Abstract
Machine Learning (ML) has been increasingly used within cardiology, particularly in the domain of cardiovascular imaging. Due to the inherent complexity and flexibility of ML algorithms, inconsistencies in the model performance and interpretation may occur. Several review articles have been recently published that introduce the fundamental principles and clinical application of ML for cardiologists. This paper builds on these introductory principles and outlines a more comprehensive list of crucial responsibilities that need to be completed when developing ML models. This paper aims to serve as a scientific foundation to aid investigators, data scientists, authors, editors, and reviewers involved in machine learning research with the intent of uniform reporting of ML investigations. An independent multidisciplinary panel of ML experts, clinicians, and statisticians worked together to review the theoretical rationale underlying 7 sets of requirements that may reduce algorithmic errors and biases. Finally, the paper summarizes a list of reporting items as an itemized checklist that highlights steps for ensuring correct application of ML models and the consistent reporting of model specifications and results. It is expected that the rapid pace of research and development and the increased availability of real-world evidence may require periodic updates to the checklist.
Keywords: artificial intelligence, cardiovascular imaging, checklist, digital health, machine learning, reporting guidelines, reproducible research
In 2016, the American College of Cardiology’s Executive Committee and Cardiovascular Imaging Section Leadership Council initiated a discussion regarding the future of cardiovascular imaging among thought leaders in the field (1). One of the goals was focused on machine learning (ML) tools and methods that embrace data-driven approaches for scientific inquiry. The 2016 paper stressed the creation and adoption of standards, the development of registries, and the use of new techniques in bioinformatics. Furthermore, the imaging community’s unfamiliarity with the approach was cited as a potential barrier to widespread adoption. Recently, the field of cardiac imaging has seen a remarkable burst of innovation with the use of ML, demonstrating powerful algorithms that start impacting the ways clinical and translational research is designed and executed (2-15).
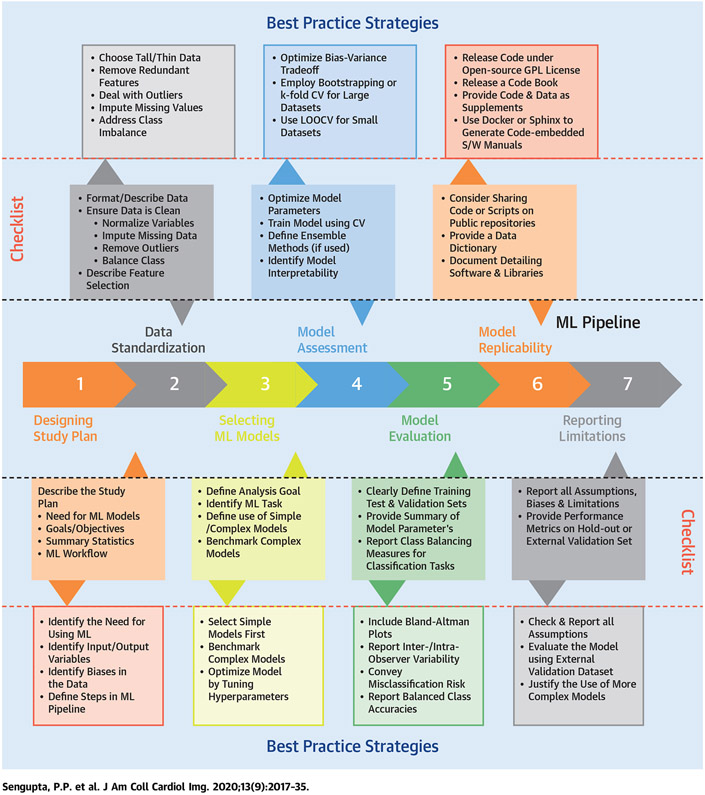
ML is a subfield of artificial intelligence (AI) where an algorithm automatically discovers patterns of data in the datasets without using explicit instructions. Several recent state-of-the-art review articles have focused on providing introductory concepts regarding ML algorithm applications for cardiologists (3,8,16,17). Although ML is creating headlines in medical journals, congress, and on the Web, considerable uncertainty and debate have arisen around topics such as problems with real-world data sources, the inconsistent availability of labeled data and outcome information, bias injection, inaccurate measurements, reproducibility, lack of external validation, and insufficient reporting, which hinder the reliable assessment of prediction model studies and reliable interpretations of the results by clinicians. This Proposed Recommendations for Cardiovascular Imaging-Related Machine Learning Evaluation (PRIME) checklist aims to provide a general framework as a reference in guiding scientific work for investigators, data scientists, authors, editors, and reviewers involved in machine learning research in cardiovascular imaging (Central Illustration). The goal of the PRIME checklist paper was to standardize the application of AI and ML, including data preparation, model selection, and performance assessment. The paper provides a set of strategic steps toward developing a pragmatic checklist (Table 1) which allows consistent reporting of ML models in cardiovascular imaging studies. To further determine its ease of use and applicability, the application of the checklist was developed in a PRIME checklist for 2 recent articles that developed ML models in cardiovascular imaging (Supplemental Table 1).
CENTRAL ILLUSTRATION. Steps for Building a Machine Learning Pipeline and the Reporting Items in a Checklist.
This illustration provides the principal requirements for building a checklist (at every step of the model building process) to enable the precise application of predictive modeling, consistent reporting of model specifications, and results in the field of cardiovascular imaging. CV = cross validation; GPL = general public license; LOOCV = leave 1 out cross validation; ML = machine learning; S/W = software.
TABLE 1.
Checklist for Standardized Reporting of Machine Learning Investigations
| Section | Checklist item | |
|---|---|---|
| 1 | Designing the Study Plan | |
| 1.1 | Describe the need for the application of machine learning to the dataset | |
| 1.2 | Describe the objectives of the machine learning analysis | |
| 1.3 | Define the study plan | |
| 1.4 | Describe the summary statistics of baseline data | |
| 1.5 | Describe the overall steps of the machine learning workflow | |
| 2 | Data Standardization, Feature Engineering, and Learning | |
| 2.1 | Describe how the data were processed in order to make it clean, uniform, and consistent | |
| 2.2 | Describe whether variables were normalized and if so, how this was done | |
| 2.3 | Provide details on the fraction of missing values (if any) and imputation methods | |
| 2.4 | Describe any feature selection processes applied | |
| 2.5 | Identify and describe the process to handle outliers if any | |
| 2.6 | Describe whether class imbalance existed, and which method was applied to deal with it | |
| 3 | Selection of Machine Learning Models | |
| 3.1 | Explicitly define the goal of the analysis e.g., regression, classification, clustering | |
| 3.2 | Identify the proper learning method used (e.g., supervised, reinforcement learning etc.) to address the problem | |
| 3.3 | Provide explicit details on the use of simpler, complex, or ensemble models | |
| 3.4 | Provide the comparison of complex models against simpler models if possible | |
| 3.5 | Define ensemble methods, if used | |
| 3.6 | Provide details on whether the model is interpretable | |
| 4 | Model Assessment | |
| 4.1 | Provide a clear description of data used for training, validation, and testing | |
| 4.2 | Describe how the model parameters were optimized (e.g., optimization technique, number of model parameters etc.) | |
| 5 | Model Evaluation | |
| 5.1 | Provide the metric(s) used to evaluate the performance of the model | |
| 5.2 | Define the prevalence of disease and the choice of the scoring rule used | |
| 5.3 | Report any methods used to balance the numbers of subjects in each class | |
| 5.4 | Discuss the risk associated to misclassification | |
| 6 | Best Practices for Model Replicability | |
| 6.1 | Consider sharing code or scripts on a public repository with appropriate copyright protection steps for further development and non-commercial use | |
| 6.2 | Release a data dictionary with appropriate explanation of the variables | |
| 6.3 | Document the version of all software and external libraries used | |
| 7 | Reporting Limitations, Biases and Alternatives | |
| 7.1 | Identify and report the relevant model assumptions and findings | |
| 7.2 | If well performing models were tested on a hold-out validation dataset, detail the data of that validation set with the same rigor as that of training dataset (see section 2 above) |
1. DESIGNING THE STUDY PLAN
Defining the goal of the analysis is a key first step that informs many downstream decisions whether to use ML at all, including the presence or absence of labeled data for supervised or unsupervised learning. These informed decisions can alter the approach to model training, model selection, development, and tuning.
DETERMINING THE APPROPRIATENESS OF MACHINE LEARNING TO THE DATASET.
