Abstract
Relevance
Human growth hormone (hGH) is synthesized, stored, and secreted by somatotroph cells in the pituitary gland, and promotes human growth and metabolism. Compared to a normal pituitary, a GH-secreting pituitary adenoma can secrete excessive GH to cause pathological changes in body tissues. GH proteoform changes would be associated with GH-related disease pathogenesis.
Purpose
This study aimed to elucidate changes in GH proteoforms between GH-secreting pituitary adenomas and control pituitaries for the predictive diagnostics, targeted prevention, and personalization of medical services.
Methods
The isoelectric point (pI) and relative molecular mass (Mr) are two basic features of a proteoform that can be used to effectively array and detect proteoforms with two-dimensional gel electrophoresis (2DGE) and 2DGE-based western blot. GH proteoforms were characterized with liquid chromatography (LC) and mass spectrometry (MS). Phosphoproteomics, ubiquitinomics, acetylomics, and bioinformatics were used to analyze post-translational modifications (PTMs) of GH proteoforms in GH-secreting pituitary adenoma tissues and control pituitaries.
Results
Sixty-six 2D gel spots were found to contain hGH, including 46 spots (46 GH proteoforms) in GH-secreting pituitary adenomas and 35 spots (35 GH proteoforms) in control pituitaries. Further, 35 GH proteoforms in control pituitary tissues were matched with 35 of 46 GH proteoforms in GH-secreting pituitary adenoma tissues; and 11 GH proteoforms were presented in only GH-secreting pituitary adenoma tissues but not in control pituitary tissues. The matched 35 GH proteoforms showed quantitative changes in GH-secreting pituitary adenomas compared to the controls. The quantitative levels of those 46 GH proteoforms in GH-secreting pituitary adenomas were significantly different from those 35 GH proteoforms in control pituitaries. Meanwhile, different types of PTMs were identified among those GH proteoforms. Phosphoproteomics identified phosphorylation at residues Ser77, Ser132, Ser134, Thr174, and Ser176 in hGH. Ubiquitinomics identified ubiquitination at residue Lys96 in hGH. Acetylomics identified acetylation at reside Lys171 in hGH. Deamination was identified at residue Asn178 in hGH.
Conclusion
These findings provide the first hGH proteoform pattern changes in GH-secreting pituitary adenoma tissues compared to control pituitary tissues, and the status of partial PTMs in hGH proteoforms. Those data provide in-depth insights into biological roles of hGH in GH-related diseases, and identify hGH proteoform pattern biomarkers for treatment of a GH-secreting pituitary adenoma in the context of 3P medicine –predictive diagnostics, targeted prevention, and personalization of medical services.
Supplementary information
The online version contains supplementary material available at 10.1007/s13167-021-00232-7.
Keywords: Growth hormone-secreting pituitary adenoma; Human growth hormone (hGH); Two-dimensional gel electrophoresis (2DGE); Liquid chromatography (LC); Mass spectrometry (MS); 2DGE-based western blot; Bioinformatics; Post-translational modifications (PTMs); Phosphorylation; Acetylation; Ubiquitination; Phosphoproteomics; Acetylomics; Ubiquitinomics; Patient stratification; Prediction/prognostic assessment; Predictive, preventive, and personalized medicine (PPPM/3P medicine)
Introduction
The structure of growth hormone
Human growth hormone (hGH) is secreted by GH-secreting cells that contain eosinophilic granules in the anterior pituitary gland [1]. hGH is encoded by the GHN gene located in the gene cluster of chromosome 17q23.3, and consists of five exons and four introns, which contains five closely related genes [2, 3]. According to the encoding rules from 5′ to 3′, these five related genes are GHN (GH1 gene), CSL (chorionic somatomammotropin-like gene), CSA, GHV (GH2 gene), and CSB. These five genes share more than 90 percent sequence identity, and are expressed in mutually exclusive tissue distribution. GHN is expressed in pituitary cells, and the other four genes are expressed in placental villous syncytiotrophoblasts [3]. The GH1 cDNA encodes the GH precursor with 217 amino acids (sequence 1–217), which contains a signal peptide at amino acid sequence positions 1–26 (Table 1). The theoretical mass for the GH precursor is 24.84 kDa. Mature GH contains 191 amino acids after removal of the signal peptide (sequence 1–26), and has a theoretical mass of 22.13 kDa (Table 1) [4, 5]. Further, hGH shares substantial similar sequence to human prolactin (PRL) [6].
Table 1.
The amino acid sequence of human GH precursor (Swiss-Prot No: P01241, position 1–217; and mature GH, position 27–217)
| GH1 precursor | ||||
| 10 | 20 | 30 | 40 | 50 |
| MATGSRTSLL | LAFGLLCLPW | LQEGSAFPTI | PLSRLFDNAM | LRAHRLHQLA |
| 60 | 70 | 80 | 90 | 100 |
| FDTYQEFEEA | YIPKEQKYSF | LQNPQTSLCF | SESIPTPSNR | EETQQKSNLE |
| 110 | 120 | 130 | 140 | 150 |
| LLRISLLLIQ | SWLEPVQFLR | SVFANSLVYG | ASDSNVYDLL | KDLEEGIQTL |
| 160 | 170 | 180 | 190 | 200 |
| MGRLEDGSPR | TGQIFKQTYS | KFDTNSHNDD | ALLKNYGLLY | CFRKDMDKVE |
| 210 | 220 | |||
| TFLRIVQCRS | VEGSCGF |
Signal peptide is position 1–26 in the underlined bold letters
The functions of growth hormone
hGH has catabolic and anabolic effects on organ or tissues systems [7], and has positive physiological effects: (i) It boosts metabolism, such as promote protein synthesis, elevate blood glucose, and lower adipose tissue; (ii) It promotes growth, such as skeletal growth, muscle growth, and tissue cell proliferation division; (iii) It increases GH secretion when involved in a stress response. In additional, it regulates carbohydrate and lipid metabolism [7]. In the endocrine system, GH is positively regulated by the secretion of GH-releasing hormone (GHRH), and negatively regulated by somatostatin (SST); both are released from the hypothalamus. hGH is secreted into body fluids and transported to the target tissues such as the liver for its physiological functions [8]. hGH induces intracellular signals through the GH receptor (GHR), and hGH in blood circulation binds to GHR in a target organ to activate signal pathways such as the Janus Kinase 2 (JAK2) signal transducer and activator of transcription 5 (STAT5) signal pathways [9, 10]. hGH can produce many growth-promoting effects through IGF-1 mediation; hGH generates IGF-1 at various target tissues in autocrine and paracrine manners, and the liver is the main tissue that synthesizes IGF-1 [11–13]. IGF-1 promotes growth in various tissues; for example, skeletal, muscle, and differentiation into proliferation of myoblasts and protein synthesis [14].
Growth hormone-related diseases
Although normal GH secretion levels are essential to maintain normal human growth, abnormal secretion of GH causes GH-related diseases. In childhood, excessive GH levels have adverse effects on skeletal architecture and strength [15] that lead to excessive growth of skeleton and result in an abnormally large skeleton to form gigantism [16]. However, hyposecretion of GH is the cause of dwarfism characterized by clinical symptoms such as short stature, delayed skeleton maturation, and sexual development disorders [17–19]. In the adult, excessive GH secretion that is derived from a pituitary adenoma leads to acromegaly with hypertrophy of the hands and feet, typical coarsening of facial features, soft-tissue swelling, and a series of clinical manifestations [20, 21].
Growth hormone variants
hGH variants are mainly derived from related genes through transcription, alternative splicing, post-translational modification (PTM), redistribution, spatial conformation, and pathway network systems [22]. GH variants are a family because more than 100 different GH variants are known to exist in the human body [23], which includes two main variants- 22 kDa GH and 20 kDa GH. Although 22 kDa GH is dominant in the blood circulation, 20 kDa GH only accounts for 5–10% [24]. Meanwhile, five forms of alternative splicing isoforms of GH are listed in the UniProtKB of public protein database number P01241. The specific biological activities of these GH variants have not yet been fully elucidated.
Growth hormone proteoforms analyzed with 2DGE-MS
Any given "protein" is thought to consist of several proteoforms. The basic unit of a proteome is a proteoform, and each proteoform has a specific isoelectric point (pI) and relative mass (Mr) [25]. Thus, GH proteoforms are derived from different alternative splicing, modification, and proteolysis processes. Two-dimensional gel electrophoresis (2DGE) can be used to array those hGH proteoforms with different pI and Mr values according to pI in the first dimension (isoelectric focusing – IEF) and Mr in the second dimension (sodium dodecyl sulfate polyacrylamide gel electrophoresis – SDS-PAGE) [26]. MS can be used to characterize 2DGE-separated GH proteoforms and to identify PTMs in the hGH proteoforms [27, 28]. Human pituitary adenoma proteomes and normal pituitary proteomes have been analyzed with 2DGE coupled with mass spectrometry (2DGE-MS) [29, 30]. Therefore, in this study, 2DGE, 2DGE-matrix assisted laser desorption ionization (MALDI)-time of flight (TOF)-peptide mass fingerprint (PMF), 2DGE-MALDI-tandem mass spectrometry (MS/MS), and 2DGE-liquid chromatography (LC)-electrospray ionization (ESI)-MS/MS were used to analyze GH-secreting pituitary adenomas and control pituitaries. A total of 46 GH proteoforms in GH-secreting pituitary adenomas and 35 GH proteoforms in normal pituitaries were identified, and their signal peptide, alternative splicing and PTMs were also analyzed. These results elucidated hGH differences between GH-secreting pituitary adenomas and control pituitaries.
Post-translational modifications of growth hormone
Protein PTMs play important roles in the regulation of different body functions. Phosphorylation is the most-extensive protein PTM- at least 10,000 phosphorylated proteins exist [31–33]. Protein phosphorylation plays a key role in tumorigenesis and development. The phosphoproteome of GH-secreting pituitary adenomas is one of the important causes of tumorigenesis [34]. In recent years, protein acetylation has been recognized as a basic PTM that plays an important role in biological regulation [35, 36]. Protein acetylation was positively correlated with the severity of a pituitary adenoma [37]. It is well-known that protein ubiquitination is associated with many diseases, and it has been reported that the occurrence of a pituitary adenoma is closely related to protein ubiquitination [38]. All of those diverse findings clearly demonstrate the importance to study phosphorylation, acetylation, and ubiquitination in the occurrence of pituitary adenomas.
Working hypotheses
We hypothesize that a GH proteoform pattern is significantly different in GH-secreting pituitary adenomas relative to control pituitaries, and that the altered GH proteoform pattern provides potential biomarkers for patient stratification, predictive diagnostics, targeted prevention, and personalization of medical services of GH-related diseases.
Study design
2DGE, 2DGE-based western blot against a GH antibody, and MS were used to array and identify GH proteoforms in GH-secreting pituitary adenomas and control pituitaries. Quantitative phosphoproteomics, ubiquitinomics, and acetylomics were used to identify and quantify phosphorylation, ubiquitination, and acetylation in hGH from whole pituitary adenoma and control tissues. The signal peptide and splicing variants of GH proteoforms were analyzed with MS. The identified phosphorylation, ubiquitination, and acetylation sites were manually compared with each MS spectrum among 46 MS spectra from GH-secreting pituitary adenomas and 35 MS spectra from control pituitaries. These findings provide the GH proteoform pattern alteration in GH-secreting pituitary adenomas relative to controls.
Expected impacts in the context of 3P medicine
The identified GH proteoform pattern alteration will help to elucidate the role of GH in GH-secreting pituitary adenomas, and might elucidate GH proteoform pattern biomarkers for effective treatment of GH-related diseases in the context of 3P medicine – patient stratification, predictive diagnostics, targeted prevention, and personalized treatment of GH-related diseases [39–41].
Materials and Methods
Pituitary tissue samples
All human pituitary adenoma tissues (n = 7, including 4 nonfunctional pituitary adenoma tissues and 3 GH-secreting pituitary adenoma tissues) were obtained from the Department of Neurosurgery of Xiangya Hospital; acquisition was approved by the Xiangya Hospital Medical Ethics Committee of Central South University in China. The post-mortem control pituitary tissue samples (n = 7) were obtained from: 1) the University of Tennessee Health Science Center (n = 4), with approval from the UTHSC Institutional Review Board, and 2) from the Department of Forensic Medicine, Tongji Medical College, Huazhong University of Science and Technology (n = 3), with approval from the Tongji Medical Ethics Committee of Huazhong University of Science and Technology in China. These pituitary adenoma tissues were removed from patients with surgery for pituitary-related diseases; all the purposes and properties of the tissue were fully explained, and the consent of each patient or family member was obtained. All tissues were removed, frozen immediately in liquid nitrogen, and stored at − 80 °C until processing. Each frozen pituitary tissue was homogenized and lyophilized, and the lyophilized solution was stored at − 80 °C until extraction analysis. The proteins extracted from 4 nonfunctional pituitary adenoma tissues were combined as tumor protein sample and the proteins extracted from 4 control pituitary tissues were combined as control protein sample, which were used for quantitative phosphoproteomics, ubiquitinomics, and acetylomics analyses. The proteins extracted from 3 GH-secreting pituitary adenoma tissues and from another 3 control pituitary tissues were combined as GH-secreting tumor proteins and control proteins, respectively, for 2DGE analysis.
Protein extraction
Tissue samples were removed from liquid nitrogen, and thawed slowly at room temperature. The tissue sample (about 500 mg) was washed with 0.9% NaCl (5 ml, 5x) to gently remove blood from the tissue surface, and was fully cut into small pieces (about 1 mm3; on ice) with clean scissors. A volume (4 ml) of protein extraction buffer that contained 7 mol/L urea, 2 mol/L thiourea, 60 mmol/L dithiothreitol (DTT), 4%(w/v)3-[(3-Cholamidopropyl)dimethylammonio] propanesulfonate (CHAPS), 0.5% v/v immobilized pH gradient (IPG) buffer (add prior to use), and a trace of bromophenol blue was added; that solution was mixed thoroughly (2 h; ice). The samples were centrifuged (15,000 × g, 15 min, 4 °C), and the supernatant was collected as the protein sample solution.
