Abstract
Diffusion MRI (dMRI) represents one of the few methods for mapping brain fiber orientations non-invasively. Unfortunately, dMRI fiber mapping is an indirect method that relies on inference from measured diffusion patterns. Comparing dMRI results with other modalities is a way to improve the interpretation of dMRI data and help advance dMRI technologies. Here, we present methods for comparing dMRI fiber orientation estimates with optical imaging of fluorescently labeled neurofilaments and vasculature in 3D human and primate brain tissue cuboids cleared using CLARITY. The recent advancements in tissue clearing provide a new opportunity to histologically map fibers projecting in 3D, which represents a captivating complement to dMRI measurements. In this work, we demonstrate the capability to directly compare dMRI and CLARITY in the same human brain tissue and assess multiple approaches for extracting fiber orientation estimates from CLARITY data. We estimate the three-dimensional neuronal fiber and vasculature orientations from neurofilament and vasculature stained CLARITY images by calculating the tertiary eigenvector of structure tensors. We then extend CLARITY orientation estimates to an orientation distribution function (ODF) formalism by summing multiple sub-voxel structure tensor orientation estimates. In a sample containing part of the human thalamus, there is a mean angular difference of 19° ± 15° between the primary eigenvectors of the dMRI tensors and the tertiary eigenvectors from the CLARITY neurofilament stain. We also demonstrate evidence that vascular compartments do not affect the dMRI orientation estimates by showing an apparent lack of correspondence (mean angular difference = 49° ± 23°) between the orientation of the dMRI tensors and the structure tensors in the vasculature stained CLARITY images. In a macaque brain dataset, we examine how the CLARITY feature extraction depends on the chosen feature extraction parameters. By varying the volume of tissue over which the structure tensor estimates are derived, we show that orientation estimates are noisier with more spurious ODF peaks for sub-voxels below 30 μm3 and that, for our data, the optimal gray matter sub-voxel size is between 62.5 μm 3 and 125 μm 3. The example experiments presented here represent an important advancement towards robust multi-modal MRI-CLARITY comparisons.
1. Introduction
Diffusion-weighted imaging (DWI) is sensitive to the diffusive motion of water molecules. Within tissue, water diffusion patterns are influenced by the presence of membranes and macromolecules. Therefore, the geometric pattern of water diffusion can be used to infer the orientation of fibrous structures such as axonal bundles and subsequently the 3D trajectories of fiber pathways, known as diffusion MRI (dMRI) fiber tracking (or tractography) (Basser et al., 2000; Conturo et al., 1999; Mori et al., 1999). Thus far, however, only the scalar measures of diffusion MRI, such as the apparent diffusion coefficient, are commonly used in the clinic (Baird and Warach, 1998), while diffusion orientation measurements have not yet become a standard clinical tool. Unfortunately, the accuracy of the voxel-wise diffusion orientation, as well as the subsequent fiber tracking, strongly depend on several different factors, including the quality and quantity of the data, efficacy of the preprocessing steps, adequacy of the model, ambiguity in intra-voxel crossings, and the choice of regions to tract between (Jones et al., 2013). This can lead to false negatives and false positives in the reconstructed diffusion fiber tracts that are difficult to identify (Maier-Hein et al., 2017). False negatives can be the result of fiber pathways that are too small, not sufficiently myelinated, with not enough fiber density or overlapping with too many other fiber pathways to produce a detectable anisotropic diffusion pattern (Canales-Rodríguez et al., 2019; Jones et al., 2013). False positives can result from dMRI fiber tracking that starts on one pathway but erroneously jumps to an unconnected, adjacent crossing or parallel pathway because the distance between the two pathways is too small for them to be uniquely resolved (O’Donnell and Westin, 2011). Further, it is not well understood whether dMRI can reliably identify fiber pathways entering and exiting gray matter.
Due to these known inaccuracies and questions about dMRI specificity (false positives) and sensitivity (false negatives), there are ongoing efforts to compare dMRI measurements against other modalities that also allow visualization of neuronal fibers (Dyrby et al., 2018; Innocenti et al., 2018). Tracer studies in animals, which inject a tracer and then sacrifice the animal to identify propagation of the tracer along neuronal fibers can be compared with diffusion fiber pathways (Knösche et al., 2015; Oh et al., 2014; Schilling et al., 2019) but is limited to animals. One-to-one comparisons with optical techniques in fixed brain tissue allow direct visualization of fiber pathways and may reveal the underlying tissue structure within the individual voxel (Kleinnijenhuis et al., 2011; Leuze et al., 2014; Seehaus et al., 2013). Unfortunately, capturing the full 3D projection of fiber pathways using these optical techniques remains a challenge. One approach is to section the brain tissue and use polarized light imaging to map the orientation at each location in the image (Axer et al., 2016; Leuze et al., 2014; Mollink et al., 2017). Another approach is to use tissue staining of 2D slices and subsequently stack the slices (Schilling et al., 2016, 2018). Yet another method involves the use of optical coherence tomography (OCT) to map microscopic structures at a depth of several millimeters and then use a vibratome to repeatedly remove the top layer of the tissue specimen (Magnain et al., 2015) to build a map of the full thickness of the specimen.
The adoption of 3D histology via tissue clearing enables optical microscopy of tissue cuboids, up to about the size of a whole rodent brain (~1 cm3) (Chung et al., 2013; Dodt et al., 2007; Ertürk and Bradke, 2013; Spalteholz, 1911). Brain clearing combined with lipophilic dyes such as long-chain dialkylcarbocyanines (Hou et al., 2015; Murray et al., 2015) or immunohistochemistry of myelin-specific proteins (Murray et al., 2015; Tomer et al., 2014) then offers a way to label neuronal fibers within intact tissue. Compared to conventional brain slicing, tissue clearing provides access to the intact 3D neuronal network without the need for further image stitching or slice alignment and is well-suited for comparisons to 3D neuroimaging data (Aswendt et al., 2017). Thus far, the development of tissue clearing has primarily focused on optimization for mouse (brain) tissue, where transgenic models allow cell-specific targeting with fluorescent probes (Chung et al., 2013; Dodt et al., 2007; Leuze et al., 2016; Renier et al., 2014; Susaki et al., 2014).
More recently, tissue clearing was improved for application to non-transgenic animals and human brain tissue through the use of active clearing and immunostaining, e.g., electrical gradients, perfusion force, or specific chemicals (Epp et al., 2015; Lee et al., 2016; Murray et al., 2015; Park et al., 2019; Zhao et al., 2020). While limited to small tissue cuboids (~3 × 3 × 0.5 cm3) (Morawski et al., 2017), these 3D histology methods potentially provide new insights into the sensitivity and specificity of diffusion MRI measurements. An initial study has shown that scalar dMRI measurements like mean diffusivity or fractional anisotropy correlate well with the density of cellular features visualized in CLARITY processed mouse brains (Stolp et al., 2018).
Here we aim to show the potential for improved interpretation of dMRI fiber orientation measurements by enabling a close correspondence with 3D histological images that directly show relevant microstructures. For this purpose, we present an example experimental protocol that enables a robust comparison of dMRI and CLARITY data in the same human brain tissue sample. We expect the neurofilament-stained CLARITY images to depict the orientation of neuronal fibers and therefore show correspondence with the dMRI orientation estimates. To examine the potential influence of tissue vasculature on dMRI measurements, we determine whether orientation estimates from vasculature-stained CLARITY images correspond with dMRI orientation estimates. Further, we examine how the CLARITY-derived orientation estimates depend on the volume of tissue over which the structure tensor estimates are derived.
There are several challenges associated with comparisons across such different imaging techniques. Anatomical orientation must be thoroughly recorded during tissue cutting to be able to later locate the origin of the specimen within the whole brain or tissue slab. Tissue deformations during the clearing process complicate a co-registration of CLARITY and MRI data. Further, the different contrast mechanisms (diffusion patterns vs. immunohistochemistry) and resolutions (2.5 μm vs. 1 mm isotropic) require a carefully considered feature extraction approach. To address the latter point, we use macaque brain data to examine the effect of calculating the structure tensors from differently sized tissue volumes.
While a complete optimization of all the methods described in the MRI-CLARITY comparison pipeline is well beyond the scope of a single manuscript, we demonstrate a highly tractable approach to perform a direct comparison in the same human tissue. This represents a critical step towards MRI-CLARITY comparisons, which have the potential to push the dMRI field forward by aiding the development of robust and interpretable dMRI fiber tracking and microstructure mapping for a wide range of applications.
2. Methods
Experiments were performed in both human brain and macaque brain samples. The multi-step process for comparing MRI and CLARITY data in the same tissue sample included tissue collection, tissue fixation in a hydrogel mixture, MRI scanning, cuboid cutting, clearing, staining, light microscopy imaging, co-registration of MRI and CLARITY images, feature extraction, comparison, and crossing fiber analysis. The human data demonstrate a comparison pipeline between diffusion MRI and CLARITY data in the same tissue sample. The macaque data demonstrate parameters that affect the CLARITY feature extraction method. The macaque experiments benefit from very high-resolution, high SNR CLARITY images but compare to an existing high-resolution dMRI dataset (Reveley et al., 2015; Schilling et al., 2019; Thomas et al., 2014) from a separate macaque.
