Abstract
Intratumor heterogeneity is an important mediator of poor outcomes in many cancers, including breast cancer. Genetic subclones frequently contribute to this heterogeneity; however, their growth dynamics and interactions remain poorly understood. PIK3CA and HER2 alterations are known to coexist in breast and other cancers. Herein, we present data that describe the ability of oncogenic PIK3CA mutant cells to induce the proliferation of quiescent HER2 mutant cells through a cell contact–mediated mechanism. Interestingly, the HER2 cells proliferated to become the major subclone over PIK3CA counterparts both in vitro and in vivo. Furthermore, this phenotype was observed in both hormone receptor–positive and –negative cell lines, and was dependent on the expression of fibronectin from mutant PIK3CA cells. Analysis of human tumors demonstrated similar HER2:PIK3CA clonal dynamics and fibronectin expression. Our study provides insight into nonrandom subclonal architecture of heterogenous tumors, which may aid the understanding of tumor evolution and inform future strategies for personalized medicine.
Keywords: Cell Biology, Oncology
Keywords: Breast cancer, Molecular biology, Molecular genetics
Introduction
Breast cancer is the most common cancer among women worldwide and the second leading cause of cancer related deaths (1). Like many other cancers, breast cancer has been shown to be extremely heterogenous. The presence of multiple clones within a single tumor is common and may be functionally important (2–5). Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA) and Erb-B2 receptor tyrosine kinase/human epidermal growth factor receptor 2 (ERBB2/HER2) are altered in approximately 30% and 20% of breast cancers, respectively (6, 7). In some patients, PIK3CA mutations have been shown to cooccur with HER2 alterations, either with amplification/overexpression of WT HER2 or less commonly with HER2 somatic mutations (3, 8, 9). Prior studies have shown that PIK3CA and HER2 missense mutations within the same cell cooperate and lead to increased biochemical signaling (10). Additionally, past reports have demonstrated that in some human breast cancers, subclonal populations with genetic alterations in PIK3CA or HER2 are present in separate cancer cells (3). Other studies have suggested that intercellular interactions between distinct tumor cells may lead to growth advantage for the tumor as an overall mass. However, common human breast cancer mutations were not queried in these studies and soluble mediators of signaling were implicated as possible mechanisms (11–17). We hypothesized that gene-specific interactions between cells carrying a PIK3CA mutation and those carrying a HER2 mutation could result in phenotypic transformations and may have important implications for tumor evolution.
Results
Clonal dynamics of HER2:PIK3CA cocultures.
To test this hypothesis, we used MCF-10A, a spontaneously immortalized nontumorigenic breast epithelial cell line, which is hormone receptor and HER2 receptor negative and epidermal growth factor–dependent (EGF-dependent) for proliferation (18). Our lab has previously demonstrated that isogenic incorporation of PIK3CA mutations into MCF-10A results in EGF growth independence, a property that is associated with increased malignant potential (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI143557DS1) (19, 20). Conversely, MCF-10A cells with an isogenic HER2 L755S missense mutation retain the parental characteristic of EGF dependence (10). These properties allowed us to design a system in which we could evaluate interactions between 2 genetically distinct subclones — one that is quiescent in the absence of exogenous growth factor and one that proliferates in the same conditions.
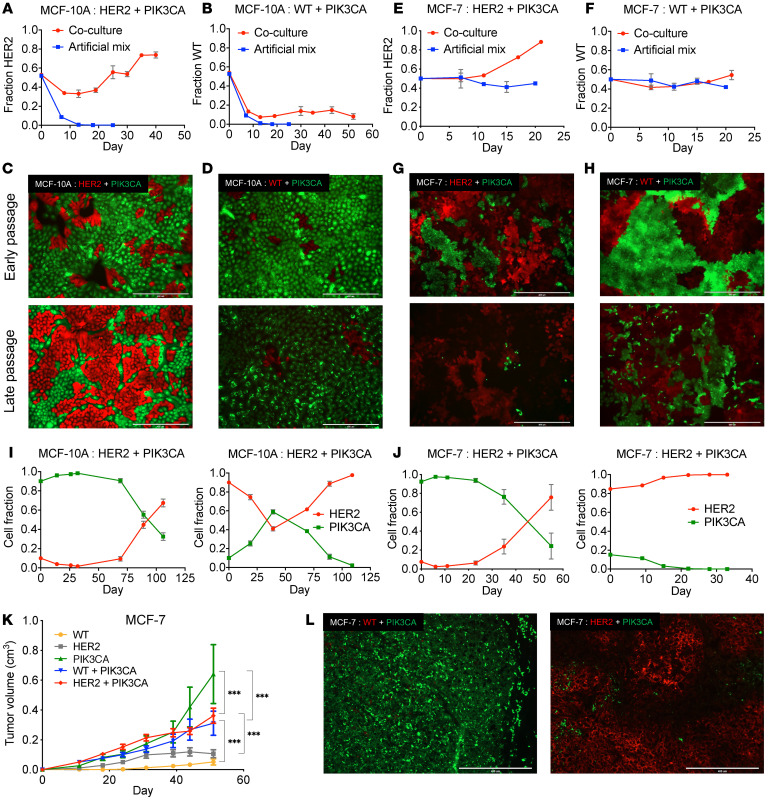
To study these interactions, we cocultured MCF-10A cells containing a HER2 L755S mutation with isogenic cells harboring a PIK3CA E545K mutation in the absence of exogenous EGF, as described in the experimental design (Supplemental Figure 1). Both the HER2 L755S and PIK3CA E545K variants are activating mutations that have oncogenic properties in the absence of amplification. Droplet digital PCR (ddPCR) and stable integration of fluorescent proteins were used to quantify and visualize the differences in growth patterns. HER2 mutant cells, which normally undergo G1 cell-cycle arrest in the absence of EGF, actively divided when cocultured with the PIK3CA mutant cells. Intriguingly, with continued passage HER2 cells outgrew their PIK3CA counterparts and became the dominant population in coculture despite the absence of EGF. This effect appeared gene specific, as neither WT MCF-10As control cell lines carrying a silent mutation (21), nor AKT1 E17K mutants, another EGF-dependent cell line (22), gained EGF independence when cocultured with the PIK3CA mutants (Figure 1, A–D and Supplemental Figure 2A). Similarly, MCF-10As containing the NOTCH1 P448fs mutation, which has also been shown to lead to EGF independence (our unpublished observations), failed to induce EGF independence in the HER2 mutants (Supplemental Figure 2B). Strikingly, this phenomenon was also observed when skewing coculture seeding ratios in favor of, or against, the HER2 mutant cells (90% or 10%, respectively, Figure 1I). These results suggest that subclonal architecture is nonrandom and selective pressures on a heterogenous population of cancer cells will establish a consistent hierarchy of genetic subclones.
