Abstract
Aims:
Comparative genomics analyses indicated that the Flavobacterium columnare genome has unique denitrification genes relative to Flavobacterium psychrophilum and Flavobacterium johnsoniae, including nasA (nitrate reductase), nirS (nitrite reductase), norB (nitric oxide reductase), and nosZ (nitrous oxide reductase). The current study determines the roles of nasA, nirS, norB, and nosZ in anaerobic growth, nitrate reduction, biofilm formation, and virulence.
Methods and Results:
Four in-frame deletion mutants in virulent F. columnare strain 94–081 were constructed by allelic exchange using pCP29 plasmid. Compared with parent strain 94–081, FcΔnasA, FcΔnirS, and FcΔnosZ mutants did not grow as well anaerobically, whereas the growth of FcΔnorB strain was similar to the parent strain (FcWT). Exogenous nitrate was not significantly consumed under anaerobic conditions in FcΔnasA, FcΔnirS, and FcΔnosZ compared to parent strain 94–081. Under anaerobic conditions, FcΔnasA, FcΔnorB, and FcΔnosZ formed significantly less biofilm than the wild type strain at 24 and 96 hours, but FcΔnirS was not significantly affected. The nitrite reductase mutant FcΔnirS was highly attenuated in catfish, whereas FcΔnasA, FcΔnorB, and FcΔnosZ had similar virulence to FcWT.
Conclusions:
These results show, for the first time, that denitrification genes enable F. columnare to grow anaerobically using nitrate as an electron acceptor, and nitrite reductase contributes to F. columnare virulence.
Significance and Impact of the Study:
These findings indicate potential for F. columnare to grow in nitrate-rich anaerobic zones in catfish production ponds, and they suggest that a FcΔnirS strain could be useful as a safe live vaccine if it protects catfish against columnaris disease.
Keywords: Flavobacterium columnare, denitrification, anaerobic respiration, catfish, columnaris disease, aquaculture, virulence
Introduction
Flavobacterium columnare is a long Gram-negative rod causing columnaris disease, which affects numerous fish species worldwide (Wagner et al. 2006). Six genomovar types have been described among F. columnare isolates (I, II, II-B, III, I/II, and II-A) (LaFrentz et al. 2014; Garcia et al. 2018). More recently, F. columnare isolates were reclassified into four genetic groups based on 16S rRNA gene sequences (LaFrentz et al. 2018). Genetic group 2 is the most virulent for channel catfish (Ictalurus punctatus) (Triyanto and Wakabayashi 1999; Shoemaker et al. 2008). In the United States, F. columnare is responsible for significant economic losses in channel catfish aquaculture (Wagner et al. 2002). Infected catfish often exhibit external lesions on the body surface, gills, and fins. F. columnare may cause chronic infection with low-level mortalities, or it may cause acute infection with mortalities occurring within a few days (Declercq et al. 2013; Mohammed and Arias 2014). Host stress (e.g., low oxygen, high nitrite and ammonia, elevated water temperature, mechanical injury, or crowding) enhances the occurrence and severity of columnaris disease. Minimizing fish stress helps prevent columnaris disease outbreaks, but the ubiquitous presence of this pathogen in aquatic environments makes eradication of the disease in aquaculture systems difficult.
F. columnare is capable of surviving for extended periods in pond water, sediments, and mud slurry (Declercq et al. 2013; Mohammed and Arias 2014). It is common for pond sediment and water at the bottom of ponds to be anaerobic. It is also common for commercial production ponds to be rich in nitrogenous compounds due to stocking density and feeding. Also, in natural habitats, F. columnare often grow as biofilms, which create oxygen-limited conditions (Cai et al. 2013). In addition, during infection and ulcer formation in fish tissue, bacteria encounter microenvironments of reduced oxygen concentration. When oxygen is not available, alternative electron acceptors, including nitrate, nitrite, or nitrous oxide, can be used by some bacteria (Filiatrault et al. 2006).
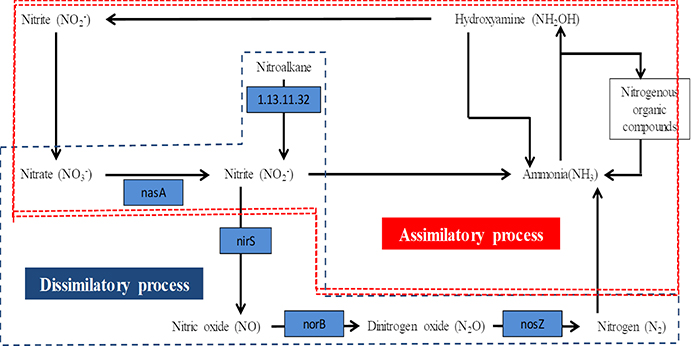
Comparison of the F. columnare genome with F. psychrophilum and F. johnsoniae showed that F. columnare encodes four enzymes capable of reducing nitrate to nitrogen gas through the denitrification pathway (Figure 1). In sequential order, the four enzymes are nitrate reductase (encoded by nasA), which reduces nitrate to nitrite; nitrite reductase (nirS), which reduces nitrite to nitric oxide (Kawasaki et al. 1997); nitric oxide reductase (norB), which reduces nitric oxide to nitrous oxide (Schreiber et al. 2007); and nitrous oxide reductase (nosZ) for reduction of nitrous oxide to inert gaseous nitrogen (Zumft et al. 1990). F. columnare is capable of anaerobic growth in the presence of sodium nitrate (Tekedar et al. 2017). This suggests that F. columnare is capable of using nitrogenous compounds as alternate electron acceptors for anaerobic respiration, which would enable F. columnare to replicate in the nitrogen-rich anaerobic pond sediments of catfish production ponds, potentially serving as an environmental source of infection in catfish ponds.
Figure 1.
Scheme for denitrification pathway from nitrate to nitrogen. Genes encoding enzymes that mediate the denitrification steps of F. columnare 94–081 include those for nitrate reductase (nasA), nitrite reductase (nirS), nitric oxide reductase (norB), and nitrous oxide reductase (nosZ). Blue highlights show enzymes encoded by F. columnare 94–081 strain 94–081 genes.
Some pathogenic bacteria are capable of denitrification (Philippot 2005). The denitrification pathway has been linked to virulence of several species such as Mycobacterium bovis, Brucella melitensis, Neisseria meningitidis, and Pseudomonas aeruginosa (Weber et al. 2000). Furthermore, inactivation of the denitrification pathway hindered biofilm formation in P. aeruginosa and Neisseria gonorrhoeae (Van Alst et al. 2007; Falsetta et al. 2009). In N. meningitidis, active denitrification was linked to modulation of host cytokine responses, enhanced intracellular survival, and inhibition of apoptosis in a macrophage model (Stevanin et al. 2005; Stevanin et al. 2007), and denitrification prevented the establishment of anti-inflammatory nitric oxide steady-state levels (Barth and Clark 2008).
