Abstract
Deep learning methods exhibit promising performance for predictive modeling in healthcare, but two important challenges remain:
Data insufficiency: Often in healthcare predictive modeling, the sample size is insufficient for deep learning methods to achieve satisfactory results.
Interpretation: The representations learned by deep learning methods should align with medical knowledge.
To address these challenges, we propose GRaph-based Attention Model (GRAM) that supplements electronic health records (EHR) with hierarchical information inherent to medical ontologies. Based on the data volume and the ontology structure, GRAM represents a medical concept as a combination of its ancestors in the ontology via an attention mechanism.
We compared predictive performance (i.e. accuracy, data needs, interpretability) of GRAM to various methods including the recurrent neural network (RNN) in two sequential diagnoses prediction tasks and one heart failure prediction task. Compared to the basic RNN, GRAM achieved 10% higher accuracy for predicting diseases rarely observed in the training data and 3% improved area under the ROC curve for predicting heart failure using an order of magnitude less training data. Additionally, unlike other methods, the medical concept representations learned by GRAM are well aligned with the medical ontology. Finally, GRAM exhibits intuitive attention behaviors by adaptively generalizing to higher level concepts when facing data insufficiency at the lower level concepts.
Keywords: Graph, Attention Model, Predictive Healthcare, Electronic Health Records
1. INTRODUCTION
The rapid growth in volume and diversity of healthcare data from electronic health records (EHR) and other sources is motivating the use of predictive modeling to improve care for individual patients. In particular, novel applications are emerging that use deep learning methods such as word embedding [11, 13], recurrent neural networks (RNN) [7, 9, 10, 25], convolutional neural networks (CNN) [30] or stacked denoising autoencoders (SDA) [6, 29], demonstrating signihcant performance enhancement for diverse prediction tasks. Deep learning models appear to perform signihcantly better than logistic regression or multilayer perceptron (MLP) models that depend, to some degree, on expert feature construction [24, 34].
Training deep learning models typically requires large amounts of data that often cannot be met by a single health system or provider organization. Sub-optimal model performance can be particularly challenging when the focus of interest is predicting onset of a rare disease. For example, using Doctor AI [9], we discovered that RNN alone was ineffective at predicting the onset of diseases such as cerebral degenerations (e.g. Leukodystrophy, Cerebral lipidoses) or developmental disorders (e.g. autistic disorder, Heller’s syndrome). In part, the low incidence of these diseases in the training data provided little learning opportunity to the flexible models like RNN.
Deep learning models require high volumes of data because of the exponential number of feature combinations that must be assessed for the model to learn. The demand for high volume can be reduced by exploiting medical ontologies to encode hierarchical clinical constructs and relationships among medical concepts, effectively reducing the search space without loss of information. Fortunately, there are many well-organized ontologies in healthcare such as the International Classification of Diseases (ICD), Clinical Classifications Software (CCS) [36] or Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) [33]. Nodes (i.e. medical concepts) close to one another in medical ontologies are likely to be associated with similar patients, allowing us to transfer knowledge among them. Use of medical ontologies are likely to be helpful when data volume is insufficient to train deep learning models, and possibly even when data volume is sufficient as a means to improve model parsimony without loss of information and by learning more interpretable representations that are consistent with the ontology structure.
In this work, we propose GRAM, a method that infuses information from medical ontologies into deep learning models via neural attention. Considering the frequency of a medical concept in the EHR data and its ancestors in the ontology, GRAM optimizes the medical concept by adaptively combining its ancestors via attention mechanism (i.e. weighted sum of the representations of ancestors). The attention mechanism is trained in an end-to-end fashion with the neural network model that predicts the onset of disease(s). We also propose an effective initialization technique to better guide the representation learning process.
We compare predictive performance (i.e. accuracy, data needs, interpretability) of GRAM to various models including the recurrent neural network (RNN) in two sequential diagnoses prediction tasks and one heart failure (HF) prediction task. We demonstrate that GRAM is up to 10% more accurate than the basic RNN for predicting diseases less observed in the training data. After discussing GRAM’s scalability, we visualize the representations learned from various models, where GRAM provides more intuitive representations by grouping similar medical concepts close to one another. Finally, we show GRAM’s attention mechanism can be interpreted to understand how it assigns the right amount of attention to the ancestors of each medical concept by considering the data availability and the ontology structure.
2. METHODOLOGY
We first define the notations describing EHR data and medical ontologies, followed by a description of GRAM (Section 2.2), the end-to-end training of the attention generation and predictive modeling (Section 2.3), and the efficient initialization scheme (Section 2.4).
2.1. Basic Notation
We denote the set of entire medical codes from the EHR as with the vocabulary size . The clinical record of each patient can be viewed as a sequence of visits where each visit contains a subset of medical codes can be represented as a binary vector where the i-th element is 1 only if Vt contains the code ci. To avoid clutter, all algorithms will be presented for a single patient.
We assume that a given medical ontology typically expresses the hierarchy of various medical concepts in the form of a parent-child relationship, where the medical codes form the leaf nodes. Ontology is represented as a directed acyclic graph (DAG) whosenodes form a set . The set consists of all non-leaf nodes {i.e. ancestors of the leaf nodes), where represents the number of all non-leaf nodes. We use knowledge DAG to refer to . A parent in the knowledge DAG represents a related but more general concept over its children. Therefore, provides a multi-resolution view of medical concepts with different degrees of specificity. While some ontologies are exclusively expressed as parent-child hierarchies (e.g. ICD-9, CCS), others are not. For example, in some instances SNOMED-CT also links medical concepts to causal or treatment relationships, but a majority of the relationships in SNOMED-CT are still parent-child. Therefore, we focus on the parent-child relationships in this work.
2.2. Knowledge DAG and the Attention Mechanism
GRAM leverages the parent-child relationship of to learn robust representations when data volume is constrained. GRAM balances the use of ontology information in relation to data volume in determining the level of specificity for a medical concept. When a medical concept is less frequent in the data, more weight is given to its ancestors as they can be learned more accurately and offer general (coarse-grained) information about their children. The process of resorting to the parent concepts can be automated via the attention mechanism and the end-to-end training as described in Figure 1.
