Abstract
Muscarinic toxins (MTs) are natural toxins produced by mamba snakes that primarily bind to muscarinic acetylcholine receptors (MAChRs) and modulate their function. Despite their similar primary and tertiary structures, MTs show distinct binding selectivity toward different MAChRs. The molecular details of how MTs distinguish MAChRs are not well understood. Here, we present the crystal structure of M1AChR in complex with MT7, a subtype-selective anti-M1AChR snake venom toxin. The structure reveals the molecular basis of the extreme subtype specificity of MT7 for M1AChR and the mechanism by which it regulates receptor function. Through in vitro engineering of MT7 finger regions that was guided by the structure, we have converted the selectivity from M1AChR toward M2AChR, suggesting that the three-finger fold is a promising scaffold for developing G protein–coupled receptor modulators.
Selective targeting of a specific subtype of a G protein–coupled receptor (GPCR) among its family members is a major challenge in developing receptor-specific drugs with minimal undesired effects (1). Efforts to design compounds with strict specificity toward target GPCRs are often hampered by high conservation of the orthosteric binding site. Allosteric binding sites, on the other hand, share less homology between family members and thus have been targeted as alternative sites for drug discovery (2,3). Muscarinic toxins (MTs) are small protein toxins consisting of 65 to 66 amino acid residues derived from the venoms of African mambas. They belong to a large superfamily, the three-finger toxin (3FT) family (4–6). Despite their high similarity in the sequence and structure, 3FTs exhibit distinct interaction profiles against the five muscarinic acetylcholine receptor (MAChR) subtypes (M1AChR to M5AChR) (7–11). Among the subtypes, MT7 has the highest specificity toward M1AChR over other muscarinic receptors, with a difference in affinity of more than five orders of magnitude (12–16). Studies have shown that MT7 binds M1AChR at subnanomolar affinity with a very slow dissociation rate (17), inhibits agonist-mediated guanosine 5′-O-(3′-thiotriphosphate)(GTP-γ-S) binding and downstream signaling (16,18– 20), and decreases the dissociation rate of orthosteric antagonists {[3H]N-methylscopolamine ([3H]NMS) or [3H]pirenzepine}(20). Thus, MT7 is a potent negative allosteric modulator (NAM) for M1AChR activation and a positive allosteric modulator (PAM) for antagonist binding. Because of its extremely high specificity toward M1AChR, MT7 has attracted research interest into understanding the structural basis for its mode of action. Here, we present the crystal structure of the MAChR-MT7 complex bound to the orthosteric antagonist. The structure reveals the molecular mechanism of the allosteric regulation by MT7 as well as the specific interactions that dictate subtype selectivity. On the basis of this structural information, we engineered MT7 in vitro to redirect its selectivity, yielding a modulator specific for M2AChR. More broadly, this work shows the utility of the 3FT fold for solving the difficult problem of generating specific high-affinity binding proteins to GPCR extracellular loops.
Preparation and structure determination of the M1AChR-MT7 complex
The M1AChR-MT7 complex was formed by using recombinantly expressed M1AChR from Sf9 insect cells and MT7 from Hi5 insect cells as described in the supplementary materials (fig. S1A). Although we were able to form a stable complex between M1AChR bound to the antagonist atropine and MT7 (fig. S1B), our initial attempts to crystallize this complex in lipidic cubic phase (LCP), a widely used method for crystallizing GPCRs, were unsuccessful, yielding crystals made of M1AChR alone. It has been shown that low–molecular weight polyethylene glycols (PEGs), reagents commonly used as precipitants in LCP crystallography, can occupy the extracellular vestibule of M4AChR (21). We determined that low–molecular weight PEGs could compete with MT7 for binding to M1AChR (fig. S1, C and D), supporting the notion that MT7 is displaced by the PEG molecules during crystallogenesis. Through the screening of alternative precipitant reagents, we finally succeeded in obtaining crystals of the M1AChR-MT7 complex by using the hanging drop vapor diffusion method in a condition with no PEG, and the structure was determined at 2.55-Å resolution (fig. S2 and table S1).