Therefore, the current study was undertaken to determine the role of four genes (nasA, nirS, norB, and nosZ) in F. columnare anaerobic growth, nitrate respiration, and biofilm formation. We also determined whether these genes are required for F. columnare virulence in catfish fingerlings. F. columnare denitrification may be physiologically relevant to its survival and growth in aquaculture ponds or during infection of fish tissue.
MATERIALS AND METHODS
Ethics statement
All fish disease challenges were conducted in compliance with protocol #17–288 approved by the Mississippi State University Institutional Animal Care and Use Committee (IACUC). The approved protocol included humane endpoints, and when morbid fish met established criteria, they were immediately euthanized by immersion in tricaine methane sulfonate (MS-222). Criteria for euthanasia were loss of balance, hanging at the water surface, or non-responsiveness to external stimuli. Some of the fish died during the study as a result of the experimentally induced systemic infection due to its rapid progression. All personnel on this experiment received IACUC-approved training in animal care and welfare by the University Laboratory Animal Veterinarian.
Bacterial strains, growth conditions, and plasmid
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strain DH5αMCR was used for cloning, and E. coli BW19851 was used for transfer of pCP29 into F. columnare strain 94–081 (genetic group 2) (Kumru et al. 2016). E. coli strains were cultured in Luria Broth (LB) broth or agar at 37 °C. F. columnare strain 94–081 was grown at 30 °C in Flavobacterium columnare growth medium (FCGM) broth [tryptone (8.00 g), yeast extract (0.80 g), MgSO4 7 H2O (1.00 g), CaCl2 2 H2O (0.74 g), NaCl (5.00 g), and sodium citrate (1.50 g) per liter] or on FCGM agar (Dumpala et al. 2010). F. columnare strains were cultured aerobically with 5% CO2 or anaerobically in a jar with GasPak system (Fisher Scientific, PA, USA) at 30 °C. The pCP29 plasmid was modified and used to construct in-frame gene deletions by allelic exchange (Kempf and McBride 2000). Plasmid pCP29 carries cefoxitin (cfxA) and erythromycin (ermF) resistance genes that function in Flavobacterium (Kempf and McBride 2000). It also carries a beta lactamase gene (bla) that is used to maintain the plasmid in E. coli. Ampicillin was used at 100 μg/ml for plasmid maintenance in E. coli DH5αMCR during cloning steps. For selection of F. columnare transconjugants, cefoxitin was used at 10 μg/ml. Colistin (12.5 μg/ml) was used for counterselection against E. coli BW19851 following conjugation. pCP29 was converted into a homologous recombination-deletion mutagenesis vector by removal of the pCP1 fragment containing the origin that allows the plasmid to replicate in Flavobacterium species (Staroscik et al. 2008). This was accomplished by digesting with a combination of two restriction endonucleases (either SmaI and SphI or SmaI and PstI). The resulting 8 kb fragment was isolated by gel purification and used for ligation with overlap extension PCR fragments (described in the next section).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| E. coli strain | ||
| DH5αMCR | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA mcrA Δ(mrr-hsdRMS-mcrBC) | (Grant et al. 1990) |
| BW19851 | RecA TraRP4+ Tpr Strr Pir+ | (Metcalf et al. 1994) |
| F. columnare strain | ||
| 94–081 | Wild-type | (Soto et al. 2008) |
| FcΔnasA | 94–081 derivative, ΔnasA | This study |
| FcΔnirS | 94–081 derivative, ΔnirS | This study |
| FcΔnorB | 94–081 derivative, ΔnorB | This study |
| FcΔnosZ | 94–081 derivative, nosZ | This sutdy |
| Plasmid | ||
| pCP29 | E. coli-F. columnare shuttle plasmid; Apr (Cfr Emr) | (Kempf and McBride 2000) |
| pCP29S | Suicide plasmid derived from pCP29; Apr (Cfr Emr) | This study |
| pCP29S-nasA | pCP39S, ΔnasA | This study |
| pCP29S-nirS | pCP39S, ΔnirS | This study |
| pCP29S-norB | pCP39S, ΔnorB | This study |
| pCP29S-nosZ | pCP39S, ΔnosZ | This study |
Antibiotic resistance phenotypes: Apr, ampicillin resistant; Cfr, cefoxitin resistant; Emr, erythromycin resistant. The antibiotic resistance phenotypes in parentheses are those expressed in F. columnare but not in E. coli.
Construction of F. columnare nasA, nirS, norB, and nosZ mutants
Four in-frame deletion strains were constructed using the overlap extension PCR method as previously described (Horton et al. 1989). In brief, the nucleotide sequences of nasA (AWN65_05950), nirS (AWN65_04985), norB (AWN65_04995), and nosZ (AWN65_09420) from the F. columnare strain 94–081 genome (GenBank accession: CP013992.1) (Kumru et al. 2016) were used to design four primers (A, B, C, D) for each gene using Primer3 (Table 2). Restriction endonuclease recognition sequences were added to A and D primers for cloning, and the reverse complement of primer B was added to the 5’ end of primer C to enable overlap extension PCR. Amplicons flanking the target genes using AB and CD primers were amplified by PCR from F. columnare 94–081 genomic DNA. Then both fragments were mixed equally and used as a template in subsequent overlap extension PCR using A and D primers. The resulting fusion fragments for each individual gene were cleaned, digested with restriction endonucleases (SmaI and SphI for nasA and norB, and SmaI and PstI for nirS and nosZ), ligated into the 8 kb fragment isolated from pCP29, and transformed into E. coli DH5αMCR to generate pCP29S-nasA, pCP29S-nirS, pCP29S-norB, and pCP29S-nosZ.
Table 2.
