Chromophore perturbations in the sodium transport intermediate states of KR2 are revealed by DNP-enhanced ssNMR.
Abstract
The functional mechanism of the light-driven sodium pump Krokinobacter eikastus rhodopsin 2 (KR2) raises fundamental questions since the transfer of cations must differ from the better-known principles of rhodopsin-based proton pumps. Addressing these questions must involve a better understanding of its photointermediates. Here, dynamic nuclear polarization–enhanced solid-state nuclear magnetic resonance spectroscopy on cryo-trapped photointermediates shows that the K-state with 13-cis retinal directly interconverts into the subsequent L-state with distinct retinal carbon chemical shift differences and an increased out-of-plane twist around the C14-C15 bond. The retinal converts back into an all-trans conformation in the O-intermediate, which is the key state for sodium transport. However, retinal carbon and Schiff base nitrogen chemical shifts differ from those observed in the KR2 dark state all-trans conformation, indicating a perturbation through the nearby bound sodium ion. Our findings are supplemented by optical and infrared spectroscopy and are discussed in the context of known three-dimensional structures.
INTRODUCTION
KR2 (Krokinobacter eikastus rhodopsin 2) is a light-driven sodium pump found in the marine bacteria K. eikastus (1, 2). It belongs to the family of microbial rhodopsins, characterized by seven transmembrane helices and a retinal cofactor linked via a Schiff base (SB) to a lysine (K255) in helix 7. If the sodium concentration is low, KR2 has been found to be able to translocate protons and lithium ions as well (1, 3). Engineering of the protein structure in the uptake cavity region leads to an expanded ion selectivity including potassium or cesium (4–6). As for other microbial rhodopsins, light-driven ion translocation by KR2 creates an electrochemical gradient and a membrane potential. KR2 has therefore attracted considerable interest because it is the first microbial rhodopsin that shows a cation pumping activity other than proton pumping, making it a promising tool for optogenetics. The potential application of KR2 as an effective optogenetic silencer would have the advantage that it could hyperpolarize the membrane potential without having an influence on the pH balance of cells (7).
Sodium pumps contain a signature NDQ motif (N112, D116, and Q123) in transmembrane helix 3 (1), which differs from the DTD [bacteriorhodopsin (BR) (8)] or DTE [proteorhodopsin (PR) (9)] motif found in proton pumps. In KR2, D116 is the retinal SB (RSB) counterion forming an H-bond with the protonated SB (pSB). The highly conserved proton donor and acceptor residues found in outward proton pumps are replaced by glutamine (Q123) and asparagine (N112) residues, which do not have such a functionality.
The discovery of KR2 was unexpected because the translocation of a cation such as Na+ across the positively charged RSB appeared energetically unfavorable due to the expected repulsion from the SB proton. Furthermore, sodium transport and its link to the photocycle raise many questions because mechanisms derived from well-studied proton pumps involving, for example, Grotthuss transfer steps do not apply.
Extensive efforts have been undertaken to resolve the structure and functional mechanism of KR2, making it the most intensively studied sodium pumping rhodopsin. Methods applied include femtosecond optical, ultrafast infrared, Raman, and solid-state nuclear magnetic resonance (NMR) spectroscopy (10–15) as well as molecular dynamics simulations (16). They were often combined with site-directed mutagenesis and transport assays to resolve the importance of key residues such as the NDQ motif as well as those affecting sodium binding, ion selectivity, or the ion transport pathway (e.g., H30, S70, D102, R109, R243, D251, N61, G263, E11, and E160) (1, 2, 4–6, 17).
Optical flash photolysis experiments revealed four distinct photointermediates named K, L, M, and O (Fig. 1A) (1, 18). After light activation, the retinal isomerizes from all-trans to 13-cis, resulting in the red-shifted K-state as known from BR and other microbial rhodopsins (10, 19). Subsequently, a blue-shifted L-state is formed, during which the pSB is prepared to transfer a proton to D116 in the even further blue-shifted M-state (1, 20). Uptake of sodium is believed to take place between the M-state and the red-shifted O-state (1, 3). The population of the O-state has been found to depend strongly on the sodium concentration (5, 21). The retinal conformation has been discussed controversially as 13-cis (20, 22) or all-trans (23, 24). The protein then relaxes back to the dark state to complete the photocycle.
Fig. 1. Experimental approach.
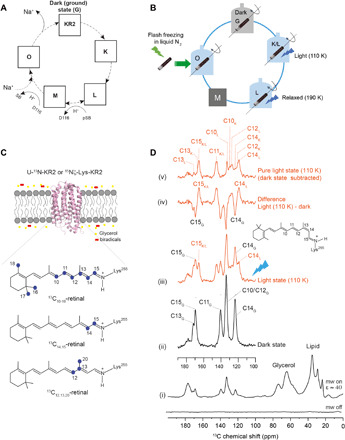
(A) Schematic photocycle of KR2. (B) Light-induced cryo-trapping for dynamic nuclear polarization (DNP)–enhanced magic angle spinning (MAS) NMR. (C) Retinal and KR2 isotope-labeling schemes. KR2 proteoliposomes were incubated with AMUPol in D2O/H2O/glycerol (see Materials and Methods). (D) DNP-enhanced 13C spectra of 13C10–18-KR2. (i) 13C CP spectra reveal a DNP signal enhancement of 40. (ii) 13C DQF spectra in the dark state and (iii) after illumination at 110 K. (iv) Difference spectrum (light dark). (v) Pure light-induced spectrum obtained by subtracting the remaining dark state contribution (see fig. S1). Retinal peak assignment is based on data in Fig. 2. mw, microwave.
Monomeric (5, 6) and pentameric (17) KR2 dark state crystal structures have been reported. In contrast to proton or chloride pumping microbial rhodopsins, no bound ion substrate has been found inside dark state KR2. The only bound sodium ion found in the resting state is at the extracellular side of the protomer interface in the pentameric crystal structure, but its binding site seems to be not directly related to the function of KR2 (6). The crystal structures of monomeric KR2 adopt a more “compact” conformation in which the large water-filled cavity close to the RSB found in the “expanded” pentameric state is missing (7).
Very recently, a serial crystallography study using an x-ray free-electron laser on monomeric KR2 has been reported (25). Two sodium-binding events have been observed, one near N112 and D251 (“O1 state”), followed by a second binding between E11, N106, and E160 (“O2 state”). In contrast, the crystal structure of the O-intermediate of pentameric KR2 (23) reveals a bound sodium ion near the RSB coordinated by N112 and D116 (23). These studies represent excellent progress in resolving structures of resting and relevant photointermediate states. However, the resolution is still limited, and fine electrostatic details may not be observable.
To link three-dimensional (3D) structures with optical properties and functional data, NMR parameters such as chemical shifts, internuclear distances, or torsion angles within the chromophore can be extremely useful, especially in the context of model building assisted by quantum chemical approaches. In particular, solid-state NMR (ssNMR) has been very strong in the field as it offers unique possibilities to obtain structural details at atomic resolution for membrane proteins directly in lipid bilayer environments (26). It has been intensively used for the study of the retinal chromophore (27–32). The sensitivity of NMR can be markedly improved by dynamic nuclear polarization (DNP), where polarization is transferred from paramagnetic dopants to nuclear spins (33). DNP, therefore, offers an excellent way to probe light-induced, cryo-trapped photointermediate states. The method was applied previously to the microbial rhodopsins BR (28, 34, 35), channelrhodopsin-2 (ChR2) (36), and PR (32, 37, 38) but has not been used for sodium pumps such as KR2.
Here, we applied DNP-enhanced ssNMR to monitor chemical shifts of retinal carbons and SB nitrogen in the dark state and K-, L-, and O-states of KR2 in its physiological pentameric form reconstituted into lipid bilayers. The light-induced photointermediates were stabilized by various cryo-trapping protocols such as illumination at 110 K, thermal relaxation, or flash freezing (Fig. 1B). Cryo- trapping is highly compatible with the low-temperature requirements of DNP (39). The applied procedures were validated by recording time-resolved as well as cryo-trapped ultraviolet/visible (UV/Vis) absorption spectra of photointermediates. Our data reveal an equilibrium between the early K- and L-states with distinct 13C chemical shift differences. The 15N chemical shift of the SB nitrogen gradually increases from the dark state via K to L, indicating a stronger H-bond to D116. Furthermore, an increased out-of-plane twist at the end of the polyene chain in the L-state is observed. In the O-state, where sodium is bound inside KR2, the chromophore adopts an all-trans conformation that, however, differs from the dark state. All-trans retinal has been suggested to aid sodium release (16), and the observed 13C and 15N chemical shift differences reflect slight structural rearrangement within the retinal binding pocket and altered SB interactions. The obtained data are complemented by time-resolved IR measurements, compared with those reported for proton pumping microbial rhodopsins, and discussed in the context of known x-ray data.
