Abstract
Biofilms are complex microbial architectures that encase microbial cells in a matrix comprising self-produced extracellular polymeric substances. Microorganisms living in biofilms are much more resistant to hostile environments than their planktonic counterparts and exhibit enhanced resistance against the microbicides. From the human perspective, biofilms can be classified into beneficial, neutral, and harmful. Harmful biofilms impact food safety, course plant and animal diseases, and threaten medical fields, making it urgent to develop effective and robust strategies to control harmful biofilms. In this review, we discuss various strategies to control biofilm formation on infected tissues, implants, and medical devices. We classify the current strategies into three main categories: (i) changing the properties of susceptible surfaces to prevent biofilm formation; (ii) regulating signaling pathways to inhibit biofilm formation; (iii) applying external forces to eradicate the biofilm. We hope this review would motivate the development of innovative and effective strategies for controlling harmful biofilms.
Keywords: Harmful biofilm, Infected tissue, Tissue implant, Medical device, Biofilm control
1. Introduction
Biofilms are microbial communities encased within a self-produced matrix of extracellular polymeric substances (EPS) that attach to biotic or abiotic surfaces (Watnick and Kolter 2000; Lohse et al. 2018). About 20-80% of microorganisms on Earth exist as biofilms (Flemming and Wuertz 2019). They can protect the microorganisms inside them, mostly bacteria, from hostile environments, by acting as a layer of “protective clothing” (Yin et al. 2019). Biofilms can comprise single or multiple species (Wolcott et al. 2013; Lohse et al. 2018), and a typical biofilm life-cycle usually includes five stages (Sauer et al. 2002; Stoodley et al. 2002; Monds and O'Toole 2009; Koo et al. 2017; Rumbaugh and Sauer 2020) (Figure 1): (i) Reversible attachment on surfaces. At this first stage, initial attachment of cells occurs via non-covalent interactions, such as hydrogen-bonding or the van der Waals’ force; (ii) Irreversible attachment. At the second stage, microbial cells become robustly attached to the surface via bacterial appendages such as flagella, pili, adhesive proteins, or exopolysaccharides (Serra et al. 2015); (iii) Development of early biofilms. After stable attachment, cells actively proliferate and produce abundant EPS; (iv) Maturation of structured biofilms. At this stage, stable biofilm forms via constructing a three-dimensional architecture; (v) Active dispersal. Finally, cells are disseminated from the biofilm and re-enter into the planktonic phase upon receiving environmental cues, waiting for a new life-cycle.
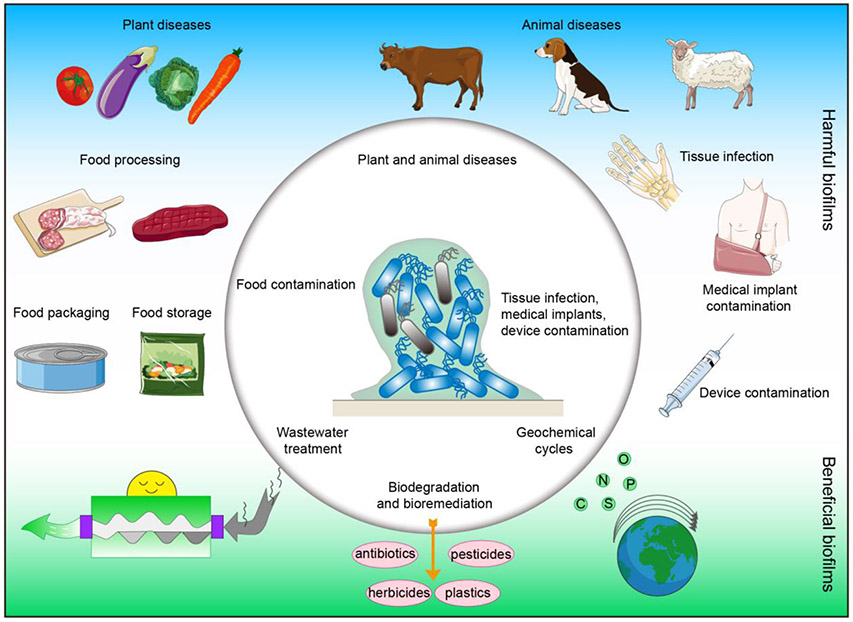
Figure 1. Schematic diagram of biofilm effects.
On one hand, biofilms play beneficial roles in wastewater treatment, biodegradation and bioremediation, and geochemical cycles of various elements. On the other hand, biofilms affect human life and health, contaminating medical implants and devices and causing a variety of infections.
Microorganisms in biofilms are distinct from their planktonic counterparts (Hathroubi et al. 2017); they usually show increased resistance to hostile environments including chemical biocides (Gupta et al. 2016), bacteriophages (Costerton et al. 1999), antibiotics (Wood 2017; Roy et al. 2018; Wolfmeier et al. 2018), and antibodies (Müsken et al. 2018). Here, according to the microbial properties in the biofilm, biofilms are classified into three main types, beneficial, neutral, and harmful biofilms, depending on their effects on humans, e.g. environment, food safety, plant and animal production, and medical fields.
Beneficial biofilms play key roles in many processes, such as wastewater treatment (Lin et al. 2019), biodegradation and bioremediation (Shukla et al. 2020), and geochemical cycles (Boer et al. 1991; Edwards et al. 2000) (Figure 2). During wastewater treatment, biofilms are massively formed in the bioreactor and participate in organic matter removal, adsorption of suspended solids, and purification of raw sewage (Huang et al. 2018) (Figure 2). In the processes of biodegradation and bioremediation, biofilms form unique microbial communities to degrade herbicides (Li et al. 2019b), pesticides (Zhang et al. 2019), antibiotics (Zhang et al. 2017), and plastics (Shah et al. 2008) and remediate contaminated water and soil (Wang et al. 2019a; Zhao et al. 2019) (Figure 2). In terms of geochemical cycles, biofilms comprising appropriate bacteria are able to decompose a wide variety of organic compounds via redox, cleavage, and hydrolysis reactions to complete the geochemical recycling of carbon, nitrogen, oxygen, phosphorus, sulfur, and other elements (Cui et al. 2018; Meyer-Dombard et al. 2018; Zhang et al. 2020) (Figure 2). For example, nitrifying and denitrifying bacteria in the biofilm play important roles in the nitrogen cycle in the soil environment (Di Trapani et al. 2011; Han et al. 2018).
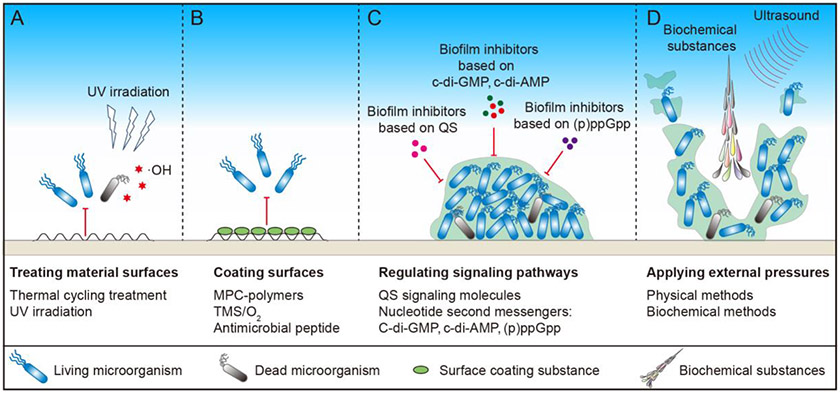
Figure 2. Strategies for controlling harmful biofilms.
(A) Material surfaces can be treated through thermal cycling and UV irradiation. (B) Surfaces can be coated by 2-methacryloyloxyethyl phosphorylcholine (MPC)-polymers, trimethylsilane (TMS)/O2, and antimicrobial peptides. (C) Biofilm formation can be inhibited by chemical agents that influence quorum sensing (QS), c-di-GMP, c-di-AMP, and (p)ppGpp related pathways. (D) External force, including biochemical substances and ultrasound, can also be applied to eradicate mature biofilms.
Harmful biofilms threaten food safety, cause plant and animal diseases (Figure 2). They are widespread in food processing (Garcia-Sanchez et al. 2019), packaging (Ripolles-Avila et al. 2019), and storage (Sternisa et al. 2019) that can significantly affect food quality (Ng et al. 2017) (Figure 2). Meanwhile, many plant and animal diseases resulting from harmful biofilms are quite difficult to control, which can significantly reduce the yield and quality of the crops (Yaron and Römling 2014) and animal products (Wang et al. 2013; Merino et al. 2019) (Figure 2). Due to the space limits, we will not discuss these aspects further in this review.
Harmful biofilms greatly threaten medical fields causing multiple infections and contaminating tissue implants and medical devices (Figure 2). Compared with the infections caused by planktonic microorganisms, biofilm-related infections are more difficult to cure and pose greater damage to human health (Abusrewil et al. 2020). For example, biofilm is present in most patients with chronic rhinosinusitis, and when these patients undergo surgery, the presence of biofilm will cause more severe diseases and result in worse surgical outcomes (Hamilos 2014). Dental plaque biofilm is a key factor for dental caries and periodontal disease (Seneviratne et al. 2011), and biofilms in the urinary tract often produce intractable chronic infections (Hola et al. 2010). Thus, biofilm typically acts as a “protective clothing” for bacteria and aggravates many tissue infection diseases. Biofilms are also a major source of contamination in tissue implants and medical devices (Blackledge et al. 2013) (Figure 2). Approximately 5% of implanted internal fractured bone fixation devices and almost 30% of open fracture devices are reported to be infected by biofilms (Rahim et al. 2019). When such biofilm-harboring medical devices contact human tissues, the bacteria will survive and proliferate, leading to harmful infection (Ribeiro et al. 2012). In fact, biofilms formed on medical devices have caused many human diseases, and the strongly attached biofilms are very difficult to eradicate from the medical devices with ordinary disinfection and antibiotic treatments (Zheng et al. 2018). The National Institutes of Health (NIH) in the USA report that 80% of all known human infections are associated with biofilms. Similarly, the U.S. Centers for Disease Control and Prevention (CDC) report that over 65% of all hospital-acquired infections are attributable to biofilms (Percival 2017). The existence of the harmful biofilms thus not only causes human diseases directly but also induces many hospital-acquired infections indirectly via contaminated tissue implants and medical devices. In addition to causing infections, biofilms can corrode metal surfaces, including those of tissue implants and prostheses (Beech and Sunner 2004; Beech et al. 2006). Therefore, effectively controlling harmful biofilms is important and urgent.
There is also a third type of biofilms, which is neither harmful nor beneficial to environment, food safety, plant and animal production, and medical fields. Such biofilms exist widely in natural environments such as mountains (Hotaling et al. 2019), wetlands (Yan et al. 2018), and marines (Kviatkovski et al. 2018; Angelova et al. 2019) that are far from human activities. Others exist in human daily life but do not exhibit any harm or benefit. Due to the absence of either harmful or beneficial effects, few reports regarding these biofilms are available. Hence we temporarily classify them as neutral biofilms. However, these biofilms can occasionally be transformed into useful biofilms under unique conditions, such as those for wastewater treatment, environmental remediation, and waste degradation. Due to the space limit, we will not describe this type of biofilms in more detail here.
In this review, we have tried to comprehensively discuss the current strategies used for preventing, inhibiting, and eradicating harmful biofilms. We aim at inspiring further developments of innovative and effective strategies for controlling harmful biofilms in medical fields, including those in infected tissues, tissue implants, and medical devices.
2. Strategies to control harmful biofilms
According to the different approaches to the task, we have divided the various strategies to control harmful biofilms into three main categories (Figures 1 and 3): (i) changing abiotic surface characteristics to prevent biofilm formation; (ii) regulating the signaling pathways to inhibit biofilm formation and stimulate biofilm dispersal, and (iii) applying external forces to eradicate the biofilm.
