Pds5 works with Rec8 to regulate chromosome axis length and crossover frequency in a dosage-dependent manner.
Abstract
Meiotic chromosomes have a loop/axis architecture, with axis length determining crossover frequency. Meiosis-specific Pds5 depletion mutants have shorter chromosome axes and lower homologous chromosome pairing and recombination frequency. However, it is poorly understood how Pds5 coordinately regulates these processes. In this study, we show that only ~20% of wild-type level of Pds5 is required for homolog pairing and that higher levels of Pds5 dosage-dependently regulate axis length and crossover frequency. Moderate changes in Pds5 protein levels do not explicitly impair the basic recombination process. Further investigations show that Pds5 does not regulate chromosome axes by altering Rec8 abundance. Conversely, Rec8 regulates chromosome axis length by modulating Pds5. These findings highlight the important role of Pds5 in regulating meiosis and its relationship with Rec8 to regulate chromosome axis length and crossover frequency with implications for evolutionary adaptation.
INTRODUCTION
Diploid progenitor cells undergo meiosis, resulting in the haploid gametes needed for sexual reproduction. A central feature of meiosis is the repair of programmed DNA double-strand breaks (DSBs) with homologous chromosomes (homologs) (1, 2). Only a small fraction of DSBs are selected as crossovers (COs), which results in the reciprocal exchange of genetic information between homologs. Most DSBs develop into non-COs, which results in the unidirectional transfer of genetic information from the intact homolog to the broken one (1). COs promote the reshuffling of genetic diversity, which is helpful for evolutionary adaptation (3–5). COs also work with sister chromatid cohesion to establish physical connections between homologs, allowing proper segregation (1, 2).
CO patterns are tightly regulated with obligatory CO and CO interference. These phenomena establish the minimum and maximum CO numbers on chromosomes, respectively (6–8). The obligatory CO is required for faithful chromosome segregation. The third feature, CO homeostasis, maintains a relatively constant CO number in the face of altered DSB frequency (7–9). These features have been proposed to arise from a single CO patterning process, where the obligatory CO is ensured and CO homeostasis depends on CO interference (10).
COs are tightly regulated by the architecture of the chromosome [e.g., (1, 2, 11–13)]. Meiotic chromosomes are proposed to be organized as linear arrays of loops, the bases of which form chromosome axes (14). Current evidence indicates that meiotic chromosome axis length determines CO frequency (15–17). First, strong positive correlations between axis lengths and the number of both DSB and COs have been observed in several organisms under various conditions (15, 17–19). Second, axis length determines DSB and CO frequencies, but DSB and CO frequencies do not determine axis length for the following reasons: (i) Alterations in axis length are always accompanied by coordinate changes in DSB and CO levels [e.g., (6, 20)]. (ii) Axis length can be maintained as is, regardless of changes in DSB level or the absence of DSBs [e.g., (6, 7, 21)]. Moreover, within individual meiotic nuclei, the axis lengths of different chromosomes tend to vary coordinately, which results in per-nucleus CO covariation (17). Therefore, understanding how meiotic chromosome organization is regulated is of great interest.
Studies suggest that meiotic cohesin is a prerequisite for proper chromosome axis formation (22, 23). Moreover, decreased cohesin levels are associated with short meiotic chromosome axes [e.g., (20, 24, 25)]. These studies suggest that cohesin regulates chromosome axis length by modulating loop size (22, 26). Pds5, an important cohesin regulator, localizes on chromosome axes during meiotic prophase I (22, 27–29). Meiosis-specific depletion of Pds5 during meiosis results in markedly short chromosome axes in budding yeast, fission yeast, and male mouse (22, 27, 28). Currently, the prevailing view is that Pds5 regulates the chromosome axis by modulating cohesin (27, 30). However, whether and how Pds5 works with cohesin to modulate the chromosome axis is poorly understood. Moreover, meiotic depletion of Pds5 results in decreased homolog pairing and recombination frequencies (22, 23, 27, 28). We are also interested to investigate how Pds5 regulates these processes.
Here, we show that ~20% of wild-type (WT) level of Pds5 can fulfill homolog pairing. However, high Pds5 levels are required for normal chromosome axis length. Both Pds5 and Rec8 regulate chromosome axis length, depending on their dosage. Our study demonstrates that Pds5 does not regulate axis length by altering Rec8 abundance. Rec8 regulates axis length mainly by altering Pds5 abundance. Changes in Pds5 protein levels result in altered chromosome axis length and, consequently, DSB and CO frequencies in the same direction without impairing the basic recombination process per se. This study clarifies the role of Pds5 in regulating homolog pairing, axis length, and CO frequency; addresses the complicated interplay between Pds5 and Rec8 in regulating chromosome axis length; and suggests the possible role of Pds5 in evolutionary adaption.
RESULTS
Homologous chromosome pairing requires low Pds5 levels
The role of Pds5 in budding yeast meiosis was investigated using two types of Pds5 depletion mutants, pCLB2-PDS5 and PDS5-AID (23, 27). For pCLB2-PDS5, the native promoter of PDS5 was replaced by the B-type cyclin CLB2 promoter, which is expressed in mitosis but not in meiosis (31). For PDS5-AID, an auxin-induced degron (AID) was fused to the C terminus of Pds5. In the presence of auxin [indole-3-acetic acid (IAA)] and the F-box TIR1 protein, this fusion protein is recognized by an auxin-dependent E3 ubiquitin ligase complex, SCFTIR1, and degraded by proteasomes (31). These studies have shown that Pds5 depletion mutants have short chromosome axes and defects in homolog pairing (Fig. 1 and fig. S1) (23, 27).
Fig. 1. Pds5 removal disrupts homolog pairing during meiosis.

(A) Representative images of Rec8 staining. Scale bar, 4 μm. (B to E) Quantification of the numbers of Rec8 lines (B), the total length of Rec8 lines per cell and corrected length with unpaired homologs (numbers in parentheses) (C), the average length of each Rec8 line per cell (D), and Pds5 fluorescence intensity (E) in WT and mutants. The numbers of nuclei measured from left to right, n = 105, 103, 104, 102, and 101, separately (B to E). pCLB2-PDS5-AID with 2 or 10 mM IAA compared with 0 mM IAA, P < 0.0001 (B, C, and E), P > 0.05 (D), t test. (F and G) Illustration of the tetO/TetR-GFP (F) and lacO/LacI-GFP (G) assays. (H) Representative images of the assay for homolog pairing and sister cohesion in the ndt80Δ background. A single GFP spot and two GFP spots indicate homologs are paired and unpaired, respectively. Sister cohesion defects are reflected by three or four GFP spots. (I and J) Quantification of homolog pairing and sister cohesion detected by tetO/TetR-GFP (H; n = 200, 203, 202, 205, and 200, separately) or LacO/LacI-GFP assay (I; n = 204, 200, 205, 201, and 201, separately). (K to N) Quantification of the number of Rec8 lines per cell (K), the corrected (in parentheses) and uncorrected total length of Rec8 lines per cell (L), the average length of each Rec8 line in individual cells (M), and the axis length of chromosome II (N), in cells with paired (one GFP focus) and unpaired (two GFP foci) chromosome II. Sample size, n = 58 (one GFP focus) and 68 (two GFP foci) (K to N). Error bar, means ± SD (B to E and K to N). P < 0.0001, t test (K to N).
There were ∼23 Zip1/Rec8 lines (dots in some cases) per pachytene nucleus in the pCLB2-PDS5 mutant, which was between ~14.9 Rec8 lines per WT pachytene nucleus when all homologs were paired and ~29.8 (14.9 × 2 = 29.8) Rec8 lines expected when all homologs were unpaired [Fig. 1, A and B, and fig. S1A; Materials and Methods; (23)]. This indicates that approximately half of the homologs are unpaired in the pCLB2-PDS5 mutant. To confirm the pairing defects, we integrated the TetO or LacO array into a specific chromosome position on both homologs. When TetR–green fluorescent protein (GFP) or LacI-GFP was expressed, they would bind to the TetO or LacO array, respectively, forming a bright spot (Fig. 1, F to H). One and two GFP spots were observed when the homologs were paired and unpaired, respectively, and three or four GFP spots indicated sister cohesion defects (Fig. 1H). In WT, ~95% of pachytene nuclei have only one GFP spot. However, the frequency of one GFP spot nuclei markedly decreased to ~40% in the pCLB2-PDS5 mutant. These results confirmed the prediction that only half of the homologs in this mutant are paired (Fig. 1, I and J) (23, 27).