Figure 1:
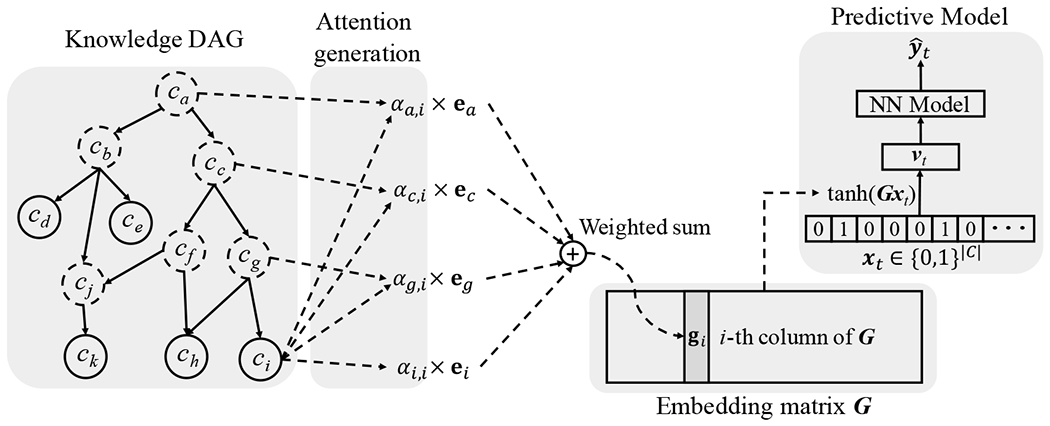
The illustration of GRAM. Leaf nodes (solid circles) represents a medical concept in the EHR, while the non-leaf nodes (dotted circles) represent more general concepts. The final representation gi of the leaf concept ci is computed by combining the basic embeddings ei of ci and eg, ec and ea of its ancestors cg, cc and ca via an attention mechanism. The final representations form the embedding matrix G for all leaf concepts. After that, we use G to embed patient visit vector xt to a visit representation vt, which is then fed to a neural network model to make the final prediction .
In the knowledge DAG, each node ci is assigned a basic embedding vector , where m represents the dimensionality. Then are the basic embeddings of the codes while represent the basic embeddings of the internal nodes . The initialization of these basic embeddings is described in Section 2.4. We formulate a leaf node’s final representation as a convex combination of the basic embeddings of itself and its ancestors:
| (1) |
where denotes the final representation of the code the indices of the code ci and ci’s ancestors, ej, the basic embedding of the code cj and the attention weight on the embedding ej when calculating gi. The attention weight in Eq. (1) is calculated by the following Softmax function,
| (2) |
f(ei, ej) is a scalar value representing the compatibility between the basic embeddings of ei and ek. We compute f(ei, ej) via the following feed-forward network with a single hidden layer (MLP),
| (3) |
where is the weight matrix for the concatenation of ei and ej, the bias vector, and the weight vector for generating the scalar value. The constant l represents the dimension size of the hidden layer of f(·, ·). We concatenate ei and ej in the child-ancestor order. Note that the compatibility function f is an MLP, because MLP is well known to be a sufficient approximator for an arbitrary function, and we empirically found that our formulation performed better in our use cases than alternatives such as inner product and Bahdanau et al.’s [2].
Remarks: The example in Figure 1 is derived based on a single path from ci to ca. However, the same mechanism can be applicable to multiple paths as well. For example, code ck has two paths to the root ca, containing hve ancestors in total. Another scenario is where the EHR data contain both leaf codes and some ancestor codes. We can move those ancestors present in EHR data from the set to and apply the same process as Eq. (1) to obtain the final representations for them.
2.3. End-to-End Training with a Predictive Model
We train the attention mechanism together with a predictive model such that the attention mechanism improves the predictive performance. By concatenating final representation of all medical codes, we have the embedding matrix where gi is the i-th column of G. As shown in the right side of Figure 1, we can convert a visit Vt to a visit representation vt by multiplying the embedding matrix G with a multi-hot (i.e. multi-label binary) vector xt indicating the clinical events in the visit Vt, followed by a nonlinear activation via tanh. Finally the visit representation vt will be used as an input to the neural network model for predicting the target label yt. In this work, we use RNN as the choice of the NN model to perform sequential diagnoses prediction [9, 10]. That is, we are interested in predicting the disease codes of the next visit Vt+1 given the visit records up to the current timestep V1, V2, …, Vt, which can be expressed as follows,
| (4) |
where denotes the multi-hot vector for the t-th visit; the t-th visit representation; the RNN’s hidden layer at the t-th time step (i.e. t-th visit); θr RNN’s parameters; and the weight matrices and the bias vector of the final Softmax function (r denotes the dimension size of the hidden layer). Note that we use Softmax instead of dimension-wise sigmoid for predicting multiple disease codes in the next visit Vt+1 because it showed better performance. Here we use “RNN” to denote any recurrent neural network variants that can cope with the vanishing gradient problem [3], such as LSTM [18], GRU [8], and IRNN [21]. The prediction loss for all time steps is calculated using the binary cross entropy as follows, ***
| (5) |
where we sum the cross entropy errors from all timestamps of , T denotes the number of timestamps of the visit sequence. Note that the above loss is dehned for a single patient. In actual implementation, we will take the average of the individual loss for multiple patients. Algorithm 1 describes the overall GRAM training procedure assuming that we are performing the sequential diagnoses prediction task using an RNN. Note that Algorithm 1 describes stochastic gradient update to avoid clutter, but it can be easily extended to other gradient based optimization such as mini-batch gradient update.
Algorithm 1.
GRAM Optimization
| Randomly initialize basic embedding matrix E, attention parameters ua, Wa, ba, RNN parameter θr, softmax parameters W, b. |
| repeat |
| Update E with GloVe objective function (see Section 2.4) |
| until convergence |
| repeat |
| X ← random patient from dataset |
| for visit Vt in X do |
| for code ci in Vt do |
| Refer to find ci’s ancestors C′ |
| for code cj in C′ do |
| Calculate attention weight αij using Eq. (2). |
| end for |
| Obtain final representation gi; using Eq. (1). |
| end for |
| vt ← tanh(∑i:ci∈Vt gi) |
| Make prediction using Eq. (4) |
| end for |
| Calculate prediction loss using Eq. (5) |
| Update parameters according to the gradient of |
| until convergence |
2.4. Initializing Basic Embeddings
The attention generation mechanism in Section 2.2 requires basic embeddings ei of each node in the knowledge DAG. The basic embeddings of ancestors, however, are not usually observed in the data. To properly initialize them, we use co-occurrence information to learn the basic embeddings of medical codes and their ancestors. Co-occurrence has proven to be an important source of information when learning representations of words or medical concepts [11, 13, 27]. To train the basic embeddings, we employ GloVe [31], which uses the global co-occurrence matrix of words to learn their representations. In our case, the co-occurrence matrix of the codes and the ancestors was generated by counting co-occurrences within each visit Vt, where we then augment each visit with the ancestors of the codes in the visit. We describe the initialization algorithm with an example knowledge DAG of Figure 1. Given a visit Vt,
we augment the leaf codes with their ancestors to obtain the augmented visit ,
where the augmented ancestors are underlined. Note that a single ancestor can appear multiple times in . In fact, the higher the ancestor is in the knowledge DAG, the more times it is likely to appear in . Co-occurrence of two codes in are counted as follows,
where is the number of times the code ci appears in the augmented visit . For example, the co-occurrence between the leaf code ci and the root ca is 3. However, the co-occurrence between the ancestor cc and the root ca is 6. Therefore our algorithm will make the higher ancestor codes more likely to be involved in all medical events (i.e. visits), which is natural in healthcare applications as those general concepts are often reliable. We repeat this calculation for all pairs of codes in all augmented visits of all patients to obtain the co-occurrence matrix depicted by Figure 2. For training the embedding vectors ei’s using M, we minimize the following loss function as described in Pennington et al. [31],
and the hyperparameters xmax and α are respectively set to 100 and 0.75 as the original paper [31]. Note that, after the initialization, the basic embeddings ei’s of both leaf nodes (i.e. medical codes) and non-leaf nodes (i.e. ancestors) are fine-tuned during model training via backpropagation.
