Abstract
Water pollution is a major global environmental problem, and it poses a great environmental risk to public health and biological diversity. This work is motivated by assessing the potential environmental threat of coal mining through increased sulfate concentrations in river networks, which do not belong to any simple parametric distribution. However, existing network models mainly focus on binary or discrete networks and weighted networks with known parametric weight distributions. We propose a principled nonparametric weighted network model based on exponential-family random graph models and local likelihood estimation, and study its model-based clustering with application to large-scale water pollution network analysis. We do not require any parametric distribution assumption on network weights. The proposed method greatly extends the methodology and applicability of statistical network models. Furthermore, it is scalable to large and complex networks in large-scale environmental studies. The power of our proposed methods is demonstrated in simulation studies and a real application to sulfate pollution network analysis in Ohio watershed located in Pennsylvania, United States.
Keywords: Exponential-Family Random Graphical Model, Local Likelihood, Variational Inference, Environmental Studies
1. Introduction
Water pollution is the leading cause of deaths and diseases, and it is a major global problem. It is known that nearly 80% of the world’s population lives in areas exposed to high levels of threat to water security (Vörösmarty et al., 2010). The recent national report on water quality by EPA (2017) pointed out that 46% of rivers, 21% of lakes, 18% of coastal waters, and 32% of wetlands in the United States are in poor biological condition or rated poor based on a water quality index. The major pollutant sources include agriculture, atmospheric deposition, construction, industrial production, municipal sewage, resource extraction, spills, and urban runoff. They pose severe health hazards like cancer, cardiovascular, respiratory, neurologic, and developmental damage. This work is motivated by assessing the potential environmental threat of coal mining in Ohio watershed of Pennsylvania through increased sulfate concentrations in the surface water, which is an important scientific problem in geoscience. Bernhardt et al. (2012) mapped surface coal mining of southern West Virginia and linked these maps with water quality and biological data of 223 streams. When these coal mines occupy > 5.4% of their contributing watershed area, the sulfate concentrations within catchments could exceed 50 mg/L (Niu et al., 2017). Residential proximity to heavy coal production is associated with higher risk for cardiopulmonary disease, chronic lung disease, hypertension, and kidney disease (Hendryx and Ahern, 2008). The study of water-quality risks will help the whole society manage them now and in future.
With advances in data collection, there are more and more modern statistical research on environmental studies using nonparametric regression, causal inference, mixture model, network analysis, and variable selection, for instance, Ebenstein (2012); Liang et al. (2015); Li et al. (2017); Lin et al. (2017); Wen et al. (2019) among many others. Especially, network analysis becomes increasingly important in large-scale environmental studies and geoscientific research to assess environmental impacts and risks for water pollution (Smith et al., 1987; Lienert et al., 2013; Ruzol et al., 2017). For example,, Gianessi and Peskin (1981) proposed a water network model to explore the impact of cropland sediment controls on improved water quality, and Montgomery (1972) and Anastasiadis et al. (2016) studies weighted pollution networks where the weights measure the pollution diminishing transition. However, none of aforementioned network models took into account the spatial heterogeneity and the hub structure of river networks. Without exploring spatial heterogeneity, these models could fail to differentiate polluted regions from less polluted but well connected regions in river networks. The hubs in river networks usually determine the flow of pollutants, and they may help identify polluted and well connected regions.
In this work, we introduce a principled model-based clustering of networks to effectively deal with the spatial heterogeneity of river pollution and efficiently identify the hub structure in river networks. Model-based clustering of networks based on stochastic block models (SBMs) and exponential-family random graph models (ERGMs) have received considerable attention in recent literature, including Snijders and Nowicki (1997); Nowicki and Snijders (2001); Girvan and Newman (2002); Airoldi et al. (2008); Karrer and Newman (2011); Zhao et al. (2012); Vu et al. (2013); Saldana et al. (2017); Wang and Bickel (2017); Lee et al. (2017), among many others. It is worth pointing out that existing research mainly focuses on the model-based clustering of networks with binary or discrete edges. In a recent paper by Ambroise and Matias (2012), parametric distributions are incorporated into a stochastic block model to model continuous network weights. Alternatively, Bayesian variational methods have been proposed by Aicher et al. (2014) to approximate posterior distribution of weights over latent block structures. However, from our motivating example, sulfate concentrations in the surface river network do not belong to any simple parametric distribution, which will be illustrated in Section 5. Thus, we need to relax the parametric assumption in network models to account for the unknown distribution of continuous network weights. To address this issue, we propose a new nonparametric weighted network model based on ERGMs and local likelihood estimation, and study its model-based clustering with application to large-scale water pollution network analysis. The proposed method greatly extends the methodology and applicability of statistical network models. Furthermore, it is scalable to large and complex networks in real-world applications.
The rest of this paper is organized as follows. Section 2 presents the methodology of our proposed nonparametric weighted network model. In Section 3, we introduce a novel variational expectation-maximization algorithm to solve the approximate maximum likelihood estimation. Section 4 demonstrates the numerical performances of our proposed methods and algorithms in simulation studies. In Section 5, we apply the proposed method to analyze the large-scale water pollution network of sulfate concentrations in the Ohio watershed of Pennsylvania.
2. Methodology
We define some necessary notation before presenting our proposed method. Let n be the number of nodes in the observed network. Let Y = (Yij )1≤i, j≤n be the corresponding weighted network such that Yij = (Eij,Wij) where Eij is a binary indicator denoting the existence of an edge in the network and Wij is the corresponding weight when Eij = 1. The weight matrix W = (Wij)1≤i,j≤n, consists of continuous weights in the network. Further we assume that the edge distribution of each network belongs to an exponential family (Besag, 1974; Frank and Strauss, 1986). We write the distribution of edge indicator matrix E as
| (1) |
where is the log of the normalizing constant, θ ∈ ℝp are canonical network parameters of interest and is the sufficient statistic. Here is the space for E consisting of 2n possible binary edge structures.
