ABSTRACT
Albumin has a serum half-life of 3 weeks in humans. This feature can be used to improve the pharmacokinetics of shorter-lived biologics. For instance, an albumin-binding domain (ABD) can be used to recruit albumin. A prerequisite for such design is that the ABD-albumin interaction does not interfere with pH-dependent binding of albumin to the human neonatal Fc receptor (FcRn), as FcRn acts as the principal regulator of the half-life of albumin. Thus, there is a need to know how ABDs act in the context of fusion partners and human FcRn. Here, we studied the binding and transport properties of human immunoglobulin A1 (IgA1), fused to a Streptococcus protein G-derived engineered ABD, in in vitro and in vivo systems harboring human FcRn. IgA has great potential as a therapeutic protein, but its short half-life is a major drawback. We demonstrate that ABD-fused IgA1 binds human FcRn pH-dependently and is rescued from cellular degradation in a receptor-specific manner in the presence of albumin. This occurs when ABD is fused to either the light or the heavy chain. In human FcRn transgenic mice, IgA1-ABD in complex with human albumin, gave 4-6-fold extended half-life compared to unmodified IgA1, where the light chain fusion showed the longest half-life. When the heavy chain-fused protein was pre-incubated with an engineered human albumin with improved FcRn binding, cellular rescue and half-life was further enhanced. Our study reveals how an ABD, which does not interfere with albumin binding to human FcRn, may be used to extend the half-life of IgA.
Abbreviations: ABD - Albumin binding domain, ADA – anti-drug-antibodies, ADCC - Antibody-dependent cellular cytotoxicity, ELISA - Enzyme-linked Immunosorbent assay, FcαRI - Fcα receptor, FcγR - Fcγ receptor, FcRn - The neonatal Fc receptor, GST - Glutathione S-transferase, HC - Heavy chain, HERA - Human endothelial cell-based recycling assay, Her2 - Human epidermal growth factor 2, HMEC - Human microvascular endothelial cells, IgG - Immunoglobulin G, IgA - Immunoglobulin A, LC - Light chain, QMP - E505Q/T527M/K573P, WT - Wild type
KEYWORDS: Immunoglobulin A (IgA), albumin-binding-domain (ABD), the neonatal Fc receptor (FcRn), human serum albumin (HSA), half-life, human FcRn transgenic mice
Introduction
Albumin is produced by hepatocytes and has a concentration of 30–50 mg/ml in both mice and humans. In addition, it has a long plasma half-life of three weeks in humans,1–5 a feature that it shares with the antibody isotype immunoglobulin G (IgG).6–8 The longevity of albumin and IgG is due to the interactions of these molecules with the neonatal Fc receptor (FcRn),5,9 which is broadly expressed throughout the body where it rescues its ligands from intracellular degradation via a cellular recycling process.10,11 This occurs as a result of noncooperative pH-dependent binding of the ligands to structurally distinct binding sites on FcRn, with binding at acidic pH and no binding and release at neutral pH. After IgG and albumin are taken up by fluid-phase pinocytosis and enter FcRn-positive endosomes, the receptor engages both its ligands. The complexes are then recycled back to the cell surface where the ligands are released upon exposure to the neutral pH of the extracellular space.11–18 This rescue-from-degradation mechanism takes place in both non-hematopoietic cells and hematopoietic cells.19,20 Proteins that are not rescued by FcRn will undergo degradation in the lysosomes. This biological mechanism is used in the design of IgG antibody variants21–28 and albumin fusions29–31 with tailored FcRn binding and transport properties. Such molecular design is clinically attractive, as the pharmacokinetic (PK) properties have a major impact on size and frequency of dosing, as well as patient compliance.
As an alternative to albumin itself, small albumin-binding modalities that can associate with endogenous albumin upon administration, such as albumin-binding peptides,32,33 albumin-binding antibody fragments34–36 and albumin-binding domains (ABDs) derived from bacteria, can be included in fusion protein drugs.37–45 Examples are Nanobodies derived from camelids,32 and designed ankyrin repeat proteins (DARPins),33 both selected for binding with nM affinity to albumin from different species. Other examples are single variable antibody domains with albumin specificity (AlbudAb),34–36 and ABDs derived from gram-positive bacteria displaying such proteins on their surface to camouflage and evade the immune system of the host.37–41 ABDs that bind albumin from different species,42–45 have been explored as biotechnological tools for purification and interaction analyses. Importantly, they have also been explored as fusion partners to extend the plasma half-life of fused proteins by indirect targeting of FcRn.46–51 The most-studied ABD, derived from Streptococcal protein G,37,52–55 has been engineered to strongly bind to human albumin at both neutral and mildly acidic pH.56
Previously, we reported on how the Streptococcal protein G-derived high affinity engineered ABD,56 fused to affibody molecules targeting human epidermal growth factor 2 (Her2), can be used to target rat and human albumin without affecting pH-dependent binding to rat/human FcRn.51 Thus, this particular engineered ABD binds the albumin species in such a fashion that FcRn engagement is not hindered, which resulted in extended half-life in a rat model.51 This ABD concept has been extended to other fusion protein designs,51,57–59 including more complex multi-domain proteins such as full-length human immunoglobulin A (IgA) antibodies.49
IgA is an attractive therapeutic protein, but its short plasma half-life of only 5 days in humans60 could be a limitation for clinical use. IgA-mediated killing of cancer cells is dependent on binding to Fcα receptor (FcαRI) expressed by myeloid cells.61 Importantly, a large body of evidence has proven that IgA-induced cross-linking of FcαRI, expressed by polymorphonuclear leukocytes (PMNs) upon binding to tumor antigens, is much more efficient in the elimination of cancer cells compared with that of IgG cross-linking to Fcγ receptors (FcγRs).49,61–71 In addition, this killing potential can be further enhanced by simultaneously blocking the CD47-SIRPα checkpoint axis.72 As such, IgA would be an attractive antibody in tumor targeting. A strategy to solve the short half-life of IgA may be to fuse IgA to an ABD to hijack human albumin in blood.49 Extended half-life combined with the potent ability of human IgA to induce antibody-dependent cell-mediated cytotoxicity (ADCC) will make this antibody isotype even more attractive as a therapeutic modality.
So far, design of ABD-fused human IgA has been studied in conventional mice expressing mouse albumin and mouse FcRn.49 However, we have demonstrated that there are great cross-species differences in FcRn-albumin interactions between mice and human that compromise studies on human albumin in conventional mice, and thus may also affect studies on ABD-strategies.73–75 Here, we conducted a comprehensive study on IgA1 fused to ABD, derived from Streptococcal protein G, in both in vitro and in vivo systems harboring human albumin and human FcRn.
Results
Design and production of ABD fused to Her2-IgA1 antibodies
Previously, we reported on the construction of ABD-fused anti-Her2 IgA variants harboring the variable domains derived from trastuzumab where IgA1 showed stronger anti-tumor effect than IgA2 in a multi-compartment xenograft model performed in human Fcα receptor transgenic mice.49 As such, we focused here on characterization of two IgA1 variants, one where the ABD was fused to the C-terminal end of the light chains (IgA1-LCABD) and another where it was fused to the heavy chains (IgA1-HCABD) (Figure 1(a)). In both cases, ABD was fused via a flexible glycine-serine linker (9GS). The ABD used was previously engineered to femtomolar affinity for human albumin, binding strongly at both acidic and neutral pH.56 The concept of indirect targeting of human FcRn is based on the ability of ABD-fused antibodies to associate with human albumin in the blood followed by cellular uptake and pH-dependent engagement of the receptor and exocytosis back at the cell surface for release (Figure 1(b)), which may happen both in the presence of wild-type human albumin and albumin engineered for improved human FcRn binding (i.e., QMP) (Figure 1(c)).
Figure 1.
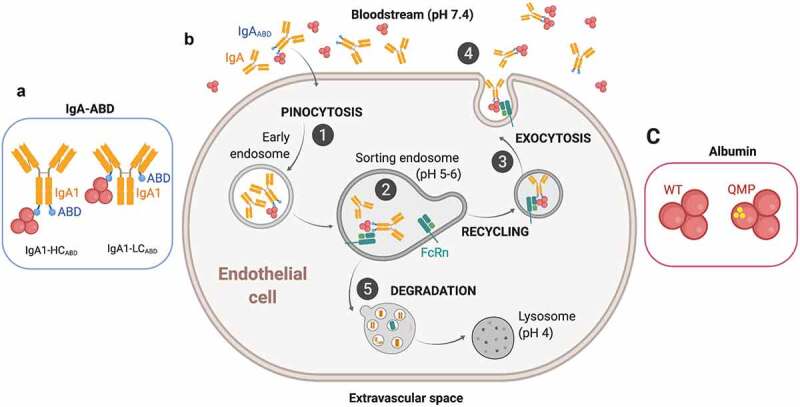
Recycling model of IgAABD through binding to albumin and indirect rescue by FcRn. A) Illustration of the two ABD-fused human IgA1 formats studied; IgA1-LCABD and IgA1-HCABD. B) (1) IgA1-LCABD and IgA1-HCABD will be taken up in complex with albumin via fluid-phase pinocytosis and enter early endosomes. (2) FcRn predominantly resides within acidified endosomes, where the lower pH will trigger binding of albumin bound IgA1ABD to the receptor. As such, the IgA1 fusions will be indirectly bound to FcRn. (3–4) The ligands bound to the receptor will then be recycled back to the cell membrane, where the physiological pH of blood (pH 7.4) results in release of the ligands back into the circulation. (5) Proteins that do not bind FcRn in the sorting endosomes, such as naked IgA, will end in lysosomes for degradation. C) Illustrations of wild-type (WT) and engineered (QMP) human albumin variants. The figure was made with BioRender
ABD-fused human IgA1 binds human albumin and engages human FcRn
In order to use the Streptococcal protein G-derived ABD as a human IgA fusion partner to achieve long half-life, it must bind human albumin in a pH-independent manner. It is a prerequisite that the IgA1ABD-albumin complex stays associated throughout the pH-gradient of the cellular recycling pathway. The mixtures of LC- and HC-fused IgA1ABD with albumin were added to an enzyme-linked immunosorbent assay (ELISA) plate coated with anti-IgA antibody, followed by extensive washing at either pH 5.5 or 7.4. Bound albumin was then detected with anti-human albumin alkaline phosphatase (ALP)-conjugated antibody (Figure 2(a)). The results clearly showed that both IgA1ABD variants bound human albumin at both pH conditions (Figure 2(b)).
