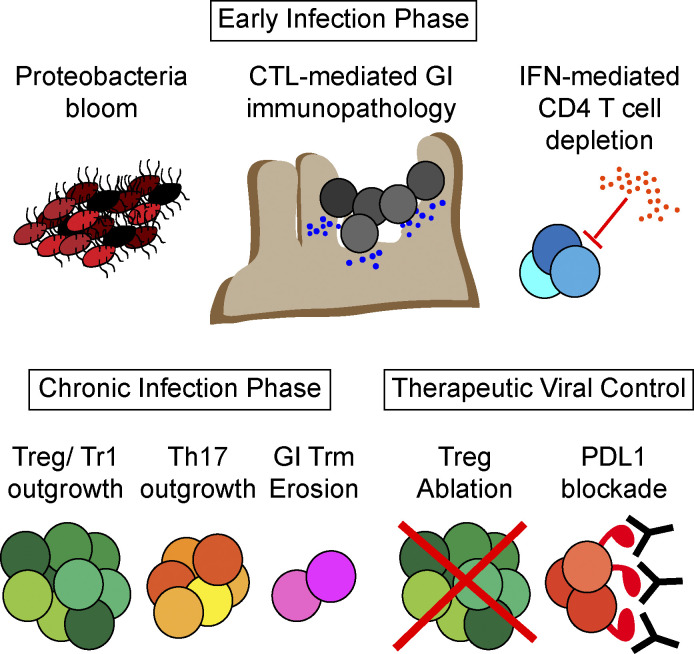
Macleod et al. uncover how chronic viral infection modulates gastrointestinal immune and microbial composition to foster persistence and long-term disease susceptibilities. The authors also identify the central role of regulatory T cells to maintain viral sanctuary in the intestinal tract.
Abstract
Many pathogens subvert intestinal immunity to persist within the gastrointestinal tract (GIT); yet, the underlying mechanisms that enable sanctuary specifically in this reservoir are unclear. Using mass cytometry and network analysis, we demonstrate that chronic LCMV infection of the GIT leads to dysregulated microbial composition, a cascade of metabolic alterations, increased susceptibility to GI disease, and a system-wide recalibration of immune composition that defines viral persistence. Chronic infection led to outgrowth of activated Tbet–expressing T reg cell populations unique to the GIT and the rapid erosion of pathogen-specific CD8 tissue-resident memory T cells. Mechanistically, T reg cells and coinhibitory receptors maintained long-term viral sanctuary within the GIT, and their targeting reactivated T cells and eliminated this viral reservoir. Thus, our data provide a high-dimensional definition of the mechanisms of immune regulation that chronic viruses implement to exploit the unique microenvironment of the GIT and identify T reg cells as key modulators of viral persistence in the intestinal tract.
Graphical Abstract
Introduction
The gastrointestinal tract (GIT) exists at the interface between the internal and external environments. As a result of this positioning, intestinal immune cells must strike an extremely fine balance between tolerance to ingested nutrients and beneficial microbiota and the generation of immunity to pathogenic organisms. The GIT contains the largest amount of immune cells in the body as well as trillions of commensal bacteria that continually shape lymphoid development and fine-tune effector responses. In the absence of excessive pathogen- or danger-associated signals, GIT APCs tend to be tolerogenic, inducing anergy and T regulatory (T reg) cell differentiation (Faria et al., 2017). At the same time, the GIT is an inflammatory environment, enriched with phenotypically activated T cells, plasma B cells, and innate lymphoid cells (ILCs) that contribute to barrier integrity and prevent disease (Mowat and Agace, 2014; Sonnenberg, 2014). Thus, perturbations in suppressive, inflammatory, or antimicrobial functions in the GIT could have dramatic consequences for host immunity and promote disease.
Although most viruses trigger a potent immune response that resolves infection, a number of viruses overcome the immune response to establish chronic infection. During chronic infection, an immunosuppressive program is initiated that actively suppresses antiviral T cell activity, leading to attenuated function, exhaustion, or deletion. In addition to immunosuppressive factors such as IL-10 and PDL1/PD1, chronic viral infections are also characterized by sustained inflammation that further potentiates immune exhaustion (Snell and Brooks, 2015). Because it must continually balance activation and suppression, the GIT is taken advantage of by many pathogens as a site of preferential infection and to generate long-lived viral sanctuaries (Jubelt and Lipton, 2014; Estes et al., 2017; OʼHara et al., 2017; Woodward et al., 2017). For example, HIV extensively depletes GIT CD4 T cells and, in contrast to peripheral blood cells, the intestinal CD4 T cell compartment often fails to fully reconstitute, even after initiation of antiretroviral therapy (Brenchley et al., 2006). Similarly, suboptimal antigen presentation to CD8 T cells and interactions with the host microbiota permit norovirus persistence in the GIT (Tomov et al., 2017).
In addition to altered immunity, chronic infection has been associated with changes in microbial composition and gastrointestinal (GI) system–associated disease susceptibilities, although the extent and direction of the microbial alterations remain highly controversial (Dubourg, 2016; Sullender and Baldridge, 2018). Colitis and GI symptoms are hallmarks of primary infection with many chronic viruses (Baldridge et al., 2016; Brenchley et al., 2006), which could be underscored by dysbiosis or redirection of immune subset composition from a suppressive to an inflammatory nature (Kmieć et al., 2017). Further adding complexity is the potential immunological diversity within different intestinal compartments and how isolated changes in the small intestine (SI) or large intestine (LI) might alter adjacent tissues.
Herein, we demonstrate that chronic lymphocytic choriomeningitis virus (LCMV) infection finds sanctuary throughout the GIT, generating lifelong persistence and causing short- and long-term pathological and dysbiotic sequelae for the host. Using high-dimensional mass cytometry (CyTOF), transcriptomics, and microbiomics in naive and acute and chronically infected mice, we provide a system-wide definition of intestinal immunity during viral clearance or persistence specifically within the GIT. Chronic, but not acute, infection induced massive reorganization of all immune subsets throughout the intestinal tract, leading to rapid IFN-I–mediated depletion of CD4 T cells and induction of colitis, a metabolic switch to oxidative and fatty acid metabolism, and highly divergent T cell differentiation among the different intestinal compartments. Comparative transcriptional analysis identified shared networks and mechanisms regulating LCMV persistence in the murine intestinal tract and HIV and simian immunodeficiency virus (SIV) intestinal persistence, establishing a common thread across chronic viral infections in the intestinal reservoir. Coupled with altered virus-specific T cell differentiation, chronic infection led to specific amplification of bystander CD4 T helper type 17 (Th17) and T reg cells and a striking reduction in preexisting pathogen-specific CD8 tissue-resident memory T (Trm) cells, creating a hole in protective immunity against previously encountered pathogens. PDL1 inhibition promoted virus control, albeit with the T cell reinvigoration simultaneously reinducing immunopathology and colitis, as is often observed following blockade in the clinic. Mechanistically, chronic infection led to the differentiation of distinct activated T reg cell populations within the GIT that sustained viral persistence through inhibiting multiple T cell populations and that could be targeted to control infection. Thus, our data provide a high-dimensional definition of immunological and microbial alterations in the acute and chronically infected GIT and define mechanisms that enable the preferential sanctuary, consequent disease susceptibility, and new targets to eliminate chronic viruses from the GIT.
Results
The GIT is a long-lived reservoir of chronic LCMV infection
To investigate how chronic viruses interact with and remodel the GI immune environment, we used the LCMV system. Infection of mice with the LCMV-Armstrong (Arm) variant induces robust T cell responses that rapidly clear the virus within 8–12 d, generating long-lived immune memory. On the other hand, infection with LCMV clone 13 (Cl13) generates a chronic infection, leading to T cell exhaustion and broad changes in innate immune cells. Cl13 infection is resolved from the majority of peripheral tissues by ∼60 d after infection, thereafter forming reservoirs in the brain for ∼200 d and the kidney for the life of the animal (Lauterbach et al., 2007; Moskophidis et al., 1995). Yet, what allows long-term sanctuary in certain tissues is ill defined.
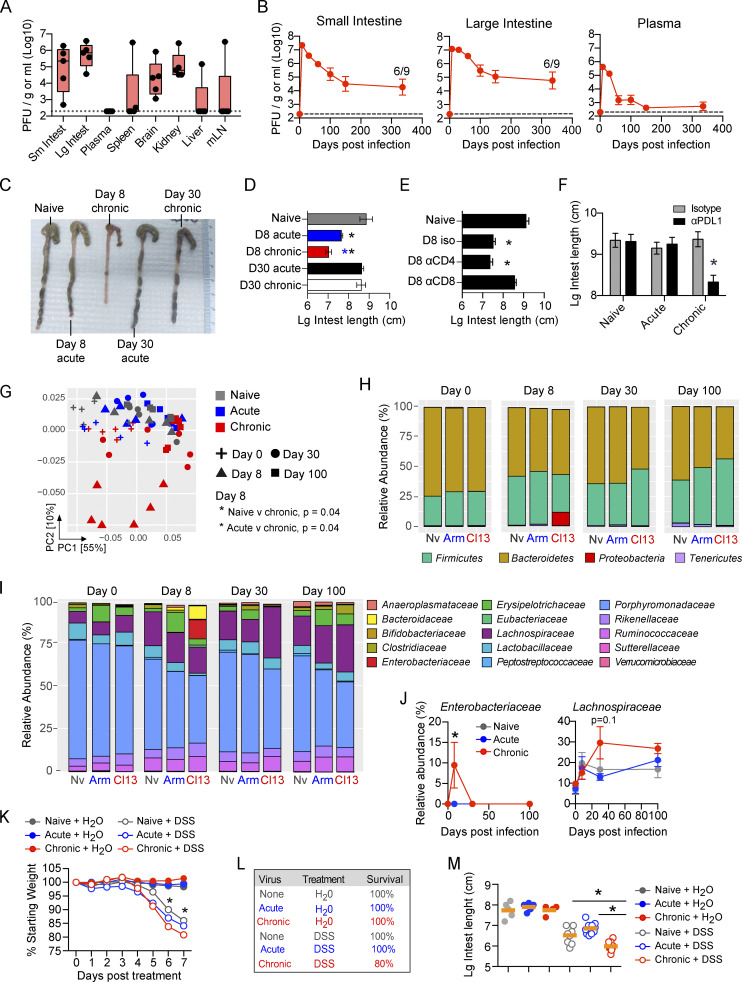
Within the SI and LI, both acute and chronic LCMV rapidly established high titer infection (Fig. S1 A); however, while by day 8, acute LCMV-Arm infection had largely resolved in all tissues, virus titers remained high following LCMV-Cl13 infection (Fig. S1 A). Interestingly, whereas virus was controlled in the plasma, spleen, liver, and mesenteric lymph nodes of most mice after 150 d, virus titers remained high in the SI and LI, persisting at levels similar to those in other viral reservoirs, the brain and kidney (Fig. 1 A). Strikingly, six of nine mice retained high-titer chronic infection in their SI and LI for >300 d after infection (Fig. 1 B), thus identifying the GIT as a previously unrecognized reservoir of chronic LCMV infection, providing a new model to mechanistically tease apart mechanisms of long-term intestinal viral persistence.
Figure S1.
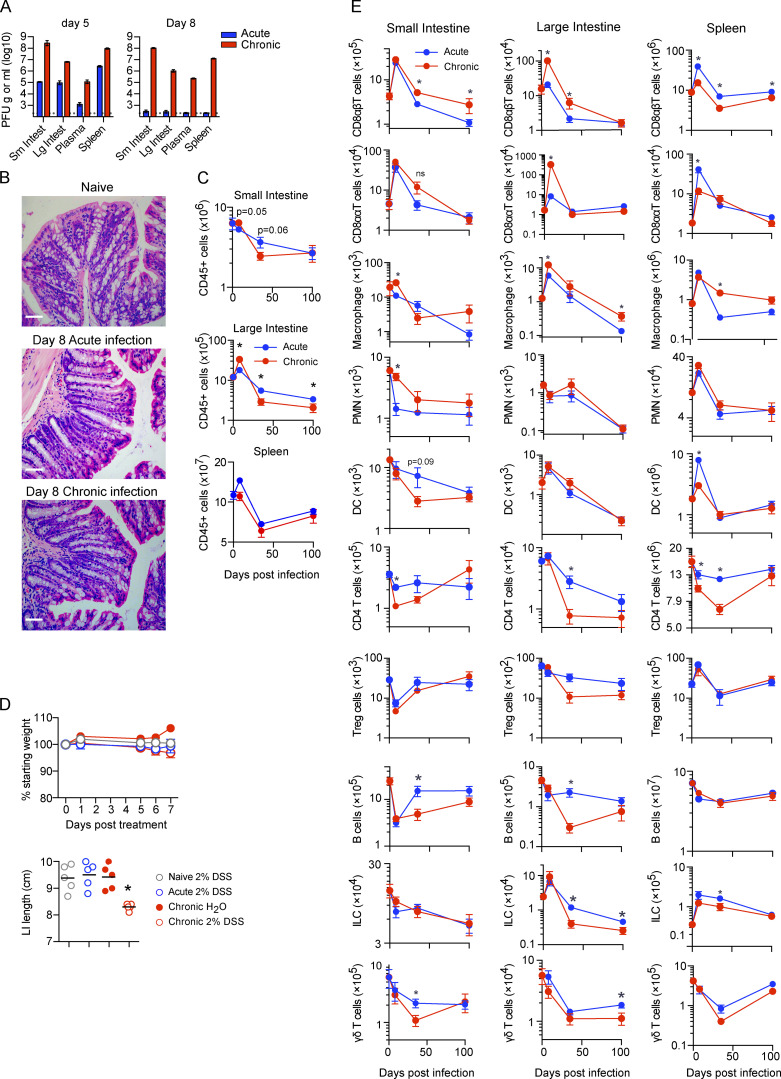
The GIT is a long-lived reservoir of LCMV-Cl13 infection. (A and B) Mice were infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic), and 5 and 8 dpi, PFU of virus per gram of tissue or per milliliter of blood plasma was determined. n = 4–5 mice per group from one experiment. *, P < 0.05 by ANOVA test. (B) H&E staining was performed on LI tissue from naive mice or 8 dpi after acute LCMV-Arm or chronic LCMV-Cl13 infection, showing a lack of epithelial or villus pathology during infection. Representative pictures from n = 5 mice per group from one experiment. Scale bars = 50 µm. (C and D) Mice were infected with LCMV-Arm or LCMV-Cl13, and 0, 8, 35, and 100 dpi, the SI, LI, and spleen were analyzed by CyTOF. Graphs show total number of (C) CD45+ viable cells and (D) the indicated immune cell subsets: CD8αβT cells (TCRβ+CD8α+CD8β+), CD8ααT cells (TCRβ+CD8α+CD8β−), monocytes/macrophages (CD11b+SiglecF−TCR−B220-CD11c−), polymorphonuclear cells (CD11b+Ly6G+TCR−B220−CD11c−), DCs (CD11chiMHC-IIhiTCR−B220−), CD4 T cells (TCRβ+CD4+), B cells (B220+MHC-II+), ILCs (lin−Thy1.2+), and γδT cells (TCRγδ+TCRβ−B220−) in the SI, LI, and spleen. *, P < 0.05 (Mann-Whitney test of log-transformed data). n = 10 mice per group. Error bars indicate SEM (A–D).
