Abstract
Background
Proteases are important for hydrolysis of proteins to generate peptides with many bioactivities. Thus, the development of novel proteases with high activities is meaningful to discover bioactive peptides. Because natural isolation from animal, plant and microbial sources is impractical to produce large quantities of proteases, gene cloning and expression of target protease are preferred.
Results
In this study, an alkaline serine protease gene (GsProS8) from Geobacillus stearothermophilus was successfully cloned and expressed in Bacillus subtilis. The recombinant GsProS8 was produced with high protease activity of 3807 U/mL after high cell density fermentation. GsProS8 was then purified through ammonium sulfate precipitation and a two-step chromatographic method to obtain the homogeneous protease. The molecular mass of GsProS8 was estimated to be 27.2 kDa by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and 28.3 kDa by gel filtration. The optimal activity of GsProS8 was found to be pH 8.5 and 50 °C, respectively. The protease exhibited a broad substrate specificity and different kinetic parameters to casein and whey protein. Furthermore, the hydrolysis of whey protein using GsProS8 resulted in a large amount of peptides with high angiotensin-I-converting enzyme (ACE) inhibitory activity (IC50 of 0.129 mg/mL).
Conclusions
GsProS8 could be a potential candidate for industrial applications, especially the preparation of antihypertensive peptides.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12896-021-00678-7.
Keywords: Alkaline serine protease, Geobacillus stearothermophilus, Bacillus subtilis, Whey protein hydrolysate, ACE inhibitory activity
Background
Microbial serine proteases (EC 3.4.21.14) are one of most important proteases to be applied in different food fields, such as meat tenderization, cheese ripening, flavor development, baking, and preparation of bioactive peptides [1]. They are composed of serine residue forming a catalytic triad with aspartic acid and histidine in the active site, which can be inactivated by phenylmethylsulfonylfluoride (PMSF), diodopropyl fluorophosphate (DFP) and chymostatin [2]. Alkaline serine proteases have mainly been produced by Bacillus species, which have abilities to tolerate pH variance to secrete much amount of proteases (> 20 g/L protein) [1]. Serine proteases from Bacillus gibsonii, Bacillus subtilis KJ-21 and Bacillus licheniformis have been patented to be suitable for use in cleaning fabrics, producing fermented food, and hydrolyzing milk protein to prepare hypoallergenic formula, respectively [3–5].
Geobacillus stearothermophilus is extensively distributed in the soil and hot spring, and is a rich source of proteases [6–8]. An alkaline serine protease from G. stearothermophilus F1 (presented in composed oil palm branches) was firstly reported by Rahman and coworkers [9], who had further cloned and expressed the gene in Escherichia coli [7]. Thereafter, Pichia pastoris was used for secretory expression of the protease gene from G. stearothermophilus F1, to solve the formation of inclusion bodies and incorrect protein folding induced by E. coli expression system, but the expression level was still low (protease activity of 4.13 U/mL) [10]. Meanwhile, another alkaline serine protease from G. stearothermophilus strains B-1172 has also been cloned and expressed in E. coli to exhibit protease activities of 69 U/mL [8]. As high protease activity and pH stability are important for industrial applications, a proper expression system will be essential to improve the expression level of the target protease genes from G. stearothermophilus [1].
Bacillus subtilis, a generally recognized as safe (GRAS) bacterium, is widely used as an expression host to secrete foreign proteases directly into culture medium. It is considered as an efficient expression system with several advantages, such as rapid growth rate to result in short fermentation cycles, distinguished ability to secrete significant amounts of proteins into the extracellular medium, easy cultivation and genetic manipulation [1]. A variety of proteases have been successfully expressed in B. subtilis, including an alkaline protease (AprE) from Bacillus licheniformis 2709 [11, 12], a serine protease (AprB) from Bacillus sp. strain B001 [13], a neutral protease (NprT) from G. stearothermophilus [14], and alkaline serine proteases from Bacillus clausii (aprE) [15, 16]. It has been reported that an alkaline protease from B. clausii yielded a protease activity of 1020 U/mL in B. subtilis as compared to 347 U/mL in the wild type strain [15]. A thermolabile alkaline protease from B. licheniformis 2709 showed a high protease production of 6280 U/mL when it was expressed in B. subtilis [11]. However, some of Bacillus species directly secreted alkaline serine proteases to present high protease activities without expression in B. subtilis. Two proteases (SAPB and SAPRH) were hyper-produced (6500 U/mL and 9000 U/mL) from Bacillus pumilus strain CBS and Bacillus safensis RH12 under optimized fermentations, as compared with crude protease activities of 310 U/mL and 450 U/mL, respectively [17, 18].
Protein hydrolysis to yield bioactive hydrolysates and peptides is an interesting field with much attentions. Antioxidant, antihypertensive, and antidiabetic potentials are most commonly reported for protein hydrolysates, which may be applied as nutraceuticals and functional food ingredients, potentially contributing to food quality and promoting human health [19]. Particularly, antihypertensive hydrolysates inhibiting angiotensin-I-converting enzyme (ACE), which is very relevant in the regulation of the cardiovascular function and blood pressure, have been abundantly investigated [1]. Recent studies have been focused on the production of antihypertensive hydrolysates using non-commercial proteases, such as alkaline serine proteases from Bacillus sp. CL18 [20], Maclura pomifera [21], and Cucurbita ficifolia [22].
The objective of this study was to clone and express a serine protease gene from G. stearothermophilus CAU209 (GsProS8) in B. subtilis to improve its activity (as compared with 69 U/mL in E. coli) [8]. The recombinant protease was then purified and characterized to evaluate its potential application in the preparation of antihypertensive hydrolysates.
Results
Expression and purification of GsProS8
The open reading frame (ORF) of GsProS8 with 1149 bp encoding 383 amino acids was cloned (Fig. S1). The mature protein had a predicted molecular mass of 39.0 kDa. There was no signal peptide in the deduced amino acid sequence based on the analysis by SignalP 4.0. According to the molecular mass determination, a pro-peptide of 107 amino acids was predicted. Thus, the amino acid sequence of GsProS8 consisted of the pro-peptide sequence (1–107) and the mature peptide sequence (108–383). It was found the amino acid sequence of GsProS8 was same as an alkaline serine protease from G. stearothermophilus B-1172 (GenBank: EU181368) that was expressed in E. coli BL21 [8].
Thereafter, GsProS8 was successfully expressed in B. subtilis WB600 under the control of P43 promotor [14]. B. subtilis WB600 transformant was then cultivated in a 5 L fermentor. The protease activity and protein content showed a continuous increment with time, up to a maximum of 3807 U/mL and 2.08 mg/mL at 114 h, respectively (Fig. 1a). By sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), a major protein band around 27 kDa was detected in the fermented medium (Fig. 1b).
Fig. 1.
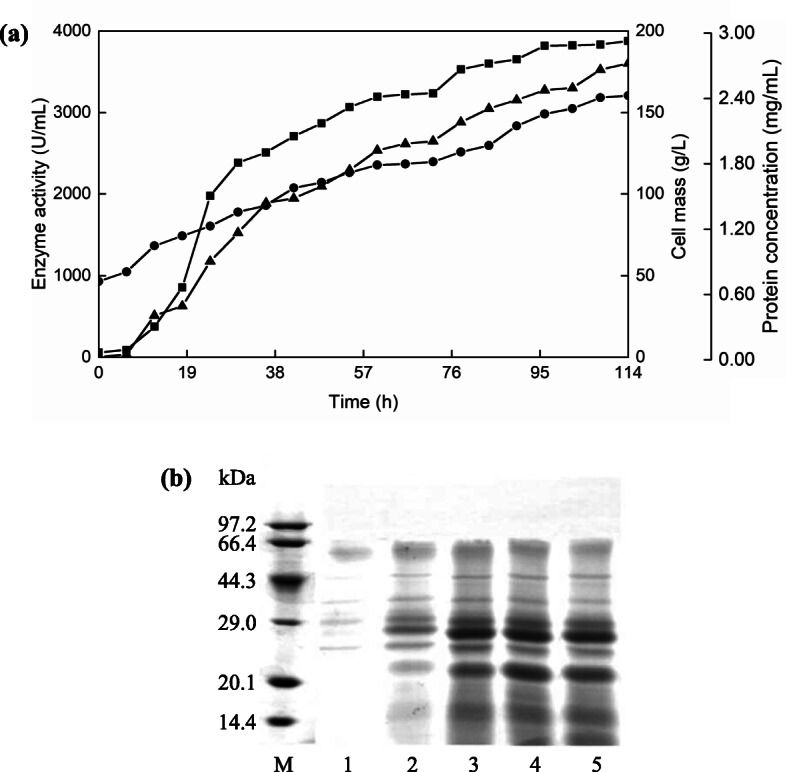
High cell density fermentation of GsProS8 expressed in B. subtilis. a enzyme activity (  ), protein concentration (
), protein concentration (  ), and cell mass (
), and cell mass (  ) of the supernatants. b extracellular protein analysis by SDS-PAGE during fermentation. Lane M, the protein marker; lanes 1–5, the supernatants collected at 24 h, 48 h, 72 h, 96 h, and 114 h, respectively. The full-length gel was presented in Fig. S2
) of the supernatants. b extracellular protein analysis by SDS-PAGE during fermentation. Lane M, the protein marker; lanes 1–5, the supernatants collected at 24 h, 48 h, 72 h, 96 h, and 114 h, respectively. The full-length gel was presented in Fig. S2
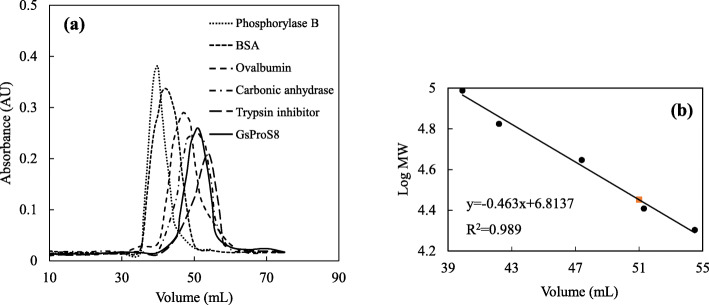
After the fermentation, GsProS8 was purified 2.1-fold to homogeneity with a yield of 23.6%. The specific activity of the protease was increased from 1679.1 U/mg to 3559.7 U/mg (Table 1). The homogeneity of purified GsProS8 was confirmed via a single smeared band at 27.2 kDa by SDS-PAGE (Fig. 2), while the native molecular mass was determined to be 28.3 kDa by gel filtration (Fig. 3). This indicated that GsProS8 is a monomer. Because the conversion process of the primary gene product into the mature enzyme can be mediated by active subtilisin to cleave off the pro-peptide sequence in the extracellular medium during the secretion [11], the molecular mass of purified GsProS8 was lower than the predicted value of 39.0 kDa. Moreover, the purified GsProS8 was evaluated and a clear band of proteolytic activity was observed in the zymogram (Fig. 2).
