer accessible amino acids on the trimer than on the monomer. A plausible explanation is that the Spike protein monomer could be exposed to the immune system at a certain, yet-to-be discovered stage. In addition, except for S1-77/78 on RBD, no big difference of the accessible amino acid numbers was observed for other epitopes in open and closed state of Spike trimer (data not shown). For the immunogenic epitopes (Table S3), the solubility (Kyte and Doolittle, 1982) and isoelectric point (pI) range from −1.97 to 1.06 and 3.01 to 11.16, respectively, and the hydrophilicity and pI of the epitopes are not correlated with response frequency (Figures S1D and S1E).
It was speculated that the N-glycosylation may interfere with antibody responses (Sikora et al., 2020). To test this speculation, we divided all the peptides into two groups: with or without an N-glycosylation site. We found that there was no significant decrease in response frequency for the group with glycosylation compared to the group without glycosylation (Figure 1C), suggesting that the distribution of the linear epitope is not or is only subtly related to N-glycosylation.
It is interesting to examine whether there is any correlation among different epitopes and epitopes versus the S protein. To answer this question, we tested the independence of IgG response among the significant epitopes. Taking S1-93 as an example, we found significant correlations for almost all of the epitopes to S1-93, including S1 protein (Figure 1D). Similar results were observed for other epitopes (Figures S3A and S3B). These results suggest that the IgG responses among the epitopes are correlated. Interestingly, although the epitopes with high response frequencies are significantly correlated with each other, the epitopes derived from the CTD region tend to have higher correlations with each other, although this is not the case for the epitopes derived from the FP region (Figure 1E). Consistently, the heatmap of hierarchical-cluster analysis of IgG response signatures in COVID-19 patients revealed associations among the significant epitopes (Figure S3C). It also demonstrated high variability among individuals, which is consistent with the concept that immune responses vary greatly from person to person (Boyd and Jackson, 2015; Li et al., 2020b).
Responsive epitopes correlate with the IgG response of the S protein and disease severity
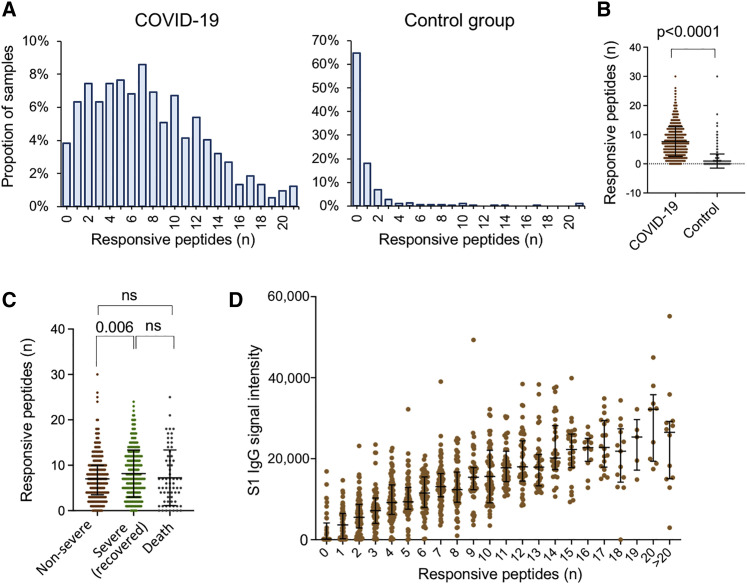
To further investigate how SARS-CoV-2 S-protein-specific IgG responses vary among COVID-19 patients, we counted the total numbers of responsive epitopes in each sample. It is known that the antibody response against SARS-CoV-2 reaches a plateau approximately 2 weeks after symptom onset (Long et al., 2020a). To maintain consistency in antibody responses, only samples collected 15 days or more after symptom onset were analyzed. The average number of responsive epitopes in the patient group is greater than those in the control group, i.e., 7.6 versus 1.2, respectively. However, the numbers are highly variable among individuals (Figures 2A and 2B). We next tested whether the responsive epitopes are related to severity. We divided the patients into three groups concerning severity and final outcome, i.e., non-severe (mild and moderate), severe survivors (severe and critical), and severe non-survivors (death). The numbers in the severe groups are significantly higher than those in the non-severe group (8.2 versus 7.1 in average), although there is no significant difference between non-survivors and severe survivors (Figure 2C). In addition, the number of responsive epitopes highly correlates with S protein IgG (Figure 2D). This observation demonstrates that S-protein-derived responsive epitopes contribute to immune responses to the S protein, which is expected.
Figure 2.
Epitope numbers in patients are related with severity
(A and B) The distribution of the numbers of the responsive epitopes in both the COVID-19 (n = 1,004) and control groups (n = 528). For the COVID-19 group, samples collected less than 15 days after symptom onset were excluded.
(C) The numbers of the responsive epitopes in the three groups with regard to severity and outcome.
(D) The S1 IgG signal intensity of the samples with different numbers of the responsive epitopes.
For (B)–(D), the data were presented by mean + SD. p values were calculated using the two-sided t test for others. For (C), the p value was adjusted for multiple comparisons by BH (Benjamini and Hochberg) method.
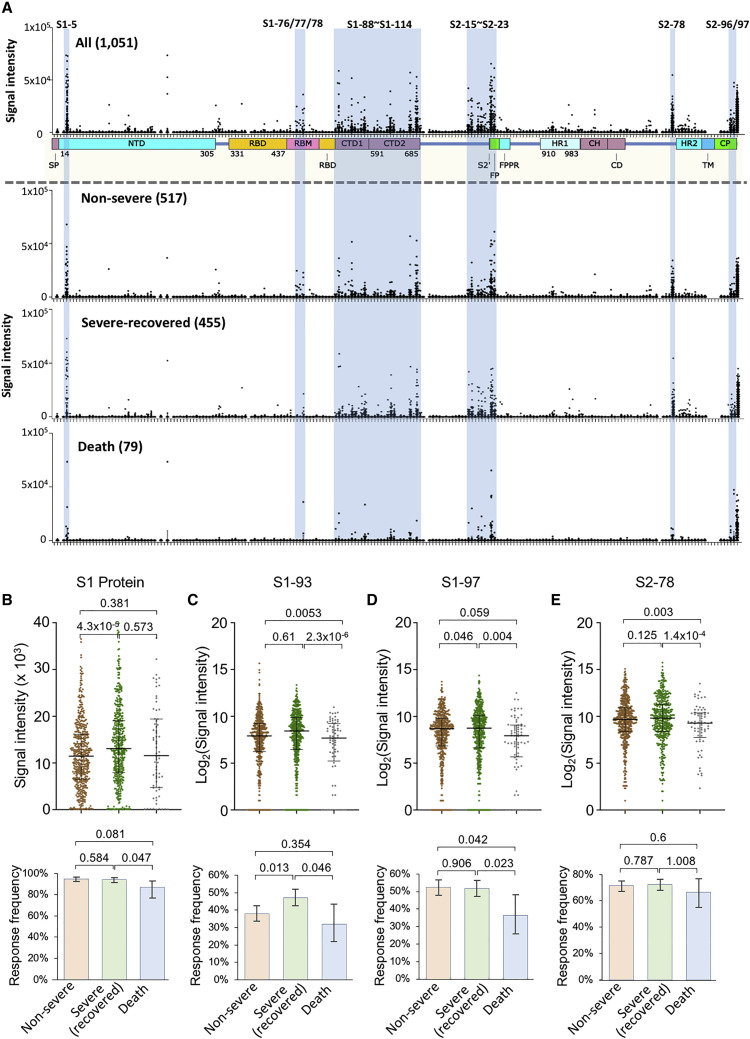
We next analyzed the possible correlation between the epitopes and disease severity. In general, there is no obvious difference among the three groups (Figure 3 A), except some epitopes and the S1 protein, which correlates with severity and/or outcome (Figures 3B–3E; Table S5). Particularly, for S1-93, S1-97, and S2-78, a statistically significant decrease in the IgG response in the non-survivor group was observed (Figures 3C–3E), suggesting protective roles for the corresponding antibodies. Consistently, the IgG response to the epitopes correlated with clinical parameters related to disease severity (Huang et al., 2020; Wu et al., 2020a), such as LDH (lactate dehydrogenase), C reactive protein, and lymphocyte percentage (Table S6). There were a few epitopes that were related to gender (Table S5) or age (Table S5). In particular, S1-5 and S1-97, as well as S1 protein, were significantly associated with age, which was consistent with our previous study with a small cohort (Jiang et al., 2020a). Although NAT (nuclear acid test) positivity is the gold standard for diagnosis, antibody tests or other approaches might be essential and complementary (Long et al., 2020a, 2020b). In our cohort, there are 248 clinically confirmed cases that were NAT negative. For this group of cases, the S1 IgG response prevalence is also high, though it is lower than that of the NAT-positive group (Table S5), although the antibody response levels against the significant epitopes were similar for both the NAT-positive and negative groups (Table S5).