2.1. Tissue collection
2.1.1. Human
In accordance with the local IRB, a whole human brain with no known pathology was extracted 24 hours after death and placed in 10% formalin for one day before the coronal slab shown in Fig. 1 was cut. A 30 × 15 × 10 mm3 tissue block covering part of the thalamus was cut from the coronal slab. Photographs with anatomical annotations were acquired during the iterative cutting process to retain perspective on the sample origin within the whole brain. For CLARITY processing, a smaller tissue cuboid (10 × 10 × 0.5 mm3) covering gray matter was cut from the tissue block as described below in Section 2.4 “Tissue Cutting ”. Photographs and MRI scans were taken during various stages of the cutting process (Fig. 1).
Fig. 1.

Photographs of the multiple stages of the tissue cutting. A A coronal slab of human brain tissue fixed for one day in 10% formalin and a sample cut from the coronal slab (red dashed box). The sample was embedded in a hydrogel solution and polymerized. A structural MRI scan (0.3 mm isotropic resolution) and a DTI scan (1 mm isotropic resolution) were performed on the sample. The arrows indicate the anatomical orientation, S – superior, R – right, A – anterior. B The tissue sample cut in half, with labels depicting the location of the lateral thalamus (LT), anterior thalamic nucleus (ATN), medial thalamic tract (MTT), red nucleus (RN), subthalamic nucleus (STN), and substantia nigra (SN). The rounded blue arrow denotes that the direction the sample is rotated (from S-R to S-A) for the photos shown in C and D. C The lateral part of the tissue sample was scanned on a small-bore scanner with a structural MRI scan at 0.1 mm isotropic resolution. D The superior part of the sample shown in C, which encompasses mostly gray matter and many visible vessels (black dots) that under-went the rest of the CLARITY pipeline (clearing, staining, and microscopy).
2.1.2. Macaque
In accordance with the local APLAC protocol, a tissue block of the occipital lobe of a macaque brain fixed in 10% formalin was extracted (Fig. 2). The tissue block was directly immersed in the CLARITY hydrogel solution after extraction. Photographs with anatomical annotations were acquired during the iterative cutting process to retain perspective on the sample origin within the whole brain.
Fig. 2.

A A photograph of the left part of the occipital lobe of a macaque brain with green lines indicating the location of the excised slab. B A photograph of the 3 mm thick slab excised from the occipital lobe in the coronal plane. C The cleared brain sample. D-G Images indicating the brain location of the extracted tissue used for high-resolution two-photon CLARITY. The confocal microscopy image in G provides an intermediate field-of-view that enables co-registration of the detailed two-photon microscopy image in H with the MRI (D-F).
2.2. Tissue fixation in hydrogel
Both human and macaque tissue samples were embedded in a hydrogel monomer solution consisting of 4% acrylamide, 0.05% bisacry-lamide, 4% paraformaldehyde (PFA) and 0.25% VA-044 thermal initiator in PBS (Tomer et al., 2014) for one week. The samples were degassed and polymerized at 37 °C for 2.5 h. Excess hydrogel was removed.
2.3. MRI
2.3.1. Human
The polymerized human brain sample (Fig. 1 B) was scanned on a 7T large bore GE human scanner, using a Nova-32 channel head coil in a large container filled with fomblin, a non-protonated fluid. While only a single sample was examined in this study, a plastic box with multiple compartments was used to simultaneously scan multiple tissue samples of similar size that were used to test variations in the clearing and staining protocol (results of other clearing and staining variations are presented in the supplemental material). The tissue samples were matched in size to the individual box dividers such that they were stabilized and did not touch each other during the scan, which simplified image segmentation of each piece after the experiment. The plastic box with dividers was placed inside a larger plastic container filled with fomblin, and holes were created inside each of the plastic box divider lids such that no air bubbles would be trapped in the individual spaces. The tissue samples were removed from the refrigerator 12 h prior to the scan such that they would equilibrate with room temperature before imaging. The parameters of the MRI sequences can be seen in Table 1.
Table 1.
MRI scan parameters for the human sample.
| Specimen | Sequence | Scanner | Resolution | TR/TE | |
|---|---|---|---|---|---|
| Extracted thalamus (Fig. 1B). | Balanced steady state free precession | 7T human | 350 × 350 × 300 um | 12.6/6.2 | 4 averages |
| Extracted thalamus (Fig. 1B). | Diffusion-weighted spin echo echo planar imaging | 7T human | 1 mm isotropic | 3000/60 | 447 directions, |
| b = 2000 s/mm2 | |||||
| 30 images at b = 0 s/mm2 | |||||
| Extracted thalamus cut in half (Fig. 1C). | FLASH | 7T Bruker small bore | 0.1 mm isotropic | 39.6/19.6 | Flipangle = 22° |
To examine the dataset with reference to a whole brain in vivo dMRI scan, the tissue sample data was compared against a single subject dMRI dataset from the Human Connectome Project (HCP) (Vu et al., 2015) acquired with a 7T scanner at 1.05 mm isotropic resolution with TE = 41 ms, b = 1000 & 2000 s/mm2 and 130 diffusion directions with two opposite phase encodings per direction.
2.3.2. Macaque
A structural scan of an ex-vivo whole macaque brain was acquired on a 7T small bore Bruker scanner with a Turbo RARE T2-weighted sequence at a resolution 0.15 × 0.15 × 0.5 mm3, and TR/TE = 2500/53.4.
A 3 mm thick slab of the occipital lobe of the macaque brain was extracted and scanned on the same 7T small bore Bruker scanner with a FLASH sequence with TR/TE = 42.9/20 and 0.2 mm isotropic spatial resolution.
The macaque brain CLARITY images were compared against a high-resolution (0.25 mm isotropic) ex vivo macaque dMRI dataset acquired on a 7-T horizontal Bruker scanner at the Bethesda National Institute of Health (NIH) and presented previously (Reveley et al., 2015; Schilling et al., 2019; Thomas et al., 2014). The NIH dMRI protocol consists of 114 directions with b = 4900 s/mm2 and 7 volumes of b = 0 s/mm2. The TORTOISE tool (Calabrese et al., 2015; Pierpaoli et al., 2010) was used to preprocess the data for eddy currents distortions and frequency drift.
2.4. Cutting tissue cuboids for CLARITY processing
2.4.1. Human
The human thalamus sample used for the FLASH MRI scan (Fig. 1 C) was cut in half, separating the superior from the inferior portion. The superior part of the thalamus sample (yellow region in Fig. 1 C) was then cut again along the sagittal orientation so that the sample’s sagittal plane was parallel to the read-slice directions of the diffusion MRI scan to produce a 10 × 10 × 0.5 mm3 section containing deep gray matter for clearing (Fig. 1 D).
2.4.2. Macaque
For the macaque brain, the tissue block from the occipital lobe was cut into 3 mm thick coronal slabs. A slab from the center of the occipital lobe (Fig. 2 B) was then used for further CLARITY processing.
2.5. Tissue clearing
2.5.1. Human and macaque brain
The human and macaque tissue cuboids were passively cleared according to the advanced CLARITY protocol by Tomer et al. (2014). Briefly, the sample was placed in a 4% SDS/borate clearing buffer (SBC) at 37 °C for five weeks (human thalamus) and at 60 °C for two weeks (macaque). The SBC solution was exchanged every two days for the first week and every week after that. After clearing, the sample was washed overnight in 0.2 M boric acid buffer/0.1% Triton X-100 at 37 °C.
2.6. Staining
For staining, the SWITCH protocol (Murray et al., 2015) was adapted to stain neurofilaments (NF) for visualization of neural fiber pathways.
2.6.1. Human
The human brain sample was incubated in SWITCH OFF (with SDS) antibody solution (NF-H chicken antibody (AB5539, Merck, Germany) (1:250) for 14 days at 37 °C, followed by a quick wash in 1x PBST (PBS + 0.1% TritonX) and an incubation in SWITCH ON (without SDS) antibody solution (1:500) for six days at 37 °C. Excess primary antibody was washed off with sequential steps in 1x PBST (10 min, 30 min, 2 days). The same labeling procedure was pursued with the secondary antibody dilution of 1:500 (donkey x chicken AlexaFluor647, A21449, Life Technologies, Germany). The stained sample was kept in PBST and transferred to FocusClear (CellExplorer Labs) 2 days before imaging.
2.6.2. Macaque
The macaque brain sample was incubated in SWITCH OFF (with SDS) antibody solution (NF-H chicken antibody (AB_2313552 AvesLab, USA)) (1:100) for 28 days at 37 °C, followed by a quick wash in 1x PBST (PBS + 0.1% TritonX) and incubation in SWITCH ON (without SDS) antibody solution (1:100) for 14 days at 37 °C. Excess primary antibody was washed off with sequential steps in 1x PBST (10 min, 30 min, 2 days). The same labeling procedure was pursued with the secondary antibody dilution of 1:100 (Goat anti-Chicken IgY (H + L) AlexaFluor647, A-21449, Thermo Fischer Scientific, USA). The stained sample was kept in PBST and transferred to FocusClear (CellExplorer Labs) 2 days before imaging.
