IFN-I provide a rapid antiviral defense, but they limit antigen availability during priming. Here, the authors demonstrate that short-term IFN-I blockade after primary viral infection improves adaptive immunity and protection against reinfection. This “adjuvant” effect is generalizable to viral vaccines.
Abstract
Type I interferons (IFN-I) are a major antiviral defense and are critical for the activation of the adaptive immune system. However, early viral clearance by IFN-I could limit antigen availability, which could in turn impinge upon the priming of the adaptive immune system. In this study, we hypothesized that transient IFN-I blockade could increase antigen presentation after acute viral infection. To test this hypothesis, we infected mice with viruses coadministered with a single dose of IFN-I receptor–blocking antibody to induce a short-term blockade of the IFN-I pathway. This resulted in a transient “spike” in antigen levels, followed by rapid antigen clearance. Interestingly, short-term IFN-I blockade after coronavirus, flavivirus, rhabdovirus, or arenavirus infection induced a long-lasting enhancement of immunological memory that conferred improved protection upon subsequent reinfections. Short-term IFN-I blockade also improved the efficacy of viral vaccines. These findings demonstrate a novel mechanism by which IFN-I regulate immunological memory and provide insights for rational vaccine design.
Graphical Abstract
Introduction
Type I interferons (IFN-I) are a first line of defense during viral infection. Absence of IFN-I results in disseminated viral infections and impaired priming of adaptive immune responses (Burns et al., 2016; Ciancanelli et al., 2015; Duncan et al., 2015; Dupuis et al., 2003; Erickson and Pfeiffer, 2013; Hambleton et al., 2013; Hernandez et al., 2019; Hernandez et al., 2018; Hoyos-Bachiloglu et al., 2017; Kolumam et al., 2005; Kreins et al., 2015; Le Bon et al., 2003; Minegishi et al., 2006; Moens et al., 2017; Sandler et al., 2014; Shahni et al., 2015; Stark et al., 1998; Teijaro et al., 2013; Wilson et al., 2013). Robust IFN-I responses are also a hallmark of effective vaccines (Gaucher et al., 2008; Li et al., 2017; Pulendran and Ahmed, 2011; Pulendran et al., 2010; Querec et al., 2009). However, there is an overlooked paradox between IFN-I and the elicitation of long-lived antiviral immunity: IFN-I can restrict initial viral replication, which in turn could limit antigen availability at a critical time when the adaptive immune system is getting primed. This conundrum motivated us to analyze the effects of transiently blocking IFN-I early after viral infection, with the goal of augmenting hyperacute antigen levels and subsequent antigen priming.
Here, we show that short-term IFN-I blockade during an initial viral infection results in a profound improvement of immunological memory, rendering the host better protected against subsequent reinfections with similar or more virulent pathogens. These findings are important for two reasons. First, they highlight an interesting mechanism by which innate immunity regulates long-lived immunological memory. Second, they may have important implications for rational vaccine design.
Results
Short-term IFN-I blockade during acute viral infection improves immunological memory
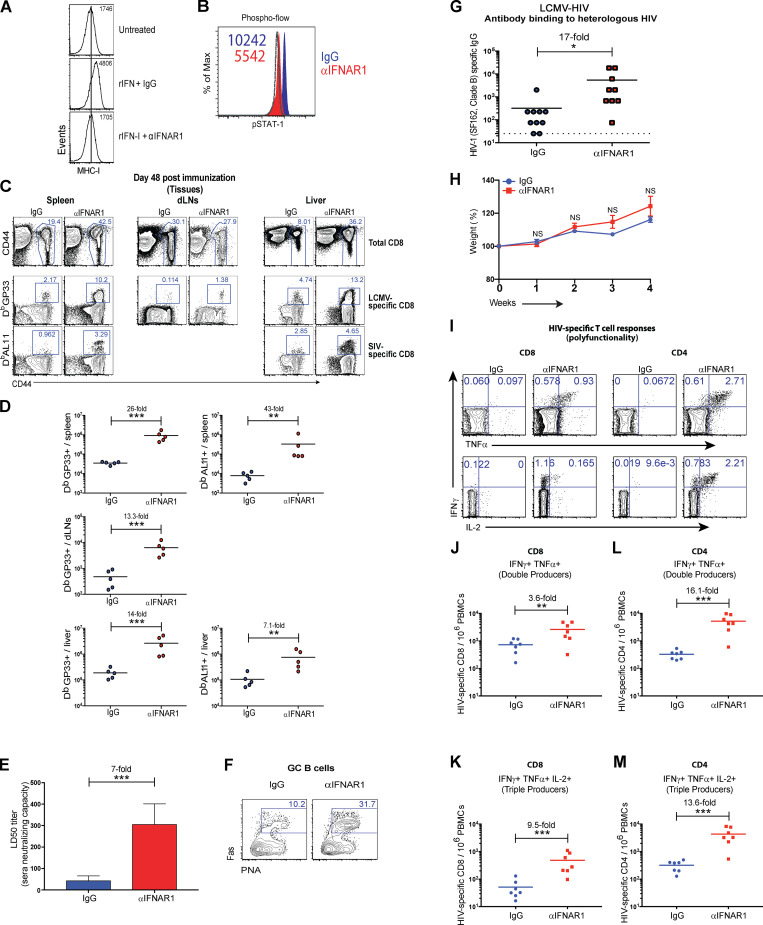
Discovered more than six decades ago, IFN-I have been shown to play an indispensable role in antiviral immunity. Long-term defects in the IFN-I pathway result in impairment of immune responses following acute viral infections or vaccinations. However, it is currently unclear whether a short-term blockade of IFN-I would have a similar effect. To transiently block the IFN-I pathway, we used an IFN-I receptor–blocking antibody (αIFNAR1, clone MAR1-5A3) that has been used in prior studies (Bhattacharyya et al., 2017; Teijaro et al., 2013; Wang et al., 2019; Wilson et al., 2013). We first corroborated that this antibody blocks IFN-I signaling in vitro (Fig. S1 A). We then immunized mice with different viruses, which were coadministered with control IgG or αIFNAR1 to induce a short-term blockade of the IFN-I pathway (Fig. 1 A). This single dose of αIFNAR1 resulted in reduced STAT1 phosphorylation (Fig. S1 B) and a short-term blockade of the IFN-I receptor lasting 96 h (Fig. 1 B). In these experiments, we coadministered the αIFNAR1 antibody locally (intramuscularly) together with the virus. As shown in Fig. 1 B, a fraction of the antibody goes systemic, since there is blockade of IFNAR1 in the blood.
Figure S1.
Effects of short-term IFN-I blockade on experimental SIV/HIV vaccines. (A) Validation of the IFNAR1 antibody (MAR1-5A3) in vitro. CT2A cells were incubated with 20 µg of IgG1 (MOPC-21) or IFNAR1-blocking antibody (MAR1-5A3) for 30 min before treatment with IFN-I (1,000 units) overnight. Representative histograms of MHC-I expression are shown. Note that MHC-I expression is induced by IFN-I signaling. (B) C57BL/6 mice were immunized intramuscularly with 104 PFU of LCMV-SIV mixed with 100 µg of control antibodies or IFNAR1-blocking antibodies, similar to Fig. 1 A. Representative histograms of phosphorylated STAT1 (pSTAT1) in whole PBMCs 1 d after infection (naive levels represented by dashed histogram, which overlapped with the αIFNAR1 group). Note that pSTAT1 is induced by IFN-I signaling. (C) Representative FACS plots showing the frequencies of LCMV- and SIV-specific CD8 T cells (gated from live CD8+ lymphocytes) in spleen, lymph nodes, and liver. (D) Summary of LCMV- and SIV-specific CD8 T cells in spleen, lymph nodes, and liver. (E) In vitro SIV neutralization in LCMV-SIV–immunized mice; sera from IgG1- or αIFNAR1-treated mice were tested for their ability to neutralize SIV in vitro. Neutralization was measured by TZM-bl assay. (F) Representative FACS plots showing the frequencies of germinal center (GC) B cells in draining lymph nodes (gated from live CD3− B220+ IgM− IgD− lymphocytes) at day 14. PNA, peanut agglutinin. (G) Levels of antibody that bind to heterologous HIV-1 (SF162, clade B envelope) at day 14. (H) Weight of mice following LCMV-HIV vaccination. (I) Representative FACS plots showing the frequencies of HIV-specific CD8 (left) and CD4 (right) T cells (gated from live CD8+ or CD4+ lymphocytes). (J) Summary of HIV-specific CD8 T cells that are double (IFNγ+ TNFα+) producers. (K) Summary of HIV-specific CD8 T cells that are triple (IFNγ+ TNFα+ IL-2) producers. (L) Summary of HIV-specific CD4 T cells that are double (IFNγ+ TNFα+) producers. (M) Summary of HIV-specific CD4 T cells that are triple (IFNγ+ TNFα+ IL-2) producers. Data in panels C and D are from day 48 after infection. All other panels are from day 14 after infection. All experiments were performed at least twice with n = 5–7 mice per group per experiment; data are from one representative experiment. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM.
Figure 1.
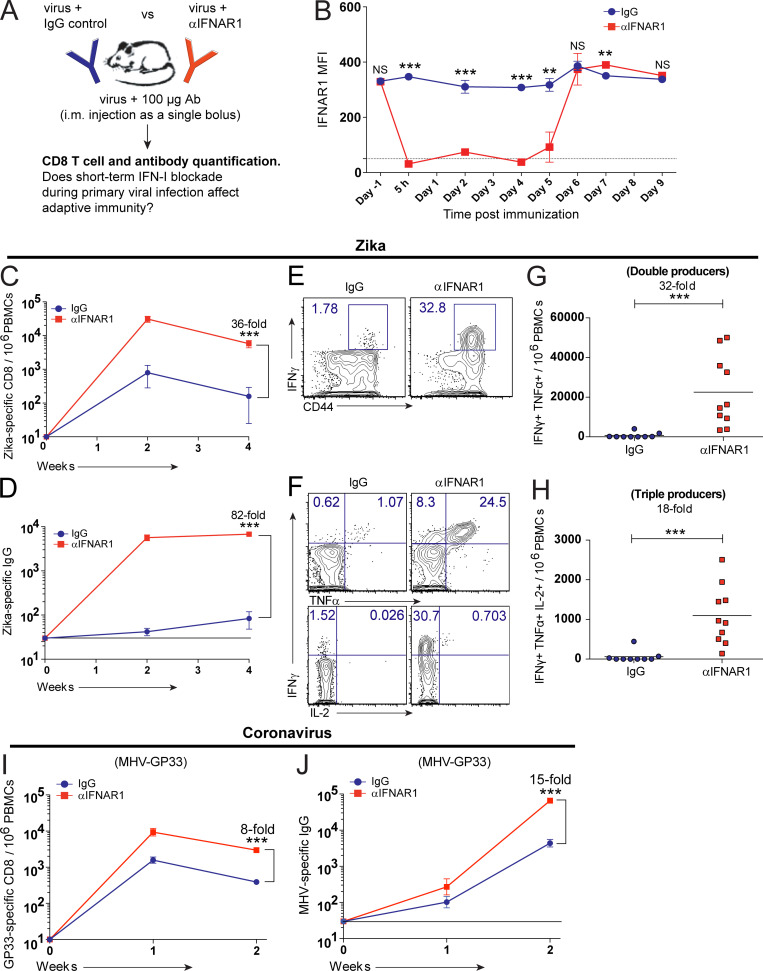
IFN-I blockade improves immunological memory after acute viral infection. (A) Experimental approach for inducing a short-term blockade of IFN-I after acute viral infection in C57BL/6 mice. (B) Duration of IFNAR1 blockade. PBMCs were stained with a fluorescently labeled αIFNAR1 antibody to evaluate competitive binding. Dotted line represents the limit of detection (IFNAR1 staining in Ifnar1−/− mice). MFI, mean fluorescence intensity. (C) Summary of Zika-specific CD8 T cells. (D) Summary of Zika-specific antibody responses. (E) Representative FACS plots showing the frequencies of IFNγ+ CD8 T cells after stimulation with Zika peptide. (F) Representative FACS plots showing the frequencies of polyfunctional CD8 T cells after stimulation with Zika peptide. (G) Summary of double cytokine producer CD8 T cells after stimulation with Zika peptide. (H) Summary of triple cytokine producer CD8 T cells after stimulation with Zika peptide. Data in panels E–H are from week 2 after infection with Zika PRV strain. Zika-specific CD8 T cells were enumerated after 5-h stimulation with a Zika-specific peptide (IGVSNRDFV). (I) Summary of GP33-specific CD8 T cells after coronavirus infection (MHV-GP33). (J) Summary of coronavirus (MHV)-specific antibody responses. Bottom lines in panels D and J indicate limit of detection. Data represent two or more combined experiments; n = 4–5 per each independent experiment. Zika infections were with 104 PFU/mouse, and coronavirus infections were with 102 PFU/mouse. **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM.
