Abstract
Oxylipins are lipid peroxidation products that participate in nociceptive, inflammatory, and vascular responses to injury. Effects of oxylipins depend on tissue-specific differences in accumulation of precursor polyunsaturated fatty acids and the expression of specific enzymes to transform the precursors. The study of oxylipins in nociception has presented technical challenges leading to critical knowledge gaps in the way these molecules operate in nociception. We applied a systems-based approach to characterize oxylipin precursor fatty acids, and expression of genes coding for proteins involved in biosynthesis, transport, signaling and inactivation of pro- and anti-nociceptive oxylipins in pain circuit tissues. We further linked these pathways to nociception by demonstrating intraplantar carrageenan injection induced gene expression changes in oxylipin biosynthetic pathways. We determined functional-biochemical relevance of the proposed pathways in rat hind paw and dorsal spinal cord by measuring basal and stimulated levels of oxylipins throughout the time-course of carrageenan-induced inflammation. Finally, when oxylipins were administered by intradermal injection we observed modulation of nociceptive thermal hypersensitivity, providing a functional-behavioral link between oxylipins, their molecular biosynthetic pathways, and involvement in pain and nociception. Together, these findings advance our understanding of molecular lipidomic systems linking oxylipins and their precursors to nociceptive and inflammatory signaling pathways in rats.
Keywords: Oxylipin, LC-MS/MS, Lipids, Pain, RNASeq
Perspective:
We applied a systems approach to characterize molecular pathways linking precursor lipids and oxylipins to nociceptive signaling. This systematic, quantitative evaluation of the molecular pathways linking oxylipins to nociception provides a framework for future basic and clinical research investigating the role of oxylipins in pain.
Introduction
The question of how pain begins must involve generation of biochemical mediators that interface with the nerve terminals of nociceptive sensory neurons. A corollary of this hypothesis is that the necessary molecular apparatus for generating such algogenic mediators is present or can be induced in tissue in response to damage or pathological conditions. Framed by the view that tissue damage involves membrane damage, the present investigation examines these questions from the perspective of bioactive lipids.
Omega-6 (n-6) and omega-3 (n-3) polyunsaturated fatty acids (PUFAs) are biosynthetic precursors to several families of bioactive lipids (oxylipins) with pro- and anti-nociceptive properties, including oxidized linoleic acid metabolites (OXLAMs) 30, prostanoids 97, lipoxins44, resolvins58, protectins 57, maresins 79, and n-3 monoepoxides 106. The majority of oxylipins derived from the n-6 PUFAs linoleic acid (LA) and arachidonic acid (AA) have pro-nociceptive properties; while oxylipins synthesized from n-3 PUFAs eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) tend to have anti-nociceptive properties 69. Since mammals cannot synthesize n-3 or n-6 PUFAs de novo, targeted changes in dietary n-6 and n-3 PUFAs may be able to change oxylipins in a manner that reduces pain. We recently established proof of principle for this hypothesis in a randomized trial of 67 subjects with severe headaches 68.
The Chronic Daily Headache trial and gaps in the proposed pathways linking dietary fatty acids to pain
Targeted dietary manipulation—increasing n-3 PUFAs with concurrent reduction in n-6 LA acid (the H3-L6 intervention)—produced statistically significant, clinically relevant reductions in headache frequency and severity among subjects with Chronic Daily Headache 67,68. These clinical improvements were accompanied by marked increases in anti-nociceptive and pro-resolving n-3 derived oxylipins, and reductions in pro-nociceptive n-6 derived mediators, in circulation 68. Reduction in plasma n-6 LA, and increases in plasma n-3 DHA, closely correlated with clinical pain reduction. Moreover, we recently demonstrated in rats that increasing LA as a controlled variable increased tissue levels of pro-nociceptive mediators and reduced n-3 derived monoepoxides91.
Despite these compelling relationships, important gaps remain in our understanding of the molecular pathways linking precursor lipids and their autacoid derivatives to nociceptive signaling pathways. For example, the tissue distributions of PUFAs (biosynthetic precursors to oxylipins), and the expression of genes coding for proteins mediating oxylipin biosynthesis, signaling and inactivation, have not yet been reported in depth and quantitatively. Nociceptive peripheral nerve endings, axons, neurons in the dorsal root ganglia (DRG), and second order spinal circuits can all be influenced by oxylipins in the event of tissue trauma or damage.
In this report, we take key first steps toward characterizing molecular pathways linking lipids to nociceptive signaling and examine their potential modulation by an inflammatory stimulus in rats. The specific objectives of the present paper are: (1) To determine the oxylipin precursor fatty acid concentrations in pain circuit tissues; (2) To characterize basal and carrageenan (CG)-stimulated expression levels of genes coding for proteins involved in biosynthesis, signaling and inactivation of pro- and anti-nociceptive oxylipins; (3) To confirm that the proposed molecular pathways (i.e. the precursor PUFA and related enzymes) are active in rat hind paw and dorsal spinal cord, by measuring basal and stimulated concentrations of pro- and anti-nociceptive oxylipins; and (4) To demonstrate the ability of oxylipins to modulate nociception in rodent behavior models. In aggregate, these findings provide an integrated and comprehensive investigation of known molecular pathways linking pro- and anti-nociceptive oxylipins and their precursor PUFA to pain signaling pathways.
Results
Characterizing and organizing molecular pathways related to oxylipins
The following sections provide quantitative data, some of which is expanded for this report based on previously published results 66, on oxylipin precursor fatty acids (i.e. PUFAs). The data are expressed as median % of total fatty acids (FA) ± interquartile range (Quartile 3-Quartile 1). Expression levels of genes involved in biosynthesis, signaling pathways and inactivation of oxylipins in nociceptive (pain) circuit tissues—including skin, peripheral nerves, sensory ganglia and dorsal spinal cord are also presented (reported as mean sFPKM). The precursor levels and transcriptomic data are accompanied by Liquid Chromatography Tandem-Mass Spectrometry (LC-MS/MS) measured concentrations of oxylipins in hind paw and spinal cord dorsal horn collected from rats in the basal state as well as in an experimental inflammation state (intraplantar CG injection; a widely used inflammatory chronic pain model) 35. Lipids exist, broadly, in both unesterified (free acid) forms, as well as esterified forms, as a component of multiple complex lipid species. We measured oxylipin concentrations in both the unesterified and esterified pools. Classically, unesterified lipids are considered to be the biologically active forms 55, although bioactivity of esterified lipids has recently been reported 38.
The results for nociception-related oxylipins are organized into four numbered sections of nociceptive oxylipins families—1. OXLAMs, 2. Prostanoids, 3. Hydroxy-eicosatetraenoic acids (HETEs), leukotrienes/lipoxins, and 4. Monohydroxy-DHA derivatives (HDHAs), monoepoxy-DHA derivatives (EDPs), and specialized pro-resolving lipid mediators (SPLMs). Each of these oxylipin families share a common precursor and biosynthetic pathways. Additionally, the effects of CG on gene expression and oxylipin concentrations are presented in section 5 and behavioral findings of intradermal injections of selected oxylipins on nociception-related sensitivity are presented in section 6 of the results.
1. Oxidized linoleic acid metabolites (OXLAMs)
1.1. OXLAM precursor concentrations
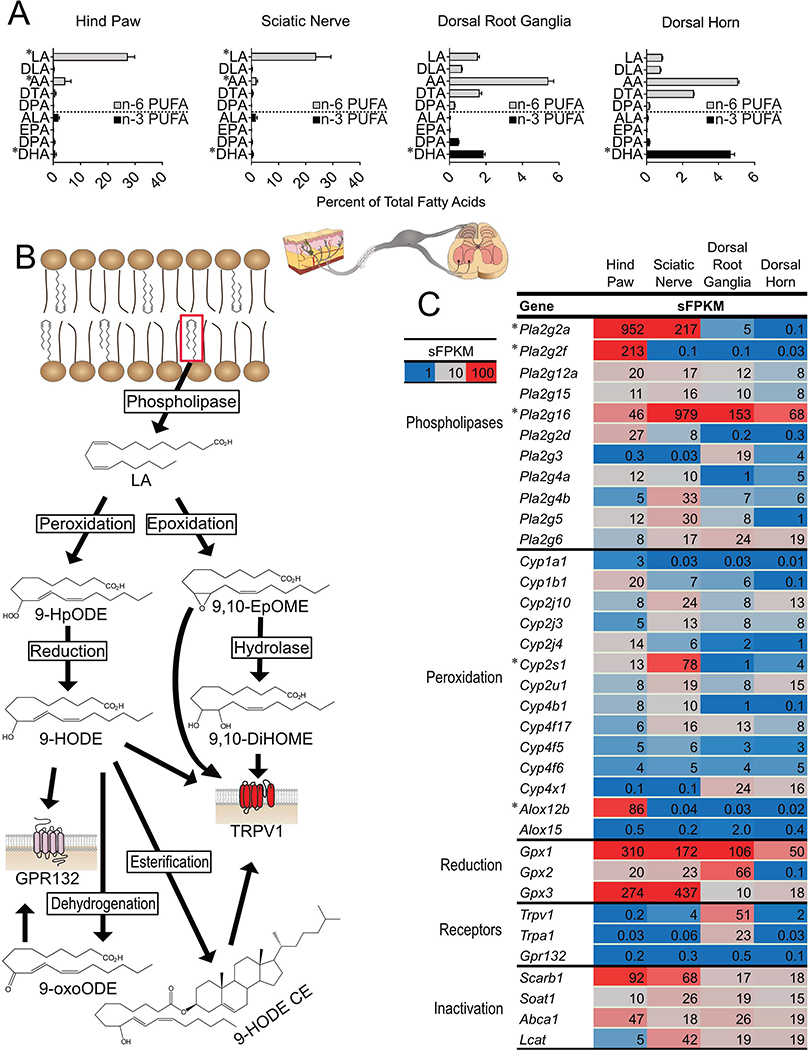
OXLAMs are oxylipins synthesized from LA [hydroperoxy- (HpODEs), mono-(HODEs), di-hydroxy- (DiHOMEs) or epoxy (EpOMEs)-LA derivatives] which have been previously reported to be pro-nociceptive 30,66. LA was most abundant in hind paw (27.3 ± 3.5%) and sciatic nerve (23.9 ± 4.9%) and comparatively scarce in DRG and spinal cord (≤1.6% of FA) (Figure 1A). In tissues where LA was abundant (hind paw and sciatic nerve) LA was approximately 3-fold more abundant as compared to the next most abundant PUFA (AA). Therefore, of the four pain-circuit tissues sampled, peripheral tissues such as hind paw and nerve trunk (which contains axons and Schwann cells, but not DRG neuronal perikarya) 75 are rich sources of the precursor (LA) for conversion to OXLAMS.
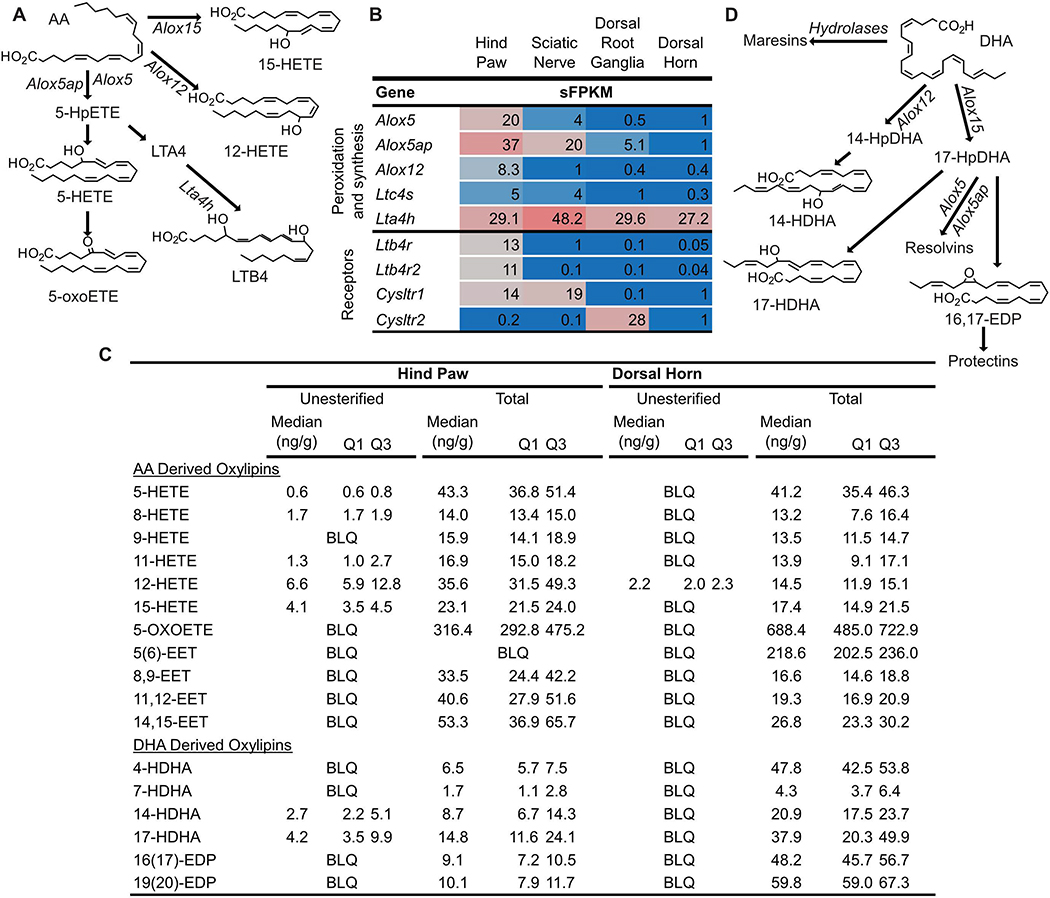
Figure 1: Linoleic acid (LA) and genes involved in oxidized linoleic acid metabolite (OXLAM) biosynthesis and inactivation are abundant in hind paw.
(A) Polyunsaturated fatty acid (PUFA) concentrations in pain circuit tissues (% of total fatty acids, n=3) (B) OXLAMs are synthesized by a multi-step process involving the release of LA from lipid membranes by phospholipases followed by peroxidation (by Arachidonate lipoxygenase enzymes or cytochrome p450 monooxygenases) or epoxidation (via the action of cytochrome p450 enzyme). The product of LA peroxidation is 9- or 13-hydroperoxy-octadecenoate (9- or 13-HpODE) while LA epoxidation results in 9,10- or 12,13-epoxy-octadecenoate (9,10- or 12,13-EpOME). HpODEs are reduced either non-enzymatically or enzymatically (by Glutathione peroxidase enzymes) to 9- or 13-hydroxy-octadecenoate (9- or 13-HODE) while EpOMEs can be hydrolyzed to 9,10- or 12,13-dihydroxy-oxtadecenoate (9,10 or 12,13-DiHOME). HODEs can be oxidized to 9- or 13-keto-octadecenoate (9-or 13-oxoODE). Unesterified OXLAMs have been reported to impact nociception via Transient Receptor Potential receptors (TRPV1 or TRPA1) as well as through G-Protein Coupled Receptors (GPR132). Lipids (including OXLAMs) are hypothesized to be inactivated by esterification into complex lipid species such as phospholipids or cholesteryl esters. (C) Expression of genes involved in OXLAM biosynthesis, signaling and deactivation were measured in pain circuit tissue using whole genome RNA Sequencing. Fatty acid compositions (A) are presented as percent of total fatty acids (median and interquartile range) n=4 per tissue. Gene expression (C) is presented as sFPKM (mean, n=3). * refers to data previously published by our group 66. DLA, Dihomo-gamma-linolenic acid; AA, Arachidonic acid; DTA, docosatetraenoic acid; DPA, docosapentaenoic acid n-6; ALA, alpha-linolenic acid; EPA, eicosapentaenoic acid; DPA n-3, docosapentaenoic acid n-3; DHA, docosahexaenoic acid; Pla2g, Phospholipase A2 Group; Cyp, Cytochrome P450; Alox, Arachidonate Lipoxygenase; Gpx, Glutathione Peroxidase; Scarb1, Scavenger Receptor Class B1; Soat1, Sterol O-Acyltransferase 1; Abca1, ATP Binding Cassette Subfamily A Member 1; Lcat, Lecithin-Cholesterol Acyltransferase.