2.7. Optical microscopy imaging
The cleared samples were mounted on a cover slide within a ring of blue tag to provide spacing between the cover slide and prevent the FocusClear from flowing out.
2.7.1. Human
The human brain sample was imaged using a confocal microscope, covering 200 slices with a 10x objective at 2.5 μm isotropic resolution using mosaic stitching to cover the whole sample. Two channels were acquired, 488 nm for the autofluorescence to show the vasculature and 648 nm to capture the NF antibody staining.
2.7.2. Macaque
The macaque brain sample was imaged using a confocal microscope with a 5x objective to obtain an overview image, including 17 slices at 264 um in-plane resolution. Two channels were acquired, 488 nm for the autofluorescence to show the vasculature and 648 nm to capture the NF antibody staining. A part of the macaque sample that contained both white and gray matter was then further imaged with a two-photon microscope at 1.5 μm isotropic resolution over a region 2.6 × 2.8 × 0.25 mm3 in size and covering 176 slices.
The signal-to-noise ratio (SNR) of each sample’s CLARITY data was measured by selecting an ROI covering the sample and an ROI covering the empty background and measuring both mean signal intensity and standard deviation of the ROI. SNR was then calculated as:
where μ is the mean signal over the ROI, and σ is the standard deviation of the background ROI.
2.8. Co-registration of MRI and CLARITY images
2.8.1. Human
Prescribing MR slices parallel to the sample surface and then cutting the cuboid and optically imaging it within the same (or very similar) plane simplified the co-registration procedure. The human CLARITY dataset was co-registered to the high-resolution 100 μm MRI scan by manually identifying vessels and the tissue boundaries as common landmarks between the two sets of images and manually placing registration markers on these landmarks. Using the landmark-based registration toolbox in 3Dslicer, a similarity transform relating the CLARITY and MR images was computed using these registration markers. The high-resolution scan from the preclinical Bruker MRI scanner was then linearly co-registered to the 300 μm structural scan from the human 7T MRI scanner and, in turn, to the dMRI scan.
2.8.2. Macaque
The portion of the macaque brain section scanned with high-resolution two-photon microscopy (Fig. 2 H) was visually identified on the confocal microscope overview (Fig. 2 G) image based on fluorescent bleaching and is delineated in Fig. 2 G by a yellow patch. To compare the two-photon microscopy images (Fig. 2 H) with a dMRI of a whole, macaque brain (Fig. 2 D), the overview confocal image (Fig. 2 G) was first manually aligned and then linearly co-registered to the structural MRI of the tissue block (Fig. 2 F). The structural MRI of the tissue block (Fig. 2 F) was co-registered to the T2-weighted whole-brain acquisition (Fig. 2 E). Finally, the T2-weighted whole macaque brain (Fig. 2 E) was co-registered to the b = 0 s/mm2 image of the NIH whole macaque brain (Fig. 2 D). All manual macaque brain co-registrations were performed with 3D Slicer (Fedorov et al., 2012) and linear co-registrations with Elastix (Klein et al., 2010).
To refine the initial point co-registration between the NIH macaque brain diffusion tensor and CLARITY structure tensor, a vector field rigid registration algorithm was developed. The optimization algorithm used rotations and translations until the angular vector distance was minimized.
All the co-registration matrices were concatenated, and a final intensity-based linear co-registration with Elastix was used, to avoid unnecessary blurring.
2.9. Feature extraction and comparison
2.9.1. Human and macaque brain
Structure tensor analysis was performed on the CLARITY data to extract fiber orientation estimates that could be compared with dMRI orientation estimates. The primary fiber orientation was estimated as the tertiary eigenvector (i.e., with the smallest eigenvalue) of a structure tensor (Budde and Frank, 2012; Kass and Witkin, 1987; Khan et al., 2015; Wang et al., 2015; Ye et al., 2016), which was defined as:
where Ix, Iy, and Iz are the gradients of image volumes I along each of the x, y, and z axes (a marker of the edges of fiber tracts), computed by convolving I with three 3-dimensional 1st order derivative of Gaussian filters with a standard deviation σ = 5 μm for the NF stain visible in the 648 nm channel of the human brain sample, σ = 12.5 μm for the vasculature visible in the 488 nm channel of the human brain sample and σ = 1 μm for the NF stain in the macaque brain (Canny, 1986). In the human sample, the structure tensors were averaged over each diffusion MRI voxel (w = 1.0 × 1.0 × 0.5 mm3). In the macaque brain, the structure tensor was computed over voxel sizes of 250 μm3. The angle (0–90°) between the primary CLARITY fiber orientation, estimated from the tertiary orientation of the structure tensor, and the primary dMRI tensor orientation was calculated in MATLAB. Note that the structure tensor’s primary eigenvector is oriented along the direction of the highest signal intensity gradient, which represents the edges of the fibers and therefore is orthogonal to their long axis. For visualization and comparison purposes, the orientation along the nerve fibers thus corresponds to the structure tensors’ tertiary orientation.
2.10. Crossing fiber analysis
2.10.1. Human brain
Diffusion data crossing fiber estimates were derived using the FSL Bayesian Estimation of Diffusion Parameters toolbox (BedpostX). The primary fiber orientation and any additional fiber orientations with an anisotropic fiber fraction > 0.05 were estimated.
CLARITY data structure tensor orientation estimates were derived from 0.09 mm isotropic regions using Gaussian filters with a standard deviation of σ = 5 μm. CLARITY data crossing fiber estimates were derived from the distribution of all the structure tensor orientation estimates within each diffusion human brain MRI voxel (w = 1.0 × 1.0 × 0.5 mm3) and represented as a polar Fiber Orientation Distribution (FOD) histogram with angle increments of 4.5°.
2.10.2. Macaque brain
The macaque brain data was used to study the effect of changing the region size over which the CLARITY crossing fiber estimates were derived. The 250 μm3 CLARITY voxel corresponding to a matched MRI voxel of the same size was subdivided into: 2 × 2 × 2, 4 × 4 × 4, 8 × 8 × 8, and 16 × 16 × 16 sub-cubes. The structure tensor was calculated for each of these sub-cubes, corresponding to 125 × 125 × 125 μm3, 61.5 × 61.5 × 61.5 μm3, 30 × 30 × 30 μm3, and 15 × 15 × 15 μm3, respectively. All primary orientations of the structure tensors in each sub-cube were then summed to estimate the FODs representative of the full 250 μm3 voxel. Due to the high number of crossings detected in the macaque data and to allow a visualization that is familiar to the diffusion MRI community, the polar histograms were fitted with spherical harmonics with order lmax = 8 with the MrTrix3 software (Tournier et al., 2019) to generate structure orientation distribution functions (sODF) such that only the dominant orientations of the histogram are visualized.
3. Results
3.1. Human
3.1.1. MRI
Fig. 3 displays the MRI scans of the human brain tissue block. Fig. 3 A is a whole-brain 1.05 mm isotropic diffusion MRI scan of a different brain from the HCP dataset. Fig. 3 B displays a zoomed view of the region of interest. The bottom of Fig. 3 displays from left to right the 1 mm isotropic diffusion MRI scan (Fig. 3 C), the 300 μm isotropic structural MRI scan (Fig. 3 D), and the 100 μm isotropic structural scan (Fig. 3 E) of the excised specimen.
Fig. 3.