Primers used in this study.
| Primer ID | Primer Sequence (5’ to 3’)a | REb | |
| nasAEF | A | CACCCGGGAAACGGCACAAGATAAATGG | SmaI |
| nasAIR | B | TCAGCGTGCAAACTAGAACC | |
| nasAIF | C | GGTTCTAGTTTGCACGCTGATAGATGGGGACCAAAATCTTC | |
| nasAER | D | CAGCATGCGGAATTGTGTATCGCACCTC | SphI |
| nasASeq | GCATCGGCTGGACATACATTT | ||
| nirSEF | A | AACCCGGGACTGGACAATGCAGGCTAAA | SmaI |
| nirSIR | B | GGCTAAAGAGGCCGAGAATA | |
| nirSIF | C | TATTCTCGGCCTCTTTAGCCATGGTTGCAGCTGTTAGAGC | |
| nirSER | D | AACTGCAGTCCATTGGCTGGTTAGGTTA | PstI |
| nirSSeq | GGCTAGACGTTTGCCTTGAC | ||
| norBEF | A | CACCCGGGCAAAAAGTTCTGGGTGCTTC | SmaI |
| norBIR | B | AAACCAATAGGCTACTTTTTGTG | |
| norBIF | C | CACAAAAAGTAGCCTATTGGTTTGGTAAAACGAACGATGAGG | |
| norBER | D | CAGCATGCTTATGATTTGTTCCATACAACGG | SphI |
| norBSeq | AATACCTCCCATGATTAATTC | ||
| nosZEF | A | AACCCGGGTTTCCATTCATTGGGCTTTT | SmaI |
| nosZIR | B | GTATCTCCCGCCAATAGTAAAG | |
| nosZIF | C | CTTTACTATTGGCGGGAGATACATCACCACTTACAGCATCGC | |
| nosZER | D | AACTGCAGCAAGCATTAAATGAAGACCCTG | PstI |
| nosZSeq | AGGGTACTGAGGGGCATCTA |
Bold letters at the 5’ end represent restriction enzyme (RE) recognition sequences. Bases preceding the RE recognition sequences were added to increase the RE efficiency. Underlined bases in primer C is the reverse complement of primer B.
RE stands for restriction enzyme recognition sequence sites incorporated into the 5’ end of the primer sequence.
These four plasmids were transferred from donor E. coli BW19851 into recipient F. columnare strain 94–081 by conjugation (Alvarez et al. 2004). Briefly, donor E. coli and recipient F. columnare were grown to mid-log phase, concentrated by centrifugation at 8,000 rpm for 10 min, and washed twice with FCGM medium. Concentrated donor and recipient bacteria were mixed at a ratio of 1:4 (based on pellet weight) and placed on a sterile nitrocellulose filter (Fisher Scientific) on an FCGM agar plate. After incubation overnight at 30 °C, the bacterial mixture was washed off the filter and resuspended in FCGM broth with a 1 ml syringe. The resuspended bacteria mixture was spread on FCGM agar containing 10 μg/ml of cefoxitin to select F. columnare transconjugants and 12.5 μg/ml colistin for counterselection. Isolated colonies were propagated in FCGM broth and incubated with shaking at 30°C overnight, and 1 ml was spread on FCGM agar with colistin to allow for a second allelic exchange event. Colonies were screened for cefoxitin sensitivity, and putative cefoxitin sensitive mutant colonies were tested by PCR using A and D primers to discriminate between mutant and wild type alleles. PCR amplicons with size consistent with a gene deletion were confirmed by DNA sequencing. FcΔnasA had a deletion of 2,085 bp out of 2,214 bp (94.17%), FcΔnirS had a deletion of 1,374 bp out of 1,437 bp (95.54%), FcΔnorB had a deletion of 1,263 bp out of 1,329 bp (95.03%), and FcΔnosZ had a deletion of 1,806 bp out of 1,971 bp (91.62%) (Table 3).
Table 3.
The sizes of upstream (AB), downstream (CD), and deletion fragments.
| Gene | Locus tag | Protein name | UF (AB) (bp)a | DF (CD) (bp) | DR (bp / aa) | 5’ UD (bp / aa) | 3’ UD (bp / aa) |
|---|---|---|---|---|---|---|---|
| nasA | AWN65_05950 | Nitrate reductase | 1101 | 969 | 2085 / 695 | 72 / 24 | 57 / 19 |
| nirS | AWN65_04985 | nitrite reductase, copper-containing | 951 | 1029 | 1374 / 458 | 36 / 12 | 27 / 9 |
| norB | AWN65_05000 | Nitric oxide reductase | 1035 | 975 | 1263/421 | 36/12 | 30/10 |
| nosZ | AWN65_09420 | Nitrous oxide reductase | 1062 | 1092 | 1806 / 602 | 66/ 22 | 99/ 33 |
UF = Upstream fragment; DF = Downstream fragment; DR = Deleted region; UD = Undeleted region; bp = base pair; aa = amino acid.
Growth of mutants
Anaerobic and aerobic growth of wild-type F. columnare (FcWT) and mutants (FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ) were compared in FCGM supplemented with 10 mM sodium nitrate (FCGM-N). Starter cultures were prepared by inoculating FCGM broth from agar plates and aerobic incubation at 30°C with shaking (220 rpm). After reaching stationary phase, cultures were added to anaerobically preconditioned FCGM-N broth in a 10% v/v proportion. FCGM-N broth without bacteria was included as a negative control. Cultures were incubated under anaerobic conditions at 30°C using a GasPak anaerobic system in a standard anaerobic jar. Bacterial growth was measured by monitoring optical density (OD600) in triplicate at 1, 7, and 14 days post-inoculation. Aerobic growth was compared using FCGM-N broth with shaking.
Addition of extracellular nitrate and nitrite under anaerobic conditions
Nitrate and nitrite concentrations were measured during anaerobic growth of the FcWT and mutant strains to determine the amount of anaerobic respiration that had occurred. For each strain, triplicate anaerobic cultures were inoculated in FCGM-N as described above. FCGM-N broth without bacterial inoculation was included as control. For each sample, both nitrate and nitrite were determined using a commercial kit (Colorimetric Nitrite/Nitrate Assay Kit, Sigma Aldrich) that is based on the Griess reaction (Griess 1879) with some modification. Serially diluted sodium nitrite was used to obtain a standard curve.
To determine nitrate concentration, the nitrate+nitrite concentration was determined, and nitrite concentration was then subtracted. For nitrate+nitrite concentration, supernatant was collected from cultured FcWT and mutants (FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ) following centrifugation, and 80 μl was transferred to a 96-well plate in triplicate (Corning, NY, USA). After adding culture supernatants, 10 μl of enzyme cofactor and 10 μl of nitrate reductase (Cayman chemical) were added to each sample and incubated for one hour at room temperature (protected from light). Then 50 μl of sulfanilamide solution (1% sulfanilamide in 5% phosphoric acid) was added to each well and incubated in the dark for 10 min at room temperature. Following incubation, 50 μl of NED solution (0.1% N-1-naphthyl ethylenediamine dihydrochloride in water) was dispensed to each well and incubated for an additional 10 min at room temperature without light. Absorbance was measured at 540 nM using a SpectraMax M5 ELISA reader (Molecular Devices, Sunnyvale, CA, USA).
For nitrite concentration, 100 μl of diluted FcWT and mutant strain culture supernatant was added to triplicate wells, and 50 μl of sulfanilamide solution and 50 μl of NED solution was subsequently dispensed to each well. Color was allowed to develop for 10 min at room temperature, and absorbance at 540 nm was measured using a SpectraMax M5 ELISA reader.