RESULTS
To monitor the retinal conformation and SB-counterion interaction by ssNMR, we used three different isotope-labeling schemes (see Fig. 1C): 10–18-13C9 or 14,15-13C2 all-trans retinal was incorporated into U-15N-KR2. In addition, 12,13,20-13C3 all-trans retinal was reconstituted into 15Nζ-Lys-KR2. We refer in the following to these samples as 13C10–18-KR2, 13C14,15-KR2, and 13C12,13,20-KR2, respectively.
KR2 was reconstituted into lipid bilayers. All experiments were carried out in the presence of NaCl (30 to 100 mM) and with AMUPol as polarizing agent (see Materials and Methods for further details). Typically, a 40-fold signal enhancement (microwave on/off) was achieved by DNP in 1H-13C cross-polarization (CP) experiments as shown for 13C10–18-KR2 [spectrum (i) in Fig. 1D]. Such an enhancement has been shown to be essential for in situ illumination and cryo-trapping experiments of photoreceptors (36, 37, 39). For 13C10–18 retinal, six of nine signals could be directly detected in double-quantum filtered (DQF) spectra (40) in which the natural abundance 13C background has been suppressed [spectrum (ii) in Fig. 1D]. Four of these signals were well resolved and two overlapped (C10 and C12). The β-ionone ring methyl resonances C16, C17, and C18 were not considered here as they could not be detected under our DNP conditions (15, 32, 41). The assignment of all other carbons was derived from double-quantum single-quantum (13C-13C DQSQ) (42) and carbon-carbon proton-driven spin diffusion (PDSD) spectra (43) (Fig. 2, A and B). The obtained chemical shifts (table S1) in the dark state were identical to those reported by us before for KR2 in the proton-pumping mode in the absence of sodium (15). The C20 resonance was determined from a 13C DQF spectrum of 13C12,13,20-KR2, which also confirmed the C12 and C13 assignments (Fig. 2E). The C14 and C15 resonance assignments were confirmed by a 13C DQF spectrum of 13C14,15-KR2 (Fig. 3B).
Fig. 2. Retinal resonance assignment.
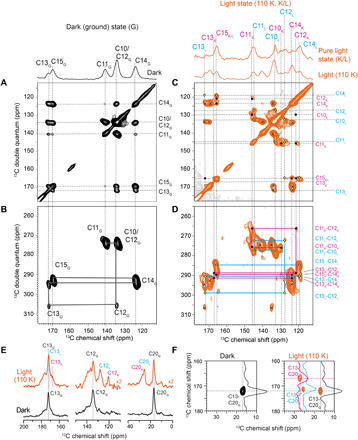
13C-13C PDSD and DQSQ spectra of 13C10–18-KR2 in the dark state (G) (A and B) and after illumination at 110 K (C and D). In the light state, two separate spin systems corresponding to the K-state (•) and L-state (◊) and the residual dark state (x) population are detected. Cross peaks belonging to the same spin system are connected in the DQSQ spectra (B and D). (E) 13C DQF spectra of 13C12,13,20-KR2 in the dark state (G) and after illumination at 110 K (G, K, and L) show resonances for C13, C12, and C20. (F) PDSD spectra of 13C12,13,20-KR2 reveal a C13-C20 cross peak in the dark, which splits into two additional signals assigned to C13K/L and C20K/L upon illumination.
Fig. 3. Thermal relaxation experiments for the identification of the K- and L-state spin system.
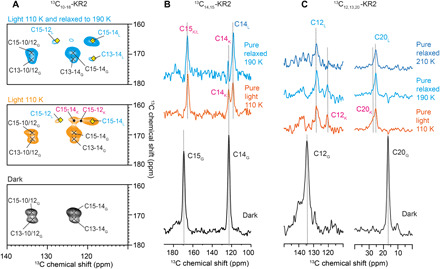
(A) 13C-13C PDSD spectrum of 13C10–18-KR2 in the dark (bottom). Upon illumination at 110 K (middle), two sets of C15-14 and C14-12 cross peaks occur in addition to the residual dark state resonances (x). Under thermal relaxation (top), only one set of these cross peaks remains and is assigned to the L-state (◊). The disappearing resonances belong to the K-state (•). (B) 13C DQF spectra of 13C14,15-KR2. Upon illumination at 110 K, C15 shifts, while C14 splits into two resonances. The latter disappears upon thermal relaxation, which confirms the assignment of both resonances to K and L. The residual dark state contribution has been subtracted from the pure light-induced spectra. (C) Same experiment as in (B) but on 13C12,13,20-KR2. Here, C20 shifts, and C12 shows a pronounced splitting, which reduces to just one peak after thermal relaxation, confirming here the resonance assignments to K- and L-states as well. See table S1 for chemical shift. An extensive temperate scan is shown in fig. S2.
Cryo-trapping and assignment of the K- and L-intermediates
Sample illumination and cryo-trapping were carried out as described previously for green PR (GPR) (37). A high-power light-emitting diode (LED) was used for in situ illumination directly within the NMR spectrometer at 110 K for 60 min. As a result, new resonances occur in the 13C DQF spectrum [spectrum (iii) in Fig. 1D]. The light-dark difference spectrum [spectrum (iv) in Fig. 1D] demonstrates the depletion of the dark state and the creation of light-induced states. A conversion of approximately 45% can be achieved as estimated from the C11 resonance (fig. S1). A pure light-induced spectrum is obtained by subtracting the remaining dark state population [spectrum (v) in Fig. 1D].
To resolve and assign all new resonances, we recorded 13C-13C PDSD and DQSQ spectra (Fig. 2, C and D). In these spectra, all signals from the residual dark state population could be identified. The newly appearing cross peaks correlate with each other and describe two separate retinal signal sets, which can be assigned by combining the DQSQ assignment walk with the correlations observed in the PDSD spectrum. Unfortunately, the coexistence of dark and apparently two photointermediate states in our cryo-illuminated samples causes severe peak overlap in the 2D spectra of 13C10–18-KR2. To unambiguously assign these new cross peaks, we also cryo-trapped 13C12,13,20- and 13C14,15-KR2 by illumination at 110 K. The C12 signal splits into two resonances (Fig. 2E). Furthermore, two additional C13-C20 cross peaks occur (Fig. 2F). Likewise, the C15 resonance shifts, and C14 splits into two signals (Fig. 3B). Using these spectra, two separate spin systems can be assigned, and it is therefore concluded that two early intermediates, tentatively assigned as K and L, have been trapped (table S1).
To identify which of the signals belongs to K and which belongs to L, we illuminated the sample at 110 K followed by thermal relaxation at various higher temperatures and subsequent detection by DNP–magic angle spinning (MAS) NMR after cooling down again to 110 K (36). Illumination under cryogenic conditions traps the protein in the earliest photointermediate state. By raising the temperature, subsequent photointermediates can be thermally reached before the system returns to the dark state. Upon illumination of 13C10–18-KR2 at 110 K, two sets of C15-C14 and C15-C12 cross peaks occur in addition to the remaining dark state signals as described above (Fig. 3A). After thermal relaxation, two of the light-induced cross peaks disappear. They are therefore assigned to the K-intermediate, while the remaining peaks belong to the L-state. Additional support is provided by 13C DQF spectra of 13C14,15-KR2, which show for C14 that under thermal relaxation at 190 K, the K-state decreases and the L-state increases (Fig. 3B). Similarly, we observe for 13C12,13,20-KR2 that one of the C12 resonances increases, which allows us to assign the K- and L-state signals (Fig. 3C). The same effects are observed for C13 (fig. S2B) and C20 (Fig. 3C). Thermal relaxation experiments over a wider temperature range are shown in fig. S2A.
The assignments to K and L have been further validated by comparing UV/Vis absorbance spectra of photointermediates extracted from time-resolved flash photolysis experiments with those obtained under cryo-trapping conditions as used for DNP ssNMR (Fig. S4). Such an approach has been used before successfully to validate cryo-trapping of ChR2 (36) and PR (37). The difference spectra recorded at 110 and 160 K correspond well with the spectral signatures of the K- and L-state. The observation of both K- and L-photointermediates at temperatures as low as 100 K is unexpected as usually only the K-state can be produced in other microbial rhodopsins at such low temperatures (28, 34, 37).
Characterization of the K- and L-intermediates
Chemical shift differences between dark and K/L-states are plotted in Fig. 4A. The 13-cis retinal isomer in the K-state causes largest effects for C12 with a shielding of 12.8 parts per million (ppm) and for C20 with a deshielding of 10.3 ppm. These values reduce in the L-state to 6.4 and 8.5 ppm for C12 and C20, respectively (table S2). The chemical shift changes are compared to those observed for the trans-cis isomerization in model compounds (gray bars in Fig. 4A) (44, 45). For the K-state, they deviate by more than 4 ppm for C10 to C13 and C20 between KR2 and model compounds, which therefore suggests a protein-induced chromophore distortion around these carbons in the K-state. For the L-state, these differences are less pronounced. The chemical shift differences between K and L are shown in Fig. 4B. C12 (6.4 ppm) and C13 (4.1 ppm) are less shielded, while C14 displays an additional shielding (4.6 ppm), suggesting a slight movement toward a 15-syn conformation (46).