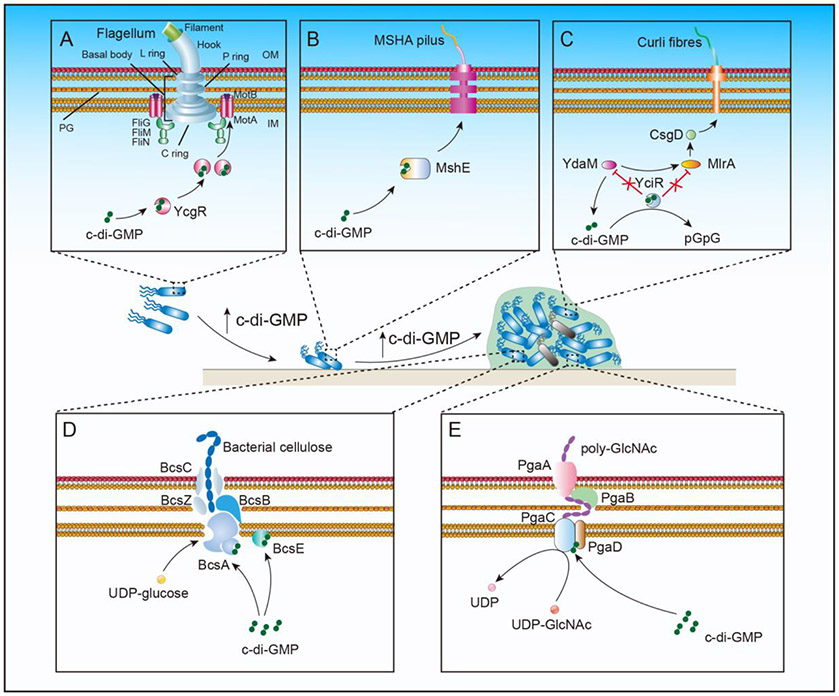
Figure 3. Cyclic di-GMP regulates biofilm formation by inhibiting bacterial motility and increasing EPS production.
(A) Binding of c-di-GMP to the bacterial flagellar brake protein YcgR inhibits the rotation of the flagellar motor, reduces cell motility, and promotes bacterial attachment to the solid surface. (B) Binding of c-di-GMP to MshE promotes the assembly of mannose-sensitive haemagglutinin pilus and helps the bacterial attachment to the solid surface. (C-E) C-di-GMP promotes the synthesis of bacterial EPS and solidifies biofilm formation: (C) When intracellular c-di-GMP concentration reaches a certain threshold, the inhibition of YdaM and MlrA proteins by YciR is relieved. YdaM can activate MlrA to interact with the central curli regulator CsgD, which induces the transcription of curli genes and facilitates curli formation; (D) Bacterial cellulose synthetase catalytic subunit BcsA is anchored on the inner membrane. When c-di-GMP binds to BcsA, its glycosyltransferase domain is activated to assemble the nascent polysaccharide with the help of BcsB/BcsC/BcsZ complex to form extracellular cellulose; (E) The PgaABCD complex promotes formation of the exopolysaccharide poly-GlcNAc. PgaC and PgaD interact with the help of c-di-GMP to form the PgaCD glycosyltransferase complex, which is used for the polymerization and extension of poly-GlcNAc. PgaA and PgaB are responsible for the transport of poly-GlcNAc outside the cell. IM, inner or plasma membrane; PG, peptidoglycan; OM, outer membrane.
2.1. Changing abiotic surface characteristics to prevent biofilm formation
It is often said that “Prevention is better than a cure” (Ling et al. 2020). Preventing biofilm formation usually requires less effort than inhibition and eradication of the biofilm. The most common strategies for preventing biofilm formation are treating and coating natural abiotic surfaces as described below.
2.1.1. Treating abiotic surfaces
Biofilm formation depends upon the physicochemical characteristics of the adsorptive surfaces (Renner and Weibel 2011). Therefore, changing the characteristics of the material surfaces, such as their smoothness, wettability, or hydrophilicity, could help in preventing biofilm formation.
Bacterial surface attachment is the first step of biofilm formation. Compared with a smooth surface, rough surface is more susceptible to microbial attachment and accumulation (Mei et al. 2011; García et al. 2016). For example, after clinical intraoral polishing, dental ceramic materials usually result in greater surface roughness and lead to increased biofilm formation (Kim et al. 2017). Vital bleaching is a popular treatment option for discoloring teeth. However, after bleaching, the roughness of the resin composite and resin-modified glass-ionomer cement can be greatly increased, leading to a significant streptococcal biofilm formation (Wongpraparatana et al. 2018) (Figure 3A). These may be due to the fact that such rough surface has more micropits, which can easily trap microorganisms and increase their interception within the rough surface to enhance biofilm formation (Kim et al. 2017).
The thermal cycling treatment of the material can improve the smoothness of the surface and inhibit the microorganism attachment. Thermal cycling is a temperature modulation process that can improve the performance, longevity, and strength of a variety of materials, which enables orthopedic implants to exhibit smoother, more uniform surfaces via molecular reorganization (Akens et al. 2018). In one example of the thermal cycling process, the material was exposed to the liquid nitrogen vapor and slowly cooled down to −148.9 °C in 2 h, followed by maintaining at this temperature for another 2 h before increasing it to 20°C over a period of 16 h (Akens et al. 2018). Staphylococcus aureus is an opportunistic pathogen and arguably the most common cause of infection in hospitalized patients. The biofilm formed by S. aureus on medical implants makes it even more difficult to eradicate and can trigger recalcitrant bacterial infections (Figueiredo et al. 2017). Thermal cycling of orthopedic stainless steel and titanium implants can prevent S. aureus from forming biofilm because of the increased smoothness (Akens et al. 2018) (Figure 3A).
Ultraviolet irradiation is another approach to provide titanium surfaces with enhanced wettability and generate reactive oxygen species such as hydroxyl radicals to destroy the microbial cell membrane and cell wall (Foster et al. 2011) (Figure 3A). Indeed, it was found that a 12-min ultraviolet treatment of titanium can transform its surface property from hydrophobic to superhydrophilic, significantly preventing oral microbial attachment and biofilm formation on the titanium implant material (de Avila et al. 2015).
The above data indicate that enhanced smoothness, wettability, and hydrophilicity are helpful in preventing biofilm formation. In the future, hopefully, more methods will be devised to alter the physicochemical properties of the solid surface for inhibiting microbial attachment, thereby preventing biofilm formation. Changing the surface properties of the material is often effective, and could play a preemptive role in controlling biofilm formation, resulting in considerably less trouble later on.
2.1.2. Coating surfaces
Coating the attachment surfaces is another way to prevent microbial attachment and biofilm formation (Figure 3B).
The surface free energy (SFE) of the attached material, which is commonly referred to as wettability (Teughels et al. 2006), can influence the microbial attachment (Nakamura et al. 2016). One typical case is when denture materials are coated with salivary and/or blood plasma proteins to change their SFE. This can prevent mature Candida albicans attachment and biofilm formation (da Silva et al. 2015).
It is difficult for microorganisms to colonize surperhydrophilic surfaces (Almaguer-Flores et al. 2012). For example, the biocompatible 2-methacryloyloxyethyl phosphorylcholine (MPC)-polymers have a phospholipid polar group that mimics the biological membrane. Due to their decent “super-hydrophilicity”, such MPC-polymers coated on saliva-coated hydroxyapatite and oral epithelial cells can markedly decrease pathogenic microbial attachment and prevent biofilm formation (Hirota et al. 2005; Hirota et al. 2011) (Figure 3B).
The coating of small molecules can also change the attachment properties of surface materials. For example, coating the surfaces of silicone rubber with monomeric trimethylsilane (TMS)/O2, which is often used in the fabrication of tissue implants, can significantly change the microbial surface proteins adsorption, and prevent S. aureus from forming biofilms (Xu et al. 2015) (Figure 3B). Since TMS/O2 coating is an environmentally friendly and efficient method, it has great potential for clinical applications.
Antimicrobial peptide coating is another valuable method for preventing biofilm formation (Figure 3B). Since antimicrobial peptides have a broad spectrum of bactericidal activity and low risk of microbial resistance, they have been studied widely in many different instances (Kazemzadeh-Narbat et al. 2010; Di Somma et al. 2020). For example, bactericidal cationic peptide GL13K was derived from the human parotid secretory protein BPIFA2 (Hirt and Gorr 2013), and people found that titanium disks that are covalently-immobilized with GL13K exhibit excellent antimicrobial activity when exposed to Streptococcus gordonii cultures and prevents the attachment of S. gordonii on the coated surface (Chen et al. 2014). GL13K coating of titanium surfaces also effectively decreases the growth of Fusobacterium nucleatum and Porphyromonas gingivali and prevents biofilm formation by these organisms (Li et al. 2017).
Thus, coating tissue implants and medical devices with specific materials can decrease microbial attachment to the material surface, prevent biofilm formation, and thereby reduce bacterial infection. The materials for surface coating are easy to synthesize and modify. Therefore, this method is simple and easy to develop and is well suited for the effective control of harmful biofilms in tissue implants and medical devices. Development of efficient, environmentally friendly, and stable coating materials for preventing the formation of harmful biofilms is a major direction of biofilm control at present and in the future.
2.2. By regulating the signaling pathways to inhibit biofilm
The biofilm formation is regulated by several signaling pathways, such as quorum sensing (QS) and nucleotide second messenger systems. Therefore, biofilm inhibitors can be designed by targeting relevant proteins in the respective regulatory systems to block their signaling pathways and thereby inhibit biofilm formation.
2.2.1. Biofilm inhibitors based on QS
2.2.1.1. Mechanism of QS
QS is a widespread mechanism of cell-cell communication in bacteria. Bacteria can communicate with each other by secreting various signaling molecules, called autoinducers (AIs). When these signaling molecules bind to their corresponding receptors, they can trigger a cascade of intracellular signaling events, ultimately regulating different physiological phenotypes (Padder et al. 2018; Kalia et al. 2019).
To date, quite a few of QS signaling molecules have been discovered, including autoinducer peptide (AIP), N-acyl homoserine lactones (AHLs), autoinducer 2 (AI-2), Pseudomonas quinolone signal (PQS), diffusible signal factor (DSF), and others (Machado et al. 2020). Among them, AIP, AHLs, and AI-2 are arguably the most widely studied.
AIPs are commonly exploited molecules in the Gram-positive bacteria for significant signal transduction. The most typical example of AIP-mediated QS is the Accessory gene regulator (Agr) system, in which the AgrD peptide is expressed in the cells, modified by AgrB on the membrane, and secreted out of cells to become mature AIP. The concentration of extracellular AIP proportionally increases with the increase in cell density, leading to a high concentration of AIP that activates the AgrC-AgrA two-component system to regulate the physiological functions of bacteria. The activated Agr system inhibits the expression of AtlE, which is an important adhesion protein for biofilm formation; inhibition of AtlE expression would inhibit biofilm formation (Yang et al. 2016). Accordingly, addition of exogenous AIP has been found to activate the agr-mediated S. aureus biofilm detachment (Boles and Horswill 2008).
Gram-negative bacteria, on the contrary, mostly use AHLs as signaling molecules, with the LuxI/LuxR system being perhaps the best well-studied signaling system among them (Galloway et al. 2011). In the LuxI/LuxR regulatory network, the LuxI protein is responsible for the AHL production, while LuxR serves as a receptor for the AHL molecule, which activates LuxR to regulate the expression of downstream genes. Indeed, a LuxI/LuxR homolog consisting of a conserved AHL synthase gene (luxI) and a transcriptional regulator gene (luxR) was identified in Pseudomonas fluorescens PF07, and in the deletion mutants ΔluxI and ΔluxR, the biofilm biomass and EPS production were significantly decreased, leading to thinner and looser biofilm structures. This result demonstrates that the LuxI/LuxR system plays a crucial regulatory role in the process of biofilm formation in P. fluorescens (Tang et al. 2019).
Another QS system that is mediated by LuxS/AI-2 was found in both Gram-positive and Gram-negative bacteria and is involved in the mixed-species biofilm regulation (Wang et al. 2019c). LuxS is involved in the production of AI-2, which is a furanosyl borate diester that functions as a universal signal for the bacterial species interaction. Streptococcus mutans that forms mixed-species biofilms with S. gordonii is related to the development of dental caries. The LuxS/AI-2 QS system of S. gordonii modulates the dual-species biofilm formation with S. mutans. luxS disruption in S. gordonii can alter the dual-species biofilm formation, architecture, and composition. Besides, it was reported to influence the S. mutans biofilm formation when the synthesized AI-2 was added in vitro by incubating the cells with both recombinant LuxS and methylthioadenosine/S-adenosylhomocysteine nucleosidase from S. gordonii (Bao et al. 2015; Wang et al. 2017).
A large number of previous studies have shown that formation, development, and functional regulation of most biofilms require the participation of QS signaling molecules. A great number of researches have convincingly shown that QS inhibitors can indeed inhibit biofilm development (Ouyang et al. 2016; Chen et al. 2018b) (Figure 3C). These data suggested that controlling the QS signaling system could serve as an effective method of controlling biofilm formation.
2.2.1.2. Biofilm inhibitors based on QS
Interfering with the QS signaling pathway to inhibit the expression of biofilm-related genes is thus one of the good methods to inhibit biofilm formation (Figure 3C and Table 1).
Table 1.