Figure 3.
The IgG response of the epitopes is associated with severity
(A) The linear epitope landscape was divided into two sub-landscapes according to severity and outcome, i.e., 517 non-severe, 455 severe (survivors), and 79 severe (non-survivors). The data are presented as the means ± SD.
(B–H) IgG signal intensities (top) and response frequencies (bottom) of the three groups, i.e., non-severe, severe (survivors), and severe (non-survivors) for the S1 protein (E), S1-93 (F), S1-97 (G), and S2-78 (H). The data were presented using the median with interquartile for the top part and response frequency with a 95% confidential interval for the bottom part. p values were calculated using the two-sided t test for the top part and the χ2 test for the bottom part for (B)–(E).
RBD lacks linear epitopes, although it is highly immunogenic
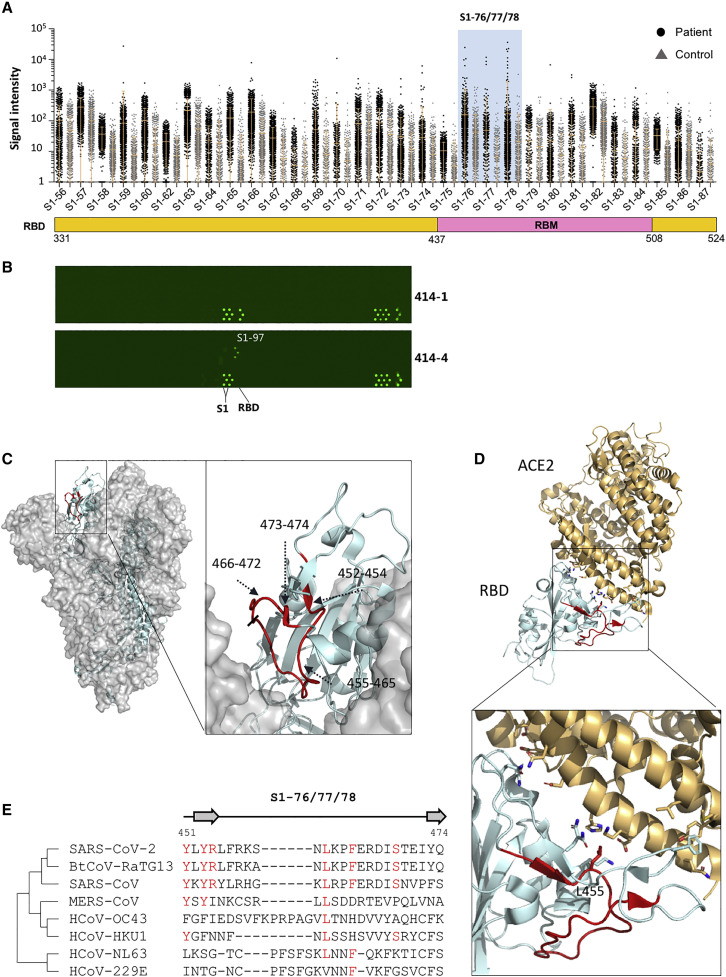
It is well known that the RBD is highly immunogenic (Jiang et al., 2020a; Premkumar et al., 2020). However, according to our selection criteria, no highly immunogenic epitope was obtained from the RBD. Only when we lowered the selection criteria were 3 peptides selected, S1-76, S1-77, and S1-78 (Figure 4 A). When the RBD is compared to other regions of the Spike protein, it is obvious that the RBD is very poor in linear epitopes. This seems contradictory to the knowledge that the RBD is highly immunogenic. It is possible that most of the epitopes of the RBD region are conformational. To test this possibility, we collected a set of 9 high-affinity monoclonal antibodies for the RBD or the Spike protein (see STAR Methods). These antibodies were obtained through memory B cell isolation from COVID-19-recovered patients (Wan et al., 2020). We analyzed these antibodies individually on the Spike protein peptide microarray (Li et al., 2020a; Figure 4B). Among these antibodies, 414-1 has the highest affinity (2.96 nM) to the RBD. As expected, strong bindings were observed for both the S1 protein and the RBD; however, negative signals were obtained for all the peptides, including the RBD peptides from aas 331–524. Additionally, no peptide bindings were observed for the rest of the RBD-specific antibodies (data not shown). For antibody 414-4, strong binding was obtained for the S1 protein, but not for the RBD. Interestingly, 414-4 binds S1-97 with high affinity, indicating the epitope that 414-4 recognizes is near aas 577–588.
Figure 4.
RBD lacks highly immunogenic linear epitopes
(A) The RBD region of the linear epitope landscape.
(B) The peptide microarray results of two Spike-protein-specific monoclonal human antibodies (from Active motif), one (414-1) is specific for the RBD and the other (414-4) is not.
(C) Detailed structures of the significant epitopes (S1-76/77/78; aas 451–474, red) on the RBD of the closed-state Spike protein trimer, side view (PDB: 6X6P).
(D) The locations of the significant epitopes s (S1-76/77/78; aas 451–474, red) on the co-crystal structure of the RBD and ACE2 (PDB: 6M0J).
(E) Homology analysis of the significant epitopes, s, among the 7 known human coronaviruses and bat coronavirus BtCoV-RaTG13, which is highly homologous to SARS-CoV-2. Amino acids with consistencies ≥50% among the 8 coronaviruses are marked in red, and the loop and β strand region are shown as a line and an arrow above the sequences, respectively.
