Arnold et al. report that eosinophils in intestinal tumors are conditioned by GM-CSF to promote antitumor immunity through the activation of Th1 and CD8+ T cell responses. GM-CSF activates IRF5 and can be administered recombinantly to reduce tumor growth. Colorectal cancer patients exhibiting high intratumoral eosinophil infiltration also have better prognosis.
Abstract
The depletion of eosinophils represents an efficient strategy to alleviate allergic asthma, but the consequences of prolonged eosinophil deficiency for human health remain poorly understood. We show here that the ablation of eosinophils severely compromises antitumor immunity in syngeneic and genetic models of colorectal cancer (CRC), which can be attributed to defective Th1 and CD8+ T cell responses. The specific loss of GM-CSF signaling or IRF5 expression in the eosinophil compartment phenocopies the loss of the entire lineage. GM-CSF activates IRF5 in vitro and in vivo and can be administered recombinantly to improve tumor immunity. IL-10 counterregulates IRF5 activation by GM-CSF. CRC patients whose tumors are infiltrated by large numbers of eosinophils also exhibit robust CD8 T cell infiltrates and have a better prognosis than patients with eosinophillow tumors. The combined results demonstrate a critical role of eosinophils in tumor control in CRC and introduce the GM-CSF–IRF5 axis as a critical driver of the antitumor activities of this versatile cell type.
Introduction
Eosinophils are granulocytes arising in the bone marrow from granulocyte-monocyte progenitors; they are released into the peripheral blood as terminally differentiated cells and rapidly migrate to their target tissues. Eosinophils are classically associated with type 2 inflammation that is characteristic of parasite infections and further are known to contribute critically to the pathogenesis of allergic asthma (Lee et al., 2004). In asthma, eosinophils drive multiple hallmarks of the disease, including mucus production, smooth muscle cell hyperplasia, angiogenesis, and fibrosis (Bergeron et al., 2009), and thereby contribute to asthma exacerbations. Targeting eosinophils is now a well-established strategy for the treatment of patients with severe eosinophilic asthma that are refractory to standard of care (i.e., steroid-based treatments; Castro et al., 2011). In the steady state, only small numbers of eosinophils are released from the bone marrow; these numbers increase dramatically during type 2 inflammation (Travers and Rothenberg, 2015). Eosinopoiesis during inflammation and in the steady state is dependent on the cytokine IL-5 (Kopf et al., 1996). IL-5 signaling requires the common β-chain, which is shared with the cytokines IL-3 and GM-CSF. IL-5 acts on eosinophils at multiple time points during their lifespan. In addition to stimulating the differentiation and maturation of eosinophil-committed progenitors in the bone marrow, contributing to eosinophil egress from the bone marrow, IL-5 synergizes with chemotactic factors such as eotaxin-1 (CCL11) to attract eosinophils to tissues, primes eosinophils for activation in response to various mediators, and extends the eosinophil lifespan by blocking apoptosis (Jung and Rothenberg, 2014; Travers and Rothenberg, 2015). IL-5 overexpression is sufficient to induce massive eosinophilia but alone does not induce tissue damage (Dent et al., 1990). IL-5 neutralization or overexpression thus represents a convenient tool to study the contribution of eosinophils to health and disease.
Eosinophils constitute an abundant cellular infiltrate of solid tumors (Lotfi et al., 2007). Interestingly, in historical surveys, tumor-associated tissue eosinophilia tends to be associated with improved prognosis in solid cancers, in particular in malignancies of the gastrointestinal tract such as gastric cancer (Cuschieri et al., 2002; Iwasaki et al., 1986) and colorectal cancer (CRC; Fernández-Aceñero et al., 2000; Nielsen et al., 1999). However, only very few experimental studies have mechanistically addressed eosinophil functions in models of carcinogenesis. In a recent study, eosinophils have been reported to enhance antitumor immune responses by normalizing the tumor vasculature, promoting macrophage polarization toward an inflammatory phenotype and enhancing the infiltration of CD8+ T cells through the release of CCL5, CXCL9, and CXCL10 (Carretero et al., 2015). Eosinophils were also shown to restrict melanoma growth upon IL-33 treatment through the recruitment and activation of cytotoxic T cells and natural killer (NK) cells (Lucarini et al., 2017). In addition, eosinophils have been suggested to exert direct tumoricidal properties by releasing their granular content. To examine possible beneficial or detrimental functions of eosinophils in syngeneic and genetic CRC models, we took advantage of various constitutive and inducible models of eosinophil deficiency or overproduction. We found that CRC cells grow more rapidly and form larger tumors in mice that lack eosinophils; this critical role of eosinophils could be linked to their ability to drive CD4+ and CD8+ T cell responses within the tumor microenvironment (TME). The antitumor activities of eosinophils were found to be activated by GM-CSF signaling through the transcription factor IRF-5 and to be counterregulated by IL-10. The administration of recombinant GM-CSF effectively stimulates antitumor immunity in an eosinophil-dependent manner. The prognostic value of eosinophil infiltration and link between the presence of eosinophils and intratumoral T cell responses could further be confirmed in a large cohort of CRC patients, implicating this cell type in tumor immunity and making it an attractive target in immunotherapy against cancer.
Results
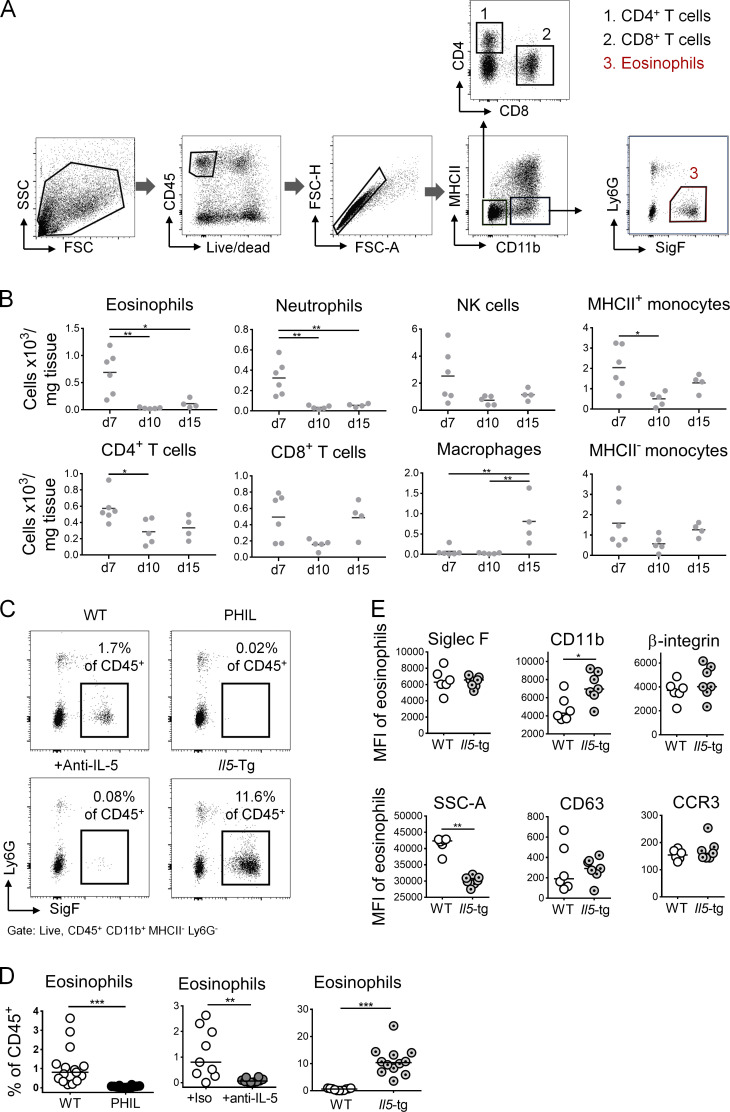
Eosinophils infiltrate subcutaneously growing MC38 tumors, and eosinophil frequencies are inversely associated with tumor growth
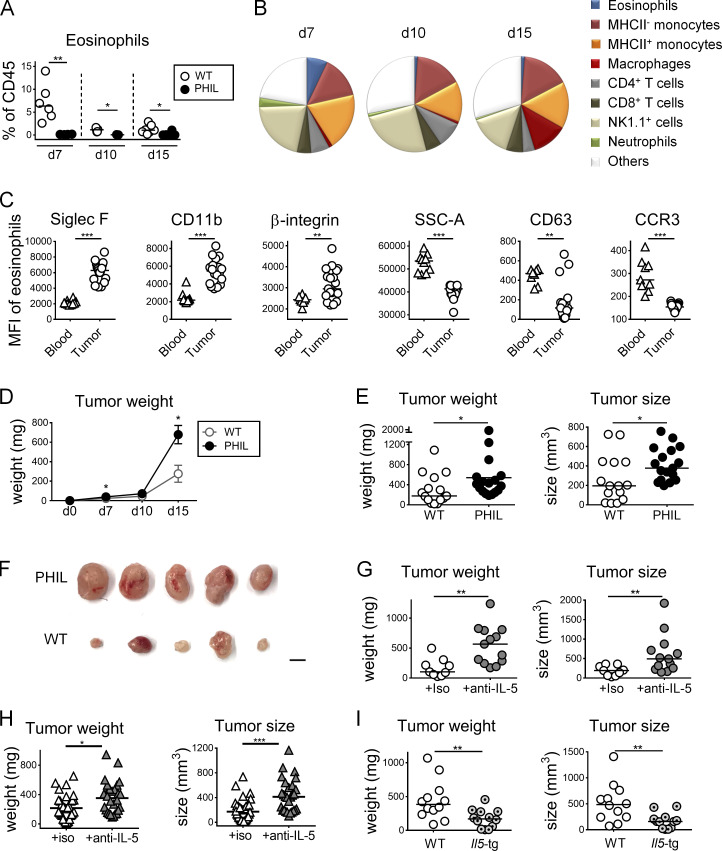
Eosinophils are known to infiltrate various solid tumors, including CRC. To examine their recruitment to and activation in the TME, we profiled the immune cell infiltrate of the subcutaneously growing CRC cell line MC38 at days 7, 10, and 15 after inoculation into the flanks of C57/BL6 mice. Eosinophils were identified as CD45+CD11b+MHCII−Ly6G− and Siglec F+ cells (see gating strategy in Fig. S1 A) and were clearly distinguishable from all other leukocyte populations in the TME. The recruitment of eosinophils was an early event in the course of MC38 growth, with eosinophils constituting one of several dominant leukocyte populations at day 7 after inoculation alongside monocytes, NK cells, and T cells, as assessed with respect to both their frequencies (Fig. 1, A and B) and absolute numbers (Fig. S1 B). At the later time points, eosinophil frequencies dwindled as tumors began to grow out and macrophages became more dominant (Fig. 1, A and B; and Fig. S1 B). The identity of eosinophils was confirmed by the specific loss of this, but not other, leukocyte populations in the TME of PHIL mice (Fig. 1 A), which transgenically express diphtheria toxin under the eosinophil peroxidase (EPO) promoter and are known to be entirely devoid of eosinophils in all tissues of the body (Lee et al., 2004). Eosinophils in the TME were more activated than their counterparts in the circulation, as judged by their expression of CD11b and Siglec F (Arnold et al., 2018; Griseri et al., 2015), but exhibited reduced granularity and concomitant reduced expression of the degranulation marker CD63 and the chemokine receptor CCR3 (Fig. 1 C). To address whether eosinophils contribute functionally to antitumor immune responses and tumor control, we compared the subcutaneous growth of MC38 cells in WT and eosinophil-deficient PHIL mice. Eosinophil deficiency led to larger and heavier tumors over time and at the study endpoint (Fig. 1, D–F; and Fig. S1, C and D). The depletion of eosinophils by another strategy (i.e., administration of a neutralizing antibody against IL-5) was effective at reducing eosinophil infiltration in the TME (Fig. S1, C and D) and had similar effects on MC38 tumor size and weight at the study endpoint as genetic eosinophil deficiency (Fig. 1 G). In both settings of eosinophil deficiency, we observed an increase in tumor-infiltrating macrophages, but not monocytes (not shown). Larger and heavier tumors were further also observed upon eosinophil depletion in a second murine CRC model (i.e., CT26 colon carcinoma cells transplanted onto the flanks of BALB/c mice; Fig. 1 H). To examine whether an excess of eosinophils would have the opposite effect on tumor growth than eosinophil deficiency, we inoculated mice that transgenically and constitutively overexpress IL-5 in T cells under the control of the CD2 promoter (Dent et al., 1990) with MC38 cells. These mice developed extreme eosinophilia, which was evident not only in the bone marrow, blood, and spleen, as reported previously (Dent et al., 1990; data not shown), but also in the TME (Fig. S1, C and D); eosinophils in the tumors of IL-5–transgenic mice were at least as mature and activated, as judged by their Siglec F and CD11b expression, as eosinophils in WT mice (Fig. S1 E). Interestingly, MC38 tumors of IL-5–transgenic mice were significantly smaller and weighed less at the study endpoint than those of WT littermates (Fig. 1 I). The combined data indicate that the frequency of tumor-infiltrating eosinophils is inversely associated with tumor growth in this model.
Figure S1.
Eosinophil depletion and overproduction by IL-5 neutralization or transgenic expression. (A) Gating strategy to identify eosinophils and CD4+ and CD8+ T cells among tumor-infiltrating CD45+ leukocytes. (B) Absolute numbers of the indicated tumor-infiltrating leukocyte populations, as quantified per mg of tumor tissue, of the tumors shown in Fig. 1 B. (C and D) Intratumoral eosinophil frequencies of WT, PHIL, anti–IL-5 antibody–treated and IL-5–transgenic mice; representative plots are shown in C along with summary plots in D of all the mice shown in the main Fig. 1. (E) Expression of the indicated surface markers on intratumoral eosinophils from IL-5–transgenic mice and their WT littermates. Data are from one representative study (n = 6–7 mice per genotype). *, P < 0.05; **, P < 0.01; ***, P < 0.001; as calculated by Mann–Whitney test (D and E) or by one-way ANOVA with Tukey’s post-test (B).
Figure 1.
Eosinophils are recruited to the TME and promote tumor control in an ectopic model of colon cancer. (A–C) PHIL mice and their WT littermates were subcutaneously injected in both flanks with 5 × 105 MC38 colon cancer cells. Tumors were analyzed at 7, 10, and 15 d after injection (n = 14–18 tumors per genotype) with respect to intratumoral eosinophil frequencies (in percentage of all CD45+ leukocytes; A), the composition of the overall leukocyte compartment (WT only; B), and the expression of the indicated surface markers (along with granularity, in corresponding blood, n = 9 versus tumor, n = 18) in the eosinophil compartment of WT mice (C). (D–F) PHIL mice and their WT littermates were injected with MC38 cells and analyzed over time (D) and at the study endpoint (day 15; E) with respect to tumor weights and volumes. Macroscopic images of representative tumors are shown in F; scale bar represents 0.5 cm (n = 14–15 tumors per genotype). (G) C57BL/6 mice were injected with MC38 cells and treated twice weekly with 250 µg/dose of isotype control (iso) or anti–IL-5 antibody. Tumor weights and volumes at the endpoint are shown (n = 9–13 tumors per condition). (H) BALB/c mice were injected with 5 × 105 CT26 colon cancer cells and received twice-weekly injections of 250 µg/dose of isotype control (iso) or anti–IL-5 antibody. Tumor weights and volumes at the study endpoint (day 20) are plotted (n = 28–32 tumors per condition). (I) IL-5–transgenic mice and their WT littermates were injected with MC38 cells and analyzed with respect to tumor weights and volumes at the study endpoint (n = 12 tumors per genotype). Data from at least two and up to three independent experiments are pooled throughout. Symbols represent individual tumors; horizontal lines indicate medians. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; as calculated by Mann–Whitney test.