The first question researchers should address is whether the ML approach would be applicable and beneficial to their study. There is an overlap between conventional statistics and ML, but they differ regarding the extent of the assumptions and the formulation of the methods to either predict or make inferences. If the dataset is relatively small (i.e., fewer than hundreds of sample per class for “average” modeling problems), then overfitting becomes a much bigger concern, resulting in models that fail to generalize well for unseen instances (18). Similarly, if variables that are important to modeling the data are missing or if the model is too simple, the resulting ML model may underfit the data, thus producing less than optimal results (18). Statistical analyses rely on simpler models that are not necessarily optimized to the specific data under observation and are therefore less prone to overfitting at the price of lower performance if the problem is complex (19). In contrast, datasets with large numbers of features, on the order of thousands or those having many irrelevant or redundant features with regard to a given task may just as easily lean toward overfitting. ML is especially useful when the data are unstructured, feature selection, or exploratory analyses are preferred to identify meaningful insights. As such, the learning algorithm may find patterns in the data to generate a homogenous fraction and identify relationships in a data-driven manner, beyond the a priori knowledge or existing hypotheses (3). Although the use of advanced ML algorithms may be better suited for handling big or heterogeneous datasets, that comes at the cost of the interpretability, complexity, and the ability to draw a causal inference. Caution should be taken against causally interpreting results derived from models designed primarily for prediction. For tasks where the goal is to establish causality, the techniques that are commonly used in “conventional” biostatistics, including statistical analyses methods such as propensity score matching or Bayesian inference, may be better suited; however, newer methods involving ML algorithms are being developed for causal inferences (20,21).
UNDERSTANDING AND DESCRIBING THE DATA.
Regardless of whether ML tools or statistical analysis methods are used, it is crucial to understand and describe the data available for analysis to draw appropriate conclusions, whether data are tabular, images, time-series data, or a combination. Important considerations about the data include the availability of data that is representative of the target population, the method used to obtain data, and the resultant biases that may influence the conclusions that can be drawn from the data. Describing the data can also help understand the relevance to the target population. The method of data collection, including the sampling method, is also important, as bias may be introduced from systematic error, coverage error, or selection. Various guidelines and associated checklists for medical research have been established to aid in the reporting of relevant details about the data, depending on the study design (22). Clearly describing the data preprocessing or data cleaning methods used is essential to enable reproducibility.
It should be acknowledged that all ML or statistical algorithms are guided by basic data assumptions. An independent and identical distribution is an important assumption where the random variables are mutually independent and have the same statistical distribution and properties. Methods to check for the model assumptions, such as learning curves (23) and diagnosis of bias and variance (24) or error analyses, may be required.
DEFINING THE PROCESS.
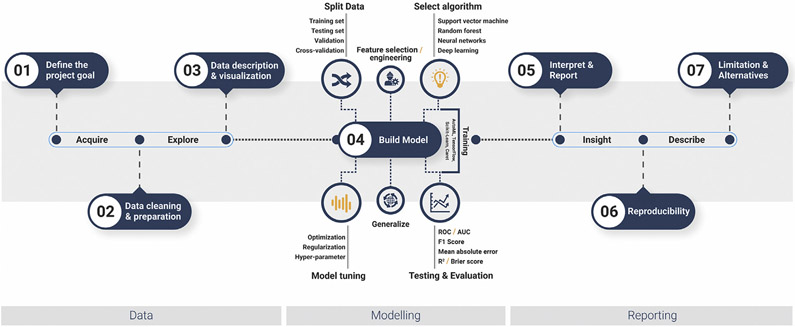
When building ML models, it is crucial to specify the inputs (e.g., pixels in images, a set of parameter values, and patient information) and desired outputs (e.g., object categories and the presence or absence of disease, an integer representing each category, the probability for each category, the prediction of a continuous outcome measurement, transformed pixel data) that are required. Although defining outputs is essential for supervised learning approaches, unsupervised learning approaches may also benefit from defining the output that is desired for the task to select an appropriate model. Some tasks, once well defined, can only be achieved using certain types of algorithms. For example, image recognition tasks from raw pixel/image data may require the extraction of the optimal features from the data, which is intrinsically performed by deep neural networks and goes beyond the use of hand-crafted features as input. At the conceptual level, deep learning works by breaking a complex task (e.g., identifying a tumor or other abnormalities in organs or tissue) into simple fewer abstract tasks. For example, if the task is to identify a square from other geometric shapes in an image, this task can be divided further into smaller, nested substeps (e.g., by first checking if there are 4 lines associated with a shape or not. Alternatively, one could check if lines are interconnected and perpendicular to each other and whether they are closed or not and so on in a step-by-step hierarchical fashion. After the consecutive hierarchical identification of complex task, deep-learning approaches automatically find out the features that are important for solving the problem. It is generally preferable to start by defining the overall broader analysis necessary to accomplish the task of interest and dividing it into several subtasks. Defining the problem or task as precisely as possible can also help guide the data annotation strategy and model selection. Once the data analysis objective has been identified and the inputs/outputs have been defined for each task, it is easier to determine the appropriate models for the analysis pipeline (Figure 1).
FIGURE 1. Machine Learning Pipeline.
Schematic diagram of a general machine learning pipeline. The data section consists of project planning, data collection, cleaning, and exploration. The modelling section describes the model building, in which hyperparameter tuning and the dimensionality reduction process, such as feature selection and engineering, model optimization and selection, and evaluation, are included. Finally, the reporting segment consists of the reporting mechanisms of the analysis, including reproducibility and maintenance, and a description of the limitations and alternatives.
Strategic steps for developing the checklist:
Identify and assess if machine learning could be appropriate.
Define the objectives of machine learning to achieve the overall goal.
Understand and describe the data.
Identify input and target variables.
Describe the baseline data and understand biases that may exist
2. DATA STANDARDIZATION, FEATURE ENGINEERING, AND LEARNING
Data preparation, standardization, and feature extraction are keys to the success of model development. They ensure that the data format is appropriate for ML, the variables used carry relevant information for solving the problem at hand, and the learning system is not biased toward a subset of the variables or categories in the database.
DATA FORMAT.
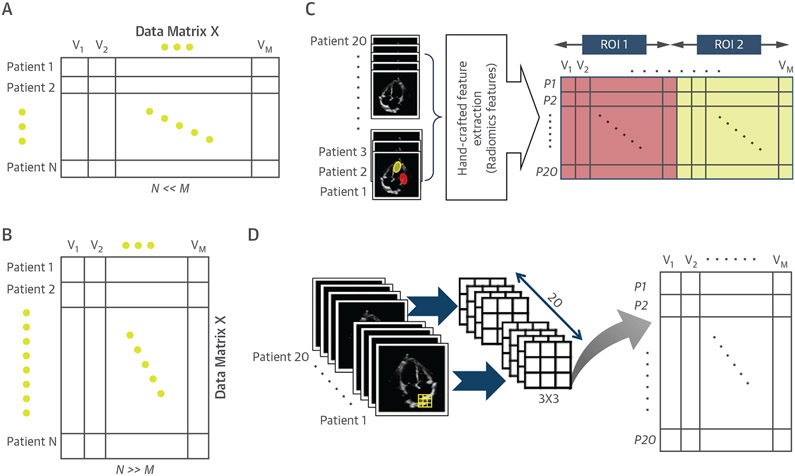
To analyze the data of N patients (also called “observations”), each with M different measurements (also called “variables” or “dimensions”), for example, ejection fraction, body mass index, and image pixels or voxels, using an ML algorithm, a data matrix X should first be constructed such that the rows of this data matrix correspond to the observations and that the columns correspond to the variables (Figure 2). Depending on the database and the problem at hand, X can be either a “wide” (Figure 2A) or a “tall” (Figure 2B) data matrix. In the former, the number of observations is much smaller than the number of variables (Figure 2C) (N << M), whereas in the latter, there is a large group of observations, but each observation has only a few variables (Figure 2D) (N >> M).
FIGURE 2. Schematic Demonstration of Short/Wide and Tall/Thin Data Matrices and the Way They Can Be Created From Image Data.
In a short and wide data matrix (A), the number of observations is much smaller than the number of variables (N << M). Considering different regions of interest (ROI) on the whole image of a given patient, the extraction of hand-crafted features (e.g., radiomics features) may lead to a short and wide data matrix, as the number of features extracted for each ROI per image is typically larger than the number of samples or patients (C). To make a tall and thin data matrix from the image data (B), an image can be divided to many (overlapping) ROIs or patches, each with a small number of pixels (D). The extraction of pixel data as features from each patch per patient may be much smaller in size than the total number of patients.
Generating a data matrix from cardiac images can be performed either on entire images or on selected regions of the images, depending on the learning purpose. When the goal is to use a learning algorithm for modeling the global characteristics of the images, hand-crafted features (e.g., radiomics) extracted based on all the pixels of selected regions of a given image are considered the variables of 1 observation (Figure 2C), which typically leads to a wide data matrix. On the contrary, to model regional image characteristics, however, a region of interest (ROI) or patch consisting of a small group of pixels, thus more easily yielding a tall data matrix (Figure 2D). Furthermore, a series of techniques (e.g., maximum pooling, or patch-based methods) can further be applied in reducing the size of the image-data matrix to highly informative elements, which often results in a tall, thin matrix, either for classification or for a pattern recognition task to identify key regions of interest (7,25). For example, a data matrix generated by using a single frame from longitudinal, 4-chamber or short-axis view of the heart that is 512 × 512 pixels in size for a total of N patients would result in creating an N × [5122] sized matrix for input into deep learning algorithms.
DATA PREPARATION.
