Abstract
Background
Plasmodium cynomolgi is a simian malaria parasite that has been reported as a naturally acquired human infection. The present study aims to systematically review reports on naturally acquired P. cynomolgi in humans, mosquitoes, and macaques to provide relevant data for pre-emptive surveillance and preparation in the event of an outbreak of zoonotic malaria in Southeast Asia.
Methods
The protocol of the systematic review was registered at PROSPERO with approval ID CRD42020203046. Three databases (Web of Science, Scopus, and MEDLINE) were searched for studies reporting the prevalence of P. cynomolgi infections in Southeast Asian countries between 1946 and 2020. The pooled prevalence or pooled proportion of P. cynomolgi parasitemia in humans, mosquitoes, and macaques was estimated using a random-effects model. Differences in the clinical characteristics of P. cynomolgi infections were also estimated using a random-effects model and presented as pooled odds ratios (ORs) or mean differences (MDs) with 95% confidence intervals (CIs).
Results
Thirteen studies reporting on the prevalence of naturally acquired P. cynomolgi in humans (3 studies, 21 cases), mosquitoes (3 studies, 28 cases), and macaques (7 studies, 334 cases) were included. The results demonstrated that the pooled proportion of naturally acquired P. cynomolgi in humans was 1% (95% CI, 0.1%, I2, 0%), while the pooled proportion of P. cynomolgi infecting mosquitoes was 18% (95% CI, 10–26%, I2, 32.7%). The pooled prevalence of naturally acquired P. cynomolgi in macaques was 47% (95% CI, 27–67%, I2, 98.3%). Most of the cases of naturally acquired P. cynomolgi in humans were reported in Cambodia (62%) and Malaysia (38%), while cases of P. cynomolgi in macaques were reported in Malaysia (35.4%), Singapore (23.2%), Indonesia (17.3%), Philippines (8.5%), Laos (7.93%), and Cambodia (7.65%). Cases of P. cynomolgi in mosquitoes were reported in Vietnam (76.9%) and Malaysia (23.1%).
Conclusions
This study demonstrated the occurrence of naturally acquired P. cynomolgi infection in humans, mosquitoes, and macaques. Further studies of P. cynomolgi in asymptomatic human cases in areas where vectors and natural hosts are endemic are extensively needed if human infections with P. cynomolgi do become public health problems.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-021-05941-y.
Keywords: Plasmodium, Malaria, Zoonoses, Mosquitoes, Monkey, Macaques
Background
Plasmodium spp. infections in humans have been identified with Plasmodium falciparum, P. malariae, P. vivax, P. ovale, and P. knowlesi [1]. Although P. falciparum and P. vivax are the two major malaria species that cause malaria in humans worldwide, the infection of P. knowlesi is a major cause of simian malaria in Malaysian Borneo [1–4] and is also reported as a cause of simian malaria in other parts of Southeast Asia, including Indonesia [5], Laos [6], Vietnam [7, 8], and Thailand [9]. P. knowlesi and other simian malaria parasites, including P. fieldi, P. coatneyi, P. cynomolgi, and P. inui, mainly infect long-tailed (Macaca fascicularis) and pig-tailed macaques (Macaca nemestrina) [2].
P. cynomolgi has accidentally and experimentally been reported as a cause of human malaria [3–5], but there was a case report showing this parasite as a naturally acquired human infection in a Malay woman from the east coast of Peninsular Malaysia who lives in an area where long-tailed macaques are present [6]. P. cynomolgi was first observed in 1907 in Macaca fascicularis collected in Java [7]. P. cynomolgi has morphological features similar to P. vivax, as shown by microscopy, including the asexual cycle (48 h), prepatent periods, and presence of hypnozoites, which can initiate relapses [6]. The identification of P. cynomolgi relies on the amplification of the 18S ribosomal RNA (rRNA) gene by nested polymerase chain reaction (PCR) sequencing. As P. cynomolgi is recorded as the most recently discovered simian malaria parasite infecting humans, its prevalence, proportion, geographical distribution, and characteristics remain unclear. The present study aimed to systematically review reports on naturally acquired P. cynomolgi in humans, mosquitoes, and macaques to provide relevant data for pre-emptive surveillance and preparation in the event of an outbreak of zoonotic malaria in Southeast Asia.
Methods
Protocol for reporting the systematic review and meta-analysis
The systematic review and meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [8]. The protocol of the systematic review was registered at the PROSPERO International Prospective Register of Systematic Reviews with approval ID CRD42020203046.
Search strategy
Studies reporting the prevalence of P. cynomolgi parasitemia in humans, mosquitoes, and macaques in Southeast Asian countries between 1946 and 2020 were systematically searched in three databases: Web of Science, Scopus, and MEDLINE. The search term used to find the potentially relevant studies was “cynomolgi”. The searches of the reference list in the included studies or the review articles and the searches of the additional source(s) such as Google Scholar were performed to maximize the number of the included studies and to prevent any missing studies during the searches of the main databases.
The eligibility criteria and study selection
The inclusion criterion for selection in the present study was that any included studies must be primary studies reporting the prevalence or incidence of P. cynomolgi parasitemia in humans, mosquitoes, and macaques. The exclusion criteria were (1) studies without data of interest; (2) studies including populations outside Southeast Asian countries where sufficient reports on P. cynomolgi could be retrieved; (3) studies without full text; and (4) studies published as case reports or case series, reviews, editorials, letters to the editor, in vitro/experimental studies, mosquito experiments, animal experiments, human experiments, or studies identifying P. cynomolgi in the same participants. Two authors (MK, FRM) independently screened the studies according to the eligibility criteria for potentially relevant studies. Any disagreement between the two authors was resolved through discussion or consultation with the third author (KUK) for the finalization of the study inclusion.
Data extraction
Data from the included studies were extracted for pilot standardization. The following data of each study were extracted: author name, year of publication, year of study, study site, types of participant or sample (humans, monkeys, or mosquitoes), age, gender ratio, types of PCR used for P. cynomolgi identification, target gene for PCR, number of Plasmodium species identified, and parasite density. Data extraction was performed by one author (MK) and cross-checked by two authors (FRM and GDM).
Quality of the included studies
The methodological quality of the included studies was assessed using the adapted version of the Newcastle-Ottawa Scale (NOS) [9] with a maximum of three scores (Is the Case Definition Adequate?, Representativeness of the Cases, and Ascertainment of Exposure). For the present study, studies achieving a NOS score of two or greater were considered high-quality studies.
Data synthesis
The pooled prevalence or the pooled proportion of P. cynomolgi parasitemia compared to all Plasmodium species or all cases enrolling humans, mosquitoes, and macaques was estimated using a random-effects model. In the case of a very low prevalence of P. cynomolgi infection compared to the number of participants enrolled, the pooled proportion instead of the pooled prevalence was used to estimate the proportion of P. cynomolgi parasitemia per Plasmodium species identified in the same participants. The difference in the geographical distribution was visualized by mapping the location(s) of the P. cynomolgi parasitemia provided by the included studies. Differences in the clinical characteristics of P. cynomolgi infections between the included studies were estimated using a random-effects model and demonstrated as pooled odds ratios (ORs) or mean differences (MDs) with 95% confidence intervals (CIs). The heterogeneity across the included studies was assessed using Cochrane’s Q and I2 (inconsistency) statistics.
