INTRODUCTION:
Strong evidence indicates that multiple genetic and environmental risk factors play a role in the pathogenesis of nonalcoholic steatohepatitis (NASH). We aimed to develop and validate a novel nomogram, incorporating both genetic and clinical factors, for predicting NASH.
METHODS:
A total of 1,070 Asian individuals with biopsy-confirmed nonalcoholic fatty liver disease (NAFLD) from 2 countries (China and South Korea) were recruited. The histological spectrum of NAFLD was classified according to the NASH clinical research network scoring system. The nomogram was developed in the Chinese training set (n = 402), and then, it was validated in both the Chinese internal validation set (n = 136) and the external Korean validation cohort (n = 532), respectively.
RESULTS:
Sex, metabolic syndrome, insulin resistance, serum aspartate aminotransferase levels, and PNPLA3 (rs738409) and HSD17B13 (rs72613567) genetic variants were strongly associated with NASH. Based on their regression coefficients, we developed a nomogram with good discriminatory ability (area under the receiver operating characteristic curve: 0.81, 95% confidence interval [CI] 0.77–0.85) and good calibration (Hosmer-Lemeshow test, P = 0.794) for identifying NASH. In the 2 validation cohorts, the nomogram showed high area under the receiver operating characteristic curves (internal validation set: 0.80, 95% CI 0.72–0.88; external validation cohort: 0.76, 95% CI 0.72–0.80) and good calibration.
DISCUSSION:
Our newly developed and externally validated nomogram, incorporating both genetic and clinical risk factors, may be conveniently used to predict NASH. Further validation studies in other ethnic groups are warranted to confirm its diagnostic utility to identify NASH, among patients with biopsy-proven NAFLD.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) has become the most common cause of chronic liver disease in many parts of the world, affecting up to a quarter of the general adult population (1). The histopathological spectrum of NAFLD ranges from simple steatosis (NAFL) to nonalcoholic steatohepatitis (NASH), advanced fibrosis, and cirrhosis (2). NASH is also becoming one of the main indications for liver transplantation (LT) among the registrants on LT waiting lists in both the United States and Europe (3,4). Approximately 20% of patients with NASH can progress to cirrhosis and hepatocellular carcinoma requiring LT (5). In addition, NASH is significantly associated with an increased risk of developing important extrahepatic complications, such as cardiovascular disease (which represents the leading cause of death in this patient population) and chronic kidney disease (6,7).
The correct identification of patients at increased risk of NASH is a critical step in the assessment of NAFLD (8). Treatment of NASH is a major focus of drug development worldwide (9,10). Although, currently, there are no therapies approved by the US Food and Drug Administration for NASH, there are ∼196 drugs being evaluated for the treatment of NASH and many ongoing phase 2 and phase 3 randomized controlled trials (11). To date, liver biopsy, and histological examination of liver tissue remains the reference method for diagnosing NASH. However, liver biopsy is an invasive method that cannot be used for screening the general population. Therefore, a major challenge is how to accurately and noninvasively identify patients with NASH, who may potentially benefit from early lifestyle intervention and future pharmacological treatment.
Metabolic disorders, such as obesity, type 2 diabetes mellitus (T2DM), and metabolic syndrome (MetS), are important clinical risk factors for NASH (12), but not all individuals with these risk factors have NASH. Familial clustering of NAFLD suggests that this disease is also strongly influenced by heritable genetic factors (13). Genome-wide association studies (GWASs) have also showed that some genetic variants play an important role in the development and progression of NAFLD (14–16). Patatin-like phospholipase domain-containing protein 3 (PNPLA3) genetic variant is the strongest genetic risk factor for the development of NASH (17). Indeed, studies have shown that individuals carrying the PNPLA3 (rs738409) variant have about a threefold increased likelihood of having NASH (18–20). Moreover, single-nucleotide polymorphisms (SNPs) in the transmembrane 6 superfamily member 2 (TM6SF2 rs58542926), membrane-bound O-acyltransferase domain containing 7 (MBOAT7 rs641738), and 17-beta-hydroxysteroid dehydrogenase 13 (HSD17B13 rs72613567) genetic variants are also associated with greater susceptibility to NASH (21–23).
Strong evidence indicates that the interaction between the genetic background and metabolic risk factors plays an important role in the pathogenesis of and disease progression in NAFLD (24). For example, the PNPLA3 (rs738409) variant has a stronger effect on liver injury in obese individuals than in lean individuals (25). Moreover, polygenic risk scores adjusted for conventional clinical risk factors may have the potential to guide and inform the care of patients with NAFLD (24). On this background of evidence, the 2 major aims of our study were as follows: (i) to identify relevant genetic and clinical risk factors associated with NASH and (ii) to develop and validate a novel nomogram for predicting NASH in a large multinational cohort of Asian patients with biopsy-proven NAFLD.
METHODS
Study population and design
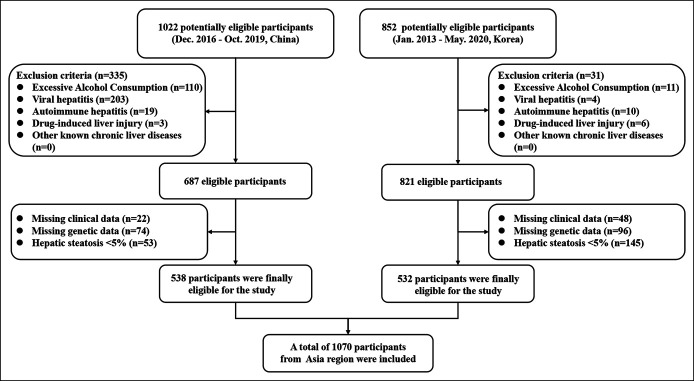
We conducted a cross-sectional study involving 2 cohorts of adult patients with biopsy-proven NAFLD from China and South Korea. The primary cohort comprised 1,022 potentially eligible Chinese patients diagnosed with suspected NAFLD (based on the presence of hepatic steatosis on imaging methods and/or elevated serum liver enzymes) between December 2016 and October 2019 at the First Affiliated Hospital of Wenzhou Medical University in Wenzhou (China). Exclusion criteria were (i) significant alcohol consumption (≥140 g/wk in men or ≥70 g/wk in women); (ii) presence of viral hepatitis, autoimmune hepatitis, drug-induced liver injury, or other known chronic liver diseases; (iii) incomplete clinical or genetic data; and (iv) hepatic steatosis <5% on liver histology. Between January 2013 and May 2020, an independent validation cohort of 852 potentially eligible patients with NAFLD from Seoul National University Seoul Metropolitan Government Boramae Medical Center in Seoul, South Korea, was also recruited. The inclusion and exclusion criteria were consistent with those of the primary Chinese cohort. As a result of the aforementioned exclusion criteria, a total of 1,070 patients with NAFLD with complete data were included in the study. More detailed information about the 2 patient cohorts is shown in Supplementary Table 1 (see Supplementary Digital Content 2, http://links.lww.com/CTG/A523).
The study protocol was approved by the local ethics committees of the 2 hospitals. All procedures involving the participants were performed in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration. Written informed consent was obtained from each subject after full explanation of the purpose and nature of all procedures.