To analyze cardiac images in an ML framework, some preprocessing stages are usually carried out. The irrelevant areas of the images can be removed in a “cropping” stage to focus on learning from useful regions and to prevent learning from extraneous regions (which can also contribute to leakage, as discussed below). If the images that are acquired from a group of subjects have different sizes, they typically should be “resized” first (26) to a reference image size to construct a data matrix with the same number of variables. More advanced techniques from computational atlases are also necessary to align the anatomy-based data of each subject to a common geometry and temporal dynamics, as spatiotemporal misalignment of input images will increase variance in the input of neurons of a neural network thereby slowing its ability to learn relevant features (27,28). Another common pre-processing stage is “noise removal,” which helps a learning algorithm to better model the essential characteristics of the images. When the acquired images have poor contrast, a “histogram equalization” (26) technique can be used to adjust the intensities of the pixels and to increase the contrast of a low-contrast region, thus facilitating its interpretation and analysis. The pixel intensities can also be manually adjusted during image acquisition. An example is the changing of the dynamic range of echocardiographic images by an operator. Techniques may also be applied to correct the differences in slice thickness, gray level distribution or even image resolution and different imaging protocols (contrast vs. noncontrast; low dose vs. high dose). Deep learning techniques, which are robust with regard to image quality may need to be used. For computed tomography and cardiac magnetic resonance images, a particular window and level setting may be applied before the deep learning training and normalized to the full intensity ranges.
In cardiovascular imaging, data samples are often represented in 3-dimensional (3D) and 4D formats, which currently may present challenges for the deep learning techniques due to the constraints posed by computing resources related to image sampling as well as large number of input variables. Nevertheless, efficient deep learning techniques have been developed for video analysis, and these can be potentially adapted to the direct interrogation of 3D or 4D data often seen in echocardiography, nuclear cardiology, or computed tomography (29). An alternative approach to deal with complex multidimensional data is to provide an intermediate simplified image representation. In cardiac imaging, often a bull’s eye representation (aka “polar map”) is used for the projection of 3D or 4D image data into a simple 2D (or 2D + time) format. For example, a bull’s eye representation approach has successfully been implemented for deep learning of nuclear cardiology studies (6, 30, 31). It allows data normalization from multiple scans and disparate sources such as motion/thickening, perfusion or flow. Such approaches could also be applied to other cardiovascular modalities.
Thus, the data preparation step aids in normalizing and compressing image, with respect to their intensities, anatomic representation, viewpoint, and so forth, across a given study prior to their entry into the ML framework. This process ensures that the algorithm spends more time and capacity learning the features that are important, rather than trying to rectify issues related to intensities or noise levels in the image data. Size of the images can also be reduced in order decrease memory requirements.
FEATURE ENGINEERING AND LEARNING.
The next stage after data preparation is extracting a set of “features” from the data matrix to be used later as the input to the learning methods. Feature extraction helps to overcome the following 2 main problems that can limit the efficient performance of a learning framework:
(i). Curse of dimensionality.
When the data matrix is wide, the variable or feature space of the data can be referred to as “high dimensional.” This may lead to an algorithm which fails to learn essential characteristics of the data due to its complexity and poor generalization power when dealing with unseen data, a phenomenon referred to as the “curse of dimensionality” (32, 33). To tackle these problems, the number of observations should increase significantly with the data dimensionality. For example, in pattern recognition, a typical rule of thumb is that there should be at least 5 training examples for each uncorrelated dimension in the representation (34). Moreover, it has been previously suggested that the sample-to-feature ratio should be between 5 and 10 depending upon the complexity of the classifier (35,36). However, a significant increase in the number of observations is not always possible, especially for medical data or studies, given that it necessitates the collection of data from a large group of patients. This curse of dimensionality is one of the main reasons why having a large database is desirable to build an efficient learning algorithm.
(ii). Correlated variables.
When a database includes correlated variables, a subset of the variables that are mutually uncorrelated may be sufficient to learn the data characteristics effectively (32). Indeed, adding correlated variables to a database will only bring redundant information and does not help the learning algorithm to achieve a better understanding of the data. For the imaging data for example, neighboring pixels typically have similar values and are highly correlated (37).
Given the curse of dimensionality, the learning process of algorithms cannot work effectively in data with too many features. Techniques to reduce the number of variables while retaining the most relevant information are critically important, a process called “feature extraction” and “dimensionality reduction.” Feature extraction can be performed either manually, using expert knowledge, or by algorithms such as principal component analysis (PCA) or multifactor dimensionality reduction (32-34). The result of the feature extraction process should be a compact set of (ideally uncorrelated) features or variables in the form of a tall matrix that encodes the essential characteristics of the data.
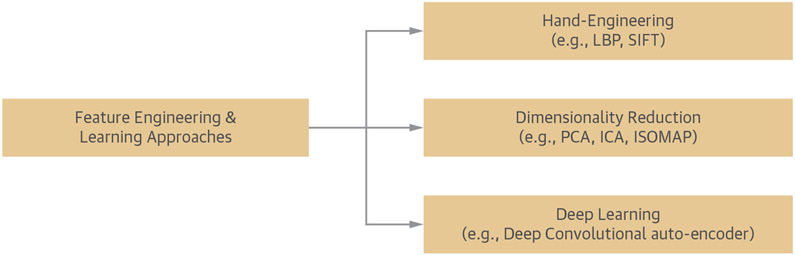
The approaches available for extracting features from the image data can be divided into the following 3 broad categories (Figure 3): 1) handcrafted methods (e.g., local binary patterns) (38) and scale-invariant feature transform (39); 2) classical ML methods for dimensionality reduction (e.g., PCA) (40), independent component analysis (41), or ISOMAP (42); and 3) deep learning methods (43). The methods in the first 2 categories are manually designed to extract specific types of features from the data, whereas in the last one, the features are learned from the database itself. Moreover, in the case of deep learning, techniques such as sub-sampling provide effective ways of down-sampling the image size in each layer, eliminating the need for feature extraction and improving overall training performance (25). Nevertheless, the classical feature learning algorithms have some limitations in the data modeling approaches such as linearity, sparsity, or lack of hierarchical representation. The deep learning techniques, on the other hand, can learn complex features from the data at multiple levels and do not have the limitations of the classical algorithms. However, they need a large-scale database to achieve efficient learning of the data characteristics. To train a deep learning algorithm with a smaller database, the following 2 main strategies can be used: 1) data augmentation (e.g., by using different types of data/image transformations) (44); and 2) transfer learning, which works by fine-tuning a deep network that has been pretrained with a different large database (e.g., natural images) (44,45). However, the field of transfer learning for medical imaging data is currently in its early stages, and more critical insights into its actual relevance will be known in the coming years.
FIGURE 3. Main Approaches for Feature Engineering and Learning.
The manually engineered approaches are manually designed to extract certain types of features from the data. For example, local binary pattern and scale-invariant feature transform derive the properties from the image such as object recognition or edge detection. The classic learning techniques use data samples to learn their characteristics for dimensionality reduction, but they have limitations in their data modeling techniques, such as linearity, sparsity, and lack of hierarchical representation. Principal component analysis (PCA) applies orthogonal transformation to produce linear combination of uncorrelated variables that best explains the variability of the data, whereas independent component analysis transforms the dataset into independent components to reduce dimensionality. Deep learning methods, however, can learn complex features from the data at multiple levels in various hidden layers.
VARIABLE NORMALIZATION.
For a database composed of several variables of different nature (e.g., anthropometric or image-derived measurements), the values of the variables lie in different ranges. Direct use of these variables may bias the learning system toward the characteristics of the variables with larger values, despite the usefulness of the variables with smaller values in solving a given problem. To deal with such challenges a “variable normalization” approach can be used to transform the variables such that they all lie in the same range prior to entering the learning phase (33,34). Variable normalization is especially helpful for a deep learning algorithm, as it helps achieve faster convergence of a deep neural network (46).
MISSING VARIABLE ESTIMATION.
ML algorithms often need complete datasets. If data are missing, the options are to exclude those subjects, to encode them as missing, or to impute missing values (45,47,48). In cardiovascular imaging, 2D images are normally collected from multiple views, for example, images can also be acquired throughout the cardiac cycle. When some of the 2D views are not accessible or when a group of 2D images at some points during the cardiac cycle or in a 3D volume are artifactual or missing, an imputation technique can estimate these images or the parameters extracted from them (48). Thanks to development of the new deep learning algorithms, such as generative adversarial networks (GAN) (49), missing images can often be estimated (50) based on the available data, although the physiological relevance of their content is not fully guaranteed. However, it should be acknowledged that most of the imputation methods assume that the missing observations occur at random, are missing completely at random, or are missing not at random (51).
Researchers should consider whether the missing observations carry any specific biases (e.g., selection bias or immortal time bias). Although there is no clear guidance or cutoff for what proportion of missing data warrants the use of data imputation techniques (52), and given that this answer depends on the complexity of the addressed problem, missing value imputation is best used whenever possible as it provides evidence about the robustness of the learned models regardless of its impact on model performance (53).
FEATURE SELECTION.