Figure 1.
The GIT is a long-lived reservoir of LCMV-Cl13 infection, which causes dysbiosis and increased susceptibility to colitis. (A) Mice were infected with LCMV-Cl13, and 150 dpi, PFUs of virus per gram of tissue or per milliliter of blood plasma were determined. mLN, mesenteric lymph node. n = 5 mice per group from one of two independent experiments. (B) Time course of LCMV-Cl13 titers in the SI, the LI, and blood plasma. Numbers on the graph indicate the number of mice with detectable virus at 336 dpi. n = 5–10 mice per time point from two independent experiments. *, P < 0.05, ANOVA test. (C and D) Representative pictures (C) and graphs (D) show LI length of naive, LCMV-Arm–infected (acute), and LCMV-Cl13–infected (chronic) mice at 8 and 30 dpi. Black asterisk, P < 0.05 naive versus day 8 chronic, ANOVA test; blue asterisk, P < 0.05 day 8 acute versus day 8 chronic, ANOVA test. n = 5–10 mice per group from two independent experiments. (E) Mice were left naive or infected with LCMV-Cl13 and treated every 2 d with CD4- or CD8-depleting antibodies. LI length was determined at 8 dpi. n = 5–10 mice per group from two independent experiments. *, P < 0.05, ANOVA test. (F) Mice were left naive or infected with LCMV-Arm or LCMV-Cl13 and treated with anti-PDL1 or isotype control antibodies beginning 25 dpi. The graph shows the LI length in isotype- or PDL1-treated mice after three treatments. n = 9–10 mice per group from two independent experiments. (G–I) 16S ribosomal RNA-seq was performed on the pellets of naive (Nv), LCMV-Arm–, and LCMV-Cl13–infected mice at 0, 8, 30, and 100 dpi. (G) PC analysis of β-diversity. By Adonis test naive versus day 8 chronic, P = 0.04; acute versus day 8 chronic, P = 0.04. Relative abundance of (H) bacterial phyla and (I) bacterial families merged by time and sample group. (J) Graphs show relative abundance of Enterobacteriaceae and Lachnospiraceae. *, P < 0.05, relative abundance means are significantly different from naive at this time point by ANOVA test. (G–I) One of two independent experiments with n = 5–6 littermate mice per group. (K–M) Naive mice or mice infected with LCMV-Arm or LCMV-Cl13 70 d earlier were treated with DSS or H2O control for 7 d. (K) Graph shows average percentage body weight change from commencement of DSS treatment. *, DSS-treated group’s weight is significantly different from that of other DSS-treated groups. (L) Table indicates survival of treated mice. (M) Graph shows LI length 8 d after the indicated treatment. n = 10 mice per group from two independent experiments. Error bars indicate SEM (A–M).
Chronic viral infection potentiates microbial dysbiosis and susceptibility to colitis-like disease
Acute colitis and GI symptoms are hallmarks of primary infection with chronic viruses (Downs, 2010). Interestingly, primary infection with both acute and chronic viruses rapidly led to reduced LI length, a surrogate marker of colitis, with the chronic virus inducing a much more severe contraction (Fig. 1, C and D). Histological examination revealed a lack of villus or epithelial damage, suggesting that inflammation induced immune pathology in the absence of major tissue damage (Fig. S1 B). The facts that the LI contraction was observed in both acute and particularly chronic infection and that its resolution by day 30 corresponded to immune exhaustion in chronic infection suggested a common immunological mechanism. Depletion of CD8 T cells, but not CD4 T cells, at the onset of chronic infection prevented the LI contraction (Fig. 1 E). Interestingly, reinvigoration of exhausted CD8 T cells following anti-PDL1 therapy in chronic infection (but not in naive mice or in mice that had previously been acutely infected with LCMV-Arm) reinduced the colitis (Fig. 1 F), indicating a prominent role for CD8 T cells in induction of GI pathology.
To identify microbial changes in viral infection, we performed longitudinal 16S ribosomal RNA sequencing (RNA-seq) using coweaned littermate mice. Despite acute LCMV-Arm replicating in the GIT and inducing moderate LI contraction, principal component (PC) analysis of β-diversity indicated no significant alteration in microbial composition in acutely infected compared with naive mice (Fig. 1 G). On the other hand, concurrent with the colitis, the microbiome of chronically infected mice substantially diverged from naive and acute virus infected mice (P = 0.04 naive versus chronic; P = 0.04 acute versus chronic; pairwise Adonis test). Chronic infection rapidly increased the abundance of the facultative anaerobes Proteobacteria, a phylum associated with pathology in inflammatory bowel disease. The Proteobacteria bloom at the onset of chronic infection was primarily due to an increase in the Enterobacteriaceae family, which now comprised ∼10% of the microbiome by day 8, while remaining negligible in naive or acutely infected groups (Fig. 1, H–J). Proteobacterial outgrowth subsided with transition to immunological exhaustion 30 d postinfection (dpi). The Bacteroidetes-to-Firmicutes ratio was reduced in chronic compared with naive or acutely infected mice. As chronic infection progressed, the Lachnospiraceae family, a family associated with short-chain fatty acid production and intestinal homeostasis (Donaldson et al., 2016), increased in prevalence (Fig. 1, I and J). Thus, viral persistence (unlike acute infection) leads to a rapid and temporally changing microbial community, with a transition from more pathogenic to wound-healing microbiomes as viral persistence progresses.
To determine how prolonged viral replication affects long-term disease susceptibilities in the GIT, we measured colitis induction in naive mice, mice previously infected with acute LCMV-Arm, or mice that had been infected for 70 d with chronic LCMV-Cl13. Mice were treated with 4% dextran sulfate sodium (DSS) to induce colitis or with vehicle control. As expected, all DSS-treated mice developed colitis (Fig. 1 K). However, chronically infected mice exhibited substantially more severe disease symptoms in terms of both increased weight loss and reduced LI size (Fig. 1, K–M). Furthermore, although all mice in the naive and acute infection groups survived, 20% of chronically infected mice ultimately succumbed to colitis (Fig. 1 L). Even lower levels of DSS (2%) that do not induce colitis in naive mice or mice previously infected with acute LCMV-Arm do induce pathology (reduced LI length) in mice chronically infected with LCMV-Cl13 (Fig. S1 D). Thus, prolonged viral replication in the GIT does not continuously manifest in colitis; however, the ongoing replication increases host susceptibility to GI inflammation, disease induction, and pathology.
Immune redirection in the GIT during chronic viral infection
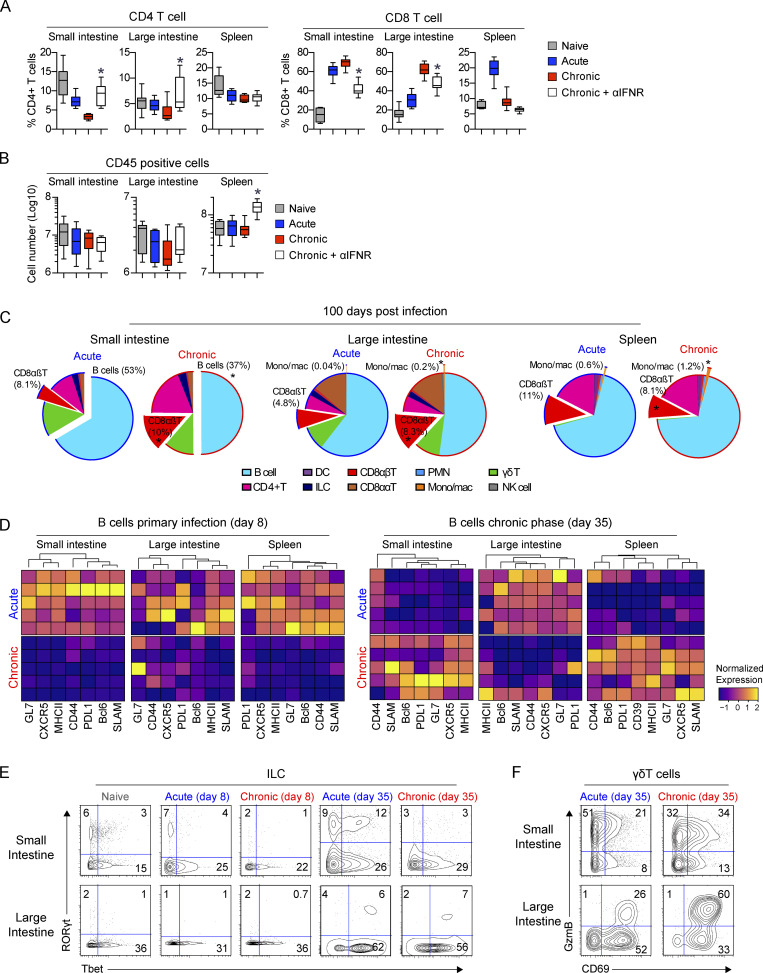
To determine how viral persistence alters immune dynamics and function in the GIT, we designed a CyTOF panel to identify major (and most minor) immune cell populations. Total immune cellularity increased in both the SI and the LI in the initial stage (8 dpi) of LCMV-Cl13 compared with LCMV-Arm infection, which was in direct contrast to the spleen, wherein these ratios were inverted and rapid leukopenia occurred following LCMV-Cl13 infection (Fig. S1 C). However, the increased GIT cellularity was not maintained, and cell numbers in the SI and the LI of chronically infected mice were decreased by 35 dpi (Fig. S1 C). In all organs, total immune cell numbers following resolution of acute infection also remained decreased relative to those in naive mice (Fig. S1 C), indicating an overall tissue remodeling evident after initial infection, with long-term consequences on immune composition.
Within 8 d, chronic infection in the LI resulted in massive expansion in the CD8 αβ T cell compartment (Fig. 2 A and Fig. S1 E). In contrast, CD8 T cell accumulation in the SI was similarly robust following acute or chronic infection, and CD8 T cell expansion in the spleen was highly attenuated by chronic compared with acute infection. These differences in CD8 T cell dynamics demonstrate the compartmentalization of antiviral responses within the GIT and their continued divergence from the spleen. By 8 d after LCMV-Cl13 infection, CD11b-expressing macrophages were increased in the GIT LI compared with acute LCMV-Arm infection, and polymorphonuclear neutrophils were increased in the SI (Fig. 2 A and Fig. S1 E). This suggested that primary infection with chronic virus increased recruitment and infiltration by inflammatory myeloid cells, potentially contributing to the observed colitis. Dendritic cells (DCs) were only minimally detected within the intestinal tract, and, unlike in the spleen, their numbers did not significantly differ between acute and chronic infection in the GIT (Fig. S1 E).
Figure 2.
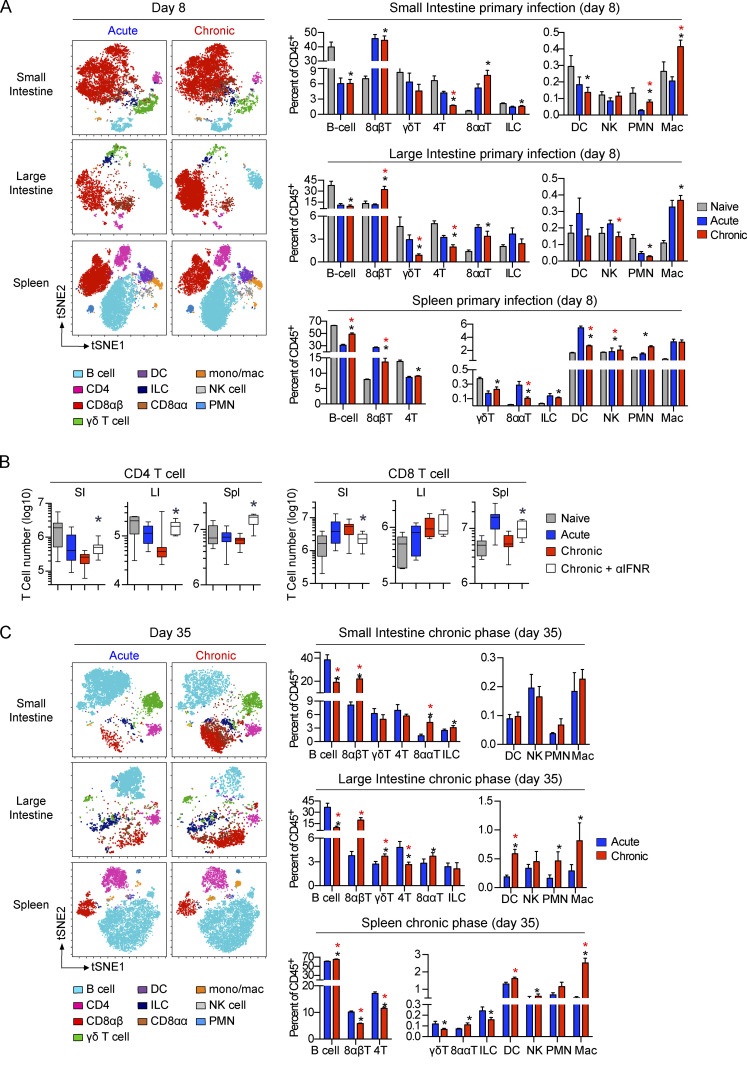
Global immune population changes in the GIT during primary infection with a persistent virus. (A and C) Mice were infected with LCMV-Arm or LCMV-Cl13, and 0, 8, 35, and 100 dpi, the SI, LI, and spleen tissue was analyzed by CyTOF. (A) Representative tSNE plots (5,000 iterations; perplexity = 30; theta = 0.5) and bar graphs at 0 and 8 dpi show the frequency and high-dimensional location of B cells (B220+MHC-II+), γδT cells (TCRγδ+ TCRβ−B220−), CD8αβT cells (TCRβ+CD8α+CD8b+), CD4 T cells (TCRβ+CD4+), ILCs (lin−Thy1.2+), DCs (CD11c+MHC-II+TCR−B220−), natural killer (NK) cells (NK1.1+TCR-B220-), CD8ααT cells (TCRβ+CD8α+CD8β-), polymorphonuclear neutrophils (PMNs; CD11b+Ly6G+TCR−B220−CD11c−), and monocytes/macrophages (CD11b+SiglecF−TCR−B220−CD11c−) among total CD45+ cells in the SI, LI, and spleen. Black asterisk, chronic versus naive, P < 0.05 by ANOVA test; red asterisk, chronic versus acute, P < 0.05 by ANOVA test. tSNE plots are concatenated data of five pooled mice per group from one of two independent experiments. Bar graphs, n = 10 mice per group. (B) Mice were left naive or infected with LCMV-Arm or LCMV-Cl13. LCMV-Cl13–infected mice were either treated with anti–IFNR-blocking antibody or isotype control antibody. The number of CD4 and CD8 T cells was analyzed in the SI, LI, and spleen (Spl) at 8 dpi by flow cytometry. *, LCMV-Cl13 + αIFNR versus LCMV-Cl13 + isotype, P < 0.05 by ANOVA test. n = 13–15 mice per group from three independent experiments. (C) Representative tSNE plots (5,000 iterations; perplexity = 30; theta = 0.5) and bar graphs at 35 dpi show the frequency and high-dimensional location of the immune populations described in A among total CD45+ cells in the SI, LI, and spleen. Black asterisk, chronic versus naive, P < 0.05 by ANOVA test; red asterisk, chronic versus acute, P < 0.05 by ANOVA test. tSNE plots are concatenated data of five pooled mice per group from one of two independent experiments. Bar graphs, n = 10 mice per group. Error bars indicate SEM (A–C).