Interactions between M1AChR and MT7
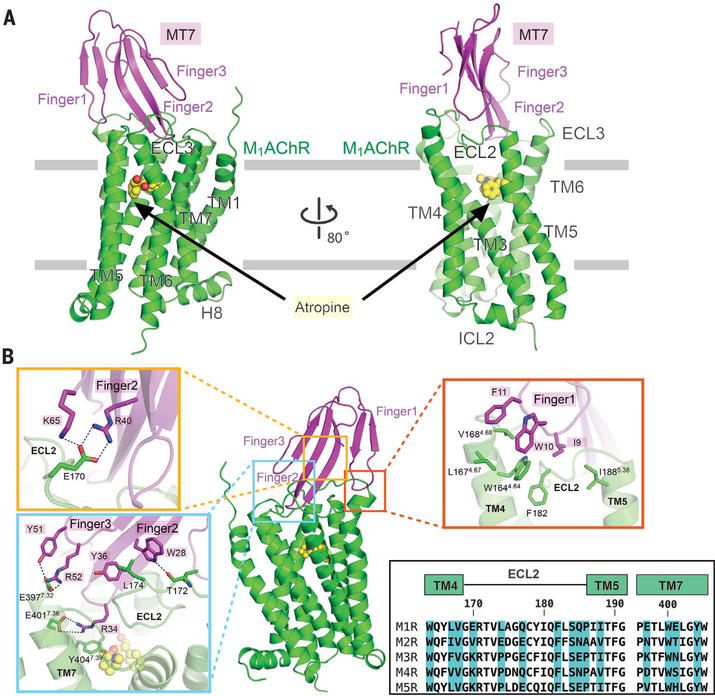
The structure shows that the extracellular vestibule is occupied, with finger loop 2 blocking access to the orthosteric site (Fig. 1A and fig. S3), and this explains the reported slow dissociation of [3H]NMS from the orthosteric site when bound to MT7 (18). Comparison of showed that finger loops 1 and 3 undergo large showed that finger loops 1 and 3 undergo large structural rearrangements upon binding to M1AChR, facilitating extensive interactions with the receptor, while finger loop 2 is mostly unchanged (22) (fig. S4). The large displacement of finger loops 1 and 3 is consistent with molecular dynamics simulations on another snake-derived 3FT, toxin α from Naja nigiicollis (23), which revealed finger loops 1 and 3 to be highly dynamic. The flexibility of finger loops 1 and 3 may explain why some 3FTs can bind to multiple targets (24). In agreement with previous studies, the interactions between M1AChR and MT7 occur predominantly with extracellular loop 2 (ECL2) M1AChR (Fig. 1B) (25–27). Finger loop 1 of MT7 forms extensive hydrophobic interactions with residues in transmembrane helix 4 (TM4) and ECL2 (Fig. 1B, red rectangle). Because these hydrophobic residues are conserved in finger loop 1 of other MT members and in TM4 and ECL2 of other muscarinic receptor subtypes (Fig. 1B and fig. S5), these interactions are not likely to contribute to MT7 subtype selectivity. Finger loop 2 forms the most extensive interactions with ECL2 and TM7, largely consisting of polar contacts. Residues in finger loops 2 and 3 form specific interactions with M1Clh residues E170 and L174 in ECL2 and E3977.32 and E4017.36 at the top of TM7 [superscripts correspond to Ballesteros-Weinstein numbering (28)] (Fig. 1B, yellow and cyan rectangles). Sequence alignment of MAChRs shows that E170, L174, E3977.32, and E4017.36 are not conserved in other MAChRs, suggesting that these M1AChR-specific interactions likely dictate the subtype selectivity of MT7 (Fig. 1B). This conclusion is supported by studies where substitution of these residues of M1AChR into the equivalent positions in M3AChR or M5AChR yields receptors that bind MT7 (25,26,29). Not surprisingly, these subtype-specific residues engaged in MT7 binding also form part of the binding pocket of M1AChR-selective small-molecule allosteric modulators (30–32).
Fig. 1. Structure of the M1AChR-MT7 complex.
(A) Side view of the overall structure of M1AChR-MT7. For clarity, T4L fused to M1AChR was removed from the structure. MT7 and M1AChR are colored magenta and green, respectively. The orthosteric antagonist atropine is colored yellow. Finger loops 1 to 3 of MT7 and ECLs and TM helices of M1AChR are labeled. (B) Detailed interactions between finger loops of MT7 and M1AChR. Interactions with finger loop 1 (red rectangle) and finger loops 2 and 3 (cyan and orange rectangles) are featured in enlarged views. Side chains making interactions are depicted as sticks. Sequence alignments of TM4-ECL2-TM5 and ECL3-TM7 from the five MAChR subtypes are shown in the bottom box. Residue numbers and TM helices of M1AChR are represented above the alignment. Amino acid residues interacting with MT7 in M1AChR are highlighted in blue. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Structural changes in M1AChR stabilized by MT7
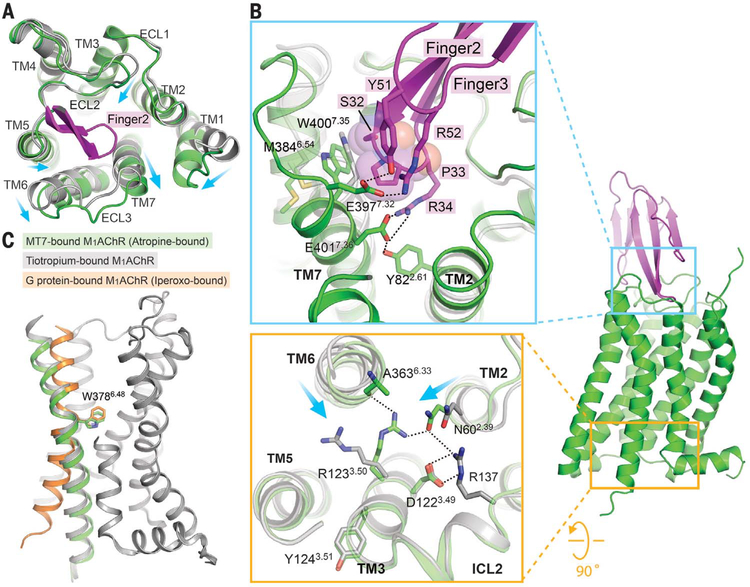