An important phase in designing a classical ML system is to determine the optimal number of preserved features. This determination can be performed by using a “feature selection” technique, where a set of features that is larger than required is first extracted and then a subset with discriminative information is selected (34,54). When a deep learning algorithm is used, the optimal features are automatically learned during the end-to-end training of the algorithm, and use of an independent feature selection method is often not required (43,55).
OUTLIERS.
An observation is considered an outlier if its values deviate substantially (or significantly, if looked through a statistical test) from the average values of a database, which may be attributed to measurement error, variability in the measurement, or abnormalities due to disease (33,34). Even though outliers can negatively influence and mislead the training process, resulting in less accurate models, they may carry relevant information related to the given task. Thus, outliers should be carefully examined to see if this comes from improper measurements that should be repeated, or not. With the existence of outliers, learning algorithms and/or a performance metrics that are robust to outliers can be useful alternatives. Methods robust to outliers including but not limited to decision trees and k-nearest neighbor (KNN) should be used as much as possible (33). If no other solution exits, the removal of outliers, using an outlier detection approach (56), may be considered and the selection criterion along with the proportion of samples removed should be reported.
CLASS IMBALANCE.
A significant imbalance in data classes (e.g., healthy vs. diseased) is quite common in medical datasets because, on the one hand, most subjects in a database are usually healthy, and on the other hand, because collecting patient data for some rare diseases is difficult and is not always possible. As a result, the performance of the learning algorithm may be skewed, as it only learns the characteristics of the larger sized categories. This problem is referred to as “class imbalance” and can be dealt with in the following 3 established ways: 1) rebalancing the categories using “under sampling” or “over sampling” (i.e., making the different classes similarly sized by omitting samples from the larger class or by up-sampling the data in the smaller class); 2) giving more importance (i.e., weight) to the samples of smaller categories during the learning process (34); or 3) using synthetic data generation methods, such as the synthetic minority over-sampling technique (SMOTE) (57). While the random over-sampling generates new data by duplicating some of the original samples of the minority class or category, SMOTE interpolates values using a k-nearest neighbor technique to synthesize new data instances (58). Recent advances in deep generative techniques, such as GAN or variational autoencoders or the use of loss functions that are robust to data imbalance (59), have made it possible to tackle complicated imbalanced data based on the learning strategies.
DATA SHIFT.
Data shift is a common problem that afflicts the ML models in cardiovascular imaging in which the distribution of the database used for testing the performance of the learning models or systems may differ from the distribution of the training data. This may occur when the data acquisition conditions or the systems that are used for collecting the test data change from when the training dataset was acquired, and could induce: 1) a covariate shift, that is, a shift in the distribution in the covariates; 2) a prior probability shift, that is, a difference in the distribution of the target variable; or 3) a domain shift, that is, a change in measurement systems or methods. It is imperative to assess and treat the shifts that may occur in the dataset prior to evaluating a model (60).
DATA LEAKAGE.
Data leakage is a major problem in ML, in which data outside of the training set seeps into the model while building the model. This event could lead to an error-prone or invalid ML model. Data leakage could occur if the same patient’s data are used in the training and testing sets and is generally a problem in complex datasets, such as time series, audio, and images, or graph problems.
Strategic steps for developing the checklist:
The data format for training a machine learning algorithm should be large and the ratio of the observations/measurements (i.e., N/M) should be at least 5.
When the data matrix is wide, a feature extraction/learning algorithm or dimensionality reduction technique should be used.
Redundant features should be removed, and variables should be normalized.
Outliers should be addressed/removed
Missing features should be imputed using relevant methods.
Testing data should never be used for preprocessing or training.
Duplicate data should be removed and ensured that the same data is not shared in the testing and training sets.
3. SELECTION OF MACHINE LEARNING MODELS
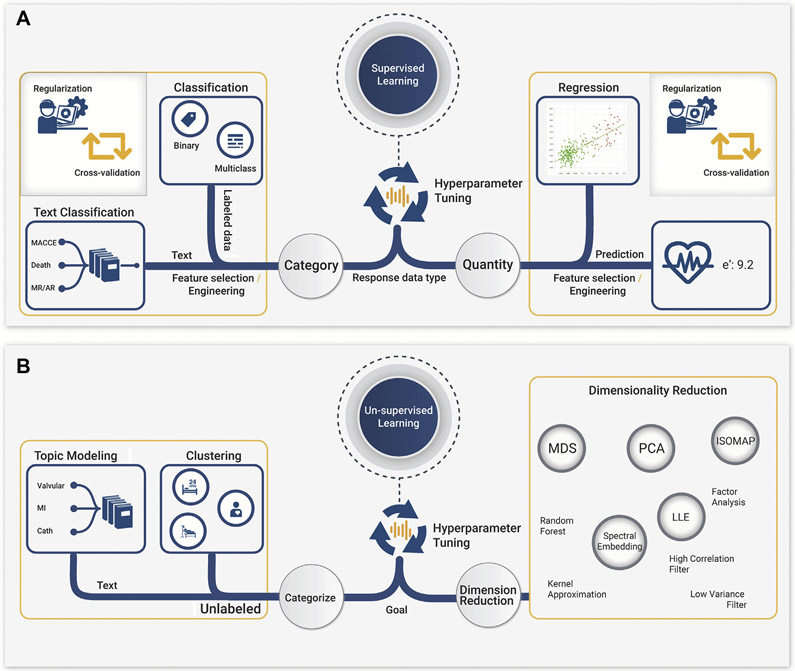
Model selection is the process of identifying the model that yields the best resolution and generalizability for the project and can be defined at multiple levels (i.e., learning methods, algorithms, and tuning hyperparameters). Learning methods include supervised, unsupervised, and reinforcement learning. Importantly, supervised learning is a method that learns from labeled data (i.e., data with outcome information to develop a prediction model), whereas unsupervised learning aims to find patterns and association rules in data that do not have labels (Figure 4).
FIGURE 4. Model Selection Process.
Illustration of the model selection process, which consists of identifying the 2 classes of ML. (A) In supervised learning method, after the hyperparameter tuning, data can be applied to 2 different tasks: classification or regression, depending on the type of the outcome. If the outcome is a category, then classification can be performed, whereas if the outcome variable is a numeric value, regression may be applied for prediction. (B) Unsupervised learning, the data in which the data are either used for clustering, topical modeling, or representing the data distribution while reducing the dimensionality of the data according to the problem to be solved. ML = machine learning.
Common algorithms, such as regression or instance-based learning, often handle high-dimensional data well and tend to perform better or equivalent to complex algorithms on small datasets while retaining the interpretability of the model. To achieve better performance, simple algorithms or weak learners may be combined in various ways using ensemble methods, such as boosting, bagging, and stacking, which sacrifice the interpretability. More complex algorithms that are also difficult to interpret, including neural networks, can outperform simpler models given an adequate amount of data. A subset of neural networks, known as deep convolutional neural networks (7,25), are particularly useful for finding patterns in image data without the need for feature extraction (61-64). The implementation of an algorithm can vary significantly in terms of the size and complexity (e.g., the size and number of features in a random forest decision tree, the number and complexity of kernels applied in a support vector machine, and the number and type of nodes and layers in a neural network) of the algorithms.
Regardless of the choice of the algorithm, it is imperative to perform hyperparameter tuning and model regularization to produce the optimal performance (65,66). These processes may be more important than selecting the types of algorithms that could impact the interpretability, simplicity, and accuracy. When performing model selection (especially involving large-scale data and/or deep learning methods), hardware constraints (e.g., memory size, cache, parallelism, and so forth) are often a key limiting factor beyond model performance. However, recent advances toward developing hardware accelerators (e.g., graphic processing units, tensor processing units) and growing convenience or abstraction by cloud computing could improve the overall process of model training, selection, and optimization. Moreover, selecting the best model often involves the relative comparison of performance between different models. Therefore, the purposeful selection of loss function and the metric that represents it (e.g., absolute error, mean-squared error etc.), which in turn is heavily influenced by the model choice, dataset, and particular problem/task to solve, becomes fundamental in selecting an appropriate model.
The size and complexity of algorithms should be chosen carefully to minimize the bias, the model error on the training dataset, and the variance, the model error on the validation dataset. Simpler models may underfit the data; they may generalize better (lower variance) at the cost of lower accuracy (higher bias). Furthermore, overfitting (high variance and low bias) may come from a model that is too complex or from insufficient representative training samples. Several considerations including size, complexity and dimensionality, number of features, and nonlinear relationships among variables in the dataset guides the choice of the initial algorithm and its complexity, but the final algorithm design (including the choice of hyperparameters) is determined empirically or by specific optimization and cross-validation.
Finally, an essential factor in algorithm selection is the need for the interpretability of the model’s decisions (i.e., an understanding of which input features caused the model to make the decision it made). Interpretability may be extremely important for certain learning tasks and less important for others. Regression, decision trees, and instance-based learning methods are generally highly interpretable, whereas methods to interpret the function of deep neural networks are still evolving; saliency mapping, class activation, and attention mapping are some examples for neural network interpretation and visualization (67,68). New mechanisms for understanding the workings of ML models (69-72), and approaches for probabilistic deep learning (73,74) further provide an opportunity to develop models to balance between both inference and prediction.
