Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is spreading around the globe since December 2019. There is an urgent need to develop sensitive and online methods for on-site diagnosing and monitoring of suspected COVID-19 patients. With the huge development of Internet of Things (IoT), the impact of Internet of Medical Things (IoMT) provides an impressive solution to this problem. In this paper, we proposed a 5G-enabled fluorescence sensor for quantitative detection of spike protein and nucleocapsid protein of SARS-CoV-2 by using mesoporous silica encapsulated up-conversion nanoparticles (UCNPs@mSiO2) labeled lateral flow immunoassay (LFIA). The sensor can detect spike protein (SP) with a detection of limit (LOD) 1.6 ng/mL and nucleocapsid protein (NP) with an LOD of 2.2 ng/mL. The feasibility of the sensor in clinical use was further demonstrated by utilizing virus culture as real clinical samples. Moreover, the proposed fluorescence sensor is IoMT enabled, which is accessible to edge hardware devices (personal computers, 5G smartphones, IPTV, etc.) through Bluetooth. Medical data can be transmitted to the fog layer of the network and 5G cloud server with ultra-low latency and high reliably for edge computing and big data analysis. Furthermore, a COVID-19 monitoring module working with the proposed the system is developed on a smartphone application (App), which endows patients and their families to record their medical data and daily conditions remotely, releasing the burdens of going to central hospitals. We believe that the proposed system will be highly practical in the future treatment and prevention of COVID-19 and other mass infectious diseases.
Keywords: Proactive prognosis of COVID-19, 5G-enabled fluorescence sensor, Lateral flow immunoassay, Internet of medical things, 5G communication
1. Introduction
The outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at the end of 2019 has posed a huge threat to global health and economy (Huang et al., 2020a). In February 2020, the World Health Organization named the coronavirus-infected pneumonia "COVID-19" (Wang et al., 2020), and up till now, this pneumonia has affected more than 200 countries and 30 million people. Coronavirus (CoV) is a positive-stranded RNA virus with a diameter of 80–120 nm, which can cause a variety of acute and chronic diseases. Before the outbreak of SARS-CoV-2, six CoV strains have been confirmed to infect humans (Velavan and Meyer, 2020; Chiolero, 2020). Among them, Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) have caused severe acute respiratory diseases worldwide (Cheng et al., 2007; Chan et al., 2015). SARS-CoV-2 discovered this time is a novel coronavirus and considered to be the seventh coronavirus that can infect humans. It enters the human cell by recognizing the corresponding receptor on the target cell through the spike protein (SP) on its surface and replicates and synthesizes new virus inside the cell to cause infection (Hoffmannet al., 2020). The most common symptoms of an infected person are fever, dry cough, and muscle pain. A small number of SARS-CoV-2 patients have intestinal signs and symptoms (such as diarrhea, nausea, and vomiting) (Chenet al., 2020).
To diagnose suspected patients of COVID-19, the routinely used medical methods include chest CT scan, molecular diagnosis and immunological detection technology (Chen et al., 2020; Won et al., 2019; Li et al., 2020a; Xiang et al., 2020). A chest CT scan uses a series of X-ray images taken from the chest and create a cross-sectional image of the lungs by computer processing to reflect radiologic features. However, this method requires bulky medical equipment, which can only be used in central hospitals (Cui and Zhou, 2019). Molecular diagnosis includes gene sequencing technology, reverse transcription real-time fluorescent quantitative PCR (RT-qPCR) (Won et al., 2019), digital PCR and loop-mediated isothermal amplification (LAMP) technology (Yu et al., 2020), etc. They identify or amplify nucleic acid molecules of SARS-CoV-2 in patient samples, and apply specific tracers, such as fluorescent particles, in the reaction process to realize quantitative diagnosis. Although these approaches are accurate and sensitive, the time-consuming and difficult operation of RNA extraction makes each test costly. Immunological detection techniques mainly include chemiluminescence, electrochemcial method (Mavrikou et al., 2020), enzyme linked immunosorbent assay (ELISA) (Liu et al., 2020) and lateral flow immunoassays (LFIA) (Li et al., 2020a; Xiang et al., 2020). Among them, LFIA shows great superiority in point-of-care tests of SARS-CoV-2 because it needs no professional operation and has rapid detection speed (Huang et al., 2020b). Traditional LFIA suffers from low sensitivity since colloidal gold is routinely served as the label. When the analyte concentration drops to a certain level low, the red color appeared on the test strip cannot be distinguished by the naked eyes, which can be detrimental to the limit of detection (LOD) (Guo et al., 2021). Grant et al. (2020) demonstrate a half-strip LFIA for the semi-quantitative detection of nucleocapsid antigen of SARS-CoV-2. In their work, an LOD of 0.65 ng/mL with recombinant antigen was achieved with the use of an optical reader. However, most of the LFIAs are usually performed off-line, which may impede real-time and effective monitoring of the patient's condition. Thus, it is critical to develop sensitive and smart detection methods of SARS-CoV-2 to release the burden on diagnosing COVID-19 and promote intelligent monitoring of suspected patients at remote place.
