Abstract
Assessing Erb-b2 receptor tyrosine kinase 2 (ERBB2) amplification status in breast and gastric cancer is necessary for deciding the best therapeutic strategy. Immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) are currently used for assessing protein levels and gene copy number (CN), respectively. The use of next-generation sequencing (NGS) to measure ERBB2 CN in breast cancer is approved by the United States Federal Drug Administration as a companion diagnostic. However, a CN of less than 8 is evaluated as “equivocal”, which means that some ERBB2 amplification cases diagnosed as “HER2 negative” might be false-negative cases. We reviewed the results of gene profiling targeting 160 cancer-related genes in breast (N = 90) and non-breast (N = 19) cancer tissue, and compared the ERBB2 CN results with the IHC/FISH scores. We obtained an estimated CN from the measured CN by factoring in the histological proportion of tumor cells and found that an ERBB2-estimated CN of 3.2 or higher was concordant with the combined IHC/FISH outcome in 98.4% (88/90) of breast cancer cases, while this was not always evident among non-breast cancer cases. Therefore, NGS-estimated ERBB2 CN could be considered a diagnostic test for anti-HER2 therapy after the completion of adequate prospective clinical trials.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12032-021-01482-1.
Keywords: Breast cancer, ERBB2, Gene copy number, Immunohistochemistry, Next-generation sequencing
Introduction
Human epidermal growth factor receptor 2 (HER2), the protein encoded by the Erb-b2 receptor tyrosine kinase 2 (ERBB2) gene, is one of the main therapeutic targets in human cancers [1]. HER2 is a transmembrane receptor tyrosine kinase that belongs to the HER family, and its overexpression leads to homodimerization and heterodimerization with other HER family-member proteins [2], triggering activation of the phosphoinositide-3-kinase/Akt and mitogen-activated protein kinase pathways [3], resulting in tumor proliferation, differentiation, apoptosis regulation, angiogenesis, and invasion [4].
HER2 overexpression is used as a biomarker in breast and gastric cancers to predict the response to anti-HER2 monoclonal antibody therapies (trastuzumab and pertuzumab) and small-molecule HER2 kinase inhibitors (lapatinib) [5, 6]. Clinical laboratories stain for HER2 protein by immunohistochemistry (IHC), or detect ERBB2 amplification by FISH or chromogenic in situ hybridization. Amplification of ERBB2 is a potential therapeutic target in other cancer types, including lung, bladder, endometrial, ovary, colorectal, esophageal, and bile duct cancers [7–14]. There are consensus scoring guidelines for these techniques for breast and gastric cancers [15, 16]; however, these have not been established for other cancer types.
Next-generation sequencing (NGS) is being increasingly adopted in clinical settings for the identification of multiple genomic changes, such as single-nucleotide variants and copy number (CN) changes. According to the current guidelines, hybrid capture-based NGS detection of ERBB2 CN amplification (CN higher than 8) is approved as a companion diagnostic for breast and upper gastrointestinal carcinomas by the United States Federal Drug Administration. However, according to their guidelines, a slight degree of amplification (CN over 3 but lower than 8) is diagnosed as ‘equivocal’, and the diagnostic value of an NGS equivocal CN status for ERBB2 has not yet been determined, as the accurate ERBB2 CN therapeutic cutoff value has not been established. In the present study, we aimed to compare targeted NGS-based ERBB2 CN estimation with HER2 IHC or FISH for breast and other cancer types, and to define the accurate ERBB2 CN therapeutic cutoff value.
Results
Patients and clinicopathological features
The present study group comprised 90 patients with breast cancer and 19 patients with other cancer types, and their characteristics (procedures performed and sample type) are listed in Table 1. The details of their clinical and pathological features are summarized in Table S1 (breast cancer) and Table S2 (non-breast cancer). Among the 90 breast cancer samples, 13 were derived from core needle biopsies and 77 from surgical resections, and the age of the patients ranged from 37 to 81 years (median 55). Eighty-four percent of patients (N = 76) had invasive ductal carcinoma, followed by 11% (N = 10) with ductal carcinoma in situ. According to the International Union Against Cancer staging system 8th edition, 94.4% (N = 85) had Tis/T1/T2, while 5.6% (N = 5) had T3/T4. Regarding the estrogen receptor (ER), progesterone receptor (PgR), and Ki67 IHC status of the cohort, 49% (N = 44) had ER-positive tumors, while 40% (N = 36) had PgR-positive, and 50% (N = 45) had Ki67-positive tumors. The percentage of tumor cells was estimated independently by three pathologists on hematoxylin and eosin-stained slides, where the tumor area was marked (histological proportions of tumor cells in Tables S1 and S2).
