Liu et al. compare epigenetic and expression profiles in isogenic ESC-derived human microglia-like cells (hMGLs) comprising Alzheimer’s disease (AD)–associated mutations. Using this model, convergent dysregulation at the APOE locus is observed with AD-associated mutations in SORL1 and TREM2.
Abstract
Variations in many genes linked to sporadic Alzheimer’s disease (AD) show abundant expression in microglia, but relationships among these genes remain largely elusive. Here, we establish isogenic human ESC–derived microglia-like cell lines (hMGLs) harboring AD variants in CD33, INPP5D, SORL1, and TREM2 loci and curate a comprehensive atlas comprising ATAC-seq, ChIP-seq, RNA-seq, and proteomics datasets. AD-like expression signatures are observed in AD mutant SORL1 and TREM2 hMGLs, while integrative multi-omic analysis of combined epigenetic and expression datasets indicates up-regulation of APOE as a convergent pathogenic node. We also observe cross-regulatory relationships between SORL1 and TREM2, in which SORL1R744X hMGLs induce TREM2 expression to enhance APOE expression. AD-associated SORL1 and TREM2 mutations also impaired hMGL Aβ uptake in an APOE-dependent manner in vitro and attenuated Aβ uptake/clearance in mouse AD brain xenotransplants. Using this modeling and analysis platform for human microglia, we provide new insight into epistatic interactions in AD genes and demonstrate convergence of microglial AD genes at the APOE locus.
Graphical Abstract
Introduction
Alzheimer’s disease (AD) is associated with cognitive decline; and while Aβ metabolism and neuronal dysfunction are etiologically fundamental events in AD pathogenesis, contributing events from other cell types such as astrocytes and microglia are also crucial. Interestingly, genome-wide association studies have identified numerous polymorphisms linked to elevated AD risk in genes that are highly expressed in microglia, including noncoding polymorphisms in CD33 (rs3865444) and INPP5D (rs35349669), as well as TREM2R47H (rs75932628) and SORL1 R744X and A528T (rs2298813) coding mutations (Harold et al., 2009; Jansen et al., 2019; Kunkle et al., 2019; Lambert et al., 2009, 2013; Raghavan et al., 2018; Vardarajan et al., 2015). Additionally, many AD-associated loci are enriched in microglia-specific enhancer regions, further implicating a vital role for microglia in AD pathogenesis (Nott et al., 2019). How microglia-associated AD risk variants contribute to AD remains unclear.
Numerous AD risk genes feature overlapping signaling pathways in microglia. TREM2 ligand engagement results in recruitment and phosphorylation of immunoreceptor tyrosine-based activation motifs within the TREM2 coreceptor, DAP12 (Bouchon et al., 2001), resulting in downstream activation of Syk and phosphoinositide 3-kinase (PI3K) to facilitate phagocytic actin rearrangement and cytokine expression (Bakker et al., 1999; Paloneva et al., 2000; Rougerie et al., 2013). SHIP1 (SH2-containing inositol 5′ phosphatase, encoded by INPP5D) and CD33, a glycosylated transmembrane protein, can both antagonize TREM2 signaling through the inactivation of Syk/PI3K (Del-Aguila et al., 2019; Malik et al., 2015). SORLA (sortilin-related receptor with A-type repeats; encoded by SORL1) is a transmembrane receptor originally identified as a potential blood biomarker in AD patients (Scherzer et al., 2004), which limits amyloidogenic APP (Aβ precursor protein) processing in neurons by trafficking APP to the Golgi (Andersen et al., 2005) or cell surface (Huang et al., 2016). Although a role for SORLA has been extensively characterized in neurons, SORLA expression has been found to be comparatively high in microglia compared with other cell types in the central nervous system (CNS) and is enriched in aged human microglia (Olah et al., 2018; Zhang et al., 2016), suggesting that SORLA may also be vital to microglia function. The APOEε4 variant is currently the strongest genetic-risk factor for sporadic, late-onset AD described thus far. Pathogenic induction of APOE in microglia has been shown to enhance AD progression (Shi et al., 2019). Importantly, APOE status has been shown to clinically alter TREM2R47H-dependent AD pathogenesis; in some instances, presence of the APOEε4 allele may be required to trigger AD onset in TREM2R47H carriers (Kunkle et al., 2019; Murray et al., 2019). In further support of APOE as a pathogenic driver in AD, viral-mediated expression of APOEε4 can increase extracellular amyloid β (Aβ) levels and enhance Aβ plaque load (Hudry et al., 2013; Zhao et al., 2014), whereas APOE deletion can reduce plaque burden in AD mouse brain (Irizarry et al., 2000; Ulrich et al., 2018).
To date, characterizing the contribution of microglia to AD pathogenesis has been problematic, as microglia in mouse models may not accurately model microglial behavior and function in humans. Given that human and mouse TREM2 features only ∼77% sequence identity, nonconserved regions in TREM2 could potentially modulate ligand repertoire, ligand binding affinity, and confer differing effects in homologous human and mouse AD mutations (Song et al., 2018). For example, the TREM2R47H mutation was previously observed to down-regulate TREM2 mRNA levels in mouse microglia, but not in human induced pluripotent stem cell (iPSC)–derived microglia models and AD cases carrying the TREM2R47H mutant allele (Xiang et al., 2018). Although, iPSC-derived microglia-like cells may be useful in modeling human microglia (Abud et al., 2017; McQuade et al., 2018; Speicher et al., 2019), variables derived from genetic variation and epigenetic memory may render complexities in comparing effects of AD mutations in nonisogenic settings (Kim et al., 2010; Polo et al., 2010). Further, characterization of microglia so far has been largely limited to singular profiles at transcriptomic or epigenetic levels (Ayata et al., 2018; Gosselin et al., 2017), thus potentially limiting our analyses to either expression or epigenetic datasets.
To address limitations with respect to nonisogenic variation and singular analyses, we describe a systematic analysis pipeline using a human embryonic stem cell (ESC) line (H9) to isogenically integrate AD-associated gene variants in CD33, INPP5D, TREM2, and SORL1 by CRISPR-Cas9 editing and characterize effects of these variants in mature human ESC–derived microglia-like cells (hMGLs). Integrated analysis of assay for transposase-accessible chromatin sequencing (ATAC-seq), chromatin immunoprecipitation sequencing (ChIP-seq), RNA sequencing (RNA-seq), and proteomic datasets from the mutant hMGL panel reveal convergence at APOE in TREM2 and SORL1 mutant hMGLs. Transcriptomic analysis indicates that TREM2R47H and SORL1R744X (“KO”) hMGLs manifest AD-like microglial signatures (including up-regulation of APOE, LPL, GPNMB, and SPP1), indicating that these mutations may confer an “AD-primed” microglial state, while TREM2 KO hMGLs show reversed expression trends. Expression analysis indicates cross-regulation between SORL1, TREM2, and APOE, where SORL1 KO and TREM2R47H hMGLs show induced APOE expression. Epistasis modeling and validation suggests that SORL1 KO up-regulates TREM2 to up-regulate APOE expression. Further, AD-primed TREM2R47H and SORL1 KO hMGLs feature defects in Aβ uptake in vitro in an APOE-dependent manner, while xenotransplantation of AD-primed hMGLs in AD mouse brain revealed impaired Aβ uptake/clearance in vivo. Together, our results demonstrate APOE up-regulation as a convergence point for AD-priming mutations such as TREM2R47H and SORL1 KO. Use of this isogenic human ESC–derived modeling platform, AD mutant lines, and the multi-omic dataset provided here will expand our capacity to understand how AD mutations interact epistatically, modulate microglial function, and enhance AD pathogenesis.
Results
Establishing isogenic hMGLs harboring AD-associated genetic variants
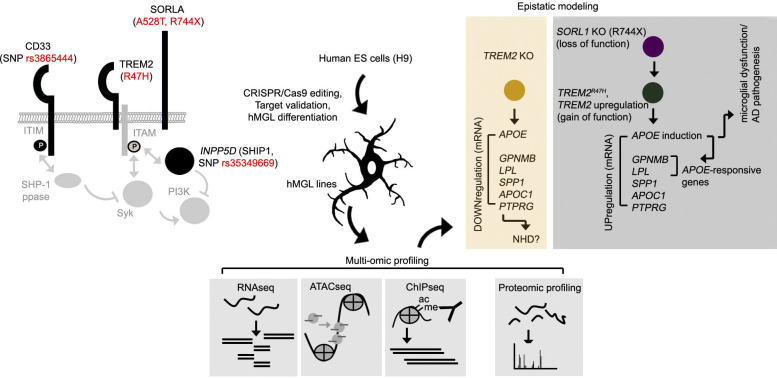
CD33, TREM2, and SORLA are all type I transmembrane components highly expressed in microglia, where CD33, TREM2, and the microglial cytosolic protein INPP5D converge at the Syk–PI3K signaling axis (Malik et al., 2015; Fig. 1 A). To characterize effects of these AD-associated variants in microglia, we integrated these mutations into the human H9 ESC line by CRISPR-Cas9 editing (Fig. S1 A), and designed an analysis scheme to characterize epigenetic and expression profiles of the hMGL panel, verify functional characteristics of mature hMGLs in vitro, and compare potential effects of AD mutations on hMGL behavior in vivo (Fig. 1 B and Fig. S1 B). Integrity of potential CRISPR-Cas9 off-target sites were validated by Sanger sequencing for each edited ESC line (Table S1 and Fig. S1, C and D). Edited H9 cell lines were then differentiated into mature hMGLs through a ∼38-d differentiation protocol (Fig. 1 C; McQuade et al., 2018), resulting in CD43/TREM2/PU.1/TMEM119/CX3CR1/Iba1-positive hMGLs (Fig. 1 C and Fig. S1 E). Transcriptomic profiles of WT hMGLs closely resemble mRNA expression in iPSC-derived microglia-like cells (iMGLs; GSE117829; Abud et al., 2017) as well as acutely isolated human microglia (Gosselin et al., 2017; Fig. 1 D). Principal-component analysis (PCA) revealed that hMGLs (purple) resemble iMGLs (dark and turquoise blue), fetal microglia (green), and adult microglia (light blue) but appear markedly different from other myeloid cells (light yellow or gold; Fig. 1 E).
Figure 1.
Integrating AD-associated genetic variations into an hMGL model. (A) Schematic depiction of AD-associated risk variants characterized. SNP variants for CD33 and INPP5D, as well as R47H, A528T, and R744X coding variants for TREM2 and SORL1 (SORLA) are marked in red. ITIM, immunoreceptor tyrosine-based inhibitory motif. (B) Workflow pipeline, for generating and characterizing AD-associated mutations in hMGLs. AD-associated coding or noncoding SNPs are introduced into corresponding genomic loci in human H9 ESC lines by CRISPR-Cas9 editing. Each line was characterized for targeted mutations and off-targeting variation before differentiation and maturation into hMGLs. hMGLs were subjected to multi-omic (RNA-seq, ATAC-seq, ChIP-seq, and label-free proteome) analysis, and functional characterization as indicated. (C) Isogenic microglial differentiation scheme used in this study. ESCs were differentiated into HPCs for 10 d, where CD43+ iHPCs are sorted (FACS plots) and cultured in serum-free media with MCSF, IL-34, TGF-β, and insulin; CD43 (green), CX3CR1 (red), Iba1 (purple), and DAPI (blue) staining is shown for HPCs at 10 d in vitro (DIV). Cells were differentiated to microglia for an additional 25 d, whereby maturation was induced by the addition of CD200 and CX3CL1. hMGLs were stained for TREM2 (red), CD43 (green), Iba1 (purple), and DAPI (blue) and compared with HPCs (bottom panels), or TMEM119 in hMGLs (red, bottom right) as indicated. Scale bars represent 100 µm (H9, left panel), 50 µm (mature hMGLs, right panel), and 20 µm (all fluorescence images). (D) Heatmap depicting RNA-seq profiles from human microglia (red; Galatro et al., 2017; GSE99074, red), hMGLs from this study (purple), iMGLs (Abud et al., 2017; GSE117829, green). (E) 3D PCA of hMGLs (this study, purple), iMGLs (GSE117829, turquoise; GSE89189, dark blue), human fetal microglia (GSE89189, green), human adult microglia (GSE89189, light blue), myeloid dendritic cells (GSE89189, light yellow), monocytes (GSE89189, gold). PCA reveals that hMGLs cluster closely with iMGLs and human adult/fetal microglia, and are distinct from myeloid cells.
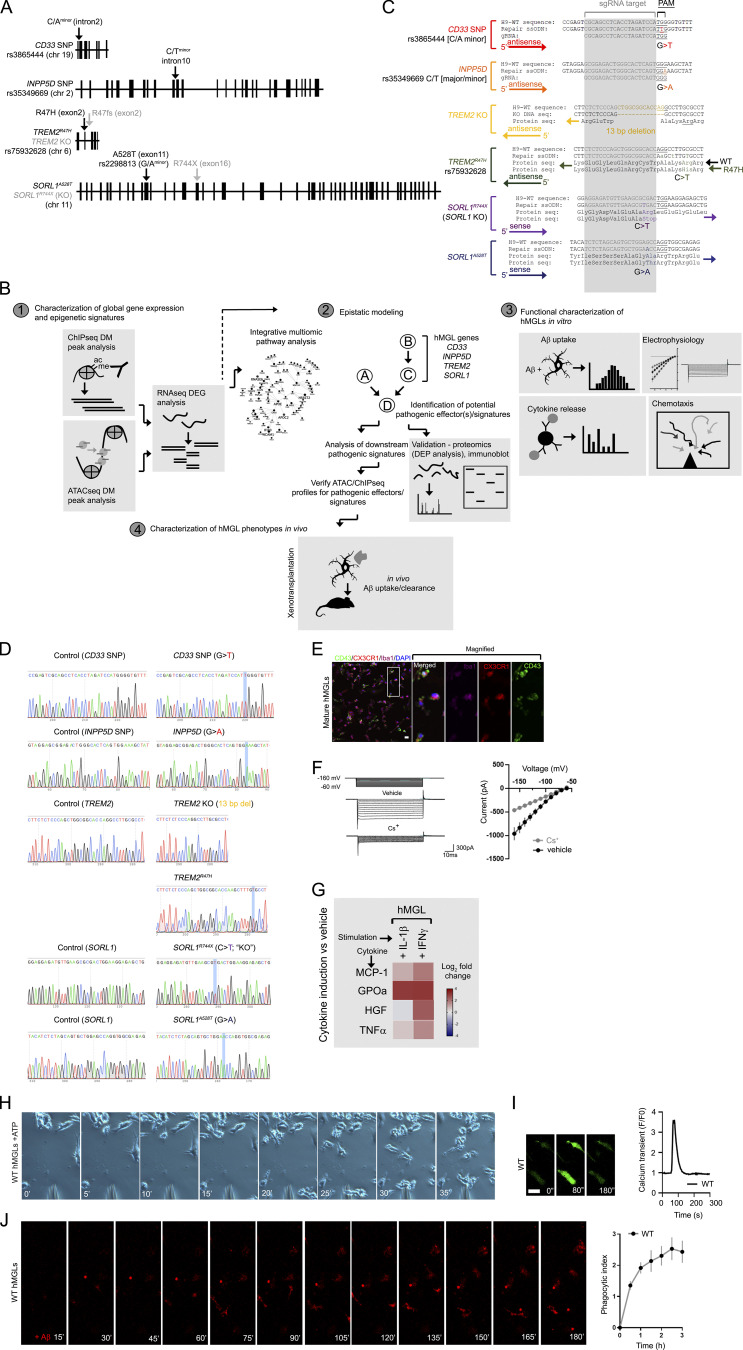
Figure S1.
Gene targeting and experimental strategy for hMGL differentiation and characterization. (A) Schematic representation of the genomic location and intron/exon schematic of AD risk SNPs CD33, INPP5D, TREM2, and SORL1 in this study. (B) Schematic diagram of the analytical workflow for this study. RNA-seq datasets from the hMGL lines (1) are analyzed for cross-regulatory interactions to generate an epistatic model (2) and identify potential pathogenic effectors or signatures. hMGL lines are characterized for physiological microglial function (3) and interactions with Aβ in immunodeficient human MCSF knockin mouse brain xenotransplants (4). (C) Representative sequences of various isogenic clones in AD-associated mutant ESC lines and H9-WT sequences. Repair single-strand donor (ssODN) templates, sgRNA (gray), corresponding amino acids, DNA directionality (arrow, 5′ to 3′) and nucleotide substitutions are shown. For TREM2R47H, two synonymous mutations were introduced in the repair ssODN, generating a new HindIII restriction site (lowercase) for consequent clone screening. (D) Sanger sequencing and validation of CD33 SNP, INPP5D SNP, TREM2 KO, TREM2R47H, SORL1 KO, and SORL1A528T lines and isogenic controls (nontargeting sgRNA). The WT H9 ESC line is heterozygous for G/A INPP5D SNPs; CRISPR-Cas9 editing was performed to convert H9 homozygously to the INPP5D “A” allele. All other modifications were converted homozygously in the H9 ESC lines. (E) After maturation induced by exposure to CD200 and CX3CL1, hMGLs were stained for CX3CR1 (red), CD43 (green), Iba1 (purple), and DAPI (blue) as indicated. Scale bar, 20 µm. (F) Representative inward currents from WT hMGLs; hyperpolarizing voltage steps from −160 mV to −60 mV were applied in the absence (top) or presence of Cs+ (bottom). At right panel, quantification of inward currents as measured in the absence (black) or presence of Cs+ (gray). (G) Induction of cytokines and chemokines in WT hMGLs stimulated with IL-1β (20 ng/ml) and IFN-γ (20 ng/ml) as determined by ELISA multiplex assay. Heatmaps indicate log2 fold change of cytokines/chemokines indicated (MCP-1, GPOa, HGF, TNFα) above vehicle treatment. Results are from three replicate cultures in three independent experiments. (H) Representative time-lapse images showing WT hMGL migration toward to ATP source (a pipette tip). (I) Representative images of calcium imaging over the time periods as indicated with 100 µM ATP stimulation. Scale bar, 25 µm. Graphs (right) depict Ca2+ traces depicting changes in Fluo-4 fluorescence over the baseline (ΔF/F0) in response to 100 µM ATP in the WT hMGLs. Results are derived from averaged values in three replicate cultures and three experiments. (J) Representative time-lapse images of fluorescent Aβ1-42 oligomers (red) bound to WT hMGLs, imaged by automated live-cell microscopy. In the adjacent graph, phagocytosis of Aβ1-42 oligomers in WT hMGLs over time was quantified, as depicted on the left. PI was determined by measuring average fluorescence intensity at each time point in comparison to the 15-min time point (set to 1.0). Images in E–J are representative of three independent experiments. Values represent mean ± SEM from n = 3 independent experiments.
