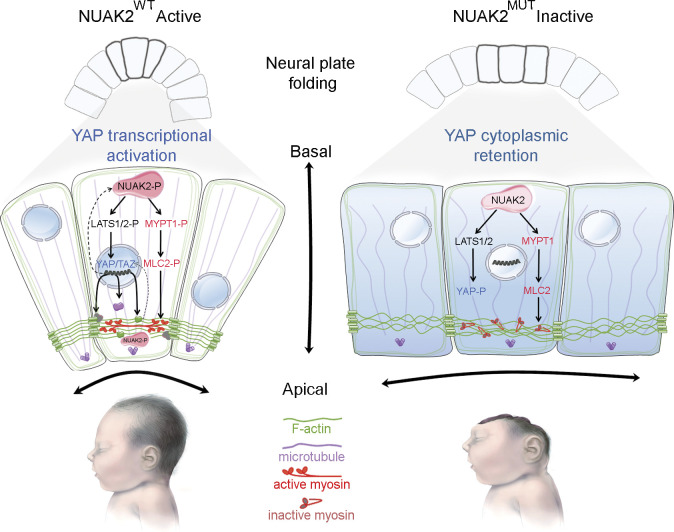
A germline homozygous loss-of-function mutation in NUAK2 causes human fetal death due to anencephaly. Our functional tests establish that NUAK2, operating on the Hippo signaling axis, is an essential kinase that regulates cytoskeletal processes that govern cell shape during neural tube closure.
Abstract
Failure of neural tube closure during embryonic development can result in anencephaly, one of the most common birth defects in humans. A family with recurrent anencephalic fetuses was investigated to understand its etiology and pathogenesis. Exome sequencing revealed a recessive germline 21-bp in-frame deletion in NUAK2 segregating with the disease. In vitro kinase assays demonstrated that the 7–amino acid truncation in NUAK2, a serine/threonine kinase, completely abrogated its catalytic activity. Patient-derived disease models including neural progenitor cells and cerebral organoids showed that loss of NUAK2 activity led to decreased Hippo signaling via cytoplasmic YAP retention. In neural tube–like structures, endogenous NUAK2 colocalized apically with the actomyosin network, which was disrupted in patient cells, causing impaired nucleokinesis and apical constriction. Our results establish NUAK2 as an indispensable kinase for brain development in humans and suggest that a NUAK2-Hippo signaling axis regulates cytoskeletal processes that govern cell shape during neural tube closure.
Graphical Abstract
Introduction
The human brain develops during the third week of gestation, when a median groove is initiated on the neural plate and cranial neural folds are generated. The neuroepithelium continues to expand and bend until the paired neural folds meet in the dorsal midline and fuse to form a neural tube along the anterior–posterior axis (Greene and Copp, 2009; Nikolopoulou et al., 2017). Failure in neural tube closure (NTC) anteriorly can give rise to anencephaly, which is the most common birth defect observed in humans, affecting 0.5 to 2 per 1,000 established pregnancies worldwide, with an occurrence as high as 6 per 1,000 live births in certain regions of China (Copp et al., 2015; Li et al., 2006). Based on animal models and epidemiological studies, most neural tube defects (NTDs) are sporadic and may be caused by genetic mutations or environmental factors (Harris and Juriloff, 1999, 2010). More than 200 candidate genes have been associated with exencephaly in mice, while in human fetuses, de novo mutations in the genes VANGL2, SHROOM3, U2SURP, and ANKRD32 have been linked to anencephaly (Lei et al., 2010; Lemay et al., 2015). Environmental factors such as deficiency of folic acid and vitamins have been shown to be critical, and supplementation before and in early phases of pregnancy has been shown to reduce NTDs by 50–70% (Czeizel and Dudás, 1992; Daly et al., 1995; Milunsky et al., 1989).
Because neurulation-stage human embryos are not easily accessible, vertebrate animal models have been used instead to understand the key molecular and cellular pathways underlying neural plate bending and subsequent NTC (Nikolopoulou et al., 2017). A number of functional studies have revealed different pathomechanisms underlying NTDs, such as disruption of actin function, premature differentiation, faulty regulation of cell proliferation, and abnormal regulation of apoptosis in the neuroepithelium (Cecconi et al., 2008; Chen et al., 1996; Herrera et al., 1999; Kim et al., 2007). Changes in cell morphology in restricted areas of the neuroepithelium, such as the median hinge point and paired dorsolateral hinge points, are primordial for the neural folds to arise (Shum and Copp, 1996). Apicobasal microtubule formation drives cell elongation, and apical concentration of actomyosin triggers cell constriction, giving the neuroepithelial cells (NECs) a wedge shape that forces the neural plate to fold onto itself. Several lines of knockout mice lacking actin cytoskeleton-associated proteins, such as SHROOM3, MARCKS, and P190RhoGAP, display NTDs (Brouns et al., 2000; Hildebrand and Soriano, 1999; Stumpo et al., 1995). Because anencephaly is caused by defective closure of the rostral end of the neural tube, it is believed that cranial neurulation must rely on specific factors that are not involved in spinal neurulation. For instance, deletion of n-Cofilin and Vinculin in mice causes exencephaly, while the spinal tube properly closes (Gurniak et al., 2005; Xu et al., 1998).
Although the morphological and cellular bases of NTC have become relatively well understood in animal models, the etiology of NTDs in humans remains largely unknown. Here, we describe a deleterious recessive mutation in NUAK2, encoding the SNF1/5′-adenosine monophosphate–activated protein kinase (AMPK)–related kinase (also known as SNARK), that caused anencephaly in three fetuses born to consanguineous parents. Using in vitro kinase assays, we show that the 7–amino acid deletion in the kinase domain causes a loss of NUAK2 phosphorylation as well as a net reduction in its enzymatic activity toward known substrates. Using in vitro models, including induced pluripotent stem cell (iPSC)–derived neural progenitor cells (NPCs) and cerebral organoids (COs), we show that loss-of-function mutation in the kinase domain of NUAK2 alters the actomyosin network via the YAP/TAZ signaling pathway. Our data provide a new pathomechanism of anencephaly in humans that involves NUAK2-mediated regulation of cytoskeleton components that drive cell contraction and elongation for proper NTC.
Results and discussion
Consanguineous Turkish family presenting with three consecutive anencephalic fetuses
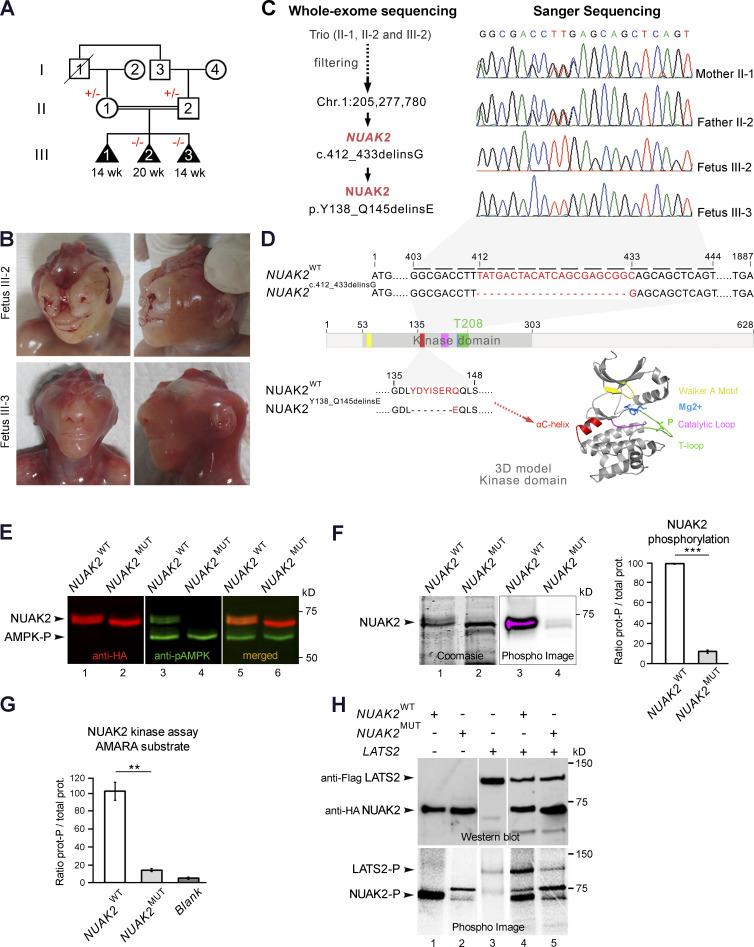
We report here a family from Turkey in which first-degree cousin parents had three consecutive fetuses with anencephaly (ANPH, MIM206500), suggesting a possible genetic etiology (Fig. 1, A and B). The parents do not have healthy living children, and no similarly affected fetuses have been reported in the extended family. The first pregnancy was medically terminated at 14 wk of gestation due to anencephaly, and no sample was available from fetus III-1. The second pregnancy was medically terminated at 20 wk of gestation due to antenatal ultrasound findings of anencephaly. Postmortem examination showed a male fetus, III-2, with severe frontonasal dysplasia findings, with midline cleft of the upper lip and alveolar ridge and total bifid nose, in addition to anencephaly (Fig. 1 B). He also had very short palpebral fissures with clinical anophthalmia, features that do not commonly accompany anencephaly. Fetal karyotype and chromosomal array in cultured amniocytes yielded normal results. The third pregnancy was also medically terminated at 14 wk due to antenatal ultrasound findings of anencephaly. Postmortem examination showed a female fetus, III-3, with anencephaly and no sign of frontonasal dysplasia or anophthalmia (Fig. 1 B). The mother had taken periconceptional folic acid supplementation for all three pregnancies, 400 μg/d for the first pregnancy and 5 mg/d for the last two.
Figure 1.