Clinical and biochemical data
Clinical and biochemical data were obtained from all participants within 48 hours from liver biopsy. Blood samples were taken in fasting conditions. Body mass index (BMI) was calculated using the formula weight (kilograms) divided by height (meters) squared. Central obesity was defined as waist circumference ≥90 cm in men and ≥80 cm in women in the Asian population (26). Insulin resistance was estimated using the homoeostasis model assessment of insulin resistance (HOMA-IR) and defined as HOMA-IR >2.5 (27). T2DM was diagnosed as either self-reported history of disease, a fasting glucose level ≥7.0 mmol/L, hemoglobin A1c ≥6.5% (≥48 mmol/mol), or use of any antihyperglycemic drugs. Hypertension and dyslipidemia were diagnosed according to consensus criteria (28). MetS was defined as having at least 3 of the following metabolic risk factors: central obesity, increased blood pressure (systolic blood pressure ≥130 mm Hg or diastolic blood pressure ≥85 mm Hg or use of any antihypertensive drugs), increased fasting glucose (≥5.6 mmol/L or use of any antihyperglycemic agents), high triglycerides (>1.7 mmol/L or use of any lipid-lowering drugs), and low high-density lipoprotein cholesterol levels (<1.03 mmol/L in men and <1.29 mmol/L in women or use of any lipid-lowering drugs) (26,29). Methodological details for measurement of plasma cytokeratin-18 fragments (cytokeratin-18 [CK-18] neoepitope M30) levels have been reported previously (30). Fibrosis-4 (FIB-4) and NAFLD fibrosis score (NFS) were calculated using the published formulas (8). Controlled attenuation parameter (CAP) and liver stiffness measurement (LSM) were measured by 2 experienced operators using vibration-controlled transient elastography (FibroScan; Echosens, Paris, France), according to the manufacturer's recommendations.
Genetic analysis
Genotyping assays for PNPLA3 (rs738409), HSD17B13 (rs72613567), TM6SF2 (rs58542926), and MBOAT7 (rs641738) variants on human peripheral blood leukocytes were carried out using the MassARRAY, Sanger sequencing, or TaqMan assays platform according to the manufacturer's protocol (31,32).
Liver histology
Percutaneous liver biopsy was performed under ultrasound guidance. Liver histology assessment was undertaken by experienced liver histopathologists (who were blinded to the clinical and genetic data of participants) according to the NASH-Clinical Research Network Scoring System (33). The NAFLD activity score was calculated as the sum of 3 histological components, including liver steatosis (0–3), ballooning (0–2), and lobular inflammation (0–3). Liver fibrosis was staged as 0 to 4 according to the Brunt's histologic criteria (34). NAFLD was defined as the presence of hepatic steatosis in more than 5% of hepatocytes. NASH was diagnosed based on an overall pattern of histological hepatic injury consisting of macrovesicular steatosis, inflammation, and hepatocellular ballooning (33,35).
Statistical analysis
Continuous variables were expressed as means ± SD or medians with interquartile ranges (IQRs), and compared using either the unpaired Student t tests or the Mann–Whitney U tests as appropriate. Categorical variables were expressed as numbers (percentages) and compared using the χ2 tests or the Fisher exact tests as appropriate.
For the development of our nomogram, the primary Chinese cohort was randomly assigned in a 3:1 ratio to training and internal validation sets, using a split-sample method by an experienced statistician. Multivariable logistic regression analysis began with the variables selected from univariable analysis (P < 0.10). Stepwise selection was applied by using the likelihood ratio test with Akaike information criterion as the stopping rule. To provide the clinician with a quantitative tool to determine the individual probability of NASH, we built the nomogram on the basis of multivariable logistic analysis results obtained in the training set. The accuracy of this novel diagnostic model was subsequently evaluated in both the internal validation set and an independent external validation cohort. The diagnostic cutoffs for the nomogram, corresponding to the 90% sensitivity and 90% specificity thresholds for NASH, were calculated in the training set. The sensitivity, specificity, negative predictive value (NPV), positive predictive value (PPV), and the gray zone of the model were calculated at each cutoff. The discrimination of the model was evaluated by calculating the area under the receiver operating characteristic curve (AUROC). The model calibration was assessed by the calibration curve and the Hosmer-Lemeshow goodness of fit test. Statistical analyses were 2 sided, and significance was set at P < 0.05. All statistical tests were performed using R (Version 3.3.1 The R Foundation).
RESULTS
Baseline characteristics of patients
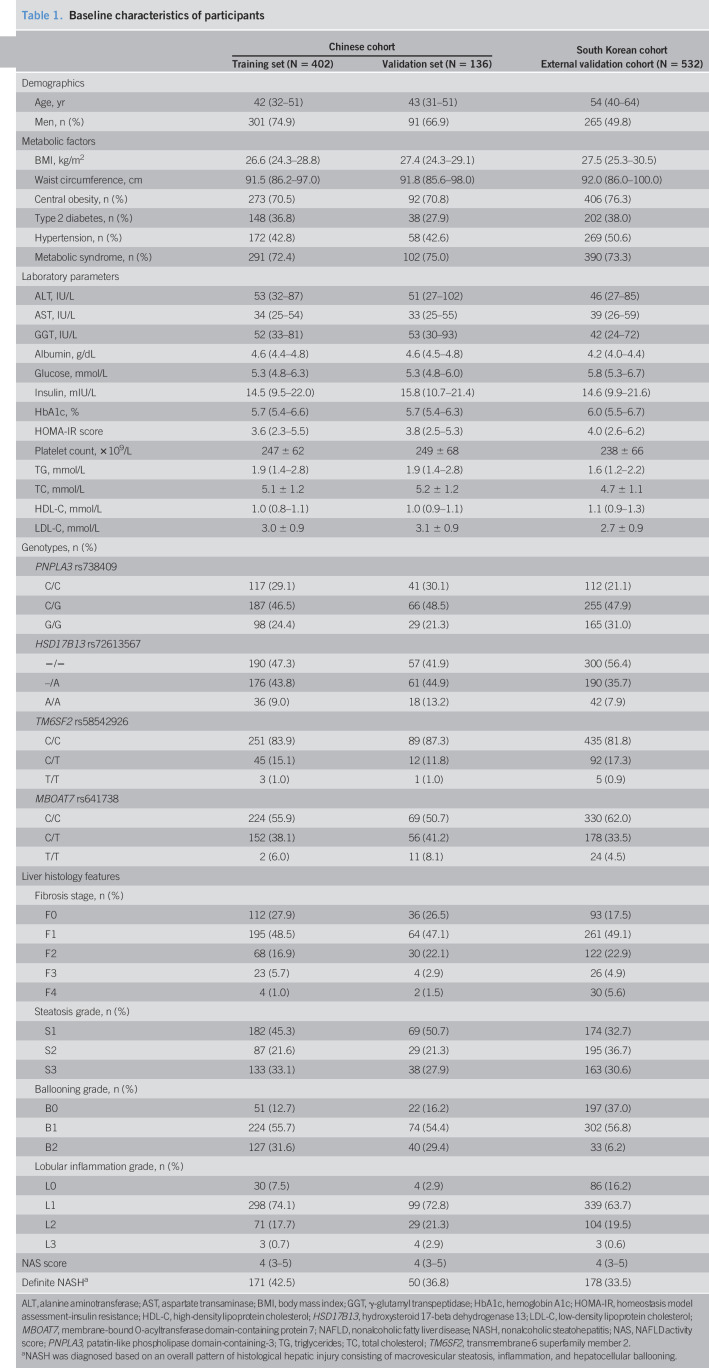
A total of 1,070 patients with biopsy-confirmed NAFLD from 2 tertiary hepatology centers were included in the study (Figure 1). In the primary Chinese cohort (n = 538), patients were randomly assembled into a “training set” (n = 402) and a “validation set” (n = 136). At the time of liver biopsy, patients had a median age of 42 years in the training set and a median age of 43 years in the validation set. The prevalence of NASH was 42.5% in the training set and 36.8% in the validation set, respectively. In the external validation cohort (involving 532 South Korean patients), the median age was 54 years and the prevalence of NASH was 33.5%. The baseline characteristics of the primary and validation cohorts are summarized in Table 1. The characteristics of the patients according to NASH status in the training set are shown in Supplementary Table 2 (see Supplementary Digital Content 3, http://links.lww.com/CTG/A524).