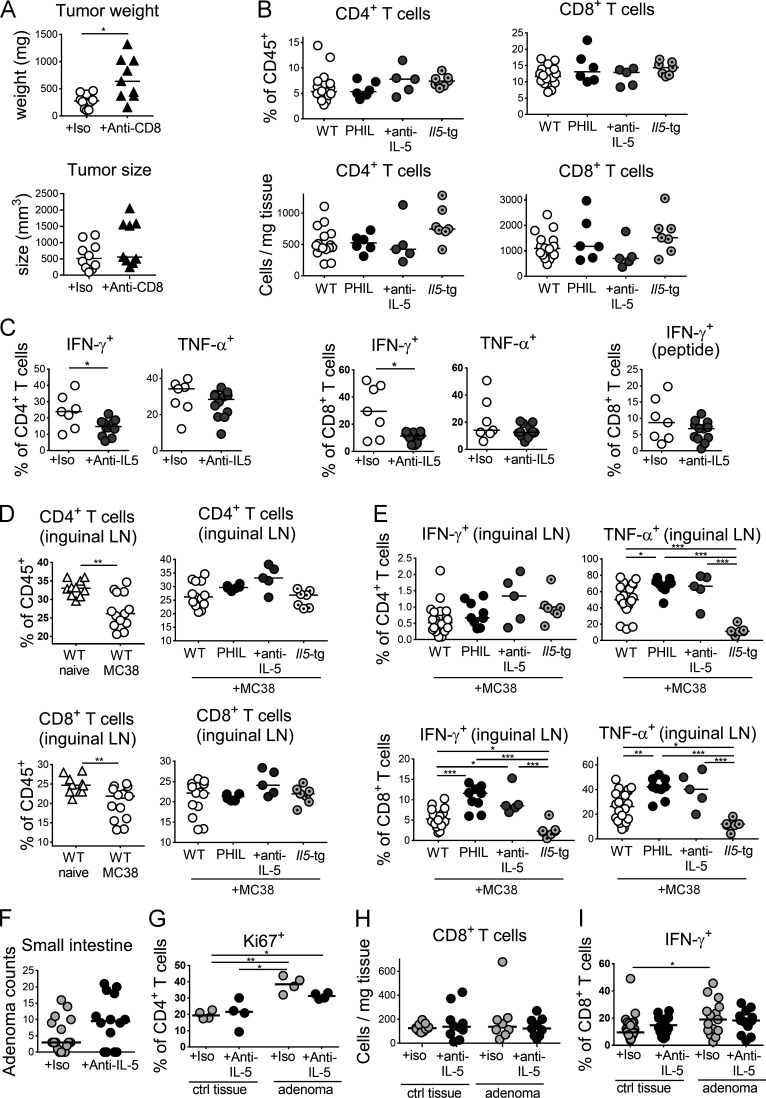
Eosinophils are required for T cell activation in the TME
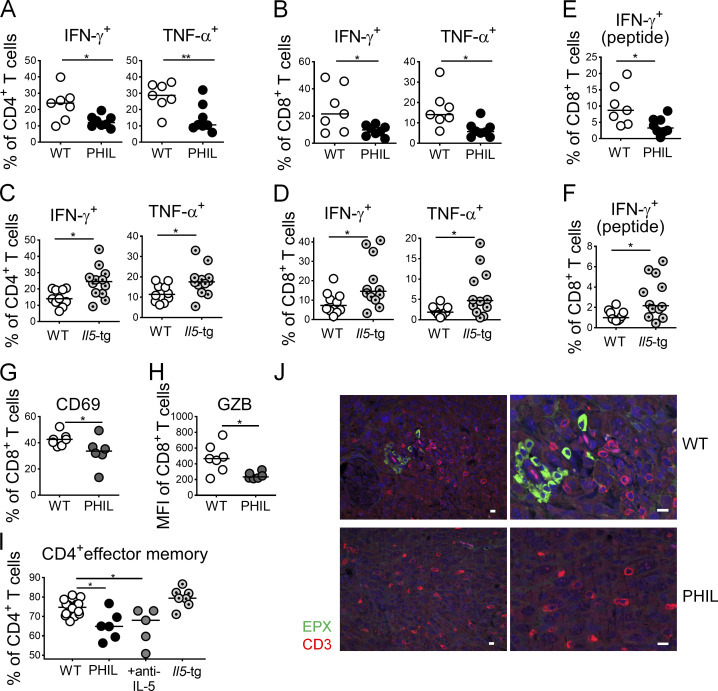
To address the mechanistic basis of the role that eosinophils play in tumor control, we examined antitumor immune responses in the TME of MC38 tumors growing on WT, eosinophil-deficient PHIL and IL-5–transgenic mice. Both CD4+ and CD8+ T cells are known to be critically involved in tumor control in this model, which we verified for CD8+ T cells by applying several doses of a CD8+ T cell–depleting antibody (Fig. S2 A). Intratumoral CD4+ and CD8+ T cell frequencies and absolute numbers were similar in PHIL mice, IL-5–depleted mice, and their WT littermates and also in IL-5–transgenic mice and their littermates (Fig. S2 B). However, we observed strong and consistent differences in IFN-γ and TNF-α production by both CD4+ and CD8+ T cells upon restimulation with PMA and ionomycin, with reduced cytokine production in the absence of eosinophils (Fig. 2, A and B; and Fig. S2 C) and increased cytokine production in their excessive presence (Fig. 2, C and D). The effects on cytokine production were also seen in CD8+ T cells that were restimulated with a tumor-specific peptide (Fig. 2, E and F; and Fig. S2 C) and were mirrored by corresponding changes in granzyme B expression and expression of the activation marker CD69 by CD8+ T cells (Fig. 2, G and H). The effects of eosinophil loss or overproduction on T cell activation were specific to the TME and were not recapitulated in the draining inguinal LNs of tumor-bearing mice (Fig. S2, D and E; data not shown). Quite to the contrary, whereas in WT mice, ectopic tumor growth led to emigration of T cells from the LNs (presumably to the tumor), this was not the case for PHIL or IL-5–depleted mice (Fig. S2, D and E); rather, we found evidence of retention of activated T cells in the tumor-draining LNs under conditions of eosinophil deficiency, indicating that T cells fail to exit the LN and migrate to their target tissues if eosinophils are absent (Fig. S2, D and E). The effects of eosinophil deficiency on T cell activation or the local expansion of activated T cells specifically affected the CD62LloCD44+ effector memory pool of CD4+ T cells (Fig. 2 I) and were not observed for naive CD62LhiCD44− T cells and CD62LhiCD44+ central memory T cells, the frequencies of which were normal (data not shown). The combined results suggest a critical role of eosinophils during the local activation or expansion of effector T cells rather than during their priming in LNs. Consistent with this model, T cells were found clustered and in close proximity to eosinophils in WT mice, whereas they appeared to be more dispersed in PHIL mice (Fig. 2 J). The combined results indicate that eosinophils promote the recruitment of cytokine-producing CD4+ and CD8+ T cells to the TME.
Figure S2.
The manipulation of the eosinophil compartment modulates T cell responses in the tumor and draining LNs. (A) Tumor weights and volumes at the study endpoint of WT mice injected with MC38 cells and treated twice weekly with 250 µg/dose of a CD8+ T cell–depleting antibody or its isotype control (n = 9–10 tumors per group). (B) CD4+ and CD8+ T cell frequencies among all leukocytes (upper panels) and absolute numbers per milligram of tumor tissue (lower panels) of PHIL, anti–IL-5 antibody–treated, and IL-5–transgenic mice shown in main Fig. 2 relative to their WT littermates. (C) Intratumoral frequencies of IFN-γ+ and TNF-α+ CD4+ and CD8+ T cells as assessed by intracellular cytokine staining upon restimulation with PMA/ionomycin and of CD8+ T cells upon restimulation with MC-38–specific peptide of the anti–IL-5–treated mice shown in Fig. 1 G. (D) Frequencies of CD4+ and CD8+ T cells among all leukocytes in the tumor-draining inguinal LNs of MC38 tumor-bearing relative to naive mice (left panels, n = 10–13 LNs per group) and of tumor-bearing PHIL, IL-5–transgenic, and anti–IL-5–treated mice and their WT littermates presented in main Figs. 1 and 2 (right panels). (E) Frequencies of IFN-γ+ and TNF-α+ CD4+ and CD8+ T cells as assessed by intracellular staining upon restimulation with PMA/ionomycin, in the inguinal LNs of tumor-bearing PHIL, IL-5–transgenic, and anti–IL-5–treated mice and their WT littermates (n = 5–21 LNs per group). (F–I) ApcMin/+ mice were treated twice weekly with 250 µg of anti–IL-5 or isotype control antibody for 3 wk beginning at 12 wk of age. At the study endpoint, small intestinal and colonic adenomas were harvested along with adjacent normal (tumor-free) colonic tissue per mouse for flow cytometric analysis. Small intestinal adenoma counts are shown in F (n = 14–17 mice per condition); frequencies of Ki67+ cells among all CD4+ T cells and CD8+ T cell counts are shown for colonic adenomas and adjacent colonic lamina propria in G and H. Frequencies of IFN-γ+ cells among all CD8+ T cells of tumor and adjacent normal tissue are shown in I. Data in D and G are from one representative study in E and H and from two pooled studies and in F and I from three pooled studies. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 2.
Eosinophils drive tumor-specific T cell responses in the TME. (A–H) PHIL (A, B, E, G, and H) and IL-5-transgenic mice (C, D, and F) and their littermates (WT) were subcutaneously injected with 5 × 105 MC38 cells and analyzed after 15 d with respect to the frequencies of intratumoral IFN-γ+ and TNF-α+ CD4+ T cells and of IFN-γ+ and TNF-α+ CD8+ T cells as assessed by intracellular cytokine staining upon restimulation with PMA/ionomycin and of IFN-γ+ CD8+ T cells upon stimulation with MC38-specific peptide where indicated (“peptide”; A–F). Littermates were used throughout (n = 7–12 tumors per genotype); note that the IL-5–transgenic line generally mounts weaker intratumoral T cell responses than the PHIL line, which is reflected in the generally lower frequencies in C, D, and F relative to A, B, and E. Tumors were further stained for expression of the activation marker CD69 and granzyme B in the CD8 compartment (G and H, n = 6–7 tumors per genotype). (I) CD4+ effector memory cells were identified by staining for CD62L and CD44, of the mice shown in A–F as well as tumor-bearing WT mice treated with anti–IL-5 neutralizing antibody (n = 5–16 tumors per condition). (J) Representative micrographs of eosinophil peroxidase (EPX)– and CD3-stained formalin-fixed sections of MC38 tumors growing on WT and PHIL mice. Scale bars represent 10 µm. Data from one representative of at least two independent experiments are shown throughout. *, P < 0.05; **, P < 0.01; as calculated by Mann–Whitney test (A–H) or by one-way ANOVA with Tukey’s post-test (I).
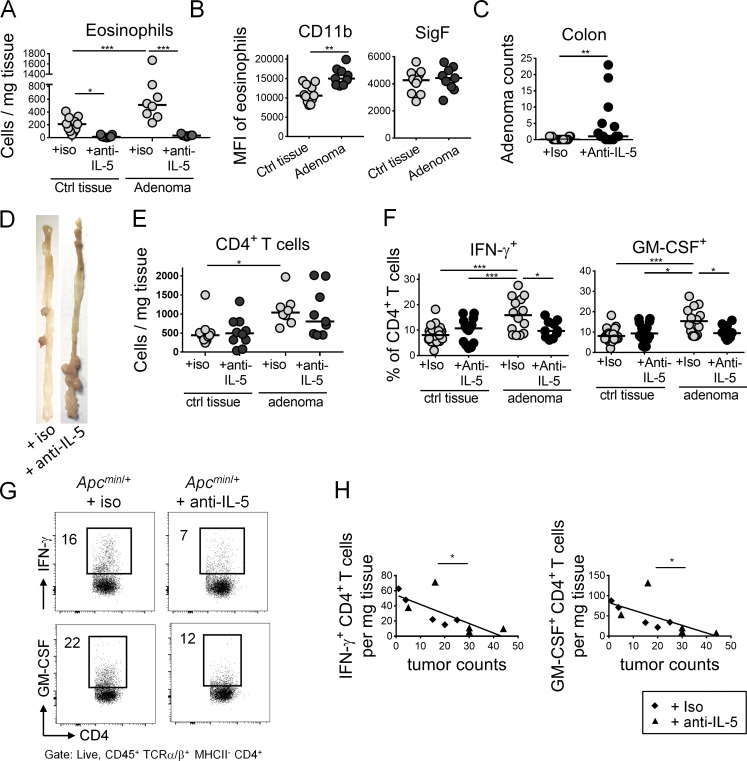
Eosinophil depletion impairs T helper type 1 (Th1) responses and enhances the tumor burden in the ApcMin/+ model of intestinal tumorigenesis
To confirm our findings in a genetic model of intestinal tumorigenesis, we depleted eosinophils for three consecutive weeks in ApcMin/+ mice (Min, multiple intestinal neoplasia) through regular twice-weekly injection of anti–IL-5 antibody beginning at 12 wk of age. ApcMin/+ mice carry a heterozygous germline mutation at codon 850 of the Apc gene (Su et al., 1992), and loss of the second (WT) Apc allele in intestinal epithelium precedes adenoma formation (Luongo et al., 1994). ApcMin/+ mice are known to develop adenomas along the length of the small intestine and colon (Su et al., 1992). Eosinophil depletion by anti–IL-5 treatment was highly efficient also in this strain, as determined in colonic tumor tissue and adjacent normal colonic lamina propria (Fig. 3 A). Eosinophils were enriched in the TME of eosinophil-replete ApcMin/+ mice treated with isotype control antibody relative to adjacent normal colon tissue (Fig. 3 A) and expressed higher levels of the activation marker CD11b, but not Siglec F (Fig. 3 B). Eosinophil depletion resulted in a higher adenoma burden in the colon (Fig. 3 C) and small intestine (Fig. S2 F) of ApcMin/+ mice. Adenomas in ApcMin/+ mice developed predominantly in the ileum and distal colon (Fig. 3 D), which was true irrespective of the eosinophil status and confirmed previous studies in ApcMin/+ mice (Su et al., 1992). Interestingly, colonic adenomas harvested from control antibody–treated mice exhibited a higher abundance of CD4+ T cells (Fig. 3 E), higher frequencies of CD4+ T cells expressing IFN-γ and GM-CSF than adjacent normal colonic tissue, and higher frequencies than tumors from eosinophil-depleted mice (Fig. 3, F and G). More CD4+ T cells were actively cycling (i.e., stained positive for Ki67) in eosinophil-replete tumors relative to adjacent tissue or eosinophil-depleted tumors (Fig. S2 G). In contrast, CD8+ T cell numbers and the frequencies of IFN-γ–expressing CD8+ T cells did not differ as a consequence of eosinophil ablation (Fig. S2, H and I). Interestingly, the absolute numbers of intratumoral CD4+ T cells expressing IFN-γ and GM-CSF per gram of tissue not only showed the same trend but also were inversely correlated with adenoma multiplicity across mice in the study (Fig. 3 H). The combined results suggest that eosinophils contribute to tumor control in this genetic tumor model through the recruitment and/or activation of tumor-infiltrating CD4+ T cells.