To functionally characterize hMGLs in vitro, we assayed hMGL response to various physiological and pathological stimuli. We applied an increasing voltage cell ramp (−160 mV to −60 mV) in the presence or absence of Cs+ to block K+-mediated inward currents and characterized changes in inward current at varying holding voltage steps in whole-cell patch-clamp recordings (Fig. S1 F). We observed a proportional increase in inward current in hMGLs, which was largely blocked with Cs+ (Fig. S1 F). Stimulation of hMGLs with proinflammatory cytokines such as IL-1β or IFN-γ showed significant cytokine induction (Fig. S1 G). Further, hMGLs also demonstrated positive purinergic chemotactic response toward an ATP source (Fig. S1 H and Video 1) and showed a typical calcium transient response in response to purinergic stimulation (Fig. S1 I; Brawek et al., 2014; Konttinen et al., 2019). We observed robust uptake of Aβ (fluorescently labeled HiLyte Fluor 555-Aβ or 555-Aβ hereon) in hMGLs; Fig. S1 J); cumulative phagocytic index (PI) was calculated by progressive uptake with 555-Aβ exposure as a function of normalized fluorescence intensity (Fig. S1 J, right graph). Together, these results demonstrate utility of hMGLs as a versatile cellular model that can recapitulate physiological and pathological microglial behavior.
Video 1.
Chemotactic migration of WT hMGLs toward an ATP gradient (1 mM) leaking from a micropipette. hMGLs were exposed to 1 mM ATPγS from a pipette tip. Images were acquired every minute for 30 min, and the video shown depicts migration at 6 frames/s.
Comparison of epigenetic and expression landscapes in AD mutant hMGLs reveal APOE as a convergent pathogenic node
To establish and compare epigenetic and expression profiles in AD mutant hMGLs, we performed RNA-seq, ATAC-seq, H3K27 acetylation (H3K27ac), and H3K4 trimethylation (H3K4me3) by ChIP-seq and label-free mass spectrometry analysis of our hMGL panel. TREM2R47H hMGLs featured the highest degree of change at both epigenetic (ATAC- and ChIP-seq; Fig. S2 A) and transcriptomic levels compared to WT (Fig. S2 B). However, proteomic profiles from SORL1A528T hMGLs specifically showed the highest degree of change compared with other hMGL lines (Fig. S2 B). While significant overlap in differentially expressed proteins (DEPs) in SORL1 KO and A528T lines was observed (Fig. S2, C and D), unique DEPs in SORL1A528T may suggest abnormalities in protein stability or turnover associated with SORL1A528T hMGLs. As a component of the endosomal retromer complex (Fjorback et al., 2012; Huang et al., 2016), which regulates trafficking of various protein substrates from the endosome to the lysosome (Van Acker et al., 2019), mutations in the cargo-binding SORLA VPS10 domain (such as A528T) may impair SORLA-dependent protein homeostasis. In agreement with this, Gene Ontology (GO) cellular component (CC) analysis of up-regulated DEPs in the SORL1A528T hMGL proteome indicates enrichment of components at the cell surface (CC: 0009986, “Cell surface”) and early endosome (CC: 0005769, “Early endosome”; Fig. S2 E), while down-regulated DEPs showed enrichment in KEGG (Kyoto Encyclopedia of Genes and Genomes) lysosome pathways (KEGG hsa04142: “Lysosome”; Fig. S2 F) and CC endosome categories (CC: 0005768, “Endosome”; Fig. S2 G). These results suggest that SORL1A528T mutation manifests accumulation of cell surface/early endosomal components, while down-regulating lysosome and endosomal components.
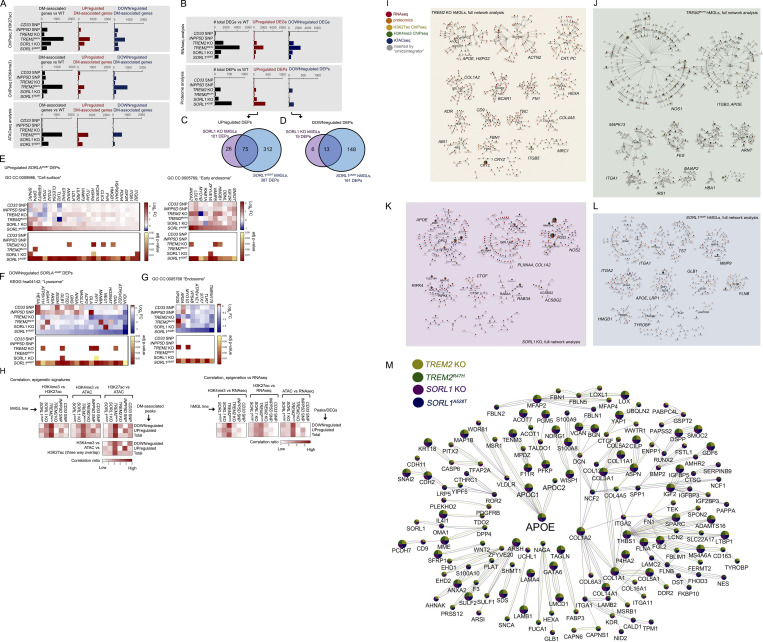
Figure S2.
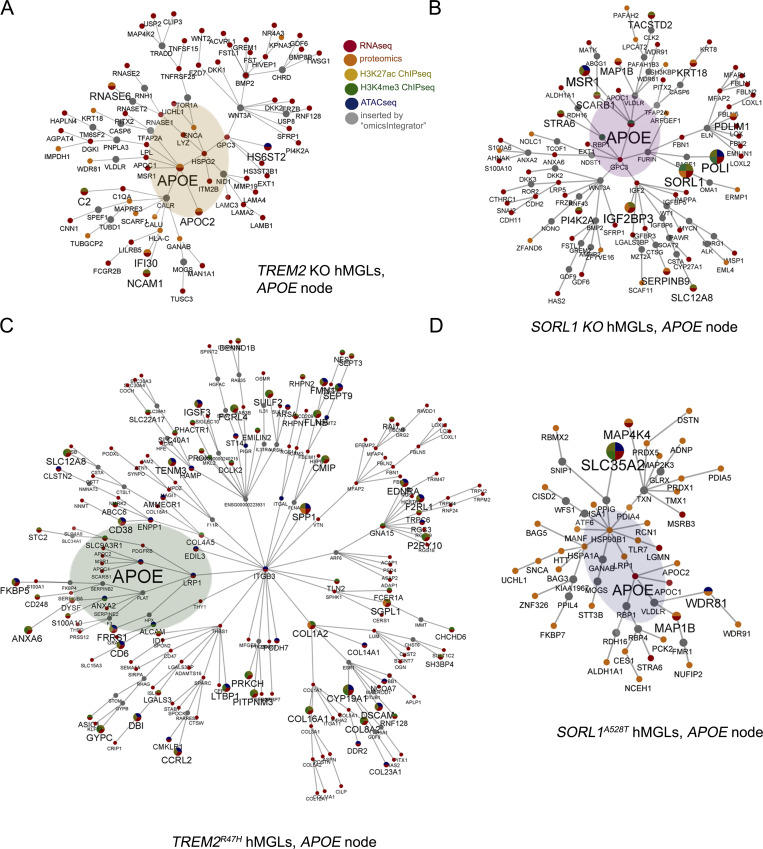
Overview of epigenetic, transcriptomic, and proteomic changes; correlation between epigenetic and transcriptomic signatures; and integrative multi-omic analysis of gene landscapes in mutant hMGLs. (A and B) Overall total number of DM gene-associated peak signals by ATAC-seq, and ChIP-seq (A) DEGs (q < 0.1), and DEPs by RNA-seq/proteomic analysis, respectively (B), in hMGLs generated and characterized in this study (black bars). Numbers of up-regulated (middle graphs, red bars) and down-regulated (right graphs, blue bars) DEGs, DEPs, and DM-associated genes are also presented as indicated. (C–G) Analysis of DEPs in SORL1A528T hMGLs, showing Venn overlap analysis between up-regulated (C) and down-regulated (D) DEPs in SORL1 KO (purple) and A528T hMGLs (blue). GO DAVID analysis of up-regulated SORL1A528T DEPs show enrichment in “cell surface” and “early endosome” CCs (E), whereas down-regulated SORL1A528T DEPs feature enrichment in lysosome-associated KEGG pathways (F) and endosome components (G). (H) Correlation of overlapping DM-associated epigenetic signatures (heatmaps, left). Heatmap depicting relative overlap between down-regulated and up-regulated ChIP-seq and ATAC-seq DM-associated peak signatures in hMGL lines, as calculated from overlapping components from ChIP-seq and ATAC-seq datasets (left heatmaps). Correlation of overlapping DM-associated epigenetic and transcriptomic signatures (heatmaps, right). Heatmap depicting relative overlap between down-regulated and up-regulated DEGs (RNA-seq) and epigenetic DM-associated peak profiles. Pairwise correlation ratios in Venn overlap diagrams (see H in Data S1; top heatmaps) are ratios of overlapping up-regulated, down-regulated, or total signatures over the total signatures (for each pairwise category) observed in each line (XhMGL line value for each hMGL line); ratios are normalized by dividing each XhMGL line value over the sum of all XhMGL line values for up-regulated, down-regulated, or total signatures. Heatmaps for three-way overlap (bottom heatmap) are ratios of down-regulated, up-regulated, or total overlapping signatures observed in all three epigenetic datasets over the total number of signatures for each hMGL line (YhMGL line value). Normalized values were established by dividing YhMGL line values from each line over the total of all YhMGL line values. Normalized correlation ratios were calculated for epigenetic/RNA-seq heatmaps in right heatmaps, as described for left heatmaps (ChIP-seq/ATAC-seq correlation). (I–L) Full molecular pathway analyses of TREM2 KO, TREM2R47H, SORL1 KO, and SORL1A528T hMGLs through integrative multi-omic analysis (OmicsIntegrator). Complete map of genes dysregulated in the mutant TREM2 KO (I), TREM2R47H (J), SORL1 KO (K), and SORL1A528T (L) hMGL lines compared to WT. Branches were defined in the OmicsIntegrator network by removing the root node. Resulting branches are displayed in a grid layout, with the most central gene highlighted with large font and labeled adjacently for each cytoscape network group with significantly centralized nodes. Node color schemes defined by various analytical platforms are indicated in I as red (RNA-seq), orange (proteomics), yellow (H3K27ac ChIP-seq), green (H3K4me3 ChIP-seq), blue (ATAC-seq), and gray (inserted by the OmicsIntegrator algorithm). See extended Data S1 for enlarged network diagrams for I–L. (M) The network of gene commonly altered across multiple cell lines. Integrative analysis implicate dysregulation of APOE in TREM2 KO and R47H, and SORL1 KO and A528T hMGL lines. Genes depicted within the Cytoscape network map were affected in one or more analytical dataset by at least three out of the four TREM2/SORL1 mutant hMGL cell lines compared to WT. Colored pie charts comprising network nodes indicate hMGL lines dysregulated at particular nodes; larger nodes indicate genes modulated in all four hMGL lines. AD-related genes of particular interest (APOE, APOC1, and APOC2) were intentionally highlighted with an enlarged font size. Edge colors indicate an edge identified within the multi-omic network for a particular hMGL line; a color scheme for nodes and edges within the network is shown (TREM2 KO, yellow; TREM2R47H, green; SORL1 KO, purple; SORL1A528T, blue).
Although we observed correlative epigenetic (H3K27ac ChIP-seq, H3K4me3 ChIP-seq, and ATAC-seq) and transcriptional (RNA-seq) overlap in most lines, we found that due to marked elevation in epigenetic and transcriptomic gene signatures, TREM2R47H hMGLs showed the greatest degree of overlap (Fig. S2 H). Overall, these results indicate that TREM2R47H hMGLs exhibit significant alterations in epigenetic and transcriptional signatures. We used the OmicsIntegrator software package (Tuncbag et al., 2016) to perform combinatorial analyses using epigenetic and expression datasets to identify potential overlapping pathways or convergent hubs within the mutant hMGL panel. As mild differences were observed with CD33 and INPP5D single-nucleotide polymorphisms (SNP; in comparison to WT) within the hMGL panel, we excluded datasets from these hMGL lines in our analysis. First, we constructed individual integrated multi-omic network maps for TREM2 KO, TREM2R47H, SORL1 KO, and SORL1A528T hMGLs (datasets compared with WT; Fig. S2, I–L). To further elucidate and identify commonly affected pathways and processes by each cell line, we combined individual OmicsIntegrator network nodes and edges (Fig. S2, I–L) and identified APOE as a shared network node in these hMGL lines (Fig. 2, A–D). Comparison of networks identified in TREM2 KO, TREM2R47H, SORL1 KO, and SORL1A528T hMGLs reveal significant convergent intersection at the APOE locus reiterated in one or more “-omics” dataset (Fig. S2 M), further implicating APOE dysregulation in TREM2 and SORL1 mutant hMGLs.
Figure 2.
Integrative multi-omic analysis demonstrates convergence at the APOE locus in mutant hMGLs. (A–D) APOE pathway network in individual hMGL lines. OmicsIntegrator network branches containing APOE is depicted from multi-omic TREM2 KO (A), TREM2R47H (C), SORL1 KO (B), and SORL1A528T (D) hMGL datasets. Color scheme for nodes identified by a particular analytical dataset is shown in A; node and font size are scaled according to identification across multi-omic datasets. APOE is intentionally marked with an increased font size, and adjacent nodes are highlighted. Gray nodes depict node inserts applied by the OmicsIntegrator algorithm to connect adjacent networks.
