The APOBEC mutation signature is abundant in cancer, but its relationship to the multistep process of carcinogenesis is unclear. Studies by Law and colleagues demonstrate that APOBEC3A catalyzes mutagenesis and promotes tumorigenesis in two different murine systems in vivo.
Abstract
The APOBEC3 family of antiviral DNA cytosine deaminases is implicated as the second largest source of mutation in cancer. This mutational process may be a causal driver or inconsequential passenger to the overall tumor phenotype. We show that human APOBEC3A expression in murine colon and liver tissues increases tumorigenesis. All other APOBEC3 family members, including APOBEC3B, fail to promote liver tumor formation. Tumor DNA sequences from APOBEC3A-expressing animals display hallmark APOBEC signature mutations in TCA/T motifs. Bioinformatic comparisons of the observed APOBEC3A mutation signature in murine tumors, previously reported APOBEC3A and APOBEC3B mutation signatures in yeast, and reanalyzed APOBEC mutation signatures in human tumor datasets support cause-and-effect relationships for APOBEC3A-catalyzed deamination and mutagenesis in driving multiple human cancers.
Graphical Abstract
Introduction
DNA damage and mutation are fundamental hallmarks of cancer (Hanahan and Weinberg, 2011). Most tumor types show phenotypic and genotypic heterogeneity attributable in large part to different mutational events accumulating throughout the existence of each tumor. The tumor mutation landscape is due to a combination of endogenous and exogenous sources with marked differences between cancer types. Knowledge of the intrinsic (bio)chemical preferences of mutagens, together with more recent advances in DNA sequencing and bioinformatics, has made it possible to break complex mutation landscapes into simpler individual components (reviewed by Helleday et al., 2014; Roberts and Gordenin, 2014; Venkatesan et al., 2018). In some cases, cause-and-effect relationships may be inferred because a given mutation spectrum or signature is only attributable to a single mutagen. The mutation landscape of melanoma, for instance, is often dominated by a UV light mutation signature, C-to-T mutations in dipyrimidine motifs due to UV-induced nucleobase cross-linking, and adenine insertion opposite cross-linked lesions during DNA replication. In comparison, cytosine-to-thymine (C-to-T) mutations in CG dinucleotide motifs in many different cancer types are attributable to water-mediated deamination of methylated cytosines followed by DNA replication. The former process is avoidable and only accrues with UV light exposure, whereas the latter is unavoidable and accumulates stochastically over time (thus associated with biological age and referred to as an aging signature; Alexandrov et al., 2015).
Another large source of C-to-T mutations in the genomes of many different tumor types is apolipoprotein B mRNA editing catalytic subunit-like (APOBEC3 [A3])–catalyzed DNA cytosine deamination (Alexandrov et al., 2013; Burns et al., 2013a, 2013b; Nik-Zainal et al., 2012; Roberts et al., 2013). Human cells have the potential to express up to seven different A3 family DNA cytosine deaminases (A3A–A3D and A3F–A3H). These enzymes function normally as an integral arm of the innate immune response to viral infections (reviewed by Harris and Dudley, 2015; Simon et al., 2015). However, A3-mediated deamination of genomic cytosines to uracils (C-to-U) can lead directly through DNA replication or indirectly through uracil excision repair and DNA replication to C-to-T mutations. In comparison to the aforementioned signatures, the primary APOBEC mutation signature is defined by C-to-T mutations in TC dinucleotides followed by adenine or thymine nucleobases to distinguish it from the aging signature (i.e., TCA and TCT motifs). A secondary APOBEC mutation signature is cytosine-to-guanine (C-to-G) transversion in the same trinucleotide motifs, which is likely due to REV1-catalyzed cytosine insertion opposite abasic lesions created by the combined action of cytosine deamination and uracil excision (Chan et al., 2013). The primary and secondary APOBEC mutation signatures are also called SBS2 and SBS13, respectively (Alexandrov et al., 2013). The overall APOBEC mutation signature is evident in over half of all cancer types and, importantly, often dominates the mutation landscape of cervical, bladder, breast, head/neck, and lung tumors (reviewed by Helleday et al., 2014; Roberts and Gordenin, 2014; Venkatesan et al., 2018).
A major factor hindering broad appreciation of the APOBEC mutation process and efforts for clinical translation is a clear mechanistic determination of whether it is a causal driver or an inconsequential passenger in the overall tumor phenotype (drivers being potentially actionable and passengers being biomarkers at best). In support of a driver mechanism, first, individual A3 enzymes as well as the overall APOBEC mutation signature have been associated with poor clinical outcomes, including drug resistance and metastasis (Cescon et al., 2015; Chen et al., 2017; Glaser et al., 2017; Law et al., 2016; Sieuwerts et al., 2014, 2017; Walker et al., 2015; Xu et al., 2015; Yan et al., 2016). Second, a handful of bona fide driver mutations occur in APOBEC signature motifs, most prominently TCA to TTA mutations resulting in E542K and E545K changes in PIK3CA (Angus et al., 2019; Bertucci et al., 2019; Cannataro et al., 2019; Faden et al., 2017; Gillison et al., 2019; Henderson et al., 2014; Lefebvre et al., 2016). Third, high-risk human papillomavirus (HPV) infections cause up-regulation of A3A and A3B gene expression, and HPV-positive tumors have stronger APOBEC mutation signatures than virus-negative tumors (Henderson et al., 2014; Mori et al., 2015, 2017; Vieira et al., 2014; Warren et al., 2015). In contrast and in support of a passenger model, first, A3A-mediated deamination of the single-stranded loop region of stem-loop structures (a mesoscale genomic feature) almost exclusively occurs in passenger genes (Buisson et al., 2019). Second, tumor genomes can contain hundreds to thousands of APOBEC signature mutations, and almost all are unique to different patients, implying little or no selective advantage (Angus et al., 2019; Bertucci et al., 2019; Nik-Zainal et al., 2016). Third, A3A and A3B expression is induced by interferon and inflammatory signals, respectively, suggesting up-regulation may be a consequence (not a cause) of other events in the overall tumor microenvironment (Koning et al., 2011; Leonard et al., 2015; Lucifora et al., 2014; Maruyama et al., 2016; Refsland et al., 2010; Siriwardena et al., 2018; Stenglein et al., 2010; Thielen et al., 2010). Fourth, the driver versus passenger question is complicated by the likelihood that these need not be mutually exclusive processes such that driver mutations could be rare and/or individually weakly penetrant and therefore difficult to distinguish from large excesses of passenger mutations. Additionally, there are challenges assigning cause-and-effect relationships for which an A3 enzyme or enzymes is the causal source of the APOBEC mutation signature in human cancer, with A3A, A3B, and A3H as leading candidates (e.g., Buisson et al., 2019; Burns et al., 2013a, 2013b; Caval et al., 2014, 2015; Chan et al., 2015; Cortez et al., 2019; de Bruin et al., 2014; Middlebrooks et al., 2016; Nik-Zainal et al., 2014; Petljak et al., 2019; Serebrenik et al., 2019; Starrett et al., 2016; Yamazaki et al., 2019).
Here, two different models for tumorigenesis in mice are used to distinguish between driver and passenger roles for A3-catalyzed genomic DNA deamination in cancer. In the first system, the adenomatous polyposis coli multiple intestinal neoplasia (ApcMin) model for colorectal carcinogenesis (Moser et al., 1990; Munteanu and Mastalier, 2014), transgenic expression of human A3A, but not A3G, causes elevated frequencies of polyp formation and inflicts C-to-T mutations in APOBEC signature trinucleotide motifs. In the second system, the fumaryl-acetoacetate hydrolase (Fah) model for hepatocellular carcinoma (Keng et al., 2009; Wangensteen et al., 2008), all seven human A3 enzymes are tested individually, including A3A, A3B, and A3H. However, only human A3A is able to significantly elevate hepatocellular tumor frequencies above control levels. In addition, A3A inflicts both primary (C-to-T) and secondary (C-to-G) single-base substitution (SBS) mutations in APOBEC signature trinucleotide motifs, indicating Rev1 functionality in hepatocytes during the transformation process. Additional bioinformatic analyses strongly indicate that the A3A mutation signature induced in these murine cancer models closely resembles the overall APOBEC mutation signature in several different human tumors. These studies combine to demonstrate that A3A is capable of catalyzing mutation and actively driving carcinogenesis in vivo and that a significant proportion of the overall APOBEC mutation signature in human cancer is attributable to this enzyme.
Results
Human A3–expressing mice
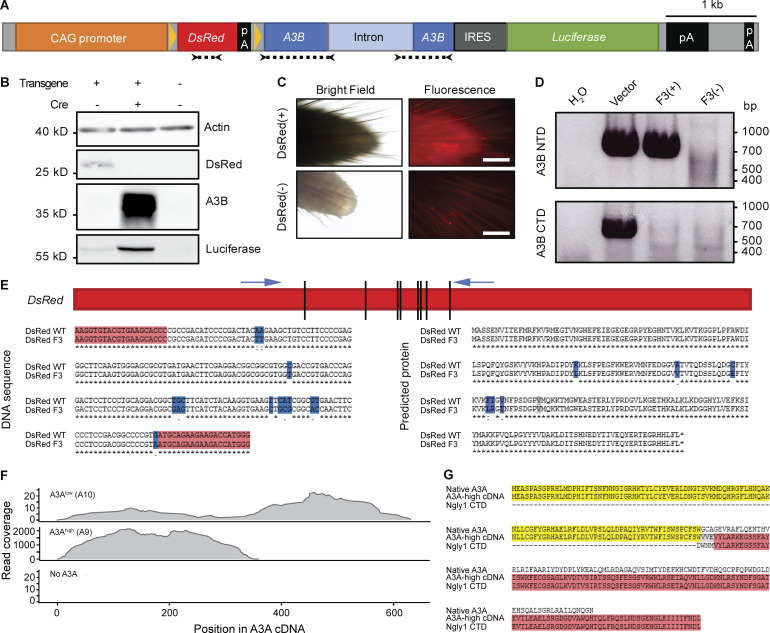
In contrast to humans with seven different A3 enzymes, mice encode only a single orthologous APOBEC3 protein that manifests lower catalytic activity and localizes to the cytoplasmic compartment (MacMillan et al., 2013). Accordingly, APOBEC signature mutations have not been reported in any murine cancer model. Two strategies were used to create animals capable of expressing human APOBEC enzymes. The first was conditional expression of A3B from a multicopy transgene (Fig. S1, A and B). This approach appeared to work initially, as evidenced by at least two generations of pups visibly expressing a linked DsRed marker (Fig. S1 C). However, both A3B functionality and DsRed expression were lost within three generations, potentially due to leaky deaminase expression from the transgene itself. Molecular analyses revealed that the A3B minigene was truncated and functionally inactivated and the linked DsRed cassette had accumulated 13 de novo base substitution mutations (Fig. S1, D and E).
Figure S1.
Cautionary tales from a conditional A3B transgenic line and an independent constitutive A3A transgenic line (A9). (A) Schematic of A3B transgene. A floxed DsRed gene is under the control of the CAG promoter and blocks transcription of an A3B minigene. Cre expression excises DsRed and allows expression of both A3B and firefly luciferase. Arrows and dotted lines indicate locations of primers used in D and E. (B) Immunoblots from HEK-293T cells either untransfected or transfected with the transgene plasmid described in A, with or without the addition of a Cre expression plasmid. (C) Representative images of DsRed expression in tail tips of transgenic mice compared with no fluorescence in nontransgenic mice (scale bars = 0.5 mm). (D) Genotyping PCR results for representative third generation animals. Mouse F3(+) was PCR positive for the 5′ end of A3B (encoding the N-terminal domain) and negative for the 3′ end of A3B (encoding the C-terminal domain). This animal also failed to show fluorescence and the DsRed cassette was later found to harbor 13-point mutations (E). Mouse F3(−) was a transgene-negative littermate. Transgene vector DNA diluted in HEK-293T genomic DNA was used as a positive control for PCR amplification. (E) Mutations detected in DsRed fragment in F3(+) mouse described in D. These 13-point mutations are predicted to change six amino acids (blue). (F) Read coverage across A3A cDNA from RNA sequencing of polyps from representative A3Alow (A10) ApcMin, A3Ahigh (A9) ApcMin, and ApcMin mice. (G) Predicted protein encoded by A3A-Ngly1 fusion in A3Ahigh (A9) transgenic animals. Amino acid prediction based upon results of 3′ RACE and Sanger sequencing.
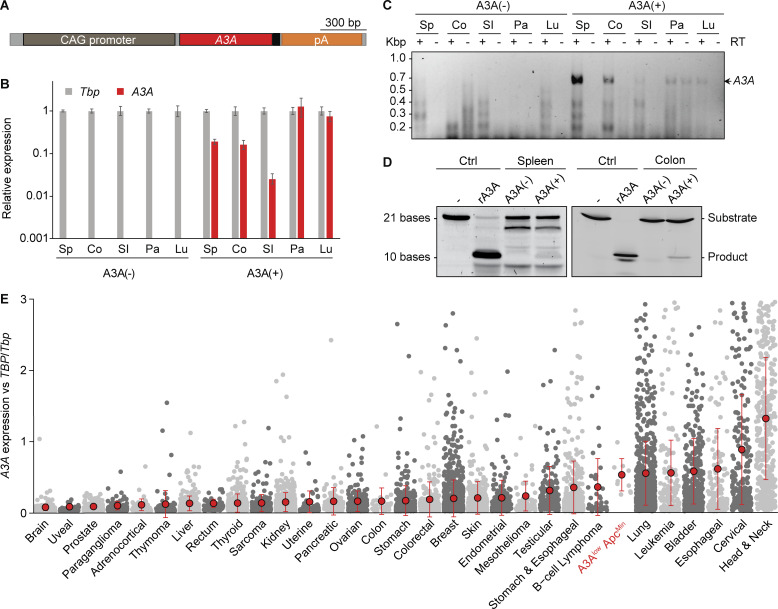
The second approach was standard transgenesis, as reported initially in the context of antiviral studies (Stavrou et al., 2014). After 10 blastocyst injections and transgenesis attempts with a CMV immediate enhancer/β-actin (CAG) promoter–driven A3A cDNA expression construct, two independent A3A lines were established: A3Ahigh with high levels of expression and A3Alow with lower levels (Fig. 1 A; Stavrou et al., 2014). However, similar to what occurred with the A3B transgenic mice, after multiple generations of breeding, we discovered through RNA sequencing that the A3Ahigh strain lost the C-terminal half of the A3A gene (Fig. S1 F). Sanger sequencing of cDNA generated from the A3Ahigh line revealed a deletion that fused A3A in-frame to the endogenous Ngly1 gene (Fig. S1 G). The encoded chimeric protein is unlikely to be a functional DNA deaminase due to loss of essential A3A structural elements (Bohn et al., 2015; Kouno et al., 2017; Shi et al., 2017). In contrast, A3A expression in the A3Alow animals was confirmed in multiple tissues, including the small intestine and colon by RT-PCR and quantitative RT-PCR, as well as by enzymatic activity in spleen and colon extracts using a modified single-stranded DNA deamination assay (Fig. 1, B–D). To compare A3Alow expression levels in mice to those in humans, mRNA expression was normalized to the housekeeping gene encoding TATA-binding protein and shown to be within the range observed across human cancers (Fig. 1 E). However, despite clear mRNA expression, the A3Alow protein did not accumulate to high enough levels to detect by immunoblotting or immunohistochemistry (IHC) using a custom mAb and validated protocols (Brown et al., 2019). A3Alow-expressing animals showed no overt cancer or other disease phenotypes, normal life spans, and normal Mendelian inheritance in three different animal facilities. These observations indicated that low levels of A3A are alone insufficient for tumorigenesis.
