Abstract
Several automated high-throughput immunoassays for detecting anti-SARS-CoV-2 antibodies by a semi-quantitative approach have been commercialized. In this study, we describe the timeline of the antibody response in patients with RT-PCR-confirmed COVID-19. A total of 292 sequential serum samples from 33 Japanese patients were retrospectively analyzed using four test kits for SARS-CoV-2: the Abbott SARS-CoV-2 IgG assay (Abbott), Elecsys® Anti-SARS-CoV-2 assay (Roche Diagnostic), and VITROS® Anti-SARS-CoV-2 Total and IgG assays (Ortho Clinical Diagnostics). All automated immunoassays could equivalently identify positive sera collected within 2 weeks after symptom onset (99.3%–100%). In addition, the S protein-based automated immunoassay, the VITROS® Anti-SARS-CoV-2 Total assay, may play a complementary role in evaluating passive antibody therapies or vaccines against SARS-CoV-2, although further research is required.
Keywords: SARS-cov-2, COVID-19, Antibodies, Immunoassay, CLIA, ECLIA
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which emerged in Wuhan, China, and is still having an enormous impact worldwide (Phelan et al., 2020; WHO 2020). The infection has now spread to 222 countries, with more than 84,000,00 confirmed cases and over 1,800,000 confirmed deaths as of January 6, 2021 (WHO 2020). Additionally, the global pandemic is still expanding due to the existence of asymptomatic carriers with high viral shedding and the long incubation period of this disease (Zhou et al., 2020).
The main in vitro diagnostic assay used for COVID-19 involves reverse transcription-polymerase chain reaction (RT-PCR)-based detection of SARS-CoV-2 RNA. It is considered the gold standard for screening and diagnosis in the early clinical phase of the infection. However, its sensitivity varies according to the duration of infection and the suitability of both the sampling technique and anatomical site. Additionally, the RT-PCR protocols are still unfortunately less automated, requiring substantial laboratory equipment, reagents, and expertise (Loeffelholz and Tang, 2020; World Health Organization 2020; Zou et al., 2020). Recently, novel high-throughput SARS-CoV-2 immunoassays that detect IgM, IgA, IgG, and total antibody by a semi-quantitative approach have been commercialized; these assays are easier to perform in clinical settings than RT-PCR, the gold standard for diagnosis, and could help to identify patients who have been exposed to SARS-CoV-2 (Theel et al., 2020). However, few peer-reviewed studies have cross-sectionally evaluated their serological response and performance (Hörber et al., 2020).
Commercially available automated high-throughput immunoassays differ not only in their target antibodies (i.e., IgA, IgM, IgG, or total antibody) and targeted SARS-CoV-2 antigens (i.e., the S1 subunit of the spike protein [S], the nucleocapsid protein [N], or the receptor binding domain [RBD]), but also in the principles of the serological assays (i.e., chemiluminescent immunoassay [CLIA], electrochemiluminescence immunoassay [ECLIA], and lateral flow assay [LFA]): the Abbott SARS-CoV-2 IgG assay (CLIA: IgG for N protein) (Abbott; Abbot Park, IL), Elecsys® Anti-SARS-CoV-2 assay (ECLIA: IgM and IgG for N protein) (Roche Diagnostic Scandinavia AB; Solna, Sweden), and VITROS® Anti-SARS-CoV-2 Total and IgG assays (CLIA: IgA, IgM, and IgG for S protein) (Ortho Clinical Diagnostics; Rochester, NY).
In this study, we describe the timeline of the antibody response and the results of cross-sectional evaluations with the above automated high-throughput immunoassays in patients with RT-PCR-confirmed COVID-19 using serially collected serum samples.
2. Materials and methods
2.1. Patients with COVID-19 and their clinical specimens
This study involved Japanese patients with laboratory-confirmed COVID-19 who were referred to Saitama Medical University Hospital in Japan from February 11 to December 31, 2020. All patients were confirmed to have COVID-19 by RT-PCR for SARS-CoV-2 using nasopharyngeal swab specimens in accordance with the nationally recommended protocol in Japan (National Institute of Infectious Diseases, Japan 2021). Briefly, RNA was extracted from each swab using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and was amplified by conventional RT-PCR using N2 gene-specific primers and TaqMan-based QuantStudioTM 5 Real-Time PCR (Thermo Fisher Scientific; Waltham, MA). All serum specimens used in the study were collected at different time points after symptom onset during hospitalization for patient diagnosis and not following a predefined research protocol. Patients who could not be evaluated via an automated immunoassay using sera collected within 7 days of symptom onset were excluded from this study. All samples were stored at –80°C until use.
