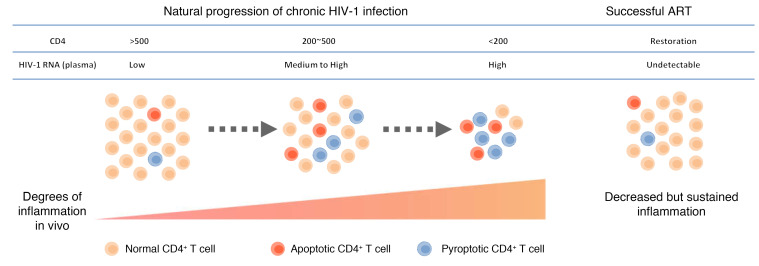
Abstract
Chronic HIV-1 infection is generally characterized by progressive CD4+ T cell depletion due to direct and bystander death that is closely associated with persistent HIV-1 replication and an inflammatory environment in vivo. The mechanisms underlying the loss of CD4+ T cells in patients with chronic HIV-1 infection are incompletely understood. In this study, we simultaneously monitored caspase-1 and caspase-3 activation in circulating CD4+ T cells, which revealed that pyroptotic and apoptotic CD4+ T cells are distinct cell populations with different phenotypic characteristics. Levels of pyroptosis and apoptosis in CD4+ T cells were significantly elevated during chronic HIV-1 infection, and decreased following effective antiretroviral therapy. Notably, the occurrence of pyroptosis was further confirmed by elevated gasdermin D activation in lymph nodes of HIV-1–infected individuals. Mechanistically, caspase-1 activation closely correlated with the inflammatory marker expression and was shown to occur through NLRP3 inflammasome activation driven by virus-dependent and/or -independent ROS production, while caspase-3 activation in CD4+ T cells was more closely related to T cell activation status. Hence, our findings show that NLRP3-dependent pyroptosis plays an essential role in CD4+ T cell loss in HIV-1–infected patients and implicate pyroptosis signaling as a target for anti–HIV-1 treatment.
Keywords: AIDS/HIV, Inflammation
Keywords: Caspases and caspase substrates, T cells
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection leads to a progressive loss of CD4+ T cells, which has long been recognized as central to HIV-1 pathogenesis but remains to be fully clarified (1). Since HIV-1 is known to primarily infect CD4+ T cells, it was believed that depletion reflects a viral cytopathic effect occurring in productively infected CD4+ T cells via direct virus-induced apoptosis and immune cell–mediated elimination. However, mounting evidence suggests that a broader and more profound depletion occurs in uninfected CD4+ T cells via a phenomenon known as the bystander effect (2). Nevertheless, the mechanism(s) by which bystander CD4+ T cells are depleted during HIV-1 infection remains a subject for debate (3, 4).
Great advances in our understanding of cell death pathways have been made in recent years (5, 6). Caspases are a family of cysteine proteases that mediate regulated cell death processes, including caspase-3–dependent apoptosis and caspase-1–dependent pyroptosis (6). Both apoptosis and pyroptosis have been proposed to contribute to the bystander death of CD4+ T cells in HIV-1 infection. Apoptosis in HIV-1 infection has been reported since the earliest days of HIV-1 research (7, 8), not only in HIV-1–infected T cells, but also in bystander CD4+ T cells (4). More recently, by using an ex vivo human lymphoid aggregate culture (HLAC) system, Doitsh et al. found that both apoptosis and pyroptosis induce the depletion of CD4+ T cells (9). Subsequent studies showed that approximately 5% of CD4+ T cell depletion occurs through apoptosis, while the remaining 95% of quiescent lymphoid CD4+ T cells die as a result of caspase-1–mediated pyroptosis triggered through abortive infection induced by cell-to-cell viral spread (10–12). The critical roles of abortive infection and pyroptosis in vivo were further supported by fitting of mathematical models with longitudinal clinical data obtained from simian immunodeficiency virus–infected (SIV-infected) macaques or HIV-1–infected patients (13, 14). However, only a few studies have described pyroptosis in humans with chronic HIV-1 infection (15, 16). Therefore, the potential mechanisms and clinical significance of CD4+ T cell loss through pyroptosis and apoptosis during the different stages of HIV-1 disease remain to be elucidated.
In the present study, we enrolled viremic HIV-1–infected patients (VIR patients), HIV-1–infected patients receiving successful antiretroviral therapy (ART patients), and elite controllers (ECs). We determined the frequencies of apoptotic and pyroptotic CD4+ T cells from both peripheral blood mononuclear cells (PBMCs) and lymph nodes (LNs) by monitoring caspase-1 activation, caspase-3 activation, and gasdermin D (GSDMD) cleavage. In addition to apoptosis, our findings also revealed a pathogenic role of NLRP3 inflammasome–dependent pyroptosis in CD4+ T cell loss and clinical progression.
Results
Increased activation of caspases in CD4+ T cells during chronic HIV-1 infection.
To analyze the distribution of apoptotic and pyroptotic cells in HIV-1–infected patients, we used fluorochrome-labeled inhibitors of caspases (FLICA) and intracellular staining with antibodies specifically recognizing the active forms of caspase-1 and caspase-3 (17, 18). Stimulation of pyroptosis by nigericin selectively induced FLICA–caspase-1 activation, whereas staurosporine stimulation resulted in preferential activation of caspase-3 (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI138861DS1).
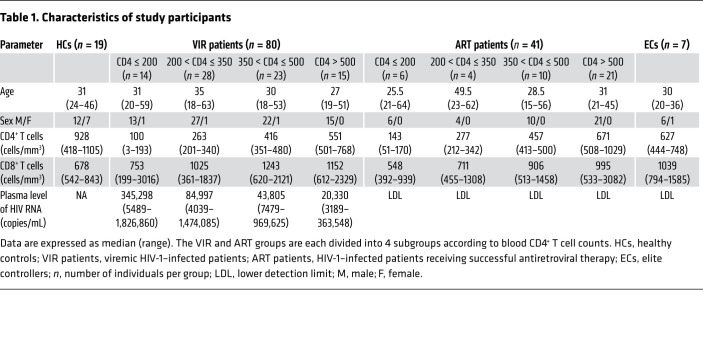
We then compared the frequencies of CD4+ T cells expressing activated caspase-1 and caspase-3 in healthy control subjects (HCs; n = 19), VIR patients (n = 80), ART patients (n = 41), and ECs (n = 7) (Table 1 and Supplemental Table 1). As shown in Figure 1, the proportion of caspase-1–activated CD4+ T cells (Figure 1, A and B; see Supplemental Figure 1B for gating strategy) was significantly increased in VIR patients compared with that in HCs (P < 0.001). The frequencies of caspase-1–activated CD4+ T cells in ART patients were lower than the frequencies in VIR patients but higher than those in HCs (P < 0.01 and P < 0.001, respectively). Notably, ECs maintained low levels of caspase-1 activation in CD4+ T cells comparable to those in HCs (P > 0.05). The activation of caspase-3 was also elevated in VIR patients but partially restored in ART patients (Figure 1C and Supplemental Figure 1C).
Table 1. Characteristics of study participants.
Figure 1. Increased activation of caspase-1 and caspase-3 in CD4+ T cells during chronic HIV-1 infection.