Biofilm inhibitors based on quorum sensing
| Compounds | Structures | Target microorganisms | References |
|---|---|---|---|
| Natural compounds | |||
| Curcumin | 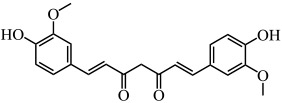 |
E. coli, P. aeruginosa, Proteus mirabilis, Serratia marcescens | (Packiavathy et al. 2014) |
| Resveratrol | 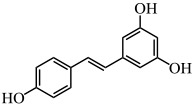 |
P. aeruginosa | (Vasavi et al. 2017) |
| Carvacrol | 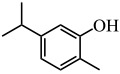 |
P. aeruginosa | (Tapia-Rodriguez et al. 2017) |
| Furocoumarins | 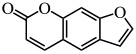 |
E. coli, S. typhimurium, P. aeruginosa | (Girennavar et al. 2008) |
| Synthetic compounds | |||
| Furanone C-30 | 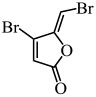 |
S. mutans | (He et al. 2012) |
| 2(5H)-Furanone | 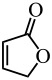 |
C. jejuni | (Castillo et al. 2015) |
| Meta-bromo-thiolactone | 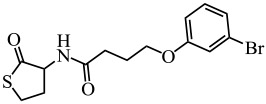 |
P. aeruginosa | (O'Loughlin et al. 2013) |
There are many natural biofilm inhibitors based on the QS phenomenon, with most of them produced by plants. For example, curcumin from Curcuma longa (turmeric) inhibited the biofilm formation in several uropathogens, such as Pseudomonas aeruginosa PAO1, Escherichia coli, Serratia marcescens, and Proteus mirabilis, via reducing QS-dependent production of exopolysaccharide (such as alginate) and slowing the bacterial motility (Packiavathy et al. 2014) (Table 1). Similarly, resveratrol, another natural product, interfered with the QS signal by binding to the protein receptor LasR of P. aeruginosa PAO1 to inhibit its biofilm formation and expression of virulence factors (Vasavi et al. 2017) (Table 1). This was also the case for carvacrol, which inhibited P. aeruginosa biofilm formation and pyocyanin production (Tapia-Rodriguez et al. 2017) (Table 1). Naturally occurring furocoumarins from grapefruit showed a >95% inhibition of AI-1 and AI-2 activities and also inhibited the biofilm formation by E. coli O157:H7, P. aeruginosa, and Salmonella enterica serovar Typhimurium (Girennavar et al. 2008) (Table 1). Clove bud oil could both inhibit biofilm formation and disrupt the preformed biofilms of P. aeruginosa. At a concentration of 1%, it can reduce biofilm formation by 85.3% while promoting biofilm dispersal by 50.4% (Kalia et al. 2019). These natural biofilm inhibitors are non-toxic and appear harmless for both human body and the environment; therefore, they exhibit good potential for application in various fields.
In addition to these natural compounds, synthetic compounds can also inhibit the QS signaling pathway. For example, synthetic furanone C-30 was found to significantly inhibit biofilm formation by S. mutans and its luxS mutant strain via a dose-dependent effect but did not affect the growth rate of planktonic cells (He et al. 2012) (Table 1). 2(5H)-Furanone reduced Campylobacter jejuni motility and biofilm formation by disturbing its QS activity (Castillo et al. 2015) (Table 1). Meta-bromo-thiolactone inhibited the pyocyanin production and biofilm formation of P. aeruginosa by binding to two QS signal receptors, LasR and RhlR (O'Loughlin et al. 2013) (Table 1).
Biofilm inhibitors based on QS have been widely used in inhibiting various biofilms, but many more QS inhibitors are still under development to treat infections caused by biofilms (Yu et al. 2018) or to eradicate biofilms on tissue implants (Luo et al. 2017). There are also additional approaches to using biofilm inhibitors based on QS, such as supplementing QS inhibitors with antibiotics for better biofilm control (Thomann et al. 2016).
2.2.2. Biofilms inhibitors based on nucleotide second messenger molecules
Nucleotide second messenger molecule-based signaling plays an important role in regulating various physiological functions of bacteria (Römling et al. 2013; Chou and Galperin 2016; He et al. 2020). The concentrations of these second messengers in the cells are maintained through dedicated synthesizing and degrading enzymes, which are activated by specific environmental cues. In the following sections, we mainly discuss the regulation of biofilm formation by c-di-GMP, c-di-AMP, and (p)ppGpp, and the corresponding inhibitors that have been developed to control the respective regulatory processes (Figure 4).
Figure 4. Applying external pressure to eradicate mature biofilm.
(A) Ultrasound directly eradicates biofilm from the solid surface. (B) Phage lysins effectively kill bacteria in the biofilm through cleaving bacterial peptidoglycan. (C) Degradative enzymes eradicate biofilm by degrading the polysaccharide and eDNA in the EPS matrix. (D) Microbial metabolites disrupt bacterial cell membrane or regulate bacterial physiological activity to eradicate the biofilm.
2.2.2.1. C-di-GMP signaling pathway and biofilm inhibitors based on c-di-GMP
C-di-GMP is synthesized from two molecules of GTP by diguanylate cyclases (DGCs) (Su et al. 2016) and degraded into GMP or pGpG by c-di-GMP-specific phosphodiesterases (PDEs) (Fu et al. 2018). C-di-GMP is a highly versatile secondary messenger in bacteria, and through combining with a wide variety of different receptors, such as PliZ (Pratt et al. 2007; Wang et al. 2016a), transcriptional regulators (Krasteva et al. 2010), MshE protein (Wang et al. 2016b), or c-di-GMP riboswitch (Tang et al. 2016; Zhou et al. 2016), it can regulate numerous bacterial physiological functions such as motility-to-sessility transition (Christen et al. 2007; Baraquet and Harwood 2013), cell cycle control (Abel et al. 2011), virulence factor production (Tang et al. 2016; Fu et al. 2018), and biofilm formation (Boyd and O'Toole 2012) or dispersal (Su et al. 2016).
C-di-GMP mainly regulates biofilm formation through the following three mechanisms: (i) Diminishing bacterial mobility to promote bacterial attachment onto a solid surface. Transition of bacteria from motility to sessility is a necessary stage during biofilm formation. In E. coli, the c-di-GMP-bound form of the flagellar brake protein YcgR interacts with the flagellar motor protein MotA, thus controlling motor output in a brake-like fashion (Hou et al. 2020). Clearly, c-di-GMP plays a role in suppressing bacterial motility and promoting bacterial surface attachment (Figure 4A). (ii) Regulating pilus formation. In Vibrio cholerae, the mannose-sensitive haemagglutinin (MSHA) pilus helps bacterial cells attach onto solid surfaces in the early stages of biofilm formation (Jones et al. 2015). It is regulated by MshE, which is an ATPase responsible for pilus polymerization (Wang et al. 2016b). When c-di-GMP binds to MshE, it promotes the assembly of MSHA pilus and increases biofilm formation (Jones et al. 2015) (Figure 4B). With the increase of intracellular c-di-GMP concentration, the number of MSHA pili on the bacterial surface also increases, leading to rapid biofilm formation. (iii) Regulating the production of various biofilm components. In E. coli K-12 strain W3110, production of biofilm matrix components curli fibers is regulated by the DGC YdaM and the PDE YciR through controlling the c-di-GMP concentrations (Lindenberg et al. 2013). YdaM can synthesize c-di-GMP or activate transcription factor MlrA, whereas YciR can degrade c-di-GMP or inhibit the activity of MlrA and YdaM. When c-di-GMP concentration reaches a certain threshold, YciR starts to exert its PDE function to release the inhibition of YdaM and MlrA. At the same time, YdaM can activate MlrA to enhance the central curli regulator CsgD, thereby inducing the transcription of curli genes and facilitating the curli formation (Lindenberg et al. 2013) (Figure 4C). Production of bacterial cellulose, another important component of the biofilm matrix, is also regulated by c-di-GMP (Römling and Galperin 2015; Galperin and Shalaeva 2018). The bacterial cellulose synthase (BcsA), which is anchored in the inner membrane, contains a catalytic glycosyltransferase domain and a c-di-GMP-binding PilZ domain in its intracellular part (Morgan et al. 2013) (Figure 4D). When c-di-GMP binds to the PilZ domain of BcsA, it activates the adjacent glycosyltransferase domain, allowing the bacterial cell to assemble the nascent polysaccharide with the help of the BcsB/BcsC/BcsZ complex to form extracellular cellulose (Figure 4D). Poly-β−1,6-N-acetylglucosamine (poly-GlcNAc) is another exopolysaccharide that is widely present in biofilms. The polymerization and secretion of poly-GlcNAc are carried out by the Pga machinery, which is encoded by the pgaABCD operon. Among them, PgaA and PgaB are responsible for transporting poly-GlcNAc outside the cell, while PgaC and PgaD form a PgaCD glycosyltransferase complex that produces poly-GlcNAc under the control of c-di-GMP. When the c-di-GMP concentration is low, PgaD cannot combine with PgaC to form the PgaCD complex, and is degraded by protease, thereby stopping the polymerization of poly-GlcNAc and leading to an eventual decrease in extracellular poly-GlcNAc levels (Steiner et al. 2013) (Figure 4E). These and other results convincingly show that high concentrations of c-di-GMP promote biofilm formation (Römling et al. 2005; Valentini and Filloux 2016; Mukherjee et al. 2018).
Designing DGC inhibitors to decrease intracellular c-di-GMP concentrations, or screening compounds that would inhibit the interaction between c-di-GMP and its receptors, are both effective approaches for controlling biofilm formation (Cho et al. 2020). For example, seven small molecules that antagonize the activity of VC2370 DGC have been found to inhibit biofilm formation by V. cholerae (Sambanthamoorthy et al. 2012), and four small molecules that could antagonize the DGCs from Thermotoga maritima and WspR from P. aeruginosa inhibited biofilm formation by P. aeruginosa and Acinetobacter baumannii (Sambanthamoorthy et al. 2014). At present, c-di-GMP signaling inhibitors are mostly c-di-GMP analogs or non-nucleotide small molecules that inhibit DGCs. They can significantly reduce c-di-GMP concentrations to inhibit biofilm formation (Jakobsen et al. 2017). Up till now, these inhibitors were mostly tested in vitro, and validating new biofilm inhibitors based on interfering with the c-di-GMP signaling pathway will require more studies in the near future.
2.2.2.2. C-di-AMP signaling pathway and biofilm inhibitors based on c-di-AMP
C-di-AMP is another second messenger molecule widely present in bacteria. It is synthesized from two molecules of ATP by diadenylyl cyclases (DACs) (Zheng et al. 2015) and degraded to pApA or AMP by distinct c-di-AMP-specific phosphodiesterases (PDEs) (Corrigan and Gründling 2013; Tang et al. 2015; Stülke and Kruger 2020; Yin et al. 2020). C-di-AMP also binds to specific receptors such as TetR family transcription factor DarR (Zhang et al. 2013), histidine kinase KdpD (Moscoso et al. 2016), pyruvate carboxylase (Choi et al. 2017), and the c-di-AMP-specific riboswitch ydaO (Wang et al. 2019b), to affect bacterial growth, regulate cellular morphology, and control biofilm formation.
In some strains, high concentrations of c-di-AMP were found to promote biofilm formation, which could be due to the role of c-di-AMP in regulating EPS formation. A typical example is that from Streptococcus pyogenes HSC5, in which the c-di-AMP synthetase DacA and the degradation enzyme Pde2 encoding genes were separately knocked out to construct a low-concentration and high-concentration c-di-AMP mutants, respectively. It was later discovered that the low-concentration c-di-AMP mutant does not produce biofilm, while the high-concentration c-di-AMP strain produces a large amount of biofilm (Fahmi et al. 2019).
In S. mutans, when c-di-AMP concentration is high, it binds to its protein receptor CabP to regulate the activity of transcription factor VicR, which then promotes the expression of GtfB that is responsible for the production of water-insoluble glucan, another important extracellular polymer. Therefore, c-di-AMP at high concentrations can enhance biofilm formation by S. mutans through signal transduction and multiple regulatory processes (Peng et al. 2016). Meanwhile, a small molecule inhibitor named ST056083 was found to significantly reduce the activity of DisA (the main DAC) in Enterococcus faecalis, thereby reducing the intracellular c-di-AMP concentration and inhibiting the exopolysaccharide production and biofilm formation (Chen et al. 2018a) (Figure 3C).
At the same time, there are reports that high concentrations of c-di-AMP in certain bacteria can inhibit biofilm formation. For example, in Bacillus subtilis, accumulated c-di-AMP affected the activity of SinR, which controls the biofilm gene expression, subsequently inhibiting biofilm formation (Gundlach et al. 2016). Similarly, knocking out c-di-AMP phosphodiesterase gdpP in Strepotoccus gallolyticus produced a strain with a higher c-di-AMP level but reduced biofilm formation and lower attachment to the intestinal cells (Teh et al. 2019).
Although the detailed mechanisms by which c-di-AMP affects biofilm formation remain to be further studied, regulating the bacterial c-di-AMP concentration has been proved to be an effective and feasible way to control the harmful biofilm formation. Since c-di-AMP in different strains has different effects on biofilm formation, studying the regulation mechanism of c-di-AMP signaling in various pathogens is a pre-requisite to using it to control biofilm formation in practice.