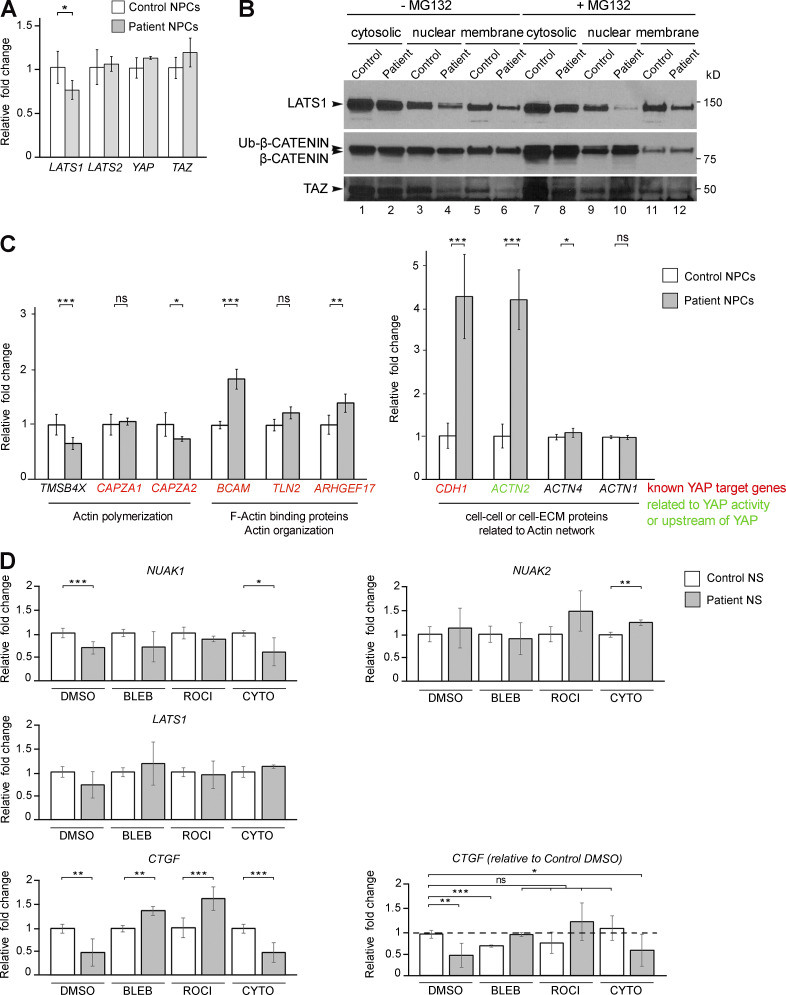
Abrogation of NUAK2 kinase activity causes anencephaly in three fetuses. (A) Pedigree of a consanguineous family from Turkey presenting with three consecutive anencephalic fetuses. Black triangle, abortion, affected state; diagonal line, deceased; +/−, heterozygous for NUAK2 mutation; −/−, homozygous for NUAK2 germline mutation. (B) Facial and profile images of affected fetuses III-2 at 20 wk and III-3 at 14 wk, both showing anencephaly, and fetus III-2, also displaying frontonasal dysplasia and clinical anophthalmia. (C) Exome sequencing of both parents and fetus III-2 delineated an in-frame deletion/insertion c.412_433delinsG in the NUAK2 gene. Sanger sequencing confirmed that NUAK2 mutation segregated with anencephaly in all family members. (D) Schematic representation of NUAK2 variant at the RNA and protein levels. The mutation causes a 7–amino acid deletion together with a 1–amino acid alteration annotated as p.Y138_Q145delinsE. Protein 3D structure prediction showed that mutated residues lie in the universally conserved αC-helix (red) of the NUAK2 kinase domain. Kinase domain is composed of a Walker-A motif (yellow) that binds to the γ-phosphate of ATP, a T-loop (green) that contains a threonine that is phosphorylated for kinase activation, a DFG motif (blue) that binds Mg2+ in catalysis, and a catalytic loop (pink) that interacts with substrate and catalytic residues. (E) Western blot of overexpressed HA-tagged NUAK2WT and NUAK2MUT showed that NUAK2MUT was not phosphorylated compared with NUAK2WT. Anti-pAMPK (T172) antibodies recognize the phosphorylated threonine T208 of NUAK2. (F) Phosphorylation assay using radioactive [32P]-ATP (Phospho Image) confirmed phosphorylation of NUAK2WT but not NUAK2MUT. Coomassie stain showed equal amount of WT and MUT NUAK2 proteins (prot.). (G) Kinase activity assay with radioactive [32P]-ATP showed significant decrease of phosphorylated AMARA peptide in presence of NUAK2MUT protein, as low as when kinase was not added in the reaction mixture (Blank). (H) Kinase activity assay repeated with LATS2, a NUAK2 substrate, showed decreased LATS2 phosphorylation in the presence of NUAK2MUT. Western blot showed equal amount of LATS2 and NUAK2 proteins. Phosphorylation and kinase assays were performed in duplicate and repeated in two independent experiments (E–H). **, P < 0.02; ***, P < 0.005.
Mutation in NUAK2 causes the first recessive form of anencephaly in humans
Exome sequencing of both parents and fetus III-2 revealed a total of 296 germline variants across protein-coding exons, UTRs, splice sites, and flanking introns. Additional filtering was applied to prioritize conserved, rare (minor allele frequency <0.1%), and deleterious variants (Mendelian clinically applicable pathogenicity score >0.025) that were homozygous in the proband and heterozygous in both parents. A deletion of 22 bp (TATGACTACATCAGCGAGCGGC) and insertion of one nucleotide in the NUAK2 gene (GRCh37/hg19; Chr1: 205,277,780; c.412_433delinsG) was the only candidate variant remaining (Table S1). Sanger sequencing of both parents and fetuses III-2 and III-3 confirmed that this germline mutation segregated with anencephaly (Fig. 1, A and C). Homozygous loss-of-function mutations in NUAK2 have never been reported in the human population (based on public databases such as gnomAD and UK10K), suggesting that this private biallelic mutation in NUAK2 could be pathogenic. NUAK2 encodes an SNF1/AMPK-related kinase (also known as SNARK), which is often amplified in human cancers (Namiki et al., 2011; Sun et al., 2013). In animal studies, it was shown that Nuak2 is specifically expressed in neural folds of the nascent head of chick and mouse embryos (Bekri et al., 2014; Hirano et al., 2006; Ohmura et al., 2012). Local deficit or accumulation of Nuak2 mRNA in chick neural folds correlated with the curvature direction of the neuroectoderm at the median hinge point and dorsolateral hinge point (Bekri et al., 2014). Importantly, Nuak2 knockout mice show NTDs including exencephaly in 40% of live births, which lends strong support for a potential role for this kinase during NTC in mammals (Ohmura et al., 2012).
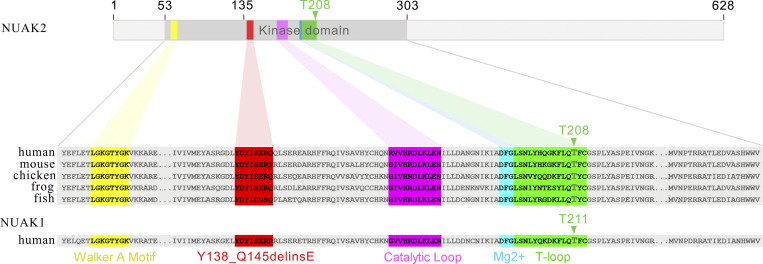
The identified germline mutation causes a 7–amino acid deletion and 1–amino acid alteration annotated as p.Y138_Q145delinsE in the kinase domain of NUAK2 (Fig. 1 D). Amino acid alignment of NUAK orthologs and paralog showed that these eight residues are phylogenetically invariant (Fig. S1). In silico three-dimensional (3D) modeling of the NUAK2 kinase domain revealed that this in-frame deletion is localized in the universally conserved αC-helix (Fig. 1 D). The αC-helix plays a key structural and functional role in protein kinases, linking the C and N lobes of kinase domains, and serves as a dynamic “signal integration motif” (Taylor and Kornev, 2011). This hinge region controls the active and inactive conformation of the kinase, allowing connections between the different subdomains and interaction with ATP and Mg2+ that are essential for catalysis. We therefore hypothesized that this internal truncation of seven critical amino acids in the αC-helix of NUAK2 would lead to major structural changes and most likely impinge on its catalytic activity.
Figure S1.
Amino acid alignments of NUAK orthologs and paralog show high conservation of the mutated residues.
Mutation in the NUAK2 kinase domain abrogates its enzymatic activity
NUAK2 activation requires phosphorylation by upstream kinases or by itself (Lefebvre et al., 2001; Lizcano et al., 2004). To investigate the effect of the mutation on NUAK2 phosphorylation, WT NUAK2 (NUAK2WT) and mutated NUAK2Y138_Q145delinsE (NUAK2MUT) were overexpressed in HEK293T cells. Although a similar amount of WT and MUT NUAK2 proteins were detected by Western blot, only NUAK2WT could be revealed using a phosphospecific antibody that recognizes phosphorylated threonine in the T-loop of AMPK (Fig. 1 E). This result suggests that although the mutation does not impact NUAK2 overall protein stability, it affects its phosphorylation. [32P]ATP phosphorylation assay also showed significant reduction of NUAK2MUT phosphorylation compared with that of NUAK2WT (Fig. 1 F), confirming that the 7–amino acid deletion in the αC-helix affects phosphorylation at residue T208, which can be achieved by NUAK2 itself or by upstream kinases. To verify the catalytic activity of NUAK2MUT, phosphorylation levels of NUAK2 targets were measured. A synthetic AMARA substrate as well as a known NUAK2 endogenous target, LATS2, could not become phosphorylated in the presence of NUAK2MUT, suggesting that unphosphorylated NUAK2MUT is enzymatically inactive (Fig. 1, G and H; Humbert et al., 2010). Taken together, these in vitro results confirmed that the p.Y138_Q145delinsE mutation in the kinase domain of NUAK2 dramatically reduces NUAK2 phosphorylation and abrogates its enzymatic activity toward known substrates, which is consistent with it being an enzymatic loss-of-function mutation.
NUAK2 controls endogenous NUAK1 expression in patient NPCs
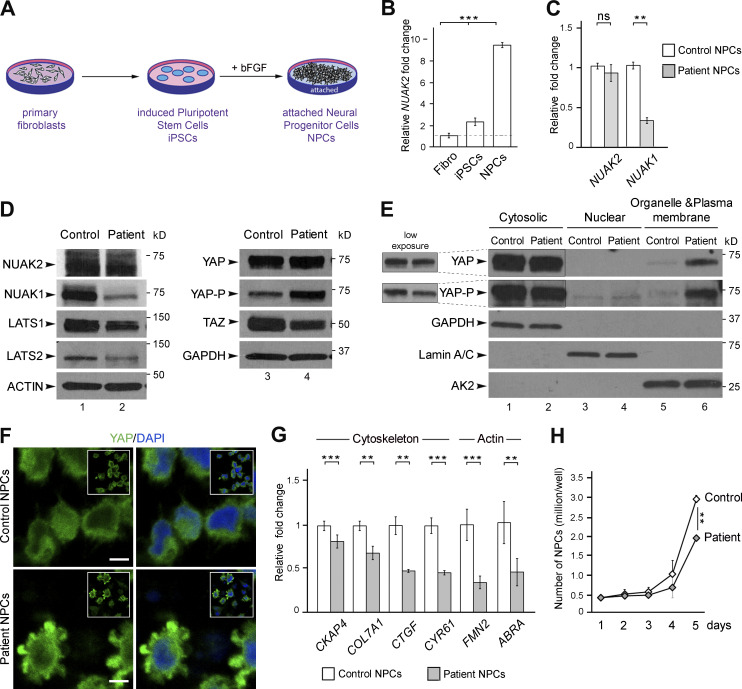
To investigate the downstream effects of this NUAK2 loss-of-function mutation, primary dermal fibroblasts were propagated from a skin biopsy of fetus III-3 and a healthy ethnicity- and sex-matched donor (control). Fibroblasts were reprogrammed into iPSCs, which were differentiated into NPCs (Fig. 2 A). Quantitative RT-PCR (Q-PCR) in control cells revealed that endogenous NUAK2 was approximately ninefold more abundant in NPCs than in dermal fibroblasts, confirming that neural cells are the likely cell-of-origin of the disease and therefore represent a disease-relevant lineage (Fig. 2 B). Although NUAK2 expression was unchanged, the levels of paralogous NUAK1 transcript were reduced by 60% in patient NPCs compared with control cells (Fig. 2 C). A dramatic decrease in the levels of endogenous NUAK1 protein in mutant cells was validated by Western blot while NUAK2 level remained unchanged, suggesting that NUAK2 activity positively regulates NUAK1 expression (Fig. 2 D, lanes 1 and 2). These results raised the possibility that, in humans at least, NUAK2 activity is needed for full NUAK1 function. This interdependence could perhaps explain why NUAK2 knockout humans show a more severe and penetrant phenotype than do single Nuak2 knockout mice (Ohmura et al., 2012).