Figure 1.
The flowchart for the study.
Table 1.
Baseline characteristics of participants
| Chinese cohort | South Korean cohort | ||
| Training set (N = 402) | Validation set (N = 136) | External validation cohort (N = 532) | |
| Demographics | |||
| Age, yr | 42 (32–51) | 43 (31–51) | 54 (40–64) |
| Men, n (%) | 301 (74.9) | 91 (66.9) | 265 (49.8) |
| Metabolic factors | |||
| BMI, kg/m2 | 26.6 (24.3–28.8) | 27.4 (24.3–29.1) | 27.5 (25.3–30.5) |
| Waist circumference, cm | 91.5 (86.2–97.0) | 91.8 (85.6–98.0) | 92.0 (86.0–100.0) |
| Central obesity, n (%) | 273 (70.5) | 92 (70.8) | 406 (76.3) |
| Type 2 diabetes, n (%) | 148 (36.8) | 38 (27.9) | 202 (38.0) |
| Hypertension, n (%) | 172 (42.8) | 58 (42.6) | 269 (50.6) |
| Metabolic syndrome, n (%) | 291 (72.4) | 102 (75.0) | 390 (73.3) |
| Laboratory parameters | |||
| ALT, IU/L | 53 (32–87) | 51 (27–102) | 46 (27–85) |
| AST, IU/L | 34 (25–54) | 33 (25–55) | 39 (26–59) |
| GGT, IU/L | 52 (33–81) | 53 (30–93) | 42 (24–72) |
| Albumin, g/dL | 4.6 (4.4–4.8) | 4.6 (4.5–4.8) | 4.2 (4.0–4.4) |
| Glucose, mmol/L | 5.3 (4.8–6.3) | 5.3 (4.8–6.0) | 5.8 (5.3–6.7) |
| Insulin, mIU/L | 14.5 (9.5–22.0) | 15.8 (10.7–21.4) | 14.6 (9.9–21.6) |
| HbA1c, % | 5.7 (5.4–6.6) | 5.7 (5.4–6.3) | 6.0 (5.5–6.7) |
| HOMA-IR score | 3.6 (2.3–5.5) | 3.8 (2.5–5.3) | 4.0 (2.6–6.2) |
| Platelet count, ×109/L | 247 ± 62 | 249 ± 68 | 238 ± 66 |
| TG, mmol/L | 1.9 (1.4–2.8) | 1.9 (1.4–2.8) | 1.6 (1.2–2.2) |
| TC, mmol/L | 5.1 ± 1.2 | 5.2 ± 1.2 | 4.7 ± 1.1 |
| HDL-C, mmol/L | 1.0 (0.8–1.1) | 1.0 (0.9–1.1) | 1.1 (0.9–1.3) |
| LDL-C, mmol/L | 3.0 ± 0.9 | 3.1 ± 0.9 | 2.7 ± 0.9 |
| Genotypes, n (%) | |||
| PNPLA3 rs738409 | |||
| C/C | 117 (29.1) | 41 (30.1) | 112 (21.1) |
| C/G | 187 (46.5) | 66 (48.5) | 255 (47.9) |
| G/G | 98 (24.4) | 29 (21.3) | 165 (31.0) |
| HSD17B13 rs72613567 | |||
| −/− | 190 (47.3) | 57 (41.9) | 300 (56.4) |
| –/A | 176 (43.8) | 61 (44.9) | 190 (35.7) |
| A/A | 36 (9.0) | 18 (13.2) | 42 (7.9) |
| TM6SF2 rs58542926 | |||
| C/C | 251 (83.9) | 89 (87.3) | 435 (81.8) |
| C/T | 45 (15.1) | 12 (11.8) | 92 (17.3) |
| T/T | 3 (1.0) | 1 (1.0) | 5 (0.9) |
| MBOAT7 rs641738 | |||
| C/C | 224 (55.9) | 69 (50.7) | 330 (62.0) |
| C/T | 152 (38.1) | 56 (41.2) | 178 (33.5) |
| T/T | 2 (6.0) | 11 (8.1) | 24 (4.5) |
| Liver histology features | |||
| Fibrosis stage, n (%) | |||
| F0 | 112 (27.9) | 36 (26.5) | 93 (17.5) |
| F1 | 195 (48.5) | 64 (47.1) | 261 (49.1) |
| F2 | 68 (16.9) | 30 (22.1) | 122 (22.9) |
| F3 | 23 (5.7) | 4 (2.9) | 26 (4.9) |
| F4 | 4 (1.0) | 2 (1.5) | 30 (5.6) |
| Steatosis grade, n (%) | |||
| S1 | 182 (45.3) | 69 (50.7) | 174 (32.7) |
| S2 | 87 (21.6) | 29 (21.3) | 195 (36.7) |
| S3 | 133 (33.1) | 38 (27.9) | 163 (30.6) |
| Ballooning grade, n (%) | |||
| B0 | 51 (12.7) | 22 (16.2) | 197 (37.0) |
| B1 | 224 (55.7) | 74 (54.4) | 302 (56.8) |
| B2 | 127 (31.6) | 40 (29.4) | 33 (6.2) |
| Lobular inflammation grade, n (%) | |||
| L0 | 30 (7.5) | 4 (2.9) | 86 (16.2) |
| L1 | 298 (74.1) | 99 (72.8) | 339 (63.7) |
| L2 | 71 (17.7) | 29 (21.3) | 104 (19.5) |
| L3 | 3 (0.7) | 4 (2.9) | 3 (0.6) |
| NAS score | 4 (3–5) | 4 (3–5) | 4 (3–5) |
| Definite NASHa | 171 (42.5) | 50 (36.8) | 178 (33.5) |
ALT, alanine aminotransferase; AST, aspartate transaminase; BMI, body mass index; GGT, γ-glutamyl transpeptidase; HbA1c, hemoglobin A1c; HOMA-IR, homeostasis model assessment-insulin resistance; HDL-C, high-density lipoprotein cholesterol; HSD17B13, hydroxysteroid 17-beta dehydrogenase 13; LDL-C, low-density lipoprotein cholesterol; MBOAT7, membrane-bound O-acyltransferase domain-containing protein 7; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NAS, NAFLD activity score; PNPLA3, patatin-like phospholipase domain-containing-3; TG, triglycerides; TC, total cholesterol; TM6SF2, transmembrane 6 superfamily member 2.
NASH was diagnosed based on an overall pattern of histological hepatic injury consisting of macrovesicular steatosis, inflammation, and hepatocellular ballooning.
The frequency distributions of PNPLA3 (rs738409), HSD17B13 (rs72613567), TM6SF2 (rs58542926), and MBOAT7 (rs641738) genotypes were all in Hardy-Weinberg equilibrium (training set: P = 0.201, 0.664, 0.781 and 0.755, respectively, and external validation cohort: P = 0.464, 0.128, 0.956 and 0.999).