Figure 2.
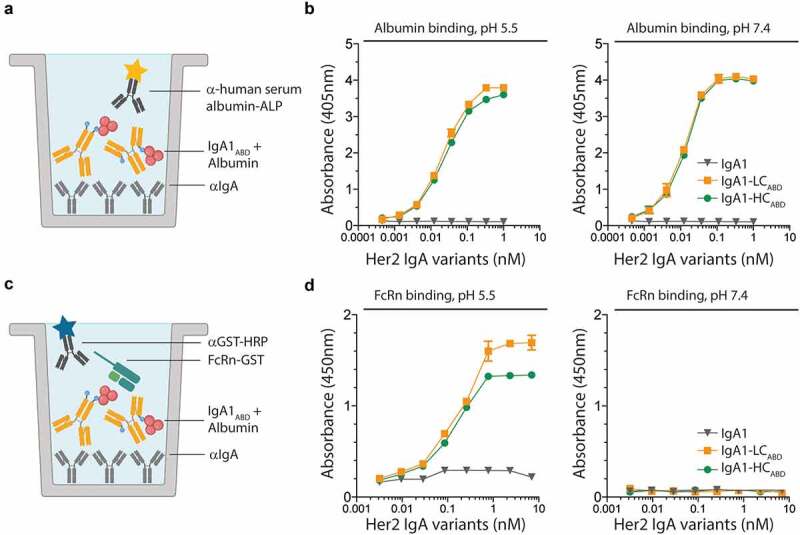
Human albumin binds IgA1-fused ABD in a pH-independent manner but human FcRn in a pH-dependent manner. A-B) ELISA showing binding of titrated amounts of ABD-fused IgA1 variants to human albumin at both pH 5.5 and 7.4. The numbers represent the mean±s.d. of duplicates. C-D) ELISA showing pH-dependent binding of ABD-fused IgA1 in complex with human albumin to human FcRn. The numbers represent the mean±s.d. of duplicates. Figure A and C were made with bioRender
To address the ability of human albumin bound IgA1ABD to engage human FcRn, ELISA was performed as above and compared with that of naked IgA1. Glutathione S-transferase (GST)-tagged recombinant soluble human FcRn was added at neutral or acidic pH, and receptor binding was detected using an anti-GST horseradish peroxidase (HRP)-conjugated antibody (Figure 2(c)). The results showed that the receptor bound human albumin when associated with the IgA1ABD variants but not naked IgA1. Binding occurred in a pH-dependent manner, with binding at acidic pH and no binding at neutral pH. At higher concentrations, IgA1-LCABD bound with slightly greater capacity to human FcRn than IgA1-HCABD, which may hint to some steric hindrance (Figure 2(d)).
To further visualize the binding events, surface plasmon resonance (SPR) experiments were performed. IgA1-HCABD was immobilized followed by injections of human albumin until saturation was reached. Then, a constant amount of soluble human FcRn was injected at pH 5.5 (Figure 3(a)). The sensorgrams demonstrated in real-time that the receptor can engage albumin in a reversible manner when bound to IgA1ABD (Figure 3(b-c)).
Figure 3.
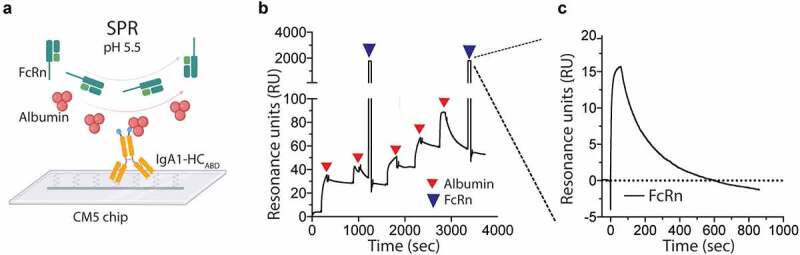
Reversible human FcRn binding to albumin in complex with ABD-fused IgA1. A) SPR setup with immobilized IgA1-HCABD, followed by injection of human albumin and soluble monomeric human FcRn at pH 5.5. The figure was made with bioRender. B) Sensorgram showing binding of human albumin after multiple injections over immobilized IgA1-HCABD, followed by injections of human FcRn. C) Close-up of the sensorgram showing binding of monomeric human FcRn to human albumin captured on ABD-fused IgA1-HCABD.
Indirect FcRn-mediated cellular rescue and extended in vivo half-life
To investigate whether the IgA1ABD variants would be rescued from intracellular degradation, a human endothelial cell-based recycling assay (HERA) was used.18 This assay is based on a human microvascular endothelial cells (HMEC)-1 cell line overexpressing human FcRn (Figure 4(a)). IgA1-LCABD and IgA1-HCABD were compared with naked IgA1 and pre-incubated with human albumin before addition to the cell medium. Cells were incubated for 4 hours to allow protein uptake. Subsequently, the cells were washed and fresh medium was added. After overnight (ON) incubation, medium was collected and the amounts of IgA1ABD variants present were quantified by ELISA. The HERA results showed that both the heavy and light chain fused IgA1ABD variants were recycled more efficiently than naked IgA1, as about 3-5-fold more of the fusions were detected in the medium. Thus, membrane-bound human FcRn can engage human albumin when bound to the IgA1ABD variants in acidified endosomes to the effect that both are rescued from degradation (Figure 4b). The light chain fused variant was recycled more efficiently than the heavy chain fusion. Notably, in the presence of albumin, ABD-fused IgA1 variants were rescued from degradation to the same extend as anti-Her2-IgG1 (supplementary Figure 1a).
Figure 4.
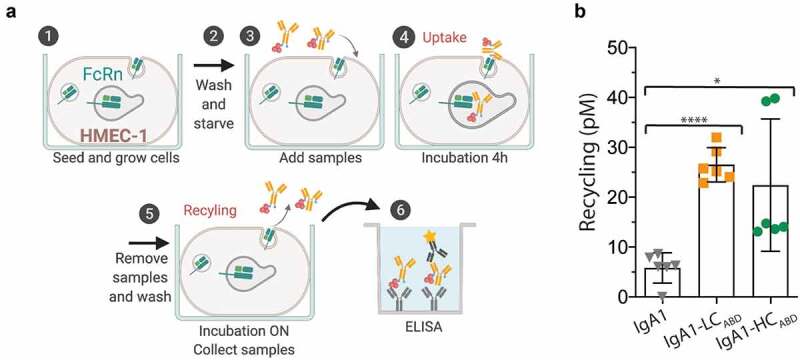
FcRn-mediated recycling in a cellular system. A) A schematic illustration of the HERA protocol. Briefly, HMEC-1 cells overexpressing human FcRn are seeded one day prior to the experiment. Cells are then washed and starved before samples are added and incubated for 4 hours. Before incubation ON, the medium is replaced. Recycling was defined as amount of protein taken up after 4 hours of incubation and further released back into the medium after ON incubation . The amounts of proteins present in the medium are then quantified in ELISA. The figure was made with BioRender. B) IgA1, IgA1-LCABD and IgA1-HCABD pre-incubated with human albumin were added to the cells. The amounts of the human IgA1 variants in collected medium samples were quantified by ELISA. The numbers given represent duplicates in ELISA of triplicates in HERA (SD). ns > 0.05, * = p 0.0136, **** = p < .0001, by two-tailed analysis using unpaired T-test (df = 10, t = 2.99, 11.05)
To investigate whether the enhanced FcRn-mediated rescue from degradation detected in HERA would translate into extended in vivo half-life, we used a state-of-the-art humanized mouse model, which is engineered to lack mouse FcRn and mouse albumin, and is transgenic for human FcRn.76 This strain has been shown to be more suitable for studies of human albumin in the context of human FcRn,77,78 due to cross-species differences.73–75,79
IgA1-HCABD and IgA1-LCABD were intravenously injected either alone or together with human albumin, and blood samples were collected after a period up to 23 days from the retrobulbar venous sinus (Figure 5(a)). When given alone, the IgA1ABD variants were rapidly cleared from blood, with half-lives of around 1 day, similar to that of naked IgA1 (Figure 5b-c). However, when mice received the IgA1ABD variants in the presence of human albumin, the half-lives of the ABD-IgA1 fusions were considerably extended, by 3.8-fold for IgA1-HCABD and 5-fold for IgA1-LCABD (Figure 5(b-c)). The longer half-life of the LC-fused IgA1 variant is in line with the in vitro data, showing better binding and recycling of the LC-fused version than the HC-fused verison. Notably, ABD-IgA1 in complex with albumin achieved a half-life in the same range as that of Her2-IgG1 in human FcRn-expressing mice (supplementary Figure 1b).
Figure 5.