Characterizing AD-associated microglial gene signatures in mutant hMGLs
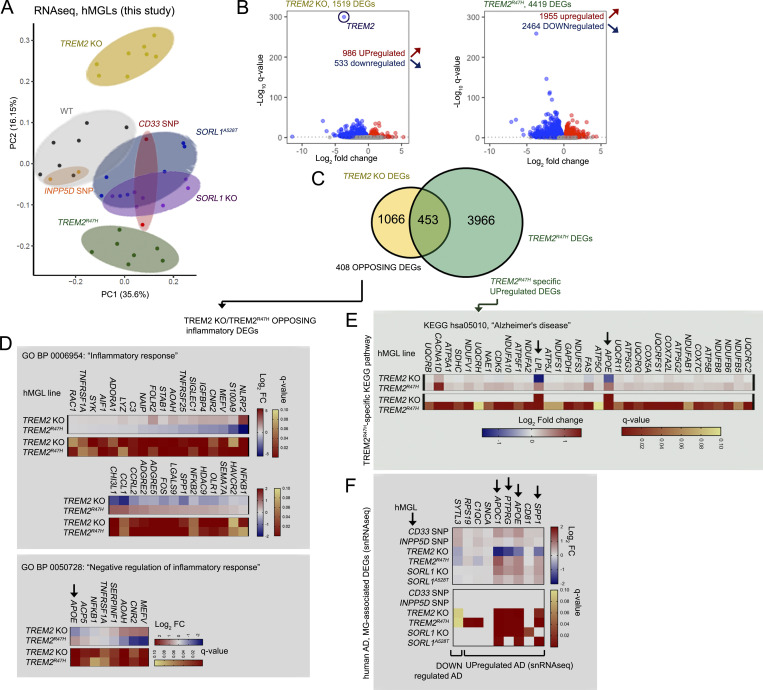
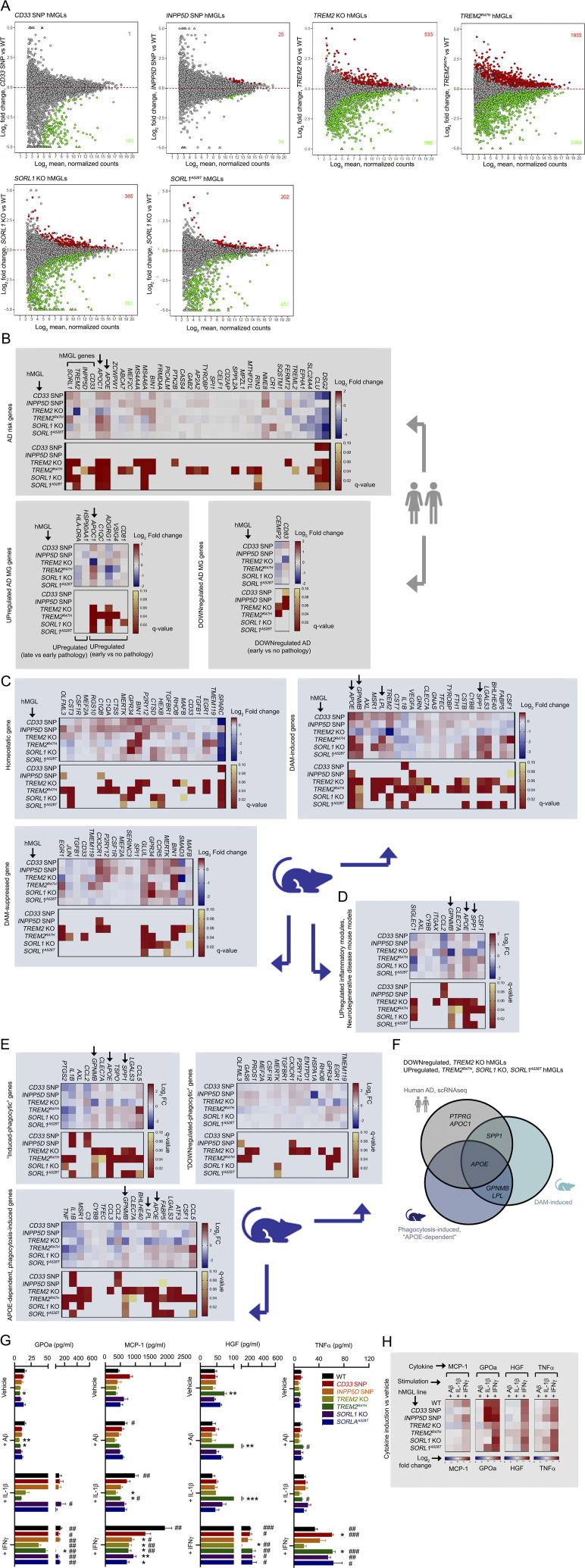
PCA of the hMGL panel demonstrated separation of WT hMGLs from CD33 SNP, TREM2 KO, TREM2R47H, SORL1 KO, and SORL1A528T hMGLs (Fig. 3 A and Fig. S3 A). Surprisingly, we observed a high degree of separation between TREM2 KO and TREM2R47H, which formed clusters distinct from WT, INPP5D SNP, CD33 SNP, SORL1 KO, and A528T hMGLs (Fig. 3 A and Fig. S3 A). Given the separation between TREM2 KO and TREM2R47H hMGLs (Fig. 3 A), we compared and characterized up-regulated and down-regulated differentially expressed genes (DEGs) in TREM2 KO and TREM2R47H RNA-seq datasets (Fig. 3, B and C). We observed some overlap between up-regulated/down-regulated TREM2 KO and TREM2R47H DEGs; 453 DEGs were found to overlap between TREM2 KO (1,519 DEGs) and TREM2R47H (4,419 DEGs) RNA-seq datasets (Fig. 3, B and C). Although APOE nodes were consistently present in multi-omic network maps in TREM2 KO, TREM2R47H, SORL1 KO, and SORL1A528T hMGLs, it remains unclear whether APOE and its associated pathways are up-regulated or down-regulated in these cell lines. GO analysis of genes with opposing up-regulatory/down-regulatory trends in TREM2 KO and TREM2R47H hMGLs showed enrichment of GO biological process (BP) components in “inflammatory response” (GO BP: 0006954) and “negative regulation of inflammatory response” (GO BP: 0050728) categories (Fig. 3 D). Interestingly, APOE was found to be down-regulated in TREM2 KO and up-regulated in TREM2R47H hMGLs (Fig. 3 D). KEGG pathway analysis also revealed enrichment of up-regulated AD-associated gene components, including APOE and LPL in TREM2R47H DEGs (Fig. 3 E); SORL1 KO and SORL1A528T also increased APOE and APOC1 (Fig. 3 F). Comparing DEGs identified from bulk or single-cell transcriptomic profiles in AD (Abud et al., 2017; Gosselin et al., 2017; Mathys et al., 2019; McQuade et al., 2018) and disease-associated microglial (DAM) signatures (Hickman et al., 2013; Kamphuis et al., 2016; Keren-Shaul et al., 2017; Krasemann et al., 2017; Yin et al., 2017), we found that genes such as APOE, APOC1, SPP1, and PTPRG were down-regulated in TREM2 KO and significantly up-regulated in TREM2R47H, SORL1 KO, and A528T (Fig. 3 F and Fig. S3 B; Harold et al., 2009; Hollingworth et al., 2011; Jansen et al., 2019; Kunkle et al., 2019; Lambert et al., 2009, 2013). Similarly, numerous DAM-induced genes previously described in AD mouse brain such as APOE, GPNMB, LPL, and SPP1 were also down-regulated in TREM2 KO and up-regulated in TREM2R47H hMGLs (Fig. S3, C and D). Interestingly, GPNMB and LPL were previously shown to be induced in microglia in an APOE-dependent manner (Krasemann et al., 2017; Fig. S3, E and F). These results suggest that within this hMGL panel, two distinct transcriptomic profiles are apparent: an AD-primed hMGL group in which induced AD-genes are up-regulated (TREM2R47H, SORL1 KO, and A528T) and TREM2 KO, which conversely shows silencing of these induced AD-related gene signatures. Importantly, microglial APOE up-regulation is observed in human AD (Mathys et al., 2019), mouse microglia models of neurodegeneration (Krasemann et al., 2017), and mouse AD brain (Keren-Shaul et al., 2017; Fig. S3 F). Thus, reciprocal expression of AD-associated microglial signatures in TREM2 KO and TREM2R47H hMGLs implicates induction of an AD-primed state in TREM2R47H, SORL1 KO, and A528T, potentially triggering pathogenic effects through APOE induction.
Figure 3.
AD-related transcriptomic signatures are altered in mutant hMGLs. (A) PCA plots derived from RNA-seq profiles in mature CD33 SNP (n = 2, red), INPP5D SNP (n = 2, orange), TREM2 KO (n = 8, yellow), TREM2R47H (n = 7, green), SORL1 KO (n = 7, purple), and SORL1A528T (n = 7, blue) and WT (n = 7, black) hMGLs. Log2-transformed transcripts per million values were used in the PCA after adjusting for batch effect. (B–F) Transcriptomic analysis reveals divergence of TREM2 KO and R47H hMGLs. (B) Volcano plot depicting significantly up-regulated (red) and down-regulated (blue) differentially regulated genes (DEGs, q < 0.1) in TREM2 KO (yellow) and TREM2R47H hMGLs; log10 q-values (y axis) and log2 fold change (x axis) are plotted for individual DEGs. (C) Venn diagram depicts overlapping DEGs in TREM2 KO and R47H hMGL lines with matched up- or down-regulatory trends. GO analysis of 408 DEGs showing opposing up- or down-regulatory expression trends demonstrate enrichment in BP categories including “inflammatory response” (D, top) and “negative regulation of inflammatory response” (D, bottom). GO analysis of TREM2R47H hMGLs indicate enrichment of DEGs related to KEGG “Alzheimer’s disease” pathways (E). (F) Analysis of RNA-seq trends in microglial genes previously shown to be altered in human AD in hMGLs. Heatmaps in D–F indicate log2 fold change (blue/red; top) and q-value (red/yellow; bottom); arrows indicate AD-associated DEGs with opposing trends in TREM2 KO and R47H hMGLs.
Figure S3.
Transcriptomic analysis of mutant hMGLs. (A) MA plots depicting differential gene expression in the different hMGL lines characterized by RNA-seq analysis. Log2 fold change (y axis) and log2 mean normalized counts (x axis) are shown. Red (up-regulated) and green (down-regulated) components, and DEG counts are indicated with at a q-value cutoff of q < 0.1. (B) Characterizing human AD-associated transcriptomic gene profiles in hMGLs. Heatmap profiles depicting log2 fold change (red/blue) and q-value (red/yellow) of DEGs related to microglial genes potentially linked AD risk (upper graph), and differentially up-regulated (lower left) or down-regulated (lower right) in human AD previously characterized by single cell RNA-seq (Mathys et al., 2019). (C) Homeostatic or DAM transcriptomic gene profiles of hMGLs. Up-regulated or down-regulated DEG profiles and q-values for the hMGL panel were plotted as indicated for homeostatic, DAM-induced, or DAM-suppressed microglial DEG profiles (RNA-seq) previously characterized in AD mice. Color scale represents log2 fold change (blue/red) or q-value (red/yellow). (D) hMGL RNA-seq profiles were characterized for up-regulated inflammatory components identified in various neurodegenerative mouse disease models (Krasemann et al., 2017). Arrows in B–D mark genes down-regulated in TREM2 KO and up-regulated in TREM2R47H hMGLs. (E) hMGL RNA-seq profiles were characterized for genes induced (upper left) and suppressed (upper right) in phagocytically active microglia. hMGL RNA-seq profiles were also characterized for microglia genes induced by phagocytosis only in the presence of APOE (APOE-dependent genes induced by phagocytosis; Krasemann et al., 2017). Genes down-regulated in TREM2 KO and up-regulated in TREM2R47H hMGLs are marked by arrows. (F) Genes from human AD and AD/phagocytic microglia mouse models down-regulated in TREM2 KO and up-regulated in TREM2R47H hMGLs; APOE is shared among all three groups. (G) Secretion of cytokines and chemokines in hMGLs stimulated with Aβ1-42 oligomers (1 µM), IL-1β (20 ng/ml), and IFN-γ (20 ng/ml) as determined by ELISA multiplex assay (mean± SEM). Statistical analyses were determined by two-way ANOVA with Dunnett's multiple comparison. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 were derived by comparing each genotype vs. WT under different stimuli. #, P < 0.05; ##, P < 0.01; and ###, P < 0.001 were derived by comparing different stimuli vs. vehicle within the same genotype. (H) Induction of cytokines and chemokines in hMGLs stimulated with Aβ1-42 oligomers (1 µM), IL-1β (20 ng/ml), and IFN-γ (20 ng/ml) as determined by ELISA multiplex assay. Heatmaps indicate log2 fold change of cytokines/chemokines indicated (MCP-1, GPOa, HGF, and TNF-α) above vehicle treatment. Results from G and H are derived from three replicate cultures in n = 3 independent experiments.
We also note interesting differences in cytokine induction profiles in TREM2 KO (non-AD) and AD-primed (TREM2R47H, SORL1 KO, and A528T) hMGLs. Specifically, we observed that AD-primed TREM2R47H, SORL1 KO, and A528T hMGLs showed enhanced IFN-γ–induced TNF-α secretion compared with WT, while TREM2 KO hMGLs showed little or no TNF-α induction in response to IFN-γ (Fig. S3, G and H). Similarly, we also see attenuated IFN-γ–induced hepatocyte growth factor (HGF) induction in TREM2 KO hMGLs, but not in other hMGL lines (Fig. S3, G and H). This suggests that alterations in DAM AD signatures are potentially associated with changes in IFN-γ–induced cytokine response; TREM2 KO hMGLs feature an impaired response, whereas AD-primed microglia show a normal (HGF) or enhanced (TNFα) cytokine response compared to WT hMGLs.
A SORL1-TREM2-APOE regulatory axis regulates microglial phagocytosis
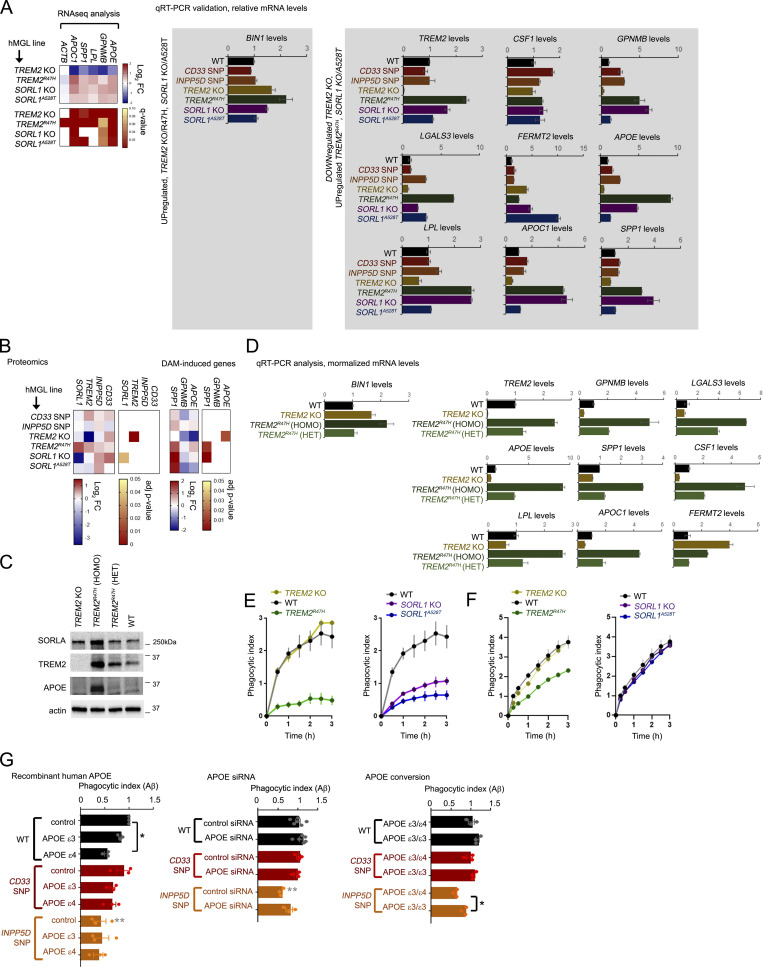
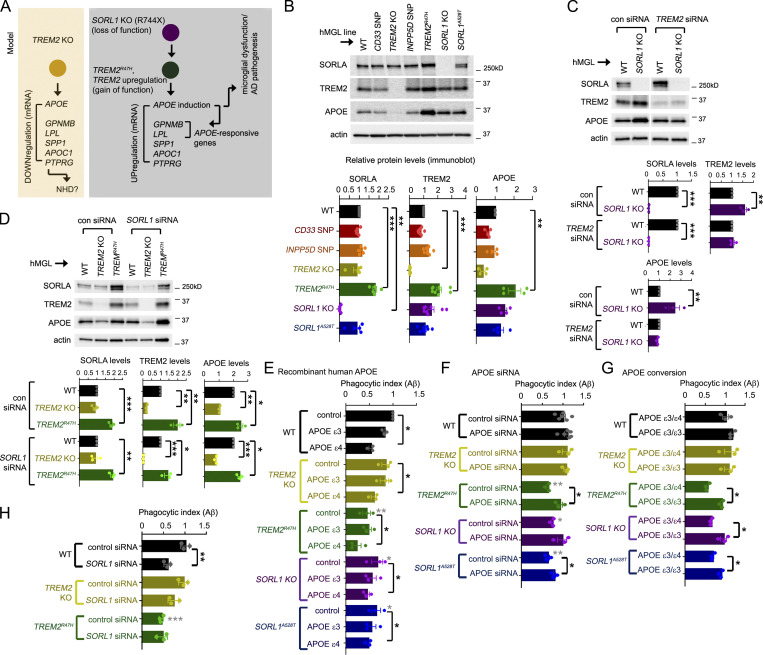
Next, we searched for cross-regulatory trends that may indicate epistatic relationships between genes within the hMGL panel; SORL1 levels were significantly up-regulated in TREM2R47H hMGLs, while TREM2 levels were also up-regulated in SORL1 KO and A528T hMGLs (Fig. S3 B). This suggests that either SORL1 modulation (KO or A528T) may up-regulate TREM2, or alternatively, TREM2 modulation (R47H) may up-regulate SORL1 to induce APOE/DAM signatures. It is unlikely that SORL1 up-regulation downstream of TREM2 KO/R47H alterations up-regulates AD signatures such as APOE, since SORL1 deletion rather than induction drives APOE up-regulation (Fig. 3 F). Given that SORL1 KO and TREM2R47H hMGLs both showed significant up-regulation of AD/DAM signatures such as APOE, GPNMB, LPL, APOC1, and SPP1 by RNA-seq and quantitative RT-PCR analysis (qRT-PCR; Fig. 3 F; Fig. S3, A–E; and Fig. S4 A), we propose an epistasis model where SORL1 deficiency up-regulates TREM2 to induce AD-associated gene signatures such as APOE (Fig. 4 A).
Figure S4.