Figure 2:
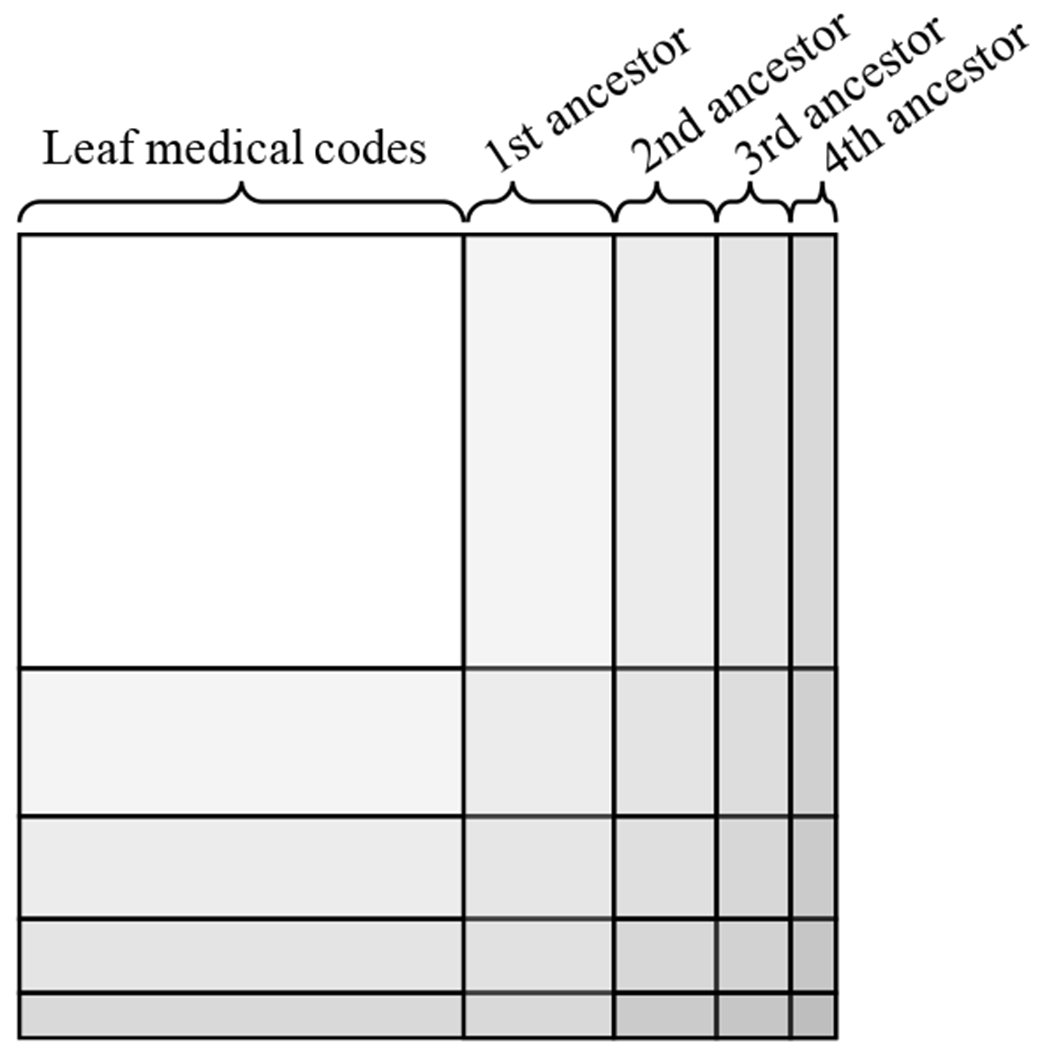
Creating the co-occurrence matrix together with the ancestors. The n-th ancestors are the group of nodes that are n hops away from any leaf node in . Here we exclude the root node, which will be just a single row (column).
3. EXPERIMENTS
We conducted three experiments to determine if GRAM offered superior prediction performance when facing data insufficiency. We first describe the experimental setup followed by results comparing predictive performance of GRAM with various baseline models. Then we present GRAM’s scalability results. Finally, we qualitatively show the intuitive interpretation of GRAM. The source code of GRAM is publicly available at https://github.com/mp2893/gram.
3.1. Experiment Setup
Prediction tasks and source of data:
We conducted two sequential diagnoses prediction (SDP) tasks using two different datasets. The overall aim of the experiments was to use all information prior to a next visit to predict all diagnosis codes that would be in that next visit. The first dataset was from the Sutter Palo Alto Medical Foundation (PAMF), which consisted of 10-years longitudinal medical records of 258K primary care patients between 50 to 89 years of age. This will determine GRAM’s performance for general adult population with many hospital visits. The second dataset was MIMIC-III [15, 19], which is a publicly available dataset consisting of medical records of 7.5K intensive care unit (ICU) patients over 11 years. This will determine GRAM’s performance for high-risk patients with veiy few hospital visits. We utilized all the patients with at least 2 visits. We prepared the true labels yt by grouping the ICD9 codes into 283 groups using CCS single-level diagnosis grouper1. This is to improve the training speed and predictive performance for easier analysis, while preserving sufficient granularity for each diagnosis. Each diagnosis code’s varying frequency in the training data can be viewed as different degrees of data insufficiency. Model performance was assessed by Accuracy@k for each of CCS single-level diagnosis codes such that, given a visit Vt, we get 1 if the target diagnosis is in the top k guesses and 0 otherwise.