In this study, days from symptom onset in patients with COVID-19 were determined by a review of the electronic medical records by a research physician. Days from symptom onset were calculated if either of the following could be confirmed from the medical records: (a) an explicit definition of "date since symptom onset with COVID-19" written by a physician or (b) a nursing record explaining acute-onset symptoms associated with COVID-19. Disease severity was classified according to the Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (China National Health Commission 2021).
2.2. Non-COVID-19 patients and their clinical specimens
To evaluate the analytical specificity of the serological assays, 110 residual serum samples randomly recruited under a previous research protocol as negative controls from Japanese patients admitted to Saitama Medical University Hospital from April to October 2019 were used as negative controls. All samples were assumed to be negative for SARS-CoV-2 antibodies because they were collected prior to December 2019, which is when SARS-CoV-2 was first reported in Wuhan, China. In addition, 36 residual sera collected from hospitalized patients with respiratory symptoms and/or fever consistent with COVID-19 but with a negative RT-PCR result for SARS-CoV-2 were also evaluated to validate the specificity of the assay (median duration from onset, 1 day; range, 0–26 days).
2.3. Detection of SARS-CoV-2 antibodies
This study used the following commercially available automated high-throughput immunoassays on the corresponding platforms for the detection of anti-SARS-CoV-2 antibodies.
1. Abbott SARS-CoV-2 IgG assay
The Abbott SARS-CoV-2 IgG assay was automatically performed according to the manufacturer's instructions on the ARCHITECT i2000 (Abbott). This CLIA detects IgG antibodies against the N protein of SARS-CoV-2. The analysis can simultaneously report a signal/cutoff (S/CO) ratio and qualitative results indicating non-reactive (S/CO < 1.4; negative) or reactive (S/CO ≥ 1.4; positive) for IgG antibodies. This assay requires a minimum of 100 μL serum per assay.
2. Elecsys® Anti-SARS-CoV-2
We estimated anti-SARS-CoV-2 antibodies using the Elecsys® Anti-SARS-CoV-2 assay on the Cobas 8000 e801 (Roche) according to the manufacturer's instructions. This ECLIA is based on a modified double-antigen sandwich immunoassay using recombinant N protein and is used for the specific detection of total SARS-CoV-2 antibodies, including IgM and IgG. Results are reported as signal sample/cutoff (cutoff index [COI]) values and as qualitative results indicating non-reactive (COI < 1.0; negative) or reactive (COI ≥ 1.0; positive). This assay requires a minimum of 100 μL serum per assay.
3. VITROS® Anti-SARS-CoV-2 Total and IgG assays
Both the VITROS® Anti-SARS-CoV-2 Total and IgG assays (Ortho Clinical Diagnostics) are based on CLIA using luminol-horseradish peroxidase (HRP)-mediated chemiluminescence. Both of the assays were performed on the VITROS 3600 automated immunoassay analyzer (Ortho Clinical Diagnostics) according to the manufacturer's instructions. In these assays, the specific antibodies against the recombinant S1 subunit of the S protein of SARS-CoV-2 were automatically analyzed. Results are reported as signal/cutoff (S/C) values and as qualitative results indicating non-reactive (S/C < 1.0; negative) or reactive (S/C ≥ 1.0; positive). The VITROS® Anti-SARS-CoV-2 Total assay can detect total antibodies (IgA, IgM, and IgG) against SARS-CoV-2 S protein. These assays also require a minimum of 100 μL serum per assay.
2.4. Ethics statement
The study design and protocol were reviewed and approved by the Institutional Review Board of Saitama Medical University Hospital (Approval Nos. 20065.01, 19136 and 20001).