Figure 2.
NUAK2 mutation affects Hippo-YAP signaling in patient NPCs. (A) Schematic representation of the different cell types generated from control and fetus III-3 skin biopsies. (B) Q-PCR showed higher level of NUAK2 transcripts in control NPCs compared with fibroblasts and iPSCs. Relative NUAK2 expression was normalized against the values for fibroblasts and set to 1 (indicated by a dashed line). (C) Q-PCR showed that while NUAK2 expression was unchanged in patient NPCs compared with control, transcript level of its paralog NUAK1 was significantly reduced. Relative NUAK2 and NUAK1 expression was normalized against the values for control NPCs and set to 1. Q-PCR was repeated in three independent experiments, using two sets of primers. (D) Western blot confirmed similar level of endogenous NUAK2, while NUAK1 was strikingly reduced in patient cells compared with control. LATS1 and LATS2 were reduced in patient cells compared with control, with more extent for LATS1. Phosphorylation of YAP (YAP-P), the main downstream effector of LATS1/2, was significantly increased in patient cells, while YAP level remained the same. TAZ, a paralog of YAP, was significantly decreased in patient cells compared with control. Protein extracts were performed on two control NPC clones and two patient NPC clones. For each NPC clone, at least three biological samples were revealed by Western blot. The same control and patient protein extracts were loaded on two 4–20% gels and revealed with different antibodies as shown in the figure. (E) Subcellular protein fractionation showed increased cytoplasmic localization of YAP in patient cells compared with control, especially in organelle and plasma membranes. Three independent fractionations were performed. (F) YAP immunofluorescence revealed increased level of YAP in the cytoplasm of mutant NPC compared with control. Two antibodies, mouse anti-YAP (Santa Cruz) and rabbit anti-YAP (Cell Signaling) gave the same result. Scale bar = 5 µm. (G) Q-PCR analysis of well-known YAP target genes showed significant reduction of YAP transcriptional activity in patient NPCs compared with control. All YAP targets encode proteins that control the cytoskeleton network including actin. For each gene, relative expression was normalized against the value for control NPCs and set to 1. Q-PCR was repeated at in two independent experiments. (H) Live-cell counting over 5 d in culture showed slower growth of NUAK2-mutated NPCs compared with control. Cells were harvested from three wells (of a 12-well plate). The values shown are the average ± SD of three technical replicates. **, P < 0.02; ***, P < 0.005; ns, not significant.
Loss of NUAK2 activity down-regulates Hippo-YAP signaling in patient NPCs
Because NUAK2MUT failed to phosphorylate LATS2 in vitro, we next investigated the effect of the loss of NUAK2 on other Hippo-YAP signaling components directly in patient cells. LATS1 and LATS2 are upstream kinases that control the transcriptional activities of coactivators YAP and TAZ (Hansen et al., 2015; Piccolo et al., 2014; Varelas, 2014). The phosphorylation of YAP by LATS1/2 serves to inhibit its nuclear translocation and downstream transcriptional activity. Conversely, NUAK2-mediated inactivation of LATS proteins causes unphosphorylated YAP to translocate into the cell nucleus and trigger downstream gene expression. We first evaluated the endogenous expression level of LATS1/2 in patient cells. Although LATS1/2 transcript levels were similar in both NPC lines (Fig. S2 A), decreased LATS1/2 proteins were observed in patient cells (Fig. 2 D, lanes 1 and 2). As previously reported (Humbert et al., 2010), we suggest that changes in LATS1/2 phosphorylation, due to inactive NUAK2, may trigger their degradation through a process that cannot be blocked by the ubiquitin proteasome inhibitor MG132 (Fig. S2 B). Next, while transcript and protein levels of YAP were comparable in patient and control NPCs, phospho-YAP was significantly increased in mutant cells (Fig. S2 A and Fig. 2 D, lanes 3 and 4), suggestive of YAP cytoplasmic retention. Cellular fractionations validated a net increase in phospho-YAP in plasma membrane fraction of mutant NPCs compared with control (Fig. 2 E, lanes 5 and 6). Immunostaining confirmed YAP cytoplasmic accumulation in NUAK2-defective cells, while also revealing altered cell shape with increased protrusions (Fig. 2 F). A robust decrease of endogenous TAZ proteins in patient NPCs was also observed (Fig. 2 D, lanes 3 and 4), confirming that YAP/TAZ signaling was compromised in the absence of NUAK2 activity.
Figure S2.
Transcriptional changes in NUAK2 mutant cells. (A) Q-PCR shows similar transcript level of Hippo components in both control and patient NPCs. (B) While the level of TAZ proteins was partly rescued in MG132-treated cells (lanes 4 and 6 vs. lanes 10 and 12), LATS2 remained lower in mutant cells. Results were confirmed in three independent Western blots. (C) Q-PCR showing transcriptional change in patient NPCs compared with control, especially in genes that are involved in actin polymerization, architecture, and dynamics. Transcription of YAP-targeted genes was significantly deregulated in patient cells compared with control. (D) Q-PCR showing transcriptional rescue of NUAK1 and CTGF in patient NS treated with BLEB and ROCK-I compared with control. Relative gene expression was normalized against the values for nontreated and treated control NS and set to 1. Q-PCR data represent the average ± SD of three technical replicates from two independent experiments. *, P < 0.05; **, P < 0.02; ***, P < 0.005; ns, not significant.
To assess the transcriptional activities of YAP and TAZ, the expression level of established YAP/TAZ target genes, including CKAP4, COL7A1, CTGF, CYR61, FMN2, and ABRA, were measured by Q-PCR. All showed a significant decrease in patient NPCs compared with control. Altogether, these results suggest that catalytically inactive NUAK2 leads to sustained YAP phosphorylation and sequestration in the cytoplasm, reducing the transcriptional output of Hippo signaling. This down-regulation of YAP signaling may explain slower growth or higher apoptosis in patient NPCs compared with that of control. After several days in culture, despite seeding the same number of NPCs, patient cells were consistently less abundant than control cells (Fig. 2 H). To conclude, our results are in line with previous studies identifying NUAK2 as a YAP/TAZ activator in diverse cancer contexts (Gill et al., 2018; Yuan et al., 2018). While YAP activation induced NUAK2 expression in a feed-forward loop to maintain robust Hippo signaling during tumorigenesis, decreased YAP signaling in patient NPCs did not affect NUAK2 transcription but did impact that of NUAK1. The observed decrease in endogenous NUAK1 suggests that it could be a YAP/TAZ target in NPCs. Finally, loss of Hippo signaling in Wnt1-Cre–driven Yap/Taz-depleted mice caused opened anterior neural tubes at embryonic day 10.5 (Wang et al., 2016). Together, these results support that NUAK2 promotes NTC through inhibition of LATS, which otherwise serve to block YAP signaling.
Loss of NUAK2 activity alters the actin network in NPCs
A major role of the Hippo-YAP pathway is to modulate cell proliferation and migration in developing organs and to restrict cell growth in adults by regulating the actin cytoskeleton architecture and dynamics (Dai et al., 2013; Gaspar and Tapon, 2014). YAP signaling and the cytoskeleton network are mutually regulated (Aragona et al., 2013; Mason et al., 2019; Morikawa et al., 2015). Because several YAP-targeted genes, which are key players in cytoskeleton organization, were down-regulated in patient cells (Fig. 2 G), we next examined the actin network in NPCs. As expected, patient NPCs had a significant increase in F-actin network and filopodia-like cellular protrusions compared with control cells, with colocalization of F-actin and NUAK2 in restricted areas of the cells (Fig. 3 A). This result is in line with previous observation that Nuak1−/− MEFs have significantly more F-actin stress fibers relative to WT counterparts (Zagórska et al., 2010). A fractionation of soluble G-actin to insoluble F-actin revealed that patient cells had higher F-actin:G-actin ratio compared with control NPCs (Fig. 3 B). This increase in actin filaments when NUAK2 is mutated echoes a previous study that demonstrated that depolymerization of F-actin into G-actin is regulated by the active form of NUAK2 (Suzuki et al., 2003). Moreover, Q-PCR analysis showed a significant transcriptional down-regulation of TMSB4X, which is involved in G-actin sequestering, and CAPZA2, which caps growing actin filaments to limit their growth (Fig. S2 C). In contrast, a series of genes coding for F-actin binding proteins were up-regulated in patient NPCs compared with control cells, consistent with a skewed F-actin accumulation in NUAK2 mutant cells (Fig. S2 C). Altogether, these results suggest that catalytically inactive NUAK2 induces inactivation of YAP signaling, causing aberrant actin cytoskeletal changes in patient cells.
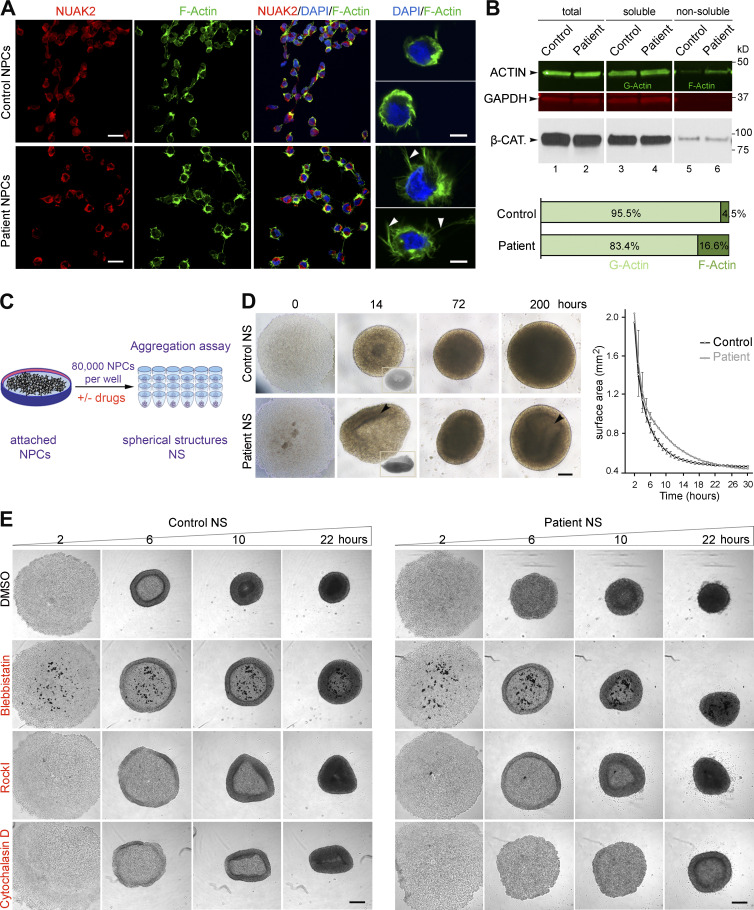
Figure 3.
