Mast cells (MCs) comprise two lineages that differ in tissue localization and ontologic origin. Derakhshan et al. define the transcriptomes of these lineages during airway inflammation, identifying distinct programs associated with each lineage and a role for TGF-β signaling in inducible MC development.
Abstract
Murine mast cells (MCs) contain two lineages: inducible bone marrow–derived mucosal MCs (MMCs) and constitutive embryonic-derived connective tissue MCs (CTMCs). Here, we use RNA sequencing, flow cytometry, and genetic deletion in two allergic lung inflammation models to define these two lineages. We found that inducible MCs, marked by β7 integrin expression, are highly distinct from airway CTMCs at rest and during inflammation and unaffected by targeted CTMC deletion. β7High MCs expand and mature during lung inflammation as part of a TGF-β–inducible transcriptional program that includes the MMC-associated proteases Mcpt1 and Mcpt2, the basophil-associated protease Mcpt8, granule components, and the epithelial-binding αE integrin. In vitro studies using bone marrow–derived MCs (BMMCs) identified a requirement for SCF in this TGF-β–mediated development and found that epithelial cells directly elicit TGF-β–dependent BMMC up-regulation of mMCP-1 and αE integrin. Thus, our findings characterize the expansion of a distinct inducible MC subset in C57BL/6 mice and highlight the potential for epithelium to direct MMC development.
Introduction
Mast cells (MCs) are tissue-resident granulocytes that fall into two distinct lineages. In mice, fetal-derived progenitors expressing β7 integrin seed virtually all peripheral tissues in multiple waves during the neonatal period (Gentek et al., 2018; Li et al., 2018) and mature into long-lived connective tissue MCs (CTMCs) with distinct transcriptional signatures modulated by their tissue of residence (Dwyer et al., 2016). Inducible mucosal MCs (MMCs) arise from the bone marrow–derived MC progenitors (MCps) that also express β7 integrin and are recruited to mucosal tissues in response to T cell–mediated type 2 inflammation (Abonia et al., 2006; Bankova et al., 2015; Gurish et al., 2001; Li et al., 2018). Mouse studies using genetic deletion of CTMCs have established their pathobiologic role in IgE-mediated ear swelling and monosodium urate–induced ankle swelling, both protective and pathogenic roles in contact hypersensitivity (Dudeck et al., 2011; Lilla et al., 2011; Otsuka et al., 2011; Reber et al., 2014), and a protective role for their proteases in detoxifying xenobiotic venoms (Tsai et al., 2015). MCs also play a central role in the development of food allergy (Leyva-Castillo et al., 2019), participate in helminth expulsion (Crowle, 1983; Knight et al., 2000), and mediate many features of experimental allergic asthma (Yu et al., 2006). While these latter models are all characterized by expansion of inducible MMCs, functional distinctions between constitutive and inducible MCs are lacking.
MMCs and CTMCs have been primarily studied histologically, identifying specific granule structure and protease expression profiles associated with each. CTMC granules contain both heparin sulfate and chondroitin sulfate and express the protease carboxypeptidase A3 (CPA3) along with murine MC protease (mMCP)-4, -5, and -6. Following maturation, induced MMCs in the intestine and airway develop granules that contain chondroitin sulfate but lack heparin and express the proteases mMCP-1 and -2 (Friend et al., 1996; Xing et al., 2011). Soluble mMCP-1 can elicit tracheal ring contraction through direct effects on the epithelium, and mice lacking mMCP-1 show impaired helminth clearance (Knight et al., 2000; Wastling et al., 1998), while in vitro studies indicate an antimicrobial role for mMCP-2 (Orinska et al., 2007). In vitro, a broad range of signals have been shown to induce these proteases in bone marrow–derived MCs (BMMCs), including IL-9 (Eklund et al., 1993; Miller et al., 1999), IL-10 (Ghildyal et al., 1992a, 1992b), TGF-β (Miller et al., 1999), and notch signaling (Honjo et al., 2017). The degree to which these signals influence MC development in vivo remains unclear.
In addition to proteases, MCs contain the biosynthetic enzymes required for rapid synthesis of proinflammatory mediators, including the lipid mediators prostaglandin D2 (PGD2), leukotriene (LT) B4 (LTB4), and LTC4 (Razin et al., 1984), and the neurotransmitters histamine and serotonin. Rat CTMCs release increased histamine but decreased LTB4 and LTC4 in response to activation relative to inducible MMCs (Heavey et al., 1988), while similar studies have not been conducted in mice. MCs are also biologically relevant sources of cytokines, with conditional deletion studies establishing CTMCs as important sources of TNF-α (Dudeck et al., 2015) and IL-10 (Reber et al., 2017), while inducible MMCs influence intestinal inflammation through production of IL-9 (Chen et al., 2015; Leyva-Castillo et al., 2019). These inflammatory mediators can be released in response to a range of exogenous signals, including cross-linking of the high-affinity IgE receptor FcɛR1α, innate signals such as the cytokine IL-33 or the complement cascade products C3a and C5a (Mukai et al., 2018), or neuron-derived signals such as substance P, recently shown to act through the receptor Mrgprb2 (Meixiong et al., 2019). However, the full extent to which the two MC lineages may differ in their capacity to generate inflammatory mediators or respond to exogenous signals is not yet understood.
Here, we use flow cytometry to identify two MC populations within the lungs of C57BL/6 mice that differ based on β7 integrin expression both at rest and in two models of allergic airway inflammation. Constitutive CTMCs, characterized by low β7 expression and dense granularity (β7Low), remain static during inflammation and are ablated in mice with mMCP-5–directed diphtheria toxin α (DTA) expression (MCpt5/DTA). Inducible MCs, characterized by high β7 expression (β7High), dynamically expand in response to pulmonary type 2 inflammation and are retained in MCpt5/DTA mice. Through sequencing analysis, we find that these two MC subsets are highly distinct transcriptionally, expressing divergent patterns of genes encoding proteases, granule components, and cell surface receptors. β7Low MCs are enriched for CTMC signature genes (Dwyer et al., 2016), including members of the mas-related G protein–coupled receptor (MRGPR) family and the canonical CTMC-associated proteases mMCP-4 (Mcpt4), mMCP-5 (Cma1), and mMCP-6 (Tpsb2). β7High MCs up-regulate a distinct transcriptional program that includes Mcpt8, a transcript previously described as basophil specific and used to target basophils in several mouse strains (Sullivan et al., 2011; Wada et al., 2010), the canonical MMC protease Mcpt1 (encoding mMCP-1), and Itgae, encoding epithelial-associated αE integrin. This up-regulated program includes canonical TGF-β target genes (Skil and Ldlrad4) and can be induced in vitro in BMMCs by TGF-β in a stem cell factor (SCF)-dependent manner. We further use an existing single-cell RNA sequencing (scRNA-seq) dataset (Travaglini et al., 2020) to provide evidence for a similar TGF-β “fingerprint” in human distal lung MCs and establish the capacity for primary epithelial cells to directly elicit TGF-β–dependent mMCP-1 synthesis in BMMCs following coculture. Thus, our findings establish that the distinctions between constitutive CTMCs and inducible MMCs go far beyond proteases and identify a key signaling axis that drives inducible MMC development in vivo.
Results
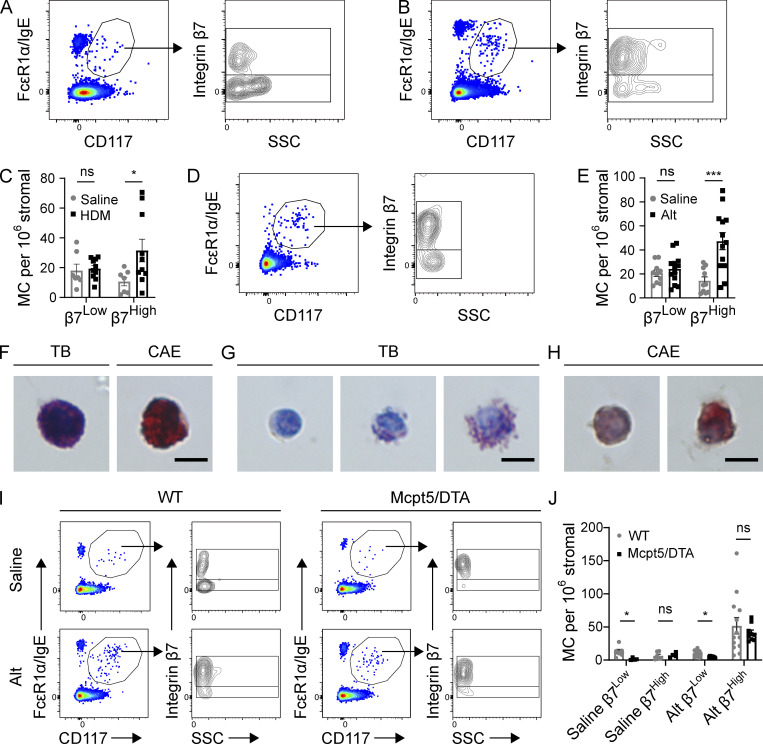
Flow cytometric assessment of pulmonary MCs identified two distinct subpopulations of MCs in naive C57BL/6 mice: one characterized by high β7 integrin expression and low side scatter (SSC; β7High) and one with low β7 integrin expression and higher SSC (β7Low; Fig. 1 A and Fig. S1). Repeated challenges with house dust mite extract (HDM) over 3 wk elicited a substantial overall increase in total airway MC concentration, driven entirely by expansion of β7High MCs (Fig. 1 B). β7High MC concentration increased fourfold in response to inflammation, in relation to the stromal (CD45−) compartment, while β7Low MC concentration remained unchanged (Fig. 1 C). Similarly, expansion of the β7High MC compartment was observed following sensitization and challenge with culture filtrate from the mold Alternaria alternata (Alt; Fig. 1, D and E), indicating that diverse type 2 inflammatory signals could induce β7High MC expansion. Upon isolation from the lungs of HDM-challenged mice, β7Low MCs were morphologically uniform cells with dense metachromatic granules visible following toluidine blue (TB) staining and a granular staining pattern with the protease stain chloroacetate esterase (CAE; Fig. 1 F). β7High MCs were more heterogeneous in morphology, ranging from agranular to granular (Fig. 1 G), although none were as heavily granulated as β7Low MCs. β7High MCs were similarly heterogeneous for CAE reactivity with a cytoplasmic staining pattern (Fig. 1 H), in contrast to the granular staining pattern observed in β7Low MCs (Fig. 1 F). To determine whether β7High MCs were a distinct lineage from the β7Low MCs, we assessed lung MCs in Alt-sensitized and -challenged WT and MCpt5/DTA mice, in which cells expressing the CTMC protease mMCP-5, encoded by Cma1, are deleted (Dudeck et al., 2011). Naive MCpt5/DTA mice lacked β7Low CTMCs but contained a small β7High MC compartment. Following antigen sensitization and challenge, WT and MCpt5/DTA mice exhibited similar expansion of β7High MCs, while β7Low MCs were observed only in WT control mice (Fig. 1, I and J). Together, these findings identify the β7High MCs as a dynamic, inducible pool of largely hypogranular MCs, and β7Low MCs as a constitutive population of heavily granulated MCs.
Figure 1.