1.2. Expression of genes related to OXLAMs
1.2.1. Expression of genes coding for enzymes involved in biosynthesis of OXLAMs
Each of the 4 pain circuit tissues examined here expressed a set of genes capable of OXLAM biosynthesis (Figure 1B and C). Generally, at least 2 phospholipases were strongly expressed (sFPKM>19) in each tissue, indicating capability to release unesterified LA (and other PUFAs) from membrane phospholipids (i.e. the esterified pool), although there were clear quantitative differences in the composition of the paralogs between the tissues. OXLAMs are generated via the action of arachidonate lipoxygenase (Alox), which generate HpODEs and cytochrome p450 monoepoxygenases (Cyp), which generate EpOMEs (Figure 1B) 53,70. Enzymes capable of lipid mediator synthesis generally exhibited tissue specific expression. For example, Alox12b and Alox15 have been reported to catalyze the synthesis of 9- and 13-hydroperoxyoctadecenoate from LA (9- and 13-HpODE, respectively) from which other, more stable OXLAMs (i.e. HODEs or EpOMEs) are synthesized 8,70. Several reports have shown roles for these more stable HODEs and EpOMEs in nociception 1,56,59 although the role of the relatively unstable and more reactive HpODEs in nociception is still being clarified in part due to the difficulty in quantifying HpODE concentrations in tissue49. We previously reported that Alox12b was expressed selectively in hind paw (sFPKM: 86) 66 and here we report that expression of Alox15 was very low in all 4 pain circuit tissues (highest expression in DRG with sFPKM=2). Low expression of Alox15 is consistent with body-wide transcriptomic datasets showing a high degree of specificity for expression of this gene in visceral adipose tissue 27 (Supplementary Figure 1). Moreover, CYP enzymes and glutahione peroxidases (Gpxs), which code for enzymes that catalyze the conversion of LA (for Cyps) or HpODEs into hydroxy- and epoxy-derivatives were expressed in most tissues (Figure 1B) 47,54,70. Cyp isoforms exhibited tissue specific expression and at least one Cyp isoform was expressed, at sFPKM>10, in each pain circuit tissue (Figure 1C).
Gpx enzymes, which catalyze the reduction of lipid peroxides to more stable hydroxy-derivatives (Figures 1B and 2), exhibit abundant expression broadly in pain circuit tissues as well as more focused tissue-specific isoform expression (Figures 1C and 2). Transcripts of Gpx1 and Gpx4 were high in all tissues examined (sFPKM ≥ 90 in all tissues, Figure 1C and 2). In particular, Gpx4, which has been demonstrated to specifically reduce esterified lipid-peroxides 94,103 exhibited greatest expression in DRG (sFPKM=406), although it was ubiquitously expressed at high levels in all tissues examined indicating very high capacity for the reduction of esterified lipid peroxides to more stable, esterified hydroxy-lipids in the pain circuit. In contrast, Gpx3 was expressed at 300–400 sFPKM in the two peripheral tissues (hind paw and sciatic nerve) vs. 10–20 sFPKM in DRG and dorsal horn (Figure 1C). This indicates that Gpx3 has more variation in expression by tissue, perhaps pointing to a unique function of this enzyme, protecting against oxidative stress by generating hydroxy-lipids from less stable lipid peroxides. Together, these data demonstrate that genes coding for enzymes that catalyze the synthesis of OXLAMs are expressed in pain circuit tissues suggesting a capacity for OXLAM synthesis in each of these tissues contingent on precursor availability. Because GPX enzymes are non-specific, this also suggests peripheral neuronal cell bodies and Schwann cells 75 have a high capacity to synthesize hydroxy-PUFA derivatives, which have been implicated in nociception and inflammation 96,99, possibly to protect against oxidative damage, and potentially cell death, induced by lipid peroxidation 103,104.
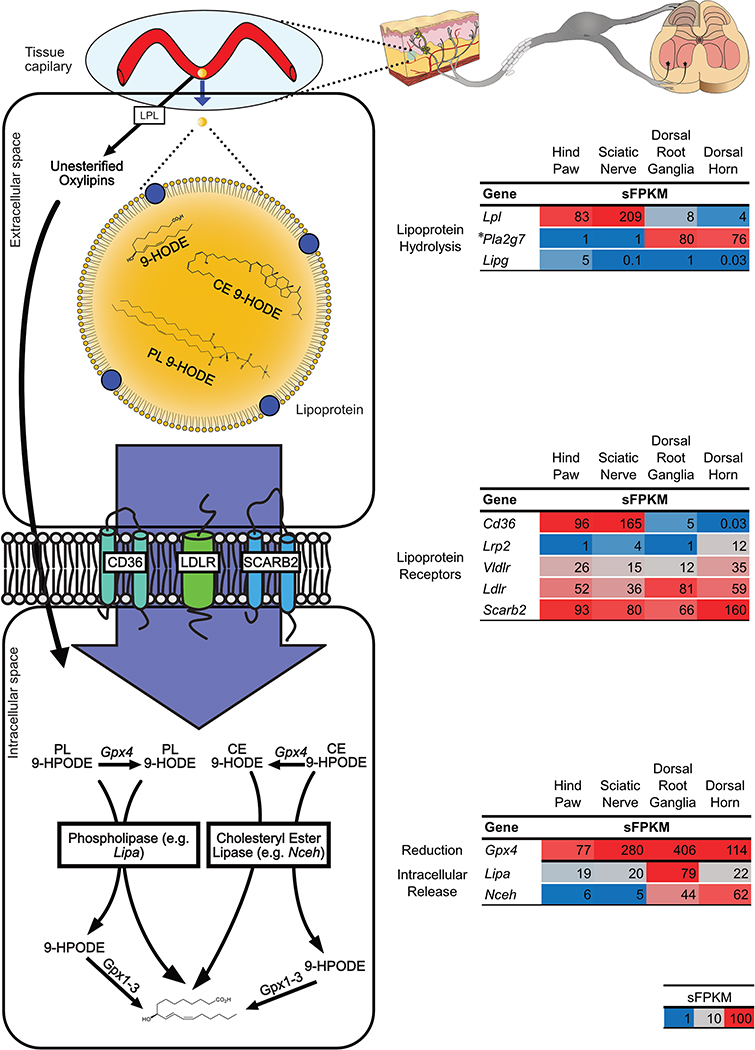
Figure 2: Oxylipins can be delivered to pain circuit tissue via circulation.
(Left) Oxylipins in circulating lipoproteins can reach tissue capillary beds and be released as unesterified oxylipins by lipoprotein hydrolysis. Unesterified oxylipins can then enter cells via passive diffusion. Additionally, lipoprotein receptors on cell membranes can mediate the uptake of lipoproteins (and oxylipins carried in them) into cells. Within cells oxylipins can be released from complex lipid species via phospholipase or cholesteryl ester lipases. Additionally, esterified hydroperoxy lipids such as HpODE can be reduced to esterified HODE by the action of Glutathione Peroxidase 4 (Gpx4). (Right) Expression of genes involved in the delivery of preformed oxylipins from circulation were measured in pain circuit tissues by whole genome RNASeq (sFPKM n=3). * refers to data previously published by our group 66. Lpl, Lipoprotein Lipase; Pla2g7, Phospholipase A2 Group VII; Lipg, Endothelial Cell-Derived Lipase; Cd36, Scavenger Receptor Class B Member 3; Lrp2, Low Density Lipoprotein Receptor Related Protein 2; Vldlr, Very Low Density Lipoprotein Receptor; Ldlr, Low Density Lipoprotein Receptor; Scarb2, Scavenger Receptor Class B Member 2; Lipa, Lipase A Lysosomal Acid Type; Nceh, Neutral Cholesterol Ester Hydrolase.
1.2.2. Expression of genes encoding enzymes involved in delivery of preformed oxylipins to pain-circuit tissues
Here we propose that circulating OXLAMs (consumed in the diet or formed and/or esterified in remote tissues such as liver) can be a source of OXLAMs in pain circuit tissue. We focus specifically on OXLAMs because LA and OXLAMs are the most abundant PUFA and oxylipins, respectively, in circulation 68,90, however, the concept described below is likely applicable to all oxylipins.
Rat pain-circuit tissues expressed genes encoding enzymes required for local delivery of preformed OXLAMs from circulation in both their unesterified and esterified forms (Figure. 2). Circulating OXLAMs, which are primarily esterified in lipoproteins [e.g. very low density lipoprotein (VLDL) and low-density lipoprotein (LDL)] as triacylglycerols, cholesteryl-esters and phospholipids 82, could potentially be delivered to pain-circuit tissues via the actions of lipoprotein lipase (Lpl), or other enzymes that mediate hydrolysis of esterified lipid species from circulating lipoproteins (i.e. Pla2g7 or Lipg) 81. We observed Lpl expression in all pain circuit tissues with greatest expression in sciatic nerve and hind paw (sFPKM=209 and 83, respectively) (Figure 2). Platelet-activating factor acetylhydrolase (Pla2g7), hydrolyzes phospholipids and has been implicated in several inflammatory disease processes in humans 89. This phospholipase, which travels with lipoproteins 87, also is expressed tissue-specifically in rat, with high expression levels in DRG and dorsal horn (sFPKM ≈ 80), and at least 76-fold less expression in hind paw and sciatic nerve (sFPKM=1 in both tissues). However, while Pla2g7 is moderately conserved at the protein level between rat and human, our observed expression pattern is divergent from what has been reported for Human PLA2G7 expression (Supplementary Figure 2).
Figure 2 also illustrates an alternate mechanism where OXLAMs (or other oxylipins) in circulating lipoproteins could be delivered to pain-circuit tissues via receptor-mediated endocytosis 12. Consistent with this hypothesis, we observed abundant expression of genes coding for traditional LDL receptors and scavenger receptors (Vldlr, Ldlr, Scarb2) in all pain circuit tissues. Interestingly, the scavenger receptor Cd36 was expressed in a tissue specific manner with strong expression in hind paw and sciatic nerve, and low expression in DRG and dorsal horn (Figure 2). Additionally, the gene encoding the scavenging receptor class B, member 2 (Scarb2), which has been reported to be fairly ubiquitous, was found at high levels throughout the pain circuit, especially the dorsal horn (sFPKM =160). We also examined the multifunctional endocytic LDLR-like protein, LDL receptor-related protein 2 (Lrp2, also known as Megalin). Megalin was expressed in dorsal horn and sciatic nerve (sFPKM=4–12), consistent with its role in CNS development and expression in myelin-producing cells 88.
Additionally, genes coding for lysosomal acid lipase (Lipa) and neutral cholesterol ester hydrolase 1 (Nceh1/Aadacl1), enzymes involved in lysosomal and endoplasmic reticulum cholesteryl ester hydrolysis 17,52, were ubiquitously expressed, providing a mechanism for intracellular release of unesterified oxylipins, initially delivered as cholesteryl esters via lipoprotein or scavenger receptors (Figure 2). Taken together, our findings indicate that all pain circuit tissues contain the enzymatic machinery requisite for the delivery of preformed OXLAMs from the circulation, but with tissue-specific profiles. Delivery of preformed OXLAMs (or other oxylipins) bypasses the need for local biosynthesis. This pathway could potentially help explain the link between systemic oxidative and inflammatory conditions and hyperalgesia 34,83.
1.2.3. Expression of genes encoding receptors involved in OXLAM signaling
Hydroxy-linoleic acid derivatives such as 9-hydroxy-octadecenoate (9-HODE) and other OXLAMs are reported to enhance nociception and immune activation via activation of Trpv1 60, Trpa1 24 and Gpr132 (G2A) receptors 32,50,51. The genes encoding transient receptor potential cation channels V1 (Trpv1) and A1 (Trpa1) were expressed in sensory ganglia in rats (sFPKM=51 and 23, respectively), and much lower (sFPKM=1.0–0.03) in other tissues in the circuit, which is consistent with previous observations that they are relatively specific for sensory ganglia 9,77.
1.2.4. Expression of genes encoding enzymes that inactivate oxylipins
Oxylipins are hypothesized to be biologically active in their unesterified (free) forms and thus can be inactivated by esterification into complex lipids, including triglycerides, cholesteryl esters, and phospholipid species that form the lipid bilayers of cellular and subcellular membranes. Enzymes that potentially inactivate oxylipins by mediating esterification of OXLAMs and other oxylipins into cholesteryl esters as part of lipoprotein synthesis [i.e. Lecithin-cholesterol acyltransferase (Lcat), sterol o-acyltransferase 1 (Soat1)] were broadly expressed throughout the pain circuit (Figure 1C). This also implies lipid turnover in the pain circuit is potentially rapid as lipids sequestered by esterification the lipids can be reutilized by cleavage from lipases (section 1.2.1).
1.3. Concentrations of OXLAMs
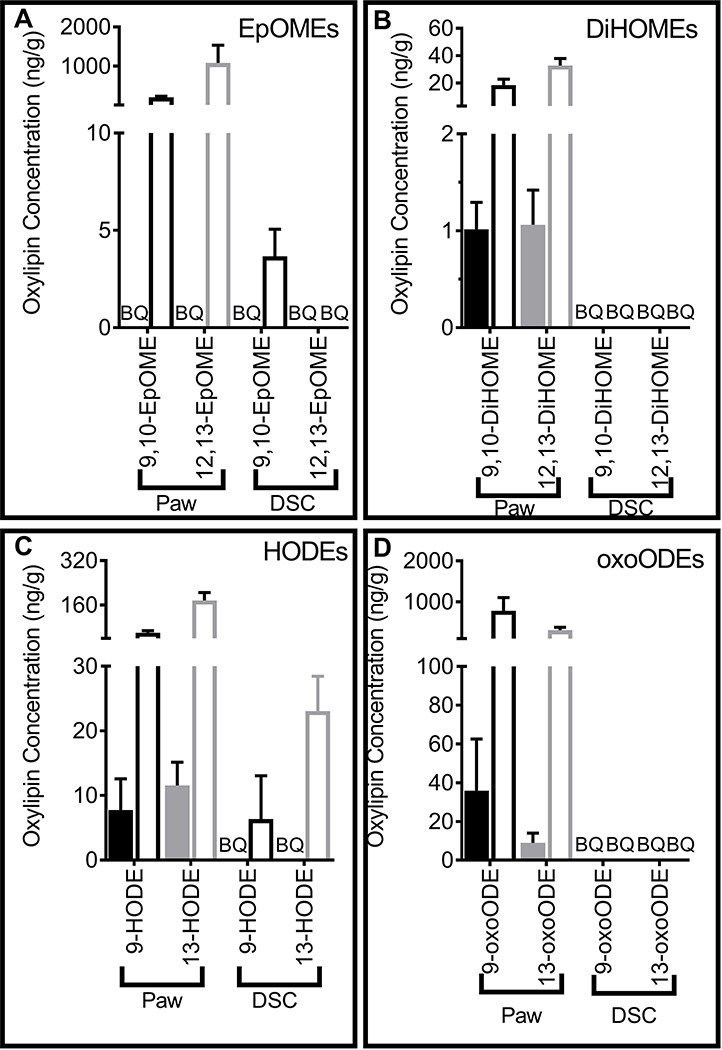
The tissue distribution of LA combined with tissue specific gene expression patterns of enzymes involved in OXLAM biosynthesis and metabolism suggested that OXLAMs would be relatively abundant in hind paw and released from esterified lipids in response to inflammatory stimuli. We performed LC-MS/MS analysis on hind paw and dorsal horn in rats to measure the concentrations of oxylipins in these two pain circuit tissues. Several OXLAM species including HODEs, oxo-octadecanoates (oxo-ODEs), epoxy-octadecenoates (EpOMEs), and dihydroxy-octadecenoates (DiHOMEs) were present at relatively high concentrations (1–30 and 18–780 ng/g, free and total pool, respectively) in hind paw (Figure 3A–D). Notably, of the unesterified oxylipins measured in the present study, 9-oxoODE and 13-HODE were the 1st and 3rd most concentrated in hind paw, respectively (Supplementary Table 1), while 12,13-EpOME and 9-oxoODE were the two most concentrated oxylipins in the total lipid pool of hind paw (Supplementary Table 2). Unesterified OXLAM concentrations tended to be less than 15% of total OXLAM pool (unesterified + esterified lipids) indicating that OXLAMs were mostly present as a population of esterified lipids (Supplementary Table 3). As predicted based on the relatively low concentration of LA and expression of biosynthetic enzymes in CNS tissue, fewer OXLAMs were detected in dorsal horn of spinal cord in both unesterified and total lipid pools. In fact, no OXLAMs were detected in the unesterified pool of the dorsal horn (Figure 3–A–D and Supplementary Tables 4–6).