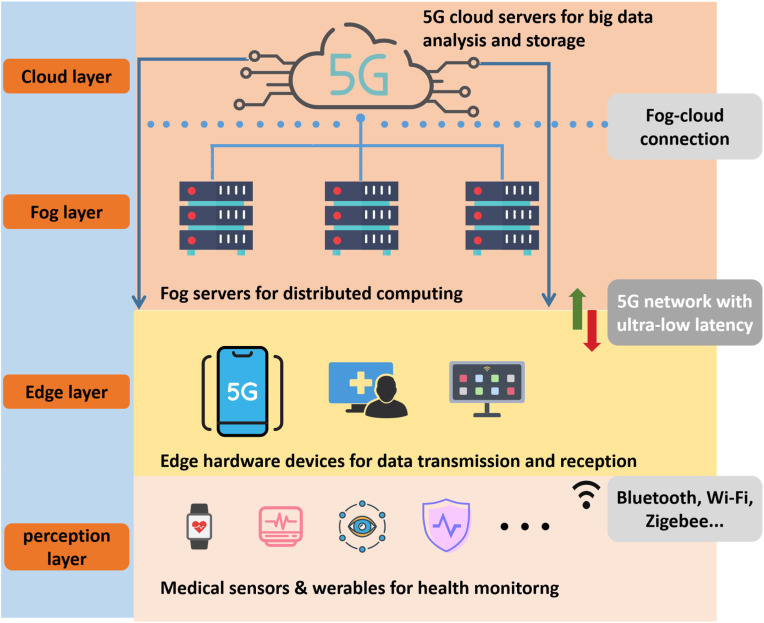
The Internet of Medical Things (IoMT) provides insight into proactive tele-healthcare of suspected COVID-19 patients. Evolved from the Internet of Things (IoT), IoMT focuses on medical applications in IoT and stands for a platform for interconnection of medical-grade devices and their integration to health networks (Catarinucciet al., 2015; Qadri et al., 2020). With the advancing of the fifth generation (5G) networks, and edge computing (Pan and McElhannon, 2018), varieties of medial sensors have reliable and massive connection to hospital systems or personal mobile devices due to the strategies of Massive Machine Type Communications (mMTC) and Ultra-reliable and Low-latency Communications (uRLLC), which further boosts IoMT growth (Sachset al., 2019; Ji et al., 2018; Kapassa, 2019). As shown in Fig. 1 , these medical sensors located in the perception layer can monitor and receive patient information through physical signals such as light, electricity, pressure, temperature, etc., and transmit it to the edge and fog layer through wireless communication (such as Wi-Fi, Bluetooth, Zigbee, etc.) with ultra-low latency and high reliability, and finally reach the cloud layer, providing hospitals and other medical institutions with the latest medical conditions. On the one hand, IoMT based smart medical system can maximize the limited medical and human resources of the hospital, and facilitate patients to obtain faster and more convenient medical services. On the other hand, the integration of mass medical data are essential to prevention and prediction of pandemics that may occur in the future. Furthermore, suspected patients can be kept informed of their health status and monitored effectively with the use of IoMT based sensors. They can stay up to date on critical health information by conducting remote consultations with specialist doctors (Guoet al., 1109).
Fig. 1.
Architecture of the 5G-enabled internet of medical things.
In this paper, we demonstrated a 5G-enabled fluorescence sensor for rapid detection and tele-monitoring of COVID-19 patients. A paper-based lateral flow assay using up-converting nanoparticles with mesoporous silica encapsulated nano-shell structure (UCNPs@mSiO2) as fluorescence labels is developed to quantitatively detect SP and Nucleocapsid Protein (NP) of SARS-CoV-2. UCNPs are a kind of rare-earth-doped luminescent nanomaterials which can convert near-infrared (NIR) excitation into visible light. Compared with other traditional nanoprobes, such as colloidal gold or quantum dots, UCNPs have unique advantages of low toxicity, anti-Stokes shifts, narrow emission spectrum, and not being interfered by biological autofluorescence (Guo et al., 2021). By encapsulating with mesoporous silica as shell outside, UCNPs@mSiO2 show great water solubility, good biocompatibility and stability, which prove to be an excellent candidate for biological probe in LFIA. SP is the membrane surface protein of SARS-CoV-2 and is an important site of action for host-neutralizing antibodies. It is also a key target for vaccine design. NP is a significant structural protein of SARS-CoV-2, which plays a critical role in the process of virus packaging, replication and protein translation. The measurement of NP has a high diagnostic value for suspected patients before the antibody appears and shortens the window period of serological diagnosis (Li et al., 2020b). Thus, by detecting the SP and NP content in the patient's respiratory tract specimens, early diagnosis of COVID-19 can be achieved. The proposed system is designed to read the test strip in 10 min and can connect to edge hardware devices (personal computers, smartphones, IPTV, etc.) and fog layer of the network to perform reliable data transmission with low latency and high security. Through the gateway devices and fog servers, the medical data are finally transmitted to the cloud for big data storage and analysis, which benefits for disease control and medical statistics. Furthermore, a COVID-19 monitoring module working with the proposed the system is developed on a smartphone application (App), which endows patients and their families to record their real-time medical data and daily conditions remotely, releasing the burdens of going to central hospitals.