A weak Pds5 signal was detected by Western blot and immunostaining assays in pCLB2-PDS5 during meiosis (Fig. 1E and figs. S1 and S2) (23). This raised the possibility that the low levels of homolog pairing are due to residual Pds5 proteins. To explore this possibility, we used a pCLB2-PDS5-AID strain to remove any residual Pds5 during meiosis (Fig. 1E; Materials and Methods). When 2 mM IAA was added, the number of Rec8 lines increased significantly, from 23.7 to 27.1 (P < 0.001, t test). The homolog pairing level also decreased significantly, from 39.5 to 12.5%, by the spot assay (P < 0.0001, t test; Fig. 1, A, B, I, and J). When 10 mM IAA was used, Pds5 was barely detected in spread nuclei (fig. S1C), the level of homolog pairing decreased to 9.0% (Fig. 1, A, B, I, and J), and the number of Rec8 lines increased to 28.4, which is close to the level of all homologs unpaired (~29.8, above). In addition, no axis breaks or gaps were observed in these mutants. This conclusion is supported by the following evidence from pCLB2-PDS5 mutant: (i) Chromosome axes can be continuously marked by either Rec8 or Red1 (fig. S3) (27); (ii) chromosome axis lengths measured by Rec8 and Red1, respectively, and the combination of both markers gave comparable results (fig. S3); and (iii) each Rec8 line has one Sgo1 focus that labels kinetochores, indicating that all Rec8 lines have centromeres (fig. S1D). These results indicate that the complete removal of Pds5 impairs homolog pairing. In the pCLB2-PDS5 mutant, the synaptonemal complexes (SCs) form between sister chromatids (27). It is likely the intersister SCs lead to the homolog pairing/synapsis defects. We also noted that Pds5 removal only slightly decreased sister cohesion (Fig. 1, H to J; nuclei with three or four GFP spots). However, we could not exclude the possibility that a globally looser association of sisters was not detected by this assay (23).
These results indicate that Pds5 dosage regulates homolog pairing levels. To explore the levels of Pds5 required for full homolog pairing, we constructed pCLB2-PDS5/pCUP1-PDS5, where Pds5 can be induced by copper. Even without copper addition, the level of homolog pairing in this mutant was higher than in pCLB2-PDS5/pCLB2-PDS5 because of the leakage expression of the CUP1 promoter (Fig. 2 and fig. S2). Adding low concentrations of copper (0.02 μM) decreased the number of Rec8 lines to nearly WT levels (16.0 versus 14.9 Rec8 lines; Fig. 2), indicating homolog pairings close to WT levels. On the basis of the results of Western blot and immunostaining analysis, we estimate that ~20% of WT level of Pds5 is required for complete homolog pairing (Fig. 2 and fig. S2, A and B). When more Pds5 is expressed, chromosome axis length, meiotic nuclear division frequency, and spore viability all gradually increased (Fig. 2 and fig. S2).
Fig. 2. Higher Pds5 levels are required for proper axis length and CO level than for homolog pairing.

(A) Representative images of Rec8 and Zip3 staining. Scale bar, 2 μm. (B to F) Quantification of the number of Rec8 lines per cell (B), the corrected (in parentheses) and uncorrected total length of Rec8 lines per cell (C), the total number of Zip3 foci per cell (D), the average length of each Rec8 line in individual cells (E), and the average number of Zip3 foci per Rec8 line from individual cells (F). Sample size, n = 103, 103, 103, 103, 101, 145, and 105, separately. Error bar, means ± SD (B to F).
Unpaired chromosomes have short chromosome axes
Meiotic chromosome axis length can be reliably measured by immunostaining Rec8 as validated by costaining it with Red1 (another axis component) or Zip1 (an SC central element) in WT and mutants studied here (Materials and Methods; fig. S3). In pCLB2-PDS5, the average chromosome axis length was 22.6 μm per pachytene nucleus, which is approximately half of the WT axis length (42.7 μm) (Fig. 1, A and C). When residual Pds5 was removed by IAA-induced degradation, the average axial length increased to 26.9 μm (Fig. 1, A and C). However, the average length per Rec8 line did not increase (0.98 μm versus 0.95 μm) (Fig. 1D). Under an epifluorescence microscope, when homologs are paired, the two axes of one homolog pair are observed and thus measured as a single Rec8 line. However, when they are unpaired, the two axes of one homolog pair are observed and thus measured as two individual Rec8 lines. To eliminate the influence of unpaired homolog axes (measured twice) on the real axis length, we corrected the measured axis length per cell as the product of the average length per Rec8 line and the average number of Rec8 lines per WT nucleus, resulting in very slightly decreased (at least not increased) ‶corrected″ axis lengths in strains with more unpaired homologs (Fig. 1C, numbers in parenthesis). This suggests that increases in the ‶measured″ axis length per nucleus (from 22.6 to 26.9 μm) come from increased number of Rec8 lines counted in nuclei that might be caused by intersister SCs (27, 32).
To confirm this prediction, we measured axis lengths of paired (a single TetO/TetR-GFP spot) and unpaired (two GFP spots) chromosome II in pCLB2-PDS5, where roughly half of the homologs are paired (Fig. 1). The results were as expected. (i) Nuclei with paired chromosome II tended to have fewer Rec8 lines and higher homolog pairing levels than nuclei with unpaired chromosome II (19.3 versus 25.4 Rec8 lines; Fig. 1K). (ii) Nuclei with paired chromosome II have significantly shorter axes than nuclei with unpaired chromosome II (20.7 μm versus 23.7 μm, P < 0.0001; Fig. 1L). However, when axis length was corrected with homolog pairing levels, nuclei with paired chromosome II have longer axes than nuclei with unpaired chromosome II (16.0 μm versus 13.9 μm, P < 0.0001; Fig. 1L). (iii) The average length of each Rec8 line is significantly shorter in nuclei with unpaired chromosomes than paired chromosomes (0.94 μm versus 1.08 μm, P < 0.0001; Fig. 1M; Materials and Methods). (iv) The axis length of unpaired chromosome II is shorter and constant with a very small coefficient of variation (CV) among nuclei (CV = 0.088, mean = 0.99, and SD = 0.087; Fig. 1N). However, the axis length of paired chromosome II is longer and more variable, with a large CV among nuclei (CV = 0.22, mean = 1.21, and SD = 0.26; Fig. 1N). This indicates that the axis length of paired homologs is likely regulated by Pds5 levels and may be also related to the pairing status. Consistently, the axis of paired chromosome II is longer in nuclei with more homologs paired (chromosome II axis length versus the number of Rec8 lines, r = −0.53). Therefore, the axis of unpaired homologs is shorter and of a more constant length in pCLB2-PDS5.
A high Pds5 level is required to maintain proper chromosome axis length
In pCLB2-PDS5/pCUP1-PDS5, adding copper induced up to ~60% of WT levels of Pds5 protein (fig. S2, A and B). Consequently, the chromosome axis length (marked by Rec8 line) gradually increased to ~85% of WT level, while the frequency of meiotic nucleus division, the sporulation efficiency, and spore viability gradually increased, almost reaching WT levels (Fig. 2C and fig. S2). To produce a higher level of Pds5 protein, we made pCUP1-PDS5/pCUP1-PDS5 and PDS5/pCUP1-PDS5 strains. More copper induced a longer chromosome axis in these strains (Fig. 3, A and B). In particular, in PDS5/pCUP1-PDS5, chromosome axis length could reach ~93% of WT level (Fig. 3, A and B). Further investigations demonstrated that Pds5 overexpression (PDS5::pCUP1-PDS5/PDS5::pCUP1-PDS5) could result in axes that exceeded WT (Fig. 3 and fig. S4). These results support that chromosome axis length is regulated by Pds5 in a dosage-dependent manner and that high levels of Pds5 are required to maintain WT axis length.
Fig. 3. Pds5 dosage-dependently regulates axis length.