2.2.2.3. (p)ppGpp signaling pathway and biofilm inhibitors based on (p)ppGpp
(p)ppGpp is synthesized from GTP and GDP by the RelA/SpoT homolog (RSH) enzymes, generating AMP as a by-product, by GppA (also known as pppGpp phosphohydrolase) and other GTPases to catalyze the interconversion of pppGpp to ppGpp. pppGpp and ppGpp are degraded by SpoT to form GTP and GDP, respectively (Hauryliuk et al. 2015). The functions of (p)ppGpp are also exerted through binding to various target enzymes, such as polyphosphate kinase, translational GTPases, DNA primase, to regulate many different bacterial physiological functions, including biofilm formation.
Although (p)ppGpp can affect biofilm formation, the specific mechanism of this regulation remains unclear. In Bordetella pertussis, accumulation of (p)ppGpp accelerates biofilm formation (Sugisaki et al. 2013). However, in Actinobacillus pleuropneumoniae S8, low-concentration (p)ppGpp was found to contribute to biofilm formation (Li et al. 2015). Such data indicate that in different organisms, (p)ppGpp concentration can exhibit different effects on biofilm formation. Furthermore, a potent anti-biofilm peptide 1018 that targets (p)ppGpp could inhibit biofilm formation in both Gram-negative and Gram-positive bacteria (de la Fuente-Nunez et al. 2014) (Figure 3C). With increasing research in this aspect, the mystery of the relationship between (p)ppGpp and biofilm will eventually be solved, and more anti-biofilm compounds will probably be found.
In general, second messenger molecules play important regulatory roles in biofilm formation. Many potential small molecule inhibitors of DGCs (Sambanthamoorthy et al. 2012; Fernicola et al. 2016) or DACs (Opoku-Temeng and Sintim 2016a; b) have been identified and more can be expected in the future. Although the efficacies of these compounds against biofilms await further validation, interference with the second messenger-based signaling pathways can be a powerful strategy for inhibiting biofilm formation.
Since biofilm inhibitors based on the above signaling systems do not directly kill bacteria, the drug resistance phenomenon can be potentially reduced (Scoffone et al. 2016; Galperin 2018). Therefore, biofilm inhibitors can serve as adjuvants and alternatives to the conventional antibiotic therapy. At the same time, they can also work synergistically with antibiotics to enhance their efficacy (Rajput et al. 2018). Hence, these biofilm inhibitors have certain advantages in controlling biofilms.
2.3. By applying external pressures to eradicate biofilm
To eradicate an already formed biofilm, one could use several different physical and biochemical approaches (Figure 5).
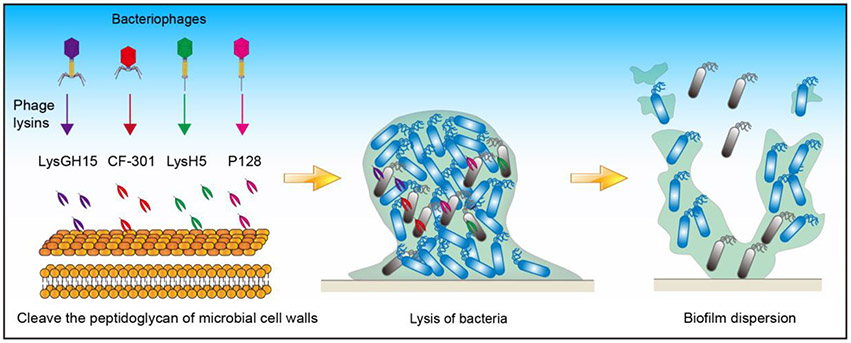
Figure 5. Applying phage lysins to eradicate mature biofilm.
Phage lysins such as LysGH15, CF-301, LysH5, and P128 can cleave the peptidoglycan of microbial cell walls enzymatically and therefore promote biofilm dispersal.
2.3.1. Physical methods
Many physical methods, such as treatment with ultrasound and magnetic fields, can be applied to effectively eradicate biofilms (Figure 3D).
Ultrasonic eradication of biofilms mainly depends on the forces generated from cavitation bubbles and fluid flow to trigger biofilm dispersion (Erriu et al. 2014) (Figure 5A). In a recent experiment, when an ultrasonic scaler tip was held 2 mm away from the Streptococcus sanguinis biofilm for 2 s, the process of cavitation bubbles contacting the surface was captured by a high-speed camera, which revealed that most of the biofilm was eradicated within 2 s (Vyas et al. 2020).
The magnetic iron oxide nanoparticles in combination with magnetic fields can also cause significant mechanical damage to the biofilm matrix and lead to biofilm eradication. Using a rotating magnetic field (Li et al. 2019a), found that nanoparticles placed on top of the biofilm were dragged across the biofilm to pull away microbial cells and matrix, thus leading to an effective biofilm eradication. Under a treatment force of 30 mg mL−1 and a rotating magnetic field with 11 nm nanoparticles, the S. aureus biofilm was reduced by nearly five orders of magnitude (Li et al. 2019a). Similarly, under a low-intensity rotating magnetic field, magneto-responsive gallium-based liquid metal droplets can also disrupt the biofilm matrix and result in bacterial cell lysis through both the nanosharp edges developed from shape transform and mechanical shear force generated from the movement of droplets (Elbourne et al. 2020).
There are also many other physical methods that can effectively eradicate mature biofilms, such as using high-velocity spray or jet irrigator to brush biofilms, or using photodynamic or photothermal therapy to destroy biofilms and kill the bacteria in them directly (Koo et al. 2017; Karygianni et al. 2020). Meanwhile, the weak electric field produced by Ag/Zn can eradicate the biofilm through electrical stimulation to accelerate wound healing (Barki et al. 2019).
Therefore, external physical factors can eradicate biofilm to a certain extent. These are simple, rough, and effective methods that are suitable for decontamination in the food industry, for biofilm eradication in the dental field and removal of biofilms adsorbed on tissue implants and medical devices. However, these methods are not suitable for infected tissues, which require more gentle approaches, such as the application of phage lysins or degradative enzymes. These approaches are described below.
2.3.2. Biochemical methods
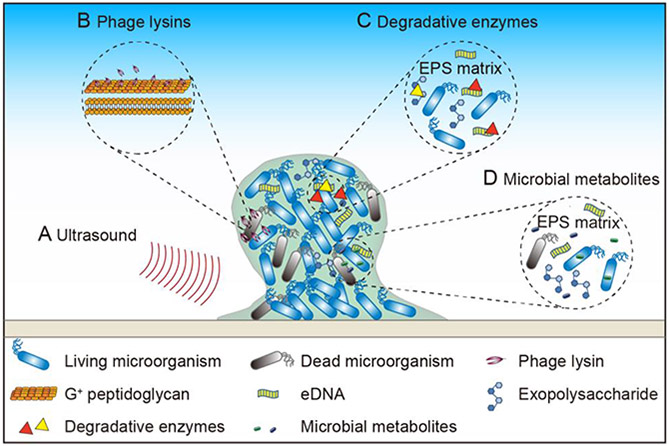
For planktonic microorganisms, antibiotics are the main means of inhibiting bacterial growth, but for those living in biofilms, they are much less useful. Therefore, for effective eradication of microorganisms in biofilms, many biochemical methods have been applied, such as application of phage lysins, degradative enzymes, and microbial metabolites (Figure 3D and Table 2).
Table 2.
Biochemical substances for eradicating biofilm
| Factors | Target microorganisms | Mechanisms | References |
|---|---|---|---|
| Phage lysins | |||
| LysGH15 | Staphylococcus | Cleaves Staphylococcus peptidoglycan | (Zhang et al. 2018) |
| CF-301 | S. aureus | Cleaves S. aureus peptidoglycan | (Schuch et al. 2017) |
| LysH5 | S. aureus | Cleaves S. aureus peptidoglycan | (Gutierrez et al. 2014) |
| P128 | Staphylococcus | Cleaves Staphylococcus peptidoglycan | (Poonacha et al. 2017) |
| Degradative enzymes | |||
| Mannosidase, Glucanase | C. auris | Degrades C. auris mannan and glucan in EPS | (Dominguez et al. 2019) |
| Alginate lyases | P. aeruginosa | Degrades exopolysaccharide alginate | (Jang et al. 2016) |
| DNase | B. licheniformis, B. subtilis, E. coli, Micrococcus luteus | Degrades eDNA | (Nijland et al. 2010) |
| DNase | Gardnerella vaginalis | Degrades eDNA | (Hymes et al. 2013) |
| Metabolites | |||
| Carolacton | Sorangium cellulosum | Causes S. mutans biofilm cells death, cell chains elongation, and cell morphology changes | (Kunze et al. 2010) |
| Rhamnolipid | S. aureus, S. enteritidis, L. monocytogenes | Possesses anti-biofilm and antimicrobial activities | (E Silva et al. 2017; Khalid et al. 2019) |
| D-amino acids | E. coli | Contribute to the cell-cell repulsion | (Kolodkin-Gal et al. 2010; Xing et al. 2015) |
2.3.2.1. Phage lysins
Pathogenic bacteria are the enemies of humans, and bacteriophages are the enemies of bacteria. The enemy’s enemy could be a friend, and phage lysins, which are the bacteriophages’ weapons, could be utilized to fight multidrug-resistant microorganisms and harmful biofilms (Drulis-Kawa and Maciejewska 2015; Melo et al. 2018; Sharma et al. 2018; Łusiak-Szelachowska et al. 2020) (Figures 5B and 6). Phage lysins are peptidoglycan hydrolases expressed by bacteriophages in the later stage of infection. They can cleave the peptidoglycan layer of the cell wall, thereby killing the bacteria (Schuch et al. 2017; Zhang et al. 2018).
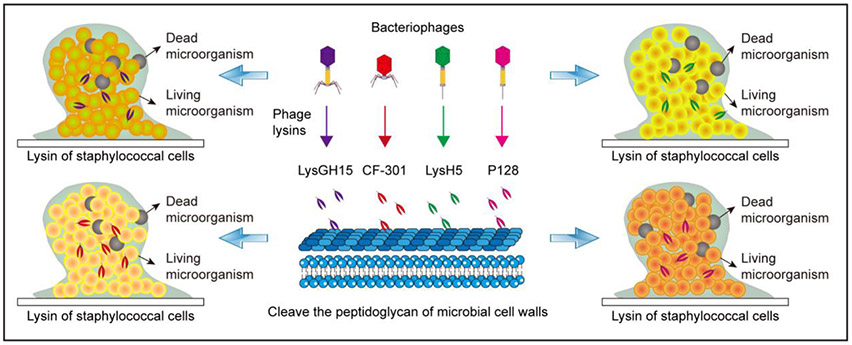
Figure 6. Applying phage lysins to eradicate mature biofilm.
Phage lysins such as LysGH15 (Zhang et al. 2018), CF-301 (Schuch et al. 2017), LysH5 (Gutierrez et al. 2014), and P128 (Poonacha et al. 2017) can enzymatically cleave the peptidoglycan of cell walls to kill staphylococci.
LysGH15, a phage lysin derived from the staphylococcal bacteriophage GH15, did not only eliminate staphylococcal planktonic cells of S. aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus hominis, but also eradicated preformed biofilms (Zhang et al. 2018) (Table 2). Phage lysin CF-301 was highly effective in eradicating S. aureus biofilms and killing microorganisms in the biofilm and has been shown to be a potential anti-biofilm agent to cure staphylococcal infections (Schuch et al. 2017) (Table 2).
Phage lysins are effective not only against growing bacteria but also against persisters, which are certain phenotypic variants that can withstand lethal doses of antibiotics (Orman and Brynildsen 2016). In fact, formation of persisters has been considered the cause of biofilm-related microbial recurrence in tissue implants and medical devices (Yan and Bassler 2019). Diminishing or killing persisters is thus crucial for biofilm eradication and infection prevention. Some phage lysins have the ability to kill persisters (Sharma et al. 2018). For example, phage lysin LysH5 was active against staphylococcal persisters both in the planktonic state and in biofilms (Gutierrez et al. 2014) (Table 2). Similarly, a chimeric recombinant phage lysin P128 showed good activity against coagulase-negative staphylococci (Poonacha et al. 2017) (Table 2). These data demonstrate that phage lysins could effectively fight against persisters in the planktonic state and biofilm state.
Phage lysins also have a species-targeted bactericidal ability and exhibit certain advantages over conventional chemotherapeutic agents in controlling biofilm formation. Therefore, using phage lysins to eradicate biofilms, especially in destroying persisters, has certain unique advantages. On the one hand, due to the species-specific bactericidal ability, a given phage lysin may only eradicate specific biofilm infections, and may not be able to treat infection of mixed species. To achieve a better therapeutic effect, several phage lysins could be mixed together in a cocktail or used in combination with other methods.