Figure 3: Oxidized Linoleic Acid metabolites (OXLAMs) were measured in hind paw and dorsal horn of the spinal cord (DSC).
Liquid chromatography-tandem mass spectrometry was utilized to measure (A) Epoxy-LA derivatives (EpOMEs) (B) di-hydroxy-LA metabolites (DiHOMEs) (C) hydroxy-LA metabolites (HODEs) (D) LA-ketone derivatives (oxoODEs) in the unesterified (filled bars and symbols) and total lipid pools (open bars and symbols) of the hind paw and DSC. Oxylipin concentrations are presented as ng/g tissues (median ± interquartile range) (n=4). BQ = Below the limit of quantitation.
2. Arachidonic acid derived prostanoids
2.1. Prostanoid precursor fatty acid concentrations
Prostanoids are derived by the action of cyclooxygenase enzymes such as cyclooxygenase 1 and 2 (Prostaglandin-endoperoxide synthase 1 and 2, Ptgs1 and Ptgs2, respectively) on arachidonic acid (AA). Prostanoids have a prominent role in mediating inflammatory, vasoconstrictive, and anaphylactic reactions, as well as other functions throughout the body 6,61 and are induced in many inflammation models 23,36,64. In the pain circuit tissues examined, AA was most abundant in DRG (5.4 ± 0.3% of FA) and lumbosacral spinal cord (5.1 ± 0.1% of FA). AA accounted for 4.4 ± 3.1 and 1.9 ± 0.6% of FA in hind paw and sciatic nerve (Figure 1A).
2.2. Expression of genes related to prostanoids
2.2.1. Expression of genes encoding enzymes that synthesize prostanoids
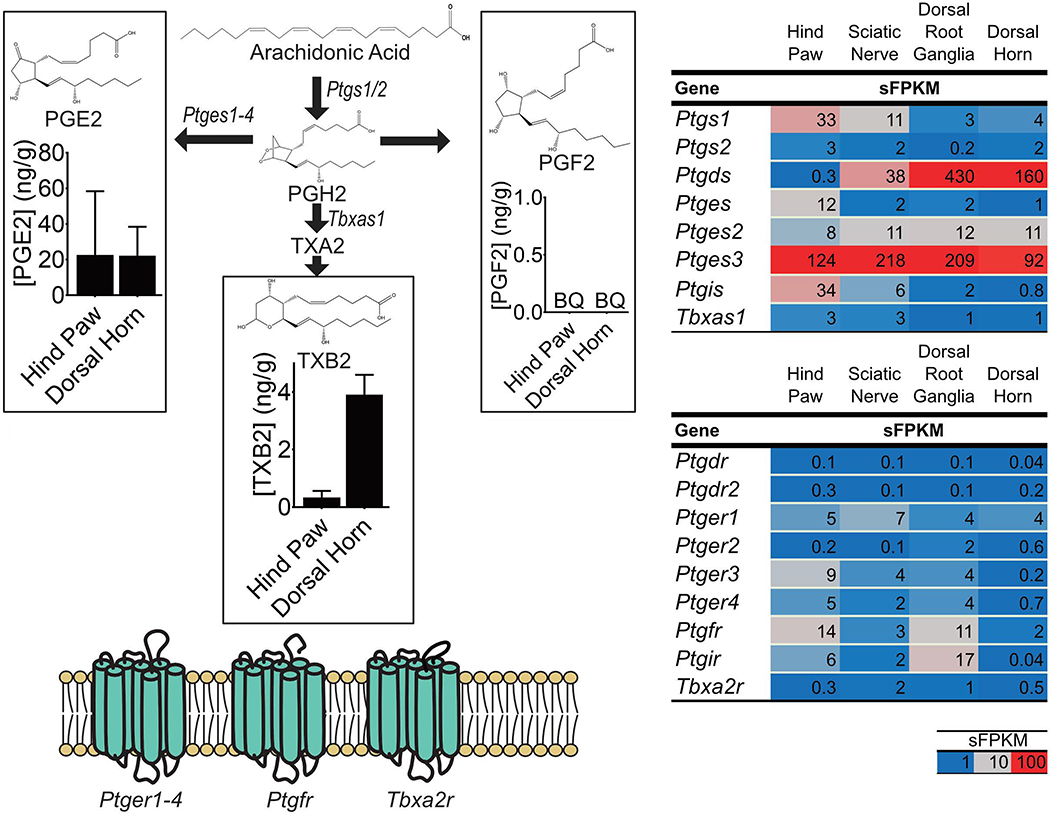
Many of the biosynthetic enzymes required for prostanoid synthesis act on AA in the unesterified lipid pool 3,71. As mentioned above (section 1.2.1) phospholipases were well expressed in each tissue indicating capability to release AA from membrane phospholipids (Figure 1C). As illustrated in Figure 4, the first step in prostanoid biosynthesis is conversion of unesterified AA to prostaglandin H2 by enzymes encoded by Ptgs1 and, under inflammatory conditions, Ptgs2 (Cox2)97. Expression of the Ptgs2 gene was low in the basal state across all pain circuit tissues examined (Figure 4), consistent with the view that Ptgs2 is inducible in response to inflammation 97. On the contrary Ptgs1 exhibited a tissue-specific expression pattern with generally low levels of expression in CNS and peripheral ganglia and 3 and10-fold greater expression in sciatic nerve and hind paw, respectively (Figure 4). The second enzymatic step(s) in prostanoid synthesis is conversion of PGH2 to specific prostaglandins or thromboxanes 97 (Figure 4). PGH2 is a precursor to several downstream mediators depending on enzymatic conversion. Three genes code for prostaglandin E synthetases, which catalyze conversion of PGH2 to PGE2 (Ptges, Ptges2, Ptges3). Of these, Ptges3 was highly expressed and ubiquitous in the pain circuit (sFPKM ≤ 92). Prostaglandin I synthetase (Ptgis), which catalyzes the conversion of PGH2 to PGI2, was mostly restricted to peripheral pain circuit tissue (especially skin, sFPKM=34). Prostaglandin D2 synthase (Ptgds), which converts PGH2 to PGD2, was >100-fold more highly expressed in rat sensory ganglia and dorsal horn (sFPKM > 100), compared to hind paw (sFPKM < 1) suggesting these tissues are uniquely situated to synthesize PGD2. In addition to conversion to other prostaglandins, PGH2 can also be converted into thromboxanes via an initial enzymatic conversion by thromboxane A synthase (Tbxas1), which converts PGH2 to thromboxane A2 (TXA2). TXA2 is a short-lived unstable intermediate which is non-enzymatically converted to thromboxane B2 (TXB2) 97. Tbxas1 is expressed relatively less than prostaglandin synthetases (1–3 sFPKM) in pain-circuit tissues.
Figure 4: Concentrations of specific prostanoids and expression of genes involved in their synthesis and signaling.
(Left Side) Prostanoids are synthesized by a series of steps starting with the action of Prostaglandin-endoperoxide synthases (Ptgs1 and 2) on unesterified arachidonic acid (AA). The product of this reaction is Prostaglandin H2 (PGH2) which is subsequently converted, by specific enzymes to prostanoids such as Prostaglandin E2 (PGE2), Prostaglandin F2 (PGF2) and Thromboxane A2 (TXA2). TXA2 is rapidly converted to Thromboxane B2 (TXB2). (Right Side) Expression of genes coding for enzymes involved in prostanoid synthesis as well as for prostanoid receptors, in pain circuit tissue, are presented as sFPKM (mean, n=3). (Left Side Graphs) Concentrations of prostanoid that were detected using liquid chromatography-tandem mass spectrometry are presented as median ± interquartile range (ng/g tissue, n=4). Prostanoids were only measured in the unesterified pool.
2.2.2. Expression of genes that code for prostanoid receptors
Genes coding for E-prostanoid receptors (Ptger1, 3 and 4) were expressed in all pain circuit tissues, with Ptger2 expression almost exclusively in the DRG. While AA concentration and gene expression data (Section 2.2.1) suggest a high capacity to for PGD2 synthesis in DRG and dorsal horn, expression of genes coding for receptors for PGD2 (Ptgdr) were low in all pain circuit tissue (sFPKM<0.3) although this gene has been detected in human DRG 21 and in human tibial nerve 27. Similarly, the receptor for TXA2 (Tbxa2r) was highest in the sciatic nerve and dorsal root ganglion (1–2 sFPKM). Notably, genes encoding the receptors for PGF2 (Ptgfr) and PGI2 (Ptgir) exhibited a tissue-specific expression pattern with relatively greater expression in skin and DRG as compared to other pain circuit tissues. Notably, Ptgfr has been reported in tibial nerve in humans 27, and in endometrium 31 while Ptgir has been observed mainly in blood vessels 19,27.
2.2.3. Expression of genes coding for enzymes that deactivate prostanoids
One of the major pathways by which PGE2 is degraded is via the oxidation of the 15(S)-hydroxyl group of prostaglandin E2 by 15-hydroxyprostaglandin dehydrogenase, reducing this mediator to less reactive metabolites 29. This enzyme has been reported in human skin 25 and we found here that this transcript was enriched in rat hind paw (sFPKM=19) relative to other pain circuit tissues (sFPKM<1.3), pointing towards a basal reservoir of enzyme capable of degrading prostanoids, which are generally produced on-demand.
2.3. Concentrations of prostanoids and thromboxanes
PGE2 and TXB2 were present in hind paw and spinal cord (Figure 4). Despite an 8-fold lower concentration in hind paw of AA compared to LA, PGE2 was the 2nd most abundant unesterified oxylipin in this tissue (Supplementary Table 1). In hind paw and dorsal horn, PGE2 was, 50- and 5-fold more abundant than TXB2, respectively. Higher concentration of PGE2 compared to TXB2 is consistent with the observed 40- and 90-fold higher expression (hind paw and dorsal horn, respectively) of Ptges3 as compared to the TXA2 synthase transcript Tbxas1. While PGE2 was concentrated at similar levels in hind paw and spinal cord, TXB2 was approximately 6-fold more concentrated in the spinal cord. Despite large differences in AA concentrations in dorsal horn compared to hind paw, TXB2 was the only prostanoid concentrated more in dorsal horn versus hind paw.
3. Arachidonic acid derived HETEs, leukotrienes and lipoxins
3.1. HETEs, leukotrienes and lipoxin precursor concentrations
HETEs comprise a group of monohydroxy-AA derivatives that are thought to play a role in inflammation 14. Similarly, leukotrienes are a group of oxylipins derived from AA via the action of Alox5 and also play a role in inflammation 14. AA derived lipoxins, a class of oxylipins containing 3 hydroxyl moieties and are considered specialized pro-resolving lipid mediators (SPMs) which are involved in the resolution of inflammation 14. Tissue distribution of AA is described above in results 2.1 and Figure 1A.
3.3. Expression of genes related to HETEs, leukotrienes and lipoxins
3.2.1. Expression of genes encoding enzymes involved in biosynthesis of HETEs, leukotrienes and lipoxins
The aforementioned tissue-specific expression of genes coding for phospholipases (Results 1.2.1) indicate the capability to generate unesterified AA from esterified lipids in each tissue (Figure 1C). Arachidonate lipoxygenase enzymes (Aloxs) catalyze the peroxidation of AA at the carbon number corresponding to the enzyme name (i.e. Alox5 adds a hydroperoxide moiety to carbon 5 of AA while Alox12 adds a hydroperoxide moiety to carbon 12 of AA) 63. These hydroperoxy-AA can then be rapidly reduced enzymatically 63, by Glutathione Peroxidases or non-enzymatically, 85 to form HETEs (Figure 5). Leukotrienes and lipoxins are formed from hydroperoxy-AA derivatives (HpETEs) via specific enzymatic reactions (Figure 5) 74. Specifically, 5-HpETE (which is formed by activity of Alox5 on AA) can be converted to both 5-HETE and leukotriene A4 (LTA4) (Figure 5A) 18. Subsequently, Leukotriene A4 hydrolase (Lta4h) converts LTA4 into LTB4. Alternatively, leukotriene C4 synthetase can convert LTA4 to cysteinyl leukotrienes, which are important because of their involvement in various physiologic functions such as asthmatic inflammation and itch 62. For simplicity, only some of the processing pathways are displayed in Figure 5A, although we note that alternatively, activity of Cyp-monooxygenase enzymes on AA directly catalyzes the conversion of AA to a series of HETEs or epoxy-arachidonic acid mediators (EETs) 102.
Figure 5: Synthesis pathways and concentrations of Arachidonic acid (AA) and Docosahexaenoic acid (DHA) derived oxylipins.
(A) Arachidonate lipoxygenase (Alox) enzymes act on AA to generate hydroperoxy-AA derivatives (HpETEs) which can be rapidly reduced to monohydroxy-AA derivatives (HETEs). Subsequent action of Alox enzymes on HpETEs leads to the generation of leukotrienes (i.e. Leukotriene B4, LTB4) and lipoxins (not shown). Additionally, HETEs can be oxidized to form keto-AA derivatives such as 5-oxo-eicosatetraenoic acid (5-oxoETE). (B) Genes coding for Alox enzymes as well as other enzymes involved in specific mediator synthesis (i.e. Leukotriene A 4 hydrolase, Lta4h) were expressed in tissue specific profiles in pain circuit tissue (mean sFPKM). (C) Several unesterified HETEs and HDHAs were detected in rat hind paw, but not rat dorsal horn. (D) The same enzymes that synthesize AA derived oxylipins can act on DHA to form hydroxy- and epoxy-DHA derivatives (HDHAs and EDPs, respectively) as well as specialized pro-resolving lipid mediators (Resolvins, Maresins and Protectins). (C) Almost all AA and DHA derived oxylipins measured, including epoxy-AA derivatives (EETs) that are generated by the action of cytochrome p450 enzymes on AA (pathway not shown) were present in the total lipid pool of hind paw and dorsal horn. Baseline oxylipins concentrations are expressed as median with quartile 1 (Q1) and quartile 3 (Q3) in ng/g tissue (n=4). BLQ indicates the concentration of that oxylipin was below the limit of detection.
Genes coding for enzymes capable of biosynthesizing HETEs, leukotrienes and lipoxins (i.e. Alox5 and 12, Ltc4s and Lta4h) tended to exhibit tissue specific gene expression patterns (Figure 5B). For example, Alox5 and Alox5-activating protein (Alox5ap) were expressed at a higher level in hind paw, compared to sciatic nerve and minimally expressed or absent in sensory ganglia and dorsal horn (Figure 5B), and both have some degree of enrichment in immune cells (Supplementary Figure 4). Additionally, Alox12 and Alox12b66 were almost exclusively expressed in rat hind paw while Alox15 was expressed only in DRG, albeit at low levels (Figure 1C and 5B, note the expression of Alox15 in adipose, Supplementary Figure 1). Notably leukotriene synthase (Ltc4s) was detectable in the pain circuit, where it was expressed at low levels in hind paw and sciatic nerve (sFPKM<5) and even lower expression levels were found in DRG and CNS (sFPKM<1). By comparison, leukotriene A4 hydrolase (Lta4h), which converts LTA4 into LTB4 (Figure 5B) expression was ≥27 sFPKM in all rat pain circuit tissues.