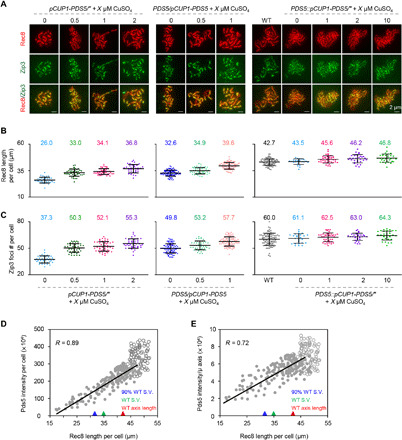
(A) Representative images of Rec8 and Zip3 staining. Scale bar, 2 μm. (B and C) Quantification of the total length of Rec8 lines per cell (B) and the total number of Zip3 foci per cell (C). Sample size, n = 39, 44, 49, 41, 97, 47, 84, 105, 25, 41, 31, and 32, separately. Error bar, means ± SD (B and C). (D and E) Nuclei with longer axis have more Pds5 per nucleus (D) and per micrometer of the axis (E) as shown by Pearson’s correlation analysis. The trend lines and correlation coefficient were calculated on the basis of presented data (n = 268), except for data points from the strain with overexpressed Pds5 (circles). Triangles indicate the axis length and Pds5 intensity per nucleus (D) and per micrometer of the axis (E) in strains with 90% of WT level of spore viability (S.V.) (blue triangle), WT level of spore viability (green triangle), and WT axis length (red triangle).
The relationship between Pds5 abundance (Pds5 fluorescence intensity) and chromosome axis length was analyzed using Pearson’s correlation analysis (Fig. 3 and fig. S5). As expected, nuclei with higher Pds5 fluorescence intensity (more Pds5) had longer chromosome axes (Fig. 3D). Nuclei with longer axes also have higher Pds5 density (Pds5 abundance per unit length of axis) (Fig. 3E). In WT, chromosome axis length varies among different nuclei (17). Pds5 fluorescence intensity was positively correlated with chromosome axis length (r = 0.57), although Pds5 density was constant (fig. S5, A and B).
However, when chromosome axis length approached WT level, further increases of Pds5 protein level were accompanied by subtle increases of chromosome axis length (as indicated by a weak correlation, r = 0.3; fig. S5C), while Pds5 density was relatively constant among nuclei with different axis lengths (fig. S5D). This suggests that Pds5 dosage reaches a plateau when regulating chromosome axis length, which could be due to limited amounts of Rec8 cohesin.
It is widely accepted that meiotic chromosomes are organized as a loop/axis architecture, where loop size is negatively correlated with axis length (14–16). Loop size can be estimated by chromosome width (20). The pCLB2-PDS5 mutant has a shorter axis and a larger chromosome width compared to WT (~2.23 μm versus ~1.82 μm at pachytene; fig. S6). This suggests that Pds5 regulates chromosome axis length by modulating loop size.
Pds5 does not regulate axis length by altering Rec8 abundance
Cohesin likely plays a critical role in regulating chromosome loop/axis organization (22). As an important regulator of cohesin, Pds5 likely regulates axis length by modulating cohesin (27, 30). However, results from the Western blot and immunostaining analyses of spread chromosomes showed that Rec8 abundance only slightly decreased in the pCLB2-PDS5 mutant (Fig. 4, A and B, and fig. S7, A, B, and E) (23, 27). Consistently, the short chromosome axis combined with nearly WT level of Rec8 protein resulted in a higher Rec8 density (amount of Rec8 per micrometer of axis), as demonstrated by the strong negative correlation between Rec8 densities and axis lengths at per-cell level (fig. S7F). These results suggest that Pds5 does not regulate chromosome axis length by directly altering Rec8 abundance.
Fig. 4. The interplay between Pds5 and Rec8 in regulating axis length.
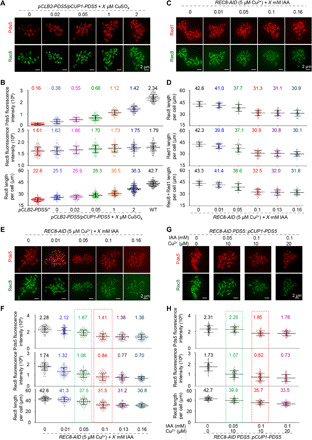
(A and B) The representative images (A) and quantifications (B) to show alterations in Pds5 abundance alter axis length in the same direction but only slightly alter Rec8 abundance. n = 80, 61, 62, 70, 67, 65, and 73, separately. (C and D) The representative images (C) and quantifications (D) showing chromosome axis can be reliably labeled and measured by either Rec8 or Red1 in Rec8 depletion mutants (Rec8 abundance is >40% of WT level) used in this study. n = 32, 30, 31, 44, 37, and 31, separately. (E and F) The representative images (E) and quantifications (F) show that decreasing Rec8 results in decreased Pds5 and axis length. n = 66, 66, 71, 67, 70, and 73, separately. (G and H) The representative images (G) and quantifications (H) show that increases in Pds5 increase axis length in Rec8 depletion mutants. n = 59, 60, 59, and 59, separately. Error bar, means ± SD (B, D, F, and H).
Rec8 regulates axis length by altering Pds5 abundance
Cohesin is required for Pds5 to regulate axis length (23, 27), so we investigated whether and how alterations in Rec8 abundance could affect axis length. Comparable chromosome axis lengths were observed when measured with either Rec8, Red1, or a combination of both markers in REC8-AID (Fig. 4, C and D). When Rec8 abundance decreased to ~75% of WT level with 0.01 mM IAA induction in REC8-AID strain, chromosome axis length showed little alteration (~96% of WT; Fig. 4, E and F). Hence, Pds5 does not regulate axis length by altering Rec8 abundance, because complete Pds5 removal only leads to ~10% Rec8 reduction (fig. S7, A and B); however, up to ~25% of Rec8 reduction does not cause significant alterations in chromosome axis length (Fig. 4, E and F; 0.01 mM IAA).
Further investigation found that more IAA resulted in further Rec8 reduction and significant decreases in chromosome axis length (Fig. 4, E and F). For example, 0.1 mM IAA induction reduced Rec8 to ~48% of WT, decreasing chromosome axis length to ~74% of WT (31.5 μm versus 42.6 μm). Therefore, the reduction of Rec8 also decreases chromosome axis length in a dosage-dependent manner.
Rec8 reduction decreased Pds5 abundance, depending on the dosage (Fig. 4, E and F, and fig. S7, C, D, and G). In a rec8Δ mutant, Pds5 is barely detected on chromosomes (27). This indicates that the localization of Pds5 depends on Rec8 and raises an interesting point: whether Rec8 completely regulates chromosome axis length by altering Pds5 abundance. To answer this question, we tried to restore Pds5 to WT levels in strains with decreased Rec8 protein levels (Fig. 4, G and H). Without affecting Rec8 levels, copper induction increased Pds5 levels, increasing chromosome axis length (Fig. 4H, the second column versus Fig. 4F, the third column; Fig. 4H, the third column versus Fig. 4F, the fourth column). When 0.05 mM IAA and 10 μM copper were added, the strain expressed Pds5 levels approaching WT but still had a lower Rec8 and shorter axis than WT (Fig. 4H, the second versus the first column). This implies that Rec8 can regulate chromosome axis length without altering Pds5. This was confirmed by the comparison: At similar Pds5 levels, strains with lower Rec8 have shorter chromosome axes (Fig. 4H, the third column versus Fig. 4F, the third column). Additional experiments performed on pCLB2-PDS5 REC8-AID strains also confirmed that alterations in Rec8 can directly regulate axis length without altering Pds5 abundance (fig. S8). Together, Rec8 can regulate chromosome axis length directly and also indirectly by modulating Pds5.
Quantification of the contributions of Pds5 and Rec8 abundance to axis length
Both Pds5 and Rec8 dosage-dependently regulate chromosome axis length, and each contributes to ~50% of the axis length in WT (fig. S7, H and I). Pds5 regulates axis length without altering Rec8 abundance. However, Rec8 regulates axis length mainly by modulating Pds5. Therefore, the altered axis length in Rec8 depletion mutants is a result of both altered Pds5 and Rec8 abundance. To further understand the respective and direct contributions of Pds5 and Rec8 abundance to chromosome axis length, we performed a regression analysis with data from strains without homolog pairing defects. Axis length could be described as a simple function of Pds5 and Rec8 abundance: axis length = 15.63 + 9.26*[Pds5] + 3.42*[Rec8] (Fig. 5B and fig. S9, A to F; Materials and Methods). Here, [Pds5] and [Rec8] represent Pds5 and Rec8 abundance (measured by fluorescence intensities), respectively. The axis lengths predicted by this function closely matched the observed axis lengths (R = 0.99; Fig. 5B and fig. S9, A to F). No synergistic effect between Pds5 and Rec8 was required (fig. S9, A to F).
