Abstract
Mitoribosomes catalyze essential protein synthesis within mitochondria. Mitoribosome biogenesis is assisted by an increasing number of assembly factors, among which guanosine triphosphate hydrolases (GTPases) are the most abundant class. Here, we will review the recent progress made about the known mitoribosome assembly GTPases. We will describe their shared and specific features and mechanisms of action, compare them with their bacterial counterparts and discuss their possible roles in the assembly of small or large mitoribosomal subunits, and formation of the monosome by establishing quality control checkpoints during these processes. Furthermore, following the recent unification of the nomenclature for the mitoribosomal proteins, we will also aim to propose a unified nomenclature for mitoribosome assembly GTPases that is currently used in the literature.
Keywords: Mitochondrial ribosome, Mitoribosome assembly GTPase, GTPBP, Quality control of mitoribosome maturation, OXPHOS deficiency, Mitochondrial diseases
The biogenesis of the mitochondrial ribosome
The mammalian mitochondrial 55S ribosome (mitoribosome) is essential for mitochondrial and cellular functions as it synthesizes the thirteen mitochondrial DNA (mtDNA)-encoded subunits of the oxidative phosphorylation (OXPHOS) system. It is composed of a 39S large ribosomal subunit (mtLSU) containing 52 mitoribosomal proteins (MRPs), the 16S rRNA and a tRNA moiety (tRNAVal or tRNAPhe) in the central protuberance, and of a 28S small ribosomal subunit (mtSSU) comprising 30 MRPs and the 12S rRNA [1–4]. While the RNA components are encoded by the mtDNA (Figure 1), all the 82 MRPs are encoded by the nuclear genome, synthesized in the cytosol and imported into mitochondria. Although the mitochondrial and bacterial ribosome derived from a common ancestor, substantial differences in structure and composition became evident from recent high-resolution structural analyses [1–5]. How the mammalian mitoribosome is assembled is poorly understood. However, defects in 55S ribosome biogenesis or function are associated with OXPHOS deficiency and severe human diseases (Box 1, [6–30]), thereby revealing the importance of understanding the complex process of mitoribosome assembly in more detail.
Figure 1. Mitoribosome assembly: compartmentalization and assembly factors.
The mtDNA (mitochondrial DNA) and proteins involved in mtDNA metabolism form a nucleoprotein complex called the mitochondrial nucleoid. Upon transcription, mitoribosome mtLSU (large mitoribosomal subunit; shown in blue) assembly starts at the nucleoid in the 16S/12S rRNA precursor. At a certain early assembly stage, the pre-particle is sorted at the RNA granule, where the rRNA precursor is processed, and mitoribosome mtSSU (small mitoribosomal subunit; shown in yellow) and mtLSU biogenesis proceeds until its completion. The process is catalyzed by several classes of assembly factors involved in RNA processing and maturation, rRNA modification, and formation of assembly intermediates.
Box 1: Biomedical relevance of the mitoribosome and associated GTPases.
Mitoribosomes are relevant biomedically since mutations in its components and assembly factors are responsible for mitochondrial disorders associated with decreased activities of multiple OXPHOS enzymes. Defects in mitoribosome assembly and mitochondrial protein synthesis lead to multisystemic mitochondrial diseases such as Leigh syndrome, cardio- and encephalo-myopathies, although other organs may also be affected. Up to date 13 mtSSU proteins (bS1m, uS2m, bS6m, uS7m, uS9m, uS11m, uS14m, bS16m, mS22, mS23, mS25, mS34 and mS39) and 4 mtLSU proteins (uL3m, bL12m, uL24m and mL44) have been implicated in mitochondrial disorders [6–11,13–28]. It has been noted that many of these proteins are recruited at early stages in mitoribosome assembly, suggesting that defects at these stages may have severe consequences on the mitoribosome structure and abolish subsequent assembly steps [9]. Multiple mutations have also been identified in the 12S rRNA, associated with non-syndromic antibiotic-induced hearing loss, the 16S rRNA, associated with cardio- and encephalo-myopathy, and the tRNAVal, associated with a variety of mitochondrial disorders.
Mitoribosome assembly factors have also been linked to human disease [91]. Among them, the cases most relevant to this review include the GTPases ERAL1, associated to Perrault syndrome (ovarian dysgenesis and sensorineuronal syndrome) [12]; GTPBP5, associated to syndromic tracheo-esophageal fistula, cardiac defects, and genitourinary anomalies [29]; and GTPBP10, for which single nucleotide polymorphisms have been associated with increased risk of prostate cancer [30].
In addition to their involvement in mitoribosome structure and function, several MRPs appear to perform important extra-ribosomal roles. Some of them, including the GTPase mS29, have been suggested to regulate apoptosis, and therefore, it is not surprising that they have been connected to numerous cancers, a topic that has been extensively reviewed elsewhere [92].
Mitoribosome biogenesis is confined to specific compartments within the mitochondrial matrix. Early human mitoribosome assembly steps occur at or near the mtDNA nucleoids [31,32], in a co-transcriptional manner. Upon transcription, RNAs are sorted at the RNA granule, a membrane-less compartment that condenses and progressively separates from the nucleoids upon transcription, where the bulk of ribosome assembly occurs [33–37] (Figure 1). Mitoribosome assembly requires a growing number of auxiliary factors acting at every step of the process, comprising RNA processing and modification enzymes, rRNA chaperones, guanosine triphosphatases (GTPases), DEAD-box RNA helicases, and kinases [38–40]. Their specific roles and the assembly stage at which they act are beginning to emerge, thanks to a combination of biochemical and structural studies (Figure 1).
Among the several classes of essential ribosome assembly factors, GTPases are the largest class in bacteria and cytoplasmic ribosomes, suggesting their evolutionarily conserved requirement for ribosome assembly regulation. In bacteria, they might couple ribosome assembly with growth control pathways by sensing the cellular GTP/GDP ratio [41]. Recent investigations have revealed that in bacteria and mitochondria, a set of GTPases belonging to several conserved families [39,42–49] (Figure 2a) can act as rRNA chaperones by recruiting MRPs and assembly factors to the assembly pathway and establishing quality control mechanisms to ensure that only the mature mtSSU and mtLSU are assembled into functional monosomes [39,41,44,46,50]. Especially the maturation of the mtLSU requires the assistance of several GTPases at late stages when they act as anti-association or quality control factors. Apparently, these final steps in mtLSU maturation are critical, and loss of these GTPases abolishes ribosome formation and leads to translation deficiency. Recent findings regarding the possible functions of mitoribosome assembly GTPases have been illustrated in Figure 3a and are the subject of this review.