Development of an individualized risk score
Through univariable analyses, age, sex, BMI, MetS, serum liver enzymes (alanine aminotransferase [ALT], aspartate transaminase [AST], and γ-glutamyl transpeptidase), albumin, HOMA-estimated insulin resistance, and the PNPLA3 rs738409 and HSD17B13 rs72613567 genetic variants were selected for developing an individualized risk score to identify NASH (all P < 0.1) (see Supplementary Table 2, Supplementary Digital Content 3, http://links.lww.com/CTG/A524). In multivariable regression analyses, there was a strong association between NASH and sex, presence of MetS, HOMA-IR >2.5, increased AST levels (≥40 U/L), and the PNPLA3 rs738409 and HSD17B13 rs72613567 genetic variants. Finally, an individualized risk score for the noninvasive identification of NASH was developed based on the regression coefficients (Figure 2). The formula for the risk score was as follows: 0.548 × sex (female = 1; male = 0) + 0.467 × MetS (yes = 1; no = 0) + 1.909 × elevated AST levels (AST ≥40 = 1; AST <40 U/L = 0) + 1.074 × insulin resistance (HOMA-IR >2.5 = 1; HOMA-IR ≤2.5 = 0) + 0.581 × PNPLA3 (rs738409) genotype (GC = 1; CC or GG = 0) + 1.228 × PNPLA3 (rs738409) genotype (GG = 1; CC or GC = 0) + 0.607 × HSD17B13 (rs72613567) genotype (AA or -/A = 1; −/− = 0). For example, for a woman whose serum AST level was 100 U/L, HOMA-IR level was 3.0, and had MetS, PNPLA3 (rs738409) GG genotype, and HSD17B13 (72613567) AA genotype, her total points score was 6 and her probability of having NASH was 94%.
Figure 2.
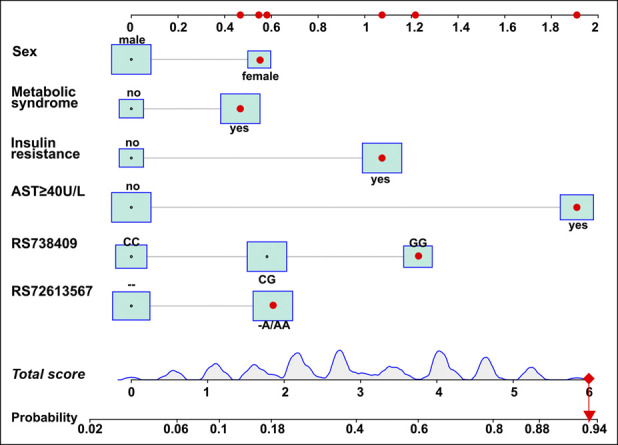
Nomogram to identify the presence of nonalcoholic steatohepatitis (NASH). To calculate the probability of having NASH, trace a vertical line from each of the predictors' axis to the first line. Add the total points and trace a vertical line from the total score axis to the risk axis to calculate the probability of having NASH. AST, aspartate transaminase.
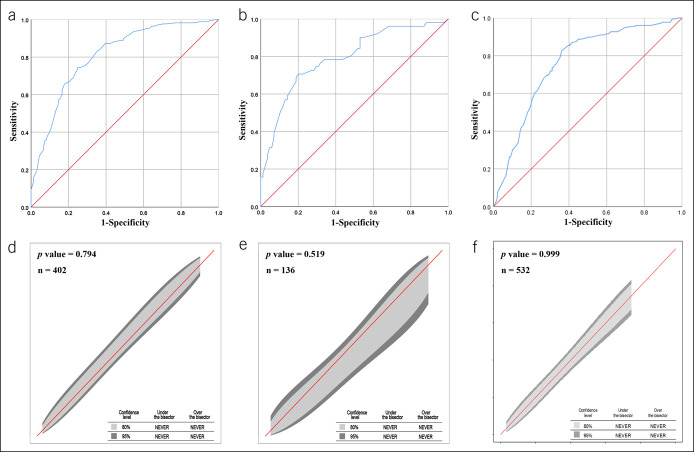
Diagnostic performance of the nomogram in the primary Chinese cohort
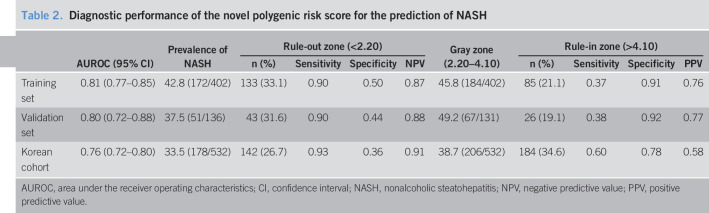
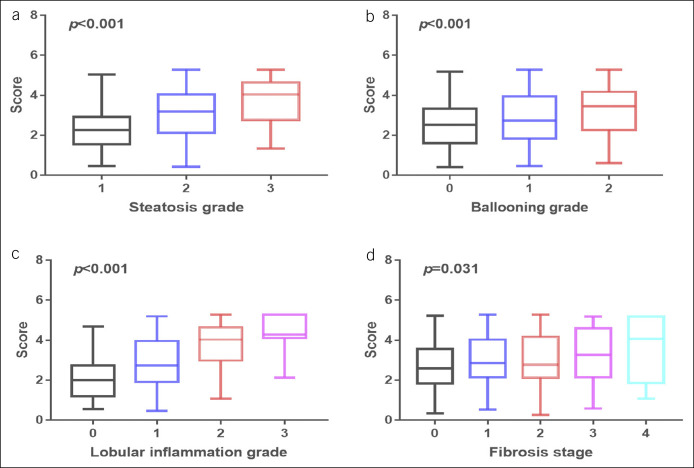
The AUROCs for the nomogram were 0.81 (95% confidence interval [CI] 0.77–0.85) for the training set (Figure 3a) and 0.80 (95% CI 0.72–0.88) for the validation set (Figure 3b). The calibration curve of the nomogram for the probability of NASH showed good agreement between prediction and observation in both the training and validation sets (Figure 3d,e). The Hosmer-Lemeshow test showed a nonsignificant statistic (training set: P = 0.794; validation set: P = 0.519), indicating that there was no departure from perfect fit. With the specific aim of identifying the most accurate nomogram cutoff values for diagnosing NASH, we used dual cutoff values of <2.20 (sensitivity = 0.90 in the training set) and >4.10 (specificity = 0.91 in the training set), respectively. In the training set, the cutoff value <2.20 had a NPV of 0.87 to rule out NASH, whereas the cutoff value >4.10 had a PPV of 0.76 to rule in NASH. Similarly, in the internal validation set, the cutoff value <2.20 had an NPV of 0.88 to rule out NASH, whereas the cutoff value >4.10 had a PPV of 0.77 to rule in NASH (Table 2). In addition, we found that the nomogram scores increased significantly across the histologic grades of steatosis, ballooning, lobular inflammation, and fibrosis (Figure 4).
Figure 3.
Diagnostic performance of the nomogram for the diagnostic of nonalcoholic steatohepatitis. (a) Area under the receiver operating characteristic curve (AUROC) of the training set, (b) AUROC of the internal validation set, (c) AUROC of the external validation cohort, (d) calibration curve of the training set, (e) calibration curve of the internal validation set, and (f) calibration curve of the external validation cohort.
Table 2.