Quantification of biofilm formation
The ability of FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ to form biofilm was compared to FcWT using a crystal violet staining technique as previously described with minor modifications (Cai et al. 2013). Briefly, FcWT and the mutant strains were grown overnight in FCGM at 30 °C before being subcultured by diluting at 1:100 into the wells of a microtiter plate (Costar, USA) under aerobic and anaerobic conditions for 24 and 96 hours. Wells with uninoculated FCGM medium were included as a negative control. Plates were incubated at 30 °C for 24 or 96 hours. Then wells were gently washed two times with PBS to remove unbound bacteria and subsequently stained with 0.01% crystal violet (Sigma-Aldrich) for 10 min at room temperature. After incubation, plates were rinsed with PBS and dried at room temperature followed by elution with 70% ethanol. The absorbance at OD538 nm was measured using a spectrophotometer (Biotek Synergy Mx, USA). The level of the biofilm formation was determined by subtracting the mean OD538 value of the negative control from the value of the test samples. The experiment was performed in four replicates for each strain and repeated two independent time.
Virulence of FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ in catfish
Approximately 480 eight-month-old specific pathogen free (SPF) channel catfish fingerlings (20 to 25 g) were stocked into 24 40 L flow-through tanks (flow rate: 1-l/ min) with a stocking density of 20 fish/tank and acclimated for one week. Fingerlings were obtained from the Mississippi State University College of Veterinary Medicine SPF Laboratory. Catfish in the SPF Laboratory are free from known obligate pathogens and parasites. Fish are hatched from disinfected eggs and maintained in indoor facilities with appropriate biosecurity protocols and monitoring for pathogens and parasites. Fish were fed to satiation twice daily and monitored three times daily for morbidity and mortality. Chlorine, dissolved oxygen, and temperature were monitored daily. Each treatment had four replicate tanks. Treatments consisted of FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ, FcWT (positive control), and FCGM (sham control). Bacterial strains were cultivated in FCGM broth overnight at 30°C. Absorbance at 600 nm (OD600) was measured, and culture densities were adjusted with sterile medium as needed to a standard absorbance. Water volume in tanks was adjusted to 10 L, and then 100 ml of appropriate bacterial culture was added to each tank (final dose of 2.45×107 CFU/ml of water). Channel catfish were infected by immersion for 6 hours, followed by gradual removal of bacteria. Mortalities were recorded daily for 21 days following challenge. F. columnare was confirmed as cause of death based on clinical signs and by culturing gills and skin swabs on FCGM agar from each mortality.
Statistical analysis
Linear regression analysis in PROC MIXED of SAS for Windows 9.4 (SAS Institute, Inc., Cary, NC, USA) was used for the anaerobic growth experiment, extracellular nitrate concentration, and biofilm formation assays where mutant strains were compared to FcWT. Separate models were developed for each analytical test using manual forward selection, and each included mutant type and time point as fixed effects. Replicates were included as a random effect. Adjustment for multiple comparisons were made using the SIMULATE option. Results are reported as least square means ± standard error. In the fish challenge experiments, mean percent mortality data were arcsine transformed, and analysis of variance (ANOVA) was applied using PROC GLM in SAS for Windows v9.4 (SAS Institute, Inc., Cary, NC) to assess significance. An alpha level of 0.05 was used in all analyses.
Results
Anaerobic growth characteristics of FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ
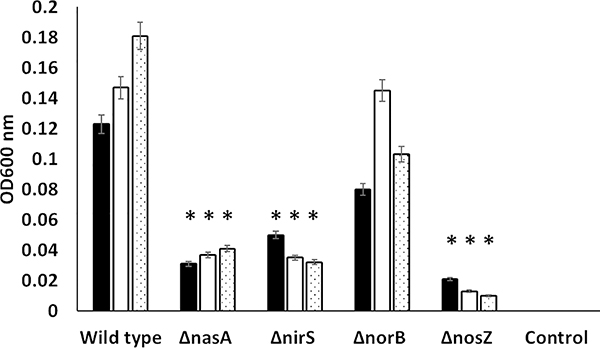
The growth of F. columnare wild-type and four mutant strains in FCGM broth supplemented with nitrate was compared under anaerobic conditions at 1, 7, and 14 days post-inoculation (Fig. 2). At all tested time points, significantly lower growth was observed in FcΔnasA, FcΔnirS, and FcΔnosZ as compared with FcWT. By contrast, anaerobic growth of FcΔnorB was not significantly decreased compared to FcWT strain with nitrate supplementation. There was no significant difference in the growth between mutant and wild type strains under aerobic condition (data not shown).
Figure 2.
Growth of FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ compared to FcWT in FCGM supplemented with 10 mM potassium nitrate (FCGM-N) under anaerobic conditions. Anaerobic conditions were generated in a jar using a standard anaerobic gas pack, and growth was measured on days 1, 7, and 14. Data points represent the mean of three replicates. Significant differences are indicated with asterisks (P < 0.05). Control group includes FCGM-N broth without bacterial inoculation.
Nitrate accumulation by FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ
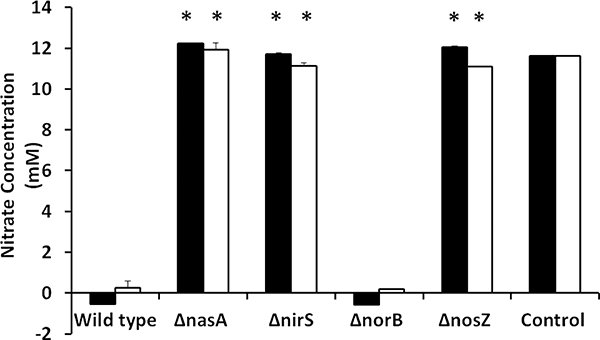
To determine whether F. columnare uses nitrate as an electron acceptor during anaerobic growth, we measured nitrate consumption during anaerobic growth. We also compared the ability of denitrification mutants to utilize nitrate during anaerobic growth (Fig. 3). At both 7 days and 14 days of anaerobic growth, FcΔnasA, FcΔnirS, and FcΔnosZ had significantly (P < 0.0001) higher nitrate concentration compared with FcWT. In contrast, the FcΔnorB mutant (encoding nitric oxide reductase) showed no significant difference in nitrate concentration compared to FcWT at 7 and 14 days.
Figure 3.
Concentration of extracellular nitrate in FcWT, FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ cultures grown anaerobically in FCGM-N medium supplemented with 10 mM nitrate (measured at 7 and 14 days post-inoculation). Data points represent the mean of six replicates. Significant differences are indicated with asterisks (P < 0.05). Control group includes FCGM-N broth without bacterial inoculation.