Publication bias
Publication bias across the included studies was assessed using the funnel plot and Egger’s test. In the absence of publication bias, the funnel plot should approximately resemble a symmetrical funnel, while an asymmetrical appearance indicates a small-study effect that will overestimate or underestimate the pooled effect [10].
Results
Search results
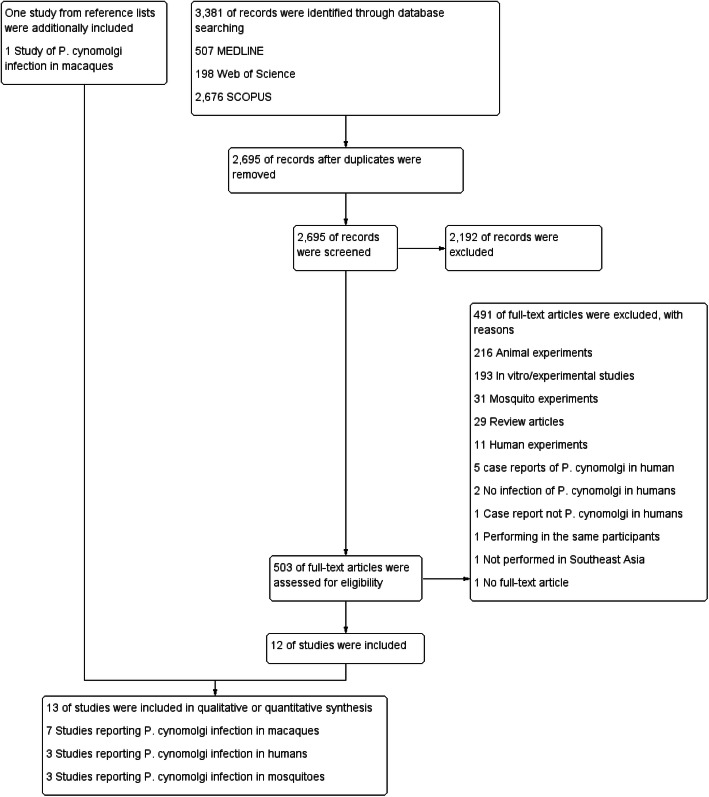
The initial search yielded 3381 articles (Fig. 1). Out of those articles, 686 articles were removed as they were duplicates. After screening the titles and abstracts of the remaining 2695 articles, 2192 articles were excluded as they did not meet the inclusion criterion. Following a full-text review of 503 articles, a further 491 articles were excluded with reason: 216 were animal experiments, 193 were in vitro/experimental studies, 31 were mosquito experiments, 29 were review articles, 11 were human experiments, 5 were case reports of P. cynomolgi in humans, 2 had no infection of P. cynomolgi in humans, 1 was a case report not on P. cynomolgi in humans, 1 involved performing identification in the same participants, 1 was not performed in Southeast Asia, and 1 was not a full-text article. Therefore, 12 articles were included in the study. Another relevant article was retrieved from the searches of the reference lists of included articles or review articles. Finally, 13 studies were included in the present study. Out of the 13 studies selected, 7 studies [11–17] reporting on P. cynomolgi infection in macaques, 3 studies [18–20] reporting on P. cynomolgi infection in humans, and 3 studies [21–23] reporting on P. cynomolgi infection in mosquitoes were included in the final analysis.
Fig. 1.
The study flow diagram. Twelve studies were from the database searches, and one study was from the searches of review articles
Characteristics of the included studies
The characteristics of the included studies are shown in Table 1. Thirteen studies [11–23] reporting the prevalence or incidence of naturally acquired P. cynomolgi in humans (3 studies, 21 cases), mosquitoes (3 studies, 26 cases), and macaques (7 studies, 334 cases) were included for qualitative and quantitative syntheses. Details of P. cynomolgi mono and mixed infections in humans, Anopheles, and macaques are shown in Table 2. Among the 3 studies reporting on P. cynomolgi in humans between 2013 and 2017, one study was conducted in Northern Sabah of Malaysia [18], one in Pailin and Battambang provinces of Cambodia [19], and one in Sarawak of Malaysia [20]. Two studies [18, 19] enrolled participants in the communities, while another study [20] enrolled malaria-positive cases for their studies. All studies used semi-nested or nested PCR with sequencing to identify Plasmodium species infection. All three studies [18–20] used semi-nested or nested PCR for amplification of the SSU rRNA gene. PCR amplification of the cytochrome C oxidase gene was used for DNA sequencing to confirm P. cynomolgi and P. knowlesi coinfections in one study [20]. Of the 21 cases of people who were infected with P. cynomolgi, 16 cases (76.2%) were male. The age of patients was 43 and 63 years in a study by Grignard et al. [18], median 28 years (range 7–64 years) in a study by Imwong et al. [19], and median 43 years (range 17–65 years) in a study by Raja et al. [20]. P. cynomolgi mono-infection was in 13 cases (61.9%), while P. cynomolgi mixed infection with other Plasmodium species was in 8 cases (38.1%). Mixed infections included P. cynomolgi with P. knowlesi (6 cases, 75%) and with P. vivax (2 cases, 25%). Only one study [18] reported on the occupation of patients and also observed monkeys. Naturally acquired P. cynomolgi in humans was reported in Cambodia (62%) and Malaysia (38%). Details of P. cynomolgi infections in humans are shown in Table 3.
Table 1.