Fig. 4. Light-induced 13C retinal carbon and SB chemical shift changes.
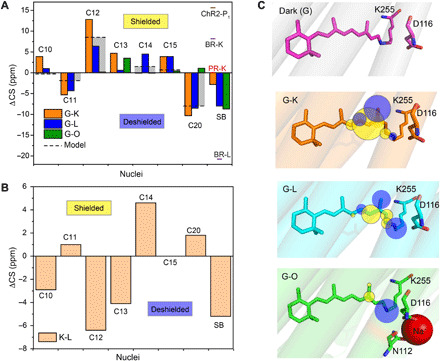
(A) Chemical shift differences between dark state (G) and photointermediates K, L, and O are plotted for carbon atoms C10 to C15, C20, and SB. The gray bars illustrate the effect of the all-trans 13-cis isomerization as observed for the model compounds N-retinylidenebutylamine (C10 to C15) and retinal (C20) (44, 45). The 15N chemical shift of KR2 in the dark and intermediate states is compared to GPR (37), BR (28), and ChR2 (36). (B) Chemical shift difference between the K- and L-state. (C) Illustration of chemical shift changes observed at retinal carbons and SB nitrogen in the dark state [Protein Data Bank (PDB): 6REW], K-state (PDB: 6TK5), L-state (PDB: 6TK4), and O-state (PDB: 6XYT). The yellow and blue spheres indicate shielding and deshielding at the nuclei, respectively.
These observations suggest a fine adjustment of the chromophore upon illumination and a distortion at least near the SB region in early photointermediates as also indicated by previous time-resolved IR studies (10, 20). Such a distortion could be, for example, an out-of-plane twist of the polyene chain. This was further studied by time-resolved IR measurements, which span the spectral range of such hydrogen out-of-plane (HOOP) modes (Fig. 5A). The highlighted transients at 960, 980, and 992 cm−1 show a biphasic decay, which correlates with the K/L-to-M transition and the decay of M, indicating a strong change of retinal distortion during the early intermediates in the photocycle. We therefore measured the HCCH torsion angle around the C14-C15 bond in 13C14,15-KR2 by double-quantum heteronuclear local field experiments (47). Unfortunately, the K-intermediate could not be analyzed unambiguously because the C14 resonances of the dark and K-state as well as the C15 peaks of K- and L-states fully overlap (Fig. 3B and fig. S3A). However, for the L-state, dephasing curves could be obtained by deconvoluting the C14L peak from the C14G,K resonance. A comparison with the dark state is shown in Fig. 5B. The curves display a substantial deviation from planarity in the dark state (150° ± 2°), which becomes larger in the L-state (136° ± 6°; see also table S1). For comparison, we also performed light-induced cryo-trapping experiments in the absence of sodium. The chemical shifts of C14 and C15 do not change in K and L and also the same HCCH torsion angle is obtained (fig. S3).
Fig. 5. Twist of the retinal polyene chain.
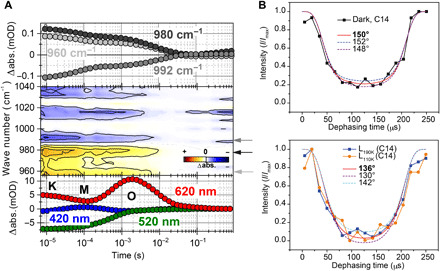
(A) Transients of the UV/Vis measurements representing the intermediates of the photocycle (bottom), contour plot of the HOOP region (middle), and picked IR transients (top). The HOOP modes at 960, 980, and 992 cm−1 show a biphasic decay, with the strongest amplitudes during the K/L-to-M transition and the decay of M. This biphasic behavior underlines the strongly twisted character of the retinal chromophore in the early photocycle of KR2, which relaxes back to a ground state–like conformation during the formation of O. (B) HCCH dephasing curves of the C14-C15 spin system in 13C14,15-KR2 reporting on the H-C14-C15-H dihedral angle. (i) Dark state and (ii) L-intermediate were obtained by illumination at 110 K and followed by thermal relaxation at 190 K. Curves were obtained by analyzing the C14 signal intensity (see fig. S3 for further details). abs, absorbance; mOD, optical density/1000.
The SB nitrogen 15N chemical shift is affected by its protonation state, H-bond formation and characteristics, its electrostatic environment, and the torsion around the C15═N bond. This NMR parameter has been extensively used for monitoring interactions between the SB and the counterion/proton acceptor during the photocycle (28, 34). The dark state signal for the pSB shifts and broadens upon illumination and cryo-trapping (fig. S2C). To resolve the K- and L-state contributions to the observed line shapes, 15N-13C transferred echo double resonance (TEDOR) (48, 49) spectra of 13C10–18-KR2 were recorded. In these spectra, cross peaks between the SB nitrogen and retinal carbons C14 and C15 are observed (Fig. 6). In the dark state, the pSB nitrogen chemical shift at 173.3 ppm is as reported before (Fig. 6A) (13, 15). Upon illumination at 110 K (Fig. 6B), cross peaks for the K- and L-state subpopulations occur. After thermal relaxation at 190 K, only the L-state cross peaks remain (Fig. 6C). These spectra reveal the pSB nitrogen chemical shift for the K-state at 176.1 ppm and for the L-state at 181.3 ppm. The SB nitrogen becomes deshielded in the K- and L-states compared to the dark state, which indicates a stronger H-bond caused by stronger interactions with the counterion (D116) (50, 51). This effect is more pronounced in the L-state, which prepares the protein for the subsequent proton transfer step in the M-state. We also tried to trap the M-state by a further thermal relaxation step. However, the de-pSB signal, which is expected at around 320 ppm, could not be detected. One reason might be a very short lifetime/low population of the M-state (1).
Fig. 6. DNP-enhanced 15N-13C TEDOR spectra of 13C10–18-KR2 recorded under different trapping conditions.
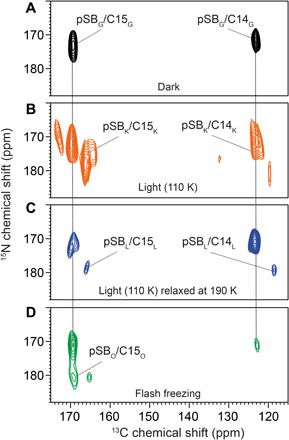
(A) In the dark state, cross peaks between the pSB nitrogen (173.3 ppm) and C14/C15 occur. (B) Upon illumination at 110 K, a mixture of new cross peaks and residual dark state populations arises. The main new population stems from the K-state. (C) A subsequent thermal relaxation step at 190 K depopulates the K-state and increases the L-state. The K- and L-state 13C chemical shifts agree with those described above (Fig. 3B). They correlate with pSB signals at 176.1 ppm (K-state) and 181.3 ppm (L-state). (D) Trapping the O-state by illumination at room temperature followed by flash freezing does not alter the C14/C15 chemical shifts significantly, but an additional cross peak between C15 and a new pSB nitrogen resonance at 182 ppm is observed. The signal at 181/175 ppm could be from a remaining L-state population but is not considered further because of its low signal-to-noise ratio. Spectra were recorded with a TEDOR mixing time of 6 ms.
Cryo-trapping and analyzing the O-intermediate
The O-state is the last photointermediate in the KR2 photocycle (1). It becomes only populated in the presence of sodium ions and has the longest lifetime of all intermediates (18). Trapping is therefore based on continuous illumination outside the NMR spectrometer, followed by rapid flash freezing in liquid nitrogen and subsequent transfer into the DNP probe. This trapping procedure was first validated by cryo-UV/Vis spectroscopy, which demonstrated the feasibility of this method (fig. S4).
The O-state trapping procedure was then applied to 13C12,13,20-KR2. A comparison of 1D 13C DQF spectra obtained after flash freezing with those recorded in the dark, after illumination at 110 K and after thermal relaxation, is shown in fig. S2B. No obvious chemical shift difference is detected between the ground and the O-state. To improve resolution, a 13C-13C PDSD spectrum was recorded after flash freezing. Here, an additional correlation between C13 and C20 due to small chemical shift differences for both resonances is observed (Fig. 7A). C13 and C20 are slightly more shielded with respect to the dark state by around 3 and 1 ppm, respectively. Similarly, an additional C13-C12 cross peak occurs. The C12 resonance is especially sensitive to the retinal isomerization. It is almost identical in the dark state and the O-state, suggesting a recovery of the all-trans retinal conformation.
