Summary
Many proteins that are needed for progression through S-phase are produced from transcripts that peak in S-phase, linking temporal expression of those proteins to the time that they are required in cell cycle. Here we explore the potential roles of long non-coding RNAs in cell cycle progression. We used a sensitive click chemistry approach to isolate nascent RNAs in a human cell line, we identified over 900 long non-coding RNAs (lncRNAs) whose synthesis peak during S-phase. Over 200 of these were long intergenic non-coding RNAs (lincRNAs) with S-phase specific expression. We characterized three of these lincRNAs by knockdown and found that all three lincRNAs were required for appropriate S-phase progression. We infer that non-coding RNAs are key regulatory effectors during the cell cycle that act on distinct regulatory networks and herein we provide a large catalogue of candidate cell cycle regulatory RNAs.
Introduction
Over 80% of the human genome is expressed as RNA (Consortium et al., 2012) yet less then 3% of the genome encodes for proteins (Bertone et al., 2004; Guttman et al., 2009; Kapranov et al., 2007). The remaining non-coding transcripts represent a major fraction of the cellular RNA pool with functions yet to be discovered. One subgroup of these noncoding transcripts is long non-coding RNAs (lncRNAs). LncRNAs are defined as 200 nucleotides or longer transcripts with no or minimal protein coding potential. They contribute to regulation via diverse molecular functions that include chromatin regulation (Pandey et al., 2008; K. C. Wang et al., 2011), higher order chromatin organization (Engreitz et al., 2013; Giorgetti et al., 2016; Hacisuleyman et al., 2014), genome stability (S. Lee et al., 2016), DNA repair (Michelini et al., 2017), DNA modification (Frank et al., 2019), subcellular/nuclear compartmentalization (Clemson et al., 2009; Sasaki, 2009; West et al., 2014), and mRNA stability, modification and splicing (Kopp and Mendell, 2018; Ransohoff et al., 2018).
DNA replication occurs at various stages of S-phase, depending upon the locus involved, and must generate daughter strands that have appropriate chromatin structure and regulatory potential. It is well established that chromatin associated proteins are required for the tight regulation of accurate replication of both the DNA and the associated chromatin structure (Alabert and Groth, 2012; Budhavarapu et al., 2013; Probst et al., 2009). Given the numerous possible roles for lncRNAs in regulation of chromatin-templated processes, we reasoned that lncRNAs might also play key roles in S-phase. The mRNA of the proteins important to S-phase transition and progression are tightly regulated and expressed at specific points in S-phase, raising the possibility that non-coding RNAs important for this process might also be transcribed specifically during S-phase.
Potential roles for lncRNAs in cell cycle progression have been indicated previously. For example, the well-studied and abundant long intergenic non-coding RNA (lincRNA) MALAT1 functions in numerous settings including G1/S transition as well as mitotic progression through p53 regulated checkpoint activation and alternative splicing of cell cycle regulatory factors (Tripathi et al., 2013). A MYC regulated lncRNA, CONCR, is highly expressed in cancer cells and is required for proper cell cycle progression and sister chromatid cohesion through the modulation of DNA helicase DDX11(Marchese et al., 2016). In addition, non-coding RNAs have been implicated in maintenance of epigenetically silenced regions in daughter cells in S. pombe (Verdel et al., 2004; Volpe et al., 2002) and similarly, in mammals, intergenic spacer originated long non-coding RNAs are essential for maintenance and inheritance of silent rDNA chromatin during the cell cycle (Guetg et al., 2012; Mayer et al., 2006). A systematic identification of lncRNAs that might be involved in cell cycle progression has not been done, thus there is limited data concerning the frequency of potential cell cycle regulatory RNA species. One hurdle in this analysis is that many lncRNAs, including the subset of intergenic lincRNAs are expressed at low to moderate levels in cells making detection difficult.
To identify lncRNAs that are candidates for S-phase regulatory function we developed a protocol to isolate nascent RNAs that are expressed during S-phase. We reasoned that lncRNAs important to regulation during S-phase would have peak in expression at this stage of the cell cycle, analogous to transcription of S-phase specific proteins that play role in cell cycle progression. We focused subsequent analysis on the subgroup of long intergenic non-coding RNAs, as the potential function of those non-coding RNAs can be assessed without the complexities introduced by an overlapping or nearby protein coding gene. Herein, we identified 909 nuclear enriched lncRNAs that peaked during S-phase, of which 229 were lincRNAs. RNA-sequencing (RNA-seq), LNA knockdown and flow cytometry were used to characterize three of these lincRNAs for their impacts on cell cycle and gene expression. These three lincRNAs are found to be frequently mutated in different cancers (“TCGA Research Network,” n.d.). We found that each lincRNA is essential to traverse S-phase and that depletion of each lincRNA resulted in perturbation of different cellular pathways in a manner specific to each lincRNA. These results indicate that long non-coding RNAs that have cell cycle specific expression are candidates for regulation of cell cycle progression and that they have distinct functions. There are a large number of these RNAs, each expressed at a specific time point in S-phase, that might facilitate the maintenance of gene expression programs in human cells.
RESULTS:
Nascent RNA profiles across S-phase of human cell cycle:
To examine whether some lncRNAs are enriched at specific stages of the cell cycle we analyzed newly synthesized transcripts. We reasoned that lncRNAs meant to function at a specific stage in cell cycle are likely to be expressed at that stage. Although on average lncRNAs are less stable than protein coding RNAs, they can have a wide range of half-lives (Clark et al., 2012). Hence detection of these lncRNAs is expected be more sensitive if nascent transcripts are characterized.
We developed a protocol to label nascent RNAs at specific stages of the cell cycle in a manner that allowed for their efficient isolation. To do this we used 5-ethynyl uridine (EU) metabolic nascent RNA labeling. To isolate the EU-labeled RNAs genome-wide we synthesized a multifunctional probe composed of an azido benzoin ester (ABE) and biotin, which we refer as ABE-biotin (Figure 1A, Supp. Figure 1A). This probe can conjugate to the EU-labeled RNA via a copper(I) catalyzed azide-alkyne cycloaddition (CuAAC) reaction (often referred to as click chemistry) (Kolb et al., 2001). The RNA is then released through photolysis by near-visible UV (350nm) irradiation (Supp. Figure 1B). This is an efficient capture and release because ABE-biotin is stable to visible-light (400 – 750 nm) during preparation of samples but undergoes rapid photocleavage when exposed to near visible UV (at 350 nm) with a half-life of ca. 5 min (Supp. Figure 1C). Thus, photolysis of the samples that were conjugated with ABE-biotin allowed efficient elution of the EU labelled molecules from biotin-streptavidin beads. The materials non-specifically bound to the beads were not released during the photocleavage based elution of specifically bound RNAs, which lowered background and increased specificity (Supp. Figure 1D, E).
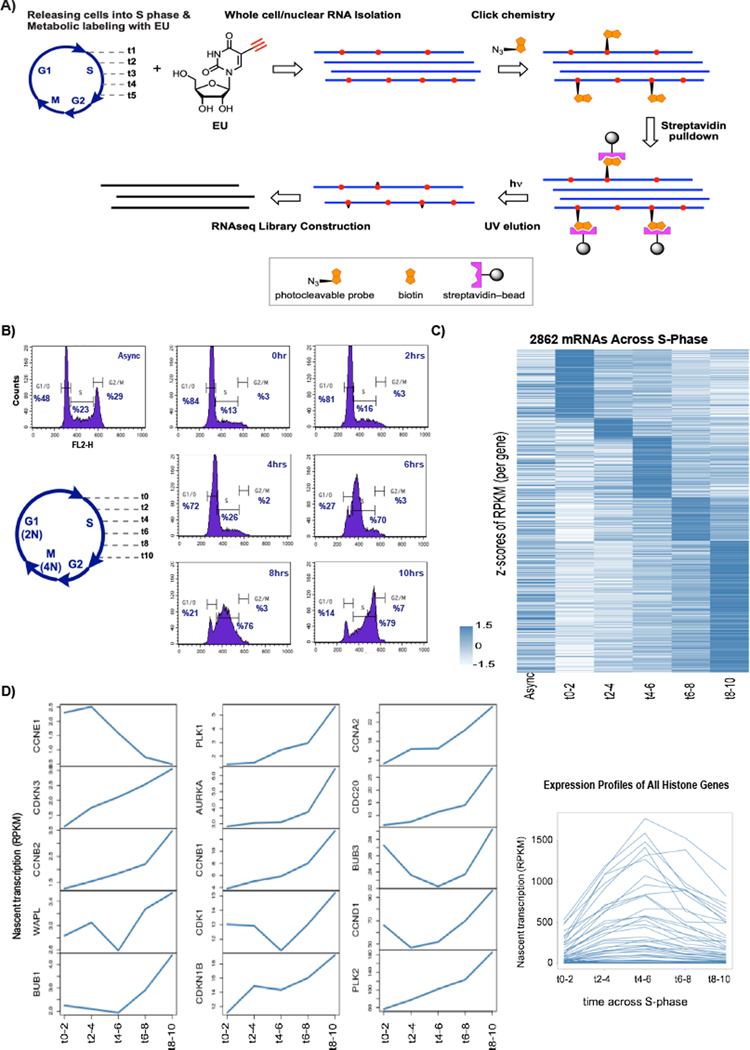
Figure 1. Nascent RNA profiles of RPE1 S-phase.
A) Schematic of experimental design for the genome wide nascent RNA pulldown after metabolic labeling with EU during S-phase (see also See also Supp. Figure 1 and Methods S1). B) Propidium iodide (PI) stained flow cytometry profiles of hTERT-RPE1 before and after mimosine synchronization and during their progress in S-phase time course. C) Whole cell nascent mRNA expression profiles for differentially expressed genes across S-phase time course (See also Supp. Figure 2D). Average of two replicates per time point was calculated and a cut-off of 1.5-fold change of maximum average RPKM across time points over minimum average RPKM was applied to find differential genes. Z-scores of average RPKMs (including Async) are plotted. Genes are clustered based on the time point of the maximum average RPKM (See Supp. Figure 2B for analyses with more stringent criteria). D) Nascent cyclin mRNAs and Histone cluster RPKM/Time line-plots showing the expression dynamics of differential cycling gene regulation across S-phase. Average of two replicates for each time point is plotted.