Strikingly, compared with acute infection, chronic infection resulted in a rapid decline in the number and proportion of CD4 T cells within the GIT (Fig. 2 A). In the SI, CD4 T cells decreased proportionally and in terms of absolute number 8 dpi, indicating a specific decline, whereas in the LI, CD4 T cells were greatly diminished in frequency in chronic infection but numerically were similar (Fig. 2 A and Fig. S1 E). Because direct infection of CD4 T cells is minimally observed following LCMV infection (Oldstone and Campbell, 2011), their decrease suggested that factors induced by chronic infection would be major drivers of CD4 culling. Interestingly, treating LCMV-Cl13–infected mice with anti–IFN receptor (anti-IFNR)–blocking antibody prevented depletion and maintained CD4 T cells at the number and frequency observed following acute infection (Fig. 2 B and Fig. S2 A), indicating that the rapid GI CD4 depletion in chronic infection is due to excessive IFN-I signaling. The IFN-mediated culling was specific to the helper T cell lineage because blockade did not increase GIT CD8 T cells, instead resulting in a reduction or no change in CD8 T cells (Fig. 2 B). At the population level, IFN-I blockade did not alter overall immune cellularity in the GIT of infected mice; yet, it resulted in massive expansion in total CD45+ cells in the spleen (Fig. S2 B), indicating that the role of IFN-I was very focused to CD4 T cell culling in the GIT while inducing broad-spectrum cell suppression in the spleen. Thus, primary chronic infection induces a robust reorganization of the GI immune system, leading to massive LI CD8 T cell infiltration, enhanced myeloid recruitment into the GIT, and IFN-I–induced CD4 T cell culling in the SI.
Figure S2.
Immune changes in the GIT during chronic viral infection. (A and B) Mice were left naive or infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic). LCMV-Cl13–infected mice were treated with either IFNR-blocking antibody or isotype control antibody between −1 to 8 dpi. (A) Graphs show the proportion of CD4 T cells and CD8 T cells in the SI, LI, and spleen. *, P < 0.05 for LCMV-Cl13 + αIFNR versus LCMV-Cl13 + isotype by ANOVA test. n = 13–15 mice per group from three independent experiments. (B) Graphs show the number of live CD45+ cells in LCMV-Cl13–infected isotype and anti–IFNR-treated mice. *, P < 0.05 by t test. n = 13–15 mice per group from three independent experiments. (C) Pie charts indicate the average proportion of the immune populations based on CyTOF analysis in the SI, LI, and spleen 100 dpi after LCMV-Arm or LCMV-Cl13 infection. Populations that significantly differ (*, P < 0.05 by Mann-Whitney test of log-transformed data) are offset and their proportion labeled. n = 9 mice per group from two independent experiments. (D–F) Mice were infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic) and at 0 (naive), 8, and 35 dpi, and the SI, LI, and spleen were analyzed by CyTOF. (D) Heatmaps show column-normalized protein expression on B cells (B220+MHC-II+) 8 and 35 dpi after LCMV-Arm or LCMV-Cl13 infection. Each row represents an individual mouse. Contour plots of n = 5 concatenated mice show average expression of the indicated proteins in (E) ILC (lineage-Thy1.2+) and (F) γδT cells (TCRγδ+ TCRβ−B220−). Heatmaps and contour plots are from one of two independent experiments with n = 5 mice per group.
After resolution of acute infection, CD8 T cells contracted and B cells increased to become the most abundant immune population by 35 dpi (Fig. 2 C and Fig. S1 E). In contrast, at day 35 following chronic infection, CD8 T cell numbers and proportions remained elevated in both the SI and the LI, whereas they were further diminished in the spleen, again demonstrating the difference between the GIT and splenic environments. The increased proportion of GI CD8 T cells and decrease in B cells continued to 100 dpi, reflective of long-term immune reorganization associated with chronic infection (Fig. S2 C). Within the LI, CD4 T cell numbers remained severely depressed through 100 dpi, whereas they rebounded in the SI with the progression of chronic infection (Fig. 2 C and Fig. S1 E).
B cells were significantly depleted throughout the GIT but not the spleen at day 35 in chronically infected mice (Fig. 2 C), indicating that sustained viral replication in the GIT increases B cell death and/or inhibits B cell survival. Phenotypically, acute infection initially induced more robust expression of activation-induced proteins (GL7, CXCR5, MHC-II, CD44, PDL1, Bcl6, and SLAM) in the SI and spleen at 8 dpi (Fig. S2 D). Yet, by day 35, there was an inversion, with B cells being more activated and differentiated, suggesting that chronic infection induced concurrent hyperactivation and SI B cell depletion, as observed during HIV infection (Moir and Fauci, 2009). In contrast to the SI and spleen, B cells in the LI were in general hypoactivated at both 8 and 35 dpi, suggesting that the humoral response to chronic infection in the GIT differed between the SI and LI and specifically that LI B cell function was inhibited.
ILC numbers were not significantly different between acute and chronic infection in the SI, whereas in the LI, ILCs were reduced with chronic infection (Fig. S1 E). However, prolonged viral replication induced clear shifts in the functional phenotype of ILCs. In the SI, there was a striking loss of RORγt+Tbet+ and RORγt+Tbet− ILC3s during primary infection with chronic virus, and this was maintained to day 35 after chronic infection (Fig. S2 E). Furthermore, γδT cells were not proportionally altered in the SI, nor were striking phenotypic changes observed (Fig. 2 and Fig. S2 F). However, in the LI at 8 and 35 dpi, chronic infection resulted in increased expression of activation markers (CD69, CD11c, granzyme B [GzmB]), indicating that chronic infection increased γδT cell activation specifically in the LI. Thus, viral persistence provokes a network of immediate and sustained alterations in cellular abundances and functional phenotypes throughout GI compartments, which differ widely from the spleen, and could profoundly impact the host’s ability to control chronic infection.
Transcriptional profiling reveals distinct environmental, metabolic, and activation states in the GIT during chronic viral infection
To gain further insight into the mechanisms that regulate GI immunity in chronic infection, we performed RNA-seq on sorted CD45+ cells from the GIT of naive mice or mice that had been infected 30 d earlier with LCMV-Arm or LCMV-Cl13. Cumulative changes in gene expression captured by PC analysis revealed that differences could be described predominantly by PC1 “infection genes” that were similarly differentially expressed following acute and chronic viral infection (Fig. 3 A). Thus, despite LCMV-Cl13 infection causing greater pathology and replicating for >300 d longer than LCMV-Arm in the GIT, both infections induced similar immune reprogramming, consistent with shared mechanisms of viral recognition and antiviral response. On the other hand, PC2 accounted for less of the variance overall but distinctly separated acute and chronic infections. There were 7,438 and 6,331 “chronic infection–associated genes” differentially expressed between LCMV-Arm– and LCMV-Cl13–infected mice in the SI and the LI, respectively. Pathway analyses revealed that the chronically infected GIT was dominated by gene signatures associated with T cell signaling and costimulation (CD28, OX40, inducible T cell costimulator signaling; Fig. 3 B). Many of the upregulated pathways mediated chronic inflammation, such as IFN signaling, T cell cytotoxicity, death receptor signaling, and cell cycling (Fig. 3 B). Interestingly, blockade of IFNR signaling to inhibit the IFN signaling network during late chronic infection led to increased virus titers (Fig. 3 C), indicating that the ongoing IFN-I signaling was critical to constraining chronic virus replication and differentially affected virus control in the GIT compared with other peripheral organs (Teijaro et al., 2013; Wilson et al., 2013; Zhen et al., 2017).
Figure 3.
Global changes in gene expression in the GI immune compartment during chronic virus infection. (A, B, and D) RNA-seq was performed on sorted CD45+ cells from the SI and LI of naive mice and mice 30 d after LCMV-Arm (acute) or LCMV-Cl13 (chronic) infection. (A) PC analysis plots of global gene expression. n = 3 mice per group from one experiment. (B) Heatmaps of activation z-scores of canonical pathways significantly upregulated and downregulated (P < 0.05, Fisher’s exact test) in LCMV-Cl13–infected mice compared with LCMV-Arm–infected mice using Ingenuity Pathway Analysis (IPA). n = 3 mice per group from one experiment. (C) Mice were infected with LCMV-Cl13, and at 60–80 dpi, they were treated with αIFNR-blocking or isotype control antibodies. Box plots show virus titers in the SI and LI. Error bars indicate the highest and lowest values. n = 8–10 mice per group. *, P < 0.05 by t test. (D) Top six subcellular compartment locations of genes upregulated in LCMV-Cl13–infected immune cells compared with LCMV-Arm–infected cells. Labeled numbers indicate the number of upregulated genes/total number of genes associated with that compartment. n = 3 mice per group from one experiment. (E) Heatmaps of relative expression of key immune cytokines and TFs in the GIT of naive, LCMV-Arm–infected, and LCMV-Cl13–infected mice. Heatmap values are row-normalized average values of n = 3 samples. Gray asterisk, P < 0.05, chronic versus naive; red asterisk, P < 0.05, chronic versus naive by ANOVA test. (F) Gene set enrichment analysis was used to compare genes upregulated (UP) in the infected LI compared with colon biopsies of HIV-infected versus uninfected patients (GSE28177). Shown are enrichment plot and leading-edge genes. *, nominal P < 0.05 using empirical phenotype-based permutation (Subramanian et al., 2005). (G) IPA comparison was performed on differentially expressed genes in LCMV-Cl13–infected versus uninfected LI samples, colon biopsies of SIV-infected versus uninfected macaques (GSE29980), and colon biopsies of HIV-infected versus uninfected patients (GSE28177). Heatmap shows activation z-scores of IPA predicted upstream regulator molecules. CoA, coenzyme A; CTL, cytotoxic T lymphocyte; EIF2, eukaryotic initiation factor 2; iCOS, inducible T cell costimulator; iCOSL, inducible T cell costimulatory ligand; NOS, nitric oxide synthetase; PI3K, phosphoinositide 3-kinase.
Pathways involved in B cell activation were consistently downregulated, likely reflecting the depletion of GI B cells during chronic infection (Fig. 3 B). Chronic infection also reduced the immune-sustaining IL-7 signaling pathway and multiple pathways involved in innate immune function (nitric oxide synthetase/ROS/IL-1). Genes mediating APC maturation were decreased (some of which overlapped with the downmodulated B cell genes), including MHC-II, CD40, CD83 CD86, IL-12, CD83, and CCR7 (Fig. S3), reflecting a diminished APC function in the GIT, as occurs in the splenic compartment during chronic LCMV infection (Cunningham et al., 2016; Snell et al., 2018).
Figure S3.

Global changes in gene expression in the GI immune compartment during chronic viral infection. RNA-seq was performed on sorted CD45+ cells from the SI and LI of naive mice and mice 30 dpi after LCMV-Arm (acute) and LCMV-Cl13 (chronic) infection. Heatmap shows relative expression of genes that make up the DC maturation pathway (IPA) in the SI and LI of LCMV-Arm– versus LCMV-Cl13–infected mice. Heatmap values are row-normalized average values of n = 3 samples.
Though SI and LI immune cells exhibited similar inflammatory and immunosuppressive pathways, their metabolism was highly distinct. Most significantly, the oxidative phosphorylation (OxPhos) pathway was upregulated in SI immune cells during chronic infection (P = 4 × 10−27), as was fatty acid β-oxidation, glutaryl–coenzyme A degradation, and TCA cycle and EIF2 signaling (Fig. 3 B), reflecting catabolic processes and consequent generation of ATP to fuel protein synthesis. Supportively, compartment analysis indicated extreme enrichment in mitochondrial and ribosomal localization among genes upregulated in the chronic SI (Fig. 3 D), key sites of OxPhos, ATP generation, and protein synthesis. Chronic infection further enhanced lipid production/signaling in the SI because genes involved in cholesterol biosynthesis and ceramide signaling were increased (Fig. 3 B). This could reflect immune cell membrane and lipid raft signaling and/or the propagation of lipid-mediated signaling intermediates (Maceyka and Spiegel, 2014). In stark contrast, chronic infection in the LI did not increase aerobic respiration or lipid metabolism. Compartment analysis revealed that genes significantly increased in the chronically infected LI were enriched inside the nucleoplasm, cytolytic granules, and (associated with the mitotic) spindle, reflecting a highly proliferative and cytotoxic phenotype consistent with CD8 T cell accumulation. Thus, chronic viral infection induced highly compartmentalized metabolic programs in the GIT, reflecting the regional immune functions.
To determine changes in key lineage-defining transcription factors (TFs), signaling molecules, effector proteins, and cytokines that shaped the chronically infected GI microenvironment, we compared differentially expressed genes (Fig. 3 E). Exhaustion-associated TFs and cytokines were expectedly upregulated during chronic viral infection, including IL-10, IL-21, eomesodermin (Eomes), Batf, Prdm1/Blimp1, Tox, and Id2 (Snell et al., 2017). Interestingly, in the LI, chronic viral infection significantly increased HIF-1α expression, and this correlated with the decreased OxPhos in the LI. (OxPhos is generally observed in aerobic conditions.) By contrast, in the chronically infected SI, where OxPhos was increased, HIF-1α was significantly decreased. Furthermore, not all cytokines typically associated with chronic inflammation were increased; for example, neither TGF-β nor IL-1, IL-12, or IL-6 was elevated. Furthermore, the conventional DC TF Zbtb46 was reduced in the SI following viral infection compared with in naive mice, but it was particularly low in chronic infection (Fig. 3 E), which, in conjunction with the reduced levels of IL-1 and IL-12 (typically considered markers of innate inflammation) and reduced DC/myeloid activation pathways, again indicated inhibition of APC function in the GIT. Interestingly, the myeloid- and T cell–attracting chemokines CCL6, CCL7, CCL8, CCL9, and CXCL12 were downmodulated in the chronically infected SI and were undetectable in the LI, further explaining the diminished myeloid activation. Conversely, the IFN-I–induced chemokines CCL3, CCL4, CCL5, and CXCL10 and signaling molecules STAT1, STAT2, IRF7, and IRF9 were increased in chronic infection, consistent with continued viral replication inducing an IFN-I–regulated environment. There were also differences between the intestinal compartments. Fos and Jun, which together form the cellular stress/activation factor AP-1, were selectively increased in the chronically infected SI but not the LI. Conversely, STAT3 and STAT5b, Notch1, and IRF2 were all selectively upregulated in the LI during chronic infection.