Figure 3.
Eosinophils are recruited to adenomas of ApcMin/+ mice and promote antitumor CD4+ T cell responses. ApcMin/+ mice were treated twice weekly with 250 µg of anti–IL-5 or isotype control antibody for 3 wk beginning at 12 wk of age (n = 14–17 mice per condition). At the study endpoint, adenoma formation in the colon was quantified by counting individual polyps with diameters of >1 mm; colonic adenomas were harvested along with adjacent normal (tumor-free) colonic tissue per mouse for flow cytometric analysis of the TME relative to normal colonic lamina propria. (A) Eosinophil numbers per milligram of tumor and adjacent normal tissue of mice treated with anti–IL-5 or isotype control antibody. (B) Eosinophil activation in tumor and normal tissue of isotype control antibody–treated mice, as assessed by flow cytometric analysis of CD11b and Siglec F (SigF) expression. (C) Colonic adenoma counts of anti–IL-5 or isotype control antibody–treated mice. (D) Representative macroscopic images of the colon of an ApcMin/+ mouse treated with anti–IL-5 or isotype control antibody. (E) CD4+ T cell numbers per milligram of colonic adenoma and adjacent normal tissue of mice treated with anti–IL-5 or isotype control antibody. (F and G) Intratumoral frequencies of IFN-γ+ and GM-CSF+ CD4+ T cells in tumor and normal colon tissue of the mice shown in A–C. Summary plots are shown in F alongside representative FACS plots in G. Numbers indicate frequencies (%) in respective gates. (H) Intratumoral counts per mg of tissue, of IFN-γ+ CD4+ T cells (ρ, correlation coefficient = −0.7368) and GM-CSF+ CD4+ T cells (ρ = −0.6507) relative to tumor counts. Data in A, B, E, and H are pooled from two (n = 8–11 samples per condition), in C and F from three independent experiments (n = 10–24 samples per experiment). Horizontal lines indicate medians. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; as calculated by Mann–Whitney test (B and C), by one-way ANOVA with Tukey’s post-test (A, E, and F) or by linear regression (H).
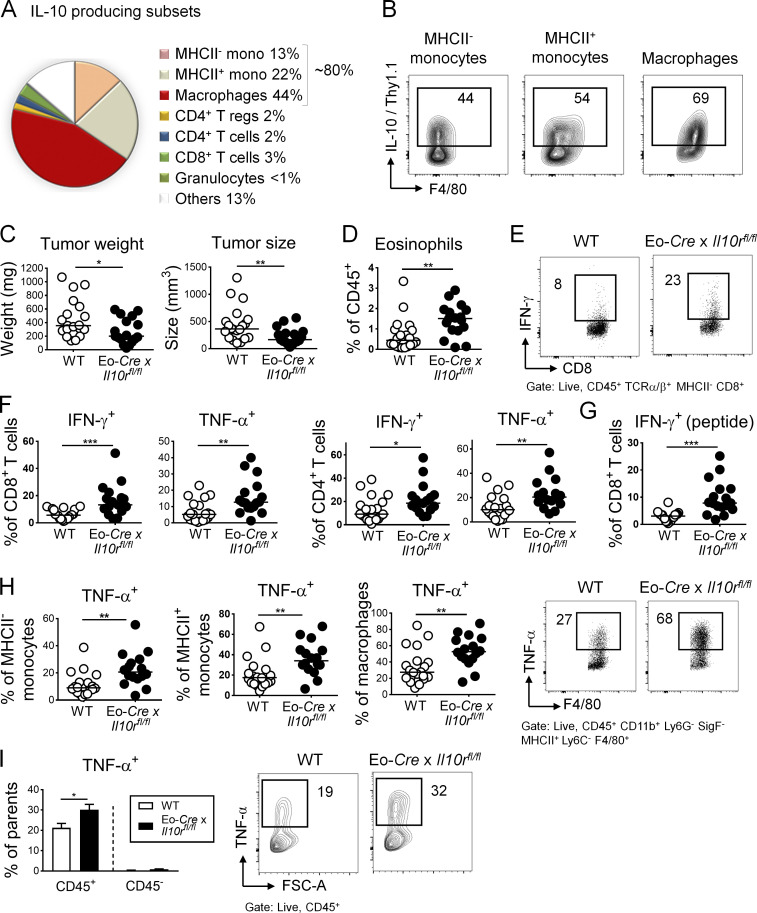
Eosinophils in the TME are negatively regulated by IL-10
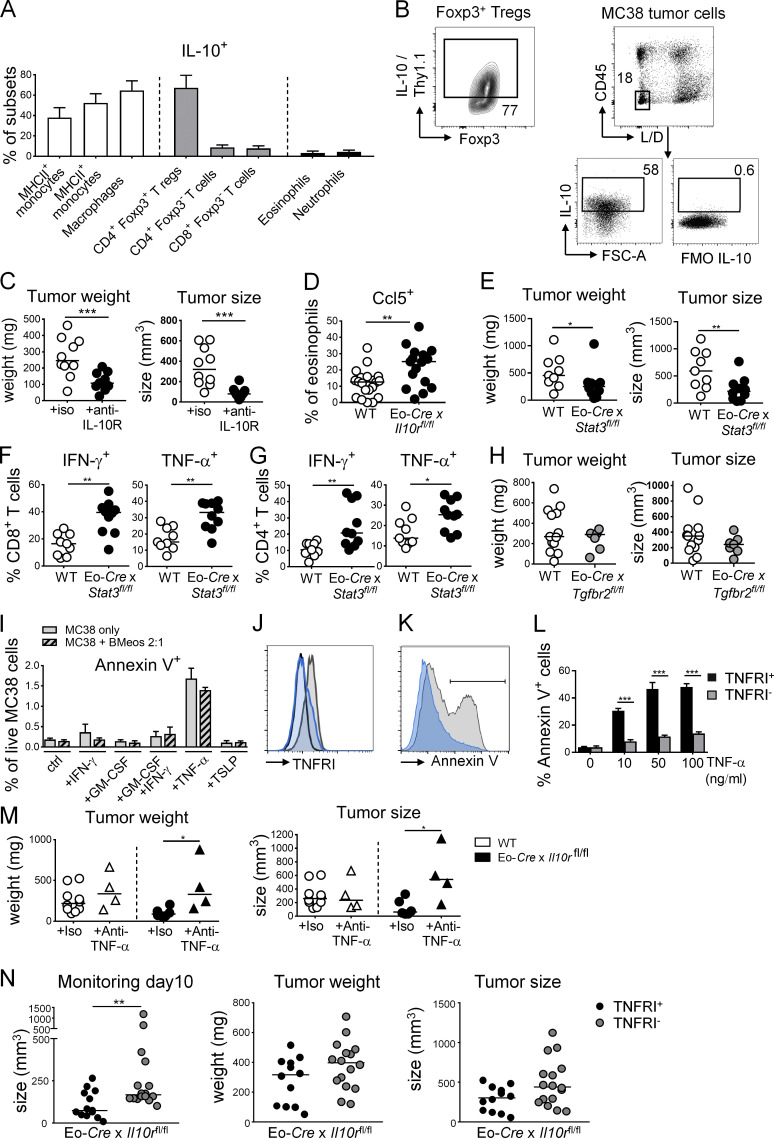
The regulatory cytokine IL-10 is known to dampen antitumor immune responses in the TME, with effects described on a variety of tumor-infiltrating cells of lymphoid and myeloid origins. We speculated that eosinophil activities in the TME might be regulated by IL-10. We first assessed which cell types produce IL-10 in MC38 tumors using a dual Il10/Foxp3 reporter in which the expression of Thy1.1 from a BAC transgene is driven by Il10 regulatory elements and GFP is expressed under the control of the Foxp3 promoter (Maynard et al., 2007). Of all IL-10–expressing cells in the TME, the vast majority were CD45+ leukocytes of myeloid origin (∼80% of Thy1.1+ cells), with most Thy1.1+ cells expressing the monocyte and macrophage markers Ly6C and F4/80, respectively, in addition to CD11b (Fig. 4 A). One half to two thirds of intratumoral monocytes and macrophages were positive for IL-10/Thy1.1 (Fig. 4 A); such frequencies were comparable to (macrophages) or slightly higher (monocytes) than IL-10/Thy1.1+ frequencies of their cellular counterparts in the colonic lamina propria of the same mice (Fig. S3 A). One of the few nonmyeloid cellular sources of intratumoral IL-10 were Foxp3+ regulatory T cells (Fig. 4 A and Fig. S3 B), which however contributed little (2%) to the overall IL-10+ leukocyte pool. In addition to the various leukocyte sources of IL-10, MC38 cells also express copious amounts of this cytokine (Fig. S3 B). To address whether IL-10 in the TME affects eosinophil activities, we crossed Il10rafl/fl mice with a strain that expresses Cre under the eosinophil peroxidase promoter (Eo-Cre; Doyle et al., 2013) to generate mice in which only the eosinophil compartment lacks the IL-10 receptor (IL-10R), and transplanted them subcutaneously with MC38 cells. Eo-Cre × Il10rafl/fl mice exhibited a phenotype that was in stark contrast to the phenotypes of eosinophil-deficient mice, as they showed improved control of the tumor burden (Fig. 4 C), with an effect size that was similar to the global neutralization of the IL-10R with a neutralizing antibody (Fig. S3 C). Eo-Cre × Il10rafl/fl mice showed strongly enhanced numbers of tumor-infiltrating eosinophils (Fig. 4 D), and intratumoral IL-10R–deficient eosinophils expressed copious amounts of the chemokine CCL5 (Fig. S3 D). CCL5 is required for eosinophil recruitment into target tissues; therefore, its excessive production provides an explanation for the overrepresentation of eosinophils in tumors of Eo-Cre × Il10rafl/fl mice. The improved tumor control observed in this model correlated with higher frequencies of IFN-γ– and TNF-α–expressing CD4+ and CD8+ T cells (Fig. 4, E–G) and with higher frequencies of intratumoral TNF-α+ monocytes and macrophages (Fig. 4 H). Indeed, the entire intratumoral production of TNF-α increased in the tumors of Eo-Cre × Il10rafl/fl mice, which could be traced to CD45+ leukocytes (Fig. 4 I). Interestingly, the eosinophil-specific loss of STAT3, a critical transcription factor mediating IL-10 signaling, in Eo-Cre × Stat3fl/fl mice phenocopied the effects of IL-10R loss in eosinophils (Fig. S3, E–G). In contrast, the eosinophil-specific ablation of TGF-β signaling by crossing of Tgfbr2fl/fl with Eo-Cre mice had no effect on the tumor burden (Fig. S3 H). The combined results indicate that eosinophils have antitumor activities that are negatively regulated by the IL-10–IL-10R–STAT3 signaling axis, but not by TGF-β; one potentially relevant feature of the TME of tumors growing in the absence of IL-10 signaling in eosinophils is the increased production of TNF-α.
Figure 4.
Eosinophil activities in the TME are suppressed by IL-10. (A and B) IL-10 reporter (10BiT) mice were subcutaneously injected with 5 × 105 MC38 cells and analyzed after 15 d with respect to their intratumoral frequencies of Thy1.1 (IL-10)+ myeloid cells, granulocytes, and T cells. Average frequencies as assessed in independent tumors are shown in A alongside representative FACS plots for the indicated major IL-10–producing myeloid populations in B (n = 8 tumors). (C–I) Eo-Cre × Il10rafl/fl mice and their Cre-negative littermates were subcutaneously injected with 5 × 105 MC38 cells and analyzed after 15 d with respect to their tumor weights and volumes (C), their intratumoral frequencies of eosinophils (D), and their intratumoral frequencies of IFN-γ+ and TNF-α+ CD4+ and CD8+ T cells (F, representative FACS plots in E; PMA and ionomycin) and IFN-γ+ CD8+ T cells upon restimulation with MC38-specific peptide (G). (H) Frequencies of TNF-α+ monocytes and macrophages among their respective parent populations, shown alongside representative FACS plots for macrophages. (I) Frequencies of TNF-α+ cells among CD45+ leukocytes and among all CD45− cells in the tumor. Data in C–I are pooled from three independent studies, n = 17–20 tumors per genotype. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; as calculated by Mann–Whitney test.
Figure S3.
The IL-10–STAT3 signaling axis in eosinophils suppresses their antitumor properties. (A) Frequencies of Thy1.1 (IL-10)+ cells in the indicated cellular compartments of the colonic lamina propria in (naive) IL-10 reporter (10BiT) mice (n = 8 mice). Means + SD are shown. (B) Representative FACS plot of Thy1.1 (IL-10) expression by intratumoral Foxp3+ T reg cells and IL-10 expression by MC38 tumor cells growing subcutaneously in mice as determined by intracellular cytokine staining relative to FMO (fluorescence minus one control). FSC-A, forward scatter area; L/D, live/dead. (C) WT C57BL/6 mice were subcutaneously injected with 5 × 105 MC38 cells, treated twice weekly with anti–IL-10R or control antibody for the duration of the experiment, and analyzed after 15 d with respect to their tumor weights and volumes (n = 10 tumors per group). (D) Ccl5 expression by intratumoral eosinophils in Eo-Cre × Il10rafl/fl mice relative to their WT littermates (n = 17–20 tumors per genotype). (E–G) Eo-Cre × Stat3fl/fl mice and their WT littermates were subcutaneously injected with 5 × 105 MC38 cells and analyzed after 15 d with respect to their tumor weights and volumes (E) and their intratumoral frequencies of IFN-γ+ and TNF-α+ CD4+ and CD8+ T cells as assessed by intracellular cytokine staining upon restimulation with PMA/ionomycin (n = 9–10 tumors per genotype). (H) Eo-Cre × Tgfbr2fl/fl mice and their Cre-negative littermates were subcutaneously injected with 5 × 105 MC38 cells and analyzed after 15 d with respect to their tumor weights and volumes (n = 7–12 tumors per genotype). Data in C and H are from one experiment, data in D are pooled from three independent experiments, and data in E–G are pooled from two independent experiments. (I) Apoptosis rate as determined by Annexin V staining followed by flow cytometry of MC38 cells cultured overnight in the presence or absence of 10 ng/ml of the indicated recombinant cytokines and in the presence or absence of bone marrow–derived eosinophils at a ratio of 2:1 (tumor/eosinophil). (J) TNFRI expression of MC38 cells subjected to genomic editing with a TNFRI-specific guide RNA (in blue) or an irrelevant guide RNA (gray); cells were stained with a TNFRI-specific (gray, blue) or control antibody (black). (K and L) Apoptosis as determined by Annexin V staining followed by flow cytometry, of MC38 cells described in J cultured o/n in the presence or absence of the indicated concentrations of TNF-α. A representative Annexin V histogram is shown in K and a summary plot of four replicate samples per condition from two independent experiments is shown in L. (M) Tumor weights and volumes at the study endpoint of Eo-Cre × Il10ra fl/fl mice and their Cre-negative littermates injected with MC38 cells, and treated twice weekly with 250 µg/dose of a TNF-α–neutralizing antibody or its isotype control (n = 4–10 tumors). (N) Tumor weights and volumes at the study endpoint of Eo-Cre × Il10ra fl/fl mice that have been subcutaneously injected with MC38 cells that express either normal (TNFRI+) or low amounts (TNFRI−) as shown in J due to CRISPR-mediated deletion of the TNFR1 locus (n = 12–16 tumors per group). The tumor size on day 10 is shown alongside tumor sizes and weights at the study endpoint (day 15). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Due to their much more efficient tumor control, Eo-Cre × Il10rafl/fl mice represent a model in which the contribution of eosinophils to tumor control can be studied mechanistically. We first examined the possibility that tumor-infiltrating eosinophils themselves have tumoricidal properties. To this end, we co-cultured eosinophils derived from bone marrow by differentiation with FLT3 ligand (FLT3L), stem cell factor, and IL-5 with MC38 cells in vitro at 1:2 ratios, with and without the addition of proinflammatory cytokines that are abundant in the TME. Eosinophils showed no cytotoxic activity toward MC38 cells under these conditions; in contrast, exposure to TNF-α, but not to GM-CSF, IFN-γ, or TSLP (Thymic stromal lymphopoietin), killed a small but robust fraction of tumor cells (Fig. S3 I). The cytotoxic effect of TNF-α was dose dependent and required TNF-α receptor I (TNFRI) expression on MC38 cells, as MC38 cells in which we had deleted the TNFRI gene by genomic editing were resistant to TNF-α–induced apoptosis (Fig. S3, J–L). To examine whether TNF-α contributes to antitumor immunity in Eo-Cre × Il10rafl/fl mice, we depleted TNF-α using a neutralizing antibody for the duration of the experiment. TNF-α–depleted tumors were larger and heavier than those depleted with a control antibody; this effect was specific to Eo-Cre × Il10rafl/fl mice (Fig. S3 M). To address whether TNF-α acts by killing tumor cells in vivo, we compared the growth of TNFRI-positive and -negative cells in Eo-Cre × Il10rafl/fl mice and observed a trend toward larger and heavier tumors produced by TNFRI-negative cells at day 10 after tumor cell injection and at the study endpoint (Fig. S3 N). The combined results indicate that (1) eosinophils exhibit no obvious cytotoxic activity toward MC38 tumor cells in vitro, (2) TNF-α is a critical cytokine in MC38 immune control with direct cytotoxic activity on tumor cells, and (3) IL-10–STAT3 signaling in eosinophils controls the intratumoral availability of TNF-α.