Table 1.
Purification summary of GsProS8
| Purification step | Total activity (U)a | Total protein (mg)b | Specific activity (U/mg) | Purification factor | Yield (%) |
|---|---|---|---|---|---|
| Crude protease | 116,250 ± 2089.3 | 68.5 ± 4.1 | 1697.1 ± 30.2 | 1.0 | 100.0 |
| Ammonium sulfate precipitation | 86,460 ± 1965.2 | 41.6 ± 1.9 | 2078.4 ± 47.5 | 1.2 | 74.4 |
| SPFF | 50,806 ± 190.5 | 22.4 ± 5.4 | 2268.1 ± 9.1 | 1.3 | 43.7 |
| QSFF | 27,409 ± 373.9 | 7.7 ± 1.2 | 3559.7 ± 48.4 | 2.1 | 23.6 |
aEnzymatic reaction was carried out using casein as a substrate at 50 °C in 0.05 mol/L MOPS buffer (pH 8.5)
bTotal protein was measured using BSA as standard by the Lowry method
Fig. 2.
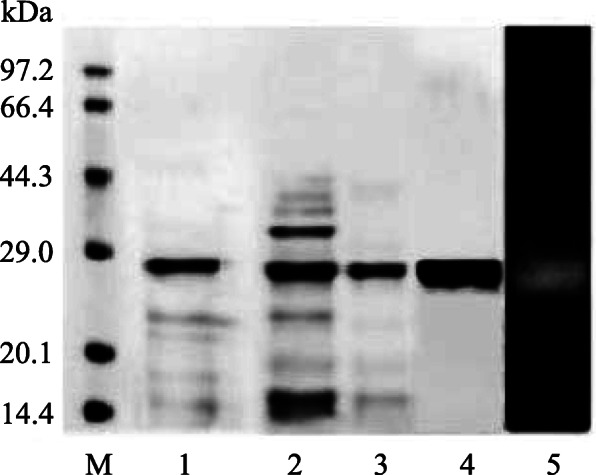
SDS-PAGE analysis of GsProS8 before and after purification. Lane M, the protein marker; lane 1, the crude proteins; lane 2, the proteins purified by ammonium sulphate precipitation; lane 3, the proteins purified by SPFF treatment; lane 4, the proteins purified by QSFF treatment; lane 5, the zymogram of the protease. The full-length gel was presented in Fig. S3 and Fig. S4
Fig. 3.
Molecular mass estimation of GsProS8 (  ) by gel filtration chromatography. a: the ultraviolet (UV) chromatogram; b: the calibration curve to calculate molecular mass of GsProS8. Standard proteins (●), including trypsin inhibitor (20.1 kDa), carbonic anhydrase (29.0 kDa), ovalbumin (44.3 kDa), bovine serum albumin (BSA, 66.4 kDa) and phosphorylase B (97.2 kDa)
) by gel filtration chromatography. a: the ultraviolet (UV) chromatogram; b: the calibration curve to calculate molecular mass of GsProS8. Standard proteins (●), including trypsin inhibitor (20.1 kDa), carbonic anhydrase (29.0 kDa), ovalbumin (44.3 kDa), bovine serum albumin (BSA, 66.4 kDa) and phosphorylase B (97.2 kDa)
Biochemical properties and kinetic parameters of GsProS8
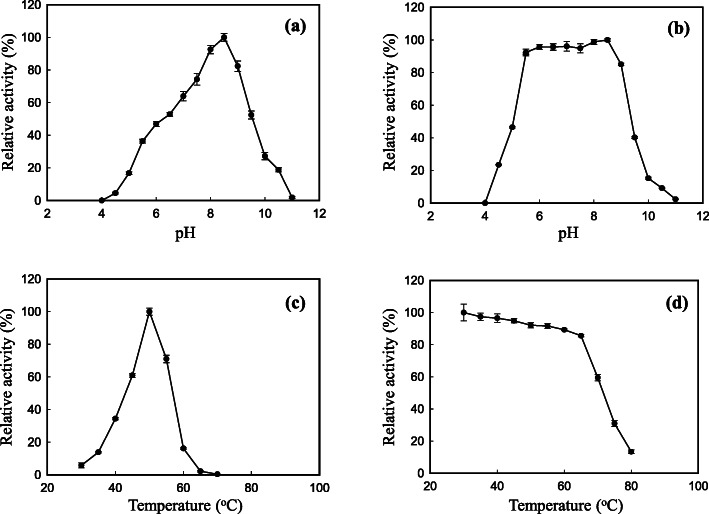
The highest activity of GsProS8 was observed at pH 8.5 (Fig. 4a). The protease was stable at pH 5.5–8.5 to retain more than 90% of the activity (Fig. 4b). The optimal temperature of GsProS8 was found to be 50 °C (Fig. 4c). It was stable up to 65 °C, retaining more than 80% of the activity (Fig. 4d). Protease inhibitors were subjected to identify groups at the active site of GsProS8. Pepstatin A (aspartic protease inhibitor) and iodoacetamide (cysteine protease inhibitor) slightly affected the protease activity, indicating that aspartate residue and -SH group did not work in the protease activity of GsProS8. The presence of ethylenediaminetetraacetic acid (EDTA, metalloenzyme inhibitor) at 1 mM and 4 mM inhibited 30.5 and 40.5% of the protease activity, respectively, revealing that some ions may be important for the stability and activity of GsProS8. The strong inhibition of 95.4 and 99.7% at 1 mM and 4 mM of PMSF (serine protease inhibitor) suggested that GsProS8 belongs to the serine protease class (Table 2).
Fig. 4.
Optimal pH (a), pH stability (b), optimal temperature (c), and thermostability (d) of GsProS8. The influence of pH on GsProS8 activity was measured in 0.05 mol/L of different buffers at 40 °C. For the optimal pH, the relative activity (%) was determined against the specific activity of 3916 U/mg. To determine pH stability against the optimal activity (2631 U/mg, defined as 100% relative activity), GsProS8 was incubated in various buffers (0.05 mol/L) at 40 °C for 30 min. For optimal temperature, GsProS8 activity was tested at different temperatures in MOPS buffer (0.05 mol/L, at pH 8.5) in contrast to the specific activity of 3916 U/mg. The thermostability of GsProS8 was assayed against the optimal activity (3915 U/mg, defined as 100% relative activity) after incubation at different temperatures for 30 min
Table 2.