To identify RNAs expressed during S-phase we used an hTERT immortalized human retinal pigment epithelium cell line, hTERT-RPE1. These are non-transformed near diploid cells which can be synchronized at the G1/S border with high (>80%) efficiency when incubated in medium containing mimosine for 22hrs. We tried several synchronization methods in different cell lines and decided on mimosine as synchronization method in RPE1 cells, due to the synchronization efficiency at G1/S and efficient S-phase progression after release. After synchronizing cells at the G1/S border we replaced mimosine containing medium with normal growth medium to release the cells from the block. We analyzed cells using FACS analysis to show their progression in S-phase. We incubated cells with EU containing medium in five two-hour windows to isolate the nascent RNAs from each time window (Figure 1A). We collected cells at release from G1 block (t0–2, labeled from 0 to 2hrs), in early S-phase (t2–4), in early/mid S-phase (t4–6), in mid to late S-phase (t6–8), and in late S-phase (t8–10) (Figure 1B). We sequenced nascent RNAs at each time point from duplicate experiments and identified RNAs that showed averaged expression levels above asynchronous expression levels at some time point during S-phase. To validate these data further we compared these results to standard RNA seq of cells at the same time points in S phase, thereby verifying that synthesis rates correlated with steady state levels (Supp. Figure 2A). From the nascent RNA data set we identified RNAs that each had at least a 1.5-fold difference between their maximum and minimum average expression during S-phase, indicating that they displayed dynamic expression during S-phase. This resulted in a list of nearly 3000 mRNAs (Figure 1C) and over 900 long non-coding RNAs (see below). A more stringent cutoff, a 2.0-fold change in at least one time point when comparing maxima to minima, lowered the number of RNAs identified in each class (Supp. Figure 2B, C) but did not change any of the conclusions that we reach. The majority of RNAs identified with either cutoff showed a single time point during S-phase where they had their most significant increase in expression (see groupings across time points in (Figure 1C and Supp. Figure 2B, C). In the analysis described below we used the 1.5-fold cutoff as we wanted to be inclusive; synchronization of cells across S-phase is not perfect which introduces noise that can diminish fold differences.
To validate the methodology, we analyzed mRNAs whose expression patterns across S-phase are well studied. Consistent with the flow cytometry profile and the timing of expression during specific phases of the cell cycle, the most enriched gene ontology (GO) terms for the nascent mRNAs that peaked during early/early-mid S-phase (t2–4 and t4–6) were DNA-protein complex, histone core and nucleosome assembly. Cell cycle progression and M phase specific genes predominated in the mRNAs enriched in mid/mid-late S-phase (t6–8 and t8–10) (Supp. Figure 2D). The transcription kinetics of the genes that are known to regulate the G1/S transition and S-phase progression, such as cyclin and cyclin dependent kinase genes and histone gene clusters (Bertoli et al., 2013; Orlando et al., 2008; Zhao et al., 2000) were as expected and provided further validation of our experimental approach (Figure 1D).
Coordinated Expression Patterns of LncRNAs Suggest Functional Roles in Cell Cycle
We identified 909 previously annotated nuclear lncRNAs that are differentially expressed across the S-phase using a 1.5-fold cutoff (Supp. Fig Figure 2E). We classified S-phase enriched lncRNAs by comparing them to mRNAs that had similar expression patterns and identified three coordinated expression profiles of mRNAs whose synthesis peaked at early, mid and late S-phase. The first cluster was enriched with mRNAs involved in developmental processes, the second in mRNAs involved in DNA packaging and nucleosome assembly and the third cluster was enriched for transcripts involved in M-phase and cell division (Figure 2A).
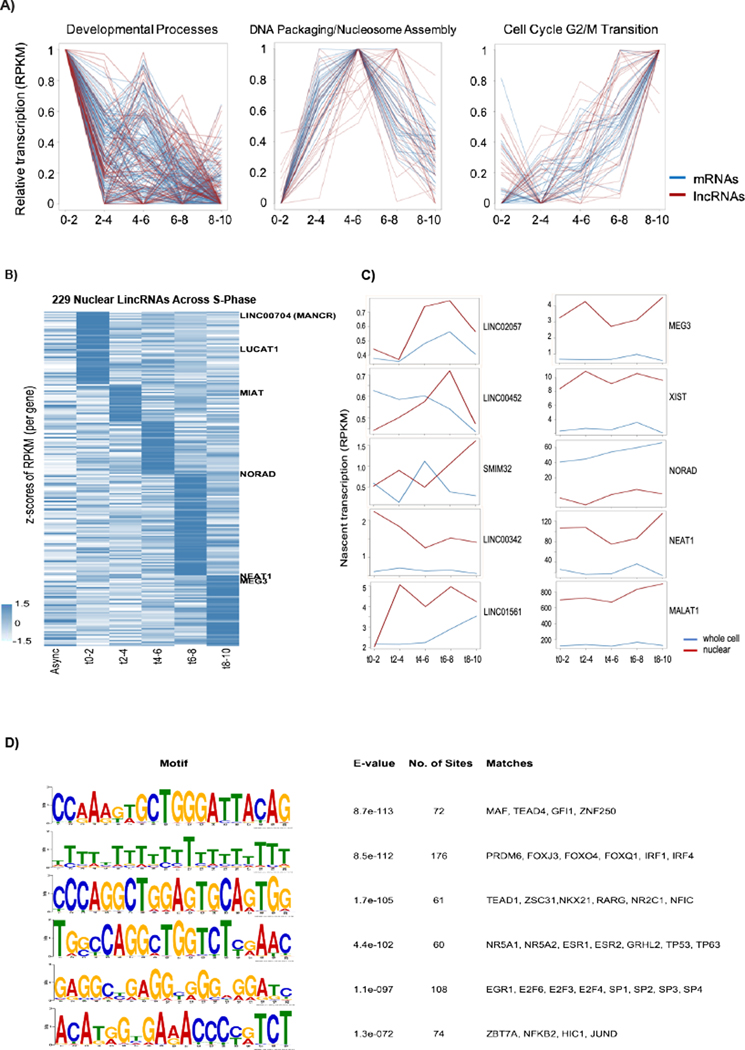
Figure 2. Coordinated expression of cell cycle regulated mRNAs and lncRNAs suggest functional significance.
A) mRNAs (blue line) and lncRNAs (red line) with similar expression patterns through S-phase (line plots). These have been divided into three categories shown left to right, in the panel 1) Peaking at early S-phase: enriched for mRNAs of proteins involved in differentiation and development processes. 2) Peaking at mid S-phase: enriched for nucleosome assembly and DNA packaging related mRNAs. 3) Peaking at late S-phase: enriched for mRNAs of proteins involved in M phase and Nuclear division. Average of two replicates for each time point is plotted. RPKMs are normalized for scale 0–1 with lowest expression as 0 and highest expression as 1 (See methods for more details on normalization). B) Nuclear nascent lincRNA expression profiles for differentially expressed genes across S-phase time course. Average of two replicates per time point was calculated and a cut-off of 1.5-fold change of maximum average RPKM across time points over minimum average RPKM was applied to find differential genes. Z-scores of average RPKMs (including Async) are plotted. lincRNAs are clustered based on the time point of the maximum average RPKM (See Supp. Figure 2E for analyses with more stringent criteria). C) RPKM/Time line-plots of individual lincRNA expression profiles (as labeled) compared in whole cell and nuclear enriched data sets. Average RPKM of two replicates per time point is shown. D) MEME results of top discovered promoter motifs in 1200 bp surrounding (1000 bp upstream and 200 bp downstream) transcription start site of nuclear lincRNA genes with S- phase expression peak. Common binding motifs of this cluster include binding sites for factors that control mitogenesis, DNA replication and cell cycle regulation. Top 6 motifs by E-value (statistical significance as calculated by MEME) and their matches to known transcription factor motifs are shown.
We chose to study three of these long non-coding RNAs in greater detail to test the hypothesis that they might be needed for S-phase progression. To minimize the confounding effects resulting from perturbation of neighboring protein coding genes on lncRNA function (Kopp and Mendell, 2018) we chose to study long intergenic lincRNAs. These were defined in the Ensembl genome database as being at least 5 kb away from other coding or non-coding gene classes, thereby excluding anti-sense and genic long non-coding RNAs. This avoids complications that arise from perturbing neighboring and/or overlapping genes. We restricted these analyses to the nuclear fraction of lincRNAs, as these have the potential to regulate gene expression and chromatin structure during replication and shortly thereafter. Of the 909 initially characterized lncRNAs we identified 229 lincRNAs that met our filtering criteria (Figure 2B). Each had at least a 1.5-fold difference between the minima and maxima of averaged expression across S-phase and most of these lincRNAs had a significant increase at a specific time point during S-phase (Figure 2B, see also Supp. Figure 2C for more stringent criteria in which we identified 112 lincRNAs that had significant peaks in both replicates). Individual lincRNAs whose expression peaks at varying time points during S-phase are shown in Figure 2C. We compare these newly identified S-phase lincRNAs with previously characterized nuclear (XIST, MEG3, NEAT1 and MALAT1) and cytoplasmic (NORAD) RNAS (S. Lee et al., 2016) for their expression timing and for whether the RNA was more abundant in nuclear fractions or whole cell extracts. We queried the Cancer Genome Atlas (TCGA) database and found 68 of the S-phase specific nuclear lincRNAs were included in that database and that 34 of the 68 were mutated or had copy number variations in different cancers (Supp. Fig 2F, Supp. Table 1).
To further validate this set of lincRNAs are bona fide S-phase transcripts we asked whether they shared promoter elements known to be involved in regulating S-phase specific transcription of mRNA genes. We queried promoter element enrichment within 1200 base pairs (bp) of the transcription start sites of the 229 nuclear enriched, S-phase lincRNAs (1000bp upstream and 200bp downstream of TSS). This analysis identified several high-confidence motifs (Figure 2D). The majority of the S-phase peaking lincRNAs had at least one of these transcription-factor binding site motifs, including the three lincRNAs we characterized further below, LINC00704, LUCAT1 and MIAT. Transcription factors with binding sites matching these motifs have roles in DNA replication, DNA damage, cell cycle regulation and mitogenesis (e.g. EGR1, several members of E2F transcription factor family, SP1–4, TP53, GIF1 TEAD1&4). Also, multiple other transcription factors with roles in growth and differentiation bind to these sequences, such as several members of the forkhead transcription factor family, GRGHL2 (epithelial cell differentiation), MAF (lens fiber differentiation), hormone receptors and retinoic acid regulated factors (Figure 2D). We conclude that these lincRNAs share expression patterns and regulatory elements with mRNAs important for the cell cycle.
LINC00704, LUCAT1 and MIAT are Essential for Proper S-Phase Progression.
We chose three lincRNAs for further analysis, LINC00704, LUCAT1 and MIAT because their upregulation in early S-phase was consistent with a potential role in the transition through S-phase (Figure 3A). We focused on transcripts peaking early in S-phase because mRNA transcripts expressed in late G1/early S phase encode proteins that regulate commitment to cell cycle entry, DNA replication and downstream events (van der Meijden et al., 2002). LINC00704 and LUCAT1 were significantly upregulated within the first two hours after release into S-phase and MIAT was upregulated two to four hours after release into S-phase. For these three RNAs, whole cell and nuclear nascent RNA pull-down sequencing and qPCR analysis confirmed both transcriptional peak timing and subcellular localization (Figure 3A, Supp. Figure 3A, B). We further verified that the peak of S-phase expression for each of these lincRNAs was higher than expression seen in asynchronous cells (Supp. Figure 3A). Each of the genic regions that encode these three RNAs were found to have elevated levels of H3K27 acetylation and low levels of H3K4 monomethylation at the transcription start sites, consistent with active expression (Supp. Figure 3C). To determine whether these RNAs might encode protein products, we assessed their protein coding potential using the CPC, which utilizes sequence alignment (Kong et al., 2007) and CPAT which is an alignment-free algorithm that utilizes regression models of sequence features (L. Wang et al., 2013). All three lincRNAs have no protein coding capacity according to CPAT; while CPC suggested minimal coding capacity for MIAT.