3.2.2. Expression of genes encoding receptors for HETEs, leukotrienes and lipoxins
Cysteinyl leukotrienes, particularly LTC4 have two known receptors (Cysltr1 and Cysltr2). These two receptors were expressed in a tissue-specific pattern with Cysltr1 being most highly expressed in hind paw and sciatic nerve (Figure 5B). Notably, Cysltr2 was almost exclusively expressed in DRG (Figure 5B) which is consistent with previous reports of Cysltr2 expression in a specific subpopulation of itch-responsive rodent DRG neurons 95. Interestingly, both Cysltr isoforms were expressed at low levels in dorsal horn. There are also two known LTB4 receptors (Ltb4r and Ltb4r2), remarkably expression of these genes, in pain circuit tissue, was exclusively observed in hind paw (sFPKM =13 and 11, respectively vs. sFPKM<1 in all other pain circuit tissues). While oxoeicosanoid receptor 1 (Oxer1) is also a receptor of 5-oxoETE in humans, there is no clear rodent ortholog, and hence it could not be included in our analysis 28.
3.3. Concentrations of HETEs, leukotrienes and lipoxins
The products of 5-, 12- and 15-arachidonate lipoxygenases, 5-HETE and 5-oxoETE, 12-HETE and 15-HETE respectively, were present in rat hind paw and spinal cord (Figure 5A and C). LTB4 and lipoxin A4 and B4 (LXA4 and LXB4, respectively) concentrations in both tissues were below the limit of quantitation in our assay (<0.06 and 0.12 ng/sample, respectively). However, except for 12-HETE, all HETEs, EETs, leukotrienes and lipoxins were only above our limit of detection in the total pool in dorsal horn.
4. n-3 mono-hydroxides, mono-epoxides and specialized pro-resolving mediators
4.1. Precursor concentrations
DHA was most abundant in lumbosacral spinal cord (4.7 ± 0.3% of FA), less abundant in sensory ganglia (1.9 ± 0.1% of FA), and comparatively scarce in sciatic nerve (0.6 ± 0.2% of FA) and hind paw (0.8 ± 0.3 % of FA)(Figure 1A). Using the same enzymes described above in Section 3.2.1 DHA can be converted to hydroperoxy-DHA (HpDHA) derivatives (Figure 5D). As summarized in Figure 5D, HpDHA can subsequently be reduced to hydroxy-DHA derivatives, isomerized to form epoxy-DHA derivatives (EDPs) or subsequently oxidized to form resolvins. Additionally, EDPs can undergo epoxide hydrolysis to form maresins and protectins which, along with resolvins are SPMs (see 3.1). Oxylipins derived from DHA are generally described as anti-inflammatory and pro-resolving 7.
4.2. Expression of genes related to n-3 mono-hydroxides, mono-epoxides and specialized pro-resolving lipid mediators
4.2.1. Genes involved in n-3 mono-hydroxides, mono-epoxides and specialized pro-resolving lipid mediators biosynthesis
Figure 5D depicts synthesis of oxylipins from DHA. Synthesis of oxylipins derived from DHA is catalyzed by the same enzymes that synthesize HETEs, EETs, lipoxins and leukotrienes and were, therefore, described above (Section 3.2.1). As described above, rat pain-circuit tissues exhibit tissue-specific expression patterns of genes coding for the cellular machinery required for n-3 monoepoxide and SPM biosynthesis (Figure 5B) such as Alox and Cyp enzymes. As mentioned above (Section 1.2.1 and Figure 1C), there was strong tissue-specific expression of Cyp genes, which convert DHA to different regio- and stereoisomers of monoepoxides and SPMs. Each pain circuit tissue had at least one Cyp isoform that was expressed at sFPKM>15 (Figure1 C), and these expression patterns have important implications for defining the specific biosynthetic capacity for each tissue.
4.2.2. Expression of genes encoding receptors for n-3 mono-hydroxides, mono-epoxides and specialized pro-resolving lipid mediators
To our knowledge, specific receptor targets for n-3 monoepoxides have not yet been identified. The gene coding for the Resolvin D1 (RvD1) receptor (Gpr32) in humans has been reported to be a pseudogene in rodents 28,41. Additionally, the gene coding for the Resolvin D2 (RvD2) receptor (Gpr18) had very low (sFPKM<0.2 in all tissue) expression in rat. However, SPMs have been reported to inhibit capsaicin and mustard oil induced TRPV1 and TRPA1 currents in mouse DRG neurons in vitro 58 and genes coding for these channels were most expressed in sensory ganglia (Section 1.2.3 and Figure 1C).
4.3. Concentrations of n-3 monohydroxides, monoepoxides and specialized pro-resolving lipid mediators
Several HDHA and EDPs were present in rat hind paw and spinal cord, indicating Cyp- and Alox-mediated metabolism of DHA in these tissues (Figure 5C). In both tissues, SPMs were below the limit of quantitation for our assay (~0.012–0.2ng/sample). However, the SPM precursors in the biosynthetic pathway for maresins (i.e. 14-hydroxy-DHA) and D-series resolvins/protectins (i.e. 16,17-EDP and 17-HDHA), were present in substantial quantities (>20 ng/g) in the total pool of both hind paw and cord. DHA derived oxylipins were 2–10-fold more concentrated in dorsal horn of the spinal cord compared to hind paw (Figure 5C), which is consistent with our finding of DHA being relatively abundant in dorsal horn (Figure 1A). Notably, DHA derived oxylipins in the free pool were only observed in hind paw (Figure 5C).
5. Effects of intraplantar carrageenan on oxylipin-related molecular pathways
Since it is possible that inflammation may alter the complement of lipid-related enzymes we assessed the potential role of oxylipins in inflammatory pain. The following sections describe results of RNA sequencing and multiplex in situ hybridization analyses in rat hind paw as well as LC-MS/MS oxylipin analyses in rat hind paw and dorsal horn of spinal cord after intraplantar injection of 100 μL of 1% CG (a commonly used, rodent inflammatory pain model, Figure 6A) 20,35. As previously reported, CG injection evoked pronounced edema (Supplementary Figure 3A) and nociceptive hypersensitivity in the hind paw persisting for up to 24 hours (Supplementary Figure 3 B–D) 76.
Figure 6: Intraplantar injection of carrageenan (CG) induced expression changes of genes encoding for proteins involved in oxylipin release, synthesis, delivery signaling and inactivation.
(A) CG was injected into the plantar surface of rat hind paw and tissue was collected at 0, 1 4, 8, 24, 48, 27 hours post injection for whole genome RNA sequencing. All genes that were statistically significantly impacted by CG injection are presented and grouped based on their function. (B) Phospholipases, (C) genes involved in oxylipin biosynthesis, (D) genes involved in delivery of preformed oxylipins to pain circuit tissues, (E) genes involved in oxylipin signaling and (F) genes involved in oxylipin esterification were all modified by CG. Gene expression is presented as mean sFPKM (n=3)
5.1. Effect of intraplantar carrageenan on oxylipin-related gene expression
Expression of all the oxylipin related genes in hind paw that were significantly modified by CG injection are presented in Figure 6B–F. CG injection significantly increased the expression, in hind paw, of genes coding for several phospholipases (Pla2g15, Pla2g2d, Pla2g2a) that can release PUFA and possibly oxylipins from membrane phospholipids (Figure 6B). Notably, expression of Pla2g5 was decreased 24 hours post-CG injection reaching a minimum at 48 hours post-CG injection (sFPKM=5 and 3 at baseline and 48 hours post-injection, respectively).
Expression of genes that encode enzymes involved in synthesis of several oxylipins were altered by CG (Figure 6C). Expression of Cytochrome P450 4B1 and 1A1 (Cyp4b1 and Cyp1a1, respectively), which encode enzymes that catalyze the conversion of PUFAs to hydroxy- and epoxy- oxylipins were increased and decreased by CG (respectively) indicating that expression of cytochrome p450 enzymes were not modified uniformly by CG injection. Additionally, Alox15, which encodes an enzyme that catalyzes the peroxidation LA, to synthesize 13-HODE (Figure 1), AA, to produce 15-HETE, LXA4 and EETs or DHA to produce 17-HDHA and several SPMs (Figure 5), was expressed at very low levels at baseline but increased to ~1 sFPKM at 1hr post-CG injection and subsequently returned to baseline expression (Figure 6C, Supplementary Figure 1). Conversely, Alox5 and Alox5ap (which encodes FLAP, required for Alox5 activation), which are involved in biosynthesis of 5-HETE, LTB4, LXA4, and DHA-derived resolvins were initially decreased but subsequently exhibited an increase above baseline at 24–48 hours post CG injection.
The above described CG-evoked changes in expression of genes that form lipid peroxides (Aloxs and Cyps) were accompanied by marked increases in expression of genes coding for enzymes that counter free-radical mediated lipid peroxidation (e.g. Gpx1 and 2) (Figure 6C) suggesting an increased capacity for lipid peroxide reduction to hydroxy-oxylipins. Additionally, prostaglandin endoperoxidases and synthases, which catalyze synthesis of PGE2, PGI2 and TXA2 were elevated by CG injection (Figure 6C). Remarkably, Ptgs2 expression increased approximately 10-fold within the first hour post injection.
Intraplantar CG injection also increased local expression of genes coding for enzymes involved in the delivery and local release of preformed OXLAMs including Pla2g7, lipoprotein lipase (Lpl), and lysosomal acid lipase (Lipa) (Figure 3D). Upregulation of these genes suggests an increased capacity for circulating oxylipins transport to tissue during inflammation.
The transcript encoding the G-protein coupled receptor (GPR) 132, Gpr132, which was originally identified as a proton sensing receptor 32,39,56,100 but has more recently been reported to respond to oxylipins (particularly OXLAMs) 51, had relatively low expression in pain circuit tissues in the baseline (sFPKM<1 in all tissue) (Figure. 1E), consistent with its predominant expression in visceral adipose and granulocytes (Supplementary Figure 1). During CG inflammation, Gpr132 expression was increased to 0.98 sFPKM (Figure 3E) which may reflect its broad expression in several classes of immune cells such as granulocytes, monocytes, and especially macrophages (Supplementary Figure 1) 11. These infiltrating cell populations enter the hind paw after inflammation but remain a small percentage of the overall hind paw transcriptome, which may explain the low expression of this transcript. Intraplantar CG injection reduced expression of cysteinyl leukotriene receptor 1 (Cysltr1) between 1- and 8-hours post CG injection (Figure 6E). Moreover, CG evoked increased expression of genes coding for prostanoid receptors for PGE2 and PGI2 (Figure 6E), some of which were sustained, suggesting an overall increase in prostanoid signaling following CG injection that coincides with the onset of behavioral hypersensitivity. The combination of increased expression of genes encoding enzymes that biosynthesize prostanoids, and prostanoid receptors is consistent with increased prostanoid signaling during inflammation.
Additionally, one gene coding for an enzyme involved in esterification of unesterified lipids into esterified lipid pools (Soat1) and two others involved in efflux, transport and clearance of esterified lipids (Abca1, Scarb1), were increased by CG (Figure 6F). These results combined with the above described increased expression of Pla2g7, Lpl and Lipa, indicate that the cellular machinery involved in the release, delivery and esterification of unesterified PUFA and preformed oxylipins (Figure 2) is enhanced with inflammation.
It should be noted that a brief review of human tissue expression of proteins encoded by genes in Figure 6B–F demonstrated that a substantial portion of these induced lipid pathway genes were identified in human immune cells and human tissues containing immune cells, although aside from Alox15, most were not strongly enriched for immune cell populations (Supplementary Figure 4). This is consistent with immune cell infiltration being a strong contributor to the CG induced lipid pathway gene expression changes 7,80.
5.2. Multiplex in situ hybridization
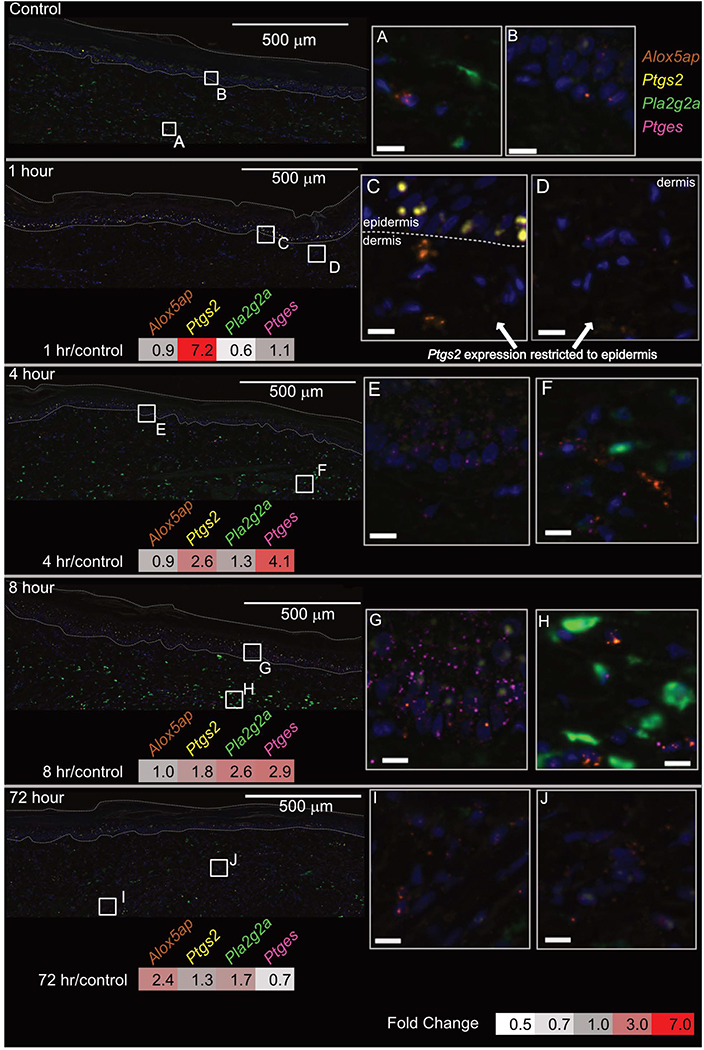
Several lipid biosynthetic enzymes were selected for further analysis. Multiplex in situ hybridization allowed us to examine the tissue localization patterns of Ptgs2, Ptges, Pla2g2a, and Alox5ap in the hind paw throughout the time course of CG induced inflammation. These genes were selected based on their expression levels (determined by RNASeq, above), as well as the magnitude and temporal patterns of their CG induced changes in expression, and relevance to lipid biosynthesis. Hence, Ptgs2 and Ptges were selected because both are rapidly induced, and strongly detected in the hind paw, each being a critical step in prostanoid biosynthesis as described in section 2.2.1. As hydrolysis of unesterified fatty acids and oxylipins is crucial for biosynthesis and activities of numerous oxylipins, we further investigated Pla2g2a since this phospholipase highly expressed and markedly altered by CG injection. The Alox5ap transcript encodes the activating protein (Five-lox activating protein, FLAP) of 5-lox (Alox5). The role of these enzymes is described in section 3.2.1, and they were selected due to their induction in the hind paw dataset between 24–72 hrs.