Interestingly, short-term IFN-I blockade after a Zika virus infection resulted in a 36-fold improvement in CD8 T cell responses and an 82-fold improvement in antibody responses (Fig. 1, C–E). Short-term IFN-I blockade also resulted in a substantial increase in cytokine coexpressing T cells relative to control immunized mice (Fig. 1, F–H). Short-term IFN-I blockade also improved adaptive immune responses following an acute coronavirus infection with mouse hepatitis virus (MHV; Fig. 1, I and J), which is in the same genera as SARS-CoV (Betacoronaviridae). These data showed that short-term or hyperacute blockade of IFN-I improves the immunogenicity of acute viral infections.
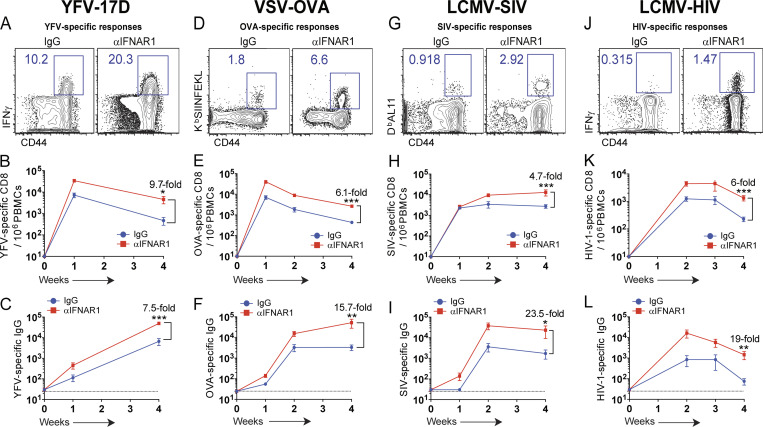
We then evaluated whether these same effects could apply to viral vaccines. Interestingly, IFN-I blockade improved the immunogenicity of the clinically approved yellow fever virus vaccine (YFV-17D; Fig. 2, A–C). Rhabdovirus- and arenavirus-based vaccines were also significantly improved after short-term IFN-I blockade (Fig. 2, D–L; and Fig. S1, C and D). IFN-I blockade during lymphocytic choriomeningitis virus (LCMV)–simian immunodeficiency virus (SIV) vaccination significantly increased antibody-mediated SIV neutralization compared with control, as measured by in vitro neutralization assays (Fig. S1 E). In the context of LCMV-HIV vaccination, short-term IFN-I blockade facilitated germinal center B cell responses (Fig. S1 F), which suggested that antibody diversification was improved. A main challenge for developing vaccines against highly evolving viruses, such as HIV, is that vaccines based on a specific viral strain may not confer substantial cross-reactive humoral immunity against heterologous viral strains. However, IFN-I blockade resulted in a 17-fold improvement in cross-reactive humoral immunity after vaccination (Fig. S1 G). Overall, our short-term IFN-I blockade regimen seemed well tolerated, and mice did not exhibit overt weight loss after infection (Fig. S1 H). In addition, IFN-I blockade improved cytokine coexpression by HIV-specific T cells compared with control vaccination (Fig. S1, I–M). Altogether, these data with different viral vaccines suggest that short-term IFN-I blockade paradoxically acts as a potent adjuvant.
Figure 2.
IFN-I blockade improves immunological memory after viral vaccination. C57BL/6 mice were immunized intramuscularly with 104 PFU of the indicated vaccines mixed with control antibodies or IFNAR1-blocking antibodies, similar to Fig. 1 A. (A) Representative FACS plots showing frequencies of YFV-specific (IFNγ+) CD8 T cells after 5-h peptide stimulation (IGITDRDFI). (B) Summary of YFV-specific CD8 T cells. (C) Summary of YFV-specific antibody responses. (D) Representative FACS plots showing the frequencies of OVA-specific (KbSIINFEKL+) CD8 T cells. (E) Summary of OVA-specific CD8 T cells. (F) Summary of OVA-specific antibody responses. (G) Representative FACS plots showing the frequencies of SIV-specific (DbAL11+) CD8 T cells. (H) Summary of SIV-specific (DbAL11+) CD8 T cells. (I) Summary of SIV-specific antibody responses. (J) Representative FACS plots showing the frequencies of HIV-specific (IFNγ+) CD8 T cells after 5-h peptide stimulation (BG505 envelope peptide pools). (K) Summary of HIV-specific CD8 T cells. (L) Summary of HIV-specific antibody responses (BG505, clade A envelope). FACS plots are gated from total CD8 T cells at week 2 after infection. Dotted lines indicate limit of detection. Experiments were performed two to four times; n = 5 mice per experiment (data from all experiments are shown). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM.
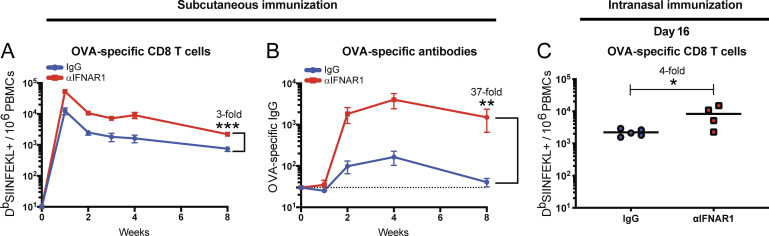
The data above involved intramuscular infection/vaccination, so we investigated whether these effects were dependent on the route. We observed a similar increase in immune responses when mice were injected via the subcutaneous or intranasal routes (Fig. S2), suggesting that our observations were not dependent on the route of infection.
Figure S2.
IFN-I blockade also improves immunological memory following subcutaneous or intranasal infection. C57BL/6 mice were immunized subcutaneously (A and B) or intranasally (C) with 104 PFU of VSV-OVA mixed with control antibodies or αIFNAR1. (A) Summary of OVA-specific CD8 T cells in PBMCs. (B) Summary of OVA-specific antibody responses in sera. (C) Summary of OVA-specific CD8 T cells in PBMCs at day 16 after infection. Data from panels A and B are combined from two experiments, with a total of eight to nine mice per group; data from panel C is from one experiment with four to five mice per group. Dotted line represents limit of detection. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM.
Transcriptional and virological analyses
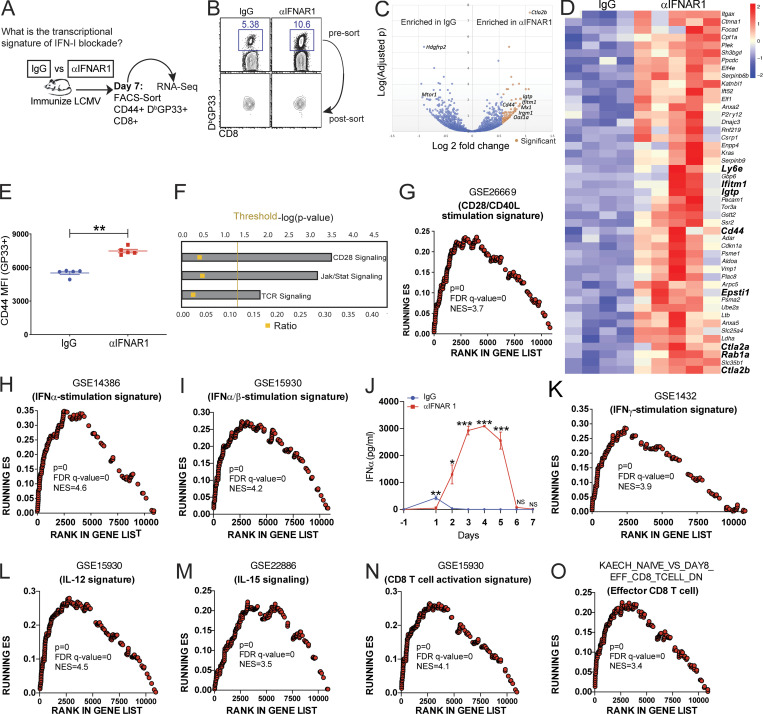
We analyzed gene expression on virus-specific CD8 T cells at the peak of the response, 7 d after acute infection with LCMV (Fig. 3, A and B). Short-term IFN-I blockade induced increased CD44 expression at the gene level (Fig. 3, C and D) and at the protein level (Fig. 3 E) relative to control. IFN-I blockade also induced enriched expression of TCR signaling genes by Ingenuity Pathway Analyses (IPA; Fig. 3 F) and costimulation genes by IPA and gene set enrichment analyses (GSEA; Fig. 3, F and G). Paradoxically, IFN-I–driven genes and IFN-I levels were increased in mice that received IFN-I blockade earlier during infection (Fig. 3, H–J), suggesting a “compensatory” IFN-I response. There was also enrichment in IFNγ, IL-12, and IL-15 signaling genes in mice that received IFN-I blockade (Fig. 3, K–M), and these mice also exhibited increased activation and effector signatures (Fig. 3, N and O). These data suggest that IFN-I blockade during the priming phase improved TCR and costimulation signaling later during the effector phase. TCR triggering and costimulation are two main signals required for activation of the adaptive immune system. Furthermore, our data showing increases in IFN-I, IFNγ, IL-12, and IL-15 cytokine signatures also suggested an increase in the so-called “third signal” needed for the activation of T cell responses.
Figure 3.
Transcriptional analyses of virus-specific CD8 T cells. C57BL/6 mice were immunized intramuscularly with 104 PFU of acute LCMV mixed with control antibodies or αIFNAR1 antibodies similar to Fig. 1 A, and gene expression was assessed on virus-specific CD8 T cells after 7 d. (A) Experimental approach for performing RNA-Seq. Splenic CD8 T cells were MACS purified by negative selection, followed by FACS sorting of live, CD8+, CD44+, and DbGP33+ cells. This resulted in ∼97% pure population of virus-specific CD8 T cells for transcriptional analyses. (B) DbGP33+ CD8 T cell purity test. (C) Volcano plot showing differentially expressed genes between mice that received IgG or αIFNAR1 at the time of infection. (D) Heat map showing the top genes up-regulated in αIFNAR1. (E) Validation of CD44 expression at the protein level by flow cytometry. MFI, mean fluorescence intensity. (F) IPA. (G) GSEA of T cell costimulation–driven genes. The GSE26669 signature contains genes that are normally increased after T cell costimulation. (H) GSEA of IFNα stimulation signature. (I) GSEA of dual IFNα/β stimulation signature. (J) Validation of IFN-I levels at the protein level using ELISA. (K) GSEA of IFNγ stimulation signature. (L) GSEA of IL-12 stimulation signature. (M) GSEA of IL-15 stimulation signature. (N) GSEA of CD8 T cell activation signature. (O) GSEA of effector CD8 T cell signature. RNA-Seq experiments were performed once, using four mice in the control group and five mice in the αIFNAR1 group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM. FDR, false discovery rate; NES, normalized enrichment score; ES, enrichment score.