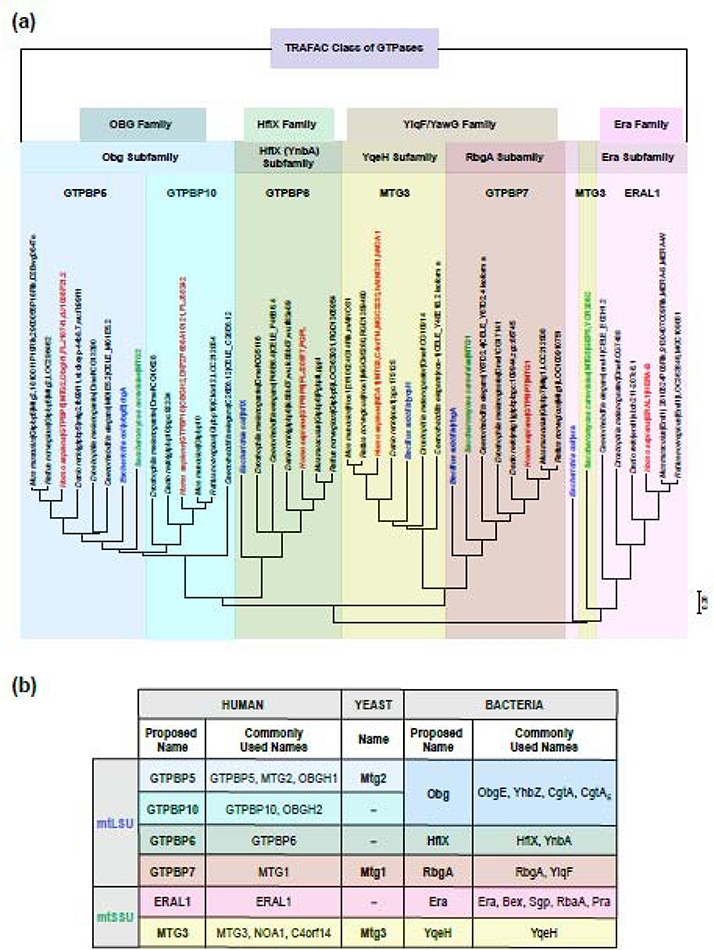
Figure 2. Evolutionary classification and phylogenetic trees of GTPases involved in mitoribosome biogenesis.
(a) On the top, we present the classification of GTPases (family and subfamily) based on the review articles [51,52]. On the bottom, the evolutionary relationships of taxa are presented. The evolutionary history was inferred using the Neighbor-Joining method [98]. The optimal tree with the sum of branch length = 22.4487 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [99] and are in the units of the number of amino acid substitutions per site. This analysis involved 44 amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 902 positions in the final dataset. Evolutionary analyses were conducted using MEGA X software (https://www.megasoftware.net/) [100,101]. (b) Proposed new nomenclature for mitoribosome biogenesis GTPases. The commonly used names have been mentioned for different species. The new nomenclature for the GTPases has been suggested under the “proposed name”.
Figure 3. Roles of GTPases during mitoribosome assembly.
(a) Proposed model for specific roles of GTPases involved at different stages of the mtLSU (large mitoribosomal subunit) and mtSSU (small mitoribosomal subunit) assembly pathway based on the biochemical studies in mammalian system. The GTPases discussed in this review are highlighted in orange. MALSU1-uL14m forms a pre-assembly complex thereby making the binding site accessible for the immediate binding of GTPBP7, which further enables the binding of uL16m. All of these assembly factors including GTPBP5 and GTPBP10 incorporates into the growing mtLSU and seems to remain bound to the particle with GTPBP6 being the last one to join. Incorporation of GTPBP6, which may do the final surveillance job to ensure that the catalytic center is correctly folded. Finally assembly factors can dissociate, with GTPBP7 might be the last one to be released when mtSSU joins and the GEF (GTP exchange factor) activity of mS27 mediates its release. CP, central protuberance of mtLSU; L1 and L12 mark the position of the corresponding stalks of mtLSU; mS29 and mS27 mark the position of the corresponding head and body of the mtSSU. (b) Role of GTPBP5 in mtLSU assembly. GTPBP5 binds subsequently to other known assembly factors (GTPBP10, MALSU1, GTPBP7, MTERF4-NSUN4) to the mtLSU assembly intermediate with partially matured PTC (peptidyl transferase center) and lacking bL36m, and facilitates the methylation of 16S rRNA at U1368 residue by MRM2 (rRNA methyltransferase 2). Once rRNA modification is completed, bL36m is recruited, followed by the joining of GTPBP6 which does the final quality control check, ensuring final maturation and might cause the release of other late-stage mtLSU assembly factors, thereby securing subunit joining and monosome formation. The order of release of all the last-stage mtLSU assembly factors is not clear yet. Figure adapted from [47]. PTC, peptidyl transferase center of mtLSU; uL16m, uL14m, bL36m mark the positions within the mitoribosome; the relative position of all the mentioned assembly factors is purely for illustration since the binding positions except for MALSU1 are unknown. (c) Role of GTPBP7 in the assembly of mtLSU. GTPBP7 interacts with 16S rRNA and bL19m of the mtLSU assembly intermediate, induces a conformational change and facilitates the incorporation of bL36m and bL35m. GTPBP7 remains bound to the mtLSU until the maturation of mtLSU is completed. Subsequently, it interacts with mS27 whose GEF (GTP exchange factor) activity enables the release of GTPBP7, thereby facilitating the formation of mB6 bridge between bL19m and mS27, and then preventing premature subunit joining. Figure adapted from [44]. bL19m and mS27 mark the positions within the mitoribosome.
We used the surface representation of the structural profile of the human mitoribosome (PDB 3J9M) [4]. The homology-based molecular modeling for the assembly factor GTPBP7 in Panel c was performed using SWISS-MODEL server. Figures in panels a, b, and c were prepared using PYMOL and Adobe Illustrator software.
Mitoribosome assembly GTPases
Ribosome assembly GTPases (RA-GTPases) are envisioned to have evolved from the ancestral GTPase along with the translation machinery [51,52]. The current list of mitoribosome assembly GTPases is presented in Figure 2a, and their general structural features are summarized in Box 2.
Box 2: Features of mitoribosome assembly GTPases.