In the basal condition, all four of these genes are expressed to some extent and showed at least some level of fluorescence signal. After staining, relatively large blocks of hind paw tissues were scanned for visualization. To facilitate orientation within these sections, two dashed lines are drawn, with the area between them indicating the epidermis. The most highly expressed baseline gene, Pla2g2a was primarily expressed in dermis (Figure 7 control and A), with dense expression in sparse cells throughout this area of the section. By contrast, Alox5ap, sparsely labeled cells throughout dermis and epidermis, although this pattern of staining was more sporadic, with the positive cells showing fewer granules of reactivity (seen in Panel B). This is consistent with a low expression level in one or more cell-types, and consistent with the proposed expression of this enzyme in adipocytes and immune cells, which would be minor cell populations in basal state hind paw. The expression of genes encoding prostanoid synthetic enzymes (Ptgs2 and Ptges) were difficult to detect at baseline, and were more readily detected after CG inflammation, as described below. At 1 hour after CG inflammation, Ptgs2 was approximately 7.2-fold induced over baseline, and showed prominent induction in the stratum basale of epidermis (Figure 7, yellow). This induction was confined to epidermis, and no Ptgs2 reactivity was detected in dermis (Figure 7D), although sporadic Alox5ap reactivity was noted (Figure 7C, orange). At 4 hours post CG injection, Ptges appeared to increase expression in epidermis (Figure 7E), whereas other markers remained unaltered (Figure 7F). By 8 hours, Ptges reactivity was prominent in epidermis, showing broad swaths of punctate reactivity, presumably corresponding to expression in a major cell type in this area, as the staining was diffuse throughout this region. Note that all transcripts except Alox5ap were upregulated transcriptionally at this time point, and the signal for Pla2g2a appeared to increase correspondingly in dermis, showing brightly filled cells in approximately the same pattern as in control (Figure 7H). By the 72-hour time point, induction of prostanoid synthetic and Pla2g2a enzymatic transcripts had largely resolved, but induction of Alox5ap was at its maximum transcriptionally. Staining of Alox5ap was faint, as in previous time points, and was inconclusive (Figure 7I, J). Note that the RNA-Seq analysis included more tissue than what was stained for, and Alox5ap induction may have occurred in another part of the tissue.
Figure 7: Spatial and temporal changes in expression of lipid related genes induced by carrageenan injection.
Multi-plex in situ hybridization revealed the spatial representation of CG induced gene expression changes. A large representative section of hind paw, at low magnification, is presented for one rat at 0, 1, 4, 8 and 72 hours post-CG injection along with two high magnification inserts. The heat map for each time point displays expression fold change vs. control (determined from RNASeq experiment, Figure 4). Notably, the in situ hybridization experiments clearly illustrate the spatial expression pattern of each gene at its highest expression time point. Cell nuclei (blue) were counterstained using a DNA-binding dye 4′,6-diamidino-2-phenylindole (DAPI). Scale bars in inserts are 10 μm. Dashed lines in panorama images outline the epidermis.
5.3. Effects of CG on oxylipin concentrations
The pronounced hind paw edema after intraplantar CG injection (Supplementary Figure 3) increased the volume and weight of the paw samples collected, therefore, impacting interpretation of oxylipin concentrations per gram of tissue. To control for edema we corrected oxylipin concentrations for total oxylipin content in hind paw (i.e. [oxylipin]/sum of all [oxylipins] x 100%). Without this correction for edema, almost all oxylipins we measured were decreased after injection of CG, on a ng/g tissue basis, indicating that the increase in amount of tissue caused by CG injection was not attributable to lipids. The following section reports on the CG evoked changes in oxylipin concentrations as a percent of total oxylipins.
We observed that intraplantar CG injection increased percent of unesterified 9-HODE, 9,10-DiHOME and 12, 13-DiHOMEs 2, 4 and 6-fold, respectively in hind paw (Supplementary Table 1). Additionally, in the total pool (unesterified + esterified), percent of 12,13-DiHOME increased almost 2-fold in hind paw, while percent of 9-oxoODE decreased in this tissue, after CG injection (Supplementary Table 2). In contrast, oxylipin concentrations were unchanged by CG injection in spinal cord.
Since it has been reported that the oxylipins are bioactive in their unesterified form and deactivated by esterification, we also measured, for each oxylipin, the percentage contribution of the unesterified pool to the total concentration of that oxylipin. In general, we found CG injection increased the proportion of almost all OXLAMs measured in the unesterified fatty acid pool of hind paw (Supplementary Table 3). The finding of increased proportion of unesterified OXLAMs is consistent with gene expression data showing enhanced expression of enzymes responsible for the hydrolysis of esterified oxylipins, such as Pla2g2a which increased 3-fold after CG injection (Figure 3B).
As predicted based on the gene expression changes outlined in section 5.2, intraplantar CG injection increased concentrations of TXB2 approximately 3-fold (Supplementary Table 1) in hind paw while changes in PGE2 concentrations were not significantly altered (Supplementary Table 1). Intraplantar CG decreased the concentration of TXB2 and had no effect on PGE2 in spinal cord (Supplementary Tables 1 and 4).
The only oxylipin to be increased in hind paw by CG injection, when expressed as ng/g tissue (i.e. without correcting for edema), was 5-HETE. Additionally, 5-HETE was the only non-LA derived lipoxygenase product to increase in the unesterified lipid pool after CG injection (Supplementary Table 1). Intraplantar CG injection also increased the adjusted concentrations of 5-oxoETE, and 15-HETE in the total lipid pool in hind paw but did not affect concentrations in dorsal horn. Contrary to what we observed with lipoxygenase derived AA products, several Cyp products (EETs) were increased in the dorsal horn by hind paw injection of CG (Figure 6C). Intraplantar CG injection broadly increased adjusted concentrations of EDPs and HDHAs in the total lipid pool of both hind paw and dorsal horn (Supplementary Tables 1–6).
6. In vivo effects of oxylipins on thermal nociceptive sensitivity
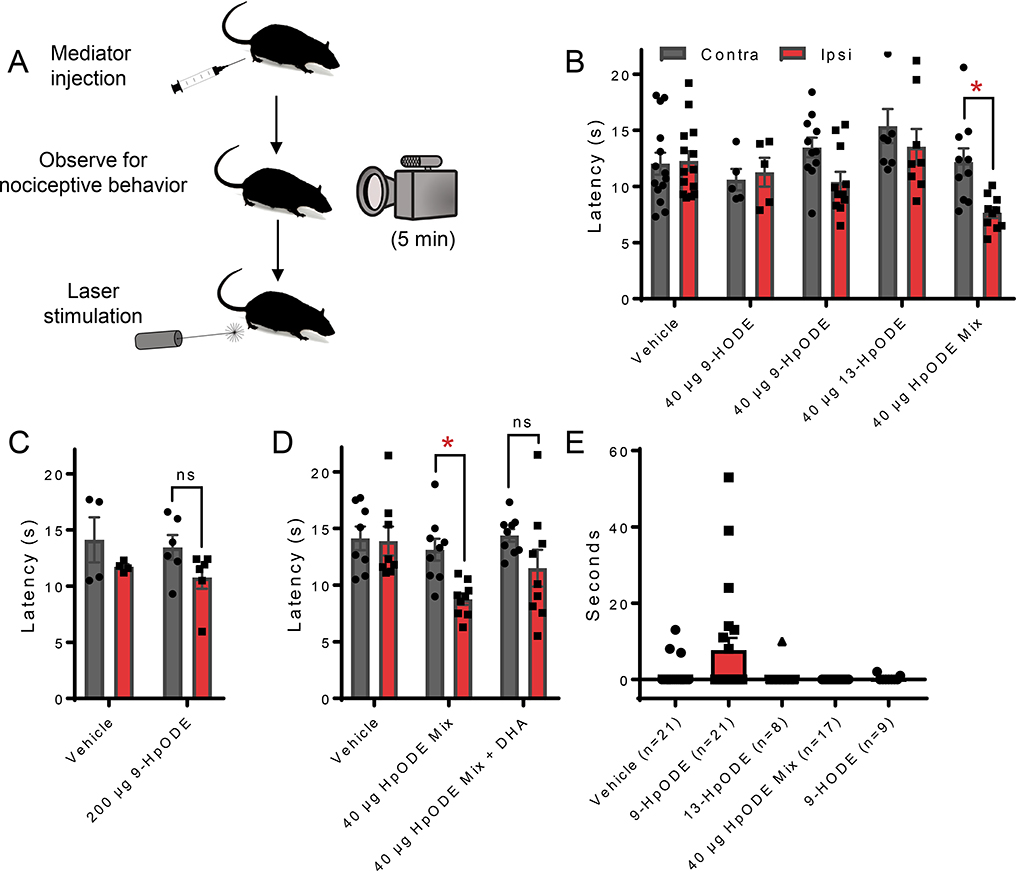
Based on the findings summarized above we investigated potential roles that oxylipins play, with respect to nociception, in vivo. Our gene expression data showed pronounced increases in the expression of phospholipase as well as several Alox and Cyp genes in hind paw in response to inflammatory stimulus. As LA was the most abundant precursor PUFA in hind paw and we observed increases in adjusted concentrations of several OXLAMs (which were among the most abundant class of oxylipins in this tissue), we decided to examine the effect of a mixture of HpODEs (the precursor to several OXLAMs). A 40 μg mixture of 9- and 13-HpODE, injected intradermally into the hind paw, evoked significantly lower relative withdrawal hind paw latencies (injected paw vs. non-injected paw) in response to a laser delivered heat stimulus that preferentially activated C-Fiber sensory neurons (Figure 8A). Injecting 40 μg of 9- or 13-HpODE or 200 μg of the 9-HpODE mixture did not significantly modify the relative thermal withdrawal latency (Figure 8A and B). Additionally, we examined the effects of injecting the HpODE mixture in combination with other classes of oxylipins and observed that the combination of the HpODE mixture and several monohydroxy DHA derivatives did not lead to significant changes in relative hind paw withdrawal latency (Figure 8C).
Figure 8: Intradermal injection of oxylipins impacts nociception in vivo.
(A) Schematic detailing oxylipin injection experiments. Rats were observed for nocifensive behaviors (i.e. guarding, licking, shaking) for at least 5 minutes following oxylipin injection and thermal hyperalgesia was assessed by withdrawal from laser evoked heat stimulus. (B) Intradermal injection, into the hind paw, of a mixture of 18 μg 9- and 22 μg 13-Hydroperoxy-octadecenoate (9- and 13-HpODE, respectively) evoked statistically significant decreased withdrawal latency from a laser delivered temperature ramp in the injected paw compared to the contralateral paw (p<0.05). This effect was not observed when only 1 HpODE was injected, 40 μg of 9-hydroxy-octadecenoate (9-HODE) was injected or when vehicle was injected. (C) An increased dose of 9-HpODE (200 μg) also did not evoke changes in withdraw latency in the injected paw compared to the contralateral paw. (D) No differences in withdrawal latency between injected and contralateral paws were observed upon intradermal injection of the above described HpODE mixture in combination with 26 μg 16,17-EDP and 13 μg 4-HDHA. (E) Oxylipin injection did not evoke strong nociceptive behavioral responses and total time that animals displayed nociceptive responses was not statistically different between vehicle or any oxylipin group. Statistical significance for thermal hyperalgesia was assessed by two-tailed, paired t-test. * signifies that means were significantly different (p<0.05). Statistical significance for nociceptive behavior observations was assessed by a Kruskal-Wallis test followed by Dunn’s multiple comparison test (comparing each oxylipin group to vehicle).
To assess if oxylipins play a role in pain modalities other than thermal hyperalgesia, we quantified the total amount of nocifensive behaviors (i.e. guarding, licking, shaking) for at least 5 minutes after injection of several different oxylipins/mixtures described above. Overall, rats exhibited very little nociceptive behavior in response to vehicle, or oxylipin injection. The majority of responses were sporadic and short-lived, and occurred only after 9-HpODE injection (Figure 8E). Note that of 21 animals tested, 14 in the 9-HpODE group showed no nocifensive behaviors over the 5 minutes of observation. None of these changes were significant compared to vehicle injection (P=0.25).
Discussion
Here we systematically investigate potential links between lipids and nociception and inflammation using quantitative transcriptomic and lipidomic measurements. We measured the concentrations of PUFA precursors and the expression of genes encoding enzymes implicated in oxylipin synthesis, delivery, release, signaling and inactivation throughout the pain circuit. We then examined the impact of intraplantar CG inflammation on transcriptional regulation, both temporally and spatially, in hind paw. Furthermore, we utilized LC-MS/MS to measure the concentrations of oxylipins before and after CG injection. Together this work provides an empirical and conceptual framework linking oxylipins and their precursor PUFAs to local biosynthetic potential, extrapolated from gene expression. This framework is a critical step towards explaining the role of lipid autacoid signaling in nociception and provides an in-depth reference for investigating the role of lipids in nociceptive circuits, and the contributions of these lipid mediators to peripheral pain pathways.
Skin is the largest sensory organ in the body and LA and OXLAMs are the most abundant oxylipins in this tissue. The most highly concentrated oxylipin in skin was 9-oxoODE which is synthesized from 9-HODE supporting our previous finding of high Alox12b expression in skin66. We and others have reported on the role of OXLAMs in nociception in skin25,30,59,66. Unexpectedly, PGE2 was among the most abundant oxylipins in skin, despite relatively low levels of its precursor (AA) in that tissue. The disproportionately high concentration of PGE2 in skin can be explained by our finding of relatively high expression of Ptgs1 and Ptges3 (which encode for enzymes that synthesize PGE2 from AA), compared to Alox enzymes in skin. While concentrations of LA as well as expression levels of Alox12b are high in basal state skin, their levels are unaffected by CG inflammation, consistent with the proposed structural barrier role of LA and OXLAMs in skin5. Similarly, Ptgs1 was well expressed in skin and Ptgs2 expression increased acutely in epidermis after CG injection and is not highly enriched in human immune cells suggesting it is not derived from these infiltrating cells. The rapid induction of Ptgs2 (a sharp peak at 1 hour after inflammation) is also inconsistent with the arrival of neutrophils and macrophages7,43, partially supporting the role of a local tissue source such as the skin. These considerations support our finding of Ptgs2 induction in the keratinocyte layer of the epidermis, which is also consistent with previous reports examining spatial-temporal distribution of Ptgs2 transcripts and COX2 protein in inflammatory models2,4,48,92. An overall perspective of the inflammatory process incorporates a dual response, whereby genes are either induced locally (as appears to be the case for Ptgs2) or imported from infiltrating immune cells, which generally comprise a subpopulation of the total cellular complement in inflamed tissue and occur in distinct temporal sequence. These incoming cells import and release potent mediators that support inflammation and nociceptive processes7,13. In general, the findings of the present report indicate that immune cell infiltration is not the primary mode of synthesis for the most highly concentrated oxylipins, such as PGE2 and OXLAMs in skin. This hypothesis does not exclude a role of immune cells in the synthesis of other oxylipins, as has previously been reported7,80. Nonetheless, many lipid biosynthetic enzymes are enriched in immune cells, and as such we do not rule out the involvement of these cells in contributing to either the inflammatory process, or the hyperalgesia driven by the peripheral inflammatory state.
As oxylipins such as PGE2 are known to be sharply induced by inflammation, and are indeed mediators of many inflammatory processes7,10,16,26,58,66,73,84, we investigated the response of oxylipins to CG-induced intraplantar inflammation. It should be stressed that the presence of inflammatory edema causes tissue distortion and hypertrophy which complicated the interpretation of oxylipin concentration measurements. We observed that concentrations of all oxylipins, except 5-HETE, decreased after CG injection, likely as a result of edema, affecting normalization by gross weight of tissue. This is consistent with the idea that the source of oxylipins is local, as opposed to being delivered via circulation or infiltrating cells, and hence is diluted by the increase in tissue mass after inflammation. Our finding of increased hind paw concentration of 5-HETE after CG injection warrants further investigation as 5-HETE has been demonstrated to activate the capsaicin receptor (Trpv1) in cultured rat DRG neurons33 suggesting a substantial role of 5-HETE in CG-induced pain. An explanation for the large increase in 5-HETE concentration is that it is imported from infiltrating immune cells, and thus goes up sharply from a very low baseline. This observation is in line with previous reports demonstrating 5-lipoxygenase (Alox5) activity in infiltrating immune cells101 and human ALOX5 and ALOX5AP expression in macrophages and granulocytes. Furthermore, our in situ hybridization results showing Alox5ap expression primarily in deep dermal layers, potentially consistent with expression in a cellular constituent of dermal tissue such as adipocytes or a resident immune cells37,86.