Diagnostic performance of the novel polygenic risk score for the prediction of NASH
| AUROC (95% CI) | Prevalence of NASH | Rule-out zone (<2.20) | Gray zone (2.20–4.10) | Rule-in zone (>4.10) | |||||||
| n (%) | Sensitivity | Specificity | NPV | n (%) | Sensitivity | Specificity | PPV | ||||
| Training set | 0.81 (0.77–0.85) | 42.8 (172/402) | 133 (33.1) | 0.90 | 0.50 | 0.87 | 45.8 (184/402) | 85 (21.1) | 0.37 | 0.91 | 0.76 |
| Validation set | 0.80 (0.72–0.88) | 37.5 (51/136) | 43 (31.6) | 0.90 | 0.44 | 0.88 | 49.2 (67/131) | 26 (19.1) | 0.38 | 0.92 | 0.77 |
| Korean cohort | 0.76 (0.72–0.80) | 33.5 (178/532) | 142 (26.7) | 0.93 | 0.36 | 0.91 | 38.7 (206/532) | 184 (34.6) | 0.60 | 0.78 | 0.58 |
AUROC, area under the receiver operating characteristics; CI, confidence interval; NASH, nonalcoholic steatohepatitis; NPV, negative predictive value; PPV, positive predictive value.
Figure 4.
Boxplot of the score versus histopathological severity of the primary Chinese cohort: (a) steatosis grade, (b) lobular inflammation grade, (c) ballooning grade, and (d) fibrosis stage.
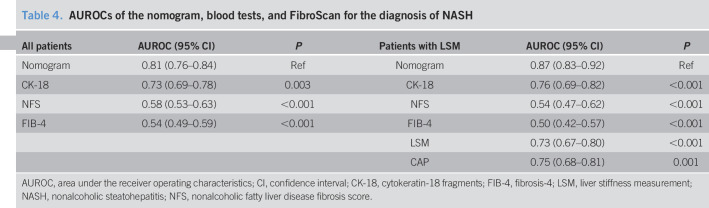
FIB-4 and NFS are widely used as noninvasive diagnostic tests for diagnosing liver fibrosis in patients with NAFLD, and plasma CK-18 levels are one of the most widely studied modalities for diagnosing NASH. We compared the performance of our nomogram with FIB-4, NFS, and plasma CK-18 for diagnosing NASH. As shown in Table 4, the discriminatory ability of our nomogram was superior to plasma CK-18, NFS, and FIB-4 scores. LSM and CAP values were measured using FibroScan in a subset of 357 patients with NAFLD. We have also compared the performance of our nomogram with that of LSM and CAP values. The AUROC of the nomogram was higher than those of LSM and CAP (Table 4).
Table 4.
AUROCs of the nomogram, blood tests, and FibroScan for the diagnosis of NASH
| All patients | AUROC (95% CI) | P | Patients with LSM | AUROC (95% CI) | P |
| Nomogram | 0.81 (0.76–0.84) | Ref | Nomogram | 0.87 (0.83–0.92) | Ref |
| CK-18 | 0.73 (0.69–0.78) | 0.003 | CK-18 | 0.76 (0.69–0.82) | <0.001 |
| NFS | 0.58 (0.53–0.63) | <0.001 | NFS | 0.54 (0.47–0.62) | <0.001 |
| FIB-4 | 0.54 (0.49–0.59) | <0.001 | FIB-4 | 0.50 (0.42–0.57) | <0.001 |
| LSM | 0.73 (0.67–0.80) | <0.001 | |||
| CAP | 0.75 (0.68–0.81) | 0.001 |
AUROC, area under the receiver operating characteristics; CI, confidence interval; CK-18, cytokeratin-18 fragments; FIB-4, fibrosis-4; LSM, liver stiffness measurement; NASH, nonalcoholic steatohepatitis; NFS, nonalcoholic fatty liver disease fibrosis score.
External validation of the nomogram
The nomogram yielded an AUROC of 0.76 (95% CI 0.72–0.80) in the external validation cohort from South Korea (Figure 3c). Good calibration was observed for the probability of NASH (Figure 3f), and the Hosmer-Lemeshow test showed a nonsignificant statistic (P = 0.999). Using the aforementioned dual cutoff approach, the NPV in the validation cohort was of 0.91, and 38.7% of the patients were in the gray zone between the 2 cutoff points (Table 2). Similar to our results in the primary Chinese cohort, the nomogram scores increased progressively across the histologic grades of hepatic steatosis, ballooning, lobular inflammation, and fibrosis in the external validation cohort (see Supplementary Figure 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A522).
Subgroup analysis
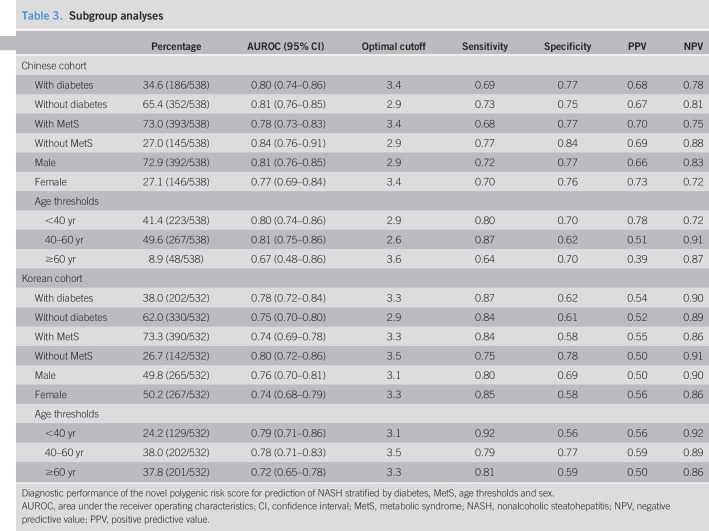
We also tested the performance of our novel nomogram by subgroup analyses (stratified by sex and age thresholds) both in the primary Chinese cohort and in the South Korean validation cohort. As shown in Table 3, the nomogram performed well in patients with and without pre-existing T2DM and in those with and without MetS. Among men with NAFLD, the AUROCs of the nomogram were 0.81 (0.76–0.85) in the Chinese cohort and 0.76 (0.70–0.81) in the Korean cohort, respectively. Among women with NAFLD, the AUROCs of the nomogram were 0.77 (0.69–0.84) in the Chinese cohort and 0.74 (0.68–0.79) in the Korean cohort. Stratifying by age groups, the AUROCs of the nomogram for patients younger than 40 years were 0.80 (0.74–0.86) in the Chinese cohort and 0.79 (0.71–0.86) in the Korean cohort, the AUROCs for patients aged between 40 years and 60 years were 0.81 (0.75–0.86) in the Chinese cohort and 0.78 (0.71–0.83) in the Korean cohort, and the AUROCs for patients older than 60 years were 0.67 (0.48–0.86) in the Chinese cohort and 0.72 (0.65–0.78) in the Korean cohort, respectively. The diagnostic performance of our nomogram was slightly diminished in women and in older (>60 years) patients in both the Chinese and the Korean cohorts.
Table 3.