Figure 1. Interactions between HER2 and PIK3CA mutant cells result in altered, nonrandom growth dynamics.
(A) Coculture of MCF-10A cells containing a HER2 mutation and those containing a PIK3CA mutation. Genomic DNA was collected upon confluency for analysis by ddPCR. Artificial mixes were obtained as described in Supplemental Figure 1. The fraction of HER2 cells was plotted as the mean ± SEM (n = 3). (B) Coculture of MCF-10A WT cells and those containing a PIK3CA mutation carried out as described in A. (C) Representative images of cocultures using fluorescently labeled HER2 mutants (red) and PIK3CA mutants (green) and (D) cocultures using fluorescently labeled MCF-10A WT cell (red) and PIK3CA mutants (green). Top image is early passage (approximately 10 days) and bottom image is late passage (approximately 40 days for MCF-10A and 25 days for MCF-7). Scale bars: 400 μM. (E) Coculture assays of MCF-7 background cells containing a HER2 mutation or (F) corrected WT MCF-7 cells, as performed in A. (G) Representative images of cocultures using fluorescently labeled MCF-7 HER2 mutants (red) and PIK3CA mutants (green) and (H) cocultures using fluorescently labeled MCF-7 WT cells (red) and PIK3CA mutants (green). Scale bars: 400 μM. (I) Cocultures of HER2 and PIK3CA mutant cells with skewed starting ratios in the MCF-10A and (J) MCF-7 backgrounds. Cellular fractions were determined by ddPCR (mean ± SEM, n = 2). (K) Xenograft study of MCF-7 panel (WT, HER2, and PIK3CA mutation). Subcutaneous injection of athymic nude mice with estrogen supplementation. Cells were inoculated individually (n = 5) and in coinoculations of HER2:PIK3CA or WT:PIK3CA (n = 10). ***P < 0.001 by 2-way ANOVA followed by Bonferroni multiple comparisons test. (L) Representative fluorescence images of cryosectioned coinoculated tumors. Left, WT (red) and PIK3CA mutant (green); right, HER2 mutant (red) and PIK3CA mutant (green). Scale bars: 400 μM.
We sought to validate these findings and examine the HER2:PIK3CA clonal dynamics in vivo by using MCF-7, a tumorigenic hormone receptor–positive, HER2-negative human breast cancer epithelial cell line (23) that natively harbors a PIK3CA E545K mutation. To create an analogous system to the MCF-10A cell line, the HER2 L755S mutation was incorporated into an isogenically PIK3CA corrected MCF-7 cell line (WT for PIK3CA) (24). The MCF-7 cells are not growth factor dependent, therefore the HER2 and PIK3CA mutant cells proliferate at similar rates in all growth conditions. However, consistent with the MCF-10A cells, the HER2 mutant cells showed increased proliferation and became the predominant population in coculture when grown with parental MCF-7 cells containing the PIK3CA mutation. This was in stark contrast to the corrected PIK3CA WT MCF-7 cells, which showed similar patterns of growth in coculture or in isolation (Figure 1, E–H). Consistent with prior results, regardless of initial seeding ratios, the HER2 mutants evolved as the predominant population when cocultured with mutant PIK3CA cells (Figure 1J).
In order to evaluate these interactions in vivo, mice were inoculated with each cell line separately or coinoculated with an equal number of either the HER2:PIK3CA mutants or the WT:PIK3CA mutants. Coinoculums of both HER2:PIK3CA and WT:PIK3CA grew better than respective HER2 and WT controls that were grown in isolation. The overall tumor volume for the PIK3CA mutant control exceeded those of the other groups (Figure 1K). On average, the WT:PIK3CA tumors were composed predominantly of PIK3CA mutant cells. Conversely, the HER2:PIK3CA coinoculated tumors were predominately HER2 mutant cells (Supplemental Figure 3). Fluorescence imaging of the tumors validated these findings (Figure 1L). These in vivo results provided additional evidence that corroborated our in vitro findings.
Cell contact interactions mediated by fibronectin.
We next addressed how mechanistically these 2 subclonal populations interacted on a cellular level. Live-cell fluorescence time-lapse imaging was used to provide an initial understanding. The videos (Supplemental Video 1) and representative images (Figure 2A) revealed the requirement of cell-cell contact for this phenomenon to occur. HER2 mutant cells (labeled red) that were in contact with the PIK3CA mutant cells (labeled green) underwent cell division (white triangles), while those devoid of contact remained quiescent (yellow triangles) despite being within a few microns of the nearest PIK3CA mutant cells. We observed similar findings when cells were cultured in 3D basement membrane (Supplemental Figure 4 and Supplemental Video 2). Consistent with these results, we did not find significant contributions from paracrine factors in trans-well coculture experiments (Supplemental Figure 5). This suggested that the mechanism driving this phenotype required extremely close proximity or direct contact between cells.
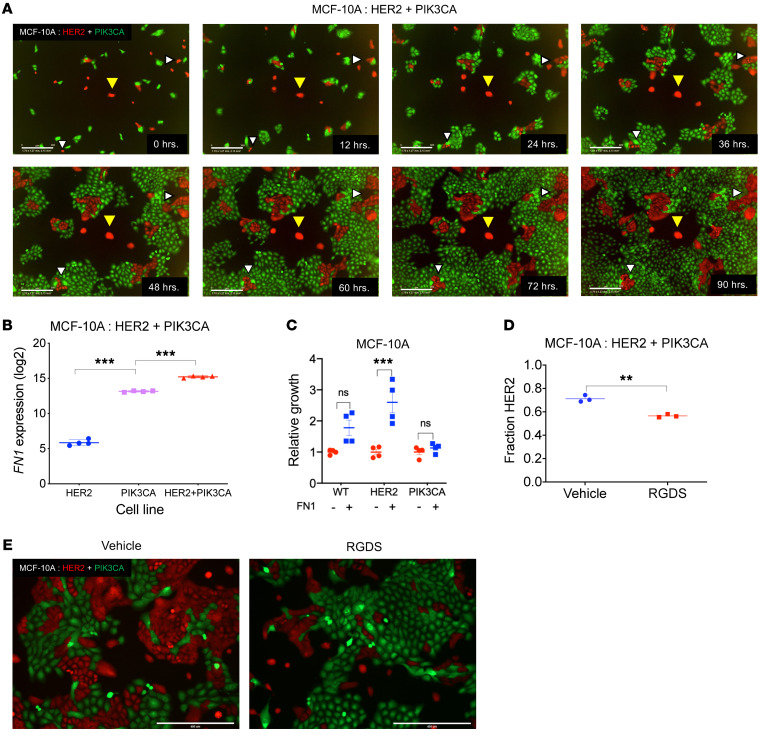
Figure 2. A contact-mediated mechanism involving fibronectin drives interactions between HER2 and PIK3CA mutant cells.
