Abstract
Purpose
Driver mutations are typically absent in esophageal adenocarcinoma (EAC). Mostly, oncogenes are amplified as driving molecular events (including GATA6-amplification in 14% of cases). However, only little is known about its biological function and clinical relevance.
Methods
We examined a large number of EAC (n = 496) for their GATA6 amplification by fluorescence in situ hybridization (FISH) analyzing both primary resected (n = 219) and neoadjuvant treated EAC (n = 277). Results were correlated to clinicopathological data and known mutations/amplifications in our EAC-cohort.
Results
GATA6 amplification was detectable in 49 (9.9%) EACs of our cohort. We observed an enrichment of GATA6-positive tumors among patients after neoadjuvant treatment (12,3% amplified tumors versus 6,8% in the primary resected group; p = 0.044). Additionally, there was a simultaneous amplification of PIK3CA and GATA6 (p < 0.001) not detectable when analyzing other genes such as EGFR, ERBB2, KRAS or MDM2. Although we did not identify a survival difference depending on GATA6 in the entire cohort (p = 0.212), GATA6 amplification was associated with prolonged overall survival among patients with primary surgery (median overall-survival 121.1 vs. 41.4 months, p = 0.032). Multivariate cox-regression analysis did not confirm GATA6 as an independent prognostic marker, neither in the entire cohort (p = 0.210), nor in the subgroup with (p = 0.655) or without pretreatment (p = 0.961).
Conclusions
Our study investigates the relevance of GATA6 amplification on a large tumor collective, which includes primary resected tumors and the clinically relevant group of neoadjuvant treated EACs. Especially in the pretreated group, we found an accumulation of GATA6-amplified tumors (12.3%) and a frequent co-amplification of PIK3CA. Our data suggest an increased resistance to radio-chemotherapy in GATA6-amplified tumors.
Keywords: GATA6, PIK3CA, Esophageal adenocarcinoma, EAC, Prognosis, Biomarker, Neoadjuvant therapy, Treatment response, Neoadjuvant treatment
Introduction
Even today, esophageal adenocarcinoma (EAC) is a devastating gastrointestinal malignancy with an overall five-years survival ranging from 15 to 20% (DeSantis et al. 2014; Rustgi and El-Serag 2014; Coleman et al. 2018) and still increasing incidences (Arnold et al. 2017). In the recent past, efforts focused on developing more effective multimodal treatment concepts including neoadjuvant chemoradiation or perioperative chemotherapy (Al-Batran et al. 2008; van Hagen et al. 2012). Therapeutic decisions are based on mere clinical parameters deriving from staging examinations and success of neoadjuvant therapy is evaluated depending on the degree of therapeutic response towards this treatment (Shapiro et al. 2015; Al-Batran et al. 2019). However, not all patients benefit from this still very standardized treatment routines, developing only significant toxic side effects. This is the case in 35% of patients undergoing chemoradiation and 39% of patients under chemotherapy (Ronellenfitsch et al. 2016; den Bakker et al. 2017). This clinical dilemma is due to the fact that EAC is a genetic extremely heterogenous disease. Its mutational burden is enormous (Mourikis et al. 2019; von Loga et al. 2020) and EAC is often associated with a high chromosomal instability (Frankell et al. 2019). Whole exome sequencing revealed TP53, CDKN2A, SMAD4, ARID1A, VEGFA, CCNE1 and PIK3CA to be among those genes most frequently affected (Dulak et al. 2013; Cancer Genome Atlas Research Network et al. 2017). Another common phenomenon is the principle of genetical amplifications [also known as copy number alterations (CNAs)]. According to recent data analyzing the genetic landscape of 551 EACs these amplifications mostly occur in KRAS (19%), c-MYC (19%), HER2 (18%), CCND1 (14%) and GATA6 (14%) (Frankell et al. 2019). However, only little is known about the function of GATA6 amplification within this entity.
GATA binding protein 6 (GATA6) belongs to the GATA family comprising of the members GATA1-6 and its gene is located on chromosome 18 (q11.1 ~ q11.2) within the human genome (Suzuki et al. 1996). During embryogenesis, GATA6 is highly expressed within the endoderm and mesoderm (Carrasco et al. 2012) as it is essential for the development of different tissues such as adrenal gland and the central nervous system (Jimenez et al. 2003; Kamnasaran and Guha 2005). Being a transcriptional factor, dysregulation of GATA6 can also result in pathological changes and it was demonstrated that GATA6 alterations implicated in several malignancies such as non-small lung cancer (NSCLC), gastric cancer, cholangiocarcinoma, pancreatic adenocarcinoma or colorectal adenocarcinoma (Zhong et al. 2011; Shen et al. 2013; Tian et al. 2013; Van Baal et al. 2013; Ma et al. 2019). For esophageal adenocarcinoma, it has been shown in a study including 85 EACs that gene amplification of GATA6 affected the patients’ survival in a negative manner (Lin et al. 2012). During the development of Barrett’s esophagus and the following malignant transformation, the expression of GATA6 is successively increasing resembling its impact on the progression of the disease (Pavlov et al. 2015).
Aim of the current study was to analyze the relevance and frequency of GATA6 amplification in a large cohort of EAC patients and the consecutively correlation with clinical, pathological and molecular parameters as well as the patients’ survival.