(A) Representative flow cytometry plot shows FLICA–caspase-1 and active caspase-3 expression in CD4+ T cells in peripheral blood of healthy controls (HC), viremic patients (VIR), antiretroviral therapy–treated patients (ART), and elite controllers (EC). (B and C) The frequencies of FLICA–caspase-1+ CD4+ T cells (B) and active caspase-3+ CD4+ T cells (C) from HCs (n = 19), VIR patients (n = 80), ART patients (n = 27), and ECs (n = 7). (D and E) Association of the frequencies of FLICA–caspase-1+ CD4+ T cells (D) and active caspase-3+ CD4+ T cells (E) with the absolute CD4+ T cell counts in VIR patients. Each dot represents a single individual, and associations were evaluated using Spearman’s correlation test. P and Spearman’s ρ values are presented. (F) CD4+ T lymphocytes from 6 VIR patients were downsampled, concatenated, and mapped by t-SNE. Each point represents a single cell. Different light colors represent different subsets. The FLICA–caspase-1+ CD4+ T cells (blue) and active caspase-3+ CD4+ T cells (red) were overlaid onto the overall t-SNE map. (G) The proportion of naive T (Tn), Tcm, Ttm, Tem, and Ttd cells in the total, FLICA–caspase-1+, and active caspase-3+ CD4+ T cell population from VIR patients (n = 6). Different symbols represent different individuals. (H) The Tcm/Tem ratio was calculated. Mann-Whitney U test (B and C); Wilcoxon’s signed-rank test (G and H); *P < 0.05, **P < 0.01, ***P < 0.001. pos, positive.
Caspase-1 activation is a marker of the formation of inflammasome and the occurrence of pyroptosis (19). We further characterized the caspase-activating CD4+ T cells by imaging flow cytometry (Supplemental Figure 1, D and E). By gating on morphologically intact cells, we detected FLICA–caspase-1+ or active caspase-3+ CD4+ T cells from VIR patients in a proportion that was comparable to that detected by flow cytometry (Supplemental Figure 1F). Strikingly, a much higher proportion of FLICA+ CD4+ T cells than negative control cells harbored a misshapen membrane (29.00% ± 8.35% vs. 5.36% ± 2.88%, P < 0.01), suggesting the occurrence of membrane rupture during pyroptosis (Supplemental Figure 1G and ref. 20). This increase in proportion of FLICA+ CD4+ T cells was not observed in caspase-3–activating CD4+ T cells (Supplemental Figure 1G).
Next, we determined whether there was a direct relationship between the increased caspase activation and CD4+ T cell loss. To this end, the statistical correlations between CD4+ T cell counts and caspase-1 and caspase-3 activation were analyzed in matched measurements performed in VIR patients. Both caspase-1 activation (Figure 1D; P < 0.001, r = –0.5823) and caspase-3 activation (Figure 1E; P < 0.001, r = –0.5369) showed significant negative correlations with CD4+ T cell counts.
When VIR patients were further stratified by CD4+ T cell count, patients with CD4+ T cell counts of ≥500 cells/μL manifested the lowest levels of caspase-1 and caspase-3 activation in CD4+ T cells, while the highest levels of caspase-1 and caspase-3 activation were detected in patients with CD4+ T cell counts of ≤200 cells/μL (Supplemental Figure 2A). ART patients showed a similar tendency to that observed in VIR patients when subgrouped by CD4+ T cell counts (Supplemental Figure 2B). Remarkably, the levels of caspase-1 activation correlated positively with caspase-3 activation levels in CD4+ T cells (Supplemental Figure 2C; P < 0.001, r = 0.9164), suggesting a close connection between pyroptosis and apoptosis, both of which might be critically involved in CD4+ T cell loss in chronically HIV-1–infected patients.
We also investigated caspase-1 and caspase-3 activation in CD8+ T cells. Although similar changes were observed among different groups (Supplemental Figure 3A), caspase-1 activation (Supplemental Figure 3B; P = 0.042, r = –0.2338) and caspase-3 activation (Supplemental Figure 3B; P = 0.0219, r = –0.2009) in CD8+ T cells showed weak negative correlation with CD4+ T cell counts. Furthermore, no significant relationship was found between caspase activation and CD8+ T cell counts (Supplemental Figure 3C). These data highlight the important pathological significance of caspase activation in CD4+ T cells during chronic HIV-1 infection.
To characterize apoptotic and pyroptotic T cells in VIR patients in vivo, we subsequently analyzed the memory status of these cells with a focus on enrichment of such cells in specific subsets during chronic HIV-1 infection. Defined by the markers CD45RA, CCR7, and CD27, naive and memory CD4+ and CD8+ T cell subsets were visualized using t-distributed stochastic neighbor embedding (t-SNE) maps in HIV-1–infected individuals (Supplemental Figure 4A). CD4+ T cells from 6 different VIR patients coclustered (Supplemental Figure 4B). In CD4+ T cells, the caspase-1–activated clusters were enriched in the effector memory (Tem) subset, while the ratio decreased in the naive, central memory (Tcm), and transient memory (Ttm) subsets, and was unchanged in the terminally differentiated (Ttd) subset (Figure 1, F and G). This is different from the in vitro data, in which pyroptosis mainly occurred in resting T cells, such as in naive and Tcm subsets. The caspase-3–activated CD4+ T cells were also enriched in the Tem subset, while the decrease in caspase-3–activated CD4+ T cells in the naive subset was less marked than the decrease in caspase-1–activated cells (Figure 1, F and G). Among the CD8+ T cells, the caspase-1–activated fractions were significantly increased in the Tcm and Ttm and decreased in the Ttd subsets (Supplemental Figure 4C). The caspase-3–activated CD8+ T cells showed a significant increase in the Tcm subset and a significant decrease in the Ttm subset compared with the caspase-1–activated CD8+ T cell fractions (Supplemental Figure 4C). Nevertheless, the Tcm/Tem ratio of caspase-activated CD4+ T cells was decreased (Figure 1H). In contrast, the Tcm/Tem ratio was increased in caspase-activated CD8+ T cells (Supplemental Figure 4D). The heterogeneity of CD4+ and CD8+ T cells suggested the existence of different mechanisms for caspase activation.
Activation of caspase-1 in CD4+ T cells correlates with both viral load and inflammation.
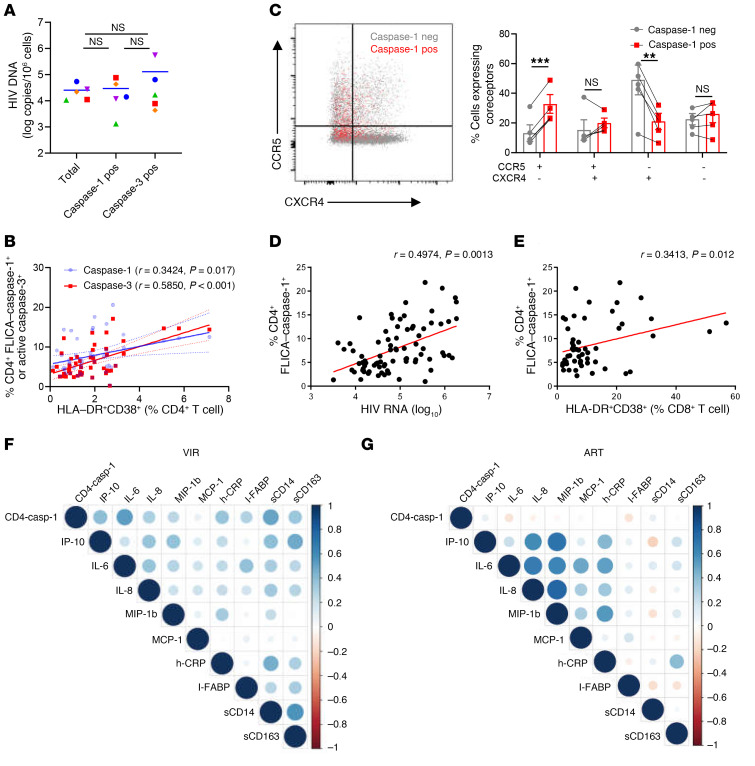
Previous ex vivo studies using the HLAC system showed that pyroptosis occurs mainly in resting CD4+ T cells, which undergo caspase-1 activation due to innate sensing of incomplete DNA fragments and are nonpermissive for HIV-1 infection (10, 11). To investigate whether caspase activation in CD4+ T cells is directly dependent on HIV-1 infection, we analyzed the presence and levels of HIV DNA in caspase-activated cells sorted from the PBMCs of VIR patients. The results showed that the frequencies of HIV DNA–containing cells were extremely low (Figure 2A). Surprisingly, neither caspase-1– nor caspase-3–activated cells showed enrichment of HIV DNA compared with control cells (Figure 2A). These data indicated that HIV-1 infection, either by a productive or an abortive mechanism, is not the main cause of caspase activation in peripheral CD4+ T cells in VIR patients. Activation-induced cell death has also been proposed as an important mechanism of CD4+ T cell loss in HIV infections (21). In this study, the activation status of CD4+ T cells (HLA-DR+CD38+) showed a stronger positive correlation with caspase-3 activation (P < 0.001, r = 0.5850) than with caspase-1 activation (P = 0.017, r = 0.3424; Figure 2B), suggesting that activation-induced cell death occurs via apoptosis, while the mechanism responsible for caspase-1 activation in CD4+ T cells remains to be clarified. Strikingly, when we assessed the expression of coreceptors recognized by HIV-1 on caspase-activating CD4+ T cells, the caspase-1–activated CD4+ T cell population was enriched in CCR5+CXCR4– cells, whereas the fraction of CCR5–CXCR4– cells was increased in caspase-3–activated CD4+ T cells (Figure 2C and Supplemental Figure 4E). Taken together, these results suggested that caspase-1 activation and caspase-3 activation in CD4+ T cells are driven by different factors.
