Abstract
Background
Toxoplasma gondii is a parasite that primarily infects through the oral route. Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) play crucial roles in the immune responses generated during parasitic infection and also drive the inflammatory response against invading parasites. However, little is known about the regulation of NLRs and inflammasome activation in T. gondii-infected human small intestinal epithelial (FHs 74 Int) cells.
Methods
FHs 74 Int cells infected with T. gondii were subsequently evaluated for morphological changes, cytotoxicity, expression profiles of NLRs, inflammasome components, caspase-cleaved interleukins (ILs), and the mechanisms of NLRP3 and NLRP6 inflammasome activation. Immunocytochemistry, lactate dehydrogenase assay, reverse transcription polymerase chain reaction (RT-PCR), real-time quantitative RT-PCR, and western blotting techniques were utilized for analysis.
Results
Under normal and T. gondii-infected conditions, members of the NLRs, inflammasome components and caspase-cleaved ILs were expressed in the FHs Int 74 cells, except for NLRC3, NLRP5, and NLRP9. Among the NLRs, mRNA expression of NOD2, NLRP3, NLRP6, and NAIP1 was significantly increased in T. gondii-infected cells, whereas that of NLRP2, NLRP7, and CIITA mRNAs decreased significantly in a time-dependent manner. In addition, T. gondii infection induced NLRP3, NLRP6 and NLRC4 inflammasome activation and production of IL-1β, IL-18, and IL-33 in FHs 74 Int cells. T. gondii-induced NLRP3 inflammasome activation was strongly associated with the phosphorylation of p38 MAPK; however, JNK1/2 had a weak effect. NLRP6 inflammasome activation was not related to the MAPK pathway in FHs 74 Int cells.
Conclusions
This study highlighted the expression profiles of NLRs and unraveled the underlying mechanisms of NLRP3 inflammasome activation in T. gondii-infected FHs 74 Int cells. These findings may contribute to understanding of the mucosal and innate immune responses induced by the NLRs and inflammasomes during T. gondii infection in FHs 74 Int cells.
Keywords: Toxoplasma gondii, Human small intestinal epithelial cells, NOD-like receptors, Inflammasome, Caspase-cleaved interleukins
Background
Toxoplasma gondii is an obligate intracellular protozoan parasite that infects one-third of the world’s population [1]. Infection is most commonly acquired through the ingestion of raw or undercooked meat containing the cystic bradyzoite form of T. gondii or through the ingestion of materials contaminated with cat feces that may contain T. gondii oocysts. Once inside the body, the parasite breaches the intestinal epithelial barrier and spreads from the lamina propria to other organs [2]. Intestinal epithelial cells can sense and respond to the invading microbial stimuli to reinforce their barrier function. They also participate in the coordination of appropriate immune responses [3]. The innate immune system plays a significant role in sensing pathogens and triggering biological mechanisms to control infection and eliminate pathogens [4, 5]. It is activated when pattern recognition receptor proteins, such as Toll-like receptors (TLRs) or nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), detect the presence of pathogens, their products, or the danger signals [5–7].
NLRs are a large group of cytosolic sensors that have diverse functions in innate immunity and inflammation. Based on the type of N-terminal domain, NLRs are classified into four subfamilies, NLRA, NLRB, NLRC, and NLRP, and an additional subfamily, NLRX1 [7, 8]. Several NLR molecules remain associated with the T. gondii-infection mediated immune responses in the infected hosts. It has been reported that NOD2-deficient mice are unable to clear T. gondii and fail to induce an appropriate adaptive immune response [9]. In addition to NOD2, NLRP1b and NLRP3 are also involved in rendering protection against T. gondii infection [10, 11]. In human acute monocytic leukemia cell line macrophages, the messenger RNA (mRNA) levels of NLRC4, NLRP6, NLRP8, NLRP13, AIM2, and NAIP are significantly elevated because of T. gondii infection, in a time-dependent manner [12]. Although some studies involving mice or cell lines have reported the involvement of NLR members in T. gondii infection protection [9–12], little information is available about the regulation of NLR activation in gut epithelial cells.
Ligand recognition by the NLR family members, such as NLRP1, NLRP3, NLRP6, NLRP12, and NLRC4, leads to the activation of inflammasome, a multiprotein complex, which cleaves interleukin (IL)-1β, IL-18, IL-33, and IL-37 (IL-17A) by caspases, the effector components of inflammasomes [8, 10–14]. T. gondii infection in cells with NLRP1 knockdown fails to induce the production of inflammatory cytokines including IL-1β, IL-18, and IL-12 compared to control cells [10]. The broad range of pathogens that act on NLRP3 in several kinds of epithelial cells include Plasmodium sp., Trypanosoma cruzi, Leishmania sp., and T. gondii [15]. The P2X7R/NLRP3 pathway plays an important role in IL-1β secretion and inhibition of T. gondii proliferation in small intestinal epithelial cells [16]. While reports have revealed NLR activation by T. gondii infection in various cells, information on inflammasome activation in gut epithelial cells infected with T. gondii is very scarce.
NLRs play a crucial role in inducing immune responses during parasitic infection and driving the inflammatory responses against invading parasites [17]. However, little is known about the regulation of NLRs and NLR-related inflammasome activation in T. gondii-infected human small intestinal epithelial (FHs 74 Int) cells. Therefore, this study evaluated the expression profiles of NLRs, inflammasome components, and caspase-cleaved ILs and investigated the mechanisms of some popular inflammasomes’ activation in T. gondii-infected FHs 74 Int cells.
Methods
Cell culture
A non-transformed human fetal small intestinal epithelial cell line (FHs 74 Int cells) was purchased from ATCC (ATCC, Manassas, VA, USA) and cultured in DMEM with 10% (v/v) heat-inactivated fetal bovine serum (FBS), an antibiotic–antimycotic solution, and 30 ng/ml human epidermal growth factor (all from Gibco, Grand Island, NY, USA) at 37 °C in a humidified atmosphere at 5% (v/v) CO2. The medium was changed every 2–3 days.
Maintenance of T. gondii
Tachyzoites of the T. gondii RFP-RH or RH strain were maintained as described previously [16]. Briefly, human retinal pigment epithelial cells (ARPE-19 cells) (ATCC) were cultured in a 1:1 (v/v) mixture of DMEM/F12 supplemented with 10% (v/v) FBS and an antibiotic–antimycotic solution (all from Gibco). ARPE-19 cells were infected with T. gondii at a multiplicity of infection (MOI) of 5 for 2–3 days. After spontaneous host cell rupture, parasites and cellular debris were pelleted by centrifugation and washed in cold PBS. The final pellet was resuspended and passed through a 26-gauge needle fitted with a 5.0 μm pore-sized filter (Millipore, Billerica, MA, USA).