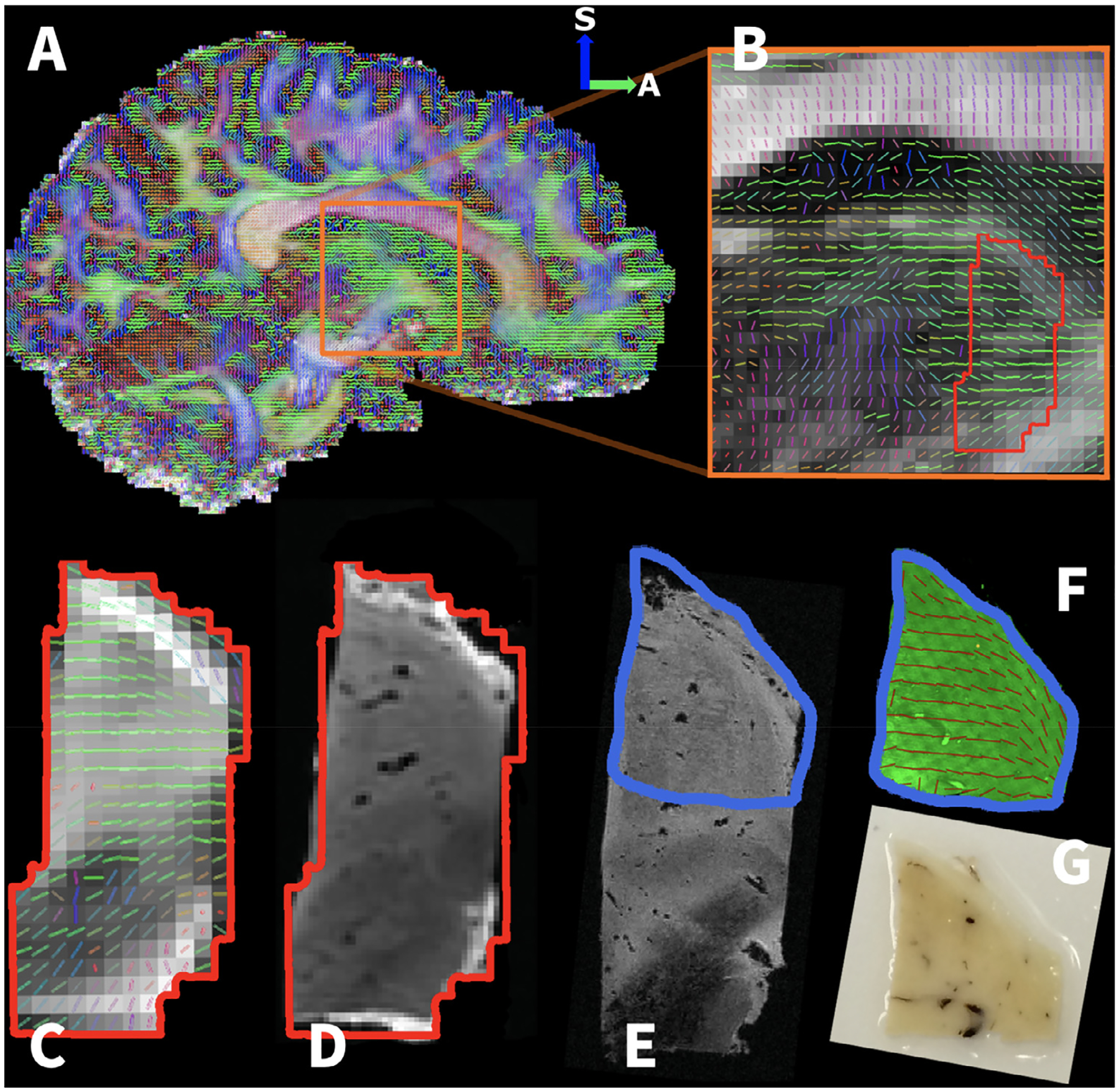
Human thalamus tissue sample relative to the whole-brain dataset. A A whole brain in vivo 1.05 mm isotropic resolution DTI scan taken from the human connectome project (HCP). B Zoomed image of the HCP DTI scan depicting the region that a specimen was obtained (indicated by red outline) for an MRI-CLARITY comparison. Ex vivo MRI of the specimen included: C 1 mm isotropic DTI, D 0.3 mm isotropic structural MRI, and E 0.1 mm isotropic structural MRI. The tissue specimen has been aligned to the whole brain HCP scan shown in A to provide whole-brain perspective on its anatomical location. F The cleared sample stained for neurofilament with extracted fiber orientations. G Photographs of the cut 10 × 10 × 0.5 mm3 tissue cuboid used for the CLARITY procedure.
3.1.2. CLARITY (clearing, staining, optical microscopy imaging)
Passive clearing led to high sample transparency while preserving the tissue’s stability throughout the subsequent staining and optical microscopy imaging experiments. Using the SWITCH protocol, the NF antibody stain penetrated the sample homogeneously throughout the whole thickness (Fig. 4). For confocal imaging, the sample was immersed in a refractive index matched solution and very carefully placed on the microscope stage such that it was as flat as possible but not compressed.
Fig. 4.

Human thalamus CLARITY data. A A cleared tissue cuboid showing (in pseudo colors) neurofilament stain (green) and autofluorescence (red), which visualizes mainly the vessels in the tissue. The right inset shows a magnification of the underlying tissue structure. B 3D rendering of the specimen showing the homogeneous stain penetration of the sample.
Fig. 4 shows the neurofilament antibody (green) and autofluorescence (red) channel of the cleared human brain sample in pseudo colors. The grid pattern that can be seen covering the sample arises from mosaic stitching of different tiles acquired with the confocal microscope. The NF stain demonstrates the overall fiber orientation in the sample, with most fibers on the posterior side of the sample traversing along the A-P orientation and on the anterior side along the S-I orientation. Note that it is only possible to infer the orientation and not the directionality of the fiber patterns. The SNR of the CLARITY image for the human sample was 3.6.
3.1.3. Comparison
Distinct vessels can be observed both in the autofluorescence channel of the CLARITY sample and the 100 μm T2* -weighted MRI data (Fig. 5 A). The root mean square error of the co-registration based on the 15 manually placed registration markers depicted in Fig. 5 A was 0.261 mm. The largest error between registration markers was 0.360 mm for registration marker #5 (Fig. 5 A, top left corner).
Fig. 5.

Human thalamus comparison of CLARITY and MRI-derived orientation estimates. A T2* -weighted structural (100 μm) MRI and the autofluorescence CLARITY image of the thalamus both depict vessels, which served as co-registration markers for linear co-registration of the CLARITY data to the MRI scan. Markers in the fused image depict the manually placed co-registration markers for multiple image slices of the CLARITY (red) and MRI (yellow) projected onto the same 2D plane. Based on the MRI and CLARITY co-registration markers, the co-registration root mean square error was RMS = 0.26 mm. The co-registered MRI and CLARITY dataset was then co-registered to the 300 μm resolution structural MRI and the 1 mm diffusion MRI. B Overlays of the CLARITY-derived structure tensors and MRI-derived diffusion tensors, on their respective images (left) and combined (right). C Overlays of the CLARITY and MRI-derived dominant orientations, on their respective images (left) and combined (right). D A map of the 3D angular difference between the CLARITY and MRI-derived dominant orientations displaying a mean angular difference and standard deviation of 19 ± 15° inside the dashed white line. The histogram shows the distribution of angular differences between the CLARITY and MRI-derived orientation estimates. E Comparison of the estimated diffusion tensor MRI orientation with the structure tensor orientations estimated for the CLARITY image of the vasculature shows a mean angular difference and standard deviation of 49 ± 23° inside the dashed white line.
The three-dimensional structure tensor estimates are superimposed on the source microscopy images (2.5 um isotropic neurofilament-stained confocal microscopy) to enable visual assessment of how well they align with visually observed fibers in the image (Fig. 5 B).
The structure tensor orientation estimates were not affected by the image stitching artifacts (grid pattern visible in Fig. 4 A, B), because these artifacts were present at a much larger scale compared to the size of the Gaussian filters used in the structure tensor estimates. Both diffusion and structure tensors estimated mainly an A-P fiber orientation, especially towards the center of the sample. The dominant fiber orientation estimated by diffusion and structure tensors was extracted for a quantitative comparison (Fig. 5 C), which yielded a mean angular difference of 19 ± 15° inside the dashed white line in Fig. 5 D. The boundaries were excluded from the analysis to avoid edge effects that lead to incorrect structure tensor estimations in that region.
The same structure tensor analysis with a slightly bigger Gaussian filter (σ= 12.5 μm) to account for the larger vasculature size compared to the neurofilament stain was applied to the 488 nm channel, which mainly highlighted the tissue vasculature (Fig. 5 E). The mean angular difference between the dominant orientation estimates of the structure tensor for the 488 nm channel and the diffusion tensor was 49 ± 23° inside the dashed white line.
Fig. 6 depicts the crossing fiber analysis of the CLARITY and the diffusion MRI data. The majority of the FODs in the CLARITY data depict a uniform single orientation with 87% of the fibers (excluding the boundaries of the sample) having a voxel-wise spread of less than 22.5° for the individual orientation estimates that compose each FOD (Fig. 6 A). The majority of the diffusion MRI voxels also support the existence of only a single orientation, although there are a few voxels where the anisotropic fiber fraction of a second fiber orientation is above the 5% threshold typically used for tractography (Fig. 6 B). A magnification of the region containing crossing fibers in the middle of the sample and comparison with the CLARITY FOD highlights a few voxels where the secondary diffusion orientation estimate and the CLARITY FOD results are not in agreement (Fig. 6 C).
Fig. 6.

Human thalamus comparison of CLARITY and MRI-derived crossing fiber estimates. A Voxel-wise fiber orientation distribution functions (FOD) are estimated from the structure tensor analysis of the CLARITY data. A CLARITY-derived FOD corresponding to each diffusion MRI voxel (1 mm isotropic) is estimated by calculating multiple structure tensors over 0.09 mm isotropic regions and then representing all the structure tensors estimated within the region of a given MRI voxel (although limited through-plane by the thickness of the CLARITY specimen) as a histogram of orientation estimates. B Crossing fibers are estimated from the diffusion MRI data. The presence of secondary fiber orientation (green) is supported by the data in only a few voxels. C A magnification of four MRI voxels highlights some disagreement between the FOD and diffusion crossing fiber analysis.