Two-dimensional gel electrophoresis and western blot
IEF
IEF was performed on an IPGphor Isoelectric Focusing System. An amount (500 μg) of prepared pituitary adenoma or control protein sample was loaded onto an 18 cm IPG strip (avoid bubbles), and 3 ml of mineral oil was added to cover the IPG strip, which was rehydrated about 18 h. IEF was performed at room temperature with the following parameters: fixed maximum current per strip = 30 µA; temperature = 20 °C; Step 1: 250 V, 1 h, 125 Vh, step and hold; Step 2: 1000 V, 1 h, 500 Vh, gradient; Step 3: 8000 V, 1 h, 4000 Vh, gradient; Step 4: 8000 V, 4 h, 32,000 Vh; and Step 5: 500 V, 0.5 h, 250 Vh, step and hold. The total time was 7.5 h and 36,875 Vh for IEF. After IEF, the IPG strips were removed and laid on its plastic back to remove the mineral oil from the surface of the IPG strips.
SDS-PAGE
A vertical Cell Electrophoresis System was used to separate the IEF-separated proteins. The 12% PAGE resolving gels (n = 12) were made by mixing 180 ml of 40% w/v acrylamide/bis-acrylamide stock solution (29:1, w:w), 150 ml of 1.5 mol/L Tris–HCl pH 8.8, 3 ml of 10% of ammonium persulfate, 270 ml deionized distilled water, and 150 µl of tetramethylethylenediamine (TEMED). The mixture was slowly poured into the multicasting chamber until the solution reached 1 cm away from the upper end, and about 3 ml deionized distilled water was added immediately to cover the upper end of the gels. The gels were left at room temperature for 1 h. The IPG strips with the focused proteins were removed and slightly rocked about 15 min in 25 ml of reducing equilibrium buffer that consisted of 375 mmol/L Tris–HCl pH 8.8, 20% v/v glycerol, 2% w/v SDS, 6 mol/L urea, a trace of bromophenol blue, and 2% w/v DTT (prior to use). The IPG strips were gently rocked about 15 min in 25 ml of alkylation equilibrium buffer that consisted of 375 mmol/L Tris–HCl pH 8.8, 20% v/v glycerol, 2% w/v SDS, 6 mol/L urea, a trace of bromophenol blue, and 2.5% w/v iodoacetamide (added prior to use). Each equilibrated IPG strip was put onto the top of the resolving SDS-PAGE gel, 30 ml of boiled 1% agarose in SDS electrophoresis buffer was poured quickly to cover the SDS-PAGE gel (avoid bubbles), and the IPGstrip with IEF-separated proteins was evenly and quickly pushed into the upper-end agarose solution, which was transferred to the electrophoresis tank that contained 10 L of electrophoresis buffer (25 mmol/L Tris, 192 mmol/L glycerin, and 0.1% w/v SDS). Electrophoresis at 200 V constant voltage for about 370 min. was performed.
2DGE-based western blot
The 2D gel between two glass plates was removed after electrophoresis and marked with a small cut at the negative end of the left-upper corner. The proteins in the gel were transferred to a polyvinylidene fluoride (PVDF) membrane with an Amersham Multiphor II semi-dry electro-transfer system with the following steps: The anode electrode plate was saturated with deionized distilled water after placement into the buffer tank; six sheets of filter paper were immersed in the anode transfer buffer R and placed onto the anode plate; three sheets of filter paper were immersed in the transfer buffer T and placed onto the six sheets of filter papers; the PVDF membrane was immersed in the anode transfer buffer and placed onto the three sheets of filter paper; the 2DE gel was placed on the PVDF membrane; and nine sheets of filter paper were immersed in the transfer buffer S and placed on the top of the 2D gel. Electro-transfer occurred at a constant current of 0.8 mA/cm2 for 90 min. The PVDF membrane with binding proteins was immersed into 100 ml of 0.3% bovine serum albumin/phosphate-buffered saline with Tween-20 (BSA/PBST) solution, blocked (gently shaking, room temperature, 60 min), and rinsed twice with deionized distilled water (200 ml). The proteins on PVDF membrane were incubated (slightly shaking, room temperature, 1 h) with 100 ml of diluted primary antibody–100 µl Rabbit anti-hGH antibody that was diluted with 100 ml of 0.3% BSA/PBST, washed with 200 ml PBST solution (15 min × 4), and rinsed twice with deionized distilled water. The proteins on PVDF membrane were incubated with the secondary antibody–20 µl goat anti-rabbit alkaline phosphatase-conjugated IgG that was diluted in 100 ml of 0.3% BSA/PBST, washed with 200 ml PBST solution (15 min × 3), and rinsed thrice with deionized distilled water. The proteins on the PVDF membrane were visualized with 1-Step nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Thermo Product no. 3404). A parallel negative-control experiment (the primary antibody hGH antibody was not added) was carried out to detect any cross-reactivity of the second antibody. The PVDF membrane with visualized protein was dried and stored with two sheets of filter papers.
Protein staining and image analysis of visualized 2D gel
The 2D gel that contained proteins was immersed (slowly shaking about 2–3 h) in a Coomassie brilliant blue staining solution that consisted of 0.75 g of Coomassie brilliant blue G250, 30 ml of glacial acetic acid, 135 ml of methanol, and 30 ml of deionized distilled water; rinsed twice with deionized distilled water; faded with 20% v/v ethanol (gently shaking until the background become nearly colorless); and rinsed twice with deionized distilled water. The visualized PVDF membrane and the corresponding Coomassie brilliant blue-stained 2D gels were scanned. The digitized images were imported into Bio-Rad PDQuest 2D gel image analysis software (version 7.0) to quantify spot volume, and the immune-positive western blotting spots were matched to the corresponding Coomassie brilliant blue-stained 2D gel spots.
MS identification of hGH
The three types of MS methods used to analyze 2DGE-separated proteins included MALDI-TOF–MS, LC–ESI–MS/MS, and MALDI-TOF-TOF–MS/MS. Briefly, the 2D gel spots that corresponded to the positive western blot spot were excised, proteins were digested with trypsin, and the tryptic peptide mixture was purified with a ZipTipC18 microcolumn.
For MALDI-TOF MS analysis, a purified tryptic peptide mixture was mixed with a-cyano-4-hydroxycinnamic acid (CHCA) matrix, and analyzed with MALDI-TOF–MS on a Perceptive Biosystems MALDI-TOF Voyager DE-RP mass spectrometer (Framingham, MA, USA) [27]. The peptide mass fingerprint (PMF) data were input into the Mascot search engine to search for a protein in the UniProtKB database 091,215 (date July 2, 2019; 513,877 sequences; 180,750,753 residues; Homo sapiens 513,877 sequences). A blank control experiment was also carried out with the analysis of margin gel pieces with MALDI-TOF MS to generate a control MS spectrum derived from any contaminating substances such as keratin and trypsin.
For LC–ESI–MS/MS analysis, a purified tryptic peptide mixture was analyzed with an LC-ESI-quadrupole-ion trap (Q-IT) mass spectrometer (LCQDeca, ThermoFinnigan, San Jose, CA, USA) to obtain MS/MS data. The LCQDeca instrument parameters were: capillary probe temperature 110 °C, ESI voltage 2.0 kV, and electron multiplier -900 V [27]. The MS/MS data were input into MASCOT software to search UniProtKB and NCBInr human databases for protein identification.
For MALDI-TOF-TOF–MS/MS analysis, a purified tryptic peptide mixture was mixed with CHCA matrix, and analyzed with a Perspective Biosystems MALDI-TOF-TOF mass spectrometer, whose parameters were: reflective mode, acceleration voltage 25 kV, and spectrum range m/z 800–4000. MS and MS/MS data were used to identify proteins with MASCOT software against the UniProtKB human protein database for protein identification. In this study, all MASCOT searching assigned a score of 70, which was the statistical threshold for the identity or high degree of homology between the search sequence and the recognition sequence, with a statistically significant level of p < 0.05.
The hGH protein amino acid sequence was obtained from the UniProtKB protein database (www.expasy.ch). For accurate and reliable MS identification of hGH in pituitary adenomas and control pituitaries, the common mass peaks from the blank gel, which were introduced from common contaminants such as trypsin, keratins derived from skin and hair, matrix, and other unknown sources, were removed from MS spectra before protein-database searching. Contaminating ion mass peaks often include m/z values 842.5, 870.5, 1045.4, 1109.3, 1179.3, 1235.2, 1277.4, 1307.3, 1365.3, 1383.3, 1434.4, 1475.3, 1493.3, 1638.3, 1708.2, 1716.3, 1791.1, 1838.3, 1940.2, 1994.2, 2211.1, 2225.1, 2239.1, 2284.1, 2389.8, 2705.7, and 2871.9 [27].
Identification of splicing events of hGH
GH precursor, mature GH, and GH splicing variants 1, 2, 3, and 4 have different amino acid sequences that can be readily identified via their characteristic tryptic peptides [29]. Peptide Mass tool (http://us.expasy.org/cgi-bin/peptide-mass.pl) was used to calculate the theoretical masses of tryptic peptides from GH precursor, mature GH, and GH splicing variants 1, 2, 3, and 4. The parameters were set as enzyme trypsin (the cleaved sites of trypsin were at the C-terminal side of Lys (K) and Arg (R), the maximum number of missed cleavages = 2, all cysteines in reduced form and methionines without oxidation, peptide mass range > 500 Da, the use of monoisotopic masses for peptide amino acid sequences, and peptide ion setting as [M + H]+. These parameters were consistent with the parameters used for MALDI-TOF and MALDI-TOF-TOF PMF data analysis. The characteristic tryptic peptides that distinguished among GH precursor, mature GH, and GH splicing variants 1, 2, 3, and 4 were compared to each MS spectrum (PMF data) derived from MALDI-TOF or MALDI-TOF-TOF analysis of each GH-immunoaffinity positive spots in GH-secreting pituitary adenoma tissues and control pituitary tissues.
Characterization of hGH PTMs
Quantitative phosphoproteomics in combination with TiO2 enrichment of phosphopeptides, quantitative ubiquitinomics in combination with ubiquitin antibody enrichment of ubiquitinated peptides, and quantitative acetylomics with acetylation antibody enrichment of acetylated peptides, were used to analyze pituitary adenoma and control tissues, respectively.
For phosphoproteomic analysis, a 6-plex iTRAQ reagent kit (AB SCIEX, Cat# 4,381,662-1KT, Foster City, CA) was used to label tryptic peptides from pituitary adenomas (triplicate) and controls (triplicate). For each iTRAQ labeling, protein (200 μg) was digested with trypsin, followed by desalting with the C18 cartridges, concentration with vacuum centrifugation, and quantification via UV absorbance at 280 nm. An amount (100 μg) of the trypsin peptide mixture of each sample was labeled with an iTRAQ reagent. The detailed procedure was described [42, 43, 46]. The iTRAQ-labeled mixed peptides were mixed (1:1:1:1:1:1, concentrated with a vacuum concentrator, and diluted in 500 μl DHB buffer. TiO2 beads were added; the mixture was slightly stirred for 2 h, and centrifuged (5000 × g, 1 min) to collect beads with phosphopeptides. Beads were washed with 50 μl of washing buffer I (30% acetonitrile and 3% trifluoroacetic acid) and 50 μl of washing buffer II (80% acetonitrile and 0.3% trifluoroacetic acid) (3 x). Enriched phosphopeptides were eluted with 50 μl of elution buffer (40% acetonitrile and15%NH4OH) (3 x), the eluate was lyophilized, and peptides were analyzed with LC–MS/MS [42, 43]. MS/MS data were used to identify the amino acid sequence and phosphorylation sites of each phosphopeptide. The intensities of iTRAQ reporter ions were used to quantify the abundance of each phosphopeptide.
For acetylomics analysis, the proteins (pituitary adenomas; control pituitaries) were digested with trypsin. Each tryptic peptide mixture was incubated with anti-N-acetyllysine antibody beads (4 °C, 2 h), the mixture was centrifuged (4 °C, 1000 × g, 1 min), and the supernatant was discarded. The anti-N-acetyllysine antibody beads with acetylated peptides were washed to remove non-specific binding peptides. Acetylated peptides were eluted in 40 µL of 0.1% TFA, desalinated with C18 STAGE Tips, and analyzed with LC–MS/MS analysis. LC used a reverse phase trap Column (Thermo Scientific Acclaim PepMap100, 100 µm × 2 cm, nanoViper C18) and C18-reversed phase analytical Column (Thermo Scientific Easy Column, 10 cm long, 75 µm inner diameter, 3 µm resin) for 120 min at a flow rate of 300 nL/min in the separation gradient through mixing buffer A (0.1% formic acid) and buffer B (84% acetonitrile and 0.1% formic acid). The LC linear gradient was set as solution B in 0–90 min for the linear gradient from 0–55%, solution B in 90–105 min for the linear gradient from 55–100%, and solution B in 105–120 min for 100%. MS/MS parameters set for the Q-Exactive mass spectrometer were: positive-ion mode, scanning range of precursor ions m/z 300–1800, automatic gain control (AGC) target 3e6, resolution of MS scan 70,000 at m/z 200, and resolution of MS/MS scan 17,500 at m/z 200. MS/MS data were input into MaxQuant software to identify protein amino acid sequence and acetylated sites of acetylated proteins, and to quantify the abundance of acetylated peptides with a label-free quantification method [44].