Figure 1.
A3A expression and activity in vivo. (A) Transgene schematic with A3A in red and myc epitope in black. (B) Quantitative RT-PCR of A3A mRNA levels relative to those of the housekeeping gene Tbp (mean ± SD for three technical replicates; Co, colon; Lu, lung; Pa, pancreas; SI, small intestine; Sp, spleen). (C) Agarose gel image of full-length A3A cDNA amplified by RT-PCR from the indicated tissues. Parallel reactions excluding RT show minimal genomic DNA contamination (only pancreas). (D) Deamination activity in A3A(−) and A3A(+) spleen and intestine extracts. Negative control is substrate oligo alone. Positive control is A3A(−) lysate spiked with 1 nM recombinant (r)A3A. (E) A3A mRNA levels in polyps from A3Alow (A10) ApcMin animals are within the range reported for human tumors (gray dots represent data from individual TCGA tumors or A3Alow (A10) ApcMin polyps; a small number of outlying TCGA data points, where A3A/TBP values were higher than three, were omitted for clarity; mean ± SD shown by red dots and error bars, respectively). Kbp, kilo basepairs.
Human A3A promotes intestinal tumor formation
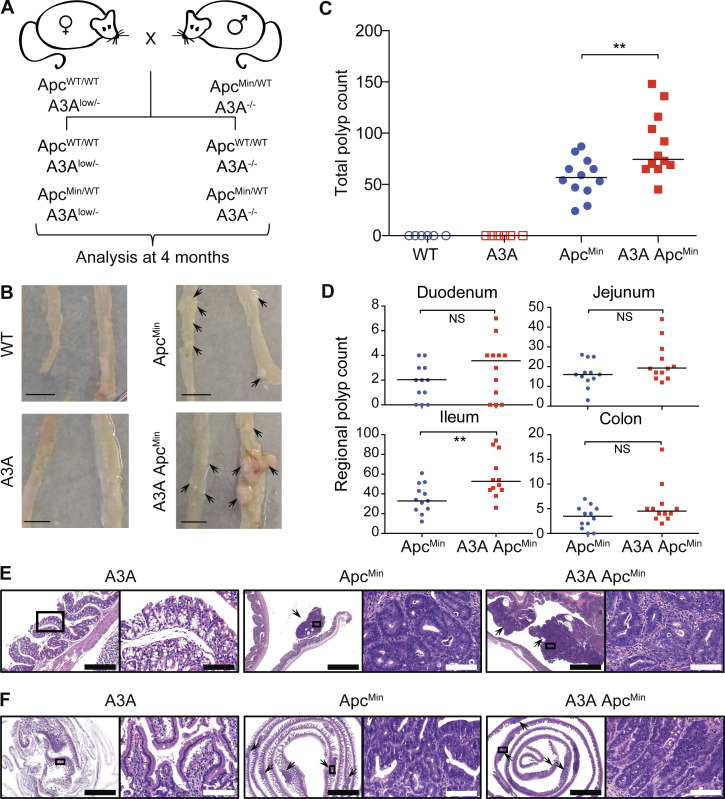
At least one initial predisposing event is likely to be required for the APOBEC3 mutagenesis program to become active and influence tumor evolution (Olson et al., 2018; Siriwardena et al., 2016; Swanton et al., 2015; Venkatesan et al., 2018). Standard breeding was therefore done to combine A3Alow (hereafter A3A) with ApcMin, which is a well-established model for intestinal tumor development (Fig. 2 A; Moser et al., 1990; Munteanu and Mastalier, 2014). This system was chosen due to high penetrance (all animals develop polyps), the quantifiable nature of polyp formation (dozens per animal), and a clearly defined analysis endpoint (sacrifice at 4 mo of age before inevitable intestinal tract blockage and death). Polyp formation in A3A ApcMin and ApcMin littermates derived from the same crosses was analyzed at 4 mo, alongside tissues from A3A and WT animals as controls. In one series of experimental crosses, two of the A3A ApcMin animals died due to intestinal blockage before the 4-mo endpoint. However, the remaining 12 animals made it to the endpoint for detailed histological and molecular analyses. The impact of A3A was immediately apparent, with a 50% increase in the median number of visible polyps in comparison to animals with ApcMin alone (Fig. 2, B and C). Polyp formation was distributed unevenly through the intestinal tract, with the greatest difference observed in the ileum (Fig. 2 D). Histopathologic examination of H&E-stained polyps in the colon and small intestine revealed multiple, exophytic, pedunculated, or sessile growths exhibiting adenomatous features; neoplastic cells were characterized by columnar or cuboidal shape and ovoid, occasionally hyperchromatic nuclei, and they formed tubular structures (Fig. 2, E and F). High-grade cytological atypia with nuclear pleomorphism, increased mitotic figures (2–5 per high-power field) and tubular necrosis were observed in adenomas of both groups. The average size of the lesions did not differ significantly between A3A ApcMin and ApcMin mice. No polyps were evident in A3A or WT littermates analyzed in parallel (Fig. 2, B and C). A second series of crosses at a different animal facility yielded similar A3A-dependent increases in polyp formation and histological features (Fig. S2, A and B). Parallel experiments with an A3G transgenic yielded no increases in polyp formation, which helped to rule-out indirect explanations for the A3A tumor phenotype (Fig. S2 A).
Figure 2.
Human A3A promotes tumorigenesis in ApcMin mice. (A) Breeding schematic and analysis timeline. (B) Representative images of small intestine (left) and distal colon (right) from WT, A3A, ApcMin, and A3A ApcMin mice (scale bars = 1 cm; arrowheads indicate polyp locations). (C) Total polyp counts in the indicated groups (n = 12 for ApcMin and A3A ApcMin and n = 6 for WT and A3A; medians shown by horizontal lines). No polyps were seen in WT or A3A mice (**, P < 0.01 by two-sided Wilcoxon rank sum test). (D) Polyp counts for the indicated intestinal regions of ApcMin and A3A ApcMin mice (subdivision of data in C (**, P < 0.01 by two-sided Wilcoxon rank sum test). (E and F) Representative low (2–10×) and high (40×) power photomicrographs of indicated H&E-stained small intestine and colon sections (arrowheads point to polyps). Right images (scale bar = 100 µm) are magnifications of the boxed regions on the left (scale bar = 2 mm, except leftmost image in E [scale bar = 400 µm]). A3A and WT H&E images are indistinguishable, and only the former are shown.
Figure S2.
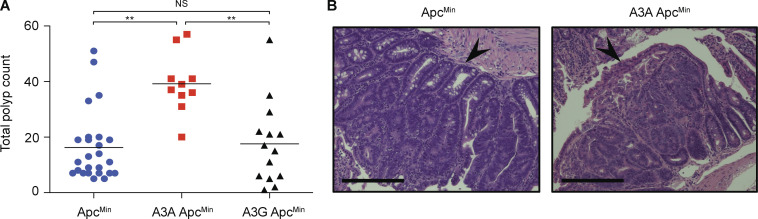
A3A ApcMin mice develop more tumors than ApcMin mice. (A) Numbers of intestinal polyps in ApcMin mice with no transgene, the A3Alow transgene, or an A3G transgene. No polyps were evident in WT animals (not shown). These experiments were performed in mouse facilities at the University of Pennsylvania independently of studies presented in Fig. 2 (**, P < 0.01 by Wilcoxon rank sum test). (B) Representative images of H&E-stained sections of small intestine from ApcMin and A3Alow ApcMin mice from the cohort in A. Arrowheads point to polyps. Scale bars = 400 µm.
Base substitution mutation signatures in human A3A expressing intestinal tumors
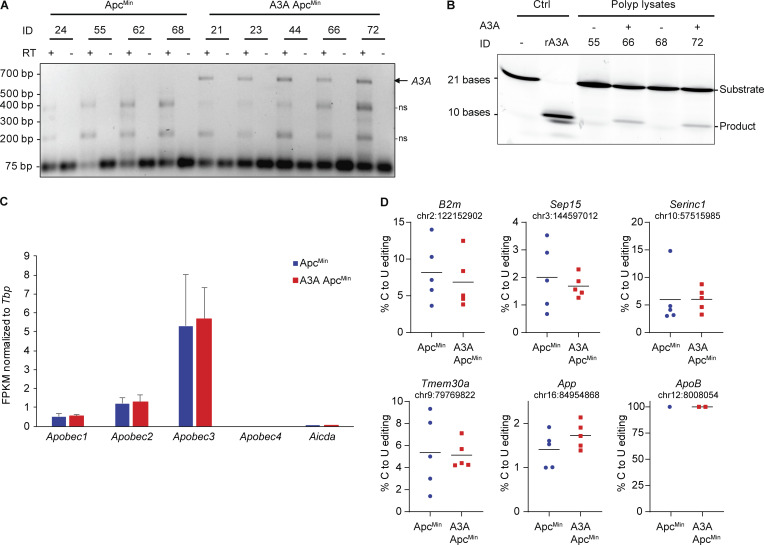
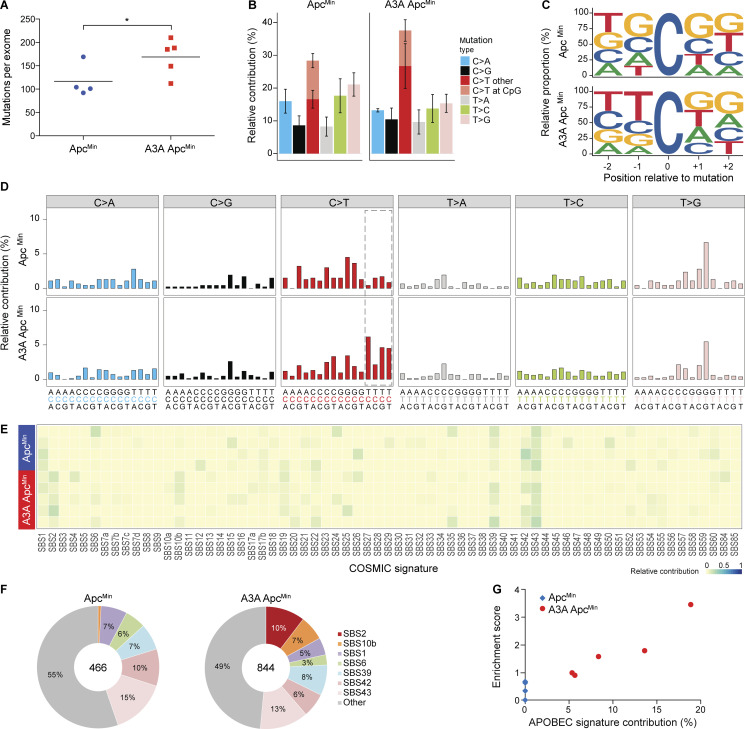
A3A mRNA expression was confirmed in individual polyps by RT-PCR and A3A protein expression by DNA deaminase activity assays (Fig. S3, A and B). Genomic DNA from polyps of A3A ApcMin and ApcMin animals was subjected to whole-exome sequencing (WES), together with matched normal tail DNA. SBS mutations present in the polyp exome profile but absent in the matched normal DNA were considered lesion-specific events. Interestingly, more mutations were detected on average in A3A-expressing tumors (Fig. 3 A). Analysis of the different SBS mutation types showed that this increase in A3A-expressing tumors is due largely to an elevated contribution of C-to-T mutations in non-CG motifs (Fig. 3 B). Evaluation of the local nucleobase contexts revealed that A3A-expressing polyps have increased C-to-T mutations within TC motifs (Fig. 3, C and D).
Figure S3.
RNA expression and editing in polyps from ApcMin and A3A ApcMin mice. (A) Agarose gel image of full-length A3A cDNA amplified from polyp mRNA from A3A ApcMin mice, but not from ApcMin animals. The smaller nonspecific (ns) bands indicate similar levels of RNA in each RT-PCR reaction. Neither the A3A-specific band nor the two smaller nonspecific bands were detected in reactions without RT (indicating no genomic DNA contamination). (B) Deamination activity in A3A(−) and A3A(+) intestinal polyps. Negative control is substrate oligo alone. Positive control is A3A(−) lysate spiked with 1 nM recombinant (r)A3A. (C) Expression levels of murine Apobec1, Apobec2, Apobec3, Apobec4, and Aicda (AID) in polyps derived from ApcMin and A3A ApcMin mice, as determined by quantifying RNA-sequencing data (n = 5 per group; mean ± SD FPKM relative to the housekeeping gene Tbp). (D) RNA C-to-U editing of the indicated mRNA sites in ApcMin and A3A ApcMin polyps, as measured by quantification of RNA-sequencing data (n = 5 per group except for ApoB). The percent editing for individual tumors is shown as colored dots and mean values are indicated by horizontal lines (no significant differences were detected). ApoB is expressed poorly in polyps and, consequently, the canonical Apobec1-catalyzed editing event in this mRNA could only be quantified in three polyps.
Figure 3.
APOBEC signature mutations in polyps from human A3A–expressing animals. (A) Dot plot of total base substitution mutations per tumor exome in ApcMin versus A3A ApcMin animals (n = 4 and n = 5, respectively; mean shown by horizontal line; *, P < 0.05 by one-sided two-sample t test). (B) Bar plot showing the percentage of each type of base substitution mutation in the tumors from A (mean ± SD). C-to-T transitions are further partitioned as shown. (C) Sequence logos for all C-to-T mutations in ApcMin versus A3A ApcMin tumors (n = 4 and n = 5, respectively). (D) Trinucleotide mutation profiles for all base substitutions in ApcMin versus A3A ApcMin tumors (n = 4 and n = 5, respectively). Dashed box highlights the trinucleotide motifs that define the APOBEC mutation signature (SBS2). (E and F) Heatmap and pie charts summarizing the relative contribution of each mutation signature to the overall landscape of mutations in ApcMin versus A3A ApcMin tumors (n = 4 and n = 5, respectively). SBS2 (APOBEC) is only evident in A3A-expressing tumors. (G) Scatterplot of APOBEC signature contribution to TCW motif enrichment score (Pearson’s correlation = 0.94, P = 0.015). COSMIC, catalogue of somatic mutations in cancer.