Materials and methods
Patients and tumor samples
Analysis was performed on 496 patients with esophageal adenocarcinoma who either underwent primary surgical resection or resection after neoadjuvant treatment between 1999 and 2017 at the Department of General, Visceral, Cancer and Transplant Surgery, University of Cologne, Germany. All patients underwent primary staging including contrast-enhanced computed tomography, esophagoduodenoscopy, endoscopic ultrasound and physical examination. Patients who qualified for multimodal treatment because of locally advanced tumors (cT > 2) or suspected locoregional lymph node metastases (cN +) received neoadjuvant chemoradiation (van Hagen et al. 2012) or chemotherapy (Donohoe and Reynolds 2017). The standardized surgical procedure was transthoracic en-bloc esophagectomy with two-field lymphadenectomy of the abdominal and mediastinal lymph nodes, reconstruction via gastric pull-up and intrathoracic anastomosis (Ivor-Lewis esophagectomy). The abdominal part was predominantly performed via laparoscopy while thoracotomy was open surgery (hybrid esophagectomy). For more technical details we refer to previous publications (Plum et al. 2018) and other authors (Mariette et al. 2019). Informed consent and ethical approval were obtained from all participating patients. This retrospective study was performed according to the criteria of the ethics committee of the University Hospital of Cologne (No. 13–091 and 10–242) and in accordance with the relevant version of the Helsinki Declaration. Clinical data was collected prospectively within the department according to a standardized protocol. During the first two years, clinical follow-up of patients was performed in the out-patient clinics every three months, followed by annual exams. These included clinical evaluation, abdominal ultrasound, chest X-ray and additional diagnostic procedures as required.
Single-spot tissue microarrays (TMA) were constructed from all surgical specimens for fluorescence in-situ hybridization (FISH) and immunohistochemical analysis. The exact procedure has been described before (Simon et al. 2005; Helbig et al. 2016). In principle, tissue cylinders with a diameter of 1.2 mm each were punched from the selected tumor tissue blocks (donor blocks) via a self-constructed semi-automated precision instrument and embedded on an empty paraffin block (recipient block). Four µm sections of the resulting TMA blocks were transferred to an adhesive coated slide system (Instrumedics Inc., Hackensack, NJ, USA) for following FISH or immunohistochemistry. Amplification of GATA6 (via FISH) was correlated with molecular profiles of these EAC samples including assessments of ARIDA 1A loss, TP53 mutations as well as ERBB2, c-MYC, KRAS and PIK3CA amplifications.
Fluorescence in-situ hybridization (FISH) of GATA6
Fluorescence in-situ hybridization (FISH) analysis for the evaluation of GATA6 gene copy numbers was performed with GATA6-20-GR Probe (Empire Genomics, New York, NY, USA) and the Zytolight centromere 18 (CEN18) Probe (Zytovision Bremerhaven, Germany) on the resulting TMA slides. For PIK3CA gene amplification analysis, the Zytolight SPEC PIK3CA/CEN3 Dual Probe Kit (Zytovision, Germany) was used according to the manufacturers' protocol. Three µm tissue sections on slides (SuperFrost Plus) were mounted by heating, followed by deparaffinization, protease digestion, washing steps (VP2000 processor system, Abbott Molecular, Wiesbaden, Germany) and hybridization at 37 °C overnight with the FISH Probe. The slides were stained with DAPI before analysis. Cases were further evaluated only when normal tissue nuclei displayed one or two clearly distinct signals of green GATA6 and orange CEN18. Tumor tissue was scanned for amplification hot spots of GATA6 signals using × 63 objective (DM5500 fluorescent microscope; Leica). This reading strategy followed that of the c-MYC-FISH probe to evaluate areas of cluster amplification. GATA6 amplification was defined as gene copy cluster > 50% of the tumor cells, respectively, gene copy number > 6 per cell. For PIK3CA reading strategy followed the recommendations of previous studies amplification such as (Essakly et al. 2020).
Immunohistochemistry
Immunohistochemistry (IHC) was performed on TMA slides using the following antibodies against MHC1, PDL1, LAG3, IDO, INI, VISTA, TP53, TIM3, TUBB3, HER2, Ki67, ARIDA 1A, BRG1, BRM, Met1 and c-MYC as already published by our group (Becker et al. 2015; Loeser et al. 2019; Plum et al. 2019; Essakly et al. 2020; Gebauer et al. 2020; Wagener-Ryczek et al. 2020; Schiffmann et al. 2020).
Statistical analysis
SPSS Statistics for Mac (Version 21, SPSS) was used for statistical analysis. Interdependence between stainings and clinical data were calculated using the chi-squared and Fisher’s exact tests, and displayed by cross-tables. Survival curves were plotted using the Kaplan–Meier method and analyzed using the log-rank test. All tests were two-sided. p values < 0.05 were considered statistically significant.
Results
Patients’ baseline characteristics
A total of 496 patients of 685 on the TMA with EAC were interpretable on the single-spot for GATA6. Reasons for non-informative cases (189 spots; 27.6%) included lack of tissue samples or absence of unequivocal cancer tissue in the TMA spot. Clinico-pathological data were summarized within Table 1. The majority of patients were male (male: n = 437; 88.1% versus female: n = 59; 11.9%). The median age was 65.2 years (range 33.6–85.6 years) at the time point of diagnosis. More than half of the patient cohort (n = 277; 55.8%) underwent multimodal treatment (including either chemoradiation or chemotherapy before surgical resection) while 219 (44.2%) patients received primary surgery.
Table 1.