Table 1.
Sample types and procedures performed in 109 cancer patients recruited for the present study
| Type of cancer | Total | Procedure | ||
|---|---|---|---|---|
| Core | Excision | |||
| Breast | 90 | 13 | 77 | |
| Non-Breast | 19 | 2 | 17 | |
| Gastric cancer | 3 | 0 | 3 | |
| Lung cancer | 2 | 0 | 2 | |
| Colorectal cancer | 2 | 0 | 2 | |
| Bladder cancer | 2 | 2 | 0 | |
| Extra-mammary Paget's disease | 2 | 0 | 2 | |
| Renal pelvic cancer | 1 | 0 | 1 | |
| Cervical cancer | 1 | 0 | 1 | |
| Endometrial cancer | 1 | 0 | 1 | |
| Ovarian cancer | 1 | 0 | 1 | |
| Gallbladder cancer | 1 | 0 | 1 | |
| Prostate cancer | 1 | 0 | 1 | |
| Uterine sarcoma | 1 | 0 | 1 | |
| Anal fistula | 1 | 0 | 1 | |
HER2 IHC, FISH, and NGS-based ERBB2 CN among patients with breast cancer
The comparison between the estimated ERBB2 CN determined by NGS and the HER2 IHC score is provided in Fig. 1. Statistically significant differences in estimated CN were found among different IHC groups. The comparison between the NGS-derived ERBB2-estimated CN and the HER2 IHC/FISH results is shown in Figure S1. Table S1 summarizes the HER2 IHC scoring, FISH results, ERBB2 CN status and measured/estimated CN by NGS, and ERBB2 mutation status in the breast cancer cohort. Among samples with amplified ERBB2, the estimated CN ranged from 3.2 to 54.3 (median 11.7). There was no significant difference in the proportion of tumor cells between the ERBB2-amplified and ERBB2 neutral CN cases (data not shown). For HER2 IHC, 23 of 90 (26%) tumors showed 3 + staining, 34 (38%) showed 2+ , 18 (20%) showed 1+ , and 15 (17%) showed no staining. FISH was performed on 34 samples, all of which showed an HER2 IHC score of 2+ , and four of which had ERBB2 amplification according to NGS. Notably, only 2 cases (5.8%) had positive results in FISH, and both had ERBB2 amplification according to NGS. Of the 27 ERBB2-amplified cases determined by NGS, 23 tumors showed strong positive HER2 staining (IHC score 3+). The four remaining ERBB2-amplified cases displayed HER2 2+ staining, two of which also had a positive FISH result, but the other two had negative FISH results. Among the four cases that met the criteria for amplification, two had a negative FISH result; one had an estimated CN = 4.8 and IHC2+ /FISH − , whereas the remaining case had an estimated of CN = 3.1 and IHC2+ /FISH − . Of the 63 cases without ERBB2 amplification according to NGS, 15 showed no HER2 IHC expression, and 48 had mild staining with an IHC score of 1+ and 2+ . Among the 30 cases with 2+ IHC staining and no ERBB2 amplification, no case was positive for FISH.
Fig. 1.