3.2. Macaque
3.2.1. Sample localization
Fig. 2 D shows the whole NIH dMRI brain and, as marked by the yellow region, the anatomical location of the macaque visual cortex brain sample processed with CLARITY. Fig. 2 H displays the two-photon microscopy image of the macaque visual cortex NF stain. The fiber orientations are easier to see in the gray matter, where the stain is less dense. The SNR of the two-photon microscopy image over both white and gray matter was 17.6.
3.2.2. Fiber orientation analysis
Fig. 7 displays the voxel-wise comparison of the primary fiber orientation estimation from the CLARITY-derived structure tensor (Fig. 7 A) and the diffusion MRI-derived tensor (Fig. 7 B). The angular difference between the CLARITY and the diffusion MRI derived orientations are shown in Fig. 7 C. There was an angle difference of 21° + /− 19° in GM and 42° + /− 16° in WM. The histograms of the angle difference in GM show 68% of voxels with an error below 20° and 40% of voxels with an error below 10°.
Fig. 7.

Macaque visual cortex comparison of CLARITY and MRI-derived orientation estimates in white matter (WM) and gray matter (GM). The dotted line provides a visual indication of the approximate gray-white matter boundary. A Structure tensor estimation from the CLARITY data. B Diffusion tensor estimation of the NIH dMRI dataset in the same region. C The angular difference between the primary orientations of the structure tensor and the diffusion tensor.
Fig. 8 displays the comparative analysis of structure tensor orientation density functions (sODFs) derived from different sizes of tissue volumes. A dominant orientation is detected for 2 × 2 × 2, 4 × 4 × 4, and 8 × 8 × 8 sub-cubes, while highly distributed orientations are detected for the 16 × 16 × 16 sub-cubes, especially in the gray matter.
Fig. 8.
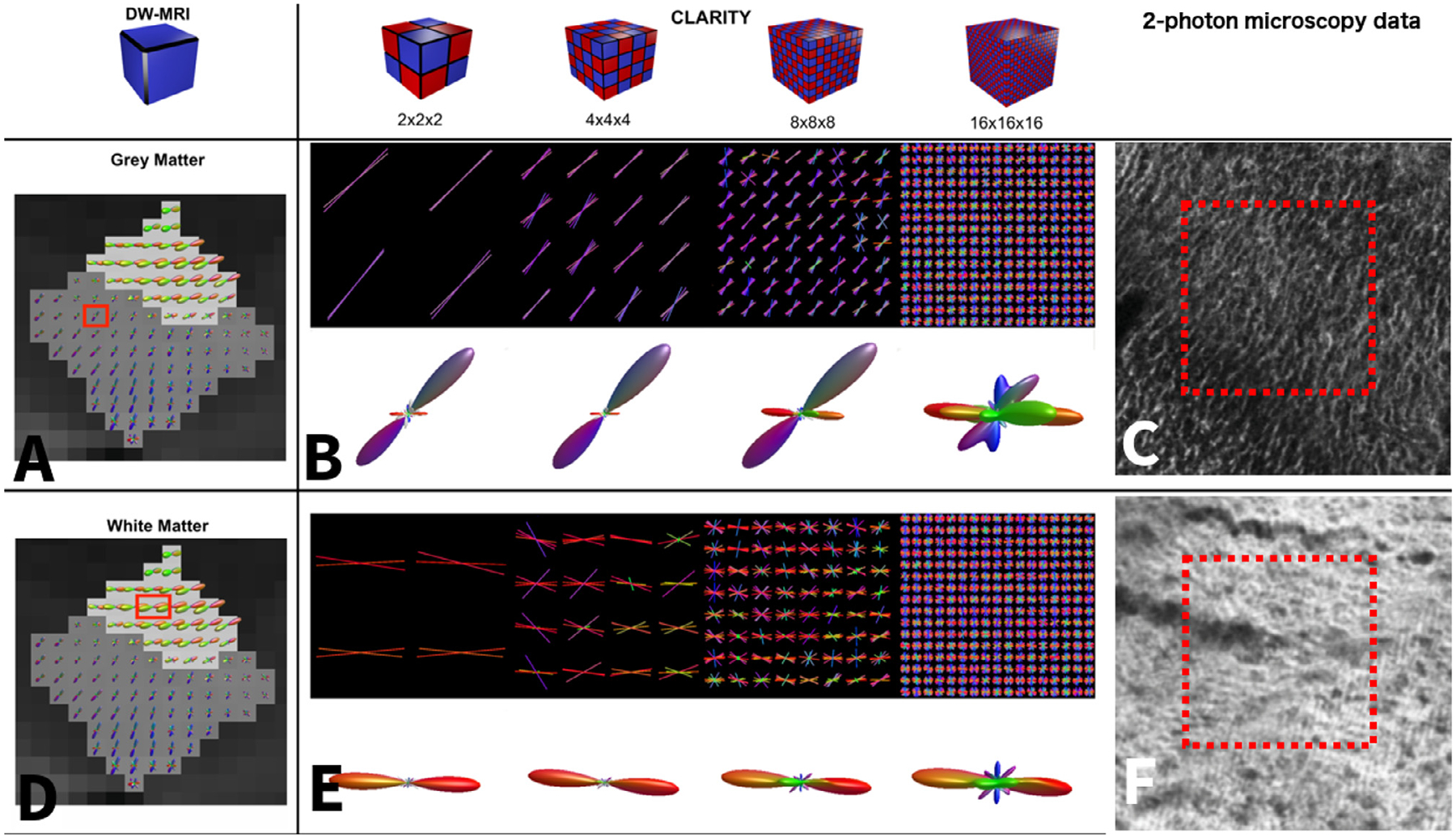
Macaque visual cortex characterization of CLARITY-derived crossing fiber estimates. A and D show fODF estimates using constrained spherical deconvolution calculated from the NIH macaque data (Thomas et al., 2014). The gray matter has a dark gray background and the white matter has a light gray background. B and E show the sODF calculated from the CLARITY two-photon microscopy data with varying sub-voxel sizes. To facilitate visual comparison with the dMRI data, the sODF was concatenated over the whole 250 μm voxel size, and a spherical harmonics function was fitted to the final sODF using MrTrix (Tournier et al., 2019). In A, a gray matter voxel is highlighted on top of the dMRI fODF reconstruction. The sODF of the corresponding voxel in the two-photon microscopy data is shown in B. The first row shows the primary structure tensor orientations for each of the sub-voxels projected on a single plane, the second row shows the corresponding sODF. In D, a white matter voxel is highlighted on top of the dMRI fODF reconstruction. The sODF of the corresponding voxel in the two-photon microscopy data is shown in E. The number of detected peaks increases for smaller sub-voxel sizes. C and F show a close-up view of the 2-photon microscopy data.
In Fig. 9, only the major peaks from each voxel-wise measure are displayed. The second and third rows show the primary peak and all the peaks with an amplitude of at least 1/3 of the primary peak amplitude. More peaks are detected in the CLARITY data than in the dMRI data, especially in the white matter. It can also be seen that the CLARITY and dMRI orientations are in better agreement for the 2 × 2 × 2 and 4 × 4 × 4 sub-cubes, especially in the gray matter.
Fig. 9.

Macaque visual cortex crossing fiber analysis of the dMRI data and CLARITY two-photon microscopy data. The first column shows the dMRI measurements. The four columns on the right show the CLARITY structure tensor results for varying sub-voxel sizes. The first row shows the spherical harmonic ODFs. The second row shows only the primary orientation of the dMRI and structure tensor measurements. The third row shows all the peaks with an amplitude of at least 0.3x the primary peak amplitude.
Table 2 lists the angular difference between the primary orientation derived from dMRI and CLARITY data for different numbers of sub-cubes averaged over the region containing ODFs shown in Fig. 9.
Table 2.
Angular differences between the primary orientation derived from dMRI and CLARITY macaque data for different numbers of sub-cubes.
| sODF | WM | GM |
|---|---|---|
| 2 × 2 × 2 | 48° + /− 15° | 28° +/− 21° |
| 4 × 4 × 4 | 40° +/− 12° | 24° +/− 16° |
| 8 × 8 × 8 | 39° +/− 14° | 36° +/− 26° |
| 16 × 16 × 16 | 35° +/− 11° | 58° +/− 11° |
4. Discussion
This study develops and demonstrates a framework for direct comparisons of tissue microstructure across MRI and CLARITY data acquired in the same human thalamus tissue sample, including the necessary data acquisition, co-registration, and automated feature extraction techniques. The MRI-CLARITY comparisons presented here demonstrate the ability to identify clear landmarks for co-registration of MRI and CLARITY images. These data also demonstrate that structure tensor estimates of the primary fiber orientation may be extracted from CLARITY confocal microscope images of a human sample with an SNR of 3.6. This study extends CLARITY fiber orientation estimates to a crossing fiber analysis. Structure tensor orientation estimates over multiple sub-cubes are used to generate fiber orientation distribution functions and structure tensor and FOD results are shown to be strongly affected by the size of the tissue volume over which the structure tensor is estimated.