3. Results
3.1. Antibody response to SARS-CoV-2
A total of 292 sequential serum samples from 33 Japanese patients (22 male, 11 female) with a median age of 67.0 (interquartile range, 42.0–78.0) years were retrospectively analyzed using the four automated high-throughput immunoassays for SARS-CoV-2. To assess the sensitivity of each assay, patients were subdivided into two groups by days from onset—7 days or less and 8 to 14 days—and analyzed for seroprevalence during each period (Table 1 ). For the first 7 days after symptom onset, the VITROS® Anti-SARS-CoV-2 Total assay (i.e., the S protein-based immunoassay) was positive in 33.3%, consistent with the results obtained with the other N protein-based immunoassays (Abbott SARS-CoV-2 IgG, 27.3%; Elecsys® Anti-SARS-CoV-2, 30.3%). However, the positive rate of the VITROS® Anti-SARS-CoV-2 IgG assay (6.1%) was significantly lower than that of the other methods. At 14 days after symptom onset, the VITROS® Anti-SARS-CoV-2 Total assay eventually reached 100%, whereas the seropositive rates of the Abbott SARS-CoV-2 IgG and Elecsys® Anti-SARS-CoV-2 assays reached 81.8% and 90.9%, respectively. No significant differences were found among the immunoassays during this period.
Table 1.
Seropositive rates of antibody responses in patients with RT-PCR-confirmed COVID-19 within 14 days of symptom onset.
| Days from onset | Seropositive rates |
P value | |||
|---|---|---|---|---|---|
| Abbotta | Rocheb | Ortho CoV2 Tc | Ortho CoV2 Gd | ||
| 1–7 days | 27.3% (9/33) | 30.3% (10/33) | 33.3% (11/33) | 6.1% (2/33) | 0.02* |
| 8–14 days | 81.8% (27/33) | 90.9% (30/33) | 100% (33/33) | 84.8% (28/33) | 0.06 |
Abbott: Abbott SARS-CoV-2 IgG assay.
Roche: Elecsys® Anti-SARS-CoV-2 assay.
Ortho CoV2 T: VITROS® Anti-SARS-CoV-2 Total assay.
Ortho CoV2 G: VITROS® Anti-SARS-CoV-2 IgG assay.
Statistical analysis was performed using Fisher's exact test.
3.2. Specificity for non-COVID-19 patients
The specificity of the automated CLIA and ECLIA systems was evaluated in 110 non-COVID-19 serum specimens collected before the emergence of SARS-CoV-2 in Japan. All samples were checked for anti-SARS-CoV-2 antibody on three machines with the four assays and all had successfully negative results. Furthermore, in a study of 38 COVID-19-negative patients with respiratory symptoms and/or fever, one specimen from an 86-year-old patient with bacterial pneumonia showed a false-positive result for antibody in the Elecsys® Anti-SARS-CoV-2 assay. Overall, the specificity of the automated CLIA and ECLIA systems was calculated to be 99.3% (147/148) for the Elecsys® Anti-SARS-CoV-2 assay and 100% for the Abbott SARS-CoV-2 IgG and VITROS® Anti-SARS-CoV-2 Total/IgG assays.
3.3. Detection timelines of anti-SARS-CoV-2 assays
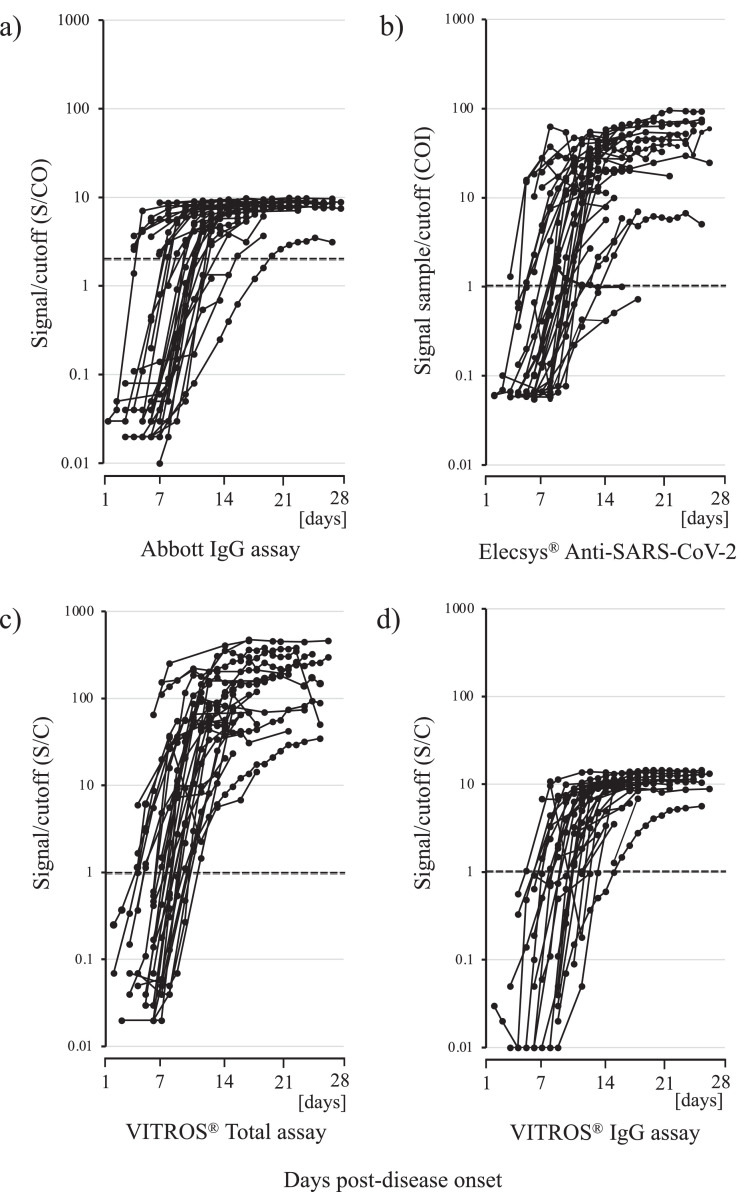