One of the major limitations of this binary network model is that it can not deal with large number of nodes due to large computational time for evaluating the likelihood function. For undirected networks, this computing time scales with node size as exp((n(n−1)log2)/2). Many estimation algorithms have been developed (Snijders, 2002; Hunter and Handcock, 2006; Møller et al., 2006; Koskinen et al., 2010; Caimo and Friel, 2011), however most of them are time-consuming and therefore unrealistic for fitting large networks. This issue of non-scalability can be resolved by the assumption of dyadic independence which assumes that all dyads are independent of each other. Note that dyad is a general term applicable for both directed and undirected edges. In the undirected weighted networks, it would imply the ties and weights are independent of each other and for all pairs of nodes, i.e.
| (2) |
This assumption facilitates both estimation and simulation of large networks as well as solves the issue of degeneracy (Strauss, 1986; Handcock et al., 2003; Schweinberger, 2011; Krivitsky, 2012). However, dyadic independence is too restrictive and most models following this assumption are either very trivial, failing to capture relational dependencies (Gilbert, 1959; Erdős and Rényi, 1959) or non-parsimonious, with a large number of parameters (Holland and Leinhardt, 1981).
We consider a model-based clustering framework to relax the dyadic independence. More specifically, we introduce the finite K-component mixture form together with a much less restrictive assumption of conditional dyadic independence (CDI) (Snijders and Nowicki, 1997; Nowicki and Snijders, 2001; Girvan and Newman, 2002; Vu et al., 2013). Under this assumption, we propose the nonparametric weighted network as
| (3) |
where Z = (Zi)1≤i≤n denote the latent cluster memberships of nodes. Here, Zi is a K × 1 vector such that zik = 1 if and only if node i lies in cluster k, otherwise zik = 0. The conditional dyadic independence strikes in a nice balance between model complexity and parsimony for model-based clustering. The number of parameters are reduced from to , thus enabling simple inter and intra community interpretations. The CDI assumption induces a block structure similar to SBMs. SBMs have been thoroughly studied in the context of social networks (Holland et al., 1983; Airoldi et al., 2008) and have a long history in multiple scientific communities (Bui et al., 1987; Dyer and Frieze, 1989; Bollobás et al., 2007).
We omit discussion of SBMs here except to point out a major difference from our current setup of ERGM. ERGMs can allow several kinds of dynamic network statistics like density, stability, transitivity (Hanneke et al., 2010), thus effectively generalizing the simple density case in SBM methodology which makes them appealing in practice. To effectively model continuous weights, for any given pair of nodes (i, j), we have
| (4) |
Here, take the parametric specification of exponential-family distributions of the network statistics as explained in (1) and network weights are assumed to be an i.i.d. sample observed from a population following an univariate nonparametric density function fkl. Note that (fkl)1≤k,l≤K do not necessarily have any parametric form. We implicitly assume that conditioning on the full Z = z is same as conditioning on just zi and zj, i.e. Eij and Wij depend on Z only via zi and zj. For ease of presentation, we assume the additive structure of pkl to be parameterized by network sparsity parameters θ as pkl = logit−1(θk + θl ). It is worth pointing out that the clusters z1,⋯,zn are determined by two sources of information in network models. On the one hand, the random block structure and also the additive structure of (pkl)1≤k,l≤K contribute to the exploration of different degrees among the clusters. On the other hand, the nonparametric network weights modulate the separation between clusters with different weight distributions.
Combining (3) and (4), the corresponding log-likelihood function given the cluster memberships can be written as:
| (5) |
where we also introduce the penalty term . Thus, (5) can be treated as a likelihood for any non-negative function fkl without imposing the additional constraint . This specification follows the similar spirit of Loader (1996).
Now we derive the localized version of the conditional log-likelihood evaluated at an arbirary grid point w as:
| (6) |
where Kh is the rescaled kernel function with a positive bandwidth h. We approximate log(fkl(u)) by , a linear combination of orthogonal basis functions (ϕm)1≤m≤p, namely, . With this approximation, the local conditional log-likelihood becomes
| (7) |
We assume that membership indicators Z = (Zi)1≤i≤n follow a multinomial distribution with a single trial and mixture proportions as π = (πk)1≤k≤K. The log-likelihood of the observed weighted network can be written as
| (8) |
In view of (5) and (7), we maximize the log-likelihood function (8) of the observed network described to estimate model parameters θ together with block densities f.
Remark 1. Allman et al. (2011) proved the identifiability of parameters for a broad class of parametric or nonparametric weighted network models. As shown in Section 4 of Allman et al. (2011), we can uniquely identify the parameters under mild conditions for parametric weighted networks while the identifiability result for nonparametric weighted networks depend on binning the values of the edge variables into a finite set. Please see Theorem 15 of Allman et al. (2011) for more details about the identifiability.
Remark 2. The proposed nonparametric weighted network model can be further extended to discrete temporally evolving weighted networks. Given the dynamic network series Y = (Yt)1≤t≤T = (Et, Wt)1≤t≤T, the dynamic nonparametric weighted network model can be derived by assuming a discrete-time Markov structure over the time (Hanneke et al., 2010; Krivitsky and Handcock, 2014; Kim et al., 2018), that is
where Pθ,f (Yt = yt | Yt−1 = yt−1, Z = z) follows the similar specification of (3) and (4). We can incorporate dynamic network statistics such as stability and transitivity in the exponential-family specification of . Hence, the interpretation of clusters will reflect the impact from dynamic network statistics. For example, the use of stability network statistic will contribute to the exploration of different levels of stability among the clusters. The dynamic nonparametric weighted network is beyond our current scope and we will study it in the future.
3. Computation
This section proposes a variational expectation maximization (EM) algorithm to approximately solve the maximum likelihood estimation. It is infeasible to directly maximize the log-likelihood function (8) due to two key challenges: (i) exponential-family form of P(Y | Z) is not scalable for large networks; (ii) the sum is over every possible assignment to Z, where for each 1≤ i ≤ n, Zi = zi can take one of K possible values.
To resolve the first challenge, the CDI assumption (3) plays a crucial role. Typically parameters in a mixture model are estimated using the classical EM algorithm (Dempster et al., 1977). The E-step proceeds by writing the complete data log-likelihood (9), assuming the network is observed while node membership indicators Z are unobserved.
| (9) |
Next we take expectation of this complete log-likelihood with the distribution ℙθ(Z | Y). Clearly the distribution ℙθ(Z | Y) = ℙθ(Zi,1≤i≤n | Y) can’t be further factored over nodes since for i ≠ j, Zi is not independent of Zj given the observed network Y. This poses a huge computational challenge. The intractability of the complete log-likelihood motivates the use of variational approximation. Variational methods are well studied in literature (Blei et al., 2017). The basic idea is to posit a tractable auxiliary distribution Aγ (z)≡ P(Z = z) for the latent variables Z and find the optimal setting for variational parameters γ that minimizes the Kullback-Liebler divergence between the approximation and true distribution. We use this auxiliary distribution to construct a tractable lower bound of the log-likelihood using Jensen’s inequality and then maximize this lower bound, yielding approximate maximum likelihood estimates.