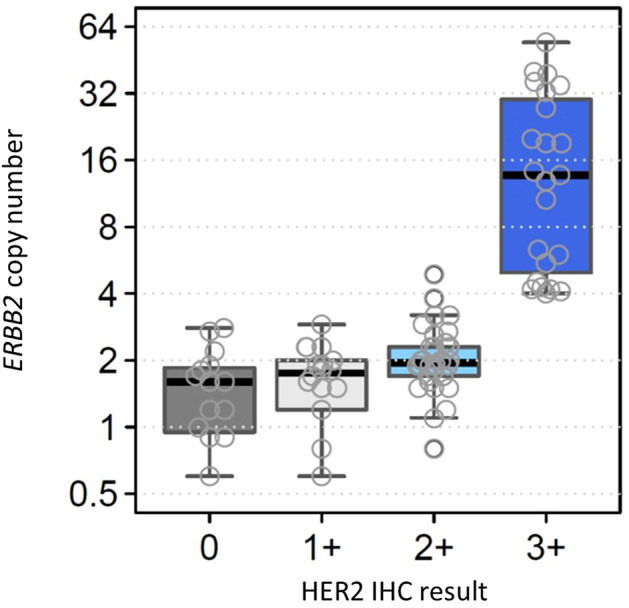
Estimated ERBB2 CN determined by NGS and HER2 IHC results among breast cancer cases. Statistically significant differences were observed between IHC 0 group (N = 15) vs. IHC 2 + group (N = 34) (p = 0.04), IHC 0 group vs. IHC 3 + group (N = 23) (p = 1.5e-06), IHC 1 + group (N = 18) vs. IHC 3 + group (p = 3.2e-07), and IHC 2 + group vs. IHC 3 + group (p = 2.2e-09). Boxplots were generated in R using default settings. Each box spans the 25th to 75th percentile range of the data, i.e., the interquartile range (IQR), and the middle line represents the median value. Whiskers extend 1.5 times the IQR from the edge of the box. ERBB2, Erb-b2 receptor tyrosine kinase 2; NGS, next-generation sequencing; HER2, Human epidermal growth factor receptor 2; IHC, immunohistochemistry
HER2 IHC, FISH, and ERBB2 CN among patients with non-breast cancer with ERBB2 amplification
Table S2 summarizes the HER2 IHC scoring, ERBB2 CN status, measured/estimated CN by NGS, and ERBB2 mutation status in non-breast cancer cases in the present study. Among 19 patients with ERBB2 amplification and an estimated CN of more than 3.0 by NGS, only 12 cases had HER2 IHC 3+ , four cases had 2+ , one case had 1+ , and two cases had no staining. The comparison between ERBB2 estimated CN and HER2 IHC results is provided in Figure S2. As a HER2 IHC scoring system has not been established for cancers other than breast and gastric cancers, we sought to evaluate HER2 protein expression by IHC in solid cancers using the breast cancer scoring system, and in luminal cancers using the gastric cancer scoring system. The IHC findings in all non-breast cancer cases are shown in Figure S3. All 19 non-breast tumors had ERBB2 amplification (estimated CN above 3.0) and varying HER2 IHC scores (Table S2).
Determining thresholds of estimated CN for NGS-based ERBB2 amplification among cases with breast cancer
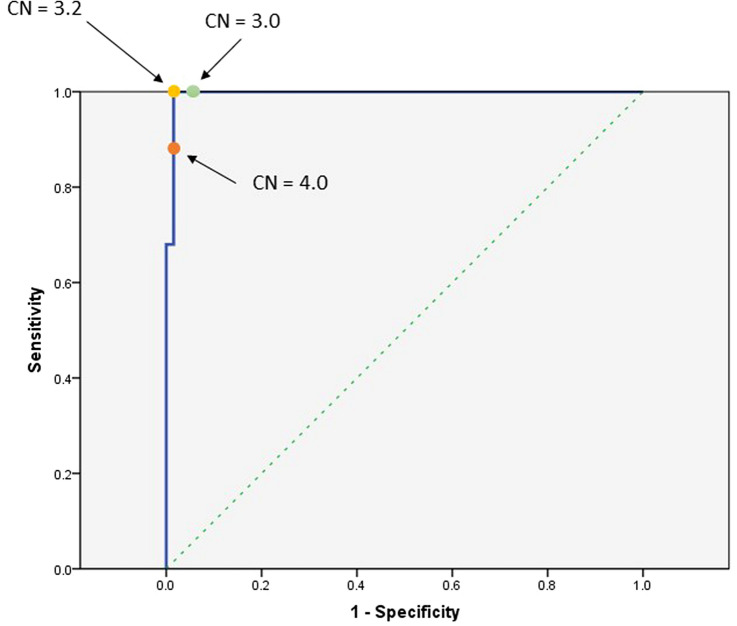
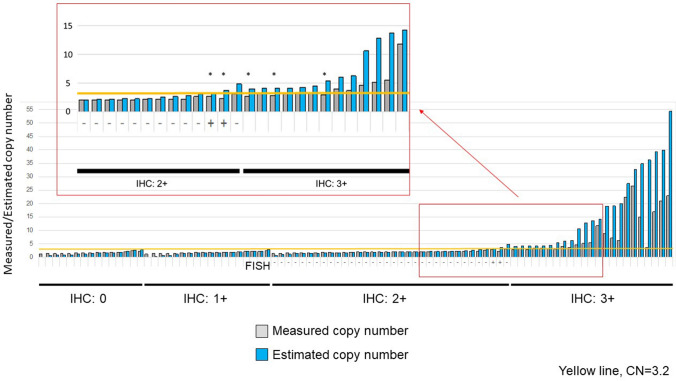
Current breast cancer treatment guidelines indicate that patients with either HER2 IHC3 + or HER2 IHC2 + /FISH + should be considered for anti-HER2 therapies. We evaluated the association between NGS-derived ERBB2 estimated CN and the IHC/FISH results. All cases with HER2 IHC3 + or IHC2 + /FISH + had ERBB2 amplification with an estimated CN of more than 3.2. The receiver operating characteristic (ROC) curve (Fig. 2) shows the relationship between sensitivity and specificity for every possible cutoff of the ERBB2 estimated CN and the HER2 IHC 3 + and IHC 2 + /FISH + outcomes. A cutoff of estimated CN = 3.2 achieved the maximum performance with 100% sensitivity and 98.5% specificity for an IHC 3 + and IHC 2 + /FISH + outcome in our breast cancer cohort. A cutoff value of estimated CN = 3.0 also achieved high sensitivity and specificity (100% and 97%, respectively), while a cutoff value of CN = 4.0 had decreased sensitivity of 88% and 98.5% specificity. Therefore, we defined ERBB2 estimated CN = 3.2 as the cutoff for therapeutic recommendation. The relationship between the NGS-derived ERBB2 measured/estimated CN and HER2 IHC and FISH results is shown in Fig. 3. Samples from the HER2 IHC 3 + group tended to have a greater discordance between measured and estimated CNs. There were five samples with measured CNs less than 3.2, while their estimated CNs were more than 3.2.
Fig. 2.
Receiver operating characteristics curve for different CN threshold values. For lower threshold values, the pipeline calls everything as amplified, which are all false positives (x axis); for higher threshold cutoff values, our ability to correctly call real events decreases. A threshold of 3.2 has the best sensitivity and specificity