2.3.2.2. Degradative enzymes
The biofilm matrix of EPS is mainly composed of nucleic acids, proteins, lipids, and exopolysaccharides (Flemming and Wingender 2010; Karygianni et al. 2020). Because biofilm serves as a “protective clothing” for microorganisms (Yin et al. 2019), one can eradicate the harmful biofilm by degrading the EPS and removing this protective clothing (Figure 5C).
Candida auris is an emerging nosocomial pathogen (Lamoth and Kontoyiannis 2018; Meis and Chowdhary 2018), and the matrix of C. auris biofilms is rich in mannan-glucan. Applying mannosidase or glucanase to hydrolyze mannan-glucan in the biofilm matrix was found to effectively cure infections caused by this organism (Dominguez et al. 2019) (Table 2). Also, Pseudomonas cells enclosed within alginate-rich biofilms have been observed in human lung tissue and sputum in cystic fibrosis patients (Bayer et al. 1992). Alginate lyase has been applied to degrade alginate into unsaturated uronic acid-containing oligosaccharides, thus removing exopolysaccharide from the P. aeruginosa cell surface and subsequently promoting biofilm eradication (Jang et al. 2016) (Table 2).
Degradation of extracellular DNA (eDNA) in the matrix by DNase is also helpful for eradicating biofilms. Since eDNA is an important component essential for biofilm formation, DNase can efficiently inhibit biofilm formation and also eradicate biofilms already formed (Whitchurch et al. 2002; Farisa Banu et al. 2019). For example, the extracellular DNase from Bacillus licheniformis was found to be capable of eradicating biofilms both from Gram-positive and Gram-negative bacteria including B. licheniformis, B. subtilis, E. coli, and Micrococcus luteus (Nijland et al. 2010) (Table 2). Also, by hydrolyzing eDNA, DNase could inhibit as well as disrupt Gardnerella vaginalis biofilms (Hymes et al. 2013) (Table 2). These results indicate that application of efficient and stable DNase could successfully degrade eDNA in biofilms and be used as a promising means of controlling biofilms.
Therefore, by careful analysis of the individual biofilm, and by treating it with enzymes capable of hydrolyzing different components of the biofilm, one could effectively eradicate different biofilms. Besides, by combining different degrading enzymes or use them as non-antibiotic assistants, we may be able to further improve the effectiveness of biofilm eradication.
2.3.2.3. Using metabolites to eradicate biofilm
Secondary metabolites have been found to serve as intercellular signals to regulate gene expression, subsequently regulating various microbial physiological functions, including biofilm formation (Dufour and Rao 2011; Yang et al. 2012). Thus, metabolites can also be used to control biofilm formation (Figure 5D).
Carolacton, a secondary metabolite isolated from Sorangium cellulosum, showed high eradication activity against S. mutans biofilms by causing cell chain elongation, cell morphology changes, and even cell death of S. mutans in biofilm (Kunze et al. 2010). Rhamnolipid, the biosurfactant produced by Pseudomonas spp. showed the ability to disrupt and eradicate S. aureus biofilms (E Silva et al. 2017) (Table 2). D-amino acids could be incorporated into the bacterial cell wall to disengage the TasA fibers that anchor to the cell wall, resulting in the release of amyloid fibers which link cells in the biofilms together, thereby eradicating B. subtilis, S. aureus, and P. aeruginosa biofilms (Kolodkin-Gal et al. 2010). Further research revealed that exogenous D-tyrosine could contribute to the repulsive nature of the E. coli cells to inhibit bacterial attachment (Xing et al. 2015) (Table 2). These characteristics may provide us a new way to control biofilm formation via adding exogenous D-amino acids to eradicate harmful biofilm. Metabolites from plants can also be used to control biofilms. A secondary metabolite of Actinidia deliciosa has been found to reduce exopolysaccharides, protein, and eDNA contents in the EPS of A. baumannii, and can be used to eradicate its biofilm (Tiwari et al. 2017).
Rhamnolipid coated silver (Ag) and iron oxide (Fe3O4) nanoparticles also show good anti-biofilm properties in preventing and inhibiting the biofilms formed by S. aureus and P. aeruginosa (Khalid et al. 2019). They are potentially useful in medical fields, e.g. for antimicrobial coating and wound dressing. These results suggest that metabolites can not only eradicate the biofilms already formed but also prevent and inhibit biofilm formation. Therefore, metabolites exhibit great potential to be used in multifunctional preparation to control biofilm formation.
2.3.2.4. Nitric oxide
Nitric oxide (NO) can also play a role in biofilm eradication (Rumbaugh and Sauer 2020). NO produced by the anaerobic respiration processes inside the P. aeruginosa biofilm can initiate biofilm dispersal at low and non-toxic concentrations (Barraud et al. 2006). Further research revealed that NO signaling in the P. aeruginosa biofilm can stimulate PDE activity, thereby decreasing the intracellular c-di-GMP levels and enhancing biofilm dispersal. In addition to P. aeruginosa, NO-induced biofilms dispersal were observed in many other bacteria including E. coli and S. aureus (Barraud et al. 2015).
Exogenous NO addition treatment can similarly induce biofilm dispersal (Barraud et al. 2009). For example, NO-releasing polymers are able to reduce the metabolic activity of various biofilms in a dose-dependent manner (Yang et al. 2020). In addition, NO-releasing cyclodextrins can eradicate P. aeruginosa biofilm irrespective of the matrix composition (Rouillard et al. 2020). By integrating NO prodrug with the glutathione-sensitive α-cyclodextrin and chlorin e6 prodrug, the generated supramolecular nanocarrier was reported to exhibit rapid NO release under the trigger of overexpressed glutathione in the biofilm, leading to effective eradication of S. aureus biofilm (Hu et al. 2020).
Due to the small size and easy penetration, NO represents a powerful potential agent to eradicate the biofilm. Therefore, development of NO-releasing carriers, such as NO prodrug-coated devices or nanoparticles might be a promising avenue for biofilm eradication.
Overall, disrupting the biofilm matrix with biochemical methods has been proven to be an efficient approach to remove EPS, disaggregate microorganisms, and eradicate the biofilm. Compared with the above-mentioned physical methods, biochemical methods are more specific, safer, and can be more widely applied for treating specific biofilm-related infections.
3. Concluding remarks
The existence of biofilms turns out to be a double-edged sword for humans. Beneficial biofilms can bring much benefit, but harmful biofilms can bring enormous damages. In this review, we discuss the currently available strategies to prevent, inhibit, and eradicate harmful biofilms in the medical fields.
There have been many strategies to control harmful biofilms. Due to the complex composition of the biofilm and its strong resistance to stress, it is difficult to completely eradicate biofilm by a single method. Therefore, using a cocktail approach by combining different strategies may be more effective in treating biofilm-related infections.
Although the research on biofilms is continuously deepening, the mechanisms of biofilm formation still need further research. A clear understanding of the details of the biofilm formation process and regulation of its signaling pathways can help one discover new targets, which can be used to develop small molecules, peptides, or protein inhibitors to produce excellent anti-biofilm effects. We can also design synthetic derivatives with structural modification to develop more effective inhibitors or changes application methods to achieve more efficient and rapid eradication of harmful biofilms.
Once the biofilm is formed, antibiotics will be less effective. Although there are many physical and biochemical methods that can be used to prevent microbial attachment to tissue implants and medical devices, which are, however, difficult to apply for infected tissues. The use of vaccines to prevent biofilm formation may be an effective strategy. Due to the diversity of microbial antigens, the proteins involved in the specific signaling pathway of biofilm formation may serve as better targets for designing vaccines to eradicate biofilms.
Bacteriophages and phage lysins have also been found to be effective as anti-biofilm agents in recent years. Using bacteriophage directly or immobilizing phage lysin as a biological coating on the surface of tissue implants and medical devices can inhibit biofilm formation; combining multiple bacteriophages or phage lysins could achieve an even more broad-spectrum antibacterial effect. At present, the clinical application of this method is still under active research, but we believe such products will appear in the near future. Compared with phage lysins, bacteriophages can not only kill bacteria directly, but also induce host bacteria to express degrading enzymes for EPS, thereby accelerating the eradication of mature biofilm. Therefore, the bacteriophage can be engineered to expand its bactericidal spectrum to better control the biofilm (Lu and Collins 2007; Pires et al. 2016; Fang et al. 2020; Melo et al. 2020).
Since some anti-biofilm agents do not kill bacteria directly, such agents possibly won’t cause bacterial drug resistance. Compared with antibiotics, anti-biofilm agents have no restriction, which has certain advantages in controlling biofilms. In the future, after research on the mechanism of biofilm life-cycle is intensified, more and stronger anti-biofilm agents may be found.
Studies have shown that the EPS produced by bacteria can maintain a high osmotic pressure in the biofilm, thus enhancing the ability of biofilm to absorb nutrients from the outside to drive biofilm expansion (Yan et al. 2017). Thus, how to modulate the osmotic pressure of biofilms to control harmful biofilm formation yet enhance beneficial biofilm formation may be a new research focus in the future.
Future research on biofilm controlling may require multidisciplinary research. In general, there seem to still have many difficulties ahead, but with the deepening of research, we believe that these difficulties will eventually be overcome. We hope this summary can motivate more innovative and effective strategies for preventing, inhibiting, and eradicating harmful biofilms.
Funding
This research was funded by the National Key Research and Development Program of China (grant 2018YFD0500204), the National Natural Science Foundation of China (grants 31770087, 31970074, and 32000055). MYG was supported by the Intramural Research Program of the U.S. National Library of Medicine at the NIH.
