Abstract
Characteristically, cells must sense and respond to environmental cues. Despite the importance of cell-cell communication, our understanding remains limited and often lacks glycans. Glycans decorate proteins and cell membranes at the cell-environment interface, and modulate intercellular communication, from development to pathogenesis. Providing further challenges, glycan biosynthesis and cellular behavior are co-regulating systems. Here, we discuss how glycosylation contributes to extracellular responses and signaling. We further organize approaches for disentangling the roles of glycans in multicellular interactions using newly available datasets and tools, including glycan biosynthesis models, omics datasets, and systems-level analyses. Thus, emerging tools in big data analytics and systems biology are facilitating novel insights on glycans and their relationship with multicellular behavior.
Keywords: Glycobiology, Systems Biology, Glycomics, Extracellular Matrix, cell-cell interaction, Bioinformatics
Extracellular glycans influence intercellular behavior
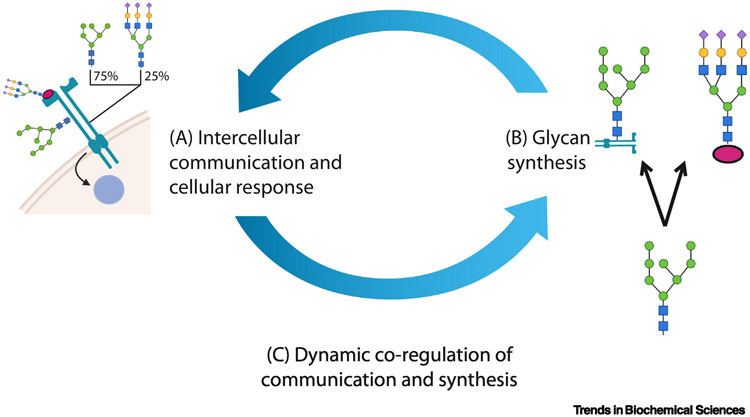
Every cell and many viruses are wrapped in a sugar-coating of functional glycan epitopes (see Glossary) and glycoconjugates that helps modulate cell-cell communication, the glycocalyx. Glycans are bound to peptide or lipid glycoconjugates and matured in the endomembrane system. They then reside on the cell-surface or diffuse into the extracellular matrix (ECM). The emergence, recycling and dispersion of glycans is often slow, variable and glycoconjugate dependent (2-1000 hours), therefore environmentally-responsive nascent glycans do not transform the cell surface immediately [1,2]. Instead, the glycocalyx constitutes a composite memory of recent and current responses to intercellular exchanges. Together, the glycocalyx, the communication-modulating glycans and glycoconjugates between conversing cells form a rich resource that can be leveraged to improve our understanding of how cells communicate (Figure 1).
Figure 1 (Key figure) -. The feedback between glycan dependent receptor sensing and glycan biosynthesis regulate cellular communication and environmental response.
Here we discuss (A) the system-level impacts of glycosylation, (B) tools to study glycan biosynthesis given various data types, and (C) databases and analytical strategies to explore the dynamic co-regulatory systems of glycan biosynthesis and intercellular communication. Diverse bioinformatics approaches and resources are now making it easier to consider glycosylation in diverse research fields.
ECM glycans are voluminous, ubiquitous and diverse, extending 2-3x the cell diameter [3]. With 4,549 of 20,365 reviewed human proteins (UniProtKB) corresponding to approximately 250,000 proteoforms per cell-type [4], there are approximately 50 glycoforms for every glycoprotein. Glycan metabolism uses 342 documented [5] human glycosyltransferases and glycosidases, each performing interdependent and distinct monosaccharide additions and removals. Borrowing from epigenetics, lectins, glycosyltransferases, and glycosidases have been described as the “readers,” “writers,” and “erasures” of the glycocalyx [6,7]. These encapsulating carbohydrates moderate many cellular responses, including development, growth, differentiation, migration, signaling, and morphogenesis (Table 1) making few biological discussions complete outside the glycan context (Figure 2) [8].
Table 1 -.
Examples of Ligand-Receptor Interactions modified by glycan.
| Mechanism | Ligand | Receptor | Type | Alt. Glycosylation | Glycan Impact | Ref. |
|---|---|---|---|---|---|---|
| Co-reception | Delta & Serrate | Notch EGF1- 10, 12, 14, 16, 17, 20, 22-32, 35 motif | O | Presence/Absence of Glycan | Boundary Formation, T-cell and marginal zone B-cell development | [25] |
| Co-reception | CD48 | 2B4 (CD244) | N & O | Differential Sialylation | Proper glycosylation necessary for binding | [162] |
| Co-reception | NMDA | NMDAR | N | Differential Mannosylation | Binding affinity | [161] |
| Structural | Antigen | IgG1 | N | Core Fucosylation | Antibody-dependent cell-mediated cytotoxicity (ADCC) | [23,163] |
| Structural | Fibronectin | Integrin | N | Removal of glycosylation sites | Integrin Assembly and Activity | [24,157] |
| Structural | ACE2 | SARS-CoV-2 Spike | N | Maturation of oligomannose | Destabilization of the Spike RBD | [15] |
| Ligand Guiding | VEGF-C | VEGFR3 | GAG | Differential Sulfation on Heparin Sulfate | Inhibition of Lymphogenesis | [159] |
| Ligand Guiding | BMP, FGF | Smad1/5, Erk1/2 | GAG | Differential Sulfation | Cartilage degradation and repair | [160] |
| Multiple | Multiple | NCAM | N | Differential Polysialylation | Cerebellum Formation & Glioblastoma Migration | [28,164] |
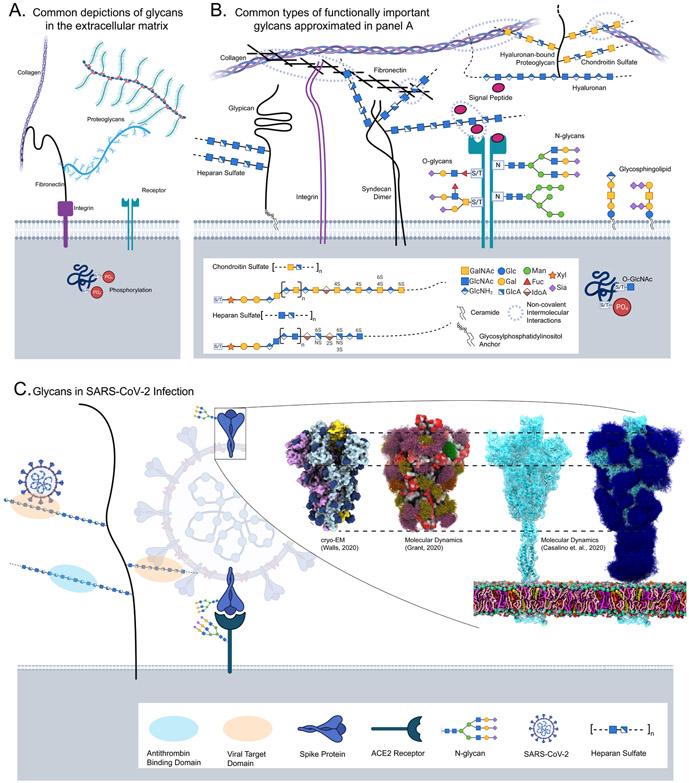
Figure 2 -. The Glycocalyx extends into the extracellular matrix.
(A) Common depictions of glycans and glycoconjugates in the extracellular matrix often censor structural and functional diversity. Panel B mirrors panel A with the structural detail unmasked glycoproteins describe proteins with branched N-or O-glycans, where carbon one of the first monosaccharide is covalently linked to the asparagine amine(N-), or serine/threonine hydroxyl(O-). Proteoglycans describe large linear glycosaminoglycans (GAGs) covalently bound to either a secretory granule protein, a protein with a transmembrane (e.g. syndecans) or glycosylphosphatidylinositol (GPI) anchor (e.g. glypicans), a pericellular protein or a Hyaluronan binding extracellular protein; GAGs like heparan and chondroitin sulfate present functional groupings of sulfation patterns. Hyaluronan-bound chondroitin sulfate proteoglycans are shown connecting two collagen fibers. Other common O-type glycans include glycolipids--mono or oligosaccharides often bound to glycerol or sphingosine backbones, and unconjugated lactation-secreted oligosaccharides. Each class is synthesized a little differently, for example, the acceptor for N-glycans, many O-glycans and GAGs is an amino acid while the conjugate for a glycolipid is a ceramide. Despite these distinctions, each of these classes follow general principles of glycan biosynthesis. Inside the cell, an O-GlcNAc modified a serine residue phosphorylation site. (C) A current example of glycans involved in SARS-CoV-2 attachment Heparan Sulfate facilitates target-cell attachment, while both the Spike protein and ACE2 are glycosylated; details cryo-EM struggles to resolve. Partially glycosylated (dark-blue) closed cryo-EM structure [19], 3D model of the open structure showing glycan range of motion (purple, green, orange, yellow) [14], and a simulation of the complete structure with and without glycans (dark-blue) [15] were adapted with permission.
Today, we are seeing glycan essentiality and neglect in the study of severe acute respiratory syndrome coronavirus 2(SARS-CoV-2), the COVID-19 causative agent (Figure 2C). Because cells and viruses are wrapped in glycans, glycans moderate the first host-pathogen interactions including immune evasion through viral glycosylation [9,10] and host-cell targeting through Heparan Sulfate (HS) binding [11]. The SARS-CoV-2 spike glycoprotein has over 20 glycosylation sites [12,13] covering nearly 50% of the protein, shielding predominantly immune targets [14]. Two sites are implicated in stabilizing the ACE2-binding receptor binding domain [15]. Considering glycosaminoglycans, tissue-specific HS sulfation patterns are necessary for SARS-CoV-2 attachment and infection thereby clarifying tissue tropism unexplainable by ACE2 and TMPRSS2 presentation alone [16]. Consistently, HS-consuming commensal bacteria can prevent SARS-CoV-2 attachment and are depleted in infected and higher-risk individuals [17]. Unfortunately, only 2 of 15 vaccine development and SARS-CoV-2 immune response manuscripts we examined considered glycosylation in their analysis [18,19].