The ongoing inflammation in the chronically infected GIT inhibited key memory programming cytokines and TFs. (IL-15, Klf2, Tcf7/Tcf1, Nfkb, and Id3 were downmodulated.) On the other hand, Th1-type immunity (IFN-γ, TNF-α, granzymes, Tbet) was increased in chronic infection, but so were gene expression patterns of Th2 (IL-4, IL-5, GATA3) and Th17 (IL-17A, RORα, Runx2) immunity (Fig. 3 E). Thus, chronic infection transcriptionally and metabolically overhauls the GIT, leading to widespread changes in inflammatory signaling pathways.
Transcriptional regulation in the GIT is conserved in human chronic infection
To investigate concordance between chronic intestinal LCMV infection and human chronic infections, we compared our RNA signatures with published colon biopsies from HIV-infected subjects (Lerner et al., 2011). Gene set enrichment analysis revealed a significant enrichment in genes upregulated in both HIV and LCMV-Cl13 infections (Fig. 3 F). Leading-edge genes that particularly contributed to the enrichment included IFN-I–induced inflammatory genes (STAT1, IRF9, CCL5, RSAD2 [viperin], PARP9, BST2 [tethrin]), CD8 T cell activation/exhaustion genes (GZMK, CD8A, NKG7, LAG3, TOX2, EOMES), antigen-processing/presentation genes (TAP1, immunoproteasome components PSMB8 and PSMB9), the proliferation-associated MKI67, leukocyte adhesion/transmigration–associated ITGAL, and the adapter protein SH2D1A (Fig. 3 F). Thus, we identified a conservation of central inflammation, immune recruitment, and effector T cell activation pathways in chronic LCMV and HIV infection of the GIT.
The similarities between LCMV and HIV infections suggested conserved upstream regulation of these shared downstream networks. Comparison of predicted upstream regulators in the GIT during chronic LCMV, HIV (Lerner et al., 2011), and SIV infections (Kwa et al., 2011) indicated a high degree of similarity among predicted upstream regulators, with highly significant overlapping regulation by multiple components within the IFN pathways (Fig. 3 G) and through networks associated with immunomodulatory cytokines that sustain T cell function (IL-2, IL-12, IL-15, IL-21, IL-27). Conversely, SOCS1 and USP18, which are negative regulators of IFN-I, were decreased. Common predicted upstream factors also included virus-responsive receptors and signaling molecules (TLR7, TLR9, Ticam1, MAVS) and the LPS pathway, potentially reflecting microbial shifts during chronic infection. Regulators predicted to be reduced included B cell/T follicular helper (Tfh) cell–associated Bcl-6, BTK, and IRF4, which, unlike other IRFs, is not directly responsive to IFN-I and broadly affects T and B cell function (Huber and Lohoff, 2014). Thus, we identify a high conservation in upstream and downstream regulatory factors that control intestinal immune responses in multiple chronic infections and across multiple species.
Chronic infection drives distinct functional CD8 T cell differentiation in the GI compartments
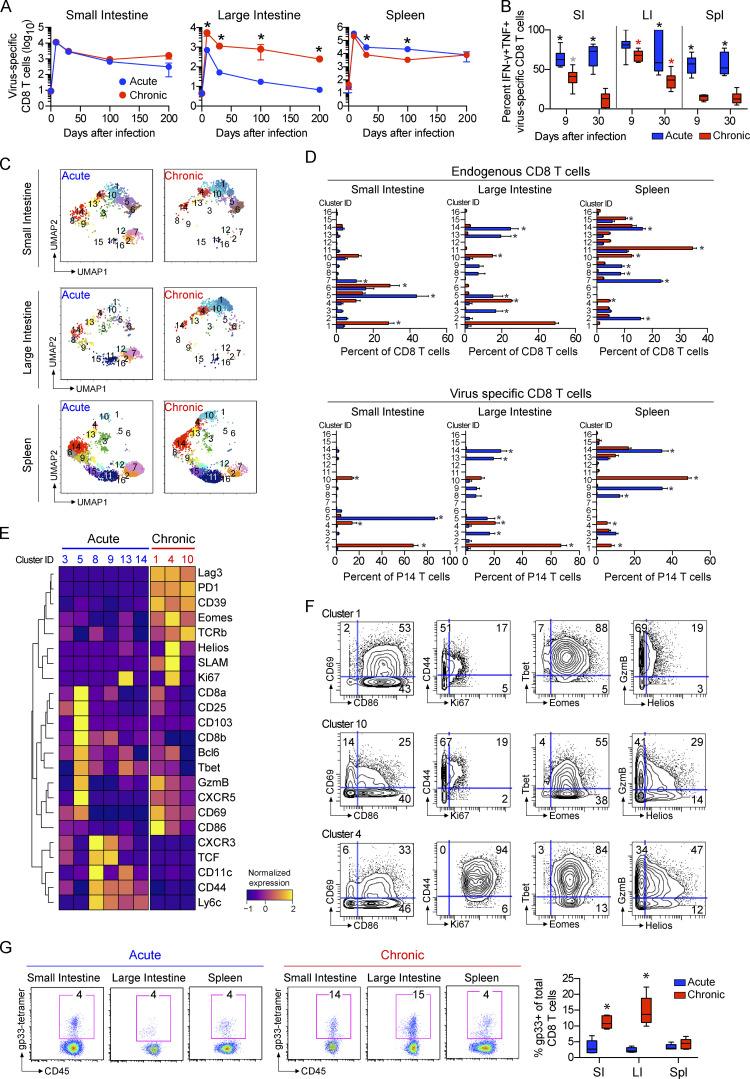
The massive CD8 T cell infiltration and transcriptomic signatures of CD8 T cell function in the LI suggested a unique niche during chronic infection. To monitor the antiviral CD8 T cell response, we tracked LCMV-specific transgenic P14 T cells throughout acute or chronic infection. Virus-specific CD8 T cells selectively accumulated and were retained in the LI during chronic infection, whereas they were maintained equally in the SI or were decreased in the spleen in chronic compared with acute infection (Fig. 4 A). Their maintenance during chronic infection corresponded with prolonged residual polyfunctional cytokine production in the SI and particularly in the LI versus the spleen, wherein rapid loss of polyfunctionality was quickly evident (Fig. 4 B).
Figure 4.
CD8 T cell accumulation and phenotype in the GIT during chronic viral infection. Mice received virus-specific CD8+ P14 T cells and were subsequently infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic). (A) Graphs show the number of virus-specific CD8 P14 T cells detected in the SI, LI, and spleen at the indicated dpi. n = 4–10 mice per group per time point from two independent experiments. *, P < 0.05 by ANOVA test. (B) 9 and 35 dpi, SI, LI, and spleen cells from LCMV-Arm– and LCMV-Cl13–infected mice were stimulated with LCMV-GP33–41 peptide. Graphs show the proportion of IFN-γ and TNF-α coproducing virus-specific CD8 P14 T cells from each compartment. Black asterisk, LCMV-Arm versus LCMV-Cl13, P < 0.05 within the tissue at that time point; gray asterisk, LCMV-Cl13 cytokine production in SI versus spleen, P < 0.05; red asterisk, LCMV-Cl13 cytokine production versus both other organs, P < 0.05 at that time point. n = 4–10 mice per group per time point from two independent experiments. (C–E) PhenoGraph clustering of total CD8 T cells and virus-specific CD8 P14 T cells analyzed by CyTOF at 35 dpi. (C) UMAP plots colored according to PhenoGraph-defined clusters. (D) Bar graphs show the relative proportion of each cluster among total CD8 T (endogenous) and P14 cells (virus specific). *, P < 0.05 acute versus chronic by two-way ANOVA. (E) Heatmap includes the clusters significantly increased (P < 0.05 by ANOVA) in virus-specific CD8 P14 T cells following LCMV-Arm (blue; acute) or LCMV-Cl13 (red; chronic) infection. (F) Contour plots show expression of the indicated markers in clusters 1, 10, and 4. Shown is one of two independent experiments with n = 5 mice per group. (G) LCMV-GP33 tetramer expression on CD8 T cells in the SI, LI, and spleen at 30 dpi after LCMV-Arm or LCMV-Cl13 infection. Box plots (left) and representative flow plots (right) show the percentage of tetramer-positive CD8 T cells in each organ. Shown is one of two independent experiments with n = 5 mice per group. *, P < 0.05 by ANOVA test. Error bars indicate highest and lowest values (A–G).
To further understand the functional impact of acute or chronic infection on CD8 T cells, CyTOF was used to assess their phenotype. Total CD8 T cells were clustered using PhenoGraph (Levine et al., 2015), generating 16 clusters, which were embedded into UMAP plots for visualization (Fig. 4, C–F; and Fig. S4 A). Further dissection identified eight clusters enriched in LCMV-Arm–infected mice and six enriched in LCMV-Cl13–infected mice. Among the virus-specific CD8 T cells, the GIT and the spleen clustered separately. There was also a large degree of overlap in cluster distribution among virus-specific and total CD8+ T cells in the GIT, likely reflecting the increased representation of virus-specific CD8 T cells among all CD8 T cells in the GIT versus the spleen (Fig. 4 G). Following acute infection, the virus-specific response had wider breadth across many clusters, except for virus-specific CD8 T cells in the SI that almost exclusively existed in a Trm cell cluster (C5) expressing CD103, CD69, Tbet, and GzmB (Fig. 4, D and E), suggesting the preferential generation of LCMV-specific Trm cells in the SI. Conversely, in chronic infection of the GIT, virus-specific CD8 T cells were primarily present in C1, whereas in the spleen, they were primarily within C10 (Fig. 4, C and D). In UMAP space, the chronic clusters (C1 and C10) localized next to each other, indicating protein expression similarity, and both expressed similar levels of the activation/exhaustion proteins PD1, Lag3, and CD39. However, within the spleen, the virus-specific cells were comparatively low for other activation markers (except CD44, Helios), indicating increased activation/effector functionality in the GIT (heightened GzmB, CD69, SLAM, CD11c, CD86; Fig. 4, E and F). Splenic virus–specific CD8 T cells in C10 contained a substantial fraction of EomeshiTbetlo cells, a differentiation state that induces exhaustion and PD1 expression (Paley et al., 2012). Interestingly, this population was not detected within the dominating cluster C1 in the GIT, where antiviral CD8 T cells instead coexpressed high levels of both Tbet and Eomes, despite high levels of PD1.
Figure S4.
T cell accumulation and phenotype in the GIT during chronic viral infection. (A) PhenoGraph clustering of total CD8 T cells analyzed by CyTOF 35 dpi after LCMV-Arm (acute) or LCMV-Cl13 (chronic) infection. Heatmap shows row-normalized protein expression. Clusters significantly increased in proportion (ANOVA, P < 0.05) between LCMV-Arm–infected and LCMV-Cl13–infected mice are labeled blue and red, respectively. Shown is one of two independent experiments with n = 5 mice per group. (B) Graphs show the number of CD103+CD69− and CD103+CD69+ CD8 T cells in the SI and LI of LCMV-Arm– and LCMV-Cl13–infected mice. *, P < 0.05 LCMV-Arm versus LCMV-Cl13 by t test at each time point. (C–F) Mice received virus-specific SMARTA CD4 T cells and were subsequently infected with LCMV-Arm or LCMV-Cl13. (C) Dot plots show the proportion of IFN-γ and TNF-α coproducing SMARTA in the SI, LI, and spleen at 9 and 30 dpi following ex vivo LCMV-GP61–80 peptide stimulation. Each dot represents an individual mouse from two independent experiments. *, P < 0.05 LCMV-Arm versus LCMV-Cl13 by ANOVA test. (D) The longitudinal frequency of virus-specific CD4 SMARTA T cells of the total CD4 T cell population in the indicated organ. n = 5–10 mice per group per time point. *, P < 0.05 LCMV-Arm versus LCMV-Cl13 by ANOVA test. (E and F) PhenoGraph was used to cluster CD4 T and SMARTA cells analyzed by CyTOF at 35 dpi. (E) Concatenated UMAP plots are colored to show individual PhenoGraph-defined clusters. n = 5 mice per group from one of two independent experiments. (F) Heatmap shows row-normalized expression of clustering markers in all PhenoGraph-defined clusters. Clusters significantly increased in frequency (P < 0.05, ANOVA) in LCMV-Arm– or LCMV-Cl13–infected mice are colored blue and red, respectively. n = 5 mice from one of two independent experiments. Error bars indicate SEM (A–F).
In the chronically infected GIT, a subdominant cluster C4 was also significantly enriched. C4 expressed the highest Ki67, activation-induced proteins (PD1, CD39, GzmB), and CXCR5 (Fig. 4, E and F), but it had low TCF1, indicating that it was distinct from the memory-like population sustaining effector cells within the spleen during chronic infection (Im et al., 2016). In the spleen, C13 was the dominant Ki67+ cluster and was absent from virus-specific CD8 T cells in the chronic GIT (Fig. 4 D), suggesting that the long-term maintenance of the effector CD8 T cells in the GIT of chronically infected mice is derived intrinsically from the cycling Ki67+, Eomeshi, Tbethi, and PD1+ population (C4) or from outside sources. Thus, unique effector differentiation states and TF coexpression patterns emerge in the GIT and diverge substantially from the spleen in response to chronic infection, yielding an activated and more polyfunctional virus-specific T cell population compared with the spleen.
Erosion of GI Trm cells in chronic infection
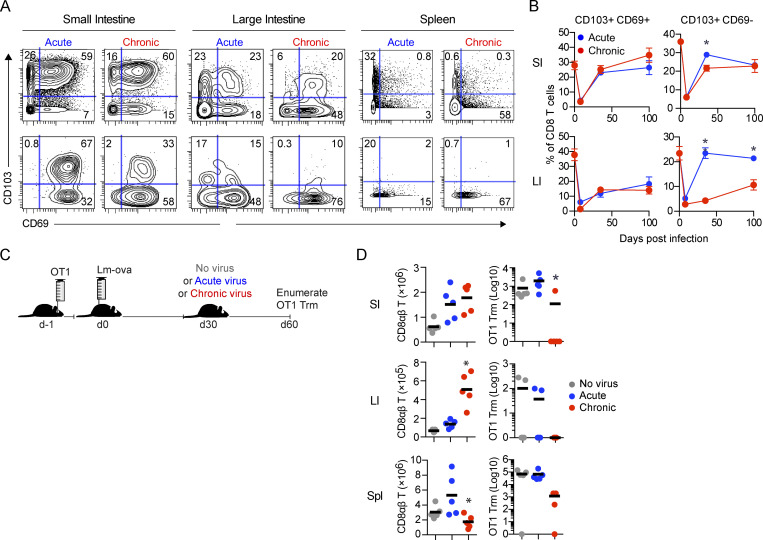
Considering the importance of Trm cells in maintaining barrier immunity (Mueller and Mackay, 2016), their loss during chronic infection could substantially alter long-term immunity and disease susceptibility. Chronic infection led to diminished frequencies of cells expressing CD8 Trm markers in both the SI (as reported by Casey et al., 2012) and in the LI, with their depletion being most prominent among CD103+CD69− CD8 Trm cells (Fig. 5, A and B; and Fig. S4 B). Interestingly, Trm cell differentiation was higher in the SI than in the LI, with increased virus-specific and endogenous CD69+CD103+ and CD103+CD69− CD8 T cells, consistent with a previous study (Steinert et al., 2015) and suggesting that the SI microenvironment preferentially promotes and retains Trm.