We also conducted a heart failure (HF) prediction task, which is a binaiy prediction task for predicting a future HF onset where the prediction is made only once at the last visit xT. The key difference between sequential diagnoses prediction and HF prediction is that the prediction target for the former can already occur in patient’s prior visits while the prediction target for the latter is a new diagnosis of HF that has not appeared before. HF prediction was conducted on Sutter heart failure (HF) cohort, which is a subset of Sutter PAMF data for a heart failure onset prediction study with 3.4K HF cases chosen by a set of criteria described in Gurwitz et al. [17], Vijayakrishnan et al. 39] and 27K matching controls chosen by a set of criteria described in Choi et al. 12]. This will determine GRAM’s performance for a different prediction task where we predict the onset of one specific condition. We randomly downsampled the training data to create different degrees of data insufficiency. We used area under the ROC curve (AUC) to measure the performance. A summary of the datasets are provided in Table 1.We used CCS multi-level diagnoses hierarchy2 as our knowledge DAG . We also tested the ICD9 code hierarchy3, but the performance was similar to using CCS multi-level hierarchy. For all three tasks, we randomly divide the dataset into the training, validation and test set by .75:.10:.15 ratio, and use the validation set to tune the hyper-parameters. Further details regarding the hyper-parameter tuning are provided below. The test set performance is reported in the paper.
Table 1:
Basic statistics of Sutter PAMF, MIMIC-III and Sutter heart failure (HF) cohort.
| Dataset | Sutter PAMF | MIMIC-III | Sutter HF cohort |
|---|---|---|---|
| # of patients | 258,555† | 7,499† | 30,727† (3,408 cases) |
| # of visits | 13,920,759 | 19,911 | 572,551 |
| Avg. # of visits per patient | 53.8 | 2.66 | 38.38 |
| # of unique ICD9 codes | 10,437 | 4,893 | 5,689 |
| Avg. # of codes per visit | 1.98 | 13.1 | 2.06 |
| Max # of codes per visit | 54 | 39 | 29 |
For all datasets, we chose patients who made at least two visits.
Implementation details:
We implemented GRAM with Theano 0.8.2 [38]. For training models, we used Adadelta [45] with a minibatch of 100 patients, on a machine equipped with Intel Xeon E5-2640, 256GB RAM, four Nvidia Titan X’s and CUDA 7.5.
Models for comparison are the following. The first two GRAM+ and GRAM are the proposed methods and the rest are baselines. Hyper-parameter tuning is configured so that the number of parameters for the baselines would be comparable to GRAM’s. Further details are provided below.
GRAM: Input sequence x1, …, xT is first transformed by the embedding matrix , then fed to the GRU with a single hidden layer, which in turn makes the prediction, as described by Eq. (4). The basic embeddings ei’s are randomly initialized.
GRAM+: We use the same setup as GRAM, but the basic embeddings ei’s are initialized according to Section 2.4.
RandomDAG: We use the same setup as GRAM, but each leaf concept has five randomly assigned ancestors from the CCS multi-level hierarchy to test the effect of correct domain knowledge.
RNN: Input xt is transformed by an embedding matrix , then fed to the GRU with a single hidden layer. The embedding size k is a hyper-parameter. Wemb is randomly initialized and trained together with the GRU.
RNN+: We use the RNN model with the same setup as before, but we initialize the embedding matrix Wemb with GloVe vectors trained only with the co-occurrence of leaf concepts. This is to compare GRAM with a similar weight initialization technique.
SimpleRollUp: We use the RNN model with the same setup as before. But for input xt, we replace all diagnosis codes with their direct parent codes in the CCS multi-level hierarchy, giving us 578, 526 and 517 input codes respectively for Sutter data, MIMIC-III and Sutter HF cohort. This is to compare the performance of GRAM with a common grouping technique.
RollUpRare: We use the RNN model with the same setup as before, but we replace any diagnosis code whose frequency is less than a certain threshold in the dataset with its direct parent. We set the threshold to 100 for Sutter data and Sutter HF cohort, and 10 for MIMIC-III, giving us 4,408, 935 and 1,538 input codes respectively for Sutter data, MIMIC-III and Sutter HF cohort. This is an intuitive way of dealing with infrequent medical codes.
Hyper-parameter Tuning:
We define five hyper-parameters for GRAM:
dimensionality m of the basic embedding ei: [100, 200, 300, 400, 500]
dimensionality r of the RNN hidden layer ht from Eq. (4): [100, 200, 300, 400, 500]
dimensionality l of Wa and ba from Eq. (3): [100, 200, 300, 400, 500]
L2 regularization coefficient for all weights except RNN weights: [0.1, 0.01, 0.001, 0.0001]
dropout rate for the dropout on the RNN hidden layer: [0.0, 0.2, 0.4, 0.6, 0.8]
We performed 100 iterations of the random search by using the above ranges for each of the three prediction experiments. In order to fairly compare the model performances, we matched the number of model parameters to be similar for all baseline methods. To facilitate reproducibility, final hyper-parameter settings we used for all models for each prediction experiments are described at the source code repository, https://github.com/mp2893/gram, along with the detailed steps we used to tune the hyper-parameters.
3.2. Prediction performance
Tables 2a and 2b show the sequential diagnoses prediction performance on Sutter data and MIMIC-III. Both tables show that GRAM+ outperforms other models when predicting labels with significant data insufficiency (i.e. less observed in the training data).The performance gain is greater for MIMIC-III, where GRAM+ outperforms the basic RNN by 10% in the 20th-40th percentile range. This seems to come from the fact that MIMIC patients on average have significantly shorter visit history than Sutter patients, with much more codes received per visit. Such short sequences make it difficult for the RNN to learn and predict diagnoses sequence. The performance difference between GRAM+ and GRAM suggests that our proposed initialization scheme of the basic embeddings ei is important for sequential diagnosis prediction.