Beyond virology, glycans are both regulators and targets in systems-level regulatory events (Figure 1 A). They modify molecules involved in intercellular communication, thereby helping to regulate cell-state (Figure 3, Table 1). Here we explore specific (Table 1) and theoretical (Figure 3) examples of glycans regulating downstream processes [8]. As evidence of their systems-level role, glycogenes are often selected or enriched in glycan-agnostic omics studies (Figure 3C.i-ii) including micro RNA (miRNA) in granulosa-hormone response [20], autoinflammatory diseases [21], and type-I interferon response [22]. Indeed, glycans can regulate many systems-level cellular phenotypes. They change protein structure and function, impacting antibody-dependent cellular cytotoxicity [23] and integrin-fibronectin-proteoglycan complex formation [24]. Glycan co-receptors facilitate receptor binding modulating notch signaling [25] and SARS-CoV-2 spike-ACE2 complex formation [13]. Glycosaminoglycans (GAGs) act as co-receptors, peptide recruiters and spatial directions in chemotaxis, cancer, and development [26]. Glycans are essential for self/non-self recognition [8,27] and inter/intra organismal coordination as they define blood group antigens, modulate neural adhesion [28], facilitate sperm egress [29], and regulate the gut microbiome [30]. O-GlcNAc regulates metabolism and competes with phosphorylation to regulate kinase activity and histone accessibility, impacting multiple cancers and neurodegenerative diseases [31]. Because of their systems-level impact, glycans show clinical promise. Glycan mimetics can inhibit influenza escape (i.e. Tamiflu® and Relenza®) [32], facilitate anticoagulation (i.e., heparin) [33] and block SARS-CoV-2 Spike-ACE2 binding [34,35]. HMOs are associated with increased survival in preterm infants [36] and decreased risk of necrotising enterocolitis [37], and glycosylation is being incorporated into vaccine development [38]. The global impact of glycosylation is undeniable.
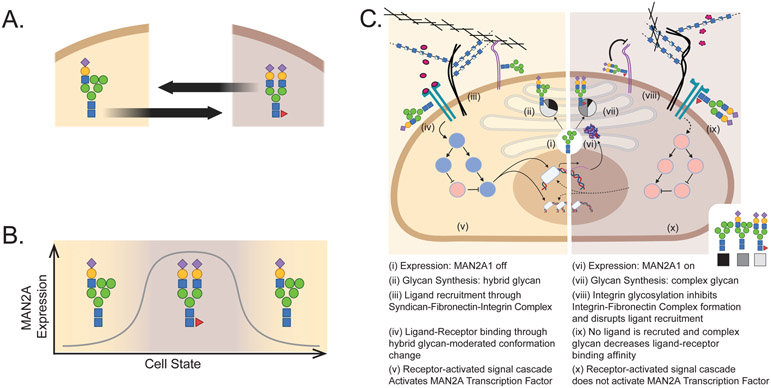
Figure 3 -. Glycans & glycoconjugates mediate multiple types of interactions modulating cell state.
(A) Transitions between cell states (tan and brown) can be modulated by differential glycosylation (e.g. cell-cycle or epithelial-mesenchymal transition [75,76,153,154]). (B) Transition between two cell states with low or high MAN2A1 (mannosidase necessary to escape the hybrid glycan) expression could result in differential abundance of hybrid and complex glycans respectively. Differential glycosylation could modulate cell state in an oscillatory fashion (C). For example, differential expression of alpha-mannosidase II (MAN2A1; i & vi) would change both mannosylation and complexity of N-linked glycans [155,156]. As a result, each cell state produces a different dominant glycan: a hybrid biantennary structure (ii) and a sialylated biantennary structure (vii). The production of different glycans could result in the differential attachment of fibronectin to the integrin [24,157] thereby facilitating (iii) or disrupting (viii) ligand recruitment [16,158-160]. Differential glycosylation of a receptor can also directly impact receptor-ligand binding by changing receptor conformation (iv & ix) [23,25,161]. Differential receptor activation can induce the activation (v) or inhibition (x) of pathways and transcription factors (red circles are activated signalling cascade elements, blue circles are inactive elements) ultimately inducing differential expression of MAN2A1 (i & vi). In this theoretical system, transcription of MAN2A1 (vi) will move the cell to the complex-glycan state (vi-x) and this state ultimately leads to the inactivation of the signal transduction (x) and the subsequent inhibition of MAN2A expression (i) moving the cell back to the hybrid state (i-v). Thus, through basic principles of cell function and glycosylation, we have constructed a theoretical glycan-modulated oscillating cell-state system.
However, the ECM is commonly depicted as a mesh of collagen fibers and fibronectin, bound to cells through cytoskeleton-associated integrins. While depictions are often brushed with the tinsel of proteoglycans (Figure 2A), it is uncommon to see a representation of glycan diversity at the cell-cell interface [39] (Figure 2B). Similarly, while technologies exist to query protein-protein interactions (PPIs) in native environments with appropriate glycosylation [40-44], many large-scale cell-to-cell and single-cell analyses use PPIs observed in non-native cellular environments with erroneous glycosylation (e.g. Yeast-2-Hybrid (Y2H)). Thus, both small-and large-scale explorations of intercellular interactions are at risk of being misled by under-descriptive or inaccurate glycosylation (Figure 2).
Here we will (1) discuss how glycans are made and how glycan biosynthesis is modelled, (Figure 1B), and (2) examine the tools, infrastructure and databases that facilitate examination of these systems in tandem (interacting systems, Figure 1C). Finally, (3) we provide initial recommendations for how to apply these tools and concepts in research to interrogate how glycan biosynthesis and intercellular communication regulate each other (Figure 1).
Models of glycan biosynthesis
The phenotypic and clinical relevance of glycans emphasizes the importance of understanding their biosynthesis. Glycosylation is a system of interrelated biosynthetic pathways that give rise to these diverse (Figure 2B) and impactful (Figure 3) macromolecules. Here, we describe general biosynthesis principles (Figure 3), and discuss the complexities of the system; how small alterations of single steps can lead to subtle or substantial compositional and functional change across the cell surface (Figure 3).
Mechanistic models of glycan biosynthesis
Mechanistic models have enhanced our understanding of glycan biosynthesis. These models organize the steps of glycosylation into explicit synthetic pathways. To build these models, information on glycosyltransferase (GT) specificity is obtained to determine reaction rules; kinetics can be tuned for each GT in multiple conditions or organisms. Using a reaction rule set, a reaction network can be outlined [45]. Depending on the method of construction, the network must be trimmed to remove irrelevant or impossible reactions then parameterized to model data and predict future behavior.
Several published models of glycan biosynthesis are detailed nonlinear kinetic models (Figure 4.i) of N-glycosylation [45-49]. These models describe reaction kinetics and therefore require multiple parameters for each reaction. These parameters are often obtained from the literature and extensive databases, which aggregate the information from diverse studies on many organisms, cell types, and cellular environments. Many kinetic reaction parameters, however, remain unmeasured and must be estimated [47,50]. Kinetic models are useful for simulating the dynamics of glycosylation to provide insights into temporal variation [46,51,52]. Small, highly parameterized kinetic models are invaluable for highlighting the dynamics and behaviors of a system, such as bistability [53] and limiting factors like nucleotide sugar availability [54,55]. Nonlinear models are challenging to construct and fit but accommodate a broader diversity of relations, a higher degree of specificity, and a multiplicity of dynamics inaccessible in other modeling approaches.
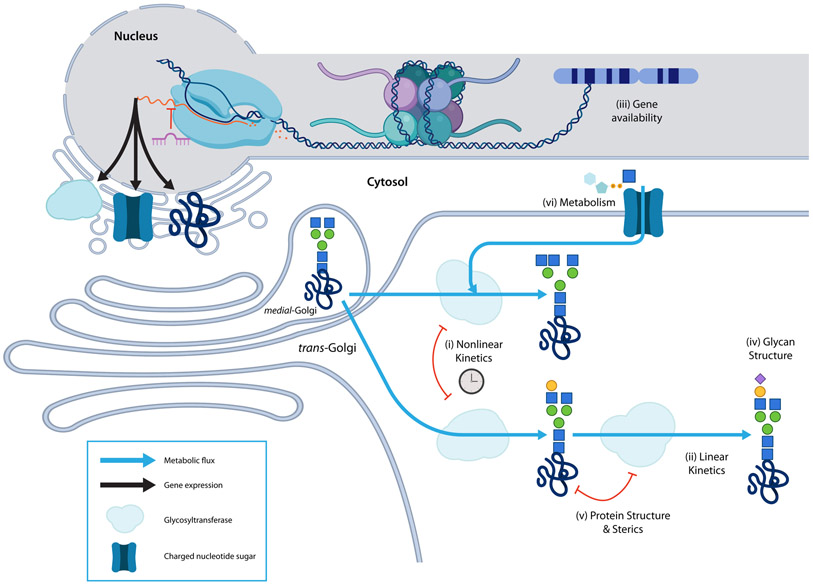
Figure 4 -. Characteristics of glycan biosynthesis.
Generally, glycans are covalently bound to a glycoconjugate and built by iterative addition, and occasional removal, of monosaccharides by highly-specific glycosyltransferases (GT), as the glycoconjugate passes through the endoplasmic reticulum (ER) and Golgi; most glycan products reactants for later reactions. GTs are retained in different endomembrane compartments thereby increasing biosynthetic diversity and control. N-glycosylation occurs in approximately 3 stages: addition and pruning of a large oligomannose structure in the ER, further pruning and GIcNAc-capping in the cis/medial Golgi, and GlcNAc-capped branch maturation. In specific models, (i) nonlinear kinetics can capture complex reaction behavior and incorporate variation over time (ii) In the absence of temporal data, simpler reaction behaviors can be adequately described with linear kinetics. (iii) Glycogenes like glycosyltransferases, nucleotide-sugar biosynthesis, and transport proteins must be present in the genome, epigenetically accessible, expressed and translated to perform their functions. Both linear and nonlinear models can be improved by including information on the availability of glycogenes. Other models can be created using only information on the availability of glycogenes. (iv) Glycan structure can also be used to supplement comprehensive modeling because the glycan structure is a complete description of every biosynthesis reaction a glycan undergoes. (v) New evidence suggests that steric interactions between glycoproteins and glycosyltransferases add additional constraints to glycan biosynthesis; sterics can decide which glycosyltransferases can access a growing glycan. (vi) Finally, metabolism can determine the availability of monosaccharide precursors thereby limiting the diversity of additions that may occur.