Characteristics of the included studies
| No. | Authors | Year of study | Study site | Types of samples/number of samples | Number of malaria cases by PCR | Number of P. cynomolgi cases by PCR | Age of patients with P. cynomolgi (years) | Number of male patients with P. cynomolgi | PCR for P. cynomolgi | Target gene for PCR |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Grignard et al., 2019 [18] | 2015 | Northern Sabah, Malaysia | Human/876 | 54 | 2 | 43, 63 | 2 (100%) | Genus specific semi-nested PCR | 18S rRNA |
| 2. | Imwong et al., 2019 [19] | 2013–2016 | Pailin and Battambang provinces, Cambodia | Human/14,732 | 1361 | 13 | Median 28, range 7–64 | 11 (84.6%) | Nested PCR | 18S rRNA |
| 3. | Raja et al., 2020 [20] | 2013–2017 | Kapit, Sarawak, Malaysia | Human/1047 (seropositive for malaria) | 845 | 6 | 36, 18, 63, 50, 17, 65 | 3 (50%) | Nested PCR | 18S rRNA |
| 4. | Chinh et al., 2019 [21] | 2005–2018 | Southern Vietnam | Mosquitoes/1386 | 40 | 9 | Nested PCR | 18S rRNA | ||
| 5. | Chua et al., 2017 [22] | 2013–2014 | Kudat district, Sabah, Malaysia | Mosquitoes/1586 | 23 | 6 | Nested PCR | 18S rRNA | ||
| 6. | Maeno et al., 2015 [23] | 2010–2013 | Vietnam | Mosquitoes/6062 | 86 | 11 | Nested PCR | 18S rRNA | ||
| 7. | Akter et al., 2015 [11] | 2014 | Selangor, Malaysia | Macaques/70 | 36 | 18 | Nested PCR | 18S rRNA | ||
| 8. | Amir et al., 2020 [12] | 2016 | Johor, Malaysia | Macaques/103 | 64 | 42 | Nested PCR | 18S rRNA | ||
| 9. | Lee et al., 2011 [15] | 2004–2008 | Sarawak, Malaysia | Macaques/108 | 101 | 61 | Nested PCR | 18S rRNA | ||
| 10. | Muehlenbein et al., 2015 [16] | Not specified | Sabah, Malaysia | Macaques/41 | 41 | 4 | Not specified type of PCR | cytochrome b | ||
| 11. | Gamalo et al., 2019 [13] | 2017 | Luzon, Philippines | Macaques/95 | 95 | 26 | Nested PCR | 18S rRNA | ||
| 12. | Zhang et al., 2016 [17] | Not specified | Batangas/Zamboanga, Philippines | Macaques/68 | 7 | 4 | Nested PCR | 18S rRNA | ||
| Southern Sumatra, Indonesia | Macaques/50 | 49 | 48 | |||||||
| Bintan Island, Indonesia | Macaques/20 | 16 | 13 | |||||||
| Singapore | Macaques/40 | 31 | 26 | |||||||
| Vanny, Cambodia | Macaques/54 | 44 | 27 | |||||||
| Guidong, Laos | Macaques/44 | 30 | 28 | |||||||
| 13. | Li Meizhi I, 2011 [14] | Not specified | Singapore | Macaques/92 | 66 | 56 | Nested PCR | 18S rRNA |
Table 2.
P. cynomolgi mono and mixed infections in humans, Anopheles, and macaques
| Infection | Case total | Location total | ||
|---|---|---|---|---|
| Mono | Mixed | |||
| Human (zoonotic) | 21 | |||
| Malaysia, Northern Sabah | Malaysia | |||
| a. Kudat | 2 | 0 | 8 | |
| b. Kota Marudu | ||||
| c. Pitas | ||||
| d. Ranau | ||||
| Malaysia, Sarawak | ||||
| e. Kapit | 0 | 6 | ||
| Cambodia | Cambodia | |||
| f. Pailin and Battambang | 11 | 2 | 13 | |
| Anopheles (vector) | 28 | |||
|
Vietnam Southern Vietnam |
Vietnam | |||
| a. Gia Lai | 6 | 3 | 20 | |
| b. Phu Yen | ||||
| c. Khanh Hoa | ||||
| d. Ninh Thuan | ||||
| e. Binh Thuan | ||||
| f. Dong Nai | ||||
| g. Binh Phuoc | ||||
|
h. Khanh Phu, Khanh Vinh district i. Khanh Hoa |
6 | 5 | ||
| Malaysia, Sabah | Malaysia | |||
| j. Kudat | 4 | 4 | 8 | |
| Macaques (natural host) | 334 | |||
| Malaysia | Malaysia | |||
| a. Hulu Selangor | 2 | 16 | 125 | |
| B. Pahang | 15 | 27 | ||
| c. Perak | ||||
| d. Johor | ||||
| e. Kapit | 1 | 60 | ||
| f. Sabah | 4 | 0 | ||
| Philippines | Philippines | |||
| g. Palawan | 0 | 23 | 27 | |
| h. Batangas | 2 | 1 | ||
| i. Zamboanga | 1 | 0 | ||
| Indonesia | Indonesia | |||
| j. South Sumatra | 39 | 9 | 61 | |
| k. Bintan Island | 9 | 4 | ||
| Singapore | Singapore | |||
| l. Singapore | 23 | 3 | (40 cases did not report species of infection) | 66 |
| Cambodia | Cambodia | |||
| m. Vanny | 24 | 3 | 27 | |
| Laos | Laos | |||
| n. Guidong | 28 | 0 | 28 | |
Table 3.
Details of P. cynomolgi infections in humans
| N = 21 (mono/mixed) | Combination | Prevalence per all malaria cases (2260 cases) | Regional trend |
|---|---|---|---|
| Mono | – | 13 (0.58) | Northern Sabah, Pailin, and Battambang |
| Dual | P. cynomolgi + P. knowlesi | 6 (0.27) | Kapit, Sarawak |
| P. cynomolgi + P. vivax | 2 (0.09) | Pailin and Battambang |
Among the 3 studies reporting the P. cynomolgi infection in mosquitoes between 2010 and 2018, 2 studies [21, 23] were conducted in South or South-Central Vietnam, while another study [22] was conducted in Malaysia. All studies used nested PCR with sequencing to identify Plasmodium species infection. One study identified Plasmodium infection in four Anopheles, including An. dirus, An. maculatus, An. aconitus, and An. minimus [21]. One study identified Plasmodium infection in An. dirus [23]. Another study identified Plasmodium infection in An. balabacensis Baisas [22]. P. cynomolgi mono-infection occurred in 16 cases (57.1%), while P. cynomolgi mixed infection with other Plasmodium species occurred in 12 cases (42.9%). Mixed infections with P. cynomolgi were dual (7 cases, 58.3%), triple (4 cases, 33.3%), and quadruple (1 case, 8.3%) infections. Most of the P. cynomolgi infected cases were mixed infections with P. inui (5 cases, 41.7%), P. vivax (5 cases, 41.7%), P. knowlesi (2 cases, 16.7%), P. coatneyi (1 case, 8.33%), and P. fieldi (1 case, 8.33%).
Among the 7 studies reporting on 334 macaques infected with P. cynomolgi between 2004 and 2019, 4 studies (57.1%) [11, 12, 15, 16] were conducted in Malaysia, 1 in the Philippines [13], and 1 in Singapore [14]. Another study [17] was conducted in five countries, including the Philippines, Indonesia, Singapore, Cambodia, and Laos. Four studies [11, 13, 14, 17] identified Plasmodium species in long-tailed macaques, while 3 studies [12, 14, 16] identified Plasmodium species in both long-tailed and pig-tailed macaques. All studies used nested PCR for amplification of the 18S rRNA gene except a study by Muehlenbein et al. [16], which used nested PCR for amplification of the cytochrome b (cytb) gene. One study did not report the status of P. cynomolgi mono or mixed infection [16]. P. cynomolgi mono-infection occurred in 148 cases (50.3%), while P. cynomolgi mixed infection with other Plasmodium species occurred in 146 cases (49.7%). Mixed infections with P. cynomolgi were dual (14 cases, 9.6%), triple (37 cases, 25.3%), quadruple (56 cases, 38.4%), and quintuple (9 cases, 6.16%) infections. Most of the P. cynomolgi cases were mixed infections with P. inui (132 cases, 90.4%), P. coatneyi (75 cases, 51.4%), P. knowlesi (67 cases, 45.9%), and P. fieldi (31 cases, 21.2%). Naturally acquired P. cynomolgi in macaques was reported in Malaysia (125/338, 37%), Philippines (30/338, 8.88%), Indonesia (61/338, 18.1%), Singapore (82/338, 24.3%), Cambodia (27/338, 7.99%), and Laos (28/338, 8.28%).