Actually, the 3 immunogenic epitopes, S1-76, S1-77, and S1-78, were also identified in other related studies (Wang et al., 2020b; Zhang et al., 2020a). These three epitopes are consecutive and located in the RBM region, which at least partially overlaps with or is close to the binding epitopes of a variety of SARS-CoV-2 neutralizing antibodies, e.g., B38 (Wu et al., 2020d), CB6 (Shi et al., 2020), and P2B-2F6 (Ju et al., 2020). To further illustrate these epitopes’ locations, we mapped them to the 3D structure of the Spike protein in both the open state and the closed state (Figure 4C). In the closed state, aas 455–465 of S1-76/77/78 is located in the contact area among the three monomers and is probably difficult to access, aas 452–454 and 473–474 form the β strand and are covered but could be accessed from both sides, and only aas 466–472 are exposed and present as a flexible sequence (Figures 4D and S4). In the open state of the Spike protein, all residues of S1-76/77/78 are exposed and highly accessible (Figure S4). To further analyze the immunogenicity of S1-76/77/78, we examined all the available neutralizing antibody-RBD complexes by the end of November 30, 2020, as far as we know (Table S7). The antibodies are CB6 (Shi et al., 2020), P2B-2F6 (Ju et al., 2020), BD23 (Cao et al., 2020), CR3022 (Yuan et al., 2020a), S309 (Pinto et al., 2020), CV30 (Hurlburt et al., 2020), BD-368-2 (Du et al., 2020), CC12.1 (Yuan et al., 2020b), H11-D4 (PDB: 6YZ5), REGN10933 (Hansen et al., 2020), Fab2-4 (Liu et al., 2020), EY6A (Zhou et al., 2020a), H014 (Lv et al., 2020), Ty1 (Hanke et al., 2020), MR17 (PDB: 7C8W), C105 (Hanke et al., 2020), COVA2-04, COVA2-39 (Wu et al., 2020c), COVA1-16 (PDB: 7JMW), S2E12, S2M11 (Tortorici et al., 2020), S2A4, S2H13, S2H14 (Piccoli et al., 2020), CV07-250, CV07-270 (Kreye et al., 2020), C110, C119, C002, C135, C121, C102, C144 (Barnes et al., 2020), Fab-52, Fab-298 (Rujas et al., 2020), Nb6 (Schoof et al., 2020), and Sb23 (Custódio et al., 2020). Among these structures, CB6, P2B-2F6, CV30, DB-368-2, CC12.1, H11-D4, REGN10933, Fab2-4, Ty1, MR17, and C105 interact directly with residues within S1-76/77/78. For the CB6-RBD complex, there are several residues within S1-76/77/78 that directly interact with the following antibodies: Y453; L455; F456; R457; K458; S459; N460; Y473; and Q474. The same interaction residues between neutralizing antibodies and the RBD can also be found in the CV30-RBD, CC12.1-RBD, and C105-RBD complexes (Table S7). Partial interaction residues of the CB6-RBD complex between S1-76/77/78 can also be found in the REGN10933-RBD, H11-D4-RBD, Fab-2-4-RBD, Ty1-RBD, and MR107-RBD complexes, and some other residues, such as L452, T470, and I472, can be found in some of these structures (Table S7). For the P2B-2F6-RBD complex, the only residue that directly interacts with the antibody is L452. In summary, most of these neutralizing antibodies interact with residues located in S1-76/77/78, residues 452–460 and 470–474 play direct roles in the interactions, and the flexible loop consisting of residues 461–469 is irrelevant to these neutralizing antibodies.
Broad neutralizing antibodies and a vaccine effective for SARS-CoV-2 and other human coronaviruses are of high interest (Jiang et al., 2020b). We performed a homology analysis for S1-76/77/78 among SARS-CoV-2, the other 6 human coronaviruses, and bat coronavirus BtCoV-RaTG13 (Zhou et al., 2020b). High homologies were observed for the all 3 epitopes among SARS-CoV-2, SARS-CoV, and BtCoV-RaTG13 although low homology level with MERS (Middle East respiratory syndrome)-CoV and other common coronavirus (Figure 4E). The high homology indicates that antibodies elicited by S1-76/77/78 may at least be effective for both SARS-CoV-2 and SARS-CoV.
These results strongly suggest that the RBD is rich in conformational epitopes, although it lacks linear epitopes. The underlying mechanism is worth further investigation.
CTD is rich in linear epitopes
The first “hot” region of linear epitopes is CTD. The whole domain is densely covered by linear epitopes (Figure 5 A). According to the selection criteria, 6 highly immunogenic epitopes, S1-93, S1-97, S1-105/106, S1-111, and S1-113, were identified. These epitopes are nearly evenly distributed across CTD. We then asked whether these 6 highly immunogenic epitopes were also revealed in other studies. It showed that S1-93 was identified by ReScan (Zamecnik et al., 2020; Table S3), as well as COVIDep (Ahmed et al., 2020), S1-97 by ReScan, and S1-111 by COVIDep.
Figure 5.
CTD is rich in significant linear epitopes
(A) The CTD region of the linear epitope landscape.
(B) The locations of the significant epitopes, s, are located on the CTD (PDB: 6X6P). Specifically, S1-93, aas 553–564, red; S1-97, aas 577–588, blue; S1-105/106, aas 625–642, yellow; S1-111, aas 661–672, green; and S1-113, S1-113, aas 673–684, orange are shown.
(C) Homology analysis of the significant epitopes, s, among the 7 known human coronaviruses and bat coronavirus BtCoV-RaTG13. Amino acids with consistencies ≥50% among the 8 coronaviruses are marked in red, and the loop, α helix, and β strand region are shown as a line, a coil, and an arrow above the sequences, respectively. An unobserved structure is shown as a dotted line.
S1-93 and S1-97 are located at CTD1, although aas 555–564 of S1-93 and aas 578–584 of S1-97 are present at the loop region and on the surface of the trimeric Spike protein. S1-105, S1-106, S1-111, and S1-113 are located at CTD2; S1-105/106 are almost at the loop and present on the surface; S1-111 is at β strand and the loop but buried inside; and only aas 667–669 on the loop region could be accessed. S1-113 is near the S1/S2 cleavage site. Although aas 677–684 are invisible in the Spike protein structure, these residues could be exposed on the surface and induce an antibody response to prevent S1/S2 hydrolysis (Figure 5B). S1-113 is also on the outer surface, although S1-111 is on the inner surface, and the underlying mechanism by which S1-111 triggers a strong IgG reaction in many patients is worth further study.
Homology analysis was performed for the 6 highly immunogenic epitopes (Figure 5C). Except for S1-113, high homologies were observed for all 5 epitopes among SARS-Cov-2, SARS-CoV, and BtCoV-RaTG13 although lower homology levels were shown among SARS-CoV-2 and MERS-CoV and the other four common coronaviruses. The high homology indicates that antibodies elicited by S1-93, S1-97, S1-105, and S1-111 may be effective for both SARS-Cov-2 and SARS-CoV. Additionally, an antibody targeting S1-113 may be specific for SARS-CoV-2.
The D614G mutant is the current dominant strain in Europe (Korber et al., 2020), which has about a 9 times higher infection efficiency in cell assays than that of the wild-type strain (Zhang et al., 2020b). D614 is within S1-102, an epitope of moderate immunogenicity, and close to the highly immunogenic S1-105. Blocking the D614 region may cause a functionally significant effect.