Introducing simplifying assumptions (e.g., assuming the system is at a steady-state) enables the development of more generalizable linear models [56]. Simplified Linear models (Figure 4.ii) can be used for studying glycosylation. These models perform well and capture many of the same predictions as nonlinear kinetic models, aside from predictions of network dynamics [52,57]. Their value, however, is seen in their ability to handle larger networks with minimal parameterization. The low parameterization and scalability of these models accommodate diverse approaches to compute fluxes, such as flux balance modeling, probabilistic learning [58,59], multi-omic comparison [60], and other linearized approaches [61]. The reduced complexity of linearized glycan biosynthesis models allows for the simulation of multiple glycogene knockouts [62], tolerance of glycan structure and reaction uncertainty [60], and analysis of many types of glycosylation including human milk oligosaccharide biosynthesis [60,63], O-linked glycans [50,64], GAGs [58], and glycolipids [65]. The simplicity of these models makes otherwise computationally taxing questions feasible.
Other models aim to approach a complete description of the systems they model and have included glycosylation. Such comprehensive models can include multiple linear and non-linear models which exchange inputs and outputs. One comprehensive model integrated processes throughout the endomembrane system including translation and translocation, post-translational modifications, metabolism, and simplified glycosylation [55]. There are many glycoconjugate-specific factors, both known and suspected, that may influence glycosylation that could be included in future comprehensive models. These include (1) glycoconjugates-specific availability and flow through the ER and Golgi compartments [59], (2) glycoconjugate-specific dependencies on transient enzymes and substrates [54,66,67], and (3) considering the impact of glycoconjugate diversity on flow and processing during glycosylation [54].
Building the highly detailed nonlinear or comprehensive models can generate invaluable insight into specific systems. However, they are hard to build and parameterize. Linear models, due to their simplicity are easier to build and can be more generalizable. However, more parsimonious frameworks for exploring glycan biosynthesis have been developed through principle-based abstraction.
Abstract glycan biosynthesis analysis
In contrast to detailed biochemical mechanistic models, more abstract approaches can model glycan biosynthesis by constraining analyses around the system principles. These approaches are more statistically driven than mechanistic models while retaining knowledge of the system through biosynthetic principles (e.g., hierarchical, or non-converging); these principle-constraints mitigate errors characteristic to variance-driven “black-box” approaches. While abstract analyses can be more error-prone, they can also be more generalizable. The tradeoff is between the high-specificity mechanistic models, which may fail due to over-specification or overfitting, and more abstract principle-driven analyses, which are less exact and more generalizable, is one that must be weighed by a user depending on their objectives. Here, we will explore those concepts and how they have been employed and explored in abstract analyses in glycan biosynthesis.
Within glycan biosynthesis, both glycosyltransferase availability and sugar-donor availability are necessary. Glycan biosynthesis can be analyzed based on gene availability (Figure 4.iii), as measured by the expression (proteome and transcriptome), accessibility (epigenome), or presence (genome) of glycogenes. Gene expression defines which reactions could be active in the biosynthetic network. Absolute and differential-expression of glycogenes have been used to predict differential glycosylation [68-71]. As expression modulators, histone methylation and acetylation can also predict glycosylation [72-74]. Similarly, expression modulating miRNAs are also a useful proxy for glycan biosynthesis [75,76]. The ability of a genome to express a glycan has been predicted from glycogene presence in minimally characterized organisms and states like bacteria and archaea [77], CHO cells [61], and cells with GT knockouts [64]. Novel glycan-glycogene relationships have been elucidated using a genome-wide association study of serum glycosylation [78]. The power of gene availability analysis is its performance in minimally characterized systems.
The glycan structure (Figure 4.iv) also provides insights into glycan biosynthesis and can be used to constrain abstract models. In a recent exploration of the glycan structure-biosynthesis dichotomy, we found that because certain glycosidic bonds implicate the activity of specific glycosyltransferases, much of the glycan biosynthesis network is encoded in the glycan structure [79]. Glycan substructure-oriented analysis has facilitated comprehensive “fingerprint” encoding [80], substructure comparison [81], substructure relations (GNOme, unpublished), glycan alignment [82], motif enrichment from datasets like glycan microarrays [78,83-86], glycoprofile reconstruction [87-90], and as a means of sterics-based glycan-receptor binding generalization [86]. Substructure-oriented computation and generalization has been codified in formal attempts to represent glycan classes using boolean logic [91] and uncertainty operators [92]. Fingerprint encodings have been combined with implicit structural alignment and glycan assembly to improve clustering and clarity while mitigating sparsity in glycoprofiles [93]. Substructure-level representations have been used with biosynthetic networks to predict novel reactions [94]. Recent work has also examined the inter-substructure distances and co-occurrence in fingerprint encodings to reveal functional similarities between glycans [95]. In our work, we combined the logic of these recent works and developed a biosynthetically cognizant substructure encoding. As a result, we were able to improve clustering, increase statistical power, discover novel reactions, and make novel insights about flux through the biosynthetic network [79]. The strength of a structural approach to biosynthesis modeling is that it limits reliance on prior knowledge and characteristics of known reactions without discarding easily accessible structural records of biosynthesis.
Metabolite availability (Figure 4.vi), also constrains monosaccharide addition. Nucleotide sugar biosynthesis and transport can affect glycosylation [96] since they are necessary substrates for growing glycans. While many mechanistic models assume sufficient nucleotide sugar availability, some models account for their impact directly. One approach defined a nucleotide-sugar metabolism and transport network wherein nucleotide sugars are synthesized and transported into the endomembrane system. This allowed the prediction of the impact of media composition on glycosylation [49]. Another model found that specifying the metabolic precursors of glycosylation allowed more accurate descriptions of glycosyltransferase dependencies [97]. Another hybrid model used nucleotide and central metabolism models to parameterize a neural network predicting glycosylation [67]. Overall, metabolism imposes a limit on the complexity and diversity achievable at a specific glycosylation site. Because of the processivity of glycan biosynthesis, the effect of a single monosaccharide shortage early in biosynthesis cascades throughout the system, as precursors may be depleted and competitive branch-points may become unbalanced. Overall, upstream metabolism is an impactful regulator of glycan structure and occupancy.
Protein structure (Figure 4.v) is another potential constraint on glycosylation. Glycoprotein structure, including secondary structure [98], can sterically interact with glycosyltransferases to promote or inhibit the addition of specific monosaccharides to a growing glycan. A meta-analysis of site-specific glycosylation revealed changes in core fucosylation and branching at low-accessibility sites [99]. Site-specific kinetics for glycosyltransferases have also been observed [47], further showing that local protein structure can inform glycosylation. It is still unclear exactly what role protein structure plays in glycosylation but its importance is evident.
Model-derived and validated principles of glycosylation
The mechanistic and statistical models provide new insights into glycan biosynthesis extending our initial understanding: (1) glycans are made by glycosyltransferases adding single monosaccharides to a growing polysaccharide in a glycosidic bond. (2) Glycan biosynthesis is predominantly hierarchical, involving the addition of one monosaccharide at a time [64,79]. The hierarchical assumption neglects, N-glycan mannosidase reactions in the cis and medial Golgi, reconvergence due to the lateral transfer of partial structures [100] and other Golgi-resident glycosidases [101]. While these limitations are important to acknowledge, they do not appear to be first-order effects and therefore models making these assumptions are still effective. (3) Linear models have demonstrated that assumptions of linearity in glycan biosynthesis are appropriate and sufficient to capture major trends [52,57,59,61,62]. (4) Considering the additional insight provided by more detailed nonlinear kinetic models, such as the bistability in galactosylation [53] and competitive inhibition [102], there are important nonlinear trends to observe within glycan biosynthesis. (5) Some analyses have demonstrated competitive inhibition [52] as a dynamic process of glycan biosynthesis. (6) Glycan biosynthesis is limited by precursors like nucleotide-sugar availability [54], protein structure and GT-accessibility [47,98,99], and enzyme presence [61,64,68-72,75-77]. (7) Segregation of glycosyltransferases and nucleotide sugar transporters into separate Golgi compartments and the rate at which glycoconjugates flow through these compartments is an important contributor to glycan diversity [59,97]. (8) glycoconjugate identity, diversity and volume can all impact glycosylation [54,66]. As these principles are further defined, they will lead to more accurate models and facilitate the interpretation of glycomics data.
Big data glycomics in glycan biosynthesis
Intuitively, “big data” should be large to reveal elusive or global trends. Glycomics datasets are small relative to RNA-Seq and negligible relative to astrophysics data. Thus, we have much to gain from modest increases in dataset size. Here, we describe the measurement, aggregation, and distribution of many glycomics datasets into “big” glycomics datasets. The structural and analytical integration of these datasets reveal invaluable insights into multicellular behaviors (Figure 1C).
High-throughput measurement of glycomes
Multiple high-throughput technologies provide temporal, spatial and systematic measurements of glycan abundance and interactions. These can identify glycans and glycan motifs that interact with a lectin of interest. Mass spectrometry (MS) based approaches can identify and quantify glycan structure. Tandem mass spectrometry can rapidly identify glycan structures and the sites of glycan linkages [103]. Highly accurate structures on glycoproteins are obtained using combined technologies such as MS, coupled with enzymatic digestion [104]. MALDI-imaging MS can recover spatial resolution of enzyme-released glycans on tissue slides [105]. Liquid Chromatography (LC) can increase the resolution of MS [106], and stand-alone LC methods [107] provide highly reproducible bulk characterization of N-glycome variability and pathology. It has been used to characterize immunoglobulin [106] and glycosylation, specifying variation in isolated populations [108], and distinguishing pathology in rheumatoid arthritis [109].
Glycan arrays can quantify glycan-protein interactions. Glycans are covalently bound to a slide and then incubated with a fluorescently-tagged protein believed to interact with the glycans. To capture diversity in under-characterized glycomes, shotgun glycomics approaches bind glycan fragments to arrays, followed by tagging with fluorescent lectins [110] and computational reassembly of targets [84]. Glycan arrays and variations on the approach can elucidate mechanisms of viral and microbial specificities and resistance [111-113]. Advances such as bead-based diffusible glycosylation probes could further provide insight into the spatiotemporal glycosylation.
Lectin arrays invert the focus of glycan arrays to identify the binding affinities of glycans to multiple lectins simultaneously. These arrays consist of lectins that are covalently bound to a slide, and fluorescent or mimetic glycans incubated on the arrays to identify specific lectin-glycan interactions [114]. Lectin arrays are notably scalable as demonstrated by the lectin-specificity characterization of glycans in the NCI-60 [75] and multiple mouse tissues [115]. This approach has been deployed on a microfluidics device to improve the reproducibility [116]. These technologies provide a high-throughput approach to the simultaneous characterization of glycans and glycan-protein interactions defining cell-cell communications.