Quality of the included studies and publication bias
The methodological quality of the included studies was assessed using the adapted version of the NOS (Table S1). Overall, all 13 studies achieving NOS scores of three were reviewed. Publication bias among the included studies could not be assessed using the funnel plot and Egger’s test due to the low number of the included studies, as the analysis required a minimum of 10 studies for each group of participants enrolled [10].
Geographical distribution of P. cynomolgi infection in humans, mosquitoes, and macaques
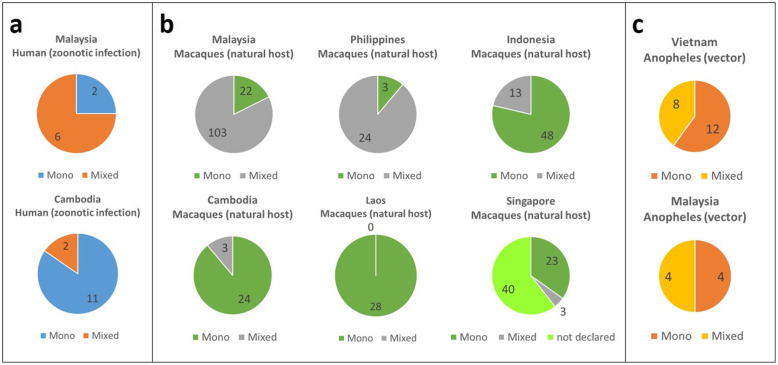
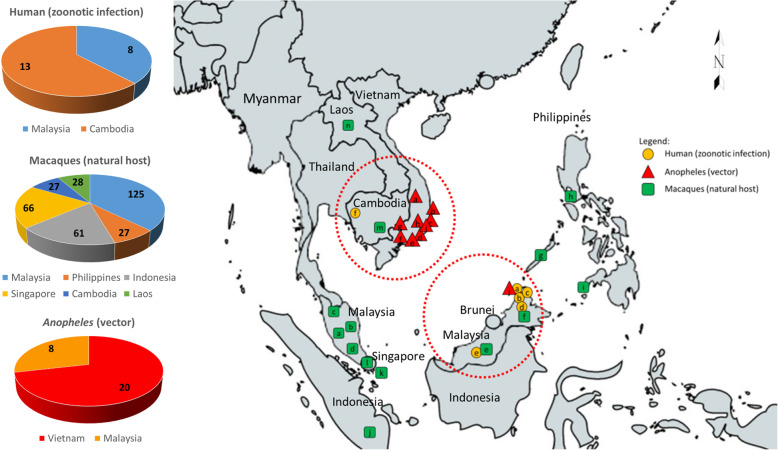
P. cynomolgi mono-infection cases were predominantly in humans and Anopheles vectors, while an almost equal number of cases were reported in macaques. The proportion of P. cynomolgi cases in humans, Anopheles, and macaques per Southeast Asian country is shown in Fig. 2. The results showed that P. cynomolgi infection in humans was demonstrated in Malaysia and Cambodia. P. cynomolgi infection in Anopheles was demonstrated in Vietnam and Malaysia. P. cynomolgi infection in macaques was demonstrated in Malaysia, the Philippines, Indonesia, Cambodia, Laos, and Singapore. Human infection with P. cynomolgi was reported in Cambodia (n = 13) and Malaysia (n = 8). Anopheles infections with P. cynomolgi were reported in Vietnam (n = 20) and Malaysia (n = 8), while the highest numbers of macaques infected with P. cynomolgi were reported in Malaysia (n = 125) and Singapore (n = 66). The majority of the reports of infected macaques in Singapore (n = 40) did not identify whether the cases were mono or mixed infections. Studies on the presence of both vectors and natural hosts also reported human infection cases, as was the case in Malaysia (Fig. 3). P. cynomolgi infections in humans were reported in Cambodia and Malaysian Borneo, while P. cynomolgi infections in macaques were reported in the Philippines, Malaysia, Laos, Singapore, and Indonesia. P. cynomolgi infections in Anopheles mosquitoes were reported in southern Vietnam and Malaysian Borneo.
Fig. 2.
The proportion of P. cynomolgi cases in; a human, b Anopheles, and c macaques per Southeast Asian country. P. cynomolgi infection in humans has been demonstrated in Malaysia and Cambodia. P. cynomolgi infection in Anopheles was demonstrated in Vietnam and Malaysia. P. cynomolgi infection in macaques was demonstrated in Malaysia, the Philippines, Indonesia, Cambodia, Laos, and Singapore. Note the higher proportion (n = 40) of undeclared speciation in Singapore compared to mono or mixed cases combined
Fig. 3.
Geographic mapping and distribution of P. cynomolgi cases in humans, Anopheles, and macaques in Southeast Asia. Regions with human cases (red circles) were observed to have the presence of both vector and natural host. Human (zoonotic infection) a Kudat, b Kota Marudu, c Pitas, and d Ranau districts, Northern Sabah, and e Kapit, Sarawak, Malaysia, f Pailin and Battambang provinces, Cambodia; Macaques (natural host) a Hulu Selangor district, Selangor b Pahang, c Perak, d Johor, e Kapit, Sarawak, and f Sabah, Malaysia, g Palawan, h Batangas, and i Zamboanga, Philippines, j Southern Sumatra, and k Bintan Island, Indonesia, l Singapore, m Vanny, Cambodia; Anopheles (vector) a Gia Lai Province, b Phu Yen Province, c Khanh Hoa Province, d Ninh Thuan Province, e Binh Thuan Province, f Dong Nai Province, g Binh Phuoc Province, Southern Vietnam, h Khanh Phu, Khanh Vinh district, i Khanh Hoa province, South-central Vietnam, j Kudat district, Sabah, Malaysia. The map was generated by authors using the map freely available at https://mapchart.net/. Authors are allowed to use, edit and modify any map created with mapchart.net for publication freely by adding the reference to mapchart.net. The project of https://mapchart.net/ is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License
The proportion of P. cynomolgi infection in humans
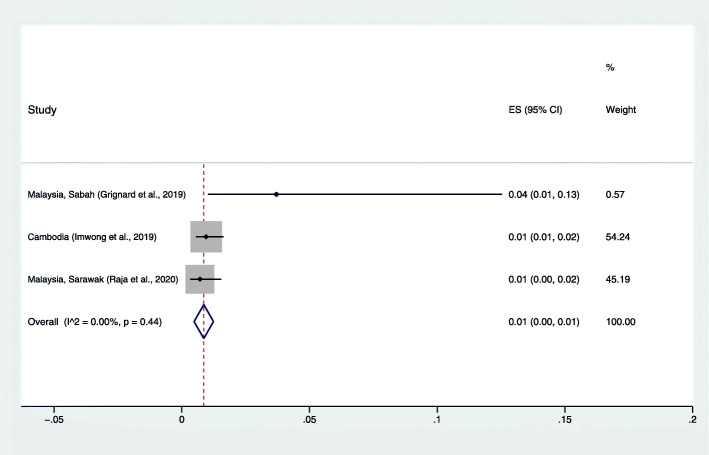
The pooled prevalence of P. cynomolgi in humans was very low (< 0.1%). Because a low prevalence of P. cynomolgi in humans was reported in the included studies, the pooled proportion of P. cynomolgi compared to Plasmodium species infections in humans was estimated. The results demonstrated that the pooled proportion of naturally acquired P. cynomolgi was 1%, with low heterogeneity (95% CI: 0.1%, I2: 0%). The highest proportion of P. cynomolgi infection in humans was demonstrated in Northern Sabah (4, 95% CI: 1–13%) [18] (Fig. 4).