The second epitope hot region spans aas 770–829, covering the S2′ cleavage site and FP
The second region with highly enriched linear epitopes spans aas 770–829 (Figure S5A). According to the selection criteria, 6 highly immunogenic epitopes were obtained: S2-15; S2-16; S2-18; S2-19; S2-22; and S2-23.
It is interesting to see whether these 6 highly immunogenic epitopes were also identified in related studies. We revealed that S2-22 was identified by a peptide microarray study (Wang et al., 2020b). Four epitopes (S2-18, S2-19, S2-22, and S2-23) were identified by ReScan (Zamecnik et al., 2020), and 2 epitopes (S2-22 and S2-23) were predicted by COVIDep (Ahmed et al., 2020). Of these epitopes, S2-22 is the only one that was identified or predicted in all these studies. Because S2-22/23 covers the S2′ cleavage site and FP, we speculate that an antibody targeting S2-22/23 may block the cleavage and disturb the function of FP; thus, it has potent neutralization activity. Interestingly, a strong S2-22-specific IgG reaction was also elicited by an mRNA vaccine study (Cai et al., 2020), which further demonstrated the high immunogenicity and high potential of the neutralization activity of S2-22.
To further illustrate these epitopes’ locations, we mapped them to the 3D structure of the Spike protein. S2-15, S2-16, S2-18, S2-19, S2-22, and S2-23 are all located near the S2′ site and the FP region. aas 770–783 of S2-15/16 form an α helix and are buried at the trimer interface but are accessible on the monomer. aas 791–805 of S2-18/19 form a loop, and aas 816–826 form an α helix and are located on the surface. The S2′ cleavage site is on S2-22 (Figure S5B).
To check the similarity of the epitopes among human coronaviruses, we performed a homology analysis for S2-22/23, S2-15/16, and S2-18/19. High homologies were observed for all these epitopes among SARS-CoV-2, SARS-CoV, and BtCoV-RaTG13, although only for S2-22/23, high homology was shown among SARS-CoV-2 and other common coronaviruses (Figure S5C). Interestingly, S2-22/23 is highly homologous among all the coronaviruses and almost identical among SARS-CoV-2, SARS-CoV, and BtCoV-RaTG13.
Other highly immunogenic linear epitopes
Except for the immunogenic epitopes that belong to the RBD and the two “hot” regions, there are other highly immunogenic epitopes dispersed across the Spike protein (Figure S6A). S1-5 is located at NTD, and part of the residues, i.e., aas 28–31, form the β strand and are on the surface of the trimeric Spike protein. aas 32–36 form a loop and are partially covered by the other region. S2-78, S2-96, and S2-97 are located in unobserved regions in the C terminus of the Spike protein. We applied a modeling structure to present these unobserved regions (Figure S6B). S2-78 is predicted to be an α helix, and S2-96/97 is predicted to be a loop. S2-96/97 are at the very C-terminal end of the Spike protein (Figure S6B), which is located in the cytoplasm of the host cell.
We checked whether these 4 highly immunogenic epitopes were also revealed in other studies. We found that S1-5 was identified by a peptide microarray study (Wang et al., 2020b). S2-78 was identified by ReScan (Zamecnik et al., 2020), and 3 epitopes (S2-78, S2-96, and S2-97) were predicted by COVIDep (Ahmed et al., 2020). The functional role of S1-5-specific antibodies may be worth further investigation. S2-78 is adjacent to HR2, and the antibody targeting this site may block the conformational change that is essential for effective virus-cell fusion (Liu et al., 2004). It is surprising to see the high immunogenicity of S2-96 and S2-97 because they are at the very C-terminal end of the Spike protein and theoretically localize in the cytoplasm. Further study is necessary to explore the underlying mechanism and functional roles of these epitopes.
High homologies were observed for all the epitopes among SARS-CoV-2, SARS-CoV, and BtCoV-RaTG13 (Figure S6C). Interestingly, S2-78 and S2-96/97 are highly homologous among all the coronaviruses and almost identical among SARS-CoV-2, SARS-CoV, and BtCoV-RaTG13. For S2-78, a high homology was also observed for MERS-CoV.
Antibody responses against most of the linear epitopes dramatically decrease after reaching the peak
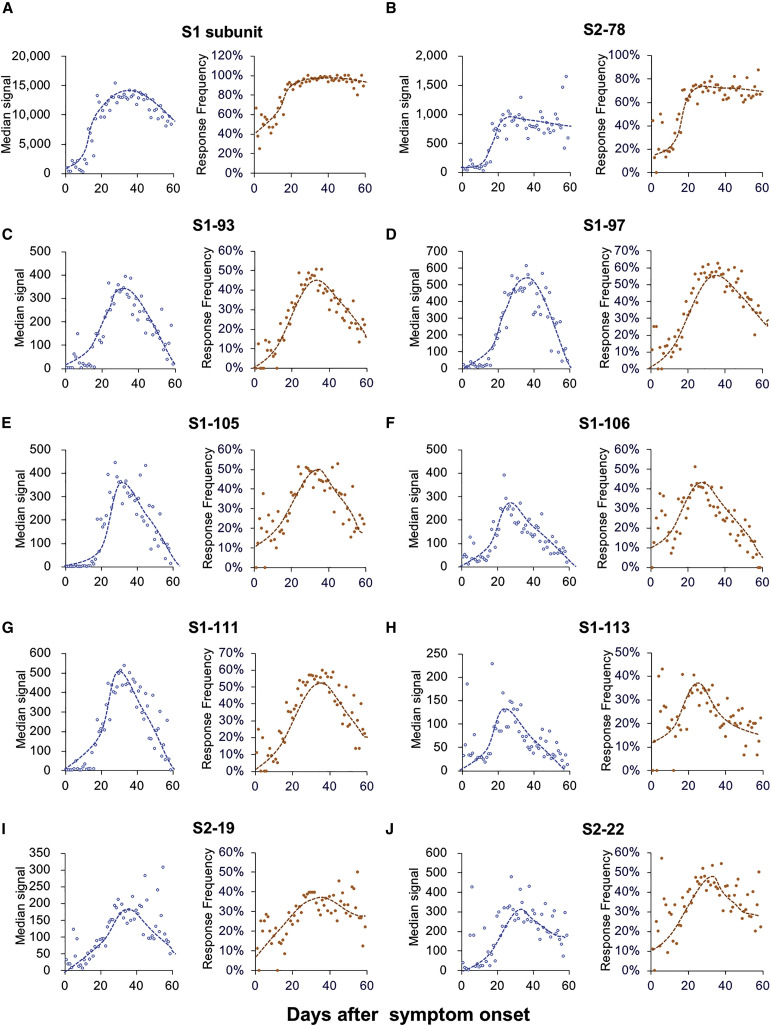
We next assessed the longevity of the antibodies elicited by the linear epitopes. For the hospitalized patients listed in Table S2, longitudinal sera samples were collected. We define collection_time as the days from symptom onset to sample collection, and the collection_time is widely distributed from 1 to 60 (Figure S7A). There are >20 samples/day for collection_time ranging from 12 to 54 days and >50 samples/day for collection_time ranging from 20 to 45 days. To fully understand the trend dynamics in the IgG to the significant epitopes, the median signal intensities and response frequencies of the epitopes were calculated and plotted according to the collection_time. The S1 protein was included as a reference for this analysis. Surprisingly, although S1 IgG slowly decreases when collection_time is approximately 40 days (Figure 6 A), the antibodies against most of the epitopes dramatically decrease when collection_time is approximately 30 days (Figures 6C–6G). However, for the antibodies against S2-78, the linear epitope of the highest frequency, no significant decrease was observed for both the median signal intensity and the response frequency (Figure 6B). This result is similar to that for S1 (Figure 6A). Consistently, the trend in the median number of responsive epitopes is similar to that of the S1 protein (Figures S7B and S7C). These observations suggest that a decrease in or disappearance of the antibodies against some epitopes may contribute to a decrease in S1 IgG antibodies, although the antibodies against some other epitopes could last longer.