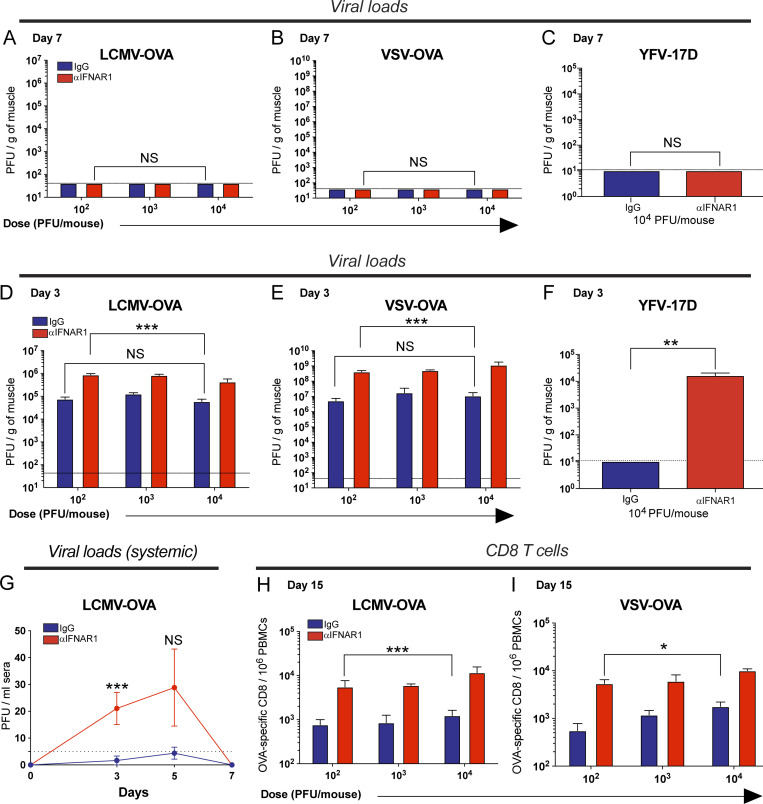
We hypothesized that our short-term IFN-I blockade regimen could give a transient advantage to the virus, increasing antigen availability. To evaluate this, muscles were harvested at day 7 after intramuscular infection, and viral loads were quantified by plaque assays. Complete viral clearance was observed at day 7 in all mice, demonstrating that short-term IFN-I blockade does not prevent the resolution of acute viral infection, which normally occurs within a week (Fig. S3, A–C). Since IFN-I plays an early antiviral effect shortly after viral encounter, we hypothesized that IFN-I blockade could increase viral burden at very early time points following infection. To test this, we performed plaque assays at earlier time points 72 h after infection. Consistent with our hypothesis, IFN-I blockade resulted in a sharp increase in viral titers (Fig. S3, D–F). These data demonstrate that short-term IFN-I blockade induces a transient hyperacute burst in viral antigen, which is then rapidly cleared from the body.
Figure S3.
Short-term IFN-I blockade transiently increases viral loads without abrogating viral clearance. C57BL/6 mice were immunized intramuscularly with escalating doses of the indicated viral vectors, mixed with control antibodies or αIFNAR1 antibodies, and viral loads were quantified at the site of infection (muscle) at different time points. (A) Viral loads at day 7 after LCMV-OVA infection. (B) Viral loads at day 7 after VSV-OVA infection. (C) Viral loads at day 7 after YFV-17D infection. (D) Viral loads at day 3 after LCMV-OVA infection. (E) Viral loads at day 3 after VSV-OVA infection. (F) Viral loads at day 3 after YFV-17D infection. Quadriceps were harvested at the indicated time points, and viral load was quantified by plaque assay. (G) Systemic viral load after intramuscular LCMV-OVA infection (104 PFU/mouse). This panel shows that very low viremia is detected after intramuscular infection. Undiluted sera were used to improve plaque assay sensitivity. (H) OVA-specific (KbSIINFEKL) CD8 T cells at day 15 after LCMV-OVA infection. (I) OVA-specific CD8 T cells at day 15 after VSV-OVA infection. Note that increasing vaccine dose does not significantly alter early viral loads and does not induce a commensurate increase in immunogenicity. Dotted lines represent limit of detection. Data are from two to three experiments, with four to five mice per group (all data are shown). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM.
A prior study in individuals receiving the YFV-17D vaccine showed a positive correlation between vaccine replication and T cell responses (Akondy et al., 2015). Namely, individuals with the highest level of vaccine replication showed the most potent T cell response following vaccination. A logical prediction from that study is that increasing the dose of the vaccine would increase antigen levels and subsequent immunogenicity. However, we show that increasing vaccine dose in control mice (from 102 to 104 PFU, blue bars) does not significantly augment early viral loads 72 h after LCMV infection (Fig. S3 D). Similar virologic effects were observed after vesicular stomatitis virus (VSV) vaccination and YFV-17D vaccination (Fig. S3, E and F). These results show that there is a strict limit on how much antigen can be expressed at the site of infection. These data also highlight that antigen levels depend mostly on the exponential replication of the virus, which critically depends on the translational machinery of infected cells. Since protein synthesis is inhibited by IFN-stimulated genes (Schoggins, 2019), this can help explain why injecting more virus did not substantially increase acute antigen loads.
It is important to highlight that, following intramuscular infection, most of the virus replicates in situ, and very low levels of virus were detected in circulation. Systemic viral load was also resolved by day 7 in all mice (Fig. S3 G). This demonstrates that short-term IFN-I blockade does not abrogate the ultimate clearance of the viral infection within a week. We also show that increasing viral dose does not significantly increase immunogenicity (Fig. S3, H and I). In summary, our findings demonstrate that short-term IFN-I blockade increases early antigen availability and immunogenicity in a way that cannot be recapitulated simply by increasing viral dose.
Mechanism: Effects of IFN-I blockade on antigen presentation and costimulation
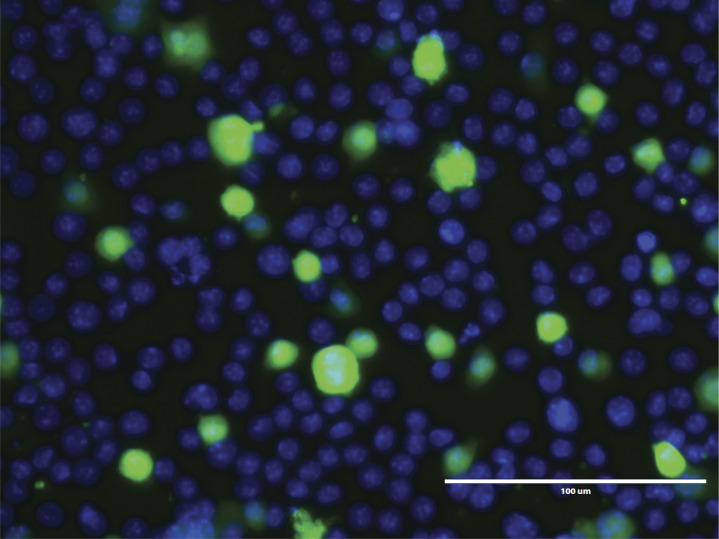
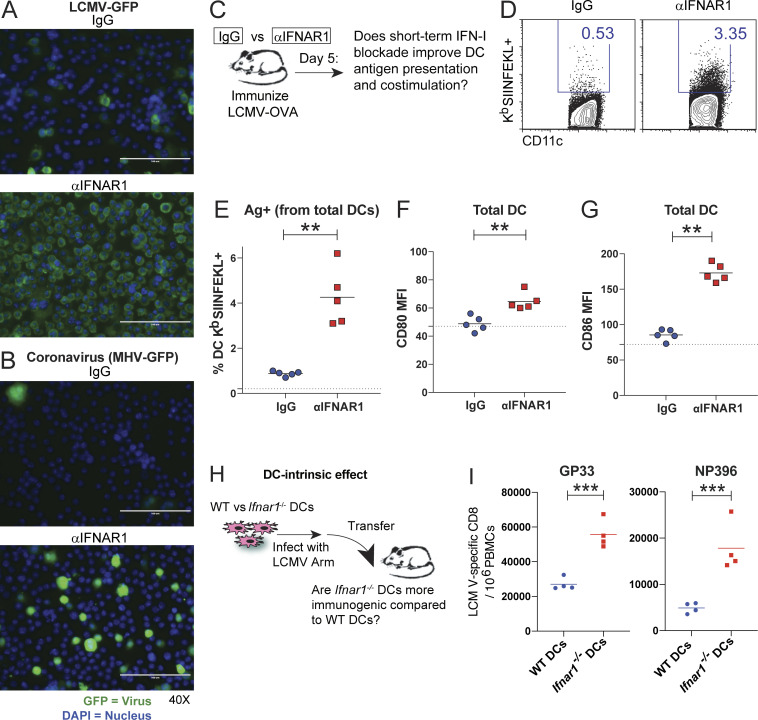
Our plaque assay data shown above demonstrate that short-term IFN-I blockade induces a transient increase in viral antigen 72 h after infection. Those experiments, however, did not specifically measure antigen loads in antigen-presenting cells, which are critical for the induction of adaptive immunity. We thus examined the effects of IFN-I blockade on dendritic cells (DCs) using an in vitro infection system. We cultured DCs with recombinant LCMV or MHV coronavirus–expressing GFP, with or without IFN-I blockade, and then we evaluated GFP expression on DCs. Our results show that IFN-I blockade resulted in more copious infection foci relative to control (Fig. 4, A and B).
Figure 4.
Phenotypic and functional analyses of DC responses. (A) Murine DCs were infected with LCMV-GFP in the presence of control antibodies or αIFNAR1 antibodies. (B) DCs infected with coronavirus (MHV-GFP) in the presence of control antibodies or αIFNAR1 antibodies. DCs were imaged by immunofluorescence 72 h after infection (see Materials and methods). Green (GFP) indicates viral foci and DAPI (blue) indicates cell nuclei. (A and B) Scale bars are 100 µm. (C) Experimental approach for analyzing DC responses in vivo. C57BL/6 mice were immunized intramuscularly with 104 PFU of LCMV-OVA mixed with control antibodies or αIFNAR1 antibodies, similar to Fig. 1 A. After 5 d, splenic DCs were characterized. (D) Representative FACS plots showing DCs that present cognate antigen in the context of MHC-I (KbSIINFEKL+). FACS plots gated on DCs (live, NK1.1−, CD3−, CD19−, Ly6G−, and CD11c+). (E) Summary of DCs that present cognate antigen. (F) Costimulatory CD80 expression on DCs. (G) Costimulatory CD86 expression on DCs. (H) Experimental approach to interrogate the DC-intrinsic effects of IFN-I. Wild-type or Ifnar1−/− DCs were infected with LCMV, and after 1 d, 2.5 × 105 DCs were intravenously transferred into naive mice to measure CD8 T cell priming. (I) Summary of LCMV DbGP33- (left panel) and DbNP396 (right panel)-specific CD8 T cell responses at day 7 after DC transfer. Experiments were performed two times; n = 4–5 mice per experiment (data from one representative experiment are shown). Dotted lines represent limit of detection based on naive mean fluorescence intensity (MFI) levels. **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test.