Biofilm assay
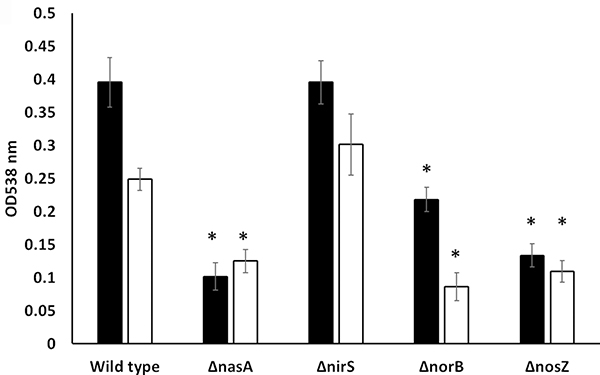
Biofilm formation of FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ was examined using a static biofilm assay at 24 and 96 hours. Under aerobic conditions, no significant (P >0.05) differences in biofilm formation between mutants and FcWT were observed (data not shown). Under anaerobic conditions, FcΔnasA, FcΔnorB, and FcΔnosZ formed significantly less biofilm than FcWT at 24 and 96 hours (Fig 4). There was no significant difference in biofilm formation between FcΔnirS and FcWT.
Figure 4.
Quantification of biofilm formation in FcWT, FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ using crystal violet staining and measured by absorbance at 538 nm after 24 and 96 hours under anerobic conditions. The data represent means of four replicates plus or minus standard error.
Virulence of FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ in catfish
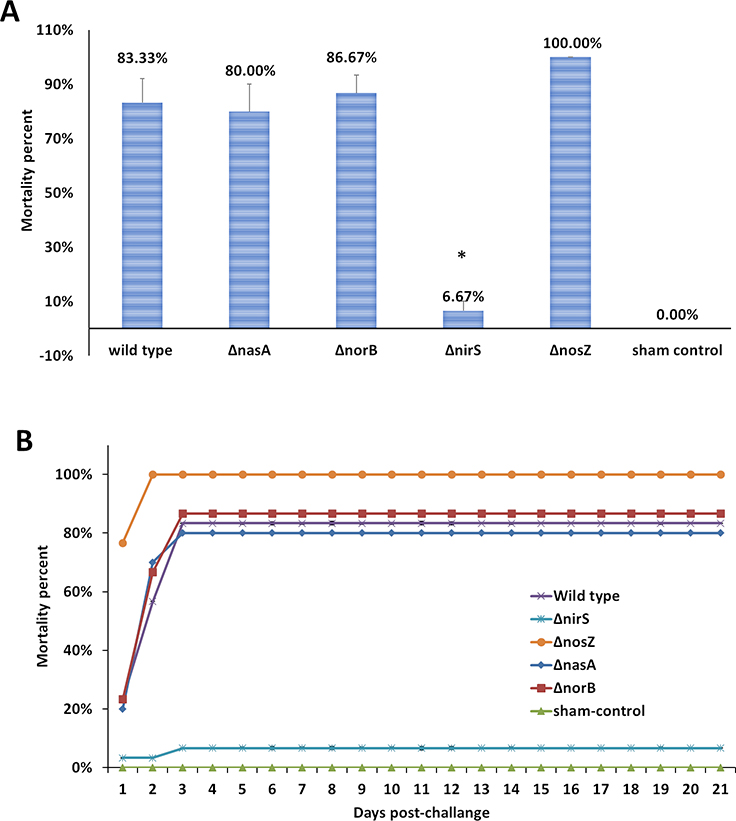
Catfish fingerlings challenged with FcΔnirS exhibited significantly fewer mortalities (6.67%) (P <0.0001) compared to those challenged with FcWT (83.33%). However, mortalities in catfish challenged with FcΔnasA (80%), FcΔnorB (86.67%), and FcΔnosZ (100%) were similar to that of FcWt challenged catfish (Fig 5 A and B).
Figure 5.
Percent mortality (A) and cumulative mortalities (B) of catfish fingerlings immersion challenged with FcWT, FcΔnasA, FcΔnirS, FcΔnorB, and FcΔnosZ. Data points represent the mean of four tanks for each treatment. Significant differences are indicated with asterisks (P < 0.05).
Discussion
Denitrification is a microbial process that reduces nitrate (NO3−) or nitrite (NO2−) to nitrogen (N2) via two obligate intermediates, nitric oxide (NO) and nitrous oxide (N2O), through anaerobic respiration. This process is linked to the respiratory electron transport chain, and denitrifying bacteria can use nitrate or nitrite as alternative electron acceptors to support respiratory growth (Zumft 1997). Each step within the denitrification pathway is catalyzed by independent metalloenzymes that are usually induced sequentially under anaerobic conditions (Zumft 1997; Tavares et al. 2006). The F. columnare genome encodes four enzymes that are likely to be involved in denitrification: nitrate reductase (encoded by nasA gene), nitrite reductase (encoded by nirS gene), nitric oxide reductase (encoded by norB gene), and nitrous oxide reductase (encoded by nosZ gene) (Tekedar et al. 2012). We have previously shown that F. columnare strain 94–081 can grow anaerobically with nitrate supplementation (Tekedar et al. 2017).
Bacterial species capable of complete denitrification such as Paracoccus denitrificans and the denitrifying pseudomonads contain up to 40 genes involved in synthesis of the denitrification apparatus (De Boer et al. 1996). On the other hand, other bacterial species that are capable of catalyzing part of the denitrification pathway such as N. meningitidis possess only a few denitrifying genes (aniA, norB, fnr, narP, narQ, and azu) (Anjum et al. 2002; Barth et al. 2009). The nasA, nirS, norB, and nosZ genes of F. columnare have significant identity with denitrification genes from other bacteria such as P. aeruginosa and N. meningitidis.
We reasoned that nasA, nirS, norB, and nosZ of F. columnare might be involved in anaerobic growth and nitrate reduction to nitrogen gas. To test this hypothesis, four mutants were constructed using plasmids derived from pCP29, which is an E. coli-F. johnsoniae shuttle vector (Kempf and McBride 2000). This vector combines the pCU19-based suicide vector pLYL03 with a cryptic plasmid (pCP1) isolated from F. psychrophilum strain D12. It carries genes encoding cefoxitin resistance and erythromycin resistance that function in F. columnare and other Bacteroidetes (Kempf and McBride 2000; Alvarez et al. 2004). pCP29 also carries a gene encoding β-lactamase that confers resistance to ampicillin in E. coli. To avoid the problem of plasmid replication in F. columnare, pCP1 was removed from pCP29, and the resulting plasmids were transferred by conjugation from E. coli to successfully construct deletion mutants in F. columnare strain 94–081.