Figure 5.
Erosion of tissue-resident memory in the chronically infected GIT. (A and B) Mice received virus-specific CD8 P14 T cells and were subsequently infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic). (A) At 35 dpi, immune cells from the SI, LI, and spleen were analyzed by CyTOF. Contour plots show representative CD69 and CD103 expression by total CD8 T cells and virus-specific CD8 P14 T cells from one of two independent experiments with n = 5 mice per group. (B) Graphs show the proportion of CD103+CD69− and CD103+CD69+ cells among total CD8 T cells in the SI and LI at the indicated time points from one of two independent experiments with n = 5 mice per group. *, P < 0.05 by t test. (C) Experimental schematic depicting mice that received OT1 cells and subsequently were infected with Listeria-OVA (Lm-ova). 30 d later, mice were challenged with no virus or LCMV-Arm or LCMV-Cl13 infection. 30 d after challenge, CD8 responses in the SI, LI, and spleen were analyzed by flow cytometry. (D) Graphs depict the number of total CD8 T cells and OT1 Trm cells (CD103+) in the SI and LI or the total number of OT1 cells in the spleen. *, P < 0.05 by Kruskal-Wallis test. Shown is one of two independent experiments with n = 5 mice per group. Error bars indicate SEM (A–D).
Chronic infections cause attrition of T cells from lymphoid organs (Stelekati et al., 2014); however, whether this influences preexisting pathogen-specific Trm cells remains unknown. To test this, we transferred OVA-specific CD8 OT1 T cells and then infected mice with Listeria-OVA to generate OT1 Trm cells (Fig. 5 C). 30 d after Listeria-OVA infection, the mice were infected with LCMV-Arm or LCMV-Cl13 or were left naive to LCMV. In the SI, LCMV-Arm infection did not alter the amount of Listeria-specific CD103+ OT1 Trm cells (Fig. 5 D). In contrast, OT1 Trm cells were almost entirely lost in the SI of subsequently chronically infected mice. This effect was not wholly specific to Trm cells, because CD103− GIT OT1 attrition was also observed, as was OT1 attrition in the spleen (which harbored few Trm cells). Listeria-specific Trm cells formed to a lesser extent in the LI; nonetheless, they were also deleted from the LI in chronically infected mice (zero of five mice) while remaining detectable in the same number of mice (two of five) that received LCMV-Arm or no virus challenge (Fig. 5 D). Thus, chronic viral replication results in a large-scale depletion of preexisting pathogen-specific Trm cells throughout the GIT.
Reduced and altered virus-specific CD4 T cells in the chronically infected GIT is coupled with bystander T reg cell and Th17 amplification
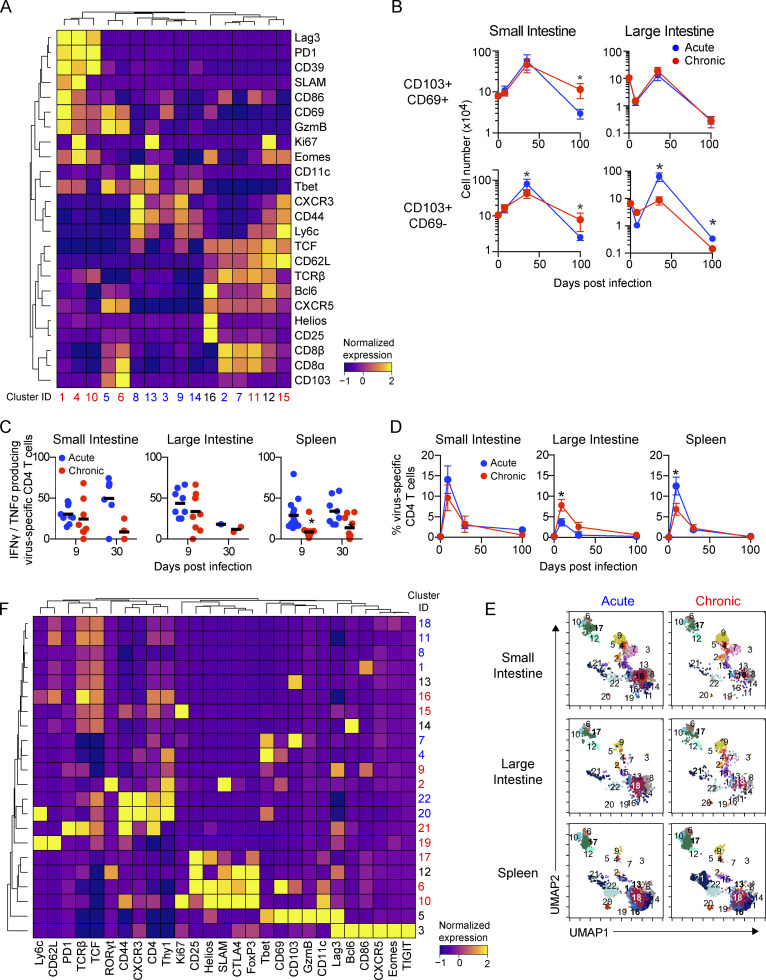
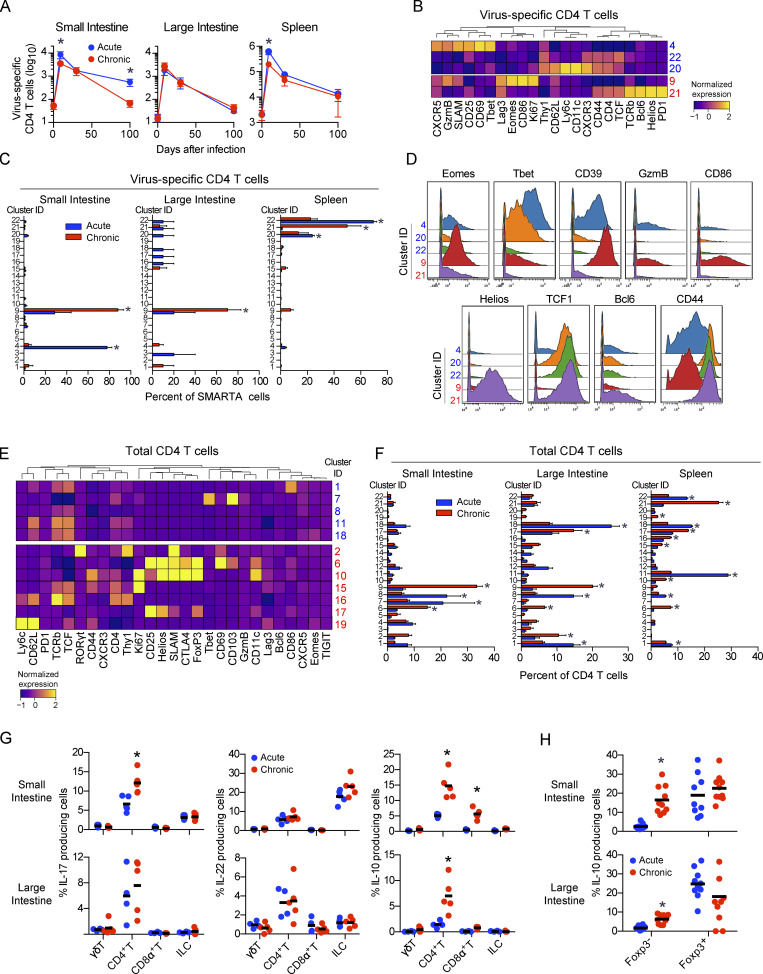
The depletion of GI CD4 T cells during establishment of chronic infection suggested that distinct subsets could emerge as the compartment was repopulated. To investigate the CD4 T cell response, we tracked total and virus-specific SMARTA transgenic CD4 T cell dynamics. During primary infection with chronic virus (day 9), virus-specific CD4 T cells in the SI and spleen were reduced, whereas they did not differ in abundance in the LI, consistent with the dynamics observed for total GI CD4 T cells (Fig. 2). Interestingly, virus-specific CD4 T cells in the GIT maintained higher frequencies of polyfunctional IFN-γ and TNF-α production than in the spleen at 9 dpi (Fig. S4 C), indicating that despite high viral titers, virus-specific CD4 T cells in the GIT retained functionality for extended periods of time. During the chronic phase of infection (30 dpi), virus-specific CD4 T cell numbers in the different tissues equilibrated between acute and chronic infection, although they continued to decline in both frequency and number with time (Fig. 6 A and Fig. S4 D) and were largely undetectable in the LI of acute or chronically infected mice and in the SI of chronically infected mice.
Figure 6.
Virus-specific CD4 T cell alterations are accompanied by T reg cell and Th17 outgrowth in the chronically infected GIT. Mice received virus-specific CD4 SMARTA T cells and were subsequently infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic). Total and virus-specific CD4 T cells were analyzed by CyTOF 35 dpi. (A) Graphs indicate the number of virus-specific CD4 SMARTA T cells in the SI, LI, and spleen at the indicated time. Data from two independent experiments with n = 9 or more mice per group. *, P < 0.05 by ANOVA test. (B) Heatmap shows PhenoGraph-defined clusters among virus-specific SMARTA cells that are significantly increased (P < 0.05 by ANOVA) in LCMV-Arm–infected mice (blue) or LCMV-Cl13–infected mice (red). (C) Bar graphs show the relative proportion of each cluster within the virus-specific CD4 SMARTA T cell population from one of two independent experiments with n = 5 mice per group. *, P < 0.05 acute versus chronic by two-way ANOVA. (D) Histograms show expression of the indicated protein by CD4 T cells in clusters significantly enriched (P < 0.05 by ANOVA) in virus-specific cells in acute (C4, C20, C22) and chronic (C9 and C21) infection. Data are pooled from n = 5 mice per group from one of two independent experiments. (E) Heatmap shows PhenoGraph-defined clusters in total CD4 T cells that are significantly increased (P < 0.05 by ANOVA test) in LCMV-Arm–infected mice (blue) or LCMV-Cl13–infected mice (red). Data are pooled from n = 5 mice per group from one of two independent experiments. (F) Bar graphs show the relative proportion of clusters that are significantly increased (P < 0.05 by ANOVA) in LCMV-Arm–infected mice (blue) or LCMV-Cl13–infected mice (red). Data from n = 5 mice per group from one of two similar experiments. *, P < 0.05 acute versus chronic by two-way ANOVA. (G and H) 45 dpi after LCMV-Arm (blue) or LCMV-Cl13 (red) infection, cells from the SI and LI were stimulated with PMA/ionomycin. (G) IL-17, IL-22, and IL-10 production was analyzed by flow cytometry in γδT cells (TCRγδ+), total CD4 T cells (CD4+), total CD8 T cells (CD8α+), and ILCs (TCR−CD8−Thy1.2+). Shown is one of two similar experiments with n = 5 mice per group. *, P < 0.05 by t test. (H) IL-10 production in total Foxp3+ and Foxp3− CD4 T cells from n = 9–10 mice per group from two independent experiments. *, P < 0.05 by t test. Error bars indicate SEM (A–G).
Clustering analysis of CD4 T cells at 35 dpi identified 22 clusters, with 5 and 10 clusters being differentially represented between the acute and chronic infections, respectively (Fig. 6, B–F; and Fig. S4, E and F). In the SI and LI after acute and chronic infections, total and virus-specific CD4 T cells predominantly fell into Th1 clusters that were only minimally present in the spleen. Spleen virus–specific CD4 T cells were dominated by a Tfh cluster (C21), as previously demonstrated (Fahey et al., 2011). Interestingly, the dominant GIT cluster, C9, in chronic infection exhibited distinct differentiation and an inverted Tbet/Eomes TF ratio from the C4 that remained following acute LCMV infection. This noncanonical Th1 cluster, C9, expressed intermediate Tbet with simultaneous high expression of Eomes and the highest activation/effector proteins (GzmB, CD86, Ki67), consistent with chronic viral replication in the GIT promoting CD4 cytotoxic T lymphocyte differentiation (Takeuchi and Saito, 2017). Counterintuitively, on the basis of low Tbet expression and heightened activation, C9 expressed CD44 and PD1 but at lower levels, suggesting decreased chronic TCR-mediated immune activation in the GIT. Thus, the intestinal environment facilitates maintenance of noncanonical virus-specific CD4 Th1 cells with a specific activation program and diminished exhaustion.
Clustering analysis revealed that, unlike the virus-specific clusters that primarily were contained in a single cluster, the total CD4 T cells were more broadly distributed (Fig. 6 F). Because the virus-specific CD4 T cells by this time comprised only a small fraction of the total CD4 T cell response (Fig. S4 D), these diverse clusters represented heterogeneity among the non–virus-specific CD4 T cells. Chronic infection simultaneously amplified activated GIT T reg (C6, C10, C17) and Th17 (C2) cells that were not observed among virus-specific cells (Fig. 6, E and F; and Fig. S4, E and F). Furthermore, a greater fraction of the total CD4 T cells from the chronically infected GIT produced IL-17 and/or IL-10 (Fig. 6 G), consistent with increased Th17 and regulatory CD4 T cells. In addition to the increased activation of protein expression on Th17 cells, chronic infection appeared to promote an inflammatory Th17 response because there was no tandem increased production of the barrier repair Th17 cell cytokine IL-22 (Fig. 6 G). Interestingly, the increased IL-10 production in chronically infected mice is observed in the FoxP3− CD4 T cells, with the percentage of FoxP3+ T reg cells that produce IL-10 being the same between acute and chronic LCMV infection, consistent with a Tr1 and not a conventional T reg cell phenotype (White and Wraith, 2016).