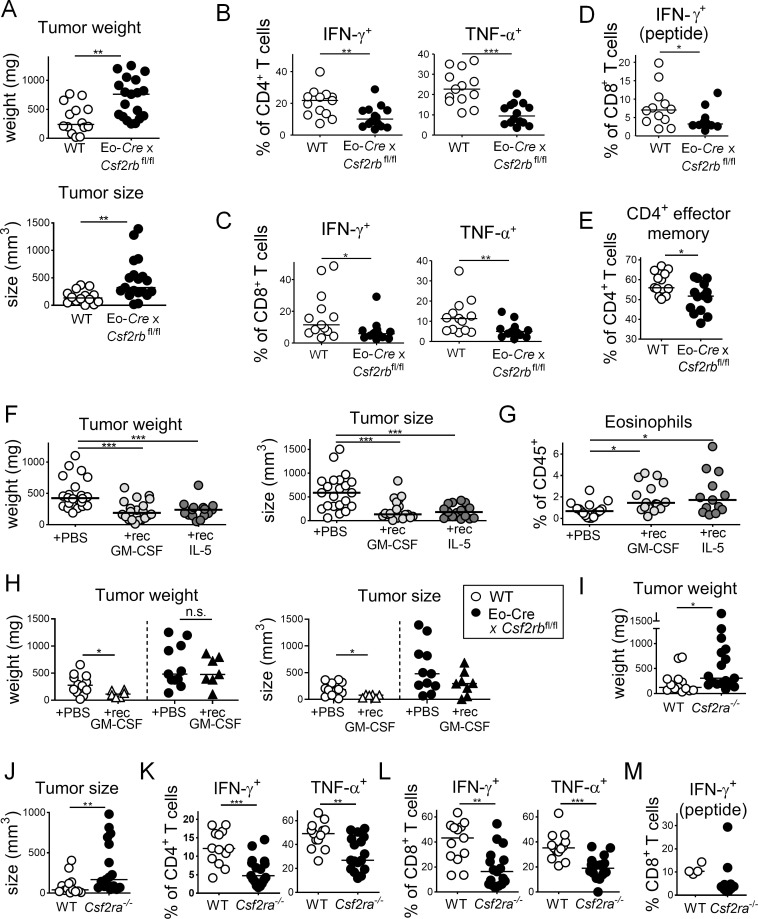
GM-CSF signaling is required and can be exploited for reducing MC38 tumor growth
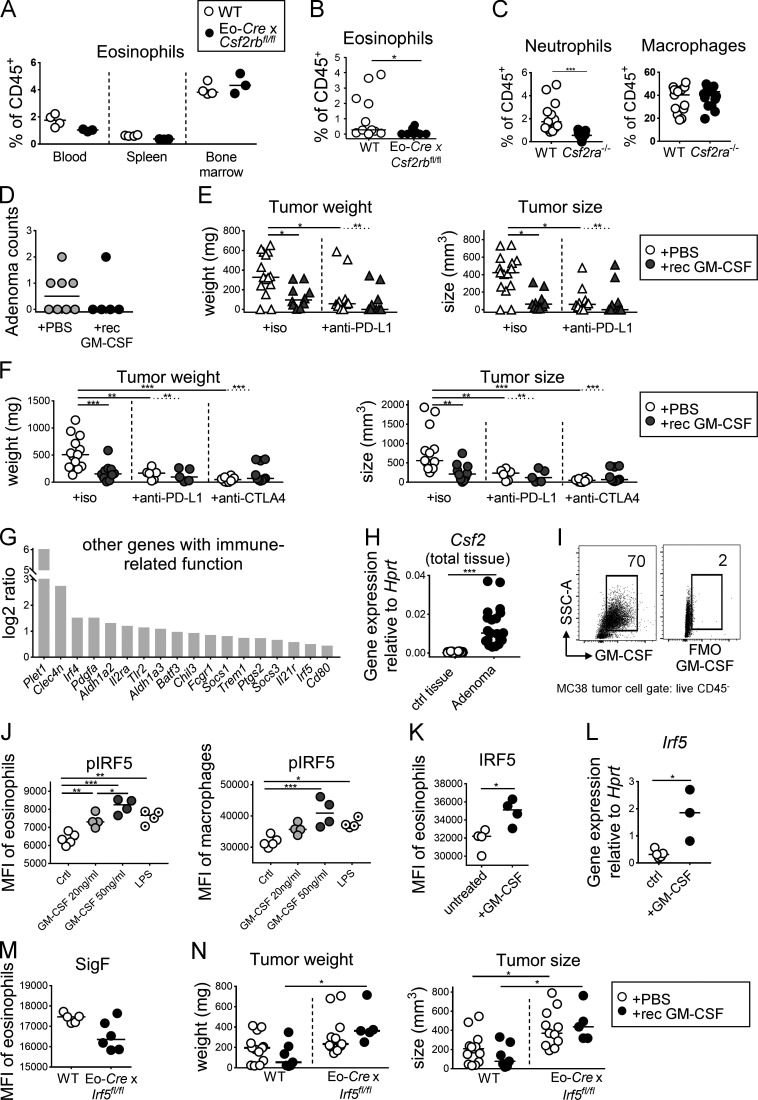
Eosinophils require the aforementioned cytokine IL-5, as well as GM-CSF, for their production in the bone marrow from myeloid progenitors, as well as their terminal differentiation and survival (Davoine and Lacy, 2014; Yamaguchi et al., 1988a, 1988b). Both cytokines bind to a distinct receptor α-chain (IL-5R-α, CSF2R-α), that upon ligand binding dimerizes with, and signals via, the shared β-chain. We generated a conditional eosinophil-specific knockout of the β-chain by crossing Csf2rbfl/fl mice with the above-mentioned Eo-Cre strain. Csf2rb deletion in the eosinophil lineage had no effect on eosinophil frequencies in the bone marrow, blood, or spleen (Fig. S4 A) and only modestly reduced the frequencies of intratumoral eosinophils (Fig. S4 B). This observation is consistent with the Eo-Cre promoter being active only in eosinophils that have egressed from the bone marrow (Arnold et al., 2018). Interestingly, MC38 cells formed larger and heavier tumors on Eo-Cre × Csf2rbfl/fl mice than on their Cre-negative littermates, as determined at the study endpoint (Fig. 5 A). Loss of GM-CSF–IL-5 signaling thus phenocopied the effect of IL-5 neutralization (Fig. 1). The larger tumors of Eo-Cre × Csf2rbfl/fl mice were infiltrated by similar absolute numbers per milligram of tissue of CD4+ and CD8+ T cells compared to their WT counterparts (data not shown) but exhibited lower frequencies of IFN-γ+ and TNF-α+ CD4+ and CD8+ T cells, as assessed by ex vivo restimulation (Fig. 5, B–D). This phenotype was mirrored by lower frequencies of CD4+ effector memory T cells (Fig. 5 E). To address whether the administration of IL-5 and/or GM-CSF in recombinant form is sufficient to promote tumor control, we i.p. administered either one or both cytokines to tumor-bearing mice once tumors were palpable. Interestingly, both cytokines effectively reduced the tumor growth and size at the study endpoint, and the combination treatment was not more efficient than either cytokine alone (Fig. 5 F). Both GM-CSF and IL-5 significantly increased eosinophil recruitment to the TME (Fig. 5 G). The tumor-suppressive effects of recombinant GM-CSF were dependent on CSF2R-β signaling in the eosinophil compartment as judged by parallel treatment of Eo-Cre × Csf2rbfl/fl mice and their WT littermates (Fig. 5 H). To obtain further proof that GM-CSF, and not IL-5, activates this signaling pathway in eosinophils, we compared tumor growth on mice that are deficient for Csf2ra. Csf2ra−/− animals phenocopied the defect of Eo-Cre × Csf2rbfl/fl mice with respect to tumor growth (Fig. 5, I and J) as well as intratumoral T cell responses (Fig. 5, K–M). While a decreased frequency of neutrophils was observed in the tumors of Csf2ra−/− mice, other myeloid populations such as macrophages were unchanged (Fig. S4 C).
Figure S4.
GM-CSF promotes tumor control through the activation of IRF5, but does not synergize with checkpoint blockade. (A) Systemic eosinophil frequencies are not affected by the deletion of Csf2rb in the eosinophil compartment. Eo-Cre × Csf2rbfl/fl mice and their Cre-negative littermates were examined with respect to their eosinophil frequencies, as determined by flow cytometry of the indicated tissues (n = 3–4 mice per genotype). (B) Eosinophil frequencies in the tumors of Eo-Cre × Csf2rbfl/fl mice and their Cre-negative littermates (n = 10–11 tumors per genotype). (C) Myeloid cell frequencies in the tumors of CSF2ra−/− mice and WT (n = 13–17 tumors per genotype). Data are pooled from two studies. (D) ApcMin/+ mice were treated three times weekly with recombinant GM-CSF during the last 3 wk of a 4-mo experiment. At the study endpoint, adenoma formation in the colon was quantified by counting individual polyps with diameters of >1 mm. Data are from one experiment and representative of two (n = 5–8 mice per group). (E) WT BALB/c mice were injected with CT26 cells and treated three times weekly with recombinant GM-CSF and/or twice weekly with a PD-L1–specific or isotype control antibody as indicated. Tumor weights and volumes are plotted for one large study (n = 10–15 tumors per group). (F) WT C57BL/6 mice were injected with MC38 cells and treated three times weekly with recombinant GM-CSF and/or twice weekly with a PD-L1–specific or CTLA4-specific antibody or isotype control as indicated. Tumor weights and volumes are pooled from two independently conducted studies (n = 6–13 tumors per group). (G) Expression of the indicated immune response–related transcripts in bone marrow–derived WT eosinophil cultures that were treated overnight with 20 ng/ml recombinant GM-CSF or vehicle control and subjected to RNA-sequencing–based transcriptome analyses, as shown in Fig. 6. The log2 ratio is shown for GM-CSF–treated cells relative to control. (H) GM-CSF (Csf2) expression, as determined by quantitative RT-PCR, in adenomas harvested from ApcMin/+ mice relative to adjacent control tissue. Each data point represents one tumor or tissue, respectively (n = 20 samples per condition). (I) GM-CSF expression, as determined by FACS, of subcutaneously growing MC38 cells at the study endpoint relative to FMO (fluorescence minus one control). (J) Splenocytes from IL-5 transgenic donors were treated for 30 min with the indicated amount of recombinant GM-CSF, or with 100 ng/ml LPS and stained for lineage markers and for phosphorylated IRF5; the p-IRF5 signal in Siglec F-positive eosinophils (left panel) and F4/80-positive macrophages (right panel) was quantified by flow cytometry. (K) Splenocytes from IL-5–transgenic donors were treated overnight with 20 ng/ml GM-CSF. Cells were stained for IRF5 and the signal in eosinophils was quantified by flow cytometry. The summary plots in J and K show the MFI (n = 4–5 parallel cultures each). (L) IRF5 expression, as determined by quantitative RT-PCR in sorted eosinophils from IL-5–transgenic splenocyte cultures that have been exposed overnight to 20 ng/ml GM-CSF (n = 3–4 samples per condition). (M) Eosinophil activation in tumors of Eo-Cre × Irf5fl/fl mice and their WT littermates, as assessed by flow cytometric analysis of Siglec F expression (n = 5–6 per genotype, P = 0.053. (N) Tumor weights and volumes of Eo-Cre × Irf5fl/fl mice and their WT littermates that have (black circles) or have not (white circles) been treated with recombinant GM-CSF three times weekly for the duration of the 2-wk experiment (n = 5–13 tumors per group). Data are pooled from two studies. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 5.
GM-CSF promotes tumor control through its activities on eosinophils. (A–E) Eo-Cre × Csf2rbfl/fl mice and their Cre-negative littermates were subcutaneously injected with 5 × 105 MC38 cells and analyzed after 15 d with respect to their tumor weights and volumes (A) as well as their intratumoral frequencies of IFN-γ+ and TNF-α+ CD4+ T cells (B), IFN-γ+ and TNF-α+ CD8+ T cells (C; PMA/ionomycin), and IFN-γ+ CD8+ T cells upon stimulation with MC38-specific peptide (D; n = 13–14 tumors per genotype). Frequencies of effector memory cells are shown as well (E). Data in A–E are pooled from two independent studies. (F–H) WT C57BL/6 mice (F and G, n = 15–22 tumors per group) and Eo-Cre × Csf2rbfl/fl mice and their Cre-negative littermates (H, n = 6–14 tumors per group) were injected with MC38 cells and treated three times weekly with recombinant GM-CSF or IL-5 as indicated. The tumor weights and volumes at the study endpoint are plotted for two (IL-5) and three (GM-CSF) independently conducted, pooled studies in F and G and a representative study of two in H. (I–M) Csf2ra−/− mice and WT controls were injected with MC38 cells and analyzed after 15 d with respect to their tumor weights and volumes (I and J) and their intratumoral frequencies of IFN-γ+ and TNF-α+ CD4+ T cells and of IFN-γ+ and TNF-α+ CD8+ T cells (K and L; PMA/ionomycin, n = 13–17), and of IFN-γ+ CD8+ T cells upon stimulation with MC38-specific peptide (M; n = 4–10). Data in I–L are pooled from three independent studies; data in M are from one representative study of three. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; n.s., not significant; as calculated by Mann–Whitney test.
