Abstract
Background
Sensitive molecular diagnostics and correct test interpretation are crucial for accurate COVID-19 diagnosis and thereby essential for good clinical practice. Furthermore, they are a key factor in outbreak control where active case finding in combination with isolation and contact tracing are crucial.
Aim
With the objective to inform the public health and laboratory responses to the pandemic, we reviewed current published knowledge on the kinetics of SARS-CoV-2 infection as assessed by RNA molecular detection in a wide range of clinical samples.
Methods
We performed an extensive search on studies published between 1 December 2019 and 15 May 2020, reporting on molecular detection and/or isolation of SARS-CoV-2 in any human laboratory specimen.
Results
We compiled a dataset of 264 studies including 32,515 COVID-19 cases, and additionally aggregated data points (n = 2,777) from sampling of 217 adults with known infection timeline. We summarised data on SARS-CoV-2 detection in the respiratory and gastrointestinal tract, blood, oral fluid, tears, cerebrospinal fluid, peritoneal fluid, semen, vaginal fluid; where provided, we also summarised specific observations on SARS-CoV-2 detection in pregnancy, infancy, children, adolescents and immunocompromised individuals.
Conclusion
Optimal SARS-CoV-2 molecular testing relies on choosing the most appropriate sample type, collected with adequate sampling technique, and with the infection timeline in mind. We outlined knowledge gaps and directions for future well-documented systematic studies.
Keywords: SARS-CoV-2, COVID-19, Laboratory Diagnosis, Polymerase Chain Reaction
Introduction
In 2019 a novel human pathogenic coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in Wuhan, China [1], leading to a worldwide outbreak, declared a public health emergency of international concern on 30 January 2020 [2] and a pandemic on 11 March 2020 [3].
SARS-CoV-2 is a positive-stranded RNA virus from the species severe acute respiratory syndrome-related coronavirus, subgenus Sarbecovirus, genus Betacoronavirus, family Coronaviridae. The species contains a wide range of bat and human viruses including SARS-CoV-1 that caused an outbreak in 2002–03. The SARS-CoV-2 origins are still unknown, but zoonotic transmission, with bats (in particular Rhinolophus spp.) as the probable primary reservoir and other yet unknown animals as intermediate hosts, is considered the most likely route [4,5].
The disease caused by SARS-CoV-2 is coronavirus disease (COVID-19). In the context of the pandemic, which is currently ongoing, rapid and reliable laboratory diagnosis is essential for detection, confirmation, and ruling out of cases, clinical management and hospital infection prevention measures, source and contact tracing, and (lifting of) isolation measures. Laboratory testing plays a critical role in surveillance to guide public health response. Nucleic acid amplification tests became the first line of testing for SARS-CoV-2 infection recommended by the World Health Organization (WHO) [6]. Serological tests are increasingly being implemented [7,8].
Here, we reviewed the current knowledge on the laboratory aspects of COVID-19 diagnostics with a focus on SARS-CoV-2 molecular assays and summarised key findings for different sample types to support the laboratory response for clinical case management and to inform public health measures to control the pandemic.
Methods
Search and selection of published reports
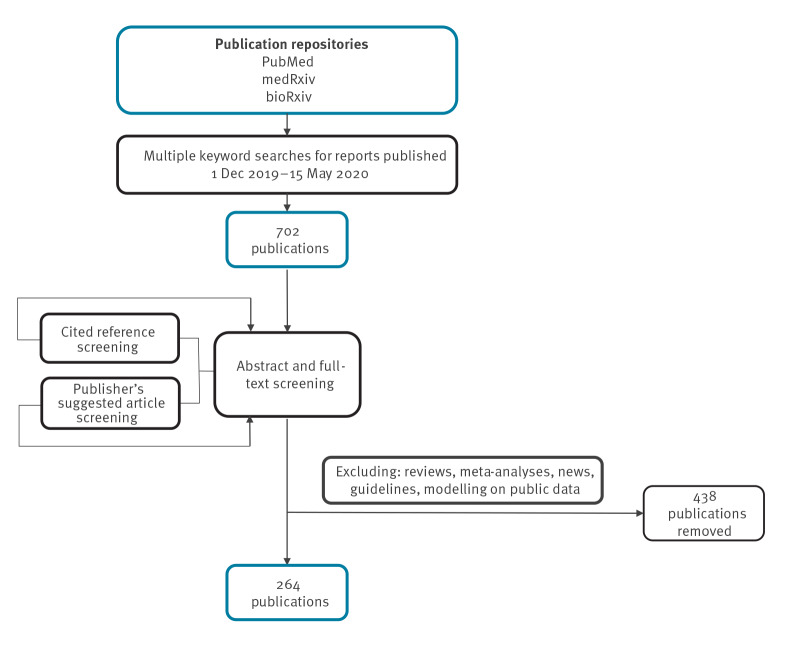
We searched repetitively PubMed, medRxiv and bioRxiv with keyword ‘coronavirus’ limiting to results published between 1 December 2019 and 15 May 2020 and screened the titles and brief descriptions of > 8,700 publications in total. We identified studies on SARS-CoV-2 (including name variations like ‘novel coronavirus’, ‘2019-nCoV’) in humans (all ages), written in English, Chinese or French, and excluded reviews, viewpoints, or news. We selected studies using the words ‘detection’, ‘testing’, ‘PCR’, ‘viral load’, ‘viral kinetics/dynamics/clearance/shedding’, ‘isolation’, ‘persistence’, ‘samples’, ‘bodily fluids’, ‘diagnosis’, ‘case report/series’, ‘case(s)’, ‘cluster’, ‘transmission’, ‘patients’, ‘neonate’, ‘child(ren)’, ‘pregnant’, ‘clinical characteristics/findings/manifestations/features/outcomes’, ‘infection’, ‘pneumonia’, ‘asymptomatic’ in the title, brief description or abstract (if available) aiming to narrow down to clinical reports. This yielded 702 publications for in-depth abstract and full-text screening. Additionally, we scanned literature cited in these articles as well as suggested similar publications and COVID-19 resource collections on the publishers’ websites. Finally, we included 264 studies reporting on SARS-CoV-2 molecular detection and/or virus isolation in any laboratory specimens of COVID-19 cases. We excluded reviews, meta-analyses, news, guidelines, or modelling studies based on public data (Figure 1).
Figure 1.
Flow-chart of the review process for publications between 1 December and 15 May 2020 on SARS-CoV-2 molecular detection and/or virus isolation in laboratory specimens
Data extraction
We aimed to summarise the current information with regard to SARS-CoV-2 infection kinetics in relation to clinical syndrome, in different bodily fluids, while also noting any specifics in some groups (pregnant women, children and immunocompromised individuals).
We extracted data on cases’ demographics such as the number of adults, children, pregnant and immunocompromised individuals as well as age, sex and severity of disease. Moreover information on specimens tested for SARS-CoV-2 was gathered, including the number of patients with collected respiratory (nasopharyngeal/oropharyngeal/other swabs, sputum, bronchoalveolar lavage fluid, endotracheal aspirate), gastrointestinal (GI) (stool, anal/rectal swabs, endoscopic samples) and blood samples, as well as those with oral fluid, tears, urine, cerebrospinal fluid, semen, vaginal fluid, breast milk and perinatal samples. All sample types tested by PCR and viral cultures, where available, were included. The number of patients with sequenced samples was also compiled.
Definitions of nasopharyngeal swabs and severe disease used for the study
Even though nasopharyngeal swabs are the routine respiratory sample for viral infections, we also included publications with nasal/midturbinate swabs or those using the term ‘nasal’ indiscriminately when describing the nasopharyngeal swab collection technique.
Due to varying disease severity definitions, we opted for a simplified approach: patients with symptoms or clinical course described as mild, moderate, common, typical, are referred to as patients with ‘mild’ disease, while patients with illnesses characterised as severe, or patients described as critically ill and/or admitted to intensive-care units (ICU), are referred to as patients with ‘severe’ disease.
Ethical statement
As the review included publicly available published data, ethical approval was not needed for the study.
Results
The compiled dataset contained 32,515 COVID-19 cases (Supplementary Dataset). Where possible, we noted duplicate case reports in different publications. Additionally, we aggregated data from 217 adults with data points from 2,777 samples with known collection day post symptom onset (dps).
Overview of SARS-CoV-2 in different specimens from adults
The distribution of different types of samples testing positive for SARS-CoV-2 as a function of dps and/or illness severity are shown in Figure 2, Figure 3, the Table and the Supplementary Table).
Figure 2.
COVID-19 aggregated RT-PCR results in different sample types (n = 2,777) by days post symptom onset in adult patients (n = 217) with mild or severe disease, data from studies published 1 December 2019–15 May 2020
COVID-19: coronavirus disease; dps: days post symptom onset; ICU: intensive care unit; NP: nasopharyngeal; OP: oropharyngeal; RT-PCR: real-time PCR; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
The number of samples with (above dps axis) and without (below dps axis) detectable SARS-CoV-2 RNA are aggregated by sample type, collection dps and disease severity. Sampling for patients with symptoms or clinical course described in the original publications as mild, moderate, common, or typical are shown in circles: a total of 1,627 samples from 173 adults. Sampling for patients with severe disease or who were critically ill and/or admitted to the ICU are shown in squares: a total of 1,150 samples from 44 adults. Asymptomatic patients were excluded. Sample types are as follows: NP swab (NP/midturbinate/nasal swab), OP swab (oropharyngeal/throat swab), NP + OP swab (both swabs collected in one tube or results published aggregated), sputum (induced/spontaneous sputum), stool, anal swab (anal/rectal swab), blood (serum, plasma, whole blood or not specified), urine. Sample collection dps are shown as stated by the authors, and if unavailable extracted from timeline descriptions.
Figure 3.
Heatmaps of laboratory-confirmed adult COVID-19 cases with mild and severe disease (n = 217) and with positive and negative RT-PCR results in different sample types (n = 2,777) by days post symptoms onset, data from studies published 1 December 2019–15 May 2020
COVID-19: coronavirus disease; ICU: intensive care unit; NP: nasopharyngeal; OP: oropharyngeal; RT-PCR: real-time PCR.
Laboratory-confirmed COVID-19 cases (n = 217) are grouped by disease severity: those (panels A and B) with symptoms or clinical course described in the original publications as mild, moderate, common, or typical (n = 173 adults) are referred to as ‘cases with mild disease’, while those (C and D) with severe disease or critically ill and/or admitted to the ICU (n = 44 adults) are referred to as ‘cases with severe disease’. Asymptomatic patients were excluded. The cumulative number of cases with respective positive (A and C) or negative (B and D) RT-PCR results on different sample types (a total of 1,627 and 1,150 samples from cases with mild and severe disease respectively) on a respective day post symptoms onset are represented using a colour gradient. Sample types are as follows: NP swab (nasopharyngeal/midturbinate/nasal swab), OP swab (oropharyngeal/throat swab), NP + OP swab (both swabs collected in one tube or results published aggregated), sputum (induced/spontaneous sputum), stool, anal swab (anal/rectal swab), blood (serum, plasma, whole blood or not specified), urine. Sample collection by days post symptoms onset are shown as stated by the authors, and if unavailable extracted from timeline descriptions.
Table. COVID-19 aggregated RT-PCR results in different sample types (n = 2,777) in adult patients (n = 217) with mild or severe disease, data from studies published 1 December 2019–15 May 2020.
| Sample type | Cases with positive results | Cases with negative results | ||||||
|---|---|---|---|---|---|---|---|---|
| Number of samples |
Number with mild disease |
Number with severe disease |
Number of samples |
Number with mild disease |
Number with severe disease |
|||
| URT | NP swab | 461 | 56 | 20 | 254 | 11 | 8 | |
| OP swab | 151 | 29 | 14 | 165 | 9 | 10 | ||
| NP + OP swaba | 197 | 64 | 1 | 296 | 3 | 0 | ||
| All URT swabs | 809 | 149 | 35 | 715 | 23 | 18 | ||
| Sputum | 376 | 36 | 23 | 131 | 0 | 2 | ||
| GIT | Stool | 172 | 14 | 11 | 112 | 11 | 11 | |
| Anal swab | 34 | 5 | 6 | 71 | 8 | 12 | ||
| All GIT samples | 206 | 19 | 17 | 183 | 19 | 23 | ||
| Blood | 26 | 3 | 6 | 222 | 25 | 24 | ||
| Urine | 3 | 1 | 1 | 106 | 26 | 20 | ||
COVID-19: coronavirus disease; GIT: gastrointestinal tract; NP: nasopharyngeal/midturbinate/nasal; OP: oropharyngeal/throat; RT-PCR: real-time PCR; URT: upper respiratory tract.
a Both swabs collected in one tube or results published aggregated.
SARS-CoV-2 kinetics and shedding in the respiratory tract
Some key points found concerning the kinetics and shedding of SARS-CoV-2 in the respiratory tract are summarised in Box 1.
Box 1. Key points with regards to shedding/kinetics of SARS-CoV-2 in the respiratory tract.
SARS-CoV-2 replicates in the throat and upper respiratory tract (URT). Respective samples collected early in the infection (regardless of the severity of symptoms) can be used for diagnostic purposes.
Viral RNA loads peak within the first infection days in the upper and later in the lower respiratory tract; viral load (VL) testing and reporting need to be standardised.
SARS-CoV-2 RNA is detectable from respiratory samples up to 6 weeks in cases with mild disease and 8 weeks in cases with severe disease, and beyond symptoms’ resolution.
There were insufficient systematic comparisons between all types of respiratory samples; higher VLs were observed in sputum than in nasopharygeal and oropharyngeal swabs. A more sensitive detection of SARS-CoV-2 detection resulted from nasopharyngeal swabs vs oropharyngeal swabs.
Computed tomography (CT) findings could sometimes precede viral RNA detection in the URT and full clinical presentation should always be evaluated.
We reviewed 262 studies with respiratory sampling for ≥ 31,957 COVID-19 cases, including data on nasopharyngeal/midturbinate/nasal (NP) swabs (≥ 13,286 cases), oropharyngeal/throat (OP) swabs (≥ 7,301 cases), combined NP and OP swabs (≥ 1,493 cases), sputum (≥ 409 cases), bronchoalveolar lavage fluid (≥ 21 cases), and other respiratory specimens (endotracheal aspirate, bronchoalveolar swab ≥ 21) (Supplementary Dataset) [1,9-268].
SARS-CoV-2 isolation from respiratory tract samples
Lower respiratory tract (LRT) bronchoalveolar lavage (BAL) sampling allowed the initial and subsequent virus culture of SARS-CoV-2 [1,11,13]. Almost all BAL specimens described in peer-reviewed literature had detectable viral RNA regardless of sampling timing, disease severity or comorbidities and were useful for ultimate confirmation of difficult cases [11,44,71,77,78].