Defects in the actomyosin network in patient-derived NPCs cause abnormal cell aggregation. (A) Immunofluorescence on NPCs revealed close proximity of NUAK2 to F-actin as well as F-actin network expansion in patient NPCs compared with control (left panels). Scale bar = 20 µm. Higher-resolution images of representative cells showed that patient NPCs were enlarged and enriched with F-actin filopodial protrusions (arrowhead) compared with control cells (right panel). Scale bar = 5 µm. (B) G- and F-actin fractionation followed by quantitative Western blot showed increased F-actin in patient NPCs compared with control. GAPDH and β-CATENIN (β-CAT) served as loading controls for each protein fraction. Two independent fractionations were performed. (C) Schematic representation of aggregation assay, in which 80,000 NPCs seeded in low-attachment U-bottom microwells formed spherical structures (NS) over time. (D) Bright-field images illustrating representative spatiotemporal assembly and organization of NPCs during aggregation assay. Patient NPC aggregation was disorganized, failing to form donut-shaped pattern (14 h, inset) and spherical structures compared with control. Heterogeneous cell density (arrowhead) was observed in patient NS compared with control, suggesting lower proliferation. Scale bar = 200 µm. NS area measurement at sequential time points showed that patient NS aggregation was delayed compared with control (right graph). All experiments were repeated three times, with two different iPSC-derived NPC clones for control and patient. Surface area was calculated using ImageJ software, and represented values are the average ± SD of eight NS. (E) Bright-field images of NS after 2, 6, 10, and 22 h in culture, showing absence of outer ring formation in patient NPCs. Treatment with BLEB rescued ring formation in patient NPCs, as well as with ROCK-I, to a lesser extent. In contrast, treatment with cytochalasin D abrogated cell aggregation in both cell lines, more so in patient NPCs. All experiments were repeated independently three times. Scale bar = 250 µm.
A defective actomyosin network in patient NPCs impairs cell aggregation
Next, we questioned whether the observed F-actin change in patient NPCs had any effects on their mechanical properties, especially with regard to cell–cell interaction and organization. In vitro assays (Fig. 3 C) showed that aggregation of NUAK2 mutant cells into neurospheres (NS) was significantly delayed and disorganized compared with control cells (NS; Fig. 3 D). Live imaging showed that, while control NPCs moved toward the center of the well by forming an outer ring that expanded and merged to create NS, NUAK2 mutant NPCs failed to form regular donut-shaped rings (Fig. 3, D and E). To obtain direct evidence that the aggregation of patient cell was affected because of F-actin accumulation, cells were treated with small molecules that diminish actin and myosin dynamics (Peng et al., 2011). Treatment with blebbistatin (BLEB), a myosin II inhibitor, entirely rescued outer ring formation in patient NPCs (Fig. 3 E). To a lesser extent, the Rho-associated protein kinase inhibitor (ROCK-I) Y-27632, which blocks myosin II activation, also partially rescued this process. In contrast, treatment with cytochalasin D, an inhibitor of actin polymerization, abrogated cell aggregation in both cell lines, with more extensive effects in patient NPCs (Fig. 3 E). Interestingly, Q-PCR analysis showed partial rescue of NUAK1 expression and that of a well-established YAP-target gene in patient NS treated with BLEB and ROCK-I compared with control NS (Fig. S2 D). This suggests that reactivation of YAP signaling, triggered by actomyosin changes, may in part rescue NUAK1 expression, which in turn could serve to compensate for the loss of NUAK2. Together, these in vitro results indicate that NUAK2 maintains an optimal level of YAP signaling and actomyosin for cultured NPCs to elicit concerted cellular movements and proliferation. Inhibition of F-actin disassembly causing actomyosin accumulation was previously reported in early-stage mouse embryos to cause defective neurulation (Escuin et al., 2015). We therefore propose that NUAK2 may regulate the actomyosin network for proper cellular shape and movements during neural plate folding.
NUAK2 mutant human COs display aberrant neural tube–like structures (NTs)
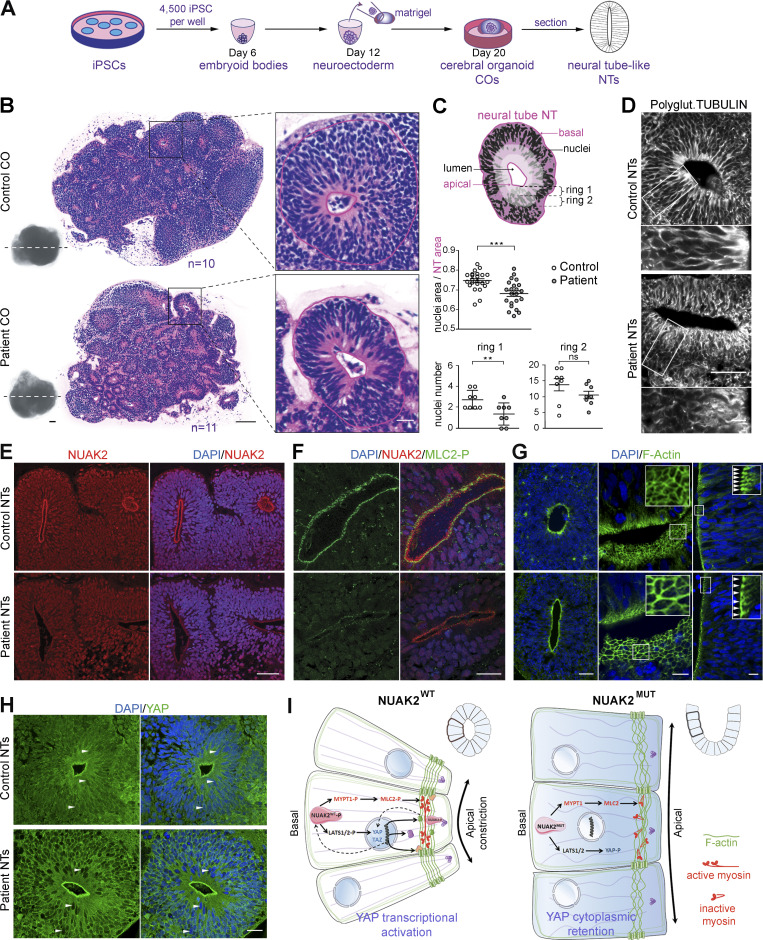
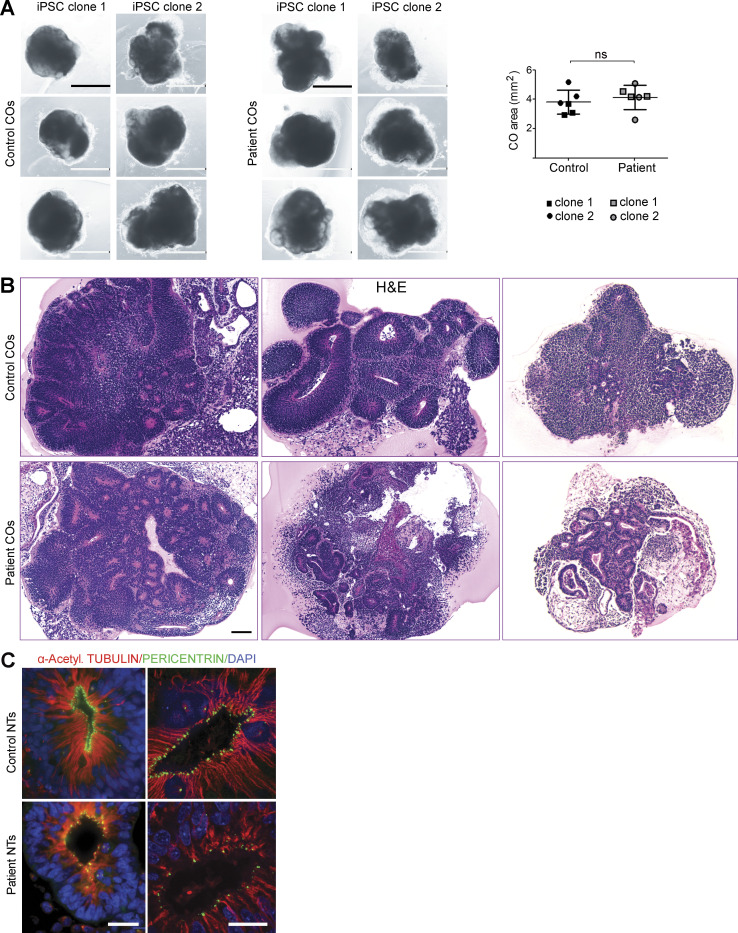
To further investigate the pathomechanisms that cause anencephaly in NUAK2 mutant human fetuses, we generated 3D COs from several clones of control- and patient-derived iPSCs (Fig. 4 A; Lancaster and Knoblich, 2014). Previous studies demonstrated the self-organizing capacity of different neural tissues within COs, including the presence of neuroectoderm composed of NECs that spontaneously establish apical-basal polarity to form NTs (Liu et al., 2019). After 3 wk in culture, the outer morphology and size of control and patient COs were similar (Fig. 4 B and Fig. S3 A). However, H&E staining of CO sections revealed that NUAK2 mutant NTs were smaller and disorganized compared with control NTs (Fig. 4 B and Fig. S3 B). While cell nuclei were distributed along the apicobasal axis of NECs in control NTs, a significantly reduced number of nuclei in the apical surface of patient NTs was observed (Fig. 4, B and C), suggestive of defective basoapical nuclear migration. As reported in the neural plate of Nuak2 knockout mice (Ohmura et al., 2012), the microtubule filaments marked by polyglutamylated and α-acetylated tubulins showed striking disorganization in NUAK2 mutant NTs, causing poor cell elongation (Fig. 4 D and Fig. S3 C) that may underlie defective nucleokinesis.
Figure 4.