For ubiquitinomics analysis, the proteins (pituitary adenomas; control pituitaries) were digested with trypsin. Each tryptic peptide mixture was incubated with anti-K-ε-GG antibody beads [PTMScan ubiquitin remnant motif (K-ε-GG) kit, Cell Signal Technology], followed by washing and centrifugation to remove non-specific binding peptides [38]. The ubiquitinated peptides on the anti-K-ε-GG antibody beads were eluted with 40 μl of 0.15% trifluoroacetic acid, desalinated with C18 STAGE Tips [38, 45], and analyzed with LC–MS/MS [38, 44]. The ubiquitinomics LC–MS/MS procedure was the same as the procedure described above for acetylomics analysis. MS/MS data were input into MaxQuant software to identify protein amino acid sequence and ubiquitination sites, and ubiquitinated peptides were quantified with a label-free quantification method [38, 44].
These quantitative phosphoproteomic, ubiquitinomic, and acetylomic data were published in other publications [38, 46]. The modification sites and levels of phosphorylation, ubiquitination, and acetylation were identified in human GH (Table 2). The PTM sites and corresponding tryptic peptides were compared to each MS spectrum of GH proteoform contained in a 2D gel spot to determine the status of phosphorylation, ubiquitination, and acetylation in each hGH proteoform in GH-secreting pituitary adenomas and control pituitaries.
Table 2.
Quantitative proteomics identified the phosphorylation, ubiquitination, and acetylation of growth hormone in pituitary adenomas compared to controls
| PTM | Accession No | Gene name | Protein name | Modified peptide | Modified Amino acid residue | Modified position | Modified Probabilities | PEP | Modified level (N) | Modified level (T) | Ratio (T/N) | P-value (t-test) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phosphorylation | P01241 | GH1 | Somatotropin | FDTNS*HNDDALLK | S | 176 | 1 | 3.20E-216 | 580,220 | 43,527 | 0.08 | 1.93E-06 |
| P01241 | GH1 | Somatotropin | FDT*NSHNDDALLK | T | 174 | 0.578 | 1.80E-03 | |||||
| P01241 | GH1 | Somatotropin | SVFANSLVYGAS*DS*NVYDLLK | S; S | 132; 134 | 0.428; 0.428 | 1.85E-10 | |||||
| Ubiquitination | B1A4G6 | GH1 | Growth hormone 1 isoform 1 | EETQQK*SNLELLR | K | 96 | 1 | 5.87E + 06 | ||||
| P01241 | GH1 | Somatotropin | EETQQK*SNLELLR | K | 96 | 1 | 4.23E + 06 | |||||
| Acetylation | P01241 | GH1 | Somatotropin | QTYSK*FDTNSHNDDALLK | K | 171 | 1 |
Results
2DGE pattern of GH proteoforms in GH-secreting pituitary adenomas and control pituitary tissues
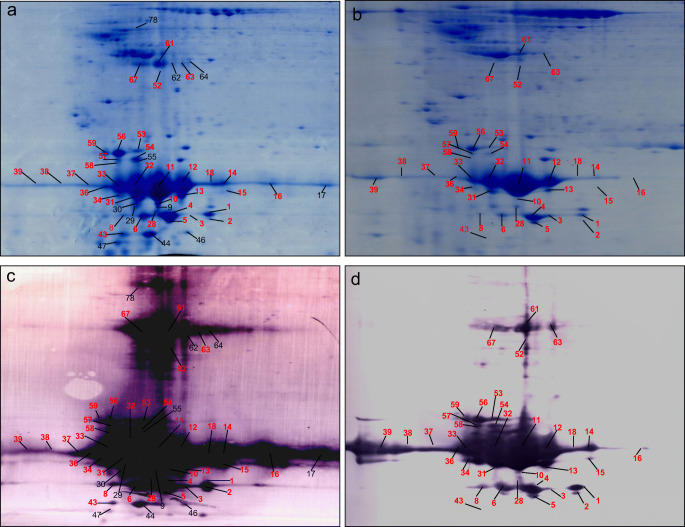
Proteoforms are the final structural and functional formats of a gene. Each proteoform has a specific pI and Mr, and can be readily arrayed with 2DGE separation according to its pI and Mr values, and can be easily detected with western immunoaffinity blot against an anti-GH antibody. A total of ~ 1,100 protein spots were detected in each 2DGE map of GH-secreting pituitary adenomas (n = 3 gels) and control pituitaries (n = 3 gels). A representative Coomassie-stained 2DGE map is shown for GH-secreting pituitary adenomas (Fig. 1A) and control pituitaries (Fig. 1B). A portion (~ 70%) of those 2DGE-separated proteins were transferred onto the PVDF membrane, and blotted with anti-hGH antibodies. A representative 2DGE-based western blot image is shown for GH-secreting pituitary adenomas (Fig. 1C) and control pituitaries (Fig. 1D). Comparative analysis of 2DGE-based western blot image and the corresponding Coomassie-stained 2DGE map identified 46 GH-immunoaffinity positive spots in GH-secreting pituitary adenomas (Figs. 1A and 1C), and 35 GH-immunoaffinity positive spots in control pituitaries (Figs. 1B and 1D). The comparative analyses of Coomassie-stained 2DGE images between GH-secreting pituitary adenomas and controls (Figs. 1A and 1B), and 2DGE-based western blot images between GH-secreting pituitary adenomas and controls (Figs. 1C and 1D) found that 35 GH-immunoaffinity positive spots in control pituitaries were all matched to the corresponding 35 of 46 spots in GH-secreting pituitary adenomas, and 11 GH-immunoaffinity positive spots only existed in GH-secreting pituitary adenomas but not in control pituitaries. The GH-immunoaffinity positive spots represented 46 GH proteoforms in GH-secreting pituitary adenomas and 35 GH proteoforms in control pituitaries (Table 3).
Fig. 1.
Two-dimensional gel electrophoresis-based GH-immunoaffinity analysis in GH-secreting pituitary adenoma and control pituitary tissues. (a) Coomassie blue-stained 2D gel image before transfer of proteins to a PVDF membrane in the GH-secreting pituitary adenoma tissue. (b) Coomassie blue-stained 2D gel image before transfer of proteins to a PVDF membrane in the control pituitary tissue. (c) 2DGE-based GH-immunoaffinity western blot image in the GH-secreting pituitary adenoma tissue. (d) 2DGE-based GH-immunoaffinity western blot image in the control pituitary tissue. GH: growth hormone. 2DGE: two-dimensional gel electrophoresis
Table 3.
MS-characterrized GH expressed in GH-serecting PA and controls tissues, and PTMs
| GH-secreting pituitary adenoma | Control pituitary | Ratio (T/N) | Control pituitary (Proteomics, 2005, 5: 1228–1241) [29] | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot No | Swiss-Prot no | Theoretical pI; Mr | Gene name | The number of matched peptides | sequence coverage (%) | Spot volume | Signal peptide removal | Splicing events | PTM | Spot No | Swiss-Prot no | Theoretical pI; Mr | Gene name | The number of matched peptides | sequence coverage (%) | Spot volume | Signal peptide removal | Splicing events | PTM | Spot No | Swiss-Prot No | Theoretical pI; Mr | Observed pI; Mr | Gene name | The number of mathed peptides | Sequence coverage (%) | Signal peptide removal | Splicing events | PTM | |
| 1 | P01241 | 5.27; 22.13 | GH1 | 10 | 41.9 | 4E + 05 | + | 2 | D178 | 1 | P01241 | 5.27; 22.13 | GH1 | 9 | 2E + 05 | + | 1 | 1.62 | 16 | P01241 | 5.27; 22.13 | 5.88; 19.37 | GH1 | 9 | 30.4 | + | 1 | P176; D178 | ||
| 2 | P01241 | 5.27; 22.13 | GH1 | 6 | 18.3 | 2E + 05 | + | 1 | 2 | P01241 | 5.27; 22.13 | GH1 | 1E + 05 | 1.79 | ||||||||||||||||
| 3 | P01241 | 5.27; 22.13 | GH1 | 9 | 30.9 | 1E + 05 | + | 1 | 3 | P01241 | 5.27; 22.13 | GH1 | 6 | 24.1 | 3E + 05 | + | 2 | 0.33 | ||||||||||||
| 4 | P01241 | 5.27; 22.13 | GH1 | 11 | 56 | 2E + 06 | + | 1 | 4 | P01241 | 5.27; 22.13 | GH1 | 15 | 9E + 05 | + | 2 | 1.64 | 9 | P01241 | 5.41; 22.99 | 5.56; 19.37 | GH1 | 15 | 59.9 | + | 2 | P176 | |||
| 5 | P01241 | 5.27; 22.13 | GH1 | 12 | 62.8 | 4E + 05 | + | 2 | D178 | 5 | P01241 | 5.27; 22.13 | GH1 | 6 | 18.8 | 3E + 05 | + | 1 | 1.33 | 10 | P01241 | 5.27; 22.13 | 5.57; 18.89 | GH1 | 11 | 41.9 | + | 1 | P176 | |
| 6 | P01241 | 5.27; 22.13 | GH1 | 11 | 53.9 | 8E + 05 | + | 1 | P174/176; D178 | 6 | P01241 | 5.27; 22.13 | GH1 | 6 | 24.1 | 3E + 05 | + | 2 | 2.96 | 1 | P01241 | 5.27; 22.13 | 5.33; 19.43 | GH1 | 10 | 33.3 | + | 1/2 | P176 | |
| 8 | P01241 | 5.27; 22.13 | GH1 | 11 | 53.9 | 3E + 05 | + | 1 | D178 | 8 | P01241 | 5.27; 22.13 | GH1 | 6 | 2E + 05 | + | 1 | 2.06 | ||||||||||||
| 9 | P01241 | 5.27; 22.13 | GH1 | 11 | 51.8 | 2E + 05 | + | 1 | P77; D178 | ± | ||||||||||||||||||||
| 10 | P01241 | 5.27; 22.13 | GH1 | 10 | 57.1 | 3E + 05 | + | 1 | P77 | 10 | P01241 | 5.27; 22.13 | GH1 | 6 | 98,039 | + | 1 | 3.08 | 12 | P01241 | 5.27; 22.13 | 5.44; 21.87 | GH1 | 14 | 66.5 | + | 1 | P176; P132 | ||
| 11 | P01241 | 5.27; 22.13 | GH1 | 7 | 49.7 | 7E + 06 | + | 1 | 11 | P01241 | 5.27; 22.13 | GH1 | 7 | 24.1 | 5E + 06 | + | 1 | P132 | 1.29 | 15 | P01241 | 5.27; 22.13 | 5.44; 22.95 | GH1 | 16 | 79.6 | + | 1 | P176; P132 | |
| 12 | P01241 | 5.27; 22.13 | GH1 | 6 | 19.4 | 3E + 06 | + | 1 | P132 | 12 | P01241 | 5.27; 22.13 | GH1 | 7 | 24.1 | 2E + 06 | + | 1 | 1.60 | 14 | P01241 | 5.27; 22.13 | 5.67; 22.79 | GH1 | 16 | 73.8 | + | 1 | P132 | |
| 13 | P01241 | 5.27; 22.13 | GH1 | 13 | 75.4 | 7E + 05 | + | 1 | P77; D178 | 13 | P01241 | 5.27; 22.13 | GH1 | 5 | 18.8 | 2E + 05 | + | 1 | 3.36 | 20 | P01241 | 5.27; 22.13 | 5.66; 21.35 | GH1 | 9 | 37.2 | + | 1 | ||
| 14 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 4E + 05 | + | 1 | D178 | 14 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 2E + 05 | + | 1 | D178 | 2.49 | 17 | P01241 | 5.27; 22.13 | 5.87; 22.65 | GH1 | 6 | 19.4 | + | 1 | |
| 15 | P01241 | 5.27; 22.13 | GH1 | 8 | 36.1 | 2E + 05 | + | 1 | 15 | P01241 | 5.27; 22.13 | GH1 | 55,144 | 3.82 | ||||||||||||||||
| 16 | P01241 | 5.27; 22.13 | GH1 | 13 | 75.4 | 3E + 05 | + | 1 | P77; D178 | 16 | P01241 | 5.27; 22.13 | GH1 | 6 | 32.5 | 56,066 | + | 1 | 5.11 | |||||||||||
| 17 | P01241 | 5.27; 22.13 | GH1 | 13 | 75.4 | 3E + 05 | + | 1 | P77; D178 | ± | ||||||||||||||||||||
| 18 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 3E + 05 | + | 1 | D178 | 18 | P01241 | 5.27; 22.13 | GH1 | 1E + 05 | 2.01 | |||||||||||||||
| 28 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 2E + 05 | + | 1 | D178 | 28 | P01241 | 5.27; 22.13 | GH1 | 7 | 24.1 | 1E + 05 | + | 1 | 1.68 | |||||||||||
| 29 | P01241 | 5.27; 22.13 | GH1 | 9 | 39.3 | 3E + 05 | + | 1 | ± | |||||||||||||||||||||
| 30 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 3E + 05 | + | 1 | AcetK171; D178 | ± | ||||||||||||||||||||
| 31 | P01241 | 5.27; 22.13 | GH1 | 11 | 57.1 | 8E + 05 | + | 1 | 31 | P01241 | 5.27; 22.13 | GH1 | 4 | 14.1 | 4E + 05 | + | 1 | P132 | 2.01 | 5 | P01241 | 5.27; 22.13 | 5.26; 21.81 | GH1 | 7 | 33 | + | 1 | P176; D178 | |
| 32 | P01241 | 5.27; 22.13 | GH1 | 10 | 66 | 4E + 06 | + | 1 | 32 | P01241 | 5.27; 22.13 | GH1 | 9 | 57.6 | 2E + 06 | + | 1 | P132 | 2.01 | 6 | P01241 | 5.27; 22.13 | 5.28; 22.95 | GH1 | 19 | 79.6 | + | 1 | P; D178 | |
| 33 | P01241 | 5.27; 22.13 | GH1 | 11 | 57.1 | 2E + 06 | + | 1 | P77 | 33 | P01241 | 5.27; 22.13 | GH1 | 8 | 31.9 | 8E + 05 | + | 1 | P132 | 2.47 | 4 | P01241 | 5.29; 24.85 | 5.14; 22.79 | GH1 | 9 | 45.8 | + | 1 | |
| 34 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 5E + 05 | + | 1 | D178 | 34 | P01241 | 5.27; 22.13 | GH1 | 12 | 2E + 05 | + | 1 | 2.47 | 8 | P01241 | 5.27; 22.13 | 5.14; 21.82 | GH1 | 12 | 49.2 | + | 1 | P176; P132 | ||
| 36 | P01241 | 5.27; 22.13 | GH1 | 13 | 75.4 | 5E + 05 | + | 1 | P132; D178 | 36 | P01241 | 5.27; 22.13 | GH1 | 7 | 36.1 | 2E + 05 | + | 1 | 2.46 | |||||||||||