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), and RNA was transcribed into cDNA using M-MLV reverse transcriptase (Invitrogen) as described by the manufacturer. Polymerase chain reaction (PCR) was performed with TaKaRa Ex Taq (Takara Bio, Shiga, Japan) in reactions containing 33.75 μl distilled water, 5 μl 10 × Ex Taq buffer, 4 μl dNTP mixture (2.5 mM each), 2 μl of each primer, 0.25 μl of TaKaRa Ex Taq, and 3 μl of template cDNA to total 50 μl. PCR amplification conditions were an initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, an annealing 60 °C for 30 s, and an extension step of 72 °C for 30 s. Finally, PCR was completed with the additional extension step for 10 min. The PCR products were analyzed on 1.5% agarose gel in 0.5 × TBE buffer and visualized using ethidium bromide and a UV transilluminator. The details of primers designed are presented in Table 1.
Table 1.
Members of the NLR family, inflammasome components and caspase-cleaved interleukins and primers used to investigate their expressions by RT-PCR and qRT-PCR
| Family | Name | Synonym | Forward 5′–3’ | Reverse 5′–3’ | Amplicon length (bp) |
|---|---|---|---|---|---|
| NLRC | NOD1 | CARD4 | ACTGAAAAGCAATCGGGAACT | ACACACAATCTCCGCATCTTC | 112 |
| NOD2 | CARD15 | GCCTGATGTTGGTCAAGAAGA | GATCCGTGAACCTGAACTTGA | 107 | |
| NLRC3 | NOD3, CLR16.2 | GGAGCCTCACCAGCTTAGATT | AGGCCACCTGGAGATAGAGAG | 117 | |
| NLRC4 | IPAF, CARD12 | GGAAAGTGCAAGGCTCTGAC | TGTCTGCTTCCTGATTGTGC | 129 | |
| NLRC5 | NOD27, CLR16.1 | CACCCTGACCAACATCCTAGA | TCTCTATCTGCCCACAGCCTA | 113 | |
| NLRP | NLRP1 | NALP1, CARD7 | ATACGAAGCCTTTGGGGACT | ACAAAGCAGAGACCCGTGTT | 148 |
| NLRP2 | NALP2 | CACCGAATGGATCTGTCTGA | GTGGTCGTTCTTTCCGTGTT | 112 | |
| NLRP3 | NALP3, CIAS1 | AAAGGAAGTGGACTGCGAGA | TTCAAACGACTCCCTGGAAC | 129 | |
| NLRP4 | NALP4 | CCAACGAGTTTGGCTGACTT | GCTGTCGATGACGAACAAGA | 105 | |
| NLRP5 | NALP5 | CTGGGGAACGAAGGTGTAAA | GCAAGTGCAAGAAAACCACA | 122 | |
| NLRP6 | NALP6 | CTGTTCTGAGCTACTGCGTGAG | AGGCTCTTCTTCTTCTTCTCCTG | 100 | |
| NLRP7 | NALP7 | TAACCCGTAGCACCTGTCATC | GGTCTTCTTCCCAATGAAAGC | 101 | |
| NLRP8 | NALP8 | CGCTGGTGTGCTTTCTACTTC | GGTCGGGTTTGGACATAATCT | 130 | |
| NLRP9 | NALP9 | CTAGCCTCTCCCAGTCTGACAT | GCGATGTCTTCACAAACTTCAC | 121 | |
| NLRP10 | NALP10 | GTCACGGTGGAGGCTCTATTT | CGAGAGTTGTCTTTCCAGTGC | 100 | |
| NLRP11 | NALP11 | GTGTTGCATGTGACGTTTCC | TTTTGTTGCTCCCAATCTCC | 157 | |
| NLRP12 | NALP12 | CGACCTTTACCTGACCAACAA | AGGTCCATCCCAAATAACCAG | 114 | |
| NLRP13 | NALP13 | ATGGTGTGTTGGACCGTATGT | GCCAAATCTACCTCTGCTGT | 140 | |
| NLRP14 | NALP14 | CCGCTTGTACTTGTCTGAAGC | GCCTCCATCTACTGGTGTGAA | 122 | |
| NLRB | NAIP1 | BIRC1, NLRB1 | AGTACTTTTTCGACCACCCAGA | TAGTTGGCACCTGTGATTTGTC | 135 |
| NLRA | CIITA | MHC2TA, C2TA | CCGACACAGACACCATCAAC | CCTCTGGGAAGGGTCTTTTC | 249 |
| NLRX | NLRX1 | NOD9 | TGGCCTTGTCTCAGCTCTTTA | CACCAGTCCAGAACCATCTTG | 121 |
| Infl. comp | Caspase 1 | IL1BC | GGGGTACAGCGTAGATGTGAA | CTTCCCGAATACCATGAGACA | 137 |
| Caspase 5 | ICH-3 | TCTGTTTGCAAGATCCACGA | GTTCTATGGTGGGCATCTGG | 223 | |
| Caspase 8 | ALPS2B | AGAAGAGGGTCATCCTGGGAGA | TCAGGACTTCCTTCAAGGCTGC | 142 | |
| ASC | PYCARD | CTGACGGATGAGCAGTACCA | CAAGTCCTTGCAGGTCCAGT | 108 | |
| Interleukins | IL-1β | IL1F2 | CCACAGACCTTCCAGGAGAA | GTGATCGTACAGGTGCATCG | 121 |
| IL-18 | IL1F4 | CACCCCGGACCATATTTATT | TCATGTCCTGGGACACTTCTC | 205 | |
| IL-33 | IL1F11 | GGTGACGGTGTTGATGGTAAG | CTGGCAGTGGTTTTTCACACT | 121 | |
| IL-37 | IL-1F7 | CAGCCTCTGCGGAGAAAGGAAGT | GTTTCTCCTTCTTCAGCTGAAGG | 120 | |
| Control | HPRT-1 | HGPRT | GACCAGTCAACAGGGGACAT | CTGCATTGTTTTGCCAGTGT | 111 |
Immunocytochemistry
FHs 74 Int cells were seeded onto coverslips in 12-well plates at a density of 1 × 104 cells/well and incubated for 24 h. The cells were infected with T. gondii at MOI 10 for 0, 4, and 8 h. Subsequently, the cells were washed with Hank's balanced salt solution (HBSS) and fixed with freshly prepared 4% paraformaldehyde for 1 h at room temperature. After washing five times with PBS containing 0.3% Triton X-100 (PBS-T) for 10 min, the cells were incubated with primary antibodies (α-tubulin, cleaved caspase-8, cleaved IL-1β, IL-33) for 2 h at room temperature. The cells were washed to remove excess primary antibody and then incubated with the appropriate fluorescently labeled secondary antibodies (anti-mouse Alexa Fluor 647, anti-rabbit Alexa Fluor 647, anti-mouse Alexa Fluor 488, and anti-rabbit Alexa Fluor 488) for 2 h at room temperature. After mounting with VECTASHIELD HardSet antifade mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA), fluorescence images were acquired using a confocal microscope (Leica, Wetzlar, German).