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. 2011. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 43(4):550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusrewil S, Alshanta OA, Albashaireh K, Alqahtani S, Nile CJ, Scott JA, McLean W. 2020. Detection, treatment and prevention of endodontic biofilm infections: what's new in 2020? Crit Rev Microbiol. 46(2):194–212. [DOI] [PubMed] [Google Scholar]
- Akens MK, Chien C, Katchky RN, Kreder HJ, Finkelstein J, Whyne CM. 2018. The impact of thermal cycling on Staphylococcus aureus biofilm growth on stainless steel and titanium orthopaedic plates. BMC Musculoskelet Disord. 19(1):260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaguer-Flores A, Olivares-Navarrete R, Wieland M, Ximénez-Fyvie LA, Schwartz Z, Boyan BD. 2012. Influence of topography and hydrophilicity on initial oral biofilm formation on microstructured titanium surfaces in vitro. Clin Oral Implan Res. 23(3):301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova AG, Ellis GA, Wijesekera HW, Vora GJ. 2019. Microbial composition and variability of natural marine planktonic and biofouling communities from the Bay of Bengal. Front Microbiol. 10:2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Zhang X, Jiang Q, Xue T, Sun B. 2015. Pfs promotes autolysis-dependent release of eDNA and biofilm formation in Staphylococcus aureus. Med Microbiol Immunol. 204(2):215–226. [DOI] [PubMed] [Google Scholar]
- Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci USA. 110(46):18478–18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barki KG, Das A, Dixith S, Ghatak PD, Mathew-Steiner S, Schwab E, Khanna S, Wozniak DJ, Roy S, Sen CK. 2019. Electric field based dressing disrupts mixed-species bacterial biofilm infection and restores functional wound healing. Ann Surg. 269(4):756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 188(21):7344–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Kelso MJ, Rice SA, Kjelleberg S. 2015. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases current pharmaceutical design. Curr Pharm Des. 21:31–42 [DOI] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 191(23):7333–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer AS, Park S, Ramos MC, Nast CC, Eftekhar F, Schiller NL. 1992. Effects of alginase on the natural history and antibiotic therapy of experimental endocarditis caused by mucoid Pseudomonas aeruginosa. Infect Immun. 60(10):3979–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech IB, Sunner J. 2004. Biocorrosion: towards understanding interactions between biofilms and metals. Curr Opin Biotechnol. 15(3):181–186. [DOI] [PubMed] [Google Scholar]
- Beech IB, Sunner JA, Arciola CR, Cristiani P. 2006. Microbially-influenced corrosion: damage to prostheses, delight for bacteria. Int J Artif Organs. 29(4):443–452. [DOI] [PubMed] [Google Scholar]
- Blackledge MS, Worthington RJ, Melander C. 2013. Biologically inspired strategies for combating bacterial biofilms. Curr Opin Pharmacol. 13(5):699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer WD, Gunnewiek PJAK, Veenhuis M, Bock E, Laanbroek HJ. 1991. Nitrification at low pH by aggregated chemolithotrophic bacteria. Appl Environ Microbiol. 57(12):3600–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4(4):e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol. 28:439–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo S, Heredia N, Garcia S. 2015. 2(5H)-Furanone, epigallocatechin gallate, and a citric-based disinfectant disturb quorum-sensing activity and reduce motility and biofilm formation of Campylobacter jejuni. Folia Microbiol (Praha). 60(1):89–95. [DOI] [PubMed] [Google Scholar]
- Chen L, Li X, Zhou X, Zeng J, Ren Z, Lei L, Kang D, Zhang K, Zou J, Li Y. 2018a. Inhibition of Enterococcus faecalis growth and biofilm formation by molecule targeting cyclic di-AMP synthetase activity. J Endod. 44(9):1381–1388. [DOI] [PubMed] [Google Scholar]
- Chen X, Hirt H, Li Y, Gorr SU, Aparicio C. 2014. Antimicrobial GL13K peptide coatings killed and ruptured the wall of Streptococcus gordonii and prevented formation and growth of biofilms. PLoS ONE. 9(11):e111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang L, Zhang M, Liu H, Lu P, Lin K. 2018b. Quorum sensing inhibitors: a patent review (2014–2018). Expert Opin Ther Pat. 28(12):849–865. [DOI] [PubMed] [Google Scholar]
- Cho KH, Tryon RG, Kim JH. 2020. Screening for diguanylate cyclase (DGC) inhibitors mitigating bacterial biofilm formation. Front Chem. 8:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PH, Vu TMN, Pham HT, Woodward JJ, Turner MS, Tong L. 2017. Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria. Proc Natl Acad Sci USA. 114(35):E7226–E7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SH, Galperin MY. 2016. Diversity of cyclic di-GMP-binding proteins and mechanisms. J Bacteriol. 198(1):32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, Jenal U. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci USA. 104(10):4112–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Gründling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 11(8):513–524. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science. 284:1318–1322. [DOI] [PubMed] [Google Scholar]
- Cui YX, Wu D, Mackey HR, Chui HK, Chen GH. 2018. Application of a moving-bed biofilm reactor for sulfur-oxidizing autotrophic denitrification. Water Sci Technol. 77(3-4):1027–1034. [DOI] [PubMed] [Google Scholar]
- da Silva WJ, Leal CM, Viu FC, Goncalves LM, Barbosa CM, Del Bel Cury AA. 2015. Influence of surface free energy of denture base and liner materials on Candida albicans biofilms. J Investig Clin Dent. 6(2):141–146. [DOI] [PubMed] [Google Scholar]
- de Avila ED, Lima BP, Sekiya T, Torii Y, Ogawa T, Shi W, Lux R. 2015. Effect of UV-photofunctionalization on oral bacterial attachment and biofilm formation to titanium implant material. Biomaterials. 67:84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Nunez C, Reffuveille F, Haney EF, Straus SK, Hancock RE. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10(5):e1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Somma A, Moretta A, Cane C, Cirillo A, Duilio A. 2020. Antimicrobial and antibiofilm peptides. Biomolecules. 10(4):652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Trapani D, Christensso M, Odegaard H. 2011. Hybrid activated sludge/biofilm process for the treatment of municipal wastewater in a cold climate region: a case study. Water Sci Technol. 63(6):1121–1129. [DOI] [PubMed] [Google Scholar]
- Domenech M, Garcia E, Moscoso M. 2011. In vitro destruction of Streptococcus pneumoniae biofilms with bacterial and phage peptidoglycan hydrolases. Antimicrob Agents Chemother. 55(9):4144–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez EG, Zarnowski R, Choy HL, Zhao M, Sanchez H, Nett JE, Andes DR. 2019. Conserved role for biofilm matrix polysaccharides in Candida auris drug resistance. mSphere. 4(1):e00680–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drulis-Kawa Z, Maciejewska GM-SB. 2015. Bacteriophages and phage-derived proteins – application approaches. Curr Med Chem. 22:1757–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour N, Rao RP. 2011. Secondary metabolites and other small molecules as intercellular pathogenic signals. FEMS Microbiol Lett. 314(1):10–17. [DOI] [PubMed] [Google Scholar]
- E Silva SS, Carvalho JWP, Aires CP, Nitschke M. 2017. Disruption of Staphylococcus aureus biofilms using rhamnolipid biosurfactants. J Dairy Sci. 100(10):7864–7873. [DOI] [PubMed] [Google Scholar]
- Edwards KJ, Bond PL, Gihring TM, Banfield JF. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science. 287(5459):1796–1799. [DOI] [PubMed] [Google Scholar]
- Elbourne A, Cheeseman S, Atkin P, Truong NP, Syed N, Zavabeti A, Mohiuddin M, Esrafilzadeh D, Cozzolino D, McConville CF, Dickey MD, Crawford RJ, Kalantar-Zadeh K, Chapman J, Daeneke T, Truong VK. 2020. Antibacterial liquid metals: biofilm treatment via magnetic activation. ACS Nano. 14(1):802–817 [DOI] [PubMed] [Google Scholar]
- Erriu M, Blus C, Szmukler-Moncler S, Buogo S, Levi R, Barbato G, Madonnaripa D, Denotti G, Piras V, Orru G. 2014. Microbial biofilm modulation by ultrasound: current concepts and controversies. Ultrason Sonochem. 21(1):15–22. [DOI] [PubMed] [Google Scholar]
- Fahmi T, Faozia S, Port GC, Cho KH, Freitag NE. 2019. The second messenger c-di-AMP regulates diverse cellular pathways involved in stress response, biofilm formation, cell wall homeostasis, SpeB expression, and virulence in Streptococcus pyogenes. Infect Immun. 87(6):e00147–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K, Park OJ, Hong SH. 2020. Controlling biofilms using synthetic biology approaches. Biotechnol Adv. 40:107518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farisa Banu S, Thamotharan S, Gowrishankar S, Karutha Pandian S, Nithyanand P. 2019. Marine bacterial DNase curtails virulence and disrupts biofilms of Candida albicans and non-albicans Candida species. Biofouling. 35(9):975–985. [DOI] [PubMed] [Google Scholar]
- Fernicola S, Paiardini A, Giardina G, Rampioni G, Leoni L, Cutruzzola F, Rinaldo S. 2016. In silico discovery and in vitro validation of catechol-containing sulfonohydrazide compounds as potent inhibitors of the diguanylate cyclase PleD. J Bacteriol. 198(1):147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo AMS, Ferreira FA, Beltrame CO, Côrtes MF. 2017. The role of biofilms in persistent infections and factors involved in ica-independent biofilm development and gene regulation in Staphylococcus aureus. Crit Rev Microbiol. 43(5):602–620. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol. 8(9):623–633. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wuertz S. 2019. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 17(4):247–260. [DOI] [PubMed] [Google Scholar]
- Foster HA, Ditta IB, Varghese S, Steele A. 2011. Photocatalytic disinfection using titanium dioxide: spectrum and mechanism of antimicrobial activity. Appl Microbiol Biotechnol. 90(6):1847–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yu Z, Liu S, Chen B, Zhu L, Li Z, Chou SH, He J. 2018. C-di-GMP regulates various phenotypes and insecticidal activity of Gram-positive Bacillus thuringiensis. Front Microbiol. 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. 2011. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 111:28–67. [DOI] [PubMed] [Google Scholar]
- Galperin MY. 2018. What bacteria want. Environ Microbiol. 20(12):4221–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Shalaeva DN. 2018. A bacterial coat that is not pure cotton. Science. 359(6373):276–277. [DOI] [PubMed] [Google Scholar]
- Garcia-Sanchez L, Melero B, Jaime I, Rossi M, Ortega I, Rovira J. 2019. Biofilm formation, virulence and antimicrobial resistance of different Campylobacter jejuni isolates from a poultry slaughterhouse. Food Microbiol. 83:193–199. [DOI] [PubMed] [Google Scholar]
- García S, Trueba A, Vega LM, Madariaga E. 2016. Impact of the surface roughness of AISI 316L stainless steel on biofilm adhesion in a seawater-cooled tubular heat exchanger-condenser. Biofouling. 32(10):1185–1193. [DOI] [PubMed] [Google Scholar]
- Girennavar B, Cepeda ML, Soni KA, Vikram A, Jesudhasan P, Jayaprakasha GK, Pillai SD, Patil BS. 2008. Grapefruit juice and its furocoumarins inhibits autoinducer signaling and biofilm formation in bacteria. Int J Food Microbiol. 125(2):204–208. [DOI] [PubMed] [Google Scholar]
- Gundlach J, Rath H, Herzberg C, Mader U, Stülke J. 2016. Second messenger signaling in Bacillus subtilis: accumulation of cyclic di-AMP inhibits biofilm formation. Front Microbiol. 7:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Sarkar S, Das B, Bhattacharjee S, Tribedi P. 2016. Biofilm, pathogenesis and prevention-a journey to break the wall: a review. Arch Microbiol. 198(1):1–15. [DOI] [PubMed] [Google Scholar]
- Gutierrez D, Ruas-Madiedo P, Martinez B, Rodriguez A, Garcia P. 2014. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE. 9(9):e107307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilos DL. 2014. Host-microbial interactions in patients with chronic rhinosinusitis. J Allergy Clin Immunol. 133(3):640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Ye W, Wei D, Xu W, Du B, Wei Q. 2018. Simultaneous nitrification-denitrification and membrane fouling alleviation in a submerged biofilm membrane bioreactor with coupling of sponge and biodegradable PBS carrier. Bioresour Technol. 270:156–165. [DOI] [PubMed] [Google Scholar]
- Hathroubi S, Mekni MA, Domenico P, Nguyen D, Jacques M. 2017. Biofilms: microbial shelters against antibiotics. Microb Drug Resist. 23(2):147–156. [DOI] [PubMed] [Google Scholar]
- Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 13(5):298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yin W, Galperin MY, Chou SH. 2020. Cyclic di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 48(6):2807–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Wang Q, Hu Y, Liang J, Jiang Y, Ma R, Tang Z, Huang Z. 2012. Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. Int J Antimicrob Agents. 40(1):30–35. [DOI] [PubMed] [Google Scholar]
- Hirota K, Murakami K, Nemoto K, Miyake Y. 2005. Coating of a surface with 2-methacryloyloxyethyl phosphorylcholine (MPC) co-polymer significantly reduces retention of human pathogenic microorganisms. FEMS Microbiol Lett. 248(1):37–45. [DOI] [PubMed] [Google Scholar]
- Hirota K, Yumoto H, Miyamoto K, Yamamoto N, Murakami K, Hoshino Y, Matsuo T, Miyake Y. 2011. MPC-polymer reduces adherence and biofilm formation by oral bacteria. J Dent Res. 90(7):900–905. [DOI] [PubMed] [Google Scholar]