Figure 6.
Dynamic changes in IgG responses to the epitopes
(A–J) Median signal intensity (left) and response frequency (right) of the samples collected at the indicated time points, i.e., collection_time or days after symptom onset for the indicated epitopes.
Antibodies from mice immunized with the linear peptides exhibit no significant neutralization activity
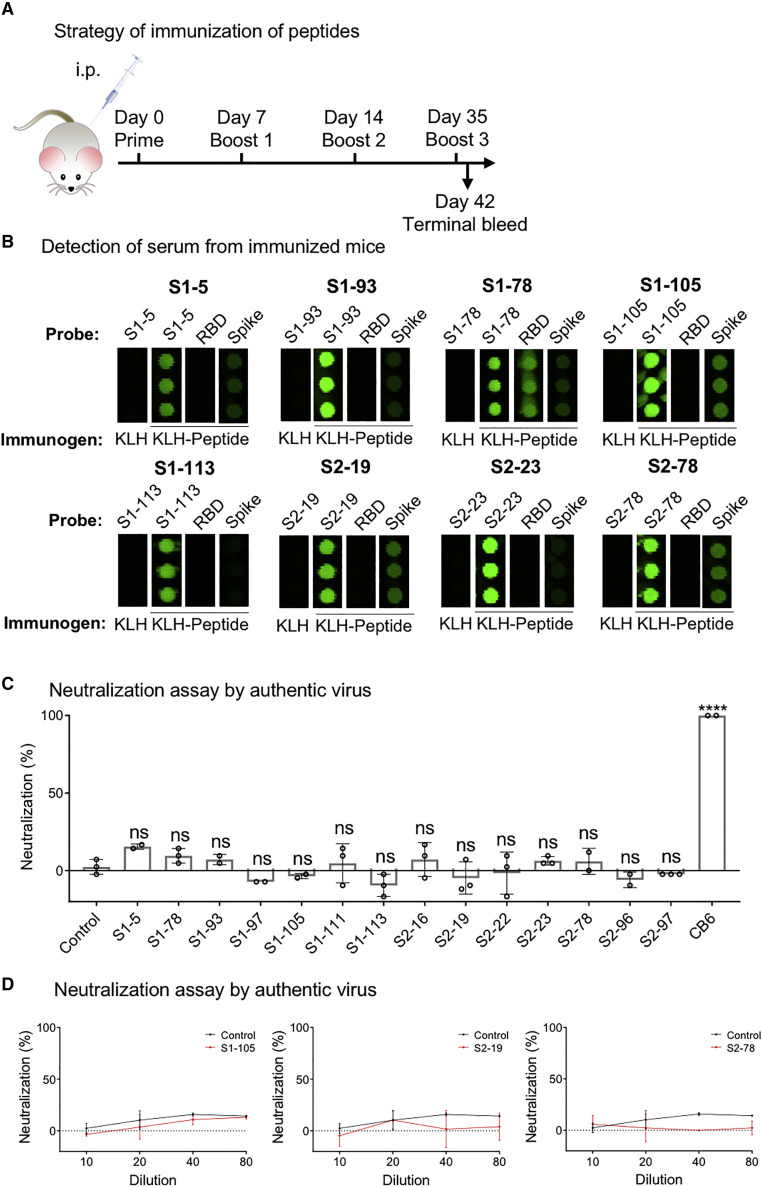
Although the RBD region is the essential part contributing to the neutralization activity of S-protein-elicited antibodies, other regions of the S protein are frequently reported to be able to elicit neutralizing antibodies (Chi et al., 2020), including linear epitopes (Li et al., 2020a; Poh et al., 2020). However, the entire spectrum showing which parts can and which parts cannot elicit neutralizing antibodies has barely been elucidated. To fully investigate the potential to elicit a neutralizing antibody using significant epitopes, the most physiologically relevant method is to enrich specific antibodies from COVID-19 sera using the significant epitopes and then run a neutralization assay (Li et al., 2020a). However, for antibody enrichment, a large volume of each serum sample is required, and it is extremely difficult to collect more COVID-19 samples in large volumes at this time. Alternatively, we tested whether the significant linear epitopes could elicit neutralizing antibodies in a mouse model. When selecting overlapping peptides, we chose one with a higher response frequency, except when both peptides were of a high response frequency. For example, S1-78 was selected to represent the RBD. In total, 14 peptides were selected. The peptides were conjugated to KLH (keyhole limpet hemocyanin), and 3 mice were immunized for each peptide (Figure 7 A). Most of the linear epitopes elicited specific antibodies (Figure 7B). Next, we performed neutralization assays on the authentic SARS-CoV-2 virus with sera collected from the immunized mice. Our results showed that some sera have marginal neutralization activities (Figures 7C and 7D), although no neutralization activity was observed for the rest of the sera. The antibodies generated by S1-78, which is located squarely in the RBM region, recognize the RBD protein (Figure 7B) and are assumed to have high neutralization activity. Unfortunately, significant neutralization activity was not observed, suggesting the lower efficacy of linear peptide immunization antibodies (Zhang et al., 2020a). These results suggest that significant epitopes may not be suitable for vaccine development because of the very limited neutralization activities; thus, our results further strengthen the central role of RBD for eliciting the neutralizing antibody.
Figure 7.
Neutralizing activities of the antibodies elicited by the linear epitopes in mice
(A) The workflow to generate and collect anti-sera from immunized mice for the peptides. KLH only was used as a control.
(B) The specificity of the sera was monitored using a peptide microarray containing the S protein, the RBD, and all the peptides that cover the entire S protein. Some representative results are shown.
(C) Summary of the neutralization activity of the peptide-immunized antibodies in mouse sera (n = 3). The sera were diluted 1:10. p values were calculated by one-way ANOVA test. ∗∗∗∗p < 0.0001.
(D) Representative results of the neutralization activity (top) and morphology of plaques (bottom). Sera were serial diluted at a range from 1:10 to 1:80.
Discussion
Herein, we aim to reveal IgG responses triggered by the SARS-CoV-2 Spike protein on a systematic level. We adopted a newly developed peptide microarray with full coverage of the Spike protein (Li et al., 2020a) and analyzed 1,051 COVID-19 sera and 528 control sera. A set of highly immunogenic epitopes were revealed, and a comprehensive IgG linear epitope landscape was constructed.