Fig. 4.
The estimated proportion of P. cynomolgi malaria in humans. ES: estimated proportion, overall: overall proportion, I2: level of heterogenicity, p: p-value less than 0.05 is statistically significant
The proportion of P. cynomolgi parasitemia in mosquitoes
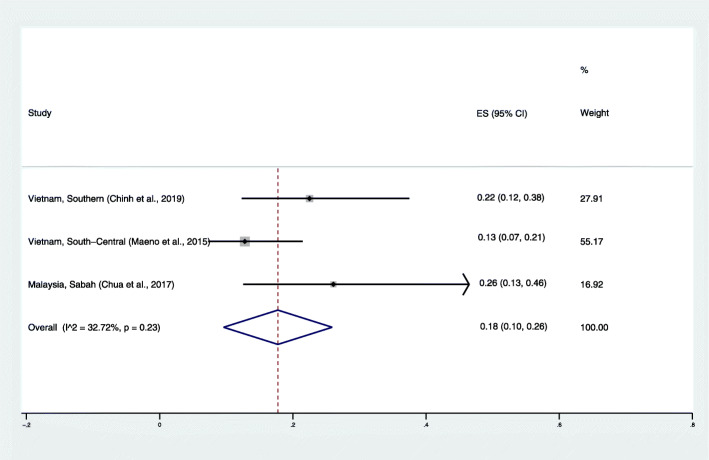
The pooled prevalence of P. cynomolgi in mosquitoes was very low at 0–1%. Because a low prevalence of P. cynomolgi in mosquitoes was reported in the included studies, the pooled proportion of P. cynomolgi compared to Plasmodium species infections in mosquitoes was estimated. The results demonstrated that the pooled proportion of P. cynomolgi infecting mosquitoes was 18%, with low heterogeneity (95% CI: 10–26%, I2: 32.7%) (Fig. 5). The highest proportion of P. cynomolgi infection in mosquitoes was demonstrated in northern Sabah (26, 95% CI: 13–46%) [22].
Fig. 5.
The estimated proportion of P. cynomolgi in vectors. ES: estimated proportion, overall: overall proportion, I2: level of heterogenicity, p: p-value less than 0.05 is statistically significant
The prevalence and proportion of P. cynomolgi infection in macaques
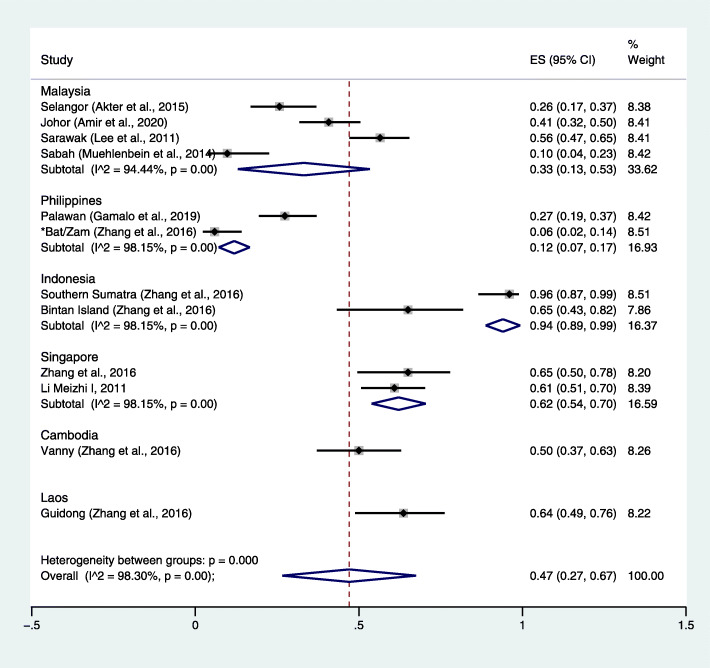
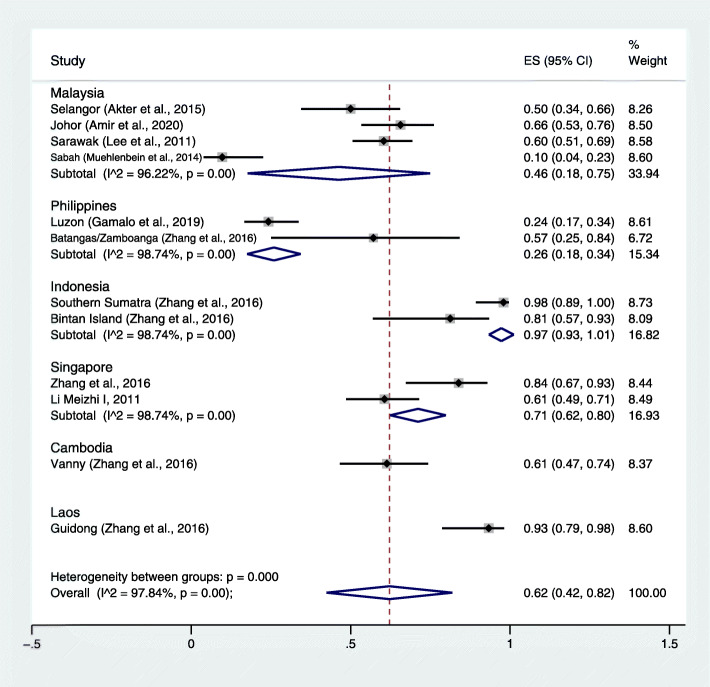
The pooled prevalence of P. cynomolgi compared to all macaques investigated was estimated. The results demonstrated that the pooled prevalence of P. cynomolgi infection in macaques was 47%, with high heterogeneity (95% CI: 27–67%, I2: 98.3%) (Fig. 6). The highest prevalence of P. cynomolgi infection in macaques was demonstrated in Indonesia (94, 95% CI: 89–99%) [17] and Singapore (62, 95% CI: 54–70%). The pooled proportion of P. cynomolgi compared to all malaria-positive samples was also estimated. The results demonstrated that the pooled proportion of P. cynomolgi infecting macaques was 67%, with high heterogeneity (95% CI: 42–82%, I2: 97.84%) (Fig. 7). The highest proportion of P. cynomolgi infection in macaques was demonstrated in Indonesia [17], while the lowest proportion of P. cynomolgi infection in macaques was demonstrated in the Philippines [13, 17].