T reg cell depletion and checkpoint blockade enhance viral clearance from the chronically infected GIT
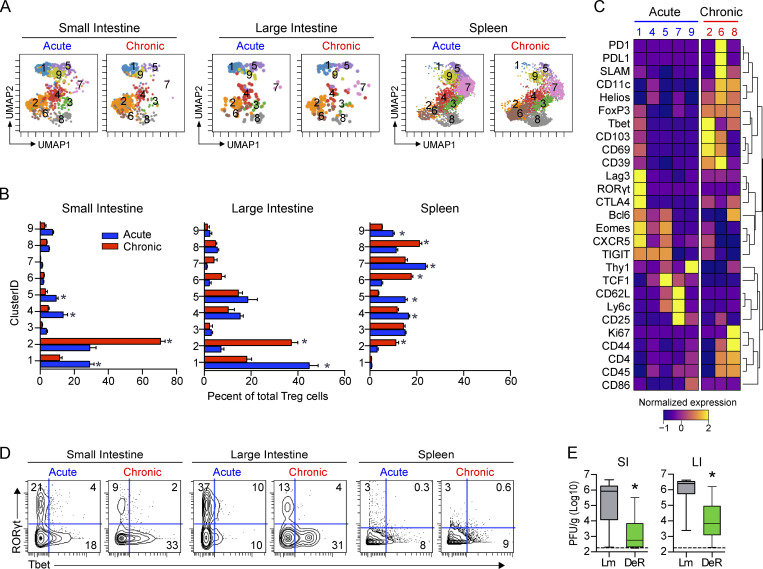
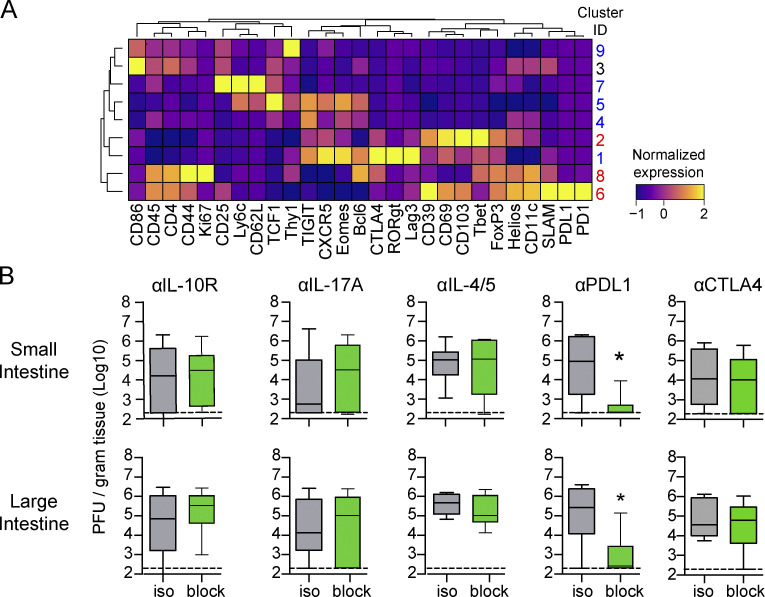
The observed increase in Foxp3-expressing clusters in the GIT during chronic infection suggested that they might be critical in maintaining the intestinal viral reservoir. To determine functional changes, we used PhenoGraph clustering and identified that FoxP3+ T reg cells distributed into nine clusters (Fig. 7, A–C; and Fig. S5). T reg cells in the GIT following acute infection were predominantly in C1, which, although positive for the canonical T reg cell markers FoxP3 and Helios, expressed them at reduced levels (Fig. 7 C). Furthermore, C1 expressed the highest RORγt, potentially suggesting a focus toward suppression of Th17 cells in the GIT following the resolution of acute infection. By contrast, T reg cells in the chronically infected GIT concentrated into C2 that exhibited elevated coexpression of multiple proteins associated with activation/immunosuppression (CD39, CD69, CD11c, Helios) and residency (CD69, CD103) and had increased Tbet with minimal RORγt (Fig. 7, B and C), indicating that chronic infection redirected GIT T reg cells to a hypersuppressive Tr1 phenotype (Levine et al., 2017). Supportively, we assessed Tbet and RORγt expression by total GIT T reg cells in acute and chronic infection and confirmed a striking loss of RORγt expression and outgrowth of Tbet-expressing T reg cells, specifically in the chronic GIT (Fig. 7 D). In the spleen following acute infection, T reg cells distributed among several clusters, expressing lower Tbet and lacking RORγt, implying a lack of Tr17 cells (Fig. 7, B and C). Instead, the dominating acute splenic clusters, C4 and C7, expressed CD62L, CXCR5, Bcl6, and TCF1, indicative of lymphoid-homing and T follicular regulatory cells. In the chronically infected spleen, T reg cells were predominantly in C6 and C8, coexpressing the highest CD44, Ki67, CD11c, and SLAM as well as PD1/PDL1, possibly reflecting a greater extent of T reg cell activation in the spleen than in the GIT. To determine how T reg cells contribute to viral persistence, we used FoxP3-DTR (DEREG) mice, in which diphtheria toxin (DT) treatment specifically depletes T reg cells (Lahl and Sparwasser, 2011). It has previously been reported that depletion 30 d into chronic LCMV infection did not alter virus titers in the spleen, liver, or kidneys (Penaloza-MacMaster et al., 2014). However, T reg cell depletion commencing 50 d after chronic infection (i.e., once the virus was nearly controlled in other peripheral tissues) significantly reduced virus titers in both the SI and the LI compared with in DT-treated littermate control mice (Fig. 7 E), indicating that T reg cells mechanistically foster viral persistence in the GIT.
Figure 7.
T reg cell depletion augments viral clearance from the chronically infected GIT. (A–D) Mice were infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic), and at 35 dpi, T reg cells (CD4+Foxp3+) were analyzed by CyTOF from SI, LI, and spleen and clustered using PhenoGraph. (A) UMAP plots colored according to PhenoGraph-defined clusters. Numbers in each UMAP plot indicate the cluster. Plots are concatenated from n = 5 mice per group from one of two independent experiments. (B) Bar graphs show the relative proportion of each cluster within the total T reg cell population. *, P < 0.05 LCMV-Arm versus LCMV-Cl13 by ANOVA test. n = 5 mice per group from one of two independent experiments. (C) Heatmap shows row-normalized expression of clustering markers in clusters where P < 0.05 by ANOVA test for LCMV-Arm infection (blue) versus LCMV-Cl13 infection (red). Heatmap was concatenated from n = 5 mice per group from one of two independent experiments. (D) Representative contour plots show Tbet and RORγt expression on total FoxP3+ cells from each organ. Plots were concatenated from n = 5 mice per group from one of two independent experiments. (E) DEREG (DeR) or littermate (Lm) control mice were infected with LCMV-Cl13 and 50–60 d later were treated with DT to deplete Foxp3+ T reg cells. Box plots show virus titers in the SI and LI 9 d after DT treatment. Error bars indicate the highest and lowest values. Data from three independent experiments with n = 14–15 mice per group. *, P < 0.05 by t test.
Figure S5.
T reg cell depletion augments virus clearance from the chronically infected GIT. (A) Mice were infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic), and 35 dpi, T reg cells (CD4+Foxp3+) from the SI and LI were analyzed by CyTOF. The heatmap shows column-normalized protein expression of PhenoGraph-defined clusters. Clusters significantly increased in frequency (ANOVA, P < 0.05) in LCMV-Arm– and LCMV-Cl13–infected mice are labeled blue and red, respectively. Shown is one of two independent experiments with n = 5 mice per group. Data represent two independent experiments with five mice per group. (B) LCMV-Cl13–infected mice were treated at 60–80 dpi with isotype control antibody or antibodies against the IL-10 receptor, IL-17, IL-4 and IL-5, PDL1, or CTLA4. Box plots show virus titers in the SI and LI after blockade. *, P < 0.05 (t test). n = 10 mice per group from two independent experiments.
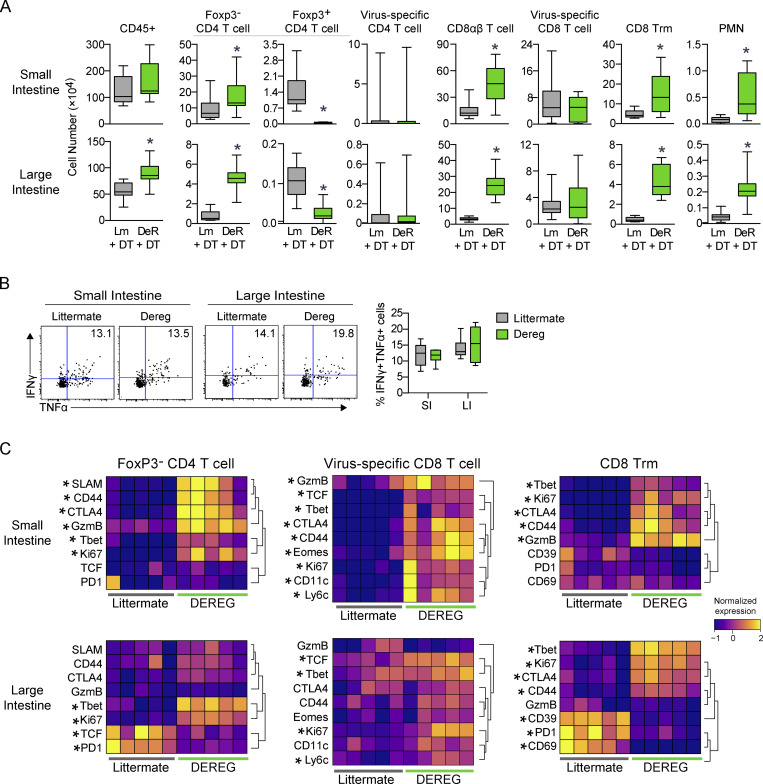
CyTOF analysis revealed that T reg cell depletion correlated with a large expansion in GIT T cells, including CD4+, total CD8αβ+, and Trm cells, as well as an increase in neutrophils (Fig. 8 A). Intriguingly, the low numbers of virus-specific CD4+ T cells did not increase, nor did virus-specific CD8+ T cells, and virus-specific CD8+ T cells showed no change in ability to produce multiple cytokines (Fig. 8 B). However, virus-specific CD8 T cells showed a striking increase in activation markers, especially in the SI (GzmB, Ki67, CD44, CD11c) following T reg cell depletion, suggesting functional reactivation of the virus-specific T cell response. Similar increases in activation of endogenous CD4 T cells and Trm cells were also observed, suggesting that depletion of GIT T reg cells enabled the broad-spectrum reinvigoration of T cell function, and this, possibly in conjunction with increases in factors produced by bystander Trm cells and non–virus-reactive T cells, mediated amelioration of the viral reservoir.
Figure 8.
Depletion of T reg cells increases virus-specific T cell activation states and increases bystander T cells in the chronically infected GIT. (A–C) DeR or Lm control mice received virus-specific CD4 SMARTA and CD8 P14 T cells and were subsequently infected with LCMV-Arm (acute) or LCMV-Cl13 (chronic). Cells from the SI and LI were analyzed by CyTOF 50 dpi. (A) The numbers of the indicated cell populations in the SI (top row) and LI (bottom row). Data from two independent experiments with n = 12 mice per group. *, P < 0.05 by t test. Error bars indicate the highest and lowest values. (B) Representative flow plots (left and center panels) and box plots (right panel) show the proportion of IFNγ+TNF+ P14 T cells following ex vivo stimulation with LCMV-GP33–41 peptide. Shown is one of three independent experiments with n = 5–6 mice per group. Error bars in box plots indicate the highest and lowest values. (C) Heatmaps show row-normalized protein expression from CyTOF analysis in the indicated immune cell populations. Each column represents an individual mouse from one of two independent experiments with n = 12 total mice per group. *, P < 0.5 (t test).
To further identify factors that impede virus control in the intestinal tract, we targeted pathways associated with immunosuppression or the redirected Th cell responses after chronic infection. Inhibition of cytokines associated with Th2 or Th17 responses (blocking IL-4, IL-5, and IL-17 at 60–80 dpi) failed to affect GI viral load (Fig. S5 B), suggesting that, although increased, Th2 and Th17 cytokines did not themselves maintain viral persistence, and further supporting the specific role of T reg cells in maintaining the GIT viral reservoir. In addition, late blockade of IL-10R, which promotes viral clearance in multiple peripheral tissues when administered at 25 dpi (Brooks et al., 2006), failed to clear the GIT chronic reservoir (Fig. S5 B). Thus, T reg cells and PDL1 are dominant suppressive mechanisms maintaining chronic infection in the GIT and providing sanctuary for long-term viral persistence.
Discussion
Knowledge of T cell responses in chronic infections is derived largely from splenic studies; yet, it remains unclear whether nonlymphoid tissues such as the GIT behave similarly, given their divergent immune landscape. Using the murine model of LCMV infection, we have identified the GIT as a long-term reservoir of chronic infection, inducing cellular, genetic, and microbial changes. Da Fonseca et al. (2015) demonstrated that bacterial infection in the GIT causes alterations in lymphoid and adipose tissue that have long-term implications for host immunity. Similarly, our studies show that chronic viral replication in the GIT had continual consequences for disease susceptibility, even after severe pathological symptoms seemingly resolved. Importantly, we identified shared immunological networks regulating LCMV, SIV, and HIV persistence, identifying a conserved regulation of the immune response in the GIT across species in times of sustained chronic viral infection in the GIT.
The temporally linked induction of antiviral immunity and LI contraction/colitis highlights the fine balance between tolerance and activation of pathogen-specific responses and the potentially beneficial role of exhaustion in minimizing GI pathology. It is interesting that the temporally increased Lachnospiraceae, which is generally associated with GIT health and physiological function (Ríos-Covián et al., 2016), was insufficient to protect mice from the increased colitis susceptibility during late chronic infection. Although colitis is a complex disease, it is possible that by maintaining increased levels of inflammatory cytokines (e.g., IFN-γ, TNF, IFN-I, cell stimulatory factor), cell polarization pathways (T cell activation, cell death, inflammasome signaling), and/or the loss of barrier-regulating ILC3s, the chronic infection overrode the impact of wound-healing bacteria and resulted in heightened colitis susceptibility. These alterations are consistent with studies of humans with inflammatory bowel disease and colorectal cancer, in whom chronic GIT inflammation and microbial shifts are associated with disease development (Nadeem et al., 2020), suggesting that by driving chronic inflammation in the GIT, even when seemingly not causing overt disease, chronic viral infection can lead to environmental changes that provide a fertile ground for disease induction.
The precise role of the microbiota in progression of chronic viral infections remains controversial, with different studies finding distinct microbial signatures of disease or no significant alterations (Dubourg, 2016). Given the different microbiome compositions we observed during primary infection and late disease, sampling different cohorts at different times after acquisition of infection and/or initiation/interruption of therapy may contribute to this variability. Multiple studies have reported an increase in Proteobacteria as a biomarker of GIT pathology (Bandera et al., 2018). Chronic LCMV infection induces substantial loss of appetite and cachexia during primary infection, and this is a known cause of dysbiosis (Bindels et al., 2018), providing a potential mechanism for the observed Proteobacteria bloom.