Fig. 7. Analyzing the retinal chromophore in the KR2 O-state trapped by illumination and flash freezing.
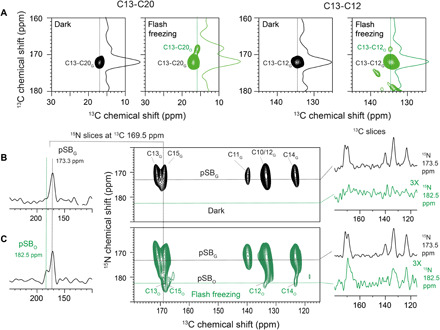
(A) 13C-13C PDSD spectra of 13C12,13,20-KR2 reveal a C13-C20 (left) and a C13-C12 cross peak (right) in the dark state. Upon flash freezing, an additional, small cross peak for C13-C20 and C13-C12 is detected. They arise from the O-state. C13 and C20 change by 3.5 and 1.1 ppm, respectively. (B) 15N-13C DCP (double cross polarization) correlation spectrum of 13C10–18-KR2 (100 mM NaCl) with an N(C15)CX polarization transfer step in the dark state and (C) upon flash freezing. The trapped O-state population is characterized by a deshielding of the SB nitrogen by 9 ppm, while the C12, C14, and C15 resonances are similar to the dark state.
To obtain O-state chemical shifts for the other retinal carbons and the SB nitrogen, we recorded 15N-13C TEDOR spectra on flash-frozen samples of 13C10–18-KR2 (Fig. 6D). A new 15N-pSB–13C15 cross peak could be detected in addition to the signals from the remaining dark state population. The retinal 13C15 chemical shift of the O-state is the same as that of the all-trans retinal in the dark state. However, the 15N chemical shift of the pSB nitrogen occurs at 182.0 ppm and is therefore deshielded with respect to the dark state (173.3 ppm). To improve the magnetization transfer along the polyene chain, we performed an N(C)CX-type experiment with an initial CP step from the SB nitrogen to C15, followed by proton-driven carbon-carbon spin diffusion (52). This N(C15)CX spectrum of 13C10–18-KR2 in the dark state shows 15N-pSB correlations at 173.3 ppm with all retinal carbons C10 to C15 (Fig. 7B). The 13C chemical shifts match those assigned by the PDSD and DQSQ spectra (Fig. 2, A and B, and table S1). Upon trapping the O-state, new cross peaks between the 15N-pSB at 182 ppm and C12, C14, and C15 occur (Fig. 7C). The C12 chemical shift in the O-state is confirmed by the experiment on 13C12,13,20-KR2 described above (Fig. 7A). The O-state C12, C14, and C15 13C chemical shifts are similar to the dark state. C13 overlaps with the C15 peak. The C10 and C11 resonances could not be observed because of their low peak intensities. The C12 and C20 O-state chemical shifts unambiguously show an all-trans conformation of the retinal chromophore similar to the dark state but with altered SB interactions since its nitrogen is deshielded by almost 9 ppm.
DISCUSSION
The KR2 retinal and SB have been probed by DNP-enhanced ssNMR after light-induced cryo-trapping in early (K and L) and late (O) photointermediate states (Fig. 1B).
Early photointermediate states
Our cryo-trapping experiments at 110 K revealed a mixture of K- and L-state populations. Very similar experimental conditions applied to BR, GPR, and ChR2 resulted just in a single trapped K-state, which hints toward differences in the energy landscape of early photointermediates between different rhodopsins. This view is also supported by the observed chemical shift changes in the K-state that are overall dominated by the retinal trans-cis isomerization in agreement with model compounds (Fig. 4). However, remarkable deviations from model compound behavior are observed for almost all labeled carbon positions, which therefore cannot solely be explained by isomerization but must partially be caused by specific interactions within the KR2 retinal binding pocket. These additional shielding/deshielding effects are protein specific as illustrated by comparing the K-state chemical shift changes of KR2 obtained with those from the same labeling scheme in the K-state of GPR (table S2). In PR, they also deviate from model compounds, but these changes are less pronounced: The overall root mean square deviation (RMSD) of the chemical shift differences (dark-K) in KR2 is larger than that in GPR (7.1 ppm versus 5.4 ppm; see table S2). Similarly, distinct protein-induced effects have also been observed for BR and other rhodopsins using a range of different labeling schemes (28). Not only the retinal 13C but also the SB 15N chemical shift shows strong photocycle-dependent changes. Factors contributing to the chemical shift are primarily the protonation state, H-bonds, and the electrostatic environment. The latter two factors are often subsumed as a measure for the SB-counterion interactions since an empirical relationship between 15N chemical shift and λmax for RSB model compounds has been found (50). In addition, the torsion around the SB bond C15═N will contribute to the observed chemical shift. Here, a deshielding in the K-state with respect to the dark state by 2.8 ppm is observed. Very different trends occur in other rhodopsin K-states although they are all red-shifted, underlining the role of the protein environment: While GPR shows no 15N chemical shift change (37), a large shielding is observed for light-adapted BR (BR568) (28) and ChR2 (36) (see Fig. 4A).
In the KR2 L-state, the overall 13C chemical shift changes with respect to the dark state are smaller compared to the K-state (13C RMSD reduces from 7.1 to 4.9 ppm; table S2). In addition, the SB nitrogen is strongly deshielded by −8 ppm and the torsion around the C14-C15 bond changes from 150° to 136°. The SB is already stronger drilled around the C15═N bond in the dark state compared to microbial proton pumps because the different position of the proton acceptor D116 in KR2 requires another orientation of the SB N─H bond vector for a hydrogen bond formation (see fig. S3) (1, 5, 6, 17, 20). This is also reflected in the C14-C15 torsion. The larger out-of-plane twist brings the SB proton into an even more favorable position with respect to D116, which would be needed for the subsequent proton transfer step during the M-state. Such a twist in the polyene chain near the SB moiety might be necessary for the inhibition of the delocalization of the positive charge along the π-conjugated system, leading to the decrease of the pKa value (where Ka is the acid dissociation constant) at the SB. This is also supported by the observed nitrogen deshielding, which is indicative of a stronger H-bond to D116. The C14 chemical shift has been found to be a sensitive marker for a 15-syn/anti conformation. In BR, the 15-anti–C14 resonance is found at 124 ppm and that for 15-syn is found at 111 ppm (28). Here, C14 shifts from 123.8 ppm in the dark and K-state to 119.2 ppm in the L-state. This means that the retinal is slightly turning toward a more compact 15-syn conformation in the L-state.
A twist along the retinal polyene chain is also supported by the HOOP modes in the range of 950 to 1000 cm−1 (Fig. 5A). The band at 992 cm−1 either stems from an in-plane bending vibration of the SB or stems from the C15-HOOP mode (53, 54). The positive signals at 960 and 980 cm−1, respectively, were also found for BR and were attributed to the coupled C15-HOOP and N-HOOP mode of the SB (53, 55). All three transients show a biphasic decay, with their strongest contributions during the K/L-to-M transition and the decay of M (fig. S5). This implies a rather strong twisting of retinal in the early photocycle compared to the ground state.
Recently published serial crystallographic x-ray data of monomeric KR2 provide structural snapshots throughout the KR2 photocycle (25). We added our chemical shift changes to an overlay plot of the dark state [Protein Data Bank (PDB): 6TK6] and K-state (PDB: 6TK5; Δt = 800 fs) structures (Fig. 8A). The largest changes stem from the isomerization. Additional protein-specific contributions to the observed chemical shift differences could be caused by altered ring-current effects due to small rearrangements of aromatic residues W113, W215, and Y218 by which the C10-C15 segment of the retinal polyene chain is surrounded. C20 tilts increasingly toward helix C in the L-state, which could also explain the C12, C13, and C14 chemical shift differences between K and L (PDB: 6TK5, Δt = 1 ns; Fig. 8B). The distance between the SB nitrogen and Oδ2 of D116 changes from 3.0 Å in the dark state via 2.9 Å in the K-state to 2.4 Å in the L-state in these x-ray structures, which would agree with a 15N deshielding due to an increasing H-bond strength.
Fig. 8. KR2 retinal binding pocket during the photocycle, structures, and chemical shifts.