Virus isolation was successful from NP swabs 2 dps in two cases with mild disease (using Vero E6 cells) [120]; NP and OP swabs 4 dps (using Vero CCL-81 cells) [76] in a mild case [63], and also at 4 dps from NP swab and nasopharyngeal aspirate in another mild case (using Vero E6 cells) [98]. SARS-CoV-2 was isolated from NP swabs of two cases with severe disease 1 and 10 dps [135]. In a German study of nine mild COVID-19 cases, SARS-CoV-2 was isolated up to 8 dps from both URT swabs (16%) and sputum samples (83%) with VL > 106 copies/mL. Furthermore, the authors detected viral subgenomic mRNAs (sgRNA) which led them to conclude there was ongoing viral replication in the throat up to 5 dps. Sequencing data also showed the continuous presence of two genotypes of SARS-CoV-2 differing by a single mutation in the throat and lungs samples of a patient [131].
La Scola and colleagues cultured 174 NP swabs and nine sputum samples testing positive via PCR (from 155 patients total) and succeeded with virus isolation from 129 samples (124 with observable cytopathic effect on Vero E6 cells). They observed a strong correlation between cycle threshold (Ct) values and virus isolation: 100% isolation rates from samples with a Ct of 13–17 decreasing to 12% at Ct 33 and no isolation from samples with Ct ≥ 34 [211].
An Indian study was successful in isolating SARS-CoV-2 (using Vero CCL-81 cells) from respiratory samples in nine of 12 samples with Ct values ranging 16–25.1 [217].
SARS-CoV-2 detection with regards to the infection timeline and disease severity
Despite the growing amount of literature, only two studies documented NP swabs, OP swabs, and sputum, collected sequentially in the same 16 [172] and 49 [240] patients respectively, while two studies described an upper respiratory specimen paired with sputum for a total of 11 cases [39,131]. SARS-CoV-2 RNA was detected in NP and OP swabs from symptoms onset up to 42 dps in cases with mild disease [158] and 50 dps in those with severe disease [172]. Sputum yielded positive results up to 27 dps in cases with mild disease [131], but 55 dps in those severely ill [172] (Figure 2 and Figure 3). Sun and colleagues aggregated data on 175 NP swabs, 88 OP swabs, and 62 sputum samples, and estimated a median/95th percentile time until loss of detection as follows: NP 22.7/46.3 dps, OP 15.6/32.8 dps, and sputum 20/43.7 dps for 43 cases with mild disease vs NP 33.5/49.4 dps, OP 33.9/53.9 dps, and sputum 30.9/44.7 dps for six cases with severe disease [240].
Nevertheless, we also aggregated the data from 31 observational studies with available infection timeline and respiratory sampling for 216 cases [9,18,23,25,29,34,37,39,42-45,58,59,63,72,85,98,111,116,120,131,134,158,160,171,172,176,187,197,206] to provide a summary (Figure 2, Figure 3, Table, and Supplementary Table).
Several studies reported SARS-CoV-2 RNA detection from the URT for a median period of 10–20 days [53,59,79,99,105,187,191] with a prolonged detection observed in cases with severe disease [99,105,191]. An aggregation of retrospective observations from 191 hospitalised adults in Wuhan showed a median duration of URT viral RNA detection of 20 (interquartile range (IQR): 16–23) dps with continuous detection until death in non-survivors and ranges 8–37 dps in survivors [79]. SARS-CoV-2 RNA was detectable in URT samples well beyond waning of respiratory symptoms [37,46,107,131].
A study in Nanchang (21 cases with mild, 10 with severe disease) observed clearance in NP swabs within 10 dps in 90% of the cases with mild disease compared with continuous RNA detection > 10 dps in all cases with severe disease [99]. Feng and colleagues detected SARS-CoV-2 RNA in NP swabs of 24 cases with mild disease for 16 ± 7 days compared with 22 ± 4 days in eight ICU patients [105].
SARS-CoV-2 remained detectable in OP swabs ≥ 2 weeks, (nine cases with mild disease) and in sputum > 3 weeks (six cases with mild disease) despite symptoms resolution [131]. Chen and colleagues described recurring SARS-CoV-2 RNA positivity in OP swabs of a patient until 30 dps (VL: 4.56×102 copies/mL), well after pneumonia resolution and hospital discharge [37]. Prolonged viral RNA detection in OP swabs (range: 5–30 dps for 22 cases) was also reported for cases with mild disease regardless of symptoms, including > 3 weeks in eight cases [199]. The study did not address if infectious virus could be detected. Another study including 66 COVID-19 cases found a median of 9.5 (6–11) dps until the first negative results in OP swabs [53]. In some cases, such as in a total of five patients with mild symptoms in Wuhan, SARS-CoV-2 RNA was re-detected 4–15 days following a last negative OP swab and hospital discharge [104]. As reported by the authors of the study [104], the COVID-19 reactivation could have been due to prolonged disease course too. Another Wuhan-based study reported re-hospitalisation with mild symptoms for 11 cases within a median of 16 (range: 6–27) days post initial hospital discharge, of whom four had viral RNA detected in OP swabs [254].
SARS-CoV-2 RNA detection considering health condition and demographic characteristics
The duration of SARS-CoV-2 shedding may be related to patient’s general health condition. In a Wuhan-based study, 27 of 56 cases with mild disease had prolonged SARS-CoV-2 RNA detection > 24 days in NP/OP swabs, associated with old age and comorbidities. The proportion of positive respiratory samples decreased from 89% to 66%, 32%, 5%, and 0% in weeks 2–6 since symptoms onset [187]. Xu and colleagues summarised respiratory samples’ data from 113 patients and observed a median time of 17 (IQR: 13–22) dps where RNA could be detected. Prolonged detection ≥ 15 dps was associated with males, old age, hypertension, severe illness upon admission, invasive mechanical ventilation, and corticosteroid treatment [150]. A Wuhan-based study on 41 discharged cases who had had severe disease reported SARS-CoV-2 RNA detection in OP swabs for a median of 31 (IQR: 24–40, range: 18–48) dps and no significant difference between male and female sex, nor between cases < 65 and ≥ 65 years of age [186].
Viral load in respiratory specimens in the context of disease severity, infection timeline and demographic characteristics
In the studies that provide quantitative results (n = 26) [9,10,25-27,34,37,39,40,59,63,99,111,119,120,122,131,158,160,161,170,172,202,206,220,248], the highest VL in URT specimens were reported in the early days of the disease [34,39,59,63,99,120,131,170,172], also before development of respiratory symptoms [39,119,170]. A study of nine cases with mild disease reported SARS-CoV-2 RNA detection from all NP and OP swabs in the first 5 dps with (average VL: 6.76×105 copies/swab and maximum 7.11×108 copies/swab), whereas detection rate in subsequent swabs was only 40% up to 28 dps (average VL: 3.44×105 copies/swab) [131].
Among four patients (two with mild, two with severe disease), VLs in NP swabs ranged from 7.4 log10 copies/1,000 cells (mild case at 2 dps) and 7.1 log10 copies/1,000 cells (case with severe disease at 6 dps) to negative at 9–14 dps, while for a critically ill case they were in the range of 4.4–6.7 log10 copies/1,000 cells and persisted 24 dps until death [120]. Among 61 healthcare workers, VLs of self-collected combined NP and OP swabs were significantly lower for 56 asymptomatic vs five symptomatic cases [248]. A study on 12 cases (nine with mild, three with severe disease) showed that Ct values from respiratory samples were correlated with disease severity scores like acute respiratory distress syndrome (ARDS) index PaO2/FiO2 ratio and lung injury Murray score, as well with biochemical indicators like albumin levels and lymphocytes and neutrophils percentages, and concluded VLs could serve as a COVID-19 severity predictor [17].
In a paediatric dialysis unit, where 12 COVID-19 cases (six asymptomatic, six mild), mainly healthcare workers, had had a NP swab, the VLs were significantly higher for the patients with symptoms [212]. Kimball and colleagues reported no significant difference between the Ct values in NP swabs collected from 10 symptomatic, 10 presymptomatic and three asymptomatic cases in a nursing facility [119].
He and colleagues reported high VL for 414 OP swabs from 94 patients in the early days of infection and gradual decrease until ca 21 dps with no difference when stratified by sex, age, or disease severity [170]. However, VLs in the sputum of 22 cases with mild disease reached a peak in week 2 after symptoms onset and were significantly lower than those of 74 cases with severe disease [191]. In a cohort of 92 cases (51 with mild disease, 11 with mild disease turning severe, 30 with severe disease) low Ct values (high VL) in sputum were correlated with severe COVID-19 and risk of progression to severe disease [202].
Comparing respiratory specimens, higher VL were reported for sputum than respiratory swabs [9,40,122,131,172]. A study using RT-PCR and droplet digital PCR found both significantly higher positive rates and average VL in sputum (66% and 17,429 ± 6,920 copies/test) compared with OP swabs (37% and 2,552 ± 1,965 copies/test) and NP swabs (16% and 651 ± 501 copies/test) [122]. For 16 critically ill patients, sputum and endotracheal aspirate samples all had detectable SARS-CoV-2 RNA at levels significantly higher than NP and OP swabs with positivity rates of 81% and 63% respectively [172]. In nine cases with mild disease up to 5 dps, the maximum SARS-CoV-2 VL in sputum (2.35×109 copies/mL) was higher compared with respiratory swabs (7.11×108 copies/swab). However, examining paired sputum and swab samples 2–4 dps in seven patients showed higher virus concentration in swabs (two cases), sputum (two cases), and similar virus concentrations in both for the remaining five cases [131]. In a cohort of 52 patients, the positivity rates of sputum samples (77%) were higher than OP swabs (44%) with a significant difference when comparing cases with positive sputum sample and negative OP swab (40%) vs those with negative sputum and positive OP swab (8%) [180].
Choice of a respiratory specimen for SARS-CoV-2 diagnostic
Choosing the most appropriate respiratory specimen depends on timing in the infection course. However, well-documented studies comparing all respiratory and other sample types collected in a known timeline were limited. A preprint study in Guangdong on 866 respiratory samples from 213 symptomatic cases (37 with severe disease), showed that apart from BAL, sputum is the most sensitive sample type for COVID-19 laboratory diagnosis, followed by NP swabs [26]. In the period 0–7 dps, the highest positivity rate was observed in sputum (89% of cases with severe disease, 82% of cases with mild disease) followed by NP swabs (73%, 72%) and OP swabs (60%, 61%). In the period 8–14 dps the same order in positivity rates was observed: sputum (83% of cases with severe disease, 74% of cases with mild disease), NP swabs (72%, 54%), and OP swabs (50%, 30%); as for ≥ 15 dps: sputum (61%, 43%), NP swabs (50%, 55%), and OP swabs (37%, 11%). Sputum samples 0–7 dps also yielded the lowest median Ct values (25 in cases with severe disease, 28.5 in cases with mild disease). BAL samples 8–14 dps were positive for SARS-CoV-2 RNA in 12 cases with severe disease and negative for three cases with mild disease. Beyond 15 dps the study showed 79% positivity in BAL in cases with severe disease [26].
Further peer-reviewed data was also in favour of sputum, or NP swabs [71,77]. A retrospective study of 4,880 cases in Wuhan found 38% positive rate for 4,818 NP and OP swabs compared with 49% for 57 sputum specimens and 80% for five BAL [71]. Another Chinese study including 205 COVID-19 cases yielded overall 1,070 samples with the following positivity rates: 93% for 15 BAL samples, 72% for 104 sputum specimens, 63% for eight NP swabs, 46% for 13 fibrobronchoscope brush biopsy samples, 32% for 398 OP swabs, 29% for 153 faeces samples, 1% for 107 blood samples [77]. Only BAL specimen sequencing, and BAL and sputum samples PCR could confirm SARS-CoV-2 co-infection with influenza A in an ICU patient, whereas repeated NP swabs were negative [78]. Although sputum might seem like a sample of choice, it was described that only a third of 1,099 COVID-19 patients had a productive cough [50], suggesting that in practice NP swabs would be preferable in most cases. Higher sensitivity for NP swabs in comparison to OP swabs was observed as well in other studies and case reports [34,52]. A study with sequential sampling in 18 patients (72 NP and 72 OP swabs) showed higher VL in the nose than the throat [34].
SARS-CoV-2 shedding potential in asymptomatic and pre-symptomatic individuals needs to be elucidated [74,94,101,119,139,144,155,156,160,167,170,224,227,269,270]. Multiple studies worldwide reported SARS-CoV-2 RNA detection in respiratory samples from cases with epidemiological link and no (189 adults, 29 children) [9,31,34,36,61,70,74,94,116,119,123,139,144,155,160,199,205,214,224,227,248] or mild/non-specific symptoms (23 adults, one child) [9,12,15,18,20,24,25,28,30,39,61,74,116,227]. One described SARS-CoV-2 RNA detection in OP swabs for 17 days in an otherwise asymptomatic patient [70]. In a series of testing campaigns in Iceland that led to SARS-CoV-2 RNA detection in combined NP and OP swabs of a total 1,321 cases, those reporting any symptoms ranged between 46% (random population screening) and 94% (targeted screening) [167].
At this stage, it was unclear whether SARS-CoV-2 affects the upper or the lower respiratory tract first, or maintains independent replication in both sites. Thus, choosing between NP and OP swabs, or sputum as sampling strategy should be done with the purpose (general population screening or confirmation of suspected cases) and potential infection timeline in mind. URT sampling would be preferable in the early infection days, especially in asymptomatic or mildly symptomatic suspected cases, whereas lower respiratory sampling provides more reliable confirmation in advanced COVID-19 with lungs’ involvement. Cases with an epidemiological link, radiological findings, and an initial negative result should be monitored further by PCR and evaluated in conjunction with their clinical presentation [10,22,35,38,41,55,56].
The discrepancy between URT and LRT test results triggered a discussion about the lack of sensitivity of PCR testing. In a study among 1,014 patients in Wuhan 59% (95% confidence interval (CI): 56–62%) of OP swabs were positive whereas 88% (95%CI: 86–90%) had chest CT findings within a median of 1 day of the PCR test, consistent with an earlier resolution of viral replication in URT than in LRT samples. Furthermore, 308 patients (75%) had negative PCR results in conjunction with radiologic findings and 14 of 15 cases with CT findings tested positive on a follow-up PCR within a mean of 5 days, a finding that may be more difficult to explain [41]. Another study that aggregated data on PCR results and CT imaging showed all 167 patients had a positive OP swab by the end of their hospitalisation [22]. A multicentre study of 80 COVID-19 cases (no critically ill) in Jiangsu reported the following positivity rates in repeated OP and/or NP swabs collection until laboratory confirmation of the infection: 51%, 38%, and 11% upon the first to third test PCR test [56]. Another Chinese, Hubei-based, study reported SARS-CoV-2 RNA detection in OP swabs of 74%, 12% and 14% of 91 cases upon the first to third test and no significant difference between the 30 cases with severe disease and 61 cases with mild diseases included [221]. A New York-based study on 5,700 COVID-19 cases (including 1,281 in ICU) reported detection upon first NP swab test in 97% (n = 5,517) of cases compared with 3% (n = 183) with repeated tests [193].
SARS-CoV-2 shedding in the gastrointestinal tract
Some key points found about shedding of SARS-CoV-2 in the GI tract are summarised in Box 2.
Box 2. Key points with regards to gastrointestinal shedding of SARS-CoV-2.