As expected, there was significant anaerobic growth deficiency in the FcΔnasA, FcΔnirS, and FcΔnosZ mutants with nitrate supplementation, indicating that nitrate reductase, nitrite reductase, and nitrous oxide reductase are needed for optimal anaerobic respiration. However, no significant decrease in anaerobic growth was observed between FcΔnorB and FcWT, indicating that nitric oxide reductase is not essential for anaerobic respiration in F. columnare. By contrast, in N. gonorrhoeae, norB gene was required for anaerobic growth, but the absence of norB did not dramatically decrease anaerobic survival (Householder et al. 2000). Similarly, mutagenesis of Paracoccus denitrificans norC, norB, norQ, and norD resulted in bacteria being unable to grow anaerobically (De Boer et al. 1996). However, Alcaligenes eutrophus mutants bearing single-site deletions in norB or norZ were shown to have no anaerobic growth defects using nitrate or nitrite. However, a mutant with double deletion of both norB and norZ failed to grow anaerobically on nitrate. Anaerobic growth was restored in the double mutant by introducing either norB or norZ on a broad-host-range plasmid (Cramm et al. 1997). Thus, it is possible that F. columnare encodes a second nitric oxide reductase that has not been identified. Indeed, the F. columnare genome has three genes currently annotated as nitric oxide reductases: norB (AWN65_04995) and two other genes (AWN65_05000 and AWN65_04960).
To further investigate if the growth defects observed in FcΔnasA, FcΔnirS, and FcΔnosZ were a result of an inability to reduce nitrate, the abilities of four mutants to consume extracellular nitrate were analyzed and compared with FcWT. Consistent with the reduced growth under anaerobic conditions, FcΔnasA, FcΔnirS, and FcΔnosZ failed to reduce nitrate to nitrite even after incubation for 14 days, as indicated by significantly more residual nitrate in the medium than was found for the wild-type. By contrast, significant residual nitrate in medium was not observed in the FcΔnorB mutant compared to FcWT, which correlates with the ability of this mutant to grow anaerobically in the presence of nitrate. Mutants of norB, norC, and nirS in P. aeruginosa are defective in nitrate and nitrite reduction activity (Borrero-de Acuna et al. 2016).
Taken together, results from the anaerobic growth experiment and nitrate consumption experiment indicate that F. columnare anaerobic growth is linked to its ability to reduce nitrate, suggesting that F. columnare can use nitrate as an alternative electron acceptor for the electron transport system during anaerobic growth. The inability of the nitrate reductase mutant to reduce nitrate was expected. Results showed that nitrite reductase and nitrous oxide reductase activities are also essential for this process, indicating that capability of the complete denitrification process to molecular nitrogen is necessary to allow use of nitrate in F. columnare. The conflicting nitric oxide reductase results are explainable by the possible presence of three genes encoding nitric oxide reductases in the F. columnare genome. In many bacterial species, reduction of either nitrate or nitrite is essential for anaerobic growth (Berks et al. 1995). Interestingly, denitrification also promotes the growth of N. meningitidis, a strictly aerobic human pathogen, under oxygen-limited conditions (Rock et al. 2005).
Biofilm formation is considered an essential feature in the pathogenicity of F. columnare (Staroscik and Nelson 2008). In the environment and during infection, F. columnare demonstrates capacity to form thick, multilayered biofilms under static and flow conditions (Cai et al. 2013). In P. aeruginosa, anaerobic respiration via denitrification is important for its formation of biofilms during anaerobic growth (Hassett et al. 2002; Worlitzsch et al. 2002). Because respiratory mucus of cystic fibrosis patients is an anaerobic environment rich in nitrate and nitrite, induction of biofilm in this environment is beneficial to the pathogen. Gene expression studies have revealed that both nirS and norCB are highly expressed in biofilms compared with planktonic P. aeruginosa (Wagner et al. 2003; Sauer et al. 2004). A P. aeruginosa norBC insertion mutant forms virtually no biofilm under anaerobic growth conditions, while a nir mutant forms biofilms similar to those of the wild-type (Yoon et al. 2002). A N. gonorrhoeae norB mutant had a more pronounced biofilm-deficient phenotype than an aniA mutant (Falsetta et al. 2009). Therefore, we investigated whether the deletion of F. columnare nasA, nirS, norB, and nosZ genes would reduce biofilm formation. Our result indicates that nasA, norB, and nosZ are necessary for biofilm formation under anaerobic growth. The role of norB in biofilm formation is noteworthy because this gene is not essential for anaerobic growth. Despite the potential existence of similar nitric oxide reductases encoded in the F. columnare genome with overlapping functions in anaerobic respiration, it appears that norB plays a unique role in biofilm formation. Taken together, F. columnare is similar to other denitrifying bacteria in that anaerobic respiration appears to be linked to biofilm formation, suggesting that biofilm formation may be a beneficial phenotype for F. columnare in anaerobic environments.
A significant contribution of anaerobic nitrate respiration to virulence has been reported in M. bovis in immunodeficient mice (Weber et al. 2000). A P. aeruginosa membrane nitrate reductase mutant was avirulent in the surrogate model host Caenorhabditis elegans, whereas nitrate sensor-response regulator mutants were fully virulent (Van Alst et al. 2007). In Brucella suis, transposon inactivation of the gene encoding nitrate reductase affected growth inside the macrophage (Kohler et al. 2002). A B. melitensis norB mutant (nitric oxide reductase deficient) is attenuated in activated macrophages but only slightly attenuated in mice at 4 weeks postinfection (Haine et al. 2006). However, in F. columnare, we found that nitrate reductase, nitric oxide reductase, and nitrous oxide reductase activities are not linked with virulence.
However, deletion of the nirS gene (nitrite reductase) significantly reduced F. columnare virulence. In Neisseria, nitrite and nitric oxide reductase are required for anaerobic growth and have a role in mitigating nitric oxide toxicity in macrophages (Anjum et al. 2002). Therefore, it is possible that reduced virulence of the F. columnare nirS mutant is due to reduced ability to inactivate reactive nitrogen species (RNS) such as nitric oxide, which is a host defense mechanism. However, this does not seem likely because nitrate reductase contributes equally to detoxification of RNS, and FcΔnasA is not attenuated. An alternative explanation is that nirS contributes to regulation of F. columnare virulence genes. In P. aeruginosa, a nirS knockout mutant was deficient in swarming motility, which involves both flagella and pili (de la Fuente-Nunez et al. 2013). Swarming motility using flagella and pili is important for initial cell-cell and cell-surface interactions in P. aeruginosa (O’Toole and Kolter 1998; Köhler et al. 2000). Furthermore, nirS expression in P. aeruginosa is required to induce the type III secretion system (T3SS) components that translocate effector proteins such as extracellular protease and elastase into host cells (Van Alst et al. 2009). Extracellular protease activity and chondroitin AC lyase activity contribute to F. columnare virulence (Stringer-Roth et al. 2002; Suomalainen et al. 2006; Li et al. 2017); it is possible that F. columnare nirS is involved in regulating expression of these or other yet-unidentified virulence factors during infection. It is interesting to note that nirS is also the only gene that is not required for biofilm formation, indicating that the ability to form biofilm anaerobically is not essential for virulence.