Lactate dehydrogenase (LDH) assay
LDH assay was performed to quantify cytotoxicity. This assay was conducted using the CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (Promega) according to the manufacturer's protocol. Briefly, 1 × 104 cells were seeded into 96-well plates and infected with T. gondii MOI 10 for 0, 4, and 8 h in an incubator (5% CO2, 90% relative humidity, 37 °C). Next, 50 µl of the supernatant was transferred into a new 96-well plate, and 50 µl of CytoTox 96 reagent was added and incubated for 30 min at room temperature. After incubation, the absorbance of the solution was measured immediately at 490 nm using a microplate reader (TECAN, Männedorf, Switzerland). LDH levels in the media were quantified and compared to control values according to the kit instructions.
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
qRT-PCR was performed using Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The primers used in this study are summarized in Table 1. All reactions were performed with an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Relative gene expression levels were quantified based on the cycle threshold (Ct) values and normalized to the reference gene hypoxanthine phosphoribosyltransferase 1 (HPRT-1). Each sample was measured in triplicate, and the gene expression levels were calculated using the 2–ΔΔCt method.
Western blotting
FHs 74 Int cells were infected with T. gondii at MOI 10 for 0, 4, and 8 h. FHs 74 Int cells were preincubated with the 30 μM of SB203580 (p38 MAPK inhibitor) and SP600125 (JNK1/2 inhibitor) for 2 h and infected with T. gondii MOI 10 for a further 8 h. Subsequently, cell lysates were collected and lysed in ice-cold radio-immunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific, Grand Island, NY, USA). Protein concentrations were determined using the Bradford assay (Bio-Rad, Hercules, CA, USA). Total protein (30 μg) was resolved on 10–12% SDS-PAGE gels and then transferred to PVDF membranes (Merck Millipore, Billerica, MA, USA). The membranes were blocked with 5% nonfat skim milk in TBS containing 0.1% Tween 20 (TBST) for 1 h and incubated with primary antibodies against NLRP1, NLRP3, NLRP6, NLRC4, NAIP1, cleaved caspase-1, ASC, cleaved IL-1β, IL-33, p-p38 MAPK, p38 MAPK, p-ERK1/2, ERK1/2, p-JNK1/2, JNK1/2, TP3, and α-tubulin overnight at 4 °C. Subsequently, the membranes were incubated with HRP-conjugated secondary antibody (Santa Cruz Biotechnology) for 2 h at room temperature. The membrane was soaked with Immobilon Western Chemiluminescent HRP Substrate (Jackson ImmunoResearch Laboratories), and chemiluminescence was detected with a Fusion Solo System (Vilber Lourmat, Collegien, France). Band intensity was quantified using ImageJ software (NIH, Bethesda, MD, USA). The result was normalized to the α-tubulin protein level and expressed as fold changes compared to the control group.
ELISA
FHs 74 Int cells were infected with T. gondii at MOI 10 for 0, 4, and 8 h. The supernatants from the mock- or T. gondii-infected FHs 74 Int cells were collected in triplicate, and IL-1β and IL-18 levels were measured using commercially available ELISA kits following the manufacturer’s instructions (R&D System, Minneapolis, MN, USA). The cytokine concentrations in the samples were calculated from standard curves obtained using recombinant cytokines.
Statistical analysis
All results are presented as the means ± standard deviations (SDs) of at least three independent experiments, unless otherwise indicated. Statistical comparisons were carried out using GraphPad Prism software (GraphPad Software Inc., San Diego, CA, USA), and multiple t-tests was used to determine one-way ANOVA procedures. Differences were considered significant at P < 0.05.
Results
Expression of NLRs, inflammasome components, and caspase-cleaved ILs in FHs 74 Int cells
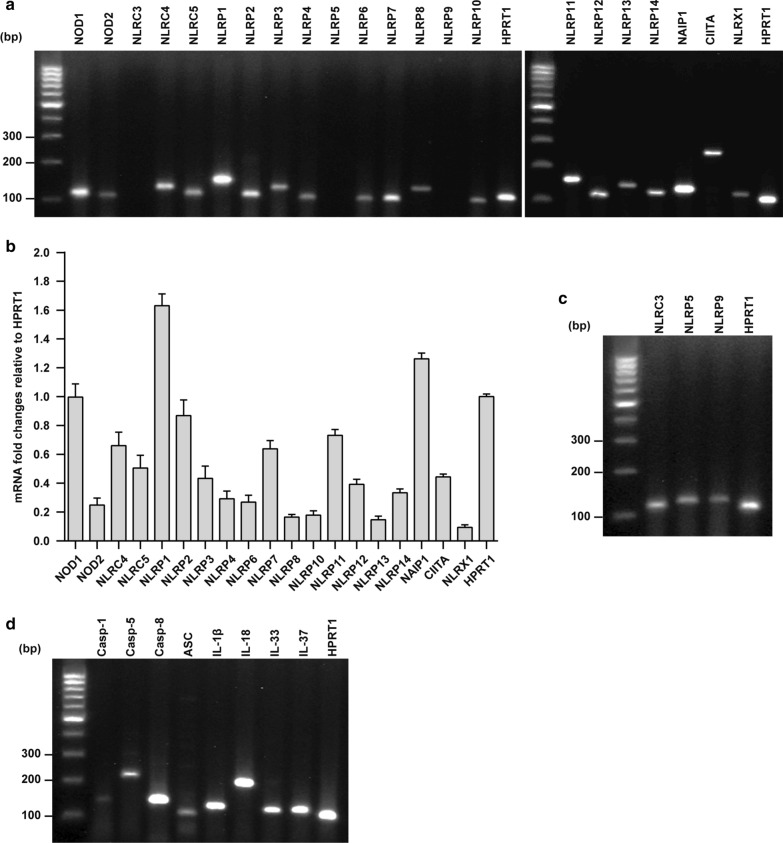
We first checked the expression of the 22 known members of the human NLR family (Table 1) in FHs 74 Int cells using RT-PCR. Our results indicated that the majority of the NLR family members, including NOD1, NOD2, NLRC4, NLRC5, NLRP1, NLRP2, NLRP3, NLRP4, NLRP6, NLRP7, NLRP8, NLRP10, NLRP11, NLRP12, NLRP13, NLRP14, NAIP1, CIITA, and NLRX1 were expressed in these cells under normal conditions (Fig. 1a). After normalization with housekeeping gene HPRT-1, NLRP1 and NOD1 were identified as the most abundantly expressed NLRs in FHs 74 Int cells (Fig. 1b). For primer functionality tests, we used different cell types of human origin that are known to express the respective NLRs and detected the expression of NLRC3, NLRP5, and NLRP9 mRNAs (Fig. 1c). Furthermore, we examined the presence of various inflammasome components and ILs cleaved by caspases. Our results revealed that caspase-1, caspase-5, caspase-8, and ASC inflammasome components and IL-1β, IL-18, IL-33, and IL-37 were expressed in the FHs 74 Int cells (Fig. 1d).
Fig. 1.