The trinucleotide mutational spectra of each tumor were assigned in relation to the current list of mutational signatures (Alexandrov et al., 2020). Strikingly, each of the A3A-expressing tumors showed clear evidence for SBS2 (APOBEC), ranging from 5–19% of total mutations, though no SBS13 was observed (Fig. 3, E and F). Control tumors lacked both SBS2 and SBS13. Enrichment scores for mutations within TCW motifs were elevated significantly in A3A-expressing tumors in relation to control tumors (t = −2.73, P = 0.043, two-sided two-sample t test) and also associated strongly with APOBEC signature contribution (Fig. 3 G). The aging signature (SBS1) was weak in polyps, likely due to the relatively young age of the animals. In further support of APOBEC signature exclusivity in A3A ApcMin tumors, a de novo deconstruction of the trinucleotide mutation data using nonnegative matrix factorization (Alexandrov et al., 2013) also led to a pattern strongly resembling SBS2 (APOBEC) in the A3A ApcMin tumors, but not the ApcMin tumors (data not shown). An even stronger SBS2 (APOBEC) emerged from analyses of whole-genome sequences generated using the same polyp genomic DNA, and again no SBS13 was detected (Fig. 4).
Figure 4.
APOBEC signature mutations in polyps from human A3A–expressing animals derived from whole-genome sequencing (WGS). (A) Composite trinucleotide mutation profiles for all base substitutions from WGS of ApcMin and A3A ApcMin tumors (n = 5 per group). The dashed box highlights the trinucleotide motifs that define the primary APOBEC mutation signature (SBS2). (B and C) Heatmap and pie charts summarizing the relative contribution of each mutation signature to the observed constellation of mutations in ApcMin versus A3A ApcMin tumor WGS (n = 5 per group). SBS2 (APOBEC) is only evident in A3A-expressing tumors. (D) Scatterplot of APOBEC signature contribution to APOBEC enrichment score in TCW motifs (Pearson’s correlation = 0.95, P = 2.98 × 10−5).
As additional controls, polyp RNA-sequencing data were used to evaluate expression levels of endogenous Apobec family members, as well as RNA editing levels for reported Apobec1 target sites (Blanc et al., 2014). Endogenous Apobec family member mRNA levels were similar between A3A-expressing and nonexpressing polyps (Fig. S3 C), making it unlikely that one of these enzymes is responsible for the exclusivity of SBS2 in A3A-expressing polyps. Moreover, Apobec1-catalyzed mRNA editing frequencies were also similar between ApcMin and A3A ApcMin polyps, indicating that the transgenic expression of human A3A is unlikely to be interfering with murine Apobec1 functionality (Fig. S3 D).
Mutational events including Apc loss of heterozygosity (LOH) in polyp formation
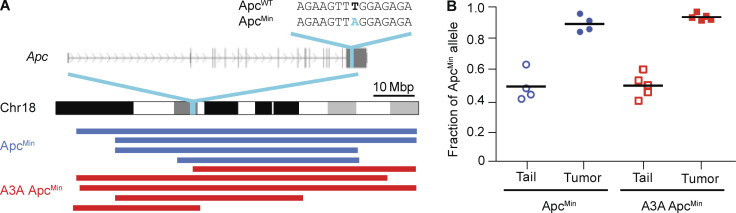
Additional mutation analyses showed that 21–30% of point mutations in each tumor were nonsynonymous. Whereas a variety of different genes were mutated, known intestinal adenocarcinoma drivers (Munteanu and Mastalier, 2014) such as Tp53, Pten, K-Ras, Tgfbr2, and Smad4 were unchanged. This is consistent with the pathological classification of the polyps as adenomas that have yet to undergo invasive transformation. However, all of the sequenced tumors for both ApcMin and A3A ApcMin showed LOH for the WT Apc allele (i.e., the thymine-to-adenine [T-to-A] base substitution mutation that defines the Min allele became homozygous; Fig. S4, A and B). Additional single-nucleotide polymorphism analyses demonstrated coincident LOH for large contiguous sections of chromosome 18 (Fig. S4 A). Apc inactivation is therefore likely to be essential for polyp formation in this system, consistent with a prior report (Starr et al., 2011). The only other recurrently mutated genes in A3A ApcMin polyps were Eif4h, Plxnd1, Rasa4, Sipa1l3, Sugp1, and Ttn (heterozygous base substitution mutations in each gene in two out of five polyps), which are not canonical intestinal cancer driver genes or part of a unified pathway. Increased polyp numbers in A3A ApcMin animals may be due therefore to a subset of A3A-catalyzed uracil lesions being processed into DNA breaks that contribute to gene conversion or break-induced replication, but additional studies will be needed to unambiguously establish mechanism (see Discussion).
Figure S4.
Apc LOH in colorectal tumors. (A) Schematic of mouse chromosome 18 highlighting the Apc gene and the ApcMin mutation. Blue and red tracts below indicate continuous regions of homozygosity (LOH) based on single-nucleotide polymorphism analysis. These tracts all include the ApcMin mutation. Mbp, mega basepair. (B) Dot plot showing the ApcMin allele frequency (chr18:34312601 T>A) in normal tail and tumor genomic DNA from ApcMin and A3A ApcMin mice. Horizontal bars represent mean values. The residual ApcWT signal in tumors is likely due to contamination by normal cells.
Human A3A promotes liver carcinogenesis
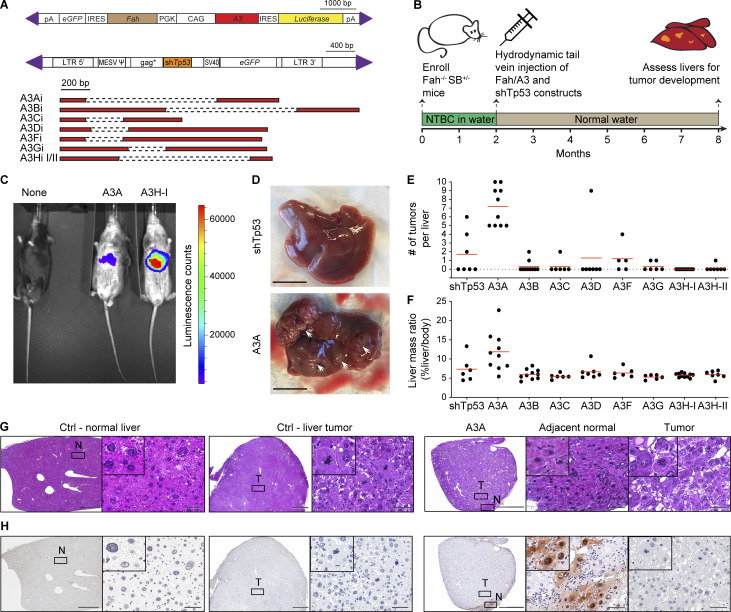
We next turned to the Fah model for liver regeneration and hepatocellular carcinoma (Grompe et al., 1998; Keng et al., 2009; Wangensteen et al., 2008), which offered the distinct advantage of enabling parallel tests of all seven human A3 enzymes. Fah-null animals are defective in tyrosine metabolism, and liver failure results from the toxic buildup of the primary Fah substrate fumaryl-acetoacetate and its precursor maleyl-acetoacetate. However, if supplemented with the drug 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC), which inhibits an enzyme upstream in the same anabolic pathway, these toxic metabolites fail to accumulate, and the liver functions normally. The essential nature of the Fah enzyme and the regenerative capacity of the liver enables genetic complementation of the deficiency upon delivery of a functional Fah transgene and withdrawal of NTBC to select for liver repopulation by complemented cells (Grompe et al., 1998). We therefore leveraged these properties and constructed a set of Fah complementation vectors for delivery by hydrodynamic transfer and heritable transmission by Sleeping Beauty (SB)–catalyzed integration into genomic DNA (schematics in Fig. 5 A). The primary SB payload has a PGK promoter driving Fah expression and a CAG promoter for direct expression of individual human A3 family members and IRES-mediated expression of luciferase. A secondary SB payload expresses a short hairpin RNA against Tp53, which promotes tumor development (Keng et al., 2009; Wangensteen et al., 2008). These genetic payloads are delivered simultaneously to the genomes of hepatocytes through SB11-mediated transposition (Keng et al., 2009; Wangensteen et al., 2008).
Figure 5.
A3A causes hepatocellular carcinogenesis in Fah liver regeneration model. (A) Schematics of Fah-A3 and shTp53 constructs injected into Fah-null SB11 animals. Purple triangles indicate IR/DR signal sequences required for transposition by SB11. Human A3 minigene schematics are depicted below with dashed lines representing introns. IRES, internal ribosome entry site; LTR, long terminal repeat; pA, poly-adenine; PGK, mouse phosphoglycerate kinase 1 promoter. (B) Overview of experimental workflow. (C) Luminescence of representative 7.5-mo-old liver-regenerated mice. (D) Representative images of livers regenerated with Fah-control (ctrl) and Fah-A3A constructs (scale bars = 1 cm; arrowheads point to tumors). (E and F) Tumor burdens and liver/body mass percentages for the indicated groups (n = 5–10 per group; mean values indicated by red lines; P < 0.0001 and P = 0.013, respectively, by one-way ANOVA). (G) Representative photomicrographs of H&E-stained sections of nonneoplastic (normal [N]) and tumorous (T) livers from Fah-control (ctrl) and Fah-A3A animals. Right images (scale bar = 60 µm) are magnifications of the boxed regions on the left (scale bar = 1 mm) and inset images are magnified an additional fourfold. (H) IHC staining of A3A in adjacent sections of the same tissues as G (scale bars are identical).
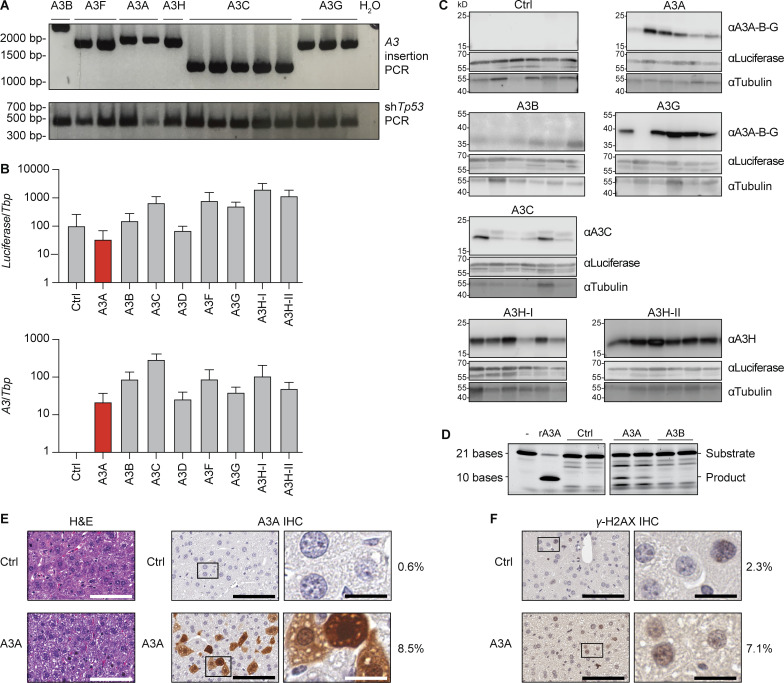
Fah-null SB11 animals were enrolled at birth and supplemented with NTBC drinking water and then, at 2 mo, provided with normal drinking water and randomized for hydrodynamic injections with the shTp53 vector and a control Fah vector (no A3) or an A3-expressing Fah vector (workflow in Fig. 5 B). Representative animals expressed luciferase in liver tissues in comparison to noninjected Fah-null SB11 animals (Fig. 5 C). At 8 mo of age (6 mo after hydrodynamic delivery), the entire cohort was sacrificed for comprehensive pathological and molecular analyses. The most visually striking result was uniformly higher tumor numbers in the livers of A3A-expressing animals (P < 0.0001 by one-way ANOVA; Fig. 5, D and E). The control (non-A3) and all other A3-expressing groups had statistically indistinguishable numbers of tumors (P value range = 0.18–0.99 by one-way ANOVA). Accordingly, livers in A3A-expressing group were an average of nearly 100% larger than those from the control or other A3-expressing groups (average liver mass of 12% body weight versus a range of 6–7% in other groups; P = 0.013 by one-way ANOVA; Fig. 5 F). The heritable integration and integrity of all A3 and shTp53 expression constructs was confirmed by diagnostic PCR assays of liver tissue genomic DNA (Fig. S5 A). Quantitative RT-PCR assays confirmed expression of the various A3 constructs at the RNA level (Fig. S5 B), and immunoblots were used to show expression of A3A, A3B, A3C, A3G, and A3H haplotypes I and II at the protein level (Fig. S5 C). Furthermore, single-stranded DNA deaminase activity assays were used to demonstrate activity of A3A and A3B in liver whole-cell extracts (Fig. S5 D). These results combined to indicate that only human A3A has the capacity to drive tumor formation in this system.
Figure S5.
A3 transposon integration and expression in livers of injected Fah−/− mice. (A) PCR amplicons from genomic DNA extracted from livers of mice hydrodynamically injected with the indicated A3 minigene constructs. This PCR assay uses common primers flanking each differently sized A3 minigene. (B) Quantitative RT-PCR results for Luciferase mRNA (identical primers for all inserts) or specific A3 mRNAs relative to those of the housekeeping gene Tbp in livers from hydrodynamically injected animals (mean ± SD across all mice injected with the control [Ctrl] or indicated A3 minigene construct). The control construct is identical apart from lacking an A3 minigene. See main text for additional details. (C) Representative immunoblots of liver protein extracts from animals of mice hydrodynamically injected with the indicated control or A3 minigene constructs. See main text for additional details. (D) Single-stranded DNA cytosine deamination activity of liver protein extracts from animals hydrodynamically injected with the indicated control or A3 minigene constructs. Deamination substrate oligo with no extract or with 1 nM purified recombinant (r)A3A are controls. A3A shows clear activity, and A3B shows weaker but still detectable activity. (E) H&E and A3A IHC staining of hepatocytes 48 h after hydrodynamic injection. A3A protein signal is cell-wide in ∼8.5% of cells (images and quantification are representative of two animals). H&E scale bars = 100 µm; A3A IHC image scale bars: left = 100 µm, right = 20 µm; A3A ApcMin image scale bars: left = 2 mm, right = 100 µm. (F) IHC staining of γ-H2AX in sections adjacent to those shown in E. The percentage of nuclei with positive staining is shown to the right (images and quantification are representative of two animals). Scale bars: left = 100 µm, right = 20 µm.