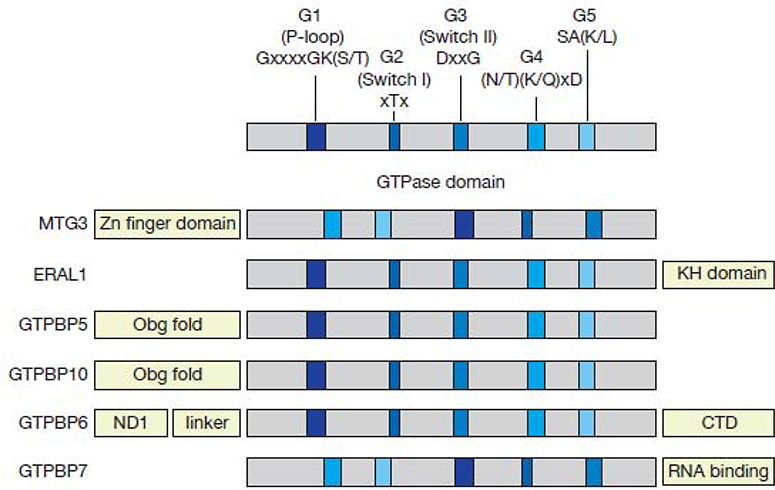
GTPases involved in ribosome maturation are P-loop NTPases and contain conserved sequence motifs, G1-G5 [93] (Figure I). The p-loop with G1, which is also called Walker A motif [GxxxxGK(S/T)] binds to the α- and β-phosphates of GTP. The loop containing G2 [xTx] termed as switch I is required for Mg2+ coordination and interacts with the β- and γ-phosphates. The loop with G3 [DxxG], also referred to as switch II or Walker B motif, interacts with the γ-phosphate and is also involved in Mg2+ coordination. G4 [(N/T)(K/Q)xD] and G5 [SA(K/L)] recognize the guanine base, however, G5 is less conserved among ribosome biogenesis GTPases. Many GTPases follow the canonical order of the signature motifs G1-G2-G3-G4-G5. However, circular permutation resulted in a distinct order with G4-G5-G1-G2-G3 as found e.g. in MTG3 or GTPBP7 [43,44]. In addition to the GTPase domain, most ribosome assembly GTPases contain an RNA-binding domain facilitating the interaction with the ribosomal RNA as demonstrated for ERAL1, GTPBP5, GTPBP7 and GTPBP10 [39,44,47,48]
Figure I (related to Box 2).
Functional domains in mitoribosome assembly GTPases
Families of conserved GTPases involved in mitoribosome SSU and LSU assembly
RA-GTPases in bacteria and mitochondria are members of the so-called TRAFAC (translation factor-related) GTPase group, which also include several translation factors, and evolutionary modern enzymes involved in signal transduction and intracellular transport [51]. A phylogenetic analysis of ribosome assembly GTPases from bacteria and mitochondria from several organisms has allowed us to group them into four families and five subfamilies (Figure 2a).
In mammalian mitochondria, two conserved GTPases are involved in mtSSU biogenesis: MTG3 (NOA1 (Nitric oxide-associated protein 1) or C4ORF14) and ERAL1 (Era G-protein-like 1). MTG3 (Mitochondrial ribosome-associated GTPase 3) is a homolog of bacterial YqeH that belongs to the YlqF/YawG family of GTPases [42,43] whose special feature is the presence of a unique circular permutation of the GTP binding domain [52]. ERAL1 is a homolog of bacterial Era (Escherichia coli Ras-like protein) that was first identified as a homolog of the eukaryotic oncogenic Ras protein [48,49]. There is no ERAL1 homolog in S. cerevisiae; however, a phylogenetic analysis places yeast Mtg3 as more related to ERAL1 than to human MTG3 or bacterial YqeH, leaving it questionable whether Mtg3 is a true homolog of bacterial YqeH. Additionally, at least four other GTPases are required for the mtLSU biogenesis: GTPBP5 (GTPBP refers to GTP-binding protein, or MTG2 (Mitochondrial ribosome-associated GTPase 2)), GTPBP6, GTPBP7 (MTG1, mitochondrial ribosome-associated GTPase 1), and GTPBP10. GTPBP7 is a homolog of bacterial RbgA (ribosome biogenesis GTPase A) [44]. They form the RbgA subfamily of GTPases that, together with the YqeH subfamily, form the YlqF/YawG family of GTPases. GTPBP5 and GTPBP10, together with GTPBP6, belong to the OBG-HflX-like superfamily [39,45–47,53]. While GTPBP6 represents the HflX family, GTPBP5 and GTPBP10 are two homologs of bacterial Obg. The Obg subfamily is a class of highly conserved and essential small monomeric P-loop GTPases found ubiquitously in prokaryotes and eukaryotes (except archaea) [51]. A fifth potential GTPase involved in mtLSU assembly is GTPBP8, the human homolog of bacterial EngB that belongs to the YihA/EngB family. Although it has not been characterized in human cells, it was recently identified in Trypanosoma brucei mtLSU assembly intermediates interacting with GTPBP7 homolog Mtg1 [54].
The role of specific mitoribosome assembly GTPases
Mitoribosome SSU assembly GTPases
MTG3
Mammalian MTG3 homologous to bacterial YqeH mainly localizes to the mitochondrial matrix where it peripherally associates with the inner membrane, although localization to the nucleus has been also suggested [55,56]. Human MTG3 specifically interacts with the mtSSU, but not with the mtLSU or the 55S ribosome suggesting a role in mtSSU assembly like YqeH in bacteria [43]. A GTPase mutant variant of MTG3 carrying point mutations in the G2, G3, and G4 motifs lost the ability to bind the mtSSU supporting the requirement of the GTPase activity for its function in mtSSU assembly. It has been shown that YqeH undergoes increased GTP hydrolysis rates in the presence of the ribosomal protein bS5, suggesting a potential role in the early stages of SSU assembly [57]. The ablation of MTG3 in murine and human cells abolishes mitochondrial translation due to reduced levels of 55S translating ribosomes. However, the mtSSU particle that is stably formed in the absence of MTG3 sedimented in sucrose gradients as the wild-type, and no assembly intermediates were identified [42,43]. Yeast mtg3 mutants are also impaired in mitochondrial translation associated with defective mtSSU assembly and accumulation of 15S-precursor [58]. However, our phylogenetic analysis (Figure 2a) does not clarify whether yeast Mtg3 is the homolog of bacterial YqeH and mammalian MTG3, although it has a circular permutation of the conserved G motifs like MTG3 (G4-G5-G1-G2-G3) and not the canonical order like ERAL1 (Box 2). Nevertheless, the molecular function of mammalian MTG3 and yeast Mtg3 in mitoribosome assembly and the precise stage of mtSSU assembly at which the proteins act require further investigations. Furthermore, human MTG3 interacts with the mitochondrial nucleoid independently of its interaction with the ribosome [43]. MTG3 depletion in human cells led to reduced mtDNA copy number [43], whereas MTG3 deletion in mice embryonic fibroblasts (MEFs) revealed normal mtDNA levels and increased mitochondrial transcription [42]. The vital role of MTG3 is demonstrated by the embryonic lethality in mice and the reduced cell viability in isolated MEFs [42]. The ablation of MTG3 results in swollen mitochondria and impaired apoptosis [42,55]. Whether compromised apoptosis in MTG3 ablated cells is directly linked to defective mtSSU assembly needs to be clarified.