When superposing M1AChR-MT7 bound to atropine and toxin-free M1AChR bound to tiotropium, engagement of MT7 stabilizes a 3- to 4-Å outward movement of TM6, ECL3, and TM7 due to the insertion of finger loop 2 into the extracellular vestibule (Fig. 2A). The outward displacement of TM7 is stabilized by a polar network involving E4017.36 and Y822.61 in M1AChR and R34 from the tip of finger loop 2 (Figs. 1B and 2B). This tyrosine residue, together with Y852.64, which is located one helical turn above Y822.61, has been reported to be one of the key elements for the binding of allosteric compounds (31, 33). In addition, Y51 and R52 in finger loop 3 form polar interactions with E3977.31 (Fig. 2B). These interactions position P33 in finger loop 2 to interact with the backbone and side chains of W4007.35 and E4017.36 of TM7, displacing it outward. The outward movement of W4007.35 at the top of TM7 in turn stabilizes the outward displacement of TM6 through interactions with M3846.54 (Fig. 2B and fig. S6). A tryptophan residue at position 7.35 (W7.35) has been identified as a critical residue for the binding of small-molecule allosteric modulators in M1AChR (30–32) and M2AChR (34). These residues in the active MAChRs undergo inward displacement, and W7.35 forms an aromatic stacking interaction with a PAM, LY2119620, in M2AChR (35, 36). Therefore, W7.35 plays a role in both the positive and negative allosteric modulation of MAChRs by stabilizing the extracellular side of TM6 either inward or outward, respectively (Fig. 2B and fig. S6). We recently reported the active-state structure of M1AChR in complex with the G protein G11 (36). In this structure, there is a 5.7-Å inward movement at the extracellular end of TM6 (at the α carbon of F390) relative to the inactive-state M1AChR, which is accompanied by an outward movement of the cytoplasmic side of TM6 (Fig. 2C). In contrast, the outward movement of the extracellular end of TM6 in the M1AChR-MT7 structure is associated with a small inward movement of the cytoplasmic end of TM6 (Fig. 2, B and C). The allosteric coupling of the extracellular and intracellular ends of TM6 can be explained by a rigid body movement which pivots around W3786.48, the “rotamer toggle switch” (37)(Fig. 2C). As a result of the inward displacement of the cytoplasmic end of TM6 in the M1AChR-MT7 complex, R1233.50 in the DRY motif forms polar interactions with N602.39 from TM2 and the backbone carbonyl of A3636.33 from TM6 (Fig. 2B). These interactions enhance the tight interhelical packing and lock the receptor in the inactive state. The extracellular end of TM5 is displaced slightly inward in the M1AChR-MT7 complex compared with its position in the tiotropium-bound state (Fig. 2A). Unlike displacements of TM6 and TM7 helices that are stabilized by MT7, the displacement of TM5 could be attributed to the different sizes of the orthosteric antagonists. Atropine has a single phenyl group that faces TM3 and TM4, whereas tiotropium has two ring systems (2-thienyl groups), with the second ring facing TM5 (fig. S7). The lack of the second ring in atropine likely allows the slight inward displacement of TM5 relative to the position in the tiotropium-bound state. Indeed, the superposition of the tiotropium into the orthosteric binding site of the atropine-bound state makes a steric clash with T1925.42 (fig. S7).
Fig. 2. Conformational changes in M1AChR stabilized by MT7 binding.
(A) Superposition of tiotropium-bound M1AChR (gray; PDB ID 5CXV) and atropine-bound M1AChR-MT7 complex (green) from the extracellular view. Part of finger loop 2 that interacts with TM7 is shown (magenta). Conformational changes are shown with arrows. (B) (Top) Insertion of finger loop 2 stabilizes conformational changes at the extracellular side of TM2, TM6, and TM7. Note that P33 from MT7, represented as spheres, stabilizes W400 outward. (Bottom) Differences in the organization of the DRY motif in the M1AChR-MT7 complex compared with that of tiotropium-bound M1AChR. Displacements of TM helices 2 and 6 are shown with arrows. (C) Conformational change of TM6 between different states of M1AChR, with W378 serving as a pivotal position: M1AChR from the M1AChR-MT7 complex (green); M1AChR from the tiotropium-bound inactive state (gray; PDB ID 5CXV), and M1AChR from the M1AChR-G11 complex (orange; PDB ID 6OIJ).