SARS-CoV-2 can be isolated from faeces and RNA can be detected regardless of GI symptoms.
Faecal sampling is not recommended for diagnostic screening (except for laboratory diagnosis of suspected cases with negative respiratory tract results).
Prolonged GI viral RNA detection can occur up to 7 weeks from either symptoms onset or hospitalisation, well after respiratory tract clearance and symptoms resolution in some patients.
We reviewed 47 studies providing data on GI sampling in ≥ 629 COVID-19 patients, including stool specimens (≥ 486 cases), anal/rectal swabs (≥ 198), and others (endoscopic samples, n = 14) (Supplementary Dataset) [9,27,39,40,42,43,48-51,53,58-60,63,64,77,84-86,89,91,93,100,102,105,109,117,120,121,126,131,137,152,158,168,171,172,178,179,191,203,206,229,237,240,271].
SARS-CoV-2 isolation from gastrointestinal samples in regard of the infection timeline
SARS-CoV-2 was isolated from stool samples 15 dps from a COVID-19 patient with severe pneumonia (using Vero cells) [271] and from two patients without diarrhoea [77]. In a study involving nine cases with mild disease, virus isolation (on Vero E6 cells) was unsuccessful in stool samples 6–12 dps from four patients, and no virus replication evidence was found through sgRNA assays despite detectable high VL [131]. SARS-CoV-2 nucleocapsid protein was detected in the cytoplasm of gastric, duodenal, and rectum glandular epithelial cells in one patient [58]. The gastric fluid samples of six of 13 critically ill patients were positive for SARS-CoV-2 RNA [172].
SARS-CoV-2 detection and presence/absence of gastrointestinal or other symptoms
GI disease has been described for some COVID-19 patients [9,10,12,14,16,63,83,110,272] and was one of the clinical signs associated with a positive SARS-CoV-2 test [272]. Though there was evidence of SARS-CoV-2 RNA detection in the GI tract, it was not necessarily in cases with GI symptoms [48,102]. Examining studies with an available timeline of sampling (Figure 2, Figure 3, Table, and Supplementary Table), SARS-CoV-2 RNA detection was reported between 3 and 50 dps in stool samples of 14 cases with mild [59,63,120,131] and 11 cases with severe disease [58,59,172], regardless of the presence of diarrhoea. Anal swabs had detectable SARS-CoV-2 RNA between 3 and 45 dps in six cases with mild disease [158,171,206] and seven cases with severe illness [42,172]. No systematic comparison between viral detection in stool and anal swabs was available.
Prolonged GI SARS-CoV-2 RNA detection after the resolution of respiratory symptoms and/or convalescence was observed in several studies [53,58,60,102,131,137,191], though infectious virus shedding is still an outstanding question. In a German study, stool samples remained RNA positive ≥ 21 dps for six cases with mild disease, including a patient with, as suggested by the authors, potential independent intestinal tract replication, based on comparison with the SARS-CoV-2 URT kinetics [131].
SARS-CoV-2 detection in stool compared to other types of specimens
Zheng and colleagues reported a 59% positivity rate in 842 stool samples from 96 patients and a median viral RNA detection duration of 22 (IQR: 17–31) dps that was significantly longer than in sputum/saliva and serum samples [191]. In a Chinese study on 98 COVID-19 cases (18 with severe disease), paired OP swabs and stool specimens for 74 cases yielded the following results: 41 with SARS-CoV-2 detection in stool for a mean 27.9 dps (standard deviation (SD): 10.7) that was 11 days after the clearance in OP swabs with mean detection 16.7 dps (SD: 6.7); while the remaining 33 patients with negative stool results had positive OP swabs for a mean 15.4 dps (SD: 6.7). A patient had detectable SARS-CoV-2 RNA in stool 47 dps and another for 33 days after respiratory clearance. Disease severity was not associated with prolonged GI shedding in this cohort [102]. In another cohort of 42 patients (11 cases with severe disease) with GI symptoms in China, 28 cases (nine with severe disease) had a median RNA detection period of 11 (IQR: 7–13) dps until first positive stool sample compared with 6.5 (IQR: 3‐7.25) dps for OP swabs. More than half (n = 18, five with severe disease) remained stool positive for a median 7 (IQR: 6‐10) days after negative OP swabs [137]. A total of 39 (53%) of 73 hospitalised patients had detectable SARS-CoV-2 RNA in stool and for 17 patients (20%), stools remained positive after respiratory samples turned negative [58]. Another Chinese study on recovering patients (n = 55) found a median of 11 (9–16) dps until the first negative results in stool: 43 patients had a 2 (1–4) days median delay in clearance in faeces compared with OP swabs, while in 12 patients both turned negative at the same time [53]. Sun and colleagues aggregated data on 165 stool samples and estimated a median/95th percentile time until loss of detection of 24.5/45.6 dps for 43 cases with mild disease and 32.5/48.9 dps for six cases with severe disease, both comparable to the estimates for NP swabs [240].
SARS-CoV-2 detection in anal swabs
Like observations for stool specimens, prolonged SARS-CoV-2 RNA detection was reported for anal swabs [42,70,158,168,171,172,206]. Presence of SARS-CoV-2 RNA in anal swabs seemed linked to disease severity in a Guangzhou cohort (two cases with severe and 16 with mild disease) [9]. Zhang and colleagues found positive anal swabs in four of 16 patients upon hospitalisation and six of 16 cases at day 5 of hospitalisation [27]. A Chinese study with discharge criteria of two consecutive negative OP swab and a negative anal swab, reported a median RNA detection duration of 12 (IQR: 9–14; range: 4–34) days between the first detection and two (consecutive) negative samples for 24 patients [158]. SARS-CoV-2 RNA detection in anal swabs ≥ 17 days was observed in an asymptomatic patient [70].
Frequency of patients with viral shedding in the gastrointestinal tract and viral loads
Large well-documented cohort studies are needed to estimate the proportion of COVID-19 cases with continuous GI shedding and the SARS-CoV-2 VL levels over time. A 29% SARS-CoV-2 RNA positivity rate in stool samples was observed in a study aggregating data on 205 patients (unclear how many provided the analysed 153 stool samples) [77]. Few studies (n = 10) provided quantitative data on SARS-CoV-2 RNA detection in stool and anal swabs for 38 COVID-19 cases [27,59,63,131,158,168,171,172,206]. SARS-CoV-2 GI VL in adults seemed to be subjectively lower (higher Ct values) than in the respiratory tract in 25 cases [59,63,131,168,171,172,206], and higher (lower Ct values) in four cases with mild disease [131,168,171], though meaningful conclusions cannot be drawn from such small sample sizes and non-systematic observations. There was no significant difference between the VL in stool between 22 cases with mild and 71 with severe disease [191].
SARS-CoV-2 detection in blood
Some key points found about SARS-CoV-2 detected in blood are summarised in Box 3.
Box 3. Key points with regards to SARS-CoV-2 detection in blood.
Blood sampling is not recommended for initial diagnostics.
There is no evidence of SARS-CoV-2 isolation from blood.
SARS-CoV-2 RNA detection in blood could be a sign of severe disease up to 4 weeks post symptoms onset and could be used as a clinical monitoring tool.
We reviewed 32 studies providing blood samples (whole blood, plasma or serum) data of ≥ 389 COVID-19 patients (Supplementary Dataset) [9,10,27,39,42,48,49,51,53,58,59,63,64,77,89,105,109,118,120,122,131,133,134,145,155,172,178,182,191,196,206,223]. A systematic comparison of SARS-CoV-2 RNA detection in different types of blood samples was lacking. No virus isolation from blood samples was reported.
SARS-CoV-2 detection with regard to the infection timeline and disease severity
Summarising data on COVID-19 patients with known infection timeline (Figure 2, Figure 3, Table, and Supplementary Table), SARS-CoV-2 RNA was detected 3–18 dps in 14 patients: 18 blood samples of 10 cases with severe disease [9,42,58,59,172] and four samples of four cases with mild disease respectively [9,39,206]. SARS-CoV-2 RNA presence in blood was linked with disease severity [42] and reported in further 54 cases with severe disease [10,27,105,109,120,172,182,191]. Additionally, viral RNA was detected in blood samples from 35 cases with mild disease [10,27,105,109,178,191,206], one asymptomatic infant [51] and three samples from unspecified cases [77]. SARS-CoV-2 RNA detection in blood might be useful as a laboratory sign of deterioration in cases with severe disease. SARS-CoV-2 RNA was detected in blood samples from 16 cases with mild disease for 10 ± 6 days and seven ICU patients for 15 ± 6 days [105]. A Chinese study reported a median viral RNA detection duration in the serum of 16 (IQR: 11–21) dps, and 27% serum positivity rates in 22 cases with mild disease compared with 45% in 74 cases with severe disease. Detection rates peaked in weeks 2–3 since symptoms onset in all patients with detectable SARS-CoV-2 RNA (17 with severe and three with mild disease) and dropped to 11% (n = 5) for cases with severe disease and 0 for cases with mild disease in week 4. However, Ct values had no significant difference between cases with severe and mild disease [191].
SARS-CoV-2 detection in blood compared to other types of specimens
A patient in critical condition had lower, but detectable SARS-CoV-2 RNA in plasma (Ct: 35.8–38.4; 7–12 dps) compared with NP swabs (6.7–4.4 log10 copies/1,000 cells; 7–24 dps) [120]. None of nine adults diagnosed with COVID-19 using OP swabs had detectable viral RNA in blood when tested with three different kits [48]. Although SARS-CoV-2 was detected and successfully isolated from respiratory samples, all 31 serum samples from nine cases with mild disease tested negative [131]. Finally, none of the serum samples from 14 convalescent patients (no respiratory symptoms and two consecutive negative OP swabs) were positive for SARS-CoV-2 RNA despite simultaneous detection in OP swabs and stool [53].
Other specimens: oral fluid, tears, urine, cerebrospinal fluid, peritoneal fluid, semen
Some key points found on SARS-CoV-2 in various other types of specimens than the ones previously considered are presented in Box 4.
Box 4. Key points with regards to SARS-CoV-2 in semen, tears, urine and oral, cerebrospinal or peritoneal fluid.
Self-collected oral fluid/saliva could be used as an alternative to respiratory sampling.
SARS-CoV-2 RNA is detectable in oral fluid/saliva up to 4 weeks.
RNA is rarely detected and SARS-CoV-2 has not been isolated in conjunctival secretions.
There is limited SARS-CoV-2 RNA detection in urine and no virus isolation.
SARS-CoV-2 can be visualised in brain tissue post mortem and viral RNA has been detected in cerebrospinal fluid.
SARS-CoV-2 RNA has been detected in semen.
SARS-CoV-2 detection in oral fluid and saliva
Fifteen studies reported on oral fluid sampling (with varying collection methods) in > 339 COVID-19 cases [21,62,89,97,105,109,148,164,178,190,191,194,208,209,265]. No study compared the different collection methods, e.g. self-collection, sampling by a healthcare worker, swabbing, stimulated secretion. Self-collected deep throat (posterior oropharyngeal) saliva was suggested as an alternative to sputum and yielded positive PCR results in 11 of 12 hospitalised COVID-19 patients in Hong Kong, as well as three positive and two negative virus cultures [21].
Further 23 patients with 173 saliva and endotracheal aspirate samples, studied by the same group, had median VL of 5.2 log10 copies/mL (IQR: 4.1–7.0) at presentation. The highest saliva VL were reported in week 1 from symptoms onset (20 patients), followed by a gradual decline, and prolonged RNA detection ≥ 20 days (seven patients) [109]. In a summary of the first cases in Hong Kong VLs in saliva were reported as 5.9×106 copies/mL compared with 3.3×106copies/mL in combined NP and OP swabs [62]. An Australian study screened 522 paired saliva and NP swabs and detected SARS-CoV-2 RNA in 33 saliva samples of 39 cases confirmed by NP swab. VLs were lower in saliva compared with NP swab with both sample types positive up to 21 dps.
Among 50 subjects with negative NP swab, one had a positive saliva sample [190]. Similarly, a Thai study screened 200 participants with paired saliva and combined NP and OP swabs and found 16 cases with matched positive saliva and swabs, two with only saliva positive and three with only NP + OP swab positive. VLs were comparable and a 97.5% agreement was observed between saliva and combined NP + OP swabs [265]. A pre-print study in the United States (later published in the New England Journal of Medicine [273]) including 44 cases reported comparable/superior sensitivity of saliva to NP swabs and higher SARS-CoV-2 saliva VL for 38 matched samples [194]. In another pre-print study saliva was collected from 31 patients (26 mild, five cases with severe disease) after stimulation of the salivary gland, paired with OP swab, and tested positive for SARS-CoV-2 RNA in four (three with severe, one with mild disease) of 13 cases with positive OP swab [97]. A study in Zhejiang confirmed COVID-19 in 96 patients by testing 668 sputum and 1,178 saliva samples but did not specify the positivity rates of the samples types separately. Taken together the latter declined from 95 to 54% in weeks 1–4 since symptoms onset with a median RNA detection duration of 18 (IQR: 13–29) days [191]. In 25 cases SARS-CoV-2 RNA detection in the saliva was reported for 13 ± 5 days in cases with mild disease and 16.5 ± 6 days in ICU patients [105]. A mild case in Wuhan had SARS-CoV-2 RNA detectable in OP swabs, saliva (Ct = 18.7), and urine sediment 54 dps, and continuous detection in OP swabs over 70 dps [208]. SARS-CoV-2 was detected in all saliva samples collected by drooling technique from 25 cases with severe disease [164], including two patients with same-day negative respiratory sampling in NP and bronchoalveolar swabs [164,209]. A comparison between throat wash with saline solution and NP swabs collected 48–57 dps in 11 cases found inconsistent results and higher positivity rates in throat wash [148].
SARS-CoV-2 detection in conjunctival secretions and tears
We reviewed six studies reporting conjunctival swab sampling in 137 COVID-19 cases [45,105,111,120,130,172]. SARS-CoV-2 RNA was detected, but virus not isolated, in the tears and conjunctival secretions of one mildly symptomatic patient with conjunctivitis [45], while samples and cultures from 46 patients without ocular symptoms were negative [45,111]. SARS-CoV-2 RNA detection in tears was also reported for one critically ill patient [172] and in five cases with unspecified disease severity [105]. Two cases with severe disease of 12 COVID-19 patients with ocular symptoms had positive conjunctival swabs [130]. No SARS-CoV-2 was detected in conjunctival swabs of four cases, including one with severe disease and conjunctivitis [120].
SARS-CoV-2 detection in urine
We reviewed 31 studies featuring urine samples from ≥ 369 patients [9,39,40,43,48,49,51,53,58,59,63,64,77,89,105,109,120,122,129,131,133,137,172,178,191,197,206,208,223,237,274]. None reported SARS-CoV-2 isolation from urine. A letter published shortly after the cut-off date of this review (not included in Supplementary Dataset) described successful isolation of SARS-CoV-2 (on Vero E6 cells) at 12 dps in a case with severe disease [275]. SARS-CoV-2 RNA was only detected in the urine of four patients (three patients with a positive sample upon OP swab turning negative) [53], including one woman with positive OP swab at 7 dps [206], one critically ill patient with suspected systemic COVID-19 infection [172], one case with urine abnormalities who later developed severe COVID-19 [274], and one neonate with mild infection 6–17 dps [178]. Urine sediments were positive for SARS-CoV-2 RNA in five COVID-19 cases [129,208].