| (10) |
The derivation in (10) uses an auxiliary distribution and also Jensen’s inequality. We choose the variational distribution A(Z) from the mean-field family as,
| (11) |
where ∀ i ∈{1,…, n}, k ∈{1,…, K}, we have 0 ≤ γik ≤ 1 with the constraint and . This class of probability distributions Aγ considers independent laws through different nodal memberships. With the definition of A(Z) in (11), we derive the following effective lower bound (ELBO).
| (12) |
Variational E-step: we maximize ELBO (θ, γ, π, f ) in (12) to obtain γ(t):
| (13) |
The direct maximization of ELBO in (13) is difficult, since the lower bound depends on the products γikγjl and, therefore the fixed-point updates of γik depend on all other γjl (Daudin et al., 2008). To separate the parameters in this maximization problem, we adopt an MM algorithm that involves constructing a surrogate (minorizing) function and optimizing it iteratively (Hunter and Lange, 2004). The surrogate function Q must satisfy the following properties to qualify as a valid minorizing function.
| (14) |
| (15) |
First we note that for all (θkl)1≤k≤l≤K we have log(pkl ) < 0 and log(1 − pkl ) < 0 which gives rise to following inequalties using the arithmetic geometric mean inequality:
| (16) |
| (17) |
with equality if and . Also the concavity of the logarithm function gives rise to following inequality (Vu et al., 2013)
| (18) |
We construct the surrogate function that satisfies (14) and (15) using the inequalities (16), (17) and (18), thus guaranteeing the ascent property of ELBO.
| (19) |
To maximize (19), we solve n separate quadratic programming problems of K variables γi under the constraints γik ≥ 0, ∀ k ∈{1,…, K} together with .
M-step: we first maximize the (12) with respect to π and θ. We have a closed-form update for π as . To update θ, we maximize (12) using the modified Newton-Raphson method with line search to guarantee the ascent property (Dennis Jr and Schnabel, 1996). Now, it remains to update block densities f. To this end, we use (6) and the approximation of log(fkl(.)). For simplicity, we use a local polynomial approximation such that log(fkl(.)) can be approximated by a low-degree polynomial ζkl in a neighborhood of the fitting point w (Loader, 1996): . With this approximation, we rewrite (7) as
and then derive the corresponding ELBO as
| (20) |
We maximize (20) over a sequence of grid points to estimate the block densities fkl.
Remark 3. We note that, the estimated densities in the M-step directly affect the optimization over variational parameters γ, which can seen by the ELBO constructed after the variational approximation. Given that cluster memberships are usually estimated by the hard clustering over these parameters, weights also affect the clusters.
4. Simulation Studies
In this section, we conduct simulation studies to examine our proposed non-parametric methods. The general procedure that we adopt to simulate entails the following steps:
First we simulate the membership indicators for all nodes from multinomial distribution with parameter vector π corresponding to uniform mixture proportions.
We simulate the binary adjacency matrix by simulating dyads in the static network given the cluster membership indicators of nodes. While simulating these dyads we use the network parameters with two settings and . The first setting corresponds to well separated clusters on the basis of density of edges while second setting considers the more extreme case when the clusters are relatively close.
For each node pair with an edge, we simulate the weight on that edge using true distribution with block parameter that depends on their cluster memberships.
We consider two distributions separately: Normal and Gamma. For space consideration, we include all results about Gamma distributions in the supplementary file.
We compare three different model-based clustering methods in each simulation, which are based on binary ERGM, proposed nonparametric weighted ERGM and “oracle” parametric weighted ERGM (Desmarais and Cranmer, 2012) with the correct specification of weight distributions. We consider different node sizes from 100 to 500 and 100 repetitions. Before proceeding, we introduce several average metrics to measure clustering and model parameters estimation performance for different simulation settings over 100 replications. First, to assess the clustering performance, we calculate the log of Rand Index (logRI). The measure calculates the proportion of pairs whose estimated labels correspond to the true labels in terms of being assigned to the same or different groups (Rand, 1971). We calculate logRI as,
Next, to assess the performance of the estimators of network parameters θ, we consider log of square root of the average squared error (logRASE),
To assess the performance of the density estimation f, we consider the Kolmogorov-Smirnov (KS) statistic,
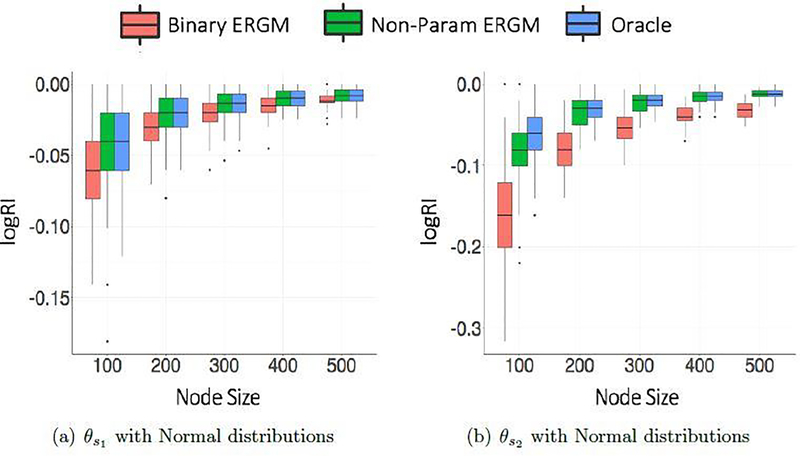
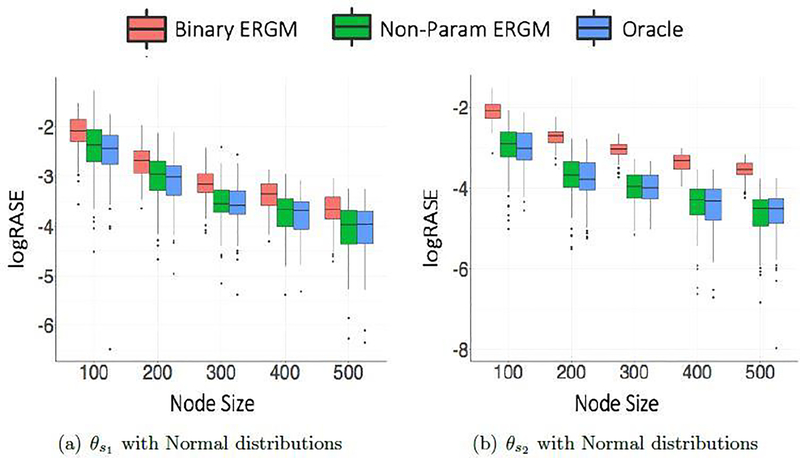
Based on the metrics defined here, Figures 1 and 2 show clustering and θ estimation performance for different sparsity parameter settings averaged over 100 simulations of graphs for normal weight distributions. Corresponding figures for Gamma weight distributions look similar and have been moved to Appendix A. The differences in logRI and logRASE for and evidently confirms the expected fact that separating two very close clusters is difficult compared to well separated clusters. It appears that the both the distributions allow a reasonable recovery of the cluster membership indicators, when the graphs considered have more than 100 nodes. As expected, the node size improves the recovery of latent structure and estimation of network parameters θ in all cases. It can be observed that our proposed nonparametric ERGM outperforms the binary ERGM by a large difference and performs competitively with the oracle method (parametric ERGM with true weight distributions) for all simulation settings. We note that our proposed strategy is best suited for real world applications when the true distributions for block pairs are unknown.