Aeroallergen challenges induce the expansion of β7High MCs in C57BL/6 mice. (A) Representative flow plots showing airway MCs in naive mice, pregated on CD45+ Lin− (CD4, CD8, CD19, CD11b, CD11c) cells (left), further subdivided by β7 integrin and SSC (right). Flow plots are representative of three independent experiments. (B) Representative flow plots showing airway MCs from the lungs of mice challenged twice per week for 3 wk with 3 µg HDM extract and sacrificed 24 h after the final challenge, pregated on CD45+ Lin− cells. Flow plots are representative of three independent experiments. (C) Quantification of β7Low and β7High MCs in mice challenged with saline ± HDM as in Fig. 1 B. Data from three independent experiments, n = 6–9 mice per group. *, P < 0.05 (t test); ns, not significant. (D) Representative flow plot showing composition of the MC compartment in the lungs of mice sensitized with 30 µg Alt on day 0, challenged twice with 3 µg Alt on days 9 and 10, and sacrificed 48 h after the final challenge. Flow plots are representative of three independent experiments. (E) Quantification of β7Low and β7High MCs in mice sensitized with either saline or 30 µg Alt and challenged twice with 3 µg Alt, as in Fig. 1 D. Data from three independent experiments, n = 6–9 mice per group. ***, P < 0.001 (t test). (F) Representative images of TB and CAE staining on β7Low airway MCs sorted from C57BL/6 mice following HDM challenge. Scale bar = 5 µm. (G) Representative images of TB staining for β7High airway MCs sorted from C57BL/6 mice following HDM challenge (multiple images shown to display heterogeneity). Scale bar = 5 µm. (H) Representative images of CAE staining for β7High MCs sorted from C57BL/6 mice following HDM challenge (multiple images shown to display heterogeneity). Scale bar = 5 µm. (I) Representative flow plots of airway MCs in littermate WT control or mMCP-5/DTA mice sensitized with either saline or 30 µg Alt, challenged twice with 3 µg Alt, and sacrificed 48 h after the final challenge. (J) Quantification of airway β7High and β7Low MCs in WT and Mcpt5/DTA mice relative to stromal cells in mice sensitized with either saline or 30 µg Alt and challenged twice with 3 µg Alt. Data from three independent experiments, n = 4–12 mice per group. *, P < 0.05 (t test).
Figure S1.
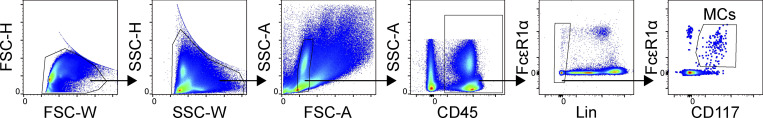
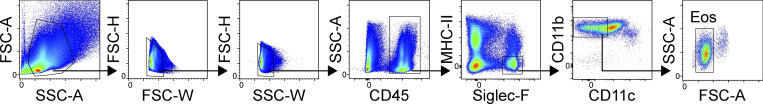
Gating strategy used for the identification of MCs within the lower airways of C57BL/6 mice. After doublet exclusion, MCs were identified as CD45 positive, Lineage (CD4, CD8, CD19, CD11b, CD11c) negative, FcεR1α positive, and CD117 positive. FSC, forward scatter; SSC, side scatter.
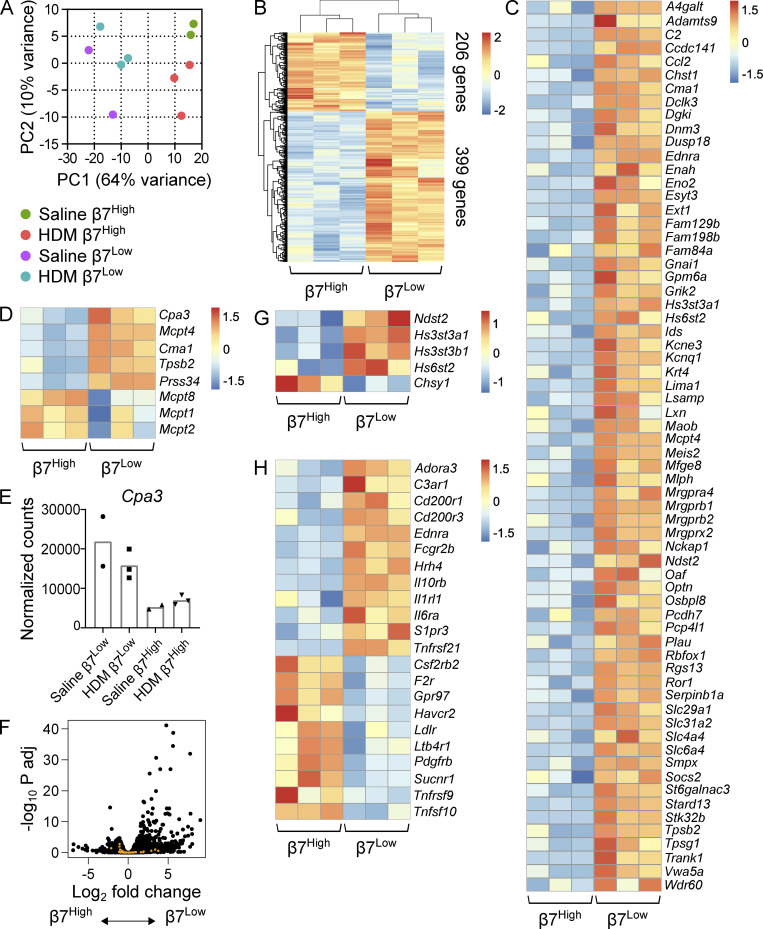
To understand the transcriptional programs underlying the histochemical and dynamic distinction between inducible β7High MCs and constitutive β7Low MCs, each population was isolated by flow sorting for RNA-seq from the lower airways of saline- and HDM-challenged mice. Principal component analysis emphasized the distinction of the two MC populations, with PC1 (64% of variance) strongly separating replicates based on population of origin (Fig. 2 A). Transcriptional analysis of β7High and β7Low MCs from HDM-challenged mice highlighted their distinction, with 605 transcripts differentially expressed (false discovery rate [FDR] < 0.1; Fig. 2 B and Table S1). Among the differentially expressed transcripts enriched in the β7Low MC subset were 66 genes from a previously identified 128-gene transcriptional signature shared by CTMCs across five murine tissues (Dwyer et al., 2016), including a number of MRGPR family members (Mrgpra4, Mrgprb1, Mrgprb2, and Mrpgrx2; Fig. 2 C). As expected, the two populations also expressed distinct protease profiles. β7Low MCs were significantly enriched for expression of transcript encoding CPA3 (Cpa3) and mMCp-4 (Mcpt4), -5 (Cma1), -6 (Tpsb2), and -11 (Prss34); however, β7High MCs still expressed CPA3 at approximately one third the level observed in β7Low MCs (Fig. 2, D and E). Unexpectedly, β7High MCs were significantly enriched for transcript encoding the basophil-associated protease mMCP-8 (Mcpt8; Fig. 2 D; Sullivan et al., 2011; Wada et al., 2010). β7High MCs showed significant enrichment for only two additional transcripts (Gpr97 and Ifitm1) from a previously identified 66-gene basophil signature (Fig. 2 F; Dwyer et al., 2016), suggesting that the observed Mcpt8 expression was not the result of contaminating basophils. β7High MCs also showed a trend toward enhanced expression of transcripts encoding mMCP-1 (Mcpt1) and -2 (Mcpt2). The two subsets also expressed distinct sets of glycose-aminoglycan biosynthetic enzymes, with β7Low MCs enriched for transcripts encoding heparin sulfate biosynthetic enzymes (Ndst2, Hs3st3a1, Hs3st3b1, and Hs6st2), while β7High MCs showed a significant increase in Chsy1 expression, encoding chondroitin sulfate synthase 1 (Fig. 2 G). Together, these findings confirmed a CTMC phenotype for β7Low MCs, including heparin sulfate-containing granules and CTMC-associated proteases, while inducible β7High MCs had a more MMC-like transcriptional phenotype, characterized by chondroitin sulfate and the MMC-associated proteases Mcpt1 and Mcpt2.
Figure 2.
β7High MCs are transcriptionally distinct from constitutive β7Low MCs. (A) Principal component plot of β7Low and β7High MCs sorted from the combined lung and trachea of mice challenged with either saline or 3 µg HDM extract twice per week for 3 wk and sacrificed 24 h after the final challenge. Percentages indicate the level of variance described by each component. Colored circles denote biological replicates (two to three per group), with each replicate consisting of cells pooled from three mice to minimize individual variability. Each replicate is the result of an independent experiment. (B) Heatmap showing all transcripts differentially expressed between β7Low and β7High MCs following repeated HDM challenge (FDR < 0.1, DESeq2). Each replicate is pooled from three mice and derived from an independent experiment; scale bar indicates z score. (C) Heatmap showing transcripts found in the ImmGen CTMC signature that were differentially expressed between β7Low and β7High MCs following HDM challenge (FDR < 0.1, DESeq2). Each replicate is pooled from three mice and derived from an independent experiment; scale bar indicates z score. (D) Heatmap showing differentially expressed MCPs in sorted β7Low and β7High subsets following HDM challenge (FDR < 0.1 with the exception of Mcpt1 and Mcpt2, DESeq2). Each replicate is pooled from three mice and derived from an independent experiment; scale bar indicates z score. (E) Normalized counts indicating Cpa3 expression in constitutive β7Low MCs and inducible β7High MCs. (F) Volcano plot of differential gene expression between β7High and β7Low MCs (black) with overlay of ImmGen basophil signature genes (orange). (G) Heatmap representation of differentially expressed glycose-aminoglycan biosynthetic enzymes in sorted β7Low and β7High subsets following HDM challenge (FDR < 0.1). Each replicate is pooled from three mice and derived from an independent experiment. (H) Heatmap indicating differentially expressed cell surface receptors in sorted β7Low and β7High subsets following HDM challenge (FDR < 0.1, DESeq2). Each replicate is pooled from three mice and derived from an independent experiment; scale bar indicates z score.
β7High and β7Low MCs further differed in their expression of transcripts encoding receptors. In addition to enhanced expression of MRGPR receptors (Fig. 2 C), β7Low MCs were enriched for transcripts encoding receptors for the C3a complement subunit and the cytokines IL-6, IL-10, and IL-33. β7High MCs were enriched for transcripts encoding receptors for LTB4 (Ltb4r1), the growth factors GM-CSF and PDGF subunit B (Csf2ra, Pdgfrb), and succinate (Sucnr1; Fig. 2 H), a compound that was previously shown to enhance IgG1-mediated release of histamine and cysteinyl LTs in guinea pig lungs (Austen and Brocklehurst, 1961). Interestingly, no significant differences were observed between the two populations in transcripts encoding cytokines or the lipid mediator biosynthetic enzymes directing synthesis of LTC4 (Alox5, Alox5ap, and Ltc4s) or PGD2 (Ptgs1, Ptgs2, and Hpgds; not depicted).