Redirecting the subtype selectivity of MT7 from M1AChR to M2AChR
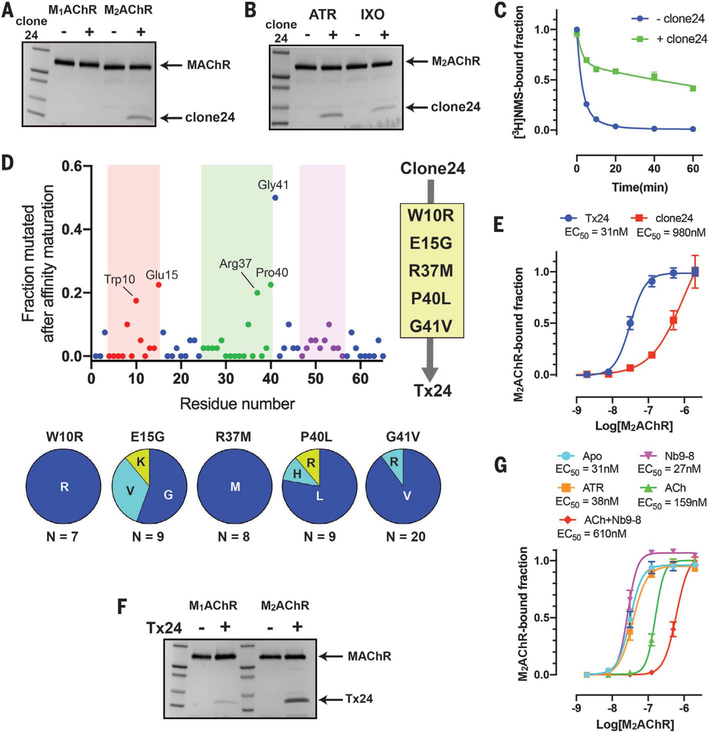
With its small size and high stability, the 3FT fold has been utilized as an alternative scaffold to generate protein binders (38, 39). Therefore, we designed a phage display library using MT7 as a template scaffold to explore whether we could redirect the specificity to other muscarinic receptor subtypes through structure-guided engineering. On the basis of the residues of MT7 in contact with M1AChR in the crystal structure, we selected residues from each finger loop to be randomized (fig. S8). For the target molecule, we used M2AChR in the presence of the antagonist atropine. After four rounds of phage panning, we identified clone 24, which binds subtype selectively toward M2AChR over M1AChR and has conformational selectivity toward the inactive state (Fig. 3, A and B). Moreover, it confers a slow dissociation of the antagonist [3H]NMS from the orthosteric binding pocket (Fig. 3C), suggesting that it binds allosterically to the extracellular vestibule. A substoichiometric amount of clone 24 in the pull-down assay suggested a weak affinity of the engineered toxin for M2AChR. We therefore performed affinity maturation by introducing random mutations throughout the toxin coding region and selection using yeast surface display to obtain a high-affinity variant (fig. S9). High-affinity binders were enriched using progressively lower concentrations of M2AChR bound with atropine during selections. After two rounds of magnetic-activated cell sorting, we further performed fluorescence-activated cell sorting (FACS) to select high-affinity clones. By combining the consensus mutations from randomly selected sequences of the affinity-maturated clones, we created a high-affinity variant, Tx24 (Fig. 3D). Tx24 gained a 30-fold enhancement in affinity for M2AChR over clone 24 (Fig. 3E) and retained the same subtype and conformation selectivity as the original clone 24 (Fig. 3, F and G). Figure S10 shows the sequence alignments for MT7, clone 24, and Tx24.
Fig. 3. Identification and affinity maturation of a subtype-selective M2AChR allosteric modulator through in vitro selection.
(A) Pull-down assay showing that clone 24 binds subtype selectively to M2AChR over M1AChR. The experiment was carried out in the presence of 10 μM atropine. (B) Pull-down assay showing the conformation preference of clone 24 toward the atropine (ATR)–bound state over the iperoxo (IXO)–bound state of M2AChR. (C) Comparison of the dissociation kinetics of the orthosteric antagonist [3H]NMS from M2AChR in the clone 24–bound and apo state. Data represent means ± SEM of three experiments performed in triplicate. (D) Sequence analysis of 50 randomly selected clones after the second FACS enrichment. The finger loop regions 1, 2, and 3 are colored red, green, and purple, respectively. The five most frequent mutations (W10R, E15G, R37M, P40L, and G41V) were combined into clone 24 to generate Tx24. (E) Comparison of the affinities of clone 24 and Tx24 to M2AChR using on-yeast affinity measurement in the presence of 10 μM atropine. Data represent means ± SEM of three independent experiments. (F) Pull-down assay of Tx24 using M1AChR or M2AChR in the presence of 10μM atropine. Tx24 prefers M2AChR over M1AChR. (G) Comparison of the affinity of Tx24 for M2AChR bound to different ligands alone or in combination with a G protein mimetic nanobody using on-yeast affinity measurement. Symbols and error bars represent means and SEM of three independent experiments.