Subgroup analyses
| Percentage | AUROC (95% CI) | Optimal cutoff | Sensitivity | Specificity | PPV | NPV | |
| Chinese cohort | |||||||
| With diabetes | 34.6 (186/538) | 0.80 (0.74–0.86) | 3.4 | 0.69 | 0.77 | 0.68 | 0.78 |
| Without diabetes | 65.4 (352/538) | 0.81 (0.76–0.85) | 2.9 | 0.73 | 0.75 | 0.67 | 0.81 |
| With MetS | 73.0 (393/538) | 0.78 (0.73–0.83) | 3.4 | 0.68 | 0.77 | 0.70 | 0.75 |
| Without MetS | 27.0 (145/538) | 0.84 (0.76–0.91) | 2.9 | 0.77 | 0.84 | 0.69 | 0.88 |
| Male | 72.9 (392/538) | 0.81 (0.76–0.85) | 2.9 | 0.72 | 0.77 | 0.66 | 0.83 |
| Female | 27.1 (146/538) | 0.77 (0.69–0.84) | 3.4 | 0.70 | 0.76 | 0.73 | 0.72 |
| Age thresholds | |||||||
| <40 yr | 41.4 (223/538) | 0.80 (0.74–0.86) | 2.9 | 0.80 | 0.70 | 0.78 | 0.72 |
| 40–60 yr | 49.6 (267/538) | 0.81 (0.75–0.86) | 2.6 | 0.87 | 0.62 | 0.51 | 0.91 |
| ≥60 yr | 8.9 (48/538) | 0.67 (0.48–0.86) | 3.6 | 0.64 | 0.70 | 0.39 | 0.87 |
| Korean cohort | |||||||
| With diabetes | 38.0 (202/532) | 0.78 (0.72–0.84) | 3.3 | 0.87 | 0.62 | 0.54 | 0.90 |
| Without diabetes | 62.0 (330/532) | 0.75 (0.70–0.80) | 2.9 | 0.84 | 0.61 | 0.52 | 0.89 |
| With MetS | 73.3 (390/532) | 0.74 (0.69–0.78) | 3.3 | 0.84 | 0.58 | 0.55 | 0.86 |
| Without MetS | 26.7 (142/532) | 0.80 (0.72–0.86) | 3.5 | 0.75 | 0.78 | 0.50 | 0.91 |
| Male | 49.8 (265/532) | 0.76 (0.70–0.81) | 3.1 | 0.80 | 0.69 | 0.50 | 0.90 |
| Female | 50.2 (267/532) | 0.74 (0.68–0.79) | 3.3 | 0.85 | 0.58 | 0.56 | 0.86 |
| Age thresholds | |||||||
| <40 yr | 24.2 (129/532) | 0.79 (0.71–0.86) | 3.1 | 0.92 | 0.56 | 0.56 | 0.92 |
| 40–60 yr | 38.0 (202/532) | 0.78 (0.71–0.83) | 3.5 | 0.79 | 0.77 | 0.59 | 0.89 |
| ≥60 yr | 37.8 (201/532) | 0.72 (0.65–0.78) | 3.3 | 0.81 | 0.59 | 0.50 | 0.86 |
Diagnostic performance of the novel polygenic risk score for prediction of NASH stratified by diabetes, MetS, age thresholds and sex.
AUROC, area under the receiver operating characteristics; CI, confidence interval; MetS, metabolic syndrome; NASH, nonalcoholic steatohepatitis; NPV, negative predictive value; PPV, positive predictive value.
DISCUSSION
In this multicenter study involving a cohort of 1,070 middle-aged individuals with biopsy-confirmed NAFLD from both China and South Korea, we developed and validated a clinical and genetic risk factors-based nomogram for identifying NASH. Our novel nomogram (including sex, MetS, HOMA-IR, serum AST level, PNPLA3, and HSD17B13 genotypes in its equation) had a good discriminatory capacity and calibration for identifying NASH in both the training and validation cohorts. This nomogram performed well in both patients with and without pre-existing T2DM and in those with and without MetS. The accuracy of the nomogram was slightly diminished in older participants. Our nomogram was positively associated with all individual histologic scores of NASH, including the fibrosis stage. These results further confirm that the interaction of genetic and metabolic risk factors plays an important role in the development and progression of NAFLD.
Because NAFLD affects up to a quarter of the general population worldwide (1), millions of patients worldwide, who are at risk of NAFLD progression, would benefit from treatment that is focused on effecting regression of NASH. Recently, a nationwide matched cohort study in Sweden showed that all histological stages of NAFLD were associated with significantly increased overall mortality, and this risk increased progressively with worsening NAFLD histology (36)—compared with matched controls, significant excess mortality risk was observed with simple steatosis (8.3/1,000 person-year [PY]) and NASH (13.4/1,000 PY); compared with those with simple steatosis, the multivariable-adjusted hazard ratio for overall mortality was increased in patients with NASH (hazard ratio 1.14; 95% CI 1.03–1.26) (36). Moreover, compared with matched controls, the mortality rate from cardiovascular causes was increased in those with NASH (absolute rate difference 2.7/1,000 PY), and compared with those with simple steatosis, the 20-year absolute excess risk of cardiovascular mortality was higher in patients with NASH (4.4%, P < 0.05) (36).
In recent years, a number of SNPs have been reported to be associated with susceptibility to NASH (24). The rs738409C>G variant in the PNPLA3 gene is the first and strongest genetic variant found to be associated with the susceptibility to NASH (17). PNPLA3 involves in lipid droplet remodeling in hepatocytes and retinol production by hepatic stellate cells (37). A recent GWAS confirmed that the rs738409 variant in the PNPLA3 gene was a risk factor across the entire histological spectrum of NAFLD (14). Our study has also confirmed that the PNPLA3 rs738409 was the most robustly associated genetic variant associated with NASH among the 4 SNPs that were tested in this study. HSD17B13 is a lipid droplet-associated protein, expressed predominantly in the liver, implying a liver-specific function (38). It has been reported that inactivating variants in the HSD17B13 gene are associated with a reduced risk of chronic liver disease among White individuals (23,39). However, we observed an inverse allelic association. As all our study participants are from East Asia, ethnic differences between patients might partly explain the results. A differential allele effect direction of genetic variants discovered by GWAS in subjects of different ethnicities is not uncommon (40). Lee et al. recently reported that the associations of apolipoprotein(a) (LPA) SNPs with size of apolipoprotein(a) isoforms, lipoprotein(a), and oxidized phospholipids on apolipoprotein B-100 levels are variable and ethnicity specific (40). In addition, HSD17B13 deficiency in mice models did not reproduce the protective effect of HSD17B13 loss-of-function mutants seen in human NAFLD (41). Interestingly, HSD17B13 deficiency induced weight gain in mice fed regular chow, which is contrary to previous findings (41).
Emerging data indicate that polygenic risk scores (PRSs) have the potential to guide and inform the care of patients with NAFLD. Costanzo et al. reported a risk score based on TM6SF2, GCKR, PNPLA3, and MBOAT7 genes could accurately identify patients with ultrasound-detected NAFLD from the general population (42). Krawczyk et al. found an increasing risk of hepatic steatosis and fibrosis with increasing number of PNPLA3, TM6SF2, and MBOAT7 risk alleles in patients with NAFLD (43). Moreover, León-Mimila et al. (44) studied 130 Mexican subjects with severe obesity undergoing bariatric surgery and found that a PRS which included the PNPLA3, LYPLAL1, PPP1R3B, and GCKR genes was associated with hepatic steatosis; however, this score did not predict NASH (AUROC = 0.56, P = 0.219). There are few studies to date that have investigated whether a PRS predicts NASH. In contrast to previous studies, we recruited Asian patients with biopsy-proven NAFLD and found that the combination of polygenic and clinical risk factors could accurately identify NASH.