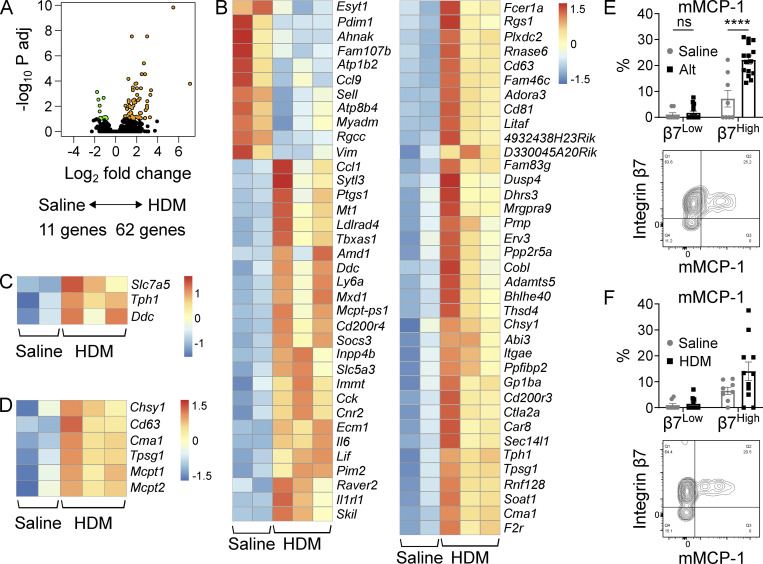
As β7High MCs exhibited a dynamic response to inflammation, we evaluated transcriptional changes elicited in this population during inflammation. In contrast to β7Low MCs, which showed few transcriptional changes in response to HDM challenge (Table S2), the β7High MC subset underwent more substantial changes in response to inflammation. β7High MCs from HDM-treated mice up-regulated 62 genes while down-regulating 9 (FDR < 0.1; Fig. 3 A and Table S2). β7High MCs up-regulated transcripts encoding IL-6 family cytokines (Il6 and Lif), chemokines (Ccl1), the lipid mediator biosynthetic enzyme cox-1 (Ptgs1), and a panel of activating receptors (Adora3, Fcer1a, Il1rl1, Cd200R3, Cd200r4, and F2r; Fig. 3 B). β7High MCs also significantly up-regulated transcripts encoding the two critical serotonin biosynthetic enzymes, Tph1 and Ddc, with a trend toward up-regulation of the L-tryptophan transporter Slc7a5 (Fig. 3 C). β7High MCs showed significant up-regulation of transcript encoding chondroitin sulfate synthase (Chsy1), the granule-associated tetraspanin CD63 (Cd63), the MCPs mMCP-5 (Cma1), and γ tryptase (Tpsg1; Fig. 3 D). The MMC-associated proteases mMCP-1 and mMCP-2 (encoded by Mcpt1 and Mcpt2, respectively) were also significantly up-regulated, but did not clear an FDR threshold of 0.1. However, this up-regulation was biologically meaningful, as following challenge with Alt, mMCP-1 protein expression was significantly up-regulated in β7High MCs, with a similar trend following repeated HDM challenges (Fig. 3, E and F). Minimal mMCP-1 protein expression was observed in β7Low MCs under either condition, identifying the expanded β7High MC pool as the predominant source of mMCP-1 within the inflamed lung.
Figure 3.
The β7High MC transcriptome dynamically changes in response to allergic inflammation. (A) Volcano plot indicating differentially expressed transcripts (FDR < 0.1) between β7High MCs isolated from the lungs of mice challenged twice per week for 3 wk with saline (n = 2) or HDM (n = 3). Transcripts highlighted in orange (62 genes) are enriched in MCs from HDM-challenged mice, and transcripts highlighted in green (11 genes) are enriched in MCs from saline-challenged mice. Adj, adjusted. (B) Heatmap showing differentially expressed transcripts between β7High MCs isolated from the lungs of mice challenged with saline or HDM (FDR < 0.1, DESeq2). Each replicate is pooled from three mice and derived from an independent experiment; scale bar indicates z score. (C) Heatmap showing differentially expressed serotonin biosynthetic components between β7High MCs isolated from the lungs of mice challenged with saline or HDM (FDR < 0.1, DESeq2). Each replicate is pooled from three mice and derived from an independent experiment; scale bar indicates z score. (D) Heatmap showing differentially expressed granule components and proteases between β7High MCs isolated from the lungs of mice challenged with saline or HDM (FDR < 0.1 with the exception of Mcpt1 and Mcpt2, DESeq2). Each replicate is pooled from three mice and derived from an independent experiment; scale bar indicates z score. (E) Quantification of mMCP-1 expression in β7Low and β7High airway MCs following sensitization with saline or 30 µg Alt on day 0 and challenge with 30 µg Alt on days 9 and 10, sacrificed 48 h after the final challenge (top), with representative flow plot (bottom). Data from three independent experiments, n = 8–15 mice per group. ****, P < 0.0001 (t test); ns, not significant. (F) Quantification of mMCP-1 expression in β7Low and β7High airway MCs following challenge with saline or 10 µg HDM twice per week for 3 wk (top) with representative flow plot (bottom). Data from four independent experiments, n = 8–11 mice per group. ns, not significant (t test).
mMCP-1 protein up-regulation was also observed among the expanded β7High MCs in Mcpt5/DTA mice at levels similar to those observed in littermate controls (Fig. 4, A and B), indicating that both development and expansion of β7High MCs were intact in this strain. Deletion of the constitutive CTMC compartment had no apparent impact on overall lung inflammation, as assessed by CD45+ cell influx, eosinophil concentration within the CD45+ cell compartment, or serum IgE increases in response to Alt sensitization or challenge (Fig. 4 and Fig. S2). These findings indicate that within the lung, Mcpt5 (Cma1)-directed cre recombinase is likely expressed only in CTMCs, while also indicating that these CTMCs do not influence pulmonary or systemic type 2 inflammation following aeroallergen challenge.
Figure 4.
CTMC deletion does not affect β7High MC development or type 2 inflammatory state. (A) Representative flow plots of intracellular mMCP-1 expression in airway MCs in WT (left) versus littermate Mcpt5/DTA (right) mice following sensitization with saline or 30 µg Alt on day 0 and challenge with 3 µg Alt on days 9 and 10, sacrificed 48 h after the final challenge. Data are representative of three independent experiments. (B) Quantification of β7High MC mMCP-1 expression in WT versus Mcpt5/DTA mice sensitized and challenged with Alt as in Fig. 4 A. Data from three independent experiments, n = 7–8 mice per group. No significant difference was observed between genotypes. ns, not significant (t test). (C) Flow cytometric quantification of lung CD45+ cell concentration as a percentage of total cells (left), lung eosinophilia as a percentage of total CD45+ cells (center), and serum IgE concentration (right) in WT versus Mcpt5/DTA mice with saline or 30 µg Alt on day 0 and challenge with 3 µg Alt on days 9 and 10, sacrificed 48 h after the final challenge. Data from two (eosinophils) or three (CD45+, IgE) independent experiments, n = 3–15 mice per group. ns, not significant (t test).
Figure S2.
Gating strategy used for the identification of eosinophils within the lower airways of WT and MCpt5/DTA mice during allergic lung inflammation. Eos, eosinophils; FSC, forward scatter; SSC, side scatter.
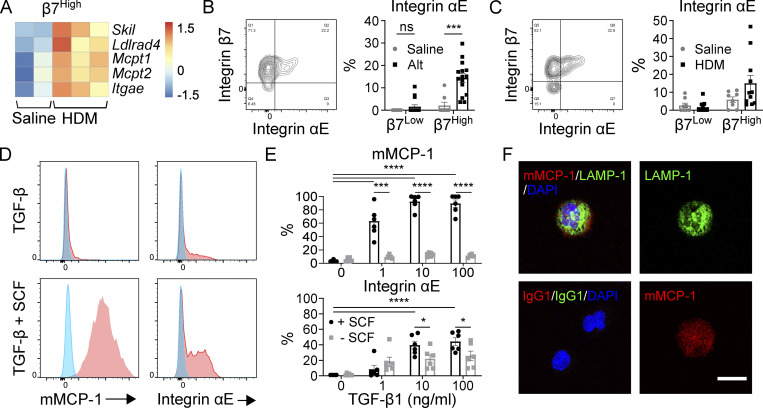
Two of the transcripts significantly up-regulated in β7High MCs following repeated HDM challenges (Skil and Ldlrad4; Fig. 5 A) are direct targets of Smad2/3 (Deheuninck and Luo, 2009; Nakano et al., 2014). Three other up-regulated transcripts, Mcpt1, Mcpt2, and Itgae, were previously identified as downstream targets of TGF-β signaling (Smith et al., 1994; Wright et al., 2002), with Mcpt1 and Mcpt2 being direct targets of Smad2 and Smad4 (Kasakura et al., 2020). Like mMCP-1, αE integrin protein (encoded by Itgae) was significantly up-regulated in β7High MCs following Alt challenge and showed a trend toward up-regulation after repeated HDM challenges (Fig. 5, B and C). Thus, we hypothesized that TGF-β signaling could play a role in regulating the induced effector program in β7High MCs during type 2 inflammation in vivo. To test this hypothesis, BMMCs grown in IL-3 were stimulated with TGF-β1 in vitro. While TGF-β1 stimulus in isolation induced only modest up-regulation of mMCP-1 and αE integrin, and SCF did not influence either protein, TGF-β1 and SCF acted synergistically to greatly enhance expression of both mMCP-1 and αE integrin in a TGF-β1 dose-dependent manner (Fig. 5, D and E). Confocal microscopy of TGF-β1– and SCF-stimulated BMMCs indicated that while some mMCP-1 colocalized with lysosome-associated membrane protein 1 (LAMP-1), a protein associated with both secretory granules and lysosomes, mMCP-1 staining was also observed in regions of the cell lacking LAMP-1 staining, suggesting extragranular localization of CAE-reactive mMCP-1 (Fig. 5 F).
Figure 5.
The β7High MC maturation phenotype can be elicited in vitro through SCF-dependent TGF-β stimulus of BMMCs. (A) Heatmap of TGF-β target genes up-regulated in β7High MCs from the lungs of saline-challenged versus HDM-challenged mice. Skil, Ldlrad4, and Itgae are significantly differentially expressed with an FDR < 0.1; all transcripts are differentially expressed with P < 0.05 (DESeq2). Each replicate is pooled from three mice and derived from an independent experiment. (B) Representative flow plot of MC surface αE integrin (left) and quantification of αE integrin expression in β7Low and β7High MCs (right) following sensitization with saline or 30 µg Alt on day 0 and challenge with 30 µg Alt on days 9 and 10, sacrificed 48 h after the final challenge. Data from three independent experiments, n = 8–15. ***, P < 0.001 (t test); ns, not significant. (C) Representative flow plot of MC surface αE integrin (left) and quantification of αE integrin expression in β7Low and β7High MCs (right) following twice-weekly challenge with saline or 10 µg HDM. Dots are from four independent experiments, n = 8–11 mice per group. (D) Representative flow plots of mMCP-1 (left) and αE integrin (right) expression in BMMCs grown and maintained in IL-3 following treatment with saline (blue) or 10 ng/ml TGF-β1 (red) in the absence of SCF (top) or in combination with 100 ng/ml SCF (bottom) for 7 d. Data are representative of three independent experiments. (E) Quantification of BMMC mMCP-1 protein expression (top) and αE integrin expression (bottom) in response to a dose range of TGF-β1 treatments with or without 100 ng/ml SCF. Data from two independent experiments, n = 6. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (Dunnett’s multiple comparisons test used to compare all doses to zero; t test used to evaluate influence of SCF at each dose of TGF-β1). (F) Confocal assessment of BMMC expression of the lysosome and granule-associated protein LAMP-1 (green) and mMCP-1 (red) with nucleus (blue) in BMMCs grown and maintained in IL-3 following treatment with 10 ng/ml TGF-β1 in combination with 100 ng/ml SCF for 7 d. Merged channels (upper left), isotype control (lower left), individual fluorescence channel for LAMP-1 (upper right), and mMCP-1 (lower right) are shown. Scale bar = 10 µm. Data representative of two independent experiments.