Fig. 1.
Clustering Performance measured using logRI against different node sizes comparing the three models for different sparsity parameter settings under Normal weight distributions
Fig. 2.
θ Estimation Performance measured using logRASE against different node sizes comparing the three models for different sparsity parameter settings under Normal weight distributions
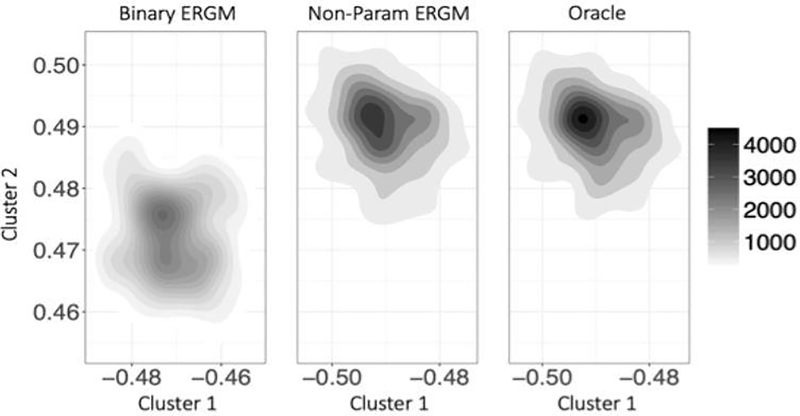
Figures 3 and 4 show the empirical distributions of network parameters θ for normal weight distributions. Corresponding figures for Gamma weight distributions have been moved to Appendix A. The proposed non-parametric model estimation again outperforms the binary ERGM uniformly for all settings. We also note that the contour plots for the proposed model seem really close to Oracle model, thus demonstrating the power of our approach.
Fig. 3.
Plots of empirical joint distributions of network parameters for Normal weight distributions over 100 simulations with 500 nodes, comparing the three models for different block distributions
Fig. 4.
Plots of empirical joint distributions of network parameters for Normal weight distributions over 100 simulations with 500 nodes, comparing the three models for different block distributions
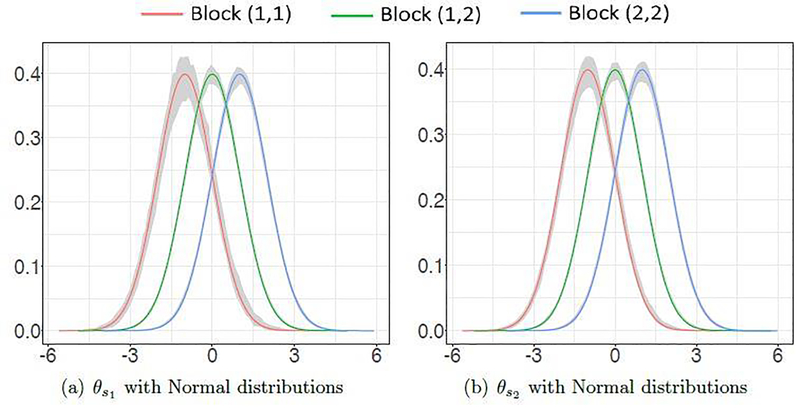
Figure 5 shows the estimated block densities within 2.5 and 97.5 percentiles for node size of 500 for normal weight distributions. Corresponding figure for Gamma weight distributions have been moved to Appendix A. Comparing and , it is evident that within cluster 1 density estimation improves substantially for . This is because cluster 1 is more sparse for compared to . Comparing the normal and gamma distributions, we observe that asymmetry of gamma distribution leads to underestimation of within cluster 1 estimated density. Cluster 1 is again most affected since it is most sparse. We point out here that for larger node sizes, these estimated densities will converge to true densities (Loader, 1996).
Fig. 5.
Estimated block densities for normal weight distributions.
Table 1 gives the summary of KS statistic for various simulation settings. We note that for sparse cluster 1, comparing and , there is a huge improvement when the true distribution is Normal. However the improvement is only minor in case of Gamma due to asymmetry. The differences are much less substantial for other blocks, however Normal uniformly outperforms Gamma for all settings.
Table 1.