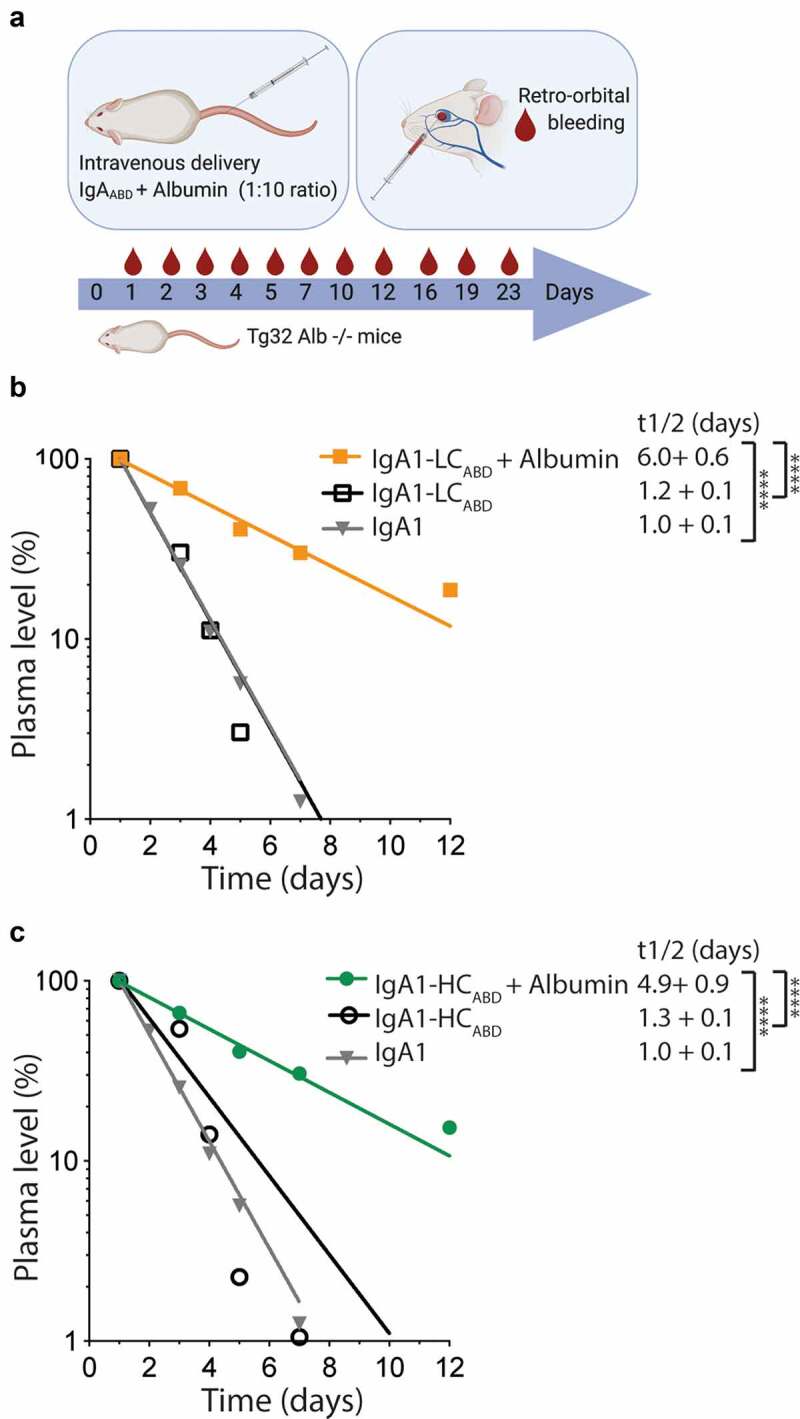
Extended plasma half-life in human FcRn transgenic mice in the presence of human albumin. A) Schematic illustration of the in vivo experimental setup, where IgA1 variants were pre-incubated with excess amounts of human albumin and given by intravenous (i.v.) injection to human FcRn transgenic mice that lack mouse albumin. Blood samples were collected up to day 23 post injection. The figure was made with BioRender. B-C) Log-linear changes in plasma levels (%) of B) IgA1 and IgA1-LCABD given with and without human albumin, and C) IgA1-HCABD given with and without human albumin. The variants were given to 5 mice per group. ns > 0.05,**** = p < .0001, by two-tailed analysis using unpaired T-test (df = 8, t = 7.45–17.17)
Engineered human albumin as a strategy to enhance recycling and half-life
ABD-fused proteins may also associate with human albumin variants that have been engineered for enhanced half-life via improved pH-dependent FcRn binding.79–81 One such variant harbors three mutations (E505Q/T527M/K573P; QMP) that was recently shown to have 2-fold extended half-life compared to wild-type (WT) albumin in human FcRn transgenic mice.80
First, we addressed if the IgA1ABD variants could bind the engineered QMP variant and subsequently engage human FcRn. This was studied using the same ELISA setups as above (Figure 2(a,c)). The results revealed that QMP and WT albumin bound equally well to the IgA1ABD variants at both acidic and neutral pH (Figure 6(a,b)), while human FcRn bound more strongly to QMP-associated complexes than to the WT albumin counterpart, in a strictly pH-dependent manner (Figure 6(c,d)). Notably, IgA1-HCABD showed slightly better binding to human FcRn than IgA1-LCABD when associated with QMP.
Figure 6.
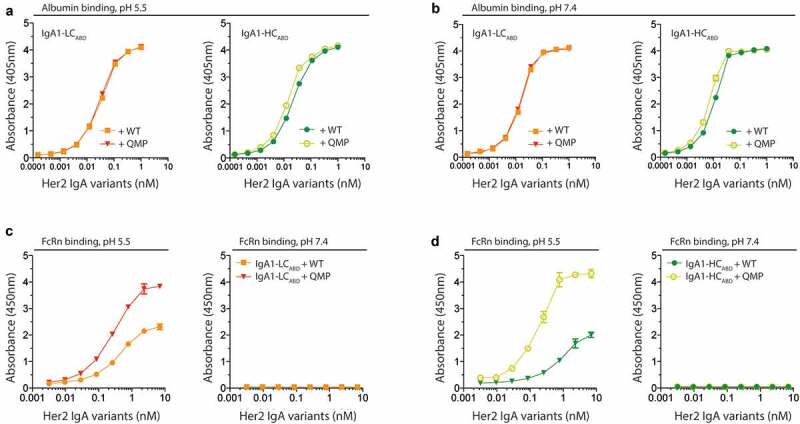
Engineered human albumin enhances human FcRn binding of ABD-fused IgA1. A-B) ELISA showing binding of titrated amounts of IgA1ABD variants to WT and QMP albumin at pH 5.5 and 7.4. The numbers represent the mean±s.d. of duplicates. C-D) ELISA showing binding of human FcRn to IgA1ABD variants pre-incubated with WT or QMP albumin at pH 5.5 and 7.4. The numbers represent the mean±s.d. of duplicates
To measure how the presence of QMP affected FcRn dependent recycling and rescue, HERA was performed as before (Figure 4(a)). IgA1-LCABD and IgA1-HCABD were pre-incubated with either WT or QMP albumin before addition to the FcRn-expressing HMEC-1 cells. Both IgA1ABD variants were recycled efficiently in the presence of QMP, as 3-5-fold more of the fusions were detected in the medium compared to in the presence of WT albumin (Figure 7(a)).
Figure 7.
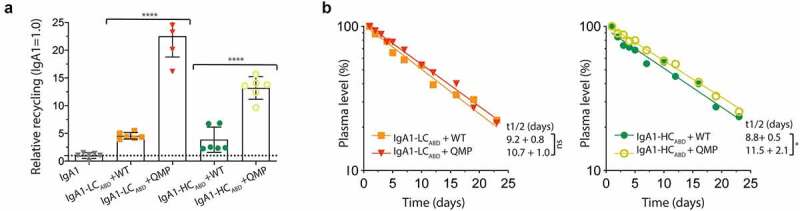
Enhanced cellular recycling and half-life of ABD-IgA1 fusion in the presence of engineered albumin. A) IgA1ABD variants were pre-incubated with QMP or WT albumin, and then given to HMEC-1 cells overexpressing human FcRn. The data is presented as relative recycling where IgA1 is set to 1.0. The numbers given represent duplicates in ELISA of triplicates in HERA (SD). ns > 0.05, **** = p < .0001, by two-tailed analysis using unpaired T-test (df = 10, t = 2.99, 11.05). B) Log-linear changes in plasma level (%) of IgA1-LCABD when given together with WT or QMP, and, IgA1-HCABD when given together with WT or QMP, in human FcRn transgenic mice lacking expression of mouse albumin. The variants were given by i.v. injections to 5 mice per group. ns > 0.05, * = p 0.0342, by two-tailed analysis using unpaired T-test (df = 8, t = 2.23, 2.55)
Finally, we measured if QMP could enhance the half-life of the two IgA1ABD variants in the human FcRn transgenic mice. The IgA1ABD variants were co-injected with QMP or WT albumin, and the clearence curves revealed that their half-life increased up to 1.3-fold in the presence of QMP. However, the gain in half-life was more pronounced and only statistically significant for the IgA1-HCABD fusion (Figure 7(b)).
Discussion
Here, we used a Streptococcal protein G-derived ABD as a model system to provide a preclinical guide on how to study indirect targeting of human FcRn in the context of human albumin. We demonstrate that human albumin in complex with an engineered version of the ABD fused to human IgA1 can engage human FcRn in a pH-dependent fashion. This translated into receptor-mediated cellular recycling in HERA and extended half-life in a humanized mouse model. The data conclusively show that the molecular design favors hijacking of soluble albumin followed by transport in and out of cells in a pH-dependent fashion. As such, the design does not sterically interfere with pH-dependent FcRn binding and release. This supports that the particular model ABD used in our study binds to a site on human albumin distal from that of human FcRn.
Human albumin consists of three homologous domains, denoted DI, DII and DIII,82 where DIII harbors the principle FcRn binding site while DI modulates binding to the receptor.83 Indeed, both DI and DIII are important for optimal receptor engagement,83 and as such, the binding site for the engineered Streptococcal protein G-derived ABD is very likely localized in DII. This is in line with data showing that the ABD homologue from Finegoldia magna binds DII.54 Thus, engaging DII may be the optimal strategy for indirect targeting to FcRn, not to interfere with the albumin:FcRn interaction.
We and others have shown that there are large differences in the FcRn-albumin interaction across species.73–75,79 In a previous study describing the design of the ABD-fused IgA variants, conventional mice and tumor-bearing SCID mice transgenic for human FcαRI were used.49 These mice express mouse FcRn and mouse albumin, which may compromise evaluation of strategies aiming for indirect targeting of human FcRn. In our case, the engineered ABD binds human albumin with femtomolar affinity, while the binding to mouse albumin is weaker.56 Notably, we recently demonstrated that, while both DI and DIII of human albumin are required for optimal interaction with human FcRn, DIII of mouse albumin plays the major role in engagement with the mouse receptor.73 This distinct difference in the mode of binding across species should be taken into consideration when exploring albumin targeting strategies. Thus, we conducted the studies in a mouse strain that is transgenic for human FcRn, but that lack mouse FcRn and mouse albumin.76 These mice can be preloaded with human albumin without any sign of immunogenicity.16,77,80 The set of in vitro and in vivo systems used in this study provides a guide to preclincical testing of drugs indirectly targeting human FcRn.