Figure 2. Activation of caspase-1 in CD4+ T cells correlates with both viral load and inflammation.
(A) Isolation of total, FLICA–caspase-1+, and active caspase-3+ CD4+ T cells from PBMCs of VIR patients (n = 5). HIV DNA in different cell fractions was detected. Wilcoxon’s matched-pairs signed-rank test was performed. (B) Correlation of HLA-DR+CD38+ CD4+ T cells with FLICA–caspase-1+ and active caspase-3+ CD4+ T cells. (C) The expression of HIV-1 coreceptor CCR5 and CXCR4 on FLICA–caspase-1+ (red dots) and FLICA–caspase-1– CD4+ T cells (gray dots) in VIR patients (n = 5). neg, negative. (D and E) Correlation of FLICA–caspase-1 frequency in CD4+ T cells with HIV-1 viral load (D) and activation of CD8+ T cells (HLA-DR+CD38+ CD8+ T cells) (E) in VIR patients (n = 80). The HIV-1 viral load is represented on a log10 scale. (F and G) Correlation matrix of FLICA–caspase-1 frequency in CD4+ T cells and plasma inflammatory markers in VIR patients and ART patients. Levels of sCD14, sCD163, I-FABP, and h-CRP were detected by ELISA, and other cytokines were measured by Luminex assay. The size and color density of circles are proportional to the correlation between 2 variables in VIR patients (D) or ART patients (E). Associations were evaluated using Spearman’s correlation test. P and Spearman’s ρ values are presented. Wilcoxon’s signed-rank test (A and C); **P < 0.01, ***P < 0.001.
The involvement of apoptosis of CD4+ T cells in HIV-1 infection has been intensively studied, while the involvement of pyroptosis is less clear. In contrast to apoptosis, pyroptosis is a proinflammatory process, which is thought to link CD4+ T cell depletion and chronic inflammation, two critical features of HIV-1 infection (3). To clarify the involvement of pyroptosis of CD4+ T cells in HIV disease progression, we investigated the potential correlation of caspase-1 activation in CD4+ T cells with clinical parameters or related biomarkers. The frequency of FLICA–caspase-1+ CD4+ T cells correlated positively with plasma HIV-1 viral load in the VIR group (Figure 2D; P = 0.0013, r = 0.4974). Moreover, the percentage of FLICA–caspase-1+ CD4+ T cells correlated positively with CD8+ T cell activation status (Figure 2E; P = 0.012, r = 0.3413). Finally, caspase-1 activation in CD4+ T cells correlated positively with plasma inflammatory markers, such as IL-6 (P < 0.001, r = 0.4997), IFN-γ–induced protein 10 (IP-10; P = 0.003, r = 0.3742), macrophage inflammatory protein 1β (MIP-1β; P < 0.001, r = 0.466), and high-sensitivity C-reactive protein (h-CRP) (P = 0.0068, r = 0.3429), and biomarkers of intestinal barrier dysfunction, such as soluble CD14 (sCD14) (P < 0.001, r = 0.4328; Figure 2F and Supplemental Figure 5). Interestingly, correlations between caspase-1 activation in CD4+ T cells and plasma inflammatory markers were not observed in ART patients (Figure 2G and Supplemental Figure 5), indicating that pyroptosis of CD4+ T cells is closely related to persistent inflammation in VIR patients, but not in ART patients.
Chronic HIV-1 infection induces GSDMD cleavage in LNs.
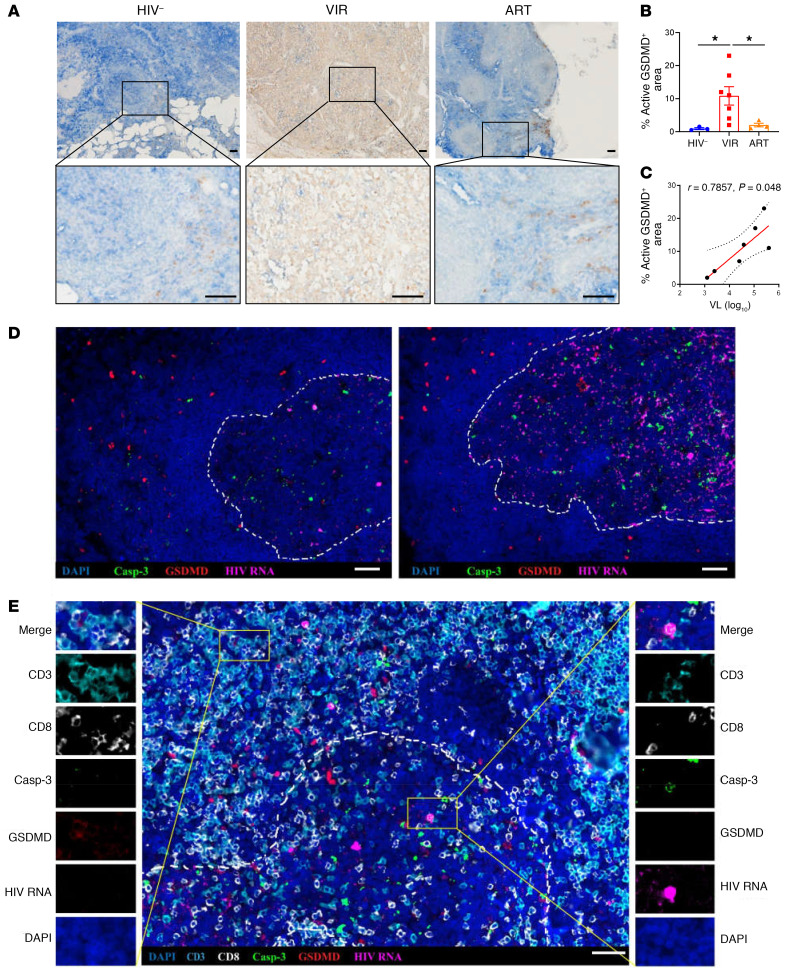
GSDMD, which has been recently identified as the central executioner of pyroptosis, functions through N-terminal insertion, oligomerization, and membrane pore formation after cleavage by activated inflammatory caspases (22–24). To identify cells with GSDMD activation, we used antibodies that target the cleavage site residues FLTD in the N-terminal region of active GSDMD; these antibodies specifically recognize the active N-terminal region, rather than the full-length form or the C-terminal region (25). To visualize the subanatomical distribution of CD4+ T cells expressing active GSDMD, LN sections from HIV-1–negative subjects, VIR patients, and ART patients were stained for active GSDMD and CD4. In 3 HIV-1–negative subjects, 0.93% ± 0.30% (mean ± SEM) of the LN sections were stained positively for active GSDMD compared with 10.8% ± 2.8% in 7 VIR patients (P = 0.0167). In the 4 ART patients, 2.0% ± 0.54% of the LN sections were stained positively for active GSDMD, which was markedly lower than the ratio observed in VIR patients (P = 0.0152) but slightly higher than that in the HIV-1–negative group (Figure 3, A and B). More importantly, for the 7 VIR patients, the percentage of active GSDMD–positive staining in LNs correlated positively with viral load (Figure 3C; P = 0.048, r = 0.7857), indicating that pyroptosis (represented by active GSDMD) might be triggered by HIV-1 replication.