We also interrogated whether DCs from mice that received short-term IFN-I blockade were more effective at presenting cognate antigen and expressing costimulatory molecules (Fig. 4 C). DCs from mice that received IFN-I blockade during primary viral infection showed higher levels of MHC-I molecules presenting cognate antigen (Fig. 4, D and E) and expressed higher levels of costimulatory molecules (Fig. 4, F and G) relative to control. At first glance, these data seemed counterintuitive, given that IFN-I signaling is a positive regulator of antigen presentation and costimulation. However, as shown earlier, blockade of the IFN-I pathway lasted only ∼96 h, and this was followed by a compensatory IFN-I response (Fig. 3, H–J), which could have explained the increase in antigen presentation and costimulation by DCs after day 5.
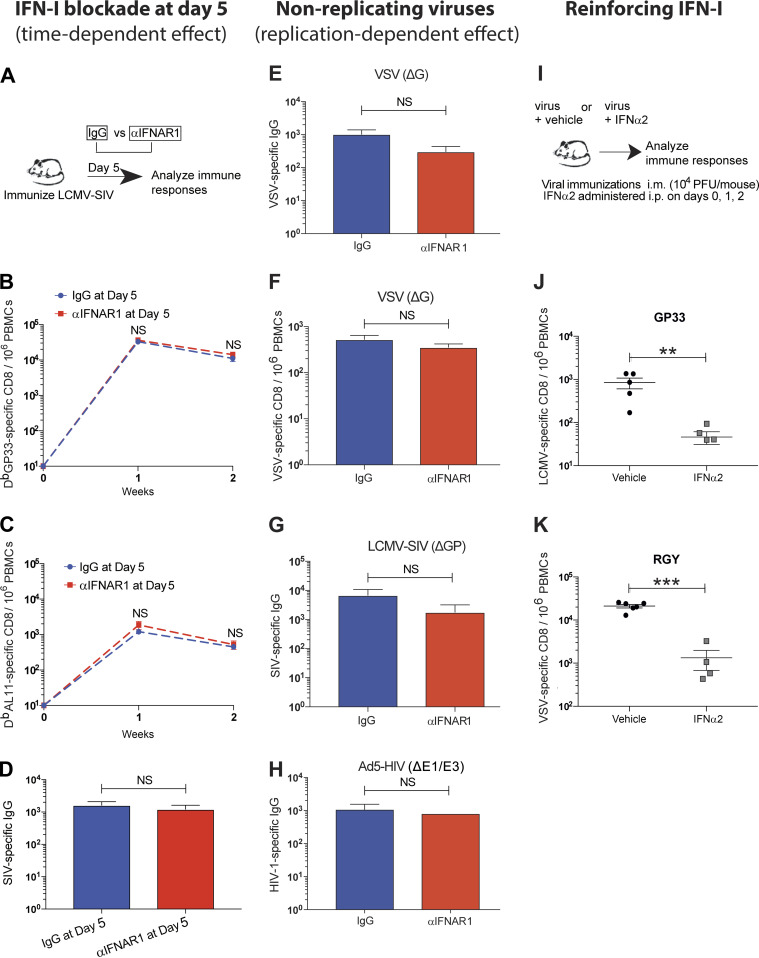
The IFN-I receptor is widely expressed on many cells, including DCs, which are also major producers of IFN-I (Reizis et al., 2011). This motivated us to study the DC-intrinsic effects of IFN-I signaling. We evaluated whether the absence of IFN-I signaling specifically on DCs would phenocopy the effect of short-term IFN-I blockade. We performed DC vaccinations using Ifnar1−/− DCs infected 24 h earlier with LCMV Armstrong (Fig. 4 H). Strikingly, transfer of Ifnar1−/− DCs resulted in greater CD8 T cell responses relative to wild-type DCs (Fig. 4 I), suggesting that IFN-I blockade modulated DC function. We also performed adoptive transfers of LCMV-specific T cells lacking IFNAR1 (Ifnar1−/−) followed by acute LCMV infection, and, consistent with prior reports (Kolumam et al., 2005; Havenar-Daughton et al., 2006), the permanent absence of IFN-I signaling on T cells resulted in long-term impairment of these responses (data not shown). According to prior studies, this could be explained by NK (Natural killer) cell–mediated killing of activated Ifnar1−/− T cells (Crouse et al., 2014; Xu et al., 2014). These earlier studies indicate that IFN-I signaling is intrinsically required for T cell immunity. Furthermore, we evaluated whether the adjuvant effect of IFN-I blockade was time dependent. We show that IFN-I blockade at day 5 after infection has no effect (Fig. S4, A–D), likely because IFN-I production normally subsides within 2–3 d of acute viral infection (Fig. 3 J; Norris et al., 2013; Zuniga et al., 2008).
Figure S4.
The adjuvant effect of short-term IFN-I blockade is time and virus replication dependent. (A–D) C57BL/6 mice were immunized intramuscularly with 104 PFU of LCMV-SIV, and at day 5 after infection, mice were treated with control antibodies or αIFNAR1 antibodies intramuscularly. (A) Experimental approach for evaluating the effect of IFN-I blockade at day 5. (B) Summary of vector-specific (DbGP33+) CD8 T cell responses. (C) Summary of SIV-specific (DbAL11+) CD8 T cell responses. (D) Summary of antibody responses in sera at day 14. (E) VSV-specific IgG in sera after infection with 104 PFU of VSV (ΔG). (F) VSV-specific CD8 T cells (KbRGY+) after infection with 104 PFU of VSV (ΔG*G). (G) SIV-specific IgG after infection with 104 PFU of LCMV-SIV (ΔGP). (H) HIV-specific IgG in sera after infection with 109 viral particle of Ad5-HIV (ΔE1/E3). Data from panels E–H are from day 14 after infection. (I) Experimental approach for evaluating immune responses after acute viral infection with IFN-I supplementation. (J) LCMV-specific (DbGP33+) CD8 T cells in blood at day 7 after infection with LCMV-HIV. (K) VSV-specific (KbRGY+) CD8 T cells in blood at day 7 after infection with VSV-OVA. All data are from PBMCs or sera. All experiments were performed at least twice with four to five mice per group per experiment; results of a representative experiment are shown. For panels J and K, IFNα2 was administered intraperitoneally on days 0, 1, and 2 (5 µg/dose). All experiments were performed at least twice with four to five mice per group per experiment. **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM.
IFN-I exert antiviral effects by regulating various biological processes; for example, by inhibiting protein translation in infected cells or by limiting a second round of infection in adjacent cells (Bailey et al., 2014; Farrell et al., 1978; McMichael et al., 2018). To determine if the adjuvant effect of short-term IFN-I blockade was dependent on the latter process, we immunized mice with single-round (nonreplicating) viruses. We first used a VSV vector that can enter cells and translate viral proteins but cannot induce a second round of infection due to genetic absence of the VSV G protein. Interestingly, short-term IFN-I blockade did not improve the immunogenicity of this virus (Fig. S4, E and F). Similar effects were reported with nonreplicating LCMV and Ad5 viruses (Fig. S4, G and H). Therefore, the adjuvant effect of IFN-I blockade is mechanistically dependent on whether the virus can cause secondary foci of infection. Until now, our experiments have involved IFN-I blockade, and we next interrogated whether IFN-I supplementation would have an opposite effect (Fig. S4 I). Systemic administration of IFN-I throughout the first 48 h of infection impaired primary immune responses (Fig. S4, J and K), consistent with a prior study that evaluated the effect of IFN-I supplementation (Honke et al., 2012). Collectively, these data suggest that the potent adjuvant effect of short-term IFN-I blockade is dependent on the timing and the ability of the virus to undergo additional rounds of infection.
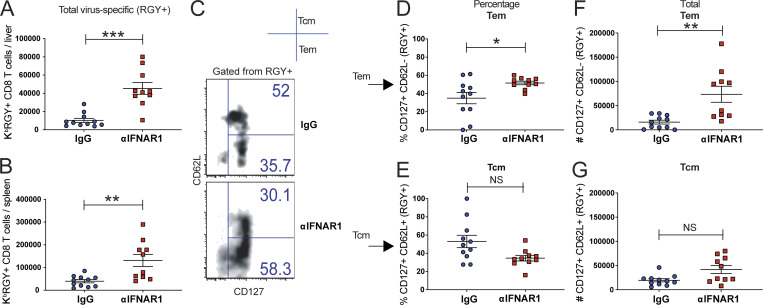
As shown earlier, short-term IFN-I blockade increases the total number of memory CD8 T cells, and the next question was if this is caused by preferential expansion of specific memory subsets. To answer this question, we immunized mice with VSV with or without IFN-I blockade, and then we immunophenotyped virus-specific CD8 T cells after 60 d. Consistent with our prior results, short-term IFN-I blockade increased the numbers of virus-specific CD8 T cells in tissues (Fig. 5, A and B). Interestingly, most of the increase in the IFN-I blockade group was due to an increase in effector memory CD8 T cells (Fig. 5, C–G). A salient feature of effector memory CD8 T cells is their response-ready state (Wherry et al., 2003), which can provide rapid sterilizing protection following subsequent reinfections, especially in the context of highly replicating pathogens. This motivated us to perform challenge studies.
Figure 5.
Memory T cell subset analyses after short-term IFN-I blockade. C57BL/6 mice were immunized intramuscularly with 104 PFU of VSV-OVA mixed with control antibodies or αIFNAR1 antibodies, similar to Fig. 1 A. Phenotypic characterization of virus-specific CD8 T cells was performed at day 60. (A) Number of VSV (KbRGY)-specific CD8 T cells in liver. (B) Number of VSV-specific CD8 T cells in spleen. (C) Representative FACS plots showing memory subsets on VSV-specific CD8 T cells from spleen. (D) Frequency of effector memory CD8 T cells (CD62L−/CD127+) in spleen. (E) Frequency of central memory CD8 T cells (CD62L+/CD127+) in spleen. (F) Number of effector memory CD8 T cells (CD62L−/CD127+) in spleen. (G) Number of central memory CD8 T cells (CD62L+/CD127+) in spleen. Experiment was performed two times (data from one representative experiment are shown). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by the Mann-Whitney U test. Error bars represent SEM.
IFN-I blockade during viral prime improves host protection following future reinfections
Does IFN-I blockade during an initial viral prime improve host protection after subsequent reinfections? To answer this simple question, we immunized mice with or without IFN-I blockade, and after several weeks we challenged mice with the same virus or a related pathogen to measure anamnestic immune protection. We used different challenge models to evaluate generalizability and to assess the contribution of different arms of the adaptive immune response in anamnestic immune protection.