NUAK2 controls apical actomyosin network in human COs. (A) Schematic representation of the culture workflow to generate COs from control and patient iPSCs. At 20 d, COs were fixed, sectioned, and used as a proxy to study NTs. Three batches of COs were independently performed using two control iPSC clones and two patient iPSC clones. For each iPSC clone, 8–16 COs were prepared (in 8–16 wells of a 96-well plate). (B) Representative image of H&E staining of control (n = 10) and patient (n = 11) CO sections shows multiple 2D NT structures that display features of the embryonic neural tube. Patient NT shows basal nucleus accumulation compared with control NT, in which nuclei were scattered along the apico-basal axis. Scale bars = 200, 100, and 20 µm (from left to right panels). (C) Schematic representation of a NT displaying total NT area (pink) and nucleus area (black), as well as two most apical subrings (rings 1 and 2, light pink). Ratio of nucleus area to NT area confirmed nucleus compaction in patient NT (n = 23) compared with control (n = 22; top graph). Number of nuclei was decreased apically (rings 1 and 2) in patient NT (n = 8) compared with control (n = 8; bottom graphs). (D) Polyglutaminated tubulin immunostaining on CO sections revealed disorganized and nonlinear microtubule network in mutant NT cells compared with control. Scale bars = 50 µm (top panel); 10 µm (lower panel). (E) Representative image of NUAK2 immunostaining on CO sections showing that, while NUAK2 localized in the nucleus of NECs, its apical concentration was significantly reduced in mutant NT compared with control. Scale bar = 50 µm. (F) Representative image of coimmunostaining of NUAK2 with phosphorylated MLC2 (MLC2-P) on CO sections, showing their proximal localization at the apical side of NT cells, with less extent in mutant NT. Scale bar = 20 µm. (G) Representative image of phalloidin staining revealing F-actin accumulation apically (left panel). Apical view of mutant NT revealed enlargement of NECs compared with control (middle panel, inset). Frontal view of patient NT confirmed enlargement of NECs compared with control (right panel, inset, white arrowheads). Scale bars = 50, 10, and 10 µm (from left to right panels). (H) Representative image of YAP immunostaining on CO sections revealing YAP cytoplasmic concentration in mutant NT cells compared with control. We also noted enrichment of YAP apically in both patient and control NTs, suggesting a possible interaction of NUAK2 and the actomyosin network during apical constriction. Scale bar = 20 µm. (I) Model showing the role of NUAK2 in regulating apical cell constriction and apicobasal cell elongation during neural tube formation. By regulating Hippo-YAP signaling, NUAK2 controls F-actin organization and dynamics. All immunostaining were confirmed at least three times using COs from different batches. **, P < 0.02; ***, P < 0.005; ns, not significant.
Figure S3.
NUAK2 controls NT-like structures in human COs. (A) Images of control and patient COs derived from two different iPSC clones. Graph representing the size (area) of control and mutant COs (right panel). Scale bar = 1 mm. (B) H&E staining of CO sections revealed either general dysplasia with significantly fewer NT structures or nucleus disorganization in patient NT compared with control. Scale bar = 100 µm. (C) Coimmunostaining using α-acetylated tubulin and pericentrin confirmed microtubule disorganization and scattered centrosomes in NUAK2 mutant NECs compared with control. Scale bar = 20 µm. ns, not significant.
NUAK2 controls the apical actomyosin network in human COs
Immunostaining analysis of CO sections revealed that endogenous NUAK2 was equally nuclear in WT and mutant NECs. Strikingly, however, mutant NUAK2 failed to be apically enriched near the lumen of NTs in patient-derived COs (Fig. 4 E). F-actin and phosphorylated MLC2 (MLC2-P), a known target of NUAK2, were found in close proximity to NUAK2 at the apical surface of WT NTs, confirming that NUAK2 interacts with the actomyosin network. (Fig. 4, F and G). The decreased signal for MLC2-P in patient NECs suggests that functional NUAK2 is required for MLC2 phosphorylation, which is instrumental for downstream actomyosin contraction. Finally, while YAP was ubiquitously found in control NECs, it was excluded from nuclei in NUAK2 mutant cells (Fig. 4 H), suggesting lower Hippo signaling activity. Accumulation of YAP was also detected apically, suggesting that it also belongs to the contractile network that controls apical cell constriction or cell–cell adhesion. A similar observation has been reported in Drosophila wing imaginal tissues, in which accumulation of Yorkie, the Drosophila homologue of YAP, in the apical junctional region at the cell cortex was shown to promote myosin activation (Xu et al., 2018). This result suggests that decreased YAP activity, which correlates with YAP cytoplasmic localization, together with apical localization of YAP, may in part contribute to the observed defective cytoskeleton architecture, causing impaired folding and growth of the neuroectoderm (Fig. 4 I).
Taken together, our study reports on the genetic, molecular, and developmental etiology of anencephaly in humans. We propose that a likely pathomechanism underlying anencephaly in human fetuses lacking NUAK2 activity is insufficient Hippo-YAP signaling. In a WT setting, the neuroepithelium expresses high levels of NUAK2, locally activating Hippo signaling, which leads to the reorganization of the actomyosin network to drive apical cell constriction. Failure to do so in a NUAK2 mutant neural plate prevents the folding and fusion of the nascent neural tube, which blocks proper brain development and results in embryonic lethal anencephaly.
Materials and methods
Human subjects
Peripheral blood samples were collected from parents (II-1 and II-2) and tissues from two fetuses (III-2 and III-3). Genomic DNA was extracted using DNeasy Blood & Tissue Kits (Qiagen). Skin biopsies from the third affected fetus (III-3) and from an ethnically matched unaffected child were collected to derive primary cells in vitro. All biospecimens were obtained after parents gave their informed consent and the local ethics commission gave its approval. All human studies were reviewed and approved by the institutional review boards of the National University of Singapore and local institutions.
Genetics
Whole-exome sequencing (WES) was performed on genomic DNA from parents II-1 and II-2 and proband III-2. The exome library was prepared on an Ion OneTouch System and sequenced on an Ion Proton instrument (Life Technologies). Sequence reads were aligned to the human GRCh37/hg19 assembly (UCSC Genome browser). WES data were deposited in the Sequence Read Archive website under accession no. PRJNA638500. Each variant was annotated with the associated gene, location, protein position, amino acid change, quality score, and coverage. Variants were filtered based on functional prediction scores including SIFT (Kumar et al., 2009), PolyPhen2 (Adzhubei et al., 2010), and Mendelian clinically applicable pathogenicity (Jagadeesh et al., 2016), as well as phyloP conservation scores (Pollard et al., 2010). Variants were further filtered for common single nucleotide polymorphisms using the National Center for Biotechnology Information’s “common and no known medical impacts” database (ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37/), the Exome Aggregation Consortium (https://gnomad.broadinstitute.org/), and the Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), as well as an in-house database of 487 sequenced individuals of Middle East origin. Variants that were predicted to be synonymous or not having a location on a coding exon, UTR, splice site junction, or flanking intron were filtered out. Finally, only variants that were homozygous in the proband while heterozygous in both parents were retained. Only one homozygous deletion/insertion variant on Chr1: 205,277,780; c.412_433delTATGACTACATCAGCGAGCGGCinsG remained (Table S1). This mutation is localized in the NUAK2 gene and has never been reported in a population. Sanger sequencing using primers flanking the mutation (forward, 5′-CAAGATCGTGATCGTCATGG-3′; reverse, 5′-CTCCTTCAGGAATCCACCAA-3′) confirmed segregation of the variant with anencephaly.
3D modeling
To model the 3D structure of NUAK2 kinase domain, the SWISS-MODEL homology modeling workspace was used (Arnold et al., 2006). We submitted the following amino acid sequence: YEFLETLGKGTYGKVKKARESSGRLVAIKSIRKDKIKDEQDLMHIRREIEIMSSLNHPHIIAIHEVFENSSKIVIVMEYASRGDLYDYISERQQLSEREARHFFRQIVSAVHYCHQNRVVHRDLKLENILLDANGNIKIADFGLSNLYHQGKFLQTFCGSPLYASPEIVNGKPYTGPEVDSWSLGVLLYILVHGTMPFDGHDHKILVKQISNGAYREPPKPSDACGLIRWLLMVNPTRRATLEDVASHWWV.
The SWISS-MODEL template library (SMTL v2015-04-15, PDB release 2015-04-10) was searched with Blast (Altschul et al., 1997) and HHBlits (Remmert et al., 2011) for evolutionarily related structures matching the target sequence. The closest x-ray 3D model of NUAK2 kinase domain was the one from the MAP/microtubule affinity-regulating kinase 3.
Plasmid constructs
Human influenza hemagglutinin (HA)–tagged human NUAK2 WT (NUAK2WT) and mutant (NUAK2MUT) cDNAs were synthesized from mRNA extracted from control and patient fibroblasts using GenScript and cloned into pCS2+ vector. pCDNA Myc-tagged LATS2 plasmid was a gift from David Bernard (Centre de Recherche en Cancérologie de Lyon, Lyon, France). The N-terminal Myc-tag was replaced with a Flag-tag and recloned into pLPC vector. For overexpression, HEK293T cells were transfected with DNA using jet polyethylenimine (PEI; Polyplus) according to manufacturer’s instruction.
Threonine phosphorylation assays
HA-tagged NUAK2WT and NUAK2MUT were transfected into HEK293T cells with JetPEI following the manufacturer’s instructions. After 36 h, cells were washed with ice-cold PBS and collected with 50 mM Hepes lysis buffer containing 150 mM NaCl and 1% Triton X-100 supplemented with protease and phosphatase inhibitors as described previously (Humbert et al., 2010). Approximately 50 μg of protein extracts was separated in a 7.5% SDS-PAGE gel and transferred overnight to polyvinylidene fluoride (PVDF) membrane in N-cyclohexyl-3-aminopropanesulfonic acid buffer. Membranes were incubated with anti-HA (mouse) and anti-phospho-AMPK (T172; rabbit) antibodies overnight. Signal was quantified using the LI-COR system.
Overexpressed NUAK2WT and NUAK2MUT proteins were also incubated at 37°C for 20 min in 25 mM Hepes, pH 7.5, and 1% Triton X-100 supplemented with protease and phosphatase inhibitors, 10% glycerol, and 0.5 μl [32P]ATP (6,000 Ci/mmol; Perkin Elmer). Reactions were stopped with SDS-PAGE sample buffer and separated on NUPAGE (Invitrogen) gels in Tris-glycine buffer. Gel was fixed in methanol:water, stained with Simply Blue stain (Invitrogen), washed in water, dried, and phosphorimaged.
Catalytic activity assays
Kinase activity assays were performed as previously described (Humbert et al., 2010). In short, equal amounts of proteins from transfected and untransfected extracts were incubated overnight with anti-HA or anti-Flag agarose resins (Sigma-Aldrich). Each protein-bound resin was then washed three times for 15 min with lysis buffer and eluted with 120 μl of elution buffer containing either HA or Flag peptide as suggested by the manufacturer. In vitro kinase assays were performed with AMARA (AMARAASAAALRRR) and LATS2 as substrates of NUAK2.
Q-PCR
Total RNA was extracted using RNAeasy Plus kit (Qiagen). cDNA was synthesized from 1 μg of RNA using iScript reverse transcription kit (Bio-Rad). Q-PCR was performed using SYBR Green Mix (Roche). Q-PCR was repeated in three biological replicates (RNA extract from different NPC subcultures). Two sets of primers were designed and tested for most of the genes as shown in Table 1. Cycle threshold values were normalized to GAPDH or ACTIN values. Data shown in figures represent the average ± SD of three technical replicates. Student’s t test (two-tailed distribution) was performed and represented as *, P < 0.05; **, P < 0.02; and ***, P < 0.005.