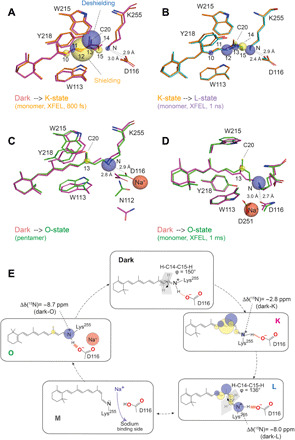
(A) Comparison between dark- and K-state serial crystallography x-ray structures (6TK6 and 6TK5) (25) with 13C and 15N chemical shift changes (see tables S1 and S2). (B) As in (A) but comparison between the K-state (6TK5) and the L-state (6TK4). Plotted chemical shift changes are with respect to the K-state. (C) The dark state and O-state of pentameric KR2 x-ray structures (6REW and 6XYT) (23) and chemical shift changes with respect to the dark state. (D) The dark state and O-state of monomeric KR2 time-resolved structures (6TK6 and 6TK2) (23) and chemical shift changes with respect to the dark state. (E) Photocycle of KR2 and illustration of retinal conformation and electrostatic changes in K, L, and O as observed by ssNMR. The chromophore is highly twisted near the SB in the L-state, and the H-bond between SB nitrogen and D116 becomes stronger in L- and O-intermediates.
BR is the only other rhodopsin for which chemical shift data for the L-state have also been obtained. As mentioned above, the 15N SB nitrogen chemical shift becomes more shielded in the K-state but is significantly deshielded in the L-state (28). In light-adapted BR568, the SB forms an indirect H-bond with the proton acceptor D85 through the structural water molecule W402. In the K-state, this interaction is disturbed because of disordering of W402, followed by the formation of a new H-bond in the L-state (28, 35). The different trend in KR2 most likely hints toward a more direct interaction between SB and D116 throughout the photocycle. Note that T89 in BR, which is at the same position as D116 in KR2, has been found to be involved in proton transfer (35). The torsion and the K- and L-state chemical shifts are sodium independent (fig. S3), which supports the view that bound sodium near the RSB only occurs in later states.
The late O-intermediate state
Structural data of microbial rhodopsin O-states are still rare. Fortunately, the KR2 O-state has a long lifetime and could be therefore trapped in proteoliposomes for ssNMR as reported here or in crystals for x-ray diffraction (23). The key difference between the KR2 O and dark states is the presence of an internally bound Na+ ion. Sodium must pass the SB region during translocation, and the O-state has been shown to be directly connected with sodium binding and transport (3, 18, 24). This additional positive charge will alter the interaction network in the retinal binding pocket and will have an effect on the chemical shifts of nearby nuclei. Our 13C chemical shift data show that the retinal in the O-state has switched back from 13-cis to all-trans but differences for the C13, C20, and of the SB nitrogen chemical shifts occur. Note that the retinal in the O-state x-ray structure is also in all-trans conformation, but the C14-C15 bond has been reported to be out of plane compared to the dark state (23). Furthermore, previous computational studies suggested that an all-trans retinal would facilitate exergonic Na+ release to the extracellular side (16). The same study also suggested an electrostatic attraction of Na+ to the negatively charged D116 resulting in a shielded interaction between D116 and the RSB with a subsequent red shift. This argument is in line with the coordination of bound Na+ by D116, N112, S70, and V67 revealed in the O-state x-ray structure (23) in which the ion is 5.4 Å away from the SB nitrogen (Fig. 8C).
For the SB nitrogen, a large deshielding by 9 ppm was observed in the O-state. As discussed before, factors contributing to the chemical shift are primarily the strength of the H-bond in which the SB nitrogen is involved, as well as electric charge effects and torsion around the C15═N bond.
An altered torsion appears less likely since the C14 chemical shift, a sensitive indicator for a 15-syn/anti isomerization, remains unchanged between the dark state and the O-state. This observation agrees with the KR2 dark state and O-state x-ray structures in which the C14-C15-N-Cε dihedral angle does not significantly change (6REW, 135°; 6XYT, 132°) and which is also supported by our IR measurements. All three HOOP modes relax back to a ground state–like conformation with the formation of O (Fig. 5A).
The observed deshielding could, however, indicate a stretched N─H bond and, hence, a stronger O…H─N hydrogen bond between D116 and the SB nitrogen in the O-state. This seems plausible when considering that the SB becomes reprotonated with a proton being transferred from D116 during the M➔O transition. This process is faster in the presence of Na+. It would require a favorable orientation of the aspartate carboxyl group and the SB with respect to each other and could result in a shorter O…H distance, which then relaxes and becomes longer again in the dark state (3, 24). On the other hand, this observation seems counterintuitive in comparison to RSB model compounds (50) for which a correlation between H-bond strength and λmax was found and which would predict a blue shift with 15N deshielding. This is usually discussed in terms of stronger counterion interactions and fits generally the observations on many rhodopsins in their dark states (13). Here, in the KR2 O-state, a red shift occurs, and it has been argued, as mentioned above, that this could be caused by a shielded counterion–SB interaction due to the bound Na+. However, the situation appears more complicated as Na+ is coordinated by D116 and three other residues (23), which will alter the polarity/electric field within the SB vicinity in a complex manner. In addition, water molecules within the proximity of the SB will contribute to the polarity since the SB cavity contains four water molecules with one of them H-bonded to D116 (17).
In the O-state x-ray structure (PDB: 6XYT) (23), KR2 adopts a structure similar to the previously mentioned compact conformation with the SB cavity disappearing. These alterations will contribute to the H-bond character. It is also important to point out that substantial alterations in the H-bond geometry could be below the usual resolution of x-ray crystallography but will cause notable chemical shift changes (56). According to the dark state and O-state x-ray structures, the distance between OD2 of D116 and the SB nitrogen is, within the resolution limits, approximately the same (2.8 Å versus 2.9 Å). The observed chemical shift change, altered H-bond character, and red shift must, therefore, be caused by synergistic interplay of factors triggered by bound Na+. If one simply considers the bound Na+ as a charge that shields D116−, then it would be interesting to know which effect a simple charge removal at the proton acceptor site would have. In both BR and GPR, upfield shifts/shieldings have been observed when replacing the proton acceptor Asp with Asn (50, 51, 57–60), which is consistent with a weaker counterion interaction. Unexpectedly, the corresponding KR2 mutant D116N shows the opposite trend with a small but notable downfield shift/deshielding of the SB nitrogen (13), which emphasizes that data interpretation in terms of a direct relationship between color/counterion interaction and SB chemical shift is in many cases too simplistic. Dissecting all contributing factors will ideally require quantum chemical computational approaches combining NMR and optical data with 3D structures.
The observed 4-ppm shielding for C13 most likely reflects an increased partial delocalization of the positive charge of the SB to C13. Furthermore, the C20 methyl group is a sensitive indicator for steric contacts between retinal chromophore and residues in the binding pocket (32, 61). Although the observed chemical shift difference is small (1 ppm), it could be caused, for example, by altered contacts with residues such as W215 within the binding pocket. An overlay of the pentameric dark state structure (PDB: 6REW) with the trapped O-state (PDB: 6XYT) and our observed chemical shift changes is shown in Fig. 8C. The x-ray structures suggest small rearrangement of the aromatic residues within the binding pocket.
So far, the O-state has been discussed solely on the basis of the x-ray structure of the cryo-trapped KR2 pentamer. However, time-resolved x-ray structures derived from serial crystallography not only cover the K- and L-states but also have been extended via the M- into two O-states (25). After 1 ms, a Na+ ion is found close to the retinal binding pocket but at a different position compared to the pentameric state. An overlay of this state with the dark state structure (Fig. 8D) shows that the structural differences should correspond to larger chemical shift changes than actually observed. One reason could be that our sample with KR2 pentamers in proteoliposomes is closer to the pentameric x-ray structure than to the monomeric preparation used for serial crystallography, a difference that has also been addressed by the authors as relevant to the O-state (25). On the other hand, the distance between SB and D116 in the serial crystallography study becomes shorter compared to the dark state (2.7 Å versus 3.0 Å), which could fit with our chemical shift changes on the SB nitrogen.
Note that the flash-freezing procedure used here for trapping the O-state has been used before successfully for long-lived states in PR and ChR2 (36, 37), but its trapping yield is limited when the lifetimes becomes shorter. A potential improvement is offered by novel fast freeze-quenching methods as recently demonstrated for DNP-enhanced ssNMR (62).
FTIR studies of trapped KR2 intermediates (22) and time-resolved Raman spectroscopy data (20) recently reported a 13-cis conformation for the retinal in the O-state. This is in contrast to our data and the pentameric O-state structure of KR2 (PDB: 6REW) (23). Data interpretation in these studies relied on spectra from all-trans and 13-cis retinal bound to BR. The C12D + C14D rocking vibrations in the IR HOOP region exhibits a band that is similar to the 13-cis conformation in BR. It might also be possible that the intermediate trapped at 240 K resembles an N-like state. The time-resolved Raman data of the C─C vibration peaks also suggested the 13-cis conformation. However, the signature interpretation was based only on the BR signature pattern, which might be divergent from KR2, and the all-trans conformation of O-intermediate might not be detected as it is very similar to the dark state.