| 37 | P01241 | 5.27; 22.13 | GH1 | 11 | 57.1 | 3E + 05 | + | 1 | 37 | P01241 | 5.27; 22.13 | GH1 | 6 | 36.1 | 1E + 05 | + | 1 | 1.96 | 22 | P01241 | 5.27; 22.13 | 4.83; 23.50 | GH1 | 5 | 25.1 | + | 1 | |||
| 38 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 2E + 05 | + | 1 | P132; D178 | 38 | P01241 | 5.27; 22.13 | GH1 | 2E + 05 | 0.92 | |||||||||||||||
| 39 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 2E + 05 | + | 1 | P174/176; D178 | 39 | P01241 | 5.27; 22.13 | GH1 | 11 | 57.1 | 4E + 05 | + | 1 | 0.39 | |||||||||||
| 43 | P01241 | 5.27; 22.13 | GH1 | 11 | 53.9 | 4E + 05 | + | 1 | P132; D178 | 43 | P01241 | 5.27; 22.13 | GH1 | 7 | 2E + 05 | + | 1 | 2.82 | 18 | P01241 | 5.27; 22.13 | 5.19; 16.25 | GH1 | 7 | 36.6 | + | 1 | P176; P132 | ||
| 44 | P01241 | 5.27; 22.13 | GH1 | 10 | 63.1 | 8E + 05 | + | 1 | P132; D178 | ± | ||||||||||||||||||||
| 46 | P01241 | 5.27; 22.13 | GH1 | 6 | 37.7 | 1E + 05 | - | 1 | ± | |||||||||||||||||||||
| 47 | P01241 | 5.27; 22.13 | GH1 | 8 | 38.7 | 3E + 05 | + | 1 | ± | |||||||||||||||||||||
| 52 | P01241 | 5.27; 22.13 | GH1 | 4 | 20.9 | 48,543 | + | 1 | 52 | P01241 | 5.27; 22.13 | GH1 | 49,575 | 0.98 | ||||||||||||||||
| 53 | P01241 | 5.27; 22.13 | GH1 | 11 | 51.8 | 2E + 05 | + | 1 | D178 | 53 | P01241 | 5.27; 22.13 | GH1 | 2E + 05 | 1.49 | |||||||||||||||
| 54 | P01241 | 5.27; 22.13 | GH1 | 9 | 36.1 | 2E + 05 | + | 1 | P132; D178 | 54 | P01241 | 5.27; 22.13 | GH1 | 2E + 05 | 1.39 | |||||||||||||||
| 55 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 2E + 05 | + | 1 | ± | |||||||||||||||||||||
| 56 | P01241 | 5.27; 22.13 | GH1 | 9 | 45 | 8E + 05 | + | 1 | 56 | P01241 | 5.27; 22.13 | GH1 | 8 | 36.1 | 5E + 05 | + | 1 | P132; D178 | 1.56 | 3 | P01241 | 5.27; 22.13 | 5.14; 25.89 | GH1 | 12 | 49.7 | + | 1 | P132 | |
| 57 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 4E + 05 | + | 1 | D178 | 57 | P01241 | 5.27; 22.13 | GH1 | 1E + 05 | 3.29 | |||||||||||||||
| 58 | P01241 | 5.27; 22.13 | GH1 | 12 | 53.9 | 1E + 05 | + | 1 | D178 | 58 | P01241 | 5.27; 22.13 | GH1 | 6 | 18.8 | 75,247 | + | 1 | 1.81 | 7 | P01241 | 5.27; 22.13 | 5.14; 25.36 | GH1 | 10 | 42.4 | + | 1 | ||
| 59 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 3E + 05 | + | 1 | D178 | 59 | P01241 | 5.27; 22.13 | GH1 | 9 | 1E + 05 | + | 1 | 1.88 | 19 | P01241 | 5.27; 22.13 | 5.00; 25.17 | GH1 | 9 | 27.2 | + | 1 | |||
| 61 | P01241 | 5.27; 22.13 | GH1 | 11 | 57.1 | 6E + 05 | + | 1 | 61 | P01241 | 5.27; 22.13 | GH1 | 2E + 05 | 2.48 | ||||||||||||||||
| 62 | P01241 | 5.27; 22.13 | GH1 | 12 | 56.5 | 47,467 | + | 1 | ± | |||||||||||||||||||||
| 63 | P01241 | 5.27; 22.13 | GH1 | 12 | 63.9 | 75,148 | + | 1 | D178 | 63 | P01241 | 5.27; 22.13 | GH1 | 80,246 | 0.94 | |||||||||||||||
| 64 | P01241 | 5.27; 22.13 | GH1 | 7 | 30.9 | 31,190 | + | 1 | ± | |||||||||||||||||||||
| 67 | P01241 | 5.27; 22.13 | GH1 | 6 | 30.9 | 4E + 05 | + | 1 | D178 | 67 | P01241 | 5.27; 22.13 | GH1 | 5 | 18.8 | 1E + 05 | + | 1 | 2.98 | 24 | P01241 | 5.27; 22.13 | 5.45; 42.42 | GH1 | 12 | 57.1 | + | 1 | P132 | |
| 78 | P01241 | 5.27; 22.13 | GH1 | 11 | 53.9 | 1E + 05 | + | 3 | P174/176; Ub-K96 | ± | ||||||||||||||||||||
For ratio (T/N), ± means that GH proteoform only existed in GH-secreting pituitary adenoma but not in control pituitary. For signal peptide remeval, + means that signal peptide (postion 1–26) was removed in GH proteoform; and—means that signal peptide (position 1–26) was not removed from GH proteoform. For splicing events, 1 means that amino acid sequence (postion 1–26) was deleted from GH precursor; 2 means that amino acid sequences (position 1–26, and 58–72) were deleted from GH precursor; 3 means that amino acid sequences (positon 1–16, and 111–148) were deleted from GH precursor; and 4 means that amino acid sequences (position 1–26, and 117–162) were deleted from GH precursor. P132 = phosphorylation at Ser132. P134 = phosphorylation at Ser134. P176 = phosphorylation at Ser176. P = phosphorylation at residues Ser77, Ser132, and Ser176. D178 = deamidation at Asn178
Our previous 2DGE-MS study revealed 24 GH proteoforms in control pituitaries [29]. A comparative analysis of 35 GH proteoforms in control pituitaries and our previously identified 24 GH proteoforms in control pituitaries [29] found that 19 GH proteoforms in this study are the same as our previous study (Table 3), including GH proteoforms 1, 4, 5, 6, 10, 11, 12, 13, 14, 31, 32, 33, 34, 37, 43, 56, 58, 59, and 67 in this study (Fig. 1B) that correspond to GH proteoforms 16, 9, 10, 1, 12, 15, 14, 20, 17, 5, 6, 4, 8, 22, 18, 3, 7, 19, and 24 in our previous study. These 19 consistent GH proteoforms in control pituitaries were all the dominant GH proteoforms. This result clearly demonstrated excellent experimental reproducibility.
Moreover, PDQuest 2D gel image analysis quantified the spot volumes of 46 Coomassie-stained spots that correspond to GH-immunoaffinity positive spots in GH-secreting pituitary adenomas (Fig. 1A) and 35 Coomassie-stained spots that correspond to GH-immunoaffinity positive spots in control pituitaries (Fig. 1B); those data reflect the abundance of each GH proteoform (Table 3, Figs. 1A and 1B). Those results indicate that (i) GH proteoforms 9, 17, 29, 30, 44, 46, 47, 55, 62, 64, and 78 (n = 11) only existed in GH-secreting pituitary adenomas but not in control pituitaries; (ii) GH proteoforms 3, 38, 39, 52, and 63 (n = 5) showed a decreased abundance in GH-secreting pituitary adenomas compared to control pituitaries; and (iii) GH proteoforms 1, 2, 4, 5, 6, 8, 10, 11, 12, 13, 14, 15, 16, 18, 28, 31, 32, 33, 34, 36, 37, 43, 53, 54, 56, 57, 58, 59, 61, and 67 (n = 30) showed an increased abundance in GH-secreting pituitary adenomas compared to control pituitaries. These extensive, detailed experimental results clearly reveal the increased abundance levels of 30 GH proteoforms, decreased abundance levels of 5 GH proteoforms, and 11 new GH proteoforms in GH-secreting pituitary adenomas compared to control pituitaries.
These findings clearly demonstrated that the hGH proteoform pattern was significantly different in GH-secreting pituitary adenomas compared to control pituitaries, and probably reflected different, potentially significant and analytically useful, underlying molecular, metabolic, and physiological processes.
MS identification of GH proteoforms in GH-secreting pituitary adenomas and control pituitary tissues
MS is an effective method to identify 2DGE-separated proteins when combined with pIand Mr information to determine proteoforms. PMF and MS/MS were used to identify each GH proteoform (Figs. 1A and 1B). A total of 46 2D gel spots– corresponding to GH-immunoaffinity positive spots in GH-secreting pituitary adenomas (Fig. 1A)– and35 2D gel spots– corresponding to GH-immunoaffinity positive spots in control pituitaries (Fig. 1B)– were excised to identify GH proteoforms. The proteins in each gel spot were subjected to in-gel digestion with trypsin, followed by the extraction and purification of tryptic peptides. The tryptic peptide mixture was analyzed with MALDI-TOF-TOF MS (PMF) and MS/MS. The PMF or MS/MS data were used to search a human protein database for protein identification. For GH-secreting pituitary adenomas (Fig. 1A), all 46 2D gel spots were identified to contain human GH (Swiss-Prot accession number: P01241) (Table 3). For control pituitaries (Fig. 1B), among 35 2D gel spots, 25 2D gel spots were identified to contain human GH (Swiss-Prot accession number: P01241). No proteins were MS-identified in 2D gel spots 2, 15, 18, 38, 52, 53, 54, 57, 61, and 63 (n = 10). However, those 10 spots (Fig. 1B) were found to contain human GH in their corresponding 2DGE spots of GH-secreting pituitary adenomas (Fig. 1A, Table 3) and their GH-immunoaffinity blot images were positive (Fig. 1D).
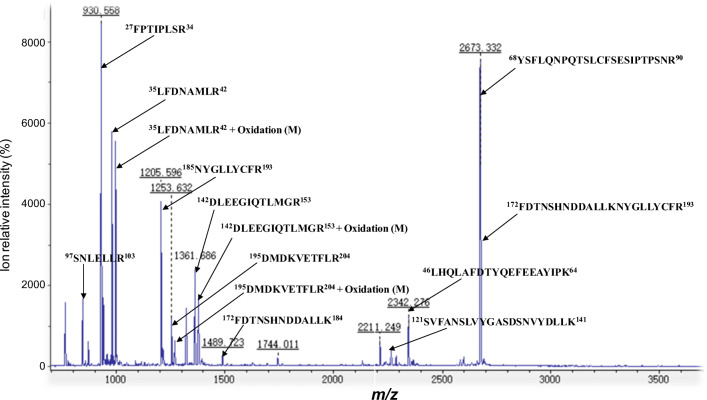
A representative MS spectrum demonstrates that hGH was contained in spot 36 in GH-secreting pituitary adenoma tissues (Fig. 2). The tryptic peptides derived from the proteins in spot 36 in GH-secreting pituitary adenomas were analyzed with MALDI-TOF-TOF–MS to obtain PMF data (Fig. 2). The PMF data used to search the UniProtKB human protein database found that 13 peptides matched with hGH (Swiss-Prot No.: P01241), including 97SNLELLR103 (m/z 844.5), 35LFDNAMLR42 (m/z 979.5), 35LFDNAM#LR42 (m/z 995.5; M# = oxidation at residue M), 185LFDNAMLR193 (m/z 1205.6), 195DMDKVETFLR204 (m/z 1253.6), 195DM#DKVETFLR204 (m/z 1269.6; M# = oxidation at residue M), 142DLEEGIQTLMGR153 (m/z 1361.7), 142DLEEGIQTLM#GR153 (m/z 1377.7; M# = oxidation at residue M), 172FDTNSHNDDALLK184 (m/z 1489.7), 121SVFANSLVYGASDSNVYDLLK141 (m/z 2262.3), 46LHQLAFDTYQEFEEAYIPK64 (m/z 2342.3), 68YSFLQNPQTSLCFSESIPTPSNR90 (m/z 2673.3), and 172FDTNSHNDDALLKNYGLLYCFR193 (m/z 2676.4) (Fig. 2; Supplemental Table 1). In addition, MALDI-TOF-TOF MS not only produced PMF data but also MS/MS data. MS/MS data elucidates the amino acid sequence to confirm PMF analysis. A representative MS/MS spectrum is shown for tryptic peptide 46LHQLAFDTYQEFEEAYIPK64 (precursor ion [M + H]+, m/z 2342.318) derived from hGH in spot 36 in GH-secreting pituitary adenomas (Fig. 3A), with a high signal-to noise (S/N) ratio and excellent b-ion and y-ion series (b1, b3, b4, b6, b7, b8, b9, b10, b11, b12, b13, b14, b15, b16, b17, y2, y5, y6, y8, y9, y10, y12, and y15). Among those 13 tryptic peptides, 7 were also found in an MS spectrum derived from proteins contained in spot 36 in control pituitary tissues, which also significantly matched to hGH (Swiss-Prot No.: P01241) in the UniProtKB human protein database (Supplemental Table 1). Thus, hGH was identified in spot 36 in GH-secreting pituitary adenomas and control pituitaries, which–combined with its observed pI and Mr in the 2DGE map (its theoretical pI and Mr were 5.27 and 22.13 kD)–characterized the GH proteoform 36. GH proteoform 36 had an increased abundance level in GH-secreting pituitary adenomas relative to control pituitary tissues with ratio (tumor/control) = 2.46 (Table 3). The same method was used to identify the other GH proteoforms in GH-secreting pituitary adenomas and control pituitary tissues (Table 3; Supplemental Table 1). Therefore, PMF data and MS/MS data clearly demonstrated that hGH was distributed in all 46 GH-immunoaffinity positive spots in GH-secreting pituitary adenomas and 35 GH-immunoaffinity positive spots in control pituitary tissues (Fig. 1 and Table 3).