Finally, we examined possible beneficial effects of GM-CSF in two additional tumor models: the CT26 subcutaneous model and the ApcMin/+ model. In ApcMin/+ mice, regular thrice-weekly GM-CSF administration during the final month of a cohort maintained for 4 mo reduced the colonic adenoma burden (Fig. S4 D); in the CT26 model, GM-CSF treatment that was initiated once tumors were palpable reduced the tumor weight and volume (Fig. S4 E). Interestingly, GM-CSF alone was as effective as checkpoint blockade with PD-L1– or CTLA4-blocking antibodies in reducing the tumor burden in both the MC38 and the CT26 tumor models (Fig. S4, E and F). Importantly, no synergy was observed between checkpoint blockade and GM-CSF treatment in either model, suggesting that eosinophils act through T cells to boost antitumor immunity. The combined results indicate that eosinophil-intrinsic GM-CSF signaling activates the tumor-suppressive functions of these granulocytes.
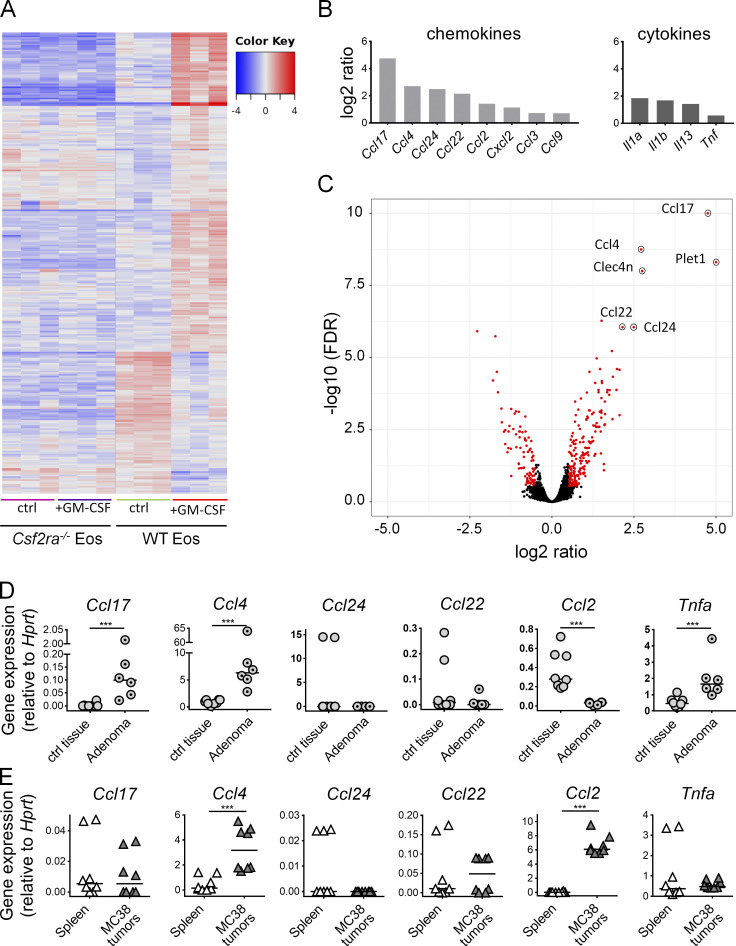
GM-CSF activates a transcriptional program in cultured eosinophils that can be recapitulated in tumor-infiltrating eosinophils
As GM-CSF has strong effects on tumor growth that are at least in part driven by its activity on eosinophils, we conducted a gene expression analysis through RNA sequencing of cultured bone marrow–derived WT or Csf2ra−/− eosinophils that were exposed or not to recombinant GM-CSF. Hierarchical clustering revealed a clear GM-CSF–specific response in WT, but not Csf2ra−/−, eosinophils (Fig. 6 A); gene ontology analysis identified the categories “inflammatory response,” “cytokine and chemokine activity,” and “CCR chemokine receptor binding” as being enriched for differentially expressed genes upon GM-CSF treatment. A volcano plot of expression changes in WT eosinophils upon GM-CSF exposure revealed the chemokines CCL4, CCL17, CCL22, CCL24, and CCL2 and to a lesser extent the cytokines IL-1α, IL-1β, IL-13, and TNF-α to be among the most obviously GM-CSF–induced, highly expressed transcripts in eosinophils (Fig. 6, B and C); none of these transcripts were induced in Csf2ra−/− eosinophils (data not shown). Several additional immune-related genes, some of them (Irf5) known to be GM-CSF target genes, were also identified as being up-regulated by GM-CSF in eosinophils (Fig. S4 G). A shared feature of most identified chemo- and cytokines is their activity in recruiting or activating T cells in the TME or other T cell–driven settings. Specifically, the processes of CD8+ T cell recruitment to sites of immune activation (CCL4; Castellino et al., 2006), activated (CCL17 and CCL22) or resting (CCL24) CD4+ T cell recruitment (Patel et al., 1997), and cytokine expression by activated CD4+ T cells (CCL22; Ushio et al., 2018) are all known to be driven by one or more of the differentially expressed eosinophil-derived chemokines. Therefore, we examined their expression in eosinophils sorted from adenomas derived from in ApcMin/+ mice relative to eosinophils sorted from adjacent tissue. We found CCL4 and CCL17, but not the other three examined CC-motif chemokines, to be up-regulated in tumor-resident eosinophils, but not control eosinophils from adjacent tissue (Fig. 6 D). The induction of CCL4 and CCL17 correlated well with the higher expression of GM-CSF in adenomas relative to adjacent tissue (Fig. S4 H). We further examined the expression of the same chemokines in eosinophils sorted from MC38 tumors relative to splenic eosinophils. In these cells, we found CCL4 and CCL2, and to some extent CCL22, to be up-regulated in the tumor context (Fig. 6 E). MC38 cells express GM-CSF, as judged by intracellular staining (Fig. S4 I), making it seem plausible that tumor-derived GM-CSF drives the eosinophil response in the TME. The combined results indicate that eosinophils respond strongly to GM-CSF, which in turn triggers T cell chemotaxis to and activation in the TME.
Figure 6.
Eosinophils respond to GM-CSF by activating the transcription of T cell–recruiting and –activating chemokines. (A–C) Triplicate bone marrow–derived WT and Csf2ra−/− eosinophil cultures were treated overnight with 20 ng/ml recombinant GM-CSF or vehicle control and subjected to RNA-sequencing–based transcriptome analyses. A heat map showing the top 500 differentially expressed genes that differed most across the 12 samples are shown in A; the log2 ratio of expression of the indicated chemokine and cytokine genes in WT eosinophils treated with GM-CSF relative to control is presented in B; the volcano plot in C shows all significantly differentially expressed transcripts (in red, significance cutoff P = 0.05, fold change >0.5), with the top transcripts annotated with their gene names. (D and E) Expression of the indicated chemokine and cytokine transcripts in eosinophils FACS-sorted from adenomas versus adjacent tissue (D; n = 6–8 samples per group) and MC38 tumors versus corresponding spleen (E; n = 8 samples per group). Each dot corresponds to sorted eosinophils from one adenoma or MC38 tumor. ***, P < 0.001, as calculated by Mann–Whitney test.
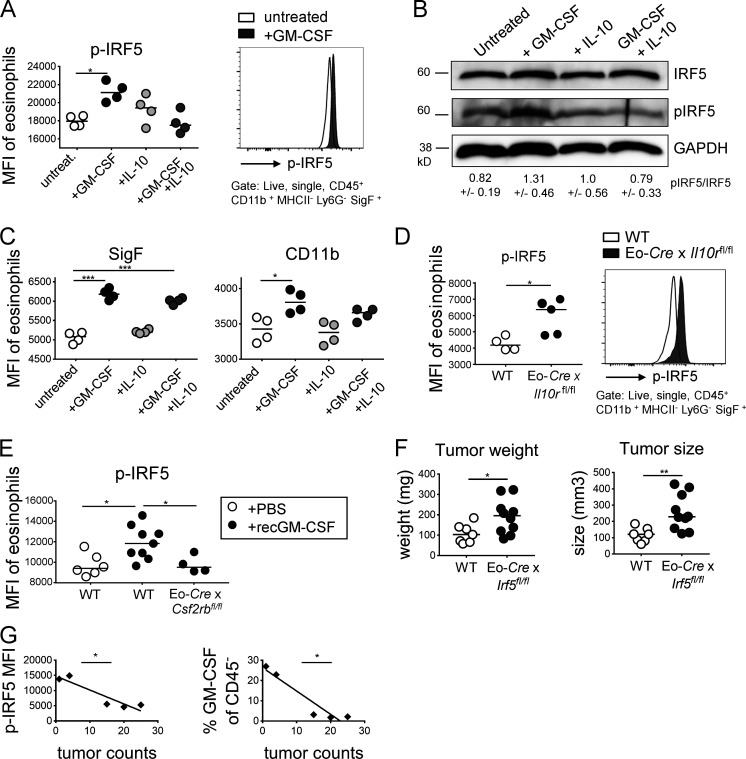
GM-CSF drives IRF5 activation in eosinophils
The interferon regulatory factor IRF5 regulates proinflammatory gene expression in myeloid cells, is involved in the polarization and function of M1 macrophages (Saliba et al., 2014; Weiss et al., 2013), and was identified as a target gene of GM-CSF signaling in cultured bone marrow–derived eosinophils in the RNA-sequencing approach described above. We speculated that eosinophils in the TME might require IRF5 for their antitumor functions. We first assessed in vitro using splenocyte preparations from IL-5–transgenic mice whether GM-CSF and/or IL-10 affect IRF5 expression or its activation by phosphorylation of Ser462. Exposure to GM-CSF induced IRF5 phosphorylation, as assessed by flow cytometry and Western blotting, which could be reversed by IL-10 (Fig. 7, A and B; and Fig. S4 J, left panel). The validity of the pIRF5 signal in eosinophils could be confirmed in macrophages (Fig. S4 J, right panel), which are well known to respond to GM-CSF or LPS exposure (Krausgruber et al., 2011). IRF5 phosphorylation was associated with enhanced IRF5 expression (Fig. S4 K), an observation that is consistent with its autoregulation and that could further be confirmed at the transcript level by quantitative RT-PCR of FACS-sorted eosinophils (Fig. S4 L). GM-CSF–induced eosinophil activation, as judged by their Siglec F, CD11b, and β-integrin expression, which also was to some extent counteracted by IL-10 (Fig. 7 C; data not shown). Interestingly, tumor-infiltrating eosinophils in MC38 tumors, which cannot sense IL-10 (in Eo-Cre × Il10rafl/fl mice), show higher levels of IRF5 phosphorylation than eosinophils infiltrating the tumors of WT littermates (Fig. 7 D), indicating that IL-10 inhibits IRF5 activation also in vivo. Strong IRF5 phosphorylation could further be detected in eosinophils infiltrating MC38 tumors that had been exposed to recombinant GM-CSF, and this signal was dependent on GM-CSF receptor expression on eosinophils (Fig. 7 E). To address whether IRF5 is required for the antitumor activities of eosinophils, we crossed Eo-Cre mice with Irf5fl/fl mice to generate a composite strain that lacks IRF5 expression exclusively in the eosinophil compartment. Interestingly, this strain was unable to control MC38 tumor growth (Fig. 7 F) and thus phenocopied the defects of GM-CSF signaling–deficient (Csf2ra−/− and Eo-Cre × Csf2rbfl/fl) mice. Eosinophils infiltrating the tumors of Eo-Cre × Irf5fl/fl mice showed lower Siglec F expression, indicating that eosinophil activation, at least as measured using this parameter, requires a functional GM-CSF–IRF5 signaling axis (Fig. S4 M). Moreover, regular doses of recombinant GM-CSF promoted tumor control in WT mice, but not their Eo-Cre × Irf5fl/fl littermates (Fig. S4 N). Finally, we found that the mean fluorescence intensity (MFI) of IRF5 phosphorylation in eosinophils, and their activation as assessed by CD11b and Siglec F expression, as well as the GM-CSF production by adenomas in ApcMin/+ mice were inversely correlated with adenoma multiplicity (Fig. 7 G). The combined results suggest that a newly identified GM-CSF–IRF5 axis in eosinophils promotes MC38 tumor control.
Figure 7.
GM-CSF-activated IRF5 is a critical regulator of eosinophil activities in the TME. (A–C) Splenocytes from IL-5–transgenic donors were treated overnight with 20 ng/ml GM-CSF, 50 ng/ml IL-10, or both cytokines. Cells were either stained for phosphorylated IRF5 and the signal in eosinophils was quantified by flow cytometry (A, summary plot of MFI and representative histogram, n = 4 technical replicates per condition) or subjected to protein extraction and Western blotting with a p-IRF5-specific antibody (B, total IRF5 and GADPH shown as loading controls). The quantification of three Western blots representing independent experiments is shown below the lanes as mean ± SEM. (C) The same cells as shown in A were also stained for Siglec F and CD11b to assess the activation state of eosinophils in the cultures. (D) IRF5 activation as assessed by p-IRF5–specific flow cytometry of eosinophils in tumors of Eo-Cre × Il10ra fl/fl mice and their Cre-negative littermates. One representative experiment of two is shown, n = 4–5 tumors per genotype. (E) IRF5 activation as assessed by p-IRF5–specific flow cytometry of eosinophils in tumors of Eo-Cre × Csf2rb fl/fl mice and their Cre-negative littermates that were treated three times weekly with recombinant GM-CSF or PBS as indicated. (F) Tumor weights and volumes at the study endpoint of Eo-Cre × Irf5 fl/fl mice and their Cre-negative littermates that had been subcutaneously injected with MC38 cells. Data are pooled from two independent experiments. (G) GM-CSF in CD45 negative leukocytes (right panel, correlation coefficient ρ = −0.948) and p-IRF5 in eosinophils infiltrating adenomas of ApcMin/+ mice treated with isotype control antibody (left panel, ρ = −0.927) as assessed flow cytometrically, relative to tumor counts (small intestine plus colon, as shown in Fig. 3). *, P < 0.05; **, P < 0.01; and ***, P < 0.001; as calculated by Mann–Whitney test (D and F), by one-way ANOVA with Tukey’s post-test (A, C, and E) or by linear regression (G).