A more comprehensive view of glycan-dependent phenotypes can be obtained by collecting multi-omic datasets that examine the interplay between molecular classes. For example, RNA-Seq data were collected in tandem with glycoprofiles to parameterize biosynthesis models on the expression of glycosyltransferases [117]. Similarly, genomic variation data were integrated with serum N-glycosylation to identify genomic elements associated with the abundance of each glycan, yielding insights into novel biosynthesis reactions [94] and regulators [78]. These large-scale multi-omic analyses have found new reactions [94] and new transcriptional regulators [78].
Glycan collections and analytics of glycan measurement are growing in diversity, magnitude, regularity, and FAIRness
The development of high-throughput glycomics methods have resulted in a rapid expansion of the amount of glycosylation data. International collaboration and computational infrastructure for the storage, integration, and distribution of these datasets have accelerated over the last decade to facilitate queries and answer fundamental questions in glycobiology. Finally, the community is becoming increasingly open-source and standardized in its nomenclature making it easier to reproduce, extend and generalize mechanistic models and biosynthetically constrained analyses. With this emerging ecosystem of Findable, Accessible, Interoperable and Reusable (FAIR) datasets and tools [118], we are closer than ever to exploring the dynamic co-regulation of glycan biosynthesis and intercellular signal transduction essential to multicellularity.
New data collections are published frequently and often integrate diverse glycosylation-related data including comprehensive glycan structures (GlyTouCan [119]), richly curated bacterial and fungal structures [120], MS glycoprofiles UniCarbDR-GlycoPost [121], and observed masses (UniCarbDB [122]). Glycoprofiles across various tissues, conditions, organisms, and cell-types can be downloaded from the Consortium for Functional Glycomics [123]. Site-specific glycosylation events can be queried through GlyConnect [124] or UniCarbKB [125]. Even glycan-mediated ECM [126] and host-pathogen [127] interactions are registered. To increase the utility and interoperability, efforts are underway to unify datasets throughout the Glycomics@ExPASy [128], GlyGen [129], and GlyCosmos [130] datahubs as the GlyspaceAliance [131]. Despite this progress, consistent formats have yet to be globally adopted hindering data sharing and collaboration within glycobiology and with adjacent collaborators in proteomics; there is still no standard format for quantitative glycoproteomics in most popular proteomics databases where glycan data is either non-canonically formatted or inaccessible. However, with the emergence and adoption of data sharing protocols like UniCarbDR-GlycoPost and mass aggregations led by the GlyspaceAliance, common formatting standards should become more prevalent making it easier for proteomics and other downstream databases to welcome glycosylation data in the near future.
Predecessors of and participants in the GlyspaceAlliance have invested considerable effort deciding formats and standards that their platforms will support and advance [121]; Minimal Information Required for a Glycoproteomics Experiment (MIRAGE) for representing metadata, and GlycoRDF for representing the data itself. The MIRAGEproject has created metadata standards for sample collection [132] and data storage for glycomics data measured in mass-spectrometry [133], glycan arrays [134] and liquid-chromatography [121,135]. Compliance with metadata standards makes data more findable but once it is found, it too must be readable. As such, the implementation of semantic web technologies--which have tamed several -omics datatypes [136], lead to the development of an ontology-based glycosylation datatype, GlycoRDF [137]; yet another major step toward defining such a standard. Though MIRAGE and GlycoRDF are well established, adoption is not yet ubiquitous posing a challenge to findability, interoperability, and reusability; each requires uniform metadata and/or common data representations.
The creation of these standards is essential to reproducibility and comparability across experiments and is essential to interoperability across platforms and analysis pipelines. Standards databases like UnicarbDB [122], common pipelines like GlycoWorkBench [138] and pGlyco [139], and open-source comparative analytics [79,140,141] allow for consistent analysis across laboratory setups to facilitate inter-lab interoperability [121]. Due to diverse methods for measuring glycans, cross platform comparison can be challenging. But, with the growing number of comparison and standardization projects [106,109,142] and the growing NIST standards project [143], we are learning about the respective power, biases, limitations and interoperability of various platforms. Increasingly standardized data, curation, and data formats [67,79,140,141] facilitate transfer of glycoprofiles to flexible yet standardized modeling platforms [45,76,79,94,144]. The regularization of data and subsequent analyses, from quantification to modeling, has positioned the community to make coordinated and sustainable progress.
A variety of high-throughput methods exist. While the diversity of methods challenges cross-platform comparisons, it is not an impassable challenge with the help of FAIR principles; rather FAIRness provides an answer to these challenges. FAIR data are a collection with rich metadata; going beyond the type of method (e.g. MALDI) and expanding into extreme detail (e.g. the exact model of mass spectrometer, the intended and true concentrations of adducts and the order of their addition). Many details are required by MIRAGE guidelines. The combination of rich metadata (MIRAGE), readable data (glycoRDF), and platform-specific effects (NIST) could provide the necessary covariates and validation data to make informed cross platform comparisons similar to those in gene expression analysis methods for cross-platform comparisons [145], and compression or removal [146] of known confounders.
The cost of glycan-naive interactome studies
Modifying one of several glycans on a protein can impact structure, function, interactions, and ultimately cell state. Since glycosylation is organism-specific, the measurement of PPIs in a non-native context can deviate substantially from the native interactions. While use of non-native Y2H is still popular, new environment-cognizant PPI methods with realistic glycosylation exist. These include proximity labeling approaches [40,41], native adaptations of Y2H [43], and Surface Plasmon Resonance (SPR). SPR is highly reproducible and goes beyond PPI to measure many other intermolecular interactions [42]. The MatrixDB project [126] used SPR, and integrated glycomics, transcriptomics and interactions from matrisome [147] to explore several phenomena including aberrant aggregation in Alzheimer's Disease [148]. The success of MatrixDB further demonstrates the importance of these molecules to extracellular questions and the power of glycan-cognizant interactome techniques.
Exploring glycan biosynthesis, cellular communication, and cellular state interactions through data integration
As glycomics increase in popularity, meaningful data-integration is essential for leveraging these datasets and furthering our understanding. Here we suggest approaches to leverage emerging tools in big-data glycomics. (Figure 1, Figure 5)
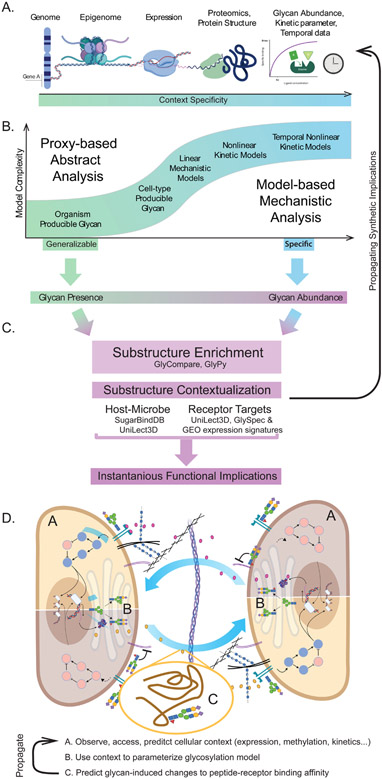
Figure 5 -. The progression of model complexity and predictions appropriate, given various common data types.
(A) Various datatypes of increasing complexity and rarity necessary to train different models. (B) The relationship between model specificity and model complexity. As the complexity increases and the magnitude and rarity of input data increases, so does the specificity of the model. Though lower complexity models are not as specific, they can be beneficially generalizable. (C) Finally, once the glycoprofile predictions are complete, they can be compared to lectin and substructure databases to predict what receptor-ligand interactions the differential glycosylation may impact. Panel D describes two theoretical cells (Figure 3C) co-regulating through differential glycosylation. If a reasonable differential expression signature can be inferred (due to differential interference or promotion of a receptor-ligand interaction), we can generate a “gene-availability” based prediction of updated glycosylation, thus exploring the sustainability of a glycosylation pattern. Labels A, B, and C correspond to methods illustrated in panels in A, B, and C respectively: (A) The omics assessment of the intracellular environment, (B) modeling of the glycan biosynthetic implications of the intracellular environment, (C) differential glycosylation of peptides and receptors and (A-C) the propagation of that information to the intercellular interface.
Synthesis Model Selection
A selected model or analysis can be mechanistic or abstract (Figure 5B). To determine which glycans can be produced in an organism, cell-type, or condition, simple models can be constrained on genome annotation or transcriptomics. Linear mechanistic models increase prediction accuracy and specificity with limited additional curation by leveraging a small set of canonical reactions extrapolated into a complete biosynthetic network. Finally, exploring biosynthesis dynamics requires curation and tuning each reaction to condition-specific kinetic parameters. Given a glycoprofile, measured or predicted, the question shifts to interpretation and relating those glycans to context-relevant insights.
Extracting important glycan substructures
Glycan substructures can be analyzed to highlight functional elements of a glycoprofile. Phenotype-associated motifs can be extracted from lectin or lectin arrays [82-86,149,150], and glycomes [79,82-86,93,140,149]. Selected substructures can be interpreted for meaning and potential impact using databases of glycan interactions and subsequent experiments.
Interpreting and generalizing selected glycan substructures
Glycoprofiles can be interpreted through selected substructure annotation using functional databases (Figure 5C). Selected substructures can be identified as virulence factors through glycan-lectin networks [151] or annotated in SugarBindDB [127]. Predictive and empirical protein-glycan databases (e.g. UniLectin [152] and MatrixDB [126]) can connect motifs to signal transduction events. A sterics-based approach, GLY-SPEC [86], can interrogate and generalize lectin-motif interaction to provide corroborative and additional glycan-lectin interactions. Ultimately, the novel insights from such analyses provide invaluable hypotheses to drive validation experiments.
Applying these tools to better understand multicellular phenotypes
With sufficient resources, we can contextualize and propagate differential glycosylation data (Figure 5D). Motifs of interest with functional relevance can generate new and generalizable insights. Differential glycosylation could become a constraint for a glycan biosynthesis model by (1) connecting differentially abundant motifs to intercellular-communication-modulating lectin-glycan interactions [86,126,152], and (2) identifying gene expression signatures corresponding to the differential activation of those lectins. Finally, (3) given regulated glycogenes, a gene availability (Figure 4.iii) analysis could predict the impact of differential glycosylation on extracellular signaling, and the subsequent impacts on glycosylation (Figure 5D). Now the analysis can be repeated with an updated glycan biosynthesis model. Once that chain of predictions is possible, interrogation of glycosylation-communication dynamics will be feasible.