Previous studies have found that the PNPLA3 variant exerted its adverse hepatic effects predominantly in obese patients compared with lean individuals (24,25). It has also been observed that the presence of insulin resistance, T2DM, or MetS affects the interaction between genetic and environmental risk factors (24). Barata et al. (45) recently found that the PNPLA3 rs738409 variant significantly interacts with insulin resistance, BMI, and plasma glucose and triglycerides levels to worsen hepatic steatosis in nondiabetic individuals carrying the G allele. The mechanism by which these modifiable metabolic traits interact with genetic variants to influence the risk of NASH remains to be clarified.
Through our multivariable logistic regression analyses, we demonstrated that female sex, MetS, HOMA-estimated insulin resistance, elevated serum AST levels, and presence of PNPLA3 (rs738409) and HSD17B13 (rs72613567) genetic variants were strongly associated with NASH. Overall, therefore, we believe that our results may contribute to better understanding of the polygenic regulation of NASH and the complex interaction between genetic and environmental risk factors. The major strengths of our study are that this is the largest study that has ever developed a polygenic risk score for identifying NASH in patients with biopsy-proven NAFLD and that we have validated the diagnostic performance of this risk score in an external validation cohort of patients with NAFLD.
The major limitation of our study is that participants were all from East Asia, and therefore, our results may not be applicable to other ethnic groups who have other metabolic risk factors. Further studies are needed to test the diagnostic accuracy of our novel nomogram in non-Asian individuals with NAFLD. Genetic testing is not widely available and not easy to perform, which makes widespread implementation difficult in clinical practice. However, genetic testing has been demonstrated to have a role in genetic counseling, prevention strategies, and treatments in other fields of medicine, including oncology, cardiology, and psychiatry (46,47). Genetic testing may also in the future have a role in genetic counseling in NAFLD. In addition, there were differences in the demographic characteristics between the Chinese and Korean cohorts, reflecting the heterogeneity of the populations with NAFLD. We have tested the performance of our nomogram in subgroup analyses and found that the diagnostic performance of our nomogram was slightly diminished in women and in older (>60 years) participants in both cohorts of patients. Finally, there is a gray zone extending from 38% to 49% when assessing the diagnostic performance of our nomogram for predicting NASH. However, almost all noninvasive tests of NAFLD are currently limited by a clear gray zone because of the use of 2 cutoff thresholds (48). Although there is a gray zone in our nomogram, liver biopsies could be correctly avoided in our study in approximately 50% of patients by using the score. In addition, a 2-step approach was recently reported to reduce indeterminate or discordant results while maintaining accuracy (49). The second test is used if a result in the gray zone is obtained from first test, and liver biopsy is performed if the result is in the gray zone for the second test. By using this 2-step approach, the need for liver biopsy would be reduced significantly without much effect on the percentage of misclassifications (50). Further studies are needed to evaluate whether the combination of our nomogram with other noninvasive scores facilitates the stratification of disease severity in NAFLD.
In conclusion, we have developed and validated a novel nomogram incorporating both genetic and clinical risk factors that accurately identifies NASH in a large cohort of Asian patients with biopsy-proven NAFLD. These results may translate into clinical practice to guide the risk stratification of NAFLD and also stimulate further research into the pathogenic role of our risk score in NASH.
CONFLICTS OF INTEREST
Guarantor of the article: Ming-Hua Zheng, MD, PhD.
Specific author contributions: F.G. and M.-H.Z.: study concept and design. S.-D.C., X.-X.W., X.-D.W., D.H.L., and Y.-P.C.: acquisition of data. F.G. and K.I.Z.: drafting of the manuscript. W.K., G.T., and C.D.B.: critical revision. F.G. and S.-D.C.: statistical analysis. M.-H.Z. and W.K.: study supervision. All authors contributed to the manuscript for important intellectual content and approved the submission.
Financial support: This work was supported by grants from the National Natural Science Foundation of China (82070588), Research Grant of Tianqing for Liver Disease (TQGB20200097), High Level Creative Talents from Department of Public Health in Zhejiang Province (S2032102600032), and Project of New Century 551 Talent Nurturing in Wenzhou. G.T. is supported in part by grants from the School of Medicine, University of Verona, Verona, Italy. C.D.B. is supported in part by the Southampton NIHR Biomedical Research Centre (IS-BRC-20004), UK. W.K. was supported by a National Research Foundation of Korea grant funded by the Korea government (2016R1D1A1B04934590), and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute funded by the Ministry of Health & Welfare, Republic of Korea (HI17C0912).
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ The correct identification of patients at increased risk of nonalcoholic steatohepatitis (NASH) is a critical step in the assessment of nonalcoholic fatty liver disease (NAFLD). There are few studies to date that have investigated whether a polygenic risk score predicts NASH.
WHAT IS NEW HERE
✓ This is the largest study that has ever developed a polygenic risk score for identifying NASH in patients with biopsy-proven NAFLD and that we have validated the diagnostic performance of this risk score in an external validation cohort of patients with NAFLD. Our results further confirm that the interaction of genetic and metabolic risk factors plays an important role in the development and progression of NAFLD.
TRANSLATIONAL IMPACT
✓ Our results may translate into clinical practice to guide the risk stratification of NAFLD and also stimulate further research into the pathogenic role of our risk score in NASH.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A522, http://links.lww.com/CTG/A523, http://links.lww.com/CTG/A524
Contributor Information
Feng Gao, Email: gaofeng@wmu.edu.cn.
Kenneth I. Zheng, Email: kenneth.zheng@wmu.edu.cn.
Sui-Dan Chen, Email: 1012032094@qq.com.
Dong Hyeon Lee, Email: donghyeonlee83@gmail.com.
Xi-Xi Wu, Email: wuxixi@wmu.edu.cn.
Xiao-Dong Wang, Email: xdwang0001@163.com.
Giovanni Targher, Email: giovanni.targher@univr.it.
Christopher D. Byrne, Email: c.d.byrne@soton.ac.uk.
Yong-Ping Chen, Email: ypchen77@hotmail.com.
Won Kim, Email: drwon1@snu.ac.kr.