Understanding how KR2 selects and pumps sodium ions requires a deep understanding of its photocycle intermediates at a level that allows one to link 3D structures with spectroscopic and functional data. ssNMR offers here a valuable insight since the relevant readout parameters are site-resolved and report not only on structural but also on electronic properties. Here, the retinal and SB-counterion interaction in the dark state and in K-, L-, and O-states of KR2 were characterized using DNP-enhanced ssNMR complemented by cryo-UV/Vis, flash photolysis, and time-resolved IR measurements. Our findings are summarized in Fig. 8E. The light-induced formation of the K-state is associated with 13C chemical shift changes due to the retinal isomerization, which also reveal strong interactions between protein and chromophore. Such a “K-state signature” of chemical shift changes is specific to individual microbial rhodopsins and reflects the energy landscape of the early photocycle steps. The chemical shift perturbations slightly relax after formation of the subsequent L-state. However, the observed large twist at the end of the retinal polyene chain further increases to prepare KR2 for the proton transfer step, which is also in line with an increased strength of the counterion interaction. These observations do not depend on the presence of sodium, which supports the view that the ion passes the protein during a later stage of the photocycle during the O-state formation. In the O-state, the chromophore relaxes back to all-trans, but in particular, the retinal C13 and SB nitrogen chemical shifts differ from the dark state and highlight a complex interaction pattern in the SB vicinity in the presence of a bound sodium ion. An all-trans retinal conformation has been suggested to be energetically favorable for sodium binding and release (16) and has also been observed by x-ray crystallography (23). Our NMR data offer an opportunity to reconcile a wealth of spectroscopic data with 3D structures if combined with future quantum mechanics/molecular mechanics calculations, by which increasingly larger systems can be addressed (63). In addition, 23Na-MAS NMR could offer a possibility to target the bound sodium ion directly.
MATERIALS AND METHODS
Sample preparation
[U-15N]-KR2 was overexpressed and purified as described previously (15). 14,15-13C2 all-trans retinal and 10–18-13C9 all-trans retinal were synthesized as described by Leeder et al. (64). 12,13,20-13C3 all-trans retinal was prepared in a similar way as published previously (32). The labeled retinals were incorporated into the membrane before the solubilization step. For 15Nζ-Lys-KR2, the preparation protocol was similar to that of the uniformly labeled sample U-15N-KR2, but NH4Cl, instead of 15NH4Cl, was used in the medium together with other unlabeled amino acids. An amount of 116 mg of 15Nζ-lysine was added to 0.5 liters of culture at the point where the optical density at 600 nm (OD600) reached a level of 0.2 and before induction with isopropyl-β-d-thiogalactopyranoside at an OD600 of 0.6. The purified protein was reconstituted into liposomes containing 1,2-dimyristoyl-sn-glycero-3-phosphocholine [90 mole percent (mol %)]/1,2-dimyristoyl-sn-glycero-3-phosphate (10 mol %) liposomes. The lipid-to-protein ratio was 0.5 w/w. The proteoliposome samples were pelleted and washed with NMR buffer containing 25 mM tris, 5 mM MgCl2, and 30 mM NaCl for 13C10–18- and 13C14,15-KR2 samples and 100 mM NaCl for 13C12,13,20-KR2. These concentrations represent a good compromise between NMR buffer requirements and a sufficient O-state formation (25). For DNP-enhanced MAS ssNMR measurements, the samples were incubated for 20 hours at 4°C with a d8-glycerol:H2O:D2O (6:3:1) solution containing 20 mM AMUPol (65), 25 mM tris (pH 8.5), 5 mM MgCl2, and 30 or 100 mM NaCl. The supernatant was removed, and samples were packed into 3.2-mm sapphire rotors. They were uniformly distributed over the rotor wall by prespinning the rotors at room temperature.
Trapping of photocycle intermediates
A high-power LED from Mightex was used to generate KR2 photocycle intermediates. The K- and L-intermediates were generated by directly illuminating KR2 inside the DNP probe within the magnet at 110 K with blue light (470 nm, 3.3 W) for 60 min as described before (37). Thermal relaxation experiments were performed by sample illumination at 110 K for 60 min, warming up to the desired temperature, followed by 2-min relaxation and subsequent cooling again to 110 K for DNP detection. The L-intermediate was best observed by relaxation at 190 or 210 K for 2 min. The O-intermediate was generated by illuminating the sample with green light (525 nm) outside the magnet for 10 s, followed by fast freezing in liquid nitrogen (flash freezing) and subsequent transfer into the precooled DNP probe (110 K) within the NMR magnet.
DNP-enhanced ssNMR experiments
All experiments were performed on a Bruker 400 DNP system consisting of a 400-MHz WB Advance II NMR spectrometer with a 3.2-mm DNP MAS probe head connected via a waveguide to a 263-GHz gyrotron. The probe head was modified with a fiber bundle for delivering light from a high-power LED directly into the MAS rotor. A MAS rate of 8 kHz was applied in all experiments, except for the spectra in Figs. 2 (E and F), 3C, and 8A, for which 10 kHz was used. The 13C and 15N chemical shifts were referenced indirectly to 2,2-dimethyl-2-silapentane-5-sulphonic acid using the 13C signal of CO-alanine at 179.85 ppm. Experimental details for CP, DQF, PDSD, DQSQ correlation, double-quantum heteronuclear local field, TEDOR, and N(C)CX experiments are summarized in table S3.
IR and UV/Vis flash photolysis
A Nd:YAG laser (SpitLight 600, InnoLas Laser GmbH) pumping an optical parametric oscillator (PreciScan, GWU-Lasertechnik) was used to generate single nanosecond pulses with a central wavelength of λmax = 525 nm to excite the sample. The power was set to 1.7 mJ cm−2 at 525-nm excitation wavelength. The IR probe light was generated by a quantum cascade laser (MIRcat 1100-U2-5086, Daylight Solutions, USA), which is tunable in the range of 950 to 1150 cm−1. The shown transients in the range from 956 to 1040 cm−1 were recorded with a spectral resolution of 4 cm−1. For a better data correlation, we also measured UV/Vis transients at 420, 520, and 620 nm of the same sample, which was achieved using a mercury-xenon lamp (LC-08, Hamamatsu, Japan) and a set of two monochromators (Photon Technology International, USA). The KR2 wild-type sample was washed three times with a buffer containing 50 mM Hepes/tris, 100 mM NaCl, and 0.05% dodecyl-β-D-maltosid detergent, which was diluted in D2O and set to pD 8.1. A concentrated pellet was then evenly spread on the center of a CaF2 window and sandwiched with a Teflon spacer of 50-μm thickness and an additional CaF2 window.