(A) Representative images taken in 12-hour intervals from time-lapse imaging of HER2 (red) and PIK3CA (green) cocultures in MCF-10A background using the IncuCyte live-cell analysis system. White triangles indicate dividing HER2 mutant cells, yellow triangles indicate quiescent HER2 mutant cells. Scale bars: 300 μM. Full video available in Supplemental Video 1. (B) Expression levels of FN1 in HER2 mutants, PIK3CA mutants and in coculture as determined by microarray. For full expression data, see Supplemental Figure 6. n = 4. ***P < 0.001 by 1-way ANOVA followed by Bonferroni multiple comparisons test. (C) Relative growth of WT, HER2 mutants, and PIK3CA mutants in the absence of EGF ± fibronectin (5 μg/mL). Cell culture plates were coated with fibronectin or vehicle and cell counts were performed after 6–8 days. n = 4. ***P < 0.001 by 1-way ANOVA followed by Bonferroni multiple comparisons test. (D) HER2:PIK3CA cocultures were treated with vehicle or an RGDS competitive inhibitor of fibronectin/integrin binding. Cellular fraction was determined by ddPCR. n = 3. **P < 0.01 by 1-way ANOVA followed by Bonferroni multiple comparisons test. (E) Representative images from each condition are shown (HER2, red; PIK3CA, green).
In pursuit of identifying the molecular agent mediating these subclonal interactions, we performed a transcriptome-wide analysis on the HER2 and PIK3CA mutant cells grown in isolation and in coculture. Genes that were differentially regulated when comparing the coculture to the HER2 mutant controls were investigated further (Supplemental Figure 6). FN1, coding for the extracellular matrix glycoprotein fibronectin, was the most upregulated gene independently and when normalized to controls (Figure 2B). Unlike other genes, whose differential expression could be attributed to the presence of PIK3CA cells in the bulk coculture, the expression of FN1 was elevated to an even greater extent in the coculture than in either control cell line. Elevated expression of fibronectin in HER2:PIK3CA cocultures was confirmed by immunofluorescence (Supplemental Figure 7). These alterations suggested that fibronectin could be playing an important role in facilitating the observed coculture phenotype.
To study the effect of extracellular fibronectin on the growth of the HER2 mutant cells, we plated WT, HER2, and PIK3CA mutant cells onto uncoated and fibronectin-coated plates in the absence of EGF. HER2 mutant cells showed a significant (> 2.5 fold) increase when plated onto fibronectin. The PIK3CA mutants and WT cells did not display any significant change in growth rate (Figure 2C). To further substantiate this finding, we transiently inhibited the cellular binding of fibronectin in cocultures by competitive inhibition of fibronectin-integrin interactions using a synthetic Arg-Gly-Asp-Ser (RGDS) peptide (25, 26). ddPCR analysis and fluorescence imaging revealed a 15% reduction (P < 0.01) in the fraction of HER2 mutant cells in cocultures that were subject to the RGDS peptide treatment (Figure 2, D and E). It is important to note that the modest, although statistically significant, reduction in the growth of HER2 mutants may be attributed to the short duration of the RGDS peptide treatment.
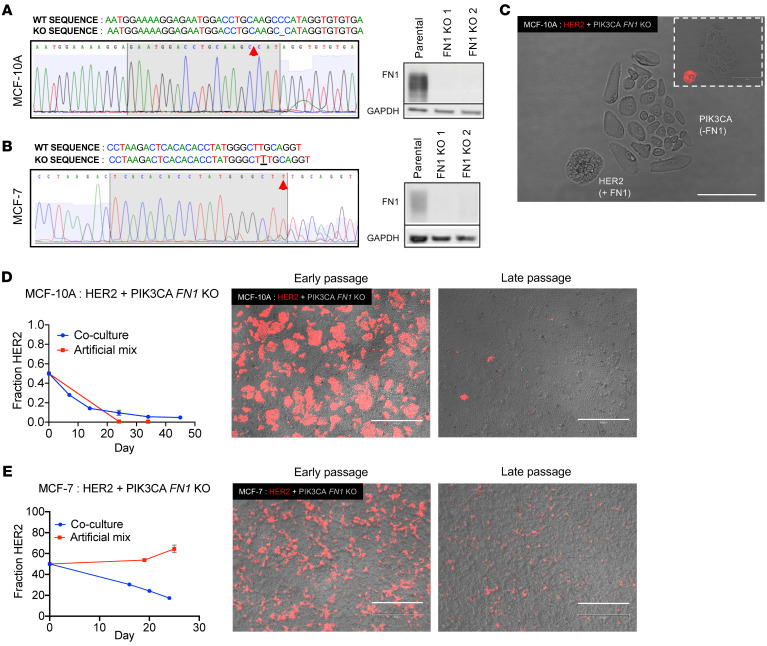
In order to definitively evaluate the role of fibronectin, we used the CRISPR-Cas 9 system to knock out FN1 in both the MCF-10A and the MCF-7 PIK3CA mutant cells. Western blots confirmed the absence of fibronectin in both cell lines (Figure 3, A and B). Disruption of FN1 did not affect the growth or morphology of the PIK3CA mutants (Supplemental Figure 8). Microscopic images showed an occasional absence of contiguous membrane contact in cells that lacked fibronectin in comparison with cells that retained expression in coculture (Figure 3C), confirming the necessity of FN1 for mediating this phenotype. Importantly, FN1-null PIK3CA mutants did not support the growth of HER2 mutants in coculture. Both MCF-10A (Figure 3D) and MCF-7 (Figure 3E) HER2 mutant cells exhibited a reversal of the growth induction phenotype when cocultured with FN1 knockout cells. These results confirmed that fibronectin is an essential component of HER2:PIK3CA interactions that lead to the observed phenotype of clonal hierarchy.