For a detailed look at the effects of the structure tensor and crossing fiber analysis and how well the findings in the human thalamus sample translate to other tissue such as white matter or a different species, an additional sample excised from a macaque brain is examined. The macaque brain data were acquired at a higher SNR (17.6) with 2-photon microscopy CLARITY images of the white and gray matters. The comparisons of the FODs with varying sub-cube sizes against a dMRI macaque brain dataset in the same region suggest that there exists an optimal sub-cube size for orientation estimation. The optimal sub-cube size likely depends on several different factors, including the SNR and resolution of the microscopy data, the density of the stain, the scale of any artifacts (e.g., stitching artifacts), and the scale of the structure of interest in the tissue (e.g., neuronal fibers). It is currently unclear whether the structure tensor analysis of the CLARITY data is sensitive to individual axons or rather the edges of striated small fiber pathways within the tissue. For the data presented here, small sub-cube sizes resulted in noisy ODFs with many spurious peaks. Large sub-cube sizes obscured intra-voxel variations between fiber orientations.
The lowest angle difference between the fiber orientation estimates derived from the macaque brain CLARITY and dMRI data in gray matter was for a sub-cube size of 61.5 μm isotropic and for the largest sub-cube size of 125 μm isotropic. This is a good indication that the 90 μm isotropic cube size for estimation of the FOD in the human dataset was chosen appropriately to probe the tissue structure in the thalamus. The higher angular differences in the macaque white matter are potentially due to the dense staining in the white matter, which does not visualize the edges of individual fibers very well. This may indicate that a sparser stain that targets a lower density component of the white matter is better for visualizing white matter orientations.
The macaque brain CLARITY data displayed higher SNR and higher spatial resolution than the human CLARITY data. Therefore, they were particularly useful for evaluating the effect of varying the volume over which the structure tensors were calculated. Nonetheless, it is important to note that the CLARITY and dMRI macaque data were from different brain specimens. Therefore some anatomical variation between animals is represented in the cross-modality comparison. In contrast, the human CLARITY data was at lower SNR and spatial resolution but featured CLARITY and dMRI in the exact same tissue and therefore represents a better test-bed for the cross-modality CLARITY-dMRI comparison.
While there was an agreement between the primary orientations of diffusion tensor and structure tensor of 20° and less in most parts of the human sample, there were some deviations with angles > 45° esp. at the boundaries of the sample leading to a mean angle of 27 ± 24° if the angle difference is measured over the whole sample. This could be due to strong gradients between tissue and background noise at the boundaries, leading to erroneous structure tensor fiber orientation estimates in these regions. This is evidenced by the lower angular difference of 19 ± 15° observed once the outer layer of voxels is excluded from the comparison. This error is still higher compared to other 3D measures such as (Schilling et al., 2016), who reported an error of 6° in voxels with a parallel fiber orientation and 10–11° in crossing fiber regions using a comparison with confocal imaging of DiI stain of 80 μm sections that were stacked to a 3D volume using block-face imaging. One source of this discrepancy could be related to the lower dMRI resolution of 1 mm isotropic used in our comparison compared to the 400 μm isotropic data used in the prior study. Future comparisons with a dMRI scan at a higher resolution would be necessary to improve this comparison step. Another limitation of the CLARITY procedure is that it relies on immunostaining to visualize fiber tracts. Since the stain only visualizes one component, in our case the neurofilaments, other structures that can contribute to tissue anisotropy are not considered. Additionally, dMRI is known to be more robust at estimating fiber orientations in white matter than gray matter. The lower anisotropy within our deep gray matter sample from the thalamus may have resulted in noisier fiber orientation estimates and a greater angular difference relative to the CLARITY fiber orientation estimates.
Crossing fiber analysis of the diffusion MRI in the human sample reveals only a small number of voxels where the anisotropic fiber fraction of a secondary orientation was estimated to be above the threshold of 0.05. The FOD analysis of the CLARITY data also revealed very few voxels with any evidence of a secondary orientation. While the close correspondence is encouraging, this dataset does not enable extensive comparisons of crossing fiber estimates.
The current study’s focus was to determine a protocol that enables a robust comparison of MRI and CLARITY 3D histology in the same tissue sample. Multiple clearing and staining protocols were tested, and therefore we employed methods that enabled multiple tissue samples to be processed simultaneously. Specifically, the diffusion MRI scanning was performed on multiple cuboids simultaneously by combining them into a single container and scanning them on a human MRI scanner. While this approach limited the achievable diffusion MRI resolution for these small samples, it was compatible with the assessment of different clearing protocols. Each sample was scanned separately on a small-bore scanner to obtain a high-resolution structural scan that facilitated the co-registration of CLARITY and MRI data and could be obtained much more quickly than a high-resolution diffusion MRI scan. In future studies, an individual high-resolution dMRI scan for each processed sample on a preclinical scanner would be desirable to allow a comparison of various diffusion models at very small voxel sizes.
As shown previously, hydrogel embedding does not significantly alter the diffusion characteristics of the dMRI results (Leuze et al., 2016). PFA percentage, fixation time, and storage solution (PFA or PBS) can affect the diffusion properties of the tissue, but it is well-established that diffusion anisotropy can still be measured in fixed tissues (Leuze et al., 2014; McNab et al., 2009; Miller et al., 2012).
Immersing tissue specimens in fomblin for MRI scans did not appear to affect the subsequent clearing or staining of the sample. Some of the samples became very soft during the clearing process, and we hypothesize that this relates to the gray versus white matter content of the sample. As expected, gray matter tends to clear faster than white matter due to its lower myelin content. For samples containing both gray and white matter, the gray matter sometimes appeared to disintegrate slightly during the extending clearing time needed to clear the white matter parts of the sample. The human tissue sample highlighted in this manuscript contained gray matter in the thalamus. It did not exhibit a strong gradient in myelination levels, and therefore differential clearing speed was not an issue.
The NF stain was selected, for the experiments presented here, because NF is the most abundant fibrillar component in axons and is organized longitudinally along the axonal wall. This makes it a valuable proxy for neuronal fiber orientation (Kandel, 1989). Staining compatibility with CLARITY cleared tissue requires special consideration. One complication is that antibodies with many binding sites (such as NF) tend to bind rapidly upon exposure to the exterior of the sample, thereby limiting staining deeper within the sample. An example of such an inhomogeneous stain can be seen in Supplemental Figure 2B. The SWITCH method tries to account for this issue by including an additional staining step in which the antibodies are temporarily prevented from binding to the sample by an SDS solution (Murray et al., 2015). Thus, the antibodies can diffuse equally throughout the sample and then, after the SDS solution is washed out, the antibodies are allowed to bind to their respective binding sites. The SWITCH protocol was applied to initially prevent the antibody from binding to the outer areas of the sample, which helped to achieve a homogeneous stain throughout the whole sample depth. In the data presented here, NF antibody was used to visualize the fiber orientations. However, Morawski et al. (2017) have previously presented within cleared tissue a range of antibodies labeling myelin basic protein (MBP), human neuronal protein C (HuC), or glial fibrillary acidic protein (GFAP) to extract myelination and specific cell types. While staining for the sample presented here was done passively, this process might be sped up by electrophoretically driving antibodies through the tissue (Li et al., 2015).
The MRI was used as the reference volume in the MRI-CLARITY co-registration because the MRI was acquired on the tissue sample before it suffered distortions from the clearing and mounting procedure. Small tissue specimens such as the one demonstrated here tend to have less severe deformations of the sample during clearing than larger samples. Similar to prior work (Stolp et al., 2018), the CLARITY images presented here show only minor distortions in the cleared sample. The low distortion levels made it easier to achieve an accurate co-registration to the MRI data (RMSE = 0.26 mm for all registration markers with a maximum error of 0.35 mm for a single registration marker after co-registration). The small RMSE (~1/4 of the dMRI voxel size) between the MRI and CLARITY registration markers indicate that the linear co-registration applied was suitable for this case (Goubran et al., 2015, 2013). Stronger deformations may demand a non-linear, deformable co-registration such as intensity-based B-spline deformation or diffeomorphic symmetric image normalization (Avants et al., 2008; Rueckert, 1999). Of note, structure tensor analyses were performed after the co-registration, thereby circumventing any need to reorient orientation estimates.
In addition to shape deformations, CLARITY tissue processing can also lead to deformations in the sample size. The sample expands when immersed in the clearing solution (SBC) but then shrinks back to approximately its original size after immersion in FocusClear. Still, the possibility for some retained expansion or contraction of the tissue sample compared to its original size should be considered. Substantial shrink-age of the tissue sample could lead to less space between stained neurofilaments and thus lower contrast between stained neurofilaments and background image noise, which would make it more challenging to estimate the dominant fiber orientation with a structure tensor analysis. Substantial expansion of the tissue sample could increase the space between stained neurofilaments, which could make fiber orientation estimates with structure tensor analysis more robust. A full analysis of the potential effect of tissue sample expansion or contraction was beyond the scope of this study.