Instead of targeting endogenous albumin, the protein drug of interest may also be directly coupled to an engineered human albumin with improved human FcRn binding.79–81 We have previously reported on such variants that have extended half-life beyond that of WT human albumin.79,80 Here, we pre-incubated the ABD-fused proteins with one such engineered variant, QMP,80 and showed that it bound equally well to IgA1ABD as WT albumin, but more strongly to human FcRn. When the complexes were compared in HERA, IgA1ABD was rescued more efficiently from degradation in the presence of QMP then when in complex with WT albumin.
Enhanced cellular rescue from degradation of ABD-fused IgA1 translated into extended half-life in human FcRn transgenic mice when given together with human albumin. Thus, human FcRn is capable of recycling a complex multi-domain protein when bound to human albumin. In addition, we could further extend the half-life of ABD-fused IgA1 when in complex with QMP. Previously, we determined the plasma half-life of WT albumin to be 16.8 days compared with 20.4 days for naked QMP in human FcRn-expressing mice lacking endogenous albumin, which gives a 1.2-fold prolonged circulation time of the engineered albumin.80 When we studied the IgA1ABD variants in this model in the absence of albumin, they were rapidly cleared with a plasma half-life of only 1 day, similar to that of IgA1. Thus, genetic fusion of ABD, to either the light or the heavy chain, does not affect plasma persistence. However, in the presence of WT human albumin, the plasma half-life was significantly extended to 4.9–9.2 days and when QMP was used, we measured an additional 1.3-fold prolongation of half-life. Thus, human FcRn engagement in vivo results in higher blood levels over time. Notably, we observed a 2-fold difference in half-life values between independent experiments using the same transgenic mouse model. As the groups of mice used were a mix of males and females at roughly the same age, we do not know the reason for this.
Despite great improvement in plasma half-life of the IgA1ABD fusions in the presence of albumin, it is still far from that of albumin, which is likely due to biophysical properties of the IgA1ABD fusions. However, the half-life measured in the human FcRn-expressing mice is considerably longer than what was measured in conventional mice, which was 1.5–1.6 days only.5 As such, the use of humanized mice preloaded with human albumin is a preclinical model that mirrors the human situation better.
With regards to molecular design, our in vitro and in vivo data indicate that fusion of ABD to IgA1-LC may be the preferred format. However, the improvement in half-life was slightly more pronounced for the HC fused version in the presence of QMP. In the previous study, LC fused IgA1 was found to bind slightly better to human FcαRI than IgA1-HC in the presence of albumin, which translated into a distinct difference in ADCC activity by human neutrophils.49 The difference observed between the LC and HC fused formats in receptor binding could be explained by the location of the FcαRI-binding site in the CH2-CH3 elbow region of IgA and possible steric hindrance of the fused ABD. However, in mice expressing human FcαRI, no difference between the ABD-fused IgA1 formats was observed, but they reduced tumor volume more than naked IgA1.49 To fully address the effect of plasma half-life extension on the tumor killing potency of human IgA in vivo, it would be attractive to construct mice expressing both human FcαRI and human FcRn, which may be pre-loaded with human albumin.
Protein-based therapeutics, including monoclonal antibodies, may be immunogenic and induce anti-drug antibody (ADA) responses in patients over time. The consequences may range from no clinical impact to reduced bioavailability and therapeutic efficacy as well as altered safety profile.84 Thus, in a clinical setting, IgA fused to the bacterial-derived ABD may induce ADAs, and we can only speculate whether the fused ABD will trigger enhanced immunogenicity. Interestingly, it has been reported that ABD binding to human albumin is used by bacteria as a strategy to hide from the immune system.37–41 In our study, we did not observe any sign of immunogenicity from the clearance curve profiles, as monophasic log-linear decay in plasma concentrations were measured. ADA responses were measured by ELISA for IgA1-HCABD from day 7 post-injection, but this was not consistent between two independent experiments (supplementary Figures 2 and 3). Although it is important to examine whether ABD modalities may induce ADA responses preclinically, caution should be taken based on our data, as we injected a human protein fused to a bacterial-derived protein into mice. Notably, prediction of B and T cell epitopes suggested that the engineered ABD contains a few potentially immunogenic peptides (supplementary Figures 4), but caution should be taken as such predictions are regarded to have largely unresolved challenges, with T cell epitope prediction being of slightly higher quality, as recently discussed.85
To further gain insight into immunogenicity towards the bacterial-derived ABD, we measured the presence of preexisting antibody responses in a commercial intravenous immunoglobulin (IVIG) preparation, representing pooled IgG from 1000–15,000 healthy human donors. While strong antibody responses were measured for an antigen derived from adenovirus 5, very low responses were detected toward the ABD-fused IgA1 variants, which were about 2-3-fold stronger than for unfused IgA1. The anti-ABD reactivity could be partly blocked in the presence of human albumin supplementary (Figure 5). Thus, some individuals may have preexisting antibody immunity against the bacterial-derived ABD, which may potentially affect treatment.
Interestingly, it has been shown that tumors use albumin as a source of nutrition.86 As such, albumin-bound cancer drugs accumulate in solid tumors and show an enhanced anti-tumor effect.86–89 This is supported by studies demonstrating that albumin accumulation is caused by the so-called “enhanced permeability and retention effect” where higher permeability of blood capillaries increases accumulation in the tumor. Based on this, it may well be that ABD-fused IgA1 in complex with albumin could be taken up by tumors more efficiently that unfused IgA1. However, in light of data showing that both the molecular size and plasma half-life greatly affect anti-tumor efficacy,90 investigating how human IgA with and without albumin-binding properties behave in mouse tumor models will be valuable.
Materials and methods
Cell culture
HEK293F cells (ThermoFisher, Catalog number: R79007) were maintained in Freestyle medium (ThermoFisher, Catalog number: 12338018), while HEK293E cells (ATCC, Catalog number: CRL-1573) were maintained in RPMI medium (Sigma, Catalog number: R2405) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Sigma, Catalog number: F7524), and 1% Penicillin-Streptomycin (Merck, Catalog number: P4458), or Freestyle medium (ThermoFisher, Catalog number: 12338018). High five cells (Invitrogen, Catalog number: B85502) were grown in Express FIVE SEF medium (ThermoFisher, Catalog number: 10486025) with 18 mM L-glutamine (ThermoFisher, Catalog number: 25030123) and 1% antibiotic–antimycotic (ThermoFisher, Catalog number: 15240096) added. The HMEC1-hFcRn cell line91 was grown in Gibco MCDB 131 medium (ThermoFisher, Catalog number:10372019) complemented with 10% heat-inactivated FCS (Sigma, Catalog number: F7524), 2 mM L-glutamine (ThermoFisher, Catalog number: 25030123), 1% Penicillin-Streptomycin (Merck, Catalog number: P4458), 10 ng/ml mouse epidermal growth factor (ThermoFisher, Catalog number: PMG8043) and 1 µg/ml hydrocortisone (Sigma, Catalog number: H0888). To ensure stable FcRn expression, the medium was supplemented with 5 µg/ml blasticidin (ThermoFisher, Catalog number: A1113903) and 100 µg/ml G418 (ThermoFisher, Catalog number: 10131027).
Her2-IgA1-ABD production and purification
Her2-IgA1, Her2-IgA1-HCABD and Her2-IgA1-LCABD variants were designed and produced as previously described.49
Albumin production and purification
cDNA encoding full-length human serum albumin, WT or the engineered albumin variant, QMP (E505Q/T527M/K573P), were expressed in pcDNA3 vectors. The vectors were transiently transfected into HEK293E cells using Polyethylenimine Max (Polysciences, Catalog number: 24885–2). The supernatant containing secreted albumin were harvested every day for up to 2 weeks and purified using a CaptureSelect human albumin affinity matrix (Life Technologies, Catalog number: 191297005) packed column (Atoll, as described by the manufacturer). Eluted proteins were up-concentrated and buffer-exchanged to phosphate-buffered saline (PBS) (Sigma, Catalog number: D8537) using Amicon Ultra-15 ml 10 K columns (Millipore, Catalog number: C7715) before purifying them in a second step using size exclusion chromatography (SEC). SEC were performed using a Superdex 200 increase 10/300GL column (GE Healthcare, Catalog number: 28–9909-44) coupled to an ÄKTA Avant instrument (GE Healthcare). Further, monomeric fractions were collected and up-concentrated using Amicon Ultra-4 ml 10 K columns (Millipore, Catalog number: C7715) before protein concentration were measured using a DS-11 spectrophotometer (Denovix Inc.) and 2 μg of the albumin variants were analyzed on a 12% SDS-PAGE (Thermo Fisher Scientific, Catalog number: NW00125BOX).