Fah tumor pathology and immunohistology were also informative. First, H&E-stained liver sections from A3A-expressing animals revealed tumorous growths with lobular architecture and marked histological heterogeneity in the neoplastic cell population. The cancer cells primarily featured a solid and/or trabecular growth pattern and showed size and shape variation, pronounced cytological atypia, nuclear pleomorphism, hyperchromatism, and increased numbers of mitotic figures. In addition, focal areas of necrosis, intracellular edema, apoptotic phenomena, and mild-to-moderate lymphocytic inflammation were seen (Fig. 5 G). Second, attempts to detect A3A at the protein level using a custom mAb and a validated IHC assay (Brown et al., 2019) revealed an additional layer of heterogeneity. In most instances, the bulk of the tumor itself showed limited/no evidence for A3A protein expression, whereas residual, adjacent to the tumor, nonneoplastic liver parenchyma often displayed aggregates of hepatocytes with moderate to strong nuclear and cytoplasmic staining (Fig. 5 H). Reconstruction experiments showed strong cell-wide A3A protein expression and elevated γ-H2AX staining in >5% of hepatocytes 48 h after hydrodynamic delivery (Fig. S5, E and F). These results are consistent with prior reports of both A3A localization in heterologous cell types (Caval et al., 2015; Chen et al., 2006; Hultquist et al., 2011; Land et al., 2013) and its capacity to exert DNA damage responses leading to cytotoxicity (Buisson et al., 2017; Caval et al., 2015; Landry et al., 2011; Stenglein et al., 2010). Thus, this remarkable heterogeneity suggests that, despite a strong positive selective pressure for Fah function, an even stronger selection against A3A may be occurring during liver tumorigenesis that results in A3A inactivation (see Discussion).
Base substitution mutation signatures in liver tumors attributable to A3A
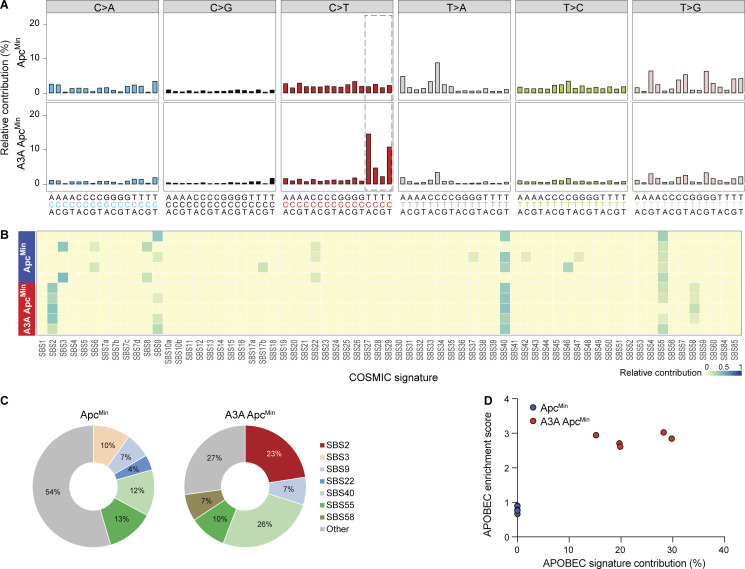
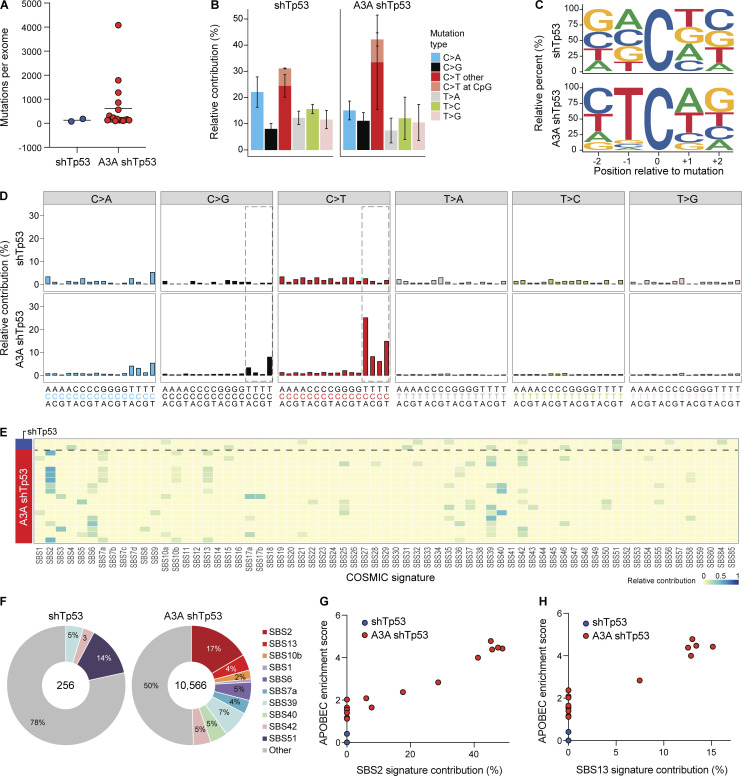
WES was done for 17 A3A shTp53 liver tumors from eight separate animals, as well as normal tissue (tail) from each animal to enable unambiguous mutation calls. Exomes from two independent shTp53 tumors and matching tail DNA were also sequenced for comparison. Mutation numbers ranged widely, even between tumors from the same animal, consistent with independent transformation events (n = 127–4,082; Fig. 6 A). The majority of base substitution mutations were C-to-T transitions in TC motifs preceded by a C or T at the −2 position and followed by an A or T at the +1 position (Fig. 6, B–D). As expected, many A3A shTp53 tumors showed a strong SBS2 (range = 0–46%). Tumors with no detectable SBS2 may have had the A3A minigene inactivated at early stages of tumor development (even before tumorigenesis). However, unlike the A3A ApcMin tumors above, several liver tumors also showed a clear SBS13 (range = 0–13%; Fig. 6, E–H; see Discussion). In contrast, liver tumors with shTp53 contained fewer mutations (n = 77, n = 179) and did not show evidence for SBS2 or SBS13 (Fig. 6). Finally, both SBS2 and SBS13 correlated positively with enrichment scores for cytosine mutation at TCW (Spearman’s rank correlation = 0.94, P = 2.49e-09; and Spearman’s rank correlation = 0.91, P = 6.38e-08, respectively; Fig. 6, G and H). These results demonstrated that A3A is capable of causing both APOBEC mutation signatures, SBS2 and SBS13, in vivo.
Figure 6.
APOBEC signature mutations in liver tumors from A3A Fah mice. (A) Dot plot of total base substitution mutations in shTp53 and A3A shTp53 liver tumor exomes (n = 2 and n = 17, respectively; mean values indicated by horizontal lines). (B) Bar plot showing the percentage of each type of base substitution mutation in the tumor groups from panel A (mean ± SD). C-to-T transitions are further partitioned as shown. (C) Logo representation of C-to-T mutation context in the tumor groups from A. (D) Composite trinucleotide mutation profiles in the tumor groups from A. Dashed boxes highlight the trinucleotide motifs that define the primary and secondary APOBEC mutation signatures (SBS2 and SBS13, respectively). (E and F) Heatmap and pie charts summarizing the relative contribution of each mutation signature to the overall landscape of mutations in the tumor groups from A. SBS2 and SBS13 (APOBEC) predominate over other signatures in A3A shTp53 tumor exomes. (G and H) Scatterplots of APOBEC enrichment scores versus SBS2 and SBS13 contributions (Pearson’s correlation = 0.96, P = 0.0002; Pearson’s correlation = 0.91, P = 0.002).
Murine and human tumor APOBEC mutation signature comparisons
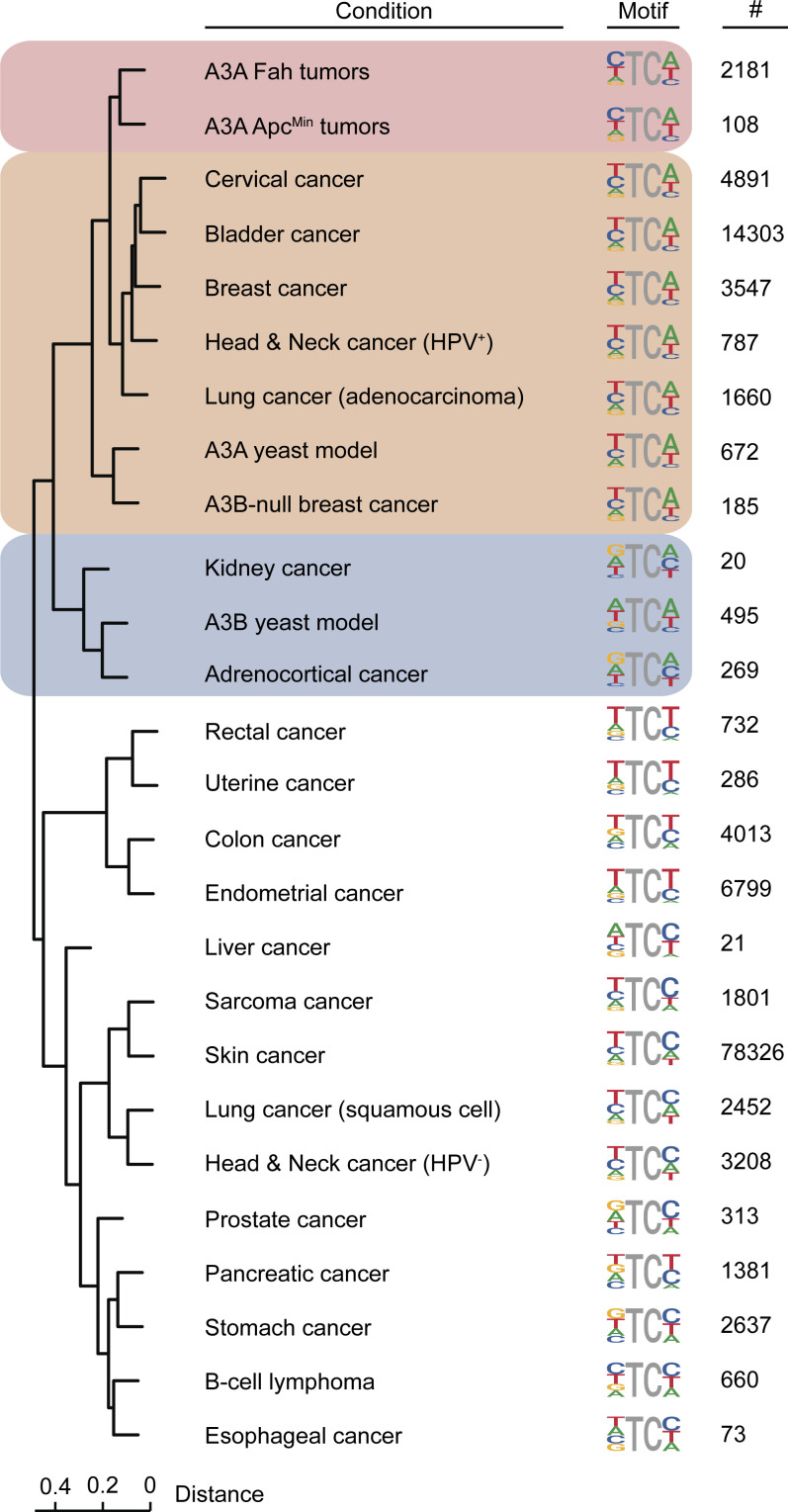
To compare the APOBEC mutation signatures observed in murine A3A tumor models and those occurring in human cancers, an unbiased Euclidean distance dendrogram was generated using the nucleobases surrounding the mutated cytosines in our murine colon and liver tumor datasets (above), The Cancer Genome Atlas (TCGA) human cancer datasets (Weinstein et al., 2013), and previously reported data for A3A and A3B mutation in yeast (Fig. 7; Chan et al., 2015). To minimize overlapping signals from other mutational processes, these analyses focused exclusively on TC-to-TT mutations in NTC (A/C/T) tetranucleotide motifs (i.e., CG motifs were excluded to avoid counting age-related spontaneous methyl-C deamination events). First, the A3A-induced signature in both the ApcMin and Fah tumors was YTCW, and these clustered tightly together at the top of the tree. Second, the A3A-induced signature in ApcMin and Fah tumors clustered adjacent to the tetranucleotide TC-to-TT mutation signatures of the largest APOBEC signature cancer types in humans (cervical, bladder, HPV-positive head/neck, and lung adenocarcinoma). Third, the tetranucleotide TC-to-TT mutation signatures from A3B-null breast cancers and A3A-expressing yeast clustered closely and as an outgroup of the aforementioned signatures. Fourth, the tetranucleotide TC-to-TT mutation signatures from A3B-expressing yeast clustered closely to those of human kidney and adrenocortical tumors but still within the APOBEC signature dominated half of the dendrogram. Last, the tetranucleotide TC-to-TT mutation signatures from all other human tumor types clustered in various ways in the other half of the dendrogram (largely non-APOBEC). These analyses provided further support to the idea that A3A is a driver of mutation in multiple human tumor types, including breast, cervix, bladder, HPV-positive head/neck, and lung.
Figure 7.
APOBEC mutation signature comparisons for murine and human tumors. Euclidian distance dendrogram depicting hierarchical relationships between TC-to-TT mutation signatures of human A3A in mouse tumors, human A3A and A3B in yeast, and the indicated human cancers. Each TC-focused tetranucleotide signature is shown next to the relevant branch tip along with the number of mutations contributing to each analysis. See text for additional details.
Discussion
The studies described here were performed with the goal of determining whether the APOBEC3 mutation process is a driver or a passenger in the overall tumorigenesis process. Prior to this, several lines of evidence have supported both models and, importantly, none have been definitive (see Introduction). Here, we showed that human A3A is able to drive tumorigenesis in two different murine systems. Low, constitutive levels of A3A from a stable transgene promoted polyp formation in the ApcMin model for colorectal cancer. Higher, likely transient levels of A3A from hydrodynamic delivery of a SB-transposable element caused hepatocellular carcinogenesis in a Tp53-depleted Fah liver regeneration system. In both systems, A3A inflicted a hallmark APOBEC3 mutation signature in the genomes of tumors. These studies are therefore the first cause-and-effect demonstrations in vivo that human A3A catalyzes mutagenesis and promotes tumorigenesis. The strong similarity of the A3A mutation signature in these murine tumor models and the overall APOBEC mutation signature in breast, cervical, bladder, HPV-positive head/neck, and lung cancers provided additional evidence for an active role of this DNA deaminase in human tumor development and evolution.