An open question that was examined with the data from this study, is whether or not water within the vascular compartments of postmortem tissue influence the measured diffusion patterns used to infer on dMRI fiber orientation. The CLARITY autofluorescence images from the human sample depict vascular compartments without coherent orientational organization. A structure tensor analysis of the CLARITY autofluorescence images confirms this incoherent orientation organization, and comparison of the vascular structure tensor orientations with the diffusion tensor MRI orientations show low correspondence (Fig. 5 E). These data inspire confidence that ex vivo diffusion MRI represents the underlying neuronal fiber structure and is not driven by water molecules that are hindered or restricted by vascular compartments. The vasculature has a different influence on the diffusion signal in vivo compared to ex vivo (Dyrby et al., 2011) because in vivo blood flow results in significant attenuation of the intra-vessel signal. The vasculature could be clearly identified as hypointensities in structural MRI of the human tissue specimen. These hypointensities may indicate the presence of a considerable amount of blood, which has a faster signal decay due to its iron content, still present inside the vessels. This lower signal level within the vessels could lower sensitivity to diffusion effects within the fixed tissue. In contrast, animal tissue, which is perfusion fixed, may have less blood remaining in the vessels and therefore be prone to the vasculature contributing diffusion anisotropy contrast.
5. Conclusion
We have demonstrated a voxel-wise comparison of CLARITY images that depict neurofilaments and vasculature with dMRI estimates of microstructural orientation in human brain tissue. Vasculature components visible in both the autofluorescence channel of the CLARITY data and the high-resolution T2* weighted MRI allowed for accurate co-registration across imaging modalities. Automated feature extraction of the fiber orientations from the CLARITY images was in good agreement with the MRI-derived diffusion tensor orientations. Crossing fiber estimation in the CLARITY data depended strongly on the chosen sub-cube size over which the structure tensor was calculated. Vasculature orientation estimates from the CLARITY images did not correspond with the MRI-derived diffusion tensor. The data presented here represents critical steps towards integrating information across multiple scales and imaging modalities such that MRI measurements may be validated, improved, and interpreted accurately.
Supplementary Material
Acknowledgements
NIH: R01 NS095985, R01 MH111444, R21 MH116484, S10 RR026351, P41 EB015891, R01 AG061120-01
The David Mahoney Neuroimaging Program from The Charles A. Dana Foundation
GE Healthcare
Stanford Radiology PHIND Center
Wu Tsai Neuroscience Institute at Stanford
Footnotes
Declaration of Competing Interest
The following authors have patents granted or pending related to the content of this manuscript: Deisseroth K., McNab J., Tian Q., Hsueh B.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2020.117692.
References
- Aswendt M, Schwarz M, Abdelmoula WM, Dijkstra J, Dedeurwaerdere S, 2017. Whole-brain microscopy meets in vivo neuroimaging: techniques, benefits, and limitations. Mol. Imaging Biol 19, 1–9. doi: 10.1007/s11307-016-0988-z. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axer M, Strohmer S, Gräßel D, Bücker O, Dohmen M, Reckfort J, Zilles K, Amunts K, 2016. Estimating fiber orientation distribution functions in 3D-polarized light imaging. Front. Neuroanat 10, 1–12. doi: 10.3389/fnana.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird AE, Warach S, 1998. Magnetic resonance imaging of acute stroke. J. Cereb. Blood Flow Metab doi: 10.1097/00004647-199806000-00001. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A, 2000. In vivo fiber tractography using DT-MRI data. Magn. Reson. Med 44, 625–632. [DOI] [PubMed] [Google Scholar]
- Budde MD, Frank JA, 2012. Examining brain microstructure using structure tensor analysis of histological sections. Neuroimage 63, 1–10. doi: 10.1016/j.neuroimage.2012.06.042. [DOI] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Shi Y, Styner MA, Johnson GA, 2015. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage doi: 10.1016/j.neuroimage.2015.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales-Rodríguez EJ, Legarreta JH, Pizzolato M, Rensonnet G, Girard G, Patino JR, Barakovic M, Romascano D, Alemán-Gómez Y, Radua J, Pomarol Clotet E, Salvador R, Thiran JP, Daducci A, 2019. Sparse wars: a survey and comparative study of spherical deconvolution algorithms for diffusion MRI. Neuroimage doi: 10.1016/j.neuroimage.2018.08.071. [DOI] [PubMed] [Google Scholar]
- Canny J, 1986. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. PAMI 8, 679–698. doi: 10.1109/TPAMI.1986.4767851. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky K.a, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K, 2013. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder a Z., Shimony JS, McK-instry RC, Burton H, Raichle ME, 1999. Tracking neuronal fiber pathways in the living human brain. Proc. Natl. Acad. Sci. U. S. A 96, 10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt H-U, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgänsberger W, Becker K, 2007. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Dyrby TB, Baare WF, Alexander DC, Jelsing J, Garde E, Sogaard LV, 2011. An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion-weighted imaging datasets. Hum. Brain Mapp 32, 544–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrby TB, Innocenti GM, Bech M, Lundell H, 2018. Validation strategies for the interpretation of microstructure imaging using diffusion MRI. Neuroimage 182, 62–79. doi: 10.1016/j.neuroimage.2018.06.049. [DOI] [PubMed] [Google Scholar]
- Epp JR, Niibori Y, Hsiang Liz, H.-L., Mercaldo V, Deisseroth K, Josselyn a., S., Frankland PW, 2015. Optimization of CLARITY for Clearing Whole Brain and Other Intact Organs. eNeuro doi: 10.1523/ENEURO.0022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Bradke F, 2013. High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO). Exp. Neurol 242, 57–64. doi: 10.1016/j.expneurol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R, 2012. 3D Slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging doi: 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubran M, Crukley C, de Ribaupierre S, Peters TM, Khan AR, 2013. Image registration of ex-vivo MRI to sparsely sectioned histology of hippocampal and neocortical temporal lobe specimens. Neuroimage doi: 10.1016/j.neuroimage.2013.07.053. [DOI] [PubMed] [Google Scholar]
- Goubran M, Hammond RR, De Ribaupierre S, Burneo JG, Mirsattari S, Steven DA, Parrent AG, Peters TM, Khan AR, 2015. Magnetic resonance imaging and histology correlation in the neocortex in temporal lobe epilepsy. Ann. Neurol doi: 10.1002/ana.24318. [DOI] [PubMed] [Google Scholar]
- Hou B, Zhang D, Zhao S, Wei M, Yang Z, Wang S, Wang J, Zhang X, Liu B, Fan L, Li Y, Qiu Z, Zhang C, Jiang T, 2015. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat 9, 1–11. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti GM, Dyrby TB, Girard G, St-Onge E, Thiran JP, Daducci A, Descoteaux M, 2018. Topological principles and developmental algorithms might refine diffusion tractography. Brain Struct. Funct doi: 10.1007/s00429-018-1759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Knoesche T, Turner R, 2013. White matter integrity, fiber count, and other fallacies: the do’s and dont’s of diffusion MRI. Neuroimage 73, 239–254. [DOI] [PubMed] [Google Scholar]
- Kandel, 1989. Principles of neuroscience. J. Chem. Inf. Model doi: 10.1017/CBO9781107415324.004. [DOI] [Google Scholar]
- Kass M, Witkin A, 1987. Analyzing oriented patterns. Comput. Vision, Graph. Image Process 37, 362–385. doi: 10.1016/0734-189X(87)90043-0. [DOI] [Google Scholar]
- Khan AR, Cornea A, Leigland LA, Kohama SG, Jespersen SN, Kroenke CD, 2015. 3D structure tensor analysis of light microscopy data for validating diffusion MRI. Neuroimage 111, 192–203. doi: 10.1016/j.neuroimage.2015.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW, 2010. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis M, Sikma KJ, Barth M, Dederen P, Zerbi V, Küsters B, Ruiter D, Slump CH, van Cappellen van Walsum A-M, 2011. Validation of diffusion weighted imaging of cortical anisotropy by means of a histological stain for myelin. Proc. Int. Soc. Magn. Reson. Med 2085. [Google Scholar]