Concluding Remarks
A protein without its glycans is like a molecule without hydrogen or a plate without food; i.e. structurally specified but functionally augmented. Though it can be challenging to find a truly glycan-independent biological process, much of biology under-represents these molecules due to the challenges of measuring and integrating glycosylation data. Fortunately, we are entering an era of FAIR and open modeling, data and functional associations in glycomics that, en masse, provide an opportunity for feasible and affordable integration of glycosylation into many analyses. Forward progress will depend on the continued construction and observance of standards (see Outstanding Questions). With FAIR and effective infrastructure we can more easily examine global trends in glycan biosynthesis, regulatory constraints imposed by and respondent to glycans and finally expose intercellular regulations regulated by and regulating glycosylation (see Outstanding Questions).
Outstanding Questions.
Within glycan biosynthesis, which glycans can be synthesized in an uncharacterized cellular state? Can the impact on different glycosylation sites be predicted? Which lectins will target new glycans thereby inducing cell-state stabilization or transformation?
How can we reach a consensus on data and model formatting to incentivize external development and tool integration? Will our journals and funders begin to require data-sharing in GlycoRDF format with MIRAGE standards met? Will our community unite behind a common open-source parser to streamline and regularize interaction with these data?
Within intercellular communication, which cellular pathways are impacted by changes in glycosylation? Which host-pathogen interactions require glycosylation? How does glycosylation change in response to a pathogenic challenge?
Within immunity, how man organisms accurately distinguish “self” from “non-self” when glycan targets are made by systems with similar glycosyltransferases? Are functional motifs prioritized? Is the processivity of glycosylation leveraged to distinguish between similar glycan biosynthetic systems?
Regarding the dynamic interdependence between inter-cellular communication and glycan synthesis, do glycans filter or modify environmental stimuli? Is their action active or passive? How extensively do glycans regulate communication, and which intercellular communications regulate glycosylation? If they co-regulate, what are the relative co-regulatory timescales?
What fundamentally challenging multicellular or multi-organism interactions could be simplified or explained by glycan augmentation? Could vaccine design become more successful with increased glycan consideration? Could neuron migratory paths become clearer?
Highlights.
Glycans are essential ECM components and are important for modulating intercellular communication.
As modulators of intercellular communication, glycans impact interaction at levels ranging from protein-protein interactions to that of whole cells, whole tissues, whole organisms, and even multi-organisms.
Glycans are synthesized by a complex system of reactions.
Glycan biosynthesis and other systems affected by intercellular communication regulate and modify each other through signal accessibility and glycogene expression.
Using big-data glycomics, we can predict new models of glycan structures and substructures to better understand the dynamic relationship between glycosylation and the broader biological systems.
Acknowledgments:
The authors thank Anne Phan and James Sorrentino for help framing the title, narrative, and figures. Emma Kellman for help developing and clarifying figures. Pascal Gagneux, Anne Phan, Daniel Sandoval, Robert Woods, Oliver Grant, Austin Chiang, David Phizicky, Bokan Bao, James Sorrentino, Claire Bergstresser, Robert Foreman, Erick Armingol, and Sarah Gasman for their feedback. Lorenzo Casalino, Zied Gaieb, Rommie Amaro, Oliver Grant, and Robert Woods for permission and support incorporating their models into the manuscript. We should also like to thank the reviewers whose feedback and insight was indispensable and notably improved this work. This work was supported by generous funding from the Novo Nordisk Foundation provided to the Center for Biosustainability at the Technical University of Denmark (NNF10CC1016517) and from NIGMS (R35 GM119850).
Glossary
- Endomembrane System
The system of membranes suspended in the cytoplasm of a eukaryotic cell. Membranes separate functional compartments often involving secretion including the nuclear envelope, endoplasmic reticulum, Golgi apparatus, secretory vesicles, lysosome, and plasma membrane.
- Epitope
A functional and/or conserved glycan structure, typically at the non-reducing end of branching glycans.
- Gene Availability
Information indicating if a gene participates in a system. Includes information in the presence of the gene in the genome, epigenetic promotion or inhibition, splicing, transcription, and translation.
- Glycan
A polymer of glycosidically-linked monosaccharides. Glycans can be linked to a protein or lipid, or unconjugated. They can be branched or linear. They are made in the Golgi by glycosyltransferases (glycan “writers”), they are typically active in the extracellular space by lectins (glycan “readers”) or through the augmentation of an extracellular element, they are broken down in the lysosome by glycosidases (glycan “erasers”) [6].
- Glycan co-receptor
Describing a receptor-bound glycan and its capacity to modify the binding affinity of the receptor with its target as a direct participant in the binding event rather than through modification of the protein structure
- Glycan substructures
combinations of connected monosaccharides found within a glycan. Ambiguous monosaccharides and linkages can be used to specify substructures where monosaccharide or linkage is not specified.
- Glycocalyx
translates to “sugar” “coat,” is the predominantly sugar encasing of one cell protruding into the extracellular matrix (ECM). Distinct from the glycocalyx of another cell which will protrude into the ECM and even touch another cell surface.
- Glycoconjugate
Typically a protein or lipid to which a glycan is bound.
- Glycogene
Genes known to regulate glycan biosynthesis or degradation including glycosyltransferases, glycosidases, and nucleotide-sugar transporters.
- Glycoprofile
The relative or absolute abundance of glycans observed on a cell surface or at a specific glycosylation site.
- Glycosidase
The “erasers” of glycosylation [6]. These enzymes are just as specific as the glycosyltransferases, removing only specific monosaccharides participating in specific linkages.
- Glycosyltransferase
The “writers” of glycosylation [6]. These Golgi-resident enzymes are highly specific to add one monosaccharide (glucose, galactose, fucose,…) at a time to a particular carbon of a monosaccharide on a growing glycan polymer. biosynthesis proceeds from the protein or lipid bound monosaccharide out.
- Lectin
An extracellular “readers,” typically a protein in the extracellular space that binds with a glycan directly or forms a co-complex with the glycan and cell-surface receptor [6].
- Linear Models
Linear models apply assumptions like temporal invariance and subsequent equilibrium, or the “steady-state” assumption.
- Mechanistic Model
A model of an event that prioritizes detailed molecular mechanisms.
- Nonlinear Models
Nonlinear models account for complex enzymatic behaviors like variation in time and complex behavior at saturation.
- Reaction Rules
A collection of generalized reactions that may, in some order, simulate glycan biosynthesis.
- Substructure-abundance
The aggregation of abundance data over substructures to determine some feature of the substructure. For example, when all substructures have a common reducing end, the sum of abundances for all glycans containing a substructure gives a substructure abundance indicative of the total number of times that substructure was synthesized [79].
- Substructure-Oriented Analysis
Using structures within a glycan to orient analysis, database query and functional analysis of a glycan. This allows greater generalization than whole-glycan analysis by avoiding distraction by irrelevant variation outside the functionally relevant substructure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Davies HW, Trotter MD. Synthesis and turnover of membrane glycoconjugates in monolayer culture of pig and human epidermal cells. Br J Dermatol. 1981; 104: 649–658. [DOI] [PubMed] [Google Scholar]
- 2.Mathieson T, Franken H, Kosinski J, Kurzawa N, Zinn N, Sweetman G, et al. Systematic analysis of protein turnover in primary cells. Nat Commun. 2018;9: 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Palomo A, Braislovsky C, Bernhard W. Ultrastructural modifications of the cell surface and intercellular contacts of some transformed cell strains. Cancer Res. 1969;29: 925–937. [PubMed] [Google Scholar]
- 4.Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, et al. How many human proteoforms are there? Nat Chem Biol. 2018; 14: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42: D490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedola S, Rugen MD, Young RJ, Field RA. Revisiting the Language of Glycoscience: Readers, Writers and Erasers in Carbohydrate Biochemistry. Chembiochem. 2019. doi: 10.1002/cbic.201900377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabius H-J. The sugar code: Why glycans are so important. Biosystems. 2018;164: 102–111. [DOI] [PubMed] [Google Scholar]
- 8.Varki A Biological roles of glycans. Glycobiology. 2017;27: 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altman MO, Angel M, Košík I, Trovão NS, Zost SJ, Gibbs JS, et al. Human Influenza A Virus Hemagglutinin Glycan Evolution Follows a Temporal Pattern to a Glycan Limit. MBio. 2019;10. doi: 10.1128/mBio.00204-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe Y, Berndsen ZT, Raghwani J, Seabright GE, Allen JD, McLellan JS, et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. bioRxiv. 2020. p. 2020.02.20.957472. doi: 10.1101/2020.02.20.957472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cagno V, Tseligka ED, Jones ST, Tapparel C. Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias? Viruses. 2019; 11. doi: 10.3390/v11070596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific analysis of the SARS-CoV-2 glycan shield, doi: 10.1101/2020.03.26.010322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T. Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. bioRxiv. 2020. Available: https://www.biorxiv.org/content/10.1101/2020.06.25.172403v1.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant OC, Montgomery D, Ito K, Woods RJ. Analysis of the SARS-CoV-2 spike protein glycan shield: implications for immune recognition. bioRxiv. 2020. p. 2020.04.07.030445. doi: 10.1101/2020.04.07.030445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casalino L, Gaieb Z, Dommer AC, Harbison AM. Shielding and Beyond: The Roles of Glycans in SARS-CoV-2 Spike Protein. bioRxiv. 2020. Available: https://www.biorxiv.org/content/10.1101/2020.06.11.146522v1.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clausen TM, Sandoval DR, Spliid CB, Pihl J, Painter CD, Thacker BE, et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. bioRxiv. 2020. p. 2020.07.14.201616. doi: 10.1101/2020.07.14.201616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martino C, Kellman BP, Sandoval DR, Clausen TM, Marotz CA, Song SJ, et al. Bacterial modification of the host glycosaminoglycan heparan sulfate modulates SARS-CoV-2 infectivity. bioRxiv. 2020. doi: 10.1101/2020.08.17.238444 [DOI] [Google Scholar]