Fig. 6.
The estimated prevalence of P. cynomolgi malaria in macaques. ES: estimated prevalence, overall: overall prevalence, I2: level of heterogenicity, p: p-value less than 0.05 is statistically significant
Fig. 7.
The estimated proportion of P. cynomolgi malaria in macaques. ES: estimated proportion, overall: overall proportion, I2: level of heterogenicity, p: p-value less than 0.05 is statistically significant
Streamlined publication years
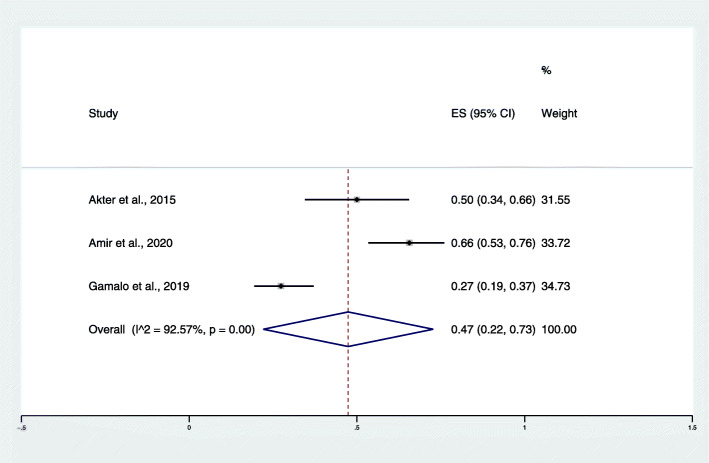
To reduce a large variance in the pooled proportion of P. cynomolgi infection between the three hosts and to describe the actual prevalence of P. cynomolgi infection in the past and present day, the cut-off point for publication years was streamlined for the purpose of this study. The year of studies was divided into studies reporting P. cynomolgi infection before 2013 and after 2013. Among the studies conducted before 2013, no studies reported the prevalence of P. cynomolgi infection in humans, while the study of Maeno et al. [23] during 2010–2013 demonstrated a pooled proportion of P. cynomolgi infection in mosquitoes of 13% (95% CI: 7–21%). The pooled proportion of P. cynomolgi infection in macaques was demonstrated by Lee et al. [15] during 2004–2008 at 56% (95% CI: 47–65%). Among studies conducted in and after 2013, the pooled proportion of P. cynomolgi infection in humans was estimated from three studies [18–20] during 2013–2017 at 1% (95% CI: 0.1%, I2: 0%). The pooled proportion of P. cynomolgi infection in mosquitoes was demonstrated by Chua et al. [22] during 2013–2014 at 26% (95% CI: 13–46%). In addition, the pooled proportion of P. cynomolgi infection in macaques during 2015–2020 was demonstrated at 47% (95% CI: 22–73%, I2: 92.57%) (Fig. 8).
Fig. 8.
The pooled proportion of P. cynomolgi infection in macaques during 2015–2020. ES: estimated proportion, overall: overall proportion, I2: level of heterogenicity, p: p-value less than 0.05 is statistically significant
Discussion
The successful transmission of zoonotic malaria is highly dependent on the bionomics and distribution of competent vectors and natural hosts. Humans and macaques live in and share overlapping spaces, particularly those who work in farming or agriculture near forests or tourists who come to visit areas where macaques are endemic. These close contacts and sharing of geographic distributions between humans and macaques and the presence of suitable vectors can lead to malarial disease transmission between host species. The increase in zoonotic malaria before the discovery of P. cynomolgi was primarily found in the occurrence of P. knowlesi malaria. P. knowlesi infection in humans is driven by multiple factors, including anthropogenic land-use factors leading to changes in the transmission pattern of the parasite between macaque reservoirs, vectors, and humans, as observed in Malaysian Borneo [24]. Deforestation for palm oil plantations or other clearing activities resulted in the loss of natural habitats for macaques, which led to their close contact with human settlements and increased incidence of P. knowlesi infection in humans [25].
The present study demonstrated a high prevalence and density of P. cynomolgi in macaques (47%), particularly in Indonesia [17] and Singapore [14, 17]. Interestingly, half of the P. cynomolgi infections in macaques were mixed infections with other Plasmodium species, and among the mixed infections, triple and quadruple infections were the most common types of mixed infections observed. It should be noted that mono-infection cases were predominantly in humans and Anopheles vectors, while an almost equal number of cases were reported in macaques. More interestingly, P. cynomolgi was likely to be a mixed infection with P. inui (90.4%), P. coatneyi (51.4%), and P. knowlesi (45.9%). The latter seems to support the similarity of transmission between P. cynomolgi and the more well-known zoonotic malaria caused by P. knowlesi. Zoonotic malaria caused by P. knowlesi infections was described and focused on in Sarawak, Malaysian Borneo, in 2004 [1], and later, it became a more common cause and was recognized as the fifth human malaria parasite throughout Southeast Asia [2]. Similar to P. knowlesi infection, naturally acquired P. cynomolgi infection in Malaysia was reported 10 years later in 2014 in endemic cases of people who lived in the same area with long-tailed macaques [6]. From that time to 2020, a tourist who traveled to Surat Thani Province, Thailand, was naturally infected with P. cynomolgi [26]. Then, naturally acquired P. cynomolgi infections in humans were detected in other parts of Southeast Asia, including Malaysia and Cambodia [18–20]. Moreover, when comparing the pooled proportion of P. cynomolgi infections in three different hosts in the studies conducted before 2013 and after 2013, the pooled proportion of P. cynomolgi infections in mosquitoes was higher in studies conducted after 2013. Nevertheless, the pooled proportion of P. cynomolgi infections in macaques was lower in studies conducted after 2013. The results of these streamlined publication years suggested that P. cynomolgi infections in humans might be due to the proximity of suitable vectors and monkeys, leading to conditions favorable for interspecies transmission, as demonstrated in cases of P. knowlesi infection [11].
Figure 3 shows that regions with the presence of both vectors and natural hosts also reported human infection cases, as was the case in Malaysia. However, human transmission in Cambodia seemed to have been caused by the presence of macaques in the country and its proximity to southern Vietnam, which reported the most cases of infected Anopheles vectors. Although cases in macaques have been reported in the Philippines, Laos, Singapore, and Indonesia, there is a curiosity as to why there is a lack of reports on infected Anopheles mosquitoes and humans. The results of the present study indicated that in areas where a high prevalence of P. cynomolgi was seen in macaques, sustained public information and advocacy in the affected areas is still necessary even if public advocacies have already been performed since 2014 [27–30].