ERAL1
ERAL1 is a homolog of bacterial Ras-like protein Era. Both share the GTPase domain and a KH-domain that facilitates RNA binding (Box 2). Bacterial Era binds via its KH-domain to the highly conserved nucleotides GAUCA and the anti-Shine-Dalgarno sequence CCUCC at the 3’end of the 16S rRNA [59–61]. Therefore, Era prohibits the binding of mRNAs with the Shine-Dalgarno sequence and the assembly of ribosomal protein bS1 to the maturing 30S SSU. In contrast, human ERAL1 binds to the stem-loop at the 3’ terminus of the 12S mt-rRNA corresponding to nucleotides 1551–1601 in the human mtDNA [48]. This stem-loop represents helix 45, which contains two highly conserved adenines that undergo methylation catalyzed by TFB1M (mitochondrial transcription factor B) in mammals and by KsgA in bacteria, respectively [62–66]. As ERAL1 mostly interacts with unmethylated 12S rRNA [67], TFB1M presumably acts downstream of ERAL1. Thus, ERAL1 might act as an RNA chaperone to protect 12S rRNA from degradation prior to maturation and assembly. ERAL1 ablation results in rapid degradation of 12S rRNA, leading to reduced mtSSU de novo assembly, whereas 16S rRNA remains stable [48]. The essential role of ERAL1 in 28S mtSSU biogenesis is demonstrated by severe cellular consequences upon loss of ERAL1, including growth retardation and apoptosis [48,49,68,69]. Mutation in the G5 motif of the GTPase domain, which is predicted to diminish GTP binding by ERAL1, cause Perrault syndrome, a recessive disorder with sensorineural deafness and ovarian dysgenesis [12]. Patients with this mutation show up to a 40% decrease in mtSSU assembly, suggesting that GTP binding is required for the function of ERAL1 in ribosome biogenesis.
Elevated ERAL1 levels are harmful to mtSSU biogenesis, as demonstrated by CLPP (Caseinolytic mitochondrial matrix peptidase proteolytic subunit) deficiency, which also causes Perrault syndrome [70,71]. CLPP, a protease located in the mitochondrial matrix, controls the levels of ERAL1 and thus coordinates mtSSU biogenesis. In the absence of CLPP, ERAL1 accumulates on the mtSSU, whereas the association of bS1m is abolished, preventing 55S ribosome formation and mitochondrial translation [70] (Figure 3a). Thus, expression and turnover of ERAL1 need to be tightly controlled to allow mtSSU assembly and, thus, mitochondrial gene expression.
Mitoribosome LSU assembly GTPases
GTPBP5
GTPBP5 is one of two Obg homologs localized in the mitochondrial matrix associated with the inner membrane [45,47,53]. GTPBP5 complements the phenotypes in ΔobgE E. coli strains suggesting a conserved function in ribosome biogenesis [53]. Loss of human GTPBP5 severely affects the formation of 55S ribosomes, thereby attenuating mitochondrial translation and OXPHOS function [47]. GTPBP5 is essential for the formation of functional mtLSU as its loss results in an immature mtLSU particle lacking bL36m and containing an excess of the assembly factors GTPBP7, GTPBP10, MALSU1 (Mitochondrial assembly of ribosomal large subunit protein 1), and MTERF4 (Mitochondrial transcription termination factor 4) [47] (Figure 3a,b).
Biochemical and proteomics studies of mitoribosome assembly intermediates accumulating in GTPBP5, GTPBP7, or GTPBP10 KO cells have allowed proposing a model for the final mtLSU maturation stages: GTPBP5 and MTERF4 are recruited to the maturing mtLSU particle after GTPBP7 and MALSU1, which would be preceded by the recruitment of GTPBP10 [47] (Figure 3b).
GTPBP5 interacts with the 16S rRNA and several mtLSU proteins and assembly factors, including MRM2 [47]. MRM2 is the methyltransferase that catalyzes the 2’-O-methylation at position U1369 of the 16S rRNA A loop, an essential component of the peptidyl transferase center (PTC), which is evolutionary conserved and responsible for the interaction of the ribosome with the aminoacyl tRNA [72,73]. The bacterial obgE mutant revealed an LSU assembly intermediate with reduced uL16, bL33, and bL34 levels, indicating a role of ObgE at late LSU maturation stages [74]. Mutations in ObgE cause defects in rRNA processing leading to the accumulation of 16S and 23S rRNA precursors and reduced levels of assembled 70S ribosomes. Additionally, ObgE was identified as a multicopy suppressor of RrmJ (bacterial homolog of MRM2), where ObgE overexpression suppresses the LSU assembly and cell growth defects in E. coli ΔrrmJ strains [75]. Similarly, overexpression of Mtg2 (yeast homolog of GTPBP5) partially suppresses the phenotype of the mrm2 mutant in S. cerevisiae [76], indicating a conserved interdependency of Obg proteins and the methyltransferase RrmJ/MRM2. Studies in human cells suggested the requirement of GTPBP5-mediated remodeling of the rRNA during the PTC maturation for efficient MRM2-mediated 16S rRNA methylation [47]. Following 16S rRNA methylation, bL36m can be recruited to the maturating mtLSU to finalize its assembly and subsequent monosome formation [47] (Figure 3b). Hence GTPBP5 plays a role in mtLSU biogenesis by facilitating the methylation of 16S rRNA residue at U1369 and securing coordinate subunit joining and release of late-stage mtLSU assembly factors. As proposed for ObgE [77], GTPBP5 might act as a quality control factor preventing premature subunit joining by monitoring the 16S rRNA modification status and folding within the PTC.
The cryo-EM (Cryogenic electron microscopy) structure of the bacterial 50S·ObgE·GMPPNP complex indicates that the conserved Obg fold of ObgE is a tRNA structural mimic, whose specific interactions with the PTC resemble those of class I release factors. In human cells, overexpression of GTPBP5 [47] and also of GTPBP10 [46] is deleterious to mitochondrial translation, presumably because the equilibrium of these proteins is shifted towards the mtLSU-bound fraction, thus preventing efficient subunit joining.