SARS-CoV-2 detection in cerebrospinal fluid
Cerebrospinal fluid (CSF) sampling for a total of 14 patients was reported in eight studies [128,138,185,189,222,230,252,257]. SARS-CoV-2 presence in the brain was evidenced post-mortem in a case who had had severe disease: viral particles were observed in the frontal lobe, moreover tissues samples, but not CSF, had detectable viral RNA [189]. SARS-CoV-2 RNA was detected (Ct > 36 for nucleocapsid protein (N) target only in a N/N2-based Japanese assay) in the CSF of a patient with meningitis 9 dps [138]. However, viral RNA was not detectable in CSF of six patients with Guillain–Barré syndrome [185,252], nor of two patients with mild respiratory symptoms and suspected viral meningoencephalitis [230], nor of two patients with intracranial haemorrhage and positive NP swabs [222], nor of a child with Kawasaki-like disease [257]. CSF was not tested in a patient with suspected COVID-19 related acute necrotising encephalopathy [128], nor in another two with Guillain–Barré syndrome [174,244].
SARS-CoV-2 detection in peritoneal fluid
SARS-CoV-2 was not detected in peritoneal fluid samples collected during an appendectomy in a patient without respiratory symptoms, but a positive NP swab [242].
SARS-CoV-2 detection in semen
We reviewed five studies reporting on semen testing in 102 COVID-19 cases [181,197,235,267,268]. SARS-CoV-2 RNA was detected in semen from six (four with acute COVID-19 and two recovering) of 38 patients [235]. SARS-CoV-2 was not detected in semen collected during the recovery of 64 cases (nine convalescent cases with severe disease, 54 with mild disease and one asymptomatic case) [181,197,267,268], nor in the testis samples from a deceased case who had had severe disease [181].
Pregnancy and infancy, vaginal sampling
We reviewed 30 studies including 400 pregnant women and their infants [19,47,54,57,64,66,81,84,92,106,110,115,118,127,146,159,162,163,173,179,201,204,210,213,223,225,226,231,234,255]. No confirmed mother-to-child transmission was reported for these babies delivered mainly via caesarean (C)-section. A single infant, delivered via C-section, isolated, and formula-fed, had a positive pharyngeal swab 36 hours after delivery. Cord blood and placenta tested negative, but it was uncertain whether it was a vertical transmission or nosocomial infection [84]. Similarly, another infant delivered via C-section and quarantined, had five NP swabs collected until 16 days of age negative for SARS-CoV-2 RNA, but serology suggestive of in utero infection. Amniotic fluid or placenta were not tested and vertical transmission could not be confirmed/excluded [115]. Buonsenso and colleagues reported on a single case of SARS-CoV-2 RNA detectable in placenta, umbilical cord blood and three of five breast milk samples collected in the first 5 days from delivery via C-section. The infant was fed with expressed breast milk that tested negative on days 14–17 and his NP swabs were negative on days 1, 3, 18 as well as the OP and anal swabs on day 18 [225]. An infant and her mother were diagnosed with COVID-19 using NP swabs on day 7 post-delivery via C-section. The infant had negative NP swabs after 14 days and horizontal transmission was considered most likely [238]. Altogether SARS-CoV-2 RNA was not detected in 41 amniotic fluid [19,54,64,92,162,179,201,223], nor in 41 umbilical cord blood [19,54,64,84,92,162,201,225], nor in 10 placental tissue samples [54,57,64,84,92,223,225]. It is unclear whether SARS-CoV-2 could be transmitted through breastmilk, so far it was not detected in samples from 39 mothers [19,51,64,84,91,92,162,178,201,225,226,237,238].
It is unclear whether shedding occurs in the female reproductive systems posing risks for vaginal delivery and sexual intercourse. In a study of 35 cases with mild disease with an age range of 37–88 years, viral RNA was not detected in exfoliated cervical cells nor vaginal fluid [226]. Vaginal fluid and vaginal swabs in further 22 women also tested negative [92,115,133,179,201,223,237] including in 10 with severe disease [133].
Children and adolescents
We reviewed 64 studies describing COVID-19 in 1,510 children and adolescents [9,17,31,32,45,49,51,56,68,74,83,84,86,87,89,91,93-95,100,101,106,108,112,114,115,121,126,140,143-145,155,156,160,166,168,177,178,183,184,193,199,203,205,212,214,225,227,229,232,233,236-238,243,246,247,249-251,253,257,264]. Some further studies included cases < 18 years old grouped with adults and/or provided incomplete stratification by age (Supplementary Dataset).
In a study of 36 cases with mild disease of ages 1–16 years it took a mean of 10 (range: 7–22) days until the first negative results in NP swabs [114]. An asymptomatic 6-month infant maintained detectable SARS-CoV-2 RNA in NP swabs until 17 days of hospitalisation and had a positive stool sample on day 9 [51]. In an Italian study, 168 paediatric cases (four asymptomatic, 131 with mild disease, 33 with severe disease) nationwide were diagnosed via NP/OP swabs [233]. Verdoni and colleagues reported an increase of Kawasaki-like disease cases in March–April 2020 with two of 10 children with positive NP swabs and eight with positive serology [257].
A case-report documented an episode of detectable SARS-CoV-2 RNA in blood and transient fever in an otherwise asymptomatic 6-month infant [51].
Prolonged GI SARS-CoV-2 RNA detection after respiratory samples clearance was observed in stool samples up to 35 dps for 20 children aged 0.15–10 years [49,89,91,100,121,126,237] and in rectal swabs ≥ 3 weeks in 15 cases aged 0.15–17 years [86,91,93,168,203]. In a cohort of 46 children and adolescents, four had positive rectal swabs within 2–12 days after recovery and hospital discharge with negative OP swabs [229]. An asymptomatic girl had SARS-CoV-2 RNA detectable in anal swabs for 42 days, but not in NP swabs [203].
Higher SARS-CoV-2 VL in stool than OP swabs were reported for > 20 days in an infant with mild COVID-19 [121]. Eight children had higher average VL in anal swabs compared with NP swabs [86]. Han and colleagues reported of a neonate with mild COVID-19 who had SARS-CoV-2 RNA detectable in respiratory swabs (NP swab VL: 1.2×1010 copies/mL; OP swab VL: 1.3×108 copies/mL to undetectable 4–17 dps), stool (VL: 1.7×106 copies/mL to 4.1×107 copies/mL 6–18 dps), saliva (6–9 dps), plasma (5–10 dps) and urine (6–17 dps) [178].
Immunocompromised individuals
We reviewed 33 studies including 317 immunocompromised individuals [16,50,80,88,95,96,103,110,136,147,151,152,154,155,165,169,188,191,193,207,218,219,221,233,245,249,251,253,257-261], however specifics for them were rarely outlined in cohorts. Data on SARS-CoV-2 shedding patterns in immunocompromised individuals are still limited and quantitative data are lacking. SARS-CoV-2 RNA was detected in NP swabs 57 and 63 dps in a kidney transplant recipient following clinical recovery and hospital discharge 35 dps [258]. In a report on two lung transplant recipients, an asymptomatic adolescent had a positive NP swab 26 days after diagnosis, while a mildly symptomatic adult’s NP swab was negative for SARS-CoV-2 RNA after 2 weeks [251]. In a study of 90 transplant recipients with COVID-19, seven (three with mild, four with severe disease) cases had an initial negative NP swab, but dps were not reported [207]. Evidence of endothelial cell infection and endothelitis was observed in three severe COVID-19 cases, including a renal transplant recipient [188].
A person living with human immunodeficiency virus (PLHIV) and hepatitis C virus (HCV) had pneumonia resolution and negative NP swabs 10 dps but delayed antibody response 42 dps [152]. Further three PLHIV had a similar mild course with negative NP and OP swabs after 1 week [218] and another two PLHIV after 2 weeks from diagnosis [80,165].
Discussion and knowledge gaps
Sensitive molecular diagnostics through PCR and correct sampling and test interpretation are crucial for accurate COVID-19 diagnosis and thereby essential for good clinical practice. Furthermore, they are a key factor in outbreak phases where active case finding in combination with quarantine and contact tracing are crucial for outbreak control. We reviewed in the early stages of the pandemic knowledge on the kinetics of SARS-CoV-2 RNA shedding in different clinical samples of 32,515 COVID-19 patients to inform clinical and public health decision making.
Our review comes with limitations like the choice of rapid instead of systematic style. Given the need for timely diagnostics information in the current pandemic and the overwhelming amount of publications indexed daily, we opted for a faster adapted systematic approach. We did not assess the included publications for biases nor could fully trace for duplicate reporting of cases. Full comparability of the assays used could not be assured due to the lack of standardised diagnostic set-up across the world with respect to e.g. sampling materials, transport conditions, cell culture, extraction and PCR platforms. These were infrequently detailed in the publications. Furthermore, we did not review separately results from PCR assays targeting different parts of SARS-CoV-2 genome, for example envelope (E) [10,276,277], RNA-dependent RNA polymerase (RdRp) [9,276,277], open reading frame (ORF)1ab [11,50,51,59,276], ORF1a [276], ORF1b [276,278], nucleaocapsid (N) [11,50,51,59,276,278,279] or spike (S) [9,51,59,276,280,281] genes, nor any effect of the different platforms used, though we tried to provide the authors’ estimations of VLs or if not Ct values, where available. However, relatively few of the reviewed articles provided such detailed data, and we could not assess the data quality. We could not estimate to what extent the genetic drift in the SARS-CoV-2 genome affects the reported PCR results, nor could summarise false-positive or false-negative rates [282] due to insufficient data. Regular monitoring of the assays performance, detailed reporting and strict quality assurance mechanisms in accredited laboratories are vital to molecular diagnostics of SARS-CoV-2.
Respiratory tract specimens are the samples of choice for widespread screening and clinical course monitoring purposes, as well as for discharge and de-isolation [283,284]. Further reports with well-documented sampling time points, comparing the different types of respiratory samples, alternative sampling like oral fluid/saliva, and their diagnostic window of use, are needed. Viral RNA concentrations in the URT peak in the early infection days, including in asymptomatic and mildly symptomatic cases. SARS-CoV-2 was successfully isolated from respiratory samples with data suggesting independent replication potential in both the upper and lower respiratory tract. Thus, respiratory sampling is the optimal strategy for both symptomatic and pre-/a-symptomatic cases and for the latter should be examined in conjunction with epidemiological evidence and clinical follow-up.
Although molecular detection on URT samples is the recommended method to diagnose a SARS-CoV-2 infection, additional assays like serology are occasionally imperative to complement RT-PCR findings, e.g. in cases of insufficient clinical sensitivity of the RT-PCR over the disease course period [285]. In addition, serology can aid decision making on clinical and infection prevention management, e.g. by confirming the presence of SARS-CoV-2 specific (neutralising) antibodies in cases with consistently low VLs (high Ct values) in URT samples [286,287]. SARS-CoV-2 RNA detection in the respiratory tract during convalescence can be prolonged. However, detection of viral genomes does not directly imply the presence of infectious virus and thereby infectivity. More information on the presence of viable SARS-CoV-2 infectivity is urgently needed, especially with de-isolation strategies in mind.
Although faecal–oral transmission has not been confirmed, it cannot be firmly excluded due to evidence of SARS-CoV-2 isolation from the GI tract [58,271]. The detection of viable SARS-CoV-2 in faecal samples also has implications for the diagnostic possibilities of faeces tests. Laboratory diagnostic and safety protocols might need adjustments, for example by implementing inactivation steps and flow cabinet work. Stool and anal swabs are not samples of choice for screening purposes, though they might play a role in clinical monitoring. Some data suggested a shift from respiratory to GI shedding in the course of COVID-19 infection or independent replication in the GI tract. More information is needed on the significance of SARS-CoV-2 detection in faecal samples and how it relates to the timing of convalescence, as well as to the viral shedding specifics in children.
SARS-CoV-2 RNA in the blood has been detected mainly in cases with severe disease and blood sampling could be useful in monitoring hospitalised patients. At this stage, there is no evidence to suggest SARS-CoV-2 could be transmitted through blood. More systematic data are needed to guide blood and organ transplantation safety protocols [288].
There were single reports on viral RNA detection in samples obtained during delivery and thus questions on the use of such reproductive tract sampling, as well as on the potential for vertical or sexual transmission of SARS-CoV-2 remain. Viral RNA was detected in semen [235], but virus isolation was not attempted. Data on SARS-CoV-2 kinetics in vulnerable groups like immunocompromised patients are insufficient. Neurological signs and syndromes associated with COVID-19 and the diagnostic/monitoring potential of CSF testing remain to be clarified.
In conclusion, using SARS-CoV-2 molecular testing by amplification techniques to the maximum of its potential is a combination of choosing the most appropriate sample type, collected with adequate sampling technique, and with the infection timeline in mind. Like all diagnostic techniques, it should be analysed considering strengths and limitations, and in conjunction with the clinical presentation and epidemiological evidence. Further well-documented systematic studies are needed to characterise fully the COVID-19 clinical course and guide public health decisions concerning optimal testing strategies.
Acknowledgements
We would like to thank Nedyalko Vasilev for script development (used for data processing and visualisation), and Jenny Deng for assisting with literature in Chinese. We also thank Silvia Herrera Leon and Kim Benschop for reviewing the manuscript. We thank GOARN-Research for the continuous partnership and helpful discussions on the work topics.
Funding statement:
KRS received fellowship funding by the European Public Health Microbiology Training Programme (EUPHEM), European Centre for Disease Prevention and Control (ECDC). An ECDC representative formally approved the manuscript before submission. MPGK received funding from the H2020 COVID-19 project RECOVER (grant agreement nr 101003589). The funders had no role in the collection, analysis, and interpretation of data, nor the writing of the review.
Supplementary Data
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: CBEMR, LMK and KRS conceived the study design. KRS searched, compiled, and organised the information, and wrote the manuscript drafts. CBEMR, LMK, AAE, AM and MPGK reviewed the evidence and agreed on the conclusions. KRS and CBEMR finalised the manuscript text. All authors reviewed and contributed to the final manuscript.