Recombinant human FcRn production and purification
A pcDNA3-GST-hβ2-microglobulin vector generated from a pcDNA3.1 vector (Invitrogen, Catalog number V79020) encoding soluble recombinant GST-tagged human FcRn was transiently transfected into HEK293E cells, and secreted receptor was purified as previously described, using a GSTrap HP column (GE Healthcare, Catalog number: 17–5282-02).92 Production of recombinant His-tagged human FcRn was done by using a Baculovirus expression vector system,93 and the viral stocks were kind gifts from Dr. Sally Ward (University of Texas, Southwestern Medical Center, Dallas, TX). The receptors were purified as previously reported using a HisTrap HP column supplied with Ni2+ ions (GE Healthcare, Catalog number: 17–5248-02).3
ELISA
To measure FcRn binding, ninety-six-well ELISA plates (Costar, Catalog number: 10544522) were coated with 1.0 μg/ml of a polyclonal IgG from goat specific for human IgA alpha chain (Abcam, Catalog number: ab97211), diluted in PBS (Sigma, Catalog number: D8537), and incubated ON 4°C. Plates were blocked with PBS containing 4% skimmed milk (S) (ITW reagents, Catalog number: A0830) for 1 hour at room temperature (RT) and washed 4 times with PBS containing 0.05% Tween 20 (T) (Sigma, Catalog number: P1379). 100 μl of titrated amounts of purified anti-Her2 IgA1ABD pre-incubated with 5 ug of albumin variants, diluted in PBS/S/T, were added to the plates and incubated for 1 hour on a shaker at RT. Wells were washed either with PBS/T pH 5.5 or PBS/T pH 7.4 before 2 μg/ml of GST-fused soluble recombinant human FcRn, diluted in PBS/S/T pH 5.5 or in pH 7.4, was added and incubated for 1 hour at RT. The plates were washed at either pH 5.5 or pH 7.4, and bound receptors were detected using HRP conjugated goat anti-GST antibody (Rockland, Catalog number: 600–102-200), diluted (1:8000) in PBS/S/T with pH 5.5 or 7.4, and incubated 1 hour at RT. The plates were washed as before and 100 μl of 3,3ʹ,5,5ʹ-tetramethybenzidine substrate solution (Merck Millipore, Catalog number: CL07) was then added to the wells. Absorbance was measured at 620 nm with a Sunrise spectrophotometer (TECAN). The reaction was stopped by adding 50 μl 1 M HCl, and then measured at 450 nm.
ABD binding to human albumin-WT and -QMP were measured by coating, blocking, washing and adding IgA1ABD+albumin variants to the plates as described above. Binding of the ABD to either human albumin-WT or -QMP were measured by adding 100 μl of anti-human albumin ALP-conjugated antibody from goat (1:4000) (Bethylx, Catalog number: A80-229AP) for 1 hour at RT. The plates were washed as above, and bound proteins were visualized by adding 100 μl of ALP substrate (1 mg/ml phosphate in diethanolamine buffer) (Sigma, Catalog number: S0942). Absorbance was measured at 405 nm with a Sunrise spectrophotometer (TECAN).
To analyze HERA samples, 96-well ELISA plates (Costar, Catalog number: 10544522) were coated with 1.0 μg/ml of a polyclonal IgG from goat specific for human IgA alpha chain (Abcam, Catalog number: ab97211), diluted in PBS (Sigma, Catalog number: D8537), and incubated ON 4°C. Plates were blocked with PBS/S for 1 hour at RT and washed 4 times with PBS/T. HERA samples were added, and binding were detected by adding 100 μl of an ALP-conjugated anti-human IgA (α-chain specific) antibody produced in goat (Sigma, Catalog number: SAB3701226) (1:2000) and incubated for 1 hour at RT. The plates were washed as above, and bound proteins were visualized by adding 100 μl of ALP substrate (1 mg/ml phosphate in diethanolamine buffer) (Sigma, Catalog number: S0942). The anti-Her2 IgG1 variant was analyzed by capture on 1.0 μg/ml human Her2 (Sino Biological, Catalog number: 10004-HCCH), and detected by using an ALP-conjugated anti-human IgG (Fc-specific) antibody from goat (1:5000) (Sigma, Catalog number: A9544). Absorbance was measured at 405 nm with a Sunrise spectrophotometer (TECAN).
To detect for ADA in the plasma samples from the PK studies, ELISA plates (Costar, Catalog number: 10544522) were coated with 1.0 μg/ml of the IgA1ABD variant of interest, with or without albumin, diluted in PBS (Sigma, Catalog number: D8537), and incubated ON 4°C. Plates were blocked and washed as described before. Plasma samples were diluted 1:200 in PBS/S/T and 100 μl was added per ELISA well. Plates were washed again, and binding was detected by adding 100 μl of an ALP-conjugated anti-mouse IgG (Fc-specific) from goat (1:4000) (Sigma, Catalog number: A2429). The plates were washed as above, and bound proteins were visualized by adding 100 μl of ALP substrate (1 mg/ml phosphate in diethanolamine buffer) (Sigma, Catalog number: S0942). Absorbance was measured at 405 nm with a Sunrise spectrophotometer (TECAN).
To study preexisting reactivity toward the ABD in IVIG, 96-well plates (Costar, Catalog number: 10544522) were either coated with 1.0 μg/ml of IgA1ABD variants directly or coated with 1.0 μg/ml Her2 (Sino Biological, Catalog number: 10004-HCCH) in PBS (Sigma, Catalog number: D8537) ON at 4°C, followed by a layer of the IgA1ABD variants in PBS/S/T. The variants were studied both in the presence and absence of albumin. Adenovirus type 5 hexon (BioRad; Catalog number:MPP002) was used as a positive control in the first setup. Plates were blocked and washed as described before. 100 μl of titrated amounts of IVIG (Octagam, Octapharma) starting at 1 mg/ml diluted in PBS/S/T was added per ELISA well. Plates were washed again. An ALP-conjugated anti-human IgA (α-chain specific) antibody (Sigma, Catalog number: SAB3701226) toward IgA1 was used as a positive control in the second setup and binding of IVIG was detected by adding 100 μl of an ALP-conjugated anti-human IgG (Fc-specific) antibody from goat (1:5000) (Sigma, Catalog number: A9544). The plates were washed, and bound proteins were visualized as before.
SPR
SPR was performed using Biacore T200 (GE Healthcare) and CM5 Series S sensor chips following the manufacturer’s protocol. IgA1-HCABD (6 μg/ml in 10 mM sodium acetate pH 4.5) were immobilized using amine coupling chemistry to ~500 RU, and unreacted moieties on the CM5 chips were blocked with 1 M ethanolamine. Phosphate buffer (155 mM phosphate buffer, 85 mM NaCl, 0.05% Tween 20) pH 5.5 and phosphate buffer (195 mM phosphate, 85 mM NaCl, 0.05% Tween20) pH 7.4 were used as running buffer and regeneration buffer, respectively. Binding measurements were performed by injecting monomeric human serum albumin with a flow rate of 30 μl/min for 320 seconds at 25°C until saturation of the chip was reached. Further, soluble monomeric human FcRn with a His-tag was injected at 1 μM at 30 μl/min for 120 seconds at 25°C.
Human endothelial cell-based recycling assay
A human endothelial recycling assay was performed as previously described.18 Briefly, 7.5 × 104 HMEC-1 cells stably expressing HA-hFcRn-EGFP were seeded into 24-well plates per well (Costar, Catalog number:10732552) and cultured for one day in growth medium. The cells were washed and starved for 1 h in Hank’s balanced salt solution (HBSS) (ThermoFisher, Catalog number: 14025100). 400 nM of IgA1ABD variants pre-incubated with 800 nM of albumin variants and IgG1 were diluted in 250 µl HBSS (pH 7.4) and added to the cells followed by 4 h incubation. The medium was removed and the cells were washed with ice cold HBSS (pH 7.4), before fresh warm HBSS (pH 7.4) or growth medium without FCS and supplemented with MEM non-essential amino acids (ThermoFisher, Catalog number: 11140050) were added. Samples were collected the next day. The amounts of anti-Her2 IgA1ABD variants and IgG1 were quantified using ELISA. The ELISA was performed as described above.
In vivo studies
Homozygote Tg32 Alb−/- mice (B6.Cg-Tg(FCGRT)32Dcr Albem12Mvw Fcgrttm1Dcr/MvwJ) from The Jackson Laboratory, expressing human FcRn and not mouse albumin, were used to determine the half-life of the IgA1 variants. Hemizygote Tg32 mice (B6.Cg-Fcgrttm1DcrTg(FCGRT)32Dcr/DcrJ) expressing human FcRn were used to determine half-life of IgG1. Female and male mice, age 7–15 weeks, weighing between 19 and 26 g (5 mice per group), received 1 mg/kg of IgA1ABD premixed with 10 times (number of molecules) more of albumin variants and 1 mg/kg IgG1 (diluted in PBS) and given by intravenous injections. Blood samples (25 µl) were drawn from the retro-orbital sinus at days 1, 2, 3, 4, 5, 7, 10, 12, 16, 19 and 23 after injection. To prevent coagulation, the blood samples were mixed with 1 µl 1% K3-EDTA and then centrifuged at 17,000 × g for 5 min at 4°C. Plasma was isolated and diluted 1:10 in 50% glycerol/PBS solution and then stored at −20°C until analysis by ELISA. Plasma samples were diluted ranging from 1:400 to 1:50 in PBS/S/T and 100 μl was added per ELISA well. The study was carried out at The Jackson Laboratory (JAX Services, Bar Harbor, ME). The experiments and procedures used were approved by the Animal Care and Use Committee at The Jackson Laboratory and performed in accordance with the approved guidelines and regulations.
Half-life calculation
The plasma concentration of the IgA1 variants is presented as percentage remaining in the circulation at indicated time points post-injection compared to the concentration on day 1. Non-linear regression analysis was performed to fit a straight line through the data using the Prism 8 software, and the β-phase half-life was calculated using the formula: t1/2 , where t1/2 is the half-life of the IgA1 variants, Ae is the amount of IgA1 remaining, A0 is the amount of IgA1 on day 1 and t is the elapsed time.
Statistical analysis
Data were analyzed using GraphPad Prism 8 for Mac (Version 8; GraphPad Software Inc.) and Microsoft Excel for Mac (version 16.35). GraphPad Prism 8 was used to perform two-tailed statistical analyses using unpaired T-test, and were performed with a 95% confidence interval and where p < 0.05 was considered a statistically significant difference.
T- and B-cell peptide epitope prediction
The WT (ABD001) and the engineered ABD (ABD035) sequence56 were aligned using Clustal Omega (1.2.4) multiple sequence alignment.
To predict T cell epitopes, we performed MHC presentation using the state-of-the-art method NetMHC4.194,95 (http://www.cbs.dtu.dk/services/NetMHCpan/) using rank threshold for strong binders: 0.5, and weak binders: 2.0.
For prediction of B-cell epitopes, the gold standard Immune Epitope Database analysis resource antibody epitope prediction tool96 (http://tools.iedb.org/bcell/) was used with a threshold of 0.500.