Expression of nucleotide-binding oligomerization domain-like receptors (NLRs), inflammasome components, and caspase-cleaved interleukins (ILs) in human small intestinal epithelial (FHs 74 Int) cells. Total RNA isolated from the untreated cells was examined by polymerase chain reaction (PCR) for mRNA expression of different genes. a Expression of NLR mRNAs in FHs 74 Int cells. b Expression of the NLR mRNAs in FHs 74 Int cells compared to HPRT-1. c Positive controls for primer functionality using human acute monocytic leukemia cell line (THP-1). d Inflammasome components and caspase-cleaved ILs expressed in FHs 74 Int cells. Images shown are representatives of five independent experiments
Effects of T. gondii infection on cell morphology and cytotoxicity of FHs 74 Int cells
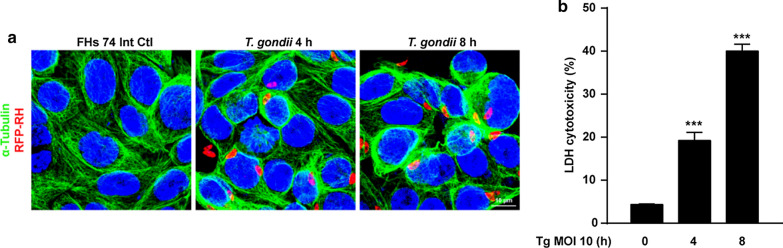
FHs 74 Int cells were incubated with T. gondii at MOI of 10 for various time periods. The integrity of the microtubule network was assessed with immunofluorescence microscopy using α-tubulin antibody and DAPI for staining cellular microtubules and DNA, respectively. As shown in Fig. 2a, the cell nucleus (blue) was wrapped with a well-developed array of hair-like microtubule networks of slim fibrous microtubules (green) in control cells. In contrast, the α-tubulin staining patterns were diffuse and disorganized in T. gondii-infected FHs 74 Int cells. The number of T. gondii-infected cells and the total number of cells were counted under a fluorescence microscope. The T. gondii infection rate significantly increased in an infection time-dependent manner (73.1% at 4 h and 89.5% at 8 h).
Fig. 2.
Effects of T. gondii infection on cell morphology and cytotoxicity of FHs 74 Int cells. FHs 74 Int cells were infected with RFP-RH T. gondii strain at a multiplicity of infection (MOI) of 10 for 0, 4, and 8 h. a Cells fixed and stained with α-tubulin (green) and nuclei stained with DAPI (blue) were observed by confocal microscopy. b Lactate dehydrogenase level in the media, which is related with cell death, was measured in FHs 74 Int cells infected with T. gondii at a MOI of 10 for 0, 4, and 8 h. The percentage of cytotoxicity was calculated. Horizontal lines in each group represent mean ± standard deviation (SD) value of at least three independent experiments. ***P < 0.001, as compared to the control group
Furthermore, to investigate T. gondii-induced cytotoxicity of FHs 74 Int cells, the cells were incubated with T. gondii at an MOI of 10 for 0, 4, and 8 h. Post-incubation, LDH assay was performed. Release of LDH significantly increased in the T. gondii-infected groups compared to that in the mock-infected control group. Cytotoxicities of FHs 74 Int cells infected with T. gondii for 0, 4, and 8 h were 4.34 ± 0.15%, 19.21 ± 1.88%, and 40.02 ± 1.57%, respectively (Fig. 2b). These data indicate that T. gondii infection induces morphological disorganization and cytotoxicity in FHs 74 Int cells in an infection time-dependent manner.
Transcriptional regulation of NLRs in FHs 74 Int cells
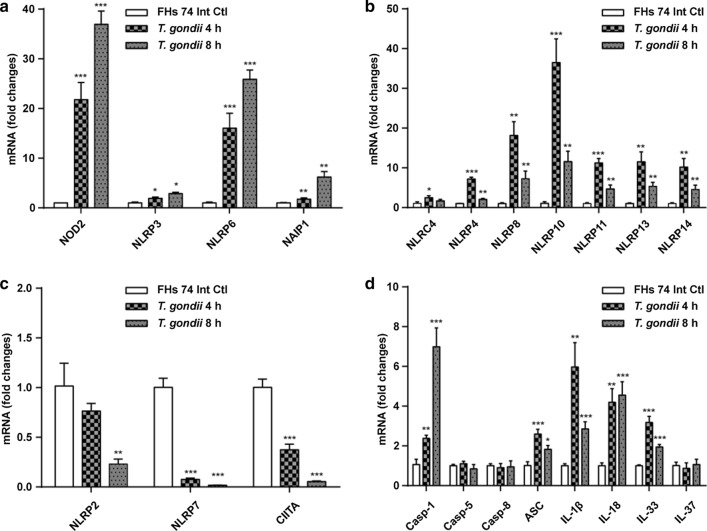
Next, we aimed to investigate the expression of the identified NLRs in response to T. gondii infection for 4 or 8 h. Real-time qRT-PCR revealed that T. gondii infection induces a significant time-dependent increase in the expression of NOD2, NLRP3, NLRP6, and NAIP1 mRNAs (Fig. 3a). Interestingly, T. gondii infection upregulated the expression of NLRC4, NLRP4, NLRP8, NLRP10, NLRP11, NLRP13, and NLRP14 mRNAs at both 4 and 8 h post-infection, but NLRP4, NLRP8, NLRP10, and NLRP11 mRNAs were significantly downregulated at 8 h post-infection compared to that at 4 h post-infection (Fig. 3b). In contrast, T. gondii infection induced a time-dependent significant decrease in the expression of NLRP2, NLRP7, and CIITA mRNAs (Fig. 3c). No significant changes in the expression of NOD1, NLRC3, NLRC5, NLRP1, NLRP9, NLRP12, and NLRX1 mRNAs were noted as a result of T. gondii infection (data not shown). Neither normal nor T. gondii-infected FHs 74 Int cells expressed NLRP5 mRNA. While T. gondii infection increased the expression of mRNAs encoding caspase-1, ASC, IL-1β, IL-18, and IL-33, it had no effect on the expression of mRNAs encoding caspase-5, caspase-8, and IL-37 (Fig. 3d). These results clearly indicate that T. gondii infection activates NLRs, but their expression patterns vary in FHs 74 Int cells.
Fig. 3.