References
- 1. Tan W, Zhao X, Ma X, Wang W, Niu P, Xu W, et al. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan, China 2019− 2020. China CDC Weekly. 2020;2(4):61-2. 10.46234/ccdcw2020.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO). Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). Geneva: WHO; 2020. Updated 30 Jan 2020. [Accessed 07 May 2020]. Available from: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov)
- 3.World Health Organization (WHO). WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. Geneva: WHO; 2020. [Accessed 9 May 2020]. Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- 4. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536-44. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181-92. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization (WHO). Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans. Geneva: WHO; 2020. [Accessed 01 May 2020]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance
- 7.World Health Organization (WHO). Coronavirus disease (COVID-2019) R&D. Geneva: WHO; 2020. [Accessed 22 Apr 2020]. Available from: https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus/en/
- 8.Foundation for Innovative New Diagnostics (FIND). SARS-COV-2 Diagnostic pipeline. Geneva: FIND; 2020. [Accessed 01 May 2020]. Available from: https://www.finddx.org/covid-19/pipeline/
- 9. Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514-23. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. China Novel Coronavirus Investigating and Research Team . A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727-33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, et al. Importation and Human-to-Human Transmission of a Novel Coronavirus in Vietnam. N Engl J Med. 2020;382(9):872-4. 10.1056/NEJMc2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199-207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507-13. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim JY, Choe PG, Oh Y, Oh KJ, Kim J, Park SJ, et al. The First Case of 2019 Novel Coronavirus Pneumonia Imported into Korea from Wuhan, China: Implication for Infection Prevention and Control Measures. J Korean Med Sci. 2020;35(5):e61. 10.3346/jkms.2020.35.e61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061-9. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364-74. 10.1007/s11427-020-1643-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bastola A, Sah R, Rodriguez-Morales AJ, Lal BK, Jha R, Ojha HC, et al. The first 2019 novel coronavirus case in Nepal. Lancet Infect Dis. 2020;20(3):279-80. 10.1016/S1473-3099(20)30067-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395(10226):809-15. 10.1016/S0140-6736(20)30360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pongpirul WA, Pongpirul K, Ratnarathon AC, Prasithsirikul W. Journey of a Thai Taxi Driver and Novel Coronavirus. N Engl J Med. 2020;382(11):1067-8. 10.1056/NEJMc2001621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. To KK-W, Tsang OT-Y, Yip CC, Chan K-H, Wu T-C, Chan JMC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841-3. 10.1093/cid/ciaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology. 2020;296(2):E41-5. 10.1148/radiol.2020200343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernard Stoecklin S, Rolland P, Silue Y, Mailles A, Campese C, Simondon A, et al. Investigation Team . First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Euro Surveill. 2020;25(6). 10.2807/1560-7917.ES.2020.25.6.2000094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silverstein WK, Stroud L, Cleghorn GE, Leis JA. First imported case of 2019 novel coronavirus in Canada, presenting as mild pneumonia. Lancet. 2020;395(10225):734. 10.1016/S0140-6736(20)30370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, Lee WJ, et al. Case of the Index Patient Who Caused Tertiary Transmission of Coronavirus Disease 2019 in Korea: the Application of Lopinavir/Ritonavir for the Treatment of COVID-19 Pneumonia Monitored by Quantitative RT-PCR. J Korean Med Sci. 2020;35(6):e79. 10.3346/jkms.2020.35.e79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Yang M, Shen C, Wang F, Yuan J, Li J, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020:2020.02.11.20021493.
- 27. Zhang W, Du R-H, Li B, Zheng X-S, Yang X-L, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386-9. 10.1080/22221751.2020.1729071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu P, Zhu J, Zhang Z, Han Y. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J Infect Dis. 2020;221(11):1757-61. 10.1093/infdis/jiaa077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han W, Quan B, Guo Y, Zhang J, Lu Y, Feng G, et al. The course of clinical diagnosis and treatment of a case infected with coronavirus disease 2019. J Med Virol. 2020;92(5):461-3. 10.1002/jmv.25711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang WH, Teng LC, Yeh TK, Chen YJ, Lo WJ, Wu MJ, et al. 2019 novel coronavirus disease (COVID-19) in Taiwan: Reports of two cases from Wuhan, China. J Microbiol Immunol Infect. 2020;53(3):481-4. 10.1016/j.jmii.2020.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan X, Chen D, Xia Y, Wu X, Li T, Ou X, et al. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect Dis. 2020;20(4):410-1. 10.1016/S1473-3099(20)30114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu X-W, Wu X-X, Jiang X-G, Xu K-J, Ying L-J, Ma C-L, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. 10.1136/bmj.m606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730-41. 10.1111/all.14238 [DOI] [PubMed] [Google Scholar]
- 34. Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382(12):1177-9. 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hao W, Li M, Huang X. First atypical case of 2019 novel coronavirus in Yan’an, China. Clin Microbiol Infect. 2020;26(7):952-3. 10.1016/j.cmi.2020.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L, et al. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323(14):1406-7. 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen D, Xu W, Lei Z, Huang Z, Liu J, Gao Z, et al. Recurrence of positive SARS-CoV-2 RNA in COVID-19: A case report. Int J Infect Dis. 2020;93:297-9. 10.1016/j.ijid.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hao W, Li M. Clinical features of atypical 2019 novel coronavirus pneumonia with an initially negative RT-PCR assay. J Infect. 2020;80(6):671-93. 10.1016/j.jinf.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, Chung YS, et al. Viral Load Kinetics of SARS-CoV-2 Infection in First Two Patients in Korea. J Korean Med Sci. 2020;35(7):e86. 10.3346/jkms.2020.35.e86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411-2. 10.1016/S1473-3099(20)30113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;296(2):E32-40. 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9(1):469-73. 10.1080/22221751.2020.1732837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng S-C, Chang Y-C, Fan Chiang Y-L, Chien Y-C, Cheng M, Yang C-H, et al. First case of Coronavirus Disease 2019 (COVID-19) pneumonia in Taiwan. J Formos Med Assoc. 2020;119(3):747-51. 10.1016/j.jfma.2020.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winichakoon P, Chaiwarith R, Liwsrisakun C, Salee P, Goonna A, Limsukon A, et al. Negative Nasopharyngeal and Oropharyngeal Swabs Do Not Rule Out COVID-19. J Clin Microbiol. 2020;58(5):e00297-20. 10.1128/JCM.00297-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xia J, Tong J, Liu M, Shen Y, Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J Med Virol. 2020;92(6):589-94. 10.1002/jmv.25725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, et al. Positive RT-PCR Test Results in Patients Recovered From COVID-19. JAMA. 2020;323(15):1502-3. 10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tian S, Hu N, Lou J, Chen K, Kang X, Xiang Z, et al. Characteristics of COVID-19 infection in Beijing. J Infect. 2020;80(4):401-6. 10.1016/j.jinf.2020.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xie C, Jiang L, Huang G, Pu H, Gong B, Lin H, et al. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264-7. 10.1016/j.ijid.2020.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiehao C, Jin X, Daojiong L, Zhi Y, Lei X, Zhenghai Q, et al. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020;71(6):1547-51. 10.1093/cid/ciaa198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, et al. China Medical Treatment Expert Group for Covid-19 . Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708-20. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kam K-Q, Yung CF, Cui L, Tzer Pin Lin R, Mak TM, Maiwald M, et al. A Well Infant With Coronavirus Disease 2019 With High Viral Load. Clin Infect Dis. 2020;71(15):847-9. 10.1093/cid/ciaa201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lillie PJ, Samson A, Li A, Adams K, Capstick R, Barlow GD, et al. Novel coronavirus disease (Covid-19): The first two patients in the UK with person to person transmission. J Infect. 2020;80(5):578-606. 10.1016/j.jinf.2020.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ling Y, Xu S-B, Lin Y-X, Tian D, Zhu Z-Q, Dai F-H, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl). 2020;133(9):1039-43. 10.1097/CM9.0000000000000774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X, Zhou Z, Zhang J, Zhu F, Tang Y, Shen X. A Case of 2019 Novel Coronavirus in a Pregnant Woman With Preterm Delivery. Clin Infect Dis. 2020;71(15):844-6. 10.1093/cid/ciaa200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu X, Yu C, Qu J, Zhang L, Jiang S, Huang D, et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47(5):1275-80. 10.1007/s00259-020-04735-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D, et al. Clinical Characteristics of Imported Cases of Coronavirus Disease 2019 (COVID-19) in Jiangsu Province: A Multicenter Descriptive Study. Clin Infect Dis. 2020;71(15):706-12. 10.1093/cid/ciaa199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen S, Huang B, Luo DJ, Li X, Yang F, Zhao Y, et al. [Pregnancy with new coronavirus infection: clinical characteristics and placental pathological analysis of three cases]. Zhonghua Bing Li Xue Za Zhi. 2020;49(5):418-23. [DOI] [PubMed] [Google Scholar]
- 58. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831-1833.e3. 10.1053/j.gastro.2020.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, et al. Singapore 2019 Novel Coronavirus Outbreak Research Team . Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488-94. 10.1001/jama.2020.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol. 2020;92(6):680-2. 10.1002/jmv.25742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63(5):706-11. 10.1007/s11427-020-1661-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cheng VCC, Wong S-C, Chen JHK, Yip CCY, Chuang VWM, Tsang OTY, et al. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect Control Hosp Epidemiol. 2020;41(5):493-8. 10.1017/ice.2020.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. Washington State 2019-nCoV Case Investigation Team . First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382(10):929-36. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li Y, Zhao R, Zheng S, Chen X, Wang J, Sheng X, et al. Lack of Vertical Transmission of Severe Acute Respiratory Syndrome Coronavirus 2, China. Emerg Infect Dis. 2020;26(6):1335-6. 10.3201/eid2606.200287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin D, Liu L, Zhang M, Hu Y, Yang Q, Guo J, et al. Co-infections of SARS-CoV-2 with multiple common respiratory pathogens in infected patients. Sci China Life Sci. 2020;63(4):606-9. 10.1007/s11427-020-1668-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu Y, Chen H, Tang K, Guo Y. Clinical manifestations and outcome of SARS-CoV-2 infection during pregnancy. J Infect. 2020. Epub ahead of print. 10.1016/j.jinf.2020.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382(10):970-1. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xia W, Shao J, Guo Y, Peng X, Li Z, Hu D. Clinical and CT features in pediatric patients with COVID-19 infection: Different points from adults. Pediatr Pulmonol. 2020;55(5):1169-74. 10.1002/ppul.24718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Arashiro T, Furukawa K, Nakamura A. COVID-19 in 2 Persons with Mild Upper Respiratory Tract Symptoms on a Cruise Ship, Japan. Emerg Infect Dis. 2020;26(6):1345-8. 10.3201/eid2606.200452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Luo S-H, Liu W, Liu Z-J, Zheng X-Y, Hong C-X, Liu Z-R, et al. A confirmed asymptomatic carrier of 2019 novel coronavirus. Chin Med J (Engl). 2020;133(9):1123-5. 10.1097/CM9.0000000000000798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172-5. 10.1016/j.cca.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Marchand-Senécal X, Kozak R, Mubareka S, Salt N, Gubbay JB, Eshaghi A, et al. Diagnosis and Management of First Case of COVID-19 in Canada: Lessons Applied From SARS-CoV-1. Clin Infect Dis. 2020;71(16):2207-10. 10.1093/cid/ciaa227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis. 2020;71(15):713-20. 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tong Z-D, Tang A, Li K-F, Li P, Wang H-L, Yi J-P, et al. Potential Presymptomatic Transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg Infect Dis. 2020;26(5):1052-4. 10.3201/eid2605.200198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen J, Qi T, Liu L, Ling Y, Qian Z, Li T, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80(5):e1-6. 10.1016/j.jinf.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg Infect Dis. 2020;26(6):1266-73. 10.3201/eid2606.200516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323(18):1843-4. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu X, Cai Y, Huang X, Yu X, Zhao L, Wang F, et al. Co-infection with SARS-CoV-2 and Influenza A Virus in Patient with Pneumonia, China. Emerg Infect Dis. 2020;26(6):1324-6. 10.3201/eid2606.200299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhu F, Cao Y, Xu S, Zhou M. Co-infection of SARS-CoV-2 and HIV in a patient in Wuhan city, China. J Med Virol. 2020;92(6):529-30. 10.1002/jmv.25732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhu H, Wang L, Fang C, Peng S, Zhang L, Chang G, et al. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Transl Pediatr. 2020;9(1):51-60. 10.21037/tp.2020.02.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cai J, Sun W, Huang J, Gamber M, Wu J, He G. Indirect Virus Transmission in Cluster of COVID-19 Cases, Wenzhou, China, 2020. Emerg Infect Dis. 2020;26(6):1343-5. 10.3201/eid2606.200412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen Q, Quan B, Li X, Gao G, Zheng W, Zhang J, et al. A report of clinical diagnosis and treatment of nine cases of coronavirus disease 2019. J Med Virol. 2020;92(6):683-7. 10.1002/jmv.25755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang S, Guo L, Chen L, Liu W, Cao Y, Zhang J, et al. A Case Report of Neonatal 2019 Coronavirus Disease in China. Clin Infect Dis. 2020;71(15):853- 7. 10.1093/cid/ciaa225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xing Y, Mo P, Xiao Y, Zhao O, Zhang Y, Wang F. Post-discharge surveillance and positive virus detection in two medical staff recovered from coronavirus disease 2019 (COVID-19), China, January to February 2020. Euro Surveill. 2020;25(10):2000191. 10.2807/1560-7917.ES.2020.25.10.2000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(4):502-5. 10.1038/s41591-020-0817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ji L-N, Chao S, Wang Y-J, Li X-J, Mu X-D, Lin M-G, et al. Clinical features of pediatric patients with COVID-19: a report of two family cluster cases. World J Pediatr. 2020;16(3):267-70. 