2. Theory
2.1. LFIA for SARS-CoV-2 detection
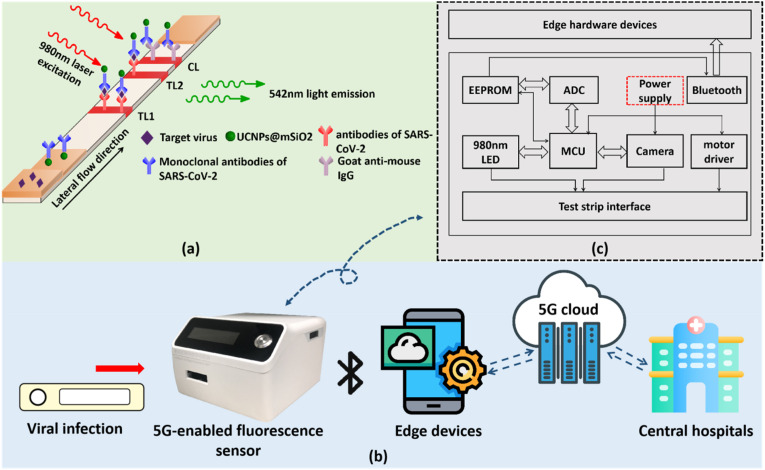
In this system, we utilize UCNPs based LFIA to quantify SP an NP of SARS-CoV-2. UCNP is a kind of rare-earth-elements-doped nanomaterial which will illuminate under near-infrared excitation. Because SP and NP both have more than two antigenic sites, we use sandwich format of LFIA in the experiment. The detection theory is illustrated in Fig. 2 a. A paper-based disposable strip is used to detect extracted SARS-CoV-2 SP and NP based on the principle of antibody-antigen specific binding. Two test lines (TLs) are drawn on the nitrocellulose (NC) membrane with antibodies of SP (for TL1) and antibodies of NP (for TL2) of SARS-CoV-2, respectively. A control line (CL) is drawn on the same membrane with goat anti-mouse IgG solution by using a scribe instrument. Fluorescent labels, UCNPs@mSiO2, are initially conjugated with monoclonal antibodies of SARS-CoV-2 and then evenly dispensed on the conjugate pad with XYZ three-dimensional dispenser in a concentration of 2.2 mg/mL. When a SARS-CoV-2 positive sample (contained SARS-CoV-2 SPs or NPs) is applied, the target proteins will initially bind to the UCNPs@mSiO2 labels and then be captured by the TLs, forming the sandwich complex of antibody-protein-antibody. As a result, the fluorescent signal appears on the TLs, whose intensity is positively correlated with SARS-CoV-2 concentration. Because antibodies on the UCNPs is murine, goat anti-mouse IgG on the CL can always capture the UCNP labels. Thus, fluorescence on the CL is stable and can be used to decide whether test strip is valid.
Fig. 2.
(a) The principle of the UCNPs based lateral flow assay in detection of SARS-CoV-2. (b) The working process of the proposed 5G-enabled fluorescence sensor. (c) The circuit configuration and hardware composition of the fluorescence sensor.
2.2. 5G-enabled fluorescence sensor
The 5G-enabled fluorescence sensor is fabricated with a size of 102 mm × 120 mm × 74 mm and a weight of 352.3 g, which proves to be compact and portable (shown in Fig. 2b and c). It is composed of a focusing lens, a 980 nm wavelength optical filter, a light emitting diode (LED) module with 980 nm wavelength, a motor driver, a high-speed camera, a Bluetooth model, and a micro controller unit (MCU) pre-embedded with control and processing modules. The sensor can be connected with computers or smartphones through Bluetooth. A certain software integrated with COVID-19 detection function is used for start the measurement. After confirming the right position of the inserted test strip, the control module of the system will automatically start the motor driver to move the test strip forward and backward. During the round trip of the test strip, the LED constantly emits 980 nm near-infrared laser on the surface of the test strip, whose light spot is shaped into a rectangular focal line of 3 mm × 1 mm due to a front cylindrical lens, to excite UCNPs on the TLs and CL. This whole process is recorded by the high-speed camera which is placed on the top of a filter. The images captured by the camera are segmented into a size of 75 × 25 pixels according to the identified fluorescence signal windows and then grayed out and de-contextualized by the data processing module. Each obtained frame is traversed pixel by pixel, and the fluorescence intensity of each frame is determined by averaging the sum of gray values of each pixel. As a result, several signal peaks will appear in the TL and CL areas during the movement of the test strip. When the measurement ends, a raw data curve is drawn in the dialog box and is subsequently processed by a smoothing algorithm and a noise filtering algorithm. Eventually, we integrate the effective area below each peak to obtain the fluorescence intensities on TLs and CL and calculate T1/C and T2/C by making ratios. The obtained T/C value is used to determine the analyte concentration because they are positively correlated. The proposed fluorescence sensor is accessible to edge hardware devices (personal computers, smartphones, etc.) through Bluetooth, and can transmit the medical data to fog servers and central cloud servers for edge computing and big data storage. Based on 5G small cells and macro base stations, medical facilities such as local hospitals can have rapid and reliable access to these critical data. To provide real-time proactive prognosis with suspected patients, an App with COVID-19 monitoring module which works with the fluorescence sensor is developed on a smartphone. For the purpose of proactive health management, in this study, fuzzy logic and deep learning algorithms has embedded into the system. According to the pre-established knowledge base of historical data and the newly delivered test data, the system can draw accurate conclusions on positive or negative detection results with the principle of maximum degree of membership after confirming the user's personal information. Since the security of patient-generated data is paramount, we are looking forward to employ complete cryptographic techniques to guarantee the privacy security during data transmission (Tiwari et al., 2019).
3. Materials & methods
3.1. Preparation of UCNPs
In the experiment, all chemicals and solvents are of analytical grade. UCNPs were synthesized according to literature protocols with slight modifications (You et al., 2019). In a typical synthesis of NaYF4: Yb,Er (Yb3+: 20 mol%, Er3+: 2 mol%), YCl3·6H2O (1.56 mmol), YbCl3·6H2O (0.40 mmol) and ErCl3·6H2O (0.04 mmol) were mixed with 12 mL oleic acid and 30 mL 1-octadecene. Under nitrogen flow protection, the mixture was heated up to 160 °C and maintained at this temperature for 2 h. Then the mixture was cooled down to room temperature, and NH4F (8 mmol) and NaOH (5 mmol) contained in 20 mL methanol was added. Subsequently, the temperature was kept at 50 °C for 30 min to remove methanol and then increased to 300 °C to react for 1 h. Finally, the solution was cooled down to room temperature and the resulting nanoparticles were isolated by centrifugation, washed twice with ethanol and dispersed in 20 mL cyclohexane. The as-prepared NaYF4: Yb,Er was then wrapped with a layer of passivation shell, NaYF4, and this particle with core-shell structure was designated as UCNP.