Table 1. Sequence of Q-PCR primers.
| Gene name | GenBank accession no. | Sequence (5′ to 3′) | ||
|---|---|---|---|---|
| Direction | First set | Second set | ||
| GAPDH | NM_002046 | Forward | TGCACCACCAACTGCTTAGC | |
| Reverse | GGCATGGACTGTGGTCATGAG | |||
| ACTB | NM_001101 | Forward | ATGTTTGAGACCTTCAACAC | |
| Reverse | AGGTAGTCAGTCAGGTCCCGGCC | |||
| NUAK1 | NM_014840 | Forward | AAGGCACCTACGGCAAAGTC | GGGAGCTGTACGATTACATCAG |
| Reverse | GTCTGATGTGAACCATGTCTTGT | ACACCGTTCTTGTGACAATAGTG | ||
| NUAK2 | NM_030952 | Forward | GAGCGCGAAGCTAGGCATT | CGCCCAAGCCCCTAATGAAG |
| Reverse | ATCTCTGGCGAGGCATAGAGG | TCCCTCCGTATGTGCATCAGA | ||
| CKAP4 | NM_006825 | Forward | CCGTGGAATCACTCCAGAAGG | TGGACAGTTTGGTTGCATACTC |
| Reverse | AGTCCTGAGCATTTTCAAGTCC | CCTCAGGTCATCTAGTAAACCCT | ||
| COL7A1 | NM_000094 | Forward | TTACGCCGCTGACATTGTGTT | GGCTGCAATTCTCCATGTGG |
| Reverse | ACCAGCCCTTCGAGAAAGC | CTGTGAGGCAACTCGCTTCA | ||
| CTGF | NM_001901 | Forward | CAGCATGGACGTTCGTCTG | ACCGACTGGAAGACACGTTTG |
| Reverse | AACCACGGTTTGGTCCTTGG | CCAGGTCAGCTTCGCAAGG | ||
| CYR61 | NM_001554 | Forward | ACCGCTCTGAAGGGGATCT | GGTCAAAGTTACCGGGCAGT |
| Reverse | ACTGATGTTTACAGTTGGGCTG | GGAGGCATCGAATCCCAGC | ||
| FMN2 | NM_020066 | Forward | CAGAGCCTCGGTGTTTTCCAA | |
| Reverse | GCACACTCGGTATCCGACAG | |||
| ABRA | NM_139166 | Forward | AGCAGTGGGCGAATGAGAAC | AGGTGTCCAAAACGGTGGTC |
| Reverse | GTGATTGGTTTAGGAGCTTGAGG | CATCCCTCTCGTACCTGTGG | ||
| LATS1 | NM_004690 | Forward | AATTTGGGACGCATCATAAAGCC | TTACCAAGATCCTCGACGAGAG |
| Reverse | TCGTCGAGGATCTTGGTAACTC | CACATTCCCTGGTTTCATGCT | ||
| LATS2 | NM_004691 | Forward | ACTTTTCCTGCCACGACTTATTC | ACCCCAAAGTTCGGACCTTAT |
| Reverse | GATGGCTGTTTTAACCCCTCA | CATTTGCCGGTTCACTTCTGC | ||
| YAP | NM_006106 | Forward | TAGCCCTGCGTAGCCAGTTA | |
| Reverse | TCATGCTTAGTCCACTGTCTGT | |||
| TAZ | NM_181313 | Forward | CACCGTGTCCAATCACCAGTC | TGGCATGTCGGAATGAATGAC |
| Reverse | TCCAACGCATCAACTTCAGGT | GCTTCCCGACAGCACAGT | ||
| TMSB4X | NM_004202 | Forward | TGACAAACCTGGTATGGCTGA | |
| Reverse | GCCTCTCCTGTTCGATAGTTTC | |||
| CAPZA1 | NM_006135 | Forward | GCCGACTTCGATGATCGTGT | TTCAATGACGTTCGGCTACTAC |
| Reverse | AGTAGCCGAACGTCATTGAATAC | GGCGTGAACTGATCCATGTTAT | ||
| CAPZA2 | NM_006136 | Forward | AAGAGAAGGTGCGTATAGCAGC | CATTACCCGAATGGAGTCTGC |
| Reverse | AAGCAGTAACCGAACATCATTGA | ACTTGAGTGGTTGAAGGAGTGA | ||
| BCAM | NM_005581 | Forward | GAGGTGCGCTTGTCTGTACC | GAATGGTTCCTTACCGACCG |
| Reverse | GCATATAATGGTCGTGGGTTCC | TCGTGCATTGTGACCTGGAG | ||
| TLN2 | NM_015059 | Forward | GCGTGTCGAGTCATTCGGG | TGACCTAAATTGGCTGGATCAC |
| Reverse | CCCTTTCCTCGGGTCTTCATC | AACTTCCGTCTAAGCAGCAAC | ||
| ARHGEF17 | NM_014786 | Forward | CAGTGACGACGACCGAGAC | CCAAGGTGAGCTTTCCCTCG |
| Reverse | TCCTGATCCAATAGGGAGCTTC | CCTACTAGACCATAGGTCGTCGT | ||
| CDH1 | NM_004360 | Forward | CGAGAGCTACACGTTCACGG | ATTTTCCCTCGACACCCGAT |
| Reverse | GGGTGTCGAGGGAAAAATAGG | TCCCAGGCGTAGACCAAGA | ||
| ACTN2 | NM_001103 | Forward | CAAACCTGACCGGGGAAAAAT | ATGGCCTTGGACTCTGTGC |
| Reverse | CTGAATAGCAAAGCGAAGGATGA | GGTGTTCACGATGTCTTCAGC | ||
| ACTN4 | NM_004924 | Forward | GCAGCATGGGCGACTACAT | TGGAGGTCATATCAGGGGGAGC |
| Reverse | TTGAGCCCGTCTCGGAAGT | GAGACCAGCTTGACGCCTTT | ||
| ACTN1 | NM_001130005 | Forward | TCCATCGGAGCCGAAGAAATC | CCCGAGCTGATTGACTACGG |
| Reverse | GTGTCGGTGGATCAAAGCACA | GCAGTTCCAACGATGTCTTCG | ||
Protein extraction, fractionation, and Western blot
Cells were lysed and protein was extracted using radioimmunoprecipitation assay buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% NP-40, and 0.5% Na-deoxycholate) supplemented with Complete Protease Inhibitor Cocktail tablets (EDTA-free, Roche; 12245300). All Western blot results were confirmed in at least three NPC subcultures derived from different control and mutant iPSC clones. For G- and F-actin biochemical fragmentation, G-actin:F-actin In Vivo Assay Kit (Cytoskeleton; BK037) was used according to the manufacturer’s instructions. Soluble proteins, which contain free monomeric actin (G-actin), were separated from insoluble cytoskeleton proteins including filamentous actin (F-actin). A cell fractionation kit (Abcam; ab109719) was also used to separate proteins from cytosol, nucleus, and organelle/plasma membranes. Proteins were prepared in 1× loading buffer with dithiothreitol, loaded on 4–20% precast polyacrylamide gel (Criterion TGX Stain-Free; Bio-Rad), and transferred on PVDF membrane, which was blocked in either 5% milk or 5% BSA Tris-buffered saline with Tween buffer. For better G- to F-actin quantification, PVDF membranes were blocked in Odyssey Blocking Buffer (LI-COR Biosciences), and bands were detected, normalized, and quantified using LI-COR Odyssey infrared Imaging System. The fractionation was duplicated using different protein extracts from control and patient NPCs.
Antibodies and dyes
Nuclei were stained with 0.5 μg/ml DAPI (Invitrogen) and F-actin with Alexa Fluor 488 phalloidin (Thermo Fisher Scientific; A12379, 1:200). For Western blotting and immunofluorescence staining, all antibodies are commercially available (Table 2).
Table 2. List of antibodies used for Western blot and immunostaining.
| Antibody | Species | Company | Catalog no. | Western blot | Immunofluorescence staining |
|---|---|---|---|---|---|
| NUAK1 | Goat | Abgent | AF2736a | 1:50 | |
| ARK5/NUAK1 | Rabbit | Cell Signaling Technology | 4458 | 1:1,000 | |
| NUAK2 | Rabbit | Abgent | AP7158a | 1:50 | |
| LATS1 | Rabbit | Bethyl | A300-477A | 1:5,000 | |
| LATS2 | Mouse | Abnova | MAB0019 | 1:500 | |
| YAP | Rabbit | Cell Signaling Technology | 14074 | 1:1,000 | |
| YAP | Mouse | Santa-Cruz | sc-101199 | 1:50 | |
| YAP-P (S127) | Rabbit | Cell Signaling Technology | 13008S | 1:1,000 | |
| TAZ (V386) | Rabbit | Cell Signaling Technology | 4883 | 1:1,000 | |
| MLC2-P (Ser19) | Mouse | Cell Signaling Technology | 3675 | 1:50 | |
| MLC2-PP (Thr18-Ser19) | Rabbit | Cell Signaling Technology | 3674S | 1:100 | |
| Polyglutamylated tubulin | Mouse | Sigma-Aldrich | T9822 | 1:1,000 | |
| α-Acetylated tubulin | Mouse | Sigma-Aldrich | T6793 | 1:1,000 | |
| γ-Tubulin | Mouse | Abcam | ab11316 | 1:200 | |
| Pericentrin | Rabbit | Abcam | ab4448 | 1:2,000 | |
| Cleaved caspase-3 | Rabbit | Cell Signaling Technology | 9664L | 1:400 | |
| Histone H3-P (Ser10) | Rabbit | Merck Millipore | 06-570 | 1:500 | |
| ACTIN | Mouse | Merck Millipore | Mab1501R | 1:2,000 | |
| GAPDH | Mouse | Santa-Cruz | sc-47724 | 1:4,000 | |
| Lamin A/C | Mouse | Merck Millipore | MAB3211 | 1:1,000 | |
| AK2 | Rabbit | Proteintech | 11014-1-AP | 1:1,000 | |
| β-Catenin | Rabbit | Sigma-Aldrich | C2206 | 1:4,000 | |
| HA-Tag | Mouse | BAbCo | 12CA5 | 1:1,000 | |
| Flag-Tag | Mouse | Sigma-Aldrich | F1804 | 1:3,000 | |
| pAMPK (T172) | Rabbit | Cell Signaling Technology | 2535 | 1:1,000 |
Culture of human cells
HEK293T cells were grown in DMEM High Glucose (Gibco) with 10% FBS (Gibco), 1% l-glutamine (Thermo Fisher Scientific; 25030-081) and 1 mM penicillin and streptomycin (Thermo Fisher Scientific; 15140-122) at 37°C in a humidified atmosphere of 5% CO2. Human primary fibroblasts were generated from skin biopsies and were expanded in the same medium as HEK293T cells.