Our transcriptomic and lipidomic results in skin indicate that OXLAMs are uniquely positioned to modulate pain and nociception because they are made in high quantities in skin. We found that intradermal injection of HpODEs evoked thermal nociceptive hypersensitivity which was in agreement with previous reports of OXLAMs and prostaglandins inducing nociceptive hypersensitivity when injected into hind paw15,40,59,66, although the receptors mediating the OXLAM effects are unknown. Oxylipin hydrolysis from esterified precursors generates local signaling molecules produced on demand, although the precise processing steps involved in making the wide variety of lipid mediators is still being delineated56. To investigate the oxylipin signaling circuit, we examined the expression of receptors for oxylipins in skin and sciatic nerve trunk, as well as in DRG cell bodies, and in the dorsal spinal cord. We detected tissue specific expression patterns for oxylipins receptors. For example, several leukotriene and prostanoid receptors were expressed in skin, while receptors through which OXLAMs are believed to signal such as Trpv1, Trpa1 and Gpr132 were expressed at very low levels in skin but rather were expressed in DRG, though it has been reported that gene expression in skin may not be reflective of actual receptor protein levels75.
The roles of oxylipins in nociceptive processes are still being elucidated56. One mechanism by which oxylipins may modulate nociceptive responses is by potentiating the release of pro-nociceptive neuropeptides such as calcitonin gene related peptide by PGE2 and OXLAMs66,98 as observed in primary DRG cultures. Oxylipin signaling has been shown to evoke or modulate nociceptive sensitivity, in vivo, in rodent models of burn or inflammation24,108. We observed that oxylipins were mostly concentrated in the esterified lipid pool (~90%) rather than the unesterified ―free‖ lipid pool that is generally considered the active pool. After CG injection the proportion of oxylipins in the free lipid pool increased, consistent with reported roles of unesterified oxylipins in pain and inflammation59,66. Besides 5-HETE, this relative increase was specifically observed for OXLAMs. Notably, unesterified concentrations of AA- and DHA- derived oxylipins were less concentrated, and relatively unchanging in dorsal horn as compared to hind paw, despite relatively high concentrations of their precursors. Taken together, this indicates unesterified oxylipin concentrations are tightly regulated in the dorsal horn implying oxylipin signaling in spinal cord, in response to a peripheral inflammatory stimulus, may be limited.
This work has several limitations, some of which are discussed. Firstly, our LC-MS/MS analysis was targeted and analysis of oxylipins was limited to those for which we were able to procure or synthesize authentic standards and resolve them with our LC-MS/MS analytical methods105. Therefore, the oxylipin concentrations reported here represent a fraction of all known oxylipins in these tissues. However, the PUFA concentrations in pain circuit tissues reported here do provide quantitative insight into the presence of oxylipin precursors and inform further studies investigating these lipids. Two unique fragment ions and HPLC were used to identify and resolve target analytes in our LC-MS/MS assay105. Nevertheless, the potential for matrix interference and failure to resolve closely related, unanticipated isomers cannot be ruled out. Additionally, not all components of the lipid synthesis and signaling pathways are conserved between rodents and humans which can impact the interpretation of the RNA-Seq measurements in several ways28,41. Even more fundamentally, not all oxylipin-related gene products examined in this study in rat have a clear human ortholog, or vice versa. Beyond this, estimation errors from various sources makes RNA-Seq only semi-quantitative in determining basal amounts, and technical considerations may lead to substantial differences in quantification. Another limitation affecting the comprehensiveness of this report is the absence of precise quantitative protein analyses, as well as precise subcellular expression. The LC-MS/MS oxylipin measurements, however, are the main focus of the paper, allowing for precise quantitative measurements of these mediators. Further, the identification of such mediators and oxylipin synthetic genes at the whole-tissue level is useful to guide future pharmacological and physiological studies where these body compartments are the likely locus of oxylipin action.
This report provides, to our knowledge, the most comprehensive analysis of oxylipins in pain circuit tissues to date. We provide a data-driven framework linking precursor PUFAs, oxylipins and genes coding for enzymes catalyzing the release, synthesis, delivery and signaling of oxylipins to pain.
Materials and Methods
Animals
The protocol was approved by the Animal Care and Use Committees of the National Institute of Dental and Craniofacial Research and National Institutes of Health (NIH) Clinical Center. Procedures followed the NIH Guide for the Care and Use of Laboratory Animals (NIH Publication No. 80–23). Male Sprague-Dawley rats (Harlan, MD) were housed in an animal facility having regulated temperature, humidity, and a 12 h light/12 h dark cycle. The rats had ad libitum access to water and Rodent NIH-31M modified formula chow (Zeigler). Food was replaced every 2 or 3 days. Three cohorts of rats were used for this report. One cohort of rats was euthanized and hind paw, sciatic nerve, dorsal root ganglia and dorsal horn of spinal cord were collected for baseline fatty acid analysis and RNASeq. The euthanization and tissue collection procedures that were performed on this cohort has been described previously 66. The remaining 2 cohorts of rats were either killed by isoflurane anesthesia followed by decapitation or injected with 100ul of 1% Carrageenan (CG) and euthanized, using the above described procedure, at set time intervals post-injection. For 1 of these rat cohorts, rats were euthanized at 0, 1, 4, 8, 24, 48 and 72 hours post injection and hind paw was collected for RNA-Seq analysis (n=3 per time point). For the other cohort rats were euthanized at 0, 1, 4, and 24 hours (n=4, 4, 4, and 6, respectively) and hind paw and dorsal horn were collected following previously published procedures 66 for oxylipin analysis. Rat DRG and sciatic nerve RNA-seq data are available under project PRJNA313202 in the Sequence Read Archive (SRA) database 75. All tissues were immediately collected, frozen and stored at −80°C.
Fatty Acid Analysis
Tissue samples were thawed, weighed, and homogenized in butylated hydroxytoluene (BHT)/methanol according to the method of Folch et al 22 and transmethylated as described by our group previously 66. Fatty acid methyl esters were analyzed with an HP-5890B gas chromatograph equipped with a flame ionization detector (Hewlett-Packard, Palo Alto, CA) and a fused silica capillary column (DB-FFAP, 30 m × 0.25 mm i.d. × 0.25 μm film thickness, J & W Scientific, Folsom, CA). The detector and injector temperatures were set to 250°C. The oven temperature program began at 130°C and increased to 175°C at the rate of 4°C/min, then at the rate of 1°C/min to 210°C, and finally increased at the rate of 30°C/min to 245°C, with a final hold for 15 min. Hydrogen was used as carrier gas at a linear velocity of 50 cm/s. A custom mixed, 30-component, quantitative methyl ester standard containing 10–24 carbons and 0–6 double bonds was used for assignment of retention times and to ensure accurate quantification (Nu Chek Prep 462, Elysian, MN). Fatty acid data were expressed as % of total peak area, which corresponded to weight% to within 5%, as demonstrated by quantitative standard mixtures. Internal standards were used to calculate tissue fatty acid concentrations.
Oxylipin extraction
Oxylipin extraction methods have been previously published 66. Briefly, hind paw and dorsal horn were transferred into FastPrep Lysing Matrix tubes on ice (MP Biomedicals, USA; Lysing Matrix A for hind paw, Lysing Matrix D for dorsal horn) and ice-cold methanol containing an antioxidant mixture were immediately added to each sample. Internal standards were added and samples were homogenized by shaking with a FastPrep-24 homogenizer (MP Bio). Proteins were precipitated by storing samples at −80°C for 1 hour followed by centrifugation at 17000g in 4°C for 10 min. Half the supernatant was transferred to a new microfuge tube. and stored in −80 °C until solid phase extraction (SPE) purification and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of unesterified oxylipins. To analyze the total oxylipin pool, the remaining half of the supernatant was saponified with methanolic sodium carbonate at 60°C for 30 min under gentle shaking. The solution was then neutralized (to pH 7) using acetic acid and stored in −80°C overnight until SPE and LC-MS/MS analysis. SPE and LC-MS/MS procedures have been described previously 66,105
RNA purification for RNA-Seq analyses
For RNA-Seq analysis, rats were deeply anesthetized with 4–5% isoflurane, decapitated and rapidly dissected. In these experiments, all animals were adult male Sprague-Dawley rats (Harlan). Spinal cords were isolated by hydraulic ejection using ice cold saline, cooled, and dissected to separate the dorsal quadrant of the spinal cord. Hind paws were dissected by cutting a square of tissue from the hind paw similarly to the dissection described above for lipid analyses. As described previously, DRGs and sciatic nerves were removed after laminectomy, and were collected as part of a previous study 66. The sciatic nerve was traced to identify L4, L5 and L6 DRGs, and L4 and L6 DRGs were collected and sequenced. Briefly, sciatic nerves were dissected by transecting the nerve to remove a section from just distal to the sciatic notch and extending to just above the sciatic trifurcation. All tissues were collected and immediately transferred to chilled tubes on dry ice containing FastPrep™ Lysing Matrix D grinding matrix (MP Biomedicals) and stored at −80°C until processed. Rat tissue samples were homogenized in Qiazol reagent (Qiagen Inc, Valencia CA) using a Fastprep 24 homogenizer (MP Biomedicals, Santa Ana, CA) followed by purification using the RNeasy Mini kit (Qiagen Inc., Valencia CA) with added DNAse digestion, according to manufacturer’s instructions. RNA integrity was assessed after gel electrophoresis using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA).
For rat hind paw and dorsal horn samples, mRNA libraries were constructed from 0.5–1 μg mRNA using the Illumina TruSeq RNA Sample Prep Kits, version 2. Samples selected for sequencing generally had RNA Integrity Numbers above 9. The resulting cDNA was fragmented using a Covaris E210. Library amplification was performed using 8–12 cycles to minimize the risk of over-amplification (8 cycles for hind paw samples, 10–12 cycles for dorsal horn samples). Unique barcode adapters were applied to each library. Libraries were pooled in equimolar ratio and sequenced together on a HiSeq 2000 with ver 3 flow cells and sequencing reagents. At least 34 million 100-base read pairs were generated for each individual library. Data was processed using RTA (version 1.12.4.2, hind paw; version 1.13.48 dorsal horn) and CASAVA 1.8.2. For rat dorsal root ganglion and sciatic nerve samples, sequencing was performed as described in the original publication in which these datasets were published 75.
Alignment, quantification, analysis and visualizations of RNA-Seq count data
Samples were aligned using the MAGIC pipeline 107 and a rat target genome built based on the Rn6 genome build as described previously 42. Quantification and normalization of gene counts was performed to produce expression estimates reported as significant fragments per kilobase per million aligned reads (sFPKM) as described previously 107. Briefly, this normalization method considers library size, gene length, insert size of the library, level of genomic contamination, and the total number of reads mapping uniquely to protein coding genes. Subsequently, several quality control metrics were examined using outputs from MAGIC with samples showing uniformity within groups. For the hind paw time course experiment significance of gene changes over time was determined by the differential expression analysis within MAGIC, which is described in greater detail in 42,78,107. For this report, we only examined a gene panel related to lipid signaling based on literature reviews. Briefly, this is a method of determining differentially expressed genes that does not assume a monomodal distribution, and determines significance based on separation of variance after noise subtraction. Significance was not tested across tissues in these analyses. Gene expression data is represented in heatmaps showing the average basal level of expression (sFPKM) according to tissue. For genes that were significant in the rat hind paw after inflammation with carrageenan, expression analysis was examined in a large dataset of tissues from the Human Protein Atlas 93. For this analysis, data were mined from the overall consensus dataset with supplementary information taken from the blood dataset to examine specific subclasses of immune cells. These expression values were taken directly from the Human Protein Atlas and clustered in R using the ward. D2 method and the hclust and heatmap.2 functions. Values are normalized such that the maximum value in each row is plotted as 1.0 and all other values are plotted as the ratio relative to that. In order to normalize for outlier expression values, 0.2 was considered a midpoint (yellow color) to more easily display a larger range of expression values without the use of a log scale. Although values were clustered in both dimensions, clusters were rearranged manually to a minor degree to keep similar tissues (by biological function) together.
Multiplex fluorescent in situ hybridization
We further interrogated CG induced RNA expression changes by utilizing multiplex fluorescent in situ hybridization to understand the localization of gene expression changes. In situ hybridization was performed on 6 μm thick sections (n=2 per time point) of paraffin blocks that contained paraformaldehyde fixed hind paw samples collected at 0, 1, 4, 8, and 72 hours post CG-injection (described above). In situ hybridization was performed using the RNAscope® Multiplex Fluorescent assays v2 (Advanced Cell Diagnostics, Newark, CA) with Tyramide Signal Amplification (Opal™ Reagent Systems; Perkin Elmer, Waltham MA) using probes for Ptgs2 (Cat#526151), Ptges (#827831), Pla2g2a (#827821), and Alox5ap (827811). Imaging of whole sections was performed using an Axio Imager.Z2 slide scanning fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a 20X/0.8 Plan-Apochromat (Phase-2) non-immersion objective (Zeiss), a high-resolution ORCA-Flash4.0 sCMOS digital camera (Hamamatsu), a 200W X-Cite 200DC broad band lamp source (Excelitas Technologies, Waltham MA) and 5 customized filter sets (Semrock, Rochester NY) optimized to detect the following fluorophores: DAPI, Opal520, Opal570, Opal620, Opal690. Image tiles (600 × 600 μm viewing area) were individually captured at 0.322 micron/pixel spatial resolution and seamlessly stitched using ZEN2 image acquisition software (Zeiss). Appropriate color tables were applied to each image channel and pseudocolored, stitched images were overlaid to create a single merged composite image. These images were analyzed and described qualitatively. To create representative images in Figure 7, the images were processed using FIJI (NIH, USA), Affinity Designer (Serif, Europe) and Adobe Photoshop (Adobe, USA First, all fluorescence channels in all the images were corrected for inter-channel bleed through. Bleeding of the yellow channel (Ptgs2) into the orange channel (Alox5ap) was observed for all scans and corrected for by converting the images of both these channels to 8-bit color format and subtracting the yellow channel image from the orange channel image in image J. Additionally, for the 1 hour image bleeding of the blue channel DAPI into the yellow channel was observed and corrected for using the process described above. In order to create large panoramic images seen in Figure 7 we had to increase the brightness of several of the channels. When the brightness was increased, we observed non- specific, blurry signal (particularly in the stratum corneum) in the yellow and magenta channels. This type of signal is not consistent with RNAScope technology. This background signal was removed from the yellow channel images in Image J by applying a Gaussian Blur (radius =10) followed by a minimum and maximum filter (radius = 10 for both) to isolate the non-specific blur. This blur was then subtracted from the original yellow channel image. The same process was performed for the magenta channel images (using radius of 5 for each step), however, magenta channel images were brightened by setting the white point of each image to 50 before performing the above described transformation. For the final figure, all color channels were brightened by setting the white point to 90 (Ptgs2), 50 (magenta), 100 (Pla2g2a), 50 (Alox5ap), 140 (DAPI) and the bright field channel was enhanced by setting the white point to 170 (figure zoom-ins do not use the enhanced bright field). Finally, after visual inspection of the panoramic figures we determined the contrast of the yellow channel was lost by the zoom out and this was corrected for by overlaying a copy of this layer.
Oxylipin injections
To assess the potential in vivo nociceptive properties of oxylipins we injected either vehicle (N-Methyl-2-pyrrolidone, ethanol, Cremophor EL, tween 20, ascorbate, 200:100:100:2:1 respectively, dissolved with 5 volumes of sterile saline), 40 μg of 9-HpODE, 40 μg of 13-HpODE, a mixture of 18 μg of 9- and 22 μg of 13-HpODE or 40 μg of 9-HODE intradermally into the plantar surface of the hind paw. Subsequent to injection animals were placed on a platform with a transparent glass floor. Animals were habituated for 10 minutes after injection, during which time behavior was monitored using a video camera for at least 5 minutes from underneath platform. Nocifensive behaviors were quantified from video playback by an observer blinded to the experimental conditions. Nocifensive behaviors were defined as guarding and/or licking of the injected foot as described previously 65. Timing was recorded manually with a stopwatch, and/or from video time stamps. Nociceptive behavior was quantified as the total time the animal spent exhibiting all nociceptive behaviors. Within 30 min of each injection C-Fiber mediated hyperalgesia was assessed (described below). Similarly, intradermal injection of a 200 μg dose of 9-HpODE as well as a combination of the HpODE mixture (described above) plus 26 μg 16,17-EDP and 13 μg 4-HDHA was performed to assess these compounds potential to impact C-Fiber mediated nociception.