Fig. 2.
MS spectrum of the tryptic peptides derived from hGH that was contained in spot 14 in GH-secreting pituitary adenoma tissues. m/z = mass to charge
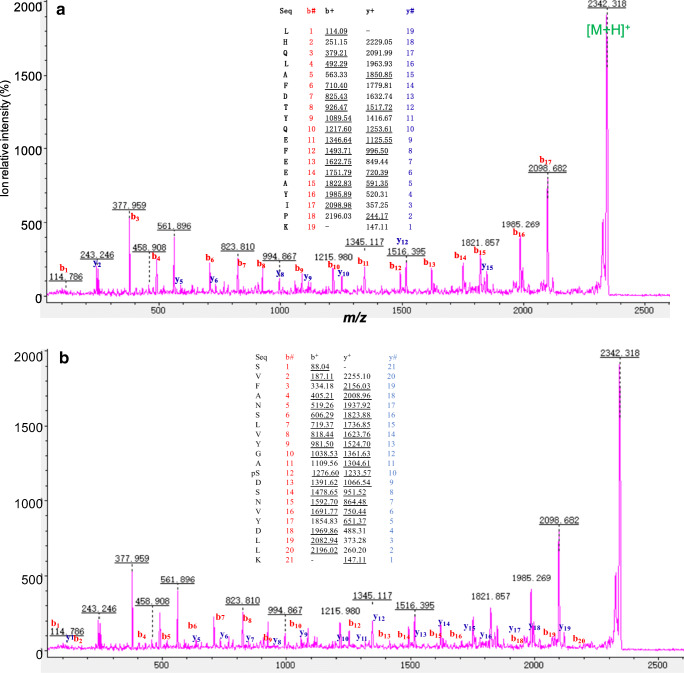
Fig. 3.
MS/MS spectra of the tryptic peptides 46LHQLAFDTYQEFEEAYIPK64 (A) and 121SVFANSLVYGAS*DSNVYDLLK141 (S* = phosphorylated Ser132) (B) derived from hGH that was contained in spot 36 in GH-secreting pituitary adenoma tissues. m/z = mass to change
Analysis of signal-peptide removal in GH proteoforms in GH-secreting pituitary adenomas and control pituitary tissues
The human GH precursor contains a signal peptide (position 1–26) that is not contained in mature GH (Table 1). Before the signal peptide is removed, theoretically there are four characteristic tryptic peptides, including 1MATGSR6 ([M + H]+ m/z 622.3), 7TSLLLAFGLLCLPWLQEGSAFPTIPLSR34 ([M + H]+ m/z 3043.7), 1MATGSRTSLLLAFGLLCLPWLQEGSAFPTIPLSR34 ([M + H]+ m/z 3646.9), and 7TSLLLAFGLLCLPWLQEGSAFPTIPLSRLFDNAMLR42 ([M + H]+m/z 4004.2) (Table 4). After the signal peptide is removed, theoretically there are two characteristic tryptic peptides, including 27FPTIPLSR34 ([M + H]+ m/z 930.5), and 27FPTIPLSRLFDNAMLR42 ([M + H]+ m/z 1891, or 1907.0 (MSO: 40)) (Table 4). These characteristic tryptic peptide ions (Table 4) were compared to the PMF data (Fig. 2) of each GH proteoform derived from GH-secreting pituitary adenoma and control samples. The results clearly demonstrated that the characteristic tryptic peptide ion 27FPTIPLSR34 ([M + H]+ m/z 930.5) appeared in all PMF spectra derived from control pituitary tissues, and from GH-secreting pituitary adenoma tissues- except for proteoform 46 of GH-secreting pituitary adenomas (Table 3). Those data indicate that these identified GH proteoforms do not contain the signal peptide (position 1–26). For proteoform 46 of GH-secreting pituitary adenomas, the characteristic tryptic peptide ion 27FPTIPLSR34 ([M + H]+ m/z 930.5) did not appear in its PMF spectrum, but the characteristic tryptic peptide ion 7TSLLLAFGLLCLPWLQEGSAFPTIPLSR34([M + H]+m/z = 3043.7) did appear (Table 3) to clearly demonstrate that the signal peptide was not removed from GH proteoform 46. Therefore, GH proteoform 46 was a GH precursor in GH-secreting pituitary adenoma tissues. All other identified GH proteoforms were not GH precursors but mature GH in GH-secreting pituitary adenoma and control pituitary tissues (Table 3).
Table 4.
Analysis of removal of signal peptide sequence (position 1–26) from hGH in GH-secreting pituitary adenomas and control pituitaries
| Before and after removal of the signal peptide 1–26 | Tryptic peptide | Position | Calculated [M + H] + ion (m/z) | Observed [M + H] + ion (m/z) with MALDI PMF data | GH-secreting pituitary adenoma (Spot number) | Control pituitary (Spot number) |
|---|---|---|---|---|---|---|
| Before | MATGSR | 1–6 | 622.3 (MSO: 1; 638.3) | |||
| TSLLLAFGLLCLPWLQEGSAFPTIPLSR | 7–34 | 3043.7 | 3042.6 | 46 | ||
| 3100.7(Cys_CAM: 17) | ||||||
| MATGSRTSLLLAFGLLCLPWLQEGSAFPTIPLSR | 1–34 | 3646.9 | ||||
| 3704.0 (Cys_CAM: 17) | ||||||
| 3662.9 (MSO: 1) | ||||||
| TSLLLAFGLLCLPWLQEGSAFPTIPLSRLFDNAMLR | 7–42 | 4004.2 | ||||
| 4061.2 (Cys_CAM: 17) | ||||||
| 4020.1 (MSO: 40) | ||||||
| After | FPTIPLSR | 27–34 | 930.5 | + | 1–6, 8–18, 28–34, 36–39, 43–44, 47, 52–59, 61–64, 67, 78 | 1, 3–6, 8, 10–14, 16, 28, 31–34, 36, 37, 39, 43, 56, 58, 59, 67 |
| FPTIPLSRLFDNAMLR | 27–42 | 1891 | ||||
| 1907.0 (MSO: 40) |
Analysis of splicing events in GH proteoforms in GH-secreting pituitary adenomas and control pituitary tissues
Alternative splicing is an important factor to produce protein diversity. Human GH has four splicing variants [29], including splicing variant 2 with removal of amino acid sequence 58–72, splicing variant 3 with removal of amino acid sequence 111–148, and splicing variant 4 with removal of amino acid sequence 117–162, from the normal GH (GH variant 1) (Table 1). Thus, before and after removal of these amino acid sequences from normal GH, there were characteristic tryptic peptides for the splicing events of human GH in human GH-secreting pituitary adenomas and control pituitary tissues (Table 5). These characteristic tryptic peptide ions were compared to the PMF data of each GH proteoform derived from GH-secreting pituitary adenoma and control pituitary tissues. The results show that, for GH-secreting pituitary adenomas, GH proteoforms 1 and 5 were splicing variant 2, GH proteoform 78 was splicing variant 3, the other 43 GH proteoforms were splicing variant 1 (normal GH), and no splicing variant 4 was identified. For control pituitaries, GH proteoforms 3, 4, and 6 were splicing variant 2, and the other GH proteoforms were splicing variant 1 (normal GH); no splicing variants 3 and 4 were identified (Tables 3 and 5).
Table 5.
Identification of splicing events of hGH in GH-secreting pituitary adenomas and control pituitaries
| GH isoform | Tryptic peptide that covers the splicing site | Calculated [M + H]+ ion (m/z) | Observed [M + H]+ ion (m/z) in PMF data | GH-secreting pituitary adenoma (Spot number) | Control pituitary (spot number) |
|---|---|---|---|---|---|
| 1 | 142DLEEGIQTLMGR153 | 1361.7 | One or two ions were detected to exclude Isoforms 3 and 4 | 2–4, 6, 8–18, 28–34, 36–39, 43–44, 46–47, 52–59, 61–64, 67 | 1, 5, 8, 10–14, 16, 28, 31–34, 36, 37, 39, 43, 56, 58, 59, 67 |
| 1377.7 (MSO: 151) | |||||
| 121SVFANSLVYGASDSNVYDLLK141 | 2262.1 | ||||
| 46LHQLA FDTYQEFEEAYIPK64 | 2342.1 |
One, two, or three ions were detected to exclude Isoform 2 |
|||
| 68YSF LQNPQTSLCFSESIPTPSNR90 | 2616.2 | ||||
| 2673.2 (Cys-CAM: 79) | |||||
| 43AHRLHQLAFDTYQ EFEEAYIPK64 | 2706.3 | ||||
| 2 | LHQLAFDTYQEF58NPQTSLCFSESIPTPSNR | 3470.6 | |||
| 3527.7 (Cys-CAM: 64) | 3527.3 | 1, 5 | |||
| 3527.5 | 3, 4, 6 | ||||
| 3529.5 | 6 | ||||
| AHRLHQLAFDTYQEF58NPQTSLCFSESIPTPSNR | 3834.8 | ||||
| 3891.9 (Cys-CAM: 64) | |||||
| LHQLAFDTYQEF58NPQTSLCFSESIPTPSNREETQQK | 4213.9 | ||||
| 4271.0 (Cys-CAM: 64) | |||||
| 3 | ISLLLIQ111TLMGR | 1357.8 | 1357.7 | 78 | |
| 1373.8 (MSO: 113) | |||||
| ISLLLIQ111TLMGRLEDGSPR | 2112.2 | ||||
| 2128.2 (MSO: 113) | |||||
| SNLE LLRISLLLIQ111TLMGR | 2183.3 | ||||
| 2199.3 (MSO: 113) | |||||
| 4 | ISLLLIQSWLEPV117QIFK | 2027.2 | |||
| ISLLLIQSWLEPV117QIFKQTYS K | 2852.7 | ||||
| SNLE LLRISLLLIQSWLEPV117QIFK | 2852.7 |
Analysis of PTMs of GH proteoforms in GH-secreting pituitary adenomas and control pituitary tissues
PTMs are another crucially important factor that produces protein diversity – namely proteoforms. Our previous quantitative phosphoproteomic, ubiquitinomic, and acetylomic studies [38, 46] determined four phosphorylation sites at residues Ser132, Ser134, Thr174, and Ser176, one ubiquitination site at residue Lys96, and one acetylation site at residue Lys171, in human GH in pituitary adenomas and control pituitary tissues (Table 2). Those PTM sites and the corresponding tryptic peptides were compared to each MS spectrum of GH proteoform that was contained in a 2DGE spot to determine the status of phosphorylation, ubiquitination, and acetylation in each GH proteoform in GH-secreting pituitary adenomas and control pituitary tissues (Table 6).
Table 6.