3.2. Preparation of UCNPs@mSiO2 with functional surface modification
Then the as-prepared UCNPs were coated with mesoporous silica through a base-catalyzed sol-gel procedure (Han et al., 2016). The as-prepared UCNPs (660 μL) were initially stirred to remove cyclohexane. Then an aqueous CTAB solution (40 mL, 0.014 M) and NaOH aqueous solution (200 μL, 0.1 M) was added. The mixture was ultrasonically dispersed for 30 min. After that, 2.7 mL TEOS/anhydrous EtOH solution (0.3 mL/2.4 mL) were dripped into the mixture using a syringe pump at a rate of 0.9 mL/h, and stirred for another 12 h. The as-synthesized UCNP@mSiO2 nanoparticles were collected by centrifugation and washed with ethanol and HCl for several times.
The modification of carboxyl groups was also according to a previous report with some modifications (Ma et al., 2016). Initially, UCNPs@mSiO2 (50 mg) was added in EtOH (10 mL) and fully dispersed. Subsequently, APTES (100 μL) was added into the solution and reacted for 24 h to attach amino group. After 3 times of centrifugation and washing with EtOH, the resulting product UCNPs@mSiO2-NH2 was dispersed in DMF, followed by an addition of SAA (50 mg) and TEA (52.1 μL), and reacted for 24 h. After three times of centrifugation and washing with EtOH and DMF, the UCNPs@mSiO2-COOH products were obtained, serving as luminescent labels for SARS-CoV-2 detection.
3.3. Preparation of UCNPs@mSiO2 labeled LFIA test strips
To link the UCNPs@mSiO2 to monoclonal antibodies of SP and NP of SARS-CoV-2, EDC/NHS activation method was carried out in the experiment (Howell et al., 1998). Initially, 2.2 mg of as-prepared UCNPs@mSiO2 is reacted with 1.2 mg NHS and 1.5 mg EDC in 1 mL MES (pH = 6.5) for 1 h. The process allows activation of the carboxyl group. Subsequently, the solution was centrifuged under a centrifugal speed of 16,000 rpm, and the buffer was changed to 1 mL PBS (pH = 7.2). After the solution was ultrasonically dispersed, 0.15 mg monoclonal antibodies of SP and 0.15 mg monoclonal antibodies NP of SARS-CoV-2 was added for labeling, and the reaction lasted for 4 h. When the labeling step is finished, the solution was centrifuged under a centrifugal speed of 16,000 rpm and dispersed in 1 mL PBS, followed by an addition of 100 μL BSA solution for sealing unconnected carboxyl groups for 1 h. Eventually, after the solution was centrifuged, the solvent was changed to 1 mL BSA solution for preservation. Once the above process was done, we utilized an XYZ three-dimensional dispenser to dispense the as-prepared UCNPs labeled solution to the glass fiber (pre-treated with treatment liquid composed of 1.21% Tris, 1% PVP, 1% S9, 5% T-casein and DI water) in a concentration of 2.2 mg/mL, and precisely scribed antibodies of SP and NP and goat anti-mouse IgG on the NC membrane as TLs (antibodies of SP on TL1, antibodies of NP on TL2) and CL in a concentration of 0.8 mg/mL, 0.8 mg/mL and 1 mg/mL, respectively. After that, the glass fiber and the substrate fixed with NC membrane were dried in the electric thermostatic drier for 8 h. The final test strip product was obtained by firmly pressing the dried glass fiber and absorbent paper on the substrate, and cutting it into pieces in a width of 3 mm.
4. Results & discussion
4.1. Preparation and characterization of UCNPs@mSiO2
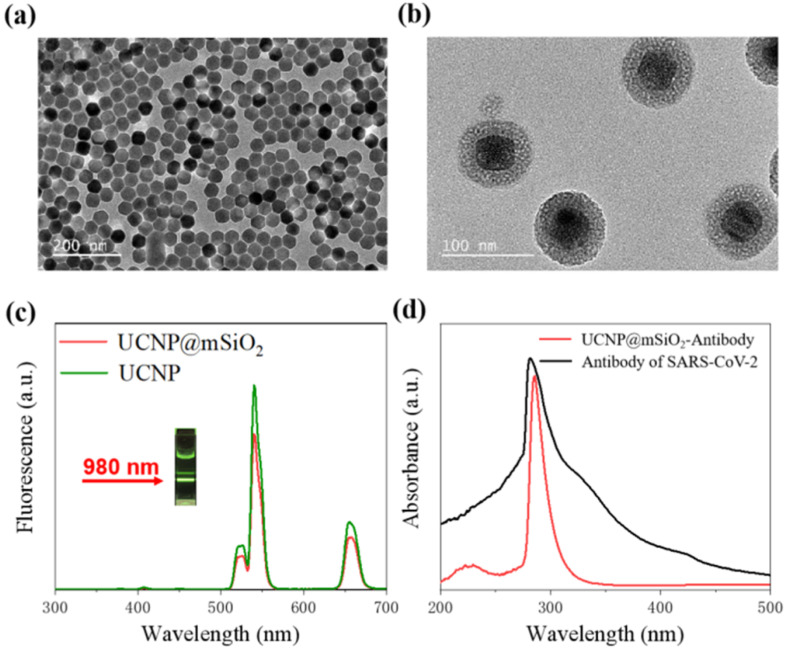
As a result, the transmission electron microscopy (TEM) proved that we fabricated UCNPs@mSiO2 with great uniformity. As illustrated in Fig. 3 a, statistical distribution results show that the synthesized core-shell UCNPs (NaYF4:Yb,Er@NaYF4) have a uniform diameter of 45.2 ± 2.7 nm (average size ± standard deviation, N = 100). After coating mSiO2, around 15 nm-thick silica shell monodispersely grows on the surface of UCNPs, which can be seen clearly from the TEM image of UCNPs@mSiO2 (Fig. 3b). Fig. 3c records the up-converting luminescence characteristics (UCL) of UCNPs@mSiO2. It can be observed that the as-prepared UCNPs@mSiO2 presents green UCL around 542 nm and red UCL around 657 nm under 980 nm NIR laser irradiation at a power of 1.3 W/cm2. The inset in Fig. 3c shows apparent green color emission from the aqueous suspension of the UCNPs@mSiO2 nanoparticles. As shown in Fig. 3d, antibodies exhibit a strong characteristic absorption peak at around 281 nm, and the label-antibody compound shows a characteristic peak at the wavelength of about 285 nm. This result indicates that antibody is successfully conjugated with UCNP@mSiO2.