In summary, we determined the role of denitrification genes nasA, nirS, norB, and nosZ in anaerobic growth, nitrate reduction, biofilm formation, and virulence in F. columnare strain 94–081. Deletion of nasA, nirS, and nosZ genes, which encode nitrate reductase, nitrite reductase, and nitrous oxide reductase made F. columnare incapable of growing anaerobically and reducing nitrate to nitrite. The nirS gene, encoding nitrite reductase, has an additional role in F. columnare virulence and may contribute to regulating expression of factors important in virulence. The FcΔnirS mutant could be useful as a safe, live vaccine against columnaris disease if it protects catfish aginst FcWT infection.
Acknowledgments
We thank Dr. Mark McBride for providing pCP29 plasmid and Dr. Adef Kordon for helping with statistical analysis. We also thank Mississippi State University Laboratory Animal Resources and Care for SPF catfish. Funding for this research was provided by a research grant from the College of Veterinary Medicine at Mississippi State University. Part of Dr. Abdelhamed’s salary was provided by the Center for Biomedical Research Excellence in Pathogen–Host Interactions, National Institute of General Medical Sciences, National Institutes of Health awarded grant number P20GM103646-07. We thank Michelle Banes for help in conducting the fish challenge experiment and proofreading the manuscript.
Footnotes
Conflict of interest
The authors have declared no conflicts of interest.
References
- Alvarez B, Secades P, McBride MJ and Guijarro JA (2004) Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum. Appl Environ Microbiol 70, 581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum MF, Stevanin TM, Read RC and Moir JW (2002) Nitric oxide metabolism in Neisseria meningitidis. J Bacteriol 184, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth K and Clark VL (2008) Differences in nitric oxide steady-states between arginine, hypoxanthine, uracil auxotrophs (AHU) and non-AHU strains of Neisseria gonorrhoeae during anaerobic respiration in the presence of nitrite. Can J Microbiol 54, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth KR, Isabella VM and Clark VL (2009) Biochemical and genomic analysis of the denitrification pathway within the genus Neisseria. Microbiology 155, 4093–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks BC, Ferguson SJ, Moir JWB and Richardson DJ (1995) Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. BBA-BIOENERGETICS 1232, 97–173. [DOI] [PubMed] [Google Scholar]
- Borrero-de Acuna JM, Rohde M, Wissing J, Jansch L, Schobert M, Molinari G, Timmis KN, Jahn M and Jahn D (2016) Protein Network of the Pseudomonas aeruginosa Denitrification Apparatus. J Bacteriol 198, 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, De La Fuente L and Arias CR (2013) Biofilm formation by the fish pathogen Flavobacterium columnare: development and parameters affecting surface attachment. Appl Environ Microbiol 79, 5633–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramm R, Siddiqui RA and Friedrich B (1997) Two isofunctional nitric oxide reductases in Alcaligenes eutrophus H16. J Bacteriol 179, 6769–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer APN, Van Der Oost J, Reijnders WNM, Westerhoff HV, Stouthamer AH and Van Spanning RJM (1996) Mutational Analysis of the Nor Gene Cluster which Encodes Nitric-Oxide Reductase from Paracoccus denitrificans. Eur J Biochem 242, 592–600. [DOI] [PubMed] [Google Scholar]
- de la Fuente-Nunez C, Reffuveille F, Fairfull-Smith KE and Hancock RE (2013) Effect of nitroxides on swarming motility and biofilm formation, multicellular behaviors in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57, 4877–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declercq AM, Haesebrouck F, Van den Broeck W, Bossier P and Decostere A (2013) Columnaris disease in fish: a review with emphasis on bacterium-host interactions. Veterinary Research 44, 27–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumpala PR, Gulsoy N, Lawrence ML and Karsi A (2010) Proteomic analysis of the fish pathogen Flavobacterium columnare. Proteome Sci 8, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Bair TB, Ku SC, Vanden Hoven RN, Steichen CT, McEwan AG, Jennings MP and Apicella MA (2009) Transcriptional profiling identifies the metabolic phenotype of gonococcal biofilms. Infect Immun 77, 3522–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiatrault MJ, Picardo KF, Ngai H, Passador L and Iglewski BH (2006) Identification of Pseudomonas aeruginosa Genes Involved in Virulence and Anaerobic Growth. Infect Immunity 74, 4237–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JC, LaFrentz BR, Waldbieser GC, Wong FS and Chang SF (2018) Characterization of atypical Flavobacterium columnare and identification of a new genomovar. J Fish Dis 41, 1159–1164. [DOI] [PubMed] [Google Scholar]
- Grant SG, Jessee J, Bloom FR and Hanahan D (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87, 4645–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griess P (1879) Bemerkungen zu der Abhandlung der HH. Weselsky und Benedikt;, Ueber einige Azoverbindungen”. Ber Dtsch Chem Ges 12, 426–428. [Google Scholar]
- Haine V, Dozot M, Dornand J, Letesson J-J and De Bolle X (2006) NnrA Is Required for Full Virulence and Regulates Several Brucella melitensis Denitrification Genes. J Bacteriol 188, 1615–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett DJ, Cuppoletti J, Trapnell B, Lymar SV, Rowe JJ, Yoon SS, Hilliard GM, Parvatiyar K, Kamani MC, Wozniak DJ, Hwang SH, McDermott TR and Ochsner UA (2002) Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv Drug Deliv Rev 54, 1425–1443. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK and Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68. [DOI] [PubMed] [Google Scholar]
- Householder TC, Fozo EM, Cardinale JA and Clark VL (2000) Gonococcal Nitric Oxide Reductase Is Encoded by a Single Gene, norB, Which Is Required for Anaerobic Growth and Is Induced by Nitric Oxide. Infect Immun 68, 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Arai H, Kodama T and Igarashi Y (1997) Gene cluster for dissimilatory nitrite reductase (nir) from Pseudomonas aeruginosa: sequencing and identification of a locus for heme d1 biosynthesis. J Bacteriol 179, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf MJ and McBride MJ (2000) Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J Bacteriol 182, 1671–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M and Liautard JP (2002) The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci U S A 99, 15711–15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, Curty LK, Barja F, van Delden C and Pechère J-C (2000) Swarming of Pseudomonas aeruginosa Is Dependent on Cell-to-Cell Signaling and Requires Flagella and Pili. J Bacteriol 182, 5990–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumru S, Tekedar HC, Waldbieser GC, Karsi A and Lawrence ML (2016) Genome Sequence of the Fish Pathogen Flavobacterium columnare Genomovar II Strain 94–081. Genome Announc 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFrentz BR, García JC, Waldbieser GC, Evenhuis JP, Loch TP, Liles MR, Wong FS and Chang SF (2018) Identification of Four Distinct Phylogenetic Groups in Flavobacterium columnare With Fish Host Associations. Front Microbiol 9, 452–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFrentz BR, Waldbieser GC, Welch TJ and Shoemaker CA (2014) Intragenomic heterogeneity in the 16S rRNA genes of Flavobacterium columnare and standard protocol for genomovar assignment. J Fish Dis 37, 657–669. [DOI] [PubMed] [Google Scholar]