Identification of phosphorylation, ubiquitination, and acetylation in growth hormone of growth hormone-secreting PA and controls
| Quantitative proteomics identified modifications in growth hormone | Modifications in growth hormone proteoforms | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene names | Swiss-Prot | Modificed peptide | Modified peptide position | Modified residue | Modified residue position | Calculated unmodified peptide (m/z) | Observed unmodified peptide (m/z; PMF data) | Calculated modified peptide (m/z) | Observed modified peptide (m/z; PMF data) | Proteoforms (2DE spot No.) | Detected PTM | Methods |
| GH1 | P01241 | SVFANSLVYGAS*DSNVYDLLK | 121–141 | S132 | P132 | 2262.13 | 2342.13 | 2342.20 | T12 | + | PMF + MS/MS | |
| 2262.30 | 2342.30 | T36 | + | PMF + MS/MS | ||||||||
| 2342.20 | T54 | + | PMF + MS/MS | |||||||||
| 2262.60 | 2342.60 | C11 | + | PMF + MS/MS | ||||||||
| 2262.60 | 2342.70 | C32 | + | PMF + MS/MS | ||||||||
| 2262.70 | 2342.80 | C33 | + | PMF + MS/MS | ||||||||
| 2262.20 | 2342.20 | C56 | + | PMF + MS/MS | ||||||||
| YSFLQNPQTS*LCFSESIPTPSNR | 68–90 | S77 | P77 | 2616.24 | 2696.24 | 2696.05 | T9 | + | PMF | |||
| 2696.18 | T10 | + | PMF | |||||||||
| 2696.27 | T13 | + | PMF | |||||||||
| 2696.14 | T16 | + | PMF | |||||||||
| 2696.17 | T17 | + | PMF | |||||||||
| 2696.44 | T33 | + | PMF | |||||||||
| 2696.11 | T38 | + | PMF | |||||||||
| 2696.14 | T43 | + | PMF | |||||||||
| 2696.27 | T44 | + | PMF | |||||||||
| FDTNS*HNDDALLK | 172–184 | S176; N178 | P176; D 178 | 1489.69 | 1489.65 | 1569.69 | 1570.81 | T6 | + | PMF | ||
| 1489.70 | 1569.85 | T39 | + | PMF | ||||||||
| 1489.71 | 1570.92 | T78 | + | PMF | ||||||||
| QTYSKFDTNS*HNDDALLK | 167–184 | 2096.99 | 2096.95 | 2176.99 | - | |||||||
| FDTNS*HNDDALLKNYGLLYCFR | 172–193 | 2619.23 | 2619.22 | 2699.23 | - | |||||||
| TGQIFKQTYSKFDTNS*HNDDALLK | 161–184 | 2771.36 | 2771.26 | 2851.36 | - | |||||||
| FDT*NSHNDDALLK | 172–184 | T174; N178 | P174; D 178 | 1489.69 | 1489.65 | 1569.69 | 1570.81 | T6 | + | PMF | ||
| 1489.70 | 1569.85 | T39 | + | PMF | ||||||||
| 1489.71 | 1570.92 | T78 | + | PMF | ||||||||
| QTYSKFDT*NSHNDDALLK | 167–184 | 2096.99 | 2096.99 | 2176.99 | - | |||||||
| FDT*NSHNDDALLKNYGLLYCFR | 172–193 | 2619.23 | 2619.22 | 2699.23 | - | |||||||
| TGQIFKQTYSKFDT*NSHNDDALLK | 161–184 | 2771.36 | 2771.36 | 2851.36 | - | |||||||
| TGQIFKQTYSK* | 161–171 | K171 | Acet-K171 | 1300.69 | 1300.18 | 1342.69 | - | |||||
| LEDGSPRTGQIFKQTYSK* | 154–171 | 2055.05 | 2055.03 | 2097.05 | - | |||||||
| TGQIFKQTYSK*FDTNSHNDDALLK | 161–184 | 2771.36 | 2771.36 | 2813.36 | - | |||||||
| QTYSK* | 167–171 | 626.31 | 626.51 | 668.31 | - | |||||||
| QTYSK*FDTNSHNDDALLK | 167–184 | 2096.99 | 2096.68 | 2138.99 | - | |||||||
| QTYSK*FDTNSHNDDALLKNY GLLYCFR | 167–193 | 3226.53 | 3226.56 | 3268.53 | 3267.79 | T30 | + | PMF | ||||
| EETQQK*SNLELLR | 91–103 | K96 | Ub-K96 | 1587.83 | 1587.81 | 1701.83 | 1701.93 | T78 | + | PMF | ||
| EETQQK* | 91–96 | 762.36 | 762.16 | 876.36 | - | |||||||
| EETQQK*SNLELLRISLLLIQSWLEPVQFLR | 91–120 | 3624.02 | 3624.13 | 3738.02 | - | |||||||
| YSFLQNPQTSLCFSESIPTPSNREETQQK* | 68–96 | 3359.59 | 3359.28 | 3473.59 | - | |||||||
| FDTNSHN*DDALLK | 172–184 | N178 | DEAM 178 | 1489.69 | 1489.67 | 1490.69 | 1490.65 | T1 | + | PMF | ||
| 1489.65 | 1490.69 | 1490.66 | T5 | + | PMF | |||||||
| 1489.65 | 1490.69 | 1490.65 | T6 | + | PMF | |||||||
| 1489.72 | 1490.69 | 1490.72 | T8 | + | PMF | |||||||
| 1489.69 | 1490.69 | 1490.67 | T9 | + | PMF | |||||||
| 1489.75 | 1490.69 | 1490.75 | T13 | + | PMF | |||||||
| 1489.71 | 1490.69 | 1490.70 | T14 | + | PMF | |||||||
| 1489.71 | 1490.69 | 1490.70 | T16 | + | PMF | |||||||
| 1489.71 | 1490.69 | 1490.72 | T17 | + | PMF | |||||||
| 1489.77 | 1490.69 | 1490.74 | T18 | + | PMF | |||||||
| 1489.72 | 1490.69 | 1490.72 | T28 | + | PMF | |||||||
| 1489.77 | 1490.69 | 1490.77 | T30 | + | PMF | |||||||
| 1489.79 | 1490.69 | 1490.80 | T34 | + | PMF | |||||||
| 1489.72 | 1490.69 | 1490.73 | T36 | + | PMF | |||||||
| 1489.67 | 1490.69 | 1490.68 | T38 | + | PMF | |||||||
| 1489.70 | 1490.69 | 1490.70 | T39 | + | PMF | |||||||
| 1489.75 | 1490.69 | 1490.71 | T43 | + | PMF | |||||||
| 1489.86 | 1490.69 | 1490.81 | T44 | + | PMF | |||||||
| 1489.75 | 1490.69 | 1490.74 | T53 | + | PMF | |||||||
| 1489.69 | 1490.69 | 1490.67 | T54 | + | PMF | |||||||
| 1489.69 | 1490.69 | 1490.78 | T57 | + | PMF | |||||||
| 1489.68 | 1490.69 | 1490.68 | T58 | + | PMF | |||||||
| 1489.74 | 1490.69 | 1490.79 | T59 | + | PMF | |||||||
| 1489.79 | 1490.69 | 1490.79 | T63 | + | PMF | |||||||
| 1489.62 | 1490.69 | 1490.60 | T67 | + | PMF | |||||||
| 1489.70 | 1490.69 | 1490.70 | C14 | + | PMF | |||||||
| 1489.68 | 1490.69 | 1490.68 | C56 | + | PMF | |||||||
+ : this PTM was detected. -: this PTM was not detected. ± : this PTM cannot be determined
For phosphorylation analysis of GH proteoforms, quantitative phosphoproteomics identified four phosphorylation sites at residues Ser132, Ser134, Thr174, and Ser176 in human GH (P02141) (Table 2) with an mass shift of + 80 Da at phosphorylated residues. NetPhos 3.1 Server software predicted potential phosphorylation sites of hGH precursor (P01241), including 18 pSer sites at residues 5, 8, 25, 33, 69, 77, 81, 88, 97, 105, 111, 121, 126, 134, 158, 170, 176, and 210, eight pThr sites at residues 3, 53, 76, 86, 93, 149, 168, and 201, and five pTyr sites at residues 54, 61, 68, 129, and 169. Those predicted phosphorylation sites assisted in the accurate identification of GH phosphorylation sites. Also, our previous study [46] found that Ser77 in peptide 68YSFLQNPQTSLCFSESIPTPSNR90, Ser176 in peptide 172FDTNSHNDDALLK184, and Ser132 in peptide 126SLVYGASDSNVYDLLK141 were phosphorylated in control pituitary tissues [29]. The tryptic peptide ions that contained these phosphorylation sites at residues Ser132, Ser134, Thr174, and Ser176 were compared with the PMF spectrum of each GH proteoform derived from GH-secreting pituitary adenomas and control pituitary tissues to determine the phosphorylation status of human GH proteoforms in GH-secreting pituitary adenomas and control pituitary tissues (Table 6). The theoretical masses were calculated for four tryptic peptide ions 121SVFANSLVYGASDSNVYDLLK141, 172FDTNSHNDDALLK184, 167QTYSKFDTNSHNDDALLK184, and 172FDTNSHNDDALLKNYGLLYCFR193, with and without phosphorylation at residues Ser132, Ser134, Thr174, or Ser176 in human GH via an increase of 80 Da for phosphorylation. Those data were compared to the corresponding observed tryptic peptide ion masses of each GH proteoform (Table 6). The phosphorylated tryptic peptide 121SVFANSLVYGASDSNVYDLLK141 (pSer132) was identified with PMF and amino acid MS/MS sequence data. For example, the tryptic peptide 121SVFANSLVYGASDSNVYDLLK141 had an [M + H]+ ion at m/z 2262.1. If Ser132 were phosphorylated, then it would have an [M + H]+ ion m/z 2342.1. However, the [M + H]+ ion of another tryptic peptide (46LHQLAFDTYQEFEEAYIPK64) derived from hGH had the same m/z value. Further, MS/MS analysis was used to determine the amino acid sequence of the peptide with an [M + H]+ ion at m/z2342.318 (Fig. 3). A careful examination of the PMF and MS/MS data derived from 46 GH proteoforms in GH-secreting pituitary adenomas and 20 GH proteoforms in controls found 7 PMF spectra (7 spots) that contained [M + H]+m/z 2342.318 with MS/MS data to determine two peptide amino acid sequences 46LHQLAFDTYQEFEEAYIPK64 (Fig. 3A) and 121SVFANSLVYGAS*DSNVYDLLK141 (S* = phosphorylated Ser132) (Fig. 3B).A total of three GH proteoforms (12, 36, and 54) in GH-pituitary adenomas and four GH proteoforms (11, 32, 33, and 56) in controls contained this phosphopeptideion121SVFANSLVYGAS*DSNVYDLLK141 (S* = phosphorylated Ser132) (Tables 3 and 6). Phosphorylated Ser77 was identified in nine GH proteoforms (9, 10, 13, 16, 17, 33, 38, 43, and 44) in GH-secreting pituitary adenoma tissues (Tables 3 and 6). Phosphorylated Thr174 or Ser176 was identified in GH proteoforms 6, 39, and 78 in GH-secreting pituitary adenoma tissues (Tables 3 and 6). These data clearly demonstrated the difference in phosphorylation status of GH proteoforms between GH-secreting pituitary adenomas and control pituitary tissues.
For ubiquitination analysis of GH proteoforms, quantitative ubiquitinomics identified one ubiquitination site at residue Lys96 (K96) in human GH (Table 2) with an mass shift of + 114 Da at residue K96. Theoretically, ubiquitinated tryptic peptides that contain the ubiquitination site K96 from human GH (P01241) have four peptides, including 68YSFLQNPQTSLCFSESIPTPSNREETQQK96, 91EETQQKSNLELLR103, 91EETQQK96, and 91EETQQKSNLELLRISLLLIQSWLEPVQFLR120 (Table 6). Only peptide 91EETQQKSNLELLR103 was found to be ubiquitinated with PMF data of GH proteoform T78 (Table 6), and was evidenced with an[M + H]+ ion at m/z 1587.8 (unmodified peptide 91EETQQKSNLELLR103) and m/z 1701.8 (ubiquitinated peptide 91EETQQKSNLELLR103 at residue K96) after careful examination of the PMF data and MS/MS data of each GH proteoformin GH-secreting pituitary adenomas and control pituitary tissues (Tables 3 and 6). Those data clearly demonstrated that ubiquitination occurred in GH proteoform T78 in GH-secreting pituitary adenomas, but not in control pituitary tissues.
For acetylation analysis of GH proteoforms, quantitative acetylomics identified one acetylation site at residue Lys171 (K171) in human GH (Table 2) with an mass shift of + 42 Da at residue K171. Also, bioinformatics analysis against a protein-modification database predicted seven potential acetylation sites at Lys residues 64, 96, 141, 166, 171, 194, and 198 in human GH (P01241) to assist in the accurate identification of acetylation at residue K171 in human GH (P012141). Theoretically, acetylated tryptic peptides that contain acetylation site K171 had six peptides, including 161TGQIFKQTYSK171, 167QTYSK171, 154LEDGSPRTGQIFKQTYSK171, 167QTYSKFDTNSHNDDALLK184, 161TGQIFKQTYSKFDTNSHNDDALLK184, and 167QTYSKFDTNSHNDDALLKNYGLLYCFR193 (Table 6). However, only peptide 167QTYSK*FDTNSHNDDALLKNYGLLYCFR193 (K* = acetylated Lys171) was detected in GH proteoform T30 in GH-secreting pituitary adenoma tissues (Tables 3 and 6).
Deamination at residue Asn178 (D178) was also detected in 25 GH proteoforms (1, 5, 6, 8, 9, 13, 14, 16, 17, 18, 28, 30, 34, 36, 38, 39, 43, 44, 53, 54, 57, 58, 59, 63, and 67) in GH-secreting pituitary adenomas, and GH proteoforms 1, 14, 31, 32, and 56 in control pituitary tissues (Tables 3 and 6). Deamidation at glutamine (Q) and asparagine (N) to form corresponding glutamate (E) and aspartate (D) with an increased mass shift of 1 Da and decreased apparent pI to a negatively charged (at pH 7.4) carboxylate anion commonly occurs with protein aging, or from basic conditions of protein sample storage [29]. Deamination clearly presents in a 2D gel a series of spots with the same Mr and different pI value; for example, spots 1, 4, 6, and 8; spots 33, 32, 11, and 12; and spots 13, 10, and 31, in that 2DE map (Fig. 1A and B).
Discussion
GH proteoform patterns in GH-secreting pituitary adenomas and control pituitaries
Human GH is synthesized in somatotroph cells in the pituitary gland, and is a very important hormone in the hypothalamus-pituitary-growth axes to maintain crucial physiological activity. However, abnormally secreted GH in human body might cause a wide-range of GH-related diseases, including dwarfism due to the reduced GH in childhood, gigantism due to the increased GH in adolescence, and acromegaly due to the increased GH in adult [47]. In the proteoform concept, a proteoform is defined by the amino acid sequence + PTMs + conformation + cofactors + binding partners + localization + function, which is the final structure and functional form encoded by a gene, and is the basic unit of a proteome [25]. Clarification of a GH proteoform pattern can help to elucidate the processes in the formation of, and rationalize molecular aspects of, GH-related diseases. Our previous study identified 24 human GH proteoforms in normal control pituitary tissues [29]. Acromegaly is commonly derived from GH-secreting pituitary adenomas. This present study investigated the GH proteoform pattern in GH-secreting pituitary adenoma tissues, compared with GH proteoforms in normal control pituitary tissues. A total of 46 GH proteoforms were identified in GH-secreting pituitary adenoma tissues, and 35 GH proteoforms in normal control pituitary tissues.