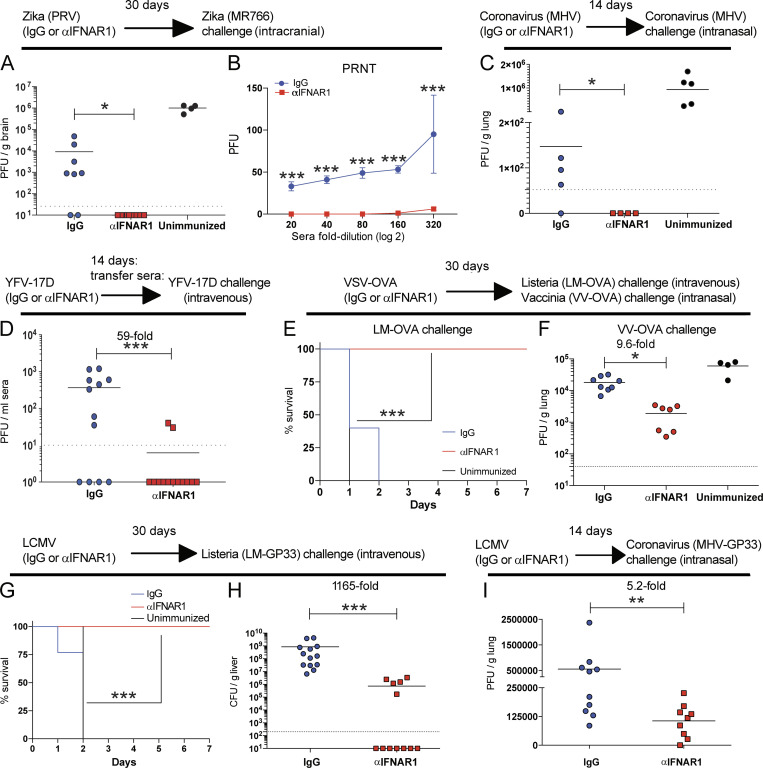
In our first challenge model, mice were immunized with Zika (Puerto Rico virus [PRV] strain) with or without IFN-I blockade. After 30 d, mice were challenged intracranially with different Zika (MR766 strain) to measure cross-protection. As expected, control Zika-immune mice showed only partial protection after heterologous Zika rechallenge when compared with unimmunized mice (Fig. 6 A). But mice that received an initial Zika infection with IFN-I blockade exhibited sterilizing immunity after subsequent heterologous Zika challenge (Fig. 6 A). These mice also exhibited improved antibody neutralization capacity by plaque reduction neutralization titer assays, with significant neutralization of heterologous Zika virus even at 320-fold sera dilution (Fig. 6 B).
Figure 6.
IFN-I blockade during an initial viral prime improves future host protection following future reinfections. C57BL/6 mice were immunized intramuscularly with the indicated viruses mixed with control antibodies or αIFNAR1 antibodies, similar to Fig. 1 A, and immune protection was assessed weeks later using various pathogen challenges. (A and B) Immune protection in Zika PRV–primed mice. (A) Viral titers in brain at day 3 following intracranial Zika MR766 challenge (104 PFU). (B) Antibody-mediated Zika MR766 neutralization in sera 14 d following Zika PRV prime by plaque reduction neutralization titer (PRNT) assays. (C) Immune protection in coronavirus (MHV)-primed mice. Viral titers in lungs at day 3 following intranasal coronavirus challenge (106 PFU). (D) Immune protection in YFV-17D–primed mice. Viral titers in sera at day 3 following intravenous YFV-17D challenge (106 PFU). In this experiment, we first vaccinated wild-type mice with YFV-17D, and after 2 wk we transferred sera from these immune mice into naive Ifnar1−/− mice (which are highly susceptible to YFV-17D). 1 d later, the recipient Ifnar1−/− mice were challenged intravenously with YFV-17D. (E and F) Immune protection in VSV-OVA–primed mice. (E) Survival following intravenous supra-lethal challenge with 107 CFU of LM-OVA. (F) Viral titers in lung at day 5 following intranasal challenge with 2 × 106 PFU of VV-OVA. (G and H) Immune protection in LCMV-immunized mice. (G) Survival following intravenous supra-lethal challenge with 107 CFU of LM-GP33. (H) Bacterial titers in liver (∼36 h after LM-GP33 challenge). (I) Viral titers in lung at day 3 following intranasal challenge with 106 PFU of coronavirus-GP33 (MHV-GP33). Limit of detection is 47 PFU. Dotted lines indicate limit of detection. Experiments were performed two times (data from all experiments are shown, except for panel C, which has data from only one experiment). The Mann-Whitney U test was used in most panels, except for panels E and G, which used the Mantel-Cox test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by indicated statistical tests. Error bars represent SEM.
In our second challenge model, we interrogated whether a primary coronavirus infection with short-term IFN-I blockade would improve protection to coronavirus reinfection. In control immune mice, prior exposure to coronavirus conferred partial protection upon coronavirus rechallenge, but only 20% of mice exhibited sterilizing immunity (Fig. 6 C). However, 100% of the mice that received IFN-I blockade during the primary coronavirus infection exhibited sterilizing immunity following subsequent coronavirus reinfections (Fig. 6 C).
In our third challenge model, we used the clinically approved YFV-17D vaccine with or without IFN-I blockade. Since immunized wild-type mice are highly resistant to YFV-17D infection, we used a passive immunization model using Ifnar1−/− recipient mice, which are highly susceptible to YFV-17D (Erickson and Pfeiffer, 2013). We first immunized wild-type mice with YFV-17D intramuscularly with or without IFN-I blockade. After 2 wk, we transferred immune sera from these mice into naive Ifnar1−/− recipient mice, followed by systemic challenge with YFV-17D. Strikingly, the sera of mice that received IFN-I blockade during prior vaccination conferred sterilizing immunity in most recipient mice (Fig. 6 D).
In our fourth challenge model, mice were immunized with a VSV-OVA vaccine and then challenged 30 d later with a supra-lethal dose of Listeria monocytogenes (LM)–expressing OVA (LM-OVA). Control immune mice succumbed to this bacterial challenge, whereas mice that had previously received IFN-I blockade during the initial VSV-OVA prime survived the subsequent LM-OVA challenge (Fig. 6 E). Enhancement of immune protection was also observed after a viral challenge with vaccinia virus (VV)–expressing OVA (Fig. 6 F).
In our fifth challenge model, mice were immunized with LCMV, and after 30 d, mice were challenged with a supra-lethal dose of LM expressing a CD8 T cell epitope derived from LCMV (LM-GP33). As expected, all naive and control immune mice succumbed rapidly to this supra-lethal bacterial challenge. However, all of the mice that received IFN-I blockade during the initial LCMV prime 30 d earlier survived the subsequent LM-GP33 challenge (Fig. 6 G), with 58% of animals exhibiting sterilizing immunity (Fig. 6 H). Finally, we used a model for coronavirus vaccination in which mice are first vaccinated with an LCMV vector, and after several weeks, they were challenged intranasally with coronavirus expressing a CD8 T cell epitope from LCMV (MHV-GP33). Interestingly, IFN-I blockade during an initial vaccination conferred a fivefold improvement in immune protection against coronavirus (Fig. 6 I). Collectively, these data show that short-term IFN-I blockade during an initial viral infection or viral vaccination renders the host better protected against subsequent infections with similar or related pathogens.
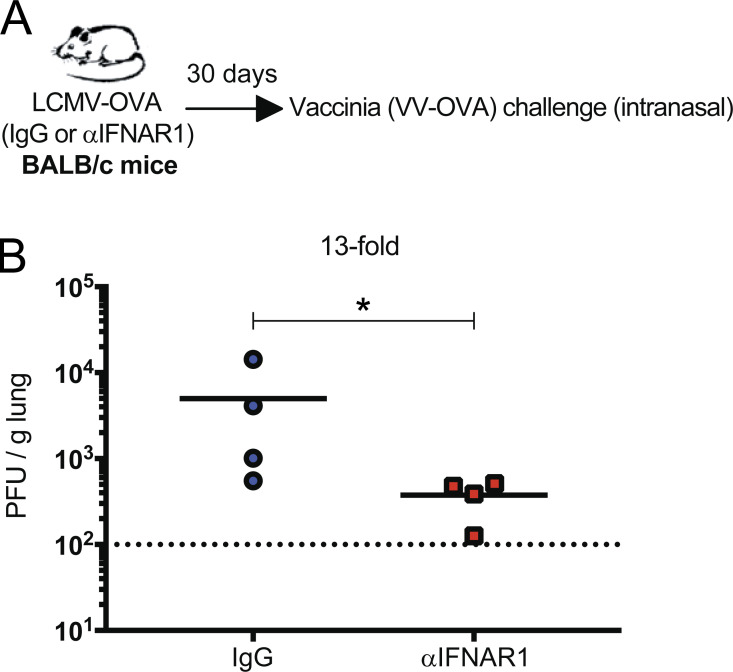
Note that most challenge models used in Fig. 6 evaluated overall protection conferred by both cellular and humoral responses. However, the experiments in Fig. 6 D specifically evaluated protection by humoral responses, whereas the experiments in Fig. 6, G–I, specifically evaluated protection by CD8 T cell responses (since only a CD8 epitope was matched between the primary and secondary infection). These results were all using C57BL/6 mice, but similarly, BALB/c mice also showed improved anamnestic protection when short-term IFN-I blockade was administered during the initial viral prime (Fig. S5). Altogether, our studies using different viruses, routes, challenges, and host genetic backgrounds show that a transient blockade of IFN-I signaling during an initial virus encounter can improve immunological memory and protection against future reinfections. Such results demonstrate a novel finding: short-term IFN-I blockade during an initial viral prime induces a long-term improvement in immunological memory. This positive effect was reproduced among different acute viral infections as well as vaccinations.
Figure S5.
Short-term IFN-I blockade during viral prime improves anamnestic protection in BALB/c mice. BALB/c mice were immunized intramuscularly with 104 PFU of LCMV-OVA mixed with control IgG antibodies or αIFNAR1. After 30 d, mice were challenged intranasally with 106 PFU of VV-OVA, and viral loads were quantified in lungs at day 5 after challenge. (A) Experimental approach for evaluating whether IFN-I blockade improves vaccine-induced protection in a different host genetic background. (B) Viral loads in lungs at day 5 after challenge. Data are from one representative experiment with four mice. Experiment was performed twice with similar results. Dotted line represents limit of detection. *, P < 0.05 by the Mann-Whitney U test.
Discussion
It is widely accepted that IFN-I play a critical role in the generation of immunological memory following acute infection or vaccination. However, most studies on IFN-I have focused on the “all-or-none” effects of IFN-I, for example, in the context of IFNAR1 genetic mutations that permanently impair IFN-I sensing or in the setting of long-term IFN-I blockade (Hernandez et al., 2019; Kolumam et al., 2005; Le Bon et al., 2003; Müller et al., 1994; Sandler et al., 2014; Teijaro et al., 2013; Wilson et al., 2013). Therefore, the effects of blocking IFN-I short term, specifically at the time of viral prime, are not well studied. Here, we demonstrate an unexpected favorable effect of blocking IFN-I short term.
Prior studies have also evaluated the effects of IFN-I blockade in the context of immune exhaustion caused by chronic infections. Elegant papers by Brooks, Oldstone, and others have shown that IFN-I blockade can ameliorate immune exhaustion during chronic LCMV infection, suggesting that IFN-I play a negative role during chronic LCMV infection. This effect is not observed in other chronic viral infections (HIV, SIV, and Hepatitis C virus) in which IFN-I are thought to play a positive role, suggesting virus-dependent effects (Torriani et al., 2004; Azzoni et al., 2013; Teijaro et al., 2013; Wilson et al., 2013; Sandler et al., 2014; Bhattacharyya et al., 2017; Cheng et al., 2017). Our study is conceptually different than prior studies because we elucidate the effects of IFN-I blockade in the context of immunological memory, focusing on how short-term modulation of this pathway, specifically at the time of prime, affects susceptibility to future reinfections. We demonstrate across multiple viral systems that short-term blockade of IFN-I improves immunological memory and anamnestic immune protection. Notably, this positive effect was also extended to clinically approved vaccines, including YFV-17D, as well as experimental HIV-1 vaccines. In this setting, we show that short-term IFN-I blockade improved the immune coverage afforded by HIV vaccines, rendering antibody responses better able to recognize variant envelopes from different HIV clades.