Fig. 3.
Characterization results of the UCNPs@mSiO2. (a) TEM images of NaYF4:Yb,Er@NaYF4 (UCNPs); (b)TEM images of UCNPs@mSiO2; (c)UCL spectrum of UCNPs@mSiO2 under 980 nm laser irradiation at a power of 1.3 W/cm2(d)UV–vis absorption spectrum of UCNP@mSiO2-COOH and UCNP@mSiO2-antibodies in water.
4.2. The feasibility of the 5G-enabled fluorescence sensor for SARS-CoV-2 prognosis
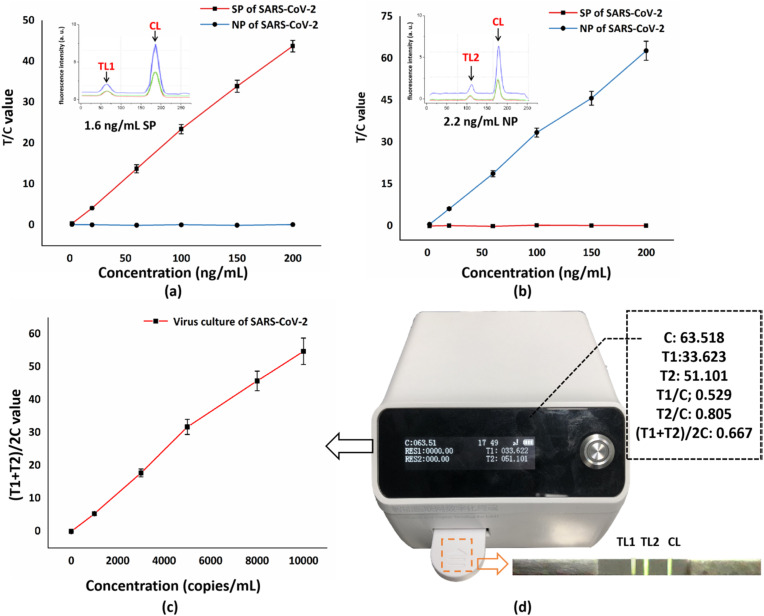
The proposed fluorescence sensor is capable of realizing quantitative detection of SP and NP of SARS-CoV-2 based on UCNPs labeled lateral flow strips and can be promisingly used for direct detection of SARS-CoV-2 from patient's nasopharyngeal samples. The detection time lasts 10 min in total, which is much shorter than that of nucleic acid detection (last for hours), benefiting much for the rapid point-of-care diagnosis. To verify its feasibility, we purchased recombinant SP and NP of SARS-CoV-2 from Acro Biosystems (Beijing, China) and obtained virus culture of SARS-CoV-2 from local hospitals to conduct a comparative experiment. We initially tested the sensor with standard solutions containing different concentrations of recombinant SP and NP. As is shown in Fig. 4 a and b, a well-established linear relationship existed between the concentrations of recombinant proteins and the fluorescence intensity. The insets in Fig. 4a and b are the fluorescence intensity curve drawn in the sensor at the LODs of the SP concentration (1.6 ng/mL) and NP concentration (2.2 ng/mL). For both recombinant proteins, the fluorescence intensity on TLs increases with the increase of SP and NP concentrations. When the concentration reaches 200 ng/mL, the fluorescence intensity becomes saturated, indicating that upper limit of detection is reached. Moreover, no crossover occurred between SP and NP detection. For recombinant SP, we reach a quantitative range from 2 ng/mL-200 ng/mL with an LOD of 1.6 ng/mL. As for recombinant NP, an LOD of 2.2 ng/mL is achieved and the detection range is from 2.2 ng/mL-200 ng/mL. We obtained the above LODs by setting a threshold above background noise. Only if the calculated T/C value is 0.3 values above the noise baseline is the signal considered valid. We applied a refined concentration gradient of analytes for testing, and it was discovered that when the analyte concentration went below the stated LODs, the fluorescence on the TLs became so faint that was drowned out by background noise (interfering fluorescence on the test strip), leading to extremely low T/C value, which could not be considered as valid. The difference in the limit of detection between two kinds of proteins is probably caused by the difference in their amino acid sequence and the different expression and renaturation in the process of recombination, which eventually leads to unequal binding ability to the antibody. It is also worthy to note that, because the murine antibodies can always bind to the goat anti-mouse IgG, the fluorescence intensity on the CL remains stable once the test strip is valid. Therefore, T/C value is used to reduce errors of test results. Compared with other LFIA reports on antigen detection, this system exhibits superiority in quantitative detection and short time consumption (Shown in Table 1 ).