Figure 3. Increased GSDMD activation in lymph nodes of HIV-1–infected patients.
(A) Representative examples of LNs from HIV-negative patients (n = 3) (left), VIR patients (n = 7) (middle), and ART patients (n = 4) (right) stained with anti–active GSDMD antibody shown at low (top panels) and high (bottom panels) magnification. Scale bars: 50 μm. (B) Quantification of active GSDMD was performed using ImageJ. Mann-Whitney U tests were used to evaluate statistically significant differences. (C) Spearman’s correlation between percentage of active GSDMD–positive area and HIV-1 viral load (VL) in VIR patients (n = 7). P and Spearman’s ρ values are presented. (D) Representative immunofluorescence microscopy images of LN sections from VIR patients hybridized with HIV-1 viral RNA and stained with anti–active caspase-3 and anti–active GSDMD antibodies, as indicated. (E) Representative immunofluorescence microscopy images of LN sections from VIR patients hybridized with HIV-1 viral RNA and stained with anti-CD3, anti-CD8, anti–active caspase-3, and anti–active GSDMD antibodies, as indicated. High-magnification fields show each fluorescence channel of representative images of pyroptotic and apoptotic cells at both sides of the main panel. Dashed lines represent B cell follicle limits. Scale bars: 50 μm. *P < 0.05.
We further used an in situ hybridization assay that combines the detection of viral RNA in fixed tissue sections with phenotypic analysis by using immunofluorescence and confocal microscopy. In VIR patients, HIV RNA–positive cells were broadly distributed in the LNs and were enriched in the follicles (Supplemental Figure 6A); however, cells expressing active GSDMD were mainly distributed in the T cell zone, in a bystander manner around HIV RNA–positive follicles (Figure 3D and Supplemental Figure 6A). In contrast, cells expressing active caspase-3 were preferentially located in the LN follicles (Figure 3D). Similar distribution patterns of active caspase-3 and active GSDMD were observed when another commercial GSDMD antibody was used (Supplemental Figure 6B). We used 6-color multiplex immunohistochemistry analysis to detect HIV RNA, active GSDMD, active caspase-3, CD3, and CD8 in LN tissue. We found that follicular CD4+ T cells with active viral replication underwent caspase-3 activation (Figure 3E, right), but pyroptotic CD4+ T cells located outside the follicle did not (Figure 3E, left). These data indicate that GSDMD activation is triggered externally by active viral replication in a bystander manner.
NLRP3 inflammasome activation contributes to caspase-1 activation in CD4+ T cells.
Subsequently, to dissect the upstream signals involved in caspase-1 activation in CD4+ T cells, we conducted quantitative reverse transcriptase PCR analysis of inflammasome-related gene expression in purified CD4+ T cells from PBMCs of HCs (n = 6), VIR patients (n = 22), ART patients (n = 11), and ECs (n = 3). Data showed that HIV-1 infection induced extensive activation of inflammasome-related genes (e.g., NLRP9, NLRP3, NLRP1, CASP1, IL1B; see Supplemental Table 4 for primer list), which was partially reversed by complete suppression of viremia in ART patients (Figure 4A). CD4+ T cells in ECs showed a pattern of inflammasome-related gene expression that was more similar to that in HCs than to that in VIR patients. Among these genes, NLRP3, CASP1, IL1B, and GSDMD showed the strongest correlation with CD4+ T cell caspase-1 activation, viral load, and CD4+ T cell counts in VIR patients (n = 22; Figure 4, B and C, and Supplemental Figure 7). In summary, these data highlight the potential role of the NLRP3 inflammasome in mediating caspase-1 activation and pyroptosis in CD4+ T cells.
Figure 4. Activation of caspase-1 is mediated by the NLRP3 inflammasome during chronic HIV-1 infection.
Peripheral blood CD4+ T cells of HCs (n = 6), VIR patients (n = 22), ART patients (n = 11), and ECs (n = 3) were enriched by magnetic cell sorting. (A) Heatmap showing inflammasome-related gene expression. (B) Heatmap of pairwise correlation of inflammasome-related gene expression with FLICA–caspase-1+ CD4+ T cell frequency, viral load, and CD4+ T cell counts. (C) Correlations of FLICA–caspase-1+ CD4+ T cell frequency with inflammasome-related gene expression. (D) PBMCs from VIR patients (n = 5) were cultured with vehicle control or MCC-950 (1 μM) for 6 hours. FLICA–caspase-1+ CD4+ T cell frequencies, IL-1β secretion, and lactate dehydrogenase (LDH) release were determined. (E) Left: Representative NLRP3 expression in CD4+ T cells of HCs (red) and VIR patients (blue). Right: MFI of NLRP3 expression in CD4+ T cells from HCs (n = 5) and VIR patients (n = 5). FMO, fluorescence minus one. (F) PBMCs from HCs (n = 4) and VIR patients (n = 4) were stimulated with 10 μM nigericin, 5 mM ATP, or HIV-1 viral particles (Bal-GFP, R5-tropic), respectively. FLICA–caspase-1+ CD4+ T cell frequencies were determined by flow cytometry, and supernatant IL-1β levels were detected by ELISA. (G) PBMCs from HCs (n = 4) were treated with conditioned medium (CM) for 48 hours; then HIV-1 viral particles (Bal-GFP, R5-tropic) were added and incubated for an additional 24 hours. (H) MFI of NLRP3 in CD4+ T cells (left) and FLICA–caspase-1+ CD4+ T cell frequencies (right). (I) Left: Representative MitoSOX (Thermo Fisher Scientific) signal of VIR patients (n = 10). Right: MFI of MitoSOX signals. (J) PBMCs from VIR patients (n = 5) were cultured in the presence of 1 mM N-acetylcysteine (NAC) or 20 μM MitoTEMPO. Frequency of FLICA–caspase-1+ CD4+ T cells determined by flow cytometry. Mann-Whitney U test (E); Wilcoxon’s signed-rank test (D, F, H, I, and J); *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether the NLRP3 inflammasome is functionally involved in caspase-1 activation in CD4+ T cells during HIV-1 infection, PBMCs isolated from VIR patients were incubated in the presence of vehicle control or MCC-950 (NLRP3 inhibitor; ref. 26). Notably, MCC-950 inhibited caspase-1 activation in CD4+ T cells, IL-1β secretion, and release of lactate dehydrogenase in the supernatant (Figure 4D).