Table 2:
Performance of three prediction tasks. The x-axis of (a) and (b) represents the labels grouped by the percentile of their frequencies in the training data in non-decreasing order. 0-20 are the most rare diagnosese while 80-100 are the most common ones. (b) uses Accuracy@20 because MIMIC-III has a large average number of codes per visit (see Table 1). For (c), we vary the size of the training data to train the models.
| (a) Accuracy@5 of sequential diagnoses prediction on Sutter data | |||||
|---|---|---|---|---|---|
| Model | 0-20 | 20-40 | 40-60 | 60-80 | 80-100 |
| GRAM+ | 0.0150 | 0.3242 | 0.4325 | 0.4238 | 0.4903 |
| GRAM | 0.0042 | 0.2987 | 0.4224 | 0.4193 | 0.4895 |
| RandomDAG | 0.0050 | 0.2700 | 0.4010 | 0.4059 | 0.4853 |
| RNN+ | 0.0069 | 0.2742 | 0.4140 | 0.4212 | 0.4959 |
| RNN | 0.0080 | 0.2691 | 0.4134 | 0.4227 | 0.4951 |
| SimpleRollUp | 0.0085 | 0.3078 | 0.4369 | 0.4330 | 0.4924 |
| RollUpRare | 0.0062 | 0.2768 | 0.4176 | 0.4226 | 0.4956 |
| (b) Accuracy@20 of sequential diagnoses prediction on MIMIC-III | |||||
|---|---|---|---|---|---|
| Model | 0-20 | 20-40 | 40-60 | 60-80 | 80-100 |
| GRAM+ | 0.0672 | 0.1787 | 0.2644 | 0.2490 | 0.6267 |
| GRAM | 0.0556 | 0.1016 | 0.1935 | 0.2296 | 0.6363 |
| RandomDAG | 0.0329 | 0.0708 | 0.1346 | 0.1512 | 0.4494 |
| RNN+ | 0.0454 | 0.0843 | 0.2080 | 0.2494 | 0.6239 |
| RNN | 0.0454 | 0.0731 | 0.1804 | 0.2371 | 0.6243 |
| SimpleRollUp | 0.0578 | 0.1328 | 0.2455 | 0.2667 | 0.6387 |
| RollUpRare | 0.0454 | 0.0653 | 0.1843 | 0.2364 | 0.6277 |
| (c) AUC of HF onset prediction on Sutter HF cohort | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model | 10% | 20% | 30% | 40% | 50% | 60% | 70% | 80% | 90% | 100% |
| GRAM+ | 0.7970 | 0.8223 | 0.8307 | 0.8332 | 0.8389 | 0.8404 | 0.8452 | 0.8456 | 0.8447 | 0.8448 |
| GRAM | 0.7981 | 0.8217 | 0.8340 | 0.8332 | 0.8372 | 0.8377 | 0.8440 | 0.8431 | 0.8430 | 0.8447 |
| RandomDAG | 0.7644 | 0.7882 | 0.7986 | 0.8070 | 0.8143 | 0.8185 | 0.8274 | 0.8312 | 0.8254 | 0.8226 |
| RNN+ | 0.7930 | 0.8117 | 0.8162 | 0.8215 | 0.8261 | 0.8333 | 0.8343 | 0.8353 | 0.8345 | 0.8335 |
| RNN | 0.7811 | 0.7942 | 0.8066 | 0.8111 | 0.8156 | 0.8207 | 0.8258 | 0.8278 | 0.8297 | 0.8314 |
| SimpleRollUp | 0.7799 | 0.8022 | 0.8108 | 0.8133 | 0.8177 | 0.8207 | 0.8223 | 0.8272 | 0.8269 | 0.8258 |
| RollUpRare | 0.7830 | 0.8067 | 0.8064 | 0.8119 | 0.8211 | 0.8202 | 0.8262 | 0.8296 | 0.8307 | 0.8291 |
Table 2c shows the HF prediction performance on Sutter HF cohort. GRAM and GRAM+ consistently outperforms other baselines (except RNN+) by 3~4% AUC, and RNN+ by maximum 1.8% AUC. These differences are quite significant given that the AUC is already in the mid-80s, a high value for HF prediction, cf. [12]. Note that, for GRAM+ and RNN+, we used the downsampled training data to initialize the basic embeddings ei’s and the embedding matrix Wemb with GloVe, respectively. The result shows that the initialization scheme of the basic embeddings in GRAM+ gives limited improvement over GRAM. This stems from the different natures of the two prediction tasks. While the goal of HF prediction is to predict a binary label for the entire visit sequence, the goal of sequential diagnosis prediction is to predict the co-occurring diagnosis codes at every visit. Therefore the co-occurrence information infused by the initialized embedding scheme is more beneficial to sequential diagnosis prediction. Additionally, this benefit is associated with the natures of the two prediction tasks than the datasets used for the prediction tasks. Because the initialized embedding shows different degrees of improvement as shown by Tables 2a and 2c, when Sutter HF cohort is a subset of Sutter PAMF, thus having similar characteristics.
Overall, GRAM showed superior predictive performance under data insufficiency in three different experiments, demonstrating its general applicability in clinical predictive modeling.
3.3. Scalability
We briehy discuss the scalability of GRAM by comparing its training time to RNN’s. Table 3 shows the number of seconds taken for the two models to train for a single epoch for each predictive modeling task. GRAM+ and RNN+ showed the similar behavior as GRAM and RNN. GRAM takes approximately 50% more time to train for a single epoch for all prediction tasks. This stems from calculating attention weights and the final representations gi for all medical codes. GRAM also generally takes about 50% more epochs to reach to the model with the lowest validation loss. This is due to optimizing an extra MLP model that generates the attention weights. Overall, use of GRAM adds a manageable amount of overhead in training time to the plain RNN.
Table 3:
Scalability result in per epoch training time in second (the number of epochs needed). SDP stands for Sequential Diagnoses Prediction
| Model | SDP (Sutter data) |
SDP (MIMIC-III) |
HF prediction (Sutter HF cohort) |
|---|---|---|---|
| GRAM | 525s (39 epochs) | 2s (11 epochs) | 12s (7 epochs) |
| RNN | 352s (24 epochs) | 1s (6 epochs) | 8s (5 epochs) |
3.4. Qualitative evaluation of interpretable representations
To qualitatively assess the interpretability, we generates the t-SNE plots [26] using the final representations gi of 2,000 randomly chosen diseases learned by GRAM+ for sequential diagnoses prediction on Sutter data4 (Figure 3a). The color of the dots represents the highest disease categories and the text annotations represent the detailed disease categories in CCS multi-level hierarchy. For comparison, we also show the t-SNE plots on the strongest results from GRAM (Figure 3b), RNN+ (Figure 3c), RNN (Figure 3d) and RandomDAG (Figure 3e). GloVe (Figure 3f) and Skip-gram (Figure 3g) were trained on the Sutter data, where a single visit Vt was used as the context window to calculate the co-occurrence of codes.