Although IFN-I–modulated vaccines seemed safe in mice, future studies are needed to evaluate safety in primates. IFN-I blockers have already been used in humans to ameliorate inflammatory diseases and have shown acceptable safety profiles (Casey et al., 2018; Felten et al., 2019; Tanaka et al., 2020). A reasonable counterargument against IFN-I–modulated vaccines, however, is that increasing vaccine replication creates a safety concern. Nevertheless, all acute viruses and vaccines were completely cleared within a week of injection. Moreover, short-term IFN-I blockade was followed by a compensatory IFN-I response, likely caused by a feedback mechanism and the transient overload in antigen. Therefore, a single dose of αIFNAR1 administered during the viral prime did not really “block” IFN-I responses per se; it just delayed the IFN-I response and actually reinforced it later throughout the effector phase of the immune response. Interestingly, a prior study showed that “pre-emptive IFN-I stimulation” before T cell priming, known as out-of-sequence signal, impairs T cell activation (Welsh et al., 2012). This suggests an additional mechanism by which delaying IFN-I signaling may improve immune responses. Furthermore, we reason that the late IFN-I response likely contributes to the improved memory response because if the IFN-I pathway is absent long term, adaptive immune responses are severely impaired and the host is unable to clear the acute viral infection (Teijaro et al., 2013; Wilson et al., 2013).
Antigen is necessary for the elicitation of adaptive immunity. However, high antigen loads for a prolonged time induces inhibitory mechanisms and is thus a main reason for immune exhaustion during chronic infection (Gallimore et al., 1998; Mueller and Ahmed, 2009; Penaloza-MacMaster et al., 2014; Penaloza-MacMaster, 2017). It is important to highlight that short-term IFN-I blockade instituted a different immune scenario: a drastic increase in antigen followed by rapid antigen control. These data suggest that a transient overload in antigen levels “raises the alarm” on the immune system, improving antigen presentation and costimulation at a critical time when the adaptive immune system is getting primed. In particular, a transient increase in antigen levels during the first 72 h of a viral infection can induce a long-term potentiation of immunological memory. These findings have not yet been validated in humans, but previous clinical studies demonstrate a positive correlation between vaccine replication and vaccine immunogenicity (Akondy et al., 2015; Lin et al., 2020). Therefore, a logical assumption is that increasing vaccine dose would proportionally increase immunogenicity, but this is not the case. We show that increasing vaccine dose above a certain level does not significantly augment immunogenicity because there is a natural limit on how much antigen can be expressed by infected cells, which is critically influenced by IFN-I and not so much by the initial virus inoculum.
Enhancing adaptive immunity by blocking an innate immune pathway seemingly violates the classical paradigm that a potent innate response gives rise to a potent adaptive response (Braciale and Hahn, 2013; Kadowaki et al., 2000; Medzhitov and Janeway, 1998; Pulendran et al., 2013). However, these two arms of the immune system do not always work in cooperation. The innate protection conferred by IFN-I has been honed over millions of years to control the initial dissemination of viruses, conferring an immediate survival benefit to the host. Nevertheless, acute IFN-I responses can extinguish viral antigen prematurely, curtailing the elicitation of immunological memory and thus limiting the future protection of the host. In conclusion, we show that coadministration of viral vaccines together with an IFN-I blocker results in profound improvement of immunological memory. Although the safety of IFN-I–modulated vaccines would need careful validation in primates, these findings provide insights for rational vaccine design, as well as a framework to understand the tug of war between innate immunity and immunological memory.
Materials and methods
Mice, treatments, infection, and challenges
For Zika infection experiments, we used 4-wk-old mice (C57BL/6) since this facilitated intracranial challenges. In all other experiments, 6–8-wk-old C57BL/6 or BALB/c mice were used. Mice were purchased from The Jackson Laboratory (approximately half males and half females). Mice were immunized intramuscularly (50 µl per quadriceps), subcutaneously (100 µl in the right flank), or intranasally (25 µl per nostril) with the indicated viral vectors. IgG isotype control (MOPC-21) or IFN-I receptor subunit 1 (IFNAR1)–blocking antibodies (MAR1-5A3) were purchased from BioXCell or Leinco and diluted in sterile PBS. 100 µg of antibody was administered, admixed together with each viral vector vaccine (as a single bolus).
Zika challenges were performed intracranially with 104 PFU. Supra-lethal bacterial challenges were performed with either LM-GP33 or LM-OVA at 107 CFU intravenously via lateral tail vein injection using a mouse restrainer. Other viral challenges consisted of 2 × 106 PFU of VV-OVA through the intranasal route, 106 PFU of YFV-17D through the intravenous route, or 106 PFU of MHV coronavirus through the intranasal route. Mice were housed at the Northwestern University Center for Comparative Medicine located in downtown Chicago. All mouse experiments were performed with approval of the Northwestern University Institutional Animal Care and Use Committee.
Adoptive cell transfers and DC analyses
For in vivo DC transfers, we generated bone marrow–derived DCs using a protocol similar to prior publications (Penaloza-MacMaster et al., 2015; Wang et al., 2019). In brief, bone marrow cells from wild-type or Ifnar1−/− mice were cultured for 5 d in GM-CSF (Sigma) at 20 ng/ml to generate DCs. On day 5, the media were aspirated and DCs were infected with LCMV Armstrong at a multiplicity of infection of 0.05 in 1% FBS RPMI, gently rocking every 10 min. After 1 h, media were replaced with 10% FBS DMEM. After 1 d of in vitro infection, DCs were washed five times and injected intravenously into naive mice (2.5 × 105 DCs/mouse). CD8 T cell responses were evaluated after specific DC transfer. In Fig. 4, A and B, we used a DC cell line (DC 2.4) for imaging.
Reagents, flow cytometry, and equipment
Single-cell suspensions were obtained from peripheral blood mononuclear cells (PBMCs) and tissues as previously described (Masopust et al., 2001). Dead cells were gated out using LIVE/DEAD fixable dead cell stain (Invitrogen). The HIV peptide pools used for intracellular cytokine staining were obtained from the AIDS Reagent Resource, and all other peptides were from AnaSpec or GenScript. MHC class I tetramers were obtained from the National Institutes of Health (NIH) tetramer facility at Emory University. Cells were stained with anti-CD8α (53–6.7 on PerCP-Cy5.5), anti-CD44 (IM7 on Pacific Blue), H-2Kb SIINFEKL (eBio25-D1.16 on APC), CD80 (16-10A1 on FITC), TNFα (MP6-XT22 on PE-Cy7), IL-2 (JES6-5H4 on PE), IFNγ (XMG1.2 on APC), peanut agglutinin (conjugated to fluorescein), Fas (Jo2 on PE), IgD (11–26 on Pacific Blue), IgM (RMM-1 on PE-Cy7), B220 (RA3-6B2 on PerCP-Cy5.5), IFNAR1 (MAR1-5A3 on PE), and CD3 (145-2c11 on FITC). Fluorescently labeled antibodies were purchased from BD PharMingen, except for CD44 (which was from Biolegend). Flow cytometry samples were acquired with a Becton Dickinson Canto II or an LSRII and analyzed using FlowJo (Treestar).
Virus-specific ELISA
Virus-specific ELISA was done as described in prior publications (Dangi et al., 2020; Wang et al., 2019). In brief, 96-well flat-bottom plates (MaxiSorp; Thermo Scientific) were coated with 100 µl/well of the respective viral lysate (e.g., Zika, YFV-17D, VSV, and MHV) diluted 1:10 in PBS for 48 h at room temperature. Plates were washed with PBS + 0.5% Tween 20. Blocking was performed for 2 h at room temperature with 200 µl of PBS + 0.2% Tween 20 + 10% FCS. 5 µl of sera were added to 145 µl of blocking solution in the first column of the plate, 1:3 serial dilutions were performed until row 12 for each sample, and plates were incubated for 90 min at room temperature. Plates were washed three times followed by addition of goat anti-mouse IgG conjugated to horseradish peroxidase (Southern Biotech) diluted in blocking solution (1:5,000) at 100 µl/well and incubated for 90 min at room temperature. Plates were washed three times, and 100 µl/well of Sure Blue substrate (SeraCare) was added for 8 min. Reaction was stopped using 100 µl/well of KPL TMB Stop Solution (SeraCare). Absorbance was measured at 450 nm using a Spectramax Plus 384 (Molecular Devices).
Protein-specific ELISA
96-well flat-bottom plates (MaxiSorp; Thermo Scientific) were coated with 0.1 µg/well of gp140 derived from SIVmac239 (ImmuneTech), HIV-1 clade A BGB505 (ImmuneTech), or HIV-1 clade B SF162 (ImmuneTech); or OVA (Worthington) for 48 h at 4°C. Plates were washed with PBS + 0.05% Tween 20. Blocking was performed for 4 h at room temperature with 200 µl of PBS + 0.05% Tween 20 + bovine serum albumin. 6 µl of sera were added to 144 µl of blocking solution in the first column of the plate, 1:3 serial dilutions were performed until row 12 for each sample, and plates were incubated for 60 min at room temperature. Plates were washed three times followed by addition of goat anti-mouse IgG conjugated to horseradish peroxidase (Southern Biotech) diluted in blocking solution (1:5,000) at 100 µl/well and incubated for 60 min at room temperature. Plates were washed three times, and 100 µl/well of Sure Blue substrate (SeraCare) was added for 8 min. Reaction was stopped using 100 µl/well of KPL TMB Stop Solution (SeraCare). Absorbance was measured at 450 nm using a Spectramax Plus 384 (Molecular Devices).
TZM-bl assays
TZM-bl cells (a HeLa cell line engineered to express human receptors and coreceptors for HIV, in addition to a Tat-inducible luciferase gene) were used to measure vaccine-induced antibody neutralization of SIV. TZM-bl cells were cultured in T75 flasks (Thermo Scientific) at 104 cell/ml density for 3 d in 10% FBS complete DMEM (GIBCO). On the day of the assay, 1:20 serial fold dilutions of mouse sera were performed. Sera were incubated with SIVmac251.TCLA pseudovirus for 30 min in low-evaporation 96-well clear plates (Corning). TZM-bl cells were detached from flasks using 0.25% trypsin-EDTA (GIBCO) and seeded at a density of 0.5 × 106/ml per well. On the following day, 10% FBS complete DMEM (GIBCO) was added to each well. At day 3, media were aspirated, and cells were lysed using luciferase cell culture lysis buffer (Promega). Luciferase reaction was performed using 30 µl of cell lysis (Promega). The reaction was added to 96-well black optiplates (Perkin Elmer). Luminescence was measured using a Perkin Elmer Victor3 luminometer.
Bacterial quantification
To quantify LM in liver, brain heart infusion agar containing 50 µg/ml streptomycin was prepared and added to 6-well plates (2 ml of agar/well). LM-GP33 and LM-OVA possess a streptomycin resistance gene. Liver was harvested and collected in 14-ml round-bottom tubes (Falcon) with 5 ml of 1% FBS DMEM (without antibiotics). Tissue was passed through a 100-µm strainer (Scientific Inc.) using 1% Triton X-100 solution. 50 µl of serial dilutions were added on each brain heart infusion agar well. Plates were incubated at 37°C overnight. CFU were counted the next day.