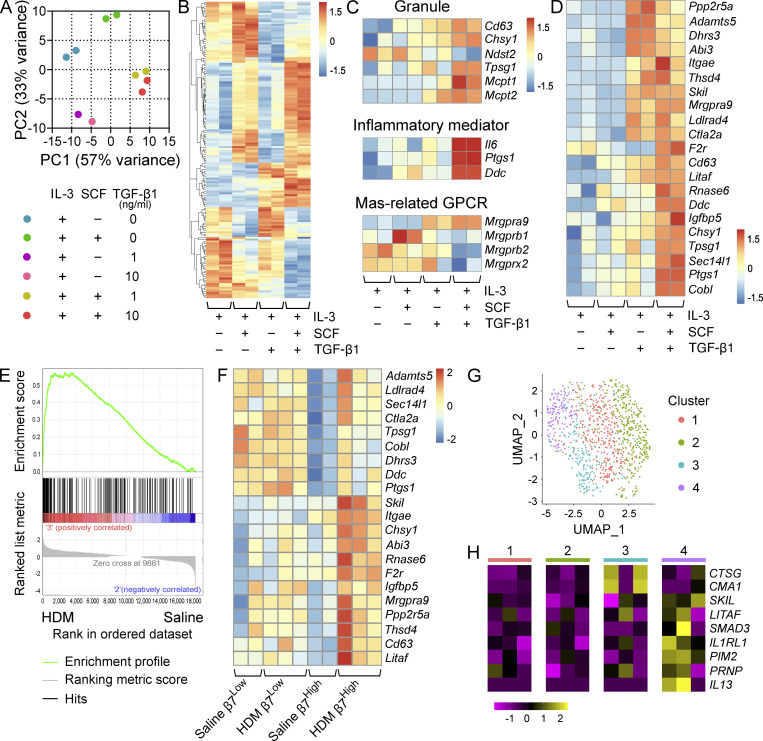
To determine whether TGF-β signaling could regulate other features of the inflammation-induced β7High MC transcriptome, BMMCs grown in IL-3 were cultured for 72 h in IL-3 alone, IL-3 plus either SCF or TGF-β1, or IL-3 plus both SCF and TGF-β1 before RNA-seq analysis. Principal component analysis indicated a strong effect of TGF-β1 signaling on the BMMC transcriptome, with TGF-β1–treated BMMCs and TGF-β1 plus SCF–treated BMMCs clustering distinctly from each other, from untreated BMMCs, and from SCF-treated BMMCs (Fig. 6 A). Analysis of the top variable transcripts showed distinct impacts of SCF, TGF-β1, and the two factors in combination on the BMMC transcriptome, with the two factors counteracting each other’s effects for some transcripts while synergizing for others (Fig. 6 B). Among these synergistically up-regulated transcripts were several associated with the MC granule that recapitulated aspects of the in vivo β7High MC transcriptome. SCF and TGF-β1 co-stimulus up-regulated Chsy1 and Cd63 relative to all other conditions, while down-regulating the heparin sulfate synthase Ndst2 (Fig. 6 C). Combined TGF-β1 and SCF signaling further induced significant up-regulation of the protease transcripts Tpsg1, Mcpt1, and Mcpt2 (Fig. 6 C). Together, TGF-β1 and SCF similarly recapitulated aspects of the inflammatory mediator profile of β7High MCs, up-regulating Il6, Ptgs1, and Ddc (Fig. 6 C). Interestingly, TGF-β signaling down-regulated expression of three Mrgpr family members (Mrgprb1, Mrgprb2, and Mrgprx2) that mark CTMCs (Fig. 6 C). In total, SCF-dependent TGF-β1 stimulation of BMMCs elicited up-regulation of a substantial fraction (21 of 62) of the in vivo HDM-elicited β7High MC transcriptome (Fig. 6 D). β7High MCs from HDM-treated mice showed a highly significant enrichment for the BMMC gene signature of TGF-β1 plus SCF relative to β7High MCs from saline-treated mice (Fig. 6 E), while β7Low MCs showed no changes in any of the TGF-β1 targets assessed (Fig. 6 F).
Figure 6.
TGF-β signaling induces a MMC-like transcriptional profile in BMMCs. (A) PCA of BMMCs grown and maintained in IL-3 and treated with SCF alone, TGF-β1 alone, or SCF in combination with TGF-β1 at the indicated concentrations for 72 h. Replicates are from two independent experiments with separate donors. (B) Heatmap showing the 100 most variable genes in BMMCs grown and maintained in IL-3 and treated with SCF alone, TGF-β1 alone, or SCF in combination with TGF-β1 at 10 ng/ml. All transcripts are significantly differentially expressed, with an FDR < 0.1 (DESeq2). Columns indicate biological replicates from two independent experiments (red, high; blue, low). (C) Heatmap analysis of MC granule-associated proteins and proteases (top) inflammatory mediator transcripts (middle) and MRGPR-family genes (bottom) differentially expressed (FDR < 0.1, DESeq2) between BMMCs grown and maintained in IL-3 and treated with SCF or SCF in combination with TGF-β1 at 10 ng/ml. Samples maintained in IL-3 alone or treated with TGF-β1 alone are shown for reference. Columns indicate biological replicates from two independent experiments (red, high; blue, low). (D) Heatmap analysis of transcripts significantly up-regulated in β7High MCs from HDM-challenged mice relative to saline treatment (FDR < 0.1, DESeq2) expressed by BMMCs grown and maintained in IL-3 and treated with SCF or SCF in combination with TGF-β1 at 10 ng/ml. Columns indicate biological replicates from two independent experiments (red, high; blue, low). Samples maintained in IL-3 alone or treated with TGF-β1 alone are shown for reference. (E) Gene set enrichment analysis plot evaluating β7High MCs from HDM-challenged versus saline-challenged mice for a set of transcripts significantly up-regulated in BMMCs treated with SCF and TGF-β1 relative to SCF alone (FDR < 0.1, DESeq2). P = 0 (below the lower limit of reporting for gene set enrichment analysis). (F) Heatmap analysis of TGF-β-target transcripts identified in D in β7High and β7Low MCs from saline or HDM-challenged mice. All transcripts are significantly up-regulated in β7High MCs (FDR < 0.1, DESeq2), while none show significant changes in β7Low MCs. Columns indicate biological replicates from two independent experiments (red, high; blue, low). (G) Uniform manifold approximation and projection embedding of human distal lung MCs computationally pooled from three healthy donors, identifying four clusters of distal lung MCs. (H) Heatmap showing expression of MCTC-associated transcripts significantly enriched (Wilcoxon test) in cluster 3 (blue; CTSG: FDR < 3 × 10−79, CMA1: FDR < 7 × 10−15); select TGF-β–induced transcripts enriched in cluster 4 (purple; LITAF: FDR < 2 × 10−8, SKIL: P < 0.001, SMAD3: FDR < 5.2 × 10−5); transcripts up-regulated in murine β7High MCs enriched in cluster 4 (purple) in relation to all other clusters (IL1RL1: FDR < 2 × 10−14, PIM2: P < 5 × 10−5, PRNP: P < 10−6); and IL13, which although not statistically significant is presented because expression was not detected outside of cluster 4. Columns indicate average expression levels for each individual donor.
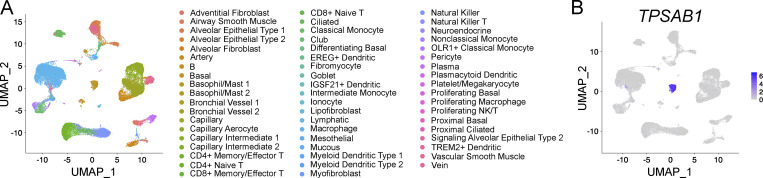
To evaluate whether TGF-β signaling may play a similar role in human MCs, we assessed a publicly available scRNA-seq dataset of healthy human distal lung tissue (Travaglini et al., 2020). MCs, previously identified as part of the dataset’s construction, were independently confirmed based on expression of tryptase β1 (TPSAB1; Fig. S3). Reclustering of pulmonary MCs identified four major clusters with distinct transcriptional profiles (Fig. 6 G and Table S3). Of these, cluster 3 was enriched for transcripts encoding cathepsin G and chymase (CTSG and CMA1), identifying cells within this cluster as MCs co-expressing tryptase and chymase (MCTC), roughly analogous to murine β7Low pulmonary MCs and identifying the remaining clusters as MCs expressing tryptase in the absence of chymase (MCT; Fig. 6 H). Of the three MCT clusters, cluster 4 exhibited enriched expression for several TGF-β signature genes (SKIL, LITAF, and SMAD3), suggesting the potential for TGF-β to also play an important role in human MC biology. While humans lack an mMCP-1 orthologue, cluster 4 was also enriched for several transcripts up-regulated by β7High MCs in response to inflammation (IL1RL1, PIM2, and PRNP). No enrichment was observed in any cluster for transcript encoding αE integrin (ITGAE; not depicted). Of note, although IL4 was not observed in any cluster, MCs within cluster 4 were the only ones with detectable IL13. Together, these data suggest a potential role for TGF-β in regulating the MMC phenotype in both mouse and human.
Figure S3.
ScRNA-seq analysis of human distal lung tissue. (A) Sequencing results for distal lung tissue from three healthy donors generated by Travaglini et al. (2020) were computationally combined using Harmony, and previously described cluster designations were applied. (B) Independent confirmation that tryptase (TPSAB1) expression was restricted to a single cluster, encompassing both previously identified MC clusters.