Analysis of expression and epigenetic profiles at AD and DAM-associated gene loci. (A) qRT-PCR validation of AD/DAM-related targets. AD-related components up-regulated in TREM2 KO/R47H and SORL1 KO/A528T (left), or down-regulated in TREM2 KO/up-regulated in TREM2R47H hMGLs by RNA-seq analysis were verified in hMGLs by qRT-PCR analysis (normalized over actin, and compared with WT, set to 1.0). One representative experiment is shown (mean ± SEM, n = 3 replicates). Heatmap of relevant targets from RNA-seq analysis is shown (right). (B) Evaluation of hMGL genes (left) and AD/DAM-related targets in the hMGL lines by label-free proteomic analysis. (C) SORLA, TREM2, and APOE levels were characterized in TREM2 KO; TREM2R47H homozygous (HOMO) and heterozygous (HET) hMGLs were compared with WT by immunoblot analysis. Western blot shown is representative of three independent experiments. (D) AD/DAM-related targets were quantified in TREM2 KO (yellow bars), TREM2R47H homozygous (HOMO, dark green), and heterozygous (HET, lighter green) hMGLs by qRT-PCR analysis. Graphed results are from n = 3 independent experiments. (E and F) Phagocytosis of Aβ1-42 (555-Aβ; E) or tau (555-tau; F) oligomers in WT and AD-variant hMGLs over time. PI was determined by measuring average fluorescence intensity in individual hMGL lines in comparison to the 15 min time point, and normalized to WT at the 15 min time point (set to 1.0). Values represent mean ± SEM from n = 3 replicate wells. Mean phagocytotic index values for the hMGL lines was quantified at the 3 h time point, and shown in the adjacent bar graph (mean ± SEM). (G) WT or mutant hMGLs were treated with 5 µg/ml recombinant human APOEε3 or APOEε4 or left untreated (control), together with 555-Aβ1-42 oligomers (left graph). Phagocytosis of 555-Aβ1-42 oligomers was measured in real time, and PI was determined by measuring average fluorescence intensity in individual hMGL lines in comparison to the 15 min time point, and normalized to WT at the 15 min time point (set to 1.0). Values represent mean ± SEM from n = 3 independent experiments. Mean PI values for the hMGL lines was quantified at the 3-h time point, and shown in the bar graph. Middle graph: mutant hMGL lines transfected with 25 nM control or APOE siRNAs were assayed for Aβ phagocytosis. Right graph: Mutant hMGL lines with endogenous APOEε3/ε4 allele combinations or edited APOEε3/ε3 alleles were assayed for Aβ phagocytosis. In G, analyses were assays were validated using two independently derived hMGL clones. Individual plots represent three replicates in n = 3 independent experiments. Graph depicts mean ± SEM of averaged values from independent experiments. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison (*, P < 0.05; **, P < 0.01). Black significance values indicate significance within treatments within one hMGL line, and gray significance values indicate differences between like treatments between different hMGL lines.
Figure 4.
Epistatic analysis of hMGL mutations and characterization of a SORL1–TREM2–APOE regulatory axis in Aβ phagocytosis in microglia. (A) Epistasis model: TREM2 KO down-regulates APOE and other induced AD-associated DEG signatures (yellow box). SORL1 KO may up-regulate TREM2; TREM2R47H may induce APOE and other APOE-responsive or AD-related microglial DEGs (gray box). (B) SORLA, TREM2, and APOE levels in the hMGL lines indicated were detected by immunoblot; relative SORLA, TREM2, and APOE levels were normalized to actin and quantified by densitometry in the lower graphs. All values were normalized to WT hMGLs (set to 1.0). (C) WT or SORL1 KO hMGLs were transfected with 25 nM control (con) or TREM2-targeting siRNAs, and lysates were immunoblotted for the components indicated. Graphs depict SORLA, TREM2, and APOE levels individually normalized to actin in WT (black bars) and SORL1 hMGLs (purple); values were compared to WT in control or TREM2 siRNA groups (set to 1.0). (D) WT, TREM2 KO or R47H hMGLs were transfected with 25 nM control or SORL1-targeting siRNAs, and lysates were subsequently immunoblotted for the components indicated. Graphs depict SORLA, TREM2, and APOE levels individually normalized to actin in WT (black bars), TREM2 KO (yellow bars), and TREM2R47H hMGLs (green bars); values were compared with WT in control or SORL1 siRNA groups (set to 1.0). (E–G) Characterizing effects of modulating APOE on hMGL-dependent Aβ binding and uptake. (E) WT or mutant hMGLs were treated with 5 µg/ml recombinant human APOEε3 or APOEε4 or left untreated (control) in the presence of 555-Aβ1-42 oligomers. Phagocytosis of 555-Aβ1-42 oligomers was measured in real time, and phagocytic index (PI) was determined by measuring average fluorescence intensity in individual hMGL lines in comparison to the 15-min time point and normalized to WT at the 15 min time point (set to 1.0). Values represent mean ± SEM from n = 3 independent experiments. Mean phagocytotic index values for each hMGL line was quantified at the 3 h time point, as shown in the bar graph. (F) Mutant hMGL lines transfected with 25 nM control or APOE siRNAs were assayed for Aβ phagocytosis as in E. (G) Mutant hMGL lines with endogenous APOEε3/ε4 allele combinations, or edited APOEε3/ε3 alleles were assayed for Aβ phagocytosis as in E. (H) WT, TREM2 KO, or R47H hMGL lines were transfected with 25 nM control or SORL1 siRNA oligos, and assayed for Aβ phagocytosis as in E. In B–H, analyses were validated using two independently derived hMGL clones; individual plots in B–D represent results from independent experiments (B, n = 4; C and D, n = 3), and plots in E–H represent phagocytosis results from averages of three replicate values from each independent experiment. All graphs depict mean ± SEM. Statistical significance in B and D were calculated using one-way ANOVA with Tukey’s multiple comparison; statistical significance in E–H was determined by two-way ANOVA with Dunnett’s multiple comparison (*, P < 0.05; **, P < 0.01; ***, P < 0.001); and statistical significance in C was determined by Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Gray asterisks in E, F, and H depict statistical differences between WT control, as determined by two-way ANOVA with Bonferroni–Dunn correction (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
We confirmed down-regulation of APOE in TREM2 KO hMGLs and up-regulation of APOE in TREM2R47H and SORL1 KO hMGLs by proteomic analysis (Fig. S4 B). We also performed immunoblot analysis of lysates from two independent clones in our hMGL panel (Fig. 4 B). Comparing APOE levels in WT, heterozygous TREM2R47H, and TREM2R47H hMGL lines, we observed dose-dependent expression of APOE in relation to TREM2R47H (Fig. S4 C). Similar dose-dependent trends were also observed for LPL, GPNMB, SPP1, APOC1, and LGALS3 (Fig. S4 D). Next, we determined whether downstream TREM2 induction was required for APOE up-regulation in SORL1 KO hMGLs. We found that TREM2 siRNA transfection abrogated APOE up-regulation in SORL1 KO hMGLs (Fig. 4 C). Conversely, we found that SORL1 siRNA had little or no effect on APOE levels in TREM2 KO and TREM2R47H hMGLs (Fig. 4 D), indicating that SORL1 is likely not a downstream (but rather an upstream) effector for TREM2 function. These results are in agreement with our proposed epistasis model, implicating APOE induction as a convergent pathogenic factor for SORL1 and TREM2 (Fig. 4 A).
The APOEε4 allele is a significant risk factor for sporadic AD, in which excessive expression of APOE has been shown to aggravate Aβ-plaque deposition in AD mouse models. Together, this suggests that up-regulation of APOEε4, and perhaps to a lesser extent, APOEε3 may impair phagocytic uptake in response to Aβ (Shi et al., 2019; Ulrich et al., 2018). To test this, we determined whether recombinant human APOEε3 or APOEε4 could alter Aβ binding/uptake in our hMGL panel. Using real-time Aβ binding/uptake assays (Fig. S1 J), we compared uptake of fluorescent Aβ oligomers in WT, TREM2, and SORL1 mutant hMGLs (Fig. S4 E). Similar to our previous observations with Trem2 KO mouse microglia (Zhao et al., 2018), TREM2 KO hMGLs showed little or no defect in Aβ binding/phagocytosis (Fig. S4 E). Interestingly, we observed that AD-primed (TREM2R47H, SORL1 KO, and A528T) hMGLs all showed defects in Aβ binding/uptake in vitro (Fig. S4 E). We also observe that TREM2R47H hMGLs feature defects in binding/uptake of oligomeric tau, while TREM2 KO, SORL1 KO, and A528T showed little or no difference in tau uptake compared to WT hMGLs (Fig. S4 F). This suggests that SORL1 deficiency causes specific defects in Aβ binding/uptake, while TREM2R47H may be associated with alterations that affect both Aβ and tau binding and/or internalization.
While the presence of APOEε3 inhibited Aβ binding/uptake to some extent, APOEε4 showed enhanced inhibition of Aβ binding/uptake in almost all hMGL lines (Fig. 4 E and Fig. S4 G). Given that APOEε4 showed equivalent effects on Aβ phagocytosis in TREM2 KO/R47H and SORL1 KO/A528T hMGLs (Fig. 4 E), it seems that APOEε4 can inhibit hMGL Aβ uptake regardless of TREM2 or SORL1 status. However, we observed that reduced Aβ uptake in TREM2R47H and SORL1 KO hMGLs appeared to be at least in part, APOE dependent; APOE siRNA restored Aβ binding/uptake in these hMGL lines (Fig. 4 F). Given that the parental H9 cell line comprises an APOEε3/ε4 genotype, we isogenically corrected each line within our mutant panel from APOEε3/ε4 to APOEε3/ε3. We observed that conversion to APOEε3/ε3 restored Aβ binding/uptake in INPP5D SNP, TREM2R47H, and SORL1 KO and A528T hMGLs (Fig. 4 G and Fig. S4 G). In agreement with our results indicating that SORL1 dysfunction (KO) can impair phagocytic uptake (Fig. 4 E), SORL1 siRNA also reduced Aβ phagocytosis in WT hMGLs (Fig. 4 H). Consistent with our epistasis model (Fig. 4 A), SORL1 siRNA transfection did not impair Aβ binding/uptake in TREM2 KO hMGLs, suggesting that defects in phagocytic uptake with SORL1 suppression are TREM2 dependent (Fig. 4 H). However, reduced Aβ binding/uptake in TREM2R47H hMGLs was not further perturbed by SORL1 siRNA (Fig. 4 H), suggesting that TREM2R47H-dependent phagocytic impairment occurs independently of SORL1. Together, these results indicated APOE induction triggers impaired Aβ binding/uptake in TREM2R47H and SORL1 KO hMGLs.
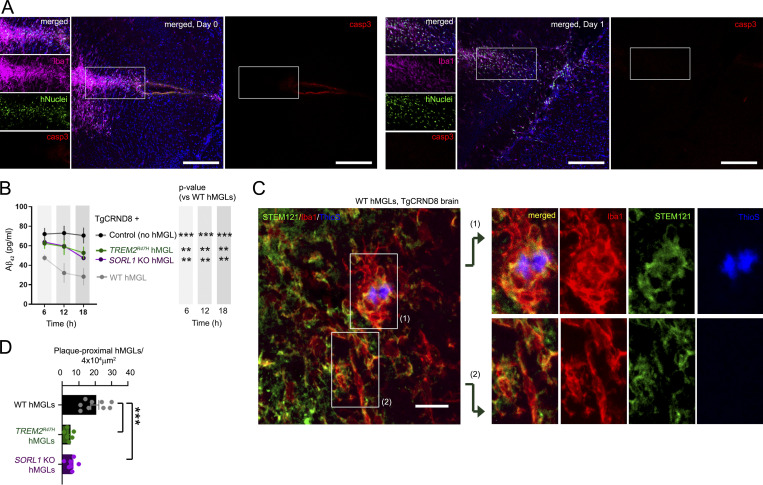
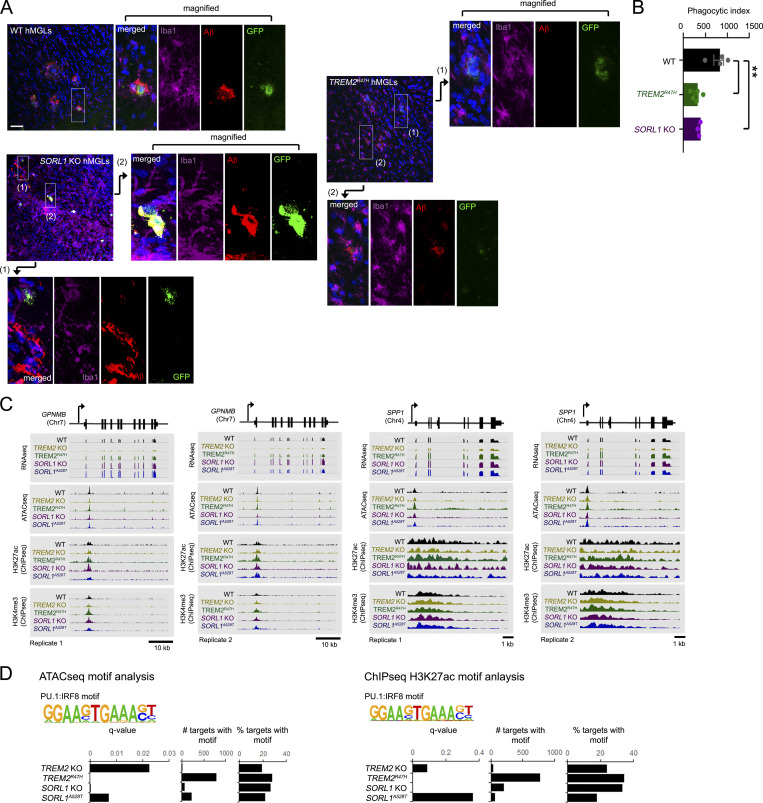
We next investigated whether TREM2R47H and SORL1 KO may affect Aβ uptake in vivo. To this end, we generated fluorescent hMGL reporter lines through insertion of a viral 2A-EGFP sequence following the last exon of CX3CR1 by homologous recombination in WT, TREM2R47H, and SORL1 KO hMGLs. Since integrity of the endogenous CX3CR1 protein is maintained through cleavage of the 2A linker, this method is preferable to GFP gene replacement strategies at the CX3CR1 gene locus (Ito et al., 2016; Jung et al., 2000; Parkhurst et al., 2013). To determine graft efficiency and survival rate of hMGLs in mouse brain, we stereotactically injected hMGLs into immunodeficient human M-CSF (hMCSF) knockin mouse brain. We observed little difference in hMGL number by hNuclei staining, or activated caspase-3 staining between 6 h and 1 d after stereotactic hMGL injection (Fig. 5 A). We then xenotransplanted hMGL reporter lines into mouse brain and tracked hMGL homing to fluorescently labeled Aβ aggregates. Over a 7-d period, we observed trends similar to our results in vitro; WT hMGLs were able to track and internalize Aβ, whereas TREM2R47H and SORL1 KO hMGLs showed reduced Aβ uptake (Fig. S5, A and B). Moreover, to examine hMGL-dependent Aβ clearance in vivo, we xenotransplanted WT, TREM2R47H, and SORL1 KO hMGLs into 3-mo-old APP transgenic (TgCRND8) mouse hippocampus (Chishti et al., 2001) and measured human Aβ42 levels in interstitial fluid (ISF) by microdialysis sampled over a course of 6–18 h (Fig. 5 D; Brody et al., 2008; Kang et al., 2009; Yan et al., 2009). Although we observed significant reduction in ISF Aβ42 levels with WT hMGL xenotransplantation, we observed significant impairment in Aβ clearance with xenotransplantation of TREM2R47H or SORL1 KO hMGLs (Fig. 5 B). Following microdialysis, TgCRND8 mice were processed for histological analysis, and we compared plaque association in WT, TREM2R47H, and SORL1 KO hMGLs (Fig. 5, C and D). We observed that significantly fewer TREM2R47H or SORL1 KO hMGLs associated with Aβ plaques in TgCRND8 animals (Fig. 5 D), further indicating that TREM2R47H or SORL1 KO mutations can render defects in microglial plaque association in vivo.
Figure 5.
Characterizing xenotransplanted hMGLs in mouse brain. (A) Histological analysis of 2 × 105 hMGLs stereotactically injected into hippocampus of human MCSF knockin mouse (Rag2−/−; γc−/−; CSF1h/h); brain tissue was processed/imaged following injection (6 h, left) or after 1 d (day 1, right). Brain tissue sections were stained for Iba1 (purple), human nuclei (hNuclei, green), active caspase-3 (casp3, red), and DAPI (blue). Scale bars, 200 µm. (B) Human Aβ1-42 levels measured in ISF from TgCRND8 mouse brain xenotransplanted with WT, TREM2R47H, and SORL1 KO hMGLs as determined by ELISA. Statistical significance was determined by two-way ANOVA with Dunnett's multiple comparison (**, P < 0.01; ***, P < 0.001); graph depicts mean ± SEM, n = 4 animals for each group with duplicate ELISA replicate readings from each animal. (C) 3-mo-old TgCRND8 mouse brain xenotransplanted with WT hMGLs into the left hippocampal hemisphere was stained with antibodies to detect human cytoplasm (STEM121, green), Iba1 (red), or ThioS to detect Aβ plaques (blue). Magnified images show that WT hMGLs (green) were able to fully encapsulate plaques in WT (top panel). Scale bar, 20 µm. (D) Phagocytosis of Aβ in xenotransplanted hMGLs is quantified in C. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison (***, P < 0.01). Data were derived from n = 3–5 animals for each group; 6–10 sagittal brain sections (30 µm) per animal were subjected to immunostaining analysis. Graph depicts mean ± SEM; data points represent averages from two or three images per animal. Images in A and C are representative images from three independent experiments.
Figure S5.