Viral quantification
Quantification of LCMV on Vero E6 cell monolayers was done as described in prior publications (Dangi et al., 2020; Wang et al., 2019). In brief, Vero E6 cells (ATCC) were grown on 6-well plates at 2 × 105 cells/ml. After cells reached ∼90% confluency, media were removed and 200 µl of 10-fold viral dilutions in 1% DMEM (GIBCO) were pipetted on top of the cell monolayers. Plates were rocked every 10 min in a 37°C, 5% CO2 incubator. After 60 min, media were aspirated, and the monolayers were overlaid with a 1:1 solution of 2 × 199 media and 1% agarose. After 4 d of culture in a 37°C, 5% CO2 incubator, a second overlay was added consisting of a 1:1 solution of 2 × 199 media and 1% agarose with neutral red. The agar overlay was removed on day 5, and plaques were counted using a transilluminator (Gradco). Quantification of VSV titers was similar to quantification of LCMV titers except that the agar overlay was removed after 1 d and 1% crystal violet was added on top of the monolayers, incubated for 1 h at room temperature, and then washed with H2O. MHV viral quantification was similar to VSV, but overlay was removed 2 d after infection. Quantification of YFV-17D titers was similar to quantification of LCMV titers except that the agar overlay was removed after 5 d and 1% crystal violet was added on top of the monolayers, incubated for 1 h at room temperature, and then washed with H2O. For quantification of vaccinia, we followed the protocol by Dr. Bernard Moss (Cotter et al., 2017). For viral load quantification in muscle, both quadriceps were harvested and collected in round-bottom tubes (Falcon) containing 3 ml of 1% FBS DMEM (GIBCO). Muscle tissue was processed using a Tissue Ruptor homogenizer (Qiagen). Lungs and brains were harvested and collected in round-bottom tubes (Falcon) containing 1 ml of 1% FBS DMEM (GIBCO) and homogenized as described above. Following homogenization, tissues were clarified using a 100-µm strainer (Scientific Inc.) to remove debris.
Viruses
LCMV-expressing SIVmac239 antigens (Gag and Env), GFP, or OVA were from Hookipa Biotech and were produced as described previously (Kallert et al., 2017). Nonreplicating LCMV vectors were also from Hookipa Biotech. The nonreplicating LCMV vectors contained the LCMV glycoprotein gene in trans, which resulted in a single round of infection (Penaloza MacMaster et al., 2017). Replicating LCMV-HIV vectors were constructed with help from the De La Torre (Scripps Research Institute, San Diego, CA) and Waggoner (University of Cincinnati College of Medicine, Cincinnati, OH) laboratories. The nonreplicating Ad5 vector is E1/E3 deleted and expresses HIV-1 gp140 V1-V3 domains from HIV-1 Bal, and was obtained from the Mascola laboratory (NIH, Bethesda, MD). The VSV-OVA used in this study was obtained from Dr. Vaiva Vezys (University of Minnesota Medical School, Minneapolis, MN) and was derived from a stock from Dr. Leo Lefrancois’ laboratory (University of Connecticut Health Center, Farmington, CT). The nonreplicating VSV vector (VSV-ΔG) that contains the G protein in trans was a gift from Dr. Connie Cepko (Harvard Medical School, Boston, MA). The acute LCMV Armstrong strain was propagated from a stock from Dr. Rafi Ahmed’s laboratory (Emory University, Atlanta, GA). Dr. Bernard Moss and Dr. Patricia Earl (NIH) provided the poxviruses. The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, and NIH (BEI Resources): YFV-17D, NR-115; Zika virus, PRVABC59 (Puerto Rico strain), NR-50240; Zika virus, MR766 (Uganda strain), NR-50065. Mouse betacoronaviruses (MHV-A59 strain) were a gift from Dr. Susan Weiss (University of Pennsylvania, Philadelphia, PA).
Murine IFNα2
293F mammalian cells were transfected with a pcDNA3.1 mIFNα2 expression vector (Addgene ID 135098). IFNα2 purification was performed by cation exchange chromatography using a HiTrap SP column (General Electric). Purified recombinant mIFNα2 was quantified by an IFNα mouse ELISA kit (Invitrogen).
Immunofluorescence staining
For immunofluorescence staining of DCs, the murine DC2.4 cell line was used. Cells were plated in clear flat-bottom 96-well plates at a density of 104 cells/well in 200 µl of 10% FBS DMEM (GIBCO), 1% L-glutamine, and 1% penicillin-streptomycin. After 2 d, cells were treated with 20 µg of IgG control or αIFNAR1 antibodies for 30 min. Media were removed from each well using a multichannel pipette, and cells were infected with LCMV-GFP or MHV-GFP (MOI 0.05) in 50 µl of 1% FBS DMEM (GIBCO) for 1 h, gently rocking every 10 min. Media were removed and replaced with 200 µl of 10% FBS DMEM (GIBCO), 1% L-glutamine, and 1% penicillin-streptomycin and incubated at 37°C and 5% CO2 for 72 h. Cells were washed once and fixed with 4% paraformaldehyde (Thermo Scientific). A Vectashield mounting medium containing DAPI (Vector Labs) was added to the wells, and images were acquired using an EVOS FL digital inverted microscope (Thermo Scientific).
RNA-Seq data acquisition and analysis
Gene expression profiling was performed as shown previously (Barnitz et al., 2013; Penaloza-MacMaster et al., 2015; Quigley et al., 2010; Wang et al., 2019). In brief, C57BL/6 mice were intramuscularly immunized with 104 PFU of LCMV, and at day 7, splenic CD8 T cells were MACS (magnetic activated cell sorting) sorted with a MACS negative selection kit (STEMCELL). Purified CD8 T cells were stained with DbGP33 tetramer, live dead stain, and flow cytometry antibodies for CD8 and CD44 to gate on activated CD8 T cells. Live, CD8+, CD44+, and DbGP33+ cells were FACS sorted to ∼97% purity on a FACS Aria cytometer (BD Biosciences) and stored at -80°C in 1 ml of TRIzol (Life Sciences). RNA extraction was performed with the RNAdvance Tissue Isolation kit (Agencourt). RNA quality and RNA-Seq downstream analyses were performed at the NUSeq core at Northwestern University. For analysis, adapters were trimmed from reads using cutadapt version 1.13 and aligned to 10 mm using STAR version 020201. For gene counting, htseq-count version 0.6.1p1 was used, and differential expression analysis was conducted using DESeq2 version 1.14.1. RNA-Seq data were uploaded into the GEO database (accession no. GSE129827) in a record titled “Gene expression comparison of splenic virus-specific CD8 T cells after infection with LCMV vector and treatment with IgG or aIFNAR1.”
Statistical analysis
Most statistical analyses used the Mann-Whitney test, unless specified otherwise in the figure legend. Survival plot analyses were performed using the Mantel-Cox test. Dashed lines in plaque assay and ELISA plots represent the limit of detection. Data were analyzed using Prism (Graphpad).
Online supplemental material
Fig. S1 shows that MAR1-5A3 blocks IFNAR1 in vitro and in vivo as well as CD8, CD4, and antibody responses with an experimental HIV vaccine (LCMV-HIV). Fig. S2 shows OVA-specific responses after VSV-OVA immunization using subcutaneous or intranasal routes. Fig. S3 shows that short-term IFNAR1 blockade increases hyperacute viral loads. Fig. S4 shows that the adjuvant effect of IFNAR1 blockade is time dependent and virus replication dependent. Fig. S5 shows immune protection in a different genetic background (BALB/c).
Acknowledgments
We thank Drs. Susan Weiss, Rafi Ahmed, Daniel Pinschewer, Chyung-Ru Wang, Richard D’Aquila, and Hank Seifert for discussions.
This work was possible with grants from the Chicago Developmental Center for AIDS Research (P30 AI117943) and National Institutes of Health (1R21AI132848-01A1 and DP2DA051912) to P. Penaloza-MacMaster and a National Science Foundation Graduate Research Fellowship Program grant (DGE-1842165) to N. Palacio. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Author contributions: P. Penaloza-MacMaster and N. Palacio designed and conducted the experiments and wrote the paper. T. Dangi, Y.R. Chung, Y. Wang, J.L. Loredo-Varela, and Z. Zhang conducted the experiments.
References
- Akondy, R.S., Johnson P.L., Nakaya H.I., Edupuganti S., Mulligan M.J., Lawson B., Miller J.D., Pulendran B., Antia R., and Ahmed R.. 2015. Initial viral load determines the magnitude of the human CD8 T cell response to yellow fever vaccination. Proc. Natl. Acad. Sci. USA. 112:3050–3055. 10.1073/pnas.1500475112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzoni, L., Foulkes A.S., Papasavvas E., Mexas A.M., Lynn K.M., Mounzer K., Tebas P., Jacobson J.M., Frank I., Busch M.P., et al. 2013. Pegylated Interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J. Infect. Dis. 207:213–222. 10.1093/infdis/jis663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, C.C., Zhong G., Huang I.C., and Farzan M.. 2014. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 1:261–283. 10.1146/annurev-virology-031413-085537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnitz, R.A., Imam S., Yates K., and Haining W.N.. 2013. Isolation of RNA and the synthesis and amplification of cDNA from antigen-specific T cells for genome-wide expression analysis. Methods Mol. Biol. 979:161–173. 10.1007/978-1-62703-290-2_13 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, M., Madden P., Henning N., Gregory S., Aid M., Martinot A.J., Barouch D.H., and Penaloza-MacMaster P.. 2017. Regulation of CD4 T cells and their effects on immunopathological inflammation following viral infection. Immunology. 152:328–343. 10.1111/imm.12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braciale, T.J., and Hahn Y.S.. 2013. Immunity to viruses. Immunol. Rev. 255:5–12. 10.1111/imr.12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, C., Cheung A., Stark Z., Choo S., Downie L., White S., Conyers R., and Cole T.. 2016. A novel presentation of homozygous loss-of-function STAT-1 mutation in an infant with hyperinflammation-A case report and review of the literature. J. Allergy Clin. Immunol. Pract. 4:777–779. 10.1016/j.jaip.2016.02.015 [DOI] [PubMed] [Google Scholar]
- Casey, K.A., Guo X., Smith M.A., Wang S., Sinibaldi D., Sanjuan M.A., Wang L., Illei G.G., and White W.I.. 2018. Type I interferon receptor blockade with anifrolumab corrects innate and adaptive immune perturbations of SLE. Lupus Sci. Med. 5. e000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L., Yu H., Li G., Li F., Ma J., Li J., Chi L., Zhang L., and Su L.. 2017. Type I interferons suppress viral replication but contribute to T cell depletion and dysfunction during chronic HIV-1 infection. JCI Insight. 2. e94366. 10.1172/jci.insight.94366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli, M.J., Huang S.X., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A., et al. 2015. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 348:448–453. 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter, C.A., Earl P.L., Wyatt L.S., and Moss B.. 2017. Preparation of Cell Cultures and Vaccinia Virus Stocks. Curr. Protoc. Mol. Biol. 117:16.16.1–16.16.18. 10.1002/cpmb.33 [DOI] [PubMed] [Google Scholar]
- Crouse, J., Bedenikovic G., Wiesel M., Ibberson M., Xenarios I., Von Laer D., Kalinke U., Vivier E., Jonjic S., and Oxenius A.. 2014. Type I interferons protect T cells against NK cell attack mediated by the activating receptor NCR1. Immunity. 40:961–973. 10.1016/j.immuni.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Dangi, T., Chung Y.R., Palacio N., and Penaloza-MacMaster P.. 2020. Interrogating Adaptive Immunity Using LCMV. Curr. Protoc. Immunol. 130. 10.1002/cpim.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, C.J., Mohamad S.M., Young D.F., Skelton A.J., Leahy T.R., Munday D.C., Butler K.M., Morfopoulou S., Brown J.R., Hubank M., et al. 2015. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci. Transl. Med. 7. 307ra154. 10.1126/scitranslmed.aac4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis, S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., et al. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391. 10.1038/ng1097 [DOI] [PubMed] [Google Scholar]