Strategic steps for developing the checklist:
For the initial model development, always select the simplest algorithm that is appropriate for the available data.
The size of the dataset and the complexity of the used algorithm should be considered to achieve a good compromise between ‘bias’ and ‘variance’ in the estimations.
Complex algorithms must be benchmarked to the performance of the initial simple model across several metrics.
Tune the hyperparameters to optimize the models and to increase performance.
4. MODEL ASSESSMENT
The next step after selecting a learning model is to evaluate the generalizability by applying it to new data (i.e., assessment of its performance on unseen data). Ideally, model assessment should be performed by randomly dividing the dataset into a “training set” for learning the data characteristics, a “validation set” for tuning the hyperparameters of the learning model, and a “test set” for estimating its generalization error, where all 3 sets have the same probability distribution (i.e., the statistical characteristics of the data in these 3 sets are identical). However, in many domains, including cardiovascular imaging, having access to a large dataset is often difficult, thus preventing model assessment using 3 independent data subsets. As mentioned in the previous section, the ratio of the training samples to the number of measured variables should be at least 5:10 (34,35), depending on the dataset and complexity of the classifier (75), to learn the data characteristics properly. If this criterion is not met, the data are called scarce. In this situation, the data may be divided into 2 subsets for training and final validation of the learning algorithm. However, the results may depend on the random selection of the samples. Therefore, the training set can then be further partitioned into 2 subsets, but this process is repeated several times by selecting different training and testing subjects to obtain a good estimate of the generalization performance of the learning algorithm (33). This method of model assessment can be performed by “cross-validation” or “bootstrapping,” as explained further below. These techniques ensure that: 1) the learning model is trained properly given that the majority of the data samples can be used in the training process; 2) the learning model is not biased toward the characteristics of a subset of the data; and 3) the optimal values of the hyperparameters of the learning model (e.g., the number of layers in a neural network and the neurons in each layer) can be determined (33).
CROSS-VALIDATION.
This technique works by dividing the data into multiple nonoverlapping training and testing subsets (also called folds) and using most of the folds for training a learning model and the remaining folds for evaluating its performance (32-34). The cross-validation process can be implemented in one of the following ways.
k-fold cross-validation.
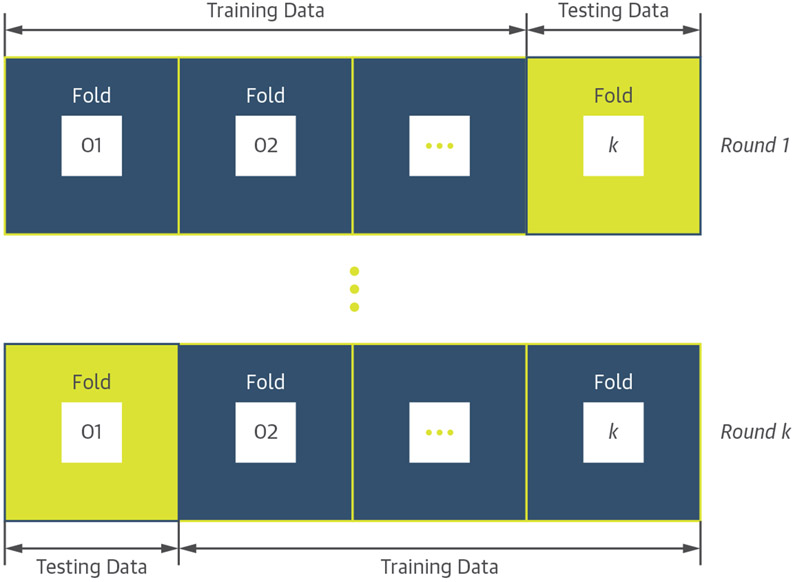
Data are randomly partitioned into k folds of roughly equal sizes, and in each round of the cross-validation process, 1 of the folds is used for testing the learning algorithm, and the rest of the folds are used for its training (Figure 5). This process is repeated k times such that all folds are used in the testing phase and the average performance on the testing folds is computed as an unbiased estimate of the overall performance of the algorithm (32,33).
FIGURE 5. Schematic Illustration of the K-Fold Cross-Validation Process.
Data are randomly partitioned into k distinct folds, and in each round (k-1) folds are used for training the learning algorithm, and the kth fold is used for testing its performance. This process is repeated k times such that all folds are used in the testing phase.
Leave-one-out cross-validation.
In this technique, the number of the folds is equal to the number of the observations in the database, and in each round, only one observation is used for testing the learning algorithm.
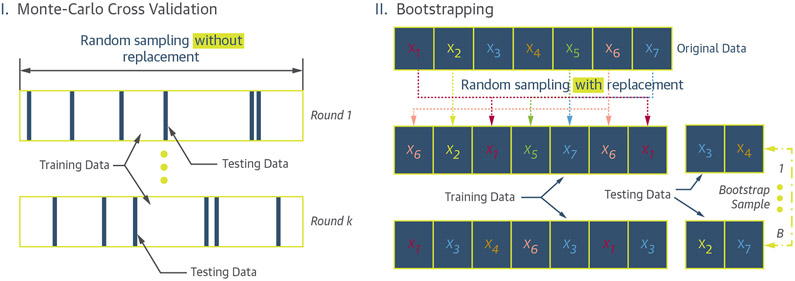
Monte Carlo cross-validation.
In this method of cross-validation, there is no limit to the number of the folds, and a database can be randomly partitioned into multiple training and testing sets. The training samples are randomly selected “without replacement,” and the remaining samples are used for the testing group (Figure 6I) (76).
FIGURE 6. Schematic Illustration of the Monte Carlo and Bootstrap Resampling Methods.
(I) Monte Carlo cross-validation performed in k rounds. In each round, the training and testing samples are randomly selected without replacement from the original data. (II) The bootstrapping process can be performed in B rounds. In each round, the training data are generated by randomly sampling from the original data with replacement. The samples that are not included in the training dataset (i.e., out-of-bag samples) form the testing dataset.
BOOTSTRAPPING.
This method works by randomly sampling observations from a database “with replacement” to form a training set whose size is equal to the original database. As a result, some of the observations can appear several times in the training set, while some may never be selected. The latter observations are called “out-of-bag” and are used to test the learning algorithm. This process is repeated multiple times to estimate the learning method’s generalization performance (Figure 6 II) (33,76). Although bootstrapping tends to drastically reduce the variance, it often tends to provide more biased results, more importantly when dealing with small sample sizes.
Strategic steps for developing the checklist:
Model assessment should be performed by randomly dividing the dataset into training, validation and testing data when applicable.
When data is inadequate or scarce, model assessments using cross-validation and/or bootstrapping techniques should be performed to obtain a good estimate of the generalization performance of the learning algorithm.
Typical numbers for k in a k-fold cross-validation should be 5 and 10.
Consider using leave-one-out cross validation as an appropriate choice when the data is small.
5. MODEL EVALUATION
The reporting of accuracy in ML is closely linked to the reporting of summary statistics, and the same background and assumptions apply. Although a review of statistical theory is out of scope for the PRIME Checklist, we encourage the readers to obtain a clear understanding of the statistics for classification and prediction (77-83). Most of the following section applies to supervised learning algorithms, for which labels are used in the definition of the performance measures. Unsupervised learning is more difficult to evaluate but should also evaluate the relevance of the output data representation and the stability of the results against the data and model parameters.
For classification tasks, the accuracy is the percentage of data that is correctly classified by the model, which could be influenced by the quality of the expert annotations. The balance of classes in the training data is also a known source of bias. As such, a prerequisite for reporting accuracy measures is to provide a clear description of the data material used for training and validation. We further suggest balancing the class data according to prevalence when possible or that balanced accuracy measures are reported (84).
The model parameters (e.g., initialization scheme, number of feature maps, and loss function), regularization strategies (e.g., smoothness and dropouts), and hyperparameters (e.g., optimizer, learning rate, and stopping criterion) also play a part in the model performance. A second prerequisite, therefore, is to provide a clear description of how the ML model was generated. It is suggested, furthermore, that the certainty of the accuracy measure is reported where applicable, for instance, by estimating the ensemble average and variance from several models generated with random initialization. Additionally, cross-validation analysis should be added to underline the robustness of the model, especially for limited training and test data (see the previous section). Furthermore, to assess the generalizability of the algorithm, it is necessary to report the accuracy of the model by testing the data from different geographic locations with similar statistical properties and distributions (85).
A report of the accuracy for ML algorithms in cardiovascular imaging will depend on the method and problem. For instance, the classification of disease from image features differs from the classification of image pixels in semantic segmentation, both in terms of the measures reported and of the risk in use.
For multiclass and label classification, it is suggested to use a statistical language close to the clinical standard. For instance, the report sensitivity, specificity, and odds ratio should be used instead of the precision, recall, and F1 score. This will also ensure that true negative outcomes are considered (86). Nonetheless, for classification tasks, the confusion matrix should normally be included but could be Supplemental Appendix. For image segmentation problems, we suggest reporting several measures to summarize both the global and local deviations, such as the mean absolute error, the Dice score to summarize the average performance, and the Hausdorff distance metric to capture local outliers.