In vivo Aβ uptake and chromatin profiles in various AD mutant hMGLs. (A) GFP-expressing WT, TREM2R47H, and SORL1 KO hMGLs lines (derived from CX3CR1-2A-GFP ES lines; see Materials and methods) xenotransplanted in human MCSF knockin mouse brain cortex and coinjected with 555-Aβ oligomers were stained with antibodies to detect GFP (green), Iba1 (purple), Aβ (red, 555-Aβ) and DAPI (blue). Magnified images indicate intracellular Aβ in WT hMGLs (green), where TREM2R47H hMGLs largely failed to show overlap in hMGL/intracellular Aβ staining, while SORL1 KO hMGLs also showed incomplete overlap. Scale bar, 50 µM. (B) Phagocytosis of Aβ in xenotransplanted hMGLs were quantified in A. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison (**, P < 0.01). Graph represents n = 3–5 animals for each group (each data point represents averages from 6–10 sagittal brain sections (30 µm) per animal; mean ± SEM) as determined by analysis of images from immunostained sections. (C) Epigenetic analysis of hMGL lines. The genome browser schematic and peak scans comparing mRNA levels and genomic epigenetic modification at GPNMB and SPP1 loci (duplicate scans) for WT, TREM2 KO, TREM2R47H, SORL1 KO, and SORL1A528T hMGLs. RNA-seq and normalized signals for ATAC-seq, H3K4me3, and H3K27ac ChIP-seq are shown in representative assays from hMGLs as indicated. (D) PU.1:IRF8 binding motifs identified by ATAC-seq (left) and anti-H3K27ac ChIP-seq (right) are quantified for significance (q-value), frequency, and percent relative frequency (% targets).
Together, these results indicate that AD-associated hMGLs show varied effects on Aβ uptake in vitro, and AD-primed hMGLs such as TREM2R47H and SORL1 feature impaired Aβ uptake and clearance in vitro and in vivo.
APOE is up-regulated through activating epigenetic pathways
To gain insight into alterations within the epigenetic landscape in our mutant hMGLs, we performed transcription factor binding motif analysis within active chromatin regions demarcated with enriched ATAC, H3K4me3, and H3K27ac signals. Interestingly, we identified enrichment of several PU.1:IRF8 binding motifs in TREM2 KO/R47H and SORL1 KO/A528T hMGLs (Fig. S5 D). This supports recent observations that increased IRF8 expression can drive microglial AD-related signatures in AD patients and in AD TREM2R47H carriers (Zhou et al., 2020).
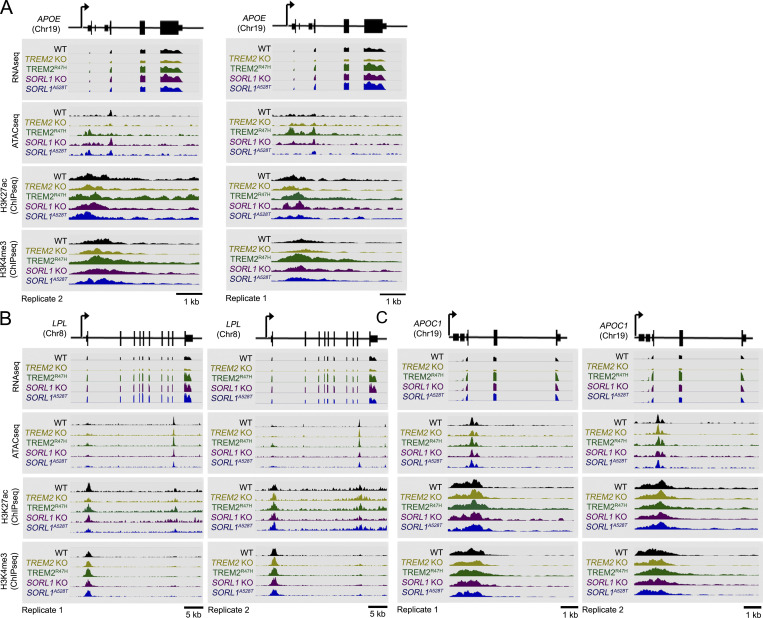
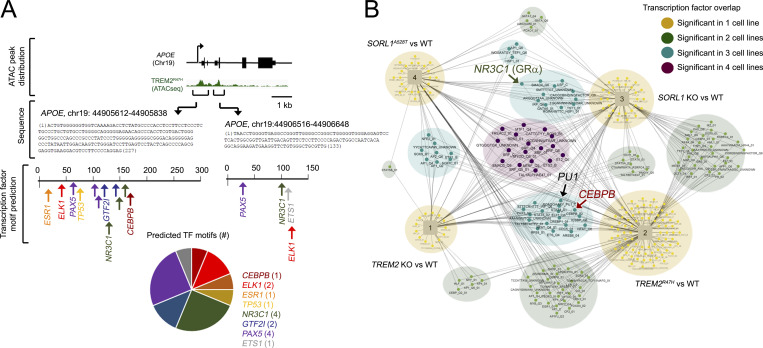
We hypothesized that opposing APOE/AD-related gene signatures in TREM2 KO, in constrast to TREM2R47H and SORL1 KO hMGLs may also be derived from chromatin-associated epigenetic/transcription activation mechanisms. We therefore characterized transcriptional/epigenomic changes at APOE, LPL, APOC1, GPNMB, and SPP1 loci using peak scans from RNA-seq or ATAC-seq/ChIP-seq datasets (Fig. 6, A–C; and Fig. S5 C). As expected, APOE, LPL, APOC1, GPNMB, and SPP1 transcripts were reduced in TREM2 KO/elevated in TREM2R47H, SORL1 KO, and A528T hMGLs compared with WT (Fig. 6, A–C; and Fig. S5 C). ATAC-seq/ChIP-seq peaks, for the most part matched RNA-seq transcripts (Fig. 6, A–C; and Fig. S5 C), where TREM2R47H hMGL peaks showed the most dramatic up-regulation at APOE (Fig. 6 A and Fig. S5 C). We searched for transcription factor motifs at the APOE locus matching up-regulated ATAC-seq peaks (APOE [Chr19]: 44905612–44905838 and 44906516–44906648) using the online transcription factor motif search tool, PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3; Fig. 7 A). We also performed upstream transcription factor analysis to characterize transcription factors that drive changes in global expression in TREM2 KO, TREM2R47H, and SORL1 KO and A528T hMGLs (Table S2 and Table S3). While numerous transcription factors were identified to overlap, we were interested to see that overlapping CEBPB (CCAAT enhancer–binding protein β [CEBPB hereon]) and NR3C1 (glucocorticoid receptor α) transcriptional pathways were observed between TREM2R47H and SORL1 KO hMGLs (Fig. 7, A and B; and Table S2). Interestingly, CEBPB appeared multiple times among TREM2 and SORL1 mutant hMGLs (Fig. 7 B and Table S2), indicating that CEBPB may play a role in driving APOE expression in TREM2R47H hMGLs. At this point, it is unclear how TREM2R47H can mediate transition between inactive and active chromatin states at loci such as APOE. Thus, further mechanistic insight will be required to define alterations in epigenetic modification mechanisms associated with TREM2R47H in microglia models and human TREM2R47H carriers.
Figure 6.
TREM2R47H enhances APOE expression through chromatin modulation. (A–C) Genome browser schematic and peak scans comparing mRNA levels and genomic epigenetic modification at the APOE (A), LPL (B), and APOC1 (C) locus; RNA-seq and normalized signals for ATAC-seq, H3K4me3, and H3K27ac ChIP-seq are shown in representative assays from hMGL as indicated. Chr, chromosome.
Figure 7.
Modulation of transcription networks in AD-associated hMGLs. (A) Two transcriptionally active regions within the APOE locus identified by ATAC-seq were characterized for transcription factor (TF) motifs using PROMO. TFs were plotted for the two ATAC-seq peaks (nucleotides [nts] 1–227, Chr19:44905612; nts 1–133, Chr19:44906516) depicted in green. Number of predicted TFs in these two regions is indicated in the pie chart. (B) Upstream transcription factors with targets significantly enriched for genes dysregulated in all four -omics analytical datasets were plotted on the network map shown. Cell line–specific transcription factors are annotated by circles, and cell lines are annotated by squares in the network diagram. Shared transcription factors are connected to each cell line in which they are enriched. Some transcription factors are enriched in a single cell line (small yellow nodes), while others are enriched in two cell lines (light green), three cell lines (teal), or all four cell lines (dark purple). PU.1, NR3C1, and CEBPB are marked in large font on the Cytoscape map.
Together, these results suggest that TREM2R47H feature enhanced chromatin activation signatures and demonstrates a unique epigenetic mechanism for APOE up-regulation through an AD-associated risk variant.
Discussion
We generated a comprehensive data resource comprising an expanded blueprint of microglial AD risk variants and their characterization at multiple genomic and biological levels that may be leveraged by the community to understand how differing risk factors can feature convergent mechanisms in AD pathogenesis. Importantly, this also provides a new research paradigm to study and compare pathogenic roles of disease-associated variants identified by genome-wide association study analyses, which allows us to surpass limitations from single -omics or function-based studies. Given that SORLA is also expressed in human astrocytes and neurons, this resource can also be repurposed to characterize AD genes in other cell types within the CNS. Further, use of an isogenic system enables an accurate comparison of genomic and functional effects derived from a single gene variant within a mutant panel; similar studies derived from human iPSCs may be complicated by variations in genetic background and are limited to isogenic correction studies. Moreover, our platform may facilitate characterization of rare AD variants, such as TREM2R47H, where tissue sources may be difficult to obtain. Lastly, identification of enriched IRF8 motifs in TREM2R47H hMGLs in our study here faithfully recapitulates in IRF8 previously observed in human AD samples (Zhou et al., 2020), demonstrating good translatability between our H9 ESC model system and human AD. At this point, use of the H9 system derived from female origin and heterozygous for APOE ε3/ε4 in our genomic/expression analysis leaves important questions as to how other fundamental factors in AD such as sex and various APOE isotypes may interact with AD risk variants in microglia. Although we caution that our global expression and epigenetic analyses were performed using a single clonal cell line, we have verified effects of APOE on AD-associated modulation of Aβ binding/uptake using two independently derived mutant hMGL cell lines. We anticipate that future studies in alternate ESC or iPSC models will confirm our findings here and elaborate how gender and APOE variants may further interact with AD risk variants in other human-derived microglia models. Altogether, the availability of these ESC lines combined with methodologies described here for differentiating hMGLs, and searchable databases comprising genomic profiles for each of these lines will greatly facilitate and enhance our understanding of how AD risk variants may alter cell function and enhance AD pathogenesis in the CNS.
Our study has also uncovered some fundamental findings with respect to striking dissimilarities in SORL1 KO, SORL1A528T, TREM2R47H, and TREM2 KO gene signatures that manifest differences in phagocytosis. Despite indication that these hMGL lines converge at the APOE locus by multi-omic analysis, these hMGL cell types are seen to segregate into two signature classes according to their AD-associated (DAM) gene profiles. Specifically, TREM2R47H and SORL1 KO render hMGLs into an AD-primed microglial state (where an AD state or environment can prime a particular microglial state as indicated by DAM-like expression profiles), while TREM2 KO shows an inverse gene profile. Loss-of-function mutations in TREM2 or its obligate coreceptor, DAP12, results in Nasu–Hakola disease (NHD), which manifests in early-onset dementia, bone cysts, and death by age ∼40 yr (Nakamagoe et al., 2011), whereas TREM2R47H carriers feature significantly increased risk of AD onset (Jonsson et al., 2013). Although NHD and AD both manifest in dementia-like symptoms, NHD results in early-onset dementia with minimal or no Aβ plaque pathology, while sporadic AD is a late-onset (age ≥65 yr) Aβ-dependent disorder with no bone pathology. This clinical divergence between NHD and AD can be partially explained by opposing expression profiles between TREM2 KO and R47H. As the TREM2R47H confers increased risk of AD onset with a potency comparable to the APOEε4 allele (Guerreiro et al., 2013; Jonsson et al., 2013; Najm et al., 2019), induced APOE levels and iteration of AD-primed gene signatures in TREM2R47H hMGLs implicate a direct association between TREM2R47H and APOE-dependent AD pathogenesis, which may explain why TREM2R47H and APOEε4 variants confer comparable clinical odds ratios. Although previous studies provide correlative evidence that the APOEε4 allele may be required to trigger AD onset in TREM2R47H carriers (Kunkle et al., 2019; Murray et al., 2019), a direct role for downstream APOE induction in TREM2R47H-dependent AD pathogenesis had not been previously demonstrated until now. Our results indicate that microglial APOE induction in TREM2R47H carriers may be critical in triggering AD onset, which suggests that suppression of a TREM2/APOE pathogenic axis may be protective.
As the R47H mutation within the TREM2 ectodomain was previously shown to impair interactions with extracellular ligands such as APOE, LDL, clusterin, and Aβ (Atagi et al., 2015; Yeh et al., 2016; Zhao et al., 2018), TREM2R47H was speculated to render TREM2 loss of function. Indeed, heterozygous expression of Trem2R47H in AD mouse models appeared to phenocopy Trem2 haploinsufficiency, where Trem2R47H expression or Trem2 heterozygous deletion resulted in impaired microglial association with Aβ plaques and reduced plaque burden (Cheng-Hathaway et al., 2018). Moreover, human TREM2R47H expression in a combined Trem2 KO/5xFAD mouse model failed to restore microglial activation and microglial plaque homing defects observed with murine Trem2 deletion (Song et al., 2018), further suggesting that Trem2R47H may primarily confer loss of function. However, we uncover striking dissimilarities in TREM2 KO and R47H gene signatures that may manifest differences in phagocytosis in mouse brain through their opposing expression profiles with respect to genes modulated with phagocytic uptake of apoptotic neurons (Krasemann et al., 2017). Opposing effects on APOE expression may be critical to TREM2 KO or R47H-dependent predisposition to NHD or AD; this suggests that APOE modulation could be a critical factor in triggering AD onset. Thus, while TREM2 KO may confer deficiencies in microglial function, lack of APOE up-regulation may drive Aβ-independent pathogenic effects in NHD.
Although APOE is primarily expressed in astrocytes in brain, microglial APOE induction has also been observed in human AD (Mathys et al., 2019), implicating a role for microglial APOE up-regulation in enhancing AD progression (Huynh et al., 2019; Shi et al., 2019). While it remains unclear at this point how an AD-induced microglial APOE pool can differ pathogenically from constitutive astrocytic APOE, it appears that APOE-immunoreactive microglia are often clustered within APOE-labeled senile plaques, while astrocytic process are often located distally to plaque cores (Uchihara et al., 1995). Thus, it may be conceivable that an induced, local microglial APOE pool potentially drives local pathogenic events at Aβ plaques. Given that both TREM2 and SORLA have been shown to interact with APOE (Yajima et al., 2015; Yeh et al., 2016), it is tempting to speculate that AD-associated defects in Aβ clearance may result from dysregulation of a SORL1–TREM2–APOE regulatory axis, thereby manifesting defects in physiological Aβ uptake/homeostasis pathways. Together, our results suggest that various risk factors for sporadic AD may demonstrate convergent overlapping mechanisms for AD pathogenesis through APOE up-regulation in microglia.
Although our hMGLs exhibit good semblance to human microglia with respect to global expression profiles and functional response, including membrane depolarization kinetics, Ca2+ homeostasis, and cytokine release, use of this analytical system may yet require further development with respect to recapitulating microglial interactions within an aged AD environment in human brain. Combined use of human astrocytes and neurons in 3D organoid co-culture models may also be useful in defining molecular events during AD onset (Gerakis and Hetz, 2019). Our xenotransplantation experiments comprised short-term integration of hMGLs in immunodeficient hMCSF mouse brain. Survivability of hMGLs may be improved in the presence of hCSF1; using these hMCSF mouse models, we are interested in characterizing long-term effects of AD mutant hMGL xenotransplantation. Moreover, current hybrid xenotransplantation models in mouse brain cannot distinguish effects from transplanted human and endogenous mouse microglia (Hasselmann et al., 2019; Mancuso et al., 2019). Thus, it will be of interest in future study to adapt our experimental platform to facilitate long-term changes in hMGL xenotransplants in the absence of mouse microglia, and monitor effects of AD-related variants on interactions with Aβ.
Epigenetic regulation is essential to many key cellular processes, and an increasing number of studies suggest that epigenetics plays an important role in AD, indicating that targeting epigenetic mechanisms may be effective therapeutically (Chatterjee et al., 2018; Dubey et al., 2018). Indeed, we show that epigenetic alterations in TREM2R47H hMGLs at the APOE locus increase chromatin accessibility to induce pathogenic APOE expression. Using WebGestalt and PROMO bioinformatic analysis platforms, our results suggest that transcription factors such as CEBPB potentially induce APOE up-regulation in microglia, which may potentiate AD onset by triggering pathogenic pathways in microglia. This may be especially relevant in the context of TREM2R47H carriers which exhibit enhanced chromatin accessibility within the APOE locus. Thus, it may be interesting in future studies to determine whether AD-associated CEBPB induction can mediate pathogenic effects in an APOE-dependent manner.
In conclusion, we provide a resource comprising an expanded blueprint of microglial AD risk variants and their characterization at multiple genomic and biological levels that may allow us to understand how differing risk factors can have convergent mechanisms in AD pathogenesis. We provide expansive bioinformatic, methodological, and characterized AD mutant ESC-based resources that will facilitate future studies focused on elucidating mechanisms underlying AD pathogenesis. Our model system may also be repurposed and adapted to drug screening platforms in future drug discovery efforts (So et al., 2019). Significantly, our results identify TREM2R47H and SORL1 KO as AD-priming mutations that induce AD-related signatures such as APOE and manifest defects in Aβ uptake. This suggests that an ordered, convergent mechanism for TREM2, SORL1, and APOE may be involved in AD pathogenesis.