Clinico-pathological parameters for the patient cohort
| Factor | Total | GATA6 | p value | ||||
|---|---|---|---|---|---|---|---|
| Negative | Positive | ||||||
| Sex | |||||||
| Female | 59 | 11.9% | 53 | 89.8% | 6 | 10.2% | |
| Male | 437 | 88.1% | 394 | 90.2% | 43 | 9.8% | 0.937 |
| Agegroup | |||||||
| < 65 yrs | 245 | 52,5% | 216 | 88.2% | 29 | 11.8% | |
| > 65 yrs | 222 | 47.5% | 202 | 91.0% | 20 | 9.0% | 0.319 |
| Tumor stage | |||||||
| pT1/2 | 125 | 25.4% | 112 | 89.6% | 13 | 10.4% | |
| pT3/4 | 368 | 74.6% | 332 | 90.2% | 36 | 9.8% | 0.842 |
| Lymph node metastasis | |||||||
| pN0 | 198 | 40.1% | 178 | 89.9% | 20 | 4.0% | |
| pN + | 296 | 59.9% | 267 | 90.2% | 29 | 9.8% | 0.912 |
| Grading | |||||||
| G1 | 5 | 1.4% | 5 | 1.4% | 0 | 0% | |
| G2 | 197 | 55.5% | 178 | 90.4% | 19 | 9.6% | |
| G3 | 151 | 42.5% | 137 | 90.7% | 14 | 9.3% | |
| G4 | 2 | 0.6% | 2 | 0.6% | 0 | 0% | 0.746 |
| UICC | |||||||
| I | 108 | 22.0% | 97 | 89.8% | 11 | 10.2% | |
| II | 106 | 21.5% | 96 | 90.6% | 10 | 9.4% | |
| III | 208 | 42.3% | 184 | 88.5% | 24 | 11.5% | |
| IV | 70 | 14.2% | 66 | 94.3% | 4 | 5.7% | 0.567 |
| Neoadjuvant therapy | |||||||
| No | 219 | 44.2% | 204 | 93.2% | 15 | 6.8% | |
| Yes | 277 | 55.8% | 243 | 87.7% | 34 | 12.3% | 0.044 |
GATA6 amplification in esophageal adenocarcinoma and correlation to clinico-pathological data
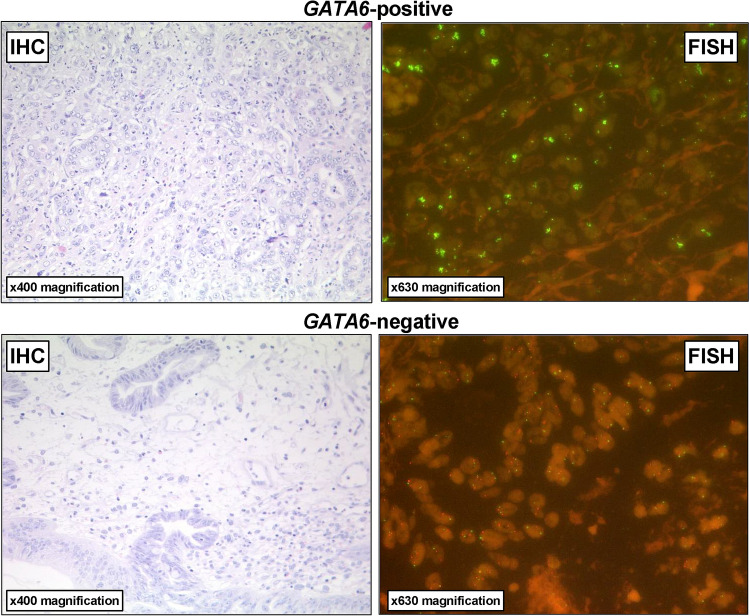
Considering the entire patient cohort, GATA6 amplification was detectable via FISH in 49 patients (9.9%) within an intranuclear pattern (compare Fig. 1). There was no significant correlation between such clinico-pathological parameters such as sex, age, grading, (y)pT-category, (y)pN-category or UICC-stage (see Table 1). However, GATA6 amplification was correlated with the status of neoadjuvant treatment (p = 0.044). Patients who had multimodal therapy showed in 12.3% an amplification in the FISH examination compared to 6.8% among those patients who had primary esophagectomy.
Fig. 1.
Representative images of immunohistochemistry (IHC) and fluorescence in-situ hybridization (FISH) analysis for the evaluation of GATA6 gene copy numbers using the GATA6-20-GR (green) and the Zytolight centromere 18 (CEN18) (red) Probe illustrating (upper row) GATA6-positive versus (lower row) GATA6-negative esophageal adenocarcinoma. GATA6 amplification was defined as gene copy cluster > 50% of the tumor cells, respectively, gene copy number > 6 per cell
GATA6 and PIK3CA co-amplification
FISH-data of GATA6 amplification was additionally correlated with other important biomarkers in EAC like other amplified oncogenes, immune checkpoint markers such as PD-L1, LAG3, IDO, INI, VISTA or the antigen-presenting protein MHC1, as well as additional proteins like the chromatin-remodeler and SWI/SNF components ARIDA 1A, BRG1, BRM and oncogene amplifications like MET, c-MYC, KRAS, ERBB2, MDM2 and PIK3CA. We observed no correlation between GATA6 and most of these other biomarkers within the cohort performing the cross-table analysis (see Table 2). However, we identified co-amplification of GATA6 together with PIK3CA in 9 (1.8%) patients of the entire cohort (p < 0.001) divided into 2 (0.3%) patients of the pretreated subgroup (p < 0.001) and 7 (1.4%) patients with primary surgery (p = 0.174). PIK3CA amplifications were seen in 24 patients (4.8%) (Essakly et al. 2020). Similar amplification rates were seen within the primary surgery group (n = 11; 5.0%) and surgery after neoadjuvant treatment (n = 13; 4.7%). All details are illustrated in Table 3.
Table 2.