- 18.Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020. doi: 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 19.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020. doi: 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velthut-Meikas A, Simm J, Tuuri T, Tapanainen JS, Metsis M, Salumets A. Research resource: small RNA-seq of human granulosa cells reveals miRNAs in FSHR and aromatase genes. Mol Endocrinol. 2013;27: 1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu F, Duan C, Zhang X, Yao D, Si G, Gao Y, et al. RNA-seq analysis reveals different gene ontologies and pathways in rheumatoid arthritis and Kashin--Beck disease. Int J Rheum Dis. 2018;21: 1686–1694. [DOI] [PubMed] [Google Scholar]
- 22.Hernáez B, Alonso G, Alonso-Lobo JM, Rastrojo A, Fischer C, Sauer S, et al. RNA-Seq Based Transcriptome Analysis of the Type I Interferon Host Response upon Vaccinia Virus Infection of Mouse Cells. J Immunol Res. 2017;2017: 5157626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, DiLillo DJ, Bournazos S, Giddens JP, Ravetch JV, Wang L-X. Modulating IgG effector function by Fc glycan engineering. Proc Natl Acad Sci U S A. 2017;114: 3485–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai X, Thinn AMM, Wang Z, Shan H, Zhu J. The importance of N-glycosylation on β3 integrin ligand binding and conformational regulation. Sci Rep. 2017;7: 4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley P, Okajima T. Chapter Four - Roles of Glycosylation in Notch Signaling. In: Kopan R, editor. Current Topics in Developmental Biology. Academic Press; 2010. pp. 131–164. [DOI] [PubMed] [Google Scholar]
- 26.Phan AQ, Lee J, Oei M, Flath C, Hwe C, Mariano R, et al. Positional information in axolotl and mouse limb extracellular matrix is mediated via heparan sulfate and fibroblast growth factor during limb regeneration in the axolotl (Ambystoma mexicanum). Regeneration (Oxf). 2015;2: 182–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinovich GA, van Kooyk Y, Cobb BA. Glycobiology of immune responses. Annals of the New York Academy of Sciences. 2012. pp. 1–15. doi: 10.1111/j.1749-6632.2012.06492.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina-Cano D, Ucuncu E, Nguyen LS, Nicouleau M, Lipecka J, Bizot J-C, et al. High N-glycan multiplicity is critical for neuronal adhesion and sensitizes the developing cerebellum to N-glycosylation defect. Elife. 2018;7. doi: 10.7554/eLife.38309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tecle E, Reynoso HS, Wang R, Gagneux P. The female reproductive tract contains multiple innate sialic acid-binding immunoglobulin-like lectins (Siglecs) that facilitate sperm survival. J Biol Chem. 2019;294: 11910–11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaramela LS, Martino C, Alisson-Silva F, Rees SD, Diaz SL, Chuzel L, et al. Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates. Nat Microbiol. 2019;4: 2082–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart GW. Nutrient regulation of signaling and transcription. Journal of Biological Chemistry. 2019. pp. 2211–2231. doi: 10.1074/jbc.aw119.003226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jefferson T, Jones MA, Doshi P, Del Mar CB, Hama R, Thompson MJ, et al. Neuraminidase inhibitors for preventing and treating influenza in adults and children. Cochrane Database Syst Rev. 2014. [cited 12 Dec 2019]. doi: 10.1002/14651858.CD008965.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9: 21–32. [DOI] [PubMed] [Google Scholar]
- 34.Kim SY, Jin W, Sood A, Montgomery DW, Grant OC, Fuster MM, et al. Glycosaminoglycan binding motif at S1/S2 proteolytic cleavage site on spike glycoprotein may facilitate novel coronavirus (SARS-CoV-2) host cell entry. bioRxiv. 2020. p. 2020.04.14.041459. doi: 10.1101/2020.04.14.041459 [DOI] [Google Scholar]
- 35.Liu L, Chopra P, Li X, Wolfert MA, Mark Tompkins S, Boons G-J. SARS-CoV-2 spike protein binds heparan sulfate in a length- and sequence-dependent manner. bioRxiv. 2020. p. 2020.05.10.087288. doi: 10.1101/2020.05.10.087288 [DOI] [Google Scholar]
- 36.Morrow AL, Meinzen-Derr J, Huang P, Schibler KR, Cahill T, Keddache M, et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. J Pediatr. 2011; 158: 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Autran CA, Kellman BP, Kim JH, Asztalos E, Blood AB, Spence ECH, et al. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut. 2018;67: 1064–1070. [DOI] [PubMed] [Google Scholar]
- 38.Bagdonaite I, Vakhrushev SY, Joshi HJ, Wandall HH. Viral glycoproteomes: technologies for characterization and outlook for vaccine design. FEBS Lett. 2018;592: 3898–3920. [DOI] [PubMed] [Google Scholar]
- 39.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010; 123: 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bar DZ, Atkatsh K, Tavarez U, Erdos MR, Gruenbaum Y, Collins FS. Biotinylation by antibody recognition—a method for proximity labeling. Nat Methods. 2018; 15: 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux KJ, Kim DI, Burke B, May DG. BioID: A Screen for Protein-Protein Interactions. Curr Protoc Protein Sci. 2018;91: 19.23.1–19.23.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drescher DG, Selvakumar D, Drescher MJ. Analysis of Protein Interactions by Surface Plasmon Resonance. Adv Protein Chem Struct Biol. 2018;110: 1–30. [DOI] [PubMed] [Google Scholar]
- 43.Stynen B, Tournu H, Tavernier J, Van Dijck P. Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol Mol Biol Rev. 2012;76: 331–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lobingier BT, Huttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, et al. An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell. 2017; 169: 350–360.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu G, Neelamegham S. A computational framework for the automated construction of glycosylation reaction networks. PLoS One. 2014;9: e100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kastelic M, Kopač D, Novak U, Likozar B. Dynamic metabolic network modeling of mammalian Chinese hamster ovary (CHO) cell cultures with continuous phase kinetics transitions. Biochem Eng J. 2019; 142: 124–134. [Google Scholar]
- 47.Losfeld M-E, Scibona E, Lin C-W, Villiger TK, Gauss R, Morbidelli M, et al. Influence of protein/glycan interaction on site-specific glycan heterogeneity. FASEB J. 2017;31: 4623–4635. [DOI] [PubMed] [Google Scholar]
- 48.Krambeck FJ, Betenbaugh MJ. A mathematical model of N-linked glycosylation. Biotechnol Bioeng. 2005;92: 711–728. [DOI] [PubMed] [Google Scholar]
- 49.Jedrzejewski PM, del Val IJ, Constantinou A, Dell A, Haslam SM, Polizzi KM, et al. Towards controlling the glycoform: a model framework linking extracellular metabolites to antibody glycosylation. Int J Mol Sci. 2014; 15: 4492–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu G, Marathe DD, Matta KL, Neelamegham S. Systems-level modeling of cellular glycosylation reaction networks: O-linked glycan formation on natural selectin ligands. Bioinformatics. 2008;24: 2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villiger TK, Scibona E, Stettler M, Broly H, Morbidelli M, Soos M. Controlling the time evolution of mAb N-linked glycosylation - Part II: Model-based predictions. Biotechnol Prog. 2016;32: 1135–1148. [DOI] [PubMed] [Google Scholar]
- 52.McDonald AG, Hayes JM, Bezak T, Głuchowska SA, Cosgrave EFJ, Struwe WB, et al. Galactosyltransferase 4 is a major control point for glycan branching in N-linked glycosylation. J Cell Sci. 2014;127: 5014–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald AG, Tipton KF, Davey GP. A mechanism for bistability in glycosylation. PLoS Comput Biol. 2018;14: e1006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Val IJ, Polizzi KM, Kontoravdi C. A theoretical estimate for nucleotide sugar demand towards Chinese Hamster Ovary cellular glycosylation. Sci Rep. 2016;6: 28547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez JM, Feizi A, Li S, Kallehauge TB, Hefzi H, Grav LM, et al. Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion. Nat Commun. 2020;11. doi: 10.1038/s41467-019-13867-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puri A, Neelamegham S. Understanding glycomechanics using mathematical modeling: a review of current approaches to simulate cellular glycosylation reaction networks. Ann Biomed Eng. 2012;40: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutter S, Villiger TK, Brühlmann D, Stettler M, Broly H, Soos M, et al. Glycosylation flux analysis reveals dynamic changes of intracellular glycosylation flux distribution in Chinese hamster ovary fed-batch cultures. Metab Eng. 2017;43: 9–20. [DOI] [PubMed] [Google Scholar]
- 58.Spencer JL, Bernanke JA, Buczek-Thomas JA, Nugent MA. A computational approach for deciphering the organization of glycosaminoglycans. PLoS One. 2010;5: e9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, Lewis NE. A Markov chain model for N-linked protein glycosylation – towards a low-parameter tool for model-driven glycoengineering. Metabolic Engineering. 2016. pp. 52–66. doi: 10.1016/j.ymben.2015.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kellman BP, Richelle A, Yang J-YE, Chapla DG, Chiang AW-T, Najera J, et al. Elucidating Human Milk Oligosaccharide biosynthetic genes through network-based multi-omics integration. bioRxiv. 2020. Available: https://www.biorxiv.org/content/10.1101/2020.09.02.278663v1.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kremkow BG, Lee KH. Glyco-Mapper: A Chinese hamster ovary (CHO) genome-specific glycosylation prediction tool. Metab Eng. 2018;47: 134–142. [DOI] [PubMed] [Google Scholar]
- 62.Spahn PN, Hansen AH, Kol S, Voldborg BG, Lewis NE. Predictive glycoengineering of biosimilars using a Markov chain glycosylation model. Biotechnol J. 2017; 12. doi: 10.1002/biot.201600489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agravat SB, Song X, Rojsajjakul T, Cummings RD, Smith DF. Computational approaches to define a human milk metaglycome. Bioinformatics. 2016;32: 1471–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDonald AG, Tipton KF, Davey GP. A Knowledge-Based System for Display and Prediction of O-Glycosylation Network Behaviour in Response to Enzyme Knockouts. PLoS Comput Biol. 2016;12: e1004844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bieberich E, Yu RK. Multi-enzyme kinetic analysis of glycolipid biosynthesis. Biochim Biophys Acta. 1999; 1432: 113–124. [DOI] [PubMed] [Google Scholar]