Fig. 4.
(a) Quantification of SP of SARS-CoV-2 from 2 to 200 ng/mL. The inset illustrates that the LOD of SP is 1.6 ng/mL. (b) Quantification of NP of SARS-CoV-2 from 2 to 200 ng/mL. The inset illustrates that the LOD of NP is 2.2 ng/mL (c) quantification of virus culture of SAES-CoV-2. (d) The real photo of the fluorescence sensor and the fluorescence image of the test strip during detection.
Table 1.
Comparison between other reports and this work on SARS-CoV-2 antigen detection.
| Detection target | Limit of detection (LOD) | Quantitative range | Detection time | Reference |
|---|---|---|---|---|
| Recombinant nucleocapsid antigen | 0.65 ng/mL | Not mentioned | 20 min | Grant et al. (2020) |
| SP | 0.1 ng/mL | 0.2–100 ng/mL | 16 min | Liu et al. (2021) |
| Antigen | Not mentioned | Not mentioned | 15 min | Lin et al. (2020) |
| NP | 2 ng antigen protein | Not mentioned | 20 min | Kim et al. (2020) |
| SP and NP | 1.6 ng/mL for SP; 2.2 ng/mL for NP | 2–200 ng/mL for both SP and NP | 10 min | This work |
4.3. Real clinical samples validation
We further tested the virus culture of SARS-CoV-2 to verify the feasibility of the sensor of detecting real samples. Inactivated virus with a concentration of 50,000 copies/mL was obtained from a P2 laboratory in Chongqing, China. In order to cleave the cell membrane so that the NP can be exposed to the outside, we used cell lysis buffer (composed of 32.9 mmol/L K2HPO4, 136.9 mmol/L NaCl, 5 g/L Casein-Na, 0.1% S9 and 0.02% NaN3, pH = 7.4) to dilute the virus sample. In dilution process, the buffer was initially mixed well with the virus sample under ambient temperature, and the obtained solution was then settled for 3 min in a centrifuge tube. After repeating the above process, a set of virus solution with concentration gradients (ranges from 1000 copies/mL to 10,000 copies/mL) was obtained. To conduct the experiment, we applied 90 μL of each well-mixed solution to the test strip for 10 min and used the proposed fluorescence senor to test the results. As is shown in Fig. 4c and d, the sensor provided two TL values of detection, representing the concentration of SP (T1) and NP (T2), respectively, and we calculated (T1+T2)/2C value to reflect the virus concentration. As a result, the newly calculated curve follows a positive linear relationship with the virus concentration. Moreover, when a negative sample is applied (containing only cell lysis buffer) to the tests strip, there are no signals detected on TLs, proving that the proposed sensor has high accuracy, which holds great potential in the medical market of diagnosing suspected COVID-19 patients. It is worth mentioning that the slope of the detection line gradually decreases as the detection concentration increases. This is mainly because the antigen-antibody binding efficiency follows an S-shaped curve (Russell et al., 1989). Excessive SARS-CoV-2 antigen contained in the sample liquid will eventually reduce its binding efficiency with the antibodies immobilized on the conjugate pad and test lines, thus resulting in a slowdown in the growth rate of the light signal intensity.
To address the stability of the proposed fluorescence sensor, we conducted three sets of repeated experiments by using different concentrations of virus culture fluid from low (1500 copies/mL) to high (8000 copies/mL). Each set of fluid sample was measured 20 times separately, and we calculated the average value, standard deviation (SD), and coefficient of variation (CV %) of each set to demonstrate the reproducibility and stability. From Table 2 we can observe that, irrespective of the analyte concentration, the result data in a same set of test fluctuates little around the average and the CV% is under 7%, indicating that the system has good stability and reproducibility in a wide range. It can be observed that the system CV% under low antigen concentration performs better than that under high antigen concentration. This is due to the nature of the binding efficiency between antibody and antigen commented above. The result proves that when the virus concentration exceeds 10,000 copies/mL, the fluorescence intensity on TL1 and TL2 will change irregularly, making it hard for quantification. However, instability in this case is acceptable because the proposed fluorescence sensor is designed for early diagnosis and proactive prognosis of suspected COVID-19 patients. On the one hand, people in real life without obvious abnormalities (such as fever, severe cough) are almost impossible to reach such a high virus concentration in the body. On the other hand, if one's SARS-CoV-2 test result is found to be slightly higher than the normal value, intervention from hospital or medical institution is required with no delay. Thus, the detection sensitivity and stability at low virus concentrations should be the key factors of this sensor to focus on.
Table 2.
The stability and reproducibility of the proposed system.