Fig. 3.
ERBB2 measured/estimated CN and HER2 IHC/FISH results. Yellow line is CN = 3.2. Five patients marked with an asterisk had estimated CN of over 3.2 but measured CN under 3.2. Erb-b2 receptor tyrosine kinase 2; HER2, Human epidermal growth factor receptor 2; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization
Genomic alterations in patients with breast cancer
Tumors with a neutral CN for ERBB2 included three cases with ERBB2 mutations. The pathogenicity of all these variants was determined to be of unknown significance by our mutation calling algorithm.
Discussion
With the advent of personalized cancer therapies, it is important to determine the expression levels of various molecular markers simultaneously in order to design effective therapeutic strategies. NGS has this capacity, as it can provide results with only a limited amount of sample material. However, clear diagnostic guidelines are necessary for the clinical use of NGS. We determined an ERBB2 estimated CN threshold of 3.2 for recommending anti-HER2 therapy in breast cancer cases. A cutoff value of estimated CN = 3.0 also achieved high concordance with IHC3+ , indicating that only hemiallelic 2 × amplification causes IHC3 +status. This threshold (estimated CN = 3.2) was capable of identifying two additional cases that had been omitted by conventional methods, suggesting that there is heterogeneity in FISH sensitivity (Figure S4), with only 50% of cases with ERBB2 amplification being FISH positive. Follow-up studies on the discordance between FISH and NGS amplification results are required to determine whether anti-HER therapy should be recommended for cases with NGS-derived amplification and IHC2+ /FISH − results. Although these cases did not meet the criteria by conventional IHC/FISH methods, they did satisfy the accepted amplification threshold using the NGS method. These cases highlight the promising capacity of NGS to offer therapeutic recommendations that could not have been possible with the IHC/FISH criteria. It is essential to observe the therapeutic response to the genotype-matched therapy in these equivocal cases, as well as to gather and observe similar cases to ascertain the validity of this new method.
All 19 non-breast tumors showed ERBB2 amplification (estimated CN above 3.0) but their HER2 IHC score varied, indicating that NGS-derived CN amplification and IHC scoring systems are not as concordant in non-breast cancers as in breast cancers. If we apply the breast or gastric IHC scoring system to our non-breast/non-gastric tumor samples, an ERBB2 estimated CN gain of 8 or more (estimated CN ≥ 8) would be equivalent to an IHC3 + score (90% sensitivity and 75% specificity). However, it is possible that establishing the cutoff point as high as 8 (instead of 3.2) would lead to the omission of patients who might have a favorable response to anti-HER2 treatment. Evaluating a large number of non-breast tumors for HER2 protein expression, including tumors without ERBB2 amplification, is necessary to examine the potential clinical use of NGS-based ERBB2 amplification status relative to the HER2 IHC score, as well as to determine whether a high estimated CN value threshold should be considered for non-breast cancers. Furthermore, given the behaviors of these marker in different tumor contexts, it is recommended that each tumor type should be analyzed separately.