Given that the GIT is a reservoir for chronic viral infection, the immunological alterations that facilitate persistence are important to understand. High-dimensional analysis denoted T cell alterations that were nearly exclusive to acute or chronic infection. The predominant virus-specific CD8 T cell clusters in the chronically infected GIT and spleen expressed exhaustion proteins, including Lag3, PD1, and CD39. Yet, virus-specific CD8 T cells in the GIT also exhibited a highly activated effector phenotype with lower levels of CD44 and Helios, the latter of which are heavily associated with T cell exhaustion in the spleen (Bengsch et al., 2018). Strikingly, the virus-specific CD8 T cells in the GIT exhibited a TbethiEomeshi coexpression pattern that does not exist in the spleen (Paley et al., 2012), depicting a unique GIT-specific differentiation pattern with dual expression of central defining TFs and diminished exhaustion/enhanced function (Bengsch et al., 2018). Interestingly, the clusters of cycling cells in the GIT expressed high Ki67 without TCF1, suggesting that GIT-resident virus-specific CD8 T cells are being maintained by de novo cycling of the effector populations themselves. Chronic infection also specifically eroded Trm cell maintenance, leading to a permanent loss of Trm cells against previously encountered pathogens, indicating a hole in the ability to fight these infections if reencountered.
Chronic infections are associated with lymphopenia and, in particular, HIV infection causes rapid GIT CD4 depletion. Yet, even during HIV infection, a relatively small fraction of CD4 T cells are infected, and thus it is believed that infection-induced/bystander cell death contributes to CD4 depletion (Doitsh and Greene, 2016). We now demonstrate that chronic LCMV infection similarly leads to massive CD4 T cell depletion in the GIT. This was not due to direct infection (few CD4 T cells are infected by LCMV); instead, it results from the large spike of IFN-I at the onset of infection, and, accordingly, blockade of IFN-I signaling prevented CD4 T cell loss. Thus, although IFN-I is important for the development of antiviral CD8 T cell responses (Kolumam et al., 2005), our studies indicate a population-wide inhibitory effect on CD4 T cells in the GIT. Furthermore, GIT-resident virus-specific CD4 T cells exhibited heightened GzmB production, multiple activation proteins (but low CD44 and PD1), and inverted Tbet:Eomes expression, indicating that, unlike the spleen, which promotes Tfh differentiation through continued TCR signaling (Fahey et al., 2011; Lindqvist et al., 2012), GIT-resident virus-specific CD4 T cells instead differentiated into a noncanonical, cytotoxic Th1 phenotype. Interestingly, chronic infection simultaneously amplified bystander T reg, Tr1, and Th17 cells in the GIT. The T reg cells were highly activated and enriched in a population of Tbet+FoxP3+ cells, associated with suppression of Th1 and CD8 T cells (Levine et al., 2017), which could specifically promote the long-term maintenance of viral persistence. Expansion of splenic T reg cells has been observed in chronic LCMV infection, although ablating T reg cells was unable to augment viral clearance (Penaloza-MacMaster et al., 2014). Yet, their activated state, coupled with increased Tbet expression, suggested that GI-resident T reg cells could be playing a more dominant suppressive role, and, indeed, late T reg cell depletion promoted viral clearance from the GIT. The molecules that T reg cells use to inhibit T cell activation/expansion to prevent control of chronic infection are likely many, considering that blockade of IL-10 or CTLA4 did not promote viral clearance. Interestingly, although T reg cell depletion increased the numbers of total CD4 T cells, CD8 T cells, and CD8 Trm cells, it did not increase the number of virus-specific CD8 T cells in the GIT. However, T reg cell depletion did lead to the multispectral upregulation of activation and antiviral factors by virus-specific T cells (particularly in the SI), suggesting that T reg cells are suppressing activation and antiviral effector functions to prevent virus control. This may further be augmented by cytokines or other factors provided by the expanded total T cell and Trm cell populations that potentially enable viral control.
The ability of PDL1 blockade to control the GI reservoir >2 mo after infection is impressive, given the challenge of reducing viral load after a set point has been established (Kelley et al., 2007). However, the reinvigoration of antiviral T cell responses by PDL1 blockade also reinduced colitis-like symptoms within these mice. Thus, although the initial colitis resolved with the exhaustion of T cell function, these chronically infected mice remained latently more susceptible to colitis-like disease. GIT pathology and disease induction are major side effects of checkpoint immunotherapy in cancer patients (Michot et al., 2016), and our results suggest that reactivation of exhausted pathogen-specific T cells against their ongoing chronic infection target in the GIT could be an important mechanism of checkpoint-associated colitis. Overall, our study provides fundamental understanding of the network of microbial, cellular, and transcriptional alterations that define chronic viral infection in the GIT and identify potential targets for eliminating this viral sanctuary.
Materials and methods
Experimental model and subject details
Mice
C57BL/6 mice were purchased from The Jackson Laboratory or the rodent breeding colony at Princess Margaret Cancer Center. LCMV glycoprotein (GP) 33–41 (LCMV-GP33–41)–specific CD8+ TCR transgenic (P14) and LCMV-GP61–80–specific CD4+ TCR transgenic (SMARTA) mice have been described previously (Brooks et al., 2006), and OVA257–264–specific CD8+ TCR transgenic (OT-I) mice and DEREG mice were purchased from The Jackson Laboratory. Experiments were performed with male and female mice (6–10 wk old) for confirmation of results. Mice were housed under specific pathogen–free conditions. Mouse handling conformed to the experimental protocols approved by the University Health Network (UHN) Animal Care Committee at the Princess Margaret Cancer Center/UHN.
Method details
T cell adoptive transfer, LCMV and Listeria infections, and DSS colitis induction
T cells were isolated from the spleens of transgenic mice by negative selection (STEMCELL Technologies), and 104 OT1, 103 P14, or 4 × 103 SMARTA cells were transferred i.v. in the retroorbital sinus of naive mice. Mice were infected i.v. via the retroorbital sinus with 2 × 106 PFU of LCMV-Arm or LCMV-Cl13. Virus stocks were prepared and viral titers quantified as described previously (Brooks et al., 2006). Mice were infected i.p. with 104 CFU of Listeria-OVA. Bacterial stocks were propagated and CFUs determined in Brain Heart Infusion media (BD Biosciences) supplemented with erythromycin (Sigma). To induce colitis, mice were treated with 2% or 4% (wt/vol) DSS (Sigma) in their drinking water. Water was replenished daily.
In vivo antibody blockade, cell depletion, and DT treatment
All antibodies were purchased from Bio X Cell. For blockade and depletion, LCMV-Cl13–infected mice were treated at the indicated time point with 250 µg anti-CD4 (GK1.5), anti-CD8 (2.43), anti-PDL1 (10F.9G2), anti–IL-10R (1B1.3A), anti-IL-4 (11B11), or anti-IL5 (TRFK5). Alternatively, mice were treated with 500 µg anti-IFNR (MAR1-5A3) and anti–IL-17A (17F3). Unless otherwise indicated, mice received five i.p. antibody treatments, with injections occurring 3 d apart. For DT-mediated depletion of FoxP3+ T reg cells, LCMV-Cl13–infected DEREG or littermate control mice were injected i.p. every second day with 1 µg DT for four treatments. Depletion was confirmed in the spleen.
Preparation of single-cell suspensions from intestine and spleen
Mice were perfused with 10 ml of PBS via intracardiac injection to remove circulating lymphocytes. Single cells were obtained from the spleen by pushing the organ through a 100-µm filter followed by RBC lysis. SI and LI tissue was sliced longitudinally, and digestive matter was expelled in cold PBS washes, and then the tissue was finely chopped with scissors in a 1.5-ml cryovial (Greiner Bio-One) in gut wash media (DMEM with 1% wt/vol FBS). Tissue was then incubated on a shaker at 37°C for 45 min in gut media (DMEM with 5% wt/vol FBS, 10 IU of penicillin and streptomycin) containing 500 mM dithiothreitol. Intraepithelial lymphocytes were isolated by pouring digested tissue through a 100-µm filter. Remaining tissue was incubated on a shaker at 37°C for 45 min in gut media containing 250 mg/ml collagenase D and 20 mg/ml DNase. Lamina propria lymphocytes were then isolated by pouring digested tissue through a 100-µm filter, and intraepithelial lymphocytes and lamina propria lymphocytes from their respective SI or LI compartment were combined. Cells were pelleted and resuspended in gut media with 40% Percoll PLUS, which was overlaid onto 80% Percoll PLUS, and cells were centrifuged at room temperature (RT) for 30 min (2,400 rpm, no brake; Sorvall ST40R centrifuge). Enriched viable intestinal leukocytes were then harvested from the buffy coat with a transfer pipette.
CyTOF and flow cytometry
The same antibody clones were used for CyTOF and flow cytometry. Vendor details for each antibody are listed in Table S1. Purified unconjugated antibodies were labeled with metal tags at the SickKids-UHN Flow and Mass Cytometry Facility using the MaxPar Antibody Labeling Kit from Fluidigm. Directly conjugated antibodies were purchased from Fluidigm.
For staining, single-cell suspensions from individual samples were first labeled for 5 min at RT with antibodies that did not perform well after fixation. The samples were then washed with PBS and pulsed with 12.5 µM cisplatin in PBS for 1 min at RT before quenching with CyTOF staining media (Mg+/Ca2+ HBSS containing 2% FBS [Multicell], 10 mM Hepes [Corning]) and FBS underlay. Cells were then fixed for 12 min at RT with TF fixative (00-5523-00; eBioscience) and permeabilized, and individual samples were barcoded according to the manufacturer’s instructions (20-Plex Pd Barcoding Kit, 201060; Fluidigm) before being combined. Combined samples were resuspended in staining media containing Fc block and then in metal-tagged surface antibodies for 30 min at 4°C. Cells were then permeabilized and stained with metal-tagged intracellular antibodies using the Transcription Factor Staining Buffer Set (00-5523-00; eBioscience) according to the manufacturer’s instructions. Cells were then incubated overnight in PBS (Multicell) containing 0.3% (wt/vol) saponin, 1.6% (vol/vol) paraformaldehyde (diluted from 16%; Polysciences Inc.), and 1 nM iridium (Fluidigm). Cells were then washed and kept in PBS with 1.6% formaldehyde in 4°C for 1–2 wk before acquisition. Cells were analyzed on a Helios or Helios2 mass cytometer (Fluidigm) at SickKids-UHN Flow and Mass Cytometry Facility. EQ Four Element Calibration Beads (Fluidigm) were used to normalize signal intensity over time on CyTOF software version 6.7. FCS files were manually debarcoded and analyzed using Cytobank 6.2 (Cytobank, Inc.).
For cytokine quantification, single-cell suspensions were restimulated for 5 h at 37°C with 2 mg/ml LCMV-GP33–40 or 5 µg/ml LCMV-GP61–80 peptide in the presence of 50 U/ml recombinant murine IL-2 and 1 mg/ml brefeldin A (Sigma). For all flow cytometry staining, single-cell suspensions were stained with a fixable viability stain, Zombie Aqua (BioLegend), and then stained with Fc block followed by cell surface binding antibodies in FACS staining media (PBS 1% [wt/vol] FBS) for 20 min at 4°C. Cells were then fixed, permeabilized, and stained with metal-tagged intracellular antibodies using the Transcription Factor Staining Buffer Set (eBioscience) or the Cytokine Staining Buffer Set (BioLegend). Samples were acquired on a FACSVerse flow cytometer (BD Biosciences), and data were analyzed using FlowJo software (FlowJo LLC).
Heatmaps were plotted in R using the blue-red, plasma, or viridis color packages and the gplots package. For CyTOF data, mean and median signal intensity values were used to generate heatmaps. T-distributed stochastic neighbor embedding (tSNE) analysis of CyTOF data was performed in Cytobank 6.2 using the indicated parameters. The R implementation of UMAP (k = 15) and the PhenoGraph (n = 80) algorithm (Levine et al., 2015) were used for cluster analysis of arcsinh-transformed CyTOF data. Arcsinh cofactors were manually determined depending on staining intensity.
16S ribosomal RNA-seq
Cohoused littermate weaned mice were used for these studies. Fecal pellets were longitudinally collected in Zymo Research DNA/RNA Shield fecal collection tubes and stored at −70°C until RNA extraction. The V4 hypervariable region of the 16S ribosomal RNA gene was amplified using KAPA2G Robust HotStart ReadyMix (Kapa Biosystems; Caporaso et al., 2012). The library was then purified using Ampure XP beads and loaded onto the Illumina MiSeq system for sequencing at the Centre for the Analysis of Genome Evolution and Function, University of Toronto. The UNOISE pipeline, available through USEARCH v10.0.240 and vsearch v2.5.0, was used for sequence analysis (Edgar, 2013; Rognes et al., 2016). The last base was removed from all sequences. Sequences were assembled and quality trimmed using fastq_mergepairs and –fastq_filter with –fastq_maxee set at 1.0. Sequences shorter than 233 bp were discarded. The trimmed data were then processed following the UNOISE pipeline. Sequences were first dereplicated and sorted to remove singletons, then denoised, and chimeras were removed using the unoise3 command. Assembled sequences were mapped back to the chimera-free denoised sequences at 97% identity operational taxonomic units. Taxonomy assignment was executed using SINTAX, available through USEARCH, and the UNOISE-compatible Ribosomal Database Project database version 16, with a minimum confidence cutoff of 0.8 (Wang et al., 2007). Operational taxonomic unit sequences were aligned using PyNast accessed through QIIME (Caporaso et al., 2010). Sequences that did not align were removed from the dataset, and a phylogenetic tree of the filtered aligned sequence data was made using FastTree (Price et al., 2009). Data analysis and visualization were done through R using Phyloseq and other open-access libraries (McMurdie and Holmes, 2013).
RNA-seq
Single-cell suspensions of intestinal cells were made, and CD45+ live (propidium iodide negative) cells were sorted on a Moflo Astrios FACS (Beckman Coulter) directly into RLT buffer (Qiagen). RNA was isolated using a single-cell RNA purification kit (Norgen Biotech Corp.) according to the manufacturer’s instructions. The SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech Laboratories) was used per the manufacturer’s instructions for amplification of RNA and subsequent cDNA synthesis. All samples proceeded through NexteraXT DNA Library Preparation (Illumina) using the NexteraXT Index Kit V1 or V2 Set A (Illumina). A portion of this library pool was sent for sequencing on an Illumina NextSeq high-throughput single read at the Princess Margaret Genomics Centre Core Facility. An average of 400 million reads were obtained per pool, with an average of 40 million reads/sample across the entire dataset. Illumina reads were aligned to the Mus musculus GRCm38 genome build 88 using HISAT2. Alignments were compressed and sorted using SAMtools. The alignments were quantified using HTSeq to obtain gene counts. Differential analysis was conducted using edgeR with modified code from the rnaseq.wiki protocol. Low-count genes were excluded from analysis if at least three samples did not have at least one count per million reads for that gene. Gene counts were normalized using trimmed mean of M-values normalization. Figures were plotted in R by using the gplots package.