Supplementary Material
Acknowledgments
We are grateful to Sathiaruby Sivaganesh and Ignacio Navas Camacho for excellent technical assistance, and we thank Dr. Wayne I. Lencer (Boston Children’s Hospital, Harvard Medical School and Harvard Digestive Diseases center) for the HMEC1 cell line stably expressing HA-hFcRn-EGFP. Funding: This work was supported in part by the Research Council of Norway through its Center of Excellence funding scheme (project no. 179573). J.T.A. and J.N. were supported by the Research Council of Norway (Grant no. 230526 and 287927), and J.T.A. and S.M. by South-Eastern Norway Regional Health Authority (Grant no. 40018). M.E. was funded by KWF (Grant no. 7650). V.G. was supported by a UiO World-Leading Research Community grant, the UiO:LifeSciences Convergence Environment Immunolingo, EU Horizon 2020 iReceptorplus (#825821), and Research Council of Norway FRIPRO project (#300740).
Funding Statement
This work was supported by the Horizon 2020 Framework Programme [825821]; KWF Kankerbestrijding [7650]; Norges Forskningsråd [230526]; Norges Forskningsråd [179573]; Norges Forskningsråd [300740]; South-Eastern Norway Regional Health Authority [40018]; Research Council of Norway [179573, 230526, 287927]; South-Eastern Norway Regional Health Authority [40018].
Disclosure of interest
M.E. is employed of TigaTx BV, and J.L. is the scientific founder and shareholder of TigaTx BV and Inc. V.G. declares an advisory board position at Enpicom B.V. I.S. and J.T.A. are co-inventors of patents, entitled “Albumin variants and uses thereof” and relate to the data described here (EP3063171B1, US10208102, and US10781245).
Data and materials availability:
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Chaudhury C, Brooks CL, Carter DC, Robinson JM, Anderson CL.. Albumin binding to FcRn: distinct from the FcRn-IgG interaction. Biochemistry. 2006;45(15):4983–13. doi: 10.1021/bi052628y. [DOI] [PubMed] [Google Scholar]
- 2.Andersen JT, Dalhus B, Cameron J, Daba MB, Plumridge A, Evans L, Brennan SO, Gunnarsen KS, Bjørås M, Sleep D, et al. Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor. Nature Communications. 2012;3(1):610. doi: 10.1038/ncomms1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sand KM, Dalhus B, Christianson GJ, Bern M, Foss S, Cameron J, Sleep D, Bjørås M, Roopenian DC, Sandlie I, et al. Dissection of the neonatal Fc receptor (FcRn)-albumin interface using mutagenesis and anti-FcRn albumin-blocking antibodies. The Journal of Biological Chemistry. 2014;289(24):17228–39. doi: 10.1074/jbc.M113.522565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters T Jr. All about albumin: biochemistry, genetics, and medical applications. Academic press, USA; 1995. [Google Scholar]
- 5.Chaudhury C, Mehnaz S, Robinson JM, Hayton WL, Pearl DK, Roopenian DC, Anderson CL.. The major histocompatibility complex-related Fc receptor for IgG (FcRn) binds albumin and prolongs its lifespan. The Journal of Experimental Medicine. 2003;197(3):315–22. doi: 10.1084/jem.20021829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West AP Jr., Bjorkman PJ. Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor. Biochemistry. 2000;39(32):9698–708. doi: 10.1021/bi000749m. [DOI] [PubMed] [Google Scholar]
- 7.Kim JK, Firan M, Radu CG, Kim CH, Ghetie V, Ward ES. Mapping the site on human IgG for binding of the MHC class I-related receptor, FcRn. European Journal of Immunology. 1999;29(9):2819–25. doi:10.1002/(sici)1521-4141(199909)29:09<2819:aid-immu2819>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Oganesyan V, Damschroder MM, Cook KE, Li Q, Gao C, Wu H, Dall’Acqua WF. Structural insights into neonatal Fc receptor-based recycling mechanisms. The Journal of Biological Chemistry. 2014;289(11):7812–24. doi: 10.1074/jbc.M113.537563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghetie V, Hubbard JG, Kim JK, Tsen MF, Lee Y, Ward ES. Abnormally short serum half-lives of IgG in beta 2-microglobulin-deficient mice. European Journal of Immunology. 1996;26(3):690–96. doi: 10.1002/eji.1830260327. [DOI] [PubMed] [Google Scholar]
- 10.Latvala S, Jacobsen B, Otteneder MB, Herrmann A, Kronenberg S. Distribution of FcRn across species and tissues. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society. 2017;65(6):321–33. doi: 10.1369/0022155417705095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward ES, Ober RJ. Chapter 4: multitasking by exploitation of intracellular transport functions the many faces of FcRn. Advances in Immunology. 2009;103:77–115. doi: 10.1016/s0065-2776(09)03004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ober RJ, Martinez C, Lai X, Zhou J, Ward ES. Exocytosis of IgG as mediated by the receptor, FcRn: an analysis at the single-molecule level. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(30):11076–81. doi: 10.1073/pnas.0402970101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ober RJ, Martinez C, Vaccaro C, Zhou J, Ward ES. Visualizing the site and dynamics of IgG salvage by the MHC class I-related receptor, FcRn. The Journal of Immunology. 2004;172(4):2021–29. doi: 10.4049/jimmunol.172.4.2021. [DOI] [PubMed] [Google Scholar]
- 14.Prabhat P, Gan Z, Chao J, et al. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(14):5889–94. doi: 10.1073/pnas.0700337104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward ES, Martinez C, Vaccaro C, Zhou J, Tang Q, Ober RJ. From sorting endosomes to exocytosis: association of Rab4 and Rab11 GTPases with the Fc receptor, FcRn, during recycling. Molecular Biology of the Cell. 2005;16(4):2028–38. doi: 10.1091/mbc.E04-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pyzik M, Rath T, Kuo TT, Win S, Baker K, Hubbard JJ, Grenha R, Gandhi A, Krämer TD, Mezo AR,et al. Hepatic FcRn regulates albumin homeostasis and susceptibility to liver injury. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(14):E2862–e2871. doi: 10.1073/pnas.1618291114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt EGW, Hvam ML, Antunes F, Cameron J, Viuff D, Andersen B, Kristensen NN, Howard KA. Direct demonstration of a neonatal Fc receptor (FcRn)-driven endosomal sorting pathway for cellular recycling of albumin. The Journal of Biological Chemistry. 2017;292(32):13312–22. doi: 10.1074/jbc.M117.794248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grevys A, Nilsen J, Sand KMK, Daba MB, Øynebråten I, Bern M, McAdam MB, Foss S, Schlothauer T, Michaelsen TE, et al. A human endothelial cell-based recycling assay for screening of FcRn targeted molecules. Nature Communications. 2018;9(1):621. doi: 10.1038/s41467-018-03061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akilesh S, Christianson GJ, Roopenian DC, Shaw AS. Neonatal FcR expression in bone marrow-derived cells functions to protect serum IgG from catabolism. The Journal of Immunology. 2007;179(7):4580–88. doi: 10.4049/jimmunol.179.7.4580. [DOI] [PubMed] [Google Scholar]
- 20.Montoyo HP, Vaccaro C, Hafner M, Ober RJ, Mueller W, Ward ES. Conditional deletion of the MHC class I-related receptor FcRn reveals the sites of IgG homeostasis in mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(8):2788–93. doi: 10.1073/pnas.0810796106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghetie V, Popov S, Borvak J, Radu C, Matesoi D, Medesan C, Ober RJ, Ward ES. Increasing the serum persistence of an IgG fragment by random mutagenesis. Nature Biotechnology. 1997;15(7):637–40. doi: 10.1038/nbt0797-637. [DOI] [PubMed] [Google Scholar]
- 22.Dall’Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). The Journal of Biological Chemistry. 2006;281(33):23514–24. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 23.Hinton PR, Johlfs MG, Xiong JM, Hanestad K, Ong KC, Bullock C, Keller S, Tang MT, Tso JY, Vásquez M, et al. Engineered human IgG antibodies with longer serum half-lives in primates. The Journal of Biological Chemistry. 2004;279(8):6213–16. doi: 10.1074/jbc.C300470200. [DOI] [PubMed] [Google Scholar]
- 24.Mi W, Wanjie S, Lo ST, Gan Z, Pickl-Herk B, Ober RJ, Ward ES. Targeting the neonatal fc receptor for antigen delivery using engineered fc fragments. The Journal of Immunology. 2008;181(11):7550–61. doi: 10.4049/jimmunol.181.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalevsky J, Chamberlain AK, Horton HM, Karki S, Leung IWL, Sproule TJ, Lazar GA, Roopenian DC, Desjarlais JR, et al. Enhanced antibody half-life improves in vivo activity. Nature Biotechnology. 2010;28(2):157–59. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbie GJ, Criste R, Dall’Acqua WF, Jensen K, Patel NK, Losonsky GA, Griffin MP. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrobial Agents and Chemotherapy. 2013;57(12):6147–53. doi: 10.1128/aac.01285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaccaro C, Zhou J, Ober RJ, Ward ES. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nature Biotechnology. 2005;23(10):1283–88. doi: 10.1038/nbt1143. [DOI] [PubMed] [Google Scholar]
- 28.Patel DA, Puig-Canto A, Challa DK, Perez Montoyo H, Ober RJ, Ward ES. Neonatal Fc receptor blockade by Fc engineering ameliorates arthritis in a murine model. The Journal of Immunology. 2011;187(2):1015–22. doi: 10.4049/jimmunol.1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rendell MS. Albiglutide: a unique GLP-1 receptor agonist. Expert Opinion on Biological Therapy. 2016;16(12):1557–69. doi: 10.1080/14712598.2016.1240780. [DOI] [PubMed] [Google Scholar]