NLRP3 inflammasome activation is a 2-step process, a priming step to license the cell and an activation step in which NLRP3 activation is triggered, followed by the assembly of the NLRP3 inflammasome and caspase-1–mediated pyroptosis (27). As shown in Figure 4A, priming of the NLRP3 inflammasome at the transcriptional level was observed in VIR patients. Additionally, intracellular staining showed that the translational level of NLRP3 was also significantly increased in VIR patients compared with the levels in HCs (Figure 4E and Supplemental Figure 8A). Thus, we hypothesized that these cells have been primed in the context of chronic HIV-1 infection, and can be readily activated by NLRP3 activators. To confirm this hypothesis, PBMCs isolated from HCs or VIR patients were stimulated with the known NLRP3 activators ATP and nigericin. Interestingly, CD4+ T cells from VIR patients, but not HCs, showed aberrant activation of caspase-1 and secretion of IL-1β following stimulation (Figure 4F). Next, we investigated the ability of viral particles to serve as an NLRP3 activator. Notably, incubation with viral particles selectively induced caspase-1 activation in CD4+ T cells (Figure 4F), but not in CD8+ T cells (Supplemental Figure 8B). Moreover, compared with PBMC-derived CD4+ T cells from HC donors primed with plasma from HCs, CD4+ T cells primed with plasma from VIR patients showed elevated NLRP3 expression and increased susceptibility to HIV particle–induced caspase-1 activation (Figure 4, G and H).
Mitochondrial dysfunction and the release of mitochondrial ROS (mtROS) are additional key upstream events implicated in NLRP3 activation (28–30). During chronic HIV-1 infection, mitochondrial depolarization and ROS production have also been described, and can be induced by Env-coreceptor interactions (31). As expected, in the setting of viral particle–induced activation, caspase-1–activated CD4+ T cells showed higher activation of mtROS (Figure 4I). Furthermore, inhibition of ROS production by N-acetylcysteine or MitoTEMPO (MilliporeSigma efficiently blocked caspase-1 activation in CD4+ T cells (Figure 4J). Overall, these data indicate that HIV-1 particle–induced ROS production drives NLRP3 inflammasome activation in CD4+ T cells during chronic HIV-1 infection.
Discussion
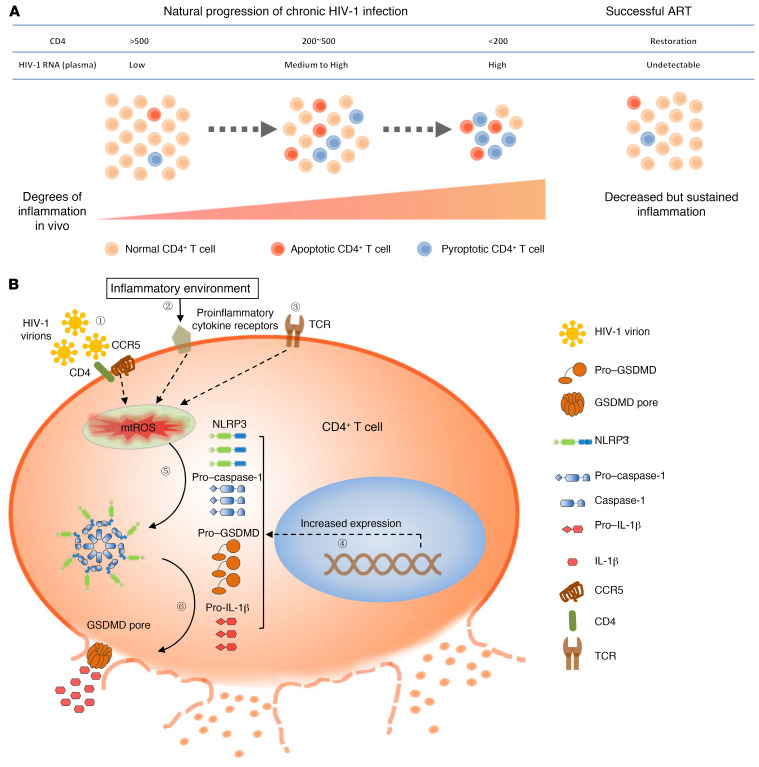
Recent studies have implicated pyroptosis in the pathogenesis of HIV-1 (3, 10, 11). In this study, we investigated the occurrence and clinical significance of pyroptosis and apoptosis in CD4+ T cells in HIV-1–infected patients. We showed that both pyroptosis and apoptosis of CD4+ T cells are associated with HIV-1 disease progression (Figure 5A). Moreover, we identified the NLRP3 inflammasome as a sensor that drives caspase-1 activation and pyroptosis in CD4+ T cells via a mechanism that depends on ROS production (Figure 5B). These findings not only provide a further understanding of the pathogenesis of HIV-1, but also highlight potential novel strategies for host immune reconstitution in HIV-1–infected patients.
Figure 5. A proposed schematic of NLRP3 inflammasome–dependent pyroptosis in CD4+ T cells in chronically HIV-1–infected patients.
(A) The pyroptotic and apoptotic levels of CD4+ T cells are elevated during chronic HIV-1 infection, showing close correlation with traditional biomarkers of disease, such as CD4+ T cell loss, increased viral load, and persistent inflammation. These symptoms are partially restored by effective ART. (B) The inflammatory milieu of the microenvironment in chronic HIV-1 infection sensitizes cells to a primed state, with upregulation of a series of inflammasome-related genes. Pyroptosis occurs through NLRP3 inflammasome activation driven by virus-dependent (signal 1) and/or -independent (signals 2 and 3) ROS production.
Different studies have indicated pyroptosis or apoptosis as the main cause of CD4+ T cell depletion during chronic HIV-1 infection (3, 4); however, the exact contribution of these processes to the clinical manifestations of CD4+ T cell loss in patients remains to be clarified. In this study, we showed that pyroptosis and apoptosis are not mutually exclusive, but rather act synergistically to orchestrate the fate of CD4+ T cells in a manner that is consistent during each stage of the disease. Notably, caspase-3 activation was preferentially restricted to follicles where there was active viral replication, whereas active GSDMD was mainly distributed around virus-producing follicles. These findings are consistent with prior studies suggesting that productively infected CD4+ T cells die via caspase-3–mediated apoptosis while bystander CD4+ T cells undergo caspase-1–mediated pyroptosis (9). Furthermore, the abundance of HIV DNA in caspase-1–activated circulating CD4+ T cells was comparable to that in the total CD4+ T cell population. These findings indicate that CD4+ T cell pyroptosis occurs in both peripheral blood and lymphoid tissues in a bystander manner.
Caspase-1–activated CD4+ T cells were detected in all memory subsets and, interestingly, were enriched in the Tem subset. Moreover, caspase-1–activated CD4+ T cells expressed higher levels of CCR5 than negative control cells. These data suggest that caspase-1–activated CD4+ T cells are highly permissive for HIV-1 infection. These data are consistent with the observation that the level of caspase-1 activation in CD4+ T cells correlated positively with viral load in VIR patients, and was significantly decreased after viral suppression in ART patients. However, other factors may also contribute to caspase-1 activation in CD4+ T cells. Despite effective viral control, ART patients showed significantly higher caspase-1 activation than HCs, and it is also worth noting that no obvious differences in caspase-1 activation were observed between patients in the VIR and ART groups with CD4+ T cell counts greater than 500 cells/μL. Since chronic inflammation and immune hyperactivation are hallmarks of HIV-1 infection (32), it is likely that these factors also contribute to caspase-1 activation in CD4+ T cells.
HIV-1 infection is characterized by development of profound immunodeficiency but sustained inflammation (32). Indeed, in VIR patients, the caspase-1 activation levels in CD4+ T cells correlated positively with inflammation and gut barrier dysfunction. Notably, an elevated level of IL-1β has been observed in tissues including cerebrospinal fluid, LNs, skin, and bronchoalveolar epithelium of HIV-infected patients (33, 34). In addition, there is evidence that inflammasome activation, which precedes IL-1β processing, occurs commonly in HIV-1 infection (35, 36). Abortive infection of resting lymphoid CD4+ T cells triggers pyroptosis through detection of incomplete viral DNA in the in vitro HLAC system, which models acute HIV-1 infection in humans (9–11). We found that NLRP3, but not other inflammasome sensors, is closely associated with caspase-1 activation in CD4+ T cells during chronic HIV-1 infection. This suggests that pyroptosis during acute infection modeled in the HLAC system may drive secondary pyroptosis through the release of intracellular mediators such as ATP and damage-associated molecular patterns (DAMPS) that are able to promote NLRP3 assembly and caspase-1 activation. It is worth noting that NLRP3 polymorphisms are associated with a higher risk for HIV-1 infection (37, 38). Studies on the potential involvement of the NLRP3 inflammasome in HIV-1 infection have focused mainly on monocytes (39–44). Here, we propose an alternative explanation for the pathological role of the CD4+ T cell–intrinsic NLRP3 inflammasome during HIV-1 infection.