Induction of NLRs, inflammasome components, and caspase-cleaved ILs in response to T. gondii infection in FHs 74 Int cells. FHs 74 Int cells were infected with RFP-RH T. gondii strain at a MOI of 10 for 0, 4, and 8 h. a–d Expression of the NLR mRNAs, inflammasome components, and caspase-cleaved ILs in FHs 74 Int cells. Each PCR was carried out with three parallels. *P < 0.05, **P < 0.01, ***P < 0.001 compared to control
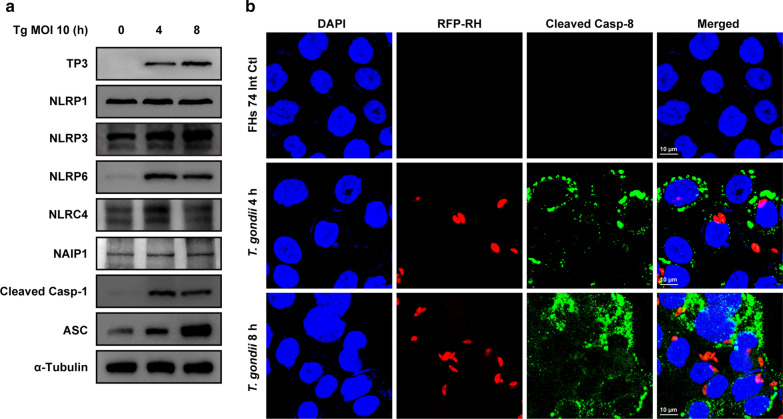
T. gondii infection induced NLRP3, NLRP6, and NLRC4 inflammasome components in FHs 74 Int cells
Until now, the most commonly studied inflammasomes in protozoan parasites were NLRP1, NLRP3, and NLRC4 [15]. Thus, we further investigated the protein levels of NLRP1, NLRP3, NLRP6, and NLRC4 inflammasome components in response to T. gondii infection. T. gondii infection time-dependently induced NLRP3 and ASC protein expression, adequately induced NLRP6 and cleaved caspase-1 expression, and moderately induced NLRC4 expression at 4 h post-infection. However, expression levels for NLRP1 and NAIP1 proteins remained unchanged in response to T. gondii infection (Fig. 4a). Confocal microscopy revealed that the expression of cleaved caspase-8 was higher in T. gondii-infected FHs 74 Int cells compared to that in mock-infected control FHs 74 Int cells (Fig. 4b). These results indicate that T. gondii infection induces NLRP3, NLRP6, and NLRC4 inflammasome activation in FHs 74 Int cells.
Fig. 4.
Expression of NLR family members, NLRP1, NLRP3, NLRP6, and NLRC4 inflammasome components in response to T. gondii infection in human small intestinal epithelial (FHs 74 Int) cells. a FHs 74 Int cells were infected with T. gondii at a MOI of 10 for 0, 4, and 8 h. Expression of NLRP1, NLRP3, NLRP6, and NLRC4 inflammasome component proteins were detected by western blot analysis; α-tubulin was used as the loading control. b FHs 74 Int cells were fixed and probed against cleaved caspase-8 (green). The cells were counterstained with 4′,6-diamidino-2-phenylindole (blue) and visualized using confocal microscopy. All data shown are representative of three independent experiments with similar results
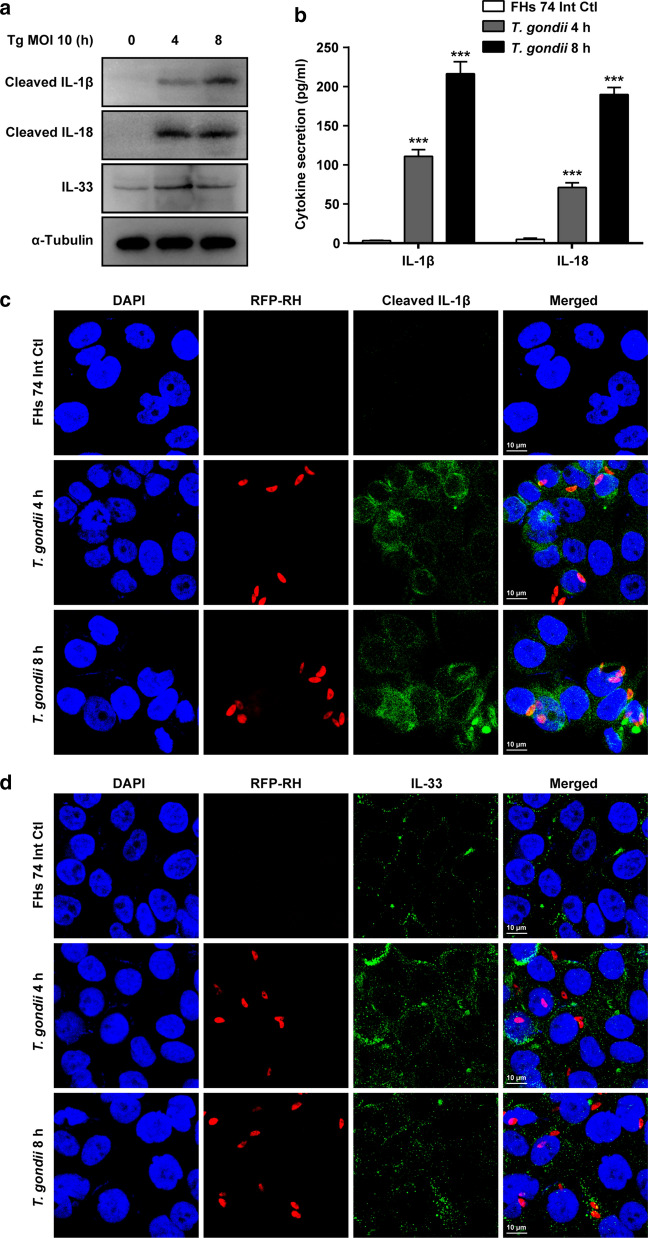
T. gondii infection upregulates ILs expression and release in FHs 74 Int cells
NLRs are a large group of cytosolic sensors whose main function is to modulate the expression of proinflammatory cytokines [8, 10–14]. Hence, we evaluated the protein expression levels of IL-1β, IL-18, IL-33, and IL-37 in T. gondii-infected FHs 74 Int cells. Western blot analysis results revealed upregulated expression of cleaved IL-1β, cleaved IL-18, and IL-33 proteins in the T. gondii-infected FHs 74 Int cell lysates (Fig. 5a). The concentrations of released IL-1β and IL-18 were measured in the T. gondii-infected FHs 74 Int cell culture medium. T. gondii infection induced a robust increase in the amount of active IL-1β and IL-18 in the culture medium (Fig. 5b). Confocal microscopy detected similar expression levels of cleaved IL-1β and IL-33. In control cells, cleaved IL-1β levels were non-detectable, while in T. gondii-infected cells, activated IL-1β and IL-33 increased significantly (Fig. 5c, d). These results indicate that T. gondii induces IL-1β, IL-18, and IL-33 production in FHs 74 Int cells.
Fig. 5.