Materials and methods
Experimental model and subject details
Generation of isogenic human ESC lines and CX3CR1-2A-EGFP reporter lines
Single-guide RNAs (sgRNAs) and primers for PCR amplification and detection of off-target sites were designed using the Zhang laboratory CRISPR design tool (http://crispr.mit.edu) or Benchling (Biology Software 2019; https://benchling.com). sgRNA sequences targeting genes or SNP loci (Table S1) were cloned into PX458 (Addgene; plasmid ID 48138) as previously described (Ran et al., 2013). 100-nt single-stranded oligodeoxynucleotide (ssODN) repair templates (PAGE purified; Integrated DNA Technology) were designed with homologous genomic sequences flanking the predicted CRISPR-Cas9 cleavage site (Table S1).
Human ESC H9 cells were transfected with PX458 containing sgRNAs and ssODNs. FACS isolation of human ESC clones, genomic DNA extraction, clone screening, and off-target analysis was performed as described previously (Ran et al., 2013). All CRISPR-Cas9 modifications converted H9 genome sequences to homozygous AD-associated variants for CD33 SNP, INPP5D SNP, TREM2 KO, TREM2R47H, SORL1R744X, and SORL1A528T. H9 was endogenously heterozygous for the INPP5D G/A polymorphism and was converted homozygously to the “A” allele. A sgRNA was designed to target a site proximal to the CX3CR1 stop codon, thereby generating a double-stranded break. Double-stranded breaks were repaired through homologous recombination of the donor plasmid, resulting in an in-frame insertion of 2A-EGFP downstream of the CX3CR1 terminal exon. Using control, isogenic TREM2R47H, and SORL1R744X lines, we created three GFP-expressing reporter lines specifically suited for live two-photon microscopy.
Cell culture and hMGL cell differentiation
Human ESC H9 cells (WA09) used in this study was purchased from the WiCell Research Institute. Undifferentiated human ESCs were maintained using the Feeder Independent Culture Platform TeSR-E8/Matrigel. Human ESC–derived hMGLs were generated using recently published protocols (McQuade et al., 2018). Differentiation of parental H9 to CD43+ primitive hematopoietic progenitor cells (HPCs) was performed using the Stem Cell Technologies STEMdiffTM Hematopoietic Kit (catalog no. 05310). H9 cells were seeded into 35-mm wells with mTeSR-E8 medium at a density of 60 100-cell colonies. On day 0, cells were switched from STEMdiff Hematopoietic Kit (Stem Cell Technologies) to medium A. On day 3, cell colonies were changed to medium B for 7 additional days, during which time HPCs separated from the colonies. At day 11, HPCs were collected and plated at a density of 10,000 cells/cm2 onto 1 mg/ml Matrigel-coated plates. Microglial differentiation was induced over 28 d by feeding the HPCs every other day with differentiation medium (DMEM/F12, 2× insulin-transferrin-selenite, 2× B27, 0.5× N2, 1× GlutaMAX, 1× nonessential amino acids, 400 mM monothioglycerol, and 5 mg/ml human insulin freshly supplemented with 100 ng/ml IL-34, 50 ng/ml TGF-β1, and 25 ng/ml M-CSF [Peprotech]). During the final 3 d of differentiation, 100 ng/ml CD200 (Novoprotein) and 100 ng/ml CX3CL1 (Peprotech) were added to mature microglia to mimic a brain-like environment.
All experimental procedures for ESCs were approved by the Sanford Burnham Prebys Medical Discovery Institute Ethics Committee.
Aβ preparation
Aβ1-42 and HiLyte Fluor (555)-Aβ1-42 (AnaSpec; AS-60480-01) oligomers or fibrils were prepared as described previously (Stine et al., 2011).
Electrophysiology
Electrophysiological recordings were taken from at least six independent microglial cultures. Data were collected and analyzed in a double-blinded fashion. 1 d before recording, hMGLs were seeded on coverslips precoated with Tropoelastin (50 μg∼125 µg/ml; Advanced Biomatrix; catalog no. 5052) and collagen I (100 µg/ml to ∼125 µg/ml; Advanced Biomatrix; catalog no. 5007) for 1 h, and screened/assayed on an ECM Select Array Kit Ultra-36 (Sigma; 5170). Cells were recorded under whole-cell patch-clamp configurations as previously described (Zhao et al., 2018). Briefly, patch pipettes with a tip resistance of 5–8 MΩ were pulled from borosilicate glass capillaries (Sutter; B150-86-10) using a laser micropipette puller (Sutter; P-2000). Patch pipettes were filled with an intracellular solution with the following composition (in mM): K-gluconate 123, KCl 6, CaCl2 1, MgCl2 4.6, EGTA 10, HEPES 10, Na2-GTP 0.4, and Na2-ATP 4.0, pH 7.3 adjusted with KOH (290 mOsm). Extracellular recording was performed in basal hMGL medium. A series of depolarizing voltage steps from −160 mV to −60 mV (Δ = 10 mV) was applied using a computer-controlled patch-clamp amplifier (Molecular Devices; Axopatch 200B) connected to an analog to digital converter (Molecular Devices; Digidata 1440A). Whole-cell capacitance was measured under voltage clamp configuration by applying a 5-mV pulse from a membrane potential of −60 mV.
Micropipette assay for ATP-induced chemotaxis
hMGLs on coverslips coated with fibronectin (Sigma-Aldrich; F4759,) were placed in a temperature-controlled stage incubator at 33°C in basal hMGL medium. A picoliter volume of 1 mM ATPγS was repetitively injected through a micropipette with an ∼2-µm tip using an electrically gated pressure application system (Parker; Picosprizer III) at pulses of 3 psi for a duration of 20 ms using clampex software. Cells were exposed to a chemotactic gradient generated by the slow release of ATPγS from the micropipette tip placed at the center of the imaging field. Serial brightfield images of cell migration toward the pipette tip were recorded every minute for up to 30 min using an Optronics MicroFire Microscope Camera (Wu et al., 2014).
Calcium imaging
Intracellular Ca2+ concentration was quantified using Fluo-4 (Thermo Fisher Scientific; F14201). In brief, hMGLs were seeded on coverslips precoated with poly-L lysine 1 d before imaging. hMGLs were incubated in basal hMGL medium with F-127 (Thermo Fisher Scientific; P6866) and 2.5 µM Fluo-4 for 30 min. Cells were washed with basal hMGL medium without Fluo-4 and incubated for an additional ∼30 min before imaging. A baseline fluorescent signal was recorded for 1 min for each well. Basal hMGL medium containing 100 µM ATP was then added, and fluorescence was recorded for an additional 9 min. Images were acquired at 2 s per frame. Traces shown represent averaged values from 20–40 single cells from at least three independent experiments. All image acquisition was performed using a Zeiss confocal microscope (LSM 880).
Multiplex cytokine and chemokine assay
hMGLs were washed and replaced with basal media for 2 h prior to stimulation with IL-1β (20 ng/ml) and IFN-γ (20 ng/ml) for 24 h, as described previously (Abud et al., 2017). Conditioned media from each treatment group were processed and analyzed using the Q-Plex Human Arrays kit (Quansys) according to the manufacturer’s specifications.
Aβ and tau binding/uptake
To image and quantify Aβ phagocytosis in hMGLs, cells were seeded at a density of 50,000 cells per well in 24-well plates. Prior to experimentation, cells were replaced with hMGL growth medium containing 555-Aβ (200 nM) or 5 µg/ml recombinant human APOEε3 or APOEε4 and incubated in a humidity- and CO2/O2-controlled chamber optimized for long-term live-cell imaging applications. A fixed area in each well was serially imaged every 15 min using a Nikon N-SIM microscope. Serial confocal images were acquired for 10 h, and Aβ intensity/area was quantified. Fluorescence intensity was normalized to microglia cell number using automated IMARIS imaging software (Bitplane); fluorescence thresholds for 555-Aβ were set to a value of 10, and individual cells were identified by differential interference contrast imaging. Fluorescence measurements normalized to cell number from hundreds of microglia/confocal imaging field were acquired from nine total confocal images from three independent batches/experiments (three independent wells per batch) at time points ranging from 15 to 180 min (for all experiments). The PI for varying time points (Fig. S1 J) or at terminal endpoints (180 min; Fig. 4, E–H) was then calculated using the following formula: PI = It / I15, where It represents averaged fluorescence intensity at various time points and I15 represents fluorescence intensity at 15 min following addition of 555-Aβ. The resulting value reflects fold change of 555-Aβ fluorescence over the 15 min time point for each hMGL line. Ratios were then normalized to the PI value in WT hMGLs under control conditions at 15 min (where WT hMGLs under control conditions/15 min will be set to 1.0) for each experiment.
To assay hMGL-dependent binding/uptake of tau oligomers, purified recombinant 2N4R human tau 1–441 (AnaSpec; 500 µg/ml, catalog no. AS-55556-50) was preincubated with 30 µM heparin (StemCell Technologies, Inc., catalog no. 07980), and allowed to oligomerize for 24 h. Tau oligomers were subsequently conjugated to Alex555 using an Alexa Fluor 555 Microscale Protein Labeling Kit (Thermo Fisher Scientific; catalog no. A30007) according to the manufacturer’s instructions. Binding/uptake of tau-Alexa Fluor 555 in hMGLs was measured in real time at a final concentration of 10 µg/ml as described for 555-Aβ above; calculations for PI and normalization to WT hMGLs at the 15-min time point were identical to methods used for 555-Aβ uptake. For experiments involving APOE or SORL1 siRNA before Aβ uptake, hMGLs were transfected with 25 nM control (Thermo Fisher Scientific; catalog no. AM4611), APOE (Thermo Fisher Scientific; catalog no. 4392420, assay ID S1495), or SORL1 siRNA (Thermo Fisher Scientific; catalog no. 4390824, assay ID S13282) using Viromer Blue (BioNTech; #VB-01LB-00) for 48 h before live-cell imaging for 555-Aβ binding/uptake.
hMGL xenotransplantation and microdialysis in mouse brain
2 × 105 hMGLs (TREM2WT/TREM2R47H/SORL1KO genotypes) or vehicle control was injected into the left hippocampus of immunodeficient human MCSF knockin (hMCSF) or TgCRND8 mice anesthetized using avertin (tribromoethanol). To facilitate injection, a small 0.8-mm hole was stereotactically drilled into the skull (coordinates, −3.1 mm bregma, +2.5 mm lateral), where the injection needle was then inserted into the hole at a 12° angle at an angular depth of 1.7 mm. Following injection, the needle was slowly removed from the mouse brain. In vivo microdialysis was performed to sample brain ISF and quantify hippocampal Aβ1–42 in freely moving mice; microdialysis procedures were performed as previously described (Kang et al., 2009). After microglia xenotransplantation, a guide cannula (Harvard Apparatus; CMA 7) was cemented into the guide hole at an angular depth of 1.2 mm. After cannulation, mice were allowed to recover for 24 h, and monitored for abnormal behavior or discomfort.
2.0-mm microdialysis probes with a 6-kD molecular weight cutoff were inserted through the guide cannula, and membranes were inserted into the hippocampus (Harvard Apparatus; CMA 7). Mice were transferred to microdialysis cages, and ISF was sampled by microdialysis at a constant flow rate of 0.5 µl/min. Samples were collected every 2 h using a refrigerated fraction collector (Amuza; FC-90) in polypropylene 96-well plates, and human Aβ1–42 was quantified by ELISA. Histological analysis of mouse brain is described below. All animal procedures described in this study, including animal husbandry and surgery, were performed under the guidelines of the Institutional Animal Care and Use Committee at Sanford Burnham Prebys Medical Discovery Institute.
Histological staining and imaging following hMGL xenotransplantation in mouse brain
GFP-expressing WT, TREM2R47H, and SORL1 KO hMGLs lines (derived from CX3CR1-2A-GFP ES lines) were xenotransplanted in hMCSF knockin mouse brain cortex and coinjected with 555-Aβ oligomers (10 µM). Mice were anesthetized with isoflurane and intracardially perfused with PBS. Brain tissues were harvested and fixed in 4% paraformaldehyde at 4°C for 48 h. Tissues were washed in PBS and cryoprotected in PBS containing 30% sucrose. Tissues were embedded in optimal cutting temperature compound containing 30% sucrose (at 1:1 vol/vol), and free-floating coronal brain cryostat slices (30 µm) were sectioned.
Brain sections were washed in PBS and counterstained using standard immunostaining procedures. For detection of Iba1 and human cells, brain slices were stained using goat anti-Iba1 (Abcam; 1:400, ab5076) and rabbit anti-GFP (Cell Signaling Technology; 1:100, 2956S). Alexa Fluor 488 or 647 secondary antibodies were used, and DAPI counterstains were applied to the sections. Aβ was visualized using an Alexa Fluor 594 anti-β-amyloid, 1–16 antibody (BioLegend; catalog no. 803019) to amplify the HiLyte Fluor 555 label. Z-stack images were acquired from multiple sections (up to 15 z-stack images/field) from each animal using a Zeiss confocal microscope (LSM 880). To quantify hMGLs in proximity to Aβ, GFP-positive cells were manually counted at a distance radius of 100 µm from Aβ. Histological analysis of TgCRND8 animals following hMGL injection/microdialysis was performed by intracardial perfusion of anesthetized animals, followed by fixation of the tissue brain tissue and sectioning as described with human MCSF mouse brain. Sections were stained with Iba1, and STEM121 (Takara; human cytoplasm, catalog no. Y40410) antibodies, as well as Thioflavin-S (Sigma-Aldrich; catalog no. T1892) to visualize plaques.
RNA extraction, RNA-seq library preparation, and analysis
After washing with cold PBS, hMGLs were lysed using TRIzol Reagent (Thermo Fisher Scientific; catalog no. 15596026) and stored at −80°C for RNA extraction. Total RNA was extracted using the RNeasy Plus Micro Kit (Qiagen; 50, #74034), following manufacturer’s recommendations with slight adjustment. Briefly, 200 µl chloroform was added to the TRIzol extraction solution/cells, mixed, and centrifuged. RNA was then extracted from the aqueous phase according to the manufacturer’s recommendations. RNA was quantified using a NanoDrop spectrophotometer, and RNA integrity numbers were assessed using an Agilent TapeStation 4200 system; all samples used for subsequent analysis showed a minimum RNA integrity number of 9 or 10. For each replicate, RNA-seq libraries were prepared from 500 ng of purified total RNA using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s recommendations and sequenced on an Illumina HiSeq platform as paired-end 150-bp reads (Genewiz). An average sequencing depth of 45.13 M (±3.6 M) was obtained from the hMGL samples, with minimum and maximum values of 39.2 and 64.6 M.
ATAC-seq library preparation and sequencing
ATAC-seq analysis was performed using the Omni-ATAC-seq protocol (Corces et al., 2017). Briefly, 80,000 viable hMGLs per line were sorted using BD FACSAriaIIu 16-color high-speed cell sorter. Cells were lysed in 50 µl ATAC-Resuspension buffer containing 0.1% NP-40, 0.1% Tween-20, and 0.01% digitonin and incubated on ice for 3 min. After washing with ATAC-RSB buffer (without NP-40), nuclei were pelleted by centrifugation and incubated in a 50-µl transposition mixture (25 µl 2 × TD buffer [Illumina; reference 15027866], 2.5 µl transposase [Illumina, reference 15027865], 16.5 µl PBS, 0.5 µl 1% digitonin, 0.5 µl 10% Tween-20, and 5 µl H2O) at 37°C for 30 min. Transposed DNA was purified using a Zymo DNA clean and concentrator-5 kit (Zymo Research; catalog no. D4014). Purified DNA was subjected to PCR amplification using the NEBNext High-Fidelity 2 × PCR Master Mix (NEB; catalog no. M0541s) system for eight cycles. Primer adapters were synthesized by Integrated DNA Technology. Libraries were purified using a Zymo DNA clean and concentrator-5 kit, size selected using AMPure XP beads, quantified using an Agilent 2100 Bioanalyzer, and pooled/sequenced using a NextSeq 500/550 High Output v2 kit using paired-end reads on a NextSeq 500 platform (Illumina).
ChIP-seq library preparation
Chromatin precipitation, library preparation, and sequencing was performed using the iDeal ChIP-seq kit for Histones (Diagenode; catalog no. C01010059), MicroPlex Library Preparation Kit v2 (Diagenode; catalog no. C05010012), and NextSeq 500/550 High Output v2 kit (75 cycles; Illumina; catalog no. FC-404-2005) kits, respectively following the manufacturer's protocols. Briefly, hMGLs were detached by accutase digestion and rinsed twice with PBS. Cells were pelleted in 1.5 ml Eppendorf tubes and fixed using 1 ml 1% formaldehyde (Sigma-Aldrich; F8775) diluted in PBS at room temperature for 10 min. Formaldehyde was quenched by adding a glycine solution to a final concentration of 125 mM and incubated for 5 min at room temperature. After washing twice with 1 ml PBS, 1 ml cell lysis buffer was added to the cell pellet, incubated for 10 min on ice, and further lysed in lysis buffer (iL2). Chromatin was resuspended in 130 µl Shearing buffer and sheared using a Covaris S220 sonicator to 200–500 bp (duty cycle 2% for 10 min) in an M220 microTUBE (Covaris; 520077). Equal aliquots of sonicated chromatin were used per immunoprecipitation reaction comprising 2 µl H3K27ac antibody (Abcam; Ab4729), H3K4me3 antibody (Millipore; 07–472), or 2 µl rabbit IgG control antibody (Diagenode) and 1 µl sheared chromatin as input. The total volume of each ChIP reaction was 300 µl and included 6 µl 5% BSA, 1.5 µl 200× protease inhibitor cocktail, 56 µl 5× buffer iC1, 20 µl magnetic beads, 2 µl antibody, and sonicated chromatin. ChIP reactions were incubated overnight at 4°C with constant rotation. Magnetic beads (conjugated with antibody-histone-DNA) were washed with 350 µl buffer iW1, iW2, iW3, and iW4. Precipitated chromatin was eluted in 100 µl elution buffer iE1 for 30 min at room temperature with constant agitation and transferred into a 200-µl tube. Chromatin cross-links were uncoupled by adding 4 µl elution buffer iE2 and incubating for 4 h at 65°C. DNA was isolated using IPure beads v2 and eluted in 25 µl elution buffer. 10 µl precipitated DNA was further processed for template preparation, library synthesis, and library amplification. Amplified products were purified, and size selected using AMPure XP beads (Beckman Coulter; A63880). All libraries were quantified using an Agilent 2100 Bioanalyzer, pooled and sequenced with a NextSeq 500/550 High Output v2 kit using single-end reads on a NextSeq 500 platform (Illumina).