The results here combined support a model in which low/ongoing or high/transient levels of A3A-catalyzed genomic DNA deamination are procarcinogenic. A direct DNA deamination mechanism is favored over indirect mechanisms, because A3A caused clear tumor phenotypes in two different murine systems, single-stranded DNA cytosine deamination is the principle activity of this enzyme, and related DNA deaminases failed to promote carcinogenesis (A3G in the ApcMin system and A3B-D/F-H in the Fah system; discussed further below). However, like many human cancers with strong APOBEC3 signature mutations, no recurring driver mutations were evident in A3A-preferred TCW motifs in either tumor system. The only recurrent driver event that may have been triggered by A3A is Apc LOH, but, as reported in prior studies, similar LOH events were found in the control ApcMin group. An analysis of larger numbers of whole-tumor genomes may be necessary to reveal additional causal events. The integration position of the human A3A transgene is unlikely to contribute to the elevated ApcMin tumor burden, because it is a single-copy insertion on chromosome 9 located over 20 kb pairs from the nearest murine gene. Future studies should also be done to compare WT A3A and a catalytic mutant to formally demonstrate a requirement for deaminase activity and rule-out indirect models such as dysregulation of an endogenous murine DNA deaminase. However, the latter possibility is unlikely because murine Apobec1 expression and editing activity were not altered by human A3A expression in polyps. Future studies should also consider the possibility that part of the tumor phenotype may be epigenetic through A3A-catalyzed RNA C-to-U editing (though levels were too low to quantify here) and/or DNA methyl-C–to–T deamination (Carpenter et al., 2012; Schutsky et al., 2017; Sharma et al., 2015; Suspène et al., 2013). However, it should be noted that these alternative mechanisms are constrained biochemically, because the catalytic rates for RNA C and DNA methyl-C deamination are much lower than those for normal C-to-U editing in single-stranded DNA (Carpenter et al., 2012; Ito et al., 2017; Schutsky et al., 2017).
None of the other human A3 enzymes, including A3B, showed an ability to accelerate tumor development in the Tp53-depleted Fah liver regeneration system. A3G also tested negative in the ApcMin system but assessing the entire A3 family was not possible due to a lack of transgenics. There are several possible, potentially interconnected explanations for these negative results. One factor may be the relatively short 6-mo timeframe from hydrodynamic delivery to analysis. A second consideration is the fact that A3A is more active biochemically than the next most potent human A3 enzymes A3B and A3H (Adolph et al., 2017; Buisson et al., 2019; Ito et al., 2017). A third consideration is expression levels. Even though each A3 cDNA was expressed using the same CAG promoter and enforced by a strong selection for Fah functionality, considerable variation in A3 mRNA levels was seen in recovered liver tissues (Fig. S5). A fourth consideration could be negative interference by an as-yet-unidentified factor. For instance, both intra-/inter-A3 oligomerization and RNA binding are known to inhibit the activity of multiple A3 family DNA deaminases, including A3B and A3H (Cortez et al., 2019; Shaban et al., 2018). However, these considerations are tempered by the fact that our original human A3B transgenic line was lost early in these studies likely due to the enzyme exerting a negative selective pressure and catalyzing self-inactivation. A fifth consideration is the possibility of anti-human A3 immune responses, which may be lower for A3A in comparison to other family members.
Indeed, it will be very important to address the potential role of immune responses in future studies. The majority of liver tumors in the Fah regeneration model showed A3A mutation signatures but little to no detectable intratumor A3A staining by IHC. For instance, the tumor with the highest mutation load in Fig. 6 A (4,082 exomic base substitutions) stained IHC negative for A3A in Fig. 5 (G and H), despite adjacent hepatocellular parenchyma showing A3A protein expression cell-wide. This apparent incongruity strongly suggests a dynamic interplay between positive selection for restoration of Fah function, negative selection against high A3A as a genotoxic and potentially cytotoxic stress (further complicated by tight linkage of A3A and Fah on the same transposable element), and additional negative selective pressure by immune responses against the foreign proteins encoded by the transposon constructs. Another related possibility is immune responses against foreign antigens (neoantigens) created by A3A-catalyzed mutation. Although reconstruction experiments demonstrated cell-wide A3A protein expression and an elevated DNA damage response in hepatocytes shortly after hydrodynamic delivery (Fig. S5, E and F), additional studies are still warranted to deconvolute these competing pressures and expand the utility of the systems described here for studying and ultimately treating the APOBEC3 mutation program in cancer.
It is curious that the A3A ApcMin colon tumors only manifest C-to-T transitions in TCA/T motifs (SBS2), whereas the A3A Fah liver tumors additionally show C-to-G and to a lesser extent C-to-A transversions in the same motifs (SBS13). This is not simply due to the lower mutation loads observed in ApcMin tumor exomes because the absence of SBS13 became even more stark upon analysis of whole-tumor genomes (Fig. 4). We therefore propose that the absence of SBS13 may be due to lower rates of uracil excision in intestinal crypt cells that ultimately give rise to polyps in the ApcMin model. Alternatively, these differences in mutation signature may be due to low (or no) Rev1 translesion DNA synthesis in colon tissue, as Rev1 is the only DNA polymerase known to insert a cytosine nucleobase opposite an abasic site (Lawrence, 2002). In support of the latter possibility, Rev1 overexpression was necessary to stimulate polyp formation and enhance mutagenesis in a chemical mutagen (N-methyl-N-nitrosourea)-dependent murine intestinal adenoma model (Sasatani et al., 2017). It is additionally interesting that structure-associated APOBEC mutagenesis events were rare in both the ApcMin and Fah systems. Specifically, strand-coordinated mutation clusters (kataegis; Nik-Zainal et al., 2012) and loop-region mutations in stem-loop (palindrome) structures (Buisson et al., 2019) were not evident in tumor exome or WGS datasets. Multiple explanations for this are possible, including differential DNA repair capabilities and/or substrate availability. An additional possibility may be that A3A is responsible for dispersed, replication-associated mutations and another A3 enzyme catalyzes kataegis and stem-loop mutagenesis (e.g., A3B in kataegis; Maciejowski et al., 2019 Preprint).
Classic studies overexpressed related deaminases in vivo (rat Apobec1, murine Aicda [activation-induced cytidine deaminase], and murine Apobec2) and reported hepatocellular carcinomas, T cell lymphomas and lung adenomas, and hepatocellular carcinomas and lung adenomas/adenocarcinomas, respectively (Okazaki et al., 2003; Okuyama et al., 2012; Yamanaka et al., 1995). The liver tumors induced by rat Apobec1 were attributed originally to dysregulated RNA editing, and, as far as we are aware, chromosomal DNA mutations were not considered. Segments of select genes, TCRβ and c-myc, were sequenced in a subset of Aicda lymphomas, and a bias toward C/G-to-T/A mutations was reported. Aicda transgenic animals also displayed shorter life spans with increasing generation times, perhaps by germline mechanisms related to the loss of the A3B and A3Ahigh transgenics described here. Similarly, human AICDA (more commonly called AID) is implicated in carcinogenesis in specific human tissues, notably B cell and myeloid cell lymphomas and gastric cancers (reviewed by Marusawa and Chiba, 2010; Robbiani and Nussenzweig, 2013). The murine Apobec2 transgenic line had tumors in liver and lung tissues but no evidence for DNA editing (Okuyama et al., 2012). However, unlike these deaminase family members, human A3A has been implicated much more broadly in carcinogenesis and, together with at least one other A3 enzyme, is arguably the second largest source of mutation across cancer (Alexandrov et al., 2020). Thus, the A3A tumor models described here provide long-awaited in vivo demonstrations that this human DNA deaminase can indeed drive mutation formation and tumor development and is not simply a passenger process. It is not clear why low levels of A3A are alone aphenotypic and, therefore, additional work will be needed to identify factors that make cells permissive for APOBEC mutagenesis and generate additional murine models for the most common APOBEC-mutated tumor types in humans (breast, cervix, bladder, head/neck, and lung).
It should be noted that mutational processes such as A3A/AID–catalyzed DNA cytosine deamination do not precisely fit the classical cancer definitions of “oncoprotein” and “driver”. These definitions may need to be expanded to include subclasses of tumor-promoting processes, exemplified by “enablers” such as A3A/AID, that provide major sources of mutational fuel for tumor development and evolution but are not themselves directly driving tumor formation. Although many exogenous and endogenous mutational processes contribute to the overall mutation landscape in human cancer, only a small number of processes such as A3A/AID are likely to be large and dominant enough to mediate measurable changes in rates of tumorigenesis and trajectories of tumor development. Such enablers have the potential to provide large proportions of the mutational fuel for many different tumor types, which, coupled with selective pressures for/against tumor development, result in the massive heterogeneity reported in the genomic DNA sequences of different human tumor types (as well as between individual tumors within single tumor types). It is therefore important to determine in vivo, as here for A3A, whether a particular source of mutation is strong enough to be defined as both a source of mutation and a driver of tumorigenesis. Such information will be critical to be able to fully investigate future diagnostic, prognostic, and therapeutic options for tumors driven by APOBEC mutagenesis.
Materials and methods
Animal care
Mice were housed at the University of Minnesota Twin Cities, University of Pennsylvania, and University of Illinois at Chicago animal facilities under specific pathogen–free conditions. All studies were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The experiments performed with mice in this study were approved by the University of Pennsylvania Institutional Animal Care and Use Committee (protocol 805690), University of Illinois at Chicago Animal Care Committee (protocol 18–168), and University of Minnesota Institutional Animal Care and Use Committee (protocols 1901-36652A, 1602-33523A, and 1808A36277).
A3A and A3G ApcMin experiments
A3Alow, A3Ahigh, and A3G transgenic mice have been described (Stavrou et al., 2014). A3Alow and A3G transgenic females were bred with ApcMin males (002020; Jackson Laboratory). Experimental animals resulting from this cross were euthanized by CO2 administration at the age of 4 mo, and intestinal tissue was analyzed for polyp formation without knowledge of genotype information (i.e., blinded). The ApcMin, A3G ApcMin, and A3A ApcMin mice shown in Fig. S2 were additionally heterozygous for a deletion in mouse APOBEC3. The A3Alow transgene conferred no selective disadvantage as evidenced by near-Mendelian ratios in crosses (e.g., A3Alow/− × A3A−/− = 70 A3Alow/− and 59 A3A−/− at University of Minnesota and 152 A3Alow/− and 185 A3A−/− at University of Chicago at Illinois.
A3B transgenesis
pTraffic was modified by replacing IRES-GFP with IRES-Firefly-luciferase using XhoI and Bsu36I cut sites. A3Bi (A3B cDNA containing L1 intron) was then amplified from pCDNA3.1-A3Bi (Hultquist et al., 2011) using primers A3B_AscI_F 5′-NNNNGGCGCGCCACCATGAATCCACAGATC-3′ and A3B_NheI_R 5′-NNNNGCTAGCTCAGTTTCCCTGATTCTGGAGAATGG-3′, digested with AscI and NheI, and inserted into pTraffic-IRES-FLuciferase. Cre-dependent expression of A3B was verified by immunoblot of HEK-293T cells transfected with pTraffic-IRES-FLuciferase-AscI-A3Bi-NheI with or without cotransfection of a Cre expressing plasmid (antibodies: β-actin [4970; CST], luciferase [NB600-307; Novus], DsRed [632392; Clontech], and A3B; Brown et al., 2019). A3B-DsRed transgenic mice were prepared by pronuclear injection of pTraffic-IRES-FLuciferase-AscI-A3Bi-NheI plasmid into a C57BL/6 background. DsRed fluorescence was visualized in tail snips on a Leica DMIL microscope. A3B-NTD, A3B-CTD, and DsRed DNA fragments were amplified with the following primer sets: NTD_F 5′-CCACAGATCAGAAATCCGATGGA-3′, NTD_R 5′-CTCGAGACTAAAGGCAACAGTGCTG-3′; CTD_F 5′-GCCAGTGACTAGTGCTGCAAG-3′, CTD_R 5′-GTTTCCCTGATTCTGGAGAATGG-3′; DsRed_F 5′-CCCATGGTCTTCTTCTGCAT-3′, DsRed_R 5′-AAGGTGTACGTGAAGCACCC-3′. A3B-NTD primers were used for genotyping. The DsRed PCR fragment was cloned using CloneJET PCR Cloning Kit (Thermo Fisher Scientific) and Sanger sequenced.
A3 Fah experiments
Fah-deficient mice expressing SB11 (Fah−/−; Rosa26-SB11Tg/WT) were generated and maintained with drinking water containing 7.5 µg/ml NTBC (Sigma Aldrich) as described previously (Keng et al., 2011) until introduction of transposon vectors by hydrodynamic tail vein injections at 8–10 wk of age. Intron-containing human A3 cDNAs were ordered as gBlocks (IDT) and cloned into the pENTR entry vector (Invitrogen) using NotI-NcoI (A3Ai, A3Bi, A3Ci, and A3Gi), MfeI-NotI (A3Di and A3Fi), or KpnI-NotI (A3Hi). The final Gateway destination plasmid coexpressing Fah, GFP, and luciferase (Keng et al., 2013) was combined with pENTR-A3 vectors using Gateway LR clonase mix (11791-020; Thermo Fisher Scientific) to generate pT2/GD-A3 delivery plasmids. The transposon vector expressing an shRNA against Tp53 (shTp53) and its validation in vivo have been described previously (Wangensteen et al., 2008).
Immediately before hydrodynamic delivery, 8–10-wk-old Fah-null SB11 mice were anesthetized by administrating 25 µl anesthetic cocktail (8 mg/ml−1 ketamine HCl, 0.1 mg/ml−1 acepromazine maleate, and 0.01 mg/ml−1 butorphanol tartrate) i.p. Each animal was injected with 20 µg of each transposon plasmid (PureLink HiPure Plasmid Filter Maxiprep kit; Invitrogen) diluted in lactated Ringer’s solution (Thermo Fisher Scientific) to an injection volume based on the weight of the mouse (10% vol/wt). Immediately following injection, animals were placed on normal drinking water to promote liver repopulation with Fah-A3 transgenic cells. Luciferase expression in repopulated transgenic livers was monitored using the Xenogen IVIS 100 (Perkin Elmer) as described previously (Wilber et al., 2005). Briefly, animals were injected with 93–150 mg luciferin/kg body weight (Xenogen) i.p. 10–15 min before imaging. Mice were anesthetized using 1–1.5% isoflurane and O2/N2O via nose cone, positioned in a custom-built cradle, and imaged for 1–5 min.
6 mo after injection, mice were euthanized and weighed, and liver tissues were harvested and weighed. Liver mass was recorded and all visible distinct nodules (>2 mm in diameter) were counted and carefully isolated from neighboring tumor-free tissue. All nodules >2 mm from were halved for DNA and RNA isolation. Genomic DNA isolations were performed using either AllPrep DNA/RNA kit or DNeasy Blood & Tissue kit per manufacturer protocols (Qiagen). RNA isolations were conducted with either the AllPrep DNA/RNA kit or RNeasy Mini Kit following manufacture protocols (Qiagen).