| Test strips #1 | Test strips #2 | Test strips #3 | |
|---|---|---|---|
| Concentrations of the virus culture (copiesl/mL) | Low concentration 1500 |
Mid concentration 4000 |
High concentration 8000 |
| Number of tests | 20 | 20 | 20 |
| Test result (μmol/L) | 1553.2 ± 21.8 | 4173.7 ± 20.3 | 8242 ± 21 |
| SD | 87.7 | 247.5 | 575.3 |
| CV% | 5.65 | 5.93 | 6.98 |
4.4. IoMT application of the 5G-enabled fluorescence sensor for online COVID-19 detection and monitoring
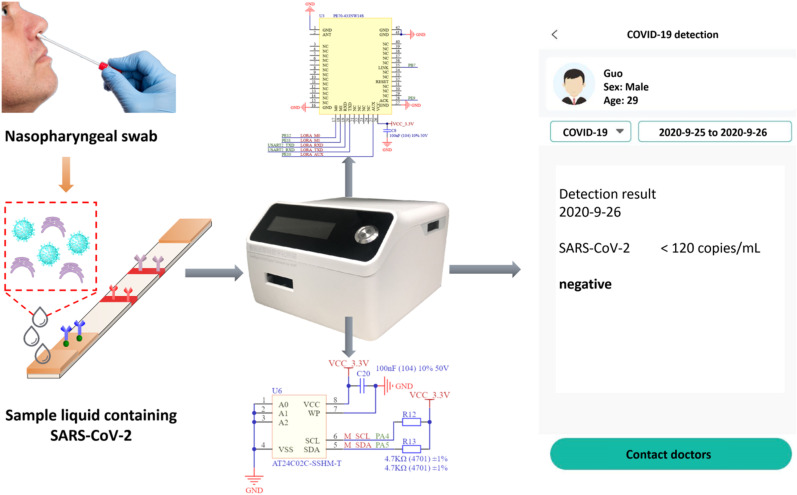
The fluorescence sensor is 5G-enabled, which is accessible to edge hardware devices (personal computers, 5G smartphones, IPTV, etc.) through integrated Bluetooth module. When the measurement is done, the vital medical data are transmitted to the fog layer and central 5G cloud server with ultra-low latency for cloud-edge computing and big data storage (Verified in the supporting information: a video of online COVID-19 detection process on a personal computer). In order to guarantee the privacy security of patients and to improve the Quality of Service (QoS) of the system, we are looking forward to employ reliable transmission protocols and well-established encryption algorithms in data transmission and storage. In this study, we also develop a COVID-19 monitoring module on a smartphone App that works with the proposed sensor to realize online detection and monitoring of suspected COVID-19 patients. As is shown in Fig. 5 , the sample solution extracted from the patient's nasopharyngeal swab is applied to the sensor, and the sensor's data storage module and 5G networking module enable biomedical information to be processed and sent out instantly. Users first log in to the module to create a personal nominee account. After performing point-of-care tests of SARS-CoV-2, they have the option of manually or automatically transferring the results to the recording log. These recordings can be dynamically and timely collected by the hospital systems due to the advancing of IoMT. Once the hospital system detects the anomaly, doctors or experts can make a quick and accurate response by generating remote consolation or face-to-face medical help. Moreover, due to the positioning function of the module, hospitals are able to rapidly locate the COVID-19 suspected patient's geographic location as soon as the positive report is uploaded. In this manner, the hospital can notify the relevant authorities to quickly screen and isolate vulnerable areas, significantly mitigating the risk and means of virus spread. Furthermore, to enable two-way monitoring, the system is endowed with functions of intelligent decision. Sudden abnormal events such as sudden server illness must reach the destination node before the deadline; otherwise, the patient's life will be threatened. Therefore, we embedded fuzzy logic and deep learning algorithms into the system to make it event-driven. Based on this method, the system can intelligently identify the urgency of the current situation according to the established data base. As long as some critical abnormal information is detected, the system will soon alert the local medical institution for a first aid, to the utmost extent to mitigate the danger from the emergency situation. Moreover, since the elderly are susceptible to SARS-CoV-2, but they are generally difficult to learn advanced smart devices without any previous related knowledge, the COVID-19 monitoring system can also connect to family healthcare centers, where adults can assist their aging relatives with SARS-CoV-2 testing and upload results to obtain appropriate medical suggestions for future treatment and prevention. In addition, the proposed system releases the burden of numerous people going to central hospitals for inspection, and consequently improving the efficiency of medical treatment and resource utilization of hospitals. As a result, people can stay up to date on critical health information anywhere and anytime, while medical facilities can obtain significant pathological data, which can be impressively beneficial for the treatment and prevention of COVID-19 and other infectious diseases in the future.
Fig. 5.
IoMT applications of the proposed 5G-enabled sensor (with a 5G networking circuit module and a data storage circuit module) on a smartphone App for online COVID-19 detection and monitoring.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.bios.2021.113160.
The following is/are the supplementary data related to this article:
5. Conclusion
In conclusion, this paper demonstrated a 5G-enabled fluorescence sensor for online SARS-CoV-2 detection. One innovation of the system is the use of UCNP@mSiO2 as probes for quantification of SP and NP of SARS-CoV-2. As a result, the proposed assay is sensitive and practical, which is capable of detecting SP of SARS-CoV-2 with an LOD of 1.6 ng/mL and NP of SARS-CoV-2 with an LOD of 2.2 ng/mL. Furthermore, the proposed sensor is IoMT enabled and is accessible to edge hardware devices through Bluetooth. The obtained medical data can be transmitted to the fog layer and 5G cloud server with ultra-low latency and high reliability. Both individuals and hospitals can stay up to date on critical state of COVID-19, benefiting much for the future treatment and prevention of SARS-CoV-2 and other infectious diseases.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors thank for the Emergency Project for COVID-19 Infection, Prevention, and Treatment, Chongqing Municipal Education Commission, KYYJ202003.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113160.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Catarinucci L., et al. vol. 2. Dec. 2015. An IoT-aware architecture for smart healthcare systems; pp. 515–526. (IEEE Internet of Things Journal). 6. [Google Scholar]
- Chan Jasper F.W., Lau Susanna K.P., Kelvin K.W., Cheng Vincent C.C., Woo Patrick C.Y., Yuen Kwok-Yung. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang G., Liu L., Li Z. 2020. Emerging biosensing technologies for improved diagnostics of COVID-19 and future pandemics; p. 121986. Talanta. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C.C., Lau S.K.P., Woo P.C.Y., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolero A. Covid-19: a digital epidemic. BMJ. 2020;368:m764. doi: 10.1136/bmj.m764. [DOI] [PubMed] [Google Scholar]
- Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease. Biosens. Bioelectron. 2019;165:2020. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant Benjamin D., Anderson Caitlin E., Williford John R., Alonzo Luis F., Glukhova Veronika A., Boyle David S., Weigl Bernhard H., Nichols Kevin P. Anal. Chem. 2020;92(16):11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- Guo J., Chen S., Guo J., Ma X. Nanomaterial labels in lateral flow immunoassays for point-of-care-testing. J. Mater. Sci. Technol. 2021;60:90–104. [Google Scholar]
- J. Guo et al., "Cyber-physical healthcare system with blood test module on broadcast television network for remote cardiovascular disease (CVD) management," in IEEE Transactions on Industrial Informatics, doi: 10.1109/TII.2020.3010280.