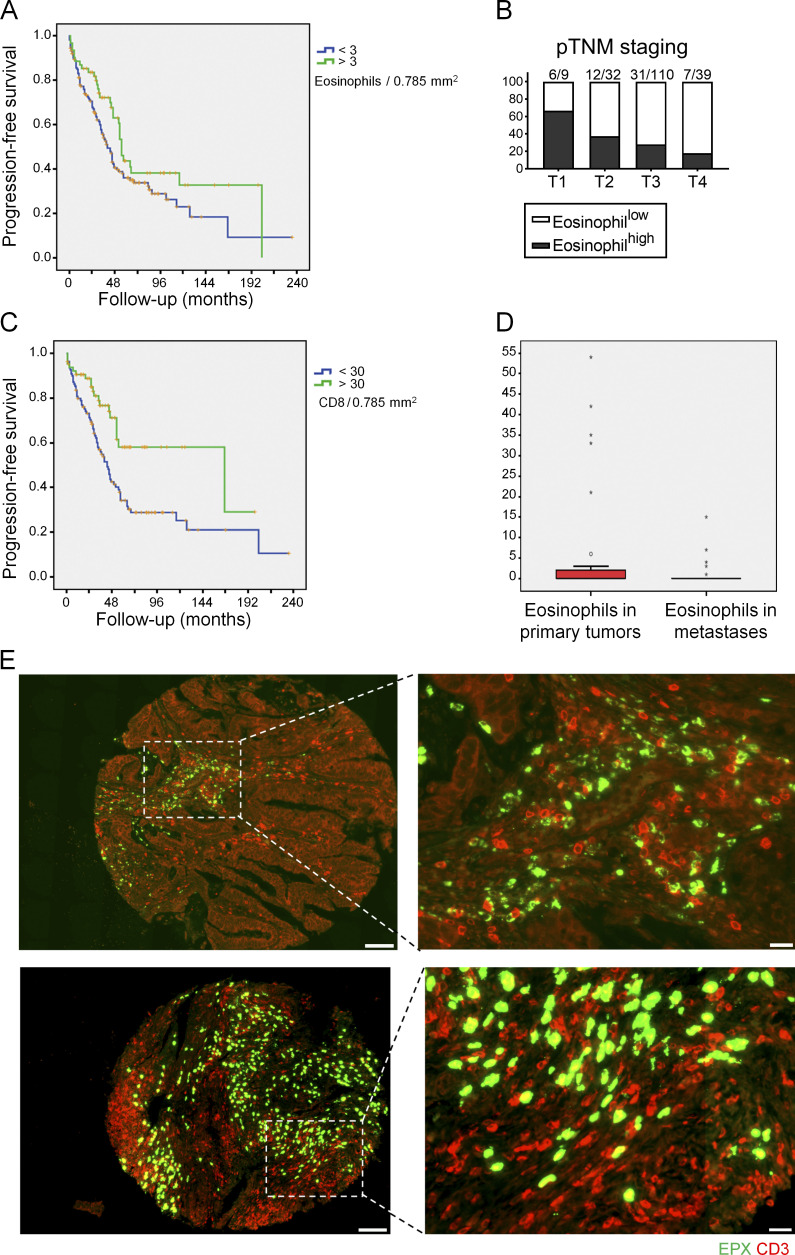
Eosinophil infiltration of human CRC is associated with superior survival and intratumoral CD8+ T cell infiltration
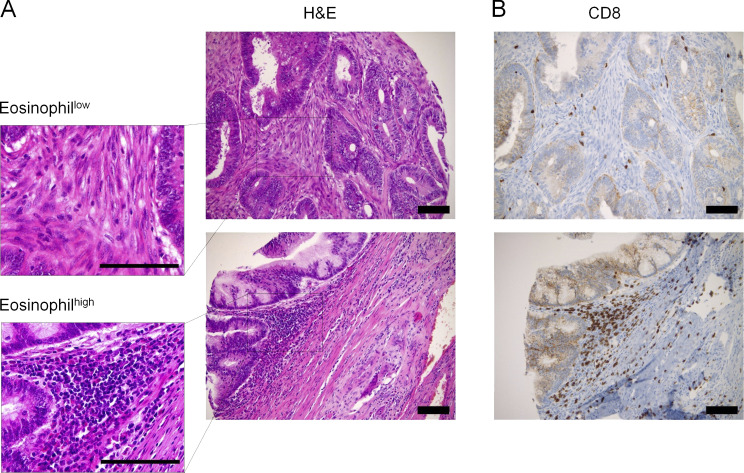
To examine the relevance of our findings for human CRC, we generated a tissue microarray (TMA) comprising 240 well-annotated cases of primary CRC treated in northwestern Switzerland. We stained the array with H&E and EPO to quantify eosinophil infiltration and with antibodies to CD3, CD8, langerin, and the mismatch repair proteins MLH1, MSH2, MSH6, and PMS2 to assess for CD8+ T cell and dendritic cell infiltration and microsatellite instability, respectively. Eosinophils were readily detectable on H&E sections, and patients could be stratified into eosinophilhigh (>3 eosinophils/0.785 mm2) and eosinophillow (<3 eosinophils/0.785 mm2) subsets (Fig. S5 A); the cutoff score was generated by means of a ROC/AUROC function to best meet prognostic significance (Tzankov et al., 2010). Of 223 evaluable primary tumors, 162 cases (73%) exhibited little or no eosinophil infiltration, whereas 61 cases (27%) were classified as eosinophilhigh. Patients with eosinophilhigh tumors had a significantly better progression-free survival (P = 0.046 according to the log rank Mantel–Cox test) than their eosinophillow counterparts (Fig. 8 A). The median progression-free survival of patients with eosinophilhigh tumors was on average 15 mo longer (55 ± 2.17 mo) than that of patients with eosinophillow tumors (40 ± 3.35 mo). Furthermore, analysis of the distribution of eosinophillow and eosinophilhigh tumors with respect to pTNM staging revealed that eosinophil infiltration was inversely correlated with pT stage, with higher proportions of eosinophilhigh instances in stage 1 and 2 patients than in stage 3 and 4 patients (Fig. 8 B). Staining for CD8 revealed that, of 174 evaluable patients, 64 cases (37%) exhibited considerable infiltration by CD8+ tumor–infiltrating T cells (>30 cells/0.785 mm2), whereas 110 cases (63%) fell under that cutoff (see representative images in Fig. S5 B). As reported in other cohorts, high CD8+ T cell infiltration was associated with superior progression-free survival (Fig. 8 C), and the difference of survival probability between CD8+ T cellhi and CD8+ T celllo patients was highly significant (P = 0.001 according to the log rank Mantel–Cox test). Eosinophilhigh tumors were further also infiltrated by larger numbers of CD8+ T cells (Fig. S5 B); the positive association between both parameters was highly significant (correlation coefficient ρ = 0.411; P = 2.02 × 10−8). Indeed, the vast majority of patients with eosinophilhigh tumors also exhibited high levels of CD8+ T cell infiltration; the reverse was not true, as ∼25% of CD8+ T cellhi tumors showed no or little eosinophil infiltration. Other examined parameters (langerin-positive Langerhans cell infiltration and microsatellite instability) had no detectable impact on survival in our cohort (data not shown), although langerin expression (and thus Langerhans cell infiltration), but not microsatellite instability, correlated with eosinophil infiltration (correlation coefficient ρ = 0.305; P = 003). To address whether eosinophils were differentially present in primary tumors and metastases, we compared 36 matched primary and metastasis pairs that had been arrayed on the TMA. The mean number of eosinophils was considerably higher at the primary sites (5.64/0.785 mm2 ± 13.47; range, 0–54) than at the metastatic sites (mean 0.83/0.785 mm2 ± 2.80; range, 0–15; P = 0.049 according to the Wilcoxon paired/signed-rank test; Fig. 8 D), suggesting that these distant sites are not efficiently targeted by eosinophils. Eosinophils and T cells infiltrated the same areas of the tumor if both cell populations were present, as shown for two representative cases at low and high magnification (Fig. 8 E). In conclusion, the observations made in our Swiss cohort of CRC patients confirms the previously noted superior survival of patients harboring tumors with high densities of eosinophils or CD8 T cells, establishes a close link between the two immune cell populations in CRC, and indicates a considerably lower presence of eosinophils at metastatic sites.
Figure S5.
Eosinophils colocalize with CD8+ T cells in human CRC biopsy specimens. (A and B) Representative consecutive H&E-stained (A) and anti-CD8–stained (B) sections of CRC biopsy specimens showing either low (upper panels) or high (lower panels) eosinophil infiltration into the tumor mass. Scale bars indicate 100 µm.
Figure 8.
Eosinophil densities in CRC have prognostic relevance and are positively correlated with T cell infiltration. (A) Progression-free survival of 61 patients whose tumors were classified as eosinophilhigh (>3 eosinophils per 0.785 mm2) versus 162 patients classified as eosinophillow (<3 eosinophils per 0.785 mm2). P = 0.046 according to the log rank Mantel–Cox test. (B) Stratification of eosinophilhigh and eosinophillow patients according to their pTNM status. (C) Progression-free survival of 64 cases whose tumors were classified as CD8high (>30 CD8+ T cells per 0.785 mm2) versus 110 patients classified as CD8low (<30 CD8+ T cells per 0.785 mm2). P = 0.001 according to the log rank Mantel–Cox test. (D) Eosinophil infiltration into the primary tumor and metastases of 36 matched pairs. P = 0.049 according to the Wilcoxon paired/signed-rank test. (E) Representative low- and high-magnification images of two stamp biopsy specimens included on our TMA that feature high eosinophil (in green) and high CD3+ T cell infiltration (in red), as assessed by immunofluorescence microscopy using antibodies for CD3 and the eosinophil peroxidase. Scale bars indicate 100 µm (left images) and 20 µm (right images), respectively.
Discussion
Eosinophils constitute a versatile and highly heterogeneous cell population with not only key functions in the settings of allergy and parasitic infection but also antibacterial and regulatory properties that are only beginning to be unraveled at the mechanistic level. The work presented here adds yet another previously understudied function to the eosinophil portfolio, i.e., their contribution to antitumor immunity. Epidemiological studies have long noted a superior prognosis of patients with cancers of the gastrointestinal tract that exhibited high levels of eosinophil infiltration, in terms of their rate of metastasis as well as overall survival probability (Lotfi et al., 2007); our own survey of 240 patients with CRC corroborated these early observations. In our cohort, eosinophil infiltration further correlated positively with CD8+ T cell infiltration and inversely with tumor stage, which supports our general hypothesis that eosinophils promote tumor control through the activation of T cells. Our experimental data suggest that eosinophils infiltrate subcutaneously growing MC38 tumors and, along with monocytes and NK cells, are among the first leukocyte populations to arrive at the tumor site. The three strategies we have used to manipulate eosinophil numbers demonstrate that eosinophils are strictly required for tumor control. In the absence of eosinophils, MC38 tumors are at least twice as large as their eosinophil-replete counterparts. Our data also show that CT26 tumors grow faster in BALB/c mice in the absence of eosinophils and that eosinophil-depleted ApcMin/+ mice develop more tumors in the small intestine and colon. The results obtained in IL-5–transgenic mice further suggest that eosinophil infiltration is a limiting factor in tumor control that can be overcome simply by the excessive bone marrow production of eosinophils under IL-5 exposure. A detailed comparison of T cell tumor infiltrates in eosinophil-replete versus depleted mice revealed lower frequencies of activated CD4+ and CD8+ T cells expressing IFN-γ, TNF-α, GM-CSF, and especially effector memory T cells, in PHIL mice and mice subjected to anti–IL-5 treatment relative to their respective controls; opposite effects were seen in tumors from IL-5–transgenic mice. The dependence of tumor-specific T cell responses on eosinophils could not be attributed to a defect in T cell priming; quite to the contrary, frequencies of activated (cytokine-expressing) T cells in the draining inguinal LNs were higher in PHIL mice than their WT counterparts. Rather, we attribute the defect of PHIL and IL-5–depleted mice in tumor control to their inability to drive the expansion of activated T cells at the tumor site. The conclusions we draw from our observations regarding the T cell dependence of the antitumor effects of eosinophils were not shared by a recent study conducted in ApcMin/+ mice, which also reported a critical role of eosinophils in controlling adenoma formation in the ApcMin/+ model but linked the reduced tumor burden of eosinophil-replete mice to direct cytotoxic activities of eosinophils on tumor cells (Reichman et al., 2019).
GM-CSF is a potent cytokine promoting the differentiation of myeloid cells, and both pro- and antitumor effects have been reported (Hong, 2016). While GM-CSF can exert stimulatory effects on tumor progression depending on the tumor type or cancer model, it can also be used as an immunostimulatory adjuvant to elicit antitumor immunity. GM-CSF is already administered in clinical trials, with clear benefit in certain cancer settings, particularly when used in combination with checkpoint inhibitors (Hodi et al., 2014). Both immune-dependent and independent mechanisms have been advanced to explain the benefit of GM-CSF therapy (Yamashita et al., 1989; Mach et al., 2000). Interestingly, GM-CSF is found to be overexpressed in one third of CRCs, and patients whose tumors express both GM-CSF and the GM-CSF receptor have excellent 5-yr survival rates (Urdinguio et al., 2013). In the TME, we have identified GM-CSF and IL-10 as critical regulators of eosinophil activity with opposing effects. Our data add to a growing body of evidence implicating GM-CSF in many aspects of eosinophil biology, including their development and survival at steady state (Willebrand and Voehringer, 2016) and their activation (Griseri et al., 2015) and migration (Liu et al., 2015) during inflammation. Studying GM-CSF is challenging due to the shared use of the common β-chain of the GM-CSF receptor by IL-5 and IL-3; we have been able to at least partly overcome this challenge through the use of a mouse strain lacking the (specific) α-chain and of a Cre recombinase that is expressed only after eosinophils have exited the bone marrow. Indeed, eosinophil counts in the bone marrow, blood and spleen are not significantly different in Eo-Cre × Csf2rbfl/fl mice and their WT littermates but rather are specifically reduced in the TME, along with the expression of activation markers by the residual intratumoral eosinophil population. Furthermore, we show that the beneficial effects of GM-CSF administration on tumor control require the expression of the GM-CSF receptor β-chain on eosinophils. The two observations combined indicate that eosinophil activation at and migration to the tumor site both require GM-CSF signaling, with GM-CSF-activated eosinophils driving CD4+ and CD8+ T cell responses.
Several observations implicate the transcription factor IRF-5 in signaling downstream of the GM-CSF receptor. On the one hand, the specific loss of IRF5 in eosinophils phenocopies the loss of the GM-CSF receptor β-chain with respect to tumor control; on the other hand, the exposure of eosinophils in vitro or in vivo to GM-CSF induces strong IRF5 phosphorylation. Our data obtained with Eo-Cre × Il10rafl/fl mice suggest that the GM-CSF–IRF5 axis is antagonized by IL-10; mice that lack IL-10R expression in the eosinophil compartment control tumors more efficiently than their WT littermates, and their eosinophil “hyperactivation” in the absence of a regulatory IL-10 signal is readily observed at the level of IRF5 phosphorylation. In vitro, the IRF5 phosphorylation upon GM-CSF stimulation is reduced by concomitant exposure to IL-10. We have begun to elucidate the major sources of GM-CSF and IL-10 in the tumor context. Myeloid cells (macrophages and monocytes) are the dominant source of IL-10, with >80% of all IL-10–positive cells expressing the myeloid cell markers CD11b, MHCII, and F4/80. Minor contributing cell types are T cells, including regulatory T cells (T reg cells). In contrast, GM-CSF is predominantly expressed by CD45-negative stromal cells and T cells. Our data are consistent with eosinophils serving as sensors and amplifiers of the environmental cues they receive from their microenvironment; in settings with high levels of GM-CSF produced by activated Th1-polarized T cells and stromal cells, eosinophils promote further T cell activation and tumor control. Conversely, under conditions of high IL-10 production, eosinophils dampen T cell responses and thereby compromise tumor control. Our data are in line with a growing body of evidence attributing both regulatory and proinflammatory properties to this versatile cell type, with microenvironmental signals tipping the balance in one or the other direction.
Targeting eosinophils through their systemic depletion by antibodies specific for IL-5 or its receptor has recently received much attention in a subset of difficult-to-treat asthma patients with steroid-resistant disease, who often present with very severe symptoms (Castro et al., 2011). The high specificity of IL-5 for eosinophils, the critical role of IL-5 in eosinophil biology and its involvement in the majority of eosinophilic conditions make it a very attractive target for the treatment of eosinophil-mediated disorders other than asthma, with clinical trials currently ongoing in patients with eosinophilic esophagitis, idiopathic hypereosinophilic syndrome, and chronic rhinosinusitis with nasal polyposis (Roufosse, 2018). The widespread use of IL-5–targeting therapies in patients with asthma, and potentially soon in patients with other eosinophil-driven conditions, means that large numbers of individuals will essentially be living without eosinophils in the very near future. The consequences of prolonged eosinophil depletion are only beginning to be debated, mostly with respect to susceptibility to parasitic infection (Gleich et al., 2013). Our recent work on the role of eosinophils in the maintenance of gastrointestinal homeostasis and bacterial infection control (Arnold et al., 2018), as well as the current study investigating eosinophil functions in the context of tumor immunity, suggests that patient populations under IL-5 treatment need to be closely monitored in phase 4 trials for possibly enhanced susceptibility to bacterial infections of the gastrointestinal tract as well as (gastrointestinal tract) malignancies. A better understanding of eosinophils functions in both health and disease, gained through the use of experimental models of eosinophil deficiency, will contribute to informed treatment choices that weigh both the benefits and risks of prolonged eosinophil depletion.