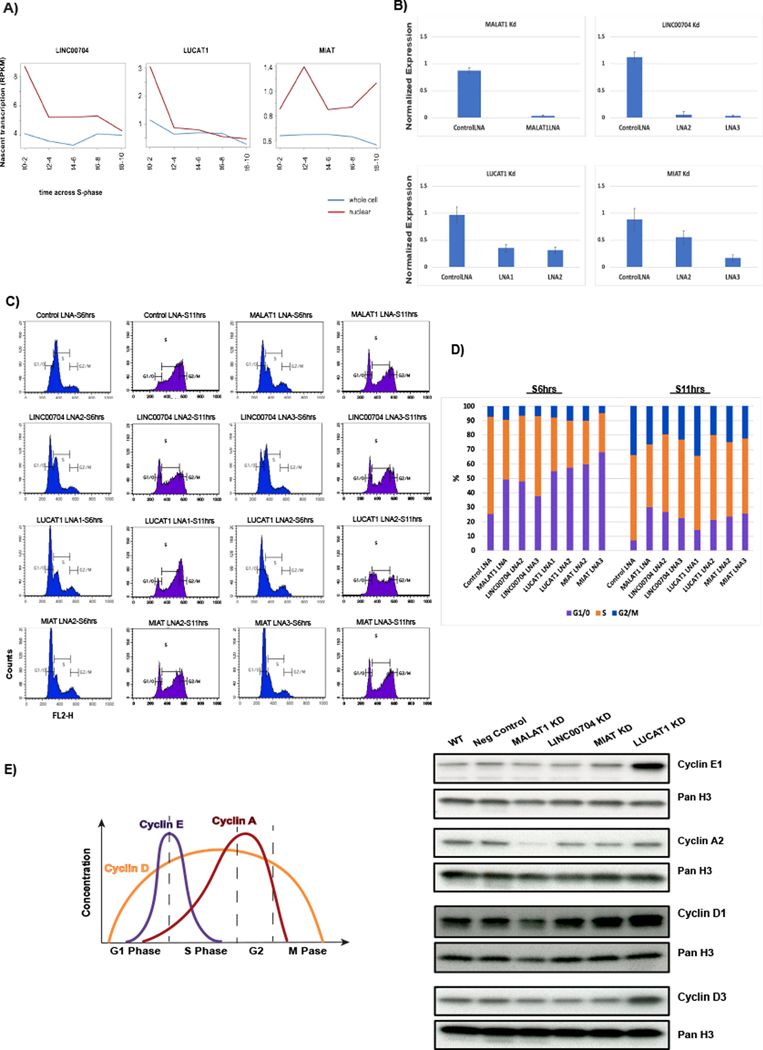
Figure 3. Depletion of early S-phase peaking lincRNAs LINC00704, LUCAT1 and MIAT causes cell cycle progression defects.
A) RPKM/Time line-plots showing expression dynamics of LINC00704, LUCAT1 and MIAT across time points in S-phase for whole cell and nuclear enriched (See also Supp. Figure 3A, B). B) RT-qPCR normalized to GAPDH showing knockdown efficiency with 2 different LNAs for LINC00704, LUCAT1 and MIAT. Cells were also treated with control LNAs D.melanogaster specific transcript targeting LNA (negative control) and MALAT1 LNA (positive control). C) Flow cytometry profiles at 6 hours and 11 hours after mimosine wash-off and release in S-phase of unperturbed (wild type) cells; control LNA transfected (negative control); MALAT1 LNA transfected cells (positive control); Linc00704 LNA treatment; LUCAT1 LNA treatment and MIAT LNA treatment. Two separate LNAs are shown for each of the experimental samples. D) Bar graph representation of the flow cytometry data. E) Western blot for different cyclin proteins that are differentially regulated at G1/S transition an S-phase progression. Alterations from normal WT levels were seen in LINC00704, LUCAT1 and MIAT knockdown cells (See also Supp. Figure 3E).
We assessed the impact of depleting these lincRNAs on cell cycle progression. We targeted each RNA of interest with LNA GapmeRs, utilizing locked antisense oligonucleotides (ASOs) that degrade complementary RNA via a ribonuclease H based mechanism. We used two different LNA GapmeRs for each lincRNA and achieved 45–90% reduction for each of them. (Figure 3B). To test the requirement for each lincRNA, RPE1 cells were transfected with LNA GapmeRs for either 42 or 47 hours using the following protocol: Approximately 14 hours after transfection of asynchronous cells mimosine was added for 22 hours to block the cells at G1/S border and then cells were released into S-phase for either 6 or 11 hours. In these experiments MALAT1 depletion was used as a positive control, as this lincRNA has previously been shown to be important in S-phase progression (Tripathi et al., 2013). Flow cytometry analysis following depletion of LINC00704, LUCAT1 and MIAT showed that each of these lincRNAs was necessary for normal progression through the cell cycle. The extent of the effect varied depending on the lincRNA and the level of knockdown, however each LNA showed significant delay of release into S-phase (Figure 3C; see remaining peaks at G1 after 6 hours of release) and incomplete progression through S-phase (Figure 3C; see 11 hours-time points). Each knockdown had effects on cell cycle similar to knockdown of MALAT1, revealing a role for all three lincRNAs in progression through the cell cycle (Figure 3C). Quantification of the percentage of cells at each stage of progression showed increased retention of cells in G1/0 and decreased numbers of cells traversing into G2/M (Figure 3D). Progression failure was apparent at all stages of S-phase (G1/S transition, S-phase and G2/M transition) but to different extents for each lincRNA depletion. These findings were further supported by defects in proliferation upon knockdown of these RNAs as measured by proliferation and cell viability measurements (Supp. Figure 3D).
Defects in traversing S-phase often correspond with differences in cyclin levels. We measured cyclin levels by Western blot after lincRNA knockdown. Cell extracts were prepared 10 hours after release into the cell cycle. Cyclin E1, which peaks at the G1/S transition, was upregulated in LUCAT1 depleted cells, whereas LINC00704 and MIAT depleted cells showed cyclin E1 levels similar to wild type cells (Figure 3E, Supp. Figure 3E). Cyclin A2 is expressed throughout S-phase with levels increasing towards late S-phase (Lim and Kaldis, 2013). Consistent with the cell cycle defects observed in LINC00704and MIAT knockdown cells, cyclin A2 levels were depleted in both cases. Surprisingly, Cyclin A2 was expressed at similar levels to wild type cells in LUCAT1 depleted cells despite a clear S-phase progression defect. Two members of cyclin D that are expressed throughout the cell cycle were also misregulated. Cyclin D1 was up regulated in MIAT and LUCAT1 depleted cells and cyclin D3 was upregulated only in LUCAT1 depleted cells, whereas they were similar to wild type cell levels in LINC00704 knockdown (Figure 3E, Supp. Figure 3E).
We conclude that three of the lincRNAs that we have identified as being transcribed primarily in S-phase are each necessary for efficient progression through S-phase. The observation that these lincRNAs displayed differences in the manner in which they impact cyclin expression, suggesting that they impact distinct regulatory programs, prompted us to look more generally at their effects on gene expression during the cell cycle.
LINC00704, LUCAT1 and MIAT Affect Different Transcriptional Programs
To dissect the differences in gene expression caused by the depletion of these three lincRNAs we performed strand specific RNA sequencing at different time points during S-phase following the knockdown of the each lincRNA. We studied time points early in S-phase to increase the likelihood that the effects reflected a direct impact of the knockdowns rather than on differences due to defects in cell cycle progression. We observed transcriptome wide patterns that were reproducible across two replicates in each of the four time points for each of the four knockdowns (Figure 4A, Supp. Figure 4A). The knockdowns resulted in significant changes in the expression of approximately 2,000–3,000 genes (depending on the time point) with LUCAT1, 1,000–2,000 genes with MIAT and 500–1,500 genes with LINC00704 depletion. Genes were more frequently up-regulated with LINC00704 or LUCAT1 knockdown and more frequently down-regulated, at the three early time points, with MIAT knockdown.
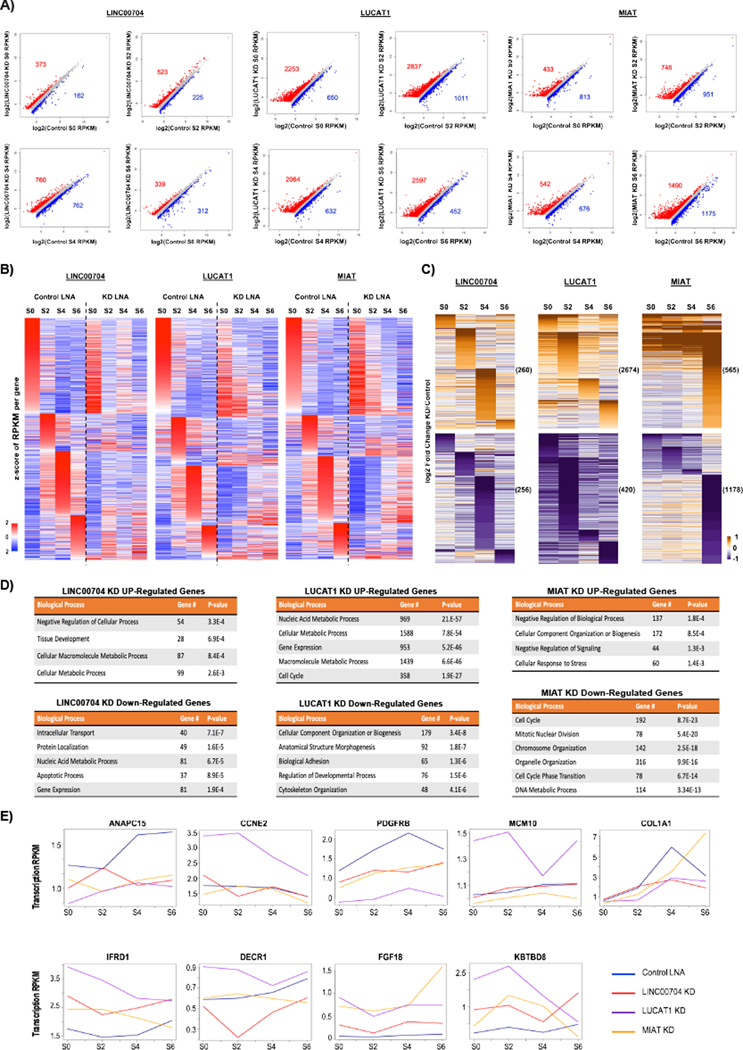
Figure 4. Different transcriptional programs are impacted by the depletion of early S-phase peaking lincRNAs LINC00704, LUCAT1 and MIAT.