Human iPSCs were derived from primary fibroblasts (Park et al., 2008). Control cells were transduced with OCT4, SOX2, KLF4, and c-MYC. After 4 d, transduced cells were seeded onto irradiated mouse embryonic fibroblasts in human ES cell medium (DMEM/F12;, Sigma; D6421) supplemented with 20% Knock Out Serum Replacement (Thermo Fisher Scientific; 10828-028), 0.1 mM 2-mercaptoethanol (Thermo Fisher Scientific; 21985-023), 2 mM l-glutamine, 0.2 mM nonessential amino acids ([NEAA] Thermo Fisher Scientific; 11140-050) and 5 ng/ml basic fibroblast growth factor (Stemgent; 03-0002). iPSC colonies were picked between days 17 and 28 and maintained in Matrigel (Corning; 354234) and mTeSR1 (Stem Cell Technologies; 85850) for expansion. Patient cells were reprogrammed using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific; A16517) in accordance with the manufacturer’s instructions. Cells were plated onto irradiated mouse embryonic feeders 7 d after viral transfection and kept in human ES cell medium as for control iPSCs.
Human NPCs were generated from iPSCs, which were dissociated into single cells and seeded in low-attachment U-bottom 96-well plates at a density of 10,000 cells per well in neural induction medium (DMEM/F12, Thermo Fisher Scientific; 10565-018) supplemented with B-27 (Thermo Fisher Scientific; 17504044), N-2 (Thermo Fisher Scientific; 17505048), 0.2 mM NEAA, 100 nM LDN 193189 (Stem Cell Technologies; 72148), 10 μM SB431542 (Stem Cell Technologies; 72234), and 10 μM ROCK-I (Stem Cell Technologies; 72304). After 6 d, cell aggregates were attached onto Matrigel-coated plates and grown in neural expansion medium (DMEM/F12; Thermo Fisher Scientific; 10565-018) supplemented with B-27, N-2, 0.2 mM NEAA, and 20 ng/ml basic fibroblast growth factor. After 3–6 d, rosette structures were manually cut and cultured in neural expansion medium attached onto Matrigel-coated plates. Living cells were counted using Trypan Blue dye and a glass hemocytometer.
Cell aggregation assay
In vitro cell aggregation assays were set up to study whether alteration observed in actin cytoskeleton impaired NPC movement and organization within a cluster of cells. NS were generated from NPCs, which were dissociated into single cells using Accutase. To form equal-sized NS, ∼80,000 cells/well were seeded in low-attachment U-bottom 96-well plates, then aggregated with 1-min spin down and maintained in neural expansion medium. Aggregation assays were repeated three times, with two control iPSC-derived NPC clones and two patient iPSC-derived NPC clones. Live imaging was started soon after centrifugation to measure the kinetics and extent of aggregation in normal conditions or under drug treatment with BLEB (50 μM), ROCK-I (10 μM), or cytochalasin D (0.1 μM). Experiments were repeated three times, and for each assay, eight wells per condition were measured. Pictures were taken every 30 min for 1–3 d. Neurosphere area was calculated using ImageJ (National Institutes of Health).
CO generation
COs were generated as described in Lancaster et al. (2013) from two different iPSC clones for mutant and control. In short, iPSCs were dissociated into single cells using Accutase (Stem Cell Technologies; 07920). Approximately 5,000 cells per well were seeded into a low-attachment U-bottom 96-well plate to form embryoid bodies (EBs) in human ES cell medium (DMEM/F12 supplemented with 20% Knock Out Serum Replacement, 0.1 mM 2-mercaptoethanol, 2 mM l-glutamine, 0.1 mM NEAA, 1 mM sodium pyruvate (Thermo Fisher Scientific; 11360-070), and 10 µM ROCK-I for 6 d. EBs were then maintained in DMEM/F12 supplemented with 2 mM l-glutamine, 1% N-2, 1% NEAA, and 1 μg/ml heparin for 5 d. On day 11, each EB was transferred to cold Matrigel droplets. When droplets were solidified at room temperature, they were transferred into low-attachment 6-well plates placed on a rocking shaker (85 rpm) and maintained in DMEM/F12 supplemented with 2 mM l-glutamine, 0.5% N-2, 1% B-27 without vitamin A, 3.5 μl/liter 2-mercaptoethanol, 1% NEAA, and 0.025% insulin (Sigma-Aldrich) for 5 d. After 5 d, B-27 was supplemented with vitamin A. After a total of 20 d in culture, COs were fixed in 4% paraformaldehyde overnight at 4°C, washed three times in PBS, and dehydrated in 70% ethanol before being embedded in paraffin blocks. Tissues were sectioned at 5 μm for H&E staining as well as immunofluorescence staining. For phalloidin staining, COs were dehydrated in 30% sucrose before cryopreservation and sectioning. Three batches of COs were independently performed using two control iPSC clones and two patient iPSC clones. For each iPSC clone, 8–16 COs were prepared (in 8–16 wells of a 96-well plate).
Immunofluorescence on cultured NPCs and CO sections
NPCs were placed in 4-well chamber slides (Merck Millipore) coated with Matrigel. Cells were fixed in 4% paraformaldehyde for 30 min at room temperature. After three washes in PBS, cells were permeabilized in 0.5% Triton X-100 for 15 min at room temperature. After three washes in PBS, cells were blocked in 1% BSA for 30 min at room temperature. CO sections were placed in an oven at 60°C for 45 min. Slides were cooled down at room temperature, followed by a deparaffinization procedure in xylene and ethanol. After being washed in water for 5 min, slides were immersed in 10% antigen retriever (citrate buffer, pH 6, Sigma-Aldrich; C9999) and placed in a pressure cooker for 30 min. After slides were cooled down at room temperature (90 min) and washed three times in PBS, they were blocked in blocking buffer (1% BSA and 0.5% Triton X-100). Primary antibodies were prepared in 1% BSA and 0.5% Triton X-100 and added accordingly for overnight incubation at 4°C. After several washes in PBS, secondary antibodies labeled with Alexa Fluor dyes (Life Technologies) were incubated for 1 h at room temperature. After three washes in PBS, cells were stained with DAPI. Slides were mounted in ProLong Gold Antifade Reagent (Life Technologies). Images were taken with bright-field and wide-field Zeiss AxioImager Z1 upright microscopes or an Olympus FV1000 upright confocal microscope.
Image analysis
ImageJ was used to analyze the bright-field and fluorescent images of NPCs and COs. To assess nucleokinesis defects observed in the NTs of mutant COs, two methodologies were used. First, each NT was manually selected (excluding lumen) to calculate NT area, and all nuclei within an NT were automatically selected to calculate surface area. To estimate the lack of nuclei in some regions of the NT, the ratio of nucleus area to NT area was calculated. Second, to show that fewer nuclei were present at the apical side of the mutant NTs, two most apical rings were drawn, and the number of nuclei was counted within each ring. For both methods, we selected NTs with similar size from control and patient COs generated from two independent iPSC clones at two different time points.
Online supplemental material
In Fig. S1, amino acid alignment of NUAK orthologs and paralog shows that the eight residues deleted in anencephalic fetuses are phylogenetically invariant. In Fig. S2, Q-PCR and Western blots compare expression levels of Hippo-YAP signaling components, as well as genes that are involved in actin polymerization, architecture, and dynamics in patient NPCs versus control cells. Additional Q-PCR on patient NSs treated with BLEB and ROCK-I shows transcriptional rescue of NUAK1 and the YAP-target gene, CTGF. In Fig. S3, pictures of control and patient COs show similar outer morphology and size, while H&E-stained CO sections show general dysplasia with significantly fewer NT structures and less nucleus disorganization in patient NTs compared with control. CO immunostaining showed striking microtubule disorganization in NUAK2 mutant NTs compared with control, which may underlie defective nucleokinesis. Table S1 shows WES of both parents and a proband that delineated two neighbor mutations causing in-frame deletion/insertion c.412_433delinsG in the NUAK2 gene.
Supplementary Material
shows WES of both parents and a proband delineated two neighbor mutations causing in-frame deletion/insertion c.412_433delinsG in the NUAK2 gene.
Acknowledgments
We are grateful to all the individuals and the family for their participation in this research. We are grateful to all members of the Reversade and Kayserili laboratories for support and constructive feedback, as well as the Microscopy Unit team at the Institute of Medical Biology, Agency for Science, Technology and Research, Singapore. OCT4, SOX2, KLF4, and c-MYC Addgene plasmids 17225, 17226, and 17227 were gifts from George Q. Daley (Harvard Stem Cell Institute, Cambridge, MA), and 18119 was a gift from John Cleveland (St. Jude Children’s Research Hospital, Memphis, TN).
N. Navaratnam and D. Carling are funded by the Medical Research Council. B. Reversade was funded by a Strategic Positioning Fund on Genetic Orphan Diseases from the Agency for Science, Technology and Research in Singapore. B. Reversade is a fellow of the Branco Weiss Fellowship–Society in Science and an Agency for Science, Technology and Research and European Molecular Biology Organization Young Investigator.
Author contributions: C. Bonnard, under the guidance of B. Reversade, directed the project, designed, and analyzed all the experiments and wrote the manuscript. N. Navaratnam and D. Carling designed and performed the kinase activity assay. K. Ghosh performed NPC and CO work, helped by O. Pomp and R. Changede. P.W. Chan and T.T. Tan generated iPSCs and derived NPCs. S. Tohari performed library preparation and exome sequencing. A.Y.J. Ng and B. Venkatesh analyzed WES results. U. Altunoglu and H. Kayserili diagnosed and provided clinical information for the Turkish family.