Effect of selective protease inhibitors on the protease activity of GsProS8. Different letters (a-f) on the same column indicated significant (p < 0.05) differences among samples*
| Inhibitor | Concentration (mM) | Specific activity (U/mg) | Relative activity (%) |
|---|---|---|---|
| Pepstatin A | 0.01 | 3201.0 ± 49.7a | 89.9 |
| 0.02 | 2993.5 ± 52.0b | 84.1 | |
| PMSF | 1 | 162.3 ± 20.3e | 4.6 |
| 4 | 10.3 ± 2.3f | 0.3 | |
| Iodoacetamide | 1 | 3277.0 ± 15.8a | 92.1 |
| 4 | 2901.0 ± 36.2b | 81.5 | |
| EDTA | 1 | 2475.7 ± 29.4c | 69.5 |
| 4 | 2118.2 ± 6.8d | 59.5 |
*Enzymatic reaction was carried out using casein as a substrate at 50 °C in 0.05 mol/L MOPS buffer (pH 8.5). Specific activity was shown as mean ± SD (n = 3). Relative activity was expressed as a percentage of the activity in the absence of inhibitors (the control)
The effects of various metal ions on the protease activity were assessed (Table 3). The protease activity of GsProS8 was not affected by Mg2+, but slightly impacted by Ca2+ (93.7%) and Na+ (91.4%). The moderate reduction in the protease activity of GsProS8 was observed in the presence of Mn2+ (78.9%), Zn2+ (78.1%), Co2+ (76.6%), Ni2+ (66.1%), and Cu2+ (40.7%). In addition, GsProS8 exhibited the highest protease activity towards casein (100%), followed by whey protein (92.1%) and skim milk powder (85.5%). The protease showed a broad substrate specificity towards hemoglobin (60.7%), soybean protein isolate (56.7%), bovine serum albumin (BSA, 45.7%), protamine (37.5%), myoglobin (36.9%), and azocasein (31.2%). However, GsProS8 displayed low activity towards gelatin (10.5%) (Table 4). The kinetic parameters of the purified GsProS8 were determined using casein and whey protein as substrates. The values of Km and Vmax were 7.37 mg/mL and 13.35 mg/mL, 231.45 μmol/mg·min and 122.96 μmol/mg·min for casein and whey protein, respectively (Table 5).
Table 3.
Effect of metal ions on the protease activity of GsProS8. Different letters (a-g) on the same column indicated significant (p < 0.05) differences among samples*
| Metal ion | Specific activity (U/mg) | Relative activity (%) |
|---|---|---|
| None | 3559.7 ± 48.4a | 100.0 |
| Mg2+ | 3593.4 ± 2.3a | 100.9 |
| Ca2+ | 3334.5 ± 15.8b | 93.7 |
| Na+ | 3254.4 ± 18.1c | 91.4 |
| Ni2+ | 2354.5 ± 9.0f | 66.1 |
| Mn2+ | 2808.6 ± 24.9d | 78.9 |
| Co2+ | 2726.4 ± 5.0e | 76.6 |
| Zn2+ | 2779.8 ± 6.8de | 78.1 |
| Cu2+ | 1450.5 ± 9.0g | 40.7 |
*Enzymatic reaction was carried out using casein as a substrate at 50 °C in 0.05 mol/L MOPS buffer (pH 8.5). Specific activity was shown as mean ± SD (n = 3). Relative activity was expressed as a percentage of the activity in the absence of metal ions (the control)
Table 4.
Substrate specificity of GsProS8. Different letters (a-h) on the same column indicated significant (p < 0.05) differences among samples*
| Substrate | Specific activity (U/mg) |
Relative activity (%) |
|---|---|---|
| Casein | 3560.5 ± 15.8a | 100 |
| Whey protein | 3277.0 ± 19.8b | 92.1 |
| Skim milk powder | 3044.8 ± 13.6c | 85.5 |
| Hemoglobin | 2161.4 ± 54.2d | 60.7 |
| Soybean protein isolate | 2025.8 ± 45.2d | 56.9 |
| Bovine serum albumin | 1627.2 ± 22.6e | 45.7 |
| Protamine | 1333.4 ± 6.8f | 37.5 |
| Myoglobin | 1312.9 ± 33.9f | 36.9 |
| Azocasein | 1109.5 ± 0.1g | 31.2 |
| Gelatin | 374.1 ± 1.6h | 10.5 |
*Enzymatic reaction was carried out using different substrates at 50 °C in 0.05 mol/L MOPS buffer (pH 8.5). Specific activity was shown as mean ± SD (n = 3). Relative activity was expressed as a percentage of the activity when casein as a substrate (the control)
Table 5.
Kinetic parameters of GsProS8 for casein and whey protein. Different letters (a-b) on the same column indicated significant (p < 0.05) differences among samples*
| Substrate |
Vmax (μmol/mg·min) |
Km (mg/mL) |
kcat (s−1) |
kcat/Km (mL/mg·s) |
|---|---|---|---|---|
| Casein | 231.50 ± 3.07a | 7.37 ± 0.24b | 0.105 | 0.014 |
| Whey protein | 122.96 ± 4.98b | 13.35 ± 1.17a | 0.056 | 0.004 |
*The measurement of kinetic parameters was carried out using casein and whey protein as substrates at 50 °C in 0.05 mol/L MOPS buffer (pH 8.5). Km and Vmax were shown as mean ± SD (n = 3)
Preparation of antihypertensive whey protein hydrolysates by GsProS8
The optimization of whey protein hydrolyzation by GsProS8 based on the orthogonal experimental design is shown in Table 6. The correlation coefficient value (R) demonstrated that whey protein concentration was the most affective factor (R = 19.1) on the peptide contents of whey protein hydrolysates, followed by temperature, pH and time. In addition, higher K value in each column showed the stronger impacts of the level (1, 2, or 3) on the peptide contents. Therefore, evaluated by K values, the optimized hydrolysis conditions to prepare whey protein hydrolysate by GsProS8 should be performed using 11% (w/v) of whey protein at pH 8.0 and 60 °C for 6 h.
Table 6.
Orthogonal experimental results and the optimization of whey protein hydrolysates by GsPros8
| Trial | Factor | Peptide content (%) |
|||
|---|---|---|---|---|---|
| pH | Temperature (°C) | Time (h) | Whey protein concentration (%) | ||
| 1 | 1 | 1 | 1 | 1 | 15.6 |
| 2 | 2 | 1 | 2 | 2 | 17.0 |
| 3 | 3 | 1 | 3 | 3 | 21.7 |
| 4 | 1 | 2 | 2 | 3 | 33.4 |
| 5 | 2 | 2 | 3 | 1 | 29.2 |
| 6 | 3 | 2 | 1 | 2 | 34.9 |
| 7 | 1 | 3 | 3 | 2 | 31.4 |
| 8 | 2 | 3 | 1 | 3 | 51.5 |
| 9 | 3 | 3 | 2 | 1 | 14.5 |
| K1a | 23.5 | 14.8 | 21.6 | 16.4 | |
| K2a | 32.6 | 32.5 | 30.7 | 27.8 | |
| K3a | 23.7 | 32.5 | 27.4 | 35.5 | |
| Rb | 9.1 | 17.7 | 9.0 | 19.1 | |
| The impact of factors | Whey protein concentration > Temperature > pH > Time | ||||
| Optimized hydrolysis condition | pH 8.0, 60 °C, 6 h, 11% of whey protein concentration | ||||
aK1, K2, and K3 indicated the sum of peptide contents corresponding to level 1, level 2, and level 3, respectively
bR was the correlation coefficient value, R = Max Ki – Min Ki (i = 1, 2, or 3)
The performance of GsProS8 was compared with commercial proteases in the preparation of antihypertensive whey protein hydrolysates (Fig. 5). The whey protein hydrolysate prepared by Flavourzyme contained the highest (p < 0.05) peptide content (52.02%) with the strongest ACE inhibitory activity (IC50 of 0.116 mg/mL), followed by the hydrolysates generated by GsProS8 (IC50 of 0.129 mg/mL) and Alcalase (IC50 of 0.143 mg/mL). Nevertheless, both of trypsin and Protamex were less (p < 0.05) suitable to prepare antihypertensive whey protein hydrolysates, because of the higher IC50 values (0.197 mg/mL and 0.192 mg/mL).
Fig. 5.
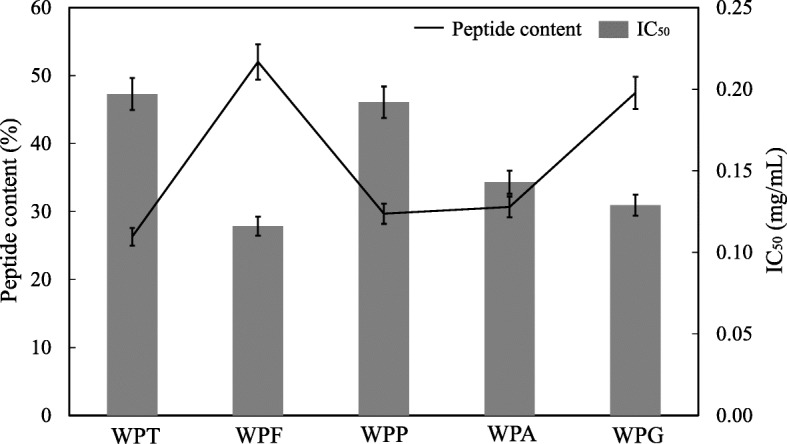
Peptide contents and IC50 values of whey protein hydrolysates prepared by GsProS8 versus commercial proteases. WPT, whey protein hydrolysate prepared by trypsin at pH 8.0 and 37 °C; WPF, whey protein hydrolysate prepared by Flavourzyme at pH 7.0 and 53 °C; WPP, whey protein hydrolysate prepared by Protamex at pH 7.5 and 40 °C; WPA, whey protein hydrolysate prepared by Alcalase at pH 8.0 and 60 °C; WPG, whey protein hydrolysate prepared by GsProS8 at pH 8.0 and 60 °C
Discussion
G. stearothermophilus can produce extracellular neutral and alkaline proteases [8, 14]. Many researches have explored alkaline serine proteases from several strains of G. stearothermophilus, such as strain F1 [7], strain B-1172 [8], and strain AP-4 [6]. In the present study, an alkaline serine protease gene (GsProS8) from G. stearothermophilus CAU209 was cloned and expressed in B. subtilis WB600. Although GsProS8 shared the same amino acid sequence with the reported protease from G. stearothermophilus B-1172 [8], the protease activity of GsProS8 (3807 U/mL) after high cell density fermentation (Fig. 1a) was much higher than the proteases from the strain AP-4 (250 U/mL) [6], the strain F1 (1500 U/mL) and the strain B-1172 (69 U/mL) that were expressed in E. coli [7, 8]. This may be attributed to the detrimental effects on the E. coli by the intracellular accumulation of the alkaline proteases [11]. On the contrary, B. subtilis is a more attractive expression host to steadily secrete extracellular proteins directly into the culture medium [23]. Moreover, the protease activity of GsProS8 was also higher than the other alkaline serine proteases from Aspergillus sojae (400 U/mL) [24], B. subtilis RD7 (607 U/mL) [2], and B. clausii (1020 U/mL) [25].