A) Scatter plots showing transcriptome wide changes of gene expression in knockdowns, regardless of expression dynamics in cell cycle progression. Genes up-regulated in the knockdowns (1.5-fold) are shown in red and down-regulated in knockdowns (1.5-fold) are shown in blue (See also Supp. Figure 4A). B) Gene expression profiles of control LNA transfected and knockdown cell mapped after strand specific RNA-seq. Z-scores of average RPKMs of two replicates across all time points are shown for genes with (maximum RPKM in Control LNA)/ (minimum RPKM in Control LNA) > 1.5 (See also Supp. Figure 4B 2-fold change). Genes are clustered based on the time point of the maximum expression level in Control LNA. Genes are ordered by increasing z-scores in control. S0 is G1/S border cells; at the time of mimosine wash-off. S2 is cells just released in S-phase; S4 is early S-phase and S6 is early-mid S-phase. LINC00704, LUCAT1 and MIAT knockdowns were compared to corresponding time points of control sample. C) Unique gene expression changes for each lincRNA knockdown compared to control LNA transfected cells at each time point. Only genes that were uniquely affected in one lincRNA knockdown at any time point, but not changing the same way in other knockdowns are shown. Log2 fold-changes knockdown over control are shown for each individual knockdown. Up-regulated genes are shown on the top followed by down-regulated genes at the bottom (See also Supp. Figure 4C). D) GO terms of gene expression changes unique to each knockdown compared to control cells in each time point. E) RPKM/Time line plots showing expression changes in control, LINC00704, LUCAT1 and MIAT knockdown cells.
To understand the impact of the depletion of each lincRNA on cell cycle progression we filtered the genes that show >1.5-fold change in expression level in at least one time point during S-phase in control cells (Figure 4B; left panels). These genes were clustered genes according to the time point of maximal expression (Figure 4B; similar conclusions were reached with a 2-fold cutoff, Supp. Figure 4B). We then compared the expression profiles of these S-phase regulated genes in control cells with the expression profiles of the same genes following lincRNA knockdowns. We observed significant differences for each of the different knockdowns (Figure 4B; right panels). The S0 point of each knockdown showed some differences with the control LNA, consistent with the observation (see below) that each of these lincRNAs impacts growth and gene expression in wild type cells. Each of the knockdowns showed greater changes with the control as the cells progressed through S-phase (Figure 4B; compare left panels with right panels). The patterns of gene expression changes were distinct for each knockdown, a finding we explored in more detail by examining genes that were uniquely regulated by each knockdown.
We identified sets of differentially regulated genes that showed at least a 1.5-fold change in expression compared to control and that were unique to each lincRNA knockdown (Fig. 4C). For MIAT knockdown at the 6-hour time point and for LUCAT1 knockdown at all time points, most of the significantly regulated genes were unique to each knockdown (Supp. Fig. 4C). A lesser, but still large, number of expression changes by LINC00704 knockdown were unique to that knockdown. LUCAT1 knockdown produced the largest effect at time point S2, whereas MIAT knockdown produced the largest number at time point at S6, timing that reflects the expression of LUCAT1 in the 0–2hour window and MIAT in the 2–4hour window of S-phase. Gene ontology (GO) analysis further supported a distinct impact of each knockdown on gene expression patterns (Figure 4D).
We infer, based upon these analyses that each lincRNA plays a specific role in S-phase progression. We conclude that knockdown of the three lincRNAs all differed from each other in substantive ways: Distinct genes were regulated, and the timing of these effects was different for each lincRNA.
Each of the analyses above was done with a protocol that used a mimosine block to generate synchronized cells, raising the possibility that the effects of the lincRNA knockdowns were specific to mimosine function and would not be seen in normally growing cells. We therefore examined the impact of lincRNA knockdown on normally growing asynchronous cells. Cells were treated with LNAs for 44 hours under normal growth conditions and RNA was extracted, ribosomal RNA depleted and strand specific RNA seq was performed. We measured the changes in expression between control LNA treated cells and lincRNA specific LNA treated cells in the asynchronous population. We then compared the expression of all RNAs in these asynchronous data sets with the average change of all RNAs across S phase in the mimosine treated cells for each lincRNA knockdown (Supp. Fig. 4D). We found good correlation indicating that lincRNA depletion impacted a similar gene set in normally growing cells. We also found good correlation of GO terms when genes uniquely regulated by each knockdown were compare between asynchronous data sets and mimosine treated data sets (compare Fig. 4D and Supp. Table 2) To further examine whether any effects were specific to mimosine, we transfected cells with LNAs and added TN-16 (3-[1-(Phenylamino)ethylidene]-5-(phenylmethyl)-2,4-pyrrolidinedione), a synthetic compound that interacts with and inhibits microtubules to synchronize cells at M-phase (Tatsumi et al., 2003). We then determined how efficiently these cells synchronized in M phase, because if depletion of a lincRNA blocked S phase progression then the cell would not progress to M phase. The wild type cells and control LNA transfected cells synchronized in M-phase with about 71 and 67% efficiency, respectively, while only about 15–18% were at G1/early S-phase. Cells depleted for one of the three lincRNAS synchronized in M phase less efficiently, varying from 35% to 55% depending on the specific LNA, and had more cells stuck in G1/early S with between 31% and 52% (Supp. Fig. 4D).
Genes dis-regulated by knockdown of each lincRNA
LINC00704 upregulation has been reported in cervical cancer (Ojesina et al., 2013) and has been associated with breast cancer recurrence (H. Liu et al., 2016). In RPE1 cells, LINC00704 depletion caused downregulation of genes associated with lipid metabolism, vesicular transport and membrane associated factors such as DECR1, SLC30A5 and EMD. In addition, RNA metabolism and RNA export related genes such as MED1, ENDOD1 and MAGOH were down regulated; while several noncoding RNAs particularly lincRNA and rRNA genes were up regulated. Examples of temporal dynamics of differentially regulated genes in knockdowns and control cells are shown as line plots of expression values and genomic tracks of the time points (Figure 4E). We conclude that LINC00704 is likely to be involved in both gene activation and repression, and that its most abundant targets are not directly related to cell cycle progression.
LUCAT1 was first identified as an elevated non-coding transcript in numerous lung cancer cell lines (Thai et al., 2013). Depletion of LUCAT1 in RPE1 cells resulted in up-regulation of genes that encode parts of the general transcription machinery (e.g. TFIID components TAF13, TAF7) and transcription factors (e.g. many members of FOX and TEAD family transcription factors). Several members of DDX family RNA helicases, RNA binding proteins (e.g. RBMs), other RNA and non-coding RNA metabolism, splicing and RNA modification related genes (e.g. SLBP, AGO2, DICER1) were also upregulated. Consistent with the observed cell cycle progression defect phenotype upon LUCAT1 depletion several proliferation and growth signaling related genes were down regulated (e.g. PDGFRB, PDGFB). More significantly, comparing the LUCAT1 knockdown to the other two lincRNA knockdowns, we identified specific downregulation of several actin/actin organization and cytoskeleton organization related genes, collagens and other cell-cell communication related genes (e.g. ACTA2, COL1A1). Despite the defective S-phase phenotype, key G1/S transition gate keeper cyclin dependent kinase inhibitors CDKN1A, CDKN1B and CDKN2B levels were reduced, while genes that are important for DNA replication and S-phase progression (e.g. MCM10, ORC1, CCNE1, CCNE2) and DNA damage genes (e.g. ATR, BRCA2, CHEK2) were upregulated. These latter findings indicate a possible early S-phase defect rather than G1/S block. Given that genes were predominantly upregulated upon LUCAT1 knockdown (Figure 4A, Supp. Figure 4C), we conclude that LUCAT1 is primarily required for repression of genes.
Myocardial Infarction Associated Transcript (MIAT) was initially identified in a large-scale association study as a susceptibility locus for myocardial infarction (Ishii et al., 2006) and was later shown to play a role in retinal cell fate specification (Rapicavoli et al., 2010). It has been linked to multiple diseases including being upregulated in several different cancers (Sun et al., 2018). Consistent with the observed strong S-phase progression defect, MIAT depleted cells showed downregulation of many genes that are directly related to DNA replication (e.g. RAD21, CDC20, CDC25A, a catalytic subunit of DNA polymerase, POLD1 and the accessory subunit POLD3). Similarly, many genes important for S-phase progression regulation were also misregulated (e.g. CDKN2A, CDKN2B, CCND1 were up regulated while CCND3, CCNE1, CCNE2 were down regulated). In addition, MIAT depletion caused misregulation of many epigenetic regulatory enzymes, both chromatin and DNA modifying enzymes (e.g. KMT2E, KMT2B, TET2, DNMT1). MIAT knockdown resulted in transcriptome wide changes that were biased towards depletion of gene expression in all time points examined, although there were also a number of significantly upregulated genes (Figure 4A, Supp. Figure 4C). We conclude that MIAT is involved in regulation of numerous genes whose functions impact cell cycle progression and chromatin dynamics and is more important for upregulation of these genes than for downregulation.
There were also common genes that were similarly impacted in all three knockdowns. The gross phenotype in S-phase progression seen in all three knockdowns was reflected via downregulation of genes related to cell cycle progression, mitosis and organelle biogenesis. Replication coordinated canonical histone genes, late S-G2/M phases specific transcripts (e.g. ANAPC15, FOXM1), actin organization and organelle function related (e.g. TGFBI, SEC61A1) genes were down regulated in all three knockdowns. In addition, forkhead family transcription factors that play important roles in retina development and morphogenesis FOXC1, FOXD1 were depleted in all 3 knockdowns. Intriguingly, some of the upregulated genes in all three knockdowns, particularly strongly upregulated in MIAT and LUCAT1 knockdowns, were neuronal lineage specific transcripts; such as neural crest specification regulator KBTBD8, dendrite branching regulator TMEM106B, neuron specific cyclin dependent kinases CDK17 and CDK5R1, and genes that plays role in early stage neuronal differentiation and signaling (e.g. DMRTA1, CEND1, NOTCH3 and FGF18). This result might be related to the maintenance of cell identity as retinal pigment epithelial cells were shown to transdifferentiate into neuronal progenitor like cells (Engelhardt et al., 2005). It is possible that gene expression changes common to all three knockdowns reflect indirect effects of perturbing normal cell cycle as opposed to direct regulatory effects of each lincRNA. Some of the differential gene expression changes upon different lincRNA knockdowns are shown in line plots and genomic tracks of the time points (Figure 4E).