HIV-1 infection induces apoptosis via different mechanisms, including the action of host factors (e.g., TNF-α, Fas ligand, and TRAIL; refs. 45, 46) and various viral factors (e.g., HIV-1 Tat, Vpr, and Nef) released from infected cells (47, 48). However, these mechanisms are thought to act on a broad spectrum of cell types and subtypes, and cannot explain the selective loss of CD4+ T cells during HIV-1 infection. The role of gp120 and gp41 Env protein in indirect cell death has also been a focus of interest (49), although the involvement of cell death pathways other than apoptosis remains to be clarified. The HIV-1 envelope (Env) glycoprotein has been shown to interact with bystander cells expressing CD4 and a coreceptor, CXCR4/CCR5. Similarly to Env glycoprotein–mediated bystander CD4+ T apoptosis, membrane damage during the hemifusion process might also induce massive mtROS production, similar to that observed in astrocytes and microglia (50, 51). Because the HIV-1 Env glycoprotein is localized on the surface of the virus and virus-infected cells, it is speculated that productively infected cells can also activate the NLRP3 inflammasome in bystander CD4+ T cells, as shown in the lymphoid tissues.
It was previously reported that peripheral blood–derived CD4+ T cells exhibit innate resistance to pyroptosis (52). Accordingly, we found that CD4+ T cells from HCs were resistant to stimulation by NLRP3 activators. However, cells from VIR patients were sensitive to stimulation of NLRP3 activators, which might be due to elevated expression of a series of inflammasome-related genes, resulting in higher pyroptosis-inducing potential (PIP). This speculation was reinforced by a recent study showing upregulation of NLRP3 and caspase-1 transcription in HIV-1–infected patients with poor immune recovery (53). This indicates that, in addition to CD4+ T cells, other cells, such as CD8+ T cells, are also susceptible to activators of the NLRP3 inflammasome. Increased susceptibility to pyroptosis has also been described for monocytes in HIV-1–infected patients (35). Considering the complexity of NLRP3 activation and regulation (27), it is highly possible that other factors also contribute to the PIP status, and this issue warrants further investigation.
The effect of antipyroptosis therapy in people living with HIV-1 is largely unknown. A recent study in monkeys demonstrated that treatment with a broad-spectrum caspase inhibitor is beneficial for immune preservation and viral control in acute SIV infection (54). Notably, the Canakinumab Antiinflammatory Thrombosis Outcome Study (CANTOS) recently demonstrated a decrease in the risk of cardiovascular events in patients with established cardiovascular disease after IL-1 inhibition with canakinumab, a monoclonal antibody targeting IL-1β (55). Moreover, canakinumab is also being investigated for effects on endothelial dysfunction and arterial inflammation (NCT02272946, ClinicalTrials.gov). However, we should be aware that IL-1β inhibition is not sufficient to prevent pyroptosis.
In summary, this study has revealed that NLRP3-dependent pyroptosis contributes as the dominant pathway in a synergistic association with apoptosis to CD4+ T cell loss in disease progression of HIV-1–infected patients. Current ART treatment can reduce, but not fully control, the pyroptosis of CD4+ T cells. Therefore, the NLRP3/caspase-1/GSDMD/IL-1β axis may provide novel therapeutic targets to improve the prognosis and outcomes of the disease.
Methods
Study patients.
In this study, we enrolled a cohort of 19 healthy controls; 80 VIR patients (with different CD4+ T cell counts) who were diagnosed with chronic HIV-1 infection at different stages of AIDS progression at Fifth Medical Center, General Hospital of the PLA (in general, the patients had started antiretroviral therapy 1 week after their diagnosis); 7 elite controllers (who were HIV antibody–positive, but with plasma HIV-1 RNA levels below the detectable limit without receiving ART); and 41 HIV-1–infected patients receiving successful ART (for more than 6 months with plasma HIV-1 RNA levels below the detectable limit) (Table 1). Exclusion criteria included coinfection with HBV and HCV, pregnancy, and moribund status. VIR patients with CD4+ T cell counts less than 250/μL are highly susceptible to opportunistic infections including tuberculosis and fungal infections. Detailed information on the patients is shown in Supplemental Table 1. LN biopsies were obtained from 7 VIR patients, 4 ART patients, and 3 HIV-negative individuals. The characteristics of the patients with LN biopsies are shown in Supplemental Table 2.
FLICA analysis of activated caspases.
Activated caspases were detected by FLICA staining using a FAM FLICA Caspase-1 kit (Bio-Rad) in accordance with the manufacturer’s instructions (17). In brief, cells were incubated with FLICA reagent for 1 hour at 37°C, and cells were washed for phenotypic staining.
Flow cytometric analysis of CD4+ T cell phenotypes.
For phenotypic staining, PBMCs were stained extracellularly using primary antibodies (1:200) for specific detection of surface markers for 30 minutes at 4°C. The cells were then washed with FACS buffer (1% FBS in PBS). For subsequent intracellular staining, the cells were permeabilized, fixed, and stained using the Permeabilization/Fixation Kit (00-5523-00, eBioscience) according to the manufacturer’s instructions. The cells were then incubated for 30 minutes at 4°C with primary antibodies (1:200) for specific detection of intracellular antigens. For GSDMD labeling, the anti–active GSDMD antibody was labeled with PE fluorochromes using commercial conjugation kits according to the manufacturer’s protocols (AB102918, Abcam). Information on the antibodies and reagents used in this study is listed in Supplemental Table 3. Samples were analyzed by flow cytometry using a BD Canto II flow cytometer (BD Biosciences) with FlowJo software (BD Biosciences). For t-distributed stochastic neighbor embedding (t-SNE) analysis, the FlowAI plugin was used to remove aberrant events. CD4+ T cells in all the samples were randomly downsampled to an equal number of cells using the DownSample plugin, and all the downsampled CD4+ T cells were concatenated into a single .fcs file. The concatenated file was exported and visualized by t-SNE plugin. t-SNE maps were generated using data for the following markers: CD45RA, CD27, and CCR7; with the following settings: iterations 300, perplexity 30, eta 7% of cell number, and theta 0.5. Other cell populations defined according to conventional gating strategy were overlaid onto the t-SNE map and color-coded as indicated. For image flow cytometry, cells were acquired using a 4-laser 12-channel imaging flow cytometer, Image StreamX MarkII (MilliporeSigma), under ×40 magnification. At least 10,000 single cells were acquired per sample, with debris and doublets excluded based on their area and aspect ratio. Data were analyzed using Inspire software. Cells in focus (using the “gradient RMS” feature for the bright-field image) and single cells (in a plot using “area” versus “aspect ratio”) were gated. Images were analyzed using image-based algorithms in ImageStream Data Exploration and Analysis Software (IDEAS 6.2.64.0, MilliporeSigma).
Luminex assays and ELISAs.
Cytokines were detected using a Luminex Bio-Plex ProTm Human Cytokine 8-Plex (including IL-6, IL-8, G-CSF, GM-CSF, IP-10, MCP-1, MIP-1β, and IFN-α2) with the Luminex 100 System (Bio-Rad) according to the manufacturer’s protocol. Plasma C-reactive protein (CRP), I-FABP, sCD14, and sCD163 were detected using ELISA kits, as described in Supplemental Table 3 (R&D Systems).