Estimated CN gain detection is impacted by the proportion of tumor cells, with low or borderline amplification being most affected. Therefore, we estimated the true CN in NGS using the measured CN and the proportion of tumor cells. If the cutoff estimated CN = 3.2 is applied to the measured CN as a diagnostic tool, then these five patients would not be eligible for anti-HER therapies, and the sensitivity would fall to 80%. As the estimated CN takes the proportion of tumor cells into account, it is a better reflection of the actual tumor context. Accordingly, the estimated CN should be used for daily clinical reporting and evaluation of CN status. NGS is based on amplicon sequencing; therefore, exact CN estimation can be achieved with a relatively low sequencing depth. In the present study, we did not use matched normal blood samples, only tumor samples. As such, the estimated CN is not influenced by the germline CN. A previous study used a log2 fold change of 1.5 as the cutoff to detect ERBB2 amplification, achieving 95% sensitivity and 100% specificity [17]. However, they used peripheral blood as the normal control and estimated the CN by comparing the CN of the tumor sample with that of the peripheral blood, without compensating for the tumor content. Therefore, especially in cases with low tumor content, the estimated CN deviated greatly from the actual CN. Furthermore, if the germline CN is lost or amplified, the estimated CN also deviates from the actual CN. Comparably, our CN cutoff is not affected by these factors, and is less likely to result in costly errors in clinical decision making. Finally, NGS also overcomes the inter- and intra-observer variabilities.
The small size of biopsy specimens may lead to false-negative results. Thus, variability in the histological features of cancers may be associated with established molecular intratumoral heterogeneity and variations in treatment response [6, 7]. In some samples, there is only a small amount of the specimen available for molecular testing, which may not be representative of the whole tumor. In cases with discrepancies between IHC and NGS results, reflex testing on the same specimen by FISH would be recommended if there is sufficient material remaining, or an alternate tumor block. This information is useful for judging ERBB2 amplification to improve precision medicine.
There were two main limitations to the present study. First, for non-breast cancers, we only evaluated tumors with an estimated CN of 3.0 or higher. Therefore, we could not draw any conclusion or inference about the possible relationship between ERBB2 estimated CN and IHC score. Second, our study did not investigate the response to anti-HER2 treatment based on ERBB2 amplification using our criteria because of the short follow-up period between the NGS testing and the manuscript preparation. In future studies we plan to evaluate the treatment response in cases using our criteria for therapeutic recommendations. Turnaround time is an additional consideration when using NGS alone to determine ERBB2 estimated CN. Comprehensive NGS assays, which yield precise CN results, are not fast, with a turnaround time of at least 14 working days. While this is longer than that expected for IHC or FISH alone, the overall time can be comparable if assays are performed sequentially, accounting for the time required for collection and transportation of material.
In conclusion, the present study showed that NGS can provide accurate ERBB2 CN status, enabling simultaneous testing for other potentially actionable genomic alterations. Our ERBB2 estimated CN cutoff for therapeutic recommendation is an acceptable solution when both the ERBB2 status and comprehensive molecular profiling are required. We believe that our cutoff value for the ERBB2 estimated CN determined by NGS can be added to the breast cancer treatment guidelines regarding anti-HER2 therapies. Although estimated CN alone could potentially be used without IHC or FISH for cases with markedly high estimated CN, confirming the results with IHC or FISH would strengthen the finding, especially when the estimated CN is marginal (around 3). An NGS approach may also be applicable to various other types of cancer for which an IHC/FISH scoring system has not been established. This method will be especially useful when biological material is limited in amount, but screening of targetable gene aberrations is needed for alternative therapeutic options. Despite the rise in NGS technology use throughout diagnostic laboratories, inherent variations among platforms, assay design, and data analysis indicate the need to update HER2 guidelines by adding criteria for NGS-based evaluation of ERBB2 status.
Materials and methods
Patients
This retrospective study was approved by the ethics committee of the Keio University (approval number: 20080015) and was conducted in accordance with the Declaration of Helsinki and Title 45, U.S. Code of Federal Regulations, Part 46, Protection of Human Subjects, effective December 13, 2001. All patients provided written informed consent. We enrolled 90 patients with breast cancer with/without ERBB2 amplification, and 19 patients with other cancer types with ERBB2 amplification (estimated CN of more than 3.0) (Table 1), who underwent surgery at Keio University. Resected specimens were used for the NGS assay. For breast cancer specimens with low histological proportion of tumor cells, we used the specimens from the core needle biopsy performed before surgery.