When the output of the regression or segmentation algorithms is linked to clinical measurements (e.g., ejection fraction), the Bland-Altman plot is suggested for conventional evaluation of the image measurements, and the importance of comparing the performance with several expert observers for both intraexpert and interexpert variability is stressed.
For the classification of disease from image features, the cost of misclassification should be clearly conveyed (e.g., rare diseases may not be properly represented in the dataset). The balance of classes should reflect the prevalence of the disease of interest, and scoring rules based on estimated probability distributions should be used for the accuracy reporting when possible, instead of direct classification. The choice of the scoring rule used for the decision (e.g., mean squared error, Brier score, and log-loss) should be rationalized. The common classification scores (sensitivity, specificity, and positive and negative predictive values) should include a full receiver-operating characteristic curve analysis to provide a more in-depth evaluation of the detection performance. It is also relevant to include benchmark results from alternative ML methods as well as more conventional techniques, such as logistic regression.
Strategic steps for developing the checklist:
Use a statistical language close to the clinical standard and introduce new measures only when needed.
Balance the classes according to prevalence where available or report balanced accuracy measures.
Estimate the accuracy certainty, e.g., from an ensemble of models, to strengthen the confidence in the values reported.
Include Bland-Altman plots when machine learning is linked to clinical measurements.
Include an inter-observer/intraobserver variability measures as a reference where possible.
The risk of misclassification should be conveyed, and appropriate scoring rules for decisions may be needed for the classification of a disease.
6. BEST PRACTICES FOR MODEL REPLICABILITY
The reproducibility of scientific results is essential to make progress in cardiac medicine. The ability to reproduce findings helps to ensure the validity and correctness, as well as enabling others to translate the results into clinical practice. However, there are several complementary definitions of reproducibility. The focus here is primarily on technical replicability (87) (i.e., the ability to independently confirm published results of a model by inspecting and executing data and code under identical conditions. Technical replicability is especially important in ML projects, which often involve customized software scripts, the use of external libraries, and intensive or expensive computations. Actions taken at any point in an ML workflow, from quality control and data preparation into suitable data structures to algorithm development to the visualization of results, are often based upon heuristic judgments, and there are potentially numerous justifiable analytical options. Ultimately, these selections may significantly alter the results and conclusions.
The first step for making ML projects reproducible could be the release of all the original code written for a project. There are several options for the publication of code. Uploading source code with software and software application version information is recommended as Supplemental Appendix alongside the manuscript. Other options include permanent archive on a laboratory Website or per-project archive on open source and public source code repositories if permitted by the investigator’s institution. Manuscripts should explicitly state where and how the code may be downloaded and under what license.
Although there are numerous open source licenses available, in most cases, either of 2 following licenses will suffice: the Massachusetts Institute of Technology/Berkeley Software Distribution (MIT/BSD) licenses (https://opensource.org/licenses/MIT; the MIT and BSD licenses are essentially equivalent) and the GNU General Public License (GNU GPLv3). The MIT/BSD licenses allow published code to be distributed, modified, and executed freely without liability or warranty; the GNU GPLv3 license allows the same with the additional restriction that all software-based upon the original code must also be freely available under the GNU GPLv3 license, meaning others cannot reuse the original code in a closed-source product.
Although the availability of code is required for technical replicability, equally important is the availability of the data used in the project (86). Clinical data should be anonymized, or if anonymization is not possible (as in the case of some genetic data), then data should be made available to other researchers with appropriate institutional review board approval. Other options include the generation of synthetic datasets with the same statistical properties as the original dataset, a field of study called differential privacy. Manuscripts must state where both the raw and manipulated or transformed data may be obtained and justify any restrictions to data availability. All data should also be accompanied by a codebook (also known as a data dictionary) containing clear and succinct explanations of all variables and class labels along with detailed description of their data types and dimensions.
Finally, even in the case of freely available data and open-source code, it can be difficult to reproduce the results of published work due to the complexities of software versions and interactions between different computing environments. Thus, it is suggested that authors make the entire analyses automatically reproducible through the use of software environments (e.g., Docker containers; https://www.docker.com/). Analyses in software containers may be freely downloaded and run from beginning to end by other scientists, greatly improving the technical replicability. Moreover, documentation generation tools (e.g., Sphinx; https://www.sphinx-doc.org/en/master/) or easy-to-launch demonstrations (e.g., through Jupiter python notebooks) should be used.
Strategic steps for developing the checklist:
Release the code and upload data as supplementary information alongside the manuscript when possible for non-commercial use; otherwise, consider making the code and data available via an academic website for non-commercial use as permissible.
Use the MIT/BSD or GPLv3 license to release open-source code.
Release a codebook (data dictionary) with clear and succinct explanations of all variables.
Document the exact version of all external libraries and software environments.
Consider the use of Docker containers or similar packaging environments such as Sphinx for straightforward technical reproducibility and to generate reliable code/software manuals.
7. REPORTING LIMITATIONS, BIASES, AND ALTERNATIVES
“All models are wrong, but some are useful” is a well-known statistical aphorism attributed to George Box. Accurate reporting and acknowledgement of limitations are required for manuscripts incorporating ML. Any statistical model or ML algorithm incorporates some assumptions regarding the data. All model assumptions should be affirmatively identified and checked with the dataset used in the manuscript, and the results should be reported in the manuscript or the Supplemental Appendix. The algorithms used in computational research efforts span a large spectrum of complexity. Generally, more basic models and algorithms should first be investigated before additional complexity is incorporated into models or different algorithms are selected. Deep learning models should be benchmarked against simpler models whenever possible, especially when applied to tabular data. Statistical or ML models incorporating large numbers of variables (e.g., polygenic risk score models) should be benchmarked against standard clinical risk prediction models using more conventional clinical variables.
Concordant findings from multiple, independent datasets dramatically increase the scientific value of manuscripts, because they decrease the likelihood that the algorithms have been erroneously overfitted to the idiosyncratic features of a certain dataset. Deep learning models are especially notorious for harnessing spurious or confounding features of the dataset to perform well. For example, Zech et al. (88) reported a case where a convolutional neural network trained on a health system’s chest radiography used the presence of a “PORTable” label on radiographic images to predict cardiomegaly with high accuracy. Furthermore, in the case of supervised ML involving human-annotated variables or outcomes, it should be noted that ML algorithms will recapitulate the underlying biases of the humans who constructed the dataset.
Strategic steps for developing the checklist:
Affirmatively identify and check relevant model assumptions and report the findings.
Benchmark complex algorithms against simpler algorithms and justify the use of more complex models.
Benchmark algorithms, incorporating high-dimensional data or novel data sources, against standard clinical risk prediction models
SUMMARY AND FUTURE DIRECTIONS
As AI and ML technologies continue to grow, 3 specific areas of opportunities will need further consideration for future standardization. First, there has been growing enthusiasm for the use of automated ML (auto-ML) platforms that democratize ML strategies. Second, using the “multiomics” approach, clinical and other data, such as smart-phone-based health data could be integrated with imaging variables to provide more algorithmic sophistication and objectivity to the existing taxonomy of risk factors and cardiac diseases (89-91). Finally, sophisticated algorithms and variations of GAN will be increasingly used to synthesize data that closely resemble the distribution of the input data (92-94). This approach may be particularly fruitful for the field of simulation and in silico clinical trials, which were recently recognized by the U.S. Food and Drug Administration (FDA) as key new directions to validate novel devices and therapies (95). In that context, recent studies have combined computational modeling with ML for synthetic data generation or tracking a disease course (96). Moreover, the ML research presents unprecedented opportunities for restructuring the industry, research, and medical alliance. This fact is well recognized by the FDA, which has mandated the standardization and applications of ML software as medical devices (97). With the advancement of organizations and cardiac medicine and imaging toward the actualization of precision medicine, the PRIME checklist would need to be updated continuously as ML algorithms continue to transform cardiovascular imaging practice over the next decade.
Supplementary Material
HIGHLIGHTS.
Algorithm complexity and flexibiLity of ML techniques can result in inconsistencies in model reporting and interpretations.
The PRIME checklist provides 7 items to be reported for reducing algorithmic errors and biases.
The checklist aims to standardize reporting on model design, data, selection, assessment, evaluation, replicability, and limitations.
As artificial intelligence and ML technologies continue to grow, the checklist will need periodic updates.
ACKNOWLEDGMENTS
The authors thank Jai K. Nahar, MD, MBA, and James E. Tcheng, MD, for reviewing and providing constructive feedback for the paper.
The views expressed in this paper do not reflect the views of the American College of Cardiology or the Journal of American College of Cardiology family. Dr. Dudley has salary support from Tempus Labs. Dr. Johnson has salary support from Tempus Labs; and has an equity interest in Tempus Labs and in Oova, Inc. Dr. Lovstakken is a consultant for GE Vingmed Ultrasound AS. Dr. Min has employment and salary support from Clearly Inc.; and serves as advisor for Arienta. Dr. Slomka has received a grant from Siemens Medical Systems; and has received royalties from Cedars-Sinai. Dr. Yanamala is member of Think2Innovate LLC. Dr. Sengupta is a consultant for Heartsciences, Ultromics, and Kencor Health; and holds equity interests in Ultromics, Kencor Health, and Heartsciences. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose. Jagat Narula, MD, was Guest Editor on this paper.