One limitation of this study is that only short peptides were involved. Though linear epitopes are nicely represented, conformational epitopes may not be. For example, for the RBD region, which is highly immunogenic, only 3 linear epitopes of moderate immunogenicity were identified. To overcome this limitation, one method is to synthesize longer peptides, which may retain some conformational information (Zhang et al., 2020a). It is necessary to note that, for the linear epitopes we identified, they are highly physiologically relevant, as all of them were revealed through analyzing sera from COVID-19 patients.
Our study presents the first IgG linear epitope landscape of the Spike protein, which could only be achieved by analyzing a large cohort of samples using a systematic approach, such as the full-coverage peptide microarray of the Spike protein. According to the landscape, it is obvious that the Spike protein is highly immunogenic, there are many epitopes on the protein, and these epitopes are not evenly distributed across the Spike protein.
Using sera from peptide-immunized mice, we examined the ability of the linear epitopes to elicit neutralization activity with the intact SARS-CoV-2 virus. However, significant neutralization activity was not observed. Our previous studies and those of other groups have identified some linear epitopes that can elicit neutralizing antibodies (Li et al., 2020a; Poh et al., 2020). One plausible reason for this inconsistency is that, although the neutralization activities of the responsive epitope-elicited antibodies are very low, a much higher concentration of antibodies is needed to demonstrate obvious neutralization activity. To support this explanation, for the three antibodies enriched from patient sera by S1-93, S1-105, and S2-78 that demonstrate neutralization activities, the estimated IC50 (half maximal inhibitory concentration) is approximately 5–20 μg/mL (Li et al., 2020a), which is much higher than that for the RBD-targeting antibodies isolated from COVID-19 patients (Cao et al., 2020; Ju et al., 2020). In addition, linear peptide-directed immunization might not effectively generate “valid” antibodies in vivo because the conformation of the peptide might differ from that in the native Spike protein. Furthermore, this result is consistent with the study in which RBD-derived peptides were used for the immunization of mice (Zhang et al., 2020a).
Our results can be used to examine the hypotheses regarding the relationship between SARS-CoV-2-specific humoral responses to linear spike peptides and the elicitation and recognition of neutralizing antibodies. In other words, the antibodies against the highly immunogenetic linear epitopes have less or very limited neutralization activities against SARS-CoV-2. Thus, when applying the S protein for vaccination, it may be necessary to block or remove some of the immunodominant linear epitopes that we identified to improve the efficacy and avoid possible side effects.
Taken together, we built a comprehensive linear epitope landscape that covers the entire sequence of the SARS-CoV-2 Spike protein by using sera from 1,051 COVID-19 patients. A set of 16 highly immunogenic epitopes outside of the RBD region were identified. The antibody responses against several epitopes are associated with severity. Little neutralization activity was observed for the antibodies against the highly immunogenic epitopes. These findings facilitate our understanding of the humoral immunity of SARS-CoV-2 on a systems level, which may provide some directions for the refinement of COVID-19 vaccine design.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cy3-Goat Anti-Human IgG | Jackson ImmunoResearch | Cat# 109-165-008 |
| Alexa Fluor 647 Donkey Anti-Human IgM | Cy3-Goat Anti-Human IgG | Cat# 709-605-073 |
| Anti-Human IgG | Sigma | Cat# I2136 |
| Anti-Human IgM | Sigma | Cat# I2386 |
| Anti-BSA antibody | Sangon Biotech | Cat# D220272-0025 |
| Biological samples | ||
| Human Sera | This paper | Table S2 |
| Chemicals, peptides, and recombinant proteins | ||
| Peptides of SARS-CoV-2 spike protein | This paper | Table S1 |
| BSA | Yeasen Biotech | Cat# B27371 |
| Sulfo-SMCC | Thermo Fisher | Cat# 22322 |
| EZ-Link Sulfo-NHS-LC-Biotin | Thermo Fisher | Cat# 21335 |
| SARS-CoV-2 Spike S1 | Hangzhou Bioeast biotech. Co.,Ltd. | N/A |
| SARS-CoV-2 Spike S1 | Sanyou biopharmaceuticals Co.,Ltd. | Cat# PNA002 |
| SARS-CoV-2 RBD | Sanyou biopharmaceuticals | Cat# PNA004 |
| SARS-CoV-2 Spike Nucleocapsid protien | VACURE Biotechnology | Cat# AG-PL-2101 |
| Human IgG Isotype Control | Thermo Fisher | Cat# 02-7102 |
| MnJ adjuvant | Zhengfan Jiang’s Lab | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 Virus (nCoV-2019BetaCoV/Wuhan/WIV04/2019) | Hongping Wei’s Lab | N/A |
| Experimental models: mouse/ cell lines | ||
| BALB/c mouse | Shanghai SLAC Laboratory Animal Co.,Ltd | N/A |
| Vero E6 Cell line | ATCC | CRL-1586 |
| Deposited data | ||
| Raw data of peptide microarray | Protein Microarray Database | http://www.proteinmicroarray.cn PMDE242 |
| Software and algorithms | ||
| SPSS | IBM | https://www.ibm.com/cn-zh/analytics/spss-statistics-software |
| ImmunomeBrowser | IEDB | http://tools.iedb.org/immunomebrowser/ |
| Pymol | Pymol | https://pymol.org/2/ |
| Pheatmap package | R | https://cran.r-project.org/web/packages/pheatmap/index.html |
| Other | ||
| PATH protein microarray slides | Grace Bio-Labs | Cat# 805025 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sheng-ce Tao (taosc@sjtu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The peptide microarray data generated during this study are entered in the Protein Microarray Database (http://www.proteinmicroarray.cn) under accession number PMDE243. Additional data related to this paper may be requested from the authors.
Experimental model and subject details
Patients and samples
The study was approved by the Ethical Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China (IRB ID:TJ-C20200128). Written informed consent was obtained from all participants enrolled in this study. COVID-19 patients were hospitalized and received treatment in Tongji Hospital from 17 February 2020 to 28 April 2020. Of the 1,051 patients, 523 are males and 528 are females and the mean age of these patients is 60.3 with 614 patients over 60-year-old (Table S2). The basic criteria to define the severity, i.e., mild, moderate, severe and critically severe, are according to the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7), released by the National Health Commission & State Administration of Traditional Chinese Medicine. Non-severe patients are those with mild or moderate symptoms, while severe patients are those with severe or critically severe symptoms. Sera were collected during hospitalization at viable time points (Table S2). Sera from the control group of healthy donors, lung cancer patients, and patients with autoimmune diseases were collected from Ruijin Hospital, Shanghai, China or Tongren Hospital, Shanghai, China. All the sera were stored at −80°C until use.
Peptide synthesis and conjugation with BSA
The N-terminal amidated peptides were synthesized using GL Biochem, Ltd. (Shanghai, China). Each peptide was individually conjugated with BSA using Sulfo-SMCC (Thermo Fisher Scientific, MA, USA), according to the manufacturer’s instructions. Briefly, BSA was activated by Sulfo-SMCC in a molar ratio of 1: 30, followed by dialysis in PBS buffer. Peptides containing cysteine were added in a w/w ratio of 1:1 and incubated for 2 h, followed by dialysis in PBS to remove free peptides. A few conjugates were randomly selected for examination by SDS-PAGE. For conjugates of the biotin-BSA-peptide, before conjugation, BSA was labeled with biotin using the NHS-LC-Biotin reagent (Thermo Fisher Scientific, MA, USA) at a molar ratio of 1: 5 and then activated using Sulfo-SMCC.