After purification, the specific activity of GsProS8 increased from 1697.1 U/mg to 3559.7 U/mg (Table 1), which is much higher than the previously reported proteases from G. stearothermophilus strain B-1172 (97.5 U/mg) [8] and strain F1 (1790 U/mg) [7]. In general, the molecular masses of alkaline serine proteases from bacteria have been reported to be 30–45 kDa. Molecular masses of alkaline serine proteases from A. sojae, B. subtilis RD7, and Bacillus lehensis JO-26 were reported to be 35.2 kDa [24], 43 kDa [2], and 34.6 kDa [26], respectively. In this study, the purified GsProS8 had a relative molecular mass of 27.2 kDa on SDS-PAGE and 28.3 kDa in gel filtration (Fig. 3), which presented a good dispersibility and permeability [27]. However, the predicted molecular mass of GsProS8 was same as the protease (100% identity of amino acid sequences) expressed in E. coli (39 kDa) [8]. The conversion of the primary gene product into the mature protease is most likely autocatalytic, and the pro-peptide sequence might be cleaved by a self-processing mechanism. Thus, the molecular mass of purified protease did not correspond with the prediction by gene sequence [11]. Tang and co-workers [11] cloned and expressed an alkaline protease gene from B. licheniformis 2709 in B. subtilis WB600. The truncation of pro-peptide was demonstrated by the molecular mass of 30.5 kDa in contrast to the predicted value of 55 kDa [28]. This truncation has also been reported in the keratinolytic protease from Thermoactinomyces sp. YT06, alkaline protease from Vibrio sp. DA1–1, and serine protease from Dichelobacter nodosus, in which the recombinant proteases were 28–37 kDa, instead of the expected 40.3–50.6 kDa [29–31]. The keratinase and alkaline protease genes from Thermoactinomyces sp. YT06 and Vibrio sp. DA1–1 were cloned and expressed in E. coli BL21, and the pro-sequences were also autoproteolytically cleaved in the periplasm [29, 30]. Therefore, the truncation was not specific to over-expression in B. subtilis WB600.
Species of Bacillus are the most representative microbial strains used in the production of alkaline serine proteases with many applications in food industry, such as hydrolyzing proteins to obtain bioactive ingredients [32] and modifying wheat gluten to improve the texture of baking products [33]. Notably, the majority alkaline serine proteases exhibit optimal pH in the range of 7 to 11 [27]. The optimal pH 8.5 of GsProS8 is committed to this range, but is slightly lower than the proteases (pH 9.0–10.6) from other strains of G. stearothermophilus [6], Bacillus sp. strain B001 [13], B. subtilis RD7 [2], and A. sojae [24]. Meanwhile, the pH stability of GsProS8 was comparable with the protease from G. stearothermophilus B-1172, in which a high residual activity (> 87%) was retained after incubation of the proteases at pH 5.0–9.0 [8]. However, the residual activities of the proteases from G. stearothermophilus F1 and AP-4 kept 95% of residual activities within pH 8.0–10.0 [6, 7].
Furthermore, GsProS8 had a maximal activity at 50 °C (Fig. 4c), which is lower than those bacterial proteases from G. stearothermophilus F1 expressed in E. coli (80 °C) and Bacillus sp. strain B001 (60 °C) [7, 13]. In theory, the expression and biochemical properties of proteases are influenced by many factors, such as the strain of expression host, culture temperature, the inducing temperature and time [34]. As the protein expressed in E. coli, the recombinant protease was secreted within cytoplasm as a soluble and active form to maintain the original living temperature of G. stearothermophilus. While expression in B. subtilis, the recombinant protease was extracellularly secreted into culture medium at 37 °C. The optimal temperature of GsProS8 expressed in B. subtilis was deduced. This was also proved by Zhang and co-workers [14], in which the protease (NprT) from G. stearothermophilus expressed in B. subtilis DB104 was different from the growth temperature of G. stearothermophilus (Table 7). In addition, the lower molecular mass of GsProS8 (27.2 kDa) was contributed to the lower optimal temperature (50 °C) as compared with the protease gene expressed in E. coli (39 kDa) [8]. A similar finding was reported by Liu and co-workers [35], who compared keratinase gene expression in E. coli, B. subtilis, and P. pastoris. However, GsProS8 was comparable and displayed an even higher optimal temperature than the proteases from B. lehensis JO-26 (50 °C) [26], A. sojae expressed in P. pastoris (40 °C) [24], and B. subtilis RD7 expressed in E. coli (40 °C) [2]. Therefore, GsProS8 can be handled for industrial applications under an alkaline condition with a relative high temperature to increase reaction rate.
Table 7.
Alkaline serine proteases from G. stearothermophilus and being used to prepare ACE inhibitory hydrolysates
| Origin of alkaline serine protease | Expression host | Molecular mass (kDa) |
Protease activity (U/mL)a |
Optimal pH and temperature | Hydrolyzed proteinb | IC50 (mg/mL) |
Reference |
|---|---|---|---|---|---|---|---|
| G. stearothermophilus CAU209 | B. subtilis WB600 | 27.2 | 3807 | pH 8.5, 50 °C | Whey protein | 0.129 | This study |
| G. stearothermophilus AP-4 | – | – | 250 | pH 9.0, 55 °C | – | – | 6 |
| G. stearothermophilus F1 | E.coli XL1-Blue | 27 | 1500 | pH 9.0, 80 °C | – | – | 7 |
| G. stearothermophilus B-1172 | E. coli BL21 | 39 | 64 | pH 9.0, 90 °C | – | – | 8 |
| G. stearothermophilus | B. subtilis DB104 | 35 | 7020 | pH 7.5, 65 °C | – | – | 14 |
| Anoxybacillus kamchatkensis M1V | E. coli BL21 | 28 | 4600 | pH 11.0, 70 °C | Shrimp protein | 0.010 | 44, 45 |
| Aeribacillus pallidus VP3 | – | 29 | 3000 | pH 10.0, 60 °C | Shrimp protein | 0.014 | 45, 46 |
| Yarrowia lipolytica | – | 35 | – | pH 9.0, 45 °C | Egg white protein | 1.229 | 47, 48 |
| Sardina pilchardus | – | 25 | 500 | pH 8.0, 60 °C | Sardinelle protein | 1.200 | 49, 50 |
| A. clavatus ES1 | E. coli BL21 | 32 | 260 | pH 8.5, 50 °C | Sardinelle protein | 7.400 | 38, 50 |
| B. licheniformis NH1 | – | 27 | 1600 | pH 10.0, 70 °C | Sardinelle protein | 2.100 | 50, 51 |
aProtease activity was measured using casein as a substrate at the optimal conditions for each protease
bThe protein was hydrolyzed under the optimal hydrolysis condition of each protease
- Not found
The protease activity of GsProS8 relatively remained constant and was not declined too much in the presence of metal ions, except Cu2+ (Table 3). Thus, the protease described in this study is not a metalloenzyme. This is similar with the subtilisins from B. subtilis RD7 [2] and B. lehensis JO-26 [26], but better than the one from Bacillus spp. B001 [13], whose activities were almost completely reduced by Cu2+, Zn2+, Ni2+, and Co2+. The protease activity of GsProS8 was affected in the presence of heavy metal Cu2+, because it might chelate with the protease causing precipitation and deactivation [36]. In addition, the reduction of protease activity in the presence of Ni2+, Mn2+, Co2+, and Zn2+ demonstrated that GsProS8 might contain a number of metal ions, so, the displacement or substitution of them compromise the catalysis [2, 8].
GsProS8 was highly specific towards casein and whey protein (Table 4). This may be resulted from the maximization of the binding energy of casein and whey protein for the protease that decided the substrate affinity [8]. The result is in accordance with the alkaline serine proteases from G. stearothermophilus B-1172 and A. sojae showing the highest activities towards casein [8, 24]. In the meantime, GsProS8 had significant hydrolysis preferences to skim milk powder, hemoglobin, and soybean protein isolate, and relative activities towards BSA, protamine, myoglobin, and azocasein (Table 4). So, it displayed broader substrate specificity than the proteases from B. subtilis RD7 [2], A. sojae [24], and G. stearothermophilus B-1172 [8]. However, azocasein was found to be the most preferred substrate for the protease from B. subtilis RD7 [2].