Correlation between GATA6 and other molecular markers within the patient cohort
| Factor | Total | GATA6 | p value | ||||
|---|---|---|---|---|---|---|---|
| Negative | Positive | ||||||
| HER2 | |||||||
| Normal | 300 | 87.7% | 268 | 89.3% | 32 | 10.7% | |
| Mutated | 42 | 12.3% | 41 | 97.6% | 1 | 2.4% | 0.089 |
| MHC1 | |||||||
| Loss | 106 | 29.4% | 101 | 95.3% | 5 | 4.7% | |
| Normal | 254 | 70.6% | 227 | 89.4% | 27 | 10.6% | 0.072 |
| ARIDA 1A | |||||||
| Loss | 45 | 9.5% | 44 | 97.8% | 1 | 2.2% | |
| Normal | 427 | 90.5% | 381 | 89.2% | 46 | 10.8% | 0.068 |
| C-myc | |||||||
| Normal | 418 | 87.8% | 379 | 90.7% | 39 | 9.3% | |
| Amplified | 58 | 12.2% | 48 | 82.8% | 10 | 17.2% | 0.063 |
| KRAS | |||||||
| Normal | 402 | 82.4% | 366 | 91.0% | 36 | 9.0% | |
| Mutated | 86 | 17.6% | 73 | 84.9% | 13 | 15.1% | 0.084 |
| PIK3CA | |||||||
| Normal | 415 | 94.5% | 379 | 91.3% | 36 | 8.7% | |
| Mutated | 24 | 5.5% | 15 | 62.5% | 9 | 37.5% | < 0.001 |
Table 3.
Correlation between GATA6 and PIK3CA within the patient cohort
| Factor | Total | GATA6 | p value | |||||
|---|---|---|---|---|---|---|---|---|
| Negative | Positive | |||||||
| Entire cohort | ||||||||
| PIK3CA | Normal | 415 | 94.5% | 379 | 91.3% | 36 | 8.7% | |
| Amplified | 24 | 5.5% | 15 | 62.5% | 9 | 37.5% | < 0.001 | |
| Patients without neoadjuvant treatment | ||||||||
| PIK3CA | Normal | 186 | 94.4% | 173 | 93.0% | 13 | 7.0% | |
| Amplified | 11 | 5.6% | 9 | 81.8% | 2 | 18.2% | 0.174 | |
| Patients with neaodjuvant treatment | ||||||||
| PIK3CA | Normal | 229 | 94.6% | 206 | 90.0% | 23 | 10.0% | |
| Amplified | 13 | 5.4% | 6 | 46.2% | 7 | 53.8% | < 0.001 | |
GATA6 amplification is associated with a prolonged survival among patients who did not receive neoadjuvant treatment
Considering the entire patient cohort of the present study, a significant difference between patients with and without GATA6 amplification could not be observed (median survival without GATA6 amplification: 26.1 months (95% CI 20.4–31.7 months) versus median survival with GATA6 amplification: 37.2 months (95% CI 29.3–45.1 months, p = 0.212) (Fig. 2a). The same was true for patients receiving neoadjuvant treatment. In this subgroup, postsurgical survival was comparable between patients with and those without GATA6 amplification (median survival without GATA6 amplification: 22.3 months (95% CI 18.2–26.4 months) versus median survival with GATA6 amplification: 31.9 months (95% CI 28.2–35.6 months, p = 0.699) (Fig. 2b). However, in patients without neoadjuvant therapy, intratumoral GATA6 amplification was associated with a prolonged overall survival (OS) compared to those tumors without this amplification (Fig. 2c) (p = 0.032). The median OS was 121.1 months (95% CI not calculable) in patients with GATA6-amplified tumors in contrast to a median OS of 41.4 months (95% CI 23.4–59.4 months, p = 0.032) in patients with normal GATA6 expression.
Fig. 2.
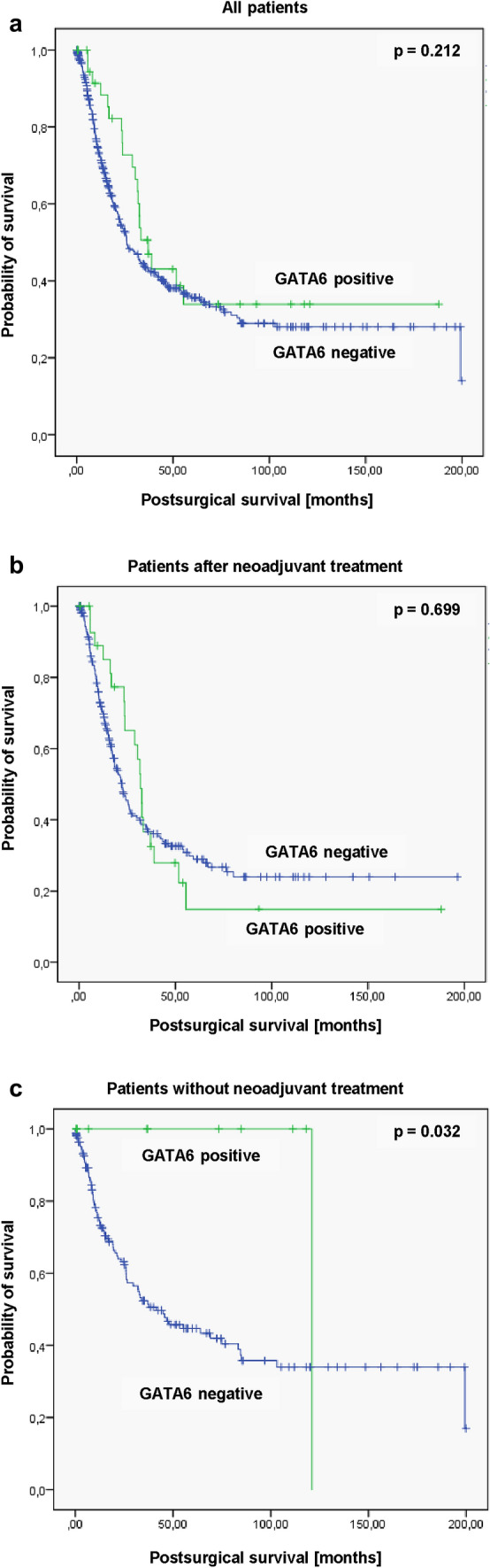
Kaplan–Meier survival analysis (log-rank test) considering the median survival depending on the GATA6 status of the patients. No significant GATA6-depending survival differences were observed within a the entire cohort (p = 0.212) as well as b those patients after neoadjuvant treatment (p = 0.699) while the subgroup of GATA6-positive patients without neoadjuvant therapy c showed a significant better postsurgical survival (p = 0.032)
Multivariate cox-regression analysis did not confirm GATA6 as an independent prognostic marker, neither in the entire cohort (p = 0.210), nor in the subgroup with (p = 0.655) or without neoadjuvant treatment (p = 0.961) (compare Table 4 for more details).