Method details
Peptide microarray fabrication
The peptide-BSA conjugates and S1 protein, the RBD protein and N protein of SARS-CoV-2, along with the negative (BSA) and positive controls (anti-human IgG and IgM antibodies), were printed in triplicate on a PATH substrate slide (Grace Bio-Labs, Oregon, USA) to generate identical arrays in a 1 × 7 subarray format using a Super Marathon printer (Arrayjet, UK). The microarrays were stored at −80°C until use.
Microarray-based serum analysis
A 14-chamber rubber gasket was mounted onto each slide to create individual chambers for the 14 identical subarrays. The microarray was used for serum profiling as previously described with minor modifications (Li et al., 2020b). Briefly, the arrays stored at −80°C were warmed to room temperature and then incubated in blocking buffer (3% BSA in 1 × PBS buffer with 0.1% Tween 20) for 3 h. A total of 200 μL of diluted sera or antibodies was incubated with each subarray for 2 h. The sera were diluted at 1:200 for most samples, and for the competition experiment, free peptides were added at a concentration of 0.25 mg/mL. For the enriched antibodies, 0.1-0.5 μg antibodies were included in the 200 μL incubation buffer. The arrays were washed with 1 × PBST, and bound antibodies were detected by incubating with Cy3-conjugated goat anti-human IgG and Alexa Fluor 647-conjugated donkey anti-human IgM (Jackson ImmunoResearch, PA, USA), which were diluted at 1: 1,000 in 1 × PBST. The incubation was carried out at room temperature for 1 h. The microarrays were then washed with 1 × PBST and dried by centrifugation at room temperature and scanned using a LuxScan 10K-A (CapitalBio Corporation, Beijing, China), with the parameters set at 95% laser power/PMT 550 and 95% laser power/PMT 480 for IgM and IgG, respectively. The fluorescent intensity was determined using GenePix Pro 6.0 software (Molecular Devices, CA, USA).
Mouse immunization
The peptides were synthesized and coupled to KLH through SMCC (Thermo Fisher Scientific, MA, USA) by GL Biochem Ltd. (Shanghai, China). For each peptide, three female mice (BALB/c) approximately 6 ~8 weeks old were immunized four times on a weekly schedule, and 50 μg KLH conjugated peptide was mixed with 60 μg manganese salt (MnJ) adjuvants (Zhang et al., 2019) for each immunization through intraperitoneal (IP) injections. Serum samples were collected 7 d after each immunization for microarray and neutralization assays.
Cell lines and viruses
Vero E6 cells (ATCC CRL-1586) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 μg/mL streptomycin and 100 U/mL penicillin, 10% FBS (Sigma-Aldrich, USA), and maintained at 37°C with 5% CO2. SARS-CoV-2 (nCoV-2019BetaCoV/Wuhan/WIV04/2019) was obtained from the National Virus Resource, Wuhan Institute of Virology, Chinese Academy of Sciences. All handling of the virus was conducted in a BSL-3 laboratory.
Plaque reduction neutralization assay
For the plaque reduction neutralization test, Vero E6 cells were grown to confluence in a 12-well plate (Corning, USA) at 37°C, 5% CO2, overnight. Additionally, 300 μL twofold serial diluted serum samples from 1:10 to 1:80 were prepared in 2.5% FBS-DMEM. The live SARS-CoV-2 virus (300 PFU/mL) was added into the serum at a 1:1 volume, mixed completely, and incubated at 37°C for 1 h. Then, cell culture media were removed from the 12-well plates, and 500 μL of serum-virus mixture was added to each cell. After incubation at 37°C for 1 h, the mixture was replaced with 1000 μL 2.5% FBS-DMEM containing 0.8% carboxymethylcellulose (Sigma-Aldrich, USA). Four days later, the plates were fixed with 8% paraformaldehyde and stained with 0.5% crystal violet. Neutralizing antibody CB6 (Shi et al., 2020) of SARS-CoV-2 was used as the positive control. The antibody was provided by Prof. Jing-Hua Yan at the Institute of Microbiology, Chinese Academy of Sciences.
Quantification and statistical analysis
Data analysis of peptide microarray
For each spot, signal intensity was defined as the mean_foreground subtracted by the mean_background. The signal intensities of the triplicate spots for each peptide or protein were averaged. The overall_mean_background and the overall_standard deviation (SD)_background of all the arrays probed with COVID-19 sera were calculated. Cutoff1 was defined as (the overall_mean_background + 2∗overall_SD_background). According to the array data, Cutoff1 was calculated as 380.7. For the control arrays, mean_foreground and SD_foreground for each peptide and protein were calculated. Cutoff2 was set as (control_mean_signal intensity + 2∗control_SD_signal intensity). For each peptide or protein, a SARS-CoV-2-specific positive response was called when the average_signal intensity was larger than both Cutoff1 and Cutoff2. Response frequency was then defined as the number of peptides with a positive response divided by the total number of peptides on the microarray.
Structure analysis
The spike protein structures (PDB: 6X6P and 6VYB), RBD-ACE2 structure (PDB: 6M0J) and antibody-RBD complex structure (PDB: 7C01, 7BWJ, 7BYR, 6W41 and 6WPT) were used to analyze the structural details of the epitopes identified from the peptide microarray. The C-terminal (1146-1273) structure of the Spike protein came from a modeling structure, QHD43416.pdb, generated by C-I-TASSER (https://zhanglab.ccmb.med.umich.edu/COVID-19/) and aligned to the C terminus of the Spike protein (PDB: 6X6P). Structural analysis was processed in Pymol. The alignment and homology analysis of 7 human coronaviruses and one bat coronavirus was generated using the ClustalW algorithm from EMBL-EBI (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Acknowledgments
We thank Dr. Daniel M. Czajkowsky for critical reading and editing. We thank Prof. Zhengfan Jiang from the Key Laboratory of Cell Proliferation and Differentiation of the Ministry of Education School of Life Science, Peking University for kindly providing the manganese salt (MnJ) adjuvant. We also thank Prof. Jing-Hua Yan at the Institute of Microbiology, Chinese Academy of Sciences for kindly providing the CB6 antibody. This work was partially supported by the National Key Research and Development Program of China grant (no. 2016YFA0500600), the Science and Technology Commission of Shanghai Municipality (no. 19441911900), the Interdisciplinary Program of Shanghai Jiao Tong University (no. YG2020YQ10), the National Natural Science Foundation of China (nos. 31970130, 31600672, 31670831, and 31370813), and China Postdoctoral Science Foundation (no. 2020M680857).
Author contributions
S.T. developed the conceptual ideas and designed the study. Z.S., F.W., H.H., Yandi Zhang, X.L., Z.Y., H.S., and J.W. collected the sera samples. X.F., Y.L., M.M., Z.X., B.Z., H.C., C.Y., J.X., X.W., Yun-xiao Zheng, D.L., H.Z., H.J., H.Q., and S.G. performed the microarray experiments and data analysis. H.W., W.H., and Yanan Zhang performed the neutralization assay. S.T., Y.L., and M.M. wrote the manuscript with suggestions from other authors.