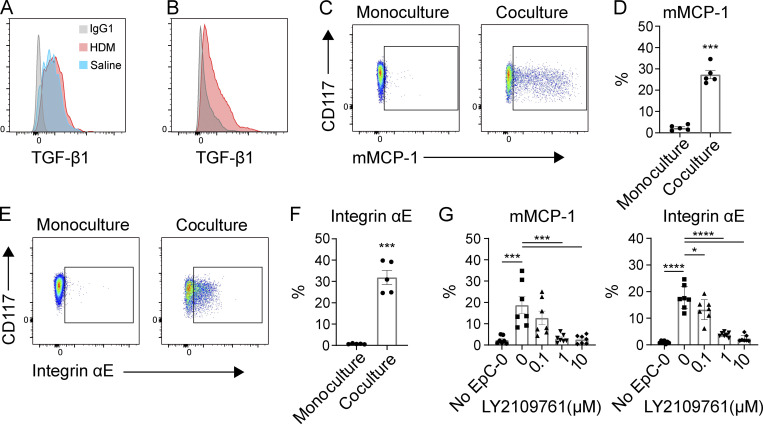
The mMCP-1 and -2 proteases are canonically associated with induced MMCs found within or adjacent to the epithelium of T helper 2 cell–inflammed murine intestine and airway (Friend et al., 1996; Xing et al., 2011), and αVβ6, a TGF-β–activating integrin restricted to epithelium (Koivisto et al., 2018), has been previously shown to regulate MMC expression of mMCP-1 in both the small intestine and airways (Knight et al., 2002; Sugimoto et al., 2012). Thus, we hypothesized that the epithelial cells could directly influence β7High MC development via TGF-β. We first evaluated TGF-β1 expression by flow cytometry in pulmonary epithelial cell adhesion molecule (EPCAM)-positive cells in vivo, noting strong expression in the lungs of both HDM- and saline-challenged mice (Fig. 7 A), and confirmed that epithelial TGF-β protein expression was maintained by primary epithelial cells expanded ex vivo (Fig. 7 B). We next tested the capacity for ex vivo expanded primary epithelial cells to directly elicit BMMC up-regulation of mMCP-1 and αE integrin. BMMCs grown in IL-3 were cultured either alone or in coculture with epithelial cells for 7 d in the presence of exogenous IL-3 to support viability. While BMMCs cultured without epithelial cells expressed minimal mMCP-1, coculture with epithelial cells led to a significant increase in intracellular mMCP-1 (Fig. 7, C and D). Similarly, αE integrin was minimally expressed by BMMCs alone, but substantially up-regulated following coculture with epithelial cells (Fig. 7, E and F). Inhibition of TGF-β receptor signaling with LY2109761, a dual inhibitor of TGF-βR1 and TGF-βR2 (Melisi et al., 2008), blocked up-regulation of both mMCP-1 and αE integrin in a dose-dependent manner (Fig. 7 G), confirming the capacity of epithelial-derived TGF-β to regulate BMMC differentiation toward an MMC phenotype.
Figure 7.
Epithelial cells induce an MMC-like phenotype through TGF-β signaling. (A) Representative flow plot of TGF-β1 expression by murine pulmonary epithelial cells (EPCAM+, Thy1−, CD90−, CD31−) in vivo from mice challenged with HDM (red) and saline (blue) relative to isotype control (gray). Results representative of three experiments with n = 2–4 mice per group in each experiment. (B) Representative flow plot of TGF-β1 expression by primary mouse tracheal epithelial cells expanded ex vivo (red) relative to isotype control (gray). Results representative of two independent experiments. (C) Representative flow plots of mMCP-1 expression by IL-3–supplemented BMMCs cultured on Matrigel alone (left) or in coculture with mouse airway epithelial cells (right) for 7 d. (D) Quantification of intracellular mMCP-1 expression by IL-3–supplemented BMMCs cultured alone or in coculture with mouse airway epithelial cells on Matrigel for 7 d. Results are from three independent experiments, with each replicate showing a unique combination of epithelial and MC biological donors. ***, P < 0.001 (t test). (E) Representative flow plots of αE integrin expression by BMMCs cultured on Matrigel alone (left) or in coculture with mouse airway epithelial cells (right) for 7 d. (F) Quantification of αE-integrin expression by BMMCs cultured on Matrigel alone or in coculture with mouse airway epithelial cells for 7 d. Results are from three independent experiments, with each replicate showing a unique biological combination of epithelial and MC cultures. ***, P < 0.001 (t test). (G) Expression of mMCP-1 and αE integrin by BMMCs cultured on Matrigel alone or in coculture with mouse airway epithelial cells for 7 d in conjunction with the indicated concentration of the TGF-β R1/R2 dual inhibitor LY2109761. Results are from three independent experiments, with each replicate showing a unique biological combination of epithelial and MC cultures. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (Dunnett’s multiple comparisons test).
Discussion
Here, we identify a population of β7High MCs in the lungs of resting C57BL/6 mice that dynamically expands in response to diverse aeroallergen challenge. β7High MCs express a distinct protease profile compared with constitutive β7Low MCs, with reduced expression of CTMC-associated proteases (Mcpt4, Cma1, Tpsb2, and Cpa3) and enhanced expression of transcripts encoding the classic MMC proteases mMCP-1 and -2, as well as mMCP-8, previously thought to be restricted to basophils and commonly used to direct basophil-specific depletion and gene deletion (Sullivan et al., 2011; Wada et al., 2010). The transcriptional differences between the two subsets extend well beyond protease expression. β7High MCs have minimal expression of many components of the previously defined CTMC transcriptional signature (Dwyer et al., 2016), including MRGPR family member receptors (Mrgprb1, Mrgprb2, and Mrgprx2) and heparin sulfate biosynthetic enzymes (Ndst2, Hs3st3a1, and Hs6st2). In contrast to CTMCs, recruited β7High MCs exhibit a dynamic response to type 2 inflammation, both expanding in number and up-regulating distinct sets of transcripts encoding the serotonin biosynthetic machinery (Slc7a5, Tph1, and Ddc), MMC proteases (Mcpt1 and Mcpt2), and activating receptors (Il1rl1, Fcer1a, Cd200r3, and Cd200r4).
The heterogeneous morphology and dynamic nature of β7High MCs during inflammation may indicate that this pool contains a range of maturation states, ranging from freshly recruited progenitors to mature granulated cells. Indeed, only a subset of β7High MCs contain metachromatic granules, indicating the presence of granule-associated proteoglycan, and intracellular protease was similarly detected in only a portion of the cells. Alternatively, the heterogeneous granularity in β7High MCs may reflect degranulation in response to allergen-specific IgE cross-linking. This morphological heterogeneity further suggests that only a fraction of these β7High MCs are likely to be recognized by conventional histology owing to their relatively poor granulation, despite this induced lineage being the dominant MC population within the lung of C57BL/6 in response to ongoing type 2 inflammation.
Of note, β7High MCs expressed abundant effector transcripts, including cytokines, chemokines, lipid mediators, and monoamine biosynthetic machinery, at levels similar to those observed in constitutive β7Low MCs, and this expression was observed even under saline challenge conditions. As transcript and protein levels do not always correlate, this raises a critical question as to the effector capacity of β7High MCs, both at rest and during inflammation. These observations could indicate that even hypogranular β7High MCs have the same capacity to synthesize histamine and lipid mediators as constitutive MCs. Alternately, β7High MC effector function may be regulated, at least in part, at the posttranscriptional level. If this is the case, the abundant transcripts observed may represent that these cells exist in a “poised” state, allowing them to rapidly translate messages in response to exogenous stimuli. This second hypothesis is supported by observations in the transgenic 4-get mouse, in which MCs, basophils, and eosinophils are known to constitutively express IL4 transcript but not translate protein until activation (Gessner et al., 2005). Importantly, the previously described committed human MCp was also found to express transcripts encoding both histadine decarboxylase (HDC) and PGD2 synthase (HPGDS) within the circulation at higher levels than mature basophils (Dahlin et al., 2016). Thus, while it is possible that human and mouse MCps enter host tissue primed for rapid histamine and lipid mediator synthesis, it is similarly possible that expression for the enzymes encoded by these transcripts are translated only in response to exogenous stimuli.
mMCP-1, prominent in β7High MCs, was initially recognized as a specific marker of intestinal MMCs that develop in response to helminth-elicited type 2 inflammation, where targeted deletion of the protease indicated an important role in helminth expulsion (Knight et al., 2000; Wastling et al., 1998). More recently, mMCP-1 has been found to elicit airway constriction through its effects on epithelial cells (Sugimoto et al., 2012), and detection of mMCP-1 in serum has become a surrogate for MC activation in helminth infection and oral food sensitization (Burton et al., 2014; Leyva-Castillo et al., 2019; Pemberton et al., 2006). Here, we find that mMCP-1 protein expression is restricted to β7High MCs within the inflamed lung. As β7High MCs contain few discrete metachromatic granules but exhibit diffuse CAE staining, we hypothesize that mMCP-1 is predominantly cytoplasmic within these cells. Interestingly, mMCP-1 is constitutively secreted by MCs in vitro (Miller et al., 1999), and intracellular storage of mMCP-1 does not require the granule core protein serglycin (Braga et al., 2007), likely because of its low net positive charge relative to the more cationic serglycin-dependent MCPs (Pejler et al., 2010). A recent study of IL-33–exacerbated ovalbumin-induced airway hyperresponsiveness specifically noted elevated bronchoalveolar fluid concentrations of mMCP-1 in the absence of histochemical detection of MMCs (Sjöberg et al., 2017). Similarly, aspirin-elicited airway hyperreactivity in Ptges−/− mice repeatedly challenged with HDM is accompanied by robust increases in bronchoalveolar fluid mMCP-1 levels (Liu et al., 2015). Thus, we speculate that β7High MCs are the dominant source of mMCP-1 in C57BL/6 allergic airway inflammation, which may be secreted from the cytoplasm rather than restricted to secretory granules.
In vivo, mMCP-1 and αE integrin expression was increased in β7High MCs in two challenge models, suggesting they were regulated by a common signal. We found that in vivo up-regulation of transcripts encoding both proteins was associated with an MC TGF-β signaling signature, including the regulatory elements Skil and Ldlrad3 (Deheuninck and Luo, 2009; Nakano et al., 2014) and the MMC-associated proteases Mcpt1 and Mcpt2. We further observed that BMMC coculture with epithelial cells directly elicited TGF-β–dependent up-regulation of mMCP-1 and αE integrin. Maturation of recruited MCp in BALB/c mice following ovalbumin sensitization and challenge is similarly associated with induction of mMCP-1 and up-regulation of αE integrin, which is retained in MMCs after maturation (Bankova et al., 2015; Xing et al., 2011). Further, MMCs from 129 mice lacking αVβ6, an epithelial cell–specific integrin that activates TGF-β (Koivisto et al., 2018), show defects in intestinal localization and mMCP-1 and mMCP-2 expression following Trichinella spiralis infection (Knight et al., 2002), while β6 integrin-deficient mice on the FvB background lack MMC expression of both mMCP-1 and mMCP-2 in the airways following ovalbumin sensitization and challenge (Sugimoto et al., 2012). Similarly, Kasakura et al. (2020) found that Mcpt1 and Mcpt2 were up-regulated by TGF-β stimulation through cooperative interactions between TGF-β–driven Smad2 and Smad4 and endogenously active GATA1 and GATA2. Of note, the αEβ7 integrin pair specifically binds to the epithelial adhesion molecule E-cadherin (Cepek et al., 1994), suggesting a role for αE integrin up-regulation in stabilizing induced MMCs within the epithelium. Thus, association of type 2 inflammation and epithelial surfaces with up-regulation of αE integrin, mMCP-1, and mMCP-2 in induced MCs across tissues and genetic backgrounds suggests that epithelial-mediated TGF-β signaling is a common regulator of MMC development. Further, our observation of a cluster of human distal lung MCTs expressing several murine TGF-β–elicited transcripts suggests that this mechanism may also play a role in human MC development.
The protease expression profile of β7High MCs has important implications for future study design using cre recombinase–mediated MC deletion. Although low in comparison to constitutive β7Low MCs, β7High MCs express robust levels of Cpa3 and could be eliminated along with CTMCs and basophils in Cpa3-DTA mice (Feyerabend et al., 2011; Lilla et al., 2011). Our finding that the β7High MC compartment is completely intact in the commonly used MCpt5/DTA model of CTMC deletion is consistent with a previous report of pulmonary MC expansion following i.v. injection of B16 melanoma cells in MCpt5/DTA on the C57BL/6 background (Öhrvik et al., 2016). Interestingly, we also observed that β7High MCs robustly express Mcpt8, suggesting that the compartment could be deleted in strains using the Mcpt8 promoter to drive DTA or DTR expression to target basophils in murine models of inflammation (Sullivan et al., 2011; Wada et al., 2010).