Cell sorting.
CD4+ T cells were isolated from PBMCs using the EasySep negative-selection Human CD4+ T Cell Enrichment Kit (STEMCELL Technologies) according to the manufacturer’s instructions. Purified CD4+ T cells were then stained with FLICA–caspase-1. Cell sorting was performed with a BD FACS Aria (BD Biosciences), and caspase-1–positive and caspase-1–negative cells were collected.
Detection of HIV DNA.
Total cellular DNA was extracted from cells using Qiagen QIAsymphony DNA Mini Kits (Qiagen). HIV DNA was quantified using a fluorescence-based real-time SUPBIO HIV Quantitative Detection Kit. The quantification range was 10 to 5 × 106 copies per 106 PBMCs.
RNAscope.
The RNAscope assay was carried out according to a previously described method (56). Briefly, after H2O2 treatment and protease digestion, LN sections were incubated overnight at 40°C with HIV clade B antisense probe (catalog 317691, Advanced Cell Diagnostics). The RNAscope Multiplex Fluorescent Detection Kit (Advanced Cell Diagnostics) was used for tyramide signaling amplification (TSA) plus Cy3 or FITC immunofluorescence detection. Images were obtained with a Leica TCS SP8 STED confocal microscope and PerkinElmer Vectra 3.0.
Immunofluorescence and immunohistochemistry assays.
Immunofluorescence and immunohistochemistry assays were carried out as previously described (57). Briefly, fresh tissues were fixed in 4% neutral-buffered formalin and embedded in paraffin. Sections (4 μm) were stained with anti-CD4, anti-GSDMD, and the appropriate secondary antibodies. Images were captured with an inverted fluorescence microscope (PerkinElmer). Quantification of GSDMD was performed using ImageJ software (NIH) after determination of the average optical density of positive staining (magnification ×400, 10 random view fields). Multiplex immunofluorescence staining of LNs was performed using a PANO 7-plex IHC kit (Panovue, Beijing, China), following the manufacturer’s protocol (58). In brief, after RNAscope assay, different primary antibodies, including anti-CD3, anti-CD8, anti–caspase-3, and anti-GSDMD, were sequentially applied, followed by incubation of HRP-conjugated secondary antibody and tyramide signal amplification reagent. Slides were counterstained with DAPI. Multispectral images were obtained on a Mantra System (PerkinElmer).
Quantitative PCR.
Total cellular RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific). RNA was then reverse-transcribed to cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). mRNA expression was examined by quantitative PCR using the SYBR Green Master Mix (Applied Biosystems) with a CF96 real-time PCR system (Bio-Rad). Primers used for amplification of the target genes are listed in Supplemental Table 4. Data represent fold changes in expression normalized to the IPO8 control.
Statistics.
Data represent the mean ± SEM, and statistical analyses were performed with GraphPad Prism version 8.0 (GraphPad Software). The correlations between variables were evaluated by a Spearman rank correlation test. Correlation matrices were generated and visualized using R with the hmisc and corrplot packages. Wilcoxon’s signed-rank test was used for matched pairs. Mann-Whitney U tests were used for comparison between 2 groups. P values less than 0.05 were considered to indicate statistical significance.
Study approval.
This study was approved by the Institutional Review Board and Ethics Committee of Fifth Medical Center of Chinese PLA General Hospital, and written informed consent was provided by each participant in accordance with the Declaration of Helsinki. Patients were managed in strict accordance with the associated guidelines and regulations.
Author contributions
FSW and CZ designed the study and wrote the manuscript. CZ, JWS, HHH, and XF designed and performed most experiments and analyzed data. The clinical samples and data were contributed by LH, JND, BT, CXY, and QWZ. KW and FS developed active GSDMD antibody. TY provided support for the experiment to quantify viral RNA. JL and MJZ performed flow cytometry on HIV samples. LFW, JYZ, RNX, YMJ, MS, FS, and RPS contributed to scientific planning. Intellectual input was provided by all authors. The order of the co–first authors was assigned on the basis of the relative contributions of the individuals.
Supplementary Material
Acknowledgments
We thank Kai Deng (Sun Yat-sen University), Lishan Su (University of North Carolina at Chapel Hill), and Liguo Zhang (Institute of Biophysics, Chinese Academy of Sciences) for the insightful critiques of the manuscript. We also thank Songshan Wang, Chunbao Zhou, Jinhong Yuan, Jiehua Jin, and Wenjing Cao for their technical support. This study was supported by the Innovation Groups of the National Natural Science Foundation of China (grant 81721002), the National Natural Science and Technology Major Project (grant 2018ZX10302104-002), and the National Natural Science Foundation of China (grants 81772185 and 81901617).
Version 1. 03/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(6):e138861.https://doi.org/10.1172/JCI138861.
Contributor Information
Chao Zhang, Email: zhangch302@163.com.
Jin-Wen Song, Email: songjinwenchina@yeah.net.
Hui-Huang Huang, Email: hhh302@163.com.
Xing Fan, Email: fanxing302@aliyun.com.
Lei Huang, Email: huangleiwa@sina.com.
Jian-Ning Deng, Email: dengjianning@126.com.
Bo Tu, Email: toby.sky@hotmail.com.
Kun Wang, Email: wangkun@nibs.ac.cn.
Jing Li, Email: jingl7820@163.com.
Ming-Ju Zhou, Email: zhoumj89@163.com.
Cui-Xian Yang, Email: ycx484@126.com.
Qi-Wen Zhao, Email: zhaoqiwen1990@hotmail.com.
Tao Yang, Email: y_t_0321@163.com.
Li-Feng Wang, Email: biowanglf@163.com.
Ji-Yuan Zhang, Email: uniquezjy@163.com.
Ruo-Nan Xu, Email: xuruonan2004@aliyun.com.
Yan-Mei Jiao, Email: jiaoyanmei@gmail.com.
Ming Shi, Email: shiming302@sina.com.
Feng Shao, Email: shaofeng@nibs.ac.cn.
Rafick-Pierre Sékaly, Email: rafick.sekaly@emory.edu.
Fu-Sheng Wang, Email: fswang302@163.com.