Fig. 1 summarizes the dynamic changes in antibodies against SARS-CoV-2 using each automated high-throughput immunoassay in 33 hospitalized patients with COVID-19; the antibody test results for each patient are detailed in Supplementary Table 1. We calculated the median and interquartile range (IQR) of the number of days from symptom onset to antibody detection. The median time to seropositivity was 10 days (IQR, I7–12 days) for the Abbott SARS-CoV-2 IgG, 9 days (IQR, 7–12 days) for the Elecsys® Anti-SARS-CoV-2 assay, 9 days (IQR, 7–10 days) for the VITROS® Anti-SARS-CoV-2 Total assay, and 11 days (IQR, 8.75–12.25 days) for the VITROS® Anti-SARS-CoV-2 IgG assay. Additional statistical analysis involving the Wilcoxon rank-sum test revealed no significant differences among the immunoassays (P > 0.05). From onset to 14 days, there were increases in qualitative results in most cases, whereas the initial antibody tests met one of the criteria for seropositivity in 11 patients (33.3%, 11 of 33) (Patients 2, 6, 7, 17, 18, 20, 21, 26, 27, 30, and 31), even though they were performed within 7 days after symptom onset (Fig. 1, Supplementary Table 1); of these, all immunoassays of the initial sera of Patients 2 and 6 were positive from the 7th day after onset. In 29 of the 33 patients (87.9%), total antibodies against SARS-CoV-2 could be detected with the VITROS® Anti-SARS-CoV-2 Total assay before or in parallel with the other immunoassays, and only 4 patients were found to have antibodies before being detected by the VITROS® Anti-SARS-CoV-2 Total assay (Patients 18, 20, 27, and 29).
Fig. 1.
Kinetics of anti-SARS-CoV-2 antibodies in 33 patients with COVID-19. a) Signal/cutoff (S/CO) values from the Abbott SARS-CoV-2 IgG assay, b) signal sample/cutoff (COI) values from the Elecsys® anti-SARS-CoV-2 assay, c) signal/cutoff (S/C) values from the VITROS® Anti-SARS-CoV-2 Total assay, d) signal/cutoff (S/C) values from the VITROS® Anti-SARS-CoV-2 IgG assay.
4. Discussion
Automated immunoassays for SARS-CoV-2 antibodies were suitable for detecting specific antibodies for epidemiological applications and could serve as alternative diagnostic tools in hospital settings, which is important due to the ever-increasing demand for the detection of SARS-CoV-2 antibodies worldwide. In a recent study of the seropositivity detected by the VITROS® Anti-SARS-CoV-2 Total assay, antibody production was found within the first 7 days and the assay had 97% sensitivity for SARS-CoV-2 at or beyond 7 days after symptom onset (Garnett et al., 2020, Qian et al., 2020). Andrea et al. (2020) have also reported a head-to-head comparison of automated immunoassays. Using serum specimens collected from Italians, they found that the Abbott SARS-CoV-2 IgG, Roche Elecsys® anti-SARS-CoV-2, and Ortho VITROS® Anti-SARS-CoV-2 Total and IgG immunoassays showed good clinical performance (sensitivity, 89.4%–95.2%; specificity, 97.6%–100%), as in our study. The sensitivity and specificity of immunoassays can depend on target population, race, and disease severity. Our study also found that these automated immunoassays demonstrated good clinical performance for detecting SARS-CoV-2 antibodies in the Japanese population.