For gene expression from RNA-seq data, transcripts per million values were used. For canonical pathway and predicted upstream regulator analysis, normalized z-scores generated by Ingenuity Pathway Analysis (Qiagen) were directly plotted on heatmaps. Subcellular locations were determined using the Jensen COMPARTMENTS database within the Enrichr analysis tool (Chen et al., 2013). Combined ranking was calculated by multiplying the log10 P value (Fisher’s exact test) by the z-score of deviation from expected rank. Gene set enrichment analysis (Subramanian et al., 2005) was based on ranked fold expression of genes in the LI of LCMV-Cl13–infected versus naive mice. This was compared with a rank file created of the top 100 (by P value) upregulated genes in colon biopsies of HIV-infected individuals from GSE28177 in the Gene Expression Omnibus database (Lerner et al., 2011). The human genes were converted to mouse genes by Ensembl BioMart, and uncharacterized loci were excluded from the comparison.
Data and software availability
The RNA-seq data generated in this paper have been deposited in the Gene Expression Omnibus database under accession number GSE142303.
Statistics
Prism 6 software (GraphPad Software) was used for statistical analysis; specific tests applied are indicated in the figure legends.
Online supplemental material
Fig. S1 shows viral titers, pathology, and immune cell kinetics in the GIT during acute and chronic LCMV infection. Fig. S2 shows the effects of anti-IFNR blockade and long-term changes in immune cells following acute and chronic LCMV infection. Fig. S3 shows the changes in DC maturation gene expression pathways. Fig. S4 further details T cell dynamics and phenotype in the GIT. Fig. S5 further phenotypically demonstrates CyTOF-based T reg cell subsets and the impact of blocking cytokines and negative inhibitory factors on chronic viral titers. Table S1 lists the vendor details for each antibody.
Supplementary Material
lists vendor details for each antibody.
Acknowledgments
The authors are supported by the Canadian Institutes of Health Research (foundation grant FDN148386), the National Institutes of Health (grant AI085043), the Scotiabank Research Chair (to D.G. Brooks), and the Fonds de la Recherche en Santé du Québec (training grant to L.M. Snell).
Author contributions: B.L. Macleod and D.G. Brooks conceived the project. B.L. Macleod, D.G. Brooks, C.J. Guidos, and T.L. McGaha designed the project and experiments. B.L. Macleod, H.J. Elsaesser, L.M. Snell, K. Hezaveh, W. Xu, and A. Kothari performed experiments. B.L. Macleod, R.J. Dickson, and M. Guo analyzed data. B.L. Macleod and D.G. Brooks wrote the manuscript. All authors reviewed the manuscript.
References
- Baldridge, M.T., Turula H., and Wobus C.E.. 2016. Norovirus regulation by host and microbe. Trends Mol. Med. 22:1047–1059. 10.1016/j.molmed.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandera, A., De Benedetto I., Bozzi G., and Gori A.. 2018. Altered gut microbiome composition in HIV infection: causes, effects and potential intervention. Curr. Opin. HIV AIDS. 13:73–80. 10.1097/COH.0000000000000429 [DOI] [PubMed] [Google Scholar]
- Bengsch, B., Ohtani T., Khan O., Setty M., Manne S., O’Brien S., Gherardini P.F., Herati R.S., Huang A.C., Chang K.M., et al. 2018. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity. 48:1029–1045.e5. 10.1016/j.immuni.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels, L.B., Neyrinck A.M., Loumaye A., Catry E., Walgrave H., Cherbuy C., Leclercq S., Van Hul M., Plovier H., Pachikian B., et al. 2018. Increased gut permeability in cancer cachexia: mechanisms and clinical relevance. Oncotarget. 9:18224–18238. 10.18632/oncotarget.24804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley, J.M., Price D.A., and Douek D.C.. 2006. HIV disease: fallout from a mucosal catastrophe? Nat. Immunol. 7:235–239. 10.1038/ni1316 [DOI] [PubMed] [Google Scholar]
- Brooks, D.G., McGavern D.B., and Oldstone M.B.A.. 2006. Reprogramming of antiviral T cells prevents inactivation and restores T cell activity during persistent viral infection. J. Clin. Invest. 116:1675–1685. 10.1172/JCI26856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188:4866–4875. 10.4049/jimmunol.1200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, C.R., Champhekar A., Tullius M.V., Dillon B.J., Zhen A., de la Fuente J.R., Herskovitz J., Elsaesser H., Snell L.M., Wilson E.B., et al. 2016. Type I and type II interferon coordinately regulate suppressive dendritic cell fate and function during viral persistence. PLoS Pathog. 12. e1005356. 10.1371/journal.ppat.1005356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Fonseca, D.M., Hand T.W., Han S.J., Gerner M.Y., Glatman Zaretsky A., Byrd A.L., Harrison O.J., Ortiz A.M., Quinones M., Trinchieri G., et al. 2015. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell. 163:354–366. 10.1016/j.cell.2015.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh, G., and Greene W.C.. 2016. Dissecting how CD4 T cells are lost during HIV infection. Cell Host Microbe. 19:280–291. 10.1016/j.chom.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, G.P., Lee S.M., and Mazmanian S.K.. 2016. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14:20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs, J.H.. 2010. The gastrointestinal tract and HIV pathogenesis. South Afr. J. Clin. Nutr. 23(sup1, Suppl 1):S65–S68. 10.1080/16070658.2010.11734275 [DOI] [Google Scholar]
- Dubourg, G.. 2016. Impact of HIV on the human gut microbiota: challenges and perspectives. Hum. Microb. J. 2:3–9. 10.1016/j.humic.2016.10.001 [DOI] [Google Scholar]
- Edgar, R.C.. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 10:996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Estes, J.D., Kityo C., Ssali F., Swainson L., Makamdop K.N., Del Prete G.Q., Deeks S.G., Luciw P.A., Chipman J.G., Beilman G.J., et al. 2017. Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 23:1271–1276. 10.1038/nm.4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey, L.M., Wilson E.B., Elsaesser H., Fistonich C.D., McGavern D.B., and Brooks D.G.. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 208:987–999. 10.1084/jem.20101773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria, A.M.C., Reis B.S., and Mucida D.. 2017. Tissue adaptation: Implications for gut immunity and tolerance. J. Exp. Med. 214:1211–1226. 10.1084/jem.20162014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, M., and Lohoff M.. 2014. IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 44:1886–1895. 10.1002/eji.201344279 [DOI] [PubMed] [Google Scholar]
- Im, S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., Shan Q., Hale J.S., Lee J., Nasti T.H., et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537:417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelt, B., and Lipton H.L.. 2014. Enterovirus/picornavirus infections. Handb. Clin. Neurol. 123:379–416. 10.1016/B978-0-444-53488-0.00018-3 [DOI] [PubMed] [Google Scholar]
- Kelley, C.F., Barbour J.D., and Hecht F.M.. 2007. The relation between symptoms, viral load, and viral load set point in primary HIV infection. J. Acquir. Immune Defic. Syndr. 45:445–448. 10.1097/QAI.0b013e318074ef6e [DOI] [PubMed] [Google Scholar]
- Kmieć, Z., Cyman M., and Ślebioda T.J.. 2017. Cells of the innate and adaptive immunity and their interactions in inflammatory bowel disease. Adv. Med. Sci. 62:1–16. 10.1016/j.advms.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Kolumam, G.A., Thomas S., Thompson L.J., Sprent J., and Murali-Krishna K.. 2005. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202:637–650. 10.1084/jem.20050821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa, S., Kannanganat S., Nigam P., Siddiqui M., Shetty R.D., Armstrong W., Ansari A., Bosinger S.E., Silvestri G., and Amara R.R.. 2011. Plasmacytoid dendritic cells are recruited to the colorectum and contribute to immune activation during pathogenic SIV infection in rhesus macaques. Blood. 118:2763–2773. 10.1182/blood-2011-02-339515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahl, K., and Sparwasser T.. 2011. In vivo depletion of FoxP3+ Tregs using the DEREG mouse model. Methods Mol. Biol. 707:157–172. 10.1007/978-1-61737-979-6_10 [DOI] [PubMed] [Google Scholar]
- Lauterbach, H., Truong P., and McGavern D.B.. 2007. Clearance of an immunosuppressive virus from the CNS coincides with immune reanimation and diversification. Virol. J. 4:53. 10.1186/1743-422X-4-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, P., Guadalupe M., Donovan R., Hung J., Flamm J., Prindiville T., Sankaran-Walters S., Syvanen M., Wong J.K., George M.D., et al. 2011. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. J. Virol. 85:4772–4782. 10.1128/JVI.02409-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., et al. 2015. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 162:184–197. 10.1016/j.cell.2015.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A.G., Mendoza A., Hemmers S., Moltedo B., Niec R.E., Schizas M., Hoyos B.E., Putintseva E.V., Chaudhry A., Dikiy S., et al. 2017. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature. 546:421–425. 10.1038/nature22360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist, M., van Lunzen J., Soghoian D.Z., Kuhl B.D., Ranasinghe S., Kranias G., Flanders M.D., Cutler S., Yudanin N., Muller M.I., et al. 2012. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J. Clin. Invest. 122:3271–3280. 10.1172/JCI64314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka, M., and Spiegel S.. 2014. Sphingolipid metabolites in inflammatory disease. Nature. 510:58–67. 10.1038/nature13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie, P.J., and Holmes S.. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 8. e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot, J.M., Bigenwald C., Champiat S., Collins M., Carbonnel F., Postel-Vinay S., Berdelou A., Varga A., Bahleda R., Hollebecque A., et al. 2016. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur. J. Cancer. 54:139–148. 10.1016/j.ejca.2015.11.016 [DOI] [PubMed] [Google Scholar]
- Moir, S., and Fauci A.S.. 2009. B cells in HIV infection and disease. Nat. Rev. Immunol. 9:235–245. 10.1038/nri2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskophidis, D., Battegay M., van den Broek M., Laine E., Hoffmann-Rohrer U., and Zinkernagel R.M.. 1995. Role of virus and host variables in virus persistence or immunopathological disease caused by a non-cytolytic virus. J. Gen. Virol. 76:381–391. 10.1099/0022-1317-76-2-381 [DOI] [PubMed] [Google Scholar]
- Mowat, A.M., and Agace W.W.. 2014. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 14:667–685. 10.1038/nri3738 [DOI] [PubMed] [Google Scholar]
- Mueller, S.N., and Mackay L.K.. 2016. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- Nadeem, M.S., Kumar V., Al-Abbasi F.A., Kamal M.A., and Anwar F.. 2020. Risk of colorectal cancer in inflammatory bowel diseases. Semin. Cancer Biol. 64:51–60. 10.1016/j.semcancer.2019.05.001 [DOI] [PubMed] [Google Scholar]
- OʼHara, K.M., Pontrelli G., and Kunstel K.L.. 2017. An introduction to gastrointestinal tract CMV disease. JAAPA. 30:48–52. 10.1097/01.JAA.0000524712.40590.76 [DOI] [PubMed] [Google Scholar]
- Oldstone, M.B., and Campbell K.P.. 2011. Decoding arenavirus pathogenesis: essential roles for alpha-dystroglycan-virus interactions and the immune response. Virology. 411:170–179. 10.1016/j.virol.2010.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley, M.A., Kroy D.C., Odorizzi P.M., Johnnidis J.B., Dolfi D.V., Barnett B.E., Bikoff E.K., Robertson E.J., Lauer G.M., Reiner S.L., et al. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 338:1220–1225. 10.1126/science.1229620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster, P., Kamphorst A.O., Wieland A., Araki K., Iyer S.S., West E.E., O’Mara L., Yang S., Konieczny B.T., Sharpe A.H., et al. 2014. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J. Exp. Med. 211:1905–1918. 10.1084/jem.20132577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M.N., Dehal P.S., and Arkin A.P.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Covián, D., Ruas-Madiedo P., Margolles A., Gueimonde M., de Los Reyes-Gavilán C.G., and Salazar N.. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7:185. 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes, T., Flouri T., Nichols B., Quince C., and Mahé F.. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4. e2584. 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, L.M., and Brooks D.G.. 2015. New insights into type I interferon and the immunopathogenesis of persistent viral infections. Curr. Opin. Immunol. 34:91–98. 10.1016/j.coi.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, L.M., McGaha T.L., and Brooks D.G.. 2017. Type I interferon in chronic virus infection and cancer. Trends Immunol. 38:542–557. 10.1016/j.it.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, L.M., MacLeod B.L., Law J.C., Osokine I., Elsaesser H.J., Hezaveh K., Dickson R.J., Gavin M.A., Guidos C.J., McGaha T.L., et al. 2018. CD8+ T cell priming in established chronic viral infection preferentially directs differentiation of memory-like cells for sustained immunity. Immunity. 49:678–694.e5. 10.1016/j.immuni.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg, G.F.. 2014. Regulation of intestinal health and disease by innate lymphoid cells. Int. Immunol. 26:501–507. 10.1093/intimm/dxu052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert, E.M., Schenkel J.M., Fraser K.A., Beura L.K., Manlove L.S., Igyártó B.Z., Southern P.J., and Masopust D.. 2015. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 161:737–749. 10.1016/j.cell.2015.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelekati, E., Shin H., Doering T.A., Dolfi D.V., Ziegler C.G., Beiting D.P., Dawson L., Liboon J., Wolski D., Ali M.A.A., et al. 2014. Bystander chronic infection negatively impacts development of CD8(+) T cell memory. Immunity. 40:801–813. 10.1016/j.immuni.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullender, M.E., and Baldridge M.T.. 2018. Norovirus interactions with the commensal microbiota. PLoS Pathog. 14. e1007183. 10.1371/journal.ppat.1007183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, A., and Saito T.. 2017. CD4 CTL, a cytotoxic subset of CD4+ T cells, their differentiation and function. Front. Immunol. 8:194. 10.3389/fimmu.2017.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro, J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C.F., Welch M., Schreiber R.D., de la Torre J.C., and Oldstone M.B.A.. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 340:207–211. 10.1126/science.1235214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomov, V.T., Palko O., Lau C.W., Pattekar A., Sun Y., Tacheva R., Bengsch B., Manne S., Cosma G.L., Eisenlohr L.C., et al. 2017. Differentiation and protective capacity of virus-specific CD8+ T cells suggest murine norovirus persistence in an immune-privileged enteric niche. Immunity. 47:723–738.e5. 10.1016/j.immuni.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Garrity G.M., Tiedje J.M., and Cole J.R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, A.M., and Wraith D.C.. 2016. Tr1-like T cells — an enigmatic regulatory T cell lineage. Front. Immunol. 7:355. 10.3389/fimmu.2016.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B.J., Karp C.L., and Brooks D.G.. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 340:202–207. 10.1126/science.1235208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, J., Gkrania-Klotsas E., and Kumararatne D.. 2017. Chronic norovirus infection and common variable immunodeficiency. Clin. Exp. Immunol. 188:363–370. 10.1111/cei.12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen, A., Rezek V., Youn C., Lam B., Chang N., Rick J., Carrillo M., Martin H., Kasparian S., Syed P., et al. 2017. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J. Clin. Invest. 127:260–268. 10.1172/JCI89488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists vendor details for each antibody.
Data Availability Statement
The RNA-seq data generated in this paper have been deposited in the Gene Expression Omnibus database under accession number GSE142303.