Fig. 5. Quantification of the respective contributions of Pds5 and Rec8 abundance to axis length.
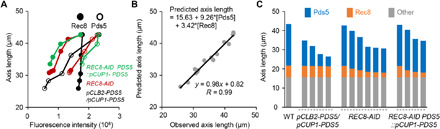
(A) Summary of Pds5 and Rec8 abundance and corresponding axis lengths in WT and various mutants. Data from pCLB2-PDS5/pCUP1-PDS5 strain without copper induction were excluded in this analysis to avoid the influence of homolog pairing defects. (B) Contributions of Pds5 and Rec8 abundance to axis length and correlation between observed and predicted axis lengths. (C) Contributions of Pds5 and Rec8 abundance to axis length in WT and various mutants.
When axis length and the intensities of Pds5 and Rec8 were normalized as “1” in WT, the function reads: relative axis length = 0.366 + 0.508*[Pds5]R + 0.144*[Rec8]R. In WT, [Pds5]R = 1 and [Rec8]R = 1. This normalized function provides valuable information. First, in WT, Rec8 and Pds5 contribute ~15% and ~50% of axis length, respectively. Second, ~37% of axis length is independent of Pds5 and Rec8. Third, in rec8 mutants, alterations in axis length primarily and indirectly result from Pds5 alteration (35%/50% = 70% of altered axis length; compare fig. S9J functions 1, 4, and 5). On the basis of this function, Pds5, Rec8, and their overall contributions to axis length could be accurately quantified (Materials and Methods; Fig. 5C and fig. S9, H and I). The results suggest that chromosome axis length can be directly and efficiently regulated by modulating Pds5.
Coordinated alterations in chromosome axis length and CO frequency
Because meiotic chromosome axis length determines CO frequency and Pds5 dosage regulates chromosome axis length, we examined whether and how alterations in axis length lead to coordinate changes in DSB and CO frequencies in Pds5 depletion mutants. During meiosis, DSB ends are resected to single-stranded DNA, which can be bound by replication protein A (RPA). Later, the RecA homologs Dmc1 and Rad51 replace RPA to form nucleoprotein filaments for homology search and DSB repair (1). As the Pds5 dosage increased, meiotic chromosome axis length and corresponding DSB levels increased, as evidenced by an increased number of RPA foci (Fig. 6, A to C).
Fig. 6. Pds5 regulates recombination frequencies but does not impair the recombination process per se.
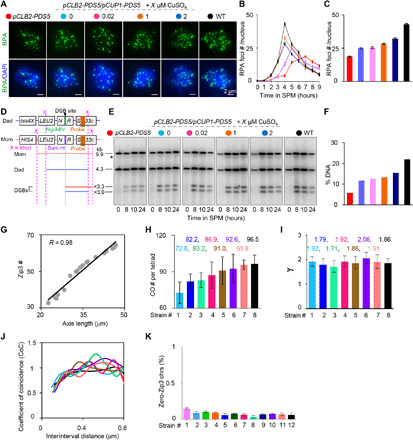
(A to C) Alterations in Pds5 protein levels alter RPA focus number. (A) Photos from the time point with maximal RPA focus number in each strain. Scale bar, 2 μm. (B) Quantification of RPA foci. (C) Maximal RPA focus number. Bar, SE (n = 3). SPM, sporulation medium. (D) Map of HIS4LEU2 hotspot locus. (E) Southern blot images for DSB assay at HIS4LEU2 locus. The asterisk, a possible DSB fragment from an additional DSB site. (F) Quantification of DSBs at 10 hours in SPM (DSBs accumulated to maximum levels in rad50S). % DNA is the percentage of total hybridizing DNA signal. (G) Zip3 focus number is highly correlated with axis length (data from Figs. 2 and 3). (H) Average CO number. Strains: pCUP1-PDS5/” with 0 μM (#1), 10 μM (#4), and 25 μM (#6) copper; PDS5/pCUP1-PDS5 with 0 μM (#2), 1 μM (#3), 5 μM (#5), and 10 μM (#7) copper; WT (#8). Bar, SD. n = 8, 6, 5, 7, 5, 7, 4, and 10 tetrads for strains #1 to #8, respectively. (I) γ values obtained from best-fit gamma distributions. Bar, 95% confidence interval. Strains and sample sizes are as in (H). (J) CoC analysis of CO interference. Data are from (H). (K) Frequencies of chromosomes absence of Zip3 focus. Strains: pCLB2-PDS5/pCUP1-PDS5 with 0.02 μM (#1), 0.05 μM (#3), 1 μM (#4), and 2 μM (#9) CuSO4; pCUP1-PDS5/” with 0 μM (#2), 0.5 μM (#6), 1 μM (#7), and 2 μM (#10) CuSO4; #5, #8, and 11# are PDS5/pCUP1-PDS5 with 0 μM (#5), 0.5 μM (#8), and 1 μM (#11) CuSO4; WT (#12). n = 100, 123, 112, 104, 98, 108, 95, 117, 104, 119, 103, and 115 nuclei, respectively. Bar, SE (n = 3). Compared with WT, P > 0.05, two-proportions test.
DSBs can be studied by a standard physical assay in combination with a Southern blot analysis at the HIS4LEU2 locus, a well-characterized DSB hotspot (33, 34). To avoid the quantification inaccuracy caused by rapid DSB turnover, the assay usually is performed in a rad50S background, where the KI81 mutation blocks the DSB end resection, accumulating DSBs. The DSB frequency was significantly reduced in pCLB2-PDS5 strain (Fig. 6, D to F) (23) and gradually increased with increases in Pds5 levels and chromosome axis length (Fig. 6, D to F). These results indicate that Pds5 regulates meiotic chromosome axis length and DSB levels.
In budding yeast, COs can be accurately defined by cytological analysis of Zip3 foci, which label future CO positions (7). Gradually increasing Pds5 led to gradual increases in the Zip3 focus number (Figs. 2D, 3C, and 6G).
The number of COs can also be assayed by resequencing the whole genome of the four haploids from the same tetrad when the two parents have proper DNA polymorphisms. To confirm the results of the Zip3 analysis, we introduced the Pds5 mutant into an S288c derivative S96 and a clinical isolate YJM789. These two strains have a 0.6% sequence difference and ~50,000 uniformly distributed single-nucleotide polymorphism (SNP) markers on chromosomes (35, 36). In agreement with previous reports, ~96.5 COs per tetrad were detected by reciprocal exchange of SNP markers in WT (35, 36). In the absence of copper, ~72.8 COs were detected in the pCUP1-PDS5/pCUP1-PDS5 strain, and when copper was added to induce more Pds5, the number of COs increased to ~92.6 per tetrad (Fig. 6H, strain 6). The positive correlation between CO number and Pds5 expression level was also observed in the PDS5/pCUP1-PDS5 mutant (Fig. 6H, strains 2, 3, 5, and 7). Therefore, gradually increasing Pds5 levels lead to gradual increases in axis length and CO frequency.
Alterations in Pds5 do not impair the recombination process per se
Pds5 regulates axis length and, consequently, DSB and CO frequencies. To investigate whether the basic CO patterning process is impaired in pds5 mutants, we investigated the strength of CO interference and the existence of obligatory CO.
CO interference distributes COs relatively evenly along chromosomes. The shape parameter (γ) of gamma distribution is often used as a measurement of the evenness of interadjacent CO distances (7, 37). The best fitting of the distribution of adjacent-CO distances to a gamma distribution showed that pds5 mutants with different CO frequencies had similar CO interference strengths as WT (γ = ~1.88 in mutants with different Pds5 levels versus 1.86 in WT; Fig. 6I). This result was further confirmed by a classical coefficient of coincidence (CoC) analysis (Fig. 6J; Materials and Methods).
The existence of obligatory CO is reflected by a very low frequency of chromosomes without even one CO [E0, usually <1%; (7)], except in human female meiosis, where E0 can be >10% because of CO maturation inefficiency (15, 16, 37). Our results demonstrated that E0 frequencies are maintained at WT levels in pds5 mutants with altered CO levels (<1%; Fig. 6K).