Figure 3. ECM protein fibronectin is a necessary mediator of HER2:PIK3CA clonal dynamics in coculture.
(A and B) Sanger sequencing traces and Western blots demonstrating the knockout of FN1 in PIK3CA mutants in (A) MCF-10A and (B) MCF-7 backgrounds. Red arrows indicate alterations. (C) Imaging of MCF-10A PIK3CA mutant cells with knocked-out fibronectin and HER2 mutant cells with WT fibronectin in coculture demonstrating differences in cellular morphology in the absence of FN1. Identification of HER2 versus PIK3CA cells was accomplished using fluorescence imaging of HER2 cells tagged with tdTomato (inset). Scale bars: 150 μM. (D) Coculture of HER2 mutant cells with FN1 knockout PIK3CA mutants in the MCF-10A and (E) MCF-7 backgrounds. Cellular fraction determined by ddPCR. n = 2. Representative fluorescence images at early and late passage are shown (HER2, red; FN1 knockout PIK3CA, untagged). Early and late passages indicate 7 and 45 days, respectively, for MCF-10As and 16 and 24 days for MCF-7s. Scale bars: 800 μM.
Fibronectin is known to interact with integrin α5β1 (27). Fibronectin-integrin interactions have been shown to stimulate the activity of MAP kinases like ERK1 and ERK2 downstream (28, 29). ERK activation, in turn, is known to induce cyclin D1 expression leading to cell cycle progression (30, 31). To test if this could be the potential downstream signaling that leads to the HER2 mutant growth in coculture, we flow sorted the HER2 mutants that were grown in coculture with the PIK3CA mutants and performed a gene expression analysis using a PCR Array (Supplemental Figure 9). In comparison to the HER2 mutants grown by themselves, those in coculture showed a significant increase in MAP2K1, which encodes a mitogen-activated protein kinase kinase known to activate ERK1 and ERK2. Additionally, they also showed a significant upregulation in the expression of CCND1, which encodes for cyclin D1. Taken together, this suggests that a MAP kinase–mediated induction of cyclin D1 results in the growth of the HER2 mutants in coculture.
Clinical evidence of clonal dynamics and the role of fibronectin.
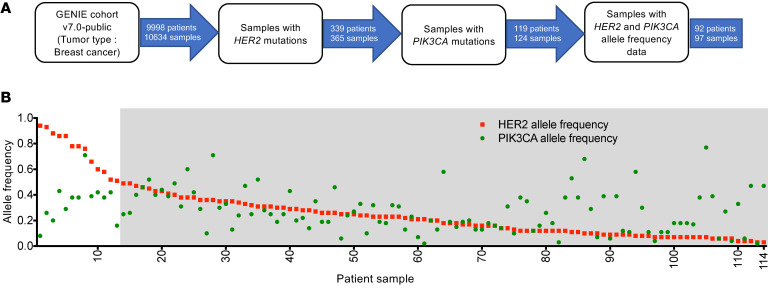
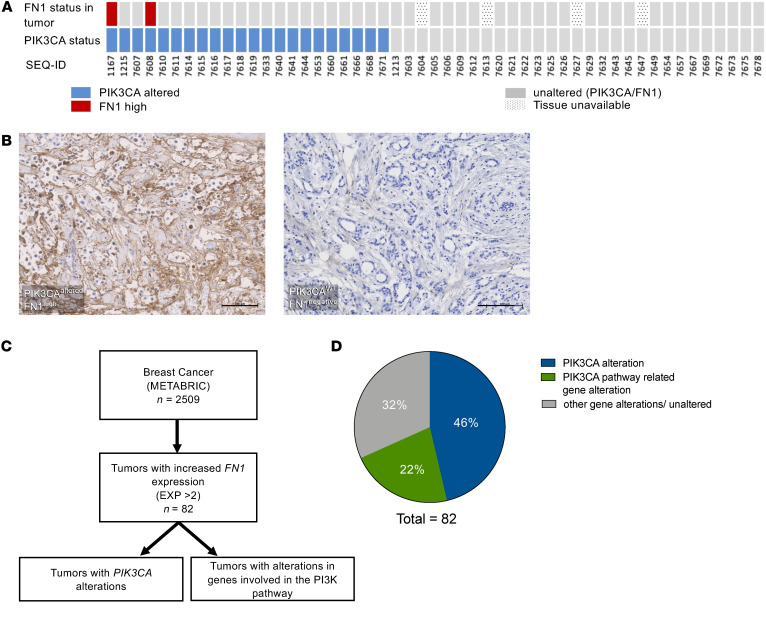
To determine the clinical relevance of this phenomenon, we retrospectively analyzed data from targeted sequencing of breast tumors (GENIE cohort v7.0) using cBioPortal (32). From a total of 9998 patients, those that had a cooccurrence of HER2 and PIK3CA mutations (n = 119 patients/124 samples) were selected and the allele frequencies of HER2 and PIK3CA were obtained (Figure 4A). Our analysis showed that all tumors with high HER2 allele frequencies (> 0.5), except one, had a PIK3CA subclonal population with dramatically lower allele frequencies (n = 12 patients/12 samples), mirroring the trend observed in our cocultures (Figure 4B and Supplemental Table 2). Additionally, patient samples 3 and 14 are serial samples obtained from the same patient (Supplemental Table 2). Sample 14 was obtained at an earlier time point and had a HER2 allele fraction of 0.49, whereas sample 3 was obtained from the same tumor at a later time point and showed a higher HER2 allele fraction of 0.88. There was also a corresponding, although minor, decrease in the PIK3CA allele fraction. These changes further mirror the dynamics seen in our in vitro system. Second, tumor tissue obtained from a cohort of patients with stage I to stage III ER-positive/HER2-negative breast cancer was analyzed for fibronectin expression (33). Of 22 tumors with a PIK3CA alteration, 2 showed robust fibronectin expression, while none of the tumors that were WT for PIK3CA had fibronectin expression (Figure 5, A and B). To expand on this finding, we retrospectively analyzed data from targeted sequencing of a large cohort of breast tumors (cBioPortal, METABRIC; Figure 5C) (34, 35). A total of 82 tumors had increased FN1 expression. Forty-six percent (n = 38) of these tumors contained a PIK3CA alteration, while an additional 22% (n = 18) had alterations in one or more other genes involved in PI3K pathway (Figure 5D). Taken together, these data provide clinical relevance to our in vitro and in vivo findings.
Figure 4. Clinical data from patients with breast cancer corroborate HER2:PIK3CA clonal dynamics.