- Han R., Yi H., Shi J., Liu Z.J., Wang H., Hou Y.F., Wang Y. PH-responsive drug release and NIR-triggered singlet oxygen generation based on a multifunctional core-shell-shell structure. Phys. Chem. Chem. Phys. 2016;18:25497–25503. doi: 10.1039/c6cp05308d. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven Protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S., Kenmore M., Kirkland M., Andy Badley R. High‐density immobilization of an antibody fragment to a carboxymethylated dextran‐linked biosensor surface. J Mol Recogn. 1998;11:1–6. doi: 10.1002/(SICI)1099-1352(199812)11:1/6<200::AID-JMR423>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Tian S., Zhao W., liu K., Ma X., Guo J. Multiplexed detection of biomarkers in lateral-flow immunoassays. Analyst. 2020:2828–2840. doi: 10.1039/c9an02485a. [DOI] [PubMed] [Google Scholar]
- Ji H., Park S., Yeo J., Kim Y., Lee J., Shim B. IEEE Wireless Communications. Vol. 25. JUNE 2018. Ultra-reliable and low-latency communications in 5G downlink: physical layer aspects; pp. 124–130. 3. [Google Scholar]
- Kapassa E., et al. Sixth International Conference on Internet of Things: Systems, Management and Security (IOTSMS), Granada, Spain, 2019. 2019. An innovative ehealth system powered by 5G network slicing; pp. 7–12. [DOI] [Google Scholar]
- Kim Bum Tae, Park Edmond Changkyun, Kim Hong Gi, Kim Seung Il. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2020:112868. doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W. Development and clinical application of A rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wang L., Wang H., Li X., Zhang S., Xu Y., Wei W. Serum SARS-COV-2 nucleocapsid protein: a sensitivity and specificity early diagnostic marker for SARS-COV-2 infection. Front Cell. Infect. Microbiol. 2020;10:470. doi: 10.3389/fcimb.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Qiuyuan, Wen Donghua, Wu Jing, Liu Liling, Wu Wenjuan, Fang Xueen, Kong Jilie. Anal. Chem. 2020;92(14):9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Dan, Ju Chenhui, Han Chao, Shi Rui, Chen Xuehui, Duan Demin, Yan Jinghua, Yan Xiyun. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;173 doi: 10.1016/j.bios.2020.112817. 112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Wang X., Hahn K., Sanchez S. Motion control of urea-powered biocompatible hollow microcapsules. ACS Nano. 2016;10:3597–3605. doi: 10.1021/acsnano.5b08067. [DOI] [PubMed] [Google Scholar]
- Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., McElhannon J. Future edge cloud and edge computing for Internet of Things applications. IEEE Internet Things J. Feb. 2018;5(1):439–449. [Google Scholar]
- Qadri Y.A., Nauman A., Zikria Y.B., Vasilakos A.V., Kim S.W. IEEE Communications Surveys & Tutorials. Vol. 22. 2020. The future of healthcare Internet of Things: a survey of emerging technologies; pp. 1121–1167. 2. Secondquarter. [Google Scholar]
- Russell Alan J., Trudel Laura J., Skipper Paul L., Groopman John D., Tannenbaum Steven R., Klibanov Alexander M. Antibody-antigen binding in organic solvents. Biochem. Biophys. Res. Commun. 1989;158(Issue 1):80–85. doi: 10.1016/s0006-291x(89)80179-2. [DOI] [PubMed] [Google Scholar]
- Sachs J., et al. “Adaptive 5G low-latency communication for tactile Internet services. Proc. IEEE. 2019;107(2):325–349. [Google Scholar]
- Tiwari A., Tripathi R.P., Bhatia D. Proc. Med. Data Security Bioengineers, Hershey, PA, USA. 2019. Advancements in data security and privacy techniques used in IoT-based hospital applications; p. 23. [Google Scholar]
- Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop. Med. Int. Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., et al. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won, J., Lee, S., Park, M., Kim, T.Y., Park, M.G., Choi, B.Y., Kim, D., Chang, H., Kim, V. N., Lee, C.J. “Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the coronavirus disease 2019 (COVID-19),” Exp. Neurobiol., 29, 2020. [DOI] [PMC free article] [PubMed]
- Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. Evaluation of enzyme-linked immunoassay and colloidal gold-immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19) medRxiv. 2020 doi: 10.1101/2020.02.27.20028787. 2020. [DOI] [Google Scholar]
- You Y.Q., Xu D.D., Pan X., Ma X. Self-propelled enzymatic nanomotors for enhancing synergetic photodynamic and starvation therapy by self-accelerated cascade reactions. Appl. Mater. Today. 2019:508–517. [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loopmediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.