Summary of KS statistic (×102) for the three block densities under different simulation settings for the proposed model, computed over 100 simulations of graphs with 500 nodes
| Summary Statistic | Normal | Gamma | Normal | Gamma | |
|---|---|---|---|---|---|
| Block (1,1) | Median | 3.94 | 4.51 | 2.77 | 4.26 |
| Mean | 4.03 | 4.70 | 2.84 | 4.36 | |
| Block (1,2) | Median | 1.40 | 1.52 | 1.46 | 1.57 |
| Mean | 1.41 | 1.56 | 1.49 | 1.61 | |
| Block (2,2) | Median | 1.51 | 1.53 | 1.63 | 1.76 |
| Mean | 1.53 | 1.60 | 1.66 | 1.76 | |
5. Application to Water Pollution Analysis
In this section, we demonstrate the power of our methodology in an environmental application to study sulfate concentrations in river networks. The dataset consists of three main parts. The first part consists of approximately 865 sulfate samples measured as concentrations in the units of parts per million (ppm) over several creeks in the Ohio watershed in Pennsylvania. The source include the following online databases: the USGS National Water Information System, the Susquehanna River Basin Commission database, the EPA STORET Data Warehouse, and the Shale Network database (doi:10.4211/his-data-shalenetwork). The second part consists of the directed geographical river network in the form of locations of all streams and creeks in Pennsylvania. The directions in the network correspond to the actual river flow. The third part consists of 93 coal mine locations which are suspected to be potential polluters of the river streams posing an environmental risk. Both the latter parts are publically available at Pennsylvania Spatial Data Access (PASDA). We map the latitude and longitude of the sampling sites onto the geographical river network. The 865 sulfate sampling sites become the nodes. Thus the nodes in the network are defined through first part of the data after mapping to nearest streams in the second part. We define the edges according to the path of river flow; for sampling sites A and B, the edge is present when river can flow either from A to B or from B to A directly or through one of its sub-tributary or some intermediary stream. The path information used to define the edges entirely comes from the second part. The spatial weights are constructed as proportional to the strength of influence of the upstream site on the downstream site, measured by difference in concentrations between the sampling locations (Peterson et al., 2007). For example if the river is flowing from A to B, with measured sulfate concentrations CA and CB, then weight on the edge between A and B would be defined as wAB = CB − CA. Note in this definition, the weights could be negative if CB < CA. In this context, we have transformed the geographically defined ‘river network’ to a weighted network defined above for the purpose of our analysis.
It is an important question to study the water pollution in river networks. Most of the existing spatial clustering methods rely on some “neighbourhood” metric that cluster the data points based on their spatial proximity. However, these approaches fail in a river network setup when the two points are very close spatially but still not connected by the river flow or vice-versa. These methods usually rely on some criteria to choose the number of clusters that is heuristic and not rigorously founded on model likelihood. There have been several parametric approaches to study spatial concentration gradients e.g. see Lawson and Denison (2002). However most methods fail in water pollution applications where the gradients tend to be asymmetric, heavy tailed and multimodal, with an unknown number of modes. We adopt skewness, kurtosis and Hartigan’s dip test of unimodality respectively to infer these properties over the whole sulfate network and individual clusters obtained using our model. The results, summarized in Table 2, clearly indicate that the gradients follow an asymmetric, leptokurtic and multimodal distribution for all block pairs. This calls for a generalized framework of clustering the weighted network while modeling the concentration gradients without making any distributional assumptions.
Table 2.
Distributional properties of sulfate concentration gradients
| Skewness | Kurtosis | Hartigan’s dip test p value | |
|---|---|---|---|
| Whole network | 0.050 | 3.885 | < 2.2 × 10−16 |
| Block (1,1) | 0.762 | 6.817 | < 2.2 × 10−16 |
| Block (1,2) | −0.011 | 3.447 | 9.37 × 10−6 |
| Block (2,2) | 0.026 | 3.672 | < 2.2 × 10−16 |
The proposed nonparametric weighted network models address all the aforementioned challenges. Now, we apply the proposed method to analyze the sulfate concentration network. In practice, we need to effectively choose the number of clusters (i.e., K). Since the likelihood is intractable (Biernacki et al., 2000), we follow (Daudin et al., 2008) to introduce a modified Integrated Classification Likelihood (ICL) criterion:
| (21) |
where the complete log-likelihood with estimated membership and densities becomes
The ICL follows the philosophy of Bayesian model selection criterion. The second term in (21) penalizes for the K − 1 free parameters in the mixture proportions π. The third term accounts for the penalization of the network parameters (Matias and Miele, 2016) given the additive structure of the specified ERGM.
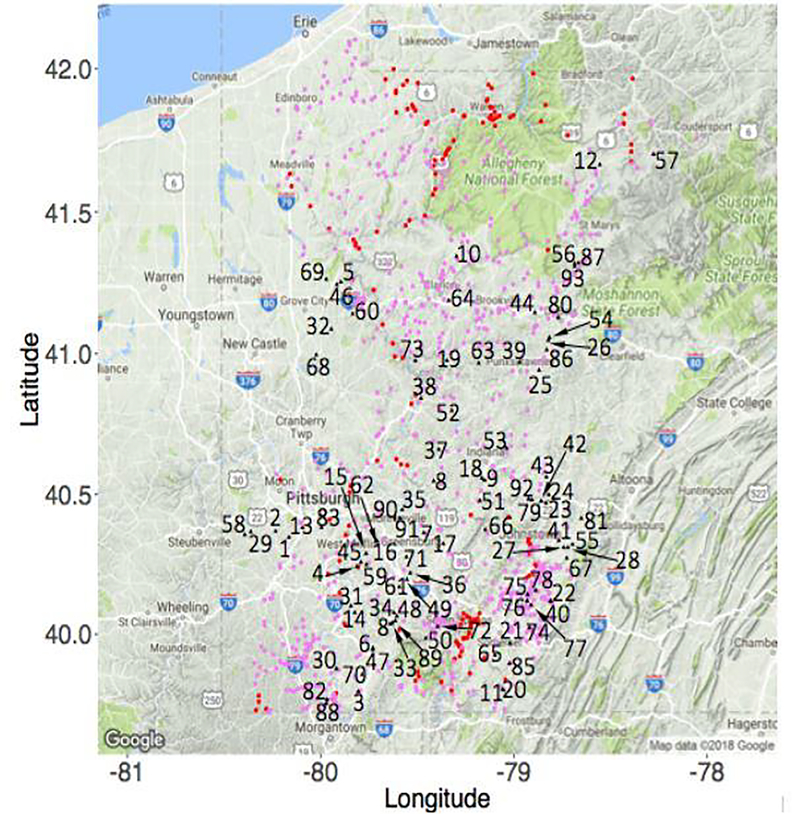
We choose the optimal number of clusters by maximizing the modified ICL criterion, which suggests K = 2. The estimated network sparsity parameters corresponding to these two clusters labelled C1 and C2 are −0.521 and −2.084 respectively indicating that C1 has higher degree in terms of the edges on average compared to C2. We plot the sampling sites belonging to the two clusters overlaying the potential polluter locations in Figure 6. It can be seen that coal mines 4, 5, 15, 20, 22, 38, 40, 42, 45, 46 73, 75 – 78, 82 and 88 lie directly either upstream or downstream of nodes in C1 suggesting they may significantly affect the sulfate concentrations. As show in Table 3, C1 consists of relatively more hubs with higher degree on average while C2 consists more nodes with lower degree on average.
Fig. 6.
Clustered sulfate sampling sites with C1 in red & C2 in pink and coal mining sites in black (numbered 1–93)
Table 3.