Although a high prevalence of P. cynomolgi in macaques was demonstrated by the included studies, the prevalence of P. cynomolgi in mosquitoes was very low at 0–1%, and the proportion of P. cynomolgi compared to all Plasmodium species detected in mosquitoes was 18%. Therefore, the transmission of P. cynomolgi from macaques to mosquitoes should be limited by the bite rates, the susceptibility of Anopheles vectors, and the parasite density of the primate host. The contrast between a high prevalence of P. cynomolgi in macaques and a lower parasitemia of Anopheles mosquitoes should be due to the limited susceptibility of Anopheles vectors to harbor a lower parasitemia of P. cynomolgi in a specific environmental condition, which can lead to disease transmission, as changes in host preference, biting behavior, and adaptation of the mosquitoes to habitat changes could affect the transmission of zoonotic malaria in Southeast Asia. Although the proportion of P. cynomolgi infecting Anopheles mosquitoes was low, the importance of transmission through the infection of human hosts by the bite of the infected Anopheles mosquitoes should be monitored. In addition, although the pooled proportion of naturally acquired P. cynomolgi in human hosts demonstrated by the included studies was 1%, the high density of infected mosquitoes, such as in Northern Sabah (26%) [22], could lead to a high prevalence of naturally acquired P. cynomolgi in humans, as demonstrated in Northern Sabah (4%) [18].
P. cynomolgi infection in humans was observed to be asymptomatic and submicroscopic [18, 19]. This implied that the mono-infection of P. cynomolgi was very low in parasite density despite the mixed infection with P. vivax as demonstrated by Imwong et al. [19]. The morphological life cycle of relapse and the genetic similarities of P. cynomolgi and P. vivax could lead to undetected or undiagnosed P. cynomolgi cases in Southeast Asia. Most of the cases with suspected malaria admitted to the hospital were identified by the microscopic method, which has a low sensitivity and specificity to differentiate P. cynomolgi from P. vivax. In addition, the misidentification or missed detection of submicroscopic parasitemia by P. cynomolgi might lead to under-reported cases. Moreover, failure to treat P. cynomolgi malaria can lead to malarial recurrences, as observed in two individuals from Charkrya and Ou Treng, Cambodia [19]. Even in symptomatic infections of P. cynomolgi, such as in the mixed infection of P. cynomolgi and P. knowlesi, as reported in the study by Raja et al. [20], the parasite density ranged from 213 to 84,299 parasites per milliliter. Moreover, the most recent study with enrolled febrile patients at malaria clinics or local hospitals in Thailand demonstrated that most of the patients with P. cynomolgi mixed infection with P. vivax, P. falciparum or both P. vivax and P. knowlesi had parasitemia less than 10,000 parasites/μL (< 0.2% parasitemia) as demonstrated by Putaporntip et al. [31]. The low proportion of P. cynomolgi detected in humans by three studies [18–20] might be due to the use of less-sensitive molecular protocols, as they used nested PCR amplifying the SSU rRNA gene for the detection of malaria parasites. PCR amplification of the malarial cytochrome oxidase gene is superior to PCR amplification of SSU rRNA, as cytochrome oxidase is a mitochondrial genome that is found at approximately 20–150 copies per parasite, whereas SSU rRNA is found at approximately 4–8 copies per parasite [32, 33].
The natural infection of P. cynomolgi in humans is similar to the infection of the natural host of long-tailed or pig-tailed macaques since mixed infections of more than one species in a single host were observed in both humans and macaques. Nevertheless, the dominance of P. knowlesi over P. cynomolgi, as demonstrated by Raja et al. [20], might be due to the differences in the developmental cycles of the two species. P. cynomolgi has both an exoerythrocytic cycle in the liver and erythrocytic cycle in red blood cells. This means that the incubation period for P. knowlesi is shorter by approximately 9–12 days than that of P. cynomolgi, which is between 15 and 37 days [7]. Furthermore, the erythrocytic cycle of P. cynomolgi (48 h) is longer than that of P. knowlesi (24 h) [34]. Therefore, P. cynomolgi mixed infection with P. knowlesi might occur simultaneously in a single mosquito bite harboring two parasite species. The simultaneous infection of P. cynomolgi with P. knowlesi in a single mosquito was demonstrated in a study by Chua et al. [22].
The present study had limitations. First, the number of studies identifying P. cynomolgi in humans and mosquitoes was limited, resulting in an imprecise estimate of the pooled prevalence or pooled proportion and of the clinical characteristics of human hosts. Second, the Northern Sabah study [18] demonstrated a lower NOS scale (2 stars) as poor comparability against the other studies which used molecular detection methods but retained in the present analysis. Therefore, the pooled proportion of P. cynomolgi infection in humans requires careful interpretation. Further studies and continued surveillance of P. cynomolgi throughout Southeast Asia and neighbouring regions may be necessary through the use of sensitive molecular protocols to obtain accurate and relevant data for the detection and reporting of this emerging zoonotic malaria. Moreover, it is essential to further explore the biology of P. cynomolgi, the possibility of relapses/recurrences, and asymptomatic infections, which have a direct impact on the epidemiology of malaria.
Conclusion
In conclusion, P. cynomolgi could continue to be a public health concern in Southeast Asian countries, as the natural habitat of the natural hosts and vectors and the evolution of the P. cynomolgi parasite itself could drive the transmission of this neglected but emerging disease. In addition, the environmental and climatic changes experienced worldwide could affect the transmission dynamics of P. cynomolgi as an emerging cause of malaria in humans. Further studies using molecular and multidisciplinary approaches to search for P. cynomolgi infection in humans, vectors, and natural hosts are necessary if human infections with P. cynomolgi do become public health concerns.
Supplementary Information
Additional file 1: Table S1. Quality of the included studies
Acknowledgments
This research was partially supported by the New Strategic Research (P2P) project, Walailak University, Thailand. The authors are also grateful to Mr. David C. Chang for editing the grammar of this manuscript.
Authors’ contributions
MK and FRM carried out the study design, study selection, data extraction, and statistical analysis and drafted the manuscript. KUK and GDM participated in the study selection and data extraction and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This research was partially supported by the New Strategic Research (P2P) project, Walailak University, Thailand. The funders had a role in the collection, analysis, and interpretation of the data.
Availability of data and materials
All data related to the present study are available in this manuscript.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest regarding the publication of this article.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Manas Kotepui, Email: manas.ko@wu.ac.th.
Frederick Ramirez Masangkay, Email: frederick_masangkay2002@yahoo.com.
Kwuntida Uthaisar Kotepui, Email: kwuntida.ut@wu.ac.th.
Giovanni De Jesus Milanez, Email: gmilanez@feu.edu.ph.