Caspase-cleaved ILs in response to T. gondii infection in FHs 74 Int cells. a FHs 74 Int cells were infected with T. gondii at a MOI of 10 for 0, 4, and 8 h. Protein levels of ILs were detected by western blot analysis; α-tubulin was used as the loading control. b IL-1β and IL-18 protein levels in the culture medium of FHs 74 Int cells after T. gondii infection were measured by ELISA. Graphs show quantified results in pg/ml as mean ± standard error. ***P < 0.001 compared to control. c, d FHs 74 Int cells were fixed and probed against cleaved IL-1β and IL-33. The cells were counterstained with 4′,6-diamidino-2-phenylindole (blue) and visualized using confocal microscopy. All data shown are representative of three independent experiments with similar results
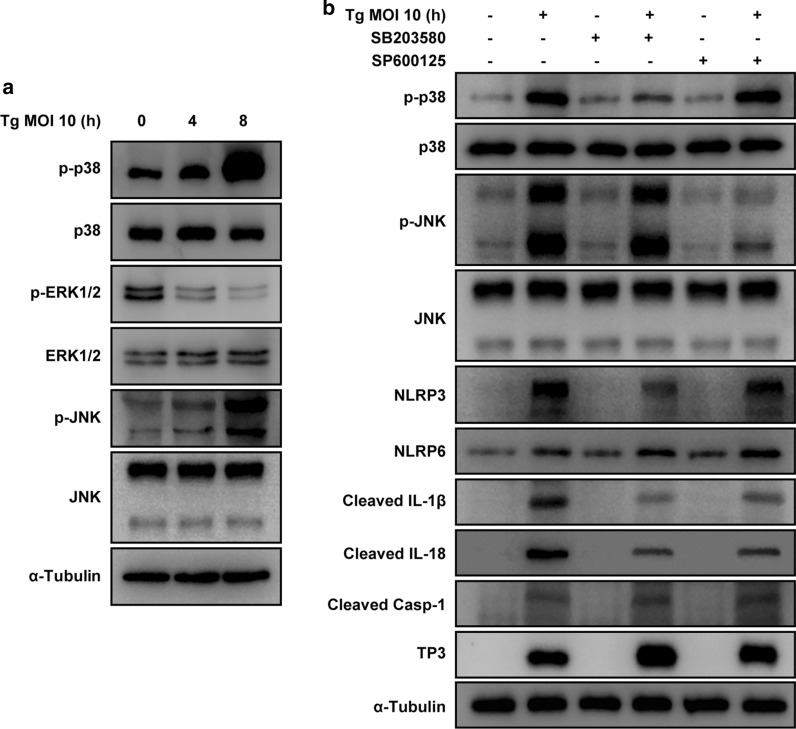
T. gondii-induced NLRP3 inflammasome activation is strongly associated with the phosphorylation of p38 MAPK
Studies have reported that the mitogen-activated protein kinase (MAPK) pathway is associated with inflammasome activation [18, 19]. Hence, we investigated the involvement of the MAPK pathway in T. gondii-induced NLRP3 and NLRP6 inflammasome activation. As shown in Fig. 6a, T. gondii infection increased the levels of phosphorylated p38 MAPK and JNK1/2; however, the level of phosphorylated ERK1/2 decreased compared with that in the control cells. There were no significant changes in the total protein levels of ERK1/2, p38 MAPK, and JNK1/2 after T. gondii infection. However, pretreatment with SB203580 (p38 inhibitor) and SP600125 (JNK inhibitor) significantly decreased the phosphorylation of p38 MAPK and JNK1/2 in T. gondii-infected cells compared with that in inhibitor-untreated T. gondii-infected cells. Interestingly, pretreatment with SB203580 significantly downregulated T. gondii infection-induced NLRP3 expression; however, the effects of SP600125 pretreatment were considerably less than those of SB203580 pretreatment. Pretreatment with SB203580 and SP600125 had no effect on the regulation of T. gondii-induced NLRP6 activation. SB203580 and SP600125 pretreatment significantly attenuated T. gondii-induced elevation in the cleaved IL-1β and cleaved IL-18 levels. Furthermore, pretreatment with SB203580 and SP600125 considerably upregulated the T. gondii antibody (TP3) level in T. gondii-infected FHs 74 Int cells compared with that in untreated T. gondii-infected cells (Fig. 6b).
Fig. 6.
Signaling pathways involved in the regulation of NLRP3 and NLRP6 inflammasome components. a FHs 74 Int cells were infected with T. gondii at a MOI of 10 for the indicated time periods. Activation of mitogen-activated protein kinase (MAPK) subsets was evaluated by western blotting. b FHs 74 Int cells were preincubated with the 30 μM of SB203580 (p38 MAPK inhibitor) and SP600125 (JNK inhibitor), respectively, for 2 h and then infected with T. gondii at a MOI of 10 for 8 h. The indicated protein levels were then evaluated by western blot. Anti-α-tubulin was used as the internal control. Similar results were obtained in three independent experiments
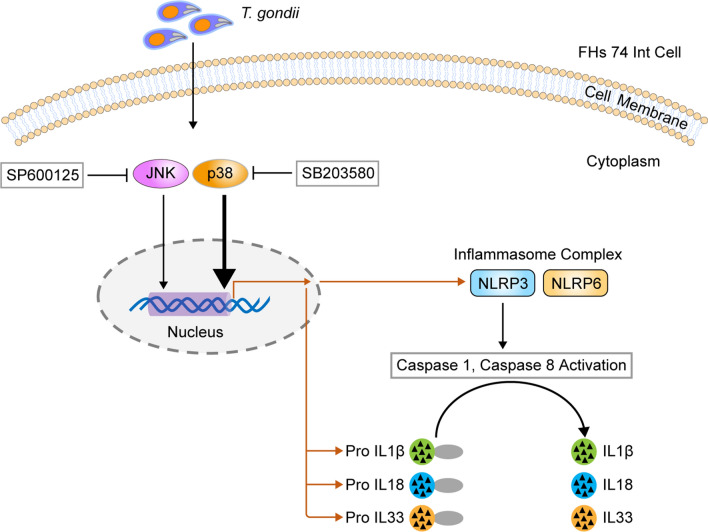
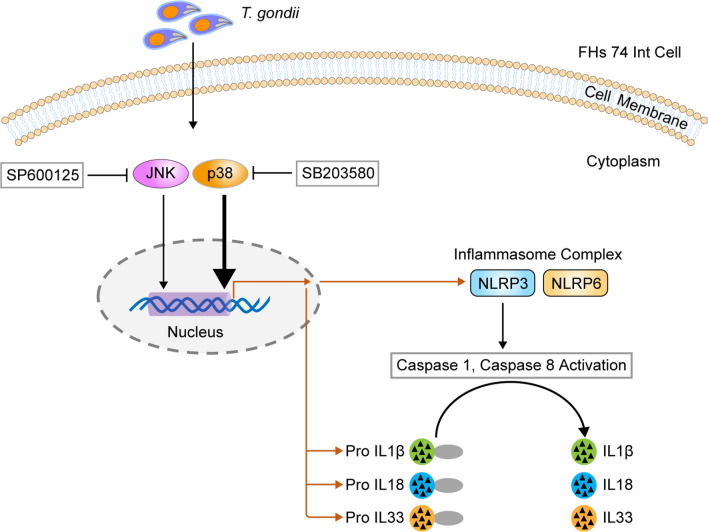
Overall, these results indicate that the p38 MAPK pathway is strongly associated with T. gondii-induced NLRP3 inflammasome production in T. gondii-infected small intestinal epithelial FHs 74 Int cells, although the p38 MAPK and JNK1/2 pathways are involved in NLRP3 activation. However, NLRP6 inflammasome activation was not related to the MAPK pathway in FHs 74 Int cells (Fig. 7).