Materials and methods
Patient cohort and TMA construction
Our cohort consisted of a total of 240 fully and irreversibly anonymized patients with therapy-naive, surgically resected CRC, of whom 98 were female (41%) and 142 were male (59%). The median follow-up of the cohort was 33 mo, with a range of 0–235 mo. Cores of primary tumors and, in 36 instances, (matched) corresponding metastases, were brought into a TMA. At the time of last follow up update, 97 patients were alive (41%), 30 were dead with/on disease (13%), 61 (25%) had died of unknown causes, and 52 had developed metachronous metastases. The pathological tumor-node-metastasis (pTNM) distribution at the time of diagnosis of our cohort was as follows: pT1, 11 patients (5%); pT2, 35 patients (17%); pT3, 118 patients (57%); pT4, 43 patients (21%); the pT stage of 33 cases could not be determined; pN0, 96 patients (47%); pN1, 77 patients (38%); pN2, 30 patients (15%); pN3, 1 patients (0.5%); the pN stage of 36 cases could not be determined; pM0, 98 patients (53%); pM1, 87 patients (47%); the pM stage of 55 cases was unknown. The pTNM staging was done according to the sixth edition of the AJCC Cancer Staging Manual (American Joint Committee on Cancer, 2002). Eosinophils were identified on H&E-stained slides based on their specific morphological appearance with large eosinophilic metachromatic granules. Immunohistochemical stainings for CD8, langerin, MLH1, MSH2, MSH6, and PMS2 were performed on an automatic platform (Benchmark Ultra; Roche/Ventana) using the following primary antibodies: CD8 (clone SP57, 790–4460 RTU-kit; Roche/Ventana), langerin (clone 12D6, NCL-langerin at a dilution of 1:100; Novocastra), MLH1 (clone M1, 790–4535 RTU-kit; Roche/Ventana), MSH2 (clone Q219-1129, 760–4265 RTU-kit; Roche/Ventana), MSH6 (clone EP49, IR08661 RTU-kit; DAKO/Agilent), and PMS2 (clone EP51 by IR08761 TRU-kit; DAKO/Agilent). Ethical approval for the TMA construction and research based on that TMA was obtained from the Ethics Committee of Northwestern Switzerland #361/12 form April 11, 2012. According to Swiss law, no informed consent is needed for retrospective archival tissue-based studies of fully and irreversibly anonymized patients, provided the tissue has been obtained before 2012, the tissue is not exhausted, and the patients have not put a veto on research with their tissues; all of these conditions applied to the cases on the TMA.
Animal experimentation
C57BL/6J (stock no. 000664), B6(SJL)-Il10ratm1.1Tlg/J (stock no. 028146), B6;129-Tgfbr2tm1Karl/J (stock no. 012603), C57BL/6-Irf5tm1Ppr/J (stock no. 017311), and C57BL/6J-ApcMin/J (stock no. 002020) mice were obtained from The Jackson Laboratory. IL-5–transgenic (Dent et al., 1990) and Csf2rbfl/fl (Croxford et al., 2015) mice have been described previously. Eosinophil-deficient mice (PHIL; Lee et al., 2004) and mice expressing Cre under the Eo-Cre (Doyle et al., 2013) were obtained from J.J. Lee (Mayo Clinic, Phoenix, AZ). IL-10 reporter (10BiT) mice (Maynard et al., 2007) and Csf2ra−/− mice (Schneider et al., 2017) were described previously. All strains were bred and maintained under specific pathogen–free conditions in accredited animal facilities at the University of Zurich. For the depletion of eosinophils, WT or ApcMin mice were i.p. injected two times per week with 0.25 mg anti–IL-5 antibody (TRFK5; BioXCell) or anti-horseradish peroxidase isotype control antibody (HRPN; BioXCell) starting from 2 d before MC38 inoculation (WT) and at 12 wk of age (ApcMin). PD-L1–blocking antibody (10F.9G2; BioXCell), CTLA4-blocking antibody (9H10; BioXCell), CD8-depleting antibody (YTS 169.4; BioXCell), and TNF-α–neutralizing antibody (XT3.11; BioXCell) were i.p. administered two times per week at doses of 0.5 mg/mouse. 100 ng recombinant GM-CSF or IL-5 (PeproTech) was i.p. administered three times per week. All animal experimentation was reviewed and approved by the Zurich Cantonal Veterinary Office (licenses ZH224/2014, ZH140/2017, and ZH021/2020 to A. Müller).
Syngeneic tumor models and genome editing by CRISPR
The murine CRC cell line MC38 is derived from C57BL/6 mice and the CT26 line is derived from BALB/c mice; both were generously provided by Prof. Lubor Borsig (University of Zurich, Zurich, Switzerland). Cells were maintained in DMEM supplemented with 100 U/ml penicillin/streptomycin and 10% heat-inactivated FCS at 37°C in 5% CO2. 5 × 105 MC38 cells were subcutaneously injected in both flanks of mice, and the tumors were analyzed after 15 d for tumor weight, volume, leukocyte infiltration, and RNA expression profile. For the generation of MC38 cells lacking TNFRI, the guide RNA for TNFRSF1A (5′-GTGTCTCACTCAGGTAGCGT-3′) was cloned into PX458 vector (#48138: pSpCas9 (BB)-2A-GFP; Addgene). 3 × 105 MC-38 cells were transfected with 2.5 µg plasmid using TransIT2020 transfection reagent (Mirus Bio). GFP-positive cells were FACS sorted 48 h after transfection. After four passages, MC-38 cells were bulk sorted based on their TNFRI expression and enriched for a TNFRIlow population and a WT population.
Leukocyte isolation and flow cytometry
For leukocyte isolation of MC-38 tumors, 80–100-mg tumor slices were cut into pieces and digested in 500 U/ml type IV collagenase and 0.05 mg/ml DNase I at 37°C for 50 min under agitation in supplemented RPMI-1640 medium, followed by pushing through a cell strainer. For leukocyte isolation of ApcMin/+ tumors or of adjacent normal colonic lamina propria, tissues were weighted, cut into pieces and incubated in HBSS with 10% FCS and 5 mM EDTA at 37°C in a shaking incubator. Tissues were then digested at 37°C for 50 min with 15 mM Hepes, 500 U/ml type IV collagenase (Sigma-Aldrich), and 0.05 mg/ml DNase I in RPMI-1640 medium supplemented with 10% FBS and 100 U/ml penicillin/streptomycin. LN cell suspensions were prepared by digesting the LNs in 500 U/ml type IV collagenase in RPMI-1640 for 15 min followed by pushing through a cell strainer using a syringe plunger. Splenocytes were prepared by mashing the spleens through a cell strainer using a syringe plunger, followed by red blood cell lysis with ACK buffer. Blood was removed by terminal cardiac puncture and subjected to ACK red blood cell lysis. Bone marrow cells were collected by flushing the mouse femur and the tibia with RPMI medium. Total leukocyte counts were determined by adding countBright Absolute Counting Beads (Life Technologies) to each sample before flow cytometry for normalization to tissue weight. Cells were stained with a fixable viability dye and a combination of the following antibodies: anti-mouse CD45 (clone 30-F11), CD11c (N418), MHCII (M5/114.15.2), F4/80 (BM8), CD11b (M1/70), Ly6G (1A8), Ly6C (HK1.4), CD4 (RM4-5), CD8 (53–6.7), NK1.1 (PK136), TCR-β (H57-597), CD44 (1M7), CD62L (MEL-14), CD69 (H1.2F3), Thy1.1 (OX-7), CD63 (NVG-2), and CCR3 (J073E5; all from BioLegend), as well as Siglec F (E50-2440; BD Biosciences) and β-integrin (TS2/16; eBioscience), CD120a (TNFR type I/p55; clone 55R-286), and Armenian hamster IgG isotype control (clone HTK888). Fc block (anti-CD16/CD32; Affymetrix) was included to minimize nonspecific antibody binding. Annexin V staining was performed with the Annexin V Apoptosis Detection Set (eBioscience) according to the manufacturer’s instructions. For intracellular cytokine staining of T cells, cells were incubated at 37°C for 3.5 h in complete IMDM containing 0.1 µM PMA and 1 µM ionomycin with 1:1,000 brefeldin A (eBioscience) and GolgiStop solutions (BD Biosciences) at 37°C in a humidified incubator with 5% CO2. To assess tumor-specific T cell responses, MC38 tumor cell suspensions were restimulated with the H-2Kb–restricted MC38 peptide KSPWFTTL at 37°C for 3.5 h in complete IMDM containing brefeldin A and GolgiStop solutions. For the detection of intracellular cytokine production by myeloid and tumor cells, cell suspensions were incubated at 37°C for 3 h in complete IMDM containing only brefeldin A. Following surface staining, cells were fixed and permeabilized with the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences) according to the manufacturer’s instructions. Cells were then stained for 50 min with antibodies to IFN-γ (XMG1.2), TNF-α (MP6-XT22), GM-CSF (MP1-22E9), IL-10 (JES5-16E3), or CCL5 (2E9). For the intracellular staining of granzyme B (QA16A02; BioLegend), p-IRF5 (p-Ser437, polyclonal; Thermo Fisher Scientific), IRF5 (polyclonal; Abcam), or Ki-67 (11F6; BioLegend), cells were surface stained followed by fixation and permeabilization with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to manufacturer’s instructions. Samples were acquired on a LSRII Fortessa (BD Biosciences) and analyzed using FlowJo software.
For quantitative PCR, RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions, and complementary DNA synthesis was performed using Superscript III reverse transcription (Qiagen). Quantitative PCR reactions for the following candidate genes were performed using TaqMan gene expression assays (Applied Biosystems by Thermo Fisher Scientific): Ccl2 (Mm00441242_m1), Ccl4 (Mm00443111_m1), Ccl17 (Mm00516136_m1), Ccl22 (Mm00436439_m1), Ccl24 (Mm00444701_m1), Csf2 (Mm01290062_m1), Hprt (Mm03024075_m1), Irf5 (Mm00496477_m1), and Tnfa (Mm00443258_m1). Complementary DNA samples were analyzed using a Light Cycler 480 detection system (Roche) and gene expression levels for each sample were normalized to HPRT expression. Mean relative gene expression was determined, and the differences were calculated using the 2ΔC(t) method.
Differentiation of bone marrow–derived eosinophils, in vitro stimulation experiments, and immunoblotting
To generate murine bone marrow–derived eosinophils, bone marrow stem cells from donor mice were flushed from the femur and the tibia and resuspended in RPMI-1640 medium upon red blood cell lysis. Cells were seeded at a density of 106 cells/ml in RPMI-1640 medium supplemented with 20% heat-inactivated FBS, 25 mM Hepes, 100 U/ml penicillin/streptomycin, 2 mM glutamine, 1× NEAA, and 1 mM sodium pyruvate, and cultured at 37°C. BM Eos were differentiated with 100 ng/ml mouse stem cell factor (PeproTech) and 100 ng/ml mouse FLT3L (PeproTech) from days 0 to 4, followed by differentiation with 10 ng/ml mouse IL-5 (PeproTech) only from day 4 onwards, as described previously (Dyer et al., 2008). At day 12, eosinophils were resuspended in DMEM supplemented with 10 ng/ml recombinant IL-5 (PeproTech), 100 U/ml penicillin/streptomycin, and 10% heat-inactivated FCS and co-cultured with MC38 cells upon stimulation with cytokines (TSLP, IFN-γ, TNF-α, GM-CSF, 20 ng/ml; PeproTech). Cells were stained with Annexin V (BioLegend) according to the manufacturer’s instructions and analyzed by flow cytometry. To analyze IRF5 and p-IRF5 expression in vitro, total splenocytes or eosinophils purified from the spleens of IL-5 transgenic mice by MACS (positive selection using anti-Siglec F-PE with anti-PE microbeads) were cultured overnight in RPMI-1640 medium supplemented with 20% heat-inactivated FBS, and 100 U/ml penicillin/streptomycin in the presence of recombinant IL-5 (10 ng/ml) and stimulated with recombinant GM-CSF and/or IL-10 (all from PeproTech) at the indicated concentrations. Cells were either stained for IRF5 and phosphorylated IRF5 and the signal in eosinophils was quantified by flow cytometry or were subjected to protein extraction and immunoblotting. Proteins were extracted in RIPA buffer (Sigma-Aldrich) supplemented with 1× complete protease inhibitor cocktail (Roche), 0.5 mM sodium orthovanadate, 1 mM benzamidine, and PMSF. Protein concentration was quantified by the Bradford method (Bio-Rad), and samples were denatured in SDS, separated by SDS-PAGE (10% gel), transferred to polyvinylidene fluoride membranes, and immunoblotted with antibodies specific for IRF5, p-IRF5, and GAPDH.
RNA sequencing and data analysis
For the RNA sequencing, live bone marrow–derived eosinophil cultures from WT and Csf2ra−/− mice were purified using the Dead Cell Removal Kit (Miltenyi Biotech), seeded at a density of 106 cells/well in RPMI-1640 medium supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin/streptomycin, and recombinant IL-5 (10 ng/ml). Cell triplicates were stimulated overnight with 20 ng/ml recombinant GM-CSF at 37°C or left untreated. RNA was isolated using the RNeasy Mini Kit (Qiagen), including an on-column DNA digestion step. Total RNA was submitted to the Functional Genomics Center Zurich. Samples were analyzed for RNA integrity and concentration using the RNA Screen Tape (Agilent were suitable for a TruSeq stranded mRNA library preparation; Illumina). After poly(A) selection using Oligo-dT beads, mRNA was reverse transcribed into cDNA. The cDNA was fragmented, end repaired, and polyadenylated before ligation of TruSeq UD Indices (IDT). The quality and quantity of the amplified sequencing libraries were validated using a Fragment Analyzer SS NGS Fragment Kit (1–6,000 bp; Agilent). The equimolar pool of samples was spiked into a NovaSeq6000 run targeting 600 million reads on a S2 FlowCell (Novaseq S2 Reagent Kit, 100 cycles; Illumina). The Bcl files were demultiplexed using bcltofastq software (Illumina). RNA-sequencing reads were aligned with the STAR aligner. As reference we used the Ensembl mouse genome build GRCm38.p6 using the gene annotation as provided by GENCODE M23 release. Gene expression values were computed with the function featureCounts from the R package Rsubread. Differential expression was computed using the Bioconductor package EdgeR. In particular, the glmQLFit function was used to fit the quasi-likelihood negative binomial generalized log-linear model to the count data and running quasi-likelihood F-test.