Figure 3:
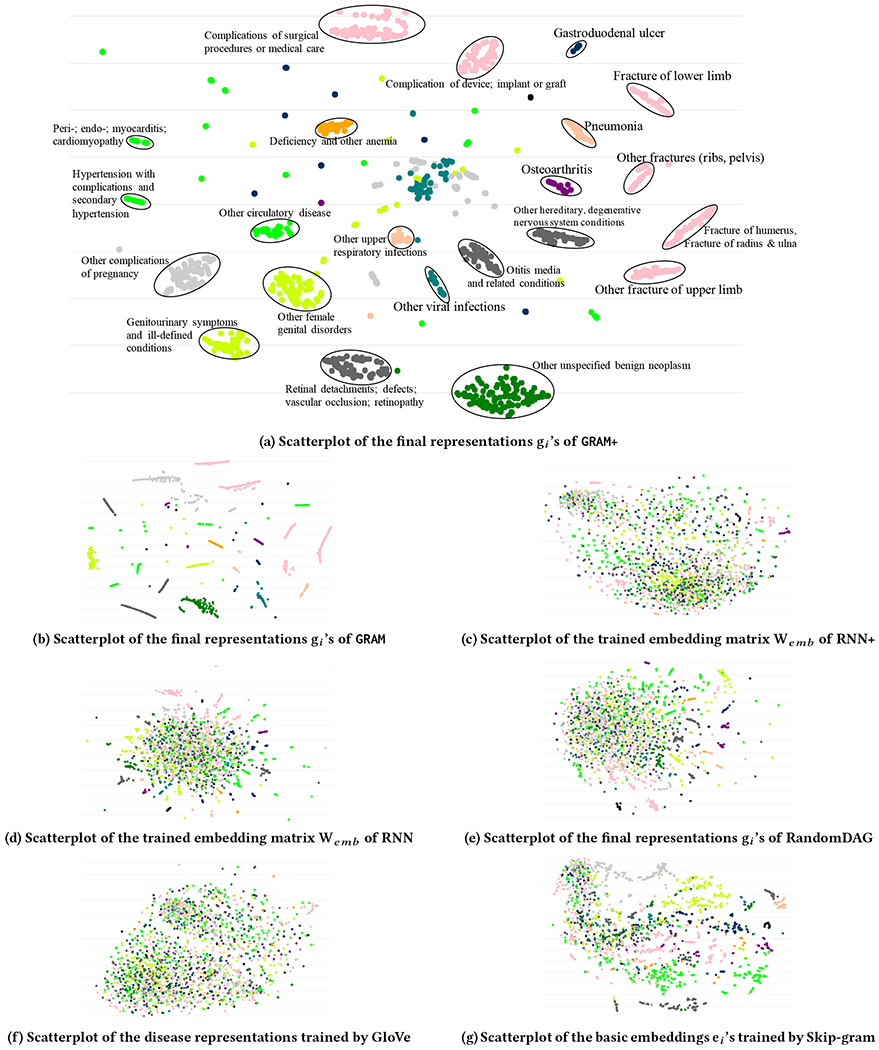
t-SNE scatterplots of medical concepts trained by GRAM+, GRAM, RNN+, RNN, RandomDAG, GloVe and Skip-gram. The color of the dots represents the highest disease categories and the text annotations represent the detailed disease categories in CCS multi-level hierarchy. It is clear that GRAM+ and GRAM exhibit interpretable embedding that are well aligned with the medical ontology.
Figures 3c and 3f conhrm that interpretable representations cannot simply be learned only by co-occurrence or supervised prediction without medical knowledge. GRAM+ and GRAM learn interpretable disease representations that are signihcantly more consistent with the given knowledge DAG . Based on the prediction performance shown by Table 2, and the fact that the representations gi’s are the final product of GRAM, we can infer that such medically meaningful representations are necessary for predictive models to cope with data insufficiency and make more accurate predictions. Figure 3b shows that the quality of the final representations gi of GRAM is quite similar to GRAM+. Compared to other baselines, GRAM demonstrates signihcantly more structured representations that align well with the given knowledge DAG. It is interesting that Skip-gram shows the most structured representation among all baselines. We used GloVe to initialize the basic embeddings ei in this work because it uses global co-occurrence information and its training time is fast as it only depends on the total number of unique concepts . Skip-gram’s training time, on the other hand, depends on both the number of patients and the number of visits each patient made, which makes the algorithm generally slower than GloVe. An interactive visualization tool can be accessed at http://www.sunlab.org/research/gram-graph-based-attention-model/.
3.5. Analysis of the attention behavior
Next we show that GRAM’s attention can be explained intuitively based on the data availability and knowledge DAG’s structure when performing a prediction task. Using Eq. (1), we can calculate the attention weights of individual disease. Figure 4 shows the attention behaviors of four representative diseases when performing HF prediction on Sutter HF cohort.
Figure 4:
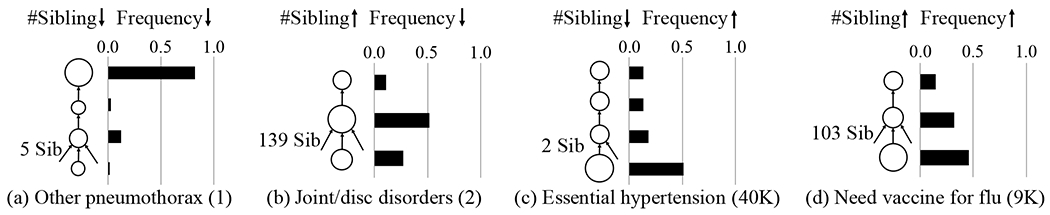
GRAM’s attention behavior during HF prediction for four representative diseases (each column). In each figure, the leaf node represents the disease and upper nodes are its ancestors. The size of the node shows the amount of attention it receives, which is also shown by the bar charts. The number in the parenthesis next to the disease is its frequency in the training data. We exclude the root of the knowledge DAG from all figures as it did not play a significant role.
Other pneumothorax (ICD9 512.89) in Figure 4a is rarely observed in the data and has only five siblings. In this case, most information is derived from the highest ancestor. Temporomandibular joint disorders & articular disc disorder (ICD9 524.63) in Figure 4b is rarely observed but has 139 siblings. In this case, its parent receives a stronger attention because it aggregates sufficient samples from all of its children to learn a more accurate representation. Note that the disease itself also receives a stronger attention to facilitate easier distinction from its large number of siblings.
Unspecified essential hypertension (ICD9 401.9) in Figure 4c is very frequently observed but has only two siblings. In this case, GRAM assigns a very strong attention to the leaf, which is logical because the more you observe a disease, the stronger your confidence becomes. Need for prophylactic vaccination and inoculation against influenza (ICD9 V04.81) in Figure 4d is quite frequently observed and also has 103 siblings. The attention behavior in this case is quite similar to the case with fewer siblings (Figure 4b) with a slight attention shift towards the leaf concept as more observations lead to higher confidence.