Fig. 7.
Schematic model of NLRP3 inflammasome activation in T. gondii-infected FHs 74 Int cells. T. gondii activates the p38 MAPK pathway in small intestinal epithelial cells, subsequently upregulating protein expression and promoting the formation of the NLRP3 inflammasome. The NLRP3 inflammasome cleaves pro-IL-1β, pro-IL-18 and pro-IL-33 to become active IL-1β, IL-18, and IL-33, which ultimately induces cytotoxicity of FHs 74 Int cells
Discussion
This study revealed that members of the NLRs, inflammasome components, and caspase-cleaved ILs are expressed differently in the FHs Int 74 cells under normal and T. gondii-infected conditions; however NLRC3, NLRP5, and NLRP9 were not expressed. The most abundantly expressed NLRs were NLRP1 and NOD1. T. gondii infection induced cytotoxicity in FHs 74 Int cells in an infection time-dependent manner. In addition, the expression of NOD2, NLRP3, NLRP6, and NAIP1 mRNAs significantly increased in T. gondii-infected cells, while that of NLRP2, NLRP7, and CIITA mRNAs decreased. T. gondii infection also induced NLRP3, NLRP6, and NLRC4 inflammasome activation and significantly produced IL-1β, IL-18, and IL-33 in FHs 74 Int cells. NLRP3 inflammasome activation was strongly associated with the p38 MAPK pathway in T. gondii-infected cells; however, no relationship was revealed between NLRP6 inflammasome activation and MAPK pathway.
Expression of the NLR family as a pattern recognition receptor is cell specific; however, little was known about the regulation of NLRs and their activation mechanisms in intestinal epithelial cells. In this study, the expression patterns of the NLRs, inflammasome components, and caspase-cleaved ILs were varied in the FHs Int 74 cells under normal and T. gondii-infected conditions. Although a previous study reported the expression patterns of NLRs in cerebral endothelial cells [8], this is the first study about the regulation of whole NLRs in the human intestinal cells after T. gondii infection. NLRs can be regulated by a wide range of cellular damages, including oxidative stress and inflammatory stimuli [5, 7]. Thus, upon evaluating the cellular changes in T. gondii-infected FHs 74 Int cells by immunofluorescence and LDH assay, we observed disorganized staining patterns for α-tubulin and significantly increased release of LDH in proportion to time. These results clearly indicated that T. gondii infection induces cellular damage and cytotoxicity in FHs 74 Int cells by activation of inflammasome-related components. Similar phenomena were observed for Schistosoma mansoni infection, wherein elicited host immune responses resulted in mitochondrial damage, generation of high levels of reactive oxygen species (ROS), and activation of apoptosis through interaction with host inflammasomes [20]. In addition, Neospora caninum-induced NADPH-dependent ROS generation plays an important role in NLRP3 inflammasome activation [21].
The present investigation in T. gondii-infected FHs 74 Int cells revealed that T. gondii infection induces the expression of NLRP3, ASC, NLRP6, NLRC4, cleaved caspase-1, and cleaved caspase-8. In addition, increased production of ILs such as IL-1β, IL-18, and IL-33 was observed in T. gondii-infected FHs 74 Int cells. These findings suggest that T. gondii infection induces the activation of NLRP3, NLRP6, and NRC4 inflammasomes via the recruitment of ASC and caspases and production of proinflammatory cytokines in FHs 74 Int cells. These findings are partially consistent with those of a previous study, that is, NLRP3, ASC, caspase-1, and IL-1β were detected in T. gondii-infected mice [11]. The findings are also consistent with those of our previous study, that is, T. gondii infection of THP-1 macrophages increased the production of IL-1β and inflammasome sensors, including NLRP1, NLRP3, NLRC4, NLRP6, NLRP8, NLRP13, AIM2, and NAIP, in a time-dependent manner [12]. Compared with the findings of previous studies, the expression patterns of inflammasome components in this study were slightly different, and this can be attributed to the cell type and culture conditions.
MAPKs, highly conserved in all eukaryotes, control a variety of cellular processes, including cell differentiation, proliferation, survival, and stress responses. It has been reported that the MAPK pathway is associated with inflammasome activation [18, 20, 22]. In the present study, we evaluated the roles of the MAPK signaling pathways in NLRP3 and NLRP6 inflammasome activation in T. gondii-infected FHs 74 Int cells by pretreatment with SB203580 and SP600125, inhibitors of p38 MAPK and JNK, respectively. While SB203580 pretreatment significantly downregulated T. gondii-induced expression of NLRP3, cleaved IL-1β, and cleaved IL-18, the effects of SP600125 pretreatment were less than those of SB203580 pretreatment. However, NLRP6 activation was not affected by pretreatment with SB203580 or SP600125 in T. gondii-infected FHs 74 Int cells. These findings suggest that p38 MAPK signaling is a more important factor than JNK1/2 signaling, although both signaling pathways were involved with NLPR3 activation in T. gondii-infected FHs 74 Int cells. However, NLRP6 inflammasome activation had no correlation with the MAPK pathway.
We also compared the parasite levels in T. gondii-infected FHs 74 Int cells after pretreatment with SB203580 or SP600125 to evaluate the host cell environment for parasite proliferation. Both inhibitors apparently increased the parasite proliferation compared with that in the untreated group of T. gondii-infected FHs 74 Int cells. However, SB203580 pretreatment induced higher TP3 expression than SP600125 pretreatment in T. gondii-infected FHs 74 Int cells, and this was opposite to the expression pattern of NLRP3 inflammasome and IL-1β and IL-18 proteins after pretreatment with SB203580 and SP600125. These data suggest that NLRP3 inflammasome is important for restricting T. gondii proliferation in T. gondii-infected FHs 74 Int cells, and the p38 MAPK pathway is a more critical factor than the JNK1/2 pathway in the regulation of T. gondii-induced NLRP3 inflammasome production. Our results are consistent with those of previous studies, that is, T. gondii triggers NLRP3 or NLRP1, which leads to parasite restriction and resistance to toxoplasmosis [11, 16], and p38 MAPK is important for the regulation of NLRP1 or NLRP3 inflammasome activation and IL-1β secretion [18, 19]. However, our results were contrary to those of Wei et al. [23], who reported that the addition of SB203580 after T. gondii infection significantly inhibited T. gondii tachyzoite replication in fibroblasts. These different effects on T. gondii growth may be a result of differences in cell type, treatment method, time of inhibitor treatment, T. gondii infection time, and host cell microenvironments.
Conclusion
We demonstrated the regulation of NLRs and NLR-related inflammasome activation in T. gondii-infected human small intestinal epithelial (FHs 74 Int) cells. T. gondii infection induced the expression of NLRs, inflammasome components, and caspase-cleaved ILs in the FHs Int 74 cells, but their expression patterns were varied. NLRP3 inflammasome activation was strongly associated with the p38 MAPK pathway; however, NLRP6 inflammasome activation had no correlation with the MAPK pathway. We believe that the study findings will contribute to the understanding of mucosal and innate immune responses induced by NLRs and inflammasomes in T. gondii-infected FHs Int 74 cells.