Functional characterization of Tx24 on M2AChR and other GPCRs
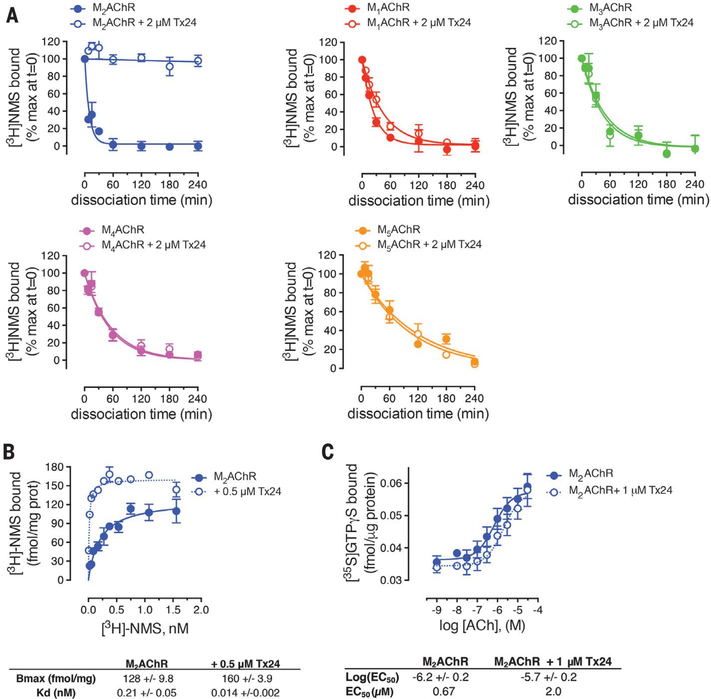
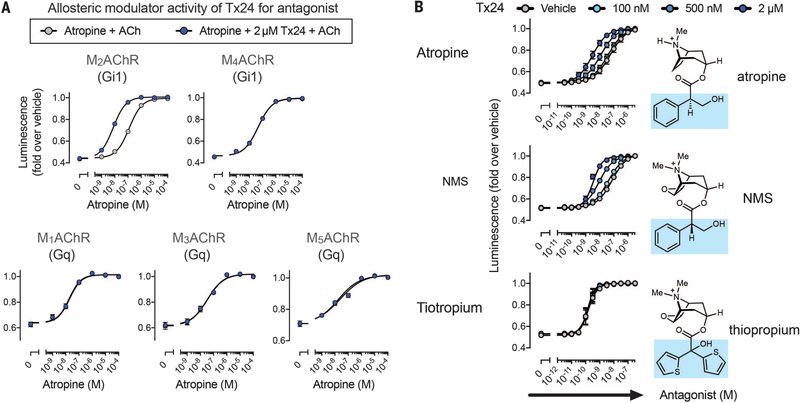
We subsequently investigated the pharmacological properties of Tx24. In the radioligand dissociation assay, Tx24 substantially decreased the dissociation rate of [3H]NMS from M2AChR, in accordance with the properties of the original clone 24 (Fig. 4). In contrast, Tx24 showed little or no impact on the dissociation rate of antagonists from other MAChR family members or from the β2-adrenergic receptor (β2AR) (Fig. 4A and fig. S11A), confirming the high subtype specificity of Tx24. A small (1.8-fold) decrease in the dissociation rate of [3H]NMS in M1AChR is likely caused by the residual binding between Tx24 and M1AChR that was observed in the pull-down assay. However, this effect was much smaller than that for M2AChR, for which a >700-fold decrease in off-rate was observed in the presence of Tx24 (fig. S11A). Moreover, the affinity of [3H]NMS for M2AChR became substantially higher in the presence of Tx24 (Fig. 4B), whereas enhancement of [3H]NMS binding was barely detectable in M1AChR and absent in M4AChR, confirming the specificity of Tx24 for M2AChR (fig. S11B). The median effective concentration of Tx24 for this effect on M2AChR is in good agreement with the on-yeast affinity measurement (Fig. 3E and fig. S11B). Apart from the substantial influence on the behavior of antagonists, Tx24 has a slight negative allosteric effect on the potency of ACh, suggesting that it is a weak NAM for agonists (Fig. 4C). Next, we investigated the effect of Tx24 on G protein activation using the NanoBiT system (40). In the NanoBiT system, MAChR-dependent cognate G protein activation is monitored through the decrease of the luminescence signal due to the heterotrimer dissociation (fig. S12A). Tx24 on its own showed no agonistic or antagonistic activity (fig. S12A). When these results were taken together, Tx24 enhanced the inhibition by the antagonist atropine and this enhancement was specific for M2AChR (Fig. 5A and table S2). In contrast, we observed no impact on G protein activation from other MAChRs or from the μ-opioid receptor or β2AR (Fig. 5A and fig. S12). These results further validate the M2AChR subtype selectivity of Tx24 in a cellular signaling context.
Fig. 4. Pharmacological impact of Tx24 for the dissociation of the orthosteric antagonist.
(A) Comparison of the [3H]NMS dissociation kinetics from M2AChR (blue), M1AChR (red), M3AChR (magenta), M4AChR (green), or M5AChR (orange) in the absence (filled symbols, solid line), or presence (empty symbol, solid line) of 2 μM Tx24. Shown are the combined results from four assays performed in duplicate. (B) Saturation binding of [3H]NMS to M2AChR in the absence (filled symbol, solid line) or presence (empty symbol, dotted line) of 0.5 μM Tx24 after 24-hour incubation. Shown are the combined results from four assays performed in duplicate. (C) ACh-stimulated [35S]GTP-γ-S binding at M2AChR expressed in CHO cells in the absence (filled symbol, solid line) or presence (empty symbol, dotted line) of 1 μM Tx24. Symbols and error bars represent means and SEM of the combined results from three or four assays performed in duplicate.