- 30.Santagostino E. Transforming the treatment for hemophilia B patients: update on the clinical development of recombinant fusion protein linking recombinant coagulation factor IX with recombinant albumin (rIX-FP). Thrombosis Research. 2016;141(Suppl 3):S5–8. doi: 10.1016/s0049-3848(16)30415-7. [DOI] [PubMed] [Google Scholar]
- 31.Elsadek B, Kratz F. Impact of albumin on drug delivery–new applications on the horizon. Journal of Controlled Release: Official Journal of the Controlled Release Society. 2012;157(1):4–28. doi: 10.1016/j.jconrel.2011.09.069. [DOI] [PubMed] [Google Scholar]
- 32.Tijink BM, Laeremans T, Budde M, Walsum MSV, Dreier T, De Haard HJ, Leemans CR, Van Dongen GAMS. Improved tumor targeting of anti–epidermal growth factor receptor Nanobodies through albumin binding: taking advantage of modular Nanobody technology. Molecular Cancer Therapeutics. 2008;7(8):2288–97. doi: 10.1158/1535-7163.Mct-07-2384. [DOI] [PubMed] [Google Scholar]
- 33.Steiner D, Merz FW, Sonderegger I, Gulotti-Georgieva M, Villemagne D, Phillips DJ, Forrer P, Stumpp MT, Zitt C, Binz HK, et al. Half-life extension using serum albumin-binding DARPin® domains. Protein Engineering, Design & Selection: PEDS. 2017;30(9):583–91. doi: 10.1093/protein/gzx022. [DOI] [PubMed] [Google Scholar]
- 34.Ward ES, Gussow D, Griffiths AD, Jones PT, Winter G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature. 1989;341(6242):544–46. doi: 10.1038/341544a0. [DOI] [PubMed] [Google Scholar]
- 35.Holt LJ, Basran A, Jones K, Chorlton J, Jespers LS, Brewis ND, Tomlinson IM. Anti-serum albumin domain antibodies for extending the half-lives of short lived drugs. Protein Engineering Design and Selection. 2008;21(5):283–88. doi: 10.1093/protein/gzm067. [DOI] [PubMed] [Google Scholar]
- 36.Jespers L, Schon O, Famm K, Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nature Biotechnology. 2004;22(9):1161–65. doi: 10.1038/nbt1000. [DOI] [PubMed] [Google Scholar]
- 37.Nilvebrant J, Hober S. The albumin-binding domain as a scaffold for protein engineering. Computational and Structural Biotechnology Journal. 2013;6(7):e201303009. doi: 10.5936/csbj.201303009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson MU, Frick IM, Nilsson H, Kraulis PJ, Hober S, Jonasson P, Linhult M, Nygren P-Å, Uhlén M, Björck L, et al. Structure, specificity, and mode of interaction for bacterial albumin-binding modules. The Journal of Biological Chemistry. 2002;277(10):8114–20. doi: 10.1074/jbc.M109943200. [DOI] [PubMed] [Google Scholar]
- 39.De Chateau M, Bjorck L. Identification of interdomain sequences promoting the intronless evolution of a bacterial protein family. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(16):8490–95. doi: 10.1073/pnas.93.16.8490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kronvall G, Simmons A, Myhre EB, Jonsson S. Specific absorption of human serum albumin, immunoglobulin A, and immunoglobulin G with selected strains of group A and G streptococci. Infection and Immunity. 1979;25(1):1–10. doi: 10.1128/IAI.25.1.1-10.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myhre EB. Surface receptors for human serum albumin in Peptococcus magnus strains. Journal of Medical Microbiology. 1984;18(2):189–95. doi: 10.1099/00222615-18-2-189. [DOI] [PubMed] [Google Scholar]
- 42.Nygren PA, Ljungquist C, Tromborg H, Nustad K, Uhlen M. Species-dependent binding of serum albumins to the streptococcal receptor protein G. European Journal of Biochemistry. 1990;193(1):143–48. doi: 10.1111/j.1432-1033.1990.tb19315.x. [DOI] [PubMed] [Google Scholar]
- 43.Falkenberg C, Bjorck L, Akerstrom B. Localization of the binding site for streptococcal protein G on human serum albumin. Identification of a 5.5-kilodalton protein G binding albumin fragment. Biochemistry. 1992;31(5):1451–57. doi: 10.1021/bi00120a023. [DOI] [PubMed] [Google Scholar]
- 44.He Y, Rozak DA, Sari N, Chen Y, Bryan P, Orban J. Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity. Biochemistry. 2006;45(33):10102–09. doi: 10.1021/bi060409m. [DOI] [PubMed] [Google Scholar]
- 45.Rozak DA, Alexander PA, He Y, Chen Y, Orban J, Bryan PN. Using offset recombinant polymerase chain reaction to identify functional determinants in a common family of bacterial albumin binding domains. Biochemistry. 2006;45(10):3263–71. doi: 10.1021/bi051926s. [DOI] [PubMed] [Google Scholar]
- 46.Hammarberg B, Nygren PA, Holmgren E, Elmblad A, Tally M, Hellman U, Moks T, Uhlén M. Dual affinity fusion approach and its use to express recombinant human insulin-like growth factor II. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(12):4367–71. doi: 10.1073/pnas.86.12.4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stahl S, Nygren PA. The use of gene fusions to protein A and protein G in immunology and biotechnology. Pathologie-biologie. 1997;45:66–76. [PubMed] [Google Scholar]
- 48.Makrides SC, Nygren PA, Andrews B, Ford PJ, Evans KS, Hayman EG, Adari H, Uhlén M, Toth CA. Extended in vivo half-life of human soluble complement receptor type 1 fused to a serum albumin-binding receptor. The Journal of Pharmacology and Experimental Therapeutics. 1996;277:534–42. [PubMed] [Google Scholar]
- 49.Meyer S, Nederend M, Jansen JH, Reiding KR, Jacobino SR, Meeldijk J, Bovenschen N, Wuhrer M, Valerius T, Ubink R, et al. Improved in vivo anti-tumor effects of IgA-Her2 antibodies through half-life extension and serum exposure enhancement by FcRn targeting. mAbs. 2016;8(1):87–98. doi: 10.1080/19420862.2015.1106658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konig T, Skerra A. Use of an albumin-binding domain for the selective immobilisation of recombinant capture antibody fragments on ELISA plates. Journal of Immunological Methods. 1998;218(1–2):73–83. doi: 10.1016/s0022-1759(98)00112-4. [DOI] [PubMed] [Google Scholar]
- 51.Andersen JT, Pehrson R, Tolmachev V, Daba MB, Abrahmsen L, Ekblad C. Extending half-life by indirect targeting of the neonatal Fc receptor (FcRn) using a minimal albumin binding domain. The Journal of Biological Chemistry. 2011;286(7):5234–41. doi: 10.1074/jbc.M110.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjorck L, Kronvall G. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. Journal of Immunology (Baltimore, Md. : 1950). 1984;133:969–74. [PubMed] [Google Scholar]
- 53.Bjorck L, Kastern W, Lindahl G, Wideback K. Streptococcal protein G, expressed by streptococci or by Escherichia coli, has separate binding sites for human albumin and IgG. Molecular Immunology. 1987;24(10):1113–22. doi: 10.1016/0161-5890(87)90080-0. [DOI] [PubMed] [Google Scholar]
- 54.Lejon S, Frick IM, Bjorck L, Wikstrom M, Svensson S. Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin. The Journal of Biological Chemistry. 2004;279(41):42924–28. doi: 10.1074/jbc.M406957200. [DOI] [PubMed] [Google Scholar]
- 55.Linhult M, Binz HK, Uhlen M, Hober S. Mutational analysis of the interaction between albumin-binding domain from streptococcal protein G and human serum albumin. Protein Science: A Publication of the Protein Society. 2002;11(2):206–13. doi: 10.1110/ps.02802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jonsson A, Dogan J, Herne N, Abrahmsen L, Nygren PA. Engineering of a femtomolar affinity binding protein to human serum albumin. Protein Engineering Design and Selection. 2008;21(8):515–27. doi: 10.1093/protein/gzn028. [DOI] [PubMed] [Google Scholar]
- 57.Stork R, Muller D, Kontermann RE. A novel tri-functional antibody fusion protein with improved pharmacokinetic properties generated by fusing a bispecific single-chain diabody with an albumin-binding domain from streptococcal protein G. Protein Engineering Design and Selection. 2007;20(11):569–76. doi: 10.1093/protein/gzm061. [DOI] [PubMed] [Google Scholar]
- 58.Hopp J, Hornig N, Zettlitz KA, Schwarz A, Fuss N, Muller D, Kontermann RE. The effects of affinity and valency of an albumin-binding domain (ABD) on the half-life of a single-chain diabody-ABD fusion protein. Protein Engineering Design and Selection. 2010;23(11):827–34. doi: 10.1093/protein/gzq058. [DOI] [PubMed] [Google Scholar]
- 59.Malm M, Bass T, Gudmundsdotter L, Lord M, Frejd FY, Ståhl S, Löfblom J. Engineering of a bispecific affibody molecule towards HER2 and HER3 by addition of an albumin-binding domain allows for affinity purification and in vivo half-life extension. Biotechnology Journal. 2014;9(9):1215–22. doi: 10.1002/biot.201400009. [DOI] [PubMed] [Google Scholar]
- 60.Delacroix DL, Elkom KB, Geubel AP, Hodgson HF, Dive C, Vaerman JP. Changes in size, subclass, and metabolic properties of serum immunoglobulin A in liver diseases and in other diseases with high serum immunoglobulin A. The Journal of Clinical Investigation. 1983;71(2):358–67. doi: 10.1172/jci110777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valerius T, Stockmeyer B, Van Spriel AB, Graziano RF, Van Den Herik-oudijk IE, Repp R, Deo YM, Lund J, Kalden JR, Gramatzki M, et al. FcalphaRI (CD89) as a novel trigger molecule for bispecific antibody therapy. Blood. 1997;90(11):4485–92. doi: 10.1182/blood.V90.11.4485. [DOI] [PubMed] [Google Scholar]