Comparative analysis of GH proteoform pattern changes in GH-secreting pituitary adenomas
Comparative analysis of GH proteoform patterns between GH-secreting pituitary adenomas and control pituitaries indicated that 11 GH proteoforms were found in only GH-secreting pituitary adenoma tissues but not in control pituitary tissues, and that 35 GH proteoforms were found in GH-secreting pituitary adenoma and control pituitary tissues, with abundance changes. Those data clearly demonstrated that the GH proteoform pattern was significantly different between GH-secreting pituitary adenoma and control pituitary tissues. This difference might be due to the following factors: (i) a certain genes such as STAT3 induce GH-secreting pituitary adenoma cell growth, and bind specifically to the hGH promoter to induce transcription to further promote GH secretion [48]; (ii) alterations in cell-cycle regulation and growth-factor signaling because of epigenetic changes lead to gene mutations for GH hypersecretion [49]; (iii) mistranslation provides a very large potential source of protein diversity, especially under the condition of stimulation by external factors; and (iv) PTMs, including glycosylation, phosphorylation, acetylation, ubiquitination, deamidation, and nitration, might have an impact on the structure and function of proteins to allow an exponentially increased number of proteoforms [29, 37, 38, 50, 51]. Similarly, these factors also contribute to the formation of GH proteoforms in pituitary tissues and GH-secreting pituitary adenoma tissues. For example, this present study found that phosphorylation, ubiquitination, acetylation, and deamination occurred in human GH in GH-secreting pituitary adenoma and control pituitary tissues. Also, alternative splicing and removal of signal peptide occurred in the human GH proteoforms. The alteration in a GH proteoform pattern between GH-secreting pituitary adenoma and control pituitary tissues not only demonstrated the altered quantity of GH, but also showed the structural differences of GH in GH-secreting pituitary adenomas relative to control pituitary tissues. Significantly, these novel experimental data demonstrate the important utility of the careful analysis of GH proteoform patterns in GH-related diseases in the context of predictive, preventive and, personalized medicine (3P medicine) [52–54].
2DGE in combination with MS is crucial to analyze GH proteoforms
2DGE in combination with MS is an effective method to detect, identify, and quantify human GH proteoforms in GH-secreting pituitary adenoma and control pituitary tissues. Also, a 2DGE-based western blot against anti-human GH antibody effectively visualizes the human GH proteoform pattern in a 2DGE map. According to the two unique properties, pI and Mr, of each GH proteoform, GH proteoforms were arrayed with 2DGE, confirmed with a two-dimensional western blot against an anti-GH antibody, and identified with mass spectrometry, in GH-secreting pituitary adenoma and control pituitary tissues. A total of 46 GH proteoforms in GH-secreting pituitary adenoma tissues and 35 GH proteoforms in control pituitary tissues were arrayed and identified in 2DGE maps (Fig. 1). Further analysis showed that the signal peptide (position 1–26) was removed from 35 GH proteoforms in control pituitaries and 45 GH proteoforms (except for GH proteoform T46) in GH-secreting pituitary adenomas. The data clearly demonstrate that all identified GH proteoforms derive from mature GH except for GH proteoform T46 (Table 3) and suggest an incomplete removal of signal peptide from GH proteoforms in GH-secreting pituitary adenomas. Alternative splicing is an important factor that contributes to GH protein diversity. This present study found that GH splicing variants 1, 2, and 3 existed in GH-secreting pituitary adenoma tissues, and that GH splicing variants 1 and 2 existed in control pituitary tissues (Table 3). The results demonstrated GH splicing variants 2 from adenomas and 1 from controls in spots 1 and 5, GH splicing variants 1 from adenomas and 2 from controls in spots 3, 4, and 6, and GH splicing variant 3 from adenoma in spot 78 (Table 3). PTMs are another crucial factor that contribute to GH proteoforms [55]. This present study clearly demonstrated phosphorylation, ubiquitination, acetylation, and deamination in human GH proteoforms in GH-secreting pituitary adenoma and control tissues.
An altered GH proteoform profile contributes to, and reflects metabolic processes that contribute to, related pathologies
Quantitative changes of human GH contribute to, and reflect metabolic processes involved in, GH-related diseases. Alterations of human GH proteoforms can be used to develop potential biomarkers to predict, diagnose, stratify patients, and prognostically assess the personalized treatment of GH-related diseases. Moreover, GH synthesized in the pituitary gland must be secreted into blood circulation to exert its functional roles. Therefore, compared to the invasive collection of human pituitary tissues, the non-invasive collection of blood of patients to detect a serum GH proteoform pattern is an effective method to develop serum GH proteoform pattern as an effective biomarker for the PPPM practice in GH-secreting pituitary adenomas and GH-related diseases [39, 53, 54].
Strength, limitations, and future studies
Human growth hormone that was synthesized, stored, and secreted by somatotroph cells in the pituitary gland plays important roles in the hypothalamus-pituitary-growth axes in the endocrine system. Its abnormal secretion is closely associated with multiple GH-related diseases, including GH-secreting pituitary adenomas that causes acromegaly. The traditional concept is that the pathologically increased GH levels secreted by GH-secreting pituitary adenomas cause the acromegaly.
For the first time, this present study: 1) identified 46 GH proteoforms in GH-secreting pituitary adenoma tissues and 35 GH proteoforms in control pituitary tissues; 2) 35 of 46 GH proteoforms in GH-secreting pituitary adenoma tissues were matched with 35 GH proteoforms in control pituitary tissues; and 3) the other 11 of 46 GH proteoforms existed only in GH-secreting pituitary adenoma tissues but not in control pituitary tissues. Also, these GH proteoforms in pituitary adenoma and control pituitary tissues might be derived from removal of signal peptides, alternative splicing, and PTMs (phosphorylation, ubiquitination, acetylation, deamination). These data, for the first time, clearly demonstrate that the quantity and type of changes of GH in GH-secreting pituitary adenomas might contribute to the initiation and progression of acromegaly.
This study emphasizes the importance, extensive information on metabolic processes, and potential clinical value of human GH proteoform profiling in GH-secreting pituitary adenoma, acromegaly, and other GH-related diseases to elucidate molecular mechanisms of GH-pituitary adenomas and acromegaly. A GH proteoform pattern indicates potential biomarkers for PPPM practice in GH-secreting pituitary adenomas and other GH-related diseases.
However, one must realize that some limitations in the current study remain to be studied further. First, for this discovery study, the sample size is very small. A significantly expanded sample size is needed to develop GH proteoform patterns as biomarker for GH-secreting pituitary adenomas, acromegaly, and other GH-related diseases. Second, GH must be secreted into the blood circulation to bind to growth hormone receptor (GHR) on its target organ to exert real functions. It is necessary to detect, identify, and quantify the serum GH proteoform pattern in patients with GH-secreting pituitary adenomas compared to normal control sera. The serum GH proteoform pattern might have more important clinical value for the development of biomarkers in the context of PPPM in GH-secreting pituitary adenoma and acromegaly. Third, serum GH must interact with GHR to exert its biological functions. Thus, different GH proteoforms must have their corresponding GHR proteoforms. It is also necessary to detect, identify, and quantify the GHR proteoform pattern on the target organ tissues/cells in patients with GH-secreting pituitary adenomas and acromegaly. Fourth, it also might be necessary to determine the binding relationship of GH proteoforms and GHR proteoforms, and also to clarify the downstream molecular signaling characterization and biological functions of different GH proteoforms through the corresponding GHR proteoforms in the targeted organ/cells.
These multiple studies on GH proteoforms at different levels of pituitary tissue, serum, and targeted organ/cell will significantly advance the understanding of human growth hormone in GH-related diseases, including GH-secreting pituitary adenomas and acromegaly, and will benefit development of a human GH proteoform pattern as biomarkers towards PPPM practice.
Conclusion and expert recommendation
The use of 2DGE and 2DGE-based Western blot in combination with MS is an effective method to identify human GH proteoforms in GH-secreting pituitary adenomas relative to control pituitaries.A total of 46 GH proteoforms was identified in GH-secreting pituitary adenoma tissues, and 35 GH proteoforms in normal control pituitary tissues. A total of 35 GH proteoforms were matched between GH-secreting pituitary adenomas and controls, and 11 GH proteoforms in GH-secreting pituitary adenomas but not in controls. Moreover, phosphorylation at residue Ser132 was found in three GH proteoforms 12, 36, and 54 in GH-secreting pituitary adenomas and four GH proteoforms 11, 32, 33, and 56 in control pituitary tissues. Ubiquitination at residue K96 was identified in GH proteoform 78 in GH-secreting pituitary adenomas. Acetylation at residue K171 was identified in GH proteoform 30 in GH-secreting pituitary adenoma tissues. Deamination at residue N178 was identified in 25 GH proteoforms in GH-secreting pituitary adenomas and 5 GH proteoforms in control pituitaries. For GH-secreting pituitary adenomas, alternative splicing variants 2 occurred in GH proteoforms 1 and 5, and alternative splicing variant 3 occurred in GH proteoform 78. For control pituitaries, alternative splicing variant 2 occurred in GH proteoforms 3, 4, and 6. The signal peptide was not removed from GH proteoform 46 in GH-secreting pituitary adenomas, but removed from all other GH proteoforms. Thereby, the GH proteoform pattern was significantly different in GH-secreting pituitary adenoma tissues relative to control pituitary tissues, with different PTMs (phosphorylation, acetylation, ubiquitination, and deamination) in GH-secreting pituitary adenomas compared to controls. These findings provide new ideas and mechanisms to effectively treat GH-secreting pituitary adenomas and GH-related diseases, and also provide the basis to develop effective GH proteoform pattern biomarkers for prediction, diagnosis, prognostic assessment, and patient stratification of GH-secreting pituitary adenomas and GH-related diseases in the context of PPPM practice.
We strongly recommend the following research activities in the context of PPPM practice. (i) It is necessary to expand the studies of human GH proteoform pattern in a significantly increased sample size of GH-secreting pituitary adenoma tissues to develop GH proteoform pattern biomarkers to personally treat GH-related diseases. (ii) The invasive and progressive characterization of pituitary adenomas is a very challenging clinical problem. It is necessary to analyze the differences in GH proteoform patterns in invasive (or aggressive) vs. noninvasive (or nonaggressive) pituitary adenomas to accurately elucidate the relationship of any proteoform and invasive (or aggressive) characterization to increase accurately the patient stratification and prognostic assessment to individualized treat pituitary adenomas. (iii) We should significantly strengthen the study of serum GH proteoform patterns, which is a noninvasive approach to better serve as effective biomarkers for prediction, diagnosis, prognostic assessment, and patient stratification of GH-secreting pituitary adenomas and GH-related diseases. (iv) We should also initiate the study of GH receptor (GHR) proteoform patterns, explore the binding relationship of GH proteoforms and GHR proteoforms to clarify and accurately analyze mechanisms and functions of GH proteoforms. These systematic studies of a GH proteoform pattern at the different levels of pituitary tissue, serum, GHR on the target cell/organ, and the binding relationship of GH proteoforms and GHR proteoforms will significantly expand, deepen, and extend our understanding of GH biological functions in GH-related diseases, provide an important data resource for the development of biomarkers and therapeutic targets for patient stratification, and allow prediction/prognostic assessment, and personalized treatment of GH-related diseases.
In sum, our greatest level of understanding a human disease always derives from an in-depth and accurate analysis of the molecules that are involved in that disease.
Supplementary information
Supplemental material provides additional information about the reactors and metagenome analysis discussed in the text and is available online on the Springer publications website at https://link.springer.com
(PDF 156 kb)
Acknowledgments
The authors acknowledge the financial support from the Shandong First Medical University Talent Introduction Funds (to X.Z.), the Hunan Provincial Hundred Talent Plan (to X.Z.), and the grants from China “863” Plan Project (Grant No. 2014AA020610-1 to XZ).
Abbreviations
- ddH2O
Deionized distilled water
- DTT
Dithiothreitol
- ESI
Electrospray ionization
- hGH
Human growth hormone
- IEF
Isoelectric focusing
- IPG
Immobilized pH gradient
- LC
Liquid chromatography
- MALDI
Matrix-assisted laser desorption ionization
- MS
Mass spectrometry
- Mr
Relative molecular mass
- PBS
Phosphate-buffered saline
- pI
Isoelectric point
- PTMs
Posttranslational modifications
- PVDF
Polyvinylidene fluoride
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- 2DGE
Two-dimensional gel electrophoresis
- TOF
Time-of-flight
Author contributions
B.L. analyzed data, carried out partial experiments, prepared figures and tables, and wrote the manuscript draft. X.W. performed 2DGE-based western blot experiment and mass spectrometry sample preparation. S.W., J.L., N.L., Y.L., and Y.M. participated in partial data analysis and experiment. J.L. assisted in 2DGE experiment and 2DGE image analysis. C.Y. provided control pituitary tissue samples and clinical diagnosis. Q.L. and X.L. obtained pituitary adenoma tissue samples and clinical diagnosis. D.M.D. provided control pituitary samples and critically reviewed the manuscript. X.Z. conceived the concept, designed experiments and manuscript, instructed experiments, analyzed data, obtained the ubiquitinated, phosphorylated, and acetylated proteomic data, supervised results, coordinated, wrote and critically revised manuscript, and was responsible for its financial supports and the corresponding works. All authors approved the final manuscript.