10.1007/s12519-020-00356-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020.ciaa270. 10.1093/cid/ciaa270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Park JY, Han MS, Park KU, Kim JY, Choi EH. First Pediatric Case of Coronavirus Disease 2019 in Korea. J Korean Med Sci. 2020;35(11):e124. 10.3346/jkms.2020.35.e124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pung R, Chiew CJ, Young BE, Chin S, Chen MIC, Clapham HE, et al. Singapore 2019 Novel Coronavirus Outbreak Research Team . Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. 2020;395(10229):1039-46. 10.1016/S0140-6736(20)30528-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cui Y, Tian M, Huang D, Wang X, Huang Y, Fan L, et al. A 55-Day-Old Female Infant Infected With 2019 Novel Coronavirus Disease: Presenting With Pneumonia, Liver Injury, and Heart Damage. J Infect Dis. 2020;221(11):1775-81. 10.1093/infdis/jiaa113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fan C, Lei D, Fang C, Li C, Wang M, Liu Y, et al. Perinatal Transmission of 2019 Coronavirus Disease–Associated Severe Acute Respiratory Syndrome Coronavirus 2: Should We Worry? Clin Infect Dis. 2020;ciaa226. 10.1093/cid/ciaa226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fan Q, Pan Y, Wu Q, Liu S, Song X, Xie Z, et al. Anal swab findings in an infant with COVID-19. Pediatr Investig. 2020;4(1):48-50. 10.1002/ped4.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang Y, Liu Y, Liu L, Wang X, Luo N, Li L. Clinical Outcomes in 55 Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Who Were Asymptomatic at Hospital Admission in Shenzhen, China. J Infect Dis. 2020;221(11):1770-4. 10.1093/infdis/jiaa119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lu X, Zhang L, Du H, Zhang J, Li YY, Qu J, et al. Chinese Pediatric Novel Coronavirus Study Team . SARS-CoV-2 Infection in Children. N Engl J Med. 2020;382(17):1663-5. 10.1056/NEJMc2005073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Arentz M, Yim E, Klaff L, Lokhandwala S, Riedo FX, Chong M, et al. Characteristics and Outcomes of 21 Critically Ill Patients With COVID-19 in Washington State. JAMA. 2020;323(16):1612-4. 10.1001/jama.2020.4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen L, Zhao J, Peng J, Li X, Deng X, Geng Z, et al. Detection of 2019-nCoV in Saliva and Characterization of Oral Symptoms in COVID-19 Patients. Preprint. SSRN Electronic Journal. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Haveri A, Smura T, Kuivanen S, Österlund P, Hepojoki J, Ikonen N, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveill. 2020;25(11):2000266. 10.2807/1560-7917.ES.2020.25.11.2000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu Y, Yan L-M, Wan L, Xiang T-X, Le A, Liu J-M, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20(6):656-7. 10.1016/S1473-3099(20)30232-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ma X, Su L, Zhang Y, Zhang X, Gai Z, Zhang Z. Do children need a longer time to shed SARS-CoV-2 in stool than adults? J Microbiol Immunol Infect. 2020;53(3):373-6. 10.1016/j.jmii.2020.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sun D, Li H, Lu X-X, Xiao H, Ren J, Zhang F-R, et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center’s observational study. World J Pediatr. 2020;16(3):251-9. 10.1007/s12519-020-00354-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(5):434-5. 10.1016/S2468-1253(20)30083-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Guillen E, Pineiro GJ, Revuelta I, Rodriguez D, Bodro M, Moreno A, et al. Case report of COVID-19 in a kidney transplant recipient: Does immunosuppression alter the clinical presentation? Am J Transplant. 2020;20(7):1875-8. 10.1111/ajt.15874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ye G, Pan Z, Pan Y, Deng Q, Chen L, Li J, et al. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80(5):e14-7. 10.1016/j.jinf.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fang Z, Zhang Y, Hang C, Ai J, Li S, Zhang W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect. 2020;81(1):147-78. 10.1016/j.jinf.2020.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu H, Liu F, Li J, Zhang T, Wang D, Lan W. Clinical and CT imaging features of the COVID-19 pneumonia: Focus on pregnant women and children. J Infect. 2020;80(5):e7-13. 10.1016/j.jinf.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chang D, Mo G, Yuan X, Tao Y, Peng X, Wang FS, et al. Time Kinetics of Viral Clearance and Resolution of Symptoms in Novel Coronavirus Infection. Am J Respir Crit Care Med. 2020;201(9):1150-2. 10.1164/rccm.202003-0524LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Le HT, Nguyen LV, Tran DM, Do HT, Tran HT, Le YT, et al. The first infant case of COVID-19 acquired from a secondary transmission in Vietnam. Lancet Child Adolesc Health. 2020;4(5):405-6. 10.1016/S2352-4642(20)30091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. To KK-W, Tsang OT-Y, Leung W-S, Tam AR, Wu T-C, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565-74. 10.1016/S1473-3099(20)30196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jin X, Lian J-S, Hu J-H, Gao J, Zheng L, Zhang Y-M, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69(6):1002-9. 10.1136/gutjnl-2020-320926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Seah IYJ, Anderson DE, Kang AEZ, Wang L, Rao P, Young BE, et al. Assessing Viral Shedding and Infectivity of Tears in Coronavirus Disease 2019 (COVID-19) Patients. Ophthalmology. 2020;127(7):977-9. 10.1016/j.ophtha.2020.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zheng F, Liao C, Fan QH, Chen HB, Zhao XG, Xie ZG, et al. Clinical Characteristics of Children with Coronavirus Disease 2019 in Hubei, China. Curr Med Sci. 2020;40(2):275-80. 10.1007/s11596-020-2172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lian J, Jin X, Hao S, Cai H, Zhang S, Zheng L, et al. Analysis of Epidemiological and Clinical Features in Older Patients With Coronavirus Disease 2019 (COVID-19) Outside Wuhan. Clin Infect Dis. 2020;71(15):740-7. 10.1093/cid/ciaa242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. 2020;20(6):689-96. 10.1016/S1473-3099(20)30198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Dong L, Tian J, He S, Zhu C, Wang J, Liu C, et al. Possible Vertical Transmission of SARS-CoV-2 From an Infected Mother to Her Newborn. JAMA. 2020;323(18):1846-8. 10.1001/jama.2020.4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hoehl S, Rabenau H, Berger A, Kortenbusch M, Cinatl J, Bojkova D, et al. Evidence of SARS-CoV-2 Infection in Returning Travelers from Wuhan, China. N Engl J Med. 2020;382(13):1278-80. 10.1056/NEJMc2001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ong SWX, Tan YK, Sutjipto S, Chia PY, Young BE, Gum M, et al. Absence of contamination of personal protective equipment (PPE) by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Infect Control Hosp Epidemiol. 2020;41(5):614-6. 10.1017/ice.2020.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zeng H, Xu C, Fan J, Tang Y, Deng Q, Zhang W, et al. Antibodies in Infants Born to Mothers With COVID-19 Pneumonia. JAMA. 2020;323(18):1848-9. 10.1001/jama.2020.4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kimball A, Hatfield KM, Arons M, James A, Taylor J, Spicer K, et al. Public Health – Seattle & King County. CDC COVID-19 Investigation Team . Asymptomatic and Presymptomatic SARS-CoV-2 Infections in Residents of a Long-Term Care Skilled Nursing Facility - King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69(13):377-81. 10.15585/mmwr.mm6913e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lescure F-X, Bouadma L, Nguyen D, Parisey M, Wicky P-H, Behillil S, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20(6):697-706. 10.1016/S1473-3099(20)30200-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Xing Y-H, Ni W, Wu Q, Li W-J, Li G-J, Wang W-D, et al. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect. 2020;53(3):473-80. 10.1016/j.jmii.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, et al. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin Infect Dis. 2020;71(15):793-8. 10.1093/cid/ciaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zhou X, Li Y, Li T, Zhang W. Follow-up of asymptomatic patients with SARS-CoV-2 infection. Clin Microbiol Infect. 2020;26(7):957-9. 10.1016/j.cmi.2020.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chu J, Yang N, Wei Y, Yue H, Zhang F, Zhao J, et al. Clinical characteristics of 54 medical staff with COVID-19: A retrospective study in a single center in Wuhan, China. J Med Virol. 2020;92(7):807-13. 10.1002/jmv.25793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yuan J, Zou R, Zeng L, Kou S, Lan J, Li X, et al. The correlation between viral clearance and biochemical outcomes of 94 COVID-19 infected discharged patients. Inflamm Res. 2020;69(6):599-606. 10.1007/s00011-020-01342-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhang T, Cui X, Zhao X, Wang J, Zheng J, Zheng G, et al. Detectable SARS-CoV-2 viral RNA in feces of three children during recovery period of COVID-19 pneumonia. J Med Virol. 2020;92(7):909-14. 10.1002/jmv.25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Li C, Ji F, Wang L, Wang L, Hao J, Dai M, et al. Asymptomatic and Human-to-Human Transmission of SARS-CoV-2 in a 2-Family Cluster, Xuzhou, China. Emerg Infect Dis. 2020;26(7):1626-8. 10.3201/eid2607.200718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B. COVID-19-associated Acute Hemorrhagic Necrotizing Encephalopathy: Imaging Features. Radiology. 2020;296(2):E119-20. 10.1148/radiol.2020201187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang L, Li X, Chen H, Yan S, Li D, Li Y, et al. Coronavirus Disease 19 Infection Does Not Result in Acute Kidney Injury: An Analysis of 116 Hospitalized Patients from Wuhan, China. Am J Nephrol. 2020;51(5):343-8. 10.1159/000507471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wu P, Duan F, Luo C, Liu Q, Qu X, Liang L, et al. Characteristics of Ocular Findings of Patients With Coronavirus Disease 2019 (COVID-19) in Hubei Province, China. JAMA Ophthalmol. 2020;138(5):575-8. 10.1001/jamaophthalmol.2020.1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465-9. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 132. Cao J, Tu W-J, Cheng W, Yu L, Liu Y-K, Hu X, et al. Clinical Features and Short-term Outcomes of 102 Patients with Coronavirus Disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71(15):748-55. 10.1093/cid/ciaa243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Qiu L, Liu X, Xiao M, Xie J, Cao W, Liu Z, et al. SARS-CoV-2 Is Not Detectable in the Vaginal Fluid of Women With Severe COVID-19 Infection. Clin Infect Dis. 2020;71(15):813-7. 10.1093/cid/ciaa375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Scott SE, Zabel K, Collins J, Hobbs KC, Kretschmer MJ, Lach M, et al. Maricopa County COVID-19 Case Investigation Team . First Mildly Ill, Nonhospitalized Case of Coronavirus Disease 2019 (COVID-19) Without Viral Transmission in the United States-Maricopa County, Arizona, 2020. Clin Infect Dis. 2020;71(15):807-12. 10.1093/cid/ciaa374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Stefanelli P, Faggioni G, Lo Presti A, Fiore S, Marchi A, Benedetti E, et al. On Behalf Of Iss Covid-Study Group . Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020;25(13):2000305. 10.2807/1560-7917.ES.2020.25.13.2000305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Beerkens F, John M, Puliafito B, Corbett V, Edwards C, Tremblay D. COVID-19 pneumonia as a cause of acute chest syndrome in an adult sickle cell patient. Am J Hematol. 2020;95(7):E154-6. 10.1002/ajh.25809 [DOI] [PubMed] [Google Scholar]
- 137. Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92(7):833-40. 10.1002/jmv.25825 [DOI] [PubMed] [Google Scholar]
- 138. Moriguchi T, Harii N, Goto J, Harada D, Sugawara H, Takamino J, et al. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int J Infect Dis. 2020;94:55-8. 10.1016/j.ijid.2020.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wan R, Mao Z-Q, He L-Y, Hu Y-C, Wei-Chen. Evidence from two cases of asymptomatic infection with SARS-CoV-2: Are 14 days of isolation sufficient? Int J Infect Dis. 2020;95:174-5. 10.1016/j.ijid.2020.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang X, Fang J, Zhu Y, Chen L, Ding F, Zhou R, et al. Clinical characteristics of non-critically ill patients with novel coronavirus infection (COVID-19) in a Fangcang Hospital. Clin Microbiol Infect. 2020;26(8):1063-8. 10.1016/j.cmi.2020.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lei S, Jiang F, Su W, Chen C, Chen J, Mei W, et al. Clinical characteristics and outcomes of patients undergoing surgeries during the incubation period of COVID-19 infection. EClinicalMedicine. 2020;21:100331. 10.1016/j.eclinm.2020.100331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wong SCY, Kwong RTS, Wu TC, Chan JWM, Chu MY, Lee SY, et al. Risk of nosocomial transmission of coronavirus disease 2019: an experience in a general ward setting in Hong Kong. J Hosp Infect. 2020;105(2):119-27. 10.1016/j.jhin.2020.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Han YN, Feng ZW, Sun LN, Ren XX, Wang H, Xue YM, et al. A comparative-descriptive analysis of clinical characteristics in 2019-coronavirus-infected children and adults. J Med Virol. 2020;92(9):1596-602. 10.1002/jmv.25835 [DOI] [PubMed] [Google Scholar]
- 144. Chen D, Li Y, Deng X, Huang H, Ou X, Lin Y, et al. Four cases from a family cluster were diagnosed as COVID-19 after 14-day of quarantine period. J Med Virol. 2020;92(10):1748-52. 10.1002/jmv.25849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z, et al. Epidemiology of COVID-19 Among Children in China. Pediatrics. 2020;145(6):e20200702. 10.1542/peds.2020-0702 [DOI] [PubMed] [Google Scholar]
- 146. Khan S, Jun L, Nawsherwan, Siddique R, Li Y, Han G, et al. Association of COVID-19 with pregnancy outcomes in health-care workers and general women. Clin Microbiol Infect. 2020;26(6):788-90. 10.1016/j.cmi.2020.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Banerjee D, Popoola J, Shah S, Ster IC, Quan V, Phanish M. COVID-19 infection in kidney transplant recipients. Kidney Int. 2020;97(6):1076-82. 10.1016/j.kint.2020.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Guo W-L, Jiang Q, Ye F, Li S-Q, Hong C, Chen L-Y, et al. Effect of throat washings on detection of 2019 novel coronavirus. Clin Infect Dis. 2020;71(8):1980-1. 10.1093/cid/ciaa416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Keeley AJ, Evans C, Colton H, Ankcorn M, Cope A, State A, et al. Roll-out of SARS-CoV-2 testing for healthcare workers at a large NHS Foundation Trust in the United Kingdom, March 2020. Euro Surveill. 2020;25(14):2000433. 10.2807/1560-7917.ES.2020.25.14.2000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Xu K, Chen Y, Yuan J, Yi P, Ding C, Wu W, et al. Factors Associated With Prolonged Viral RNA Shedding in Patients with Coronavirus Disease 2019 (COVID-19). Clin Infect Dis. 2020;71(15):799- 806. 10.1093/cid/ciaa351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhang G, Hu C, Luo L, Fang F, Chen Y, Li J, et al. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J Clin Virol. 2020;127:104364. 10.1016/j.jcv.2020.104364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhao J, Liao X, Wang H, Wei L, Xing M, Liu L, et al. Early Virus Clearance and Delayed Antibody Response in a Case of Coronavirus Disease 2019 (COVID-19) With a History of Coinfection With Human Immunodeficiency Virus Type 1 and Hepatitis C Virus. Clin Infect Dis. 2020;71(16):2233-5. 10.1093/cid/ciaa408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Du X, Yu X, Li Q, Li X, Qin T, Luo Q, et al. Duration for carrying SARS-CoV-2 in COVID-19 patients. J Infect. 2020;81(1):e78-9. 10.1016/j.jinf.2020.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J, et al. COVID-19 with Different Severities: A Multicenter Study of Clinical Features. Am J Respir Crit Care Med. 2020;201(11):1380-8. 10.1164/rccm.202002-0445OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. He G, Sun W, Fang P, Huang J, Gamber M, Cai J, et al. The clinical feature of silent infections of novel coronavirus infection (COVID-19) in Wenzhou. J Med Virol. 2020;92(10):1761-3. 10.1002/jmv.25861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Huang L, Zhang X, Zhang X, Wei Z, Zhang L, Xu J, et al. Rapid asymptomatic transmission of COVID-19 during the incubation period demonstrating strong infectivity in a cluster of youngsters aged 16-23 years outside Wuhan and characteristics of young patients with COVID-19: A prospective contact-tracing study. J Infect. 2020;80(6):e1-13. 10.1016/j.jinf.2020.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683-90. 10.1001/jamaneurol.2020.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Qian G-Q, Chen X-Q, Lv D-F, Ma AHY, Wang L-P, Yang N-B, et al. Duration of SARS-CoV-2 viral shedding during COVID-19 infection. Infect Dis (Lond). 2020;52(7):511-2. 10.1080/23744235.2020.1748705 [DOI] [PubMed] [Google Scholar]