As summarized in literature, the values of Km and Vmax of proteases presented as 0.08–5.10 mg/mL and 25–7692 U/mg, respectively [37–40]. GsProS8 exhibited higher affinity than the other alkaline serine proteases (Table 5), to indicate potential application in commercial products. Additionally, because caseins carried open structures to be susceptible for proteolysis than whey protein with a globular nature [41], the kinetic parameters of GsProS8 acting on whey protein were significantly lower than those on casein.
Proteases have been found to display many applications in the food industry, in which the preparation of protein hydrolysates with remarkable biological activities (e.g., antihypertensive, immunostimulating, antimicrobial, and antioxidant activities) have attracted much attention [1]. Due to excellent nutritional and functional properties, whey protein hydrolysates produced by protease hydrolysis, microbial fermentation, and heat treatment have been widely used in food industry [42]. ACE inhibitory activity of whey protein hydrolysates has been discovered using various proteases (e.g., trypsin, Alcalase, bromelain, papain, and pepsin) [43]. In this study, whey protein was hydrolyzed by GsProS8 and commercial proteases (e.g., trypsin, Flavourzyme, Protamex, and Alcalase), GsProS8 hydrolysate contained higher peptide content and ACE inhibitory activity (Fig. 5). In particular, the whey protein hydrolysate prepared by GsProS8 (IC50 of 0.129 mg/mL) exhibited better antihypertensive activity than the hydrolysates produced by other alkaline serine proteases from Maclura pomifera (IC50 of 0.53 mg/mL) [21] and Cucurbita ficifolia (IC50 of 0.65 mg/mL) [22]. In addition, GsProS8 was compared with other alkaline serine proteases for the preparation of ACE inhibitory hydrolysates (Table 7). GsProS8 has comparable biochemical properties as compared with alkaline serine proteases from Anoxybacillus kamchatkensis strain M1V and Aeribacillus pallidus strain VP3. Moreover, GsProS8 hydrolyzed whey protein to yield a lower IC50 value (0.129 mg/mL) than the hydrolysates prepared by the proteases from Yarrowia lipolytica, S. pilchardus, A. clavatus ES1 and B. licheniformis NH1 (1.200–7.400 mg/mL) [38, 44–51]. This revealed that GsProS8 has potential to be used for preparation of antihypertensive hydrolysates and peptides.
Conclusions
In summary, an alkaline serine protease gene from G. stearothermophilus CAU209 has been expressed in B. subtilis WB600, to exhibit high protease activity (3807 U/mL) after high cell density fermentation. GsProS8 was purified as a monomeric protein with a molecular mass of 27.2 kDa. It showed maximal protease activity at pH 8.5 and 50 °C with a broad substrate specificity. Moreover, GsProS8 displayed a good ability to hydrolyze whey protein to prepare antihypertensive hydrolysates. The properties of GsProS8 provided important insights for its applications into food industry.
Methods
Bacterial strain, plasmids and media
G. stearothermophilus CAU209 was deposited in Key Laboratory of Food Bioengineering (China National Light Industry) in Beijing, China. B. subtilis WB600 (Δbpr, Δepr, Δmpr, ΔnprB, Δvpr, ΔwprA), a six extracellular protease deficient strain gifted from Guangxi University, Nanning, China, was used as the expression host. E. coli DH5α [F− supE44 Ф80 δlacZ ΔM15 Δ (lacZYA-argF) U169 endA1 recA1 hsdR17 (rK−, mK+) deoR thi-1 λ- gyrA96 relA1] (Biomed, Beijing, China) was used as the host strain for DNA manipulation, and was cultured in Luria-Bertani (LB) medium composed of 10 g/L peptone, 5 g/L yeast extract, and 5 g/L NaCl. Plasmid pWB980 (TaKaRa Corporation, Dalian, China) was used as the expression vector. Whey protein (80% of protein content on dry basis) was obtained from Ausnutria Dairy Corporation (Changsha, China). All other chemicals and reagents were of analytical grades and commercially available from Sigma-Aldrich (St. Louis, MO, USA).
Cloning of the alkaline serine protease gene (GsProS8)
Genomic DNA from G. stearothermophilus CAU209 (GenBank: MW084977) was extracted as described previously [52]. According to the genome sequence of G. stearothermophilus CAU209, a pair of oligonucleotide primers, named GsProS8-F and GsProS8-R (Table S1), were designed to amplify the coding region of the alkaline serine protease. Polymerase chain reaction (PCR) was performed at 94 °C for 5 min, 30 cycles with denaturing at 94 °C for 30 s, annealing at 56 °C for 30 s and extension at 72 °C for 1.5 min, and the last cycle was at 72 °C for 10 min. The PCR product was ligated with pMD-19 T and transformed into E. coli DH5α for sequencing. Sequence analysis was performed using DNAMAN 6.0. The ORF was found using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi/). The signal peptide was predicted by SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/).
Plasmid construction for expression of GsProS8
To express the protease gene in B. subtilis WB600, two pairs of primers (GsProS8-IF and GsProS8-IR, pWB980-VF and pWB980-VR, Table S1) were designed to amplify the ORF region of the GsProS8 gene on the chromosome DNA and the vector, respectively. The overlapping PCR was carried out using 0.0002 mol/L each of the four dNTPs and 0.04 U Phusion DNA polymerase. P43 promoter was located at the upstream of SacB signal peptide in the plasmid pWB980. PCR conditions were as follows: 94 °C for 5 min, 30 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1.5 min (the last cycle at 72 °C for 10 min). The purified PCR product was digested with EcoRI and SmaI restriction endonucleases, and then inserted into the plasmid pWB980. The recombinant plasmid pWB980-GsProS8 was transformed into the B. subtilis WB600 by electroporation. The transformants were cultured on LB plates and incubated at 37 °C until colonies appeared. The colonies were then suspended in the LB medium for further growth in a rotary shaker at 37 °C for 18–24 h. The cultures were centrifuged at 10,000×g for 10 min to collect the supernatant. The protease activity as well as the expression in the supernatant were analyzed by the Folin-Ciocalteu reagent and SDS-PAGE, respectively.
High cell density fermentation
To scale up GsProS8 production, high cell density fermentation was carried out in a 5.0 L fermentor (Guoqiang, Shanghai, China) with a 1.5 L working volume at 37 °C. 1% (v/v) inoculum was initially incubated at 37 °C for 12 h. Subsequently, the fermentor was inoculated with 750 mL of the inoculum and cultivated at 37 °C for 114 h. The fermentation medium was composed of glucose (10 g/L), yeast extract powder (10 g/L), peptone (20 g/L), corn steep powder (5 g/L), sodium chloride (10 g/L), magnesium sulfate (0.3 g/L), sodium phosphate (6 g/L), and dipotassium phosphate (3 g/L). The cultivation was maintained at pH 7.0–7.2 with the aid of ammonium hydroxide and phosphoric acid. The dissolved oxygen level was maintained at 30% air-saturation. Samples were withdrawn at 6 h interval to analyze enzyme activity, cell mass, and protein concentration, and SDS-PAGE was also performed.
Purification of GsProS8
The fermented culture was purified through ammonium sulfate precipitation and a two-step chromatographic method. In short, the crude enzyme was initially precipitated by ammonium sulphate (40–70% w/v saturation) and dialyzed against 0.02 mol/L Tris-HCl at pH 8.0 (buffer A) for overnight. Thereafter, the dialyzed sample was subjected to a HiTrap SP Fast Flow (SPFF) column (GE Healthcare, Wuxi, China), which was pre-equilibrated with buffer A. After washing with buffer A until the absorbance (at 280, OD280) < 0.05, the bounded proteins were then eluted with a linear gradient (0–0.25 mol/L) of NaCl at a flow rate of 1 mL/min. The fraction with high protease activity was collected and dialyzed against 0.05 mol/L N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) at pH 10.3 (buffer B) for overnight. The sample was loaded onto a HiTrap Q-Sepharose Fast Flow (QSFF) column (GE Healthcare, Wuxi, China) that was pre-equilibrated with buffer B. Subsequently, the proteins were eluted with buffer B until OD280 < 0.05, followed by a gradient elution of NaCl (0–0.15 mol/L) at 1 mL/min. The purity of the fractions was verified by SDS-PAGE.
SDS-PAGE, zymogram and molecular mass determination
SDS-PAGE was used to evaluate the purity and molecular mass of the protease. The protein bands were stained by Coomassie Brillaint Blue R-250. The molecular weight markers for electrophoresis included α-lactalbumin (14.4 kDa), trypsin inhibitor (20.1 kDa), carbonic anhydrase (29.0 kDa), ovalbumin (44.3 kDa), BSA (66.4 kDa) and phosphorylase B (97.2 kDa). Zymogram of the protease was analyzed using the SDS-polyacrylamide gel co-polymerized with 0.1% gelatin as described by Sun and co-workers [53]. The native molecular mass of the purified GsProS8 was estimated by a gel filtration chromatography via a Sephacryl S-100 column (1.0 × 100 cm). The purified protease and protein markers were eluted by 20 mM Tris-HCl buffer (at pH 8.0) containing 500 mM NaCl at a flow rate of 0.33 mL/min.