The induced MC response has been predominantly studied in BALB/c mice, in which MMCs with prominent secretory granules arise within the epithelium of large airways during type 2 inflammation. In C57BL/6 mice, we find that β7High BMMCs are present within the lungs of naive mice and both expand and develop to an MMC-like phenotype following aeroallergen challenge, characterized by up-regulation of mMCP-1 and αE integrin. Following the induction of allergic airway inflammation, induced β7High MCs represent the majority of pulmonary MCs in C57BL/6 mice, while up-regulating additional effector-associated transcripts, some shared with CTMCs and some distinct. This response likely allows these cells to respond to distinct sets of exogenous stimuli within the lung microenvironment. While we observed no impact of CTMC deletion on pulmonary type 2 inflammation, a role for MCs in regulating pulmonary inflammation has been previously established by multiple groups using kit-deficient strains. As these models use reconstitution with BMMCs, their effect on pulmonary inflammation may be mediated by the infiltrated TGF-β–responding BMMCs. Prior literature has identified distinct characteristics for the progenitors for constitutive and inducible MCs (Kitamura et al., 1979; Sonoda et al., 1983); thus, our findings of distinct adult forms emphasizes the need to similarly distinguish functions of the mature cells in integrated host inflammatory responses.
Materials and methods
Flow cytometry
For detection and analysis, the following antibodies/clones were used: FcɛR1a (MAR-1; BioLegend); CD117 (2B8; BioLegend); CD45 (30-F11; BioLegend), β7 integrin (M293; BD Biosciences); CD11b (M1/70; BioLegend); CD11c (N418; BioLegend); CD4 (GK1.5; BioLegend); CD8 (53–6.7; BioLegend); CD103 (2E7; BioLegend); EPCAM (G8.8; BioLegend); Thy1.2 (53–2.1; BioLegend); CD31 (390; eBioscience); TGF-β1 (Tw7-16B4; BioLegend); and mMCP-1 (RF6.1; eBioscience). Anti-mMCP-1 and isotype control were conjugated in parallel to Alexa Fluor 647 using a conjugation kit (Life Technologies). Intracellular staining was conducted using a BD Cytofix/Cytoperm kit (BD Bioscience), according to the manufacturer-supplied protocol. For flow cytometry, the cells were collected and stained for surface markers for 45 min before fixation. Intracellular mMCP-1 staining was conducted overnight at 4°C. All cell sorting was on a BD FACSAria Fusion cell sorter using BD FACSDiva software. For all flow cytometry not involving cell sorting, data were collected on a BD LSRII Fortessa or BD CANTO-II using BD FACSDiva software. All downstream data analysis was conducted in FlowJo.
Mice
C57BL/6 were initially obtained from the Jackson Laboratory. To generate CTMC-deficient Mcpt5/DTA mice, C57BL/6-TgMcpt5-Cre mice, expressing cre recombinase under the control of the Mcpt5 (Cma1) promoter (Dudeck et al., 2011), were crossed to B6.129P2-Gt (ROSA)26Sortm1(DTA)Lky/J mice, which express the DTA chain after a floxed stop codon under the control of the Gt(ROSA)26Sor promoter, obtained from the Jackson Laboratory. All mice were bred and housed in a specific pathogen–free facility at the Dana-Farber Cancer Institute and Brigham and Women’s Hospital. Pups were weaned 19–28 d after birth. Experiments were conducted on female mice. Mice were housed in groups of four to five mice per cage and maintained with a standard 12-h light, 12-h dark cycle with food and water ad libitum. For repeated HDM challenge, mice were anesthetized through isoflurane inhalation and challenged intranasally with 3 or 10 µg Dermatophagoides farina extract (depending on lot; Greer) by protein weight in 20 µl saline or vehicle control twice per week for 3 wk and sacrificed 24 h after the final challenge. Dosages were calibrated to elicit similar levels of β7High MC recruitment between lots. All RNA-seq was conducted on mice challenged with batch. For Alt sensitization and challenge models, mice were anesthetized with an i.p. injection of ketamine and xylazine and sensitized with an intranasal dose of 30 µg Alt culture filtrate (Greer) by protein weight in 20 µl saline or saline alone on day 0. Both Alt- and saline-sensitized mice were subsequently anesthetized with i.p. ketamine and xylazine and challenged intranasally with 3 µg Alt in 20 µl on days 9 and 10. Mice were sacrificed 48 h after the final challenge. All mouse experiments were conducted in accordance with review and approval by the Animal Care and Use Committees of the Dana-Farber Cancer Institute and Brigham and Women’s Hospital.
Cell culture
BMMCs were grown from bone marrow of C57BL/6 mice in RPMI 1640 supplemented with 10% (vol/vol) FBS, 2-mercaptoethanol, and IL-3 at 5 ng/ml (R10 medium). Once mature, BMMCs were stimulated with TGF-β1 (Peprotech) at various concentrations in R10 medium and analyzed after 7 d. The effect of SCF at 100 ng/ml was also tested. Epithelial cells were grown from trachea using keratinocyte serum-free medium (Gibco) following a previous protocol and seeded on 50% (vol/vol) Matrigel (Corning) for 3 d in supplemented DMEM/F-12 medium (Gibco; Eenjes et al., 2018). BMMCs were added to the epithelial cells at a 1:10 ratio and collected after 7 d for analysis by flow cytometry. In the blocking experiments, LY2109761, a dual TGF-βRI and II inhibitor (Cayman), was added to the medium at defined concentrations at the time of BMMC addition to epithelial cells.
Cell isolation
After sacrifice, lungs were removed and digested with collagenase IV (600 U/ml; Worthington) and Dispase I (0.1% wt/vol; Gibco) with 28 µg/ml DNase 1 (Roche) for 30 min in RPMI 1640 supplemented with 10% (vol/vol) FBS. Samples were agitated on a shaker set to 500 rpm. After 30 min, samples were triturated 10 times through a syringe using a 16G needle. For RNA-seq experiments, trachea was included to ensure that enough CTMCs were collected for analysis. Samples were centrifuged at 500 relative centrifugal force for 10 min after dilution with equal parts HBSS and 2 mmol EDTA to stop enzymatic digestion. Erythrocytes were lysed with RBC lysis buffer (Sigma-Aldrich). For intracellular staining, leukocytes were further enriched by resuspension in 44% Percoll (GE Healthcare), which causes epithelial cells to float to the top and leaves leukocytes in the pellet fraction. To maximize purity of flow-sorted MCs, a two-step sorting protocol was used. MCs were identified as propidium iodide negative, CD45 positive, lineage (CD4, CD8, CD19, CD11b, and CD11c) negative, and coexpressing CD117 and FcɛR1α. On the first sort, to maximize recovery, β7High and β7Low MCs were not separated. Enriched MCs were then sorted a second time, with fresh propidium iodide was added to exclude dead cells, enriched MCs were identified using an identical gating strategy, and β7High and β7Low MCs were further separated based on β7 integrin and SSC.
Cytological analysis
For histochemical analysis of sorted CTMCs and β7High MCs, sorted cells were pelleted and resuspended in <10 µl saline. This suspension was placed on a glass slide and quickly dried using a plate warmer at the lowest setting. Cells were stained with TB solution (94.2% H2O, 5.8% concentrated HCl, and 0.5 g TB) to visualize granules or with CAE to determine protease reactivity. For microscopy, BMMCs were fixed using a Cytofix/Cytoperm kit following the manufacturer’s protocol before incubation in permeabilization buffer containing BSA (Sigma-Aldrich) for 1 h. The cells were stained for mMCP-1 (RF6.1; eBioscience) and LAMP-1 (1D4B; BioLegend) overnight. Cytospins were prepared using mounting medium containing DAPI (Life Technologies).
RNA-seq and data analysis
All sequencing sample cells were collected into TCL lysis buffer. Sorted upper-airway MCs were processed for sequencing at the Broad Technology Labs using a low-input eukaryotic Smart-seq2 protocol. Briefly, total RNA was purified using RNA-SPRI beads. Poly(A)+ mRNA was converted to cDNA, which was then amplified. cDNA was subjected to transposon-based fragmentation that used dual indexing to barcode each fragment of each converted transcript with a combination of barcodes specific to each sample. Barcoded cDNA fragments were then pooled before sequencing. Sequencing was performed as paired-end 2× 25 bp with an additional 8 cycles for each index. Data were separated by barcode and aligned using Tophat v2.0.10 with default settings (Kim et al., 2013). Transcripts were quantified by Broad Technology Labs using the Cufflinks suite v2.2.1 (Trapnell et al., 2012). Sequencing of in vitro stimulated BMMCs was conducted on the Broad Institute Genomics Platform. SmartSeq2 libraries were prepared according to the SmartSeq2 protocol described Trombetta et al. (2014). Briefly, total RNA was purified using RNA-SPRI (solid phase reversible immobilization) beads. cDNA was generated from full-length mRNA transcripts using reverse transcription with terminal transferase activity. Combined with a second “template switch” primer, the cDNA was constructed to have two universal priming sequences. After preamplification, the Nextera XT library construction kit was used to prepare 96 unique indexes specific to each sample. Barcoded cDNA fragments were then pooled before sequencing. Sequencing was performed using an Illumina NextSeq500 as paired-end 2× 38 bp to a coverage of ∼4 million reads per well. Data for each lane were separated by barcode and concatenated. Transcripts were aligned to GRCm38 (mm10) genome assembly with STAR (Dobin et al., 2013) and quantified with HTseq (Anders et al., 2015). Data analysis for all samples was conducted using Bioconductor for R. Count normalization and differential were conducted using the DESeq2 software package (Love et al., 2014). Genes were considered significantly differentially regulated based on an FDR of <0.1 using a Benjamini–Hochberg adjusted P value to correct for multiple comparisons. Heatmaps were generated using the pheatmap software package.
For human scRNA-seq analysis, we used a series of droplet-based human lung scRNA-seq datasets generated by the Krasnow Laboratory (Travaglini et al., 2020). Annotated Seurat objects generated by Travaglini et al. (2020) for three healthy donors were downloaded from GitHub (https://github.com/krasnowlab/hlca). Cells isolated from distal lung, a compartment assessed in all three donors, were computationally isolated from each donor. The resulting cells were merged using the Seurat package for R (Butler et al., 2018) and normalized using the Harmony package (Korsunsky et al., 2019). Upon confirming that MC tryptase (TPSAB1) was restricted to a population encompassing the previously defined Basophil/Mast 1 and Basophil/Mast 2 populations, these populations were computationally isolated for further analysis. Datasets were normalized using the scTransform wrapper for Seurat, and iterative reclustering was used to remove all contaminating cell populations. No cluster with a gene signature suggestive of basophils was observed in distal lung. Differential gene analysis was conducted using the Wilcoxon test implemented within Seurat.