- Hirt H, Gorr SU. 2013. Antimicrobial peptide GL13K is effective in reducing biofilms of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 57(10):4903–4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hola V, Ruzicka F, Horka M. 2010. Microbial diversity in biofilm infections of the urinary tract with the use of sonication techniques. FEMS Immunol Med Microbiol. 59(3):525–528. [DOI] [PubMed] [Google Scholar]
- Hotaling S, Foley ME, Zeglin LH, Finn DS, Tronstad LM, Giersch JJ, Muhlfeld CC, Weisrock DW. 2019. Microbial assemblages reflect environmental heterogeneity in alpine streams. Glob Chang Biol. 25(8):2576–2590. [DOI] [PubMed] [Google Scholar]
- Hou YJ, Yang WS, Hong Y, Zhang Y, Wang DC, Li DF. 2020. Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility in Escherichia coli. J Biol Chem. 295(3):808–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Deng Y, Jia F, Jin Q, Ji J. 2020. Surface charge switchable supramolecular nanocarriers for nitric oxide synergistic photodynamic eradication of biofilms. ACS Nano. 14(1):347–359. [DOI] [PubMed] [Google Scholar]
- Huang H, Peng C, Peng P, Lin Y, Zhang X, Ren H. 2018. Towards the biofilm characterization and regulation in biological wastewater treatment. Appl Microbiol Biotechnol. 103(3):1115–1129. [DOI] [PubMed] [Google Scholar]
- Hymes SR, Randis TM, Sun TY, Ratner AJ. 2013. DNase inhibits Gardnerella vaginalis biofilms in vitro and in vivo. J Infect Dis. 207(10):1491–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen TH, Tolker-Nielsen T, Givskov M. 2017. Bacterial biofilm control by perturbation of bacterial signaling processes. Int J Mol Sci. 18(9):1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CH, Piao YL, Huang X, Yoon EJ, Park SH, Lee K, Zhan CG, Cho H. 2016. Modeling and re-engineering of Azotobacter vinelandii alginate lyase to enhance its catalytic efficiency for accelerating biofilm degradation. PLoS ONE. 11(6):e0156197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Utada A, Davis KR, Thongsomboon W, Zamorano Sanchez D, Banakar V, Cegelski L, Wong GC, Yildiz FH. 2015. C-di-GMP regulates motile to sessile transition by modulating MshA pili biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS Pathog. 11(10):e1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia VC, Patel SKS, Kang YC, Lee JK. 2019. Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol Adv. 37(1):68–90. [DOI] [PubMed] [Google Scholar]
- Karygianni L, Ren Z, Koo H, Thurnheer T. 2020. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 28(8):668–681. [DOI] [PubMed] [Google Scholar]
- Kazemzadeh-Narbat M, Kindrachuk J, Duan K, Jenssen H, Hancock RE, Wang R. 2010. Antimicrobial peptides on calcium phosphate-coated titanium for the prevention of implant-associated infections. Biomaterials. 31(36):9519–9526. [DOI] [PubMed] [Google Scholar]
- Khalid HF, Tehseen B, Sarwar Y, Hussain SZ, Khan WS, Raza ZA, Bajwa SZ, Kanaras AG, Hussain I, Rehman A. 2019. Biosurfactant coated silver and iron oxide nanoparticles with enhanced anti-biofilm and anti-adhesive properties. J Hazard Mater. 364:441–448. [DOI] [PubMed] [Google Scholar]
- Kim KH, Loch C, Waddell JN, Tompkins G, Schwass D. 2017. Surface characteristics and biofilm development on selected dental ceramic materials. Int J Dent. 2017:7627945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. D-amino acids trigger biofilm disassembly. Science. 328(5978):627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. 2017. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 15(12):740–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 327(5967):866–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze B, Reck M, Dötsch A, Lemme A, Schummer D, Irschik H, Steinmetz H, Wagner-Döbler I. 2010. Damage of Streptococcus mutans biofilms by carolacton, a secondary metabolite from the myxobacterium Sorangium cellulosum. BMC Microbiol. 10:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kviatkovski I, Mamane H, Lakretz A, Sherman I, Beno-Moualem D, Minz D. 2018. Resistance of a multiple-isolate marine culture to ultraviolet C irradiation: inactivation vs biofilm formation. Lett Appl Microbiol. 67(3):278–284. [DOI] [PubMed] [Google Scholar]
- Lamoth F, Kontoyiannis DP. 2018. The Candida auris Alert: facts and perspectives. J Infect Dis. 217(4):516–520. [DOI] [PubMed] [Google Scholar]
- Li G, Xie F, Zhang Y, Bosse JT, Langford PR, Wang C. 2015. Role of (p)ppGpp in viability and biofilm formation of Actinobacillus pleuropneumoniae S8. PLoS ONE. 10(10):e0141501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nickel R, Wu J, Lin F, van Lierop J, Liu S. 2019a. A new tool to attack biofilms: driving magnetic iron-oxide nanoparticles to disrupt the matrix. Nanoscale. 11(14):6905–6915. [DOI] [PubMed] [Google Scholar]
- Li T, Wang N, Chen S, Lu R, Li H, Zhang Z. 2017. Antibacterial activity and cytocompatibility of an implant coating consisting of TiO2 nanotubes combined with a GL13K antimicrobial peptide. Int J Nanomedicine. 12:2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang X, Zhao X, Yu B, Weng L, Li Y. 2019b. Efficient removal of metolachlor and bacterial community of biofilm in bioelectrochemical reactors. Appl Biochem Biotechnol. 189(2):384–395. [DOI] [PubMed] [Google Scholar]
- Lin S, Rong K, Lamichhane KM, Babcock RW, Kirs M, Cooney MJ. 2019. Anaerobic-aerobic biofilm-based digestion of chemical contaminants of emerging concern (CEC) and pathogen indicator organisms in synthetic wastewater. Bioresour Technol. 299:122554. [DOI] [PubMed] [Google Scholar]
- Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 32(14):2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling N, Forsythe S, Wu Q, Ding Y, Zhang J, Zeng H. 2020. Insights into Cronobacter sakazakii biofilm formation and control strategies in the food industry. Engineering. 6(4):393–405. [Google Scholar]
- Lohse MB, Gulati M, Johnson AD, Nobile CJ. 2018. Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol. 16(1):19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood R, Winer BY, Pelzek AJ, Diez-Martinez R, Thandar M, Euler CW, Schuch R, Fischetti VA. 2015. Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother. 59(4):1983–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. 2007. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci USA. 104(27):11197–11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Dong B, Wang K, Cai S, Liu T, Cheng X, Lei D, Chen Y, Li Y, Kong J, Chen Y. 2017. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE. 12(4):e0176883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łusiak-Szelachowska M, Weber-Dąbrowska B, Górski A. 2020. Bacteriophages and lysins in biofilm control. Virol Sin. 35(2):125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado I, Silva LR, Giaouris ED, Melo LF, Simoes M. 2020. Quorum sensing in food spoilage and natural-based strategies for its inhibition. Food Res Int. 127:108754. [DOI] [PubMed] [Google Scholar]
- Mei L, Busscher HJ, van der Mei HC, Ren Y. 2011. Influence of surface roughness on streptococcal adhesion forces to composite resins. Dent Mater. 27(8):770–778. [DOI] [PubMed] [Google Scholar]
- Meis JF, Chowdhary A. 2018. Candida auris: a global fungal public health threat. Lancet Infect Dis. 18(12):1298–1299. [DOI] [PubMed] [Google Scholar]
- Melo LDR, Brandao A, Akturk E, Santos SB, Azeredo J. 2018. Characterization of a new Staphylococcus aureus Kayvirus harboring a lysin active against biofilms. Viruses. 10(4):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo LDR, Oliveira H, Pires DP, Dabrowska K, Azeredo J. 2020. Phage therapy efficacy: a review of the last 10 years of preclinical studies. Crit Rev Microbiol. 46(1):78–99. [DOI] [PubMed] [Google Scholar]
- Meng X, Shi Y, Ji W, Meng X, Zhang J, Wang H, Lu C, Sun J, Yan Y. 2011. Application of a Bacteriophage lysin to disrupt biofilms formed by the animal pathogen Streptococcus suis. Appl Environ Microbiol. 77(23):8272–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino L, Procura F, Trejo FM, Bueno DJ, Golowczyc MA. 2019. Biofilm formation by Salmonella sp. in the poultry industry: detection, control and eradication strategies. Food Res Int. 119:530–540. [DOI] [PubMed] [Google Scholar]
- Meyer-Dombard DR, Casar CP, Simon AG, Cardace D, Schrenk MO, Arcilla CA. 2018. Biofilm formation and potential for iron cycling in serpentinization-influenced groundwater of the Zambales and Coast Range ophiolites. Extremophiles. 22(3):407–431. [DOI] [PubMed] [Google Scholar]
- Monds RD, O'Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17(2):73–87. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Strumillo J, Zimmer J. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 493(7431):181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso JA, Schramke H, Zhang Y, Tosi T, Dehbi A, Jung K, Gründling A. 2016. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter. J Bacteriol. 198(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Hu Y, Tan CH, Rice SA, Cao B. 2018. Engineering a light-responsive, quorum quenching biofilm to mitigate biofouling on water purification membranes. Sci. Adv 4(12):eaau1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müsken M, Pawar V, Schwebs T, Bähre H, Felgner S, Weiss S, Häussle S. 2018. Breaking the vicious cycle of antibiotic killing and regrowth of biofilm-residing Pseudomonas aeruginosa. Antimicrob Agents Chemother. 62(12):e01635–01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Hori N, Ando H, Namba S, Toyama T, Nishimiya N, Yamashita K. 2016. Surface free energy predominates in cell adhesion to hydroxyapatite through wettability. Mater Sci Eng C Mater Biol Appl. 62:283–292. [DOI] [PubMed] [Google Scholar]
- Ng CG, Loke MF, Goh KL, Vadivelu J, Ho B. 2017. Biofilm formation enhances Helicobacter pylori survivability in vegetables. Food Microbiol. 62:68–76. [DOI] [PubMed] [Google Scholar]
- Nijland R, Hall MJ, Burgess JG. 2010. Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS ONE. 5(12):e15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. 2013. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA. 110(44):17981–17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opoku-Temeng C, Sintim HO. 2016a. Inhibition of cyclic diadenylate cyclase, DisA, by polyphenols. Sci Rep. 6:25445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opoku-Temeng C, Sintim HO. 2016b. Potent inhibition of cyclic diadenylate monophosphate cyclase by the antiparasitic drug, suramin. Chem Commun (Camb). 52(19):3754–3757. [DOI] [PubMed] [Google Scholar]
- Orman MA, Brynildsen MP. 2016. Persister formation in Escherichia coli can be inhibited by treatment with nitric oxide. Free Radic Biol Med. 93:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Sun F, Feng W, Sun Y, Qiu X, Xiong L, Liu Y, Chen Y. 2016. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J Appl Microbiol. 120(4):966–974. [DOI] [PubMed] [Google Scholar]
- Packiavathy IA, Priya S, Pandian SK, Ravi AV. 2014. Inhibition of biofilm development of uropathogens by curcumin - an anti-quorum sensing agent from Curcuma longa. Food Chem. 148:453–460. [DOI] [PubMed] [Google Scholar]
- Padder SA, Prasad R, Shah AH. 2018. Quorum sensing: a less known mode of communication among fungi. Microbiol Res. 210:51–58. [DOI] [PubMed] [Google Scholar]
- Peng X, Zhang Y, Bai G, Zhou X, Wu H. 2016. Cyclic di-AMP mediates biofilm formation. Mol Microbiol. 99(5):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival SL. 2017. Importance of biofilm formation in surgical infection. Br J Surg. 104(2):e85–e94. [DOI] [PubMed] [Google Scholar]
- Pires DP, Cleto S, Sillankorva S, Azeredo J, Lu TK. 2016. Genetically engineered phages: a review of advances over the last decade. Microbiol Mol Biol Rev. 80(3):523–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poonacha N, Nair S, Desai S, Tuppad D, Hiremath D, Mohan T, Vipra A, Sharma U. 2017. Efficient killing of planktonic and biofilm-embedded coagulase-negative staphylococci by bactericidal protein P128. Antimicrob Agents Chemother. 61(8):e00457–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 282(17):12860–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim MI, Szafranski SP, Ingendoh-Tsakmakidis A, Stiesch M, Mueller PP. 2019. Evidence for inoculum size and gas interfaces as critical factors in bacterial biofilm formation on magnesium implants in an animal model. Colloids Surf B Biointerfaces. 186:110684. [DOI] [PubMed] [Google Scholar]
- Rajput A, Thakur A, Sharma S, Kumar M. 2018. aBiofilm: a resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res. 46(D1):D894–D900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 68(11):5459–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. 2011. Physicochemical regulation of biofilm formation. MRS Bull. 36(5):347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M, Monteiro FJ, Ferraz MP. 2012. Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions. Biomatter. 2(4):176–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripolles-Avila C, Garcia-Hernandez N, Cervantes-Huaman BH, Mazaheri T, Rodriguez-Jerez JJ. 2019. Quantitative and compositional study of monospecies biofilms of spoilage microorganisms in the meat industry and their interaction in the development of multispecies biofilms. Microorganisms. 7(12):655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY. 2015. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23(9):545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 77(1):1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 57(3):629–639. [DOI] [PubMed] [Google Scholar]
- Rouillard KR, Markovetz MR, Bacudio LG, Hill DB, Schoenfisch MH. 2020. Pseudomonas aeruginosa biofilm eradication via nitric oxide-releasing cyclodextrins. ACS Infect Dis. 6(7):1940–1950. [DOI] [PubMed] [Google Scholar]