- 62.Boross P, Lohse S, Nederend M, Jansen JHM, Van Tetering G, Dechant M, Peipp M, Royle L, Liew LP, Boon L, et al. IgA EGFR antibodies mediate tumour killing in vivo. EMBO Molecular Medicine. 2013;5(8):1213–26. doi: 10.1002/emmm.201201929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dechant M, Beyer T, Schneider-Merck T, Weisner W, Peipp M, Van De Winkel JG, Valerius T. Effector mechanisms of recombinant IgA antibodies against epidermal growth factor receptor. The Journal of Immunology. 2007;179(5):2936–43. doi: 10.4049/jimmunol.179.5.2936. [DOI] [PubMed] [Google Scholar]
- 64.Morton HC, Van Egmond M, Van De Winkel JG. van de Winkel JG. Structure and function of human IgA Fc receptors (Fc alpha R). Crit Rev Immunol. 1996;16:423–40. [PubMed] [Google Scholar]
- 65.Huls G, Heijnen IA, Cuomo E, Van Der Linden J, Boel E, Van De Winkel JG, Logtenberg T. Antitumor immune effector mechanisms recruited by phage display-derived fully human IgG1 and IgA1 monoclonal antibodies. Cancer Research. 1999;59:5778–84. [PubMed] [Google Scholar]
- 66.Lohse S, Loew S, Kretschmer A, Jansen JHM, Meyer S, Ten Broeke T, Rösner T, Dechant M, Derer S, Klausz K, et al. Effector mechanisms of IgA antibodies against CD20 include recruitment of myeloid cells for antibody-dependent cell-mediated cytotoxicity and complement-dependent cytotoxicity. British Journal of Haematology. 2018;181(3):413–17. doi: 10.1111/bjh.14624. [DOI] [PubMed] [Google Scholar]
- 67.Breedveld A, Van Egmond M. IgA and FcalphaRI: pathological roles and therapeutic opportunities. Frontiers in Immunology. 2019;10:553. doi: 10.3389/fimmu.2019.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J, Kuroki M, Shibaguchi H, Wang L, Huo Q, Takami N, Tanaka T, Kinugasa T, Kuroki M. Recombinant human monoclonal igA antibody against CEA to recruit neutrophils to CEA-expressing cells. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2008;17(5):217–22. doi: 10.3727/096504008786111374. [DOI] [PubMed] [Google Scholar]
- 69.Lohse S, Meyer S, Meulenbroek LA, Jansen JHM, Nederend M, Kretschmer A, Klausz K, Möginger U, Derer S, Rösner T, et al. An anti-EGFR IgA that displays improved pharmacokinetics and myeloid effector cell engagement in Vivo. Cancer Res. 2016;76(2):403–17. doi: 10.1158/0008-5472.CAN-15-1232. [DOI] [PubMed] [Google Scholar]
- 70.Rouwendal GJ, Van Der Lee MM, Meyer S, Reiding KR, Schouten J, De Roo G, Egging DF, Leusen JH, Boross P, Wuhrer M, et al. A comparison of anti-HER2 IgA and IgG1 in vivo efficacy is facilitated by high N-glycan sialylation of the IgA. mAbs. 2016;8(1):74–86. doi: 10.1080/19420862.2015.1102812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lohse S, Loew S, Kretschmer A, Jansen JHM, Meyer S, Ten Broeke T, Rösner T, Dechant M, Derer S, Klausz K, et al. Effector mechanisms of IgA antibodies against CD20 include recruitment of myeloid cells for antibody-dependent cell-mediated cytotoxicity and complement-dependent cytotoxicity. British Journal of Haematology. 2017;181(3):413–17. doi: 10.1111/bjh.14624. [DOI] [PubMed] [Google Scholar]
- 72.Treffers LW, Ten Broeke T, Rösner T, Jansen JHM, Van Houdt M, Kahle S, Schornagel K, Verkuijlen PJJH, Prins JM, Franke K, et al. IgA-mediated killing of tumor cells by neutrophils is enhanced by CD47-SIRPα checkpoint inhibition. Cancer Immunology Research. 2020;8(1):120–30. doi: 10.1158/2326-6066.Cir-19-0144. [DOI] [PubMed] [Google Scholar]
- 73.Nilsen J, Bern M, Sand KMK, Grevys A, Dalhus B, Sandlie I, Andersen JT. Human and mouse albumin bind their respective neonatal Fc receptors differently. Scientific Reports. 2018;8(1):14648. doi: 10.1038/s41598-018-32817-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andersen JT, Daba MB, Berntzen G, Michaelsen TE, Sandlie I. Cross-species binding analyses of mouse and human neonatal Fc receptor show dramatic differences in immunoglobulin G and albumin binding. The Journal of Biological Chemistry. 2010;285(7):4826–36. doi: 10.1074/jbc.M109.081828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andersen JT, Cameron J, Plumridge A, Evans L, Sleep D, Sandlie I. Single-chain variable fragment albumin fusions bind the neonatal Fc receptor (FcRn) in a species-dependent manner: implications for in vivo half-life evaluation of albumin fusion therapeutics. The Journal of Biological Chemistry. 2013;288(33):24277–85. doi: 10.1074/jbc.M113.463000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roopenian DC, Low BE, Christianson GJ, Proetzel G, Sproule TJ, Wiles MV. Albumin-deficient mouse models for studying metabolism of human albumin and pharmacokinetics of albumin-based drugs. mAbs. 2015;7(2):344–51. doi: 10.1080/19420862.2015.1008345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nilsen J, Sandlie I, Roopenian DC, Andersen JT. Animal models for evaluation of albumin-based therapeutics. Current Opinion in Chemical Engineering. 2018;19:68–76. doi: 10.1016/j.coche.2017.11.007. [DOI] [Google Scholar]
- 78.Nilsen J, Trabjerg E, Grevys A, Azevedo C, Brennan SO, Stensland M, Wilson J, Sand KMK, Bern M, Dalhus B, et al. An intact C-terminal end of albumin is required for its long half-life in humans. Communications Biology. 2020;3(1):181. doi: 10.1038/s42003-020-0903-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andersen JT, Dalhus B, Viuff D, Ravn BT, Gunnarsen KS, Plumridge A, Bunting K, Antunes F, Williamson R, Athwal S, et al. Extending serum half-life of albumin by engineering neonatal Fc receptor (FcRn) binding. The Journal of Biological Chemistry. 2014;289(19):13492–502. doi: 10.1074/jbc.M114.549832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bern M, Nilsen J, Ferrarese M, Sand KMK, Gjølberg TT, Lode HE, Davidson RJ, Camire RM, Bækkevold ES, Foss S, et al. An engineered human albumin enhances half-life and transmucosal delivery when fused to protein-based biologics. Science Translational Medicine. 2020;12(565):eabb0580. doi: 10.1126/scitranslmed.abb0580. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt MM, Townson SA, Andreucci AJ, King B, Schirmer E, Murillo A, Dombrowski C, Tisdale A, Lowden P, Masci A, et al. Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface. Structure (London, England: 1993. 2013;21(11):1966–78. doi: 10.1016/j.str.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 82.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Engineering, Design and Selection. 1999;12(6):439–46. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 83.Sand KM, Bern M, Nilsen J, Dalhus B, Gunnarsen KS, Cameron J, Grevys A, Bunting K, Sandlie I, Andersen JT, et al. Interaction with both domain I and III of albumin is required for optimal pH-dependent binding to the neonatal Fc receptor (FcRn). The Journal of Biological Chemistry. 2014;289(50):34583–94. doi: 10.1074/jbc.M114.587675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carter PJ, Lazar GA. Next generation antibody drugs: pursuit of the ‘high-hanging fruit’. Nature Reviews Drug Discovery. 2018;17(3):197–223. doi: 10.1038/nrd.2017.227. [DOI] [PubMed] [Google Scholar]
- 85.Greiff V, Yaari G, Cowell LG. Mining adaptive immune receptor repertoires for biological and clinical information using machine learning. Current Opinion in Systems Biology. 2020;24:109–19. doi: 10.1016/j.coisb.2020.10.010. [DOI] [Google Scholar]
- 86.Hoogenboezem EN, Duvall CL. Harnessing albumin as a carrier for cancer therapies. Advanced Drug Delivery Reviews. 2018;130:73–89. doi: 10.1016/j.addr.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Research. 1986;46:6387–92. [PubMed] [Google Scholar]
- 88.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. Journal of Controlled Release: Official Journal of the Controlled Release Society. 2000;65(1–2):271–84. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 89.Abdallah M, Müllertz OO, Styles IK, Mörsdorf A, Quinn JF, Whittaker MR, Trevaskis NL. Lymphatic targeting by albumin-hitchhiking: applications and optimisation. Journal of Controlled Release: Official Journal of the Controlled Release Society. 2020;327:117–28. doi: 10.1016/j.jconrel.2020.07.046. [DOI] [PubMed] [Google Scholar]
- 90.Brandl F, Busslinger S, Zangemeister-Wittke U, Plückthun A. Optimizing the anti-tumor efficacy of protein-drug conjugates by engineering the molecular size and half-life. Journal of Controlled Release: Official Journal of the Controlled Release Society. 2020;327:186–97. doi: 10.1016/j.jconrel.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 91.Weflen AW, Baier N, Tang QJ, Van Den Hof M, Blumberg RS, Lencer WI, Massol RH. Multivalent immune complexes divert FcRn to lysosomes by exclusion from recycling sorting tubules. Molecular Biology of the Cell. 2013;24(15):2398–405. doi: 10.1091/mbc.E13-04-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berntzen G, Lunde E, Flobakk M, Andersen JT, Lauvrak V, Sandlie I. Prolonged and increased expression of soluble Fc receptors, IgG and a TCR-Ig fusion protein by transiently transfected adherent 293E cells. Journal of Immunological Methods. 2005;298(1–2):93–104. doi: 10.1016/j.jim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 93.Firan M, Bawdon R, Radu C, Ober RJ, Eaken D, Antohe F, Ghetie V, Ward ES. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of gamma-globulin in humans. International Immunology. 2001;13(8):993–1002. doi: 10.1093/intimm/13.8.993. [DOI] [PubMed] [Google Scholar]
- 94.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Research. 2020;48(W1):W449–w454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Research. 2017;45(W1):W24–w29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