(A) Schematic of the patient selection criteria for the cBioPortal-based analysis of the GENIE cohort v7.0. (B) Allelic frequencies of HER2 (red) and PIK3CA (green) in breast tumors that had a cooccurrence of HER2 and PIK3CA mutations, sorted by HER2 allele frequency. The gray box indicates patients with HER2 allele frequency less than 0.5. For further information on patient samples, see Supplemental Table 2.
Figure 5. Clinical data from patients with breast cancer corroborate the role of fibronectin.
(A) Immunohistochemical staining of small patient cohort for fibronectin in human breast tumors with and without PIK3CA alterations. Tumors with alterations in PIK3CA are shown in blue and those with high fibronectin are shown in red. Unaltered tumors are shown in gray. (B) Representative images of fibronectin IHC in human breast tumors with a PIK3CA (left) alteration and without a PIK3CA alteration (right). Scale bars: 100 μM. (C) Schematic of patient selection and analysis of the METABRIC data set using cBioPortal. (D) Quantification of PIK3CA alterations and alterations in other genes involved in the PI3K pathway, in breast tumors with increased fibronectin as analyzed by cBioPortal using the METABRIC data set.
Discussion
Tumor subclones have been portrayed as self-interested competitors, but theoretical hypotheses suggesting a role for subclonal interactions have gained experimental support in recent years (11–17, 36). We believe the system described herein is a powerful model to study interactions between genetically distinct subclones and allows for focused interrogation of specific subclonal contributions to clonal dynamics and heterogeneity in human cells. Our results show that interaction between subclonal populations can lead to unexpected but consistent patterns in subclonal architecture in vitro, in vivo, and across different cancerous and noncancerous human cell lines.
The vast majority of studies examining subclonal interactions have implicated paracrine factors such as TGF-α and -β, WNT, amphiregulin, and various cytokines as mediators (11–17). Our findings, however, demonstrate a contact-mediated mechanism involving fibronectin as the primary mediator of subclonal growth in mutant HER2 and mutant PIK3CA cocultures. Prior studies have shown that fibronectin is upregulated during breast cancer development (37–39), and previous work using the PIK3CA E545K mutant MCF-10A cells used in this study demonstrated that these cells produce increased fibronectin (40). Altogether, our results consistently indicate that fibronectin can drive the proliferation of HER2 cells and that there may be cooperative subclonal remodeling of the microenvironment.
Improving our understanding of tumor evolution is a critical need to advance cancer diagnosis, therapy, and personalized medicine. Here, we demonstrate that fibronectin-mediated interactions between genetically distinct subclones can result in a consistent clonal hierarchy. In the era of clinical tumor sequencing, deciphering these subclonal interactions has the potential to positively impact the management of tumors with varying degrees of heterogeneity.
Methods
Cell culture.
The MCF-10A cell line (18) and its derivatives were maintained in DMEM/F12 (1:1) (Thermo Fisher Scientific) supplemented with 5% horse serum (Life Technologies), 20 ng/mL EGF (Sigma-Aldrich), 10 μg/mL insulin (Life Technologies), 0.5 μg/mL hydrocortisone (Sigma-Aldrich), 0.1 μg/mL cholera toxin (Sigma-Aldrich), and 1% penicillin-streptomycin (Life Technologies). The exception was the MCF-10A–derived PIK3CA E545K mutant cell line, which was maintained on the same media described above, but without supplemental EGF. All assays using MCF-10A background cells were done in DMEM/F12 (1:1) media containing 1% charcoal dextran–stripped FBS (Life Technologies) supplemented with insulin, hydrocortisone, cholera toxin, penicillin-streptomycin, and lacking EGF. MCF-10A parental cells were purchased from ATCC. PIK3CA E545K knockins were created in a previous study (19), as were the HER2 L755S mutant (10) and SF3B1 R702R silent substitution cells (21). The MCF-7 cell line (23) was purchased from ATCC. Parental MCF-7 cells and its derivatives were maintained in phenol red–free DMEM/F12 (1:1) supplemented with 5% FBS (Life Technologies) and 1% penicillin-streptomycin. The same media was used for any assays involving these cells. The PIK3CA WT MCF-7 cells and the HER2 L755S knockins were previously developed in our lab (10, 24). The MCF-10A FN1 knockouts were maintained in the same medium as the parental with an addition of 1 μg/mL of puromycin (Thermo Fisher Scientific). The MCF-7 FN1 knockouts were maintained in DMEM/F12 (1:1) supplemented with 5% FBS (Life Technologies), 1% penicillin-streptomycin, and 2 μg/mL puromycin. Assays involving the knockouts were performed in the same assay media used for their corresponding parental cell line. MCF-10A and MCF-7 cell lines were verified by STR profiling.
Cocultures.
Cells were plated at the indicated ratio into T25 flasks (Corning) as shown in Supplemental Figure 1. The cocultures were grown to near confluency and then split at a 1:10 ratio. Genomic DNA (gDNA) was extracted at each split for analysis by droplet digital PCR (ddPCR). Control cultures were allowed to grow until the flasks of one of the controls (e.g., HER2 or PIK3CA) reached confluency. At this point, cells from each control were mixed with equal volumes to yield the artificial mixes.
Droplet digital PCR.
gDNA used for ddPCR was extracted using the QIAamp DNA Blood Mini Kit (Qiagen) and then digested using CviQI (New England Biolabs) restriction enzyme. The digested DNA was purified using the QIAquick PCR Purification Kit (Qiagen). ddPCR reactions were performed using the BioRad QX200 system (BioRad) according to the manufacturer’s protocol. Sequences of the primers and probes used in this study are listed in Supplemental Table 3. In the case of the dual-mutant assay, the FAM-tagged probe was designed to detect the mutant allele of one gene and the HEX-tagged probe was designed to detect the mutant allele of the other gene of interest. For dual-mutant reactions, forward and reverse primers were added for both loci. The working concentrations of the primers were doubled, and their volumes halved to maintain the same concentrations as recommended in the manufacturer protocol. Positive droplets for each channel were identified using the QuantaSoft program (BioRad) and the fraction of each cell in the coculture was calculated according to the calculations shown in the Supplemental Methods.
Lentivirus-mediated stable integration of fluorescence proteins.