Protein lysates were obtained through vortex-mediated homogenization with stainless steel beads (Qiagen) for 2 min at 4°C in protein lysis buffer (50 mM Tris-Cl, pH 7.4, 250 mM NaCl, and 0.5% Igepal NP-40) at 10-20× vol/wt tissue. Homogenized lysates were cleared of debris through centrifugation (maximum speed for 20 min at 4°C) and quantified with a NanoDrop (Thermo Fisher Scientific). Samples were treated with RNaseA (1.67 µg/µl) for 10 min at room temperature and then combined 1:1 with SDS-PAGE loading buffer (62.5 mM Tris-Cl, pH 6.8, 20% glycerol, 7.5% SDS, 5% 2-mercaptoethanol, and 250 mM DTT). Proteins were separated by a 12.5% SDS-PAGE gel and transferred to polyvinylidene Immobilon-FL membranes (Millipore). Membranes were blocked in blocking solution (5% milk + PBS supplemented with 0.1% Tween20) and then incubated with primary antibody diluted in blocking solution. Primary immunoblotting antibodies were mouse α-tubulin (T5168; Sigma Aldrich), rabbit α-human A3A/B/G mAb (mAb 5210–87-13; Brown et al., 2019), rabbit α-human A3C pAb (10591–1-AP; Proteintech), rabbit α-human A3H pAb (Novus NBP1-91682), and rabbit α-firefly luciferase pAb (Abcam ab21176). Secondary antibodies were diluted in blocking solution supplemented with 0.02% SDS. Secondary antibodies used for detection were α-rabbit 800CW (827–08365; LI-COR), α-mouse 680LT (925–68020; LI-COR), α-rabbit HRP (7074P2; Cell Signaling Technology), and α-mouse HRP (7076P2; Cell Signaling Technology). Membranes were imaged with an Odyssey Classic scanner and Odyssey Fc imager (LI-COR).
A3Ahigh transgene characterization
RNA from the intestinal polyps of ApcMin, A3Ahigh ApcMin, and A3Alow ApcMin animals was extracted, and RNA-sequencing libraries were prepared and sequenced on a HiSeq 2500 125 × 2 bp. Reads were mapped to the mouse genome with human A3A cDNA appended as an additional chromosome using tophat. Read coverage plots suggested a 3′ truncation.
For further mapping of the 3′ truncation, spleens were harvested from 3-mo-old mice. Splenic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). RNA was isolated with the use of Trizol (Invitrogen) and further processed according to the Qiagen RNA cleanup protocol (treated with DNaseI to eliminate any contaminating genomic DNA). Purified RNA was converted to cDNA using the SuperScript III First Strand Synthesis System for RT-PCR using 50 μM oligo(dT)20 as the primer (Invitrogen). Full-length cDNA was obtained by rapid amplification of cDNA ends (RACE) using A3A transgene forward primer 5′-TGGACCTGGTTCCTTCTTT-3′, with the poly(A) tail functioning as the 3′ end tag. Bands were excised, and the fragments were cloned into pCR2.1-TOPO vector as specified by the manufacturer (Invitrogen). Additionally, the transgene DNA was amplified using primers CAG promoter 5′-GGGCGGGGTTCGGCTTCTGGCGTGTGAC-3′ and CAG poly(A) tail 5′-CAGGGCATTGGCCACACCAGCCACCACC-3′. Both the cloned cDNA and amplified genomic DNA were Sanger sequenced.
RT-PCR and quantitative PCR
RNA was extracted from mouse tissues (Fah and ApcMin) with RNeasy Mini Kit (Qiagen) and from polyps with AllPrep DNA/RNA mini kit (Qiagen). cDNA was prepared using Transcriptor reverse transcription (Roche) with random hexamer priming. Full-length A3A transcript was amplified with the following primers: A3A_full_length-F 5′-ATGGAAGCCAGCCCAGCATC-3′; A3A_full_length-R 5′-GTTTCCCTGATTCTGGAGAATGG-3′. Relative transcript levels were measured by quantitative PCR with LightCycler 480 Probes Master mix (Roche) on a LightCycler 480 instrument (Roche) and the following primers: A3A (in conjunction with UPL probe 97; Roche) A3A_qPCR_F 5′-CCACACATATTCACTTCCAACT-3′, A3A_qPCR_R 5′-TGTGCTGGTCCATCTTGA-3′; Tbp (in conjunction with UPL probe 97; Roche) mouse_Tbp_F 5′-GGGGAGCTGTGATGTGAAGT-3′, mouse_Tbp_R 5′-CCAGGAAATAATTCTGGCTCA-3′. APOBEC3 mRNA was quantified with previously described primers (A3B, A3D, A3F, A3G, and TBP) or A3A (forward 5′-CGGTCAAGATGGACCAGCAC-3′; reverse 5′-GAAGGAACGCACGCACTTC-3′), A3C (forward 5′-AGCCAACGATCGGAACGAAA-3′; reverse 5′-AGGGCTCCAAGATGTGTACC-3′), A3H (forward 5′-TCAGAAGGCCTTACTACCCG-3′; reverse 5′-ATGAAGTCAACCAGCTCCCAG-3′) using SsoFast EvaGreen Supermix (Bio-Rad) per the manufacturer’s protocols. Luciferase mRNA was quantified using LightCycler 480 Probes Master Mix (Roche) with primers forward 5′-TCCATCTTGCTCCAACACCC-3′ and reverse 5′-TCGTCTTTCCGTGCTCCAAA-3′ along with UPL 70. All mRNA quantification was obtained using a LightCycler 480 instrument (Roche).
Deaminase activity assays
Spleens and colons from 4-mo-old A3Alow mice were homogenized and lysed in HED buffer (25 mM Hepes, 5 mM EDTA, 10% glycerol, 1 mM DTT, and 1× protease inhibitor [cOmplete; Roche]). Lysates were sonicated for 20 min in a water bath sonicator and cleared by centrifugation, and spleen lysates were further concentrated using Amicon Ultra 0.5 ml centrifugal filters with a molecular weight cutoff of 10 kD with two washes of 400 µl buffer (25 mM Hepes, pH 7.4, 15 mM EDTA, cOmplete EDTA-free protease inhibitors, and 10% glycerol). Protein concentration in lysates was quantified by Bradford assay (Zor and Selinger, 1996). Oligo NUP93 (6-FAM)-GCAAGCTGTTCAGCTTGCTGA (Buisson et al., 2019) was allowed to form a hairpin by heating a 10 µM stock in 1 mM Tris-Cl and 0.1 mM EDTA to 65°C for 5 min and then allowing the DNA to cool to room temperature. 75 µg of spleen and 17 µg of colon protein lysates were incubated with 800 nM NUP93 hairpin oligo at 37°C with 100 µg/ml RNase A for 24 h. 0.1 U uracil DNA glycosylase (NEB) was added to each reaction and incubated for 10 min at 37°C. NaOH was added to a final concentration of 100 mM, and samples were heated to 95°C for 10 min to cause breakage of abasic sites caused by deamination and subsequent uracil removal. Samples were separated by 15% TBE-Urea PAGE to resolve product and imaged on a Typhoon FLA 7000 biomolecular imager (GE Healthcare Life Sciences). A3A-mycHis purified from HEK-293T cells was used at 1 nM as a positive control for deamination activity (Shi et al., 2017). APOBEC3 activity in Fah liver samples was detected similarly using lysates obtained as described above.
Handling of intestinal tissue
Intestines from 4-mo-old ApcMin and A3Alow/ApcMin mice were removed from duodenum to colon and placed on PBS-soaked Bibulous paper. Intestines were divided into sections and then carefully sliced lengthwise, cleaned, and spread with lumen side upward. Select large polyps from the distal colon were excised and flash frozen for future molecular analysis. Intestines were fixed overnight in 10% buffered formalin solution. Polyp numbers were counted (blinded to mouse genotype) using a Leica S8 APO stereo microscope (at University of Minnesota) or a Nikon SMZ1500 (at University of Pennsylvania). Statistical differences between ApcMin and A3Alow/ApcMin animals were determined by two-sided Wilcoxon rank sum test using Prism 6 software.
Histology
The small intestine and colon of ApcMin and A3Alow/ApcMin mice were isolated and fixed in 10% neutral formalin. The flattened segments of intestinal tissues were rolled lengthwise into “Swiss rolls” (Moolenbeek and Ruitenberg, 1981) and embedded in paraffin (FFPE). In addition, normal livers and liver tumors generated with the A3A Fah model were collected and fixed as above. H&E staining of the FFPE specimens was performed as follows. Tissues were sectioned at 4 µm, mounted on positively charged adhesive slides, and allowed to air-dry for at least 24 h before staining. To deparaffinize and rehydrate the samples, slides were baked in a 60–62°C oven for 20 min, washed three times with xylene for 5 min each, soaked in graded alcohols (100% × 2, 95% and 80% for 3 min each), and then rinsed in running water for 5 min. The tissues were stained with hematoxylin for 5 min and rinsed in running water for 30 s, followed by two dips in acid solution and 30–90 s in ammonia water (bluing solution). After a 10-min water rinse, the slides were transferred in 80% ethanol for 1 min, counterstained with eosin for 1 min, dehydrated in graded alcohols and xylene, and cover-slipped using Permount mounting media. The H&E-stained slides were subsequently scanned at 40× magnification and visualized using the Aperio ScanScope XT system (Leica Biosystems) as described previously (Brown et al., 2019).
IHC
4-µm-thick sections of FFPE intestinal and liver tissues were mounted on positively charged, adhesive slides and allowed to air-dry for at least 24 h. To deparaffinize and rehydrate the samples, slides were baked in a 65°C oven for 20 min, washed three times with CitriSolv (1601; Decon Labs) or xylene for 5 min each, soaked in graded alcohols (100% × 2, 95% and 80% for 3 min/each), and then rinsed in running water for at least 5 min. Epitope retrieval was performed using Reveal Decloaker (RV1000M; BioCare Medical) in a steamer for 35 min, followed by a 20-min “cool-down” period. Then, slides were rinsed with running tap water for 5 min and transferred to Tris-buffered saline with 0.1% Tween20 (TBST) for 5 min. Endogenous peroxidase activity was quenched by placing the slides in 3% H2O2 in TBST for 10 min at room temperature, followed by a 5-min rinse under running water. To block nonspecific binding of primary antibody, sections were covered with Rodent Block M (RBM961; BioCare Medical) for 15 min at room temperature. After blocking, sections of each specimen were incubated overnight at 4°C with a rabbit α-human A3A/B/G mAb (5210–87-13; Brown et al., 2019) diluted 1:350 or a rabbit α-γ-H2AX mAb (9718; Cell Signaling Technology) diluted 1:200 in 10% Rodent Block M in TBST.
Following overnight incubation with primary antibody, sections were rinsed in TBST for 5 min and completely covered with anti-rabbit poly-HRP-IgG (Novolink Polymer, RE7260-K; Leica Biosystems) for 30 min at room temperature. The reaction product was developed using the Novolink DAB substrate kit (RE7230-K; Leica Biosystems) at room temperature for 3 min, rinsed in tap water for 5 min, counterstained in Mayer’s hematoxylin solution (26252–01; Electron Microscopy Sciences) for 5 min, dehydrated in graded alcohols and CitriSolv, and cover-slipped using Permount mounting media. Nuclear and cytoplasmic A3A and nuclear γ-H2AX immunoreactivities were visualized and quantified using the Aperio ScanScope XT (Leica Biosystems). Approximately 100,000 cells were analyzed from each specimen for quantification of the positively stained population.
Genomic DNA sequencing and analysis of mutational patterns
Genomic DNA was prepared from frozen colon polyp tissue using AllPrep DNA/RNA mini kit (Qiagen). Matched normal DNA was extracted from tail snips following the Gentra Puregene Tissue DNA isolation protocols (Qiagen). DNA was sheared and adapters added using SureSelectQXT Library Prep kit (Agilent), and exonic DNA fragments were captured using SureSelectXT Mouse All Exon kit (Agilent). ApcMin exome libraries were sequenced 75 × 2 bp on a NextSeq 550 instrument (Illumina) to a target depth of 75× coverage for tumor samples and 30× coverage for matched normal. ApcMin whole-genome libraries were sequenced 150 × 2 bp on a NovaSeq 6000 (Illumina) to a target read depth of 30× coverage for both tumor and matched normal samples. Fah exome libraries were sequenced 150 × 2 bp on a NovaSeq 6000 (Illumina) to a read depth similar to the ApcMin exome libraries. Resulting sequences were aligned to the mouse genome (mm10) using Burrows–Wheeler Aligner (BWA; version 0.7.17). For whole-genome analysis, reads were aligned to the mm10 reference genome using SpeedSeq (Chiang et al., 2015), which utilizes BWA. PCR duplicates were removed using Picard (version 2.18.16). Reads were locally realigned around indels using GATK3 (version 3.6.0) tools RealignerTargetCreator to create intervals, followed by IndelRealigner on the aligned bam files. The exome-sequencing alignments from the normal tail tissues were used to create a “panel of normals” background set using MuTect2 from GATK3 (version 3.6.0) in artifact detection mode. Mutations were called from the polyp samples compared with the matched normal, including the “panel of normals” as background using MuTect2 from GATK3. Single-nucleotide variants that passed the internal GATK3 filter were used for downstream analysis. Mutational patterns were analyzed in R (version 3.6.0) using the “MutationalPatterns” package (Blokzijl et al., 2018). No panel of normals was used for ApcMin whole-genome samples, and filters of a minimum read depth of 20× and 0.05 variant allele frequency were applied. Mouse 59 failed quality control for WES library construction (for unknown reasons) and the same DNA passed quality control for WGS. Aligned WES reads are available at the NCBI Sequence Read Archive through BioProject accession no. PRJNA655491.
SBS mutation signatures (v3, May 2019; https://cancer.sanger.ac.uk/cosmic/signatures/SBS/) were obtained from https://www.synapse.org/#!Synapse:syn11738319. De novo nonnegative matrix factorization of mutational signatures was performed with the “extract_signature” command, with a rank of 2 and 10 iterations. TCW mutation enrichment score was calculated as described previously (Chan et al., 2015; Jarvis et al., 2018). Sequence logos of −2 to +2 sequence surrounding C-to-T mutations was created using “ggseqlogo” package in R. Prediction of mutation effect on protein sequence was done with SnpEff (version 4.3t) using the SnpEff mm10 database. LOH of chromosome 18 was calculated by comparing tumor to matched normal alignments using Varscan (version 2.3.9) in somatic mode.
WGS reads were trimmed using TrimGalore with default settings. Trimmed reads were then aligned against mm10 using BWA. Alignments were sorted and marked for duplicates using sambamba. Somatic copy number variants from WGS alignments were determined using GATK4 CNV for each tumor-normal pair. Structural variants were determined using BreakDancer (Fan et al., 2014) and structural variants less than two times as frequent in the tumor versus the normal normalized for coverage were excluded. The integration site of the A3Alow (A10) transgene was determined using oncovirus_tools get_int.pl using the vector sequence as the input bait and the bam alignment from a representative A3A ApcMin whole-genome sequence (https://github.com/gstarrett/oncovirus_tools; Starrett et al., 2020).