ABBREVIATIONS AND ACRONYMS
- AI
artificial intelligence
- ML
machine learning
- PRIME
Proposed Recommendations for Cardiovascular Imaging-Related Machine Learning Evaluation
Footnotes
APPENDIX For supplemental tables and vocabulary, please see the online version of this paper.
REFERENCES
- 1.Douglas PS, Cerqueira MD, Berman DS, et al. The future of cardiac imaging: report of a think tank convened by the American College of Cardiology. J Am Coll Cardiol Img 2016;9:1211–23. [DOI] [PubMed] [Google Scholar]
- 2.Sengupta PP, Shrestha S. Machine learning for data-driven discovery: the rise and relevance. J Am Coll Cardiol Img 2019;12:690–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dey D, Slomka PJ, Leeson P, et al. Artificial intelligence in cardiovascular imaging: JACC State-of-the-Art Review. J Am Coll Cardiol 2019;73: 1317–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winther HB, Hundt C, Schmidt B, et al. ν-net: Deep learning for generalized biventricular mass and function parameters using multicenter cardiac MRI data. J Am Coll Cardiol Img 2018;11:1036–8. [DOI] [PubMed] [Google Scholar]
- 5.Tan LK, McLaughlin RA, Lim E, Abdul Aziz YF, Liew YM. Fully automated segmentation of the left ventricle in cine cardiac MRI using neural network regression. J Magn Reson Imaging 2018; 48:140–52. [DOI] [PubMed] [Google Scholar]
- 6.Betancur J, Commandeur F, Motlagh M, et al. Deep learning for prediction of obstructive disease from fast myocardial perfusion SPECT: a multicenter study. J Am Coll Cardiol Img 2018;11: 1654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madani A, Arnaout R, Mofrad M. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit Med 2018;1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol 2017; 69:2657–64. [DOI] [PubMed] [Google Scholar]
- 9.Zheng T, Xie W, Xu L, et al. A machine learning-based framework to identify type 2 diabetes through electronic health records. Int J Med Inform 2017;97:120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawes TJW, de Marvao A, Shi W, et al. Machine learning of three-dimensional right ventricular motion enables outcome prediction in pulmonary hypertension: a cardiac MR imaging study. Radiology 2017;283:381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang RM, Badano LP, Mor-Avi V, et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr 2015;28:1–39. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Gajjala S, Agrawal P, et al. Fully automated echocardiogram interpretation in clinical practice. Circulation 2018;138:1623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouyang D, He B, Ghorbani A, et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 2020;580:252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahmy AS, Rausch J, Neisius U, et al. Automated cardiac MR scar quantification in hypertrophic cardiomyopathy using deep convolutional neural networks. J Am Coll Cardiol Img 2018;11: 1917–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai W, Sinclair M, Tarroni G, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson 2018;20:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson KW, Torres Soto J, Glicksberg BS, et al. Artificial intelligence in cardiology. J Am Coll Cardiol 2018;71:2668–79. [DOI] [PubMed] [Google Scholar]
- 17.Al’Aref SJ, Anchouche K, Singh G, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975–86. [DOI] [PubMed] [Google Scholar]
- 18.Ghojogh B, Crowley M. The Theory Behind Overfitting, Cross Validation, Regularization, Bagging, and Boosting: Tutorial. Available at: https://arxiv.org/abs/1905.12787:arXiv. Accessed August 11, 2020.
- 19.Dhurandhar A, Shanmugam K, Luss R, Olsen PA. Improving simple models with confidence profiles. 32nd Conference on Neural Information Processing Systems. Montréal, Canada; 2018. [Google Scholar]
- 20.Blakely T, Lynch J, Simons K, Bentley R, Rose S. Reflection on modern methods: when worlds collide-prediction, machine learning and causal inference. Int J Epidemiol 2019:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng S, Xu Z, Ma H. Reconstructing directional causal networks with random forest: Causality meeting machine learning. Chaos 2019;29: 093130. [DOI] [PubMed] [Google Scholar]
- 22.Vandenbroucke JP, von Elm E, Altman DG, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. Int J Surg 2014;12: 1500–24. [DOI] [PubMed] [Google Scholar]
- 23.Cohen O, Malka O, Ringel Z. Learning Curves for Deep Neural Networks: A Gaussian Field Theory Perspective. 2019. Available at: http://arxiv.org/abs/1906.05301. Accessed August 10, 2020.
- 24.Mehta P, Wang CH, Day AGR, et al. A high-bias, low-variance introduction to Machine Learning for physicists. Phys Rep 2019;810:1–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. J Med Syst 2018;42:226. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez RC, Woods RE. Digital Image Processing. 3rd edition. Upper Saddle River, NJ: Prentice-Hall, Inc.; 2007. [Google Scholar]
- 27.Duchateau N, Craene MD, Pennec X, Merino B, Sitges M, Bijnens B. Which reorientation framework for the atlas-based comparison of motion from cardiac image sequences?. In: Durrleman S, Fletcher T, Gerig G, Niethammer M, editors. Spatio-temporal Image Analysis for Longitudinal and Time-Series Image Data. STIA 2012. Lecture Notes in Computer Science, vol 7570. Berlin: Springer, 2012; p 25–37. [Google Scholar]
- 28.Duchateau N, De Craene M, Piella G, et al. A spatiotemporal statistical atlas of motion for the quantification of abnormal myocardial tissue velocities. Med Image Anal 2011;15:316–28. [DOI] [PubMed] [Google Scholar]
- 29.Ullah A, Ahmad J, Muhammad K, Sajjad M, Baik SW. Action recognition in video sequences using deep bi-directional LSTM with CNN features. IEEE Access 2018;6:1155–66. [Google Scholar]
- 30.Betancur J, Hu LH, Commandeur F, et al. Deep learning analysis of upright-supine high-efficiency SPECT myocardial perfusion imaging for prediction of obstructive coronary artery disease: a multicenter study. J Nucl Med 2019;60:664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spier N, Nekolla S, Rupprecht C, Mustafa M, Navab N, Baust M. Classification of polar maps from cardiac perfusion imaging with graph-convolutional neural networks. Sci Rep 2019;9: 7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bishop CM. Pattern Recognition and Machine Learning. New York: Springer-Verlag; 2006. [Google Scholar]
- 33.Hastie T, Tibshirani, Robert JF. The Elements of Statistical Learning Data Mining, Inference, and Prediction. 2nd edition. New York: Springer; 2009. [Google Scholar]
- 34.Koutroumbas K, Theodoridis S. Pattern Recognition. 4th edition. Washington, DC: Academic Press; 2008. [Google Scholar]
- 35.Somorjai RL, Dolenko B, Baumgartner R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics 2003;19: 1484–91. [DOI] [PubMed] [Google Scholar]
- 36.Dernoncourt D, Hanczar B, Zucker J-D. Analysis of feature selection stability on high dimension and small sample data. Comput Stat Data Anal 2014;71:681–93. [Google Scholar]
- 37.Hyvärinen A, Hurri J, Hoyer PO. Natural Image Statistics: A Probabilistic Approach to Early Computational Vision. 1st edition. London: Springer-Verlag; 2009. [Google Scholar]
- 38.Ojala T, Pietikäinen MK, Mäenpää T. Multi-resolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 2002;24: 971–87. [Google Scholar]
- 39.Lowe DG. Object recognition from local scale-invariant features. ICCV ‘99. Proceedings of the International Conference on Computer Vision 1999;2:1150–7. [Google Scholar]
- 40.Jolliffe IT. Principal Component Analysis. 2nd edition. New York: Springer-Verlag; 2002. [Google Scholar]
- 41.Hyvärinen A, Oja E. Independent component analysis: algorithms and applications. Neural Netw 2000;13:411–30. [DOI] [PubMed] [Google Scholar]
- 42.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science 2000;290: 2319–23. [DOI] [PubMed] [Google Scholar]
- 43.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- 44.Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bengio Y Deep Learning of representations for unsupervised and transfer learning. jmlr: workshop and conference proceedings 27. Proc Mach Learn Res 2012:17–37. [Google Scholar]
- 46.Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Available at: http://arxiv.org/abs/1502.03167. Accessed August 11, 2020.