Discussion:
We isolated nascent transcripts and identified over 200 long intergenic non-coding RNAs that are expressed with specific timing during S-phase. Our data set provides insights on transcription peak time and function relation for non-coding RNAs at specific stages of cell cycle focused in detailed S phase progression. Protein factors necessary for cell cycle display a similar periodic expression pattern of their mRNAs (Grant et al., 2013; Martin and A, 2019). Consistent with these observations, the S-phase specific lincRNAs we cataloged share similar promoter elements with co-transcribed protein coding genes. Three of the S-phase expressed lincRNAs were shown to be necessary for proper regulation of numerous genes during cell cycle, with each of the three regulating distinct sets of genes. The timing of the expression of these RNAs in S-phase appeared relevant to their functions, as depletion of the three caused a widespread alteration in gene expression that occurred immediately after the expression peak time of each lincRNA. We infer that there are multiple lincRNAs in mammalian cells that play a regulatory role during S-phase and provide a list of candidates for this function.
Long non-coding RNAs have been associated with pluripotency (Guttman et al., 2011; Savić et al., 2014), lineage specification/differentiation (Frank et al., 2019; Guo et al., 2018; Schwarzer et al., 2017; X. Zhang et al., 2014), apoptosis (Hu et al., 2018; Huarte et al., 2010; A. Zhang et al., 2013) and cell cycle coordination (Hung et al., 2011; Y. Liu et al., 2017). Most of these lncRNAs were either genic (intronic or antisense) or located in the close vicinity of a promoter element of a protein coding gene or an enhancer. Many of these lncRNAs and their function in growth modification are cell type specific, perhaps due to differences in cell type specific transcriptional networks (S. J. Liu et al., 2017). Certain lncRNAs have been shown previously to have a cell-cycle function and shown to be important for proper cell cycle progression (S. Lee et al., 2016; Marchese et al., 2016; Nötzold et al., 2017; Tripathi et al., 2013). We find these latter lncRNAs in our analysis of nascent RNAs expressed during S-phase, indicating that their synthesis is consistent with their previously described function, and also identify approximately 900 other lncRNAs whose expression peaks in S-phase.
Diverse regulatory functions in controlling chromatin structure and transcriptional profiles have been attributed to long non-coding RNAs, yet very few of them have well-defined expression patterns with respect to their proposed biological function. Furthermore, the overall low abundance of many lncRNAs throws into question their ability to execute their functional roles in the cell (Cabili et al., 2015; Kopp and Mendell, 2018; Palazzo and E. S. Lee, 2015). While there have been numerous analyses of lncRNAs that are expressed in specific cell types and in malignancies, few of these studies have examined temporal regulation of lncRNAs. The ability to efficiently isolate nascent transcripts at specific stages during cell cycle has allowed us to expand upon previous studies in several important ways. Many nuclear long non-coding RNAs are relatively short-lived (Clark et al., 2012), which creates issues in abundance that are mitigated by isolating nascent RNAs in a cell cycle stage specific manner. It is possible that their lower abundance is due to their transient nature; expression at high levels at specific time points might allow sufficient abundance at those time points to allow broad functional roles. We infer from our data that the timing of synthesis of a non-coding RNA offers information on its function, and that there are a large number of non-coding RNAs with temporal regulation.
We suggest that lincRNAs play a key role in regulating S-phase progression. Molecular events, including transcription during S-phase, are tightly regulated to ensure controlled cell cycle progression and propagation of cellular integrity. For example, after replication, gene dosage homeostasis is preserved with nascent chromatin modifications (Voichek et al., 2016) while coordination with the replication fork is important so that the collision of these two DNA templated processes can be prevented and genome stability can be maintained (Duch et al., 2013; García-Muse and Aguilera, 2016; Helmrich et al., 2011; Wansink et al., 1994). Hence, there is a need for transcriptional control of non-coding RNAs during S-phase as has been seen in many instances with mRNAs. Therefore, S-phase peaking lincRNAs might be less likely to be the result of fortuitous transcriptional activity produced from junk DNA (Palazzo and E. S. Lee, 2015) or transcriptional noise byproducts (Struhl, 2007). Consistent with non-random expression, the lincRNAs we have identified share promoter elements linked to cell cycle regulation and differentiation of the mRNA genes expressed in S-phase. Each of the three lincRNAs that we investigated further were required for proper S-phase progression. Importantly, despite similar characteristics in expression timing, nuclear enrichment and knockdown phenotype, all three lincRNAs impacted different gene expression programs. Emphasizing these differences, depletion of MIAT or LUCAT1 had the opposite effect on many genes. The observation that over 200 lincRNAs display S-phase specific expression indicates a possible coordination between the regulatory effects of these lincRNAs and the regulatory effects of proteins that are expressed during S-phase to allow effective progression through cell cycle and re-establishment of regulation on both daughter strands.
Some members of the set of lincRNAs we identify here might be important in establishing the ‘memory’ of expression state on the replicated daughter strands that is essential for proper development. There are several examples of small non-coding RNAs being required for maintenance of epigenetic states in a variety of organisms. Transgenerational inheritance has been shown to involve RNA, for example through use of maternal piRNAs in flies (Brennecke et al., 2008) and paternal tRNAs in mice (Sharma et al., 2016), regulation of DNA methylation in plants (Saze et al., 2008) and paramutation in mice and maize (Alleman et al., 2006; Rassoulzadegan et al., 2006; Woodhouse et al., 2006). Pericentromeric heterochromatin is maintained through small RNAs in S. pombe (Verdel et al., 2004; Volpe et al., 2002). The most extensively studied lincRNA, Xist, is known to trigger a chromosomal memory during early differentiation of embryonic stem cells that is required for enforcement of transcriptional repression (Kohlmaier et al., 2004). Other examples include rDNA heterochromatinization during differentiation in mammals (Guetg et al., 2012; Savić et al., 2014) and mesoderm specification (Frank et al., 2019). The set of lincRNAs described here provides a set of candidates for function in epigenetic memory in addition to candidates for S-phase progression and potential biomarkers and/or therapeutic targets.
STAR METHODS:
Resource Availibilty:
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Robert E. Kingston (kingston@molbio.mgh.harvard.edu).
Materials Availability:
There are restrictions to the availability of ABE-biotin probe generated in this study due to the lack of our need to maintain the stock. A detailed synthetic method of this reagent has been provided in Method S1.
Data and Code Availability:
The accession number for the next generation sequencing data reported in this paper is GEO: GSE137448. This study did not generate code.
Experimental Model and Subject Details
The hTERT-RPE1 cell line:
Organism: Homo sapiens, human; Gender: Female; Cell Type: Epithelial cells immortalized with hTERT; Tissue: Retina, eye, pigment epithelium; Disease: Normal
Cell Culture:
Cells were cultured (37° C, 5% CO2), in DMEM-F12 + GlutaMAX™ and penicillin and streptavidin medium (Gibco, Waltham, MA, USA) supplemented with 10% fetal calf serum (FCS; Gibco) and %0.25 Na-Bicarbonate. Cells were synchronized at the G1/S border with L-Mimosine (Sigma-Aldrich, M0253) prepared as described before with slight modifications (Galgano and Schildkraut, 2006).
METHOD DETAILS
Synchronization and Flow Cytometry:
This protocol was adapted from “Cell Synchronization,” Chapter 14, in Cells (eds. Spector et al.). Cold Spring Harbor Laboratory Press). 79mg of mimosine was dissolved completely in 5 mL of 0.1N NaOH and 5 mL DMEM/F12 was added to achieve a 100X solution, final concentration of 40mM. Cells were plated at 3X106 cells/15cm plate in 1X mimosine (400 uM) containing medium and cultured for 22 hrs. To release into S-phase, they were washed with 15 mL PBS twice and plated in regular RPE1 medium.
Propidium iodine staining and Flow Cytometry adapted from Ormerod and Kubbies: Cells were trypsinized, collected and washed once with PBS. They are then resuspended in 200ul PBS and the cell suspension was transferred drop-wise fashion into a tube containing 4 mL ice cold 70% ethanol, incubated at +4°C overnight. Next day, 300,000 cells resuspended in 1 mL PBS containing 40 ug/mL Propidium iodine and 100ug/mL RNase A, incubated at 37°C for 30 minutes, filtered and FACS analyzed for their DNA content (Ormerod and Kubbies, 1992).
EU Metabolic Labeling and Nascent RNA Sequencing:
After 22 hrs of mimosine synchronization cells were washed twice with 15mL PBS and release them into S-phase in normal culture medium. At different time points of after release into S-phase, culture medium was changed to 5-Ethynyl Uridine (Cat.No. PY7563, Berr&Associates) containing medium with a final concentration of 100uM. After 2 hours of labeling nascent transcripts with EU (EU labeling has been reported as little as in 10 min in vivo (Jao and Salic, 2008)), the medium was aspirated, and cells were washed once with PBS, then collected in 1.2 mL Trizol Reagent (Cat. No. 15596–026, Life Technologies) using cell lifter. For nuclear enriched RNA seq, cells were trypsinized and the nuclear fraction was isolated using cell fractionation buffer (PARIS RNA Kit, Cat.No. AM1921, Thermo Fisher Scientific). The nuclear fraction was resuspended in 1.2mL Trizol. Samples were flash-frozen in liquid nitrogen and stored in −80°C.
RNA Extraction, Ribosomal RNA Removal: 0.4 mL chloroform was added to samples collected in 1.2 mL Trizol®. Samples were centrifuged for 15 min at 12,000 x g, 4°C. The aqueous phase was transferred to RNA fresh tubes and an equal amount of isopropanol added. RNA was precipitated by centrifugation for 15 min at 12,000 x g, 4°C and washed once with 80% Ethanol, pellets were air-dried for 5 min, then resuspended in 44 ul nuclease-free water and DNAseI treated for 1hour at 37°C. The DNAseI reaction was cleaned up with the RNA clean & concentrator kit (Zymo Research), according to the manufacturer’s protocol. For each sample (time point) 15 ug RNA was ribosome depleted using RiboZero Gold Magnetic Kit (Cat.No. MRZG12324, Illumina Inc.).