Summary of nodes, degree and edge weights of the two clusters.
| Summary | C1 | C2 |
|---|---|---|
| Number of nodes | 147 | 718 |
| Average Degree | 18.72 | 5.21 |
| Minimum | −385.50 | −389.70 |
| 1st quantile | −2.00 | −35.99 |
| Median | 8.081 | 10.24 |
| Mean | 40.41 | 23.66 |
| 3rd quantile | 66.96 | 111.80 |
| Maximum | 436.40 | 441.00 |
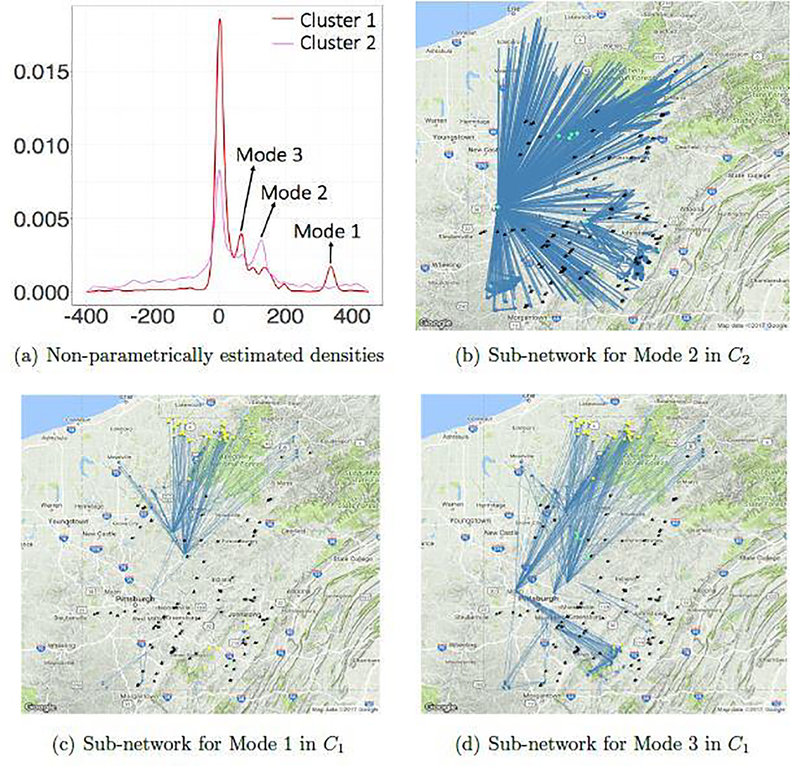
In Figure 7(a), we plot densities estimated for these clusters. From this density plot, it is clear that C1 has two modes (Mode 1 and Mode 3) and C2 has one mode (Mode 2) on the positive sulfate concentrations. Based on these modes and estimated network parameters, we identify sub-regions of interest corresponding to 3 modes:
Fig. 7.
Non-parametrically estimated densities for ties within clusters and subnetworks for different modes
Mode 1 consists of sub-regions with high degrees and higher differences of sulfate concentrations among adjacent nodes;
Mode 2 consists of sparse sub-regions with low degrees and moderate differences of sulfate concentrations among adjacent nodes;
Mode 3 consists of dense sub-regions with high degrees and low differences of sulfate concentrations among adjacent nodes.
The modes on positive weights indicate that the pollutant’s concentration increases downstream, thus pointing towards polluters making significant impact on the environment. We analyze three modes by plotting their corresponding sub-networks in Figure 7 and summarizing the descriptive statistics of edge weights in Table 4.
Table 4.
Descriptive statistics of edge weights of the three modes
| Summary | Mode 1 | Mode 2 | Mode 3 |
|---|---|---|---|
| Minimum | 303.9 | 90.18 | 42.33 |
| 1st quantile | 328.5 | 111.70 | 56.86 |
| Median | 334.60 | 126.80 | 65.46 |
| Mean | 334.80 | 128.30 | 65.42 |
| 3rd quantile | 341.9 | 140.00 | 70.92 |
| Maximum | 365.8 | 186.20 | 90.02 |
Figures 7(c) and 7(d) gives sub-networks for the positive modes in C1. We identify coal mine ids 10, 19, 64 and 73 directly lying on highly weighted edges around Mode 2 and emanating from the hubs. We define the hubs as nodes with an outlier degree. The outlier degree is defined in the usual sense of outliers, i.e. any degree which is greater than Q3 + 1.5 × IQR, where Q3 is the upper quartile of the degree distribution and IQR is the interquartile range. These nodes correspond to sampling sites usually located at junctions of several streams. The coal mines and hubs are marked black and green respectively. Since these coal mines belong to Mode 1, they are most likely to cause serious water pollution and must be investigated in a prioritized manner. Next we identify coal mine ids 2, 4, 5, 15, 32, 36, 38, 45, 49, 59 and 68 in Mode 3. These mines emanate from nodes that are connected densely to other nodes and so the affected region is larger, even though the pollution is relatively low. These mines in Mode 3 cause moderate impact over a large section of river network and must be monitored accordingly. Figure 7(b) shows the sub-network for the only positive mode in C2. This corresponds to Mode 2 with relatively moderate to low local impacts.
We also compare the binary ERGM model with weighted model and observe that binary ERGM’s cluster 1 consists of 108 nodes, a strict subset of 147 nodes of weighted model’s C1. Figures 7(c) and 7(d) shows these difference of nodes by yellow. These indicate that several edges would have been absent in C1 had we used binary model, thus missing significant coal mines that lie in Mode 1. Clearly taking weights into account while clustering helps to differentiate mines between Mode 1 and Mode 3 and hence could uncover important potential polluters.
6. Discussion
We introduce a new nonparametric model-based approach for clustering large-scale weighted networks. The ERGM specification allows the flexibility to incorporate interesting network statistics and the nonparametric density function provides the robustness to study the network weights. We illustrate the power of our proposed method in a real application to study water pollution networks.
In general, our proposed method does not require a parametric specification of network weights and thus it is robust to the model mis-specification, and it can be extended to incorporate the nonparametric mixture functions or parametric constraints (DeSarbo et al., 2017; Lee and Xue, 2018). Like most nonparametric methods, when the sample size is limited, our proposed method may not perform well. The semiparametric extension such as Xue and Zou (2012, 2014) and Fan et al. (2016) seems a promising alternative to the proposed nonparametric method. Moreover, the proposed method could be computationally intensive when the number of nodes is huge. To make the proposed methods scalable, we shall follow the stochastic variational methods (Hoffman et al., 2013) to employ the minibatch sampling scheme.