GTPBP10
GTPBP10 is another homolog to bacterial ObgE and is able to rescue obgE deleted E. coli strains [53]. Initially, GTPBP10 was localized to the nucleolus [53], but several studies have confirmed its mitochondrial matrix localization and its requirement for mitochondrial translation [39,46,78]. Like its bacterial counterpart, GTPBP10 mostly associates with the mtLSU in a GTP-dependent manner. A mutation in the G5 motif abolishes GTPBP10 association with the mtLSU, and when using a non-hydrolyzable GTP analog, GTPBP10 is trapped by the mtLSU, suggesting that GTP hydrolysis is required to dissociate GTPBP10 from the mtLSU [39,46,78]. While a deletion in the Obg fold, which compromises GTPBP10 stability, mostly results in reduced levels of selected mtLSU proteins and the 16S rRNA [46], the complete KO of GTPBP10 also causes a reduction of mtSSU proteins and the 12S rRNA and a parallel increase in the 12S-16S precursor [39,78]. It has been suggested that the assembly of the mitoribosome starts on the 12S-16S rRNA precursor, which is eventually processed at a certain mtLSU maturation stage [79] (Figure 1). Thus, the accumulation of the 12S-16S precursor in mtLSU assembly-deficient GTPBP10-KO cells might secondarily affect mtSSU maturation [39].
GTPBP10 interacts with the mtLSU at late maturation stages and might present a quality check-point inhibiting the joining of immature ribosomal subunits [39,46,78]. In agreement with such a scenario is the accumulation of GTPBP10 in a late mtLSU assembly intermediate together with other biogenesis factors, including MALSU1 and L0R8F8 in MTERF4-deficient mice [78]. Similarly, GTPBP10 protein levels increase in cells deficient for GTPBP5, GTPBP6, GTPBP7, or the late assembly protein bL36m [39,47,80]. On the contrary, mtLSU intermediates in GTPBP10-deficient cells appear to lack MTERF4, NSUN4 (5-methylcytosine rRNA methyltransferase), and GTPBP7, suggesting that MTERF4, NSUN4, and GTPBP7 act downstream to GTPBP10 [39].
Although the two human mitochondrial Obg proteins complement the phenotypes in ΔobgE E. coli strains [53], GTPBP5 and GTPBP10 display distinct functions in mtLSU assembly and cannot compensate for each other. Whereas both are required for the late mtLSU maturation stages [39,46,47], they act at different time points. While GTPBP10 acts probably upstream of MTERF4-NSUN4 and GTPBP7, the action of GTPBP5 is required at an even later step, downstream or concomitantly of MTERF4-NSUN4 (Figure 3a).
GTPBP6
GTPBP6 belongs to the universally conserved HflX GTPase family found in nearly all domains of life except fungi [81]. Initially, GTPBP6 was identified in the DDX28 (DEAD box RNA helicase 28) interactome, and later, to associate with the mitoribosome ([39,80]). In bacteria, HflX is non-essential under physiological growth conditions; however, it is required under certain stress conditions when it recycles damaged ribosomes [82,83]. The ribosome recycling activity is conserved among HflX family members as GTPBP6 also promotes ribosome dissociation into subunits with the preference of splitting vacant ribosomes and post-hydrolysis complexes containing deacylated tRNA in the P site [80]. The exact concentration of GTPBP6 within mitochondria needs to be tightly controlled as elevated levels lead to mitochondrial translation deficiency due to ribosome dissociation. Although the exact biological relevance of GTPBP6-mediated ribosome recycling in mammalian mitochondria remains to be further evaluated, it is reasonable to speculate that it involves a mechanism that differs from the canonical mtRRF-mtEFG2 (Mitochondrial ribosome recycling factor and mitochondrial elongation factor 2) recycling system as GTPBP6 activity does not rely on GTP hydrolysis but GTP binding.
In contrast to its bacterial counterpart, GTPBP6 has acquired an additional function as a ribosome biogenesis factor essential for cell growth and mitochondrial gene expression under physiological conditions [80]. GTPBP6 ablation abolishes 55S ribosome formation associated with the accumulation of 28S and 39S subunits, leading to mitochondrial translation deficiency. GTPBP6 loss stalls mtLSU biogenesis at a very late assembly stage when all of the 52 MRPs, including bL36m, are incorporated, and mtLSU biogenetic factors such as MALSU1, GTPBP5, GTPBP7, GTPBP10, and NSUN4-MTERF4 are bound to the complex. The presence of bL36m and the accumulation of GTPBP5 indicate that GTPBP6 acts downstream of GTPBP5 (Figure 3a), and it is tempting to assume that GTPBP6 is required for the very final tuning steps of mtLSU maturation, potentially acting as an anti-association factor like other GTPases to prevent premature subunit joining. Only after the surveillance action of GTPBP6, all other assembly factors are released from the matured mLSU, and subunit joining occurs.
GTPBP7
GTPBP7, homologous to bacterial RbgA, interacts with the mtLSU in a GTP dependent manner [44,45,53]. Loss of GTPBP7 affects cell growth and attenuates mitochondrial translation and OXPHOS function [44,45]. This is also true for the Δmtg1 yeast strain, whose mitochondrial translation defect was suppressed by mutations in the stem-loop V of the PTC domain of 21S rRNA, suggesting that Mtg1 might interact with this 21S rRNA fold to stabilize it or to facilitate its interaction with MRPs [84].
Depletion of human GTPBP7 perturbed the assembly of functional mtLSU and thus 55S formation [44]. GTPBP7 catalyzes a late step in mtLSU biogenesis necessary for the incorporation of bL36m and bL35m [44] (Figure 3c). This is reminiscent of bacterial RbgA, which is required to incorporate uL16, bL27, bL28, bL33, bL35, and bL36 [85–88]. Deleting RbgA in bacteria leads to the accumulation of 45S LSU intermediate with severe distortion in the key functional sites such as the central protuberance, tRNA binding sites, and GTPase associated regions [88]. Thus, the functional core of the 50S subunit is the last region to mature during the assembly, and RbgA activity is essential for this process [88]. A recent cryo-EM structure of the RbgA-bound immature LSU particle has allowed the visualization of the conformational changes induced by this GTPase and the unusual location for its catalytic residue [89] (Box 3). Similarly, GTPBP7 interacts with domain VI helices in the 16S rRNA and with bL19m, which induces a conformational change and remodeling of the bL19m-containing mtLSU domain, thereby facilitating the incorporation of the late assembly proteins bL36m and bL35m [44] (Figure 3c). GTPBP7 remains bound to the mtLSU and is not readily released from the pre-mtLSU particle until maturation is completed by GTPBP6 [80]. Only when subunit joining is about to occur, GTPBP7 interacts with the mtSSU protein mS27, a putative guanosine triphosphate exchange factor (GEF) [44], which was proposed to catalyze fast GDP-GTP exchange that enables GTPBP7 release from the ribosome and facilitates the formation of the mB6 intersubunit bridge between bL19m and mS27 [44] (Figure 3a and 3c). Thus, GTPBP7 acts as a mtLSU quality control check-point protein in mitoribosome assembly, where it regulates mitochondrial translation by linking mtLSU assembly with intersubunit bridge formation, thereby preventing premature subunit joining [44].