RNA library sequencing and analysis
RNA was prepared from frozen colon polyp tissue using the AllPrep DNA/RNA mini kit (Qiagen) simultaneously with genomic DNA. Sequencing libraries were prepared using Illumina Truseq Stranded mRNA kit (Illumina) and sequenced 2 × 150 bp on a NovaSeq 6000 instrument (Illumina). Sequencing results were aligned to the mm10 mouse genome with human A3A cDNA sequence appended as an additional chromosome using tophat (version 2.0.13). FPKM levels of various genes were extracted using cufflinks (version 2.2.1). RNA editing levels at Apobec1 target sites were extracted from aligned reads using the mpileup command.
TCGA variant and Euclidean distance analysis
All WES variant information from TCGA was downloaded from the Broad GDAC firehose database (https://gdac.broadinstitute.org/). Variant information from the A3A and A3B yeast models of mutagenesis were downloaded from previous published material (Chan et al., 2015). Only samples with >300 C-to-T mutations were included in the Euclidean distance and sequencing logo analyses. Single-nucleotide variants that were used in these analyses included only C-to-T variants in a TC dinucleotide, excluding all mutations as CG motifs. All C-to-T variants from the ApcMin, A3A yeast model, and A3B yeast model were used in this analysis. A matrix comprised of the number of mutations within a tetranucleotide across all samples within a cancer type was generated, and counts were normalized to frequency within each cancer type. This matrix was converted to a hierarchical distance matrix using the hclust function in R, which was then plotted as dendrograms. All sequences logos were again generated and visualized using “ggseqlogo” package in R. Total mutation counts per cancer type were generated by simple counting of all C-to-T mutations at TC dinucleotide, excluding all mutations as CG motifs.
Statistical analyses
GraphPad Prism (version 6) was used for statistical analyses, including Wilcoxon rank sum tests (Fig. 2, C and D; Fig. 3 A; and Fig. S2) and one-way ANOVA tests (Fig. 5, E and F). R was used to calculate Pearson’s correlations (Fig. 3 G, Fig. 4 D, and Fig. 6, G and H). Additional statistics involved Student’s t test comparisons of mean values with SDs. P values < 0.05 were considered significant.
Online supplemental material
Fig. S1 is a summary of data from human A3B and A3A transgenics that were lost potentially due to self-inactivation. Fig. S2 provides an independent demonstration of elevated polyp formation in A3A ApcMin animals. Fig. S3 provides further support that human A3A is expressed and active in A3A ApcMin polyps and also not affecting the mRNA expression levels or RNA editing functions of endogenous murine deaminases. Fig. S4 provides a visual summary of the LOH observed for murine chromosome 18, which invariably includes the Apc gene. Fig. S5 shows control experiments for the Fah liver tumor system.
Acknowledgments
We thank Hilde Nilsen and Douglas Yee for thoughtful feedback, Bobbi Tschida for training on hydrodynamic injections, Brian Dunnette for sharing expertise regarding the use of the Aperio ScanScope XT, and Kristin Blouch and David Ryan for assistance with mouse breeding.
These studies were supported in part by the National Cancer Institute (grant P01CA234228 to R.S. Harris), the National Institute of Allergy and Infectious Diseases (grant R01AI085015 to S.R. Ross), the National Institute on Aging (grant R21AG047114 to S.R. Ross), the University of Minnesota College of Biological Sciences and Academic Health Center (R.S. Harris), the Randy Shaver Cancer Research and Community Fund (R.S. Harris), the Masonic Cancer Center Translational Working Group (T.K. Starr), and the Mezin-Koats Colorectal Cancer Research Fund (T.K. Starr). National Institutes of Health training grants and career development awards provided salary support for R. Levin-Klein (T32HL007062 and F32CA232458) and M.C. Jarvis (T32CA009138 and F31CA243306). R. Levin-Klein is an awardee of the Weizmann Institute of Science National Postdoctoral Award Program for Advancing Women in Science. M.B. Burns was supported in part by a US Department of Defense Breast Cancer Research Program Predoctoral Fellowship (BC101124). R.S. Harris is the Margaret Harvey Schering Land Grant Chair for Cancer Research, a Distinguished University McKnight Professor, and an Investigator of the Howard Hughes Medical Institute.
Author contributions: R.S. Harris and S.R. Ross conceptualized and supervised the overall project. E.K. Law, R. Levin-Klein, M.C. Jarvis, H. Kim, P.P. Argyris, M.A. Carpenter, L.K. Larson, C. Durfee, M.B. Burns, S. Stavrou, A.N. Aguilera, and T.K. Starr performed experiments. R. Levin-Klein and H. Kim performed validation studies. E.K. Law, R. Levin-Klein, M.C. Jarvis, G.J. Starrett, N.A. Temiz, and R.I. Vogel did formal data analysis. E.K. Law, R. Levin-Klein, M.C. Jarvis, H. Kim, M.A. Carpenter, S. Wagner, D.A. Largaespada, and T.K. Starr contributed methodology. R. Levin-Klein, M.C. Jarvis, M.B. Burns, D.A. Largaespada, T.K. Starr, S.R. Ross, and R.S. Harris contributed to funding acquisition. R. Levin-Klein, M.C. Jarvis, E.K. Law, P.P. Argyris, S.R. Ross, and R.S. Harris drafted the manuscript, and all authors contributed to revisions.
References
- Adolph, M.B., Love R.P., Feng Y., and Chelico L.. 2017. Enzyme cycling contributes to efficient induction of genome mutagenesis by the cytidine deaminase APOBEC3B. Nucleic Acids Res. 45:11925–11940. 10.1093/nar/gkx832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., et al. ; ICGC PedBrain . 2013. Signatures of mutational processes in human cancer. Nature. 500:415–421. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, L.B., Jones P.H., Wedge D.C., Sale J.E., Campbell P.J., Nik-Zainal S., and Stratton M.R.. 2015. Clock-like mutational processes in human somatic cells. Nat. Genet. 47:1402–1407. 10.1038/ng.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov, L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., Boot A., Covington K.R., Gordenin D.A., Bergstrom E.N., et al. ; PCAWG Consortium . 2020. The repertoire of mutational signatures in human cancer. Nature. 578:94–101. 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus, L., Smid M., Wilting S.M., van Riet J., Van Hoeck A., Nguyen L., Nik-Zainal S., Steenbruggen T.G., Tjan-Heijnen V.C.G., Labots M., et al. 2019. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat. Genet. 51:1450–1458. 10.1038/s41588-019-0507-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci, F., Ng C.K.Y., Patsouris A., Droin N., Piscuoglio S., Carbuccia N., Soria J.C., Dien A.T., Adnani Y., Kamal M., et al. 2019. Genomic characterization of metastatic breast cancers. Nature. 569:560–564. 10.1038/s41586-019-1056-z [DOI] [PubMed] [Google Scholar]
- Blanc, V., Park E., Schaefer S., Miller M., Lin Y., Kennedy S., Billing A.M., Ben Hamidane H., Graumann J., Mortazavi A., et al. 2014. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 15:R79. 10.1186/gb-2014-15-6-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl, F., Janssen R., van Boxtel R., and Cuppen E.. 2018. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 10:33. 10.1186/s13073-018-0539-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn, M.F., Shandilya S.M.D., Silvas T.V., Nalivaika E.A., Kouno T., Kelch B.A., Ryder S.P., Kurt-Yilmaz N., Somasundaran M., and Schiffer C.A.. 2015. The ssDNA mutator APOBEC3A is regulated by cooperative dimerization. Structure. 23:903–911. 10.1016/j.str.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W.L., Law E.K., Argyris P.P., Carpenter M.A., Levin-Klein R., Ranum A.N., Molan A.M., Forster C.L., Anderson B.D., Lackey L., et al. 2019. A rabbit monoclonal antibody against the antiviral and cancer genomic DNA mutating enzyme APOBEC3B. Antibodies (Basel). 8. E47. 10.3390/antib8030047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson, R., Lawrence M.S., Benes C.H., and Zou L.. 2017. APOBEC3A and APOBEC3B activities render cancer cells susceptible to ATR inhibition. Cancer Res. 77:4567–4578. 10.1158/0008-5472.CAN-16-3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson, R., Langenbucher A., Bowen D., Kwan E.E., Benes C.H., Zou L., and Lawrence M.S.. 2019. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science. 364. eaaw2872. 10.1126/science.aaw2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B., et al. 2013a. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 494:366–370. 10.1038/nature11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, M.B., Temiz N.A., and Harris R.S.. 2013b. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45:977–983. 10.1038/ng.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannataro, V.L., Gaffney S.G., Sasaki T., Issaeva N., Grewal N.K.S., Grandis J.R., Yarbrough W.G., Burtness B., Anderson K.S., and Townsend J.P.. 2019. APOBEC-induced mutations and their cancer effect size in head and neck squamous cell carcinoma. Oncogene. 38:3475–3487. 10.1038/s41388-018-0657-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, M.A., Li M., Rathore A., Lackey L., Law E.K., Land A.M., Leonard B., Shandilya S.M., Bohn M.F., Schiffer C.A., et al. 2012. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 287:34801–34808. 10.1074/jbc.M112.385161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caval, V., Suspène R., Shapira M., Vartanian J.P., and Wain-Hobson S.. 2014. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage. Nat. Commun. 5:5129. 10.1038/ncomms6129 [DOI] [PubMed] [Google Scholar]
- Caval, V., Bouzidi M.S., Suspène R., Laude H., Dumargne M.C., Bashamboo A., Krey T., Vartanian J.P., and Wain-Hobson S.. 2015. Molecular basis of the attenuated phenotype of human APOBEC3B DNA mutator enzyme. Nucleic Acids Res. 43:9340–9349. 10.1093/nar/gkv935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescon, D.W., Haibe-Kains B., and Mak T.W.. 2015. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc. Natl. Acad. Sci. USA. 112:2841–2846. 10.1073/pnas.1424869112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K., Resnick M.A., and Gordenin D.A.. 2013. The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst.). 12:878–889. 10.1016/j.dnarep.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K., Roberts S.A., Klimczak L.J., Sterling J.F., Saini N., Malc E.P., Kim J., Kwiatkowski D.J., Fargo D.C., Mieczkowski P.A., et al. 2015. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 47:1067–1072. 10.1038/ng.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Lilley C.E., Yu Q., Lee D.V., Chou J., Narvaiza I., Landau N.R., and Weitzman M.D.. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480–485. 10.1016/j.cub.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Chen, T.W., Lee C.C., Liu H., Wu C.S., Pickering C.R., Huang P.J., Wang J., Chang I.Y., Yeh Y.M., Chen C.D., et al. 2017. APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism. Nat. Commun. 8:465. 10.1038/s41467-017-00493-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, C., Layer R.M., Faust G.G., Lindberg M.R., Rose D.B., Garrison E.P., Marth G.T., Quinlan A.R., and Hall I.M.. 2015. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat. Methods. 12:966–968. 10.1038/nmeth.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez, L.M., Brown A.L., Dennis M.A., Collins C.D., Brown A.J., Mitchell D., Mertz T.M., and Roberts S.A.. 2019. APOBEC3A is a prominent cytidine deaminase in breast cancer. PLoS Genet. 15. e1008545. 10.1371/journal.pgen.1008545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin, E.C., McGranahan N., Mitter R., Salm M., Wedge D.C., Yates L., Jamal-Hanjani M., Shafi S., Murugaesu N., Rowan A.J., et al. 2014. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 346:251–256. 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faden, D.L., Thomas S., Cantalupo P.G., Agrawal N., Myers J., and DeRisi J.. 2017. Multi-modality analysis supports APOBEC as a major source of mutations in head and neck squamous cell carcinoma. Oral Oncol. 74:8–14. 10.1016/j.oraloncology.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Fan, X., Abbott T.E., Larson D., and Chen K.. 2014. BreakDancer: identification of genomic structural variation from paired-end read mapping. Curr. Protoc. Bioinformatics. 45:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison, M.L., Akagi K., Xiao W., Jiang B., Pickard R.K.L., Li J., Swanson B.J., Agrawal A.D., Zucker M., Stache-Crain B., et al. 2019. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res. 29:1–17. 10.1101/gr.241141.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, A.P., Fantini D., Wang Y., Yu Y., Rimar K.J., Podojil J.R., Miller S.D., and Meeks J.J.. 2017. APOBEC-mediated mutagenesis in urothelial carcinoma is associated with improved survival, mutations in DNA damage response genes, and immune response. Oncotarget. 9:4537–4548. 10.18632/oncotarget.23344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompe, M., Overturf K., al-Dhalimy M., and Finegold M.. 1998. Therapeutic trials in the murine model of hereditary tyrosinaemia type I: a progress report. J. Inherit. Metab. Dis. 21:518–531. 10.1023/A:1005462804271 [DOI] [PubMed] [Google Scholar]
- Hanahan, D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Harris, R.S., and Dudley J.P.. 2015. APOBECs and virus restriction. Virology. 479-480:131–145. 10.1016/j.virol.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday, T., Eshtad S., and Nik-Zainal S.. 2014. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15:585–598. 10.1038/nrg3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, S., Chakravarthy A., Su X., Boshoff C., and Fenton T.R.. 2014. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 7:1833–1841. 10.1016/j.celrep.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Hultquist, J.F., Lengyel J.A., Refsland E.W., LaRue R.S., Lackey L., Brown W.L., and Harris R.S.. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 85:11220–11234. 10.1128/JVI.05238-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, F., Fu Y., Kao S.A., Yang H., and Chen X.S.. 2017. Family-wide comparative analysis of cytidine and methylcytidine deamination by eleven human APOBEC proteins. J. Mol. Biol. 429:1787–1799. 10.1016/j.jmb.2017.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, M.C., Ebrahimi D., Temiz N.A., and Harris R.S.. 2018. Mutation signatures including APOBEC in cancer cell lines. JNCI Cancer Spectr. 2. pky002. 10.1093/jncics/pky002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng, V.W., Villanueva A., Chiang D.Y., Dupuy A.J., Ryan B.J., Matise I., Silverstein K.A., Sarver A., Starr T.K., Akagi K., et al. 2009. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 27:264–274. 10.1038/nbt.1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng, V.W., Tschida B.R., Bell J.B., and Largaespada D.A.. 2011. Modeling hepatitis B virus X-induced hepatocellular carcinoma in mice with the Sleeping Beauty transposon system. Hepatology. 53:781–790. 10.1002/hep.24091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng, V.W., Sia D., Sarver A.L., Tschida B.R., Fan D., Alsinet C., Solé M., Lee W.L., Kuka T.P., Moriarity B.S., et al. 2013. Sex bias occurrence of hepatocellular carcinoma in Poly7 molecular subclass is associated with EGFR. Hepatology. 57:120–130. 10.1002/hep.26004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning, F.A., Goujon C., Bauby H., and Malim M.H.. 2011. Target cell-mediated editing of HIV-1 cDNA by APOBEC3 proteins in human macrophages. J. Virol. 85:13448–13452. 10.1128/JVI.00775-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouno, T., Silvas T.V., Hilbert B.J., Shandilya S.M.D., Bohn M.F., Kelch B.A., Royer W.E., Somasundaran M., Kurt Yilmaz N., Matsuo H., et al. 2017. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat. Commun. 8:15024. 10.1038/ncomms15024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land, A.M., Law E.K., Carpenter M.A., Lackey L., Brown W.L., and Harris R.S.. 2013. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J. Biol. Chem. 288:17253–17260. 10.1074/jbc.M113.458661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, S., Narvaiza I., Linfesty D.C., and Weitzman M.D.. 2011. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 12:444–450. 10.1038/embor.2011.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, E.K., Sieuwerts A.M., LaPara K., Leonard B., Starrett G.J., Molan A.M., Temiz N.A., Vogel R.I., Meijer-van Gelder M.E., Sweep F.C., et al. 2016. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci. Adv. 2. e1601737. 10.1126/sciadv.1601737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.W.. 2002. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst.). 1:425–435. 10.1016/S1568-7864(02)00038-1 [DOI] [PubMed] [Google Scholar]