- Roy R, Tiwari M, Donelli G, Tiwari V. 2018. Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence. 9(1):522–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh KP, Sauer K. 2020. Biofilm dispersion. Nat Rev Microbiol. 18(10):571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambanthamoorthy K, Luo C, Pattabiraman N, Feng X, Koestler B, Waters CM, Palys TJ. 2014. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling. 30(1):17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambanthamoorthy K, Sloup RE, Parashar V, Smith JM, Kim EE, Semmelhack MF, Neiditch MB, Waters CM. 2012. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob Agents Chemother. 56(10):5202–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 184(4):1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch R, Khan BK, Raz A, Rotolo JA, Wittekind M. 2017. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob Agents Chemother. 61(7):e02666–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoffone VC, Chiarelli LR, Makarov V, Brackman G, Israyilova A, Azzalin A, Forneris F, Riabova O, Savina S, Coenye T, Riccardi G, Buroni S. 2016. Discovery of new diketopiperazines inhibiting Burkholderia cenocepacia quorum sensing in vitro and in vivo. Sci Rep. 6:32487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne CJ, Zhang CF, Samaranayake LP. 2011. Dental plaque biofilm in oral health and disease. Chin J Dent Res 14(2):87–94. [PubMed] [Google Scholar]
- Serra DO, Klauck G, Hengge R. 2015. Vertical stratification of matrix production is essential for physical integrity and architecture of macrocolony biofilms of Escherichia coli. Environ Microbiol. 17(12):5073–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AA, Hasan F, Hameed A, Ahmed S. 2008. Biological degradation of plastics: a comprehensive review. Biotechnol Adv. 26:246–265. [DOI] [PubMed] [Google Scholar]
- Sharma U, Vipra A, Channabasappa S. 2018. Phage-derived lysins as potential agents for eradicating biofilms and persisters. Drug Discov Today. 23(4):848–856. [DOI] [PubMed] [Google Scholar]
- Shukla SK, Hariharan S, Rao TS. 2020. Uranium bioremediation by acid phosphatase activity of Staphylococcus aureus biofilms: can a foe turn a friend? J Hazard Mater. 384:121316. [DOI] [PubMed] [Google Scholar]
- Singh PK, Donovan DM, Kumar A. 2014. Intravitreal injection of the chimeric phage endolysin Ply187 protects mice from Staphylococcus aureus endophthalmitis. Antimicrob Agents Chemother. 58(8):4621–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son JS, Lee SJ, Jun SY, Yoon SJ, Kang SH, Paik HR, Kang JO, Choi YJ. 2010. Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Appl Microbiol Biotechnol. 86(5):1439–1449. [DOI] [PubMed] [Google Scholar]
- Steiner S, Lori C, Boehm A, Jenal U. 2013. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 32(3):354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternisa M, Klancnik A, Smole Mozina S. 2019. Spoilage Pseudomonas biofilm with Escherichia coli protection in fish meat at 5 °C. J Sci Food Agric. 99(10):4635–4641. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol. 56:187–209. [DOI] [PubMed] [Google Scholar]
- Stülke J, Kruger L. 2020. Cyclic di-AMP Signaling in Bacteria. Annu Rev Microbiol. 74:159–179. [DOI] [PubMed] [Google Scholar]
- Su J, Zou X, Huang L, Bai T, Liu S, Yuan M, Chou SH, He YW, Wang H, He J. 2016. DgcA, a diguanylate cyclase from Xanthomonas oryzae pv. oryzae regulates bacterial pathogenicity on rice. Sci Rep. 6:25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki K, Hanawa T, Yonezawa H, Osaki T, Fukutomi T, Kawakami H, Yamamoto T, Kamiya S. 2013. Role of (p)ppGpp in biofilm formation and expression of filamentous structures in Bordetella pertussis. Microbiology. 159(Pt 7):1379–1389. [DOI] [PubMed] [Google Scholar]
- Tang Q, Luo Y, Zheng C, Yin K, Ali MK, Li X, He J. 2015. Functional analysis of a c-di-AMP-specific phosphodiesterase MsPDE from Mycobacterium smegmatis. Int J Biol Sci. 11(7):813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Yin K, Qian H, Zhao Y, Wang W, Chou SH, Fu Y, He J. 2016. Cyclic di-GMP contributes to adaption and virulence of Bacillus thuringiensis through a riboswitch-regulated collagen adhesion protein. Sci Rep. 6:28807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Zhu J, Feng L, Li J, Liu X. 2019. Characterization of LuxI/LuxR and their regulation involved in biofilm formation and stress resistance in fish spoilers Pseudomonas fluorescens. Int J Food Microbiol. 297:60–71. [DOI] [PubMed] [Google Scholar]
- Tapia-Rodriguez MR, Hernandez-Mendoza A, Gonzalez-Aguilar GA, Martinez-Tellez MA, Martins CM, Ayala-Zavala JF. 2017. Carvacrol as potential quorum sensing inhibitor of Pseudomonas aeruginosa and biofilm production on stainless steel surfaces. Food Control. 75:255–261. [Google Scholar]
- Teh WK, Dramsi S, Tolker-Nielsen T, Yang L, Givskov M. 2019. Increased intracellular cyclic di-AMP levels sensitize Streptococcus gallolyticus subsp. gallolyticus to osmotic stress and reduce biofilm formation and adherence on intestinal cells. J Bacteriol. 201(6): e00597–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teughels W, Assche NV, Sliepen I, Quirynen M. 2006. Effect of material characteristics and/or surface topography on biofilm development. Clin Oral Imp Res. 17:68–81. [DOI] [PubMed] [Google Scholar]
- Thomann A, de Mello Martins AG, Brengel C, Empting M, Hartmann RW. 2016. Application of dual inhibition concept within looped autoregulatory systems toward antivirulence agents against Pseudomonas aeruginosa infections. ACS Chem Biol. 11(5):1279–1286. [DOI] [PubMed] [Google Scholar]
- Thongsomboon W, Serra DO, Possling A, Hadjineophytou C, Hengge R, Cegelski L. 2018. Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science 359(6373): 334–338. [DOI] [PubMed] [Google Scholar]
- Tiwari V, Tiwari D, Patel V, Tiwari M. 2017. Effect of secondary metabolite of Actinidia deliciosa on the biofilm and extra-cellular matrix components of Acinetobacter baumannii. Microb Pathog. 110:345–351. [DOI] [PubMed] [Google Scholar]
- Valentini M, Filloux A. 2016. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem. 291(24):12547–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasavi HS, Sudeep HV, Lingaraju HB, Shyam Prasad K. 2017. Bioavailability-enhanced resveramax modulates quorum sensing and inhibits biofilm formation in Pseudomonas aeruginosa PAO1. Microb Pathog. 104:64–71. [DOI] [PubMed] [Google Scholar]
- Vyas N, Wang QX, Manmi KA, Sammons RL, Kuehne SA, Walmsley AD. 2020. How does ultrasonic cavitation remove dental bacterial biofilm? Ultrason Sonochem. 67:105112. [DOI] [PubMed] [Google Scholar]
- Wang F, He Q, Su K, Gao F, Huang Y, Lin Z, Zhu D, Gu L. 2016a. The PilZ domain of MrkH represents a novel DNA binding motif. Protein Cell. 7(10):766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lu L, Mao D, Huang Z, Cui Y, Jin S, Zuo Y, Ren ZJ. 2019a. Dominance of electroactive microbiomes in bioelectrochemical remediation of hydrocarbon-contaminated soils with different textures. Chemosphere. 235:776–784. [DOI] [PubMed] [Google Scholar]
- Wang H, Ye K, Wei X, Cao J, Xua X, Zhou G. 2013. Occurrence, antimicrobial resistance and biofilm formation of Salmonella isolates from a chicken slaughter plant in China. Food Control. 33(2):378–384. [Google Scholar]
- Wang X, Cai X, Ma H, Yin W, Zhu L, Li X, Lim HM, Chou SH, He J. 2019b. A c-di-AMP riboswitch controlling kdpFABC operon transcription regulates the potassium transporter system in Bacillus thuringiensis. Commun Biol. 2:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li X, Ling J. 2017. Streptococcus gordonii LuxS/autoinducer-2 quorum-sensing system modulates the dual-species biofilm formation with Streptococcus mutans. J Basic Microbiol. 57(7):605–616. [DOI] [PubMed] [Google Scholar]
- Wang YC, Chin KH, Tu ZL, He J, Jones CJ, Sanchez DZ, Yildiz FH, Galperin MY, Chou SH. 2016b. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat Commun. 7:12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu B, Grenier D, Yi L. 2019c. Regulatory Mechanisms of the LuxS/AI-2 System and Bacterial Resistance. Antimicrob Agents Chemother. 63(10):e01186–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick P, Kolter R. 2000. Biofilm, city of microbes. J Bacteriol. 182(10):2675–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science. 295(5559):1487. [DOI] [PubMed] [Google Scholar]
- Wolcott R, Costerton JW, Raoult D, Cutler SJ. 2013. The polymicrobial nature of biofilm infection. Clin Microbiol Infect. 19:107–112 [DOI] [PubMed] [Google Scholar]
- Wolfmeier H, Pletzer D, Mansour SC, Hancock REW. 2018. New perspectives in biofilm eradication. ACS Infect Dis. 4(2):93–106. [DOI] [PubMed] [Google Scholar]
- Wongpraparatana I, Matangkasombut O, Thanyasrisung P, Panich M. 2018. Effect of vital tooth bleaching on surface roughness and streptococcal biofilm formation on direct tooth-colored restorative materials. Oper Dent. 43(1):51–59. [DOI] [PubMed] [Google Scholar]
- Wood TK. 2017. Strategies for combating persister cell and biofilm infections. Microb Biotechnol. 10(5):1054–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing SF, Sun XF, Taylor AA, Walker SL, Wang YF, Wang SG. 2015. D-amino acids inhibit initial bacterial adhesion: thermodynamic evidence. Biotechnol Bioeng. 112(4):696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jones JE, Yu H, Yu Q, Christensen GD, Chen M, Sun H. 2015. Nanoscale plasma coating inhibits formation of Staphylococcus aureus biofilm. Antimicrob Agents Chemother. 59(12):7308–7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Bassler BL. 2019. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe. 26(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Nadell CD, Stone HA, Wingreen NS, Bassler BL. 2017. Extracellular-matrix-mediated osmotic pressure drives Vibrio cholerae biofilm expansion and cheater exclusion. Nat Commun. 8(1):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Zhang S, Lin D, Guo C, Yan L, Wang S, He Z. 2018. Nitrogen loading affects microbes, nitrifiers and denitrifiers attached to submerged macrophyte in constructed wetlands. Sci Total Environ. 622-623:121–126. [DOI] [PubMed] [Google Scholar]
- Yan Y, Tan F, Miao H, Wang H, Cao Y. 2019. Effect of shikonin against Candida albicans biofilms. Front Microbiol. 10:1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhang H, Wang J, Yu J, Wei H. 2017. A novel chimeric lysin with robust antibacterial activity against planktonic and biofilm methicillin-resistant Staphylococcus aureus. Sci Rep. 7:40182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Y, Wu H, Song Z, Hoiby N, Molin S, Givskov M. 2012. Combating biofilms. FEMS Immunol Med Microbiol. 65(2):146–157. [DOI] [PubMed] [Google Scholar]
- Yang L, Teles F, Gong W, Dua SA, Martin L, Schoenfisch MH. 2020. Antibacterial action of nitric oxide-releasing hyperbranched polymers against ex vivo dental biofilms. Dent Mater. 36(5):635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Tal-Gan Y, Paharik AE, Horswill AR, Blackwell HE. 2016. Structure-function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC-type receptor modulation. ACS Chem Biol. 11(7):1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron S, Römling U. 2014. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb Biotechnol. 7(6):496–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Cai X, Ma H, Zhu L, Zhang Y, Chou SH, Galperin MY, He J. 2020. A decade of research on the second messenger c-di-AMP. FEMS Microbiol Rev. doi: 10.1093/femsre/fuaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Wang Y, Liu L, He J. 2019. Biofilms: the microbial "Protective clothing" in extreme environments. Int J Mol Sci. 20(14):3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhu X, Zhou J, Cai Z. 2018. Biofilm inhibition and pathogenicity attenuation in bacteria by Proteus mirabilis. R Soc Open Sci. 5(4):170702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Miao Y, Zhang Q, Sun Y, Wu L, Peng Y. 2020. Mechanism of stable sewage nitrogen removal in a partial nitrification-anammox biofilm system at low temperatures: Microbial community and EPS analysis. Bioresour Technol. 297:122459. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li W, He ZG. 2013. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 288(5):3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Li Z, Zhang L, Li D. 2019. Enhancement of fipronil degradation with eliminating its toxicity in a microbial fuel cell and the catabolic versatility of anodic biofilm. Bioresour Technol. 290:121723. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang Y, Li D. 2017. Cometabolic degradation of chloramphenicol via a meta-cleavage pathway in a microbial fuel cell and its microbial community. Bioresour Technol. 229:104–110. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng M, Zhang H, Dai J, Guo Z, Li X, Ji Y, Cai R, Xi H, Wang X, Xue Y, Sun C, Feng X, Lei L, Han W, Gu J. 2018. Antibacterial effects of phage lysin LysGH15 on planktonic cells and biofilms of diverse staphylococci. Appl Environ Microbiol. 84(15):e00886–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Liu D, Huang W, Yang Y, Ji M, Nghiem LD, Trinh QT, Tran NH. 2019. Insights into biofilm carriers for biological wastewater treatment processes: current state-of-the-art, challenges, and opportunities. Bioresour Technol. 288:121619. [DOI] [PubMed] [Google Scholar]
- Zheng C, Ma Y, Wang X, Xie Y, Ali MK, He J. 2015. Functional analysis of the sporulation-specific diadenylate cyclase CdaS in Bacillus thuringiensis. Front Microbiol. 6:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, He L, Asiamah TK, Otto M. 2018. Colonization of medical devices by staphylococci. Environ Microbiol. 20(9):3141–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zheng C, Su J, Chen B, Fu Y, Xie Y, Tang Q, Chou SH, He J. 2016. Characterization of a natural triple-tandem c-di-GMP riboswitch and application of the riboswitch-based dual-fluorescence reporter. Sci Rep. 6:20871. [DOI] [PMC free article] [PubMed] [Google Scholar]