4. RELATED WORK
The attention mechanism is a general framework for neural network learning [2], and has been since used in many areas such as speech recognition [14], computer vision [1,43] andhealthcare [10]. However, no one has designed attention model based on knowledge ontology, which is the focus of this work.
There are related works in learning the representations of graphs. Several studies focused on learning the representations of graph vertices by using the neighbor information. DeepWalk [32] and node2vec [16] use random walk while FINE [37] uses breadth-first search to find the neighbors of a vertex and learn its representation based on the neighbor information. Graph convolutional approaches [20, 44] also focus on learning the vertex representations to mainly perform vertex classification. All those works focus on solving the graph data problems whereas GRAM focuses on solving clinical predictive modeling problems using the knowledge DAG as supplementary information.
Several researchers tried to model the knowledge DAG such as WordNet [28] or Freebase [4] where two entities are connected with various types of relation, forming a set of triples. They aim to project entities and relations [5, 23, 35, 40] to the latent space based on the triples or additional information such as hierarchy of entities [42]. These works demonstrated tasks such as link prediction, triple classification or entity classification using the learned representations. More recently, Li et al. [22] learned the representations of words and Wikipedia categories by utilizing the hierarchy of Wikipedia categories. GRAM is fundamentally different from the above studies in that it aims to design intuitive attention mechanism on the knowledge DAG as a knowledge prior to cope with data insufficiency and learn medically interpretable representations to make accurate predictions.
A classical approach for incorporating side information in the predictive models is to use graph Laplacian regularization [6, 41]. However, using this approach is not straightforward as it relies on the appropriate definition of distance on graphs which is often unavailable.
5. CONCLUSION
Data insufficiency, either due to less common diseases or small datasets, is one of the key hurdles in healthcare analytics, especially when we apply deep neural networks models. To overcome this challenge, we leveraged the knowledge DAG, which provides a multi-resolution view of medical concepts. We proposed GRAM, a graph-based attention model using both a knowledge DAG and EHR to learn an accurate and interpretable representations for medical concepts. GRAM chooses a weighted average of ancestors of a medical concept and train the entire process with a predictive model in an end-to-end fashion. We conducted three predictive modeling experiments on real EHR datasets and showed signihcant improvement in the prediction performance, especially on low-frequency diseases and small datasets. Analysis of the attention behavior provided intuitive insight of GRAM. Although GRAM showed good performance, there is room for improving the way we incorporate knowledge DAG into neural networks. For future work, we plan to devise a method to systematically leverage knowledge DAG in addition to using attention-weighted embeddings.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation, award IIS-#1418511 and CCF-#1533768, Children’s Healthcare of Atlanta, Google Faculty Award, UCB and Samsung Scholarship. Dr. Stewart was supported by NIH RO1 HL116832. Dr. Song was supported in part by NSF IIS-1218749, NIH BIGDATA 1R01GM108341, NSF CAREER IIS-1350983, NSF IIS-1639792 EAGER, ONR N00014-15-1-2340, Nvidia and Intel.
Footnotes
The scatterplots of models trained for sequential diagnoses prediction on MIMIC-III and HF prediction for Sutter HF cohort were similar but less structured due to smaller data size.
REFERENCES
- [1].Ba Jimmy, Mnih Volodymyr, and Kavukcuoglu Koray. 2014. Multiple object recognition with visual attention. arXiv:1412.7755 (2014). [Google Scholar]
- [2].Bahdanau Dzmitry, Cho Kyunghyun, and Bengio Yoshua. 2014. Neural Machine Translation by Jointly Learning to Align and Translate. arXiv:1409.0473 (2014). [Google Scholar]
- [3].Bengio Yoshua, Simard Patrice, and Frasconi Paolo. 1994. Learning long-term dependencies with gradient descent is difficult. IEEE Transactions on Neural Networks 5, 2 (1994). [DOI] [PubMed] [Google Scholar]
- [4].Bollacker Kurt, Evans Colin, Paritosh Praveen, Sturge Tim, and Taylor Jamie. 2008. Freebase: a collaboratively created graph database for structuring human knowledge. In SIGMOD. [Google Scholar]
- [5].Bordes Antoine, Usunier Nicolas, Garcia-Duran Alberto, Weston Jason and Yakhnenko Oksana. 2013. Translating embeddings for modeling multi-relational data. In NIPS. [Google Scholar]
- [6].Che Zhengping, Kale David, Li Wenzhe, Mohammad Taha Bahadori, and Yan Liu. 2015. Deep Computational Phenotyping. In SIGKDD. [Google Scholar]
- [7].Che Zhengping, Purushotham Sanjay, Cho Kyunghyun, Sontag David, and Liu Yan. 2016. Recurrent Neural Networks for Multivariate Time Series with Missing Values. arXiv:1606.01865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cho Kyunghyun, Van Merriënboer Bart, Gulcehre Caglar, Bahdanau Dzmitry, Bougares Fethi, Schwenk Holger, and Bengio Yoshua. 2014. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In EMNLP. [Google Scholar]
- [9].Choi Edward, Bahadori Mohammad Taha, Schuetz Andy, Stewart Walter F., and Sun Jimeng. 2016. Doctor AI: Predicting Clinical Events via Recurrent Neural Networks. In MLHC. [PMC free article] [PubMed] [Google Scholar]
- [10].Choi Edward, Bahadori Mohammad Taha, Schuetz Andy, Stewart Walter F., and Sun Jimeng. 2016. RETAIN: Interpretable Predictive Model in Healthcare using Reverse Time Attention Mechanism. In NIPS. [Google Scholar]
- [11].Choi Edward, Bahadori Mohammad Taha, Searles Elizabeth, Coffey Catherine, Thompson Michael, Bost James, Javier T Sojo, and Jimeng Sun. 2016. Multi-layer Representation Learning for Medical Concepts. In SIGKDD. [Google Scholar]