Assessments of Hyperalgesia.
Baseline measurements were taken for all tests prior to injection with CG, as well as at 1, 4 and 24 hours post CG injection or after oxylipin injection. Additionally, each test was performed on both hind paws. Hind paw measurements were taken by measuring the diameter (dorsal-ventral) of the hind paw at the thickest cross-section. A-delta mediated paw withdrawal responses were measured by stimulation of the plantar surface of the hind paw with a 100ms laser pulse, producing a rapid withdrawal response. Laser pulses were delivered by an infrared diode laser 46 (LASS-10 M; Lasmed, Mountain View, CA, USA) and calibrated to 3500 mA at 0.5 mm diameter, and delivered from 1cm distance. Nociceptive sensitivity was assessed by scoring the response to the stimulus on a previously validated scale 45. C-fiber mediated responses were measured by delivery of a slow temperature ramp to the plantar surface of the hind paw (1000 mA, 13cm distance). Voluntary paw withdrawal latency was recorded. Pinprick withdrawal was delivered by a pin stimulus at the plantar surface of the hind paw, resulting in a withdrawal. Inflamed animals subsequently exhibit guarding behavior by lifting the paw. The duration of this behavior was timed using a stopwatch.
Statistics
Fatty acid concentrations were presented as percent of total fatty acids [median ± interquartile range (IQR)]. Baseline oxylipin concentrations were presented as ng/g tissue (median ± IQR). Oxylipin concentrations were also expressed as percent of total oxylipins when assessing the impact of CG oxylipin concentrations. Kruskal-Wallis test was used to assess the statistical significance of CG evoked changes in adjusted oxylipin concentrations. A main effect of effect of CG with p<0.05 was considered statistically significant. RNASeq data are presented as mean sFPKM and changes in gene expression evoked by CG were assessed by Differential gene expression was analyzed using the raw counts generated by MAGIC, and statistical analysis was performed using limma voom in R with a p<0.01 considered to be significant 72.Thermal withdrawal latency of injected paws were compared to non-injected paws and statistical significance was assessed by two-tailed, paired t-test (p<0.05). Nociceptive behavior was assessed by Kruskal-Wallis test followed by Dunn’s multiple comparison test, comparing vehicle to each oxylipin group.
Supplementary Material
Highlights:
The biochemical pathways related to oxylipins were outlined
Genes encoding oxylipin related enzymes are expressed in the pain-circuit
Gene expression and oxylipin concentrations were modified by inflammation
Oxylipin injections evoked thermal nociceptive hypersensitivity in rats.
Acknowledgements
We would like to thank Natalie Labinger for scoring rat behavior the videos Mary Kate Dougherty and Radhika Narasimhan for creating in situ hybridization and supplemental figures, Katherine Ness for assisting with review of the literature and Amit Ringel and Kristen Maiden for conceptualizing the design of the lipid pathway figures.
Disclosures: This work was supported by the intramural research programs of the National Institute on Aging, National Institute on Alcohol Abuse and Alcoholism, National Center for Complementary and Integrative Health and the National Institutes of Health Clinical Center. Supplemental funding was provided by the NIH Bench-to-Bedside Program to M.J.I. from NCCIH, and the Office of Behavioral and Social Sciences. The NIA (NIH) has claimed intellectual property related to analogs of oxidized lipids (PCT/US2018/041086;), with C.E.R. and G.S.K. named as inventors. All other authors declare that they have no conflicts of interest interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data and Materials Availability:
N/A
References
- 1.Alsalem M, Wong A, Millns P, Arya PH, Chan MSL, Bennett A, Barrett DA, Chapman V, Kendall DA: The contribution of the endogenous TRPV1 ligands 9-HODE and 13-HODE to nociceptive processing and their role in peripheral inflammatory pain mechanisms. Br J Pharmacol 168:1961–1974, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakry OA, El Farargy SM, El Kady NNED, Dawy HFA: Immunohistochemical Expression of Cyclo-oxygenase 2 and Liver X Receptor-α in Acne Vulgaris. J Clin Diagn Res 11:WC01–WC07, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkman DJ: The effect of aspirin and nonsteroidal anti-inflammatory drugs on prostaglandins. Am J Med 105:8S–12S, 1998. [DOI] [PubMed] [Google Scholar]
- 4.Blomme EAG, Chinn KS, Hardy MM, Casler JJ, Kim SH, Opsahl AC, Hall WA, Trajkovic D, Khan KN, Tripp CS: Selective cyclooxygenase-2 inhibition does not affect the healing of cutaneous full-thickness incisional wounds in SKH-1 mice. Br J Dermatol 148:211–223, 2003. [DOI] [PubMed] [Google Scholar]
- 5.Boeglin WE, Kim RB, Brash AR: A 12R-lipoxygenase in human skin: mechanistic evidence, molecular cloning, and expression. Proc Natl Acad Sci USA 95:6744–6749, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breyer RM, Bagdassarian CK, Myers SA, Breyer MD: Prostanoid receptors: subtypes and signaling. Annu Rev Pharmacol Toxicol 41:661–690, 2001. [DOI] [PubMed] [Google Scholar]
- 7.Buckley CD, Gilroy DW, Serhan CN: Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 40:315–327, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bürger F, Krieg P, Marks F, Fürstenberger G: Positional- and stereo-selectivity of fatty acid oxygenation catalysed by mouse (12S)-lipoxygenase isoenzymes. Biochem J 348 Pt 2:329–335, 2000. [PMC free article] [PubMed] [Google Scholar]
- 9.Cavanaugh DJ, Chesler AT, Jackson AC, Sigal YM, Yamanaka H, Grant R, O’Donnell D, Nicoll RA, Shah NM, Julius D, Basbaum AI: Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J Neurosci 31:5067–5077, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cezar TLC, Martinez RM, da Rocha C, Melo CPB, Vale DL, Borghi SM, Fattori V, Vignoli JA, Camilios-Neto D, Baracat MM, Georgetti SR, Verri WA, Casagrande R: Treatment with maresin 1, a docosahexaenoic acid-derived pro-resolution lipid, protects skin from inflammation and oxidative stress caused by UVB irradiation. Sci Rep 9:3062, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P, Zuo H, Xiong H, Kolar MJ, Chu Q, Saghatelian A, Siegwart DJ, Wan Y: Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc Natl Acad Sci USA 114:580–585, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung NS, Wasan KM: Potential role of the low-density lipoprotein receptor family as mediators of cellular drug uptake. Adv Drug Deliv Rev 56:1315–1334, 2004. [DOI] [PubMed] [Google Scholar]
- 13.Cui JG, Holmin S, Mathiesen T, Meyerson BA, Linderoth B: Possible role of inflammatory mediators in tactile hypersensitivity in rat models of mononeuropathy. Pain 88:239–248, 2000. [DOI] [PubMed] [Google Scholar]
- 14.Dennis EA, Norris PC: Eicosanoid storm in infection and inflammation. Nat Rev Immunol 15:511–523, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domenichiello AF, Wilhite BC, Keyes GS, Ramsden CE: A dose response study of the effect of prostaglandin E2 on thermal nociceptive sensitivity. Prostaglandins Leukot Essent Fatty Acids 126:20–24, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douda DN, Grasemann H, Pace-Asciak C, Palaniyar N: A lipid mediator hepoxilin A3 is a natural inducer of neutrophil extracellular traps in human neutrophils. Mediators Inflamm 2015:520871, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubland JA, Francis GA: Lysosomal acid lipase: at the crossroads of normal and atherogenic cholesterol metabolism. Front Cell Dev Biol 3:3, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Dyke TE, Serhan CN: Resolution of inflammation: a new paradigm for the pathogenesis of periodontal diseases. J Dent Res 82:82–90, 2003. [DOI] [PubMed] [Google Scholar]
- 19.Eivers SB, Kinsella BT: Regulated expression of the prostacyclin receptor (IP) gene by androgens within the vasculature: Combined role for androgens and serum cholesterol. Biochim Biophys Acta 1859:1333–1351, 2016. [DOI] [PubMed] [Google Scholar]
- 20.Fehrenbacher JC, Vasko MR, Duarte DB: Models of inflammation: Carrageenan- or complete Freund’s Adjuvant (CFA)-induced edema and hypersensitivity in the rat. Curr Protoc Pharmacol Chapter 5:Unit5.4, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flegel C, Schöbel N, Altmüller J, Becker C, Tannapfel A, Hatt H, Gisselmann G: RNA-Seq Analysis of Human Trigeminal and Dorsal Root Ganglia with a Focus on Chemoreceptors. PLoS One 10:e0128951, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folch J, Lees M, Sloane Stanley GH: A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226:497–509, 1957. [PubMed] [Google Scholar]
- 23.Goulet JL, Pace AJ, Key ML, Byrum RS, Nguyen M, Tilley SL, Morham SG, Langenbach R, Stock JL, McNeish JD, Smithies O, Coffman TM, Koller BH: E-prostanoid-3 receptors mediate the proinflammatory actions of prostaglandin E2 in acute cutaneous inflammation. J Immunol 173:1321–1326, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Green D, Ruparel S, Gao X, Ruparel N, Patil M, Akopian A, Hargreaves K: Central activation of TRPV1 and TRPA1 by novel endogenous agonists contributes to mechanical allodynia and thermal hyperalgesia after burn injury. Mol Pain 12:, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green DP, Ruparel S, Roman L, Henry MA, Hargreaves KM: Role of endogenous TRPV1 agonists in a postburn pain model of partial-thickness injury. Pain 154:2512–2520, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grubb BD, Birrell GJ, McQueen DS, Iggo A: The role of PGE2 in the sensitization of mechanoreceptors in normal and inflamed ankle joints of the rat. Exp Brain Res 84:383–392, 1991. [DOI] [PubMed] [Google Scholar]
- 27.GTEx Consortium: The Genotype-Tissue Expression (GTEx) project. Nat Genet 45:580–585, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haitina T, Fredriksson R, Foord SM, Schiöth HB, Gloriam DE: The G protein-coupled receptor subset of the dog genome is more similar to that in humans than rodents. BMC Genomics 10:24, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen HS: 15-hydroxyprostaglandin dehydrogenase. A review. Prostaglandins 12:647–679, 1976. [DOI] [PubMed] [Google Scholar]
- 30.Hargreaves KM, Ruparel S: Role of oxidized lipids and TRP channels in orofacial pain and inflammation. J Dent Res 95:1117–1123, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hay A, Wood S, Olson D, Slater DM: Labour is associated with decreased expression of the PGF2alpha receptor (PTGFR) and a novel PTGFR splice variant in human myometrium but not decidua. Mol Hum Reprod 16:752–760, 2010. [DOI] [PubMed] [Google Scholar]
- 32.Hohmann SW, Angioni C, Tunaru S, Lee S, Woolf CJ, Offermanns S, Geisslinger G, Scholich K, Sisignano M: The G2A receptor (GPR132) contributes to oxaliplatin-induced mechanical pain hypersensitivity. Sci Rep 7:446, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang SW, Cho H, Kwak J, Lee SY, Kang CJ, Jung J, Cho S, Min KH, Suh YG, Kim D, Oh U: Direct activation of capsaicin receptors by products of lipoxygenases: endogenous capsaicin-like substances. Proc Natl Acad Sci USA 97:6155–6160, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hysing E-B, Smith L, Thulin M, Karlsten R, Bothelius K, Gordh T: Detection of systemic inflammation in severely impaired chronic pain patients and effects of a multimodal pain rehabilitation program. Scand J Pain 19:235–244, 2019. [DOI] [PubMed] [Google Scholar]
- 35.Iadarola MJ, Brady LS, Draisci G, Dubner R: Enhancement of dynorphin gene expression in spinal cord following experimental inflammation: stimulus specificity, behavioral parameters and opioid receptor binding. Pain 35:313–326, 1988. [DOI] [PubMed] [Google Scholar]
- 36.Ibuki T, Matsumura K, Yamazaki Y, Nozaki T, Tanaka Y, Kobayashi S: Cyclooxygenase-2 is induced in the endothelial cells throughout the central nervous system during carrageenan-induced hind paw inflammation; its possible role in hyperalgesia. J Neurochem 86:318–328, 2003. [DOI] [PubMed] [Google Scholar]
- 37.Kaaman M, Rydén M, Axelsson T, Nordström E, Sicard A, Bouloumié A, Langin D, Arner P, Dahlman I: ALOX5AP expression, but not gene haplotypes, is associated with obesity and insulin resistance. Int J Obes 30:447–452, 2006. [DOI] [PubMed] [Google Scholar]
- 38.Kagan VE, Mao G, Qu F, Angeli JPF, Doll S, Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, Kapralov AA, Amoscato AA, Jiang J, Anthonymuthu T, Mohammadyani D, Yang Q, Proneth B, Klein-Seetharaman J, Watkins S, Bahar I, Greenberger J, Mallampalli RK, Stockwell BR, Tyurina YY, Conrad M, Bayir H: Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol 13:81–90, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kern K, Schäfer SMG, Cohnen J, Pierre S, Osthues T, Tarighi N, Hohmann S, Ferreiros N, Brüne B, Weigert A, Geisslinger G, Sisignano M, Scholich K: The G2A receptor controls polarization of macrophage by determining their localization within the inflamed tissue. Front Immunol 9:2261, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khasar SG, Green PG, Levine JD: Comparison of intradermal and subcutaneous hyperalgesic effects of inflammatory mediators in the rat. Neurosci Lett 153:215–218, 1993. [DOI] [PubMed] [Google Scholar]
- 41.Krishnamoorthy S, Recchiuti A, Chiang N, Yacoubian S, Lee C-H, Yang R, Petasis NA, Serhan CN: Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc Natl Acad Sci USA 107:1660–1665, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaPaglia DM, Sapio MR, Burbelo PD, Thierry-Mieg J, Thierry-Mieg D, Raithel SJ, Ramsden CE, Iadarola MJ, Mannes AJ: RNA-Seq investigations of human post-mortem trigeminal ganglia. Cephalalgia 38:912–932, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Dalli J, Chiang N, Baron RM, Quintana C, Serhan CN: Plasticity of leukocytic exudates in resolving acute inflammation is regulated by MicroRNA and proresolving mediators. Immunity 39:885–898, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martini AC, Berta T, Forner S, Chen G, Bento AF, Ji R-R, Rae GA: Lipoxin A4 inhibits microglial activation and reduces neuroinflammation and neuropathic pain after spinal cord hemisection. J Neuroinflammation 13:75, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell K, Bates BD, Keller JM, Lopez M, Scholl L, Navarro J, Madian N, Haspel G, Nemenov MI, Iadarola MJ: Ablation of rat TRPV1-expressing Adelta/C-fibers with resiniferatoxin: analysis of withdrawal behaviors, recovery of function and molecular correlates. Mol Pain 6:94, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell K, Lebovitz EE, Keller JM, Mannes AJ, Nemenov MI, Iadarola MJ: Nociception and inflammatory hyperalgesia evaluated in rodents using infrared laser stimulation after Trpv1 gene knockout or resiniferatoxin lesion. Pain 155:733–745, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muñoz-Garcia A, Thomas CP, Keeney DS, Zheng Y, Brash AR: The importance of the lipoxygenase-hepoxilin pathway in the mammalian epidermal barrier. Biochim Biophys Acta 1841:401–408, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nantel F, Denis D, Gordon R, Northey A, Cirino M, Metters KM, Chan CC: Distribution and regulation of cyclooxygenase-2 in carrageenan-induced inflammation. Br J Pharmacol 128:853–859, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niki E: Biomarkers of lipid peroxidation in clinical material. Biochim Biophys Acta 1840:809–817, 2014. [DOI] [PubMed] [Google Scholar]