Protease assay and protein determination
Protease activity was determined using casein as a substrate. Briefly, the protease solution (100 μL) was incubated with 2% (w/v) casein (100 μL) in 20 mM phosphate buffer (700 μL, pH 7.5) at 37 °C for 30 min. The reaction was terminated by trichloroacetic acid (10% w/v, 200 μL). After centrifugation at 10,000×g for 5 min, the supernatant (100 μL) was mixed with 600 μL alkaline reagent (0.4 mol/L Na2CO3: Folin-Ciocalteu reagent = 5:1 v/v) and incubated at 40 °C for 20 min. The absorbance was measured at 660 nm using a spectrophotometer (TU1901, Puxi General Instruments Co., Ltd., Beijing, China). The mixture without the protease was used as control. One unit of protease activity (U) was defined as the amount of the protease to liberate 1 μmol tyrosine per minute at the above conditions. Moreover, the protein concentration was measured by the Lowry method using BSA as standard. Specific activity was expressed as units per milligram of protein.
Biochemical properties of GsProS8
The optimal pH for protease activity was evaluated at 40 °C in a pH range of 4.0–11.0. The following buffers (0.05 mol/L) were used: citrate (pH 4.0–5.0), acetate (pH 5.0–5.5), 2-(N-morpholino) ethanesulfonic acid (MES, pH 5.5–7.0), 4-(N-morpholino) propanesulfonic acid (MOPS, pH 7.0–8.5), and N-cyclohexyl-2-aminoethanesulfonic acid (CHES, pH 8.5–11.0). The pH stability was investigated after incubating the protease in various buffers (0.05 mol/L) at 40 °C for 30 min. The residual activity was determined by the standard assay.
The optimal temperature was determined in 0.05 mol/L of MOPS (pH 8.5) buffer at 30-70 °C. The thermostability was measured after incubating the protease at a temperature range of 30-80 °C for 30 min. The residual activity was determined by the standard assay.
The effect of several inhibitors on the protease activity was analyzed. The following inhibitors were used: pepstain A (at 0.01 mM and 0.02 mM), EDTA (at 1 mM and 4 mM), PMSF (at 1 mM and 4 mM), and iodoacetamide (at 1 mM and 4 mM). Moreover, the effect of metal ions on protease activity was examined in the presence of magnesium sulfate, calcium chloride, sodium chloride, nickel chloride, manganese sulfate, cobalt chloride, zinc sulfate, and copper sulfate at the final concentration of 0.001 mol/L.
Substrate specificity was evaluated at pH 8.5 and 50 °C using casein, whey protein, skim milk powder, hemoglobin, soybean protein isolate, BSA, protamine, myoglobin, azocasein, or gelatin (1% w/v). The protease activity was measured as described previously. The protease activity towards casein was determined to be 100%, to calculate the relative protease activities to other substrates.
Kinetic parameters of GsProS8
The kinetic parameters of the purified GsProS8 were determined using Michaelis-Menten plot after measuring the protease activities at various concentrations of casein (4–16 mg/mL) and whey protein (8–25 mg/mL) at pH 8.5 and 50 °C for 5 min. The parameters included Km, Vmax, kcat and kcat/Km, in which Km and Vmax were obtained from non-linear regression analysis by GraphPad Prism V.8 [54].
Hydrolysis of whey protein by GsProS8
According to the preliminary test, pH, temperature, time, and whey protein concentration were identified as four critical factors to impact the hydrolysis of whey protein by GsProS8. Each factor was analyzed at three levels using an orthogonal experimental design (Table S2). In brief, whey protein (7–11%, w/v) was hydrolyzed by GsProS8 (200 U/mL) at designed temperature (40-60 °C) and pH (7.5–8.5) for 4–8 h in a temperature-control shaker (HZQ-X100, Suzhou, China) at 100 rpm, and terminated by heating at 85 °C for 10 min. The hydrolysate solutions were centrifuged (Refrigerated Centrifuge GL-20B, Shanghai, China) at 10,000×g for 10 min to obtain the supernatants for later analysis. Commercial proteases (200 U/mL, e.g., trypsin at pH 8.0 and 37 °C; Flavourzyme at pH 7.0 and 53 °C; Protamex at pH 7.5 and 40 °C; and Alcalase at pH 8.0 and 60 °C) were also applied in the hydrolysis of whey protein.
Peptide content of whey protein hydrolysate
The peptide content in the whey protein hydrolysate was determined by o-phthalaldehyde (OPA) method as described by Church and co-workers [55] with slight modifications. Concisely, the supernatant of whey protein hydrolysate (25 μL) was added into OPA mixture (1 mL), which was prepared by 100 mM sodium tetraborate, 20% (w/w) SDS, and OPA solution (containing 0.02% w/v OPA, methanol, and β-mercaptoethanol). The incubation was then processed at room temperature for 8 min, and the absorbance of resulting solution at 340 nm (TU-1800PC spectrophotometer, Persee General Instrument Co. Ltd., Beijing, China) was recorded. Both distilled water and glutathione were performed as a blank and a standard, respectively. The peptide content was calculated based on the standard curve of glutathione.
Antihypertensive activity of whey protein hydrolysate
Antihypertensive activity of whey protein hydrolysate was expressed as ACE inhibitory activity (%) and measured as described by Cushman and Cheung [56] with some modifications. Namely, the reaction of whey protein hydrolysate (2 mg/mL, 20 μL) with ACE (0.1 U/mL, 10 μL) in 120 μL of 0.1 M sodium borate buffer (containing 5 mM N-Hippuryl-His-Leu hydrate and 0.3 M NaCl at pH 8.3) was performed at 37 °C for 60 min. The reaction was then terminated by HCl (1 M, 150 μL) and ethyl acetate (1 mL), followed by centrifugation (GL-20B Refrigerated Centrifuge, Anting Scientific Instrument Co. Ltd., Shanghai, China) at 4000 rpm for 10 min at room temperature to collect the supernatant, which was dried at 105 °C for 30 min. The liberated hippuric acid was dissolved in the deionized water (0.5 mL) to record the absorbance at 228 nm (TU-1800PC, Persee General Instrument Co. Ltd., Beijing, China). The enzyme reaction without the whey protein hydrolysate and sodium borate buffer (0.1 M) containing substrate were used as a control and a blank, respectively. ACE inhibitory activity (%) was calculated as follows:
where Aa is the absorbance of the sample supernatant, Ab is the absorbance of the control, and Ac is the absorbance of the blank. ACE inhibitory activity (%) was transformed to IC50 (mg/mL, half maximal inhibitory concentration) based on a plot of relative ACE inhibitory activity (%) against different dilutions (0.01–1.00%) of the hydrolyzed supernatant using non-linear regression analysis by GraphPad Prism V.8 (Fig. S5).
Statistics
The experiments were performed in triplicate and expressed as the mean ± one standard deviation. A one-way analysis of variance (ANOVA) was carried out using Systat v10 software (San Jose, CA, USA) to analyze statistical differences on the protease activity, peptide content, and ACE inhibitory activity (IC50). p < 0.05 was considered significant.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- PMSF
Phenylmethylsulfonylfluoride
- DFP
Diodopropyl fluorophosphate
- GRAS
Generally recognized as safe
- ACE
Angiotensin-I-converting enzyme
- LB
Luria-Bertani
- PCR
Polymerase chain reaction
- ORF
Open reading frame
- SDS-PAGE
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- SPFF
SP Fast Flow
- CAPS
N-cyclohexyl-3-aminopropanesulfonic acid
- QSFF
Q-Sepharose Fast Flow
- BSA
bovine serum albumin
- MES
2-(N-morpholino) ethanesulfonic acid
- MOPS
4-(N-morpholino) propanesulfonic acid
- CHES
N-cyclohexyl-2-aminoethanesulfonic acid
- EDTA
Ethylenediaminetetraacetic acid
- OPA
O-phthaladehyde
- ANOVA
A one-way analysis of variance
Authors’ contributions
CC: Manuscript writing, revision, and data interpretation; SYG and ZPL: Performing experiments, data collection and analysis; QJY and ZQJ: Study design, manuscript editing, and supervision. All authors read and approved the final manuscript.
Authors’ information
CC: specialized on the protein science and development of bioactive peptides, Assistant Professor, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China. SYG: Master Graduate, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China. ZPL: Master Student, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China. QJY: Professor, College of Engineering, China Agricultural University, Beijing, China. ZQJ: specialized on the food enzymology, Professor, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0400200).
Availability of data and materials
The sequencing data of the alkaline serine protease gene (GsProS8) from G. stearothermophilus is presented on the NCBI database (GenBank accession number MW084977). The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Contesini FJ, de Melo RR, Sato HH. An overview of Bacillus proteases: from production to application. Crit Rev Biotechnol. 2018;38(3):321–334. doi: 10.1080/07388551.2017.1354354. [DOI] [PubMed] [Google Scholar]
- 2.Suberu Y, Akande I, Samuel T, Lawal A, Olaniran A. Cloning, expression, purification and characterization of serine alkaline protease from Bacillus subtilis RD7. Biocatal Agric Biotechnol. 2019;20:101264. doi: 10.1016/j.bcab.2019.101264. [DOI] [Google Scholar]
- 3.Kolkman M, Mejldal R, Goedegebuur F, Babe LM, Kellett-Smith AH, Mulder H, Bott RR, Scotcher MC. Serine proteases of the Bacillus gibsonii-clade: Danisco US Inc., US10533165; 2016.https://patentimages.storage.googleapis.com/7c/6b/c8/ea8c5f68ea0e1b/US10533165.pdf.