NGS
Genomic testing was performed in-house using the PleSSision testing platform (Keio University Hospital, Tokyo, Japan). Briefly, genomic DNA was extracted from 10-µm-thick formalin-fixed paraffin-embedded (FFPE) tissue sections of tumor specimens using the Maxwell RSC FFPE Plus DNA Kit (Cat.AS1720, Promega, Madison, USA) according to the manufacturer’s instructions. DNA quality was checked by calculating the DNA integrity number (DIN), using an Agilent 4200 TapeStation (Agilent Technologies, Waldbronn, Germany); all analytes had DIN ≥ 2.0. Libraries were generated from 80 (DIN ≤ 2.5) or 160 (DIN > 2.5) ng of DNA per sample using the Human Comprehensive Cancer Panel, GeneRead DNAseq Panel PCR kit V2, GeneRead DNA Library I Core Kit, and GeneRead DNA Library I Amp Kit (Qiagen, Hilden, Germany) and the library quality was assessed using the Agilent D1000 ScreenTape (Agilent Technologies). Targeted amplicon exome sequencing was performed using a 160 cancer-related gene panel as previously described [18, 19]. The targeted regions of all 160 genes were specifically enriched using oligonucleotide probes. The enriched libraries were sequenced with a paired-end (150 bp × 2) sequencing method using the NextSeq sequencing platform (Illumina, San Diego, CA, USA), resulting in a mean depth of 500. The sequencing data were analyzed using the GenomeJack bioinformatics pipeline (Mitsubishi Space Software Co., Ltd., Tokyo, Japan) (http://genomejack.net/) as previously described [20]. The proportion of tumor cells ranged from 5 to 80% (median 45%). The estimated CN of the tumor cells was calculated by the following formula: estimated CN = (measured CN − 2)/proportion of tumor cells + 2.
IHC
IHC for HER2 was performed on 4-µm thick FFPE whole-tissue sections using the PATHWAY anti-HER-2/neu rabbit monoclonal antibody (clone 4B5, Ventana Medical Systems, Tokyo, Japan) on the Leica BOND-III (Leica Microsystems, Wetzlar, Germany). External positive controls were tested with each run. To determine the level of HER2 expression, the membrane staining pattern was estimated and scored on a scale of 0 to 3 + for breast15 and gastric16 cancers. An established breast cancer HER2 IHC scoring method [15] was applied to other solid cancers including lung, bladder, renal pelvic, cervical, endometrial, ovary, and prostate cancers, uterine sarcoma, and extra-mammary Paget’s disease; for gastric cancers and cancers with clear luminal structure (colorectal, gallbladder, and fistula cancers), the gastric cancer scoring system [16] was used. Scoring was performed independently by three pathologists. The IHC for the ER and PgR was performed using the CONFIRM rabbit monoclonal antibodies (clone SP1 and IE2, respectively). Tumors with ≥ 1% of cells showing positive nuclear staining for the expression of ER and PgR were evaluated as ER/PgR-positive. IHC for Ki67 was performed using a mouse monoclonal anti-human Ki67 antibody (MIB-1, Dako). The labeling index (LI) was assessed as the percentage of tumor cells showing definite nuclear staining among > 1000 invasive tumor cells.
FISH
FFPE tissue Sects. (5-µm thick) were deparaffinized and digested using standard processing methods for FISH. The PathVysion HER2 DNA Probe Kit (Abbott Molecular/Vysis) was used for the hybridization of tissue sections. Hybridization and counterstaining were performed according to the manufacturer’s instructions. Slides were imaged using an Applied Imaging system running Ariol SL200 (Leica Biosystems, Tokyo, Japan). At least 30 nuclei were evaluated, and the results were interpreted following the 2018 American Society of Clinical Oncology and the College of American Pathologists guidelines [15].