Supplementary Material
7. Acknowledgement
The authors thank the Editor, an associate editor, and two referees for their constructive comments and suggestions. Amal Agarwal and Lingzhou Xue have been partially supported by the National Institute on Drug Abuse grant P50DA039838 and the National Science Foundation grants DMS-1505256 and DMS-1811552.
References
- Aicher C, Jacobs AZ, and Clauset A (2014). Learning latent block structure in weighted networks. Journal of Complex Networks, 3(2):221–248. [Google Scholar]
- Airoldi EM, Blei DM, Fienberg SE, and Xing EP (2008). Mixed membership stochastic blockmodels. Journal of Machine Learning Research, 9(Sep):1981–2014. [PMC free article] [PubMed] [Google Scholar]
- Allman ES, Matias C, and Rhodes JA (2011). Parameter identifiability in a class of random graph mixture models. Journal of Statistical Planning and Inference, 141(5):1719–1736. [Google Scholar]
- Ambroise C and Matias C (2012). New consistent and asymptotically normal parameter estimates for random-graph mixture models. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 74(1):3–35. [Google Scholar]
- Anastasiadis E, Deng X, Krysta P, Li M, Qiao H, and Zhang J (2016). Network pollution games. In Proceedings of the 2016 International Conference on Autonomous Agents & Multiagent Systems, pages 23–31. International Foundation for Autonomous Agents and Multiagent Systems. [Google Scholar]
- Bernhardt ES, Lutz BD, King RS, Fay JP, Carter CE, Helton AM, Campagna D, and Amos J (2012). How many mountains can we mine? Assessing the regional degradation of central Appalachian rivers by surface coal mining. Environmental Science & Technology, 46(15):8115–8122. [DOI] [PubMed] [Google Scholar]
- Besag J (1974). Spatial interaction and the statistical analysis of lattice systems. Journal of the Royal Statistical Society. Series B (Methodological), 36(2):192–236. [Google Scholar]
- Biernacki C, Celeux G, and Govaert G (2000). Assessing a mixture model for clustering with the integrated completed likelihood. IEEE Transactions on Pattern Analysis and Machine Intelligence, 22(7):719–725. [Google Scholar]
- Blei DM, Kucukelbir A, and McAuliffe JD (2017). Variational inference: A review for statisticians. Journal of the American Statistical Association, 112(518):859–877. [Google Scholar]
- Bollobás B, Janson S, and Riordan O (2007). The phase transition in inhomogeneous random graphs. Random Structures & Algorithms, 31(1):3–122. [Google Scholar]
- Bui TN, Chaudhuri S, Leighton FT, and Sipser M (1987). Graph bisection algorithms with good average case behavior. Combinatorica, 7(2):171–191. [Google Scholar]
- Caimo A and Friel N (2011). Bayesian inference for exponential random graph models. Social Networks, 33(1):41–55. [Google Scholar]
- Chang J (2015). lda: Collapsed Gibbs Sampling Methods for Topic Model. R package, version ≥ 2.10.
- Daudin J-J, Picard F, and Robin S (2008). A mixture model for random graphs. Statistics and Computing, 18(2):173–183. [Google Scholar]
- Dempster AP, Laird NM, and Rubin DB (1977). Maximum likelihood from incomplete data via the em algorithm. Journal of the Royal Statistical Society. Series B (Methodological), 39(1):1–38. [Google Scholar]
- Dennis JE Jr and Schnabel RB (1996). Numerical Methods for Unconstrained Optimization and Nonlinear Equations. SIAM, Philadelphia. [Google Scholar]
- DeSarbo WS, Chen Q, and Blank AS (2017). A parametric constrained segmentation methodology for application in sport marketing. Customer Needs and Solutions, 4(4):37–55. [Google Scholar]
- Desmarais BA and Cranmer SJ (2012). Statistical inference for valued-edge networks: The generalized exponential random graph model. PloS ONE, 7(1):e30136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer ME and Frieze AM (1989). The solution of some random NP-hard problems in polynomial expected time. Journal of Algorithms, 10(4):451–489. [Google Scholar]
- Ebenstein A (2012). The consequences of industrialization: evidence from water pollution and digestive cancers in china. Review of Economics and Statistics, 94(1):186–201. [Google Scholar]
- EPA, E. P. A. (2017). 2017 National water quality inventory report to Congress. Washington, D.C.: United States. Technical report. [Google Scholar]
- Erdős P and Rényi A (1959). On random graphs i. Publicationes Mathematicae, Debrecen, 6:290–297. [Google Scholar]
- Fan J, Xue L and Zou H (2016). Multitask quantile regression under the transnormal model. Journal of the American Statistical Association, 111(516):1726–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank O and Strauss D (1986). Markov graphs. Journal of the American Statistical Association, 81(395):832–842. [Google Scholar]
- Gianessi LP and Peskin HM (1981). Analysis of national water pollution control policies: 2. agricultural sediment control. Water Resources Research, 17(4):803–821. [Google Scholar]
- Gilbert EN (1959). Random graphs. The Annals of Mathematical Statistics, 30(4):1141–1144. [Google Scholar]
- Girvan M and Newman ME (2002). Community structure in social and biological networks. Proceedings of the National Academy of Sciences, 99(12):7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handcock MS, Robins G, Snijders TA, Moody J, and Besag J (2003). Assessing degeneracy in statistical models of social networks. Technical report, Citeseer. [Google Scholar]
- Hanneke S, Fu W, and Xing EP (2010). Discrete temporal models of social networks. Electronic Journal of Statistics, 4:585–605. [Google Scholar]
- Hendryx M and Ahern MM (2008). Relations between health indicators and residential proximity to coal mining in West Virginia. American Journal of Public Health, 98(4):669–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MD, Blei DM, Wang C, and Paisley J (2013). Stochastic variational inference. The Journal of Machine Learning Research, 14(1):1303–1347. [Google Scholar]
- Holland PW, Laskey KB, and Leinhardt S (1983). Stochastic blockmodels: First steps. Social Networks, 5(2):109–137. [Google Scholar]
- Holland PW and Leinhardt S (1981). An exponential family of probability distributions for directed graphs. Journal of the American Statistical Association, 76(373):33–50. [Google Scholar]
- Hunter DR and Handcock MS (2006). Inference in curved exponential family models for networks. Journal of Computational and Graphical Statistics, 15(3):565–583. [Google Scholar]
- Hunter DR and Lange K (2004). A tutorial on MM algorithms. The American Statistician, 58(1):30–37. [Google Scholar]