Biotinylation with Click Chemistry and UV Elution:
Ribosome-depleted, EU labeled RNAs were biotinylated using a novel UV-cleavable biotin azide probe, ABE-biotin (See detailed synthetic method in Methods S1) under previously described click reaction conditions (Yildirim, 2015). The detailed description for the synthesis of ABE-biotin, accompanied with the spectral data of new compounds, can be found in Supplementary Information. Ribozero-treated RNA samples were pooled to achieve a yield of at least 500 ng RNA, which was used for the click reaction. Nascent transcripts were then isolated using streptavidin coated magnetic beads (MyOne Streptavidin C1 magnetic beads, Cat.No. 65001, Life Technologies). Nascent RNA bound magnetic beads were then transferred to sterile, UV transparent glass tubes and released from biotin-streptavidin complex by irradiating for 15 min at 350 nm irradiation using a bench-top Rayonet UV reactor. Eluted RNA cleaned up with the RNA clean & concentrator kit (Cat.No. R1015, Zymo Research). 9 ul RNA is fragmented in 7 ul 5X First strand buffer (SuperScript III, Cat.No. 18080044, Thermo Fisher Scientific) at 94 °C for 5 min in PCR machine with a heated lid. The RNA was quickly chilled on ice. Then 2,25 ul 100mM DTT (SuperScript III, Cat.No. 18080044, Thermo Fisher Scientific), 1,25 ul Random primer (Cat.No. 48190011, Thermo Fisher Scientific) and 6ul RNAse free water were added and incubated at 65 °C for 3 min, then chilled on ice for 3min. 1,5 ul dNTP mixure (10 mM each); 0,75 ul RNase inhibitor (Cat. No. PRN2615, Promega); 0,75 ul 100 mM DTT; 1,5 ul SuperScript III enzyme (Cat.No. 18080044, Thermo Fisher Scientific) were added to chilled samples and incubated for 10 min at 25 °C; 1hr and 15 min at 50 °C and then 15 min at 70°C. 72 ul SPRI beads (Cat. No. A63882, Beckman Coulter Genomics Inc.) were used to purify and isolate the cDNA, samples were then eluted with 22 ul EB (10mM Tris·Cl, pH 8.5). Second strand synthesis was performed by addition of 3 ul 10X NEB Buffer 2; 2 ul 10 mM dNTP mixture; 0.75 ul RNaseH (NEB); 2 μl DNA polymerase I (10 U/μl, NEB); 0.5 μl 10mM DTT and incubation at 16 °C for 2.5 hrs. The reaction was cleaned up with 56 ul SPRI beads (1.6X ratio) and eluted in 38 ul dH2O. A picogram scale sequencing library protocol (Bowman et al., 2013) was then used.
LNA GapMeRs lncRNA knockdown:
hTERT-RPE1 cells were transfected with 130–150 pmol LNA GapMeRs(Qiagen) in Opti-MEM™ (Cat.No. 31985070, Thermo Fisher Scientific) via the facilitation of Lipofectamine™ RNAiMax Transfection Reagent (Cat.No. 13778150, Thermo Fisher Scientific). Knockdown efficiency was checked with rt-qPCR using primers specific to the appropriate lincRNA for each LNA.
RT-qPCR:
RNA is extracted and DNaseI treated as described before and reaction was cleaned up with the RNA clean & concentrator kit (Zymo Research), according to the manufacturer’s protocol. Reverse transcription reaction is done with SuperScript III, according to the manufacturer’s protocol (SuperScript III, Cat.No. 18080044, Thermo Fisher Scientific). RNA is degraded at 65°C with 6.5ul 0.5M EDTA + 6.5ul 1N NaOH incubation for 15 min in PCR machine with a heated lid. Reaction is neutralized with 17ul 1M HEPES pH 7.5 and cleaned with Amicon YM10 cleanup spin columns. qPCR reaction is run with 2X BioRad Sybergreen protocol.
LNA KD-Strand Specific RNAseq:
12 hrs after LNA GapMeR transfection cells were synchronized with mimosine for 21 hrs as described before and then released in S-phase. Collected time points are: At the time of release (S0), 2 hrs after release (S2), 4 hrs after release (S4) and 6hrs after release time points collected. RNA is isolated using phenol chloroform extraction and isopropanol precipitation. Turbo DNAseI treated samples cleaned with RNA clean & concentrator kit (Zymo Research).
After rRNA depletion as described. 4 ul RNA was fragmented in 4 ul 5X First strand buffer (SuperScript III, Cat.No. 18080044, Thermo Fisher Scientific) at 94 °C for 5 min in PCR machine with a heated-lid. RNA was chilled on ice, then 1.5 ul 100 mM DTT (SuperScript III, Cat.No. 18080044, Thermo Fisher Scientific) 1 ul Random primer (Cat.No. 48190011, Thermo Fisher Scientific) 4 ul RNAse free water were added and incubated at 65 °C for 3 min; then it was again chilled on ice for 3 min. Finally, 1 ul dNTP mixure (10 mM each); 0.5 ul RNase inhibitor (Cat. No. PRN2615, Promega); 0.5 ul 100 mM DTT; 1 ul SuperScript III enzyme (Cat.No. 18080044, Thermo Fisher Scientific) were added to sample and incubated for 10 min at 25 °C; 1 hr and 15 min at 50 °C and then 15 min at 70°C. 36 ul SPRI beads (Cat. No. A63882, Beckman Coulter Genomics Inc.) were used to purify the cDNA and were eluted with 22 ul EB (10 mM Tris·Cl, pH 8.5). dUTP incorporation performed with addition of 3 ul 10X NEB Buffer 2; 2 ul 10 mM dUTP mixture (Cat.No. 77330, Afftmetrix Inc.); 0.75 ul RNaseH (NEB); 2 ul DNA polymerase I (10 U/μl, NEB); 0.5 ul 10mM DTT incubated at 16 °C for 2.5 hrs. The reaction was cleaned up with 56 ul SPRI beads (1.6X ratio), eluted with 38 ul dH2O. End repair reaction was executed in 50 ul. 5 ul T4 DNA ligase buffer, 2 ul 10 mM dNTP mixture, 2.5 ul T4 DNA polymerase (NEB), 0.5 ul Klenow DNA polymerase (NEB), 2.5 ul T4PNK added and incubated at 20°C for 30 min. Reaction was cleaned up with 1.6X SPRI beads and eluted in 19 ul. End repaired samples then were A’tailed with 3 ul NEB buffer 2, 6 ul dATP(1mM), 2 ul Klenow 3’ to 5’ exo (5U/ul, NEB) and incubated at 37°C for 30 min. Reaction cleaned up with 1.6X SPRI beads and eluted in 10,5 ul dH2O. 2 ul Universal Illumina adaptors (1 uM) were ligated to A’tailed samples using 15 ul 2X Enzymatics T4 rapid ligase buffer and 2,5 ul T4 rapid ligase (Cat. No. L603-HCL-L Enzymatics Inc.) and sample was incubated for 15 min at room temperature. The reaction was cleaned up with 1.2X SPRI beads and eluted in 25 ul dH2O for UDG treatment. 2 ul UDG (Cat. No. M02280S, NEB) and 3 ul UDG buffer were added and incubated at 37°C for 30 min. Reaction was cleaned up with 54 ul SPRI beads (1.8X ratio), eluted in 38 ul dH2O. PCR amplification and barcoding were done as described previously (Bowman et al., 2013).
ChIP-Seq:
Cells were cross-linked in 1% formaldehyde at room temperature and ChIP carried out as described in (Schmiedeberg et al., 2009) Sequencing libraries were barcoded and prepared as described in Bowman et al. (Bowman et al., 2013).
Cut and Run:
H3K27me3 mapping carried out according to Cut & Run protocol. IgG used as negative control. ~3×105 cells per reaction were fixed with 1% formaldehyde at RT for 10 min. Crosslinking was quenched glycine at final concentration of 125 mM before continuing with the Cut&Run protocol as described before (Skene and Henikoff, 2017). After targeted fragments were released, crosslink reversed by incubation at 65 °C with 2 ul 10% SDS and 2.5 ul Proteinase K (20 mg/ml) overnight. DNA was phenol/chloroform extracted and EtOH precipitated in the presence of 2 ul glycogen (2 mg/mL). Samples were recovered in 25 ul 1 mM Tris-HCl pH8, 0.1 mM EDTA. Sequencing libraries were barcoded and prepared as described in Bowman et al. (Bowman et al., 2013).
Cell Viability and Proliferation Assay:
Cell proliferation was assayed for each lincRNA knockdown for 60hrs. After LNA GapMeR transfection for each lincRNA, cells were seeded in 96-well plates in RPE1 medium and assayed every 12hr. To determine the lincRNA depletion on cell proliferation CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used according to the manufacturer’s recommendations.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-seq data processing:
RNA-seq sequencing reads were aligned to the hg38 genome using STAR v2.5.3 (Dobin et al., 2013). using default parameters and --outFilterMultimapNmax 10. Gene annotations were obtained from Ensembl (Hunt et al., n.d.). Genome browser tracks were generated using Homer v4.10.3 (Heinz et al., 2010) and visualized in IGV(J. T. Robinson et al., n.d.). Reads in the whole gene body were counted using featureCounts v1.6.1 (Liao et al., 2014). Each knockdown condition was compared separately to the control condition. For each comparison, reads were normalized using edgeR’s (M. D. Robinson et al., 2009) TMM method and RPKM was calculated. All further calculations, heatmaps and figures were made using R v3.3.2 (Team, n.d.) unless otherwise noted.
All RNA-seq experiments were performed in duplicate. RNA-seq on asynchronous cell lincRNA knockdowns were performed in triplicate. RPKMs were calculated using R v3.3.2. For all heat-maps, average RPKMs were used to calculate z-score with the formula (RPKM- average of all RPKMs for gene)/standard deviation across all RPKMs.
Data for Figure 2A was obtained by using the software STEM v1.3.11 (Ernst and Bar-Joseph, 2006). Relative transcription was calculated by the formula (RPKM – minimum RPKM across all time points)/(maximum – minimum RPKM across all time points).
Motif analysis was done using the web version of MEME (Bailey et al., 2009). E-values were used to determine the top discovered motifs. E-value is defined as the expected number of sequences in a random database of the same size that would match the motifs as well as the sequence does and is equal to the combined p-value of the sequence times the number of sequences in the database.
For Figure 4A and 4B, normalized read counts (CPMs) were calculated for every knockdown condition separately with the control using edgeR’s (M. D. Robinson et al., 2009) TMM method. RPKM was then calculated using the normalized counts. Fold changes were calculated using average RPKM values and an added pseudocount of 0.1. For Supplementary Figure 7, RNA-seq data for asynchronous cells was processed the same way. For synchronous cells, average of all RPKMs was calculated for control and knockdown conditions. Pearson correlation between log2 fold-changes in synchronous and asynchronous cells was calculated using R. EdgeR’s p-value and FDR calculations were used to make volcano plots in Supplementary Figure 7.
For Figure 4C, genes that were changing 1.5-fold in one knockdown, but not in the other knockdowns were chosen. log2(fold changes) for these uniquely changing genes in each knockdown are shown, separated by if they were up-regulated in the knockdown (upper panel) or down-regulated in the knockdown (lower panel).
Gene ontology was done using DAVID (Huang et al., 2009). For Supplementary Figure 3C, each circle in the venn diagram represents the number of genes that were 1.5-fold over Async at the labeled time-point.