- 159. Xiong X, Wei H, Zhang Z, Chang J, Ma X, Gao X, et al. Vaginal delivery report of a healthy neonate born to a convalescent mother with COVID--19. J Med Virol. 2020;92(9):1657-9. 10.1002/jmv.25857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Danis K, Epaulard O, Bénet T, Gaymard A, Campoy S, Botelho-Nevers E, et al. Investigation Team . Cluster of Coronavirus Disease 2019 (COVID-19) in the French Alps, February 2020. Clin Infect Dis. 2020;71(15):825-32. 10.1093/cid/ciaa424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Xia XY, Wu J, Liu HL, Xia H, Jia B, Huang WX. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID-19. J Clin Virol. 2020;127:104360. 10.1016/j.jcv.2020.104360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Liu W, Wang J, Li W, Zhou Z, Liu S, Rong Z. Clinical characteristics of 19 neonates born to mothers with COVID-19. Front Med. 2020;14(2):193-8. 10.1007/s11684-020-0772-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Sutton D, Fuchs K, D’Alton M, Goffman D. Universal Screening for SARS-CoV-2 in Women Admitted for Delivery. N Engl J Med. 2020;382(22):2163-4. 10.1056/NEJMc2009316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45-50. 10.1016/j.jinf.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Chen J, Cheng X, Wang R, Zeng X. Computed tomography imaging of an HIV-infected patient with coronavirus disease 2019. J Med Virol. 2020;92(10):1774-6. 10.1002/jmv.25879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Chen J, Zhang Z-Z, Chen Y-K, Long Q-X, Tian W-G, Deng H-J, et al. The clinical and immunological features of pediatric COVID-19 patients in China. Genes Dis. 2020;7(4):535-41. 10.1016/j.gendis.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Gudbjartsson DF, Helgason A, Jonsson H, Magnusson OT, Melsted P, Norddahl GL, et al. Spread of SARS-CoV-2 in the Icelandic Population. N Engl J Med. 2020;382(24):2302-15. 10.1056/NEJMoa2006100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Liu J, Xiao Y, Shen Y, Shi C, Chen Y, Shi P, et al. Detection of SARS-CoV-2 by RT-PCR in anal from patients who have recovered from coronavirus disease 2019. J Med Virol. 2020;92(10):1769-71. 10.1002/jmv.25875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Blanco JL, Ambrosioni J, Garcia F, Martínez E, Soriano A, Mallolas J, et al. COVID-19 in HIV Investigators . COVID-19 in patients with HIV: clinical case series. Lancet HIV. 2020;7(5):e314-6. 10.1016/S2352-3018(20)30111-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672-5. 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 171. Hu Y, Shen L, Yao Y, Xu Z, Zhou J, Zhou H. A report of three COVID-19 cases with prolonged viral RNA detection in anal swabs. Clin Microbiol Infect. 2020;26(6):786-7. 10.1016/j.cmi.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z, et al. SARS-CoV-2 Viral Load in Clinical Samples from Critically Ill Patients. Am J Respir Crit Care Med. 2020;201(11):1435-8. 10.1164/rccm.202003-0572LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Lowe B, Bopp B. COVID-19 vaginal delivery - A case report. Aust N Z J Obstet Gynaecol. 2020;60(3):465-6. 10.1111/ajo.13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Sedaghat Z, Karimi N. Guillain Barre syndrome associated with COVID-19 infection: A case report. J Clin Neurosci. 2020;76:233-5. 10.1016/j.jocn.2020.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Yang F, Shi S, Zhu J, Shi J, Dai K, Chen X. Analysis of 92 deceased patients with COVID-19. J Med Virol. 2020;92(11):2511-5. 10.1002/jmv.25891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Baettig SJ, Parini A, Cardona I, Morand GB. Case series of coronavirus (SARS-CoV-2) in a military recruit school: clinical, sanitary and logistical implications. BMJ Mil Health. 2020:bmjmilitary-2020-001482. 10.1136/bmjmilitary-2020-001482 [DOI] [PubMed] [Google Scholar]
- 177. Du W, Yu J, Wang H, Zhang X, Zhang S, Li Q, et al. Clinical characteristics of COVID-19 in children compared with adults in Shandong Province, China. Infection. 2020;48(3):445-52. 10.1007/s15010-020-01427-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Han MS, Seong M-W, Heo EY, Park JH, Kim N, Shin S, et al. Sequential Analysis of Viral Load in a Neonate and Her Mother Infected With Severe Acute Respiratory Syndrome Coronavirus 2. Clin Infect Dis. 2020;71(16): 2236-9. 10.1093/cid/ciaa447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Iqbal SN, Overcash R, Mokhtari N, Saeed H, Gold S, Auguste T, et al. An Uncomplicated Delivery in a Patient with Covid-19 in the United States. N Engl J Med. 2020;382(16):e34. 10.1056/NEJMc2007605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Lin C, Xiang J, Yan M, Li H, Huang S, Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19). Clin Chem Lab Med. 2020;58(7):1089-94. 10.1515/cclm-2020-0187 [DOI] [PubMed] [Google Scholar]
- 181. Song C, Wang Y, Li W, Hu B, Chen G, Xia P, et al. Absence of 2019 novel coronavirus in semen and testes of COVID-19 patients. Biol Reprod. 2020;103(1):4-6. 10.1093/biolre/ioaa050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, et al. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin Infect Dis. 2020;71(8):1937-42. 10.1093/cid/ciaa449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Li W, Zhang B, Lu J, Liu S, Chang Z, Peng C, et al. Characteristics of Household Transmission of COVID-19. Clin Infect Dis. 2020;71(8):1943-6. 10.1093/cid/ciaa450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Paret M, Lighter J, Pellett Madan R, Raabe VN, Shust GF, Ratner AJ. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection in Febrile Infants Without Respiratory Distress. Clin Infect Dis. 2020;71(16): 2243-5. 10.1093/cid/ciaa452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Toscano G, Palmerini F, Ravaglia S, Ruiz L, Invernizzi P, Cuzzoni MG, et al. Guillain-Barré Syndrome Associated with SARS-CoV-2. N Engl J Med. 2020;382(26):2574-6. 10.1056/NEJMc2009191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhou B, She J, Wang Y, Ma X. Duration of Viral Shedding of Discharged Patients With Severe COVID-19. Clin Infect Dis. 2020;71(16):2240-2. 10.1093/cid/ciaa451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Xiao AT, Tong YX, Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis. 2020;71(16):2249-51. 10.1093/cid/ciaa460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417-8. 10.1016/S0140-6736(20)30937-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Paniz-Mondolfi A, Bryce C, Grimes Z, Gordon RE, Reidy J, Lednicky J, et al. Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). J Med Virol. 2020;92(7):699-702. 10.1002/jmv.25915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8):e00776-20. 10.1128/JCM.00776-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. 10.1136/bmj.m1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Blasco ML, Buesa J, Colomina J, Forner MJ, Galindo MJ, Navarro J, et al. Co-detection of respiratory pathogens in patients hospitalized with Coronavirus viral disease-2019 pneumonia. J Med Virol. 2020;92(10):1799-801. 10.1002/jmv.25922 [DOI] [PubMed] [Google Scholar]
- 193. Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al. the Northwell COVID-19 Research Consortium . Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323(20):2052-9. 10.1001/jama.2020.6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv. 2020:2020.04.16.20067835.
- 195. Colaneri M, Sacchi P, Zuccaro V, Biscarini S, Sachs M, Roda S, et al. COVID19 IRCCS San Matteo Pavia Task Force . Clinical characteristics of coronavirus disease (COVID-19) early findings from a teaching hospital in Pavia, North Italy, 21 to 28 February 2020. Euro Surveill. 2020;25(16):2000460. 10.2807/1560-7917.ES.2020.25.16.2000460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Liu J, Chen T, Yang H, Cai Y, Yu Q, Chen J, et al. Clinical and radiological changes of hospitalised patients with COVID-19 pneumonia from disease onset to acute exacerbation: a multicentre paired cohort study. Eur Radiol. 2020;30(10):5702-8. 10.1007/s00330-020-06916-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Paoli D, Pallotti F, Colangelo S, Basilico F, Mazzuti L, Turriziani O, et al. Study of SARS-CoV-2 in semen and urine samples of a volunteer with positive naso-pharyngeal swab. J Endocrinol Invest. 2020;43(12):1819-22. 10.1007/s40618-020-01261-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Park SY, Kim Y-M, Yi S, Lee S, Na B-J, Kim CB, et al. Coronavirus Disease Outbreak in Call Center, South Korea. Emerg Infect Dis. 2020;26(8):1666-70. 10.3201/eid2608.201274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Song R, Han B, Song M, Wang L, Conlon CP, Dong T, et al. Clinical and epidemiological features of COVID-19 family clusters in Beijing, China. J Infect. 2020;81(2):e26-30. 10.1016/j.jinf.2020.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Tostmann A, Bradley J, Bousema T, Yiek W-K, Holwerda M, Bleeker-Rovers C, et al. Strong associations and moderate predictive value of early symptoms for SARS-CoV-2 test positivity among healthcare workers, the Netherlands, March 2020. Euro Surveill. 2020;25(16):2000508. 10.2807/1560-7917.ES.2020.25.16.2000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Yan J, Guo J, Fan C, Juan J, Yu X, Li J, et al. Coronavirus disease 2019 in pregnant women: a report based on 116 cases. Am J Obstet Gynecol. 2020;223(1):111.e1-14. 10.1016/j.ajog.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Yu X, Sun S, Shi Y, Wang H, Zhao R, Sheng J. SARS-CoV-2 viral load in sputum correlates with risk of COVID-19 progression. Crit Care. 2020;24(1):170. 10.1186/s13054-020-02893-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol. 2020;92(10):1807-9. 10.1002/jmv.25941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Lu D, Sang L, Du S, Li T, Chang Y, Yang X-A. Asymptomatic COVID-19 infection in late pregnancy indicated no vertical transmission. J Med Virol. 2020;92(9):1660-4. 10.1002/jmv.25927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Luo Y, Trevathan E, Qian Z, Li Y, Li J, Xiao W, et al. Asymptomatic SARS-CoV-2 Infection in Household Contacts of a Healthcare Provider, Wuhan, China. Emerg Infect Dis. 2020;26(8):1930-3. 10.3201/eid2608.201016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Peng L, Liu J, Xu W, Luo Q, Chen D, Lei Z, et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol. 2020;92(9):1676-80. 10.1002/jmv.25936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Pereira MR, Mohan S, Cohen DJ, Husain SA, Dube GK, Ratner LE, et al. COVID-19 in solid organ transplant recipients: Initial report from the US epicenter. Am J Transplant. 2020;20(7):1800-8. 10.1111/ajt.15941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Yang J-R, Deng D-T, Wu N, Yang B, Li H-J, Pan X-B. Persistent viral RNA positivity during the recovery period of a patient with SARS-CoV-2 infection. J Med Virol. 2020;92(9):1681-3. 10.1002/jmv.25940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Azzi L, Carcano G, Dalla Gasperina D, Sessa F, Maurino V, Baj A. Two cases of COVID-19 with positive salivary and negative pharyngeal or respiratory swabs at hospital discharge: A rising concern. Oral Dis. 2020;odi.13368. 10.1111/odi.13368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Vintzileos WS, Muscat J, Hoffmann E, John NS, Vertichio R, Vintzileos AM, et al. Screening all pregnant women admitted to labor and delivery for the virus responsible for coronavirus disease 2019. Am J Obstet Gynecol. 2020;223(2):284-6. 10.1016/j.ajog.2020.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059-61. 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Schwierzeck V, König JC, Kühn J, Mellmann A, Correa-Martínez CL, Omran H, et al. First reported nosocomial outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a pediatric dialysis unit. Clin Infect Dis. 2020;ciaa491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Hantoushzadeh S, Shamshirsaz AA, Aleyasin A, Seferovic MD, Aski SK, Arian SE, et al. Maternal death due to COVID-19. Am J Obstet Gynecol. 2020;223(1):109.e1-16. 10.1016/j.ajog.2020.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Liu S-F, Kuo N-Y, Kuo H-C. Three Taiwan’s domestic family cluster infections of coronavirus disease 2019. J Med Virol. 2020;92(10):2011-8. 10.1002/jmv.25949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Meng Y, Wu P, Lu W, Liu K, Ma K, Huang L, et al. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: A retrospective study of 168 severe patients. PLoS Pathog. 2020;16(4):e1008520. 10.1371/journal.ppat.1008520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Potdar V, Cherian SS, Deshpande GR, Ullas PT, Yadav PD, Choudhary ML, et al. National Influenza Centre (NIC) Team . Genomic analysis of SARS-CoV-2 strains among Indians returning from Italy, Iran & China, & Italian tourists in India. Indian J Med Res. 2020;151(2 & 3):255-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Sarkale P, Patil S, Yadav PD, Nyayanit DA, Sapkal G, Baradkar S, et al. First isolation of SARS-CoV-2 from clinical samples in India. Indian J Med Res. 2020;151(2 & 3):244-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Altuntas Aydin O, Kumbasar Karaosmanoglu H, Kart Yasar K. HIV/SARS-CoV-2 coinfected patients in Istanbul, Turkey. J Med Virol. 2020;92(11):2288-90. 10.1002/jmv.25955 [DOI] [PubMed] [Google Scholar]
- 219. Nair V, Jandovitz N, Hirsch JS, Nair G, Abate M, Bhaskaran M, et al. COVID-19 in kidney transplant recipients. Am J Transplant. 2020;20(7):1819-25. 10.1111/ajt.15967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569-78. 10.1016/S0140-6736(20)31022-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Zhao X-Y, Xu X-X, Yin H-S, Hu Q-M, Xiong T, Tang Y-Y, et al. Clinical characteristics of patients with 2019 coronavirus disease in a non-Wuhan area of Hubei Province, China: a retrospective study. BMC Infect Dis. 2020;20(1):311. 10.1186/s12879-020-05010-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Al Saiegh F, Ghosh R, Leibold A, Avery MB, Schmidt RF, Theofanis T, et al. Status of SARS-CoV-2 in cerebrospinal fluid of patients with COVID-19 and stroke. J Neurol Neurosurg Psychiatry. 2020;91(8):846-8. 10.1136/jnnp-2020-323522 [DOI] [PubMed] [Google Scholar]