Fig. 5. Tx24 is a probe-selective PAM for antagonists.
(A) Allosteric modulator activity of Tx24 toward antagonist atropine. The cells expressing the NanoBiT–G protein and the test GPCR were treated with titrated atropine followed by addition of Tx24 or vehicle. Luminescent signals were measured before and after ligand stimulation (10 μM ACh). (B) HEK293 cells transiently expressing the NanoBiT-Gi1 protein and M2AChR were loaded with coelenterazine and pretreated with titrated antagonist (atropine, NMS, or tiotropium), followed by addition of Tx24 (100 nM, 500 nM, or 2 μM). Luminescent signals were measured before and after ACh treatment (10 μM). Changes in luminescent signals were normalized to those with vehicle treatment. The chemical structure of each antagonist compound is placed next to the respective curve, with the aromatic head group highlighted in blue. Symbols and error bars represent means and SEM of three to five independent experiments, each performed in duplicate (see tables S2 and S3 for statistics).
Probe-dependent positive allosteric modulation of Tx24
Probe dependency of the allosteric modulator in M2AChR has been reported with a PAM molecule for agonists that implicates a complex interaction between orthosteric and allosteric binding sites (41). To determine the probe dependency of Tx24, we assessed three orthosteric antagonist compounds: atropine, NMS, and tiotropium. Although Tx24 enhanced the inhibition of M2AChR activity by atropine or NMS, it had no impact on the inhibition by tiotropium (Fig. 5B and table S3), indicating that Tx24 is a probe-selective PAM for atropine and NMS. Recent structural dynamics analysis by nuclear magnetic resonance (NMR) spectroscopy revealed distinct conformations of M2AChR for NMS- and tiotropium-bound states (42). Atropine and NMS share a similar structure with a single ring system, whereas tiotropium has two rings (Fig. 5B). The preferential allosteric effects of Tx24 for these smaller antagonists are likely due to the fact that it was developed against atropine-bound M2AChR. Consistent with this notion, Tx24 has a higher affinity for M2AChR bound to NMS than for M2AChR bound to tiotropium (fig. S13).
Effect of Tx24 binding on conformation of M2AChR
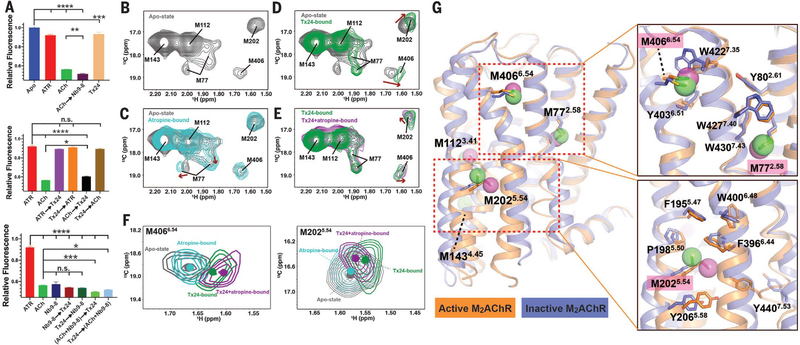
To assess the allosteric structural changes resulting from Tx24, we labeled T3866.34 at the cytoplasmic end of TM6 with monobromobimane (mBBr) and introduced a tryptophan mutation at S2105.62, which upon activation of the receptor quenched the fluorescence emitted from mBBr on TM6 (fig. S14). Compared with the apo state, the ACh-activated state had a significantly lower fluorescence intensity, which was further decreased in the presence of an intracellular G protein–mimetic nanobody, Nb9–8, whereas there was only a small decrease in fluorescence in the atropine-bound or Tx24-bound state (Fig. 6A). There was little change in the fluorescence intensity in the presence of both atropine and Tx24 regardless of the order of addition. When the receptor was first incubated with ACh, the addition of Tx24 resulted in only a small increase in fluorescence. However, when the receptor was first incubated with Tx24, the addition of ACh did not reduce the fluorescence. These results are consistent with the observation that Tx24 binds with lower affinity to ACh-bound M2AChR (Fig. 3G) and, therefore, cannot stabilize an inactive conformation in TM6. Notably, the receptor remains in the active conformation in the presence of Nb9–8 regardless of the presence of Tx24 or the order of addition of these components.
Fig. 6. Structural impact of Tx24 on M2AChR conformation dynamics.