Previous reports indicated that some patients with COVID-19 could have negative results on RT-PCR tests for SARS-CoV-2 due to sampling errors or sample transport restrictions (Guo et al., 2020; Li et al., 2020; Qian et al., 2020). Automated assays would be useful, as would high-throughput assays able to rapidly analyze many samples for antibody detection. In particular, whereas the diagnosis of active SARS-CoV-2 infection must rely on the detection of viral RNA in the hospital, the antibody test could eliminate the need to collect nasopharyngeal swab samples or sputum, which poses a risk of infection for medical staff. Combinations of automated immunoassays for SARS-CoV-2 antibodies with repeated swab tests, the gold standard for diagnosis, could also be helpful for the alternative diagnosis of patients with a high index of suspicion for COVID-19. Additionally, the S protein, located on the surface of coronavirus, which is the target of the VITROS® Anti-SARS-CoV-2 Total and IgG assays, plays a pivotal role in viral entry and is a main target for neutralizing antibodies and vaccine design against SARS-CoV-2 (Chi et al., 2020). Therefore, we should not exclude a potential role for these tests in COVID-19 diagnosis and evaluation of the effectiveness of the vaccine. However, further study is needed.
Recent work has demonstrated that the sensitivity of the S protein-based IgM immunoassay was significantly higher than that of an immunoassay based on N protein and that there was no significant difference in sensitivity between the S and N proteins for the detection of IgG and total antibodies (Liu et al., 2020). In the third week after symptom onset, the seropositive rates of IgM antibodies against both S and N proteins were maintained at 73.7%, whereas the seropositive rates of IgG antibodies against S and N proteins reached 100% (Sun et al., 2020). The level of IgA antibody increased from days 2 to 6 after symptom onset and showed higher levels compared with IgM antibody throughout the observation period (Ma et al., 2020; Padoan et al., 2020, Yu et al., 2020). However, the VITROS® Anti-SARS-CoV-2 Total assay, which detects IgA, IgM, and IgG antibodies against the S1 subunit of the S protein, was ultimately equivalent to the other immunoassays, with all patients found to have the specific antibody within 2 weeks after symptom onset in this study. Furthermore, it should be noted that the VITROS® Anti-SARS-CoV-2 IgG assays may have lower detection sensitivity in the early period after onset than the other immunoassays.
The data acquired from patients with negative COVID-19 demonstrated that these automated immunoassays were reliable due to their high specificity. In particular, the specificities of the Abbott SARS-CoV-2 IgG and VITROS® Anti-SARS-CoV-2 Total/IgG assays reached 100% while that of Elecsys® Anti-SARS-CoV-2 assay was 99.3%. Previous reports have suggested that the specificities of the Abbott and VITROS® Anti-SARS-CoV-2 IgG assays for healthy populations exceeded 99% (Bryan et al., 2020, Theel et al., 2020), in agreement with our findings.
Our study has some limitations. First, the number of patients was small (n = 33) and all patients were hospitalized with symptoms. Second, the overall serological response of individuals with SARS-CoV-2 infection cannot be determined because asymptomatic individuals were not evaluated. Third, the specificity for sera that are positive for common human coronaviruses other than SARS-CoV-2 was not evaluated.
In conclusion, the Abbott SARS-CoV-2 IgG, Elecsys® Anti-SARS-CoV-2, and VITROS® Anti-SARS-CoV-2 Total assays were able to equivalently identify positive sera collected within 2 weeks after symptom onset. In addition, S protein-based automated immunoassays may play a complementary role in the evaluation of passive antibody therapies or vaccines against SARS-CoV-2, although further research is required.
Credit author statement
Katsumi Kubota: Data curation, Writing- Original draft preparation. Yutaro Kitagawa: Data curation. Masaru Matsuoka: Conceptualization, Data curation. Kazuo Imai: Data curation. Yuta Orihara: Data curation. Rieko Kawamura: Data curation. Jun Sakai: Data curation. Ishibashi Noriomi: Data curation. Norihito Tarumoto: Supervision. Shinichi Takeuchi: Supervision. Shigefumi Maesaki: Supervision. Takuya Maeda: Conceptualization, Methodology, Writing- Reviewing and Editing.