Table 4.
Multivariate cox-regression analysis for all patients and for those with/without neoadjuvant treatment
| Factor | All patients | Patients with neoadjuvant treatment | Patients without neoadjuvant treatment | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% confidence interval | p value | Hazard ratio | 95% confidence interval | p value | Hazard ratio | 95% confidence interval | p value | ||||
| Lower | Upper | Lower | Upper | Lower | Upper | |||||||
| Sex: male versus female | 1.181 | 0.727 | 1.918 | 0.501 | 1.558 | 0.836 | 2.902 | 0.162 | 0.481 | 0.216 | 1.072 | 0.074 |
| Age groups: < 65yrs versus > 65 years | 1.273 | 0.980 | 1.655 | 0.071 | 1.170 | 0.838 | 1.632 | 0.357 | 1.662 | 1.038 | 2.662 | 0.034 |
| Tumor stage:pT1/2 versus pT3/4 | 1.434 | 0.992 | 2.073 | 0.055 | 0.874 | 0.550 | 1.391 | 0.571 | 2.629 | 1.403 | 4.924 | 0.03 |
| Lymph node metastasis: pN0 versus pN + | 2.863 | 2.106 | 3.981 | 0.001 | 2.197 | 1.504 | 3.210 | 0.001 | 4.294 | 2.509 | 7.351 | 0.001 |
| GATA6:negative versus positive | 0.745 | 0.470 | 1.181 | 0.210 | 0.897 | 0.558 | 1.443 | 0.655 | 0.001 | 0.000 | 10.000 | 0.961 |
Discussion
In the current study, we focused on the frequency and clinical relevance of GATA6 amplification within a large EAC (n = 496) cohort by performing FISH-analysis. We identified gene amplification of GATA6 in up to 12,6% of patients. However, it had no correlation to clinico-pathological parameters such as sex, age, grading, pT-category, pN-category or UICC-stage. Interestingly, there was a positive correlation between the amplification of GATA6 and multimodal treatment since patients after neoadjuvant therapy more frequently showed corresponding amplification compared to patients who primarily underwent surgical resection (p = 0.044). Additionally, distinct subgroup analysis revealed that an influence of GATA6 on the patients’ survival was present depending on a multimodal treatment concept. GATA6 amplification had no effect on the OS in those patients who received neoadjuvant treatment while in patients without neoadjuvant procedures, GATA6-positive patients had a significantly prolonged OS. Correlated with other molecular alterations/amplifications common for EAC, we observed a co-amplification of GATA6 and PIK3CA in about 1.8% of patients. This effect was detectable in both subgroups with and without neoadjuvant treatment.
Our current results considering the frequency of amplified GATA6 is consistent with previous publications by recent large genetic studies (14%) (n = 551) (Frankell et al. 2019) or the TCGA-database (12%) (n = 185) (compare http://cancergenome.nih.gov/) focusing on this malignancy. Both studies analyze primarily operated tumors (without chemoradiation) and conclude on gene amplification using a next-generation sequencing technique. Using the fluorescence in-situ technique (FISH; gold standard for determining gene amplification) we have the possibility of a direct and reliable visualization of gene copy alterations in tumor cells. In primarily operated tumors we can detect only half of GATA6-amplified EACs (6.8%). In our cohort there is an accumulation of GATA6 amplified tumors in the group of neoadjuvant treated tumors, which has not been considered in all studies so far. However, the vast majority of EACs are now treated neoadjuvantly. Therefore, our results may suggest that GATA6-amplified tumors induce an increased resistance to either radiotherapy or chemotherapy.
One study described a much higher frequency of amplification in 20.5% of patients. However, only 85 tumors were included in this work and amplification was observed by performing an array-based comparative genomic hybridization on 20 EACs and further validation via SNP-array analysis and quantitative real-time PCR (qRT-PCR) within the rest of the cohort (Lin et al. 2012). Contrary to this, we performed FISH-analysis which resembles the current gold standard for detection of gene copy number alterations within the daily pathological routine diagnostics.