Data about copy number variation frequencies were obtained from the TCGA database using the R package “cgdsr” (Jacobsen and Questions, n.d.).
ChIP-seq and Cut-n-run data processing: ChIP-seq and Cut-n-run sequencing reads were aligned to the hg38 genome using bowtie 2.3.4.3 (Langmead and Salzberg, n.d.). Genome browser tracks were generated using Homer v4.10.3 (Heinz et al., 2010) and visualized in IGV (J. T. Robinson et al., n.d.).
Supplementary Material
Supplementary Table S1, Related to Figure 2 Data from The Cancer Genome Atlas showing frequencies of mutations in lincRNAs enriched in S-phase in various cancers.
Supplementary Table S2, Related to Figure 4 Gene ontology terms for genes uniquely differential in LINC00704, MIAT and LUCAT1 knockdowns in Asynchronous cells.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CYCLIN A2 | Cell Signaling Technology | 4656 |
| CYCLIN D1 | Cell Signaling Technology | 2978 |
| CYCLIN D3 | Cell Signaling Technology | 2936 |
| CYCLIN E1 | Cell Signaling Technology | 4129 |
| H3K4me3 | E M D Millipore | 07–473 |
| H3K27me3 | Cell Signaling Technology | 9733 |
| H3K27Ac | Cell Signaling Technology | 8173 |
| H3Kme1 | Abcam | 8895 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L-Mimosine | Sigma-Aldrich | M0253 |
| 5-Ethynyl Uridine (EU) | Berry & Associates | PY7563 |
| dUTP mixture | Affymetrix Inc. | 77330 |
| RNase inhibitor | Promega | PRN2615 |
| Random primer | Thermo Fisher Scientific | 48190011 |
| RNAiMax Transfection Reagent | Thermo Fisher Scientific | 13778150 |
| My1 Streptavidin C1 magnetic beads | Life Technologies | 65001 |
| SuperScript III | Thermo Fisher Scientific | 18080044 |
| SPRI beads | Beckman Coulter Genomics Inc | A63882 |
| Deposited Data | ||
| Next Generation Sequencing | GEO | GSE137448. |
| Experimental Models: Cell Lines | ||
| hTERT-RPE1 cell line | ATCC | CRL-4000 |
| Oligonucleotides | ||
| LUCAT1 Fw | Integrated DNA Technologies | atgcctgggacagacagaga |
| LUCAT1 Rev | Integrated DNA Technologies | actgaccctggcttgcttc |
| LINC00704 Fw | Integrated DNA Technologies | atacctgtgcatctcatgctgt |
| LINC00704 Rev | Integrated DNA Technologies | cattcccagattgtggagttgg |
| MIAT Fw | Integrated DNA Technologies | gacccgagttggaggcatct |
| MIAT Rev | Integrated DNA Technologies | cttggttacccctgtgatgcc |
| Software and Algorithms | ||
| STAR v2.5.3 | https://github.com/alexdobin/STAR | |
| bowtie 2.3.4.3 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | |
| Homer v4.10.3 | http://homer.ucsd.edu/homer/index.html | |
| IGV | https://software.broadinstitute.org/software/igv/ | |
| featureCounts v1.6.1 from Subread | http://subread.sourceforge.net/ | |
| STEM v1.3.11 | https://www.ncbi.nlm.nih.gov/pubmed/16597342 | |
| MEME web application | http://meme-suite.org/tools/meme | |
| edgeR | https://bioconductor.org/packages/release/bioc/html/edgeR.html | |
| R v3.3.2 | https://www.r-project.org/ | |
| TCGA cBioPortal | https://www.cbioportal.org/ | |
| R package “cgdsr” | https://cran.r-project.org/web/packages/cgdsr/index.html | |
| Other | ||
| PARIS RNA Kit (cell fractionation) | Thermo Fisher Scientific | AM1921 |
| RNA clean & concentrator kit | Zymo Research | R1013 |
| RiboZero Gold Magnetic Kit | Illumina Inc | MRZG12324 |
Acknowledgements:
O.Y. was supported by HHMI-Damon Runyon Cancer Research Foundation, DRG 2156–13.
J.W.S. is an Investigator of the Howard Hughes Medical Institute.
This work was supported by R35-GM131743 from the NIH (R.E.K).
Footnotes
Declaration of Interests: The authors declare no competing interests
References:
- Alabert C, Groth A, 2012. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167. doi: 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL, 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442, 295–298. doi: 10.1038/nature04884 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS, 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C, Skotheim JM, de Bruin RAM, 2013. Control of cell cycle transcription during G1 and S phases. Nature Publishing Group 14, 518–528. doi: 10.1038/nrm3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M, 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246. doi: 10.1126/science.1103388 [DOI] [PubMed] [Google Scholar]
- Bowman SK, Simon MD, Deaton AM, Tolstorukov M, Borowsky ML, Kingston RE, 2013. Multiplexed Illumina sequencing libraries from picogram quantities of DNA. BMC Genomics 14, 466–7. doi: 10.1186/1471-2164-14-466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ, 2008. An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. Science 322, 1387–1392. doi: 10.1126/science.1165171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhavarapu VN, Chavez M, Tyler JK, 2013. How is epigenetic information maintained through DNA replication? Epigenetics & Chromatin 6, 1–1. doi: 10.1186/1756-8935-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A, 2015. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20. doi: 10.1186/s13059-015-0586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS, 2012. Genome-wide analysis of long noncoding RNA stability. Genome Research 22, 885–898. doi: 10.1101/gr.131037.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB, 2009. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Molecular Cell 33, 717–726. doi: 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T.E.P., Consortium, T.E.P., data analysis coordination, O.C., data production, D.P.L., data analysis, L.A., group, W., scientific management, N.P.M., steering committee, P.I., Boise State University and University of North Carolina at Chapel Hill Proteomics groups (data production and analysis), Broad Institute Group (data production and analysis), Cold Spring Harbor, University of Geneva, Center for Genomic Regulation, Barcelona, RIKEN, Sanger Institute, University of Lausanne, Genome Institute of Singapore group (data production and analysis), Data coordination center at UC Santa Cruz (production data coordination), Duke University, EBI, University of Texas, Austin, University of North Carolina-Chapel Hill group (data production and analysis), Genome Institute of Singapore group (data production and analysis), HudsonAlpha Institute, Caltech, UC Irvine, Stanford group (data production and analysis), targeted experimental validation, L.B.N.L.G., data production and analysis, N.G., Sanger Institute, Washington University, Yale University, Center for Genomic Regulation, Barcelona, UCSC, MIT, University of Lausanne, CNIO group (data production and analysis), Stanford-Yale, Harvard, University of Massachusetts Medical School, University of Southern California/UC Davis group (data production and analysis), University of Albany SUNY group (data production and analysis), University of Chicago, Stanford group (data production and analysis), University of Heidelberg group (targeted experimental validation), University of Massachusetts Medical School Bioinformatics group (data production and analysis), University of Massachusetts Medical School Genome Folding group (data production and analysis), University of Washington, University of Massachusetts Medical Center group (data production and analysis), Data Analysis Center (data analysis), 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 488, 57–74. doi: 10.1038/nature11247 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duch A, Felipe-Abrio I, Barroso S, Yaakov G, García-Rubio M, Aguilera A, de Nadal E, Posas F, 2013. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 493, 116–119. doi: 10.1038/nature11675 [DOI] [PubMed] [Google Scholar]
- Engelhardt M, Bogdahn U, Aigner L, 2005. Adult retinal pigment epithelium cells express neural progenitor properties and the neuronal precursor protein doublecortin. Brain Research 1040, 98–111. doi: 10.1016/j.brainres.2005.01.075 [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M, 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973–1237973. doi: 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z, 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191. doi: 10.1186/1471-2105-7-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Ahuja G, Bartsch D, Russ N, Yao W, Kuo JC-C, Derks J-P, Akhade VS, Kargapolova Y, Georgomanolis T, Messling J-E, Gramm M, Brant L, Rehimi R, Vargas NE, Kuroczik A, Yang T-P, Sahito RGA, Franzen J, Hescheler J, Sachinidis A, Peifer M, Rada-Iglesias A, Kanduri M, Costa IG, Kanduri C, Papantonis A, Kurian L, 2019. yylncT Defines a Class of Divergently Transcribed lncRNAs and Safeguards the T-mediated Mesodermal Commitment of Human PSCs. Cell Stem Cell 24, 318–327.e8. doi: 10.1016/j.stem.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Galgano PJ, Schildkraut CL, 2006. G1/S phase synchronization using mimosine arrest. CSH Protoc 2006, pdb.prot4488–pdb.prot4488. doi: 10.1101/pdb.prot4488 [DOI] [PubMed] [Google Scholar]
- García-Muse T, Aguilera A, 2016. Transcription-replication conflicts: how they occur and how they are resolved. Nat rev mol cell 17, 553–563. doi: 10.1038/nrm.2016.88 [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, Dekker J, 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579. doi: 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GD, Brooks L, Zhang X, Mahoney JM, Martyanov V, Wood TA, Sherlock G, Cheng C, Whitfield ML, 2013. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell 24, 3634–3650. doi: 10.1091/mbc.E13-05-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R, 2012. Inheritance of Silent rDNA Chromatin Is Mediated by PARP1 via Noncoding RNA. Molecular Cell 45, 790–800. doi: 10.1016/j.molcel.2012.01.024 [DOI] [PubMed] [Google Scholar]
- Guo X, Xu Y, Wang Z, Wu Y, Chen J, Wang G, Lu C, Jia W, Xi J, Zhu S, Jiapaer Z, Wan X, Liu Z, Gao S, Kang J, 2018. A Linc1405/Eomes Complex Promotes Cardiac Mesoderm Specification and Cardiogenesis. Stem Cell 22, 893–908.e6. doi: 10.1016/j.stem.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES, 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 457, 223–227. doi: 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES, 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. doi: 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL, 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206. doi: 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell 38, 576–589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L, 2011. Collisions between Replication and Transcription Complexes Cause Common Fragile Site Instability at the Longest Human Genes. Molecular Cell 44, 966–977. doi: 10.1016/j.molcel.2011.10.013 [DOI] [PubMed] [Google Scholar]
- Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD, Wu M, 2018. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nature Cell Biology 1–17. doi: 10.1038/s41556-018-0066-7 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL, 2010. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 142, 409–419. doi: 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerød A, Børresen-Dale A-L, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY, 2011. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genetics 43, 621–629. doi: 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SE, McLaren W, Gil L, Thormann A, Schuilenburg H, Sheppard D, Parton A, Armean IM, Trevanion SJ, Flicek P, Cunningham F, n.d. Ensembl variation resources [WWW Document]. httpsdoi.org.databasebay. URL (accessed 8.2.19). [DOI] [PMC free article] [PubMed]
- Hurst TE, Macklin TK, Becker M, Hartmann E, Kügel W, Parisienne-La Salle J-C, Batsanov AS, Marder TB, Snieckus V, 2010. Iridium-catalyzed C-H activation versus directed ortho metalation: complementary borylation of aromatics and heteroaromatics. Chemistry 16, 8155–8161. doi: 10.1002/chem.201000401 [DOI] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Susumu Saito, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Satoshi Saito, Nakamura Y, Tanaka T, 2006. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51, 1087–1099. doi: 10.1007/s10038-006-0070-9 [DOI] [PubMed] [Google Scholar]
- Jacobsen A, Questions C, n.d. cgdsr: R-Based API for Accessing the MSKCC Cancer Genomics Data Server (CGDS). R package version 1.2.10 [WWW Document]. URL https://CRAN.R-project.org/package=cgdsr (accessed 8.2.19).