Sample preparation for proteomic analysis and liquid chromatography–tandem mass spectrometry
hMGLs were resuspended in buffer containing 8 M urea and 50 mM ammonium bicarbonate. Protein concentration was determined by BCA assay (Thermo Scientific) according to the manufacturer’s instructions. Cells were lysed, and proteins were precipitated in 90% methanol (final concentration), followed by centrifugation (16,000 g for 5 min). Proteins were then reduced by the addition of 5 mM tris(2-carboxyethyl) phosphine (TCEP) at 30°C for 60 min, followed by alkylation of cysteines with 15 mM iodoacetamide for 30 min in the dark at room temperature. Urea was diluted to a final concentration of 1 M with 50 mM ammonium bicarbonate. Samples were digested overnight with Lys-C/trypsin (Promega) at room temperature with constant agitation at a 1:25 enzyme/protein ratio. Following digestion, samples were acidified using 0.1% formic acid (FA) and desalted using AssayMap C18 cartridges mounted on an Agilent AssayMap BRAVO liquid handling system. Cartridges were sequentially conditioned with 100% acetonitrile (ACN) and 0.1% FA; samples were then loaded, washed with 0.1% FA, and eluted with 60% ACN, 0.1% FA. Peptide concentration was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
Samples were subjected to mass spectrometry analysis using an EASY nanoLC system (Thermo Fisher Scientific). Buffer A consisted of H2O/0.1% FA, and buffer B consisted of 80% ACN/0.1% FA. Samples were separated over a 4 h gradient of increasing buffer B on a 25-cm column packed in-house with 1.7 µm bridged-ethylene hybrid packing material (Waters). The Orbitrap Lumos mass spectrometer was operated in positive, data-dependent mode. An Orbitrap MS1 scan was performed at 120, 000 resolution with 1e6 accumulation gain control, with a scan range of 350–1,400 m/z. MS/MS analysis was performed by high collision dissociation in the ion trap using the rapid scan rate, an accumulation gain control target of 2e4 and a maximum injection time of 18 ms. Data were acquired using a cycle time of 1 s. Dynamic exclusion was set to 20 s, with a 10-ppm high/low mass tolerance.
Multiomic data analyses with customized bioinformatic pipelines
RNA-seq analysis
Adapter remnants from sequencing reads were removed using cutadapt v1.18 (Martin, 2011). RNA-seq sequencing reads were aligned using STAR aligner version 2.7 (Dobin et al., 2013). Human genome version 38 and Ensembl gene annotation version 84 were used in the alignment and quantification. Quantification was performed based on RSEM v1.3.1 (Li and Dewey, 2011). Estimated read counts from RSEM were compared using the R Bioconductor package DESeq2 following generalized linear model based on negative binomial distribution. Genes with Benjamini-Hochberg–corrected P value <0.1 were selected as significantly differentially expressed genes.
ChIP-seq and ATAC-seq
Adapter remnants of sequencing reads were removed using cutadapt v1.18 (Martin, 2011). ChIP-seq and ATAC-seq sequencing reads were aligned using STAR aligner version 2.7 to Human genome version 38 (Dobin et al., 2013). Homer v4.10 (Heinz et al., 2010) was used to call peaks from ChIP-seq and ATAC-seq samples, annotate peaks to human genes, and quantify reads count to peaks. The raw reads count for different peaks were compared using DESeq2 (Love et al., 2014) based on a generalized linear model adjusting for batch effect. Peaks with a P value <0.05 and fold change ≥1.5 or ≤0.6667 were selected as significantly differentially marked (DM) peaks. Genes associated with any DM peaks at exon, intron, promoter, transcription termination site, and closest intergenic region were investigated for GO and pathway functional enrichment tests.
Proteomic data analysis
Raw files were searched with MaxQuant (version 1.5.5.1; Cox and Mann, 2008) using the Andromeda search engine (Cox et al., 2011). Spectra were searched against the UniProt human database, including contaminants. Precursor mass tolerance was set to 20 ppm, and product ions were searched with a mass tolerance of 0.35 daltons. Oxidation of methionine and protein N-terminal acetylation were set as variable modifications, and carbamidomethylation of cysteines was set as a fixed modification. Cleavage specificity was set to trypsin, with up to two missed cleavage sites. The target-decoy–based false discovery rate (Elias and Gygi, 2007) was set to 1% at the spectrum and protein level. Data were further processed using the MSstats software package in R (Choi et al., 2014). Proteins with Benjamini–Hochberg–corrected P value <0.05 and fold change ≥1.5 or ≤0.6667 were selected as significantly DEPs.
Network analysis methods
Individual cell line network integration
The Forest tool from the OmicsIntegrator suite (Tuncbag et al., 2016) was used to integrate data layers for each cell line individually. This tool uses an algorithm to solve the Prize Collecting Steiner Forest problem, to identify the relevant neighborhoods of altered genes to interpret underlying biological and processes affected by the genetic variant. The algorithm inputs gene sets of interest and optimally connects them using edges from an input interactome. Genes may be added to the final network which were not in the input set if they form bridges between sets of input genes. These added genes represent genes which are predicted to be functionally relevant by the algorithm. Genes which were identified as dysregulated by RNA-seq (Benjamini–Hochberg adjusted P < 0.005), or by proteomics analysis (Benjamini–Hochberg adjusted P < 0.05), when compared with WT, were used as input to the Forest tool. A more stringent P value was applied to the RNA-seq dataset due to the large number of DEGs (thousands) since the OmicsIntegrator tool optimally operates when the input gene sets are on the order of hundreds, thereby producing results with greater specificity. Input datasets comprising thousands of genes make up a large fraction of the genome, and thus are not specific to the phenotype of interest. The absolute value of the log fold change between genetic variants and WT was used as the prize value for the algorithm. This process was repeated for each of TREM2 KO vs. WT, TREM2R47H vs. WT, SORL1 KO vs. WT, and SORL1A528T vs. WT. The STRING high-confidence interactome (Szklarczyk et al., 2017), a database of known and predicted protein–protein interactions, was used as the source of gene–gene interactions for the algorithm. Parameters for Forest were optimized as recommended by the tool documentation (Kedaigle and Fraenkel, 2018; Tuncbag et al., 2016), by choosing parameters which resulted in a final graph that was not dominated by hub nodes (where a single node was connected to more than 25% of the other network nodes), and the final networks contained a large ratio of input nodes to nodes added by the algorithm. Resulting individual cell line networks were additionally integrated with the genes identified by differential ChIP-seq and ATAC-seq analysis. Nodes in the network were color-coded by data type. Nodes affected in multiple datatypes were visualized using pie charts and by mapping the node size and label font size to the number of datatypes in which they are affected. Network visualization was conducted using Cytoscape (Shannon et al., 2003).
Each individual cell line network was further analyzed by dividing the full network into branches by removing the root node, identified by betweenness centrality, resulting in ∼10 connected components for each cell line, representing different perturbed biological pathways. The branches containing APOE are visualized in Fig. 2. The full set of branches for each network are visualized in Fig. S2, I–L.
Genes commonly altered across individual cell line networks
We filtered this combined network by genes which were found in at least three out of the four networks (Fig. S2 M). A pie chart was mapped to the node color to indicate the cell line networks in which the gene was found. Similarly, edges which connect nodes in multiple cell line networks are visualized as multi-edges, using the same color scheme as the nodes. Because the OmicsIntegrator algorithm returns networks with edges optimally connecting input genes, it is possible to have two nodes present in the same sets of cell lines but with different sets of edges connecting them. In general, if a group of nodes is found in the same sets of cell line networks, and if this group of nodes has similar sets of connecting edges, it likely represents a commonly perturbed pathway or process.
Upstream transcription factor analysis
Overrepresentation analysis was used to identify transcription factors with target genes significantly enriched for genes altered by each of the cell line knockouts, using the WebGestalt R interface (Wang et al., 2017). The union of genes identified by each differential -omics analysis (RNA-seq, proteomics, ChIP-seq, and ATAC-seq) was used as the input set for the over-representation analysis. WebGestalt uses the transcription factor-target interactions defined in MSigDB (Liberzon et al., 2011). Transcription factors with targets significantly overlapping with the input sets (Benjamini–Hochberg adjusted P < 0.05) were retained for further analysis. A bipartite network was used to visualize the results, with transcription factors annotated by circles, and cell lines in which they are enriched annotated by squares in the network. Transcription factors are connected to each cell line in which they are enriched. Some transcription factors are enriched in just a single cell line (small yellow nodes), while others are enriched in two cell lines (light green), three cell lines (teal), or all four cell lines (dark purple; Fig. 7 B, Table S2, and Table S3).
Quantification and statistical analysis
All statistical analysis in this study was performed using GraphPad Prism (version 8) software and RStudio. Data were collected and analyzed in a double-blind fashion; no data were excluded. SEM was used in this study. Differences were assessed by paired or unpaired t tests, or one-way or two-way ANOVA where appropriate. P < 0.05 were considered as statistically significant. All experiments in figures (including supplemental figures) were repeated at least three times (independent experiments) unless specified otherwise in the figure legends.
Data availability
All sequencing data acquired in this study have been deposited in the Gene Expression Omnibus under accession number GSE153658. Proteomics data included in this study have been deposited in the ProteomeXchange database under accession number PXD020240.
Online supplemental material
Fig. S1 includes schematics outlining CRISPR-Cas9 targeting of the AD-associated loci, validation of targets, and characterization of differentiated WT hMGLs. Fig. S2 presents analytical summaries of changes in AD-associated hMGL mutants as detected by epigenetic, transcriptomic, and proteomic analysis, analysis of DEPs in SORL1 mutant hMGLs, overlap between up-regulated and down-regulated components at epigenetic, mRNA, and protein expression within each mutant hMGL line, and multi-omic analysis summaries using OmicsIntegrator. Fig. S3 shows MA plot distribution of DEGs in mutant hMGLs identified by RNA-seq analysis, and mRNA expression profiles of microglial genes related to AD risk, modulated in human AD brain, and modulated in AD or neurodegenerative mouse models in mutant hMGL lines; and also shows changes in cytokine profiles in hMGL lines at steady-state and under induced conditions. Fig. S4 presents qRT-PCR validation of transcripts differentially expressed in hMGLs and proteomic profiles of AD risk genes characterized in this study in the hMGL panel. It compares expression of gene targets in TREM2R47H homozygous and heterozygous hMGL lines, shows kinetics of real-time Aβ binding/uptake in TREM2 and SORL1 mutant hMGLs, and compares Aβ uptake in WT and mutant CD33, INPP5D hMGLs with APOE modulation. Fig. S5 compares Aβ uptake in WT, TREM2R47H, and SORL1 KO hMGLs following xenotransplantation in mouse brain, genomic track scans obtained from RNA-seq, ATAC-seq, and ChIP-seq analysis at GPNMB and SPP1 loci, and PU.1 motif enrichment in hMGL lines identified by ATAC-seq or H3K27ac ChIP-seq analysis. Video 1 shows chemotaxis of WT hMGLs toward an ATP gradient (1 mM) leaking from a micropipette. Table S1 summarizes primers, sgRNAs, and off-target analyses to generate mutant hMGLs described in this study. Table S2 lists shared upstream transcription factor motifs enriched in the mutant hMGL lines. Table S3 lists all upstream transcription factor motifs identified in individual hMGL lines. Data S1 is an expanded dataset related to Fig. S2, showing an overview of epigenetic, transcriptomic, and proteomic changes and the correlation between epigenetic and transcriptomic signatures and integrative multi-omic analysis of gene landscapes in mutant hMGLs.
Supplementary Material
summarizes primers, sgRNAs, and off-target analyses to generate mutant hMGLs described in this study.
lists shared upstream transcription factor motifs enriched in the mutant hMGL lines.
lists all upstream transcription factor motifs identified in individual hMGL lines.
is an expanded dataset related to Fig. S2, showing an overview of epigenetic, transcriptomic, and proteomic changes and the correlation between epigenetic and transcriptomic signatures and integrative multi-omic analysis of gene landscapes in mutant hMGLs.
Acknowledgments
We thank all the members in Huang/Xu laboratory for critical discussion. We are grateful to have support from the Sanford Burnham Prebys Animal Resources Core, Histology Core, Proteomics Core, and Genomics Core.
This work was supported by National Institutes of Health grants R01 AG061875, R01 AG056130, R01 AG044420, and R21 AG059217 (to T.Y. Huang) and grants R01 AG038710, R01 NS046673, and R01 AG056114 (to H. Xu), the Tanz Family Fund (to H. Xu), the Cure Alzheimer’s Fund (to H. Xu, T.Y. Huang, and R.E. Tanzi), and the National Cancer Institute (Cancer Center Support Grant P30CA030199 to S.B. Rosenthal).
Author contributions: T. Liu, B. Zhu, T.Y. Huang, and H. Xu conceived the project. T. Liu designed and cloned plasmid constructs, performed gene editing, screened and characterized ESC clones, performed RNA-seq, generated and maintained all isogenic lines, and provided hMGL cells and Aβ oligomers used in this study. B. Zhu performed calcium imaging, Aβ ELISA analysis, cytokine release, Aβ phagocytosis, and proteomics. Y. Liu performed electrophysiology, immunohistochemistry, microglial migration, and confocal imaging and analysis. X. Zhang (with help from T. Liu) performed ChIP-seq, ATAC-seq, quantitative PCR, and Western blot validation. X. Li executed microdialysis procedures. J. Yin, A.P. Hodges, L. Jiang, and T.Y. Huang performed all bioinformatic analyses and generated/processed all datasets. J. Yin, X. Zhang, L. Jiang, S.B. Rosenthal, and T.Y. Huang contributed to bioinformatic data interpretation. T. Liu, B. Zhu, Y. Liu, X. Zhang, J. Yancey, and L. Zhou analyzed the data. M. Blurton-Jones, A. McQuade, T. Liu, and B. Zhu developed the human microglia differentiation protocol. R.E. Tanzi provided genetic information and discussion. T.Y. Huang, T. Liu, B. Zhu, and H. Xu wrote the paper, and all authors provided feedback.