- Lefebvre, C., Bachelot T., Filleron T., Pedrero M., Campone M., Soria J.C., Massard C., Lévy C., Arnedos M., Lacroix-Triki M., et al. 2016. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 13. e1002201. 10.1371/journal.pmed.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, B., McCann J.L., Starrett G.J., Kosyakovsky L., Luengas E.M., Molan A.M., Burns M.B., McDougle R.M., Parker P.J., Brown W.L., et al. 2015. The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res. 75:4538–4547. 10.1158/0008-5472.CAN-15-2171-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifora, J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X., Sprinzl M.F., Koppensteiner H., Makowska Z., Volz T., et al. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 343:1221–1228. 10.1126/science.1243462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski, J., Chatzipli A., Dananberg A., de Lange T., and Campbell P.J.. 2019. APOBEC3B-dependent kataegis and TREX1-driven chromothripsis in telomere crisis. bioRxiv. doi: 10.1101/725366 (Preprint posted August 5, 2019) [DOI] [PMC free article] [PubMed]
- MacMillan, A.L., Kohli R.M., and Ross S.R.. 2013. APOBEC3 inhibition of mouse mammary tumor virus infection: the role of cytidine deamination versus inhibition of reverse transcription. J. Virol. 87:4808–4817. 10.1128/JVI.00112-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusawa, H., and Chiba T.. 2010. Helicobacter pylori-induced activation-induced cytidine deaminase expression and carcinogenesis. Curr. Opin. Immunol. 22:442–447. 10.1016/j.coi.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Maruyama, W., Shirakawa K., Matsui H., Matsumoto T., Yamazaki H., Sarca A.D., Kazuma Y., Kobayashi M., Shindo K., and Takaori-Kondo A.. 2016. Classical NF-κB pathway is responsible for APOBEC3B expression in cancer cells. Biochem. Biophys. Res. Commun. 478:1466–1471. 10.1016/j.bbrc.2016.08.148 [DOI] [PubMed] [Google Scholar]
- Middlebrooks, C.D., Banday A.R., Matsuda K., Udquim K.I., Onabajo O.O., Paquin A., Figueroa J.D., Zhu B., Koutros S., Kubo M., et al. 2016. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat. Genet. 48:1330–1338. 10.1038/ng.3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenbeek, C., and Ruitenberg E.J.. 1981. The “Swiss roll”: a simple technique for histological studies of the rodent intestine. Lab. Anim. 15:57–59. 10.1258/002367781780958577 [DOI] [PubMed] [Google Scholar]
- Mori, S., Takeuchi T., Ishii Y., and Kukimoto I.. 2015. Identification of APOBEC3B promoter elements responsible for activation by human papillomavirus type 16 E6. Biochem. Biophys. Res. Commun. 460:555–560. 10.1016/j.bbrc.2015.03.068 [DOI] [PubMed] [Google Scholar]
- Mori, S., Takeuchi T., Ishii Y., Yugawa T., Kiyono T., Nishina H., and Kukimoto I.. 2017. Human Papillomavirus 16 E6 upregulates APOBEC3B via the TEAD transcription factor. J. Virol. 91. e02413-16. 10.1128/JVI.02413-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, A.R., Pitot H.C., and Dove W.F.. 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 247:322–324. 10.1126/science.2296722 [DOI] [PubMed] [Google Scholar]
- Munteanu, I., and Mastalier B.. 2014. Genetics of colorectal cancer. J. Med. Life. 7:507–511. [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., et al. ; Breast Cancer Working Group of the International Cancer Genome Consortium . 2012. Mutational processes molding the genomes of 21 breast cancers. Cell. 149:979–993. 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S., Wedge D.C., Alexandrov L.B., Petljak M., Butler A.P., Bolli N., Davies H.R., Knappskog S., Martin S., Papaemmanuil E., et al. 2014. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 46:487–491. 10.1038/ng.2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal, S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. 2016. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 534:47–54. 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, I.M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., and Honjo T.. 2003. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197:1173–1181. 10.1084/jem.20030275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama, S., Marusawa H., Matsumoto T., Ueda Y., Matsumoto Y., Endo Y., Takai A., and Chiba T.. 2012. Excessive activity of apolipoprotein B mRNA editing enzyme catalytic polypeptide 2 (APOBEC2) contributes to liver and lung tumorigenesis. Int. J. Cancer. 130:1294–1301. 10.1002/ijc.26114 [DOI] [PubMed] [Google Scholar]
- Olson, M.E., Harris R.S., and Harki D.A.. 2018. APOBEC enzymes as targets for virus and cancer therapy. Cell Chem. Biol. 25:36–49. 10.1016/j.chembiol.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petljak, M., Alexandrov L.B., Brammeld J.S., Price S., Wedge D.C., Grossmann S., Dawson K.J., Ju Y.S., Iorio F., Tubio J.M.C., et al. 2019. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell. 176:1282–1294.e20. 10.1016/j.cell.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland, E.W., Stenglein M.D., Shindo K., Albin J.S., Brown W.L., and Harris R.S.. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284. 10.1093/nar/gkq174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani, D.F., and Nussenzweig M.C.. 2013. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 8:79–103. 10.1146/annurev-pathol-020712-164004 [DOI] [PubMed] [Google Scholar]
- Roberts, S.A., and Gordenin D.A.. 2014. Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer. 14:786–800. 10.1038/nrc3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G., et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45:970–976. 10.1038/ng.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasatani, M., Xi Y., Kajimura J., Kawamura T., Piao J., Masuda Y., Honda H., Kubo K., Mikamoto T., Watanabe H., et al. 2017. Overexpression of Rev1 promotes the development of carcinogen-induced intestinal adenomas via accumulation of point mutation and suppression of apoptosis proportionally to the Rev1 expression level. Carcinogenesis. 38:570–578. 10.1093/carcin/bgw208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutsky, E.K., Nabel C.S., Davis A.K.F., DeNizio J.E., and Kohli R.M.. 2017. APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA. Nucleic Acids Res. 45:7655–7665. 10.1093/nar/gkx345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebrenik, A.A., Starrett G.J., Leenen S., Jarvis M.C., Shaban N.M., Salamango D.J., Nilsen H., Brown W.L., and Harris R.S.. 2019. The deaminase APOBEC3B triggers the death of cells lacking uracil DNA glycosylase. Proc. Natl. Acad. Sci. USA. 116:22158–22163. 10.1073/pnas.1904024116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban, N.M., Shi K., Lauer K.V., Carpenter M.A., Richards C.M., Salamango D., Wang J., Lopresti M.W., Banerjee S., Levin-Klein R., et al. 2018. The antiviral and cancer genomic DNA deaminase APOBEC3H is regulated by an RNA-mediated dimerization mechanism. Mol. Cell. 69:75–86.e9. 10.1016/j.molcel.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S., Patnaik S.K., Taggart R.T., Kannisto E.D., Enriquez S.M., Gollnick P., and Baysal B.E.. 2015. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 6:6881. 10.1038/ncomms7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, K., Carpenter M.A., Banerjee S., Shaban N.M., Kurahashi K., Salamango D.J., McCann J.L., Starrett G.J., Duffy J.V., Demir Ö., et al. 2017. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 24:131–139. 10.1038/nsmb.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts, A.M., Willis S., Burns M.B., Look M.P., Meijer-Van Gelder M.E., Schlicker A., Heideman M.R., Jacobs H., Wessels L., Leyland-Jones B., et al. 2014. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer. 5:405–413. 10.1007/s12672-014-0196-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts, A.M., Schrijver W.A., Dalm S.U., de Weerd V., Moelans C.B., Ter Hoeve N., van Diest P.J., Martens J.W., and van Deurzen C.H.. 2017. Progressive APOBEC3B mRNA expression in distant breast cancer metastases. PLoS One. 12. e0171343. 10.1371/journal.pone.0171343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, V., Bloch N., and Landau N.R.. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 16:546–553. 10.1038/ni.3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena, S.U., Chen K., and Bhagwat A.S.. 2016. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 116:12688–12710. 10.1021/acs.chemrev.6b00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena, S.U., Perera M.L.W., Senevirathne V., Stewart J., and Bhagwat A.S.. 2018. A tumor-promoting phorbol ester causes a large increase in APOBEC3A expression and a moderate increase in APOBEC3B expression in a normal human keratinocyte cell line without increasing genomic uracils. Mol. Cell. Biol. 39. e00238-18. 10.1128/MCB.00238-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, T.K., Scott P.M., Marsh B.M., Zhao L., Than B.L., O’Sullivan M.G., Sarver A.L., Dupuy A.J., Largaespada D.A., and Cormier R.T.. 2011. A Sleeping Beauty transposon-mediated screen identifies murine susceptibility genes for adenomatous polyposis coli (Apc)-dependent intestinal tumorigenesis. Proc. Natl. Acad. Sci. USA. 108:5765–5770. 10.1073/pnas.1018012108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrett, G.J., Luengas E.M., McCann J.L., Ebrahimi D., Temiz N.A., Love R.P., Feng Y., Adolph M.B., Chelico L., Law E.K., et al. 2016. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat. Commun. 7:12918. 10.1038/ncomms12918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrett, G.J., Thakuria M., Chen T., Marcelus C., Cheng J., Nomburg J., Thorner A.R., Slevin M.K., Powers W., Burns R.T., et al. 2020. Clinical and molecular characterization of virus-positive and virus-negative Merkel cell carcinoma. Genome Med. 12:30. 10.1186/s13073-020-00727-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou, S., Crawford D., Blouch K., Browne E.P., Kohli R.M., and Ross S.R.. 2014. Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo. PLoS Pathog. 10. e1004145. 10.1371/journal.ppat.1004145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein, M.D., Burns M.B., Li M., Lengyel J., and Harris R.S.. 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17:222–229. 10.1038/nsmb.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène, R., Aynaud M.M., Vartanian J.P., and Wain-Hobson S.. 2013. Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase. PLoS One. 8. e63461. 10.1371/journal.pone.0063461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton, C., McGranahan N., Starrett G.J., and Harris R.S.. 2015. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5:704–712. 10.1158/2159-8290.CD-15-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen, B.K., McNevin J.P., McElrath M.J., Hunt B.V., Klein K.C., and Lingappa J.R.. 2010. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J. Biol. Chem. 285:27753–27766. 10.1074/jbc.M110.102822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan, S., Rosenthal R., Kanu N., McGranahan N., Bartek J., Quezada S.A., Hare J., Harris R.S., and Swanton C.. 2018. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 29:563–572. 10.1093/annonc/mdy003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, V.C., Leonard B., White E.A., Starrett G.J., Temiz N.A., Lorenz L.D., Lee D., Soares M.A., Lambert P.F., Howley P.M., et al. 2014. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio. 5. e02234-14. 10.1128/mBio.02234-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, B.A., Wardell C.P., Murison A., Boyle E.M., Begum D.B., Dahir N.M., Proszek P.Z., Melchor L., Pawlyn C., Kaiser M.F., et al. 2015. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat. Commun. 6:6997. 10.1038/ncomms7997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangensteen, K.J., Wilber A., Keng V.W., He Z., Matise I., Wangensteen L., Carson C.M., Chen Y., Steer C.J., McIvor R.S., et al. 2008. A facile method for somatic, lifelong manipulation of multiple genes in the mouse liver. Hepatology. 47:1714–1724. 10.1002/hep.22195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, C.J., Xu T., Guo K., Griffin L.M., Westrich J.A., Lee D., Lambert P.F., Santiago M.L., and Pyeon D.. 2015. APOBEC3A functions as a restriction factor of human papillomavirus. J. Virol. 89:688–702. 10.1128/JVI.02383-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., and Stuart J.M.; Cancer Genome Atlas Research Network . 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45:1113–1120. 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber, A., Frandsen J.L., Wangensteen K.J., Ekker S.C., Wang X., and McIvor R.S.. 2005. Dynamic gene expression after systemic delivery of plasmid DNA as determined by in vivo bioluminescence imaging. Hum. Gene Ther. 16:1325–1332. 10.1089/hum.2005.16.1325 [DOI] [PubMed] [Google Scholar]
- Xu, L., Chang Y., An H., Zhu Y., Yang Y., and Xu J.. 2015. High APOBEC3B expression is a predictor of recurrence in patients with low-risk clear cell renal cell carcinoma. Urol. Oncol. 33:340.e1-340.e8. 10.1016/j.urolonc.2015.05.009 [DOI] [PubMed] [Google Scholar]
- Yamanaka, S., Balestra M.E., Ferrell L.D., Fan J., Arnold K.S., Taylor S., Taylor J.M., and Innerarity T.L.. 1995. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. USA. 92:8483–8487. 10.1073/pnas.92.18.8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, H., Shirakawa K., Matsumoto T., Hirabayashi S., Murakawa Y., Kobayashi M., Sarca A.D., Kazuma Y., Matsui H., Maruyama W., et al. 2019. Endogenous APOBEC3B overexpression constitutively generates DNA substitutions and deletions in myeloma cells. Sci. Rep. 9:7122. 10.1038/s41598-019-43575-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, S., He F., Gao B., Wu H., Li M., Huang L., Liang J., Wu Q., and Li Y.. 2016. Increased APOBEC3B predicts worse outcomes in lung cancer: a comprehensive retrospective study. J. Cancer. 7:618–625. 10.7150/jca.14030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zor, T., and Selinger Z.. 1996. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal. Biochem. 236:302–308. 10.1006/abio.1996.0171 [DOI] [PubMed] [Google Scholar]