- 223. Baud D, Greub G, Favre G, Gengler C, Jaton K, Dubruc E, et al. Second-Trimester Miscarriage in a Pregnant Woman With SARS-CoV-2 Infection. JAMA. 2020;323(21):2198-200. 10.1001/jama.2020.7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Kim GU, Kim MJ, Ra SH, Lee J, Bae S, Jung J, et al. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clin Microbiol Infect. 2020;26(7):948.e1-3. 10.1016/j.cmi.2020.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Buonsenso D, Costa S, Sanguinetti M, Cattani P, Posteraro B, Marchetti S, et al. Neonatal Late Onset Infection with Severe Acute Respiratory Syndrome Coronavirus 2. Am J Perinatol. 2020;37(08):869-72. 10.1055/s-0040-1710541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Cui P, Chen Z, Wang T, Dai J, Zhang J, Ding T, et al. Severe acute respiratory syndrome coronavirus 2 detection in the female lower genital tract. Am J Obstet Gynecol. 2020;223(1):131-4. 10.1016/j.ajog.2020.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Kim SE, Jeong HS, Yu Y, Shin SU, Kim S, Oh TH, et al. Viral kinetics of SARS-CoV-2 in asymptomatic carriers and presymptomatic patients. Int J Infect Dis. 2020;95:441-3. 10.1016/j.ijid.2020.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Turner J, Eliot Hodgson L, Leckie T, Eade L, Ford-Dunn S. A Dual-Center Observational Review of Hospital-Based Palliative Care in Patients Dying With COVID-19. J Pain Symptom Manage. 2020;60(2):e75-8. 10.1016/j.jpainsymman.2020.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Zhang B, Liu S, Zhang J, Xiao J, Zhu S, Dong Y, et al. Children hospitalized for coronavirus disease 2019 (COVID-19): A multicenter retrospective descriptive study. J Infect. 2020;81(2):e74-5. 10.1016/j.jinf.2020.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Bernard-Valnet R, Pizzarotti B, Anichini A, Demars Y, Russo E, Schmidhauser M, et al. Two patients with acute meningoencephalitis concomitant with SARS-CoV-2 infection. Eur J Neurol. 2020;27(9):e43-4. 10.1111/ene.14298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Blitz MJ, Grünebaum A, Tekbali A, Bornstein E, Rochelson B, Nimaroff M, et al. Intensive care unit admissions for pregnant and nonpregnant women with coronavirus disease 2019. Am J Obstet Gynecol. 2020;223(2):290-1. 10.1016/j.ajog.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Coronado Munoz A, Nawaratne U, McMann D, Ellsworth M, Meliones J, Boukas K. Late-Onset Neonatal Sepsis in a Patient with Covid-19. N Engl J Med. 2020;382(19):e49. 10.1056/NEJMc2010614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Garazzino S, Montagnani C, Donà D, Meini A, Felici E, Vergine G, et al. Multicentre Italian study of SARS-CoV-2 infection in children and adolescents, preliminary data as at 10 April 2020. Euro Surveill. 2020;25(18):2000600. 10.2807/1560-7917.ES.2020.25.18.2000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Khalil A, Hill R, Ladhani S, Pattisson K, O’Brien P. Severe acute respiratory syndrome coronavirus 2 in pregnancy: symptomatic pregnant women are only the tip of the iceberg. Am J Obstet Gynecol. 2020;223(2):296-7. 10.1016/j.ajog.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Li D, Jin M, Bao P, Zhao W, Zhang S. Clinical Characteristics and Results of Semen Tests Among Men With Coronavirus Disease 2019. JAMA Netw Open. 2020;3(5):e208292. 10.1001/jamanetworkopen.2020.8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Ma Y, Xu QN, Wang FL, Ma XM, Wang XY, Zhang XG, et al. Characteristics of asymptomatic patients with SARS-CoV-2 infection in Jinan, China. Microbes Infect. 2020;22(4-5):212-7. 10.1016/j.micinf.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Mao LJ, Xu J, Xu ZH, Xia XP, Li B, He JG, et al. A child with household transmitted COVID-19. BMC Infect Dis. 2020;20(1):329. 10.1186/s12879-020-05056-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Piersigilli F, Carkeek K, Hocq C, van Grambezen B, Hubinont C, Chatzis O, et al. COVID-19 in a 26-week preterm neonate. Lancet Child Adolesc Health. 2020;4(6):476-8. 10.1016/S2352-4642(20)30140-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. De Maria A, Varese P, Dentone C, Barisione E, Bassetti M. High prevalence of olfactory and taste disorder during SARS-CoV-2 infection in outpatients. J Med Virol. 2020;92(11):2310-1. 10.1002/jmv.25995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Sun J, Xiao J, Sun R, Tang X, Liang C, Lin H, et al. Prolonged Persistence of SARS-CoV-2 RNA in Body Fluids. Emerg Infect Dis. 2020;26(8):1834-8. 10.3201/eid2608.201097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Yokoo K, Sugaya F, Matsuzaka S, Ueda K, Kamimura R, Yokoyama T, et al. The first case of COVID-19 occurring as community-acquired pneumonia in Hokkaido, Japan and our preventive measures against nosocomial infection. Respir Med Case Rep. 2020;30:101078. 10.1016/j.rmcr.2020.101078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Ngaserin SH-N, Koh FH, Ong B-C, Chew M-H. COVID-19 not detected in peritoneal fluid: a case of laparoscopic appendicectomy for acute appendicitis in a COVID-19-infected patient. Langenbecks Arch Surg. 2020;405(3):353-5. 10.1007/s00423-020-01891-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Ibrahim LF, Tosif S, McNab S, Hall S, Lee HJ, Lewena S, et al. SARS-CoV-2 testing and outcomes in the first 30 days after the first case of COVID-19 at an Australian children’s hospital. Emerg Med Australas. 2020;32(5):801-8. 10.1111/1742-6723.13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Scheidl E, Canseco DD, Hadji-Naumov A, Bereznai B. Guillain-Barré syndrome during SARS-CoV-2 pandemic: A case report and review of recent literature. J Peripher Nerv Syst. 2020;25(2):204-7. 10.1111/jns.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Anand P, Slama MCC, Kaku M, Ong C, Cervantes-Arslanian AM, Zhou L, et al. COVID-19 in patients with myasthenia gravis. Muscle Nerve. 2020;62(2):254-8. 10.1002/mus.26918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Cellina M, Orsi M, Toluian T, Valenti Pittino C, Oliva G. False negative chest X-Rays in patients affected by COVID-19 pneumonia and corresponding chest CT findings. Radiography (Lond). 2020;26(3):e189-94. 10.1016/j.radi.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Dodi I, Castellone E, Pappalardo M, Rubini M, Veronese P, Ruberto C, et al. SARS-CoV-2 infection in children in Parma. Acta Biomed. 2020;91(2):214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Rivett L, Sridhar S, Sparkes D, Routledge M, Jones NK, Forrest S, et al. CITIID-NIHR COVID-19 BioResource Collaboration . Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. eLife. 2020;9:e58728. 10.7554/eLife.58728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Shekerdemian LS, Mahmood NR, Wolfe KK, Riggs BJ, Ross CE, McKiernan CA, et al. International COVID-19 PICU Collaborative . Characteristics and Outcomes of Children With Coronavirus Disease 2019 (COVID-19) Infection Admitted to US and Canadian Pediatric Intensive Care Units. JAMA Pediatr. 2020;174(9):868-73. 10.1001/jamapediatrics.2020.1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Wei M, Yuan J, Liu Y, Fu T, Yu X, Zhang Z-J. Novel Coronavirus Infection in Hospitalized Infants Under 1 Year of Age in China. JAMA. 2020;323(13):1313-4. 10.1001/jama.2020.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Koczulla RA, Sczepanski B, Koteczki A, Kuhnert S, Hecker M, Askevold I, et al. SARS-CoV-2 infection in two patients following recent lung transplantation. Am J Transplant. 2020;20(10):2928-32. 10.1111/ajt.15998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Ottaviani D, Boso F, Tranquillini E, Gapeni I, Pedrotti G, Cozzio S, et al. Early Guillain-Barré syndrome in coronavirus disease 2019 (COVID-19): a case report from an Italian COVID-hospital. Neurol Sci. 2020;41(6):1351-4. 10.1007/s10072-020-04449-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Bush R, Johns F, Acharya R, Upadhyay K. Mild COVID-19 in a pediatric renal transplant recipient. Am J Transplant. 2020;20(10):2942-5. 10.1111/ajt.16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Chen M, An W, Xia F, Yang P, Li K, Zhou Q, et al. Clinical characteristics of rehospitalized patients with COVID-19 in China. J Med Virol. 2020;92(10):2146-51. 10.1002/jmv.26002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Huang W, Zhao Z, He Z, Liu S, Wu Q, Zhang X, et al. Unfavorable outcomes in pregnant patients with COVID-19. J Infect. 2020;81(2):e99-101. 10.1016/j.jinf.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Rodríguez-Cola M, Jiménez-Velasco I, Gutiérrez-Henares F, López-Dolado E, Gambarrutta-Malfatti C, Vargas-Baquero E, et al. Clinical features of coronavirus disease 2019 (COVID-19) in a cohort of patients with disability due to spinal cord injury. Spinal Cord Ser Cases. 2020;6(1):39. 10.1038/s41394-020-0288-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Verdoni L, Mazza A, Gervasoni A, Martelli L, Ruggeri M, Ciuffreda M, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395(10239):1771-8. 10.1016/S0140-6736(20)31103-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Man Z, Jing Z, Huibo S, Bin L, Fanjun Z. Viral shedding prolongation in a kidney transplant patient with COVID-19 pneumonia. Am J Transplant. 2020;20(9):2626-7. 10.1111/ajt.15996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Ferguson J, Rosser JI, Quintero O, Scott J, Subramanian A, Gumma M, et al. Characteristics and Outcomes of Coronavirus Disease Patients under Nonsurge Conditions, Northern California, USA, March-April 2020. Emerg Infect Dis. 2020;26(8):1679-85. 10.3201/eid2608.201776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Gervasoni C, Meraviglia P, Riva A, Giacomelli A, Oreni L, Minisci D, et al. Clinical Features and Outcomes of Patients With Human Immunodeficiency Virus With COVID-19. Clin Infect Dis. 2020;71(16):2276-8. 10.1093/cid/ciaa579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM, et al. China Medical Treatment Expert Group for COVID-19 . Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020;55(5):2000547. 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Böhmer MM, Buchholz U, Corman VM, Hoch M, Katz K, Marosevic DV, et al. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect Dis. 2020;20(8):920-8. 10.1016/S1473-3099(20)30314-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Chen W, Zheng KI, Liu S, Yan Z, Xu C, Qiao Z. Plasma CRP level is positively associated with the severity of COVID-19. Ann Clin Microbiol Antimicrob. 2020;19(1):18. 10.1186/s12941-020-00362-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Diotallevi F, Campanati A, Bianchelli T, Bobyr I, Luchetti MM, Marconi B, et al. Skin involvement in SARS-CoV-2 infection: Case series. J Med Virol. 2020;92(11):2332-4. 10.1002/jmv.26012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, Suksuwan W, et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2021;27(2):285.e1-4. 10.1016/j.cmi.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Zheng L, Wang X, Zhou C, Liu Q, Li S, Sun Q, et al. Analysis of the Infection Status of Healthcare Workers in Wuhan During the COVID-19 Outbreak: A Cross-sectional Study. Clin Infect Dis. 2020;71(16):2109-13. 10.1093/cid/ciaa588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ning J, Li W, Ruan Y, Xia Y, Wu X, Hu K, et al. Effects of 2019 Novel Coronavirus on Male Reproductive System: A Retrospective Study. Preprints. 2020. [Google Scholar]
- 268. Pan F, Xiao X, Guo J, Song Y, Li H, Patel DP, et al. No evidence of severe acute respiratory syndrome-coronavirus 2 in semen of males recovering from coronavirus disease 2019. Fertil Steril. 2020;113(6):1135-9. 10.1016/j.fertnstert.2020.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Han Y, Yang H. The transmission and diagnosis of 2019 novel coronavirus infection disease (COVID-19): A Chinese perspective. J Med Virol. 2020;92(6):639-44. 10.1002/jmv.25749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Wei WE, Li Z, Chiew CJ, Yong SE, Toh MP, Lee VJ. Presymptomatic Transmission of SARS-CoV-2 - Singapore, January 23-March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(14):411-5. 10.15585/mmwr.mm6914e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Zhang Y, Chen C, Zhu S, Shu C, Wang D, Song J, et al. Isolation of 2019-nCoV from a Stool Specimen of a Laboratory-Confirmed Case of the Coronavirus Disease 2019 (COVID-19). China CDC Weekly. 2020;2(8):123-4. 10.46234/ccdcw2020.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Sun Y, Koh V, Marimuthu K, Ng OT, Young B, Vasoo S, et al. National Centre for Infectious Diseases COVID-19 Outbreak Research Team . Epidemiological and Clinical Predictors of COVID-19. Clin Infect Dis. 2020;71(15):786-92. 10.1093/cid/ciaa322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N Engl J Med. 2020;383(13):1283-6. 10.1056/NEJMc2016359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Gross O, Moerer O, Weber M, Huber TB, Scheithauer S. COVID-19-associated nephritis: early warning for disease severity and complications? Lancet. 2020;395(10236):e87-8. 10.1016/S0140-6736(20)31041-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Sun J, Zhu A, Li H, Zheng K, Zhuang Z, Chen Z, et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg Microbes Infect. 2020;9(1):991-3. 10.1080/22221751.2020.1760144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.World Health Organization (WHO). Molecular assays to diagnose COVID-19. In-house developed molecular assays. Geneva: WHO;2020 [Accessed 14 Apr 2020]. Available from: https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2.
- 277. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020;66(4):549-55. 10.1093/clinchem/hvaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Centers for Disease Control and Prevention (CDC). 2019-Novel Coronavirus (2019-nCoV) Real-time rRT-PCR Panel Primers and Probes. Atlanta: CDC; 2020 [Accessed 9 Mar 2020]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- 280. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-3. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395(10226):809-15. 10.1016/S0140-6736(20)30360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure. Ann Intern Med. 2020;173(4):262-7. 10.7326/M20-1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.European Centre for Disease Prevention and Control (ECDC). Guidance for discharge and ending isolation in the context of widespread community transmission of COVID-19. Stockholm: ECDC; 8 Apr 2020. Available from: https://www.ecdc.europa.eu/en/publications-data/covid-19-guidance-discharge-and-ending-isolation
- 284. Tay JY, Lim PL, Marimuthu K, Sadarangani SP, Ling LM, Ang BSP, et al. De-isolating Coronavirus Disease 2019 Suspected Cases: A Continuing Challenge. Clin Infect Dis. 2020;71(15):883-4. 10.1093/cid/ciaa179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Bergmans BJM, Reusken CBEM, Van Oudheusden AJG, Godeke G-J, Bonacic Marinovic AA, de Vries E, et al. Declining SARS-CoV-2 PCR sensitivity with time and dependence on clinical features: consequences for control. medRxiv. 2020:2020.08.23.20179408.
- 286. Sethuraman N, Jeremiah SS, Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 2020;323(22):2249-51. 10.1001/jama.2020.8259 [DOI] [PubMed] [Google Scholar]
- 287.van Kampen JJA, van de Vijver DAMC, Fraaij PLA, Haagmans BL, Lamers MM, Okba N, et al. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. medRxiv. 2020:2020.06.08.20125310. [DOI] [PMC free article] [PubMed]
- 288. Chang L, Yan Y, Wang L. Coronavirus Disease 2019: Coronaviruses and Blood Safety. Transfus Med Rev. 2020;34(2):75-80. 10.1016/j.tmrv.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.