Funding
This work was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), the Hunan Provincial Hundred Talent Plan (to X.Z.), and China “863” Plan Project (Grant No. 2014AA020610-1 to XZ).
Data availability
All data and materials are provided in this article and supplemental materials, which can be available publicly.
Code availability
All protein and gene accession codes can be available in the Swiss-Prot and Genbank databases. Bio-Rad PDQuest 2D gel image analysis software (version 7.0) is commercially available.
Declarations
Competing interests
The authors declare that there is no conflict of interests regarding the publication of this article.
Ethical approval
All the patients were informed about the purposes of the study and consequently have signed their “consent of the patient”. All investigations conformed to the principles outlined in the Declaration of Helsinki and were performed with permission (Approval number: 2013030181) by the responsible Medical Ethics Committee of Xiangya Hospital, Central South University, China.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
References
- 1.Melmed S. Mechanisms for pituitary tumorigenesis: the plastic pituitary. J ClinIinvest. 2003;112:1603–18. doi: 10.1172/JCI20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Männik J, Vaas P, Rull K, Teesalu P, Rebane T, Laan M. Differential expression profile of growth hormone/chorionic somatomammotropin genes in placenta of small- and large-for-gestational-age newborns. J Clin Endocrinol Metab. 2010;95:2433–2442. doi: 10.1210/jc.2010-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho Y, Liebhaber SA, Cooke NE. Activation of the human GH gene cluster: roles for targeted chromatin modification. Trend EndocrinolMetabol. 2004;15:40–5. doi: 10.1016/j.tem.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Sedman L, Padhukasahasram B, Kelgo P, Laan M. Complex signatures of locus-specific selective pressures and gene conversion on human growth hormone/chorionic somatomammotropin genes. Hum Mutat. 2008;29:1181–1193. doi: 10.1002/humu.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankenne F, Scippo ML, Van Beeumen J, Igout A, Hennen G. Identification of placental human growth hormone as the growth hormone-V gene expression product. J ClinEndocrinolMetabol. 1990;71:15–8. doi: 10.1210/jcem-71-1-15. [DOI] [PubMed] [Google Scholar]
- 6.Qian S, Yang Y, Li N, Cheng T, Wang X, Liu J, et al. Prolactin variants in human pituitaries and pituitary adenomas identified with two-dimensional gel electrophoresis and mass spectrometry. Front Endocrinol. 2018;9:468. doi: 10.3389/fendo.2018.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vijayakumar A, Novosyadlyy R, Wu Y, Yakar S, LeRoith D. Biological effects of growth hormone on carbohydrate and lipid metabolism. Growth Horm IGF Res. 2010;20:1–7. doi: 10.1016/j.ghir.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry JK, Wu ZS, Mertani HC, Zhu T, Lobie PE. Tumour-derived human growth hormone as a therapeutic target in oncology. Trend Endocrinol Metabol. 2017;28:587–596. doi: 10.1016/j.tem.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Sos BC, Harris C, Nordstrom SM, Tran JL, Balazs M, Caplazi P, et al. Abrogation of growth hormone secretion rescues fatty liver in mice with hepatocyte-specific deletion of JAK2. J Clin Invest. 2011;121:1412–1423. doi: 10.1172/JCI42894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui Y, Hosui A, Sun R, Shen K, Gavrilova O, Chen W, et al. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration. Hepatol. 2007;46:504–513. doi: 10.1002/hep.21713. [DOI] [PubMed] [Google Scholar]
- 11.Liu JL, Yakar S, LeRoith D. Conditional knockout of mouse insulin-like growth factor-1 gene using the Cre/loxP system. Proc Soc Exp Biol Med. 2000;223:344–351. doi: 10.1046/j.1525-1373.2000.22349.x. [DOI] [PubMed] [Google Scholar]
- 12.Yakar S, Rosen CJ, Beamer WG, Ackert-Bicknell CL, Wu Y, Liu JL, et al. Circulating levels of IGF-1 directly regulate bone growth and density. J Clin Invest. 2002;110:771–781. doi: 10.1172/JCI15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rrev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- 15.Lim SV, Marenzana M, Hopkinson M, List EO, Kopchick JJ, Pereira M, et al. Excessive growth hormone expression in male GH transgenic mice adversely alters bone architecture and mechanical strength. Endocrinol. 2015;156:1362–1371. doi: 10.1210/en.2014-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beckers A, Petrossians P, Hanson J, Daly AF. The causes and consequences of pituitary gigantism. Nat Rev Endocrinol. 2018;14:705–720. doi: 10.1038/s41574-018-0114-1. [DOI] [PubMed] [Google Scholar]
- 17.Castro C, Trivin C, Souberbielle JC, Zerah M, Brauner R. Growth hormone deficiency: permanence and diagnosis in young adults. Horm Res. 2002;58:165–171. doi: 10.1159/000065489. [DOI] [PubMed] [Google Scholar]
- 18.de Boer H, Blok GJ, Van der Veen EA. Clinical aspects of growth hormone deficiency in adults. Endocr Rev. 1995;16:63–86. doi: 10.1210/edrv-16-1-63. [DOI] [PubMed] [Google Scholar]
- 19.Molitch ME, Clemmons DR, Malozowski S, Merriam GR, Vance ML, Endocrine Society Evaluation and treatment of adult growth hormone deficiency: an Endocrine Society clinical practice guideline. J ClinEndocrinolMetab. 2011;96:1587–609. doi: 10.1210/jc.2011-0179. [DOI] [PubMed] [Google Scholar]
- 20.Vilar L, Vilar CF, Lyra R, Lyra R, Naves LA. Acromegaly: clinical features at diagnosis. Pituitary. 2017;20:22–32. doi: 10.1007/s11102-016-0772-8. [DOI] [PubMed] [Google Scholar]
- 21.Clemmons DR. Role of insulin-like growth factor-I in diagnosis and management of acromegaly. Endocr Practice. 2004;10:362–371. doi: 10.4158/EP.10.4.362. [DOI] [PubMed] [Google Scholar]
- 22.Zhan X, Long Y, Lu M. Exploration of variations in proteome and metabolome for predictive diagnostics and personalized treatment algorithms: Innovative approach and examples for potential clinical application. J Proteomics. 2018;188:30–40. doi: 10.1016/j.jprot.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Baumann G. Growth hormone heterogeneity: genes, isohormones, variants, and binding proteins. Endocr Rev. 1991;12:424–449. doi: 10.1210/edrv-12-4-424. [DOI] [PubMed] [Google Scholar]
- 24.Liu YX, Chen JY, Tang XL, Chen P, Zhang M. The 20kDa and 22kDa forms of human growth hormone (hGH) exhibit different intracellular signalling profiles and properties. Gen Compar Endocrinol. 2017;248:49–54. doi: 10.1016/j.ygcen.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Zhan X, Li B, Zhan X, Schluter H, Jungblut PR, Coorssen JR. Innovating the concept and practice of two-dimensional gel electrophoresis in the analysis of proteomes at the proteoform level. Proteomes. 2019;7:1–15. doi: 10.3390/proteomes7040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhan X, Yang H, Peng F, Li J, Mu Y, Long Y, et al. How many proteins can be identified in a 2DE gel spot within an analysis of a complex human cancer tissue proteome? Electrophoresis. 2018;39:965–980. doi: 10.1002/elps.201700330. [DOI] [PubMed] [Google Scholar]
- 27.Zhan X, Desiderio DM. A reference map of a human pituitary adenoma proteome. Proteomics. 2003;3:699–713. doi: 10.1002/pmic.200300408. [DOI] [PubMed] [Google Scholar]
- 28.Wilkins MR, Gasteiger E, Gooley AA, Herbert BR, Molloy MP, Binz PA, et al. High-throughput mass spectrometric discovery of protein post-translational modifications. J Mol Biol. 1999;289:645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- 29.Zhan X, Giorgianni F, Desiderio DM. Proteomics analysis of growth hormone isoforms in the human pituitary. Proteomics. 2005;5:1228–1241. doi: 10.1002/pmic.200400987. [DOI] [PubMed] [Google Scholar]
- 30.Zhan X, Huang Y, Long Y. Two-dimensional gel electrophoresis coupled with mass spectrometry methods for an analysis of human pituitary adenoma tissue proteome. J Vis Exp. 2018;134:56739. doi: 10.3791/56739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engholm-Keller K, Larsen MR. Technologies and challenges in large-scale phosphoproteomics. Proteomics. 2013;13:910–931. doi: 10.1002/pmic.201200484. [DOI] [PubMed] [Google Scholar]
- 32.Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun Mass Spectrom. 2007;21:3635–3645. doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- 33.Beltran L, Cutillas PR. Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids. 2012;43:1009–1024. doi: 10.1007/s00726-012-1288-9. [DOI] [PubMed] [Google Scholar]
- 34.Zhao S, Feng J, Li C, Gao H, Lv P, Li J, et al. Phosphoproteome profiling revealed abnormally phosphorylated AMPK and ATF2 involved in glucose metabolism and tumorigenesis of GH-PAs. J Endocrinol Invest. 2019;42:137–148. doi: 10.1007/s40618-018-0890-4. [DOI] [PubMed] [Google Scholar]
- 35.Ezzat S, Yu S, Asa SL. Ikaros isoforms in human pituitary tumors: distinct localization, histone acetylation, and activation of the 5’ fibroblast growth factor receptor-4 promoter. Am J Pathol. 2003;163:1177–84. 10.1016/S0002-9440(10)63477-3. [DOI] [PMC free article] [PubMed]
- 36.Smith KT, Workman JL. Introducing the acetylome. Nat Biotechnol. 2009;27:917–919. doi: 10.1038/nbt1009-917. [DOI] [PubMed] [Google Scholar]
- 37.Ebrahimi A, Schittenhelm J, Honegger J, Schluesener HJ. Histone acetylation patterns of typical and atypical pituitary adenomas indicate epigenetic shift of these tumours. J Neuroendocrinol. 2011;23:525–530. doi: 10.1111/j.1365-2826.2011.02129.x. [DOI] [PubMed] [Google Scholar]
- 38.Qian S, Zhan X, Lu M, Li N, Long Y, Li X, et al. Quantitative analysis of ubiquitinated proteins in human pituitary and pituitary adenoma tissues. Front Endocrinol. 2019;10:328. doi: 10.3389/fendo.2019.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan X, Desiderio DM. The use of variations in proteomes to predict, prevent, and personalize treatment for clinically nonfunctional pituitary adenomas. EPMA J. 2010;1:439–459. doi: 10.1007/s13167-010-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu R, Wang X, Zhan X. Multi-parameter systematic strategies for predictive, preventive and personalised medicine in cancer. EPMA J. 2013;4:2. doi: 10.1186/1878-5085-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8:51–60. doi: 10.1007/s13167-017-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, Qian S, Li B, Zhan X. Quantitative analysis of the human ovarian carcinoma mitochondrial phosphoproteome. Aging (Albany NY) 2019;11:6449–68. doi: 10.18632/aging.102199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li N, Li H, Cao L, Zhan X. Quantitative analysis of the mitochondrial proteome in human ovarian carcinomas. Endocr Relat Cancer. 2018;25:909–931. doi: 10.1530/ERC-18-0243. [DOI] [PubMed] [Google Scholar]
- 44.Lu M, Chen W, Zhuang W, Zhan X. Label-free quantitative identification of abnormally ubiquitinated proteins as useful biomarkers for human lung squamous cell carcinomas. EPMA J. 2020;11:73–94. doi: 10.1007/s13167-019-00197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 46.Liu D, Li J, Li N, Lu M, Wen S, Zhan X. Integration of quantitative phosphoproteomics and transcriptomics revealed phosphorylation-mediated molecular events as useful tools for a potential patient stratification and personalized treatment of human nonfunctional pituitary adenomas. EPMA J. 2020;11:419–467. doi: 10.1007/s13167-020-00215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bello MO, Garla VV. Gigantism and acromegaly. In: StatPearls [Internet]. Treasure Island (FL), StatPearls Publishing. 2020; Bookshelf ID: NBK538261. [PubMed]
- 48.Zhou C, Jiao Y, Wang R, Ren SG, Wawrowsky K, Melmed S. STAT3 upregulation in pituitary somatotroph adenomas induces growth hormone hypersecretion. J Clin Invest. 2015;125:1692–1702. doi: 10.1172/JCI78173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caimaril F, Korbonits M. Novel genetic causes of pituitary adenomas. Clin Cancer Res. 2016;22:5030–5042. doi: 10.1158/1078-0432.CCR-16-0452. [DOI] [PubMed] [Google Scholar]
- 50.Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, et al. How many human proteoforms are there? Nat Chem Biol. 2018;14:206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan X, Wang X, Desiderio DM. Pituitary adenoma nitroproteomics: current status and perspectives. Oxid Med Cell Longev. 2013;2013:580710. doi: 10.1155/2013/580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9:77–102. doi: 10.1007/s13167-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Golubnitschaja O, Costigliola V, EPMA General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive, Preventive and Personalised Medicine. EPMA J. 2012;2012(3):14. doi: 10.1186/1878-5085-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, Vittadini G, Desiderio DM. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6:9. doi: 10.1186/s13167-015-0030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva AMN, Vitorino R, Domingues MRM, Spickett CM, Domingues P. Post-translational modifications and mass spectrometry detection. Free Radic Biol Med. 2013;65:925–941. doi: 10.1016/j.freeradbiomed.2013.08.184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 156 kb)
Data Availability Statement
All data and materials are provided in this article and supplemental materials, which can be available publicly.
All protein and gene accession codes can be available in the Swiss-Prot and Genbank databases. Bio-Rad PDQuest 2D gel image analysis software (version 7.0) is commercially available.