REFERENCES
- 1.Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73–84. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z, Anstee QM, Marietti M, et al. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11–20. [DOI] [PubMed] [Google Scholar]
- 3.Wong RJ, Aguilar M, Cheung R, et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015;148:547–55. [DOI] [PubMed] [Google Scholar]
- 4.Haldar D, Kern B, Hodson J, et al. Outcomes of liver transplantation for non-alcoholic steatohepatitis: A European Liver Transplant Registry study. J Hepatol 2019;71:313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marjot T, Moolla A, Cobbold JF, et al. Nonalcoholic fatty liver disease in adults: Current concepts in etiology, outcomes, and management. Endocr Rev 2020;41:bnz009. [DOI] [PubMed] [Google Scholar]
- 6.Targher G, Byrne CD, Tilg H. NAFLD and increased risk of cardiovascular disease: Clinical associations, pathophysiological mechanisms and pharmacological implications. Gut 2020;69:1691–705. [DOI] [PubMed] [Google Scholar]
- 7.Byrne CD, Targher G. NAFLD as a driver of chronic kidney disease. J Hepatol 2020;72:785–801. [DOI] [PubMed] [Google Scholar]
- 8.Vilar-Gomez E, Chalasani N. Non-invasive assessment of non-alcoholic fatty liver disease: Clinical prediction rules and blood-based biomarkers. J Hepatol 2018;68:305–15. [DOI] [PubMed] [Google Scholar]
- 9.Dufour JF, Caussy C, Loomba R. Combination therapy for non-alcoholic steatohepatitis: Rationale, opportunities and challenges. Gut 2020;69:1877–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newsome PN, Buchholtz K, Cusi K, et al. A placebo-controlled trial of subcutaneous semaglutide in nonalcoholic steatohepatitis. N Engl J Med. [Epub ahead of print November 13, 2020.] [DOI] [PubMed] [Google Scholar]
- 11.Cardoso AC, de Figueiredo-Mendes C, C AVN, et al. New drugs for non-alcoholic steatohepatitis. Liver Int 2020;40(Suppl 1):96–101. [DOI] [PubMed] [Google Scholar]
- 12.Zheng KI, Fan JG, Shi JP, et al. From NAFLD to MAFLD: A “redefining” moment for fatty liver disease. Chin Med J (Engl) 2020;19:2271–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loomba R, Schork N, Chen CH, et al. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology 2015;149:1784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anstee QM, Darlay R, Cockell S, et al. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol 2020;73:505–15. [DOI] [PubMed] [Google Scholar]
- 15.Chalasani N, Guo X, Loomba R, et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology 2010;139:1567–76, 1576.e1561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Namjou B, Lingren T, Huang Y, et al. GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network. BMC Med 2019;17:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salameh H, Hanayneh MA, Masadeh M, et al. PNPLA3 as a genetic determinant of risk for and severity of non-alcoholic fatty liver disease spectrum. J Clin Transl Hepatol 2016;4:175–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol 2018;68:268–79. [DOI] [PubMed] [Google Scholar]
- 20.Koo BK, Joo SK, Kim D, et al. Additive effects of PNPLA3 and TM6SF2 on the histological severity of non-alcoholic fatty liver disease. J Gastroenterol Hepatol 2018;33:1277–85. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Zhou P, De L, et al. The roles of transmembrane 6 superfamily member 2 rs58542926 polymorphism in chronic liver disease: A meta-analysis of 24,147 subjects. Mol Genet Genomic Med 2019;7:e824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology 2016;150:1219–30.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med 2018;378:1096–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlsson B, Lindén D, Brolén G, et al. Review article: The emerging role of genetics in precision medicine for patients with non-alcoholic steatohepatitis. Aliment Pharmacol Ther 2020;51:1305–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romeo S, Sentinelli F, Dash S, et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes 2010;34:190–4. [DOI] [PubMed] [Google Scholar]
- 26.Alberti KG, Eckel RH, Grundy SM, et al. Harmonizing the metabolic syndrome: A joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009;120:1640–5. [DOI] [PubMed] [Google Scholar]
- 27.Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–9. [DOI] [PubMed] [Google Scholar]
- 28.Alberti KG, Zimmet P, Shaw J. Metabolic syndrome: A new world-wide definition. A consensus statement from the International Diabetes Federation. Diabetic Med 2006;23:469–80. [DOI] [PubMed] [Google Scholar]
- 29.Executive summary of the third report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III). JAMA 2001;285:2486–97. [DOI] [PubMed] [Google Scholar]
- 30.Gao F, Huang JF, Zheng KI, et al. Development and validation of a novel non-invasive test for diagnosing fibrotic non-alcoholic steatohepatitis in patients with biopsy-proven non-alcoholic fatty liver disease. J Gastroenterol Hepatol 2020;35:1804–12. [DOI] [PubMed] [Google Scholar]
- 31.Sun DQ, Zheng KI, Xu G, et al. PNPLA3 rs738409 is associated with renal glomerular and tubular injury in NAFLD patients with persistently normal ALT levels. Liver Int 2020;40:107–19. [DOI] [PubMed] [Google Scholar]
- 32.Koo BK, Joo SK, Kim D, et al. Development and validation of a scoring system, based on genetic and clinical factors, to determine risk of steatohepatitis in Asian patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2020;18:2592–9.e2. [DOI] [PubMed] [Google Scholar]
- 33.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313–21. [DOI] [PubMed] [Google Scholar]
- 34.Brunt EM, Janney CG, Di Bisceglie AM, et al. Nonalcoholic steatohepatitis: A proposal for grading and staging the histological lesions. Am J Gastroenterol 1999;94:2467–74. [DOI] [PubMed] [Google Scholar]
- 35.Brunt EM, Kleiner DE, Wilson LA, et al. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: Distinct clinicopathologic meanings. Hepatology 2011;53:810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon TG, Roelstraete B, Khalili H, et al. Mortality in biopsy-confirmed nonalcoholic fatty liver disease: Results from a nationwide cohort. Gut. [Epub ahead of print October 9, 2020.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruschi FV, Claudel T, Tardelli M, et al. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology 2017;65:1875–90. [DOI] [PubMed] [Google Scholar]
- 38.Su W, Mao Z, Liu Y, et al. Role of HSD17B13 in the liver physiology and pathophysiology. Mol Cell Endocrinol 2019;489:119–25. [DOI] [PubMed] [Google Scholar]
- 39.Ma Y, Belyaeva OV, Brown PM, et al. 17-beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology 2019;69:1504–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SR, Prasad A, Choi YS, et al. LPA gene, ethnicity, and cardiovascular events. Circulation 2017;135:251–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y, Brown PM, Lin DD, et al. Hsd17b13 deficiency does not protect mice from obesogenic diet injury. Hepatology. [Epub ahead of print August 11, 2020.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Costanzo A, Belardinilli F, Bailetti D, et al. Evaluation of polygenic determinants of non-alcoholic fatty liver disease (NAFLD) by a candidate genes resequencing strategy. Sci Rep 2018;8:3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krawczyk M, Rau M, Schattenberg JM, et al. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: A multicenter biopsy-based study. J Lipid Res 2017;58:247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.León-Mimila P, Vega-Badillo J, Gutiérrez-Vidal R, et al. A genetic risk score is associated with hepatic triglyceride content and non-alcoholic steatohepatitis in Mexicans with morbid obesity. Exp Mol Pathol 2015;98:178–83. [DOI] [PubMed] [Google Scholar]
- 45.Barata L, Feitosa MF, Bielak LF, et al. Insulin resistance exacerbates genetic predisposition to nonalcoholic fatty liver disease in individuals without diabetes. Hepatol Commun 2019;3:894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cirino AL, Harris S, Lakdawala NK, et al. Role of genetic testing in inherited cardiovascular disease: A review. JAMA Cardiol 2017;2:1153–60. [DOI] [PubMed] [Google Scholar]
- 47.Eeltink E, van der Horst MZ, Zinkstok JR, et al. Polygenic risk scores for genetic counseling in psychiatry: Lessons learned from other fields of medicine. Neurosci Biobehav Rev 2020;121:119–27. [DOI] [PubMed] [Google Scholar]
- 48.Boursier J, Guillaume M, Leroy V, et al. New sequential combinations of non-invasive fibrosis tests provide an accurate diagnosis of advanced fibrosis in NAFLD. J Hepatol 2019;71:389–96. [DOI] [PubMed] [Google Scholar]
- 49.Chan WK, Treeprasertsuk S, Goh GB, et al. Optimizing use of nonalcoholic fatty liver disease fibrosis score, fibrosis-4 score, and liver stiffness measurement to identify patients with advanced fibrosis. Clin Gastroenterol Hepatol 2019;17:2570–80.e7. [DOI] [PubMed] [Google Scholar]
- 50.Chan WK, Nik Mustapha NR, Mahadeva S. A novel 2-step approach combining the NAFLD fibrosis score and liver stiffness measurement for predicting advanced fibrosis. Hepatol Int 2015;9:594–602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.