- Jahromi AH, Fu Y, Miller KA, Nguyen L, Luu LM, Baranger AM, Zimmerman SC, 2013. Developing bivalent ligands to target CUG triplet repeats, the causative agent of myotonic dystrophy type 1. J. Med. Chem. 56, 9471–9481. doi: 10.1021/jm400794z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, Salic A, 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U.S.A. 105, 15779–15784. doi: 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR, 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. doi: 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- Kawamoto K, Zhong M, Wang R, Olsen BD, 2015, n.d. Loops versus branch functionality in model click hydrogels. ACS Publications. doi: 10.1021/acs.macromol.5b02243 [DOI] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A, 2004. A Chromosomal Memory Triggered by Xist Regulates Histone Methylation in X Inactivation. Plos Biol 2, e171. doi: 10.1371/journal.pbio.0020171.sg005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB, 2001. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. Engl. 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, Gao G, 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–W349. doi: 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Mendell JT, 2018. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL, n.d. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357 EP –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang T-C, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT, 2016. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 164, 69–80. doi: 10.1016/j.cell.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W, 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lim S, Kaldis P, 2013. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 140, 3079–3093. doi: 10.1242/dev.091744 [DOI] [PubMed] [Google Scholar]
- Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, Wang Z, Wang C, Zhang X, Mo Y-Y, 2016. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget 7, 20584–20596. doi: 10.18632/oncotarget.7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA, 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, eaah7111–11. doi: 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen S, Wang S, Soares F, Fischer M, Meng F, Du Z, Lin C, Meyer C, DeCaprio JA, Brown M, Liu XS, He HH, 2017. Transcriptional landscape of the human cell cycle. Proc Natl Acad Sci U S A 114, 3473–3478. doi: 10.1073/pnas.1617636114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese FP, Grossi E, Marín-Béjar O, Bharti SK, Raimondi I, González J, Martínez-Herrera DJ, Athie A, Amadoz A, Brosh RM, Huarte M, 2016. A Long Noncoding RNA Regulates Sister Chromatid Cohesion. Molecular Cell 63, 397–407. doi: 10.1016/j.molcel.2016.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, A MG, 2019. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Critical Reviews in Biochemistry and Molecular Biology 0, 638–662. doi: 10.1080/10409238.2017.1360836 [DOI] [PubMed] [Google Scholar]
- Mayer C, Schmitz K-M, Li J, Grummt I, Santoro R, 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Molecular Cell 22, 351–361. doi: 10.1016/j.molcel.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, Cabrini M, Wang Y, Capozzo I, Iannelli F, Matti V, Francia S, Shivashankar GV, Walter NG, d’Adda di Fagagna F, 2017. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nature Cell Biology 19, 1400–1411. doi: 10.1038/ncb3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nötzold L, Frank L, Gandhi M, Polycarpou-Schwarz M, Groß M, Gunkel M, Beil N, Erfle H, Harder N, Rohr K, Trendel J, Krijgsveld J, Longerich T, Schirmacher P, Boutros M, Erhardt S, Diederichs S, 2017. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci. Rep. 7, 2265. doi: 10.1038/s41598-017-02357-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, Romero-Cordoba S, Treviño V, Vazquez-Santillan K, Guadarrama AS, Wright AA, Rosenberg MW, Duke F, Kaplan B, Wang R, Nickerson E, Walline HM, Lawrence MS, Stewart C, Carter SL, McKenna A, Rodriguez-Sanchez IP, Espinosa-Castilla M, Woie K, Bjorge L, Wik E, Halle MK, Hoivik EA, Krakstad C, Gabiño NB, Gómez-Macías GS, Valdez-Chapa LD, Garza-Rodríguez ML, Maytorena G, Vazquez J, Rodea C, Cravioto A, Cortes ML, Greulich H, Crum CP, Neuberg DS, Hidalgo-Miranda A, Escareno CR, Akslen LA, Carey TE, Vintermyr OK, Gabriel SB, Barrera-Saldaña HA, Melendez-Zajgla J, Getz G, Salvesen HB, Meyerson M, 2013. Landscape of genomic alterations in cervical carcinomas. Nature 506, 371–375. doi: 10.1038/nature12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JES, Iversen ES, Hartemink AJ, Haase SB, 2008. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453, 944–947. doi: 10.1038/nature06955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormerod MG, Kubbies M, 1992. Cell cycle analysis of asynchronous cell populations by flow cytometry using bromodeoxyuridine label and Hoechst-propidium iodide stain. Cytometry 13, 678–685. doi: 10.1002/cyto.990130703 [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES, 2015. Non-coding RNA: what is functional and what is junk? Front Genet 6, 2. doi: 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-DiNardo D, Kanduri C, 2008. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation. Molecular Cell 32, 232–246. doi: 10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G, 2009. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206. doi: 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, Khavari PA, 2018. The functions and unique features of long intergenic non-coding RNA. Nature Publishing Group 19, 143–157. doi: 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Blackshaw S, 2010. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 10, 49–10. doi: 10.1186/1471-213X-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F, 2006. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature Cell Biology 441, 469–474. doi: 10.1038/nature04674 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, n.d. Integrative genomics viewer. Nat Biotechnol 29, 24 EP –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK, 2009. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YTF, 2009. MEN?/? noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 106, 2525–2530. doi: 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savić N, Bär D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, Cinelli P, Santoro R, 2014. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734. doi: 10.1016/j.stem.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T, 2008. Control of Genic DNA Methylation by a jmjC Domain-Containing Protein in Arabidopsis thaliana. Science 319, 462–465. doi: 10.1126/science.1150987 [DOI] [PubMed] [Google Scholar]
- Schmiedeberg L, Skene P, Deaton A, Bird A, 2009. A Temporal Threshold for Formaldehyde Crosslinking and Fixation. PLoS ONE 4, e4636–5. doi: 10.1371/journal.pone.0004636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer A, Emmrich S, Schmidt F, Beck D, Ng M, Reimer C, Adams FF, Grasedieck S, Witte D, Käbler S, Wong JWH, Shah A, Huang Y, Jammal R, Maroz A, Jongen-Lavrencic M, Schambach A, Kuchenbauer F, Pimanda JE, Reinhardt D, Heckl D, Klusmann J-H, 2017. The non-coding RNA landscape of human hematopoiesis and leukemia. Nature Communications 1–16. doi: 10.1038/s41467-017-00212-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ, 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. doi: 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S, 2017. CUT&RUN: Targeted <em>in situ</em> genome-wide profiling with high efficiency for low cell numbers. bioRxiv 193219. [DOI] [PubMed] [Google Scholar]
- Struhl K, 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14, 103–105. doi: 10.1038/nsmb0207-103 [DOI] [PubMed] [Google Scholar]
- Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X, Cui Y, 2018. Long non-coding RNA MIAT in development and disease: a new player in an old game 1–7. doi: 10.1186/s12929-018-0427-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susumu K, Uyeda HT, Medintz IL, Pons T, Delehanty JB, Mattoussi H, 2007. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. J. Am. Chem. Soc. 129, 13987–13996. doi: 10.1021/ja0749744 [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C, 2003. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J Biol Chem 278, 41528–41534. doi: 10.1074/jbc.M307534200 [DOI] [PubMed] [Google Scholar]
- TCGA Research Network [WWW Document], n.d. TCGA Research Network [WWW Document]. httpswww.cancer.govtcga. URL httpswww.cancer.govtcga (accessed 7.30.19).
- Team, R.D.C., n.d. R: A language and environment for statistical computing. [WWW Document]. httpwww.R-project.org. URL http://www.R-project.org (accessed 8.2.19).
- Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R, 2013. Characterization of a Novel Long Noncoding RNA, SCAL1, Induced by Cigarette Smoke and Elevated in Lung Cancer Cell Lines. Am J Respir Cell Mol Biol 49, 204–211. doi: 10.1165/rcmb.2013-0159RC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV, 2013. Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. PLoS Genet 9, e1003368. doi: 10.1371/journal.pgen.1003368.s014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden CMJ, Lapointe DS, Luong MX, Peric-Hupkes D, Cho B, Stein JL, van Wijnen AJ, Stein GS, 2002. Gene profiling of cell cycle progression through S-phase reveals sequential expression of genes required for DNA replication and nucleosome assembly. Cancer Research 62, 3233–3243. [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D, 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676. doi: 10.1126/science.1093686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voichek Y, Bar-Ziv R, Barkai N, 2016. Expression homeostasis during DNA replication. Science 351, 1087–1090. doi: 10.1126/science.aad1162 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA, 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. doi: 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY, 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W, 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74–e74. doi: 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Manders EE, van der Kraan I, Aten JA, van Driel R, de Jong L, 1994. RNA polymerase II transcription is concentrated outside replication domains throughout S-phase. J. Cell. Sci. 107 ( Pt 6), 1449–1456. [DOI] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE, 2014. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Molecular Cell 55, 791–802. doi: 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Freeling M, Lisch D, 2006. Initiation, Establishment, and Maintenance of Heritable MuDR Transposon Silencing in Maize Are Mediated by Distinct Factors. Plos Biol 4, e339–11. doi: 10.1371/journal.pbio.0040339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, 2015. Isolation of Nascent Transcripts with Click Chemistry. Curr Protoc Mol Biol 111, 4.24.1–4.24.13. doi: 10.1002/0471142727.mb0424s111 [DOI] [PubMed] [Google Scholar]
- Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, Lu Z, Bai C, Watabe K, Mo Y-Y, 2013. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Research 23, 340–350. doi: 10.1038/cr.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Weissman SM, Newburger PE, 2014. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol 11, 777–787. doi: 10.4161/rna.28828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E, 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14, 2283–2297. doi: 10.1101/gad.827700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1, Related to Figure 2 Data from The Cancer Genome Atlas showing frequencies of mutations in lincRNAs enriched in S-phase in various cancers.
Supplementary Table S2, Related to Figure 4 Gene ontology terms for genes uniquely differential in LINC00704, MIAT and LUCAT1 knockdowns in Asynchronous cells.
Data Availability Statement
The accession number for the next generation sequencing data reported in this paper is GEO: GSE137448. This study did not generate code.