References
- Adzhubei, I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., and Sunyaev S.R.. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., and Lipman D.J.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona, M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., and Piccolo S.. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 154:1047–1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Arnold, K., Bordoli L., Kopp J., and Schwede T.. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22:195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Bekri, A., Billaud M., and Thélu J.. 2014. Analysis of NUAK1 and NUAK2 expression during early chick development reveals specific patterns in the developing head. Int. J. Dev. Biol. 58:379–384. 10.1387/ijdb.140024jt [DOI] [PubMed] [Google Scholar]
- Brouns, M.R., Matheson S.F., Hu K.Q., Delalle I., Caviness V.S., Silver J., Bronson R.T., and Settleman J.. 2000. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 127:4891–4903. [DOI] [PubMed] [Google Scholar]
- Cecconi, F., Piacentini M., and Fimia G.M.. 2008. The involvement of cell death and survival in neural tube defects: a distinct role for apoptosis and autophagy? Cell Death Differ. 15:1170–1177. 10.1038/cdd.2008.64 [DOI] [PubMed] [Google Scholar]
- Chen, J., Chang S., Duncan S.A., Okano H.J., Fishell G., and Aderem A.. 1996. Disruption of the MacMARCKS gene prevents cranial neural tube closure and results in anencephaly. Proc. Natl. Acad. Sci. USA. 93:6275–6279. 10.1073/pnas.93.13.6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp, A.J., Adzick N.S., Chitty L.S., Fletcher J.M., Holmbeck G.N., and Shaw G.M.. 2015. Spina bifida. Nat. Rev. Dis. Primers. 1:15007. 10.1038/nrdp.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeizel, A.E., and Dudás I.. 1992. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N. Engl. J. Med. 327:1832–1835. 10.1056/NEJM199212243272602 [DOI] [PubMed] [Google Scholar]
- Dai, X., She P., Chi F., Feng Y., Liu H., Jin D., Zhao Y., Guo X., Jiang D., Guan K.L., et al. 2013. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J. Biol. Chem. 288:34041–34051. 10.1074/jbc.M113.518019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, L.E., Kirke P.N., Molloy A., Weir D.G., and Scott J.M.. 1995. Folate levels and neural tube defects. Implications for prevention. JAMA. 274:1698–1702. 10.1001/jama.1995.03530210052030 [DOI] [PubMed] [Google Scholar]
- Escuin, S., Vernay B., Savery D., Gurniak C.B., Witke W., Greene N.D., and Copp A.J.. 2015. Rho-kinase-dependent actin turnover and actomyosin disassembly are necessary for mouse spinal neural tube closure. J. Cell Sci. 128:2468–2481. 10.1242/jcs.164574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar, P., and Tapon N.. 2014. Sensing the local environment: actin architecture and Hippo signalling. Curr. Opin. Cell Biol. 31:74–83. 10.1016/j.ceb.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Gill, M.K., Christova T., Zhang Y.Y., Gregorieff A., Zhang L., Narimatsu M., Song S., Xiong S., Couzens A.L., Tong J., et al. 2018. A feed forward loop enforces YAP/TAZ signaling during tumorigenesis. Nat. Commun. 9:3510. 10.1038/s41467-018-05939-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, N.D., and Copp A.J.. 2009. Development of the vertebrate central nervous system: formation of the neural tube. Prenat. Diagn. 29:303–311. 10.1002/pd.2206 [DOI] [PubMed] [Google Scholar]
- Gurniak, C.B., Perlas E., and Witke W.. 2005. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Dev. Biol. 278:231–241. 10.1016/j.ydbio.2004.11.010 [DOI] [PubMed] [Google Scholar]
- Hansen, C.G., Moroishi T., and Guan K.L.. 2015. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 25:499–513. 10.1016/j.tcb.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, M.J., and Juriloff D.M.. 1999. Mini-review: toward understanding mechanisms of genetic neural tube defects in mice. Teratology. 60:292–305. [DOI] [PubMed] [Google Scholar]
- Harris, M.J., and Juriloff D.M.. 2010. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res. A Clin. Mol. Teratol. 88:653–669. 10.1002/bdra.20676 [DOI] [PubMed] [Google Scholar]
- Herrera, E., Samper E., and Blasco M.A.. 1999. Telomere shortening in mTR-/- embryos is associated with failure to close the neural tube. EMBO J. 18:1172–1181. 10.1093/emboj/18.5.1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand, J.D., and Soriano P.. 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 99:485–497. 10.1016/S0092-8674(00)81537-8 [DOI] [PubMed] [Google Scholar]
- Hirano, M., Kiyonari H., Inoue A., Furushima K., Murata T., Suda Y., and Aizawa S.. 2006. A new serine/threonine protein kinase, Omphk1, essential to ventral body wall formation. Dev. Dyn. 235:2229–2237. 10.1002/dvdy.20823 [DOI] [PubMed] [Google Scholar]
- Humbert, N., Navaratnam N., Augert A., Da Costa M., Martien S., Wang J., Martinez D., Abbadie C., Carling D., de Launoit Y., et al. 2010. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 29:376–386. 10.1038/emboj.2009.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesh, K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., and Bejerano G.. 2016. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 48:1581–1586. 10.1038/ng.3703 [DOI] [PubMed] [Google Scholar]
- Kim, T.H., Goodman J., Anderson K.V., and Niswander L.. 2007. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Dev. Cell. 13:87–102. 10.1016/j.devcel.2007.04.018 [DOI] [PubMed] [Google Scholar]
- Kumar, P., Henikoff S., and Ng P.C.. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4:1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Lancaster, M.A., and Knoblich J.A.. 2014. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9:2329–2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., and Knoblich J.A.. 2013. Cerebral organoids model human brain development and microcephaly. Nature. 501:373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, D.L., Bai Y., Shahmolky N., Sharma M., Poon R., Drucker D.J., and Rosen C.F.. 2001. Identification and characterization of a novel sucrose-non-fermenting protein kinase/AMP-activated protein kinase-related protein kinase, SNARK. Biochem. J. 355:297–305. 10.1042/bj3550297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y.P., Zhang T., Li H., Wu B.L., Jin L., and Wang H.Y.. 2010. VANGL2 mutations in human cranial neural-tube defects. N. Engl. J. Med. 362:2232–2235. 10.1056/NEJMc0910820 [DOI] [PubMed] [Google Scholar]
- Lemay, P., Guyot M.C., Tremblay É., Dionne-Laporte A., Spiegelman D., Henrion É., Diallo O., De Marco P., Merello E., Massicotte C., et al. 2015. Loss-of-function de novo mutations play an important role in severe human neural tube defects. J. Med. Genet. 52:493–497. 10.1136/jmedgenet-2015-103027 [DOI] [PubMed] [Google Scholar]
- Li, Z., Ren A., Zhang L., Ye R., Li S., Zheng J., Hong S., Wang T., and Li Z.. 2006. Extremely high prevalence of neural tube defects in a 4-county area in Shanxi Province, China. Birth Defects Res. A Clin. Mol. Teratol. 76:237–240. 10.1002/bdra.20248 [DOI] [PubMed] [Google Scholar]
- Liu, F., Huang J., Zhang L., Chen J., Zeng Y., Tang Y., and Liu Z.. 2019. Advances in Cerebral Organoid Systems and their Application in Disease Modeling. Neuroscience. 399:28–38. 10.1016/j.neuroscience.2018.12.013 [DOI] [PubMed] [Google Scholar]
- Lizcano, J.M., Göransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Mäkelä T.P., Hardie D.G., et al. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833–843. 10.1038/sj.emboj.7600110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, D.E., Collins J.M., Dawahare J.H., Nguyen T.D., Lin Y., Voytik-Harbin S.L., Zorlutuna P., Yoder M.C., and Boerckel J.D.. 2019. YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J. Cell Biol. 218:1369–1389. 10.1083/jcb.201806065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky, A., Jick H., Jick S.S., Bruell C.L., MacLaughlin D.S., Rothman K.J., and Willett W.. 1989. Multivitamin/folic acid supplementation in early pregnancy reduces the prevalence of neural tube defects. JAMA. 262:2847–2852. 10.1001/jama.1989.03430200091032 [DOI] [PubMed] [Google Scholar]
- Morikawa, Y., Zhang M., Heallen T., Leach J., Tao G., Xiao Y., Bai Y., Li W., Willerson J.T., and Martin J.F.. 2015. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci. Signal. 8:ra41. 10.1126/scisignal.2005781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki, T., Tanemura A., Valencia J.C., Coelho S.G., Passeron T., Kawaguchi M., Vieira W.D., Ishikawa M., Nishijima W., Izumo T., et al. 2011. AMP kinase-related kinase NUAK2 affects tumor growth, migration, and clinical outcome of human melanoma. Proc. Natl. Acad. Sci. USA. 108:6597–6602. 10.1073/pnas.1007694108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulou, E., Galea G.L., Rolo A., Greene N.D., and Copp A.J.. 2017. Neural tube closure: cellular, molecular and biomechanical mechanisms. Development. 144:552–566. 10.1242/dev.145904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura, T., Shioi G., Hirano M., and Aizawa S.. 2012. Neural tube defects by NUAK1 and NUAK2 double mutation. Dev. Dyn. 241:1350–1364. 10.1002/dvdy.23816 [DOI] [PubMed] [Google Scholar]
- Park, I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., and Daley G.Q.. 2008. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 451:141–146. 10.1038/nature06534 [DOI] [PubMed] [Google Scholar]
- Peng, G.E., Wilson S.R., and Weiner O.D.. 2011. A pharmacological cocktail for arresting actin dynamics in living cells. Mol. Biol. Cell. 22:3986–3994. 10.1091/mbc.e11-04-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo, S., Dupont S., and Cordenonsi M.. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94:1287–1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Pollard, K.S., Hubisz M.J., Rosenbloom K.R., and Siepel A.. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20:110–121. 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert, M., Biegert A., Hauser A., and Söding J.. 2011. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 9:173–175. 10.1038/nmeth.1818 [DOI] [PubMed] [Google Scholar]
- Shum, A.S., and Copp A.J.. 1996. Regional differences in morphogenesis of the neuroepithelium suggest multiple mechanisms of spinal neurulation in the mouse. Anat. Embryol. (Berl.). 194:65–73. 10.1007/BF00196316 [DOI] [PubMed] [Google Scholar]
- Stumpo, D.J., Bock C.B., Tuttle J.S., and Blackshear P.J.. 1995. MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc. Natl. Acad. Sci. USA. 92:944–948. 10.1073/pnas.92.4.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Gao L., Chien H.Y., Li W.C., and Zhao J.. 2013. The regulation and function of the NUAK family. J. Mol. Endocrinol. 51:R15–R22. 10.1530/JME-13-0063 [DOI] [PubMed] [Google Scholar]
- Suzuki, A., Kusakai G., Kishimoto A., Minegichi Y., Ogura T., and Esumi H.. 2003. Induction of cell-cell detachment during glucose starvation through F-actin conversion by SNARK, the fourth member of the AMP-activated protein kinase catalytic subunit family. Biochem. Biophys. Res. Commun. 311:156–161. 10.1016/j.bbrc.2003.09.184 [DOI] [PubMed] [Google Scholar]
- Taylor, S.S., and Kornev A.P.. 2011. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36:65–77. 10.1016/j.tibs.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas, X.. 2014. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 141:1614–1626. 10.1242/dev.102376 [DOI] [PubMed] [Google Scholar]
- Wang, J., Xiao Y., Hsu C.W., Martinez-Traverso I.M., Zhang M., Bai Y., Ishii M., Maxson R.E., Olson E.N., Dickinson M.E., et al. 2016. Yap and Taz play a crucial role in neural crest-derived craniofacial development. Development. 143:504–515. 10.1242/dev.126920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Baribault H., and Adamson E.D.. 1998. Vinculin knockout results in heart and brain defects during embryonic development. Development. 125:327–337. [DOI] [PubMed] [Google Scholar]
- Xu, J., Vanderzalm P.J., Ludwig M., Su T., Tokamov S.A., and Fehon R.G.. 2018. Yorkie Functions at the Cell Cortex to Promote Myosin Activation in a Non-transcriptional Manner. Dev. Cell. 46:271–284,e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, W.C., Pepe-Mooney B., Galli G.G., Dill M.T., Huang H.T., Hao M., Wang Y., Liang H., Calogero R.A., and Camargo F.D.. 2018. NUAK2 is a critical YAP target in liver cancer. Nat. Commun. 9:4834. 10.1038/s41467-018-07394-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagórska, A., Deak M., Campbell D.G., Banerjee S., Hirano M., Aizawa S., Prescott A.R., and Alessi D.R.. 2010. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci. Signal. 3:ra25. 10.1126/scisignal.2000616 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows WES of both parents and a proband delineated two neighbor mutations causing in-frame deletion/insertion c.412_433delinsG in the NUAK2 gene.