Acknowledgments
Funding: This work was funded by DFG/SFB807 “Transport and communications across membranes.” DNP experiments were enabled through an equipment grant provided by DFG (GL307/4-1) and Cluster of Excellence Macromolecular Complexes Frankfurt. Author contributions: O.J. prepared samples, performed all NMR experiments, carried out data analysis, prepared figures, and wrote the paper. P.E. performed optical spectroscopy, analyzed data, and contributed to discussion and interpretation. M.A. performed IR spectroscopy, analyzed data, and contributed to discussion and interpretation. J.K. and C.N.K. contributed to sample preparation and data interpretation. J.M. contributed to retinal synthesis and data interpretation. A.J.L., L.J.B., and R.C.D.B. synthesized retinal. J.B.-B. contributed to NMR spectroscopy and data interpretation. C.B. contributed to sample preparation and discussions. J.W. designed and supervised optical and IR experiments and contributed to data interpretation, discussion, and paper writing. C.G. designed and supervised the study, carried out data interpretation, prepared figures, and wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/11/eabf4213/DC1
REFERENCES AND NOTES
- 1.Inoue K., Ono H., Abe-Yoshizumi R., Yoshizawa S., Ito H., Kogure K., Kandori H., A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Kandori H., Ion-pumping microbial rhodopsins. Front. Mol. Biosci. 2, 52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato Y., Inoue K., Kandori H., Kinetic analysis of H+-Na+ selectivity in a light-driven Na+-pumping rhodopsin. J. Phys. Chem. Lett. 6, 5111–5115 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Konno M., Kato Y., Kato H. E., Inoue K., Nureki O., Kandori H., Mutant of a light-driven sodium ion pump can transport cesium ions. J. Phys. Chem. Lett. 7, 51–55 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kato H. E., Inoue K., Abe-Yoshizumi R., Kato Y., Ono H., Konno M., Hososhima S., Ishizuka T., Hoque M. R., Kunitomo H., Ito J., Yoshizawa S., Yamashita K., Takemoto M., Nishizawa T., Taniguchi R., Kogure K., Maturana A. D., Iino Y., Yawo H., Ishitani R., Kandori H., Nureki O., Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature 521, 48–53 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Gushchin I., Shevchenko V., Polovinkin V., Kovalev K., Alekseev A., Round E., Borshchevskiy V., Balandin T., Popov A., Gensch T., Fahlke C., Bamann C., Willbold D., Büldt G., Bamberg E., Gordeliy V., Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 22, 390–395 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Gushchin I., Shevchenko V., Polovinkin V., Borshchevskiy V., Buslaev P., Bamberg E., Gordeliy V., Structure of the light-driven sodium pump KR2 and its implications for optogenetics. FEBS J. 283, 1232–1238 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Oesterhelt D., Stoeckenius W., Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233, 149–152 (1971). [DOI] [PubMed] [Google Scholar]
- 9.Béjà O., Aravind L., Koonin E. V., Suzuki M. T., Hadd A., Nguyen L. P., Jovanovich S. B., Gates C. M., Feldman R. A., Spudich J. L., Spudich E. N., De Long E. F., Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Ono H., Inoue K., Abe-Yoshizumi R., Kandori H., FTIR spectroscopy of a light-driven compatible sodium ion-proton pumping rhodopsin at 77 K. J. Phys. Chem. B 118, 4784–4792 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Tomida S., Ito S., Inoue K., Kandori H., Hydrogen-bonding network at the cytoplasmic region of a light-driven sodium pump rhodopsin KR2. Biochim. Biophys. Acta Bioenerg. 1859, 684–691 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Hontani Y., Inoue K., Kloz M., Kato Y., Kandori H., Kennis J. T. M., The photochemistry of sodium ion pump rhodopsin observed by watermarked femto- to submillisecond stimulated Raman spectroscopy. Phys. Chem. Chem. Phys. 18, 24729–24736 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Shigeta A., Ito S., Inoue K., Okitsu T., Wada A., Kandori H., Kawamura I., Solid-state nuclear magnetic resonance structural study of the retinal-binding pocket in sodium ion pump rhodopsin. Biochemistry 56, 543–550 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Shigeta A., Ito S., Kaneko R., Tomida S., Inoue K., Kandori H., Kawamura I., Long-distance perturbation on Schiff base-counterion interactions by His30 and the extracellular Na+-binding site in Krokinobacter rhodopsin 2. Phys. Chem. Chem. Phys. 20, 8450–8455 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Kaur J., Kriebel C. N., Eberhardt P., Jakdetchai O., Leeder A. J., Weber I., Brown L. J., Brown R. C. D., Becker-Baldus J., Bamann C., Wachtveitl J., Glaubitz C., Solid-state NMR analysis of the sodium pump Krokinobacter rhodopsin 2 and its H30A mutant. J. Struct. Biol. 206, 55–65 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Suomivuori C.-M., Gamiz-Hernandez A. P., Sundholm D., Kaila V. R. I., Energetics and dynamics of a light-driven sodium-pumping rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 114, 7043–7048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovalev K., Polovinkin V., Gushchin I., Alekseev A., Shevchenko V., Borshchevskiy V., Astashkin R., Balandin T., Bratanov D., Vaganova S., Popov A., Chupin V., Büldt G., Bamberg E., Gordeliy V., Structure and mechanisms of sodium-pumping KR2 rhodopsin. Sci. Adv. 5, eaav2671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asido M., Eberhardt P., Kriebel C. N., Braun M., Glaubitz C., Wachtveitl J., Time-resolved IR spectroscopy reveals mechanistic details of ion transport in the sodium pump Krokinobacter eikastus rhodopsin 2. Phys. Chem. Chem. Phys. 21, 4461–4471 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Tahara S., Takeuchi S., Abe-Yoshizumi R., Inoue K., Ohtani H., Kandori H., Tahara T., Ultrafast photoreaction dynamics of a light-driven sodium-ion-pumping retinal protein from Krokinobacter eikastus revealed by femtosecond time-resolved absorption spectroscopy. J. Phys. Chem. Lett. 6, 4481–4486 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Nishimura N., Mizuno M., Kandori H., Mizutani Y., Distortion and a strong hydrogen bond in the retinal chromophore enable sodium-ion transport by the sodium-ion pump KR2. J. Phys. Chem. B 123, 3430–3440 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Bogachev A. V., Bertsova Y. V., Verkhovskaya M. L., Mamedov M. D., Skulachev V. P., Real-time kinetics of electrogenic Na+ transport by rhodopsin from the marine flavobacterium Dokdonia sp. PRO95. Sci. Rep. 6, 21397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H.-F., Inoue K., Ono H., Abe-Yoshizumi R., Wada A., Kandori H., Time-resolved FTIR study of light-driven sodium pump rhodopsins. Phys. Chem. Chem. Phys. 20, 17694–17704 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Kovalev K., Astashkin R., Gushchin I., Orekhov P., Volkov D., Zinovev E., Marin E., Rulev M., Alekseev A., Royant A., Carpentier P., Vaganova S., Zabelskii D., Baeken C., Sergeev I., Balandin T., Bourenkov G., Carpena X., Boer R., Maliar N., Borshchevskiy V., Büldt G., Bamberg E., Gordeliy V., Molecular mechanism of light-driven sodium pumping. Nat. Commun. 11, 2137 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balashov S. P., Imasheva E. S., Dioumaev A. K., Wang J. M., Jung K. H., Lanyi J. K., Light-driven Na+ pump from Gillisia limnaea: A high-affinity Na+ binding site is formed transiently in the photocycle. Biochemistry 53, 7549–7561 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skopintsev P., Ehrenberg D., Weinert T., James D., Kar R. K., Johnson P. J. M., Ozerov D., Furrer A., Martiel I., Dworkowski F., Nass K., Knopp G., Cirelli C., Arrell C., Gashi D., Mous S., Wranik M., Gruhl T., Kekilli D., Brünle S., Deupi X., Schertler G. F. X., Benoit R. M., Panneels V., Nogly P., Schapiro I., Milne C., Heberle J., Standfuss J., Femtosecond-to-millisecond structural changes in a light-driven sodium pump. Nature 583, 314–318 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Mandala V. S., Williams J. K., Hong M., Structure and dynamics of membrane proteins from solid-state NMR. Annu. Rev. Biophys. 47, 201–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lansing J. C., Hohwy M., Jaroniec C. P., Creemers A. F., Lugtenburg J., Herzfeld J., Griffin R. G., Chromophore distortions in the bacteriorhodopsin photocycle: Evolution of the H–C14–C15–H dihedral angle measured by solid-state NMR. Biochemistry 41, 431–438 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Bajaj V. S., Mak-Jurkauskas M. L., Belenky M., Herzfeld J., Griffin R. G., Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization–enhanced solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 106, 9244–9249 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimata N., Pope A., Eilers M., Opefi C. A., Ziliox M., Hirshfeld A., Zaitseva E., Vogel R., Sheves M., Reeves P. J., Smith S. O., Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation. Nat. Commun. 