Statistics
Results are presented as mean ± SEM of data from at least three independent experiments. Student’s t test was used for pairwise comparisons, and Dunnett’s multiple comparisons test was used for repeated comparisons using Prism (GraphPad Software). A value of P < 0.05 was considered significant.
Data availability
Sequencing data have been deposited in GEO under accession no. GSE155575.
Online supplemental material
Fig. S1 displays the gating strategy used for the identification of MCs within the lower airways of C57BL/6 mice.Fig. S2 shows the gating strategy used for the identification of eosinophils within the lower airways of WT and MCpt5/DTA mice during allergic lung inflammation. Fig. S3 depicts scRNA-seq analysis of human distal lung tissue. Table S1 shows DESeq2 analysis of differential gene expression between constitutive β7Low MCs and inducible β7High MCs following sequential HDM challenges. Table S2 shows DESeq2 analysis of differential gene expression between constitutive β7Low MCs from saline or sequential HDM-challenged mice (tab 1) and inducible β7High MCs following saline or sequential HDM challenges (tab 2). Table S3 shows Seurat analysis of differentially expressed transcripts across the four human MC clusters (Wilcoxon test).
Supplementary Material
shows DESeq2 analysis of differential gene expression between constitutive β7Low MCs and inducible β7High MCs following sequential HDM challenges.
shows DESeq2 analysis of differential gene expression between constitutive β7Low MCs from saline or sequential HDM-challenged mice (tab 1) and inducible β7High MCs following saline or sequential HDM challenges (tab 2).
shows Seurat analysis of differentially expressed transcripts across the four human MC clusters (Wilcoxon test).
Acknowledgments
We thank J. Lai and H. Katz for histology support, the Brigham and Women’s Hospital Human Immunology Center Flow Cytometry Core for cell sorting assistance, and F. Al Haddad for administrative support. For RNA-seq assistance, we thank the Broad Institute’s technology labs, the Broad Genomics Platform, and Robert Chase of the Brigham and Women’s Hospital Channing Division of Network Medicine.
J.A. Boyce was supported by National Institutes of Health grants R37AI052353, R01AI136041, R01AI130109, R01HL136209, and U19 AI095219. N.A. Barrett was supported by National Institutes of Health grants R01HL120952, R01AI134989, and U19AI095219 and the Steven and Judy Kaye Young Innovators Award. D.F. Dwyer was supported by National Institutes of Health grants T32 AI007306 (to J.A. Boyce) and K22 AI146281, National Institutes of Health AADCRC Opportunity Fund Award U19AI070535, and generous contributions from the Vinik family.
Author contributions: K.F. Austen and D.F. Dwyer conceptualized the project. T. Derakhshan, S.K. Samuchiwal, L.G. Bankova, N.A. Barrett, K.F. Austen, and D.F. Dwyer designed experiments. N. Hallen adapted murine epithelial cell expansion protocols and maintained cell lines. T. Derakhshan conducted and analyzed in vitro experiments. T. Derakhshan, S.K. Samuchiwal, and D.F. Dwyer conducted and analyzed animal experiments. T. Derakhshan, S.K. Samuchiwal, and D.F. Dwyer conducted and analyzed flow cytometry experiments. T. Derakhshan and D.F. Dwyer conducted RNA-seq analysis. L.G. Bankova, N.A. Barrett, and J.A. Boyce provided experimental advice. T. Derakhshan, K.F. Austen, and D.F. Dwyer wrote the manuscript and designed figures.
References
- Abonia, J.P., Hallgren J., Jones T., Shi T., Xu Y., Koni P., Flavell R.A., Boyce J.A., Austen K.F., and Gurish M.F.. 2006. Alpha-4 integrins and VCAM-1, but not MAdCAM-1, are essential for recruitment of mast cell progenitors to the inflamed lung. Blood. 108:1588–1594. 10.1182/blood-2005-12-012781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen, K.F., and Brocklehurst W.E.. 1961. Anaphylaxis in chopped guinea pig lung. II. Enhancement of the anaphylactic release of histamine and slow reacting substance by certain dibasic aliphatic acids and inhibition by monobasic fatty acids. J. Exp. Med. 113:541–557. 10.1084/jem.113.3.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankova, L.G., Dwyer D.F., Liu A.Y., Austen K.F., and Gurish M.F.. 2015. Maturation of mast cell progenitors to mucosal mast cells during allergic pulmonary inflammation in mice. Mucosal Immunol. 8:596–606. 10.1038/mi.2014.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga, T., Grujic M., Lukinius A., Hellman L., Abrink M., and Pejler G.. 2007. Serglycin proteoglycan is required for secretory granule integrity in mucosal mast cells. Biochem. J. 403:49–57. 10.1042/BJ20061257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, O.T., Noval Rivas M., Zhou J.S., Logsdon S.L., Darling A.R., Koleoglou K.J., Roers A., Houshyar H., Crackower M.A., Chatila T.A., et al. 2014. Immunoglobulin E signal inhibition during allergen ingestion leads to reversal of established food allergy and induction of regulatory T cells. Immunity. 41:141–151. 10.1016/j.immuni.2014.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepek, K.L., Shaw S.K., Parker C.M., Russell G.J., Morrow J.S., Rimm D.L., and Brenner M.B.. 1994. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature. 372:190–193. 10.1038/372190a0 [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Lee J.B., Liu B., Ohta S., Wang P.Y., Kartashov A.V., Mugge L., Abonia J.P., Barski A., Izuhara K., et al. 2015. Induction of Interleukin-9-Producing Mucosal Mast Cells Promotes Susceptibility to IgE-Mediated Experimental Food Allergy. Immunity. 43:788–802. 10.1016/j.immuni.2015.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowle, P.K. 1983. Mucosal mast cell reconstitution and Nippostrongylus brasiliensis rejection by W/Wv mice. J. Parasitol. 69:66–69. 10.2307/3281276 [DOI] [PubMed] [Google Scholar]
- Dahlin, J.S., Malinovschi A., Öhrvik H., Sandelin M., Janson C., Alving K., and Hallgren J.. 2016. Lin- CD34hi CD117int/hi FcεRI+ cells in human blood constitute a rare population of mast cell progenitors. Blood. 127:383–391. 10.1182/blood-2015-06-650648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deheuninck, J., and Luo K.. 2009. Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res. 19:47–57. 10.1038/cr.2008.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudeck, A., Dudeck J., Scholten J., Petzold A., Surianarayanan S., Köhler A., Peschke K., Vöhringer D., Waskow C., Krieg T., et al. 2011. Mast cells are key promoters of contact allergy that mediate the adjuvant effects of haptens. Immunity. 34:973–984. 10.1016/j.immuni.2011.03.028 [DOI] [PubMed] [Google Scholar]
- Dudeck, J., Ghouse S.M., Lehmann C.H., Hoppe A., Schubert N., Nedospasov S.A., Dudziak D., and Dudeck A.. 2015. Mast-Cell-Derived TNF Amplifies CD8(+) Dendritic Cell Functionality and CD8(+) T Cell Priming. Cell Rep. 13:399–411. 10.1016/j.celrep.2015.08.078 [DOI] [PubMed] [Google Scholar]
- Dwyer, D.F., Barrett N.A., and Austen K.F.; Immunological Genome Project Consortium . 2016. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 17:878–887. 10.1038/ni.3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eenjes, E., Mertens T.C.J., Buscop-van Kempen M.J., van Wijck Y., Taube C., Rottier R.J., and Hiemstra P.S.. 2018. A novel method for expansion and differentiation of mouse tracheal epithelial cells in culture. Sci. Rep. 8:7349. 10.1038/s41598-018-25799-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund, K.K., Ghildyal N., Austen K.F., and Stevens R.L.. 1993. Induction by IL-9 and suppression by IL-3 and IL-4 of the levels of chromosome 14-derived transcripts that encode late-expressed mouse mast cell proteases. J. Immunol. 151:4266–4273. [PubMed] [Google Scholar]
- Feyerabend, T.B., Weiser A., Tietz A., Stassen M., Harris N., Kopf M., Radermacher P., Möller P., Benoist C., Mathis D., et al. 2011. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity. 35:832–844. 10.1016/j.immuni.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Friend, D.S., Ghildyal N., Austen K.F., Gurish M.F., Matsumoto R., and Stevens R.L.. 1996. Mast cells that reside at different locations in the jejunum of mice infected with Trichinella spiralis exhibit sequential changes in their granule ultrastructure and chymase phenotype. J. Cell Biol. 135:279–290. 10.1083/jcb.135.1.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek, R., Ghigo C., Hoeffel G., Bulle M.J., Msallam R., Gautier G., Launay P., Chen J., Ginhoux F., and Bajénoff M.. 2018. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity. 48:1160–1171.e5. 10.1016/j.immuni.2018.04.025 [DOI] [PubMed] [Google Scholar]
- Gessner, A., Mohrs K., and Mohrs M.. 2005. Mast cells, basophils, and eosinophils acquire constitutive IL-4 and IL-13 transcripts during lineage differentiation that are sufficient for rapid cytokine production. J. Immunol. 174:1063–1072. 10.4049/jimmunol.174.2.1063 [DOI] [PubMed] [Google Scholar]
- Ghildyal, N., McNeil H.P., Gurish M.F., Austen K.F., and Stevens R.L.. 1992a. Transcriptional regulation of the mucosal mast cell-specific protease gene, MMCP-2, by interleukin 10 and interleukin 3. J. Biol. Chem. 267:8473–8477. [PubMed] [Google Scholar]
- Ghildyal, N., McNeil H.P., Stechschulte S., Austen K.F., Silberstein D., Gurish M.F., Somerville L.L., and Stevens R.L.. 1992b. IL-10 induces transcription of the gene for mouse mast cell protease-1, a serine protease preferentially expressed in mucosal mast cells of Trichinella spiralis-infected mice. J. Immunol. 149:2123–2129. [PubMed] [Google Scholar]
- Gurish, M.F., Tao H., Abonia J.P., Arya A., Friend D.S., Parker C.M., and Austen K.F.. 2001. Intestinal mast cell progenitors require CD49dbeta7 (alpha4beta7 integrin) for tissue-specific homing. J. Exp. Med. 194:1243–1252. 10.1084/jem.194.9.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavey, D.J., Ernst P.B., Stevens R.L., Befus A.D., Bienenstock J., and Austen K.F.. 1988. Generation of leukotriene C4, leukotriene B4, and prostaglandin D2 by immunologically activated rat intestinal mucosa mast cells. J. Immunol. 140:1953–1957. [PubMed] [Google Scholar]
- Honjo, A., Nakano N., Yamazaki S., Hara M., Uchida K., Kitaura J., Nishiyama C., Yagita H., Ohtsuka Y., Ogawa H., et al. 2017. Pharmacologic inhibition of Notch signaling suppresses food antigen-induced mucosal mast cell hyperplasia. J. Allergy Clin. Immunol. 139:987–996.e10. 10.1016/j.jaci.2016.05.046 [DOI] [PubMed] [Google Scholar]