- [12].Choi Edward, Schuetz Andy, Walter F Stewart, and Jimeng Sun. 2016. Using Recurrent Neural Network Models for Early Detection of Heart Failure Onset. JAMIA (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Choi Youngduck, Chiu Chill Yi-I, and David Sontag. 2016. Learning Low-Dimensional Representations of Medical Concepts. (2016). AMIA CRI. [PMC free article] [PubMed] [Google Scholar]
- [14].Chorowski Jan, Bahdanau Dzmitry, Cho Kyunghyun, and Bengio Yoshua. 2014. End-to-end continuous speech recognition using attention-based recurrent NN: First results. arXiv:1412.1602 (2014). [Google Scholar]
- [15].Goldberger Ary and others. 2000. Physiobank, physiotoolkit, and physionet components of a new research resource for complex physiologic signals. Circulation (2000). [DOI] [PubMed] [Google Scholar]
- [16].Grover Aditya and Leskovec Jure. 2016. Node2Vec: Scalable Feature Learning for Networks. In SIGKDD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gurwitz Jerry, Magid David, Smith David, Goldberg Robert, McManus David, Allen Larry, Saczynski Jane, Thorp Micah, Hsu Grace, Sue Hee Sung, and others. 2013. Contemporary prevalence and correlates of incident heart failure with preserved ejection fraction. The American journal of medicine 126, 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hochreiter Sepp and Schmidhuber Jürgen. 1997. Long short-term memory. Neural Computation 9, 8 (1997). [DOI] [PubMed] [Google Scholar]
- [19].Johnson Alistair and others. 2016. MIMIC-III, a freely accessible critical care database. Scientific Data 3(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Thomas N Kipfand Max Welling. 2016. Semi-supervised classification with graph convolutional networks. arXiv:1609.02907 (2016). [Google Scholar]
- [21].Quoc V Le, Navdeep Jaitly, and Geoffrey E Hinton. 2015. A Simple Way to Initialize Recurrent Networks of Rectified Linear Units. arXiv:1504.00941 (2015). [Google Scholar]
- [22].Li Yuezhang, Zheng Ronghuo, Tian Tian, Hu Zhiting, Iyer Rahul, and Sycara Katia. 2016. Joint Embedding of Hierarchical Categories and Entities for Concept Categorization and Dataless Classification. (2016). [Google Scholar]
- [23].Lin Yankai, Liu Zhiyuan, Sun Maosong, Liu Yang, and Zhu Xuan. 2015. Learning Entity and Relation Embeddings for Knowledge Graph Completion. In AAAI. [Google Scholar]
- [24].Zachary C Lipton, David C Kale, Charles Elkan, and Randall Wetzell. 2015. Learning to Diagnose with LSTM Recurrent Neural Networks. arXiv:1511.03677 (2015). [Google Scholar]
- [25].Zachary C Lipton, David C Kale, and Randall Wetzel. 2016. Modeling Missing Data in Clinical Time Series with RNNs. In MLHC. [Google Scholar]
- [26].Laurens Maaten van der and Geoffrey Hinton. 2008. Visualizing data using t-SNE. JMLR 9, November (2008). [Google Scholar]
- [27].Mikolov Tomas, Sutskever Ilya, Chen Kai, Corrado Greg, and Dean Jeff. 2013. Distributed representations of words and phrases and their compositionality. In NIPS. [Google Scholar]
- [28].George A Miller. 1995. WordNet: a lexical database for English. Commun. ACM 38, 11 (1995). [Google Scholar]
- [29].Miotto Riccardo, Li Li, Brian A Kidd, and Joel T Dudley. 2016. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Scientific Reports 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nguyen Phuoc, Tran Truyen, Wickramasinghe Nilmini, and Venkatesh Svetha. 2016. Deepr: A Convolutional Net for Medical Records. arXiv:1607.07519 (2016). [DOI] [PubMed] [Google Scholar]
- [31].Pennington Jeffrey, Socher Richard, and Christopher D Manning. 2014. Glove: Global Vectors for Word Representation. In EMNLP. [Google Scholar]
- [32].Perozzi Bryan, Rami Al-Rfou, and Steven Skiena. 2014. DeepWalk: Online Learning of Social Representations. In SIGKDD. [Google Scholar]
- [33].Cost Healthcare & Project Utilization and others. 2010. Clinical classifications software (CCS) for ICD-9-CM. Rockville, MD: Agency for Healthcare Research and Quality; (2010). [Google Scholar]
- [34].Razavian Narges, Marcus Jake, and Sontag David. 2016. Multi-task Prediction of Disease Onsets from Longitudinal Lab Tests. In MLHC. [Google Scholar]
- [35].Socher Richard, Chen Danqi, Christopher D Manning, and Andrew Ng. 2013. Reasoning with neural tensor networks for knowledge base completion. In NIPS. [Google Scholar]
- [36].Michael Q Stearns, Colin Price, Kent A Spackman, and Amy Y Wang. 2001. SNOMED clinical terms: overview of the development process and project status. In AMIA. [PMC free article] [PubMed] [Google Scholar]
- [37].Tang Jian, Qu Meng, Wang Mingzhe, Zhang Ming, Yan Jun, and Mei Qiaozhu. 2015. LINE: Large-scale Information Network Embedding. In WWW. [Google Scholar]
- [38].The Theano Development Team. 2016. Theano: A Python framework for fast computation of mathematical expressions. arXiv:1605.02688 (2016). [Google Scholar]
- [39].Vijayakrishnan Rajakrishnan, Steinhubl Steven, Ng Kenney, Sun Jimeng, Byrd Roy, Daar Zahra, Williams Brent, Ebadollahi Shahram, Stewart Walter, and others. 2014. Prevalence of heart failure signs and symptoms in a large primary care population identified through the use of text and data mining of the electronic health record. Journal of cardiac failure 20, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Zhen, Zhang Jianwen, Feng Jianlin, and Chen Zheng. 2014. Knowledge Graph Embedding by Translating on Hyperplanes. In AAAI. [Google Scholar]
- [41].Weinberger Kilian Q, Sha Fei, Zhu Qihui, and Lawrence K Saul. 2006. Graph Laplacian Regularization for Large-Scale Semidefinite Programming. In NIPS. [Google Scholar]
- [42].Xie Ruobing, Liu Zhiyuan, and Sun Maosong. 2016. Representation Learning of Knowledge Graphs with Hierarchical Types. In IJCAI. [Google Scholar]
- [43].Xu Kelvin, Ba Jimmy, Kiros Ryan, Cho Kyunghyun, Courville Aaron, Salakhutdinov Ruslan, Zemel Richard, and Bengio Yoshua. Show, Attend and Tell: Neural Image Caption Generation with Visual Attention.. In ICML. [Google Scholar]
- [44].Yang Zhilin, Cohen William, and Salakhutdinov Ruslan. 2016. Revisiting Semi-Supervised Learning with Graph Embeddings. arXiv:1603.08861 (2016). [Google Scholar]
- [45].Matthew D Zeiler. 2012. ADADELTA: an adaptive learning rate method. arXiv:1212.5701 (2012). [Google Scholar]