Together, alterations in Pds5 abundance lead to altered chromosome axis length and, consequently, altered DSB and CO frequencies all in the same direction. However, both the occurrence of obligatory CO and the strength of CO interference were unchanged. These results support that alterations in Pds5 protein levels do not impair the CO patterning process.
DISCUSSION
We have demonstrated that ~20% of the WT level of Pds5 is sufficient for meiotic homolog pairing. Both Pds5 and Rec8 regulate axis length and, consequently, DSB and CO frequencies, depending on their dosage. Alterations in Pds5 protein levels directly alter axis length; however, Rec8 mainly regulates axis length by altering Pds5 abundance. Moderate alterations in Pds5 protein levels do not impair the CO patterning process per se.
The relationship between Pds5 and Rec8 in modulating chromosome axis length
Meiotic chromosomes are organized as linear arrays of loops, where the bases of these loops are the axes. Although loop sizes markedly vary among organisms, from ~20 kb in budding yeast to ~2500 kb in grasshoppers, the numbers of loops per unit length of axis are highly conserved in a variety of organisms, with ~20 loops per micrometer of axis (14). Therefore, loop size determines axis length and varies inversely under various conditions (15, 17, 18).
The depletion of cohesin results in shorter axes during mammalian meiosis (20, 24, 25). Pds5 meiotic depletion mutants show shorter chromosome axis in budding and fission yeast meiosis and also in mammal spermatogenesis (22, 27, 28). Our current study demonstrates that Pds5 and Rec8 dosage-dependently regulate meiotic chromosome axes in budding yeast. Moreover, our investigations have confirmed and provided further insights into the complicated interplay between Pds5 and cohesin in regulating axis length.
-
1)
In the absence of cohesin, meiotic chromosomes are disorganized and axes are not formed, regardless of the presence or absence of Pds5 (22, 27). This suggests that cohesin is necessary for loop/axis formation, which is consistent with the loop extrusion model (22, 38). In this aspect, Rad21/Mcd1, the mitotic ortholog of Rec8, can at least partially replace Rec8 (fig. S10) (27).
-
2)
Alterations in either Pds5 or Rec8 can regulate the chromosomal binding of the other. In both budding and fission yeasts, Pds5 meiotic depletion only slightly decreases Rec8 abundance (Fig. 4, A and B, and fig. S7, A and B) (22, 23). However, Rec8 depletion markedly decreases Pds5 abundance (Fig. 4, E and F, and fig. S7, C and D) (27). Our current study confirmed and quantified these effects: (i) An alteration in one of the two factors results in an alteration in the other one in a dosage-dependent manner. (ii) A reduction of Pds5 only slightly decreases Rec8 at ~10% of altered Pds5 abundance (Fig. 4, A and B, and fig. S7E). However, (iii) a reduction of Rec8 markedly decreases Pds5 at ~70% of altered Rec8 abundance (Fig. 4, E and F, and fig. S7G). Pds5 and Rec8 differentially affect each other’s abundance. Temporally, Rec8 is loaded onto DNA during premeiotic DNA replication, while Pds5 is recruited via interaction with cohesin (39). Therefore, the reduction of Rec8 (a component of cohesin) decreases Pds5 binding sites and Pds5 abundance on chromosomes. Functionally, Pds5 may regulate the stability of Rec8. In mitotic cells, Pds5 antagonizes the polySUMO-dependent degradation of cohesin. Therefore, Pds5 depletion accelerates cohesin degradation (40, 41). Pds5 likely plays the same role in meiosis.
-
3)
Both Pds5 and Rec8 can dosage-dependently regulate chromosome axis length (Fig. 4), and it has been proposed that Pds5 regulates chromosome axis length via Rec8 cohesin (27, 30). However, our study showed that Rec8 regulates chromosome axis length primarily by altering Pds5 abundance (fig. S9J). Rec8 reduction (~25%) barely affects Pds5 abundance or axis length, but additional Rec8 reduction does. This suggests that Rec8 exists in abundance (Pds5 is the limiting factor) in WT and Pds5 depletion mutants. However, when Pds5 is overexpressed, Rec8 may be the limiting factor in regulating axis length. Therefore, further Pds5 increases have little effect on the axis length under these conditions (fig. S5, C and D).
-
4)
Pds5 and Rec8 abundance contribute to ~15% and ~50% of chromosome axis length, respectively, and ~65% of chromosome axis length in WT (Fig. 5 and fig. S9). There are likely other reasons for additional compaction. Besides mutants of cohesin and associated regulators, several other mutants also show altered chromosome axis lengths (6). Homolog synapsis may also promote additional compaction (42). Together, all of these factors can regulate meiotic chromosome axis length.
Pds5 can regulate axis length in the absence of sister chromatids, condensin, or meiotic DSBs
Rec8 is not only a component of the meiotic chromosome axis but also a component of meiotic cohesin. Rec8 may regulate chromosome axis length by altering the number of cohesin-mediated interactions (22, 42). In the pCLB2-PDS5 rec8Δ double mutant, chromosomes were disorganized and intact chromosome axes were not observed (23, 27). This suggests that chromosome hypercompaction in pds5 mutants requires Rec8. However, neither Rec8’s role in sister cohesion nor its role in axis assembly is required, because the meiosis-specific depletion of Cdc6 required for the formation of sister chromatids or the deletion of axis component Hop1 does not affect chromosome hypercompaction in Pds5 depletion mutants (23, 27).
In rec8 deletion mutants, the induced expression of Mcd1 (the mitotic ortholog of Rec8) can reconstruct cohesin and rescue sister cohesion during meiosis, although it does not replace Rec8’s role in axis assembly (43). Mcd1 induction in the pCLB2-PDS5 rec8Δ double mutant (pCLB2-PDS5 rec8Δ pCUP1-MCD1) can at least partially rescue the phenotype of hypercompacted chromosomes. However, the chromosome axis still differs from that of pCLB2-PDS5 mutants (fig. S10) (23, 27). These results suggest that Pds5 can regulate axis length in the absence of sister chromatids or sister cohesion but requires the existence of cohesin complex and probably its role in mediating chromatin loops.
Condensin complexes are required for proper chromosome condensation (individualization and compaction) in both mitosis and meiosis [e.g., (6, 44)]. Meiotic depletion of Brn1, a subunit of condensin, results in a longer chromosome axis and more COs (fig. S11, A and E) (6). Unexpectedly, the pCLB2-PDS5 pCLB2-BRN1 double mutant showed hypercompacted chromosomes similar to pCLB2-PDS5 but not the pCLB2-BRN1 mutant (fig. S11, A and E). Moreover, Pds5 also can dosage-dependently regulate axis length in the absence of Brn1. A detailed comparison showed that the effects of Pds5’s dosage on chromosome axis length were greater in the absence of Brn1 than in its presence (fig. S11, A and E). This suggests that Pds5 can regulate axis length independent of condensin (condensin may restrict Pds5’s dosage effects). Our work and previous studies have demonstrated that the phenotypes of the Pds5 depletion mutant were independent of meiotic DSBs (spo11Y135F), Hop1 (a meiotic specific component of chromosome axis), or Zip1 (the transverse filament of synapsis complex) (fig. S11, B to E) (23, 27).
Why do organisms regulate axis length mainly by altering Pds5 but not Rec8 (cohesin) abundance?
A loop extrusion model and similar ideas have been proposed to explain the formation and regulation of chromatin loop/axis (18, 22, 38). Studies have suggested that cohesin holds two loci together along a chromatid to produce a loop. As a cohesin regulator, Pds5 modulates loop size and axis length by regulating cohesin’s processive activity in loop extrusion (26, 42, 45). Pds5 can suppress loop size in two ways: It suppresses cohesin’s translocation ability and restricts cohesin’s residence on chromatin (46, 47). This study suggests that Pds5 abundance regulates meiotic chromosome axis length; however, Rec8 regulates axis length mainly by altering Pds5 abundance. On the basis of previous reports and our current results, there are several possibilities why cells alter Pds5 and keep Rec8 on chromosomes but do not regulate axis length by directly altering Rec8 abundance.