References
- 1.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 2.White NJ. Plasmodium knowlesi: the fifth human malaria parasite. Clin Infect Dis. 2008;46(2):172–173. doi: 10.1086/524889. [DOI] [PubMed] [Google Scholar]
- 3.Cross JH, Hsu Kuo MY, Lien JC. Accidental human infection with Plasmodium cynomolgi bastianellii. Southeast Asian J Trop Med Public Health. 1973;4(4):481–483. [PubMed] [Google Scholar]
- 4.Druilhe P, Trape JF, Leroy JP, Godard C, Gentilini M. Two accidental human infections by Plasmodium cynomolgi bastianellii. Ann Soc Med Trop. 1980;60(4):349–354. [PubMed] [Google Scholar]
- 5.Most H. Plasmodium cynomolgi malaria: accidental human infection. Am J Trop Med Hyg. 1973;22(2):157–158. doi: 10.4269/ajtmh.1973.22.157. [DOI] [PubMed] [Google Scholar]
- 6.Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J. 2014;13(1):68. doi: 10.1186/1475-2875-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coatney GR, Collins WE, Warren M, Contacos PG. The primate malarias. 10. Atlanta: CDC; 2003. [Google Scholar]
- 8.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells G, Shea B, O’Connell D, Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 2013. [Google Scholar]
- 10.Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, Welch VA. Cochrane Handbook for Systematic Reviews of Interventions version 6.0 (updated March 2011): Cochrane; 2011. Available from: www.training.cochrane.org/handbook
- 11.Akter R, Vythilingam I, Khaw LT, Qvist R, Lim YA, Sitam FT, et al. Simian malaria in wild macaques: first report from Hulu Selangor district, Selangor, Malaysia. Malar J. 2015;14:386. doi: 10.1186/s12936-015-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amir A, Shahari S, Liew JWK, de Silva JR, Khan MB, Lai MY, et al. Natural Plasmodium infection in wild macaques of three states in peninsular Malaysia. Acta Trop. 2020;211:105596. doi: 10.1016/j.actatropica.2020.105596. [DOI] [PubMed] [Google Scholar]
- 13.Gamalo LE, Dimalibot J, Kadir KA, Singh B, Paller VG. Plasmodium knowlesi and other malaria parasites in long-tailed macaques from the Philippines. Malar J. 2019;18(1):1–7. doi: 10.1186/s12936-019-2780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irene LM. Identification and molecular characterisation of simian malaria parasites in wild monkeys of Singapore: National University of Singapore; 2011.
- 15.Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7(4):e1002015. doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muehlenbein MP, Pacheco MA, Taylor JE, Prall SP, Ambu L, Nathan S, et al. Accelerated diversification of nonhuman primate malarias in Southeast Asia: adaptive radiation or geographic speciation? Mol Biol Evol. 2015;32(2):422–439. doi: 10.1093/molbev/msu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Kadir KA, Quintanilla-Zariñan LF, Villano J, Houghton P, Du H, et al. Distribution and prevalence of malaria parasites among long-tailed macaques (Macaca fascicularis) in regional populations across Southeast Asia. Malar J. 2016;15(1):1–8. doi: 10.1186/s12936-015-1044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grignard L, Shah S, Chua TH, William T, Drakeley CJ, Fornace KM. Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in Sabah, Malaysia. J Infect Dis. 2019;220(12):1946–1949. doi: 10.1093/infdis/jiz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imwong M, Madmanee W, Suwannasin K, Kunasol C, Peto TJ, Tripura R, et al. Asymptomatic natural human infections with the simian malaria parasites plasmodium cynomolgi and plasmodium knowlesi. J Infect Dis. 2019;219(5):695–702. doi: 10.1093/infdis/jiy519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raja TN, Hu TH, Kadir KA, Mohamad DSA, Rosli N, Wong LL, et al. Naturally acquired human Plasmodium cynomolgi and P. knowlesi infections, Malaysian Borneo. Emerg Infect Dis. 2020;26(8):1801–1809. doi: 10.3201/eid2608.200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chinh VD, Masuda G, Hung VV, Takagi H, Kawai S, Annoura T, et al. Prevalence of human and non-human primate Plasmodium parasites in anopheline mosquitoes: A cross-sectional epidemiological study in Southern Vietnam. Trop Med Health. 2019;47(1):1–6. doi: 10.1186/s41182-019-0139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chua TH, Manin BO, Daim S, Vythilingam I, Drakeley C. Phylogenetic analysis of simian Plasmodium spp. infecting Anopheles balabacensis Baisas in Sabah, Malaysia. Plos Neglect Trop Dis. 2017;11(10):e0005991. doi: 10.1371/journal.pntd.0005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeno Y, Quang NT, Culleton R, Kawai S, Masuda G, Nakazawa S, et al. Humans frequently exposed to a range of non-human primate malaria parasite species through the bites of Anopheles dirus mosquitoes in South-central Vietnam. Parasit Vectors. 2015;8(1):1–7. doi: 10.1186/s13071-015-0995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson G, Chua TH, Cook A, Speldewinde P, Weinstein P. The role of ecological linkage mechanisms in Plasmodium knowlesi transmission and spread. Ecohealth. 2019;16(4):594–610. doi: 10.1007/s10393-019-01395-6. [DOI] [PubMed] [Google Scholar]
- 25.Fornace KM, Herman LS, Abidin TR, Chua TH, Daim S, Lorenzo PJ, et al. Exposure and infection to Plasmodium knowlesi in case study communities in northern Sabah, Malaysia and Palawan The Philippines. PLoS Negl Trop Dis. 2018;12(6):e0006432. doi: 10.1371/journal.pntd.0006432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmeyer GN, Stensvold CR, Fabricius T, Marmolin ES, Hoegh SV, Nielsen HV, et al. Plasmodium cynomolgi as cause of malaria in tourist to Southeast Asia, 2018. Emerg Infect Dis. 2019;25(10):1936–1939. doi: 10.3201/eid2510.190448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaehler N, Adhikari B, Cheah PY, von Seidlein L, Day NPJ, Paris DH, et al. Prospects and strategies for malaria elimination in the greater Mekong sub-region: a qualitative study. Malar J. 2019;18(1):203. doi: 10.1186/s12936-019-2835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Seidlein L, Peto TJ, Landier J, Nguyen TN, Tripura R, Phommasone K, et al. The impact of targeted malaria elimination with mass drug administrations on falciparum malaria in Southeast Asia: a cluster randomised trial. PLoS Med. 2019;16(2):e1002745. doi: 10.1371/journal.pmed.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Seidlein L, Peto TJ, Tripura R, Pell C, Yeung S, Kindermans JM, et al. Novel approaches to control malaria in forested areas of Southeast Asia. Trends Parasitol. 2019;35(6):388–398. doi: 10.1016/j.pt.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Scott J. Proposed intergrated control of zoonotic Plasmodium knowlesi in Southeast Asia using themes of One Health. Trop Med Infect Dis. 2020;5(4):175. [DOI] [PMC free article] [PubMed]
- 31.Putaporntip C, Kuamsab N, Pattanawong U, Yanmanee S, Seethamchai S, Jongwutiwes S. Plasmodium cynomolgi co-infections among symptomatic malaria patients, Thailand. Emerg Infect Dis. 2021;27(2):590–593. doi: 10.3201/eid2702.191660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaidya AB, Arasu P. Tandemly arranged gene clusters of malarial parasites that are highly conserved and transcribed. Mol Biochem Parasitol. 1987;22(2–3):249–257. doi: 10.1016/0166-6851(87)90056-9. [DOI] [PubMed] [Google Scholar]
- 33.Preiser PR, Wilson RJ, Moore PW, McCready S, Hajibagheri MA, Blight KJ, et al. Recombination associated with replication of malarial mitochondrial DNA. EMBO J. 1996;15(3):684–693. doi: 10.1002/j.1460-2075.1996.tb00401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 2013;26(2):165–184. doi: 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Quality of the included studies
Data Availability Statement
All data related to the present study are available in this manuscript.