Statistical analysis
For comparisons of the estimated CN among the IHC score groups, data were analyzed using a Kruskal–Wallis test and Steel–Dwass multiple comparison test. ROC analysis was conducted to evaluate the diagnostic performance of estimated CN for IHC3 + or IHC2 + /FISH( +) outcomes. P values of < 0.05 were considered to indicate a significant difference. Statistical analysis was performed using SPSS software (version 19.0 for Windows; IBM Corp., Armonk, NY, USA) and R v3.6.1.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file4 Details of the clinical and pathological features of the patients with breast cancer (XLSX 18 KB)
Supplementary file5 Table S2: Details of the clinical and pathological features of the patients with non-breast cancer (XLSX 12 KB)
Acknowledgements
The authors would like to thank Emmy Yanagida and Hiroshi Yamada for providing technical assistance, Dr. Akihito Hagihara for his statistical expertise and advice, and Dr. A. Gordon Robertson for his helpful advice on data visualization and analysis.
Abbreviations
- DIN
DNA integrity number
- ER
Estrogen receptor
- ERBB2
Erb-b2 receptor tyrosine kinase 2
- FFPE
Formalin-fixed paraffin-embedded
- HER2
Human epidermal growth factor receptor 2
- IHC
Immunochemistry
- PgR
Progesterone receptor
- ROC
Receiver operating characteristic
- CN
Copy number
- NGS
Next-generation sequencing
Authors’ contributions
Conceptualization, H.N. and S.I.; methodology, J.O. and S.T.; investigation, E.A. and H.O.; resources, T.H., T.C., T.K., T.H., H.K., M.K., K.O., T.F.; data curation, S.T.; writing—original draft preparation, K.N.; writing—review and editing, J.O. and H.N.; supervision, H.H. and H.T.; funding acquisition, S.I. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the JSPS KAKENHI (Grant-in-Aid for Young Scientists B; grant 20K18232), and the Japan Agency for Medical Research and Development (AMED) under Grant Number JP20mk0102145 and 20ck0106448h0003.
Data Availability
The data that support the findings of this study are available from the corresponding author, [author initials], upon reasonable request.
Compliance with ethical standard
Conflicts of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Ethical approval
Approval number: 20180005 (Keio University).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Written consent was obtained from the patient for publication of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kohei Nakamura, Email: knakamura320@keio.jp.
Eriko Aimono, Email: eriko0123@keio.jp.
Junna Oba, Email: joba@keio.jp.
Hideyuki Hayashi, Email: rock-hayashi-pop@keio.jp.
Shigeki Tanishima, Email: tanishima.shigeki@mss.co.jp.
Tetsu Hayashida, Email: tetsu@keio.jp.
Tatsuyuki Chiyoda, Email: chiyoda@keio.jp.
Takeo Kosaka, Email: takemduro@gmail.com.
Tomoyuki Hishida, Email: tetsu@keio.jp.
Hirohumi Kawakubo, Email: hkawakubo@z3.keio.jp.
Minoru Kitago, Email: dragonpegasus427@gmail.com.
Koji Okabayashi, Email: okabayashikoji@gmail.com.
Takeru Funakoshi, Email: takeruf@a8.keio.jp.
Hajime Okita, Email: okita-h@keio.jp.
Sadakatsu Ikeda, Email: ikeda.canc@tmd.ac.jp.
Hiromasa Takaishi, Email: takaishi@keio.jp.
Hiroshi Nishihara, Email: hnishihara1971@keio.jp.