Acknowledgements
The authors thank In-Wook Choi (Chungnam National University) for her help in performing the experiments.
Abbreviations
- BSA
Bovine serum albumin
- DAPI
4′,6-Diamidino-2-phenylindole
- ELISA
Enzyme-linked immunosorbent assay
- FBS
Fetal bovine serum
- HPRT-1
Hypoxanthine phosphoribosyltransferase 1
- ILs
Interleukins
- LDH
Lactate dehydrogenase
- MAPK
Mitogen-activated protein kinase
- MOI
Multiplicities of infection
- NLRs
Nucleotide-binding oligomerization domain (NOD)-like receptors
- NOD
Nucleotide-binding oligomerization domain
- PBS
Phosphate buffer saline
- PCR
Polymerase chain reaction
- qPCR
Quantitative polymerase chain reaction
- SD
Standard deviation
Authors' contributions
JQC, FFG, WW, JHQ and YHL designed and conceived the experiments. JQC, FFG, WW, CL, ZP, JS, HW, CH and SHL carried out the experiments. JQC, FFG, WW, JHQ and YHL performed the data analysis. JQC, JHQ, YHL and WW drafted and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2019R1A2C1088346) at Chungnam National University, and the National Natural Science Foundation of China (81771612 and 81971389), the Natural Science Foundation of Guangdong Province (2019A1515011888 and 2019A1515011715), and the Characteristic Innovation Projects of Guangdong Universities (2018KTSCX081, 2018KTSCX079).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jia-Qi Chu, Fei Fei Gao, and Weiyun Wu contributed equally to this work
Contributor Information
Juan-Hua Quan, Email: quanjuanhua@gdmu.edu.cn.
Young-Ha Lee, Email: yhalee@cnu.ac.kr.
References
- 1.Robert-Gangneux F, Dardé ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. 2012;25:264–296. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harker K, Ueno N, Lodoen M. Toxoplasma gondii dissemination: a parasite's journey through the infected host. Parasit Immunol. 2015;37:141–149. doi: 10.1111/pim.12163. [DOI] [PubMed] [Google Scholar]
- 3.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 4.Pifer R, Yarovinsky F. Innate responses to Toxoplasma gondii in mice and humans. Trends Parasitol. 2011;27:388–393. doi: 10.1016/j.pt.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olive C. Pattern recognition receptors: sentinels in innate immunity and targets of new vaccine adjuvants. Expert Rev Vaccines. 2012;11:237–256. doi: 10.1586/erv.11.189. [DOI] [PubMed] [Google Scholar]
- 6.Yarovinsky F. Toll-like receptors and their role in host resistance to Toxoplasma gondii. Immunol Letters. 2008;119:17–21. doi: 10.1016/j.imlet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Franchi L, Warner N, Viani K, Nuñez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagyőszi P, Nyúl-Tóth Á, Fazakas C, Wilhelm I, Kozma M, Molnár J, et al. Regulation of NOD-like receptors and inflammasome activation in cerebral endothelial cells. J Neurochem. 2015;135:551–564. doi: 10.1111/jnc.13197. [DOI] [PubMed] [Google Scholar]
- 9.Shaw MH, Reimer T, Sanchez-Valdepenas C, Warner N, Kim YG, Fresno M, et al. T cell-intrinsic role of Nod2 in promoting type 1 immunity to Toxoplasma gondii. Nat Immunol. 2009;10:1267–1274. doi: 10.1038/ni.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witola WH, Mui E, Hargrave A, Liu S, Hypolite M, Montpetit A, et al. NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect Immun. 2011;79:756–766. doi: 10.1128/IAI.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorfu G, Cirelli KM, Melo MB, Mayer-Barber K, Crown D, Koller BH, et al. Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. mBio. 2014;5:e01117–e1213. doi: 10.1128/mBio.01117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu JQ, Shi G, Fan YM, Choi IW, Cha GH, Zhou Y, et al. Production of IL-1beta and inflammasome with up-regulated expressions of NOD-like receptor related genes in Toxoplasma gondii-infected THP-1 macrophages. Korean J Parasitol. 2016;54:711–717. doi: 10.3347/kjp.2016.54.6.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 14.Vladimer GI, Weng D, Paquette SWM, Vanaja SK, Rathinam VA, Aune MH, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunit. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clay GM, Sutterwala FS, Wilson ME. NLR proteins and parasitic disease. Immunol Res. 2014;59:142–152. doi: 10.1007/s12026-014-8544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan JH, Huang R, Wang Z, Huang S, Choi I-W, Zhou Y, et al. P2X7 receptor mediates NLRP3-dependent IL-1β secretion and parasite proliferation in Toxoplasma gondii-infected human small intestinal epithelial cells. Parasit Vectors. 2018;11:1–10. doi: 10.1186/s13071-017-2573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Carvalho RVH, Zamboni DS. Inflammasome activation in response to intracellular protozoan parasites. Trends Parasitol. 2020;36:459–472. doi: 10.1016/j.pt.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z, Li H, Tian S, Yi W, Zhou Y, Yang H, et al. Critical roles of NLRP3 inflammasome in IL-1beta secretion induced by Corynebacterium pseudotuberculosis in vitro. Mol Immunol. 2019;116:11–17. doi: 10.1016/j.molimm.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Fenini G, Grossi S, Gehrke S, Beer H-D, Satoh TK, Contassot E, et al. The p38 mitogen-activated protein kinase critically regulates human keratinocyte inflammasome activation. J Invest Dermatol. 2018;138:1380–1390. doi: 10.1016/j.jid.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Chen TTW, Cheng PC, Chang KC, Cao JP, Feng JL, Chen CC, et al. Activation of the NLRP3 and AIM2 inflammasomes in a mouse model of Schistosoma mansoni infection. J Helminthol. 2019;94:e72. doi: 10.1017/S0022149X19000622. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Wang XC, Gong PT, Zhang N, Zhang X, Li S, et al. ROS-mediated NLRP3 inflammasome activation participates in the response against Neospora caninum infection. Parasit Vectors. 2020;13:449. doi: 10.1186/s13071-020-04331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, Wang Z, Gu Y, Li Z, Li Z, Wei F, et al. Toxoplasma gondii mitogen-activated protein kinases are associated with inflammasome activation in infected mice. Microbes Infect. 2016;18:696–700. doi: 10.1016/j.micinf.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Wei S, Daniel BJ, Brumlik MJ, Burow ME, Zou W, Khan IA, et al. Drugs designed to inhibit human p38 mitogen-activated protein kinase activation treat Toxoplasma gondii and Encephalitozoon cuniculi infection. Antimicrob Agents Chemother. 2007;51:4324–4328. doi: 10.1128/AAC.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.