Lentiviral vectors containing fluorescence proteins (#20131202, Takara Bio USA) were used to transduce cells according to the manufacturer’s protocol. Plasmid sequences were rLV-EF1a-tdTomato-IRES-Puro-WPRE for red cells, and rLV-EF1a-ZsGreen1-IRES-Puro-WPRE for green cells. Briefly, cells were seeded at subconfluent concentrations and infected after 24 hours with a mixture of the viral particles in media and polybrene (800 μg/mL). Following an overnight incubation, the cells were recovered by replacing the transduction media with normal maintenance media. Fluorescence was assessed 48 hours after recovery and the cells were then single-cell diluted to isolate individual clones. The isolated clones were then evaluated for fluorescence and their growth rates were compared with that of the parental using crystal violet staining (data not shown). Clones that most closely resembled the parental growth rates and expressed the fluorescence protein were selected. Fluorescence imaging was performed using the EVOS M7000 system (Thermo Fisher Scientific).
Xenograft assays.
A total of 35 female athymic nude mice with supplemental estrogen pellets (Harlan Laboratories) were assigned to a total of 5 groups: 5 mice times 3 control groups and 10 mice times 2 coinoculation groups. Cells (2 × 106) were resuspended in Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix (Thermo Fisher Scientific) and subcutaneously injected. Coinoculations were accomplished by mixing equal number of cells from each cell line prior to resuspension in Geltrex. Tumor volumes were periodically assessed using calipers to measure the length, width, and height for each tumor.
Xenograft processing.
Tumors were harvested upon reaching a predetermined threshold (1.5 cm3) or at the termination of the experiment. Upon extraction, each tumor was placed in 10% formalin for 1 hour and then 30% sucrose in PBS overnight. The tumors were then cut into 2 halves. One half was used for genomic DNA extraction using the DNeasy Blood & Tissue Kit (Qiagen) for ddPCR analysis. The other half was prepared for fluorescence imaging. In order to preserve the endogenous fluorescence of the xenograft cells, tumors were preserved in OCT (Thermo Fisher Scientific) and stored at –80°C. Cryosectioning was performed by the Johns Hopkins Oncology Tissue Services Core. Cryosectioned slides were stained with DAPI and mounted using a mixture of 20 mM Tris, pH 8.0, 0.5% N-propyl gallate, and 90% glycerol.
Time-lapse imaging.
Cells were plated in isolation or in coculture and were allowed to grow for 24 hours. They were then transferred to the incubator of the IncuCyte (Essen BioScience), which was kept at 37°C and 5% CO2. Periodic phase and fluorescence images were taken at ×10 magnification using the IncuCyte’s internal camera.
Microarray.
RNA was extracted from controls and cocultures in quadruplicate using the RNeasy and QiaShredder kits (Qiagen). Samples were sent to the Johns Hopkins Deep Sequencing & Microarray Core, which performed quality control measures and subjected the RNA to the Affymetrix Clariom S Array. Downstream analysis was performed using the Transcriptome Analysis Console (TAC) Software (Thermo Fisher Scientific). Transcriptome data were deposited in NCBI’s Gene Expression Omnibus (GEO) and is accessible through GEO series accession number GSE165065.
Fibronectin coating.
Fibronectin from human plasma (Sigma-Aldrich) was dissolved to 1 mg/mL in sterile water by placing in 37°C for 30 minutes. The soluble fibronectin was then diluted in a minimal volume of HBSS and applied to each plate at a concentration of 5 μg/cm. Plates were left to air-dry for at least 45 minutes before cells were added.
CRISPR Cas-9 system.
The CRISPR-Cas 9 system was used to knockout FN1 in the MCF-10A and the MCF-7 PIK3CA mutants. Guide RNA sequences were chosen from the GeCKov2 library pool (41). Vectors were created by cloning the guide RNAs into the lentiCRISPR v2 single-vector system backbone. Each vector was then cotransfected with lentiviral packaging plasmids CMV dR8.91 and VSVG into HEK293T cells. The harvested virus was used to infect the PIK3CA mutants in confluent 6-well plates for 24 hours. After infection, the transduced cells were recovered for 4 hours following which they were selected using puromycin for 3 days (1 μg/mL for MCF-10As and 2 μg/mL for MCF-7). After selection, the cells were single-cell diluted and individual colonies were sequenced at the locus to identify clones with insertions/deletions at the cut site. The selected clones were further analyzed by Western blot to verify complete knockout of FN1.
Western blot analysis.
MCF-10A cells were seeded in 6-well plates in DMEM/F12 (1:1) media containing 1% charcoal dextran–stripped FBS (Life Technologies) supplemented with insulin, hydrocortisone, cholera toxin, and penicillin-streptomycin. The MCF-7 cells were plated in DMEM/F12 (1:1) supplemented with 1% charcoal dextran stripped FBS (Life Technologies) and 1% penicillin-streptomycin. For lysate preparation, the cells were washed thoroughly with PBS to avoid serum cross-reactivity with the FN1 antibody, following which they were scrapped in PBS and centrifuged to remove any residual serum. The supernatant was discarded and the cell pellet was resuspended in Laemmli buffer. The lysates were resolved by SDS-PAGE using NuPAGE gels (Invitrogen). Since fibronectin is a high-molecular-weight protein, the gels were incubated in 20% ethanol at room temperature for 5–10 minutes prior to transfer. The proteins were transferred onto PVDF membranes using iBlot 2 (Invitrogen) and the membranes were probed with anti-fibronectin antibody (ab2413, Abcam) diluted in primary antibody dilution buffer (R2004, Kwikquant). TBST was replaced with clean wash buffer (R2001) in all steps. Images were acquired according to the manufacturer’s protocol using the Kwikquant Imager (Kindle Biosciences, LLC).
Flow sorting for PCR array.
HER2 mutants (MCF-10A background) were cocultured with the PIK3CA mutants in MCF-10A assay media in the absence of phenol red for 5 days. HER2 mutants grown in isolation were used as control. The control and cocultured cells were trypsinized and centrifuged at 200g for 5 minutes. The cell pellets were resuspended in PBS with 3% charcoal dextran–stripped FBS. Flow sorting was carried out at the Vanderbilt Flow Cytometry Shared Resource using the 100 μM flow nozzle. RFP-tagged HER2 mutants were separated using assay media with 20% charcoal dextran–stripped FBS as catch buffer.
PCR array.