References
- 1.Popescu NC, King CR, Kraus MH. Localization of the human erbB-2 gene on normal and rearranged chromosomes 17 to bands q12–21.32. Genomics. 1989;4:362–366. doi: 10.1016/0888-7543(89)90343-1. [DOI] [PubMed] [Google Scholar]
- 2.Coussens L, Yang-Feng TL, Liao YC, Chen E, Gray A, McGrath J, Seeburg PH, Libermann TA, Schlessinger J, Francke U, et al. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science. 1985;230:1132–1139. doi: 10.1126/science.2999974. [DOI] [PubMed] [Google Scholar]
- 3.Kirouac, D.C.; Du, J.; Lahdenranta, J.; Onsum, M.D.; Nielsen, U.B.; Schoeberl, B.; McDonagh, C.F. HER2+ Cancer Dependence on PI3K vs. MAPK Signaling Axes Is Determined by Expression of EGFR, ERBB3 and CDKN1B. PLoS Comput Biol 2016, 12, e1004827. [DOI] [PMC free article] [PubMed]
- 4.Gutierrez C, Schiff R. HER2: biology, detection, and clinical implications. Arch Pathol Lab Med. 2011;135:55–62. doi: 10.5858/2010-0454-RAR.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 6.Bang YJ, Van C, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomized controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 7.Santin AD, Bellone S, Roman JJ, McKenney JK, Pecorelli S. Trastuzumab treatment in patients with advanced or recurrent endometrial carcinoma overexpressing HER2/neu. Int J Gynaecol Obstet. 2008;102:128–131. doi: 10.1016/j.ijgo.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Heinmoller P, Gross C, Beyser K, Schmidtgen C, Maass G, Pedrocchi M, Rüschoff J. HER2 status in non-small cell lung cancer: results from patient screening for enrollment to a phase II study of Herceptin. Clin Cancer Res. 2003;9:5238–5243. [PubMed] [Google Scholar]
- 9.Gatzemeier U, Growth G, Butts C. Randomized phase II trial of gemcitabine-cisplatin with or without trastuzumab in HER2-positive non-small-cell lung cancer. Ann Oncol. 2004;15:19–27. doi: 10.1093/annonc/mdh031. [DOI] [PubMed] [Google Scholar]
- 10.Serrano-Olvera A, Duenas-Gozalez A, Gallardo-Ricon D, Candelaria M. Prognostic, predictive and therapeutic implications of HER2 in invasive epithelial ovarian cancer. Cancer Treat Rev. 2006;32:180–190. doi: 10.1016/j.ctrv.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su P, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellegrini C, Falleni M, Marchetti A, Cassani B, Miozzo M, Buttitta F, Roncalli M, Coggi G, Bosari S. HER2/Neu alterations in non-small cell lung cancer: a comprehensive evaluation by real time reverse transcription-PCR, fluorescence in situ hybridization, and immunohistochemistry. Clin Cancer Res. 2003;9:3645–3652. [PubMed] [Google Scholar]
- 13.Grushko TA, Filiacy VL, Mundt AJ, Ridderstrale K, Olopade OI, Fleming GF. An exploratory analysis of HER-2 amplification and overexpression in advanced endometrial carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2008;108:3–9. doi: 10.1016/j.ygyno.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAlpine JN, Wiegand KC, Vang R, Ronnett BM, Adamiak A, Kobel M, Kalloger SE, Swenerton KD, Huntsman DG, Gilks CB, et al. HER2 overexpression and amplification is present in a subset of ovarian mucinous carcinomas and can be targeted with trastuzumab therapy. BMC Cancer. 2009;9:433. doi: 10.1186/1471-2407-9-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P, Hanna W, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol. 2018;36:2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann M, Stoss O, Shi D, Buttner R. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology. 2008;52:797–805. doi: 10.1111/j.1365-2559.2008.03028.x. [DOI] [PubMed] [Google Scholar]
- 17.Ross DS, Zehir A, Cheng DT, Benayed R, Nafa K, Hechtman JF, Janjigian YY, Weigelt B, Razavi P, Hyman DM, et al. Next-Generation Assessment of Human Epidermal Growth Factor Receptor 2 (ERBB2) Amplification Status: Clinical Validation in the Context of a Hybrid Capture-Based Comprehensive Solid Tumor Genomic Profiling Assay. J Mol Diagn. 2017;19:244–254. doi: 10.1016/j.jmoldx.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura K, Aimono E, Tanishima S, Imai M, Nagatsuma AK, Hayashi H, Yoshimura Y, Nakayama K, Kyo S, Nishihara H. Intratumoral Genomic Heterogeneity May Hinder Precision Medicine Strategies in Patients with Serous Ovarian Carcinoma. Diagnostics (Basel) 2020;10:E200. doi: 10.3390/diagnostics10040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura K, Aimono E, Tanishima S, Imai M, Nagatsuma AK, Hayashi H, Yoshimura Y, Nakayama K, Kyo S, Nishihara H. Olaparib monotherapy for BRIP1-mutated high-grade serous endometrial cancer. JCO Precis Oncol. 2020;4:283–290. doi: 10.1200/PO.19.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsumura K, Arai E, Tian Y, Shibuya A, Nishihara H, Yotani T, Yamada Y, Takahashi Y, Maeshima AM, Fujimoto H, et al. Establishment of permutation for cancer risk estimation in the urothelium based on genome-wide DNA methylation analysis. Carcinogenesis. 2019;40:1308–1319. doi: 10.1093/carcin/bgz112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file4 Details of the clinical and pathological features of the patients with breast cancer (XLSX 18 KB)
Supplementary file5 Table S2: Details of the clinical and pathological features of the patients with non-breast cancer (XLSX 12 KB)
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, [author initials], upon reasonable request.