- Knösche TR, Anwander A, Liptrot M, Dyrby TB, 2015. Validation of tractography: comparison with manganese tracing. Hum. Brain Mapp 36, 4116–4134. doi: 10.1002/hbm.22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Choi J, Jo Y, Kim JY, Jang YJ, Lee HM, Kim SY, Lee HJ, Cho K, Jung N, Hur EM, Jeong SJ, Moon C, Choe Y, Rhyu IJ, Kim H, Sun W, 2016. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep 6. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuze C, Aswendt M, Ferenczi E, Liu CW, Hsueh B, Goubran M, Tian Q, Steinberg G, Zeineh MM, Deisseroth K, McNab JA, 2016. The separate effects of lipids and proteins on brain MRI contrast revealed through tissue clearing. Neuroimage 0–1. doi: 10.1016/j.neuroimage.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuze CWU, Anwander A, Bazin PL, Dhital B, Stüber C, Reimann K, Geyer S, Turner R, 2014. Layer-specific intracortical connectivity revealed with diffusion MRI. Cereb. Cortex 24, 328–339. doi: 10.1093/cercor/bhs311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Czajkowsky DM, Li X, Shao Z, 2015. Fast immuno-labeling by electrophoretically driven infiltration for intact tissue imaging. Nat. Publ. Gr 1–7. doi: 10.1038/srep10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnain C, Augustinack JC, Konukoglu E, Frosch MP, Sakadžic S, Varjabedian A, Garcia N, Wedeen VJ, Boas D.a., Fischl B, Konukoglu E, Frosch MP, Sakad ž ic S, Varjabedian A, Garcia N, Wedeen VJ, Boas D.a., Fischl B, 2015. Optical coherence tomography visualizes neurons in human entorhinal cortex. Neurophotonics 2, 015004. doi: 10.1117/1.NPh.2.1.015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein KH, Neher PF, Houde JC, Côté MA, Garyfallidis E, Zhong J, Chamberland M, Yeh FC, Lin YC, Ji Q, Reddick WE, Glass JO, Chen DQ, Feng Y, Gao C, Wu Y, Ma J, Renjie H, Li Q, Westin CF, Deslauriers-Gauthier S, González JOO, Paquette M, St-Jean S, Girard G, Rheault F, Sidhu J, Tax CMW, Guo F, Mesri HY, Dávid S, Froeling M, Heemskerk AM, Leemans A, Boré A, Pinsard B, Bedetti C, Desrosiers M, Brambati S, Doyon J, Sarica A, Vasta R, Cerasa A, Quattrone A, Yeatman J, Khan AR, Hodges W, Alexander S, Romascano D, Barakovic M, Auría A, Esteban O, Lemkaddem A, Thiran JP, Cetingul HE, Odry BL, Mailhe B, Nadar MS, Pizzagalli F, Prasad G, Villalon-Reina JE, Galvis J, Thompson PM, Requejo FDS, Laguna PL, Lacerda LM, Barrett R, Dell’Acqua F, Catani M, Petit L, Caruyer E, Daducci A, Dyrby TB, Holland-Letz T, Hilgetag CC, Stieltjes B, Descoteaux M, 2017. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun doi: 10.1038/s41467-017-01285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab J.a, Jbabdi S, Deoni SCL, Douaud G, Behrens TEJ, Miller KL, 2009. High resolution diffusion-weighted imaging in fixed human brain using diffusion-weighted steady state free precession. Neuroimage 46, 775–785. doi: 10.1016/j.neuroimage.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Miller KL, McNab J.a., Jbabdi S, Douaud G, 2012. Diffusion tractography of post-mortem human brains: optimization and comparison of spin echo and steady-state free precession techniques. Neuroimage 59, 2284–2297. doi: 10.1016/j.neuroimage.2011.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollink J, Kleinnijenhuis M, van Cappellen van Walsum AM, Sotiropoulos SN, Cottaar M, Mirfin C, Heinrich MP, Jenkinson M, Pallebage-Gamarallage M, Ansorge O, Jbabdi S, Miller KL, 2017. Evaluating fibre orientation dispersion in white matter: comparison of diffusion MRI, histology and polarized light imaging. Neuroimage 157, 561–574. doi: 10.1016/j.neuroimage.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski M, Kirilina E, Scherf N, Jäger C, Reimann K, Trampel R, Gavriilidis F, Geyer S, Biedermann B, Arendt T, Weiskopf N, 2017. Developing 3D microscopy with CLARITY on human brain tissue: towards a tool for informing and validating MRI-based histology. Neuroimage 1–12. doi: 10.1016/j.neuroimage.2017.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC, 1999. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann. Neurol 45, 265–269. doi: . [DOI] [PubMed] [Google Scholar]
- Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim S-Y, Choi H, Park Y-G, Park J-Y, Hubbert A, McCue M, Vassallo SL, Bakh N, Frosch MP, Wedeen VJ, Seung HS, Chung K, 2015. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell LJ, Westin CF, 2011. An introduction to diffusion tensor image analysis. Neurosurg. Clin. N. Am doi: 10.1016/j.nec.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, Mortrud MT, Ouellette B, Nguyen TN, Sorensen SA, Slaughter-beck CR, Wakeman W, Li Y, Feng D, Ho A, Nicholas E, Hirokawa KE, Bohn P, Joines KM, Peng H, Hawrylycz MJ, Phillips JW, Hohmann JG, Wohnoutka P, Gerfen CR, Koch C, Bernard A, Dang C, Jones AR, Zeng H, 2014. A mesoscale connectome of the mouse brain. Nature doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YG, Sohn CH, Chen R, McCue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W, Choi H, Jin X, Lilascharoen V, Wang J, Truttmann MC, Qi HW, Ploegh HL, Golub TR, Chen SC, Frosch MP, Kulik HJ, Lim BK, Chung K, 2019. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol 37, 73. doi: 10.1038/nbt.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu MO, Barnett A, Basser P, Chang L-C, Koay C, Pajevic S, Rohde G, Sarlls J, Wu M, 2010. TORTOISE: an integrated software package for processing of diffusion MRI data. In: ISMRM 18th Annu. Meet, Stock. Sweden, p. 1597. [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-lavigne M, 2014. Resource iDISCO : a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Reveley C, Seth AK, Pierpaoli C, Silva AC, Yu D, Saunders RC, Leopold DA, Ye FQ, 2015. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion MR tractography. Proc. Natl. Acad. Sci. U. S. A doi: 10.1073/pnas.1418198112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert D, 1999. Nonrigid registration using free-form deformations: Application to breast mr images. IEEE Trans. Med. Imaging doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Schilling K, Janve V, Gao Y, Stepniewska I, Landman BA, Anderson AW, 2016. Comparison of 3D orientation distribution functions measured with confocal microscopy and diffusion MRI. Neuroimage 129, 1–13. doi: 10.1016/j.neuroimage.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Janve V, Gao Y, Stepniewska I, Landman BA, Anderson AW, 2018. Histological validation of diffusion MRI fiber orientation distributions and dispersion. Neuroimage 165, 200–221. doi: 10.1016/j.neuroimage.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Nath V, Hansen C, Parvathaneni P, Blaber J, Gao Y, Neher P, Aydogan DB, Shi Y, Ocampo-Pineda M, Schiavi S, Daducci A, Girard G, Barakovic M, Rafael-Patino J, Romascano D, Rensonnet G, Pizzolato M, Bates A, Fischi E, Thiran JP, Canales-Rodríguez EJ, Huang C, Zhu H, Zhong L, Cabeen R, Toga AW, Rheault F, Theaud G, Houde JC, Sidhu J, Chamberland M, Westin CF, Dyrby TB, Verma R, Rathi Y, Irfanoglu MO, Thomas C, Pierpaoli C, Descoteaux M, Anderson AW, Landman BA, 2019. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage doi: 10.1016/j.neuroimage.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehaus AK, Roebroeck A, Chiry O, Kim DS, Ronen I, Bratzke H, Goebel R, Galuske RAW, 2013. Histological validation of DW-MRI tractography in human postmortem tissue. Cereb. Cortex 23, 442–450. doi: 10.1093/cercor/bhs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalteholz W, 1911. Über das Durchsichtigmachen von menschlichen und tierischen Prä-paraten Leipzig, Verlag von S.Hirzel. [Google Scholar]
- Stolp HB, Ball G, So P-W, Tournier J-D, Jones M, Thornton C, Edwards AD, 2018. Voxel-wise comparisons of cellular microstructure and diffusion-MRI in mouse hippocampus using 3D Bridging of Optically-clear histology with neuroimaging data (3D-BOND). Sci. Rep 8, 4011. doi: 10.1038/s41598-018-22295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki E.a, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, Kiyonari H, Shimizu Y, Miyawaki A, Yokota H, Ueda HR, 2014. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C, 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc. Natl. Acad. Sci. USA doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K, 2014. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc 9, 1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh CH, Connelly A, 2019. MRtrix3: a fast, flexible and open software framework for medical image processing and visualisation. Neuroimage doi: 10.1016/j.neuroimage.2019.116137. [DOI] [PubMed] [Google Scholar]
- Vu AT, Auerbach E, Lenglet C, Moeller S, Sotiropoulos SN, Jbabdi S, An-dersson J, Yacoub E, Ugurbil K, 2015. High resolution whole brain diffusion imaging at 7T for the human connectome project. Neuroimage 122, 318–331. doi: 10.1016/j.neuroimage.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lenglet C, Akkin T, 2015. Structure tensor analysis of serial optical coherence scanner images for mapping fiber orientations and tractography in the brain. J. Biomed. Opt 20, 036003. doi: 10.1117/1.JBO.20.3.036003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang AC, Jennings JH, Adhikari A, Halpern CH, Witten IB, Barth AL, Luo L, McNab JA, Deisseroth K, 2016. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell 165, 1776–1788. doi: 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Todorov MI, Cai R, Maskari RA, Steinke H, Kemter E, Mai H, Rong Z, Warmer M, Stanic K, Schoppe O, Paetzold JC, Gesierich B, Wong MN, Huber TB, Duering M, Bruns OT, Menze B, Lipfert J, Puelles VG, Wolf E, Bechmann I, Ertürk A, 2020. Cellular and molecular probing of intact human organs. Cell 180, 796–812. doi: 10.1016/j.cell.2020.01.030, .e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