Declaration of interests
The authors declare no competing interests.
Published: March 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108915.
Supplemental information
References
- Abbott R.K., Crotty S. Factors in B cell competition and immunodominance. Immunol. Rev. 2020;296:120–131. doi: 10.1111/imr.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.F., Quadeer A.A., McKay M.R. COVIDep: a web-based platform for real-time reporting of vaccine target recommendations for SARS-CoV-2. Nat. Protoc. 2020;15:2141–2142. doi: 10.1038/s41596-020-0358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S.D., Jackson K.J.L. Predicting vaccine responsiveness. Cell Host Microbe. 2015;17:301–307. doi: 10.1016/j.chom.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Cai Y., Yin D., Ling S., Tian X., Li Y., Xu Z., Jiang H., Zhang X., Wang X., Shi Y. A single dose SARS-CoV-2 simulating particle vaccine induces potent neutralizing activities. bioRxiv. 2020 doi: 10.1101/2020.05.14.093054. [DOI] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli K.M., Carnathan D.G., Nogal B., Martin J.T., Rodriguez O.L., Upadhyay A.A., Enemuo C.A., Gebru E.H., Choe Y., Viviano F. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell. 2019;177:1153–1171.e28. doi: 10.1016/j.cell.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio T.F., Das H., Sheward D.J., Hanke L., Pazicky S., Pieprzyk J., Sorgenfrei M., Schroer M.A., Gruzinov A.Y., Jeffries C.M. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11:5588. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini E.A., Hughes J.V., Perlow D.S., Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F., Corey L., Fernandes P., Gilbert P.B., Hotez P.J., Rao S., Santos M.R., Schuitemaker H., Watson M., Arvin A. Prospects for a safe COVID-19 vaccine. Sci. Transl. Med. 2020;12:eabe0948. doi: 10.1126/scitranslmed.abe0948. [DOI] [PubMed] [Google Scholar]
- Herrera N.G., Morano N.C., Celikgil A., Georgiev G.I., Malonis R.J., Lee J.H., Tong K., Vergnolle O., Massimi A.B., Yen L.Y. Characterization of the SARS-CoV-2 S protein: biophysical, biochemical, structural, and antigenic analysis. bioRxiv. 2020 doi: 10.1101/2020.06.14.150607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt N.K., Seydoux E., Wan Y.-H., Edara V.V., Stuart A.B., Feng J., Suthar M.S., McGuire A.T., Stamatatos L., Pancera M. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 2020;11:5413. doi: 10.1038/s41467-020-19231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020;5:eabe0367. doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.W., Li Y., Zhang H.N., Wang W., Yang X., Qi H., Li H., Men D., Zhou J., Tao S.C. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11:3581. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Sheffield COVID-19 Genomics Group Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.-C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Larman H.B., Zhao Z., Laserson U., Li M.Z., Ciccia A., Gakidis M.A.M., Church G.M., Kesari S., Leproust E.M., Solimini N.L., Elledge S.J. Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lai D.Y., Zhang H.N., Jiang H.W., Tian X., Ma M.L., Qi H., Meng Q.F., Guo S.J., Wu Y. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell. Mol. Immunol. 2020;17:1095–1097. doi: 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li C.-Q., Guo S.-J., Guo W., Jiang H.-W., Li H.-C., Tao S.-C. Longitudinal serum autoantibody repertoire profiling identifies surgery-associated biomarkers in lung adenocarcinoma. EBioMedicine. 2020;53:102674. doi: 10.1016/j.ebiom.2020.102674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., HLH Across Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 Spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv. 2020 doi: 10.1101/2020.04.07.023903. [DOI] [Google Scholar]
- Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y., Chee R.S.-L., Fong S.W., Yeo N.K., Lee W.H., Torres-Ruesta A. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., C4591001 Clinical Trial Group Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujas E., Kucharska I., Tan Y.Z., Benlekbir S., Cui H., Zhao T., Wasney G.A., Budylowski P., Guvenc F., Newton J.C. Multivalency transforms SARS-CoV-2 antibodies into broad and ultrapotent neutralizers. bioRxiv. 2020 doi: 10.1101/2020.10.15.341636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Fan X., Nie S., Kou L., Zhang X., Liu H., Ji S., Deng R., Wang A., Zhang G. Identification of a linear B-cell epitope on glycoprotein (GP) 2a of porcine reproductive and respiratory syndrome virus (PRRSV) Int. J. Biol. Macromol. 2019;139:1288–1294. doi: 10.1016/j.ijbiomac.2019.07.171. [DOI] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora M., von Bülow S., Blanc F.E.C., Gecht M., Covino R., Hummer G. Map of SARS-CoV-2 spike epitopes not shielded by glycans. bioRxiv. 2020 doi: 10.1101/2020.07.03.186825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Sinai Immunology Review Project Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Xing S., Ding L., Wang Y., Gu C., Wu Y., Rong B., Li C., Wang S., Chen K. Human-IgG-neutralizing monoclonal antibodies block the SARS-CoV-2 infection. Cell Rep. 2020;32:107918. doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., Teng F., Dai J., Duan H., Guo S. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020;6:2238–2249. doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hou X., Wu X., Liang T., Zhang X., Wang D., Teng F., Dai J., Duan H., Guo S. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. bioRxiv. 2020 doi: 10.1101/2020.03.26.994756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li Y., Hou B., Pronobis M.I., Wang M., Wang Y., Cheng G., Weng W., Wang Y., Tang Y. An array of 60,000 antibodies for proteome-scale antibody generation and target discovery. Sci. Adv. 2020;6:eaax2271. doi: 10.1126/sciadv.aax2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.-C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., van Gils M.J. An alternative binding mode of IGHV3-53 antibodies to the SARS-CoV-2 receptor binding domain. bioRxiv. 2020 doi: 10.1101/2020.07.26.222232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.J., Kula T., Xu Q., Li M.Z., Vernon S.D., Ndung’u T., Ruxrungtham K., Sanchez J., Brander C., Chung R.T. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.-C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamecnik C.R., Rajan J.V., Yamauchi K.A., Mann S.A., Sowa G.M., Zorn K.C., Alvarenga B.D., Stone M., Norris P.J., Gu W. ReScan, a multiplex diagnostic pipeline, pans human sera for SARS-CoV-2 antigens. medRxiv. 2020 doi: 10.1101/2020.05.11.20092528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang C., Guan Y., Wei X., Sha M., Jing M., Lv M., Xu J., Wan Y., Jiang Z. The manganese salt (MnJ) functions as a potent universal adjuvant. BioRxiv. 2019:783910. [Google Scholar]
- Zhang B.Z., Hu Y.F., Chen L.L., Yau T., Tong Y.G., Hu J.C., Cai J.P., Chan K.-H., Dou Y., Deng J. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res. 2020;30:702–704. doi: 10.1038/s41422-020-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- Zhou D., Duyvesteyn H.M.E., Chen C.-P., Huang C.-G., Chen T.-H., Shih S.-R., Lin Y.-C., Cheng C.-Y., Cheng S.-H., Huang Y.-C. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The peptide microarray data generated during this study are entered in the Protein Microarray Database (http://www.proteinmicroarray.cn) under accession number PMDE243. Additional data related to this paper may be requested from the authors.