Pds5 could affect the cohesin responsible for loop formation but not those for sister cohesion (48). This would regulate axis length by modulating Pds5 and maintaining sister cohesion. Because axis length regulates recombination frequency, a better possibility is that it alters Pds5 and keeps cohesin on chromosomes to maintain chromosomal organization. This ensures successful and efficient meiosis, producing viable gametes with altered recombination frequency without damaging the basic recombination process. This agrees with the fact that Rec8 (cohesin) plays multiple roles during meiosis and that alterations in Rec8 result in abnormal gametes (23, 49). Moreover, cohesin variation is linked to axis length and recombination rate differences that are linked to fertility in humans (24, 50, 51). Spore viability was more severely impaired at similar reductions of axis length by altering Rec8 than Pds5 (fig. S2G). In addition, reductions in Rec8 (but not Pds5) resulted in decreased Red1 abundance (fig. S12). However, we cannot exclude the possibility that Pds5 regulates chromosomal axis length independent of cohesin (e.g., modulating chromosome property that affects looping velocity).
Does Pds5 play a role in evolutionary adaptation?
CO recombination promotes genetic diversity by creating new features [e.g., (52)]. Some features are good because they generate favored allele combinations and/or disrupt unfavorable ones. However, some features are bad because of the combination of unfavorable alleles and/or the disruption of favorable combinations [e.g., (52)]. Therefore, COs must be tightly controlled to balance these two opposite effects [e.g., (52)].
Chromosome axis length determines the CO frequency (Introduction). Hence, modulating chromosome axis length is an efficient way of regulating evolutionary adaptation, which is supported by recent studies. (i) Efforts toward how recombination rate evolves have identified axis/SC length has an important contribution to recombination differences among and within house mice (53). (ii) Decreased chromosome axis length plays a crucial role in the evolution of Arabidopsis autotetraploids (4). In tetraploids, each chromosome has four homologous copies. One chromosome connected by more than one homolog will result in chromosome segregation errors and meiosis failures. To avoid this, the chromosome axis shortens to ensure interference, reaching chromosome ends once one CO is designated. This ensures only one CO formed between each pair of homologs (4, 54).
Another way to modulate CO frequency is to generate both hyper- and hypo-CO gametes via covariation of CO frequencies across different chromosomes within individual nuclei (17). This per-nucleus CO covariation results from the per-nucleus covariation of chromosome axis lengths (17).
This study suggests that Pds5 is a candidate regulator of axis length covariation, as discussed below. Pds5 works in a dosage-dependent manner to regulate chromosome axis length and recombination frequency. In the population of WT meiotic nuclei, Pds5 abundance is highly correlated with chromosome axis length (Fig. 3, D and E). Therefore, variations in Pds5 abundance could result in variations in recombination frequency among nuclei. Moreover, this regulation does not alter the basic aspects of the recombination process (Fig. 6, I to K). A moderate Pds5 alteration significantly changes recombination frequency but has only mild effects on sporulation efficiency and spore viability. For example, when Pds5 abundance decreases to ~50% of WT, axis length and recombination frequency decrease to 70 to 80% of WT. However, sporulation efficiency and spore viability still exceed 90% of WT (Fig. 2 and fig. S2). Therefore, alterations in Pds5 to modulate axis length are an efficient way of regulating recombination frequency. Pds5 is conserved because decreased axis length and recombination frequency have also been observed in fission yeast and mammals when Pds5 is depleted during meiosis (23, 27, 28, 55).
Alternatively, alterations in Pds5 protein levels could be selected under certain conditions by altering the expression or occurrence of new alleles with altered activity or stability. Pds5 is a top candidate in modulating chromosome structure for the evolution of Arabidopsis autotetraploids (56). Pds5 alleles in tetraploid Arabidopsis are predicted to eliminate several potential phosphorylation sites (56). It is worth further investigating whether these Pds5 alleles affect Pds5 protein abundance (either its expression or stability) or its activity.
Pds5 has a role in multiple processes during meiosis and is also related to evolution. However, it is unclear whether Pds5 actively functions in this process or is just a pleiotropic outcome of Pds5’s role in other processes.
MATERIALS AND METHODS
Yeast strains
Saccharomyces cerevisiae strains used in this study are listed in table S1. All yeast strains are of SK1 background, except for strains used for whole-genome resequencing, which were made by mating S96 and YJM789 haploids.
The HIS4LEU2 locus, the pCLB2-PDS5, and spo11Y135F strains have been described (9, 27). The strains carrying TetO/TetR-GFP and LacO/LacI-GFP on chromosomes II and XV, respectively, have also been described previously (23, 33). The pCUP1-PDS5 strain was constructed by replacing the native promoter of the PDS5 gene with the CUP1 promoter. The pCLB2-PDS5-AID strain was modified on the basis of the pCLB2-PDS5 strain with an AID tagged to the C terminus of PDS5. The AID tag was obtained by polymerase chain reaction (PCR) from a pHyg-AID71-114-9myc plasmid (57). The strain with an additional pCUP1-PDS5 was constructed by inserting a copy of pCUP1-PDS5 after the PDS5 gene locus.
Meiotic time course
The meiotic time course was performed as previously described (23). Briefly, yeast cells were grown in presporulation medium (SPS) for 16 to 18 hours and transferred to sporulation medium (SPM) to induce meiosis. To induce pCUP1-PDS5 expression, CuSO4 was added at specified time points during meiosis. To induce the degradation of Pds5 or Rec8, CuSO4 and auxin (IAA) [or dimethyl sulfoxide as a control] were added to the cultures of pCLB2-PDS5-AID or REC8-AID at the specified time points (57).
Chromosome spreading and immunofluorescence
For chromosome spreading, the yeast cells were first processed into spheroplasts with Zymolyase-100 T (Nacalai Tesque), spread on a clean slide with 1% Lipsol, and fixed with 3% paraformaldehyde containing 3.4% sucrose (6). For immunostaining, slides with spread nuclei were sequentially incubated with the proper primary and secondary antibodies. Primary antibodies used in this study include mouse monoclonal anti-Myc (sc-40, Santa Cruz Biotechnology), rat monoclonal anti–hemagglutinin (HA; 11867423001, Roche), goat polyclonal anti-Zip1 (sc-48716, Santa Cruz Biotechnology), rabbit polyclonal anti-RPA (AS07214, Agrisera), and rabbit polyclonal anti-GFP (A-11122, Thermo Fisher Scientific). The following secondary antibodies were used in this study: Alexa 488–conjugated donkey anti-mouse/goat/rabbit (A-21202/A-11055/A-21206, respectively; Thermo Fisher Scientific), Alexa 594–conjugated donkey anti-rat/goat (A-21209/A-11058, respectively; Thermo Fisher Scientific). Chromosomal DNA was stained by 4,6-diamidino-2-phenylindole. Fluorescence images were visualized and acquired using a Zeiss fluorescence microscope (Axio Imager.Z2) equipped with an Andor charge-coupled device (CCD) camera (iXon Ultra 897).
Western blot
Yeast cells were lysed in 20% trichloroacetic acid using glass beads. The resultant pellets were extracted with a Laemmli buffer and denatured in boiling water for 5 min. Proteins were separated on a 10% SDS–polyacrylamamide gel electrophoresis gel. The membrane transfer was performed at a current of 200 mA on ice for 1.5 hours. The following antibodies were used for immunoblotting: mouse monoclonal anti-HA (H9658, Sigma-Aldrich), mouse monoclonal anti-Myc (sc-40, Santa Cruz Biotechnology), and mouse monoclonal anti-phosphoglycerate kinase 1 (PGK1; ab113687, Abcam). Membranes were imaged with a Bio-Rad Imager. Quantification of protein bands was performed using ImageJ software.
Whole-genome resequencing to analyze COs and CO interference
S96 and YJM789 haploids were patched together on yeast extract, peptone, and dextrose (YPD) plates and incubated for 6 hours. The mixtures were transferred into SPM, and the diploids were sporulated. Tetrads were dissected onto YPD plates under a tetrad dissection microscope and incubated for 3 days. Tetrads with four viable spores were confirmed by colony PCR with allele-specific primers to check for aneuploidy spores. DNA was prepared from each spore colony and sequenced (150 PE) with Novogene using the Illumina HiSeq X platform. In our experiments, approximately 90% of reads could be aligned to the reference genome covered at 100× read depth. The ReCombine program (http://sourceforge.net/projects/recombine) was used to determine COs (58).