- Erickson, A.K., and Pfeiffer J.K.. 2013. Dynamic viral dissemination in mice infected with yellow fever virus strain 17D. J. Virol. 87:12392–12397. 10.1128/JVI.02149-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell, P.J., Sen G.C., Dubois M.F., Ratner L., Slattery E., and Lengyel P.. 1978. Interferon action: two distinct pathways for inhibition of protein synthesis by double-stranded RNA. Proc. Natl. Acad. Sci. USA. 75:5893–5897. 10.1073/pnas.75.12.5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten, R., Scher F., Sagez F., Chasset F., and Arnaud L.. 2019. Spotlight on anifrolumab and its potential for the treatment of moderate-to-severe systemic lupus erythematosus: evidence to date. Drug Des. Devel. Ther. 13:1535–1543. 10.2147/DDDT.S170969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore, A., Glithero A., Godkin A., Tissot A.C., Plückthun A., Elliott T., Hengartner H., and Zinkernagel R.. 1998. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187:1383–1393. 10.1084/jem.187.9.1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher, D., Therrien R., Kettaf N., Angermann B.R., Boucher G., Filali-Mouhim A., Moser J.M., Mehta R.S., D.R. Drake, III, Castro E., et al. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med. 205:3119–3131. 10.1084/jem.20082292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton, S., Goodbourn S., Young D.F., Dickinson P., Mohamad S.M., Valappil M., McGovern N., Cant A.J., Hackett S.J., Ghazal P., et al. 2013. STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. USA. 110:3053–3058. 10.1073/pnas.1220098110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton, C., Kolumam G.A., and Murali-Krishna K.. 2006. Cutting Edge: The direct action of type I IFN on CD4 T cells is critical for sustaining clonal expansion in response to a viral but not a bacterial infection. J. Immunol. 176:3315–3319. 10.4049/jimmunol.176.6.3315 [DOI] [PubMed] [Google Scholar]
- Hernandez, N., Melki I., Jing H., Habib T., Huang S.S.Y., Danielson J., Kula T., Drutman S., Belkaya S., Rattina V., et al. 2018. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 215:2567–2585. 10.1084/jem.20180628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, N., Bucciol G., Moens L., Le Pen J., Shahrooei M., Goudouris E., Shirkani A., Changi-Ashtiani M., Rokni-Zadeh H., Sayar E.H., et al. 2019. Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J. Exp. Med. 216:2057–2070. 10.1084/jem.20182295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honke, N., Shaabani N., Cadeddu G., Sorg U.R., Zhang D.E., Trilling M., Klingel K., Sauter M., Kandolf R., Gailus N., et al. 2012. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat. Immunol. 13:51–57. 10.1038/ni.2169 [DOI] [PubMed] [Google Scholar]
- Hoyos-Bachiloglu, R., Chou J., Sodroski C.N., Beano A., Bainter W., Angelova M., Al Idrissi E., Habazi M.K., Alghamdi H.A., Almanjomi F., et al. 2017. A digenic human immunodeficiency characterized by IFNAR1 and IFNGR2 mutations. J. Clin. Invest. 127:4415–4420. 10.1172/JCI93486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, N., Antonenko S., Lau J.Y., and Liu Y.J.. 2000. Natural interferon alpha/beta-producing cells link innate and adaptive immunity. J. Exp. Med. 192:219–226. 10.1084/jem.192.2.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallert, S.M., Darbre S., Bonilla W.V., Kreutzfeldt M., Page N., Müller P., Kreuzaler M., Lu M., Favre S., Kreppel F., et al. 2017. Replicating viral vector platform exploits alarmin signals for potent CD8+ T cell-mediated tumour immunotherapy. Nat. Commun. 8:15327. 10.1038/ncomms15327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolumam, G.A., Thomas S., Thompson L.J., Sprent J., and Murali-Krishna K.. 2005. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202:637–650. 10.1084/jem.20050821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreins, A.Y., Ciancanelli M.J., Okada S., Kong X.F., Ramírez-Alejo N., Kilic S.S., El Baghdadi J., Nonoyama S., Mahdaviani S.A., Ailal F., et al. 2015. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212:1641–1662. 10.1084/jem.20140280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bon, A., Etchart N., Rossmann C., Ashton M., Hou S., Gewert D., Borrow P., and Tough D.F.. 2003. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol. 4:1009–1015. 10.1038/ni978 [DOI] [PubMed] [Google Scholar]
- Li, S., Sullivan N.L., Rouphael N., Yu T., Banton S., Maddur M.S., McCausland M., Chiu C., Canniff J., Dubey S., et al. 2017. Metabolic Phenotypes of Response to Vaccination in Humans. Cell. 169:862–877.e17. 10.1016/j.cell.2017.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.W., Moran E., Adams R.J., Sievers R.E., Hauer D., Godin S., and Griffin D.E.. 2020. A durable protective immune response to wild-type measles virus infection of macaques is due to viral replication and spread in lymphoid tissues. Sci. Transl. Med. 12:eaax7799. 10.1126/scitranslmed.aax7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust, D., Vezys V., Marzo A.L., and Lefrançois L.. 2001. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 291:2413–2417. 10.1126/science.1058867 [DOI] [PubMed] [Google Scholar]
- McMichael, T.M., Zhang Y., Kenney A.D., Zhang L., Zani A., Lu M., Chemudupati M., Li J., and Yount J.S.. 2018. IFITM3 Restricts Human Metapneumovirus Infection. J. Infect. Dis. 218:1582–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov, R., and C.A. Janeway, Jr.. 1998. Innate immune recognition and control of adaptive immune responses. Semin. Immunol. 10:351–353. 10.1006/smim.1998.0136 [DOI] [PubMed] [Google Scholar]
- Minegishi, Y., Saito M., Morio T., Watanabe K., Agematsu K., Tsuchiya S., Takada H., Hara T., Kawamura N., Ariga T., et al. 2006. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 25:745–755. 10.1016/j.immuni.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Moens, L., Van Eyck L., Jochmans D., Mitera T., Frans G., Bossuyt X., Matthys P., Neyts J., Ciancanelli M., Zhang S.Y., et al. 2017. A novel kindred with inherited STAT2 deficiency and severe viral illness. J. Allergy Clin. Immunol. 139:1995–1997.e9. 10.1016/j.jaci.2016.10.033 [DOI] [PubMed] [Google Scholar]
- Mueller, S.N., and Ahmed R.. 2009. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA. 106:8623–8628. 10.1073/pnas.0809818106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., and Aguet M.. 1994. Functional role of type I and type II interferons in antiviral defense. Science. 264:1918–1921. 10.1126/science.8009221 [DOI] [PubMed] [Google Scholar]
- Norris, B.A., Uebelhoer L.S., Nakaya H.I., Price A.A., Grakoui A., and Pulendran B.. 2013. Chronic but not acute virus infection induces sustained expansion of myeloid suppressor cell numbers that inhibit viral-specific T cell immunity. Immunity. 38:309–321. 10.1016/j.immuni.2012.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster, P. 2017. CD8 T-cell regulation by T regulatory cells and the programmed cell death protein 1 pathway. Immunology. 151(2):146–153. 10.1111/imm.12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster, P., Kamphorst A.O., Wieland A., Araki K., Iyer S.S., West E.E., O’Mara L., Yang S., Konieczny B.T., Sharpe A.H., et al. 2014. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J. Exp. Med. 211(9):1905–1918. 10.1084/jem.20132577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster, P., Barber D.L., Wherry E.J., Provine N.M., Teigler J.E., Parenteau L., Blackmore S., Borducchi E.N., Larocca R.A., Yates K.B., et al. 2015. Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science. 347:278–282. 10.1126/science.aaa2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza MacMaster, P., Shields J.L., Alayo Q.A., Cabral C., Jimenez J., Mondesir J., Chandrashekar A., Cabral J.M., Lim M., Iampietro M.J., et al. 2017. Development of novel replication-defective lymphocytic choriomeningitis virus vectors expressing SIV antigens. Vaccine. 35:1–9. 10.1016/j.vaccine.2016.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran, B., and Ahmed R.. 2011. Immunological mechanisms of vaccination. Nat. Immunol. 12:509–517. 10.1038/ni.2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran, B., Li S., and Nakaya H.I.. 2010. Systems vaccinology. Immunity. 33:516–529. 10.1016/j.immuni.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran, B., Oh J.Z., Nakaya H.I., Ravindran R., and Kazmin D.A.. 2013. Immunity to viruses: learning from successful human vaccines. Immunol. Rev. 255:243–255. 10.1111/imr.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec, T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B., et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10:116–125. 10.1038/ni.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley, M., Pereyra F., Nilsson B., Porichis F., Fonseca C., Eichbaum Q., Julg B., Jesneck J.L., Brosnahan K., Imam S., et al. 2010. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 16:1147–1151. 10.1038/nm.2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis, B., Bunin A., Ghosh H.S., Lewis K.L., and Sisirak V.. 2011. Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev. Immunol. 29:163–183. 10.1146/annurev-immunol-031210-101345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler, N.G., Bosinger S.E., Estes J.D., Zhu R.T., Tharp G.K., Boritz E., Levin D., Wijeyesinghe S., Makamdop K.N., del Prete G.Q., et al. 2014. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 511:601–605. 10.1038/nature13554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins, J.W.. 2019. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 6:567–584. 10.1146/annurev-virology-092818-015756 [DOI] [PubMed] [Google Scholar]
- Shahni, R., Cale C.M., Anderson G., Osellame L.D., Hambleton S., Jacques T.S., Wedatilake Y., Taanman J.W., Chan E., Qasim W., et al. 2015. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain. 138:2834–2846. 10.1093/brain/awv182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, G.R., Kerr I.M., Williams B.R., Silverman R.H., and Schreiber R.D.. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264. 10.1146/annurev.biochem.67.1.227 [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Takeuchi T., Okada M., Ishii T., Nakajima H., Kawai S., Nagashima T., Hayashi N., Wang L., and Tummala R.. 2020. Safety and tolerability of anifrolumab, a monoclonal antibody targeting type I interferon receptor, in Japanese patients with systemic lupus erythematosus: A multicenter, phase 2, open-label study. Mod. Rheumatol. 30:101–108. 10.1080/14397595.2019.1583833 [DOI] [PubMed] [Google Scholar]
- Teijaro, J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C., Welch M., Schreiber R.D., de la Torre J.C., and Oldstone M.B.. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 340:207–211. 10.1126/science.1235214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani, F.J., Rodriguez-Torres M., Rockstroh J.K., Lissen E., Gonzalez-García J., Lazzarin A., Carosi G., Sasadeusz J., Katlama C., Montaner J., et al. ; APRICOT Study Group . 2004. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N. Engl. J. Med. 351:438–450. 10.1056/NEJMoa040842 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Chung Y.R., Eitzinger S., Palacio N., Gregory S., Bhattacharyya M., and Penaloza-MacMaster P.. 2019. TLR4 signaling improves PD-1 blockade therapy during chronic viral infection. PLoS Pathog. 15. e1007583. 10.1371/journal.ppat.1007583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh, R.M., Bahl K., Marshall H.D., and Urban S.L.. 2012. Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog. 8. e1002352. 10.1371/journal.ppat.1002352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., Teichgräber V., Becker T.C., Masopust D., Kaech S.M., Antia R., von Andrian U.H., and Ahmed R.. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4:225–234. 10.1038/ni889 [DOI] [PubMed] [Google Scholar]
- Wilson, E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B.J., Karp C.L., and Brooks D.G.. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 340:202–207. 10.1126/science.1235208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H.C., Grusdat M., Pandyra A.A., Polz R., Huang J., Sharma P., Deenen R., Köhrer K., Rahbar R., Diefenbach A., et al. 2014. Type I interferon protects antiviral CD8+ T cells from NK cell cytotoxicity. Immunity. 40:949–960. 10.1016/j.immuni.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Zuniga, E.I., Liou L.Y., Mack L., Mendoza M., and Oldstone M.B.. 2008. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe. 4:374–386. 10.1016/j.chom.2008.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]