References
- 1.Ghosn J, et al. HIV. Lancet. 2018;392(10148):685–697. doi: 10.1016/S0140-6736(18)31311-4. [DOI] [PubMed] [Google Scholar]
- 2.Finkel T, et al. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat Med. 1995;1(2):129–134. doi: 10.1038/nm0295-129. [DOI] [PubMed] [Google Scholar]
- 3.Doitsh G, Greene WC. Dissecting how CD4 T cells are lost during HIV infection. Cell Host Microbe. 2016;19(3):280–291. doi: 10.1016/j.chom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg H, Joshi A. Host and viral factors in HIV-mediated bystander apoptosis. Viruses. 2017;9(8):237. doi: 10.3390/v9080237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang D, et al. The molecular machinery of regulated cell death. Cell Res. 2019;29(5):347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantaleo G, Fauci AS. Apoptosis in HIV infection. Nat Med. 1995;1(2):118–120. doi: 10.1038/nm0295-118. [DOI] [PubMed] [Google Scholar]
- 8.Gougeon ML, Montagnier L. Apoptosis in AIDS. Science. 1993;260(5112):1269–1270. doi: 10.1126/science.8098552. [DOI] [PubMed] [Google Scholar]
- 9.Doitsh G, et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143(5):789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doitsh G, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monroe KM, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343(6169):428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galloway NL, et al. Cell-to-cell transmission of HIV-1 is required to trigger pyroptotic death of lymphoid-tissue-derived CD4 T cells. Cell Rep. 2015;12(10):1555–1563. doi: 10.1016/j.celrep.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, et al. Modeling the slow CD4+ T cell decline in HIV-infected individuals. PLoS Comput Biol. 2015;11(12):e1004665. doi: 10.1371/journal.pcbi.1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke R, et al. On the death rate of abortively infected cells: estimation from simian-human immunodeficiency virus infection. J Virol. 2017;91(18):e00352–17. doi: 10.1128/JVI.00352-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J, et al. Longitudinal changes in plasma Caspase-1 and Caspase-3 during the first 2 years of HIV-1 infection in CD4Low and CD4High patient groups. PLoS One. 2015;10(3):e0121011. doi: 10.1371/journal.pone.0121011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai R, et al. Caspase-1 activity in CD4 T cells is downregulated following antiretroviral therapy for HIV-1 infection. AIDS Res Hum Retroviruses. 2017;33(2):164–171. doi: 10.1089/aid.2016.0234. [DOI] [PubMed] [Google Scholar]
- 17.Bedner E, et al. Activation of caspases measured in situ by binding of fluorochrome-labeled inhibitors of caspases (FLICA): correlation with DNA fragmentation. Exp Cell Res. 2000;259(1):308–313. doi: 10.1006/excr.2000.4955. [DOI] [PubMed] [Google Scholar]
- 18. Mazumder S, et al. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. In: Mor G, et al., eds. Apoptosis and Cancer. Springer; 2008:13–21. [DOI] [PubMed] [Google Scholar]
- 19.Miao EA, et al. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243(1):206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Plasma membrane changes during programmed cell deaths. Cell Res. 2018;28(1):9–21. doi: 10.1038/cr.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groux H, et al. Activation-induced death by apoptosis in CD4+ T cells from human immunodeficiency virus-infected asymptomatic individuals. J Exp Med. 1992;175(2):331–340. doi: 10.1084/jem.175.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535(7610):111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 25.Wang K, et al. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell. 2020;180(5):941–955.e20. doi: 10.1016/j.cell.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 26.van Hout GP, et al. The selective NLRP3-inflammasome inhibitor MCC950 reduces infarct size and preserves cardiac function in a pig model of myocardial infarction. Eur Heart J. 2016;38(11):828–836. doi: 10.1093/eurheartj/ehw247. [DOI] [PubMed] [Google Scholar]
- 27.Swanson KV, et al. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19(8):477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi H, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol. 2016;17(3):250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10(3):210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 30.Zhou R, et al. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov AV, et al. Oxidative stress during HIV infection: mechanisms and consequences. Oxid Med Cell Longev. 2016;2016:8910396. doi: 10.1155/2016/8910396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deeks SG. HIV infection, inflammation, immunosenescence, and aging. Annu Rev Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tufa DM, et al. Brief Report: HIV-1 infection results in increased frequency of active and inflammatory SlanDCs that produce high level of IL-1β. J Acquir Immune Defic Syndr. 2016;73(1):34–38. doi: 10.1097/QAI.0000000000001082. [DOI] [PubMed] [Google Scholar]
- 34.Brabers N, Nottet H. Role of the pro-inflammatory cytokines TNF-alpha and IL-1beta in HIV-associated dementia. Eur J Clin Invest. 2006;36(7):447–458. doi: 10.1111/j.1365-2362.2006.01657.x. [DOI] [PubMed] [Google Scholar]
- 35.Ahmad F, et al. Evidence of inflammasome activation and formation of monocyte-derived ASC specks in HIV-1 positive patients. AIDS. 2018;32(3):299–307. doi: 10.1097/QAD.0000000000001693. [DOI] [PubMed] [Google Scholar]
- 36.Lisco A, et al. Identification of rare HIV-1-infected patients with extreme CD4+ T cell decline despite ART-mediated viral suppression. JCI Insight. 2019;4(8):127113. doi: 10.1172/jci.insight.127113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pontillo A, et al. A 3′ UTR SNP in NLRP3 gene is associated with susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr. 2010;54(3):236–240. doi: 10.1097/QAI.0b013e3181dd17d4. [DOI] [PubMed] [Google Scholar]
- 38.Pontillo A, et al. Polymorphisms in inflammasome’ genes and susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr. 2012;59(2):121–125. doi: 10.1097/QAI.0b013e3182392ebe. [DOI] [PubMed] [Google Scholar]
- 39.Chivero ET, et al. HIV-1 Tat primes and activates microglial NLRP3 inflammasome-mediated neuroinflammation. J Neurosci. 2017;37(13):3599–3609. doi: 10.1523/JNEUROSCI.3045-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh JG, et al. Rapid inflammasome activation in microglia contributes to brain disease in HIV/AIDS. Retrovirology. 2014;11:35. doi: 10.1186/1742-4690-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez JC, et al. HIV-1 induces the first signal to activate the NLRP3 inflammasome in monocyte-derived macrophages. Intervirology. 2014;57(1):36–42. doi: 10.1159/000353902. [DOI] [PubMed] [Google Scholar]
- 42.Mamik MK, et al. HIV-1 viral protein R activates NLRP3 inflammasome in microglia: implications for HIV-1 associated neuroinflammation. J Neuroimmune Pharmacol. 2017;12(2):233–248. doi: 10.1007/s11481-016-9708-3. [DOI] [PubMed] [Google Scholar]
- 43.Guo H, et al. HIV-1 infection induces interleukin-1β production via TLR8 protein-dependent and NLRP3 inflammasome mechanisms in human monocytes. J Biol Chem. 2014;289(31):21716–21726. doi: 10.1074/jbc.M114.566620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He X, et al. NLRP3-dependent pyroptosis is required for HIV-1 gp120-induced neuropathology. Cell Mol Immunol. 2020;17(3):283–299. doi: 10.1038/s41423-019-0260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gandhi RT, et al. HIV-1 directly kills CD4+ T cells by a Fas-independent mechanism. J Exp Med. 1998;187(7):1113–1122. doi: 10.1084/jem.187.7.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herbeuval JP, et al. CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood. 2005;106(10):3524–3531. doi: 10.1182/blood-2005-03-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindler M, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125(6):1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 48.Westendorp MO, et al. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature. 1995;375(6531):497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- 49.Perfettini J, et al. Mechanisms of apoptosis induction by the HIV-1 envelope. Cell Death Differ. 2005;12(S1):916–923. doi: 10.1038/sj.cdd.4401584. [DOI] [PubMed] [Google Scholar]
- 50.Dröge W, et al. Oxidant-antioxidant status in human immunodeficiency virus infection. Methods Enzymol. 1994;233:594–601. doi: 10.1016/s0076-6879(94)33062-x. [DOI] [PubMed] [Google Scholar]
- 51.Kaul M, et al. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ. 2005;12(suppl 1):878–892. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- 52.Muñoz-Arias I, et al. Blood-derived CD4 T cells naturally resist pyroptosis during abortive HIV-1 infection. Cell Host Microbe. 2015;18(4):463–470. doi: 10.1016/j.chom.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bandera A, et al. The NLRP3 inflammasome is upregulated in HIV-infected antiretroviral therapy-treated individuals with defective immune recovery. Front Immunol. 2018;9:214. doi: 10.3389/fimmu.2018.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laforge M, et al. The anti-caspase inhibitor Q-VD-OPH prevents AIDS disease progression in SIV-infected rhesus macaques. J Clin Invest. 2018;128(4):1627–1640. doi: 10.1172/JCI95127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ridker PM, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 56.Rose R, et al. HIV maintains an evolving and dispersed population in multiple tissues during suppressive combined antiretroviral therapy in individuals with cancer. J Virol. 2016;90(20):8984–8993. doi: 10.1128/JVI.00684-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiao Y-M, et al. Dichotomous roles of programmed cell death 1 on HIV-specific CXCR5+ and CXCR5- CD8+ T cells during chronic HIV infection. Front Immunol. 2017;8:1786. doi: 10.3389/fimmu.2017.01786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen YP, et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 2020;30(11):1024–1042. doi: 10.1038/s41422-020-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.