7, 12683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Good D. B., Wang S., Ward M. E., Struppe J., Brown L. S., Lewandowski J. R., Ladizhansky V., Conformational dynamics of a seven transmembrane helical protein Anabaena sensory rhodopsin probed by solid-state NMR. J. Am. Chem. Soc. 136, 2833–2842 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Ahuja S., Eilers M., Hirshfeld A., Yan E. C., Ziliox M., Sakmar T. P., Sheves M., Smith S. O., 6-s-cis Conformation and polar binding pocket of the retinal chromophore in the photoactivated state of rhodopsin. J. Am. Chem. Soc. 131, 15160–15169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao J., Aladin V., Jin X., Leeder A. J., Brown L. J., Brown R. C. D., He X., Corzilius B., Glaubitz C., Exploring protein structures by DNP-enhanced methyl solid-state NMR spectroscopy. J. Am. Chem. Soc. 141, 19888–19901 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Ni Q. Z., Daviso E., Can T. V., Markhasin E., Jawla S. K., Swager T. M., Temkin R. J., Herzfeld J., Griffin R. G., High frequency dynamic nuclear polarization. Acc. Chem. Res. 46, 1933–1941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mak-Jurkauskas M. L., Bajaj V. S., Hornstein M. K., Belenky M., Griffin R. G., Herzfeld J., Energy transformations early in the bacteriorhodopsin photocycle revealed by DNP-enhanced solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 105, 883–888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni Q. Z., Can T. V., Daviso E., Belenky M., Griffin R. G., Herzfeld J., Primary transfer step in the light-driven ion pump bacteriorhodopsin: An irreversible U-turn revealed by dynamic nuclear polarization-enhanced magic angle spinning NMR. J. Am. Chem. Soc. 140, 4085–4091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker-Baldus J., Bamann C., Saxena K., Gustmann H., Brown L. J., Brown R. C. D., Reiter C., Bamberg E., Wachtveitl J., Schwalbe H., Glaubitz C., Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 112, 9896–9901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehler M., Eckert C. E., Leeder A. J., Kaur J., Fischer T., Kubatova N., Brown L. J., Brown R. C. D., Becker-Baldus J., Wachtveitl J., Glaubitz C., Chromophore distortions in photointermediates of proteorhodopsin visualized by dynamic nuclear polarization-enhanced solid-state NMR. J. Am. Chem. Soc. 139, 16143–16153 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Maciejko J., Kaur J., Becker-Baldus J., Glaubitz C., Photocycle-dependent conformational changes in the proteorhodopsin cross-protomer Asp-His-Trp triad revealed by DNP-enhanced MAS-NMR. Proc. Natl. Acad. Sci. U.S.A. 116, 8342–8349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker-Baldus J., Glaubitz C., Cryo-trapped intermediates of retinal proteins studied by DNP-enhanced MAS NMR spectroscopy. Emagres 7, 79–91 (2018). [Google Scholar]
- 40.Hohwy M., Jakobsen H. J., Edén M., Levitt M. H., Nielsen N. C., Broadband dipolar recoupling in the nuclear magnetic resonance of rotating solids: A compensated C7 pulse sequence. J. Chem. Phys. 108, 2686–2694 (1998). [Google Scholar]
- 41.Ni Q. Z., Markhasin E., Can T. V., Corzilius B., Tan K. O., Barnes A. B., Daviso E., Su Y., Herzfeld J., Griffin R. G., Peptide and protein dynamics and low-temperature/DNP magic angle spinning NMR. J. Phys. Chem. B 121, 4997–5006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong M., Solid-state dipolar INADEQUATE NMR spectroscopy with a large double-quantum spectral width. J. Magn. Reson. 136, 86–91 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Szeverenyi N. M., Sullivan M. J., Maciel G. E., Observation of spin exchange by two-dimensional fourier-transform 13C cross polarization-magic-angle spinning. J. Magn. Reson. 47, 462–475 (1982). [Google Scholar]
- 44.Inoue Y., Tokito Y., Tomonoh S., Chujo R., Carbon-13 chemical-shifts of retinal isomers and their Schiff-bases as models of visual chromophores. B. Chem. Soc. Jpn. 52, 265–266 (1979). [Google Scholar]
- 45.Wang K.-W., Wang S.-W., Du Q.-Z., Complete NMR assignment of retinal and its related compounds. Magn. Reson. Chem. 51, 435–438 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Harbison G. S., Smith S. O., Pardoen J. A., Winkel C., Lugtenburg J., Herzfeld J., Mathies R., Griffin R. G., Dark-adapted bacteriorhodopsin contains 13-cis, 15-syn and all-trans, 15-anti retinal Schiff bases. Proc. Natl. Acad. Sci. U.S.A. 81, 1706–1709 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Concistre M., Johannessen O. G., McLean N., Bovee-Geurts P. H., Brown R. C., Degrip W. J., Levitt M. H., A large geometric distortion in the first photointermediate of rhodopsin, determined by double-quantum solid-state NMR. J. Biomol. NMR 53, 247–256 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Jaroniec C. P., Filip C., Griffin R. G., 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly 13C, 15N-labeled solids. J. Am. Chem. Soc. 124, 10728–10742 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Bajaj V. S., Mak-Jurkauskas M. L., Belenky M., Herzfeld J., Griffin R. G., DNP enhanced frequency-selective TEDOR experiments in bacteriorhodopsin. J. Magn. Reson. 202, 9–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu J., Griffin R. G., Herzfeld J., Synergy in the spectral tuning of retinal pigments: Complete accounting of the opsin shift in bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 91, 8880–8884 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J. G., Griffin R. G., Herzfeld J., Interactions between the protonated Schiff base and its counterion in the photointermediates of bacteriorhodopsin. J. Am. Chem. Soc. 119, 9495–9498 (1997). [Google Scholar]
- 52.Pauli J., Baldus M., van Rossum B., de Groot H., Oschkinat H., Backbone and side-chain 13C and 15N signal assignments of the α-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 tesla. Chembiochem 2, 272–281 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Maeda A., Balashov S. P., Lugtenburg J., Verhoeven M. A., Herzfeld J., Belenky M., Gennis R. B., Tomson F. L., Ebrey T. G., Interaction of internal water molecules with the schiff base in the L intermediate of the bacteriorhodopsin photocycle. Biochemistry 41, 3803–3809 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Mizuide N., Shibata M., Friedman N., Sheves M., Belenky M., Herzfeld J., Kandori H., Structural changes in bacteriorhodopsin following retinal photoisomerization from the 13-cis form. Biochemistry 45, 10674–10681 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Furutani Y., Sudo Y., Wada A., Ito M., Shimono K., Kamo N., Kandori H., Assignment of the hydrogen-out-of-plane and -in-plane vibrations of the retinal chromophore in the K intermediate of pharaonis phoborhodopsin. Biochemistry 45, 11836–11843 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Sharif S., Denisov G. S., Toney M. D., Limbach H.-H., NMR studies of coupled low- and high-barrier hydrogen bonds in pyridoxal-5′-phosphate model systems in polar solution. J. Am. Chem. Soc. 129, 6313–6327 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Harbison G. S., Herzfeld J., Griffin R. G., Solid-state nitrogen-15 nuclear magnetic resonance study of the Schiff base in bacteriorhodopsin. Biochemistry 22, 1–5 (1983). [DOI] [PubMed] [Google Scholar]
- 58.de Groot H. J., Harbison G. S., Herzfeld J., Griffin R. G., Nuclear magnetic resonance study of the Schiff base in bacteriorhodopsin: Counterion effects on the 15N shift anisotropy. Biochemistry 28, 3346–3353 (1989). [DOI] [PubMed] [Google Scholar]
- 59.Hatcher M. E., Hu J. G., Belenky M., Verdegem P., Lugtenburg J., Griffin R. G., Herzfeld J., Control of the pump cycle in bacteriorhodopsin: Mechanisms elucidated by solid-state NMR of the D85N mutant. Biophys. J. 82, 1017–1029 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfleger N., Wörner A. C., Yang J., Shastri S., Hellmich U. A., Aslimovska L., Maier M. S. M., Glaubitz C., Solid-state NMR and functional studies on proteorhodopsin. Biochim. Biophys. Acta 1787, 697–705 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Yomoda H., Makino Y., Tomonaga Y., Hidaka T., Kawamura I., Okitsu T., Wada A., Sudo Y., Naito A., Color-discriminating retinal configurations of sensory rhodopsin I by photo-irradiation solid-state NMR spectroscopy. Angew. Chem. Int. Ed. 53, 6960–6964 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Jeon J., Yau W.-M., Tycko R., Millisecond time-resolved solid-state NMR reveals a two-stage molecular mechanism for formation of complexes between calmodulin and a target peptide from myosin light chain kinase. J. Am. Chem. Soc. 142, 21220–21232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He X., Wang B., Merz K. M. Jr., Protein NMR chemical shift calculations based on the automated fragmentation QM/MM approach. J. Phys. Chem. B 113, 10380–10388 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Leeder A. J., Brown L. J., Becker-Baldus J., Mehler M., Glaubitz C., Brown R. C. D., Synthesis of isotopically labeled all-trans retinals for DNP-enhanced solid-state NMR studies of retinylidene proteins. J. Labelled Comp. Radiopharm. 61, 922–933 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Sauvée C., Rosay M., Casano G., Aussenac F., Weber R. T., Ouari O., Tordo P., Highly efficient, water-soluble polarizing agents for dynamic nuclear polarization at high frequency. Angew. Chem. Int. Ed. 52, 10858–10861 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Mao J., Do N.-N., Scholz F., Reggie L., Mehler M., Lakatos A., Ong Y.-S., Ullrich S. J., Brown L. J., Brown R. C. D., Becker-Baldus J., Wachtveitl J., Glaubitz C., Structural basis of the green-blue color switching in proteorhodopsin as determined by NMR spectroscopy. J. Am. Chem. Soc. 136, 17578–17590 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Bak M., Rasmussen J. T., Nielsen N. C., SIMPSON: A general simulation program for solid-state NMR spectroscopy. J. Magn. Reson. 147, 296–330 (2000). [DOI] [PubMed] [Google Scholar]
- 68.F. A. Carey, R. J. Sundberg, Advanced Organic Chemistry (Springer, 5th ed, 2007), p. 2. [Google Scholar]
- 69.Henrich E., Sörmann J., Eberhardt P., Peetz O., Mezhyrova J., Morgner N., Fendler K., Dötsch V., Wachtveitl J., Bernhard F., Bamann C., From gene to function: Cell-free electrophysiological and optical analysis of ion pumps in nanodiscs. Biophys. J. 113, 1331–1341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinogradov E., Madhu P. K., Vega S., High-resolution proton solid-state NMR spectroscopy by phase-modulated Lee-Goldburg experiment. Chem. Phys. Lett. 314, 443–450 (1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/11/eabf4213/DC1