Although GATA6 amplification is recurrent in EAC, little is known about the molecular mechanisms this transcriptional factor regulates. GATA6 amplification increases during the progression from normal esophageal squamous epithelia to Barrett’s metaplasia and finally to the invasive EAC (Pavlov et al. 2015). It was experimentally validated by Van Baal et al. that BMP4, a key protein within the development of Barrett’s esophagus (BE) which induces SOX9 mRNA expression and which promotor is activated by GATA6, is negatively regulated via microRNA (miR)-145 (Van Baal et al. 2013). Overexpression of miR-145 in HET-1A (an esophageal squamous cell line) and BAR-T cells (a non-neoplastic Barrett’s esophagus cell line) resulted in an inhibition of GATA6, BMP4 and SOX9 expression and in a reduced proliferation rate. This suggested that miRNA-145 might indirectly target BMP4 via GATA6 and impact the development of BE (Van Baal et al. 2013). Another in vitro study by Lin et al. demonstrated that ectopic expression of GATA6 increased anchorage-independent growth in immortalized Barrett’s esophageal cells (Lin et al. 2012). Contrary to this, GATA6 deprivation induced apoptotic (TNF-associated) pathways in EAC cells (Lin et al. 2012). Own previous data could reveal a possible connection between Dickkopf-2 (DKK2) and GATA6 in EAC (Schiffmann et al. 2020). Nevertheless, it remained unclear how these molecules interact on the molecular level. In pancreatic adenocarcinoma, GATA6 directly binds to the DKK2-promotor leading to a down-regulation of its expression and, therefore, reduces its suppressive effect on the oncogenic Wnt pathways (Zhong et al. 2011). Interestingly, a large genome-wide association study (GWAS) on EAC performed by the German Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON) including about 1065 EAC cases and 1019 controls identified variants of GATA6 to be strongly associated with the disease reflecting its central role within the tumor development (Becker et al. 2015).
The reasons for the higher frequency of GATA6 amplification among patients with neoadjuvant therapy in the current analysis are unsolved. It would be interesting to assess putative changes in the GATA6 amplification rate under therapeutic pressure. To identify dynamic alterations, prospective sample collection of initial treatment-naïve biopsies during the time point of staging followed by consecutive samples from surgical specimens of patients after neoadjuvant therapy would be necessary.
To our best knowledge, this is the first study describing a simultaneous amplification of GATA6 and PIK3CA in EAC. Confirming own previous studies (Schallenberg et al. 2020), no significant co-amplifications with other common CNAs in EAC occurred. We observed amplification of PIK3CA in 4.8% of the entire cohort with no differences between patients with or without neoadjuvant treatment as already published by our group (Essakly et al. 2020). However, 1.8% of all patients showed an amplification of both GATA6 and PIK3CA. Chromotrypsis is a recognized oncogenic mechanism of development in EAC. By this route, a synergistic co-amplification of PIK3CA and GATA6 is well conceivable (Nones et al. 2014).
In the present study, we observed a positive prognostic relevance within the subgroup of patients who did not receive neoadjuvant treatment before surgery while the prognosis of the entire cohort was not affected by GATA6 amplification. After all, the prognosis of EAC patients is still impaired and according to our results upregulation of GATA6 does not affect this in any manner. On the first sight, this seems contradictory as GATA6 has been reported to decrease the patients survival in different malignancies (Zhong et al. 2011; Shen et al. 2013, 2019; Tian et al. 2013; Rao et al. 2019). But at second glance the results for EAC are controversial. Some studies with relatively small cohorts of patients (n = 73, respectively, n = 58) reported poor prognosis in patients with GATA6 amplifications (Lin et al. 2012; Toxopeus et al. 2019) while another analysis including two separated cohorts (first cohort: 130 tissue samples of normal squamous epithelium, metaplasia, dysplasia, and esophageal adenocarcinoma; second cohort: 92 esophageal adenocarcinoma) demonstrated no association between GATA6 and overall or disease-free survival in this entity (Pavlov et al. 2015). After all, our own study based on a much larger cohort size utilizing FISH as the gold standard for the detection of copy number alteration in the current pathological routine work-flow. Additionally, there are reports from gastric cancer that suggested multiple roles of GATA6 within carcinogenesis. Recently a novel suppressive function of GATA6 has been described within gastric adenocarcinoma revealing that patients with metastatic tumors had low GATA6 expression with a negative impact on the patients’ survival (Liu et al. 2019). The authors illustrated that GATA6 directly targets the expression of miR-520b and that this microRNA again reduced its functional target cAMP-responsive element binding protein 1 (CREB1) leading to a suppressed cell migration, invasion and metastasis both in vitro and in vivo (Liu et al. 2019). Whether these mechanisms are also responsible for the prolonged survival within our study and why this is selectively within those patients with primary surgery remains unclear and needs further investigations.
In summary, our study identified GATA6 amplification to be significantly associated with multimodal treatment concepts in EAC and to be of prognostic impact for at least those patients with primary surgery. This might indicate an increased resistance to radio-chemotherapy in GATA6-amplified tumors. For the first time, simultaneous co-amplification of GATA6 and PIK3CA has been observed within this malignancy. Despite our large cohort, the resulting subgroups for further analysis are quite small (amongst others due to the low frequency of GATA6 amplification). Consequently, large prospective studies are essential for further validation. Finally, mechanistic approaches for further investigation of the biological functions/interactions related to GATA6 amplification in EAC via in-vitro, respectively, in vivo experiments should gain more knowledge about how this molecular alteration might be a target for future treatment concepts.
Author contributions
PSP, HL, FG, and AQ conceived and designed the study; PSP, TZ, HA, WS, and FG enrolled the patients and collected the clinical data while HL, AE, and AQ performed the pathological analysis; PSP, and FG carried out the statistical analyses; PSP, HL, TZ,CJB, AMH, RB, FG, and AQ contributed to the interpretation of data. PSP, FG, and AQ drafted the manuscript; All authors were involved in critically revising the manuscript for important intellectual content, and approving of the submitted version.