- Karrer B and Newman MEJ (2011). Stochastic blockmodels and community structure in networks. Physical Review E, 83(1):016107. [DOI] [PubMed] [Google Scholar]
- Kim B, Lee KH, Xue L, Niu X, et al. (2018). A review of dynamic network models with latent variables. Statistics Surveys, 12:105–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen JH, Robins GL, and Pattison PE (2010). Analysing exponential random graph (p-star) models with missing data using bayesian data augmentation. Statistical Methodology, 7(3):366–384. [Google Scholar]
- Krivitsky PN (2012). Exponential-family random graph models for valued networks. Electronic Journal of Statistics, 6:1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivitsky PN and Handcock MS (2014). A separable model for dynamic networks. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 76(1):29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson AB and Denison DG (2002). Spatial Cluster Modelling. CRC press, Florida. [Google Scholar]
- Lee KH and Xue L (2018). Nonparametric finite mixture of gaussian graphical models. Technometrics, 60(4):511–521. [Google Scholar]
- Lee KH, Xue L, and Hunter DR (2017). Model-based clustering of time-evolving networks through temporal exponential-family random graph models. arXiv preprint arXiv:1712.07325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zheng G, Agarwal A, Xue L, and Lauvaux T (2017). Discovery of causal time intervals. In Proceedings of the 2017 SIAM International Conference on Data Mining, pages 804–812. SIAM. [Google Scholar]
- Liang X, Zou T, Guo B, Li S, Zhang H, Zhang S, Huang H, and Chen SX (2015). Assessing beijing’s PM2.5 pollution: severity, weather impact, apec and winter heating. Proceedings of the Royal Society A, 471(2182):20150257. [Google Scholar]
- Lienert J, Schnetzer F, and Ingold K (2013). Stakeholder analysis combined with social network analysis provides fine-grained insights into water infrastructure planning processes. Journal of Environmental Management, 125:134–148. [DOI] [PubMed] [Google Scholar]
- Lin N, Jing R, Wang Y, Yonekura E, Fan J, and Xue L (2017). A statistical investigation of the dependence of tropical cyclone intensity change on the surrounding environment. Monthly Weather Review, 145(7):2813–2831. [Google Scholar]
- Loader CR (1996). Local likelihood density estimation. The Annals of Statistics, 24(4):1602–1618. [Google Scholar]
- Matias C and Miele V (2016). Statistical clustering of temporal networks through a dynamic stochastic block model. Journal of the Royal Statistical Society: Series B (Statistical Methodology). [Google Scholar]
- Møller J, Pettitt AN, Reeves R, and Berthelsen KK (2006). An efficient markov chain monte carlo method for distributions with intractable normalising constants. Biometrika, 93(2):451–458. [Google Scholar]
- Montgomery WD (1972). Markets in licenses and efficient pollution control programs. Journal of Economic Theory, 5(3):395–418. [Google Scholar]
- Niu X, Wendt A, Li Z, Agarwal A, Xue L, Gonzales M, and Brantley SL (2017). Detecting the effects of coal mining, acid rain, and natural gas extraction in Appalachian basin streams in Pennsylvania (USA) through analysis of barium and sulfate concentrations. Environmental Geochemistry and Health, pages 1–21. [DOI] [PubMed] [Google Scholar]
- Nowicki K and Snijders TA (2001). Estimation and prediction for stochastic blockstructures. Journal of the American Statistical Association, 96(455):1077–1087. [Google Scholar]
- Peterson EE, Theobald DM, and ver Hoef JM (2007). Geostatistical modelling on stream networks: developing valid covariance matrices based on hydrologic distance and stream flow. Freshwater Biology, 52(2):267–279. [Google Scholar]
- Rand WM (1971). Objective criteria for the evaluation of clustering methods. Journal of the American Statistical Association, 66(336):846–850. [Google Scholar]
- Ruzol C, Banzon-Cabanilla D, Ancog R, and Peralta E (2017). Understanding water pollution management: Evidence and insights from incorporating cultural theory in social network analysis. Global Environmental Change, 45:183–193. [Google Scholar]
- Saldana DF, Yu Y, and Feng Y (2017). How many communities are there? Journal of Computational and Graphical Statistics, 26(1):171–181. [Google Scholar]
- Schweinberger M (2011). Instability, sensitivity, and degeneracy of discrete exponential families. Journal of the American Statistical Association, 106(496):1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Alexander RB, and Wolman MG (1987). Water-quality trends in the nation’s rivers. Science, 235:1607–1616. [DOI] [PubMed] [Google Scholar]
- Snijders TA (2002). Markov chain monte carlo estimation of exponential random graph models. Journal of Social Structure, 3(2):1–40. [Google Scholar]
- Snijders TA and Nowicki K (1997). Estimation and prediction for stochastic blockmodels for graphs with latent block structure. Journal of Classification, 14:75–100. [Google Scholar]
- Strauss D (1986). On a general class of models for interaction. SIAM Review, 28(4):513–527. [Google Scholar]
- Vörösmarty CJ, McIntyre PB, Gessner MO, Dudgeon D, Prusevich A, Green P, Glidden S, Bunn SE, Sullivan CA, and Liermann CR (2010). Global threats to human water security and river biodiversity. Nature, 467(7315):555–561. [DOI] [PubMed] [Google Scholar]
- Vu DQ, Hunter DR, and Schweinberger M (2013). Model-based clustering of large networks. The Annals of Applied Statistics, 7(2):1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YR and Bickel PJ (2017). Likelihood-based model selection for stochastic block models. The Annals of Statistics, 45(2):500–528. [Google Scholar]
- Wen T, Agarwal A, Xue L, Chen A, Herman A, Li Z, and Brantley SL (2019). Assessing changes in groundwater chemistry in landscapes with more than 100 years of oil and gas development. Environmental Science: Processes & Impacts, 21(2):384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L and Zou H (2012). Regularized rank-based estimation of high-dimensional nonparanormal graphical models. The Annals of Statistics, 24(1):2541–2571. [Google Scholar]
- Xue L and Zou H (2014). Rank-based tapering estimation of bandable correlation matrices. Statistica Sinica, 40(5):83–100 [Google Scholar]
- Zhao Y, Levina E, and Zhu J (2012). Consistency of community detection in networks under degree-corrected stochastic block models. The Annals of Statistics, 40(4):2266–2292. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.