- 4.Soo JK, Soon WK, Seung BH, Soon JS, Ji YM. New Bacillus subtilis KJ-21 strain useful as microbial agent to produce fermented food i.e. meju or Korean miso, doenjang, seasoned miso, gochujang, seasoned gochujang, spring roll, cheonggukjang, mixed roll, and Korean-style soy sauce: Rural Dev Administration, KR1740543-B1; 2017. https://patentimages.storage.googleapis.com/e7/f4/4d/d6a38cecd0b761/KR101740543B1.pdf.
- 5.Maynard F, Salvatore D, Thevenier A, Salvato D. Methods of preparing milk protein hydrolysate for composition e.g. infant formula involves hydrolyzing milk-based proteinaceous material with microbial alkaline serine protease in combination with bromelain and protease from Aspergillus: Nestec SA, AU2020267141-A1; 2020.https://patentimages.storage.googleapis.com/c7/74/20/f20c0d21843ed6/AU2020267141A1.pdf.
- 6.Dhandapani R, Vijayaragavan R. Production of a thermophilic, extracellular alkaline protease by Bacillus stearothermophilus AP-4. World J Microbiol Biotechnol. 1994;10(1):33–35. doi: 10.1007/BF00357559. [DOI] [PubMed] [Google Scholar]
- 7.Fu ZB, Hamid SBA, Bazak CNA, Basri M, Salleh AB, Rahman RNZA. Secretory expression in Escherichia coli and single-step purification of a heat-stable alkaline protease. Protein Expr Purif. 2003;28(1):63–68. doi: 10.1016/S1046-5928(02)00637-X. [DOI] [PubMed] [Google Scholar]
- 8.Iqbal I, Aftab MN, Afzal M, Ur-Rehman A, Aftab S, Zafar A, Ud-Din Z, Khuharo AR, Iqbal J, Ul-Haq I. Purification and characterization of cloned alkaline protease gene of Geobacillus stearothermophilus. J Basic Microbiol. 2015;55(2):160–171. doi: 10.1002/jobm.201400190. [DOI] [PubMed] [Google Scholar]
- 9.Rahman RNZA, Razak CN, Ampon K, Basri M, Yunus WMZW, Salleh AB. Purification and characterization of a heat-stable alkaline protease from Bacillus stearothermophilus F1. Appl Microbiol Biotechnol. 1994;40(6):822–827. doi: 10.1007/BF00173982. [DOI] [Google Scholar]
- 10.Latiffi AA, Salleh AB, Rahman RNZRA, Oslan SN, Basri M. Secretory expression of thermostable alkaline protease from Bacillus stearothermophilus F1 by using native signal peptide and α-factor secretion signal in Pichia pastoris. Genes Genet Syst. 2013;88(2):85–91. doi: 10.1266/ggs.88.85. [DOI] [PubMed] [Google Scholar]
- 11.Tang XM, Shen W, Lakay FM, Shao WL, Wang ZX, Prior BA, Zhuge J. Cloning and over-expression of an alkaline protease from Bacillus licheniformis. Biotechnol Lett. 2004;26(12):975–979. doi: 10.1023/B:BILE.0000030042.91094.38. [DOI] [PubMed] [Google Scholar]
- 12.Zhou CX, Zhou HY, Li DK, Zhang HT, Wang HB, Lu FP. Optimized expression and enhanced production of alkaline protease by genetically modified Bacillus licheniformis 2709. Microb Cell Factories. 2020;19(1):45. doi: 10.1186/s12934-020-01307-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng A, Wu J, Zhang GQ, Wen TY. Molecular and structural characterization of a surfactant-stable high-alkaline protease AprB with a novel structural feature unique to subtilisin family. Biochimie. 2011;93(4):783–791. doi: 10.1016/j.biochi.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Zhao C, Du LX, Lu FP, Gao C. Expression, purification, and characterization of a thermophilic neutral protease from Bacillus stearothermophilus in Bacillus subtilis. Sci China C Life Sci. 2008;51(1):52–59. doi: 10.1007/s11427-008-0009-9. [DOI] [PubMed] [Google Scholar]
- 15.Du H, Lu Y, Fan Y, Fu Q, Qiao B, Du X. New Bacillus subtilis expressing foreign protein, produced by performing gene knockout of two exocrine protease encoding genes including alkaline serine protease (subtilisin E) (aprE) gene and neutral metalloprotease (nprE) gene: Genscript Nanjing Co Ltd, CN104073458-A; 2014. https://patentimages.storage.googleapis.com/8d/ff/fc/1f26e8047ada06/CN104073458A.pdf.
- 16.Lu F, Li Y, Wang X, Shi C, Yu J, Liu Y, Peng C, Liu F, Zhang H, Wang H. New genetically engineered bacterium useful for fermentation production of alkaline proteases and constructing comprising e.g. synthetic nucleotide sequence and a signal peptide combination and using Bacillus clausii genome as template: Univ Tianjin Sci & Technology, CN110144319-A; 2019. https://patentimages.storage.googleapis.com/c4/69/42/90b67eb31d4f67/CN110144319A.pdf.
- 17.Rekik H, Jaouadi NZ, Gargouri F, Bejar W, Frikha F, Jmal N, Bejar S, Jaouadi B. Production, purification and biochemical characterization of a novel detergent-stable serine alkaline protease from Bacillus safensis strain RH12. Int J Biol Macromol. 2019;121:1227–1239. doi: 10.1016/j.ijbiomac.2018.10.139. [DOI] [PubMed] [Google Scholar]
- 18.Jaouadi B, Ellouz-Chaabouni S, Ali MB, Messaoud EB, Naili B, Dhouib A, Bejar S. Excellent laundry detergent compatibility and high dehairing ability of the Bacillus pumilus CBS alkaline proteinase (SAPB) Biotechnol Bioproc E. 2009;14:503–512. doi: 10.1007/s12257-008-0244-8. [DOI] [Google Scholar]
- 19.Nwachukwu ID, Aluko RE. Structural and functional properties of food protein-derived antioxidant peptides. J Food Biochem. 2019;43(1):e12761. doi: 10.1111/jfbc.12761. [DOI] [PubMed] [Google Scholar]
- 20.Lermen AM, Clerici NJ, Daroit DJ. Biochemical properties of a partially purified protease from Bacillus sp. CL18 and its use to obtain bioactive soy protein hydrolysates. Appl Biochem Biotechnol. 2020;192(2):643–664. doi: 10.1007/s12010-020-03355-1. [DOI] [PubMed] [Google Scholar]
- 21.Bertucci JI, Liggieri CS, Colombo ML, Cavalli SEV, Bruno MA. Application of peptidases from Maclura pomifera fruit for the production of active biopeptides from whey protein. LWT Food Sci Technol. 2015;64(1):157–163. doi: 10.1016/j.lwt.2015.05.041. [DOI] [Google Scholar]
- 22.Konrad B, Anna D, Marek S, Marta P, Aleksandra Z, Jozefa C. The evaluation of dipeptidyl peptidase (DPP)-IV, α-glucosidase and angiotensin converting enzyme (ACE) inhibitory activities of whey proteins hydrolyzed with serine protease isolated from Asian pumpkin (Cucurbita ficifolia) Int J Rept Res Ther. 2014;20:483–491. doi: 10.1007/s10989-014-9413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drejer EB, Hakvag S, Irla M, Brautaset T. Genetic tools and techniques for recombinant expression in thermophilic bacillaceae. Microorganisms. 2018;6(2):1–19. doi: 10.3390/microorganisms6020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke Y, Yuan XM, Li JS, Zhou W, Huang XH, Wang T. High-level expression, purification, and enzymatic characterization of a recombinant Aspergillus sojae alkaline protease in Pichia pastoris. Protein Expr Purif. 2018;148:24–29. doi: 10.1016/j.pep.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Abbasi-Hosseini SM, Eftekhar F, Yakhchali B, Minai-Tehrani D. Cloning and enhanced expression of an extracellular alkaline protease from a soil isolate of Bacillus clausii in Bacillus subtilis. Iran J Biotechnol. 2011;9(40):275–280. [Google Scholar]
- 26.Bhatt HB, Singh SP. Cloning, expression, and structural elucidation of a biotechnologically potential alkaline serine protease from a newly isolated haloalkaliphilic Bacillus lehensis JO-26. Front Microbiol. 2020;11:941. doi: 10.3389/fmicb.2020.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimenes NC, Silveira E, Tambourgi EB. An overview of proteases: production, downstream processes and industrial applications. Sep Purif Rev. 2019;00:1–21. doi: 10.1080/15422119.2019.1677249. [DOI] [Google Scholar]
- 28.Sareen R, Bornscheuer UT, Mishra P. Cloning, functional expression and characterization of an alkaline protease from Bacillus licheniformis. Biotechnol Lett. 2005;27(23–24):1901–1907. doi: 10.1007/s10529-005-3901-4. [DOI] [PubMed] [Google Scholar]