Box 3: GTP binding and hydrolysis by mitoribosome assembly GTPases.
As for bacterial ribosome assembly GTPases, the intrinsic rate of GTP hydrolysis by GTPBP5 and GTPBP10 is very slow [45,53].
The GTPase activity of recombinant GTPBP7 is only detectable in the presence of the mtLSU or the 55S monosome [45]. Nucleotide-bound GTPBP7 interacting with the mtLSU and 55S monosomes, was only detected in the presence of non-hydrolyzable analog of GTP suggesting that GTP hydrolysis stimulate the dissociation of the proteins from the ribosome [45].
Canonical members of the Ras superfamily of GTPases contain a conserved glutamine in the switch II domain, located one amino acid C-terminal to the G3 motif essential for GTP hydrolysis. Instead, in B. subtilis RbgA (human GTPBP7) and several other ribosome assembly-GTPases, the catalytic residue is His9, at an alternate location [89].
For several mitoribosome assembly GTPases, the ribosome can serve as a GAP (GTPase-accelerating protein) and stimulate GTP hydrolysis by more than 50 fold. GTPBP5 interacts with the mtLSU in a GTP dependent manner, although only its intrinsic GTPase activity could be measured [45,53]. GTPBP10 and GTPBP7 associate with the mtLSU in a GTP-dependent manner [45,46,78].
GTPBP6-facilitated ribosome dissociation into subunits depends on GTP binding, but not GTP hydrolysis [80]. It is tempting to assume that GTP hydrolysis induces the release of GTPBP6 from the mtLSU.
A GTPBP6 variant mutated in the GTPase domain is incompetent to form translational active ribosomes indicating that GTPBP6-mediated mtLSU maturation depends on GTP binding or hydrolysis [80].
Bacterial Era exhibits low intrinsic GTPase activity, although it is stimulated by the 16S rRNA [60,61,94–96]. Also human MTG3 binds GTP and exhibit intrinsic GTPase activity like bacterial YqeH [42]. However, MTG3-facilitated GTP hydrolysis is enhanced by the mitochondrial and bacterial rRNAs, the bacterial 30S SSU and 50S LSU, but not by the 70S ribosome [42].
Ribosome assembly GTPases bind nucleotides weakly and are thus not expected to be dependent on GEFs (guanine nucleotide exchange factors) to exchange GDP for GTP. However, GTPBP7 uses the GEF activity of the mtSSU MRP mS27 leading to the dissociation of GDP-bound GTPBP7 from the mature mtLSU [44].
Two isoforms of RCC1L (RCC1LV1, and RCC1LV3), a putative GEF, interact with the mtLSU and mtSSU, respectively. GTPBP10, ERAL1, and MTG3 could be their targets [97]
The mtSSU protein mS29 is a GTPase [3,4], however the role of this activity in mitoribosome assembly and function remains unknown.
The physiological role of GTPBP7 was demonstrated by the development of early cardiovascular lesions upon GTPBP7 depletion in human cardiomyocytes and developing heart in zebrafish, highlighting its role in cardiac hypertrophy [44]. Consistently, Xu et al. have shown a protective role of GTPBP7 in the development of cardiac hypertrophy [90].
Towards a unification of the nomenclature for mitoribosome assembly GTPases
Despite the smaller number of proteins involved, the nomenclature of mammalian mitoribosome assembly GTPases has become plural in most instances. The table in Figure 2b presents the nomenclature that we are proposing for mitoribosome assembly GTPases, and we encourage the community to use it. The names of the mammalian proteins align in most cases with the most generic GTPBP (GTP binding protein) nomenclature and the most common names used in the literature so that the proposed names can be easily recognized.
Concluding Remarks and Future Perspectives
GTPases are a major class of mitoribosome assembly factors. They can act as protein and rRNA chaperones inducing conformational changes and facilitating rRNA modifications. In addition, they can prevent premature MRP binding by acting as placeholders. GTPases especially required for mtLSU maturation act as anti-association factors to prohibit premature subunit joining. As all these GTPases are critical for the biogenesis of the mtSSU or mtLSU, mostly acting as quality control factors in late maturation steps, their genes are candidates when screening for genetic causes of mitochondrial disorders associated with mitochondrial translation defects and multiple OXPHOS enzyme deficiencies.
Despite recent substantial advances in the molecular biology and biochemistry of mitoribosome assembly GTPases, an array of fundamental questions remains open, some of which are listed in the Outstanding Questions Box. The next few years are expected to witness discoveries of the precise mitoribosome subunit assembly intermediate in which each GTPase act, whether all GTPases function in a linear pathway or in cooperation among them and with other assembly factors, and the role of GTP binding and hydrolysis. At the current pace of progress in multiple laboratories, we anticipate that with a combination of biochemical studies (in vitro and in cellular/animal models) and cryo-EM reconstruction of mitochondrial ribosome assembly intermediates, we will have a comprehensive picture of the functional roles of mitoribosome assembly GTPases.
Outstanding Questions Box.
Why do so many GTPases act during the late stages of mitoribosome assembly?
What is the role of GTP binding and hydrolysis for each mitoribosome assembly GTPase? Is there a competition for GTP binding among GTPases?
Do GTPases sense the organellar and cellular energy level to coordinate mitoribosome assembly accordingly?
How do the GTPases bind to the rRNA and modify their conformation?
How do mitoribosome assembly RNA helicases and GTPases cooperate?
Highlights.
Mitoribosome assembly involves at least six GTPases belonging to several conserved families.
Mitoribosome assembly GTPases act to facilitate rRNA folding, and recruit mitoribosomal proteins and assembly factors to the assembly pathway.
Maturation of the mtLSU requires the assistance of several GTPases acting at late stages, when they function as anti-association or quality control factors to ensure joining of mature mtSSU and mtLSU into functional ribosomes.
Impaired mitoribosome assembly GTPase function leads to defective mitochondrial protein synthesis and human disease.
A novel unifying nomenclature for mitoribosome assembly GTPases is proposed.
Acknowledgements
Our research is supported by:
NIGMS-MIRA [R35GM118141 to A.B.]; Muscular Dystrophy Association Research Grant [MDA-381828 to A.B.]; American Heart Association postdoctoral fellowship [19POST34450174 to P.M.]; Deutsche Forschungsgemeinschaft by the Emmy-Noether grant [RI 2715/1-1 to R.R.-D.] and the Excellence Cluster [EXC 2067/1- 390729940 to R.R.-D.].