Total RNA was extracted from controls and cocultures using the RNeasy Mini Kit (Qiagen). The RNA was reverse transcribed using the RT2 First Strand Kit (Qiagen) according to the manufacturer’s protocol. The expression of a total of 84 genes was tested using the RT2 Profiler PCR Array Human Kit EGF/PDGF Signaling Pathway Sampler (GeneGlobe ID PAHS-040Z). The data were analyzed using the GeneGlobe data analysis center from Qiagen according to the manufacturer’s instructions. The average of 5 housekeeping genes (ACTB, B2M, GAPDH, HPRT1, RPLP0) was used for normalization. The data generated in this array were deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE165150.
Immunohistochemical staining of patient tumors.
Patient tissue, from stage 1 to stage III operable ER-positive/HER2-negative breast tumors from postmenopausal patients who received 15 to 21 days of letrozole prior to surgery were obtained from a prior Vanderbilt Ingram-Cancer center trial (BRE0776) (33). Since fibronectin is widely expressed by different cell types and is a part of the extracellular matrix, we were unable to use the matched normal tissue from the tissue microarray as a negative control. Therefore, cell pellets from the FN1 knockouts and the corresponding FN1+ parentals created in our lab for this paper were used as negative and positive controls, respectively. Pellets were sectioned by the Translational Pathology Shared Resource at Vanderbilt University. The tissues and sectioned pellet controls were stained with ab2413 anti-fibronectin primary (Abcam) and 26836 Fibronectin/FN1 (E5H6X) Rabbit mAb (Cell Signaling).
cBioPortal data analysis.
Information for in silico data analyses were obtained using the METABRIC and GENIE databases and analyzed using cBioPortal (https://www.cbioportal.org). Genomic profiles, including mutations, allelic frequencies, and mRNA expression z scores, were obtained and analyzed further using the GraphPad prism. In the METABRIC data set, samples were filtered based on increased FN1 expression (FN1: EXP > 2) prior to analysis of genetic alterations in PIK3CA, AKT1, AKT2, AKT3, PIK3C2B, and PTEN genes. In the GENIE data set (breast cancer cohort), samples with cooccurrence of HER2 (searched using the genetic nomenclature of ERBB2 in cBioPortal) and PIK3CA mutations were selected. Within the selected subset, samples which had both the HER2 and PIK3CA allele frequency data were separated and analyzed.
Transwell assays.
PIK3CA mutant cells were seeded into the wells of transwell plates (VWR). Meanwhile, HER2 or WT cells were seeded onto transwell inserts with a pore size of 0.4 μM. After allowing the cells on the insert to settle, the inserts were transferred into the transwell plates. Assay media was added such that cells on the insert were covered on the apical and basolateral sides. Media was replenished periodically. After 7–10 days, inserts were removed and both the inserts and the plated cells were trypsinized and counted using a Beckman Coulter counter.
Immunofluorescence staining of in vitro cocultures.
A 1:1 mixture of fluorescence HER2 cells (described above) and nonfluorescence PIK3CA cells (MCF-10A background) were plated onto Nunc Lab-Tek Chamber Slides (Thermo Fisher Scientific). Cocultures were grown for 7 days. Slides were fixed with 4% paraformaldehyde and stained using ab2413 anti-fibronectin primary (Abcam) and donkey anti-rabbit Alexa Fluor 405 secondary (Abcam). Prior to imaging, Fluorescence Mounting Media were added to each slide (https://nic.med.harvard.edu/resources/media/). Images were taken using a Nikon C1 Confocal Microscope.
3D matrigel assay and imaging.
Fluorescence HER2 and PIK3CA MCF-10A background cells (described above) were plated into multi-well plates containing Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix (Thermo Fisher Scientific) and assay media as previously described (42). Cells were grown for 12 days and then imaged. Images were taken using a Nikon C1 Confocal Microscope and image processing was performed using ImageJ (https://imagej.nih.gov/ij/).
Statistics.
Statistical analyses were performed using the GraphPad Prism 5 software (GraphPad Software). Unpaired t test, 1-way ANOVA, and 2-way ANOVA functions were used in the appropriate contexts to analyze our results. All statistical tests were 2-tailed. All averages and error bars represent the mean and SEM of each group. *P < 0.05, **P < 0.01, and ***P < 0.001.
Study approval.
All animal care and experimental protocols were approved by the IACUC of The Johns Hopkins University. All human samples were obtained from prior studies conducted in accordance with the IRB of Vanderbilt University Medical Center.
Author contributions
SK, IGW, and BHP conceived and designed research. BHP and SC supervised the project. SK, IGW, LD, and DC developed protocols and designed experiments. SK, IGW, LD, DHS, DMR, MVP, REB, BAD, and SCR carried out experiments. PIGE, VS, and MES carried out immunohistochemical staining of patient tumors. SK and SC carried out in silico data analysis. SK, IGW, LD, SC, and BHP analyzed and interpreted data. JD, KC, KKS, BB, DJZ, HYW, and PJH contributed to data interpretation and troubleshooting discussions. SK, IGW, SC, and BHP wrote the manuscript with comments from all coauthors. The order of the co–first authors was determined by alphabetical order of last name.
Supplementary Material
Acknowledgments
This work was supported by the Microenvironment Influences in Cancer Training Program T32 (2T32CA009592-31A1) (to BAD); The American Cancer Society 131356-RSG-17-160-01-CSM and The National Cancer Institute/National Institute of Health RO1CA211695-01A1 (to PJH); The Breast Cancer Research Foundation, Komen Foundation, and NIH CA214494, CA194024 (to BHP). We would also like to thank and acknowledge the support of The Canney Foundation, the Marcie and Ellen Foundation, Amy and Barry Baker and the Vanderbilt-Ingram Cancer Center support grant (NIH CA068485), and Breast Cancer SPORE (NIH CA098131). The authors would like to acknowledge the American Association for Cancer Research (AACR) and its financial and material support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of study authors. We dedicate this work to the loving memory of Dr. David Chu, who continues to inspire us to find cures for all cancers.
Version 1. 02/02/2021
In-Press Preview
Version 2. 03/15/2021
Print issue publication
Footnotes
Conflict of interest: BHP is a paid consultant for Jackson Labs, Casdin Capital, Pathovax, and Sermonix, and is a paid scientific advisory board member for Celcuity Inc. DJZ, PJH, and BHP are entitled to a share of royalties received by The Johns Hopkins University on sales of products, under separate licensing agreements between Horizon Discovery Ltd. and The Johns Hopkins University.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(6):e143557.https://doi.org/10.1172/JCI143557.
Contributor Information
Swathi Karthikeyan, Email: swathi.karthikeyan@Vanderbilt.Edu.
Ian G. Waters, Email: watersia@umich.edu.