- 29.Chen XJ, Zhou C, Xue YF, Shi JS, Ma YH. Cloning, expression, and characterization of an alkaline protease, AprV, from Vibrio sp. DA1-1. Bioprocess Biosyst Eng. 2018;41:1437–1447. doi: 10.1007/s00449-018-1972-6. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Zhou Y, Huang Y, Wei QS, Huang HY, Guo CB. Cloning and expression of a thermostable keratinase gene from Thermoactinomyces sp. YT06 in Escherichia coli and characterization of purified recombinant enzymes. World J Microbiol Biotechnol. 2019;35:135. doi: 10.1007/s11274-019-2710-1. [DOI] [PubMed] [Google Scholar]
- 31.Wani AH, Sharma M, Salwan R, Singh G, Chahota R, Verma S. Cloning, expression, and functional characterization of serine protease Aprv2 from virulent isolate Dichelobacter nodosus of Indian origin. Appl Biochem Biotechnol. 2016;180:576–587. doi: 10.1007/s12010-016-2117-5. [DOI] [PubMed] [Google Scholar]
- 32.Zheng LF, Yu XY, Wei CH, Qiu LY, Yu CW, Xing Q, Fan YW, Deng ZY. Production and characterization of a novel alkaline protease from a newly isolated Neurospora crassa through solid-state fermentation. LWT Food Sci Technol. 2020;122:108990. doi: 10.1016/j.lwt.2019.108990. [DOI] [Google Scholar]
- 33.Verbauwhede AE, Lambrecht MA, Fierens E, Hermans S, Shegay O, Brijs K, Delcour JA. Thermo-reversible inhibition makes aqualysin 1 from Thermus aquaticus a potent tool for studying the contribution of the wheat gluten network to the crumb texture of fresh bread. Food Chem. 2018;264:118–125. doi: 10.1016/j.foodchem.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Cui YL, Luo LL, Wang X, Lu YY, Yi YL, Shan YY, Liu BF, Zhou Y, Lv X. Mining, heterologous expression, purification, antibactericidal mechanism, and application of bacteriocins: a review. Comp Rev Food Sci Food Safety. 2020. 10.1111/1541-4337.12658. [DOI] [PubMed]
- 35.Liu BH, Zhang J, Gu L, Du GC, Chen J, Liao XR. Comparative analysis of bacterial expression systems for keratinase production. Appl Biochem Biotechnol. 2014;173:1222–1235. doi: 10.1007/s12010-014-0925-z. [DOI] [PubMed] [Google Scholar]
- 36.Pandey S, Ghosh PK, Ghosh S, De TK, Maiti TK. Role of heavy metal resistant Ochrobactrum sp. and Bacillus spp. strains in bioremediation of a rice cultivar and their PGPR like activities. J Microbiol. 2013;51(1):11–17. doi: 10.1007/s12275-013-2330-7. [DOI] [PubMed] [Google Scholar]
- 37.Li F, Yang L, Lv X, Liu D, Xia H, Chen S. Purification and characterization of a novel extracellular alkaline protease from Cellulomonas bogoriensis. Protein Expr Purif. 2016;121:125–132. doi: 10.1016/j.pep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Hajji M, Hmidet N, Jellouli K, Vallaeys T, Nasri M, Sellami-Kamoun A. Gene cloning and expression of a detergent stable alkaline protease from Aspergillus clavatus ES1. Process Biochem. 2010;45:1746–1752. doi: 10.1016/j.procbio.2010.07.005. [DOI] [Google Scholar]
- 39.Kannan K, Jasra RV. Immobilization of alkaline serine endopeptidase from Bacillus licheniformis on SBA-15 and MCF by surface covalent binding. J Mol Catal B. 2009;56(1):34–40. doi: 10.1016/j.molcatb.2008.04.007. [DOI] [Google Scholar]
- 40.Tarrahimofrad H, Meimandipour A, Arjmand S, Nassiri MB, Jahangirian E, Tavana H, Zamani J, Rahimnahal S, Aminzadeh S. Structural and biochemical characterization of a novel thermophilic Coh01147 protease. PLoS One. 2020;15(6):e0234958. doi: 10.1371/journal.pone.0234958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barros RM, Malcata FX. A kinetic model for hydrolysis of whey proteins by cardosin a extracted from Cynara cardunculus. Food Chem. 2004;88(3):351–359. doi: 10.1016/j.foodchem.2004.01.046. [DOI] [Google Scholar]
- 42.Sarmadi B, Ismail A. Antioxidative peptides from food proteins: a review. Peptides. 2010;31(10):1949–1956. doi: 10.1016/j.peptides.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 43.Shu GW, Huang J, Bao CJ, Meng JP, Chen H, Cao JL. Effect of different proteases on the degree of hydrolysis and angiotensin I-converting enzyme-inhibitory activity in goat and cow milk. Biomolecules. 2018;8(4):101. doi: 10.3390/biom8040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mechri S, Bouacem K, Zarai Jaouadi N, Rekik H, Berrouina MB, Omrane Benmrad M, Hacene H, Bejar S, Bouanane-Darenfed A, Jaouadi B. Identification of a novel protease from the thermophilic Anoxybacillus kamchatkensis M1V and its application as laundry detergent additive. Extremophiles. 2019;23:687–706. doi: 10.1007/s00792-019-01123-6. [DOI] [PubMed] [Google Scholar]
- 45.Mechri S, Sellem I, Bouacem K, Jabeur F, Chamkha M, Hacene H, Bouanane-Darenfed A, Jaouadi B. Antioxidant and enzyme inhibitory activities of Metapenaeus monoceros by-product hydrolysates elaborated by purified alkaline proteases. Waste Biomass Valor. 2020;11:6741–6755. doi: 10.1007/s12649-020-00942-5. [DOI] [Google Scholar]
- 46.Mechri S, Berrouina MB, Benmarad MO, Jaouadi NZ, Rekik H, Moujehed E, Chebbi A, Sayadi S, Chamkha M, Bejar S, Jaouadi B. Characterization of a novel protease from Aeribacillus pallidus strain VP3 with potential biotechnological interest. Int J Biol Macromol. 2017;94:221–232. doi: 10.1016/j.ijbiomac.2016.09.112. [DOI] [PubMed] [Google Scholar]
- 47.Bessadok B, Masri M, Breuck T, Sadok S. Characterization of the crude alkaline extracellular protease of Yarrowia lipolytica YITun15. J Fish Sci. 2017;11(4):19–24. [Google Scholar]
- 48.Pokora M, Zambrowicz A, Zablocka A, Dabrowska A, Szoltysik M, Babij K, Echert E, Trziszka T, Chrzanowska J. The use of serine protease from Yarrowia lipolytica yeast in the production of biopeptides from denatured egg white proteins. Acta Biochim Pol. 2017;62(2):245–253. doi: 10.18388/abp.2016_1316. [DOI] [PubMed] [Google Scholar]
- 49.Bougatef A, Souissi N, Fakhfakh N, Triki-Ellouz Y, Nasri M. Purification and characterization of trypsin from the viscera of sardine (Sardina pilchardus) Food Chem. 2007;102:343–350. doi: 10.1016/j.foodchem.2006.05.050. [DOI] [Google Scholar]
- 50.Bougatef A, Nedjar-Arroume N, Ravallec-Ple R, Leroy Y, Guillochon D, Barkia A, Nasri M. Angiotensin I-converting enzyme (ACE) inhibitory activities of sardinella (Sardinella aurita) by-products protein hydrolysates obtained by treatment with microbial and visceral fish serine proteases. Food Chem. 2008;111:350–356. doi: 10.1016/j.foodchem.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 51.El Hadj-Ali N, Agrebi R, Ghorbel-Frikha B, Sellami-Kamoun A, Kanoun S, Nasri M. Biochemical and molecular characterization of a detergent stable alkaline seine-protease from a newly isolated Bacillus licheniformis NH1. Enzyme Microb Tech. 2007;40:515–523. doi: 10.1016/j.enzmictec.2006.05.007. [DOI] [Google Scholar]
- 52.Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL. Georgenia ruanii sp nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol. 2007;57:1424–1428. doi: 10.1099/ijs.0.64749-0. [DOI] [PubMed] [Google Scholar]
- 53.Sun Q, Zhang B, Yan QJ, Jiang ZQ. Comparative analysis on the distribution of protease activities among fruits and vegetable sources. Food Chem. 2016;213:708–713. doi: 10.1016/j.foodchem.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 54.Lineweaver H, Burk D. The determination of enzyme dissociation constant. J Am Chem Sci. 1934;56:665–666. [Google Scholar]
- 55.Church FC, Swaisgood HE, Porter DH, Catignani GL. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J Dairy Sci. 1983;66(6):1219–1227. doi: 10.3168/jds.S0022-0302(83)81926-2. [DOI] [Google Scholar]
- 56.Cushman DW, Cheung HS. Spectrophotometric assay and properties of angiotensin-converting enzyme of rabbit lung. Biochem Pharmacol. 1971;20(7):1637–1648. doi: 10.1016/0006-2952(71)90292-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data of the alkaline serine protease gene (GsProS8) from G. stearothermophilus is presented on the NCBI database (GenBank accession number MW084977). The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.