- 66.Liang C, Chiang AWT, Hansen AH, Arnsdorf J, Schoffelen S, Sorrentino JT, et al. A Markov model of glycosylation elucidates isozyme specificity and glycosyltransferase interactions for glycoengineering. Curr Res Biotechnol. 2020;2: 22–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kotidis P, Kontoravdi C. Harnessing the potential of artificial neural networks for predicting protein glycosylation. Metabolic Engineering Communications. 2020; e00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin Y, Zhao L, Wang X, Tong D, Hoover C, Wu F, et al. MeCP2 regulated glycogenes contribute to proliferation and apoptosis of gastric cancer cells. Glycobiology. 2017;27: 306–317. [DOI] [PubMed] [Google Scholar]
- 69.Aco-Tlachi M, Carreño-López R, Martínez-Morales PL, Maycotte P, Aguilar-Lemarroy A, Jave-Suárez LF, et al. Glycogene expression profiles based on microarray data from cervical carcinoma HeLa cells with partially silenced E6 and E7 HPV oncogenes. Infect Agent Cancer. 2018; 13: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suga A, Yamanishi Y, Hashimoto K, Goto S, Kanehisa M. AN IMPROVED SCORING SCHEME FOR PREDICTING GLYCAN STRUCTURES FROM GENE EXPRESSION DATA. Genome Informatics 2007. 2007. doi: 10.1142/9781860949920_0023 [DOI] [PubMed] [Google Scholar]
- 71.Frenkel-Pinter M, Shmueli MD, Raz C, Yanku M, Zilberzwige S, Gazit E, et al. Interplay between protein glycosylation pathways in Alzheimer’s disease. Sci Adv. 2017;3: e1601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Greville G, McCann A, Rudd PM, Saldova R. Epigenetic regulation of glycosylation and the impact on chemo-resistance in breast and ovarian cancer. Epigenetics. 2016; 11: 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horvat T, Deželjin M, Redžić I, Barišić D, Herak Bosnar M, Lauc G, et al. Reversibility of membrane N-glycome of HeLa cells upon treatment with epigenetic inhibitors. PLoS One. 2013;8: e54672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vojta A, Samaržija I, Bočkor L, Zoldoš V. Glyco-genes change expression in cancer through aberrant methylation. Biochim Biophys Acta. 2016;1860: 1776–1785. [DOI] [PubMed] [Google Scholar]
- 75.Agrawal P, Kurcon T, Pilobello KT, Rakus JF, Koppolu S, Liu Z, et al. Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. Proc Natl Acad Sci U S A. 2014; 111: 4338–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kurcon T, Liu Z, Paradkar AV, Vaiana CA, Koppolu S, Agrawal P, et al. miRNA proxy approach reveals hidden functions of glycosylation. Proc Natl Acad Sci U S A. 2015; 112: 7327–7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eichler J, Koomey M. Sweet New Roles for Protein Glycosylation in Prokaryotes. Trends Microbiol. 2017;25: 662–672. [DOI] [PubMed] [Google Scholar]
- 78.Sharapov SZ, Tsepilov YA, Klaric L, Mangino M, Thareja G, Shadrina AS, et al. Defining the genetic control of human blood plasma N-glycome using genome-wide association study. Hum Mol Genet. 2019;28: 2062–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bao B, Kellman BP, Chiang AWT, York AK, Mohammad MA, Haymond MW, et al. Correcting for sparsity and non-independence in glycomic data through a systems biology framework. bioRxiv. 2019. p. 693507. doi: 10.1101/693507 [DOI] [Google Scholar]
- 80.Rademacher C, Paulson JC. Glycan fingerprints: calculating diversity in glycan libraries. ACS Chem Biol. 2012;7: 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robin T, Mariethoz J, Lisacek F. Examining and fine-tuning the selection of glycan compositions with GlyConnect Compozitor. Mol Cell Proteomics. 2020. doi: 10.1074/mcp.RA120.002041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hosoda M, Takahashi Y, Shiota M, Shinmachi D, Inomoto R, Higashimoto S, et al. MCAW-DB: A glycan profile database capturing the ambiguity of glycan recognition patterns. Carbohydr Res. 2018;464: 44–56. [DOI] [PubMed] [Google Scholar]
- 83.Tang H, Hsueh P, Kletter D, Bern M, Haab B. The detection and discovery of glycan motifs in biological samples using lectins and antibodies: new methods and opportunities. Adv Cancer Res. 2015;126: 167–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cholleti SR, Agravat S, Morris T, Saltz JH, Song X, Cummings RD, et al. Automated motif discovery from glycan array data. OMICS. 2012;16: 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Agravat SB, Saltz JH, Cummings RD, Smith DF. GlycoPattern: a web platform for glycan array mining. Bioinformatics. 2014;30: 3417–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grant OC, Xue X, Ra D, Khatamian A, Foley BL, Woods RJ. Gly-Spec: a webtool for predicting glycan specificity by integrating glycan array screening data and 3D structure. Glycobiology. 2016. pp. 1027–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alocci D, Ghraichy M, Barletta E, Gastaldello A, Mariethoz J, Lisacek F. Understanding the glycome: an interactive view of glycosylation from glycocompositions to glycoepitopes. Glycobiology. 2018;28: 349–362. [DOI] [PubMed] [Google Scholar]
- 88.Klein J, Carvalho L, Zaia J. Application of network smoothing to glycan LC-MS profiling. Bioinformatics. 2018;34: 3511–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choo MS, Wan C, Rudd PM, Nguyen-Khuong T. GlycopeptideGraphMS: Improved Glycopeptide Detection and Identification by Exploiting Graph Theoretical Patterns in Mass and Retention Time. Anal Chem. 2019;91: 7236–7244. [DOI] [PubMed] [Google Scholar]
- 90.Hong P, Sun H, Sha L, Pu Y, Khatri K, Yu X, et al. GlycoDeNovo – an Efficient Algorithm for Accurate de novo Glycan Topology Reconstruction from Tandem Mass Spectra. Journal of The American Society for Mass Spectrometry. 2017. pp. 2288–2301. doi: 10.1007/s13361-017-1760-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klamer Z, Staal B, Prudden AR, Liu L, Smith DF, Boons G-J, et al. Mining High-Complexity Motifs in Glycans: A New Language To Uncover the Fine Specificities of Lectins and Glycosidases. Anal Chem. 2017;89: 12342–12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kellman BP, Zhang Y, Logomasini E, Meinhardt E, Chiang AWT, Sorrentino JT, et al. A consensus-based and readable extension of Linear Code for Reaction Rules (LiCoRR). bioRxiv. bioRxiv; 2020. p. 2020.05.31.126623. doi: 10.1101/2020.05.31.126623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ashwood C, Pratt B, MacLean BX, Gundry RL, Packer NH. Standardization of PGC-LC-MS-based glycomics for sample specific glycotyping. Analyst. 2019; 144: 3601–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benedetti E, Pučić-Baković M, Keser T, Wahl A, Hassinen A, Yang J-Y, et al. Network inference from glycoproteomics data reveals new reactions in the IgG glycosylation pathway. Nat Commun. 2017;8: 1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bojar D, Camacho DM, Collins JJ. Using Natural Language Processing to Learn the Grammar of Glycans. bioRxiv. 2020. p. 2020.01.10.902114. doi: 10.1101/2020.01.10.902114 [DOI] [Google Scholar]
- 96.Wong NSC, Wati L, Nissom PM, Feng HT, Lee MM, Yap MGS. An investigation of intracellular glycosylation activities in CHO cells: effects of nucleotide sugar precursor feeding. Biotechnol Bioeng. 2010; 107: 321–336. [DOI] [PubMed] [Google Scholar]
- 97.Jimenez del Val I, Nagy JM, Kontoravdi C. A dynamic mathematical model for monoclonal antibody N-linked glycosylation and nucleotide sugar donor transport within a maturing Golgi apparatus. Biotechnol Prog. 2011;27: 1730–1743. [DOI] [PubMed] [Google Scholar]
- 98.Silverman JM, Imperiali B. Bacterial N-Glycosylation Efficiency Is Dependent on the Structural Context of Target Sequons. J Biol Chem. 2016;291: 22001–22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thaysen-Andersen M, Packer NH. Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology. 2012;22: 1440–1452. [DOI] [PubMed] [Google Scholar]
- 100.Loke I, Østergaard O, Heegaard NHH, Packer NH, Thaysen-Andersen M. Paucimannose-Rich N-glycosylation of Spatiotemporally Regulated Human Neutrophil Elastase Modulates Its Immune Functions. Mol Cell Proteomics. 2017; 16: 1507–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miura K, Hakamata W, Tanaka A, Hirano T, Nishio T. Discovery of human Golgi β-galactosidase with no identified glycosidase using a QMC substrate design platform for exo-glycosidase. Bioorg Med Chem. 2016;24: 1369–1375. [DOI] [PubMed] [Google Scholar]
- 102.Lee C-G, Oh MJ, Park S-Y, An HJ, Kim JH. Inhibition of poly-LacNAc biosynthesis with release of CMP-Neu5Ac feedback inhibition increases the sialylation of recombinant EPO produced in CHO cells. Sci Rep. 2018;8: 7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dong X, Huang Y, Cho BG, Zhong J, Gautam S, Peng W, et al. Advances in mass spectrometry-based glycomics. Electrophoresis. 2018;39: 3063–3081. [DOI] [PubMed] [Google Scholar]
- 104.Klamer Z, Hsueh P, Ayala-Talavera D, Haab B. Deciphering Protein Glycosylation by Computational Integration of On-chip Profiling, Glycan-array Data, and Mass Spectrometry. Mol Cell Proteomics. 2019; 18: 28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Angel PM, Mehta A, Norris-Caneda K, Drake RR. MALDI Imaging Mass Spectrometry of N-glycans and Tryptic Peptides from the Same Formalin-Fixed, Paraffin-Embedded Tissue Section. Methods Mol Biol. 2018; 1788: 225–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huffman JE, Pucic-Bakovic M, Klaric L, Hennig R, Selman MHJ, Vuckovic F, et al. Comparative performance of four methods for high-throughput glycosylation analysis of immunoglobulin G in genetic and epidemiological research. Mol Cell Proteomics. 2014;13: 1598–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adamczyk B, Stockmann H, O’Flaherty R, Karlsson NG, Rudd PM. High-Throughput Analysis of the Plasma N-Glycome by UHPLC. Methods Mol Biol. 2017;1503: 97–108. [DOI] [PubMed] [Google Scholar]
- 108.Pucic M, Knezevic A, Vidic J, Adamczyk B, Novokmet M, Polasek O, et al. High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol Cell Proteomics. 2011; 10: M111.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reiding KR, Bondt A, Hennig R, Gardner RA, O’Flaherty R, Trbojevic-Akmacic I, et al. High-throughput Serum N-Glycomics: Method Comparison and Application to Study Rheumatoid Arthritis and Pregnancy-associated Changes. Mol Cell Proteomics. 2019; 18: 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, et al. Shotgun glycomics: a microarray strategy for functional glycomics. Nat Methods. 2011;8: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Geissner A, Reinhardt A, Rademacher C, Johannssen T, Monteiro J, Lepenies B, et al. Microbe-focused glycan array screening platform. Proc Natl Acad Sci USA. 2019;116: 1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, et al. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell. 2014;159: 1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang ML, Cohen M, Fisher CJ, Schooley RT, Gagneux P, Godula K. Determination of receptor specificities for whole influenza viruses using multivalent glycan arrays. Chem Commun . 2015;51: 5326–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6: 985–989. [DOI] [PubMed] [Google Scholar]