- Kasakura, K., Nagata K., Miura R., Iida M., Nakaya H., Okada H., Arai T., Arai T., Kawakami Y., Kawakami T., et al. 2020. Cooperative Regulation of the Mucosal Mast Cell-Specific Protease Genes Mcpt1 and Mcpt2 by GATA and Smad Transcription Factors. J. Immunol. 204:1641–1649. 10.4049/jimmunol.1900094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, Y., Hatanaka K., Murakami M., and Shibata H.. 1979. Presence of mast cell precursors in peripheral blood of mice demonstrated by parabiosis. Blood. 53:1085–1088. 10.1182/blood.V53.6.1085.1085 [DOI] [PubMed] [Google Scholar]
- Knight, P.A., Wright S.H., Lawrence C.E., Paterson Y.Y., and Miller H.R.. 2000. Delayed expulsion of the nematode Trichinella spiralis in mice lacking the mucosal mast cell-specific granule chymase, mouse mast cell protease-1. J. Exp. Med. 192:1849–1856. 10.1084/jem.192.12.1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, P.A., Wright S.H., Brown J.K., Huang X., Sheppard D., and Miller H.R.. 2002. Enteric expression of the integrin alpha(v)beta(6) is essential for nematode-induced mucosal mast cell hyperplasia and expression of the granule chymase, mouse mast cell protease-1. Am. J. Pathol. 161:771–779. 10.1016/S0002-9440(10)64236-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivisto, L., Bi J., Häkkinen L., and Larjava H.. 2018. Integrin αvβ6: Structure, function and role in health and disease. Int. J. Biochem. Cell Biol. 99:186–196. 10.1016/j.biocel.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Korsunsky, I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., and Raychaudhuri S.. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 16:1289–1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Castillo, J.M., Galand C., Kam C., Burton O., Gurish M., Musser M.A., Goldsmith J.D., Hait E., Nurko S., Brombacher F., et al. 2019. Mechanical Skin Injury Promotes Food Anaphylaxis by Driving Intestinal Mast Cell Expansion. Immunity. 50:1262–1275.e4. 10.1016/j.immuni.2019.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Liu S., Xu J., Zhang X., Han D., Liu J., Xia M., Yi L., Shen Q., Xu S., et al. 2018. Adult Connective Tissue-Resident Mast Cells Originate from Late Erythro-Myeloid Progenitors. Immunity. 49:640–653.e5. 10.1016/j.immuni.2018.09.023 [DOI] [PubMed] [Google Scholar]
- Lilla, J.N., Chen C.C., Mukai K., BenBarak M.J., Franco C.B., Kalesnikoff J., Yu M., Tsai M., Piliponsky A.M., and Galli S.J.. 2011. Reduced mast cell and basophil numbers and function in Cpa3-Cre; Mcl-1fl/fl mice. Blood. 118:6930–6938. 10.1182/blood-2011-03-343962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T., Kanaoka Y., Barrett N.A., Feng C., Garofalo D., Lai J., Buchheit K., Bhattacharya N., Laidlaw T.M., Katz H.R., et al. 2015. Aspirin-Exacerbated Respiratory Disease Involves a Cysteinyl Leukotriene-Driven IL-33-Mediated Mast Cell Activation Pathway. J. Immunol. 195:3537–3545. 10.4049/jimmunol.1500905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixiong, J., Anderson M., Limjunyawong N., Sabbagh M.F., Hu E., Mack M.R., Oetjen L.K., Wang F., Kim B.S., and Dong X.. 2019. Activation of Mast-Cell-Expressed Mas-Related G-Protein-Coupled Receptors Drives Non-histaminergic Itch. Immunity. 50:1163–1171.e5. 10.1016/j.immuni.2019.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melisi, D., Ishiyama S., Sclabas G.M., Fleming J.B., Xia Q., Tortora G., Abbruzzese J.L., and Chiao P.J.. 2008. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol. Cancer Ther. 7:829–840. 10.1158/1535-7163.MCT-07-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, H.R., Wright S.H., Knight P.A., and Thornton E.M.. 1999. A novel function for transforming growth factor-beta1: upregulation of the expression and the IgE-independent extracellular release of a mucosal mast cell granule-specific beta-chymase, mouse mast cell protease-1. Blood. 93:3473–3486. 10.1182/blood.V93.10.3473.410k01_3473_3486 [DOI] [PubMed] [Google Scholar]
- Mukai, K., Tsai M., Saito H., and Galli S.J.. 2018. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol. Rev. 282:121–150. 10.1111/imr.12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, N., Maeyama K., Sakata N., Itoh F., Akatsu R., Nakata M., Katsu Y., Ikeno S., Togawa Y., Vo Nguyen T.T., et al. 2014. C18 ORF1, a novel negative regulator of transforming growth factor-β signaling. J. Biol. Chem. 289:12680–12692. 10.1074/jbc.M114.558981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhrvik, H., Grujic M., Waern I., Gustafson A.M., Ernst N., Roers A., Hartmann K., and Pejler G.. 2016. Mast cells promote melanoma colonization of lungs. Oncotarget. 7:68990–69001. 10.18632/oncotarget.11837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orinska, Z., Maurer M., Mirghomizadeh F., Bulanova E., Metz M., Nashkevich N., Schiemann F., Schulmistrat J., Budagian V., Giron-Michel J., et al. 2007. IL-15 constrains mast cell-dependent antibacterial defenses by suppressing chymase activities. Nat. Med. 13:927–934. 10.1038/nm1615 [DOI] [PubMed] [Google Scholar]
- Otsuka, A., Kubo M., Honda T., Egawa G., Nakajima S., Tanizaki H., Kim B., Matsuoka S., Watanabe T., Nakae S., et al. 2011. Requirement of interaction between mast cells and skin dendritic cells to establish contact hypersensitivity. PLoS One. 6. e25538. 10.1371/journal.pone.0025538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejler, G., Rönnberg E., Waern I., and Wernersson S.. 2010. Mast cell proteases: multifaceted regulators of inflammatory disease. Blood. 115:4981–4990. 10.1182/blood-2010-01-257287 [DOI] [PubMed] [Google Scholar]
- Pemberton, A.D., Wright S.H., Knight P.A., and Miller H.R.. 2006. Anaphylactic release of mucosal mast cell granule proteases: role of serpins in the differential clearance of mouse mast cell proteases-1 and -2. J. Immunol. 176:899–904. 10.4049/jimmunol.176.2.899 [DOI] [PubMed] [Google Scholar]
- Razin, E., Ihle J.N., Seldin D., Mencia-Huerta J.M., Katz H.R., LeBlanc P.A., Hein A., Caulfield J.P., Austen K.F., and Stevens R.L.. 1984. Interleukin 3: A differentiation and growth factor for the mouse mast cell that contains chondroitin sulfate E proteoglycan. J. Immunol. 132:1479–1486. [PubMed] [Google Scholar]
- Reber, L.L., Marichal T., Sokolove J., Starkl P., Gaudenzio N., Iwakura Y., Karasuyama H., Schwartz L.B., Robinson W.H., Tsai M., et al. 2014. Contribution of mast cell-derived interleukin-1β to uric acid crystal-induced acute arthritis in mice. Arthritis Rheumatol. 66:2881–2891. 10.1002/art.38747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber, L.L., Sibilano R., Starkl P., Roers A., Grimbaldeston M.A., Tsai M., Gaudenzio N., and Galli S.J.. 2017. Imaging protective mast cells in living mice during severe contact hypersensitivity. JCI Insight. 2. e92900. 10.1172/jci.insight.92900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg, L.C., Nilsson A.Z., Lei Y., Gregory J.A., Adner M., and Nilsson G.P.. 2017. Interleukin 33 exacerbates antigen driven airway hyperresponsiveness, inflammation and remodeling in a mouse model of asthma. Sci. Rep. 7:4219. 10.1038/s41598-017-03674-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.J., Ducharme L.A., Shaw S.K., Parker C.M., Brenner M.B., Kilshaw P.J., and Weis J.H.. 1994. Murine M290 integrin expression modulated by mast cell activation. Immunity. 1:393–403. 10.1016/1074-7613(94)90070-1 [DOI] [PubMed] [Google Scholar]
- Sonoda, T., Hayashi C., and Kitamura Y.. 1983. Presence of mast cell precursors in the yolk sac of mice. Dev. Biol. 97:89–94. 10.1016/0012-1606(83)90066-0 [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Kudo M., Sundaram A., Ren X., Huang K., Bernstein X., Wang Y., Raymond W.W., Erle D.J., Abrink M., et al. 2012. The αvβ6 integrin modulates airway hyperresponsiveness in mice by regulating intraepithelial mast cells. J. Clin. Invest. 122:748–758. 10.1172/JCI58815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.M., Liang H.E., Bando J.K., Wu D., Cheng L.E., McKerrow J.K., Allen C.D., and Locksley R.M.. 2011. Genetic analysis of basophil function in vivo. Nat. Immunol. 12:527–535. 10.1038/ni.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini, K.J., Nabhan A.N., Penland L., Sinha R., Gillich A., Sit R.V., Chang S., Conley S.D., Mori Y., Seita J., et al. 2020. A molecular cell atlas of the human lung from single cell RNA sequencing. bioRxiv. (Preprint posted August 27, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta, J.J., Gennert D., Lu D., Satija R., Shalek A.K., and Regev A.. 2014. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr. Protoc. Mol. Biol. 107:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, M., Starkl P., Marichal T., and Galli S.J.. 2015. Testing the ‘toxin hypothesis of allergy’: mast cells, IgE, and innate and acquired immune responses to venoms. Curr. Opin. Immunol. 36:80–87. 10.1016/j.coi.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Ishiwata K., Koseki H., Ishikura T., Ugajin T., Ohnuma N., Obata K., Ishikawa R., Yoshikawa S., Mukai K., et al. 2010. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. J. Clin. Invest. 120:2867–2875. 10.1172/JCI42680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wastling, J.M., Knight P., Ure J., Wright S., Thornton E.M., Scudamore C.L., Mason J., Smith A., and Miller H.R.. 1998. Histochemical and ultrastructural modification of mucosal mast cell granules in parasitized mice lacking the beta-chymase, mouse mast cell protease-1. Am. J. Pathol. 153:491–504. 10.1016/S0002-9440(10)65592-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S.H., Brown J., Knight P.A., Thornton E.M., Kilshaw P.J., and Miller H.R.. 2002. Transforming growth factor-beta1 mediates coexpression of the integrin subunit alphaE and the chymase mouse mast cell protease-1 during the early differentiation of bone marrow-derived mucosal mast cell homologues. Clin. Exp. Allergy. 32:315–324. 10.1046/j.1365-2222.2002.01233.x [DOI] [PubMed] [Google Scholar]
- Xing, W., Austen K.F., Gurish M.F., and Jones T.G.. 2011. Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue. Proc. Natl. Acad. Sci. USA. 108:14210–14215. 10.1073/pnas.1111048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M., Tsai M., Tam S.Y., Jones C., Zehnder J., and Galli S.J.. 2006. Mast cells can promote the development of multiple features of chronic asthma in mice. J. Clin. Invest. 116:1633–1641. 10.1172/JCI25702 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows DESeq2 analysis of differential gene expression between constitutive β7Low MCs and inducible β7High MCs following sequential HDM challenges.
shows DESeq2 analysis of differential gene expression between constitutive β7Low MCs from saline or sequential HDM-challenged mice (tab 1) and inducible β7High MCs following saline or sequential HDM challenges (tab 2).
shows Seurat analysis of differentially expressed transcripts across the four human MC clusters (Wilcoxon test).
Data Availability Statement
Sequencing data have been deposited in GEO under accession no. GSE155575.