(A) Comparison of the fluorescence intensity of the mBBr-labeled M2AChR in various conditions. Receptor was first incubated with the compound or protein listed first and then further incubated with the second molecule(s) before fluorescence spectra were obtained. Fluorescence peak intensities at 461 nm are plotted. Data represent at least three independent measurements. *P < 0.05; **P < 0.005; ***P < 0.001; ****P < 0.0001; n.s., not significant (one-way analysis of variance). (B to E) Structural impact of Tx24 on the HSQC spectra of the apo state, atropine-bound state, Tx24-bound state, and Tx24+atropine-bound state of M2AChRmini△5M. (F) Overlay of spectra for M4066.54 and M2025.54 in different conditions from panels (B) to (E): apo state (gray), atropine-bound (cyan), Tx24-bound (green), Tx24+atropine–bound (magenta). The centers of the resonances are indicated with dots. (G) Distribution of methionine probes with the structural comparison between inactive (blue; PDB ID 3UON) and active M2AChR (orange; PDB ID 4MQS). Side-chain interactions surrounding M406, M77, and M202 are detailed in the right panels.
The structural impact of Tx24 on M2AChR conformation and dynamics was further investigated by solution NMR spectroscopy using methionine residues as conformational probes (42). The labeled methionines are well-positioned to detect conformational changes in the extracellular vestibule (M4066.54), the orthosteric binding pocket (M772.58), the transduction core (M1123.41), the junction of ICL2 and TM4 (M1434.45), and the cytoplasmic ends of TM5 and TM6 (M2025.54) (fig. S15). On the basis of the structure of M1AChR-MT7, we would not expect Tx24 to directly interact with any of the methionine probes. Binding of Tx24 caused chemical shift perturbations of M772.58, M2025.54 and M4066.54 in the heteronuclear single-quantum coherence (HSQC) spectra among the five labeled methionines (Fig. 6, B to E). M772.58 underwent substantial changes in chemical shift and peak pattern upon binding to Tx24 compared with the apo or atropine-bound state, suggesting distinct local conformations and dynamics stabilized by Tx24. Co-incubation with atropine had no further influence on the M772.58 peak, which indicated that the conformation of the extracellular side of TM2 is mostly dominated by Tx24. Y802.61 and Y832.64, both located above M772.58, play pivotal roles in the allosteric transmission of structural changes in the extracellular vestibule to the orthosteric pocket (31, 33). It is possible that Tx24 triggers a conformational change around Y802.61 in M2AChR that is similar to the change we observed around the analogous Y822.61 residue in the M1AChR-MT7 complex (Fig. 2B), and this conformational change is reported by M772.58 in the HSQC spectra. Although the M2025.54 and M4066.54 peak profiles showed little change from apo state to atropine-bound state, their positions shifted markedly upon binding with Tx24 (Fig. 6F). M4066.54 is located in close contact with W4227.35, which faces the extracellular vestibule in the inactive state (Fig. 6G). Given the size of Tx24, it is likely that it interacts with W4227.35 and provides M4066.54 with a different microenvironment in the apo state and the atropine-bound state. Unlike M772.58 and M4066.54, M2025.54 is located at the TM5-TM6 interface on the intracellular side of the receptor and away from the putative Tx24 binding pocket (Fig. 6G). The spectral changes observed in M2025.54 indicate that the extracellular conformational change induced by Tx24 propagates to the intracellular region and allosterically stabilizes a distinct intracellular conformation of the TM5-TM6 interface from the apo or atropine-bound state. Both M2025.54 and M4066.54 peaks further shift when co-incubated with atropine (Fig. 6F), indicating a cooperative action between Tx24 and atropine, as evident from the pharmacological response (Fig. 5G).
Discussion
Natural organisms such as plants, bacteria, fungi, and animals have developed various molecules for their survival: for self-defense against predators and for capturing prey. These molecules have been a rich source of drugs and tool compounds for a diverse range of targets, including GPCRs (43). Whereas small molecules dominate the pharmacopeia for GPCRs, there is growing interest in peptides and small proteins because of their potential to be highly selective. The structure of M1AChR -MT7 reveals the molecular basis for its strict subtype selectivity and also how the conformational changes of M1AChR upon MT7 binding lead to inactivation of the receptor. The ability to redirect its MAChR subtype preference through in vitro protein evolution demonstrates the promise of the 3FT fold as an alternative scaffold for generating protein-based modulators.
Supplementary Material
ACKNOWLEDGMENTS
We thank N. Fastman and D. Hilger for help with phage display experiments and C. Glassman for help with yeast surface display experiments. We thank staff at Advanced Photon Source GM/CA beamlines for technical assistance and support for data collection. We thank K. Jude for technical advice in the crystal structure analysis; K. Sato, S. Nakano, and A. Inoue at Tohoku University for assistance in plasmid preparation and cell-based GPCR assays; and X. Niu and H. Li for help with NMR data collection. NMR data were collected at the Beijing NMR Center and the NMR facility of the National Center for Protein Sciences at Peking University.
Funding: The work was supported by NIH grant R01GM083118 for R.K.S. B.K.K. was funded by R01NS028471. K.C.G. was funded by NIH R01AI125320, Mathers Foundation, and Howard Hughes Medical Institute. A.I. was funded by the PRIME 18gm5910013 and the LEAP 18gm0010004 from the Japan Agency for Medical Research and Development (AMED) and the Japan Society for the Promotion of Science (JSPS) KAKENHI 17K08264. J.A. was funded by the LEAP 18gm0010004 from AMED. B.K.K. is a Chan Zuckerberg Biohub investigator.