Funding
Open Access funding enabled and organized by Projekt DEAL. The current study was not funded by any organization.
Data availability
The datasets generated and/or analyzed during this current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
Patrick Sven Plum is fellow of the Else Kröner Forschungskolleg Cologne “Clonal Evolution in Cancer” (2016-Kolleg-19). All other authors declare no conflict of interest.
Ethics approval
This retrospective study was performed according to the criteria of the ethics committee of the University Hospital of Cologne (No. 13–091 and 10–242) and in accordance with the relevant version of the Helsinki Declaration.
Consent to participate/publication
All patients declared their participation and written consent was obtained before participation in the study (No. 13–091 and 10–242). The objective of the project was primarily in the field of diagnostics and quality assurance.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Al-Batran S-E, Hartmann JT, Hofheinz R, et al. Biweekly fluorouracil, leucovorin, oxaliplatin, and docetaxel (FLOT) for patients with metastatic adenocarcinoma of the stomach or esophagogastric junction: a phase II trial of the Arbeitsgemeinschaft Internistische Onkologie. Ann Oncol Off J Eur Soc Med Oncol. 2008;19:1882–1887. doi: 10.1093/annonc/mdn403. [DOI] [PubMed] [Google Scholar]
- Al-Batran SE, Homann N, Pauligk C, et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a ra. Lancet. 2019;393:1948–1957. doi: 10.1016/S0140-6736(18)32557-1. [DOI] [PubMed] [Google Scholar]
- Arnold M, Laversanne M, Brown LM, et al. Predicting the future burden of esophageal cancer by histological subtype: international trends in incidence up to 2030. Am J Gastroenterol. 2017;112:1247–1255. doi: 10.1038/ajg.2017.155. [DOI] [PubMed] [Google Scholar]
- Becker J, May A, Gerges C, et al. Supportive evidence for FOXP1, BARX1, and FOXF1 as genetic risk loci for the development of esophageal adenocarcinoma. Cancer Med. 2015;4:1700–1704. doi: 10.1002/cam4.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer genome Atlas research network, analysis working group: asan university, BC cancer agency et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco M, Delgado I, Soria B, et al. GATA4 and GATA6 control mouse pancreas organogenesis. J Clin Invest. 2012;122:3504–3515. doi: 10.1172/JCI63240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HG, Xie SH, Lagergren J. The epidemiology of esophageal adenocarcinoma. Gastroenterology. 2018;154:390–405. doi: 10.1053/j.gastro.2017.07.046. [DOI] [PubMed] [Google Scholar]
- den Bakker CM, Smit JK, Bruynzeel AME, et al. Non responders to neoadjuvant chemoradiation for esophageal cancer: Why better prediction is necessary. J Thorac Dis. 2017;9:S843–S850. doi: 10.21037/jtd.2017.06.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- Donohoe CL, Reynolds JV. Neoadjuvant treatment of locally advanced esophageal and junctional cancer: the evidence-base, current key questions and clinical trials. J Thorac Dis. 2017;9:S697–S704. doi: 10.21037/jtd.2017.03.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essakly A, Loeser H, Kraemer M, et al. PIK3CA and KRAS amplification in esophageal adenocarcinoma and their impact on the inflammatory tumor microenvironment and prognosis. Transl Oncol. 2020;13:157–164. doi: 10.1016/j.tranon.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankell AM, Jammula SG, Li X, et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat Genet. 2019;51:506–516. doi: 10.1038/s41588-018-0331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Krämer M, Bruns C, et al. Lymphocyte activation gene-3 (LAG3) mRNA and protein expression on tumour infiltrating lymphocytes (TILs) in oesophageal adenocarcinoma. J Cancer Res Clin Oncol. 2020;146:2319–2327. doi: 10.1007/s00432-020-03295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig D, Ihle MA, Pütz K, et al. Oncogene and therapeutic target analyses in Atypical fibroxanthomas and pleomorphic dermal sarcomas. Oncotarget. 2016;7:21763–21774. doi: 10.18632/oncotarget.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez P, Saner K, Mayhew B, Rainey WE. GATA-6 is expressed in the human adrenal and regulates transcription of genes required for adrenal androgen biosynthesis. Endocrinology. 2003;144:4285–4288. doi: 10.1210/en.2003-0472. [DOI] [PubMed] [Google Scholar]
- Kamnasaran D, Guha A. Expression of GATA6 in the human and mouse central nervous system. Dev Brain Res. 2005;160:90–95. doi: 10.1016/j.devbrainres.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Lin BAJ, Lockwood WW, et al. Activation of GATA binding protein 6 (GATA6) sustains oncogenic lineage-survival in esophageal adenocarcinoma. Proc Natl Acad Sci USA. 2012;109:4251–4256. doi: 10.1073/pnas.1011989109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Du F, Sun L, et al. GATA6 suppresses migration and metastasis by regulating the miR-520b/CREB1 axis in gastric cancer. Cell Death Dis. 2019;10:35. doi: 10.1038/s41419-018-1270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeser H, Kraemer M, Gebauer F, et al. The expression of the immune checkpoint regulator VISTA correlates with improved overall survival in pT1/2 tumor stages in esophageal adenocarcinoma. Oncoimmunology. 2019;8:1–8. doi: 10.1080/2162402X.2019.1581546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R, Li X, Liu H, et al. GATA6-upregulating autophagy promotes TKI resistance in nonsmall cell lung cancer. Cancer Biol Ther. 2019;20:1206–1212. doi: 10.1080/15384047.2019.1599665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariette C, Markar SR, Dabakuyo-Yonli TS, et al. Hybrid minimally invasive esophagectomy for esophageal cancer. N Engl J Med. 2019;380:152–162. doi: 10.1056/nejmoa1805101. [DOI] [PubMed] [Google Scholar]