Footnotes
Conflict of interest statement. None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greber BJ et al. (2014) The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515, 283–286 [DOI] [PubMed] [Google Scholar]
- 2.Brown A et al. (2014) Structure of the large ribosomal subunit from human mitochondria. Science 346, 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greber BJ et al. (2015) Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 [DOI] [PubMed] [Google Scholar]
- 4.Amunts A et al. (2015) Ribosome. The structure of the human mitochondrial ribosome. Science 348, 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greber BJ and Ban N (2016) Structure and Function of the Mitochondrial Ribosome. Annu. Rev. Biochem. 85, 103–132 [DOI] [PubMed] [Google Scholar]
- 6.Pulman J et al. (2019) Mutations in the MRPS28gene encoding the small mitoribosomal subunit protein bS1m in a patient with intrauterine growth retardation, craniofacial dysmorphism and multisystemic involvement. Human Molecular Genetics 28, 1445–1462 [DOI] [PubMed] [Google Scholar]
- 7.Bugiardini E et al. (2019) MRPS25 mutations impair mitochondrial translation and cause encephalomyopathy. Human Molecular Genetics 28, 2711–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormier-Daire V et al. (1996) Craniofacial anomalies and malformations in respiratory chain deficiency. Am. J. Med. Genet. 66, 457–463 [DOI] [PubMed] [Google Scholar]
- 9.Gardeitchik T et al. (2018) Bi-allelic Mutations in the Mitochondrial Ribosomal Protein MRPS2 Cause Sensorineural Hearing Loss, Hypoglycemia, and Multiple OXPHOS Complex Deficiencies. Am. J. Hum. Genet. 102, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell S et al. (2009) Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szpakowicz A et al. (2015) The rs9982601 polymorphism of the region between the SLC5A3/MRPS6 and KCNE2 genes associated with a prevalence of myocardial infarction and subsequent long-term mortality. Pol. Arch. Med. Wewn. 125, 240–248 [DOI] [PubMed] [Google Scholar]
- 12.Chatzispyrou IA et al. (2017) A homozygous missense mutation in ERAL1, encoding a mitochondrial rRNA chaperone, causes Perrault syndrome. Human Molecular Genetics 26, 2541–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menezes MJ et al. (2015) Mutation in mitochondrial ribosomal protein S7 (MRPS7) causes congenital sensorineural deafness, progressive hepatic and renal failure and lactic acidemia. Human Molecular Genetics 24, 2297–2307 [DOI] [PubMed] [Google Scholar]
- 14.Dheedene A et al. (2014) A de novo POU3F3 Deletion in a Boy with Intellectual Disability and Dysmorphic Features. Mol Syndromol 5, 32–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YH and Song GG (2015) Meta-analysis of differentially expressed genes in ankylosing spondylitis. Genet. Mol. Res. 14, 5161–5170 [DOI] [PubMed] [Google Scholar]
- 16.Jackson CB et al. (2018) A variant in MRPS14 (uS14m) causes perinatal hypertrophic cardiomyopathy with neonatal lactic acidosis, growth retardation, dysmorphic features and neurological involvement. Human Molecular Genetics DOI: 10.1093/hmg/ddy374 [DOI] [PubMed] [Google Scholar]
- 17.Miller C et al. (2004) Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann. Neurol. 56, 734–738 [DOI] [PubMed] [Google Scholar]
- 18.Saada A et al. (2007) Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J. Med. Genet. 44, 784–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen A et al. (2018) Mutations in the mitochondrial ribosomal protein MRPS22 lead to primary ovarian insufficiency. Human Molecular Genetics 27, 1913–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smits P et al. (2011) Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 19, 394–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohda M et al. (2016) A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet 12, e1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lake NJ et al. (2017) Biallelic Mutations in MRPS34 Lead to Instability of the Small Mitoribosomal Subunit and Leigh Syndrome. Am. J. Hum. Genet. 101, 239–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galmiche L et al. (2011) Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum. Mutat. 32, 1225–1231 [DOI] [PubMed] [Google Scholar]
- 24.Serre V et al. (2013) Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neurological deterioration and mitochondrial translation deficiency. Biochim. Biophys. Acta 1832, 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carroll CJ et al. (2013) Whole-exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J. Med. Genet. 50, 151–159 [DOI] [PubMed] [Google Scholar]
- 26.Distelmaier F et al. (2015) MRPL44 mutations cause a slowly progressive multisystem disease with childhood-onset hypertrophic cardiomyopathy. Neurogenetics 16, 319–323 [DOI] [PubMed] [Google Scholar]
- 27.Borna NN et al. (2019) Mitochondrial ribosomal protein PTCD3 mutations cause oxidative phosphorylation defects with Leigh syndrome. 20, 9–25 [DOI] [PubMed] [Google Scholar]
- 28.Di Nottia M et al. (2020) A homozygous MRPL24 mutation causes a complex movement disorder and affects the mitoribosome assembly. Neurobiol. Dis. 141, 104880. [DOI] [PubMed] [Google Scholar]
- 29.Solomon BD et al. (2011) De novo deletion of chromosome 20q13.33 in a patient with tracheo-esophageal fistula, cardiac defects and genitourinary anomalies implicates GTPBP5 as a candidate gene. Birth Defects Res. Part A Clin. Mol. Teratol. 91, 862–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin H-J et al. (2016) Identification and validation of regulatory SNPs that modulate transcription factor chromatin binding and gene expression in prostate cancer. Oncotarget 7, 54616–54626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogenhagen DF et al. (2018) Kinetics and Mechanism of Mammalian Mitochondrial Ribosome Assembly. CellReports 22, 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogenhagen DF et al. (2014) Initial Steps in RNA Processing and Ribosome Assembly Occur at Mitochondrial DNA Nucleoids. Cell Metabolism 19, 618–629 [DOI] [PubMed] [Google Scholar]
- 33.Antonicka H et al. (2013) The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metabolism 17, 386–398 [DOI] [PubMed] [Google Scholar]
- 34.Antonicka H and Shoubridge EA (2015) Mitochondrial RNA Granules Are Centers for Posttranscriptional RNA Processing and Ribosome Biogenesis. Cell Reports DOI: 10.1016/j.celrep.2015.01.030 [DOI] [PubMed] [Google Scholar]
- 35.Jourdain AA et al. (2013) GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metabolism 17, 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tu Y-T and Barrientos A (2015) The Human Mitochondrial DEAD-Box Protein DDX28 Resides in RNA Granules and Functions in Mitoribosome Assembly. Cell Reports DOI: 10.1016/j.celrep.2015.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrientos A (2015) Mitochondriolus: assembling mitoribosomes. Oncotarget 6, 16800–16801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Silva D et al. (2013) The DEAD box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metabolism 18, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maiti P et al. (2018) Human GTPBP10 is required for mitoribosome maturation. Nucleic Acids Research 46, 11423–11437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jourdain AA et al. (2017) The FASTK family of proteins: emerging regulators of mitochondrial RNA biology. Nucleic Acids Research 45, 10941–10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Britton RA (2009) Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63, 155–176 [DOI] [PubMed] [Google Scholar]