References
- Abud, E.M., Ramirez R.N., Martinez E.S., Healy L.M., Nguyen C.H.H., Newman S.A., Yeromin A.V., Scarfone V.M., Marsh S.E., Fimbres C., et al. 2017. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 94:278–293.e9. 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, O.M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., von Arnim C.A., Breiderhoff T., Jansen P., Wu X., et al. 2005. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA. 102:13461–13466. 10.1073/pnas.0503689102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atagi, Y., Liu C.C., Painter M.M., Chen X.F., Verbeeck C., Zheng H., Li X., Rademakers R., Kang S.S., Xu H., et al. 2015. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 290:26043–26050. 10.1074/jbc.M115.679043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayata, P., Badimon A., Strasburger H.J., Duff M.K., Montgomery S.E., Loh Y.E., Ebert A., Pimenova A.A., Ramirez B.R., Chan A.T., et al. 2018. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 21:1049–1060. 10.1038/s41593-018-0192-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, A.B., Baker E., Sutherland G.R., Phillips J.H., and Lanier L.L.. 1999. Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc. Natl. Acad. Sci. USA. 96:9792–9796. 10.1073/pnas.96.17.9792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon, A., Hernandez-Munain C., Cella M., and Colonna M.. 2001. A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J. Exp. Med. 194:1111–1122. 10.1084/jem.194.8.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawek, B., Schwendele B., Riester K., Kohsaka S., Lerdkrai C., Liang Y., and Garaschuk O.. 2014. Impairment of in vivo calcium signaling in amyloid plaque-associated microglia. Acta Neuropathol. 127:495–505. 10.1007/s00401-013-1242-2 [DOI] [PubMed] [Google Scholar]
- Brody, D.L., Magnoni S., Schwetye K.E., Spinner M.L., Esparza T.J., Stocchetti N., Zipfel G.J., and Holtzman D.M.. 2008. Amyloid-beta dynamics correlate with neurological status in the injured human brain. Science. 321:1221–1224. 10.1126/science.1161591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, P., Roy D., and Rathi N.. 2018. Epigenetic Drug Repositioning for Alzheimer’s Disease Based on Epigenetic Targets in Human Interactome. J. Alzheimers Dis. 61:53–65. 10.3233/JAD-161104 [DOI] [PubMed] [Google Scholar]
- Cheng-Hathaway, P.J., Reed-Geaghan E.G., Jay T.R., Casali B.T., Bemiller S.M., Puntambekar S.S., von Saucken V.E., Williams R.Y., Karlo J.C., Moutinho M., et al. 2018. The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol. Neurodegener. 13:29. 10.1186/s13024-018-0262-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti, M.A., Yang D.S., Janus C., Phinney A.L., Horne P., Pearson J., Strome R., Zuker N., Loukides J., French J., et al. 2001. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J. Biol. Chem. 276:21562–21570. 10.1074/jbc.M100710200 [DOI] [PubMed] [Google Scholar]
- Choi, M., Chang C.Y., Clough T., Broudy D., Killeen T., MacLean B., and Vitek O.. 2014. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 30:2524–2526. 10.1093/bioinformatics/btu305 [DOI] [PubMed] [Google Scholar]
- Corces, M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14:959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox, J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Del-Aguila, J.L., Benitez B.A., Li Z., Dube U., Mihindukulasuriya K.A., Budde J.P., Farias F.H.G., Fernandez M.V., Ibanez L., Jiang S., et al. 2019. TREM2 brain transcript-specific studies in AD and TREM2 mutation carriers. Mol. Neurodegener. 14:18. 10.1186/s13024-019-0319-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, H., Gulati K., and Ray A.. 2018. Recent studies on cellular and molecular mechanisms in Alzheimer’s disease: focus on epigenetic factors and histone deacetylase. Rev. Neurosci. 29:241–260. 10.1515/revneuro-2017-0049 [DOI] [PubMed] [Google Scholar]
- Elias, J.E., and Gygi S.P.. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 4:207–214. 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- Fjorback, A.W., Seaman M., Gustafsen C., Mehmedbasic A., Gokool S., Wu C., Militz D., Schmidt V., Madsen P., Nyengaard J.R., et al. 2012. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J. Neurosci. 32:1467–1480. 10.1523/JNEUROSCI.2272-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro, T.F., Holtman I.R., Lerario A.M., Vainchtein I.D., Brouwer N., Sola P.R., Veras M.M., Pereira T.F., Leite R.E.P., Möller T., et al. 2017. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 20:1162–1171. 10.1038/nn.4597 [DOI] [PubMed] [Google Scholar]
- Gerakis, Y., and Hetz C.. 2019. Brain organoids: a next step for humanized Alzheimer’s disease models? Mol. Psychiatry. 24:474–478. 10.1038/s41380-018-0343-7 [DOI] [PubMed] [Google Scholar]
- Gosselin, D., Skola D., Coufal N.G., Holtman I.R., Schlachetzki J.C.M., Sajti E., Jaeger B.N., O’Connor C., Fitzpatrick C., Pasillas M.P., et al. 2017. An environment-dependent transcriptional network specifies human microglia identity. Science. 356. eaal3222. 10.1126/science.aal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro, R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold, D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. 2009. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41:1088–1093. 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann, J., Coburn M.A., England W., Figueroa Velez D.X., Kiani Shabestari S., Tu C.H., McQuade A., Kolahdouzan M., Echeverria K., Claes C., et al. 2019. Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron. 103:1016–1033.e10. 10.1016/j.neuron.2019.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L.C., Means T.K., and El Khoury J.. 2013. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16:1896–1905. 10.1038/nn.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth, P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina, V., et al. 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43:429–435. 10.1038/ng.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T.Y., Zhao Y., Li X., Wang X., Tseng I.C., Thompson R., Tu S., Willnow T.E., Zhang Y.W., and Xu H.. 2016. SNX27 and SORLA Interact to Reduce Amyloidogenic Subcellular Distribution and Processing of Amyloid Precursor Protein. J. Neurosci. 36:7996–8011. 10.1523/JNEUROSCI.0206-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudry, E., Dashkoff J., Roe A.D., Takeda S., Koffie R.M., Hashimoto T., Scheel M., Spires-Jones T., Arbel-Ornath M., Betensky R., et al. 2013. Gene transfer of human Apoe isoforms results in differential modulation of amyloid deposition and neurotoxicity in mouse brain. Sci. Transl. Med. 5. 212ra161. 10.1126/scitranslmed.3007000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, T.V., Wang C., Tran A.C., Tabor G.T., Mahan T.E., Francis C.M., Finn M.B., Spellman R., Manis M., Tanzi R.E., et al. 2019. Lack of hepatic apoE does not influence early Abeta deposition: observations from a new APOE knock-in model. Mol. Neurodegener. 14:37. 10.1186/s13024-019-0337-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, M.C., Cheung B.S., Rebeck G.W., Paul S.M., Bales K.R., and Hyman B.T.. 2000. Apolipoprotein E affects the amount, form, and anatomical distribution of amyloid beta-peptide deposition in homozygous APP(V717F) transgenic mice. Acta Neuropathol. 100:451–458. 10.1007/s004010000263 [DOI] [PubMed] [Google Scholar]
- Ito, Y., Ofengeim D., Najafov A., Das S., Saberi S., Li Y., Hitomi J., Zhu H., Chen H., Mayo L., et al. 2016. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 353:603–608. 10.1126/science.aaf6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S., Sealock J., Karlsson I.K., Hagg S., Athanasiu L., et al. 2019. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51:404–413. 10.1038/s41588-018-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368:107–116. 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis, W., Kooijman L., Schetters S., Orre M., and Hol E.M.. 2016. Transcriptional profiling of CD11c-positive microglia accumulating around amyloid plaques in a mouse model for Alzheimer’s disease. Biochim. Biophys. Acta. 1862:1847–1860. 10.1016/j.bbadis.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Kang, J.E., Lim M.M., Bateman R.J., Lee J.J., Smyth L.P., Cirrito J.R., Fujiki N., Nishino S., and Holtzman D.M.. 2009. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science. 326:1005–1007. 10.1126/science.1180962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedaigle, A.J., and Fraenkel E.. 2018. Discovering Altered Regulation and Signaling Through Network-based Integration of Transcriptomic, Epigenomic, and Proteomic Tumor Data. Methods Mol. Biol. 1711:13–26. 10.1007/978-1-4939-7493-1_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul, H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kim, K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I., et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature. 467:285–290. 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konttinen, H., Cabral-da-Silva M.E.C., Ohtonen S., Wojciechowski S., Shakirzyanova A., Caligola S., Giugno R., Ishchenko Y., Hernandez D., Fazaludeen M.F., et al. 2019. PSEN1DeltaE9, APPswe, and APOE4 Confer Disparate Phenotypes in Human iPSC-Derived Microglia. Stem Cell Reports. 13:669–683. 10.1016/j.stemcr.2019.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann, S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle, B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., Boland A., Vronskaya M., van der Lee S.J., Amlie-Wolf A., et al. 2019. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 51:414–430. 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B., et al. 2009. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41:1094–1099. 10.1038/ng.439 [DOI] [PubMed] [Google Scholar]
- Lambert, J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., and Grenier-Boley B.. 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45:1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon, A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., and Mesirov J.P.. 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, M., Parikh I., Vasquez J.B., Smith C., Tai L., Bu G., LaDu M.J., Fardo D.W., Rebeck G.W., and Estus S.. 2015. Genetics ignite focus on microglial inflammation in Alzheimer’s disease. Mol. Neurodegener. 10:52. 10.1186/s13024-015-0048-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso, R., Van Den Daele J., Fattorelli N., Wolfs L., Balusu S., Burton O., Liston A., Sierksma A., Fourne Y., Poovathingal S., et al. 2019. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci. 22:2111–2116. 10.1038/s41593-019-0525-x [DOI] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequenes from high-throughput sequencing reads. EMBnet. J. 17:10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mathys, H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., Menon M., He L., Abdurrob F., Jiang X., et al. 2019. Author Correction: Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 571. E1. 10.1038/s41586-019-1329-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade, A., Coburn M., Tu C.H., Hasselmann J., Davtyan H., and Blurton-Jones M.. 2018. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 13:67. 10.1186/s13024-018-0297-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, C.E., King A., Troakes C., Hodges A., and Lashley T.. 2019. APOE epsilon4 is also required in TREM2 R47H variant carriers for Alzheimer’s disease to develop. Neuropathol. Appl. Neurobiol. 45:183–186. 10.1111/nan.12474 [DOI] [PubMed] [Google Scholar]
- Najm, R., Jones E.A., and Huang Y.. 2019. Apolipoprotein E4, inhibitory network dysfunction, and Alzheimer’s disease. Mol. Neurodegener. 14:24. 10.1186/s13024-019-0324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamagoe, K., Shioya A., Yamaguchi T., Takahashi H., Koide R., Monzen T., Satoh J., and Tamaoka A.. 2011. A Japanese case with Nasu-Hakola disease of DAP12 gene mutation exhibiting precuneus hypoperfusion. Intern. Med. 50:2839–2844. 10.2169/internalmedicine.50.5891 [DOI] [PubMed] [Google Scholar]
- Nott, A., Holtman I.R., Coufal N.G., Schlachetzki J.C.M., Yu M., Hu R., Han C.Z., Pena M., Xiao J., Wu Y., et al. 2019. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science. 366:1134–1139. 10.1126/science.aay0793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah, M., Patrick E., Villani A.C., Xu J., White C.C., Ryan K.J., Piehowski P., Kapasi A., Nejad P., Cimpean M., et al. 2018. A transcriptomic atlas of aged human microglia. Nat. Commun. 9:539. 10.1038/s41467-018-02926-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva, J., Kestila M., Wu J., Salminen A., Bohling T., Ruotsalainen V., Hakola P., Bakker A.B., Phillips J.H., Pekkarinen P., et al. 2000. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat. Genet. 25:357–361. 10.1038/77153 [DOI] [PubMed] [Google Scholar]
- Parkhurst, C.N., Yang G., Ninan I., Savas J.N., J.R. Yates, III, Lafaille J.J., Hempstead B.L., Littman D.R., and Gan W.B.. 2013. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 155:1596–1609. 10.1016/j.cell.2013.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, J.M., Liu S., Figueroa M.E., Kulalert W., Eminli S., Tan K.Y., Apostolou E., Stadtfeld M., Li Y., Shioda T., et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28:848–855. 10.1038/nbt.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan, N.S., Brickman A.M., Andrews H., Manly J.J., Schupf N., Lantigua R., Wolock C.J., Kamalakaran S., Petrovski S., Tosto G., et al. 2018. Whole-exome sequencing in 20,197 persons for rare variants in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 5:832–842. 10.1002/acn3.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougerie, P., Miskolci V., and Cox D.. 2013. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: role and regulation of the actin cytoskeleton. Immunol. Rev. 256:222–239. 10.1111/imr.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer, C.R., Offe K., Gearing M., Rees H.D., Fang G., Heilman C.J., Schaller C., Bujo H., Levey A.I., and Lah J.J.. 2004. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch. Neurol. 61:1200–1205. 10.1001/archneur.61.8.1200 [DOI] [PubMed] [Google Scholar]
- Shannon, P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Manis M., Long J., Wang K., Sullivan P.M., Remolina Serrano J., Hoyle R., and Holtzman D.M.. 2019. Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. J. Exp. Med. 216:2546–2561. 10.1084/jem.20190980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So, R.W.L., Chung S.W., Lau H.H.C., Watts J.J., Gaudette E., Al-Azzawi Z.A.M., Bishay J., Lin L.T., Joung J., Wang X., et al. 2019. Application of CRISPR genetic screens to investigate neurological diseases. Mol. Neurodegener. 14:41. 10.1186/s13024-019-0343-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.M., Joshita S., Zhou Y., Ulland T.K., Gilfillan S., and Colonna M.. 2018. Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J. Exp. Med. 215:745–760. 10.1084/jem.20171529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher, A.M., Wiendl H., Meuth S.G., and Pawlowski M.. 2019. Generating microglia from human pluripotent stem cells: novel in vitro models for the study of neurodegeneration. Mol. Neurodegener. 14:46. 10.1186/s13024-019-0347-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine, W.B., Jungbauer L., Yu C., and LaDu M.J.. 2011. Preparing synthetic Abeta in different aggregation states. Methods Mol. Biol. 670:13–32. 10.1007/978-1-60761-744-0_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. 2017. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45(D1):D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncbag, N., Gosline S.J., Kedaigle A., Soltis A.R., Gitter A., and Fraenkel E.. 2016. Network-Based Interpretation of Diverse High-Throughput Datasets through the Omics Integrator Software Package. PLOS Comput. Biol. 12. e1004879. 10.1371/journal.pcbi.1004879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihara, T., Duyckaerts C., He Y., Kobayashi K., Seilhean D., Amouyel P., and Hauw J.J.. 1995. ApoE immunoreactivity and microglial cells in Alzheimer’s disease brain. Neurosci. Lett. 195:5–8. 10.1016/0304-3940(95)11763-M [DOI] [PubMed] [Google Scholar]
- Ulrich, J.D., Ulland T.K., Mahan T.E., Nystrom S., Nilsson K.P., Song W.M., Zhou Y., Reinartz M., Choi S., Jiang H., et al. 2018. ApoE facilitates the microglial response to amyloid plaque pathology. J. Exp. Med. 215:1047–1058. 10.1084/jem.20171265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker, Z.P., Bretou M., and Annaert W.. 2019. Endo-lysosomal dysregulations and late-onset Alzheimer’s disease: impact of genetic risk factors. Mol. Neurodegener. 14:20. 10.1186/s13024-019-0323-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardarajan, B.N., Zhang Y., Lee J.H., Cheng R., Bohm C., Ghani M., Reitz C., Reyes-Dumeyer D., Shen Y., Rogaeva E., et al. 2015. Coding mutations in SORL1 and Alzheimer disease. Ann. Neurol. 77:215–227. 10.1002/ana.24305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Vasaikar S., Shi Z., Greer M., and Zhang B.. 2017. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45(W1):W130–W137. 10.1093/nar/gkx356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H.J., Liu Y.J., Li H.Q., Chen C., Dou Y., Lou H.F., Ho M.S., Li X.M., Gao Z., and Duan S.. 2014. Analysis of microglial migration by a micropipette assay. Nat. Protoc. 9:491–500. 10.1038/nprot.2014.015 [DOI] [PubMed] [Google Scholar]
- Xiang, X., Piers T.M., Wefers B., Zhu K., Mallach A., Brunner B., Kleinberger G., Song W., Colonna M., Herms J., et al. 2018. The Trem2 R47H Alzheimer’s risk variant impairs splicing and reduces Trem2 mRNA and protein in mice but not in humans. Mol. Neurodegener. 13:49. 10.1186/s13024-018-0280-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima, R., Tokutake T., Koyama A., Kasuga K., Tezuka T., Nishizawa M., and Ikeuchi T.. 2015. ApoE-isoform-dependent cellular uptake of amyloid-beta is mediated by lipoprotein receptor LR11/SorLA. Biochem. Biophys. Res. Commun. 456:482–488. 10.1016/j.bbrc.2014.11.111 [DOI] [PubMed] [Google Scholar]
- Yan, P., Bero A.W., Cirrito J.R., Xiao Q., Hu X., Wang Y., Gonzales E., Holtzman D.M., and Lee J.M.. 2009. Characterizing the appearance and growth of amyloid plaques in APP/PS1 mice. J. Neurosci. 29:10706–10714. 10.1523/JNEUROSCI.2637-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, F.L., Wang Y., Tom I., Gonzalez L.C., and Sheng M.. 2016. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron. 91:328–340. 10.1016/j.neuron.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Yin, Z., Raj D., Saiepour N., Van Dam D., Brouwer N., Holtman I.R., Eggen B.J.L., Moller T., Tamm J.A., Abdourahman A., et al. 2017. Immune hyperreactivity of Abeta plaque-associated microglia in Alzheimer’s disease. Neurobiol. Aging. 55:115–122. 10.1016/j.neurobiolaging.2017.03.021 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G., et al. 2016. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 89:37–53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W., Dumanis S.B., Tamboli I.Y., Rodriguez G.A., Jo Ladu M., Moussa C.E., and William Rebeck G.. 2014. Human APOE genotype affects intraneuronal Abeta1-42 accumulation in a lentiviral gene transfer model. Hum. Mol. Genet. 23:1365–1375. 10.1093/hmg/ddt525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Wu X., Li X., Jiang L.L., Gui X., Liu Y., Sun Y., Zhu B., Pina-Crespo J.C., Zhang M., et al. 2018. TREM2 Is a Receptor for beta-Amyloid that Mediates Microglial Function. Neuron. 97:1023–1031.e1027. 10.1016/j.neuron.2018.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Song W.M., Andhey P.S., Swain A., Levy T., Miller K.R., Poliani P.L., Cominelli M., Grover S., Gilfillan S., et al. 2020. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26:131–142. 10.1038/s41591-019-0695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
summarizes primers, sgRNAs, and off-target analyses to generate mutant hMGLs described in this study.
lists shared upstream transcription factor motifs enriched in the mutant hMGL lines.
lists all upstream transcription factor motifs identified in individual hMGL lines.
is an expanded dataset related to Fig. S2, showing an overview of epigenetic, transcriptomic, and proteomic changes and the correlation between epigenetic and transcriptomic signatures and integrative multi-omic analysis of gene landscapes in mutant hMGLs.
Data Availability Statement
All sequencing data acquired in this study have been deposited in the Gene Expression Omnibus under accession number GSE153658. Proteomics data included in this study have been deposited in the ProteomeXchange database under accession number PXD020240.