To analyze CO interference, whole-genome resequencing data were used and gamma distribution and CoC curve were performed as previously described (7, 35). For CoC analysis, the CO data from different chromosomes were combined, though only the first 320 kb was used (excluding chromosomes I and VI, which are less than 320 kb). For the CoC curve plot, the genomic distance was converted to axis length with 320 kb = 1.2 μm in WT and scaled with the number of COs (which is scaled axis length) in mutants (7).
Physical analysis of DSBs
The HIS4LEU2 locus was used to quantify DSBs. DNA preparation and physical analysis were performed as previously described (34). Briefly, genomic DNA was digested with Xho I and separated on an agarose gel in tris/borate/EDTA (TBE) buffer. For Southern blot analysis, “Probe A” labeled with 32P-dCTP radioactive nucleotides was used for hybridization (33). Hybridizing signals were quantified using Quantity One.
Measurement of chromosome axis length
In yeast meiosis, Rec8, Red1, and Hop1 are critical chromosome axis components. At pachytene, the homologs are fully synapsed with the help of Zip1 (the central element of SC). Therefore, at pachytene, chromosome axis length can be measured by quantifying the length of either the Rec8, Red1, Hop1, or Zip1 line. Previous studies have shown that these components are continuously distributed along chromosomes, although with alternative hyper- and hypoabundant regions. Red1 and Hop1 show the same hyperabundant regions and complement Zip1 and Rec8 hyperabundant regions (33). To test whether measuring axis length by staining one component is reliable, the surface spread nuclei were double-stained with Rec8 and Zip1 or with Rec8 and Red1.
Our results show that chromosome axes can be reliably visualized and measured by immunostaining any one of those components (fig. S3). First, at pachytene, Rec8, Red1, and Zip1 were continuously localized along chromosomes. Second, Rec8 and Zip1 overlapped each other, and Rec8 and Red1 overlapped each other, although their hyperabundance differed. Third, the per-nucleus axis lengths (as measured with Rec8, Red1, or both together) were well matched and highly correlated (R > 0.9). Per-nucleus axis lengths measured with Rec8, Zip1, or both together were also well matched and were highly correlated (R > 0.9). Fourth, the average axis length as measured by Rec8, Red1, Zip1, the combination of Rec8 and Red1, or the combination of Rec8 and Zip1 displayed no significant differences.
In the Rec8 and Pds5 mutants, our measurements of chromosome axis length using Rec8 were validated by costaining Rec8 and Red1. Furthermore, no axis breaks or gaps were observed. This is supported by the following facts: (i) Chromosome axes can be marked by Rec8 or Red1 with continuous signals. (ii) Chromosome axis length measured by Rec8 and Red1 and the combination of both markers produced comparable results (fig. S3). (iii) Each Rec8 line has one Sgo1 focus that labels kinetochores, indicating that all Rec8 lines have centromeres (fig. S1D).
Quantification of immunofluorescence intensity
Pds5 and Rec8 levels were estimated by their fluorescence intensity after immunostaining in spread nuclei. For accurate quantification, mutant cells were mixed with WT cells and spread on the same slide. The left end of the WT chromosome XV was labeled by a LacO/LacI-GFP spot, making WT and mutant cells easily distinguished. The same protocol was used for all immunostaining, including the concentration of antibodies, incubation time, and buffers. After immunostaining, the same parameters were used in imaging. All images were captured using a Zeiss fluorescence microscope (Axio Imager.Z2) equipped with an Andor CCD camera (iXon Ultra 897), which was controlled by MetaMorph software (Molecular Devices). Fluorescence intensity was measured using ImageJ software.
The fluorescence intensities of WT cells obtained from different slides were comparable, meaning that fluorescence intensities between WT and different mutant cells could be directly compared. The fluorescence intensity was obtained by subtracting the background fluorescence intensity from the raw fluorescence intensity.
We randomly selected a position on a chromosome in the target nucleus (not close to chromosome ends) and drew a line perpendicular to the chromosome axis at that position. The signal intensity profile (signal intensity of each pixel) of the line is displayed as a normal distribution curve, with the peak (highest signal intensity) of the curve in the middle of the chromosome axis width. The signal intensity gradually decreases from the middle of the axis, moving out bidirectionally along the line, while the slope at each pixel position on the curve decreases. Two methods were used to define background signal intensity. First, signal intensity was selected as the background level at a certain place where the slope has a maximal change. Second, the curve is flattened as a horizontal line where the minimal background pixel intensity is defined. The real background is defined by an increase of 20% of the minimum pixel intensity. In our experience, this background signal intensity is close to the maximum slope change. Three different positions on each of three different chromosomes were randomly selected for each nucleus, resulting in a total of nine background values. The average of these values was considered the average background pixel intensity. A minimal circle was drawn to cover the target nuclear area, while each pixel intensity was displayed in the circled area. We then counted the number of pixels whose intensity exceeded the average background pixel intensity. The sum of these pixel intensities was considered the total raw fluorescence intensity [Total background intensity] = [average background pixel intensity] × [the number of pixel whose intensity exceeds background pixel intensity].
Quantification of respective contributions of Pds5 and Rec8 abundance to chromosome axis length
The chromosome axis length and the fluorescence intensities of Rec8 and Pds5 were measured in pCLB2-PDS5/pCUP1-PDS5, REC8-AID, and REC8-AID PDS5::pCUP1-PDS5 strains under various conditions. To identify the individual contributions of Rec8 and Pds5 abundance to axis length, we performed a regression analysis using GraphPad Prism (v8.0.1). Data from the pCLB2-PDS5/pCUP1-PDS5 strain without copper induction were excluded in this analysis to avoid the possible influence of homolog pairing defects.
Prediction homolog pairing level based on the number of Rec8 lines
Using an epifluorescence microscope, each pair of paired homologs is displayed as one Rec8 line, and each pair of unpaired homologs is displayed as two Rec8 lines. Diploid budding yeast has 16 pairs of homologs. When all homologs are paired during meiosis, there should be 16 Zip1/Rec8 lines observed under an epifluorescence microscope. When all homologs are unpaired, there should be 32 Rec8 lines. In the pCLB2-PDS5 mutant, an average of 23 Rec8 lines was observed, indicating that approximately half of the homologs in this mutant are paired. Homolog pairing levels can be similarly estimated in other mutants.
Statistical analysis
Data are presented as means ± SD, SEM, or 95% confidence interval. Sample sizes and n values are described in figure legends. The correlation coefficient was calculated with Pearson’s correlation analysis using EXCEL or GraphPad Prism.