- 50.Obinata H, Hattori T, Nakane S, Tatei K, Izumi T: Identification of 9-hydroxyoctadecadienoic acid and other oxidized free fatty acids as ligands of the G protein-coupled receptor G2A. J Biol Chem 280:40676–40683, 2005. [DOI] [PubMed] [Google Scholar]
- 51.Obinata H, Izumi T: G2A as a receptor for oxidized free fatty acids. Prostaglandins Other Lipid Mediat 89:66–72, 2009. [DOI] [PubMed] [Google Scholar]
- 52.Okazaki H, Igarashi M, Nishi M, Sekiya M, Tajima M, Takase S, Takanashi M, Ohta K, Tamura Y, Okazaki S, Yahagi N, Ohashi K, Amemiya-Kudo M, Nakagawa Y, Nagai R, Kadowaki T, Osuga J, Ishibashi S: Identification of neutral cholesterol ester hydrolase, a key enzyme removing cholesterol from macrophages. J Biol Chem 283:33357–33364, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliw EH: Oxygenation of linolenic and linoleic acid to novel vicinal dihydroxy acids by hepatic microsomes of the rabbit. Biochem Biophys Res Commun 111:644–651, 1983. [DOI] [PubMed] [Google Scholar]
- 54.Oliw EH: Oxygenation of polyunsaturated fatty acids by cytochrome P450 monooxygenates. Prog Lipid Res 33:329–354, 1994. [DOI] [PubMed] [Google Scholar]
- 55.Orr SK, Palumbo S, Bosetti F, Mount HT, Kang JX, Greenwood CE, Ma DWL, Serhan CN, Bazinet RP: Unesterified docosahexaenoic acid is protective in neuroinflammation. J Neurochem 127:378–393, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osthues T, Sisignano M: Oxidized lipids in persistent pain states. Front Pharmacol 10:1147, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park C-K, Lü N, Xu Z-Z, Liu T, Serhan CN, Ji R-R: Resolving TRPV1- and TNF-α-mediated spinal cord synaptic plasticity and inflammatory pain with neuroprotectin D1. J Neurosci 31:15072–15085, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park C-K, Xu Z-Z, Liu T, Lü N, Serhan CN, Ji R-R: Resolvin D2 is a potent endogenous inhibitor for transient receptor potential subtype V1/A1, inflammatory pain, and spinal cord synaptic plasticity in mice: distinct roles of resolvin D1, D2, and E1. J Neurosci 31:18433–18438, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patwardhan AM, Akopian AN, Ruparel NB, Diogenes A, Weintraub ST, Uhlson C, Murphy RC, Hargreaves KM: Heat generates oxidized linoleic acid metabolites that activate TRPV1 and produce pain in rodents. J Clin Invest 120:1617–1626, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patwardhan AM, Scotland PE, Akopian AN, Hargreaves KM: Activation of TRPV1 in the spinal cord by oxidized linoleic acid metabolites contributes to inflammatory hyperalgesia. Proc Natl Acad Sci USA 106:18820–18824, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peavy RD, Metcalfe DD: Understanding the mechanisms of anaphylaxis. Curr Opin Allergy Clin Immunol 8:310–315, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peters-Golden M, Gleason MM, Togias A: Cysteinyl leukotrienes: multi-functional mediators in allergic rhinitis. Clin Exp Allergy 36:689–703, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Powell WS, Rokach J: Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochim Biophys Acta 1851:340–355, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quan N, Whiteside M, Herkenham M: Cyclooxygenase 2 mRNA expression in rat brain after peripheral injection of lipopolysaccharide. Brain Res 802:189–197, 1998. [DOI] [PubMed] [Google Scholar]
- 65.Raithel SJ, Sapio MR, LaPaglia DM, Iadarola MJ, Mannes AJ: Transcriptional Changes in Dorsal Spinal Cord Persist after Surgical Incision Despite Preemptive Analgesia with Peripheral Resiniferatoxin. Anesthesiology 128:620–635, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramsden CE, Domenichiello AF, Yuan Z-X, Sapio MR, Keyes GS, Mishra SK, Gross JR, Majchrzak-Hong S, Zamora D, Horowitz MS, Davis JM, Sorokin AV, Dey A, LaPaglia DM, Wheeler JJ, Vasko MR, Mehta NN, Mannes AJ, Iadarola MJ: A systems approach for discovering linoleic acid derivatives that potentially mediate pain and itch. Sci Signal 10:, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramsden CE, Faurot KR, Zamora D, Palsson OS, MacIntosh BA, Gaylord S, Taha AY, Rapoport SI, Hibbeln JR, Davis JM, Mann JD: Targeted alterations in dietary n-3 and n-6 fatty acids improve life functioning and reduce psychological distress among patients with chronic headache: a secondary analysis of a randomized trial. Pain 156:587–596, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramsden CE, Faurot KR, Zamora D, Suchindran CM, Macintosh BA, Gaylord S, Ringel A, Hibbeln JR, Feldstein AE, Mori TA, Barden A, Lynch C, Coble R, Mas E, Palsson O, Barrow DA, Mann JD: Targeted alteration of dietary n-3 and n-6 fatty acids for the treatment of chronic headaches: a randomized trial. Pain 154:2441–2451, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramsden CE, Ringel A, Majchrzak-Hong SF, Yang J, Blanchard H, Zamora D, Loewke JD, Rapoport SI, Hibbeln JR, Davis JM, Hammock BD, Taha AY: Dietary linoleic acid-induced alterations in pro- and anti-nociceptive lipid autacoids: Implications for idiopathic pain syndromes? Mol Pain 12:, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reinaud O, Delaforge M, Boucher JL, Rocchiccioli F, Mansuy D: Oxidative metabolism of linoleic acid by human leukocytes. Biochem Biophys Res Commun 161:883–891, 1989. [DOI] [PubMed] [Google Scholar]
- 71.Ribardo DA, Crowe SE, Kuhl KR, Peterson JW, Chopra AK: Prostaglandin levels in stimulated macrophages are controlled by phospholipase A2-activating protein and by activation of phospholipase C and D. J Biol Chem 276:5467–5475, 2001. [DOI] [PubMed] [Google Scholar]
- 72.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK: limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruparel S, Green D, Chen P, Hargreaves KM: The cytochrome P450 inhibitor, ketoconazole, inhibits oxidized linoleic acid metabolite-mediated peripheral inflammatory pain. Mol Pain 8:73, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samuelsson B, Dahlén SE, Lindgren JA, Rouzer CA, Serhan CN: Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science 237:1171–1176, 1987. [DOI] [PubMed] [Google Scholar]
- 75.Sapio MR, Goswami SC, Gross JR, Mannes AJ, Iadarola MJ: Transcriptomic analyses of genes and tissues in inherited sensory neuropathies. Exp Neurol 283:375–395, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sapio MR, Iadarola MJ, Loydpierson AJ, Kim JJ, Thierry-Mieg D, Thierry-Mieg J, Maric D, Mannes AJ: Dynorphin and enkephalin opioid peptides and transcripts in spinal cord and dorsal root ganglion during peripheral inflammatory hyperalgesia and allodynia. J Pain, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sapio MR, Neubert JK, LaPaglia DM, Maric D, Keller JM, Raithel SJ, Rohrs EL, Anderson EM, Butman JA, Caudle RM, Brown DC, Heiss JD, Mannes AJ, Iadarola MJ: Pain control through selective chemo-axotomy of centrally projecting TRPV1+ sensory neurons. J Clin Invest 128:1657–1670, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.SEQC/MAQC-III Consortium: A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol 32:903–914, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Serhan CN, Dalli J, Karamnov S, Choi A, Park C-K, Xu Z-Z, Ji R-R, Zhu M, Petasis NA: Macrophage proresolving mediator maresin 1 stimulates tissue regeneration and controls pain. FASEB J 26:1755–1765, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Serhan CN: Pro-resolving lipid mediators are leads for resolution physiology. Nature 510:92–101, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shearer GC, Newman JW: Impact of circulating esterified eicosanoids and other oxylipins on endothelial function. Curr Atheroscler Rep 11:403–410, 2009. [DOI] [PubMed] [Google Scholar]
- 82.Shearer GC, Newman JW: Lipoprotein lipase releases esterified oxylipins from very low-density lipoproteins. Prostaglandins Leukot Essent Fatty Acids 79:215–222, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shim HS, Bae C, Wang J, Lee K-H, Hankerd KM, Kim HK, Chung JM, La J-H: Peripheral and central oxidative stress in chemotherapy-induced neuropathic pain. Mol Pain 15:1744806919840098, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shinohara M, Serhan CN: Novel Endogenous Proresolving Molecules:Essential Fatty Acid-Derived and Gaseous Mediators in the Resolution of Inflammation. J Atheroscler Thromb 23:655–664, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Skoog MT, Nichols JS, Harrison BL, Wiseman JS: Glutathione peroxidase is neither required nor kinetically competent for conversion of 5-HPETE to 5-HETE in rat PMN lysates. Prostaglandins 31:577–593, 1986. [DOI] [PubMed] [Google Scholar]
- 86.Sorgi CA, Zarini S, Martin SA, Sanchez RL, Scandiuzzi RF, Gijón MA, Guijas C, Flamand N, Murphy RC, Faccioli LH: Dormant 5-lipoxygenase in inflammatory macrophages is triggered by exogenous arachidonic acid. Sci Rep 7:10981, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stafforini DM, McIntyre TM, Carter ME, Prescott SM: Human plasma platelet-activating factor acetylhydrolase. Association with lipoprotein particles and role in the degradation of platelet-activating factor. J Biol Chem 262:4215–4222, 1987. [PubMed] [Google Scholar]
- 88.Strickland DK, Gonias SL, Argraves WS: Diverse roles for the LDL receptor family. Trends Endocrinol Metab 13:66–74, 2002. [DOI] [PubMed] [Google Scholar]
- 89.Sutton BS, Crosslin DR, Shah SH, Nelson SC, Bassil A, Hale AB, Haynes C, Goldschmidt-Clermont PJ, Vance JM, Seo D, Kraus WE, Gregory SG, Hauser ER: Comprehensive genetic analysis of the platelet activating factor acetylhydrolase (PLA2G7) gene and cardiovascular disease in case-control and family datasets. Hum Mol Genet 17:1318–1328, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taha AY, Cheon Y, Faurot KF, Macintosh B, Majchrzak-Hong SF, Mann JD, Hibbeln JR, Ringel A, Ramsden CE: Dietary omega-6 fatty acid lowering increases bioavailability of omega-3 polyunsaturated fatty acids in human plasma lipid pools. Prostaglandins Leukot Essent Fatty Acids 90:151–157, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taha AY, Hennebelle M, Yang J, Zamora D, Rapoport SI, Hammock BD, Ramsden CE: Regulation of rat plasma and cerebral cortex oxylipin concentrations with increasing levels of dietary linoleic acid. Prostaglandins Leukot Essent Fatty Acids, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tripp CS, Blomme EAG, Chinn KS, Hardy MM, LaCelle P, Pentland AP: Epidermal COX-2 induction following ultraviolet irradiation: suggested mechanism for the role of COX-2 inhibition in photoprotection. J Invest Dermatol 121:853–861, 2003. [DOI] [PubMed] [Google Scholar]
- 93.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F: Proteomics. Tissue-based map of the human proteome. Science 347:1260419, 2015. [DOI] [PubMed] [Google Scholar]
- 94.Ursini F, Maiorino M, Valente M, Ferri L, Gregolin C: Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim Biophys Acta 710:197–211, 1982. [DOI] [PubMed] [Google Scholar]
- 95.Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P: Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18:145–153, 2015. [DOI] [PubMed] [Google Scholar]
- 96.Valdes AM, Ravipati S, Menni C, Abhishek A, Metrustry S, Harris J, Nessa A, Williams FMK, Spector TD, Doherty M, Chapman V, Barrett DA: Association of the resolvin precursor 17-HDHA, but not D- or E- series resolvins, with heat pain sensitivity and osteoarthritis pain in humans. Sci Rep 7:10748, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vane JR, Bakhle YS, Botting RM: Cyclooxygenases 1 and 2. Annu Rev Pharmacol Toxicol 38:97–120, 1998. [DOI] [PubMed] [Google Scholar]
- 98.Vasko MR, Habashy Malty R, Guo C, Duarte DB, Zhang Y, Nicol GD: Nerve growth factor mediates a switch in intracellular signaling for PGE2-induced sensitization of sensory neurons from protein kinase A to Epac. PLoS One 9:e104529, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wen H, Östman J, Bubb KJ, Panayiotou C, Priestley JV, Baker MD, Ahluwalia A: 20-Hydroxyeicosatetraenoic acid (20-HETE) is a novel activator of transient receptor potential vanilloid 1 (TRPV1) channel. J Biol Chem 287:13868–13876, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weng Z, Fluckiger AC, Nisitani S, Wahl MI, Le LQ, Hunter CA, Fernal AA, Le Beau MM, Witte ON: A DNA damage and stress inducible G protein-coupled receptor blocks cells in G2/M. Proc Natl Acad Sci USA 95:12334–12339, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Werz O, Gerstmeier J, Libreros S, De la Rosa X, Werner M, Norris PC, Chiang N, Serhan CN: Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat Commun 9:59, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Willenberg I, Ostermann AI, Schebb NH: Targeted metabolomics of the arachidonic acid cascade: current state and challenges of LC-MS analysis of oxylipins. Anal Bioanal Chem 407:2675–2683, 2015. [DOI] [PubMed] [Google Scholar]
- 103.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA: The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med 34:496–502, 2003. [DOI] [PubMed] [Google Scholar]
- 104.Yoo S-E, Chen L, Na R, Liu Y, Rios C, Van Remmen H, Richardson A, Ran Q: Gpx4 ablation in adult mice results in a lethal phenotype accompanied by neuronal loss in brain. Free Radic Biol Med 52:1820–1827, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yuan Z-X, Majchrzak-Hong S, Keyes GS, Iadarola MJ, Mannes AJ, Ramsden CE: Lipidomic profiling of targeted oxylipins with ultra-performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem 410:6009–6029, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang G, Kodani S, Hammock BD: Stabilized epoxygenated fatty acids regulate inflammation, pain, angiogenesis and cancer. Prog Lipid Res 53:108–123, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang W, Yu Y, Hertwig F, Thierry-Mieg J, Zhang W, Thierry-Mieg D, Wang J, Furlanello C, Devanarayan V, Cheng J, Deng Y, Hero B, Hong H, Jia M, Li L, Lin SM, Nikolsky Y, Oberthuer A, Qing T, Su Z, Volland R, Wang C, Wang MD, Ai J, Albanese D, Asgharzadeh S, Avigad S, Bao W, Bessarabova M, Brilliant MH, Brors B, Chierici M, Chu T-M, Zhang J, Grundy RG, He MM, Hebbring S, Kaufman HL, Lababidi S, Lancashire LJ, Li Y, Lu XX, Luo H, Ma X, Ning B, Noguera R, Peifer M, Phan JH, Roels F, Rosswog C, Shao S, Shen J, Theissen J, Tonini GP, Vandesompele J, Wu P-Y, Xiao W, Xu J, Xu W, Xuan J, Yang Y, Ye Z, Dong Z, Zhang KK, Yin Y, Zhao C, Zheng Y, Wolfinger RD, Shi T, Malkas LH, Berthold F, Wang J, Tong W, Shi L, Peng Z, Fischer M: Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol 16:133, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zimmer B, Angioni C, Osthues T, Toewe A, Thomas D, Pierre SC, Geisslinger G, Scholich K, Sisignano M: The oxidized linoleic acid metabolite 12,13-DiHOME mediates thermal hyperalgesia during inflammatory pain. Biochim Biophys Acta 1863:669–678, 2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
N/A