Footnotes
Competing interests: B.K.K. is a cofounder of and consultant for ConfometRx.
SUPPLEMENTARY MATERIALS
Data and materials availability:
The atomic coordinates of M1AChR-MT7 have been deposited in the RCSB Protein Data Bank (PDB) with the identifier 6WJC. Materials for these engineered toxins are available from the corresponding authors upon request. Expression plasmids for the NanoBiT-G-protein dissociation assay are available from A.I. under a material transfer agreement with Tohoku University.
REFERENCES AND NOTES
- 1.Lee S-M, Booe JM, Pioszak AA, Eur. J. Pharmacol. 763, 196–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christopoulos A, Mol. Pharmacol. 86, 463–478 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Langmead CJ, Christopoulos A, Curr. Opin. Cell Biol. 27, 94–101 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kessler P, Marchot P, Silva M, Servent D, J. Neurochem 142 (suppl. 2), 7–18 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Ségalas I et al. , Biochemistry 34, 1248–1260 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Liang JS et al. , Toxicon 34, 1257–1267 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Bradley KN, Pharmacol. Ther. 85, 87–109 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Karlsson E, Jolkkonen M, Mulugeta E, Onali P, Adem A, Biochimie 82, 793–806 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Jerusalinsky D et al. , Toxicon 38, 747–761 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Servent D et al. , Toxicon 58, 455–463 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Servent D, Fruchart-Gaillard C, J. Neurochem. 109, 1193–1202 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Max SI, Liang JS, Potter LT, J. Neurosci. 13, 4293–4300 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Max SI, Liang JS, Potter LT, Mol. Pharmacol. 44, 1171–1175 (1993). [PubMed] [Google Scholar]
- 14.Max SI, Liang JS, Valentine HH, Potter LT, J. Pharmacol. Exp. Ther. 267, 480–485 (1993). [PubMed] [Google Scholar]
- 15.Carsi JM, Potter LT, Life Sci. 68, 2541–2547 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Näsman J, Jolkkonen M, Ammoun S, Karlsson E, Åkerman KEO, Biochem. Biophys. Res. Commun. 271, 435–439 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Krajewski JL, Dickerson IM, Potter LT, Mol. Pharmacol. 60, 725–731 (2001). [PubMed] [Google Scholar]
- 18.Olianas MC et al. , Br. J. Pharmacol. 131, 447–452 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley KN, Rowan EG, Harvey AL, Toxicon 41, 207–215 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Olianas MC, Adem A, Karlsson E, Onali P, Eur. J. Pharmacol. 487, 65–72 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Thal DM et al. , Nature 531, 335–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fruchart-Gaillard C et al. , Mol. Pharmacol. 74, 1554–1563 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Gilquin B et al. , Protein Sci. 12, 266–277 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchet G et al. , Biochimie 103, 109–117 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Kukkonen A, Peräkylä M, Åkerman KEO, Näsman J, J. Biol. Chem. 279, 50923–50929 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Rondinelli S, Näreoja K, Näsman J, Toxins 3, 1393–1404 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquer C et al. , J. Biol. Chem. 286, 31661–31675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballesteros JA, Weinstein H, in Receptor Molecular Biology, vol. 25 of Methods in Neurosciences, Sealfon SC, Ed. (Academic Press, 1995), pp. 366–428. [Google Scholar]
- 29.Xu J et al. , J. Mol. Recognit. 28, 239–252 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Abdul-Ridha A et al. , J. Biol. Chem. 289, 6067–6079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdul-Ridha A et al. , J. Biol. Chem. 289, 33701–33711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma L et al. , Proc. Natl. Acad. Sci. U.S.A. 106, 15950–15955 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollingsworth SA et al. , Nat. Commun. 10, 3289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dror RO et al. , Nature 503, 295–299 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Kruse AC et al. , Nature 504, 101–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeda S, Qu Q, Robertson MJ, Skiniotis G, Kobilka BK, Science 364, 552–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visiers I, Ballesteros JA, Weinstein H, Methods Enzymol. 343, 329–371 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Naimuddin M et al. , Mol. Brain 4, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai W et al. , J. Recept. Signal Transduct. 34, 154–161 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Inoue A et al. , Cell 177, 1933–1947.e25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valant C, Felder CC, Sexton PM, Christopoulos A, Mol. Pharmacol. 81, 41–52 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Xu J et al. , Mol. Cell 75, 53–65.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Muratspahic E, Freissmuth M, Gruber CW, Trends Pharmacol. Sci. 40, 309–326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of M1AChR-MT7 have been deposited in the RCSB Protein Data Bank (PDB) with the identifier 6WJC. Materials for these engineered toxins are available from the corresponding authors upon request. Expression plasmids for the NanoBiT-G-protein dissociation assay are available from A.I. under a material transfer agreement with Tohoku University.