- 47.Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics 2001;17:520–5. [DOI] [PubMed] [Google Scholar]
- 48.Tabassian M, Alessandrini M, Jasaityte R, Marchi LD, Masetti G, D’hooge J. Handling missing strain (rate) curves using K-nearest neighbor imputation. Tours, France: 2016. IEEE Int Ultrason Symp 2016:1–4. [Google Scholar]
- 49.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. Adv Neural Inf Process Syst 27 2014;2:2672–80. [Google Scholar]
- 50.Shang C, Palmer A, Sun J, Chen KS, Lu J, Bi J. VIGAN: missing view imputation with generative adversarial networks. Proc IEEE Int Conf Big Data 2017:766–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhodes W Improving disparity research by imputing missing data in health care records. Health Serv Res 2015;50:939–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madley-Dowd P, Hughes R, Tilling K, Heron J. The proportion of missing data should not be used to guide decisions on multiple imputation. J Clin Epidemiol 2019:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Gopalakrishnan V. An Overview and evaluation of recent machine learning imputation methods using cardiac imaging data. Data (Basel) 2017;2:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inza I, Larranaga P, Blanco R, Cerrolaza AJ. Filter versus wrapper gene selection approaches in DNA microarray domains. Artif Intell Med 2004; 31:91–103. [DOI] [PubMed] [Google Scholar]
- 55.Arnaout R, Curran L, Chinn E, Zhao Y, Moon-Grady A. Deep-learning models improve on community-level diagnosis for common congenital heart disease lesions. Available at: http://arxiv.org/abs/1809.06993. 2018. Accessed August 11, 2020.
- 56.Atkinson A, Riani M. Robust Diagnostic Regression Analysis. 1st edition. New York: Springer-Verlag; 2000. [Google Scholar]
- 57.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority oversampling technique. J Artif Intell Res 2002;16: 321–57. [Google Scholar]
- 58.Last F, Douzas G, Bacao F. Oversampling for Imbalanced Learning Based on K-Means and SMOTE. 12December2017 ed. Available at: https://arxiv.org/abs/1711.00837. 2017. Accessed August 11, 2020.
- 59.Rezende DJ, Mohamed S. Variational inference with normalizing flows. Available at: http://arxiv.org/abs/1505.05770. Accessed August 11, 2020.
- 60.Subbaswamy A, Schulam P, Saria S. Preventing failures due to dataset shift: learning predictive models that transport. Proc Mach Learn Res 2019:3118–27. [Google Scholar]
- 61.Wang J, Ding H, Bidgoli FA, et al. Detecting cardiovascular disease from mammograms with deep learning. IEEE Trans Med Imaging 2017;36: 1172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Litjens G, Ciompi F, Wolterink JM, et al. State-of-the-art deep learning in cardiovascular image analysis. J Am Coll Cardiol Img 2019;12:1549–65. [DOI] [PubMed] [Google Scholar]
- 63.Retson TA, Besser AH, Sall S, Golden D, Hsiao A. Machine learning and deep neural networks in thoracic and cardiovascular imaging. J Thorac Imaging 2019;34:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 2016;30:108–19. [DOI] [PubMed] [Google Scholar]
- 65.Kostoglou K, Robertson AD, MacIntosh BJ, Mitsis GD. A novel framework for estimating time-varying multivariate autoregressive models and application to cardiovascular responses to acute exercise. IEEE Trans Biomed Eng 2019;66: 3257–66. [DOI] [PubMed] [Google Scholar]
- 66.Al’Aref SJ, Singh G, van Rosendael AR, et al. Determinants of in-hospital mortality after percutaneous coronary intervention: a machine learning approach. J Am Heart Assoc 2019;8: e011160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilpin LH, Bau D, Yuan BZ, Bajwa A, Specter M, Kagal L. Explaining explanations: an overview of interpretability of machine learning. 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA) 2018:80–9. [Google Scholar]
- 68.Fong RC, Vedaldi A. Interpretable explanations of black boxes by meaningful perturbation. 2017 IEEE International Conference on Computer Vision (ICCV) 2017:3449–57. [Google Scholar]
- 69.Doshi-Velez F, Kim B. Towards A Rigorous Science of Interpretable Machine Learning. Available at: http://arxiv.org/abs/1702.08608. 2017. Accessed August 11, 2020.
- 70.Guidotti R, Monreale A, Ruggieri S, Turini F, Giannotti F, Pedreschi D. A survey of methods for explaining black box models. ACM Comput Surv 2018:1–42. [Google Scholar]
- 71.Olah C, Satyanarayan A, Johnson I, et al. The Building Blocks of Interpretability. 2018. Distill 2018. Available at: https://distill.pub/2018/building-blocks/. Accessed July 26, 2020.
- 72.Samek W, Wiegand T, Müller K-R. Explainable Artificial Intelligence: Understanding, Visualizing and Interpreting Deep Learning Models. August 28, 2017. Available at: https://arxiv.org/abs/1708.08296. Accessed July 26, 2020.
- 73.Lake BM, Salakhutdinov R, Tenenbaum JB. Human-level concept learning through probabilistic program induction. Science 2015;350: 1332–8. [DOI] [PubMed] [Google Scholar]
- 74.Gal Y, Ghahramani Z. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning. 33rd Int. Conf. Mach. Learn. Available at: http://arxiv.org/abs/1506.02142. Accessed on August 10, 2020 [Google Scholar]
- 75.Raudys Š. Statistical and neural classifiers: an integrated approach to design. New York: Springer; 2001. [Google Scholar]
- 76.Kuhn M, Johnson K. Applied Predictive Modeling. New York: Springer; 2013. [Google Scholar]
- 77.Wheelan C Naked statistics. stripping the dread from the data. 1st edition. Philadelphia: WW Norton; 2013; p 302. [Google Scholar]
- 78.Mlodinow L The Drunkard’s Walk: How Randomness Rules Our Lives. New York: Vintage; 2009. [Google Scholar]
- 79.Wasserman L All of Statistics: A Concise Course In Statistical Inference. 1st edition. New York: Springer-Verlag; 2004. [Google Scholar]
- 80.Urdan TC. Statistics in Plain English. 2nd edition. Sussex, UK: Psychology Press; 2005. [Google Scholar]
- 81.Cohen PR. Empirical Methods For Artificial Intelligence. Cambridge, MA: MIT Press; 1995. [Google Scholar]
- 82.Box GEP, Hunter JS, Hunter WG. Statistics for Experimenters: Design, Innovation, and Discovery. 2nd edition. New York: Wiley and Sons; 2005. [Google Scholar]
- 83.Sabo R, Boone E. Statistical Research Methods: A Guide for Non-Statisticians. 1st edition. New York: Springer-Verlag; 2013. [Google Scholar]
- 84.Wainer J, Franceschinell RA. An empirical evaluation of imbalanced data strategies from a practitioner’s point of view. October 16, 2018. Available at: https://arxiv.org/abs/1810.07168. Accessed July 26, 2020.
- 85.Abazeed M Walking the tightrope of artificial intelligence guidelines in clinical practice. The Lancet Digital Health 2019:PE100. [DOI] [PubMed] [Google Scholar]
- 86.Tharwat A Classification assessment methods. Appl. Comput. Informatics; 2020. Available at: https://www.emerald.com/insight/content/doi/10.1016/j.aci.2018.08.003/full/html. Accessed August 9, 2020.
- 87.McDermott MBA, Wang S, Marinsek N, Ranganath R, Ghassemi M, Foschini L. Reproducibility in Machine Learning for Health. July 2, 2019. Available at: https://arxiv.org/abs/1907.01463. Accessed July 26, 2020. [DOI] [PubMed]
- 88.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med 2018;15:e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Natarajan S, Jain A, Krishnan R, Rogye A, Sivaprasad S. Diagnostic accuracy of community-based diabetic retinopathy screening with an offline artificial intelligence system on a smartphone. JAMA Ophthalmol 2019;137:1182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rajalakshmi R, Subashini R, Anjana RM, Mohan V. Automated diabetic retinopathy detection in smartphone-based fundus photography using artificial intelligence 2018;32: 1138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wasserlauf J, You C, Patel R, Valys A, Albert D, Passman R. Smartwatch performance for the detection and quantification of atrial fibrillation. Circ Arrhythm Electrophysiol 2019;12:e006834. [DOI] [PubMed] [Google Scholar]
- 92.Suarez J GAN You Do the GAN GAN? April 1, 2019. Available at: https://arxiv.org/abs/1904.00724. Accessed July 26, 2020.
- 93.Wang X, He K, Hopcroft JE. AT-GAN: A Generative Attack Model for Adversarial Transferring on Generative Adversarial Nets. rev May 21. 2019. Available at: https://arxiv.org/abs/1904.07793v3. Accessed July 26, 2019.
- 94.Chang C-H, Yu C-H, Chen S-Y, Chang EY, GAN KG-. Knowledge-Guided Generative Adversarial Networks. September 23, 2019. Available at: https://arxiv.org/abs/1905.12261. Accessed July 26, 2020.
- 95.Morrison T How Simulation Can Transform Regulatory Pathways. 2018. FDA; 2018. Available at: https://www.fda.gov/science-research/about-science-research-fda/how-simulation-can-transform-regulatory-pathways. Accessed October 14, 2019. [Google Scholar]
- 96.Kagiyama N, Shrestha S, Farjo PD, Sengupta PP. Artificial intelligence: practical primer for clinical research in cardiovascular disease. J Am Heart Assoc 2019;8:e012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Software as a Medical Device (SaMD): Key Definitions. International Medical Device Regulatory Forum, 2013. Available at: https://www.fda.gov/medical-devices/digital-health/software-medical-device-samd. Accessed October 14, 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.