- Mourikis TP, Benedetti L, Foxall E, et al. Patient-specific cancer genes contribute to recurrently perturbed pathways and establish therapeutic vulnerabilities in esophageal adenocarcinoma. Nat Commun. 2019;10:3101. doi: 10.1038/s41467-019-10898-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nones K, Waddell N, Wayte N, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun. 2014;5:5224. doi: 10.1038/ncomms6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov K, Honing J, Meijer C, et al. GATA6 expression in Barrett’s oesophagus and oesophageal adenocarcinoma. Dig Liver Dis. 2015;47:73–80. doi: 10.1016/j.dld.2014.09.014. [DOI] [PubMed] [Google Scholar]
- Plum PS, Hölscher AH, Pacheco Godoy K, et al. Prognosis of patients with superficial T1 esophageal cancer who underwent endoscopic resection before esophagectomy—A propensity score-matched comparison. Surg Endosc. 2018;32:3972–3980. doi: 10.1007/s00464-018-6139-7. [DOI] [PubMed] [Google Scholar]
- Plum PS, Gebauer F, Krämer M, et al. HER2/neu (ERBB2) expression and gene amplification correlates with better survival in esophageal adenocarcinoma. BMC Cancer. 2019;19:38. doi: 10.1186/s12885-018-5242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Wan L, Jie Z, et al. Upregulated miR-27a-3p indicates a poor prognosis in pancreatic carcinoma patients and promotes the angiogenesis and migration by epigenetic silencing of GATA6 and activating VEGFA/VEGFR2 signaling pathway. Onco Targets Ther. 2019;12:11241–11254. doi: 10.2147/OTT.S220621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ronellenfitsch U, Liodaki E, Trunk MJ, et al. Association between tumor response and postoperative morbidity after neoadjuvant chemotherapy for gastroesophageal adenocarcinoma? J Unexplored Med Data. 2016;1:6–14. doi: 10.20517/2572-8180.2016.01. [DOI] [Google Scholar]
- Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–2509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- Schallenberg S, Bork J, Essakly A, et al. Loss of the SWI/SNF-ATPase subunit members SMARCF1 (ARID1A), SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1) in oesophageal adenocarcinoma. BMC Cancer. 2020;20:1–12. doi: 10.1186/s12885-019-6425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann LM, Loeser H, Jacob AS, et al. Dickkopf-2 (DKK2) as context dependent factor in patients with esophageal adenocarcinoma. Cancers (Basel) 2020 doi: 10.3390/cancers12020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J, van Lanschot JJB, Hulshof MCCM, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol. 2015;16:1090–1098. doi: 10.1016/S1470-2045(15)00040-6. [DOI] [PubMed] [Google Scholar]
- Shen F, Li J, Cai W, et al. GATA6 predicts prognosis and hepatic metastasis of colorectal cancer. Oncol Rep. 2013;30:1355–1361. doi: 10.3892/or.2013.2544. [DOI] [PubMed] [Google Scholar]
- Shen W, Niu N, Lawson B, et al. GATA6: a new predictor for prognosis in ovarian cancer. Hum Pathol. 2019;86:163–169. doi: 10.1016/j.humpath.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Simon R, Mirlacher M, Sauter G. Tissue microarrays. Methods Mol Med. 2005;114:257–268. doi: 10.1385/1-59259-923-0:257. [DOI] [PubMed] [Google Scholar]
- Suzuki E, Evans T, Lowry J, et al. The human GATA-6 gene: Structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics. 1996;38:283–290. doi: 10.1006/geno.1996.0630. [DOI] [PubMed] [Google Scholar]
- Tian F, Li D, Chen J, et al. Aberrant expression of GATA binding protein 6 correlates with poor prognosis and promotes metastasis in cholangiocarcinoma. Eur J Cancer. 2013;49:1771–1780. doi: 10.1016/j.ejca.2012.12.015. [DOI] [PubMed] [Google Scholar]
- Toxopeus ELA, Lynam-Lennon N, Biermann K, et al. Tumor microRNA-126 controls cell viability and associates with poor survival in patients with esophageal adenocarcinoma. Exp Biol Med. 2019;244:1210–1219. doi: 10.1177/1535370219868671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baal JWPM, Verbeek RE, Bus P, et al. MicroRNA-145 in Barrett’s oesophagus: regulating BMP4 signalling via GATA6. Gut. 2013;62:664–675. doi: 10.1136/gutjnl-2011-301061. [DOI] [PubMed] [Google Scholar]
- van Hagen P, Hulshof MCCM, van Lanschot JJB, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- von Loga K, Woolston A, Punta M, et al. Extreme intratumour heterogeneity and driver evolution in mismatch repair deficient gastro-oesophageal cancer. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13915-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener-Ryczek S, Schoemmel M, Kraemer M, et al. Immune profile and immunosurveillance in treatment-naive and neoadjuvantly treated esophageal adenocarcinoma. Cancer Immunol Immunother. 2020;69:523–533. doi: 10.1007/s00262-019-02475-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang Z, Fu B, et al. GATA6 activates Wnt signaling in pancreatic cancer by negatively regulating the Wnt antagonist Dickkopf-1. PLoS ONE. 2011 doi: 10.1371/journal.pone.0022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during this current study are available from the corresponding author on reasonable request.