Acknowledgments
We would like to thank J. Fung (UCSF), H.-G. Yu (Florida State University), and H. Ulrich (Institute of Molecular Biology, Germany) for strains/plasmids, as well as N. Kleckner and D. Zickler for discussions. Funding: This work was supported by the National Natural Science Foundation of China (31671293, 31890782, 31900402, 31801203, and 31771385), China Postdoctoral Science Foundation (2018 M630771), and the Taishan Scholars Youth Project of Shandong Province. Author contributions: M.S., S.W., and L.Z. conceived of the study and designed experiments. M.S., B.Z., Xiao Yang, T.T., Y.W., Xuan Yang, Y.T., T.C., Y.C., and Y.S. performed the experiments. M.S., S.W., and L.Z. analyzed the data and wrote the manuscript with inputs and edits from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The whole-genome sequencing data are available at NCBI (SRA BioProject, accession number PRJNA695084). Additional data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/11/eabe7920/DC1
REFERENCES AND NOTES
- 1.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zickler D., Kleckner N., Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole F., Keeney S., Jasin M., Preaching about the converted: How meiotic gene conversion influences genomic diversity. Ann. N. Y. Acad. Sci. 1267, 95–102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bomblies K., Jones G., Franklin C., Zickler D., Kleckner N., The challenge of evolving stable polyploidy: Could an increase in “crossover interference distance” play a central role? Chromosoma 125, 287–300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veller C., Kleckner N., Nowak M. A., A rigorous measure of genome-wide genetic shuffling that takes into account crossover positions and Mendel’s second law. Proc. Natl. Acad. Sci. U.S.A. 116, 1659–1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Wang S., Yin S., Hong S., Kim K. P., Kleckner N., Topoisomerase II mediates meiotic crossover interference. Nature 511, 551–556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., Liang Z., Hutchinson J., Kleckner N., Crossover patterning by the beam-film model: Analysis and implications. PLOS Genet. 10, e1004042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones G. H., Franklin F. C. H., Meiotic crossing-over: Obligation and interference. Cell 126, 246–248 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Martini E., Diaz R. L., Hunter N., Keeney S., Crossover homeostasis in yeast meiosis. Cell 126, 285–295 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Zickler D., Kleckner N., Zhang L., Meiotic crossover patterns: Obligatory crossover, interference and homeostasis in a single process. Cell Cycle 14, 305–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keeney S., Lange J., Mohibullah N., Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu. Rev. Genet. 48, 187–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray S., Cohen P. E., Control of meiotic crossovers: From double-strand break formation to designation. Annu. Rev. Genet. 50, 175–210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medhi D., Goldman A. S., Lichten M., Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. eLife 5, e19669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleckner N., Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Hassold T., Hunt P., White M. A., Zickler D., Kleckner N., Zhang L., Inefficient crossover maturation underlies elevated aneuploidy in human female meiosis. Cell 168, 977–989.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S., Kleckner N., Zhang L., Crossover maturation inefficiency and aneuploidy in human female meiosis. Cell Cycle 16, 1017–1019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S., Veller C., Sun F., Ruiz-Herrera A., Shang Y., Liu H., Zickler D., Chen Z., Kleckner N., Zhang L., Per-nucleus crossover covariation and implications for evolution. Cell 177, 326–338.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleckner N., Storlazzi A., Zickler D., Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet. 19, 623–628 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Morgan C. H., Zhang H., Bomblies K., Are the effects of elevated temperature on meiotic recombination and thermotolerance linked via the axis and synaptonemal complex? Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revenkova E., Eijpe M., Heyting C., Hodges C. A., Hunt P. A., Liebe B., Scherthan H., Jessberger R., Cohesin SMC1β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6, 555–562 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Storlazzi A., Tessé S., Gargano S., James F., Kleckner N., Zickler D., Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17, 2675–2687 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding D.-Q., Sakurai N., Katou Y., Itoh T., Shirahige K., Haraguchi T., Hiraoka Y., Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J. Cell Biol. 174, 499–508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong S., Joo J. H., Yun H., Kleckner N., Kim K. P., Recruitment of Rec8, Pds5 and Rad61/Wapl to meiotic homolog pairing, recombination, axis formation and S-phase. Nucleic Acids Res. 47, 11691–11708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas U., Hempel K., Llano E., Pendás A., Jessberger R., Distinct roles of meiosis-specific cohesin complexes in mammalian spermatogenesis. PLOS Genet. 12, e1006389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward A., Hopkins J., Mckay M., Murray S., Jordan P. W., Genetic interactions between the meiosis-specific cohesin components, STAG3, REC8, and RAD21L. G3 6, 1713–1724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wutz G., Várnai C., Nagasaka K., Cisneros D. A., Stocsits R. R., Tang W., Schoenfelder S., Jessberger G., Muhar M., Hossain M. J., Walther N., Koch B., Kueblbeck M., Ellenberg J., Zuber J., Fraser P., Peters J.-M., Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin H., Guacci V., Yu H.-G., Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J. Cell Biol. 186, 713–725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viera A., Berenguer I., Ruiz-Torres M., Gómez R., Guajardo A., Luis Barbero J., Losada A., Suja J., PDS5 proteins regulate the length of axial elements and telomere integrity during male mouse meiosis. EMBO Rep. 21, e49273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heemst D. V., James F., Pöggeler S., Lecellier V. B., Zickler D., Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98, 261–271 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Fukuda T., Hoog C., The mouse cohesin-associated protein PDS5B is expressed in testicular cells and is associated with the meiotic chromosome axes. Genes 3, 484–494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur H., Ahuja J. S., Lichten M., Methods for controlled protein depletion to study protein function during meiosis. Methods Enzymol. 601, 331–357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agostinho A., Manneberg O., Schendel R. V., Hernández A. H., Kouznetsova A., Blom H., Brismar H., Höög C., High density of REC8 constrains sister chromatid axes and prevents illegitimate synaptonemal complex formation. EMBO Rep. 6, 901–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim K. P., Weiner B. M., Zhang L., Jordan A., Dekker J., Kleckner N., Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143, 924–937 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter N., Kleckner N., The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Chen S. Y., Tsubouchi T., Rockmill B., Sandler J. S., Richards D. R., Vader G., Hochwagen A., Roeder G. S., Fung J. C., Global analysis of the meiotic crossover landscape. Dev. Cell 15, 401–415 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Liu Y., Shang Y., Zhai B., Yang X., Kleckner N., Zhang L., Crossover interference, crossover maturation, and human aneuploidy. Bioessays 41, e1800221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alipour E., Marko J. F., Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 40, 11202–11212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartman T., Stead K., Koshland D., Guacci V., Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151, 613–626 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan K. L., Gligoris T., Upcher W., Kato Y., Shirahige K., Nasmyth K., Beckouët F., Pds5 promotes and protects cohesin acetylation. Proc. Natl. Acad. Sci. U.S.A. 110, 13020–13025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Ambrosio L. M., Lavoie B. D., Pds5 prevents the polysumo-dependent separation of sister chromatids. Curr. Biol. 24, 361–371 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Schalbetter S. A., Fudenberg G., Baxter J., Pollard K. S., Neale M. J., Principles of meiotic chromosome assembly revealed in S. cerevisiae. Nat. Commun. 10, 4795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buonomo S. B., Clyne R. K., Fuchs J., Loidl J., Uhlmann F., Nasmyth K., Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103, 387–398 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Yu H. G., Koshland D. E., Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J. Cell Biol. 163, 937–947 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller H., Scolari V. F., Agier N., Piazza A., Thierry A., Mercy G., Descorps-Declere S., Lazar-Stefanita L., Espeli O., Llorente B., Fischer G., Mozziconacci J., Koszul R., Characterizing meiotic chromosomes’ structure and pairing using a designer sequence optimized for Hi-C. Mol. Syst. Biol. 14, e8293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanke M., Tahara E., Huis In’t Veld P. J., Nishiyama T., Cohesin acetylation and Wapl-Pds5 oppositely regulate translocation of cohesin along DNA. EMBO J. 35, 2686–2698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morales C., Ruiz-Torres M., Rodríguez-Acebes S., Lafarga V., Rodríguez-Corsino M., Megías D., Cisneros D. A., Peters J.-M., Méndez J., Losada A., PDS5 proteins are required for proper cohesin dynamics and participate in replication fork protection. J. Biol. Chem. 295, 146–157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayerova N., Cipak L., Gregan J., Cohesin biology: From passive rings to molecular motors. Trends Genet. 36, 387–389 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Brar G. A., Hochwagen A., Ee L.-s. S., Amon A., The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol. Biol. Cell 20, 1030–1047 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murdoch B., Owen N., Stevense M., Smith H., Nagaoka S., Hassold T., Kay M. M., Xu H., Fu J., Revenkova E., Jessberger R., Hunt P., Altered cohesin gene dosage affects Mammalian meiotic chromosome structure and behavior. PLOS Genet. 2, e1003241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chernus J. M., Allen E. G., Zeng Z., Hoffman E. R., Hassold T. J., Feingold E., Sherman S. L., A candidate gene analysis and GWAS for genes associated with maternal nondisjunction of chromosome 21. PLOS Genet. 12, e1008414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otto S. P., Payseur B. A., Crossover interference: Shedding light on the evolution of recombination. Annu. Rev. Genet. 53, 19–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang R. J., Dumont B. L., Jing P., Payseur B. A., A first genetic portrait of synaptonemal complex variation. PLOS Genet. 15, e1008337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgan C., Zhang H., Henry C. E., Franklin F. C. H., Bomblies K., Derived alleles of two axis proteins affect meiotic traits in autotetraploid Arabidopsis arenosa. Proc. Natl. Acad. Sci. U.S.A. 117, 8980–8988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding D.-Q., Matsuda A., Okamasa K., Nagahama Y., Haraguchi T., Hiraoka Y., Meiotic cohesin-based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe. Chromosoma 125, 205–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright K. M., Arnold B., Xue K., Šurinová M., O’Connell J., Bomblies K., Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Mol. Biol. Evol. 32, 944–955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morawska M., Ulrich H. D., An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341–351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson C. M., Oke A., Yam P., Zhuge T., Fung J. C., Reduced crossover interference and increased ZMM-independent recombination in the absence of Tel1/ATM. PLOS Genet. 11, e1005478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/11/eabe7920/DC1