- Li N, Zhu Y, LaFrentz BR, Evenhuis JP, Hunnicutt DW, Conrad RA, Barbier P, Gullstrand CW, Roets JE, Powers JL, Kulkarni SS, Erbes DH, García JC, Nie P and McBride MJ (2017) The Type IX Secretion System Is Required for Virulence of the Fish Pathogen Flavobacterium columnare. Appl Env Microbiol 83, e01769–01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Jiang W and Wanner BL (1994) Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6Kλ origin plasmids at different copy numbers. Gene 138, 1–7. [DOI] [PubMed] [Google Scholar]
- Mohammed HH and Arias CR (2014) Epidemiology of columnaris disease affecting fishes within the same watershed. Dis Aquat Organ 109, 201–211. [DOI] [PubMed] [Google Scholar]
- O’Toole GA and Kolter R (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30, 295–304. [DOI] [PubMed] [Google Scholar]
- Philippot L (2005) Denitrification in pathogenic bacteria: for better or worst? Trends Microbiol 13, 191–192. [DOI] [PubMed] [Google Scholar]
- Rock JD, Mahnane MR, Anjum MF, Shaw JG, Read RC and Moir JW (2005) The pathogen Neisseria meningitidis requires oxygen, but supplements growth by denitrification. Nitrite, nitric oxide and oxygen control respiratory flux at genetic and metabolic levels. Mol Microbiol 58, 800–809. [DOI] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG and Gilbert P (2004) Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186, 7312–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber K, Krieger R, Benkert B, Eschbach M, Arai H, Schobert M and Jahn D (2007) The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol 189, 4310–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CA, Olivares-Fuster O, Arias CR and Klesius PH (2008) Flavobacterium columnare genomovar influences mortality in channel catfish (Ictalurus punctatus). Vet Microbiol 127, 353–359. [DOI] [PubMed] [Google Scholar]
- Soto E, Mauel MJ, Karsi A and Lawrence ML (2008) Genetic and virulence characterization of Flavobacterium columnare from channel catfish (Ictalurus punctatus). J Appl Microbiol 104, 1302–1310. [DOI] [PubMed] [Google Scholar]
- Staroscik AM, Hunnicutt DW, Archibald KE and Nelson DR (2008) Development of methods for the genetic manipulation of Flavobacterium columnare. BMC Microbiol 8, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staroscik AM and Nelson DR (2008) The influence of salmon surface mucus on the growth of Flavobacterium columnare. J Fish Dis 31, 59–69. [DOI] [PubMed] [Google Scholar]
- Stevanin TM, Laver JR, Poole RK, Moir JW and Read RC (2007) Metabolism of nitric oxide by Neisseria meningitidis modifies release of NO-regulated cytokines and chemokines by human macrophages. Microbes Infect 9, 981–987. [DOI] [PubMed] [Google Scholar]
- Stevanin TM, Moir JW and Read RC (2005) Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect Immun 73, 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer-Roth KM, Yunghans W and Caslake LF (2002) Differences in chondroitin AC lyase activity of Flavobacterium columnare isolates. J Fish Dis 25, 687–691. [Google Scholar]
- Suomalainen LR, Tiirola M and Valtonen ET (2006) Chondroitin AC lyase activity is related to virulence of fish pathogenic Flavobacterium columnare. J Fish Dis 29, 757–763. [DOI] [PubMed] [Google Scholar]
- Tavares P, Pereira AS, Moura JJ and Moura I (2006) Metalloenzymes of the denitrification pathway. J Inorg Biochem 100, 2087–2100. [DOI] [PubMed] [Google Scholar]
- Tekedar HC, Karsi A, Gillaspy AF, Dyer DW, Benton NR, Zaitshik J, Vamenta S, Banes MM, Gulsoy N, Aboko-Cole M, Waldbieser GC and Lawrence ML (2012) Genome sequence of the fish pathogen Flavobacterium columnare ATCC 49512. J Bacteriol 194, 2763–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekedar HC, Karsi A, Reddy JS, Nho SW, Kalindamar S and Lawrence ML (2017) Comparative Genomics and Transcriptional Analysis of Flavobacterium columnare Strain ATCC 49512. Front Microbiol 8, 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triyanto H (1999) Genotypic Diversity of Strains of Flavobacterium columnare from Diseased Fishes. Fish Pathol 34, 65–71. [Google Scholar]
- Van Alst NE, Picardo KF, Iglewski BH and Haidaris CG (2007) Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun 75, 3780–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alst NE, Wellington M, Clark VL, Haidaris CG and Iglewski BH (2009) Nitrite reductase NirS is required for type III secretion system expression and virulence in the human monocyte cell line THP-1 by Pseudomonas aeruginosa. Infect Immun 77, 4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner BA, Wise DJ, Khoo LH and Terhune JS (2002) The Epidemiology of Bacterial Diseases in Food-Size Channel Catfish. J Aquat Anim Health 14, 263–272. [DOI] [PubMed] [Google Scholar]
- Wagner BA, Wise DJ, Khoo LH and Terhune JS (2006) The Epidemiology of Bacterial Diseases in Food-Size Channel Catfish. J Aquat Anim Health 18, 263–272. [DOI] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI and Iglewski BH (2003) Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185, 2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I, Fritz C, Ruttkowski S, Kreft A and Bange FC (2000) Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol Microbiol 35, 1017–1025. [DOI] [PubMed] [Google Scholar]
- Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC and Doring G (2002) Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC and Hassett DJ (2002) Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3, 593–603. [DOI] [PubMed] [Google Scholar]
- Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61, 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG, Viebrock-Sambale A and Braun C (1990) Nitrous oxide reductase from denitrifying Pseudomonas stutzeri. Genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. Eur J Biochem 192, 591–599. [DOI] [PubMed] [Google Scholar]