Author contributions
TM and MM designed the research; KK, YK, MM, KI, YO, RK, NI, and JS performed the research; NT, ST, and SM provided scientific guidance; KK and TM prepared the manuscript.
Conflict of interest
All authors have no conflicts of interest.
Funding
This study did not receive funding support.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.diagmicrobio.2021.115370.
Appendix. Supplementary materials
Supplementary Table 1 Detailed timeline of the antibody response in 33 patients with COVID-19 from symptom onset (day 1) according to the four automated immunoassays. Black cells indicate the date at which the specific antibody was first detected by each immunoassay. Severity was classified as follows: mild, 1; moderate, 2; severe, 3; and critical, 4.
References
- Andrea P, Francesco B, Matteo P, Alessio B, Davide N, Silvia Z, et al. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103101. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, et al. Performance characteristics of the abbott architect SARS-CoV-2 IgG assay and seroprevalence in boise, idaho. Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00941-20. Jul 23e00941-20Print 2020 Jul 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. Aug 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China National Health Commission. Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (7th edition). http://kjfy.meetingchina.org/msite/news/show/cn/3337.html. Accessed July 12, 2020.
- Coronavirus disease (COVID-19) pandemic, WHO. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed August 9, 2020.
- Garnett E, Jung J, Tam E, Rajapakshe D, Cheney S, Brown C, et al. Clinical validation and performance evaluation of the automated vitros total anti-SARS-CoV-2 antibodies assay for screening of erostatus in COVID-19. Am J Clin Pathol. 2020;154(6):742–747. doi: 10.1093/ajcp/aqaa157. Nov 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörber S, Soldo J, Relker L, Jürgens S, Guther J, Peter S, et al. Evaluation of three fully-automated SARS-CoV-2 antibody assays. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0975. Aug 4:/j/cclm.ahead-of-print/cclm-2020-0975/cclm-2020-0975.xml. [DOI] [PubMed] [Google Scholar]
- Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.00461-20. May 26e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections - the state of the art. Emerg Microbes Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zeng W, He H, Zhao D, Jiang D, Zhou P, et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol. 2020;17(7):773–775. doi: 10.1038/s41423-020-0474-z. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Infectious Diseases, Japan. Manual for the Detection of Pathogen 2019-nCoV Ver.2.6. https://www.niid.go.jp/niid/ja/diseases/ka/corona-virus/2019-ncov/2484-idsc/9403-labo-manual.html. Accessed March 17, 2021.
- Padoan A, Sciacovelli L, Basso D, Negrini D, Zuin S, Cosma C, et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin Chim Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan AL, Katz R, Gostin LO. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020 doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- Qian M, Yi Q, Qihua F, Ming G. Understanding the influencing factors of nucleic acid detection of 2019 novel coronavirus. Chin J Lab Med. 2020 doi: 10.3760/cma.j.issn.1009-8158.2020.0002. [DOI] [Google Scholar]
- Sun B, Feng Y, Mo X, Zheng P, Wang Q, Li P, et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):940–948. doi: 10.1080/22221751.2020.1762515. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theel ES, Harring J, Hilgart H, Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.01243-20. Jul 23e01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theel ES, Harring J, Hilgart H, Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/JCM.01243-20. Jun 8; JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland.: 2020. Coronavirus Disease 2019.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 Accessed March 17, 2021. [Google Scholar]
- World Health Organization. 2020. Laboratory Biosafety Guidance Related to the Novel Coronavirus (2019-nCoV). https://www.who.int/docs/default-source/coronaviruse/laboratory-biosafety-novel-coronavirus-version-1-1.pdf?sfvrsn=912a9847_2. Accessed March 17, 2021.
- Yu HQ, Sun BQ, Fang ZF, Zhao JC, Liu XY, Li YM, et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. 2020 doi: 10.1183/13993003.01526-2020. Jun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Detailed timeline of the antibody response in 33 patients with COVID-19 from symptom onset (day 1) according to the four automated immunoassays. Black cells indicate the date at which the specific antibody was first detected by each immunoassay. Severity was classified as follows: mild, 1; moderate, 2; severe, 3; and critical, 4.