- 115.Zou X, Yoshida M, Nagai-Okatani C, Iwaki J, Matsuda A, Tan B, et al. A standardized method for lectin microarray-based tissue glycome mapping. Sci Rep. 2017;7: 43560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shang Y, Zeng Y, Zeng Y. Integrated Microfluidic Lectin Barcode Platform for High-Performance Focused Glycomic Profiling. Sci Rep. 2016;6: 20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scott E, Munkley J. Glycans as Biomarkers in Prostate Cancer. Int J Mol Sci. 2019;20: 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3: 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tiemeyer M, Aoki K, Paulson J, Cummings RD, York WS, Karlsson NG, et al. GlyTouCan: an accessible glycan structure repository. Glycobiology. 2017;27: 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Toukach PV, Egorova KS. Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res. 2016;44: D1229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rojas-Macias MA, Mariethoz J, Andersson P, Jin C, Venkatakrishnan V, Aoki NP, et al. Towards a standardized bioinformatics infrastructure for N-and O-glycomics. Nat Commun. 2019; 10: 3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Campbell MP, Nguyen-Khuong T, Hayes CA, Flowers SA, Alagesan K, Kolarich D, et al. Validation of the curation pipeline of UniCarb-DB: building a global glycan reference MS/MS repository. Biochim Biophys Acta. 2014;1844: 108–116. [DOI] [PubMed] [Google Scholar]
- 123.Venkataraman M, Sasisekharan R, Raman R. Glycan array data management at Consortium for Functional Glycomics. Methods Mol Biol. 2015; 1273: 181–190. [DOI] [PubMed] [Google Scholar]
- 124.Alocci D, Mariethoz J, Gastaldello A, Gasteiger E, Karlsson NG, Kolarich D, et al. GlyConnect: Glycoproteomics Goes Visual, Interactive, and Analytical. J Proteome Res. 2019; 18: 664–677. [DOI] [PubMed] [Google Scholar]
- 125.Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, et al. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014;42: D215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Clerc O, Deniaud M, Vallet SD, Naba A, Rivet A, Perez S, et al. MatrixDB: integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2019;47: D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mariethoz J, Khatib K, Alocci D, Campbell MP, Karlsson NG, Packer NH, et al. SugarBindDB, a resource of glycan-mediated host-pathogen interactions. Nucleic Acids Res. 2016;44: D1243–D1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mariethoz J, Alocci D, Gastaldello A, Horlacher O, Gasteiger E, Rojas-Macias M, et al. Glycomics@ExPASy: Bridging the Gap. Mol Cell Proteomics. 2018; 17: 2164–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.York WS, Mazumder R, Ranzinger R, Edwards N, Kahsay R, Aoki-Kinoshita KF, et al. GlyGen: Computational and Informatics Resources for Glycoscience. Glycobiology. 2019. doi: 10.1093/glycob/cwz080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yamada I, Shiota M, Shinmachi D, Ono T, Tsuchiya S, Hosoda M, et al. The GlyCosmos Portal: a unified and comprehensive web resource for the glycosciences. Nat Methods. 2020. doi: 10.1038/s41592-020-0879-8 [DOI] [PubMed] [Google Scholar]
- 131.Aoki-Kinoshita KF, Lisacek F, Mazumder R, York WS, Packer NH. The GlySpace Alliance: towards a collaborative global glycoinformatics community. Glycobiology. 2019. doi: 10.1093/glycob/cwz078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Struwe WB, Agravat S, Aoki-Kinoshita KF, Campbell MP, Costello CE, Dell A, et al. The minimum information required for a glycomics experiment (MIRAGE) project: sample preparation guidelines for reliable reporting of glycomics datasets. Glycobiology. 2016;26: 907–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, et al. The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting mass-spectrometry-based glycoanalytic data. Mol Cell Proteomics. 2013; 12: 991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Y, McBride R, Stoll M, Palma AS, Silva L, Agravat S, et al. The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting glycan microarray-based data. Glycobiology. 2016. doi: 10.1093/glycob/cww118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Campbell MP, Abrahams JL, Rapp E, Struwe WB, Costello CE, Novotny M, et al. The minimum information required for a glycomics experiment (MIRAGE) project: LC guidelines. Glycobiology. 2019;29: 349–354. [DOI] [PubMed] [Google Scholar]
- 136.Sima AC, Mendes de Farias T, Zbinden E, Anisimova M, Gil M, Stockinger H, et al. Enabling semantic queries across federated bioinformatics databases. Database . 2019;2019. doi: 10.1093/database/baz106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ranzinger R, Aoki-Kinoshita KF, Campbell MP, Kawano S, Lütteke T, Okuda S, et al. GlycoRDF: an ontology to standardize glycomics data in RDF. Bioinformatics. 2015;31: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. Annotation of glycomics MS and MS/MS spectra using the GlycoWorkbench software tool. Methods Mol Biol. 2015;1273: 3–15. [DOI] [PubMed] [Google Scholar]
- 139.Liu M-Q, Zeng W-F, Fang P, Cao W-Q, Liu C, Yan G-Q, et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat Commun. 2017;8: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Klein J, Zaia J. glypy: An Open Source Glycoinformatics Library. J Proteome Res. 2019; 18: 3532–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu G, Cheng K, Lo CY, Li J, Qu J, Neelamegham S. A Comprehensive, Open-source Platform for Mass Spectrometry-based Glycoproteomics Data Analysis. Mol Cell Proteomics. 2017; 16: 2032–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reusch D, Haberger M, Falck D, Peter B, Maier B, Gassner J, et al. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles-Part 2: Mass spectrometric methods. MAbs. 2015;7: 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.De Leoz MLA, Duewer DL, Fung A, Liu L, Yau HK, Potter O, et al. NIST Interlaboratory Study on Glycosylation Analysis of Monoclonal Antibodies: Comparison of Results from Diverse Analytical Methods. Mol Cell Proteomics. 2020; 19: 11–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hou W, Qiu Y, Hashimoto N, Ching W-K, Aoki-Kinoshita KF. A systematic framework to derive N-glycan biosynthesis process and the automated construction of glycosylation networks. BMC Bioinformatics. 2016;17 Suppl 7: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Franks JM, Cai G, Whitfield ML. Feature specific quantile normalization enables cross-platform classification of molecular subtypes using gene expression data. Bioinformatics. 2018;34: 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32: 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016;49: 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Salza R, Lethias C, Ricard-Blum S. The Multimerization State of the Amyloid-β42 Peptide (Aβ42) Governs its Interaction Network with the Extracellular Matrix. Journal of Alzheimer’s Disease. 2017. pp. 991–1005. doi: 10.3233/jad-160751 [DOI] [PubMed] [Google Scholar]
- 149.Coff L, Chan J, Ramsland PA, Guy AJ. Identifying glycan motifs using a novel subtree mining approach. BMC Bioinformatics. 2020;21: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mehta AY, Cummings RD. GLAD: GLycan Array Dashboard, a Visual Analytics Tool for Glycan Microarrays. Bioinformatics. 2019. doi: 10.1093/bioinformatics/btz075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ielasi FS, Alioscha-Perez M, Donohue D, Claes S, Sahli H, Schols D, et al. Lectin-Glycan Interaction Network-Based Identification of Host Receptors of Microbial Pathogenic Adhesins. MBio. 2016;7. doi: 10.1128/mBio.00584-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bonnardel F, Mariethoz J, Salentin S, Robin X, Schroeder M, Perez S, et al. UniLectin3D, a database of carbohydrate binding proteins with curated information on 3D structures and interacting ligands. Nucleic Acids Res. 2019;47: D1236–D1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Naticchia MR, Laubach LK, Tota EM, Lucas TM, Huang ML, Godula K. Embryonic Stem Cell Engineering with a Glycomimetic FGF2/BMP4 Co-Receptor Drives Mesodermal Differentiation in a Three-Dimensional Culture. ACS Chem Biol. 2018; 13: 2880–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huang ML, Tota EM, Lucas TM, Godula K. Influencing Early Stages of Neuromuscular Junction Formation through Glycocalyx Engineering. ACS Chem Neurosci. 2018;9: 3086–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Moremen KW. Golgi α-mannosidase II deficiency in vertebrate systems: implications for asparagine-linked oligosaccharide processing in mammals. Biochimica et Biophysica Acta (BBA) - General Subjects. 2002; 1573: 225–235. [DOI] [PubMed] [Google Scholar]
- 156.Crispin M, Chang VT, Harvey DJ, Dwek RA. A human embryonic kidney 293T cell line mutated at the Golgi α-mannosidase II locus. Journal of Biological. 2009. Available: http://www.jbc.org/content/284/32/21684.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hsiao C-T, Cheng H-W, Huang C-M, Li H-R, Ou M-H, Huang J-R, et al. Fibronectin in cell adhesion and migration via N-glycosylation. Oncotarget. 2017;8: 70653–70668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weiss RJ, Esko JD, Tor Y. Targeting heparin and heparan sulfate protein interactions. Org Biomol Chem. 2017; 15: 5656–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Johns SC, Yin X, Jeltsch M, Bishop JR, Schuksz M, El Ghazal R, et al. Functional Importance of a Proteoglycan Coreceptor in Pathologic Lymphangiogenesis. Circ Res. 2016;119: 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Otsuki S, Hanson SR, Miyaki S, Grogan SP, Kinoshita M, Asahara H, et al. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc Natl Acad Sci U S A. 2010; 107: 10202–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sinitskiy AV, Pande VS. Simulated Dynamics of Glycans on Ligand-Binding Domain of NMDA Receptors Reveals Strong Dynamic Coupling between Glycans and Protein Core. J Chem Theory Comput. 2017;13: 5496–5505. [DOI] [PubMed] [Google Scholar]
- 162.Margraf-Schönfeld S, Böhm C, Watzl C. Glycosylation affects ligand binding and function of the activating natural killer cell receptor 2B4 (CD244) protein. J Biol Chem. 2011. ;286: 24142–24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, et al. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human FcγRIII and Antibody-dependent Cellular Toxicity. J Biol Chem. 2002;277: 26733–26740. [DOI] [PubMed] [Google Scholar]
- 164.Seifert A, Glanz D, Glaubitz N, Horstkorte R, Bork K. Polysialylation of the neural cell adhesion molecule: interfering with polysialylation and migration in neuroblastoma cells. Arch Biochem Biophys. 2012;524: 56–63. [DOI] [PubMed] [Google Scholar]