Lauren Dennison, Email: ldenni13@jhmi.edu.
David Chu, Email: dchu21@jhmi.edu.
Joshua Donaldson, Email: joshua.donaldson@vumc.org.
Dong Ho Shin, Email: donghoshin@mednet.ucla.edu.
D. Marc Rosen, Email: rosenma@jhmi.edu.
Paula I. Gonzalez-Ericsson, Email: paula.i.gonzalez.ericsson@vumc.org.
Violeta Sanchez, Email: violeta.sanchez@vumc.org.
Melinda E. Sanders, Email: melinda.sanders@vanderbilt.edu.
Morgan V. Pantone, Email: morgan.v.pantone@vumc.org.
Riley E. Bergman, Email: riley.e.bergman@vanderbilt.edu.
Brad A. Davidson, Email: brad.a.davidson@vanderbilt.edu.
Sarah C. Reed, Email: sarah.c.reed@vanderbilt.edu.
Daniel J. Zabransky, Email: dzabran1@jhmi.edu.
Karen Cravero, Email: kcravero@ufl.edu.
Kelly Kyker-Snowman, Email: k.kykersnowman@gmail.com.
Berry Button, Email: bcbutton@gmail.com.
Hong Yuen Wong, Email: hongyuen.wong@vumc.org.
Paula J. Hurley, Email: paula.hurley@vumc.org.
Sarah Croessmann, Email: sarah.croessmann@vanderbilt.edu.
References
- 1.Siegel RL, et al. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janiszewska M, et al. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet. 2015;47(10):1212–1219. doi: 10.1038/ng.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstung M, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578(7793):122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salcedo A, et al. A community effort to create standards for evaluating tumor subclonal reconstruction. Nat Biotechnol. 2020;38(1):97–107. doi: 10.1038/s41587-019-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukohara T. PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast Cancer (Dove Med Press) 2015;7:111–123. doi: 10.2147/BCTT.S60696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zabransky DJ, et al. HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc Natl Acad Sci U S A. 2015;112(45):E6205–E6214. doi: 10.1073/pnas.1516853112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu M, et al. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463(7280):545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inda MM, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calbo J, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19(2):244–256. doi: 10.1016/j.ccr.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Cleary AS, et al. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508(7494):113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514(7520):54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, et al. Intratumoral heterogeneity in a Trp53-null mouse model of human breast cancer. Cancer Discov. 2015;5(5):520–533. doi: 10.1158/2159-8290.CD-14-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franco OE, et al. Altered TGF-α/β signaling drives cooperation between breast cancer cell populations. FASEB J. 2016;30(10):3441–3452. doi: 10.1096/fj.201500187RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soule HD, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–6086. [PubMed] [Google Scholar]
- 19.Gustin JP, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proc Natl Acad Sci U S A. 2009;106(8):2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ignatoski KM, et al. erbB-2 overexpression in human mammary epithelial cells confers growth factor independence. Endocrinology. 1999;140(8):3615–3622. doi: 10.1210/endo.140.8.6939. [DOI] [PubMed] [Google Scholar]
- 21.Dalton WB, et al. Hotspot SF3B1 mutations induce metabolic reprogramming and vulnerability to serine deprivation. J Clin Invest. 2019;129(11):4708–4723. doi: 10.1172/JCI125022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauring J, et al. Knock in of the AKT1 E17K mutation in human breast epithelial cells does not recapitulate oncogenic PIK3CA mutations. Oncogene. 2010;29(16):2337–2345. doi: 10.1038/onc.2009.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soule HD, et al. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51(5):1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 24.Beaver JA, et al. PIK3CA and AKT1 mutations have distinct effects on sensitivity to targeted pathway inhibitors in an isogenic luminal breast cancer model system. Clin Cancer Res. 2013;19(19):5413–5422. doi: 10.1158/1078-0432.CCR-13-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309(5963):30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 26.Moon C, et al. Synthetic RGDS peptide attenuates lipopolysaccharide-induced pulmonary inflammation by inhibiting integrin signaled MAP kinase pathways. Respir Res. 2009;10(1):18. doi: 10.1186/1465-9921-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, et al. Force measurements of the α5β1 integrin-fibronectin interaction. Biophys J. 2003;84(2):1252–1262. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, Assoian RK. Integrin-dependent activation of MAP kinase: a link to shape-dependent cell proliferation. Mol Biol Cell. 1995;6(3):273–282. doi: 10.1091/mbc.6.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, et al. Integrin-mediated cell adhesion activates mitogen-activated protein kinases. J Biol Chem. 1994;269(43):26602–26605. doi: 10.1016/S0021-9258(18)47058-5. [DOI] [PubMed] [Google Scholar]
- 30.Lavoie JN, et al. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271(34):20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 31.Weber JD, et al. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D1 in G1 phase. Biochem J. 1997;326(Pt 1):61–68. doi: 10.1042/bj3260061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium APG. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giltnane JM, et al. Genomic profiling of ER+ breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Sci Transl Med. 2017;9(402):eaai7993. doi: 10.1126/scitranslmed.aai7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Axelrod R, et al. Evolution of cooperation among tumor cells. Proc Natl Acad Sci U S A. 2006;103(36):13474–13479. doi: 10.1073/pnas.0606053103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasaturo F, et al. Comparison of extracellular matrix and apoptotic markers between benign lesions and carcinomas in human breast. Int J Oncol. 2005;27(4):1005–1011. [PubMed] [Google Scholar]
- 38.Hao X, et al. Differential gene and protein expression in primary breast malignancies and their lymph node metastases as revealed by combined cDNA microarray and tissue microarray analysis. Cancer. 2004;100(6):1110–1122. doi: 10.1002/cncr.20095. [DOI] [PubMed] [Google Scholar]
- 39.Woodward TL, et al. Fibronectin and the alpha(5)beta(1) integrin are under developmental and ovarian steroid regulation in the normal mouse mammary gland. Endocrinology. 2001;142(7):3214–3222. doi: 10.1210/endo.142.7.8273. [DOI] [PubMed] [Google Scholar]
- 40.Young CD, et al. Activating PIK3CA mutations induce an epidermal growth factor receptor (EGFR)/extracellular signal-regulated kinase (ERK) paracrine signaling axis in basal-like breast cancer. Mol Cell Proteomics. 2015;14(7):1959–1976. doi: 10.1074/mcp.M115.049783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanjana NE, et al. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debnath J, et al. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30(3):256–268. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.