Confocal immunofluorescence microscopy
Paraffin-embedded mouse tissue section or human TMA slides were deparaffinized and rehydrated with graded ethanol dilutions. After antigen retrieval in a pressure cooker using EDTA-Tris buffer (10 mM Tris base and 1 mM EDTA, pH 9.0), nonspecific binding was prevented by preincubation of tissue sections with a blocking buffer (PBS containing human immunoglobulins, normal goat serum, and 7.5% BSA) at room temperature for 1 h. Indirect immunofluorescence staining was performed by incubating the paraffin sections with mouse monoclonal anti-EPX antibody (1:200; clone MM25-82 2; obtained from Lee Laboratories, Mayo Clinic) and rabbit monoclonal anti-CD3 antibody (1:20; clone SP7; Thermo Fisher Scientific, distributed by LuBioScience), in blocking solution, at 4°C, overnight. After several washes in PBS, secondary Alexa Fluor 488–conjugated goat anti-mouse (1:400) and Alexa Fluor 532–conjugated goat anti-rabbit (1:400) antibodies (Thermo Fisher Scientific) were applied and tissue samples incubated at room temperature for 1 h. Samples were washed and mounted in Prolong Gold mounting medium (Thermo Fisher Scientific), and image acquisition was performed using confocal laser scanning microscopy LSM 700 (Carl Zeiss Micro Imaging) with a 20× or 40×/1.40 oil DIC objective and analyzed with IMARIS software (Bitplane AG).
Statistical analysis
Statistical analysis was performed with Prism 6.0 (GraphPad Software) and with SPSS 22.0. The nonparametric Mann–Whitney U, Spearman rank correlation, and the log rank Mantel–Cox tests in combination with Kaplan–Meier curves and one-way ANOVA with Tukey’s post-test were used, where appropriate, for statistical analyses. Differences were considered statistically significant when P < 0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Online supplemental material
Fig. S1 shows the gating strategy and absolute numbers of the tumor-infiltrating leukocyte populations as well as eosinophil frequencies of WT, PHIL, anti–IL-5 antibody–treated and IL-5–transgenic mice. Fig. S2 shows CD4+ and CD8+ T-cell responses in tumors and tumor-draining lymph nodes upon manipulation of the eosinophil compartment. Fig. S3 shows the effect of manipulating the IL-10/STAT3 signaling axis on subcutaneous MC38 tumor growth and T cell responses. Fig. S4 shows eosinophil and myeloid cell frequencies in the tumors and tissues of Eo-Cre × Csf2rbfl/fl and CSF2ra−/− mice. Fig. S5 shows representative H&E- and anti-CD8–stained sections of CRC biopsy specimens.
Acknowledgments
We thank Maries van den Broek, Paulino Tallon de Lara, Lubor Borsig, and Jesus Glaus-Garzon for helpful discussions.
This work was supported by the Swiss National Science Foundation (grant BSCGIO_157841/1 to A. Müller). The sponsor had no role in the study design, data analysis, or any other part of the research and manuscript submission.
Author contributions: I.C. Arnold, M. Artola-Boran, A. Gurtner, K. Bertram, and M. Bauer conducted experiments; B. Becher, M. Kopf, and H.-U. Simon provided critical mouse strains; Z. Frangez, S. Yousefi, and H.-U. Simon stained TMAs and analyzed data; A. Tzankov generated TMAs and analyzed patient data; and I.C. Arnold and A. Müller supervised the study.
References
- American Joint Committee on Cancer . 2002. AJCC Cancer Staging Manual. Sixth edition. Springer, New York. [Google Scholar]
- Arnold, I.C., Artola-Borán M., Tallón de Lara P., Kyburz A., Taube C., Ottemann K., van den Broek M., Yousefi S., Simon H.U., and Müller A.. 2018. Eosinophils suppress Th1 responses and restrict bacterially induced gastrointestinal inflammation. J. Exp. Med. 215:2055–2072. 10.1084/jem.20172049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron, C., Al-Ramli W., and Hamid Q.. 2009. Remodeling in asthma. Proc. Am. Thorac. Soc. 6:301–305. 10.1513/pats.200808-089RM [DOI] [PubMed] [Google Scholar]
- Carretero, R., Sektioglu I.M., Garbi N., Salgado O.C., Beckhove P., and Hämmerling G.J.. 2015. Eosinophils orchestrate cancer rejection by normalizing tumor vessels and enhancing infiltration of CD8(+) T cells. Nat. Immunol. 16:609–617. 10.1038/ni.3159 [DOI] [PubMed] [Google Scholar]
- Castellino, F., Huang A.Y., Altan-Bonnet G., Stoll S., Scheinecker C., and Germain R.N.. 2006. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 440:890–895. 10.1038/nature04651 [DOI] [PubMed] [Google Scholar]
- Castro, M., Mathur S., Hargreave F., Boulet L.P., Xie F., Young J., Wilkins H.J., Henkel T., and Nair P.; Res-5-0010 Study Group . 2011. Reslizumab for poorly controlled, eosinophilic asthma: a randomized, placebo-controlled study. Am. J. Respir. Crit. Care Med. 184:1125–1132. 10.1164/rccm.201103-0396OC [DOI] [PubMed] [Google Scholar]
- Croxford, A.L., Lanzinger M., Hartmann F.J., Schreiner B., Mair F., Pelczar P., Clausen B.E., Jung S., Greter M., and Becher B.. 2015. The Cytokine GM-CSF Drives the Inflammatory Signature of CCR2+ Monocytes and Licenses Autoimmunity. Immunity. 43:502–514. 10.1016/j.immuni.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Cuschieri, A., Talbot I.C., and Weeden S.; MRC Upper GI Cancer Working Party . 2002. Influence of pathological tumour variables on long-term survival in resectable gastric cancer. Br. J. Cancer. 86:674–679. 10.1038/sj.bjc.6600161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoine, F., and Lacy P.. 2014. Eosinophil cytokines, chemokines, and growth factors: emerging roles in immunity. Front. Immunol. 5:570. 10.3389/fimmu.2014.00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, L.A., Strath M., Mellor A.L., and Sanderson C.J.. 1990. Eosinophilia in transgenic mice expressing interleukin 5. J. Exp. Med. 172:1425–1431. 10.1084/jem.172.5.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, A.D., Jacobsen E.A., Ochkur S.I., Willetts L., Shim K., Neely J., Kloeber J., Lesuer W.E., Pero R.S., Lacy P., et al. 2013. Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J. Leukoc. Biol. 94:17–24. 10.1189/jlb.0213089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, K.D., Moser J.M., Czapiga M., Siegel S.J., Percopo C.M., and Rosenberg H.F.. 2008. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J. Immunol. 181:4004–4009. 10.4049/jimmunol.181.6.4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aceñero, M.J., Galindo-Gallego M., Sanz J., and Aljama A.. 2000. Prognostic influence of tumor-associated eosinophilic infiltrate in colorectal carcinoma. Cancer. 88:1544–1548. [DOI] [PubMed] [Google Scholar]
- Gleich, G.J., Klion A.D., Lee J.J., and Weller P.F.. 2013. The consequences of not having eosinophils. Allergy. 68:829–835. 10.1111/all.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griseri, T., Arnold I.C., Pearson C., Krausgruber T., Schiering C., Franchini F., Schulthess J., McKenzie B.S., Crocker P.R., and Powrie F.. 2015. Granulocyte Macrophage Colony-Stimulating Factor-Activated Eosinophils Promote Interleukin-23 Driven Chronic Colitis. Immunity. 43:187–199. 10.1016/j.immuni.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi, F.S., Lee S., McDermott D.F., Rao U.N., Butterfield L.H., Tarhini A.A., Leming P., Puzanov I., Shin D., and Kirkwood J.M.. 2014. Ipilimumab plus sargramostim vs ipilimumab alone for treatment of metastatic melanoma: a randomized clinical trial. JAMA. 312:1744–1753. 10.1001/jama.2014.13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, I.S. 2016. Stimulatory versus suppressive effects of GM-CSF on tumor progression in multiple cancer types. Exp. Mol. Med. 48. e242. 10.1038/emm.2016.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, K., Torisu M., and Fujimura T.. 1986. Malignant tumor and eosinophils. I. Prognostic significance in gastric cancer. Cancer. 58:1321–1327. [DOI] [PubMed] [Google Scholar]
- Jung, Y., and Rothenberg M.E.. 2014. Roles and regulation of gastrointestinal eosinophils in immunity and disease. J. Immunol. 193:999–1005. 10.4049/jimmunol.1400413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf, M., Brombacher F., Hodgkin P.D., Ramsay A.J., Milbourne E.A., Dai W.J., Ovington K.S., Behm C.A., Köhler G., Young I.G., et al. 1996. IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity. 4:15–24. 10.1016/S1074-7613(00)80294-0 [DOI] [PubMed] [Google Scholar]
- Krausgruber, T., Blazek K., Smallie T., Alzabin S., Lockstone H., Sahgal N., Hussell T., Feldmann M., and Udalova I.A.. 2011. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 12:231–238. 10.1038/ni.1990 [DOI] [PubMed] [Google Scholar]
- Lee, J.J., Dimina D., Macias M.P., Ochkur S.I., McGarry M.P., O’Neill K.R., Protheroe C., Pero R., Nguyen T., Cormier S.A., et al. 2004. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 305:1773–1776. 10.1126/science.1099472 [DOI] [PubMed] [Google Scholar]
- Liu, L.Y., Wang H., Xenakis J.J., and Spencer L.A.. 2015. Notch signaling mediates granulocyte-macrophage colony-stimulating factor priming-induced transendothelial migration of human eosinophils. Allergy. 70:805–812. 10.1111/all.12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi, R., Lee J.J., and Lotze M.T.. 2007. Eosinophilic granulocytes and damage-associated molecular pattern molecules (DAMPs): role in the inflammatory response within tumors. J. Immunother. 30:16–28. 10.1097/01.cji.0000211324.53396.f6 [DOI] [PubMed] [Google Scholar]
- Lucarini, V., Ziccheddu G., Macchia I., La Sorsa V., Peschiaroli F., Buccione C., Sistigu A., Sanchez M., Andreone S., D’Urso M.T., et al. 2017. IL-33 restricts tumor growth and inhibits pulmonary metastasis in melanoma-bearing mice through eosinophils. OncoImmunology. 6. e1317420. 10.1080/2162402X.2017.1317420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo, C., Moser A.R., Gledhill S., and Dove W.F.. 1994. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 54:5947–5952. [PubMed] [Google Scholar]
- Mach, N., Gillessen S., Wilson S.B., Sheehan C., Mihm M., and Dranoff G.. 2000. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 60:3239–3246. [PubMed] [Google Scholar]
- Maynard, C.L., Harrington L.E., Janowski K.M., Oliver J.R., Zindl C.L., Rudensky A.Y., and Weaver C.T.. 2007. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat. Immunol. 8:931–941. 10.1038/ni1504 [DOI] [PubMed] [Google Scholar]
- Nielsen, H.J., Hansen U., Christensen I.J., Reimert C.M., Brünner N., and Moesgaard F.. 1999. Independent prognostic value of eosinophil and mast cell infiltration in colorectal cancer tissue. J. Pathol. 189:487–495. [DOI] [PubMed] [Google Scholar]
- Patel, V.P., Kreider B.L., Li Y., Li H., Leung K., Salcedo T., Nardelli B., Pippalla V., Gentz S., Thotakura R., et al. 1997. Molecular and functional characterization of two novel human C-C chemokines as inhibitors of two distinct classes of myeloid progenitors. J. Exp. Med. 185:1163–1172. 10.1084/jem.185.7.1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman, H., Itan M., Rozenberg P., Yarmolovski T., Brazowski E., Varol C., Gluck N., Shapira S., Arber N., Qimron U., et al. 2019. Activated Eosinophils Exert Antitumorigenic Activities in Colorectal Cancer. Cancer Immunol. Res. 7:388–400. 10.1158/2326-6066.CIR-18-0494 [DOI] [PubMed] [Google Scholar]
- Roufosse, F. 2018. Targeting the Interleukin-5 Pathway for Treatment of Eosinophilic Conditions Other than Asthma. Front. Med. (Lausanne). 5:49. 10.3389/fmed.2018.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba, D.G., Heger A., Eames H.L., Oikonomopoulos S., Teixeira A., Blazek K., Androulidaki A., Wong D., Goh F.G., Weiss M., et al. 2014. IRF5:RelA interaction targets inflammatory genes in macrophages. Cell Rep. 8:1308–1317. 10.1016/j.celrep.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., Nobs S.P., Heer A.K., Hirsch E., Penninger J., Siggs O.M., and Kopf M.. 2017. Frontline Science: Coincidental null mutation of Csf2rα in a colony of PI3Kγ-/- mice causes alveolar macrophage deficiency and fatal respiratory viral infection. J. Leukoc. Biol. 101:367–376. 10.1189/jlb.4HI0316-157R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, L.K., Kinzler K.W., Vogelstein B., Preisinger A.C., Moser A.R., Luongo C., Gould K.A., and Dove W.F.. 1992. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 256:668–670. 10.1126/science.1350108 [DOI] [PubMed] [Google Scholar]
- Travers, J., and Rothenberg M.E.. 2015. Eosinophils in mucosal immune responses. Mucosal Immunol. 8:464–475. 10.1038/mi.2015.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzankov, A., Zlobec I., Went P., Robl H., Hoeller S., and Dirnhofer S.. 2010. Prognostic immunophenotypic biomarker studies in diffuse large B cell lymphoma with special emphasis on rational determination of cut-off scores. Leuk. Lymphoma. 51:199–212. 10.3109/10428190903370338 [DOI] [PubMed] [Google Scholar]
- Urdinguio, R.G., Fernandez A.F., Moncada-Pazos A., Huidobro C., Rodriguez R.M., Ferrero C., Martinez-Camblor P., Obaya A.J., Bernal T., Parra-Blanco A., et al. 2013. Immune-dependent and independent antitumor activity of GM-CSF aberrantly expressed by mouse and human colorectal tumors. Cancer Res. 73:395–405. 10.1158/0008-5472.CAN-12-0806 [DOI] [PubMed] [Google Scholar]
- Ushio, A., Arakaki R., Otsuka K., Yamada A., Tsunematsu T., Kudo Y., Aota K., Azuma M., and Ishimaru N.. 2018. CCL22-Producing Resident Macrophages Enhance T Cell Response in Sjögren’s Syndrome. Front. Immunol. 9:2594. 10.3389/fimmu.2018.02594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, M., Blazek K., Byrne A.J., Perocheau D.P., and Udalova I.A.. 2013. IRF5 is a specific marker of inflammatory macrophages in vivo. Mediators Inflamm. 2013. 245804. 10.1155/2013/245804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willebrand, R., and Voehringer D.. 2016. IL-33-Induced Cytokine Secretion and Survival of Mouse Eosinophils Is Promoted by Autocrine GM-CSF. PLoS One. 11. e0163751. 10.1371/journal.pone.0163751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y., Hayashi Y., Sugama Y., Miura Y., Kasahara T., Kitamura S., Torisu M., Mita S., Tominaga A., and Takatsu K.. 1988a. Highly purified murine interleukin 5 (IL-5) stimulates eosinophil function and prolongs in vitro survival. IL-5 as an eosinophil chemotactic factor. J. Exp. Med. 167:1737–1742. 10.1084/jem.167.5.1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y., Suda T., Suda J., Eguchi M., Miura Y., Harada N., Tominaga A., and Takatsu K.. 1988b. Purified interleukin 5 supports the terminal differentiation and proliferation of murine eosinophilic precursors. J. Exp. Med. 167:43–56. 10.1084/jem.167.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, Y., Nara N., and Aoki N.. 1989. Antiproliferative and differentiative effect of granulocyte-macrophage colony-stimulating factor on a variant human small cell lung cancer cell line. Cancer Res. 49:5334–5338. [PubMed] [Google Scholar]