- 42.Kolanczyk M et al. (2011) NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell 22, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He J et al. (2012) Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Research 40, 6097–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim H-J and Barrientos A (2018) MTG1 couples mitoribosome large subunit assembly with intersubunit bridge formation. Nucleic Acids Research 46, 8435–8453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kotani T et al. (2013) Human G-proteins, ObgH1 and Mtg1, associate with the large mitochondrial ribosome subunit and are involved in translation and assembly of respiratory complexes. Nucleic Acids Research 41, 3713–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavdovskaia E et al. (2018) The human Obg protein GTPBP10 is involved in mitoribosomal biogenesis. Nucleic Acids Research 46, 8471–8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maiti P et al. (2020) Human GTPBP5 (MTG2) fuels mitoribosome large subunit maturation by facilitating 16S rRNA methylation. Nucleic Acids Research 348, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennerlein S et al. (2010) Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem. J. 430, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchiumi T et al. (2010) ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Research 38, 5554–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metodiev MD et al. (2014) NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly. PLoS Genet 10, e1004110–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leipe DD et al. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 52.Goto S et al. (2013) GTPases involved in bacterial ribosome maturation. J. Biochem. 153, 403–414 [DOI] [PubMed] [Google Scholar]
- 53.Hirano Y et al. (2006) Human small G proteins, ObgH1, and ObgH2, participate in the maintenance of mitochondria and nucleolar architectures. Genes to Cells 11, 1295–1304 [DOI] [PubMed] [Google Scholar]
- 54.Jaskolowski M et al. (2020) Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis. Mol. Cell DOI: 10.1016/j.molcel.2020.06.030 [DOI] [PubMed] [Google Scholar]
- 55.Tang T et al. (2009) hNOA1 interacts with complex I and DAP3 and regulates mitochondrial respiration and apoptosis. J. Biol. Chem. 284, 5414–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Furoukh N et al. (2014) NOA1, a novel ClpXP substrate, takes an unexpected nuclear detour prior to mitochondrial import. PLoS ONE 9, e103141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anand B et al. (2009) Circularly permuted GTPase YqeH binds 30S ribosomal subunit: Implications for its role in ribosome assembly. Biochem. Biophys. Res. Commun. 386, 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paul M-F et al. (2012) The putative GTPase encoded by MTG3 functions in a novel pathway for regulating assembly of the small subunit of yeast mitochondrial ribosomes. Journal of Biological Chemistry 287, 24346–24355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma MR et al. (2005) Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol. Cell 18, 319–329 [DOI] [PubMed] [Google Scholar]
- 60.Tu C et al. (2009) Structure of ERA in complex with the 3’ end of 16S rRNA: implications for ribosome biogenesis. Proceedings of the National Academy of Sciences 106, 14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tu C et al. (2011) The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3’ end of 16S rRNA. Proceedings of the National Academy of Sciences 108, 10156–10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seidel-Rogol BL et al. (2003) Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 33, 23–24 [DOI] [PubMed] [Google Scholar]
- 63.Metodiev MD et al. (2009) Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metabolism 9, 386–397 [DOI] [PubMed] [Google Scholar]
- 64.Formenoy LJ et al. (1994) Methylation of the conserved A1518-A1519 in Escherichia coli 16S ribosomal RNA by the ksgA methyltransferase is influenced by methylations around the similarly conserved U1512.G1523 base pair in the 3’ terminal hairpin. Biochimie 76, 1123–1128 [DOI] [PubMed] [Google Scholar]
- 65.Helser TL et al. (1971) Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nature New Biol. 233, 12–14 [DOI] [PubMed] [Google Scholar]
- 66.Van Buul CP et al. (1983) Kasugamycin resistant mutants of Bacillus stearothermophilus lacking the enzyme for the methylation of two adjacent adenosines in 16S ribosomal RNA. Mol. Gen. Genet. 189, 475–478 [DOI] [PubMed] [Google Scholar]
- 67.Rozanska A et al. (2017) The human RNA-binding protein RBFA promotes the maturation of the mitochondrial ribosome. Biochem. J. 474, 2145–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akiyama T et al. (2001) Mammalian homologue of E. coli Ras-like GTPase (ERA) is a possible apoptosis regulator with RNA binding activity. Genes Cells 6, 987–1001 [DOI] [PubMed] [Google Scholar]
- 69.Gohda J et al. (2003) Elimination of the vertebrate Escherichia coli Ras-like protein homologue leads to cell cycle arrest at G1 phase and apoptosis. Oncogene 22, 1340–1348 [DOI] [PubMed] [Google Scholar]
- 70.Szczepanowska K et al. (2016) CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J. 35, 2566–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jenkinson EM et al. (2013) Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet. 92, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee K-W and Bogenhagen DF (2014) Assignment of 2’-O-Methyltransferases to Modification Sites on the Mammalian Mitochondrial Large Subunit 16 S Ribosomal RNA (rRNA). Journal of Biological Chemistry 289, 24936–24942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rorbach J et al. (2014) MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol. Biol. Cell 25, 2542–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang M et al. (2006) The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 188, 6757–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan J et al. (2002) Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184, 2692–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Datta K et al. (2005) The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol. Biol. Cell 16, 954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng B et al. (2014) Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 12, e1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Busch JD et al. (2019) MitoRibo-Tag Mice Provide a Tool for In Vivo Studies of Mitoribosome Composition. CellReports 29, 1728–1738.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rackham O et al. (2016) Hierarchical RNA Processing Is Required for Mitochondrial Ribosome Assembly. Cell Reports 16, 1874–1890 [DOI] [PubMed] [Google Scholar]
- 80.Lavdovskaia E et al. (2020) Dual function of GTPBP6 in biogenesis and recycling of human mitochondrial ribosomes. Nucleic Acids Research DOI: 10.1093/nar/gkaa1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Srinivasan K et al. (2019) Structural modules of the stress-induced protein HflX: an outlook on its evolution and biological role. Curr. Genet. 65, 363–370 [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y et al. (2015) HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat. Struct. Mol. Biol. 22, 906–913 [DOI] [PubMed] [Google Scholar]
- 83.Dey S et al. (2018) The universally conserved GTPase HflX is an RNA helicase that restores heat-damaged Escherichia coli ribosomes. J. Cell Biol. 217, 2519–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barrientos A et al. (2003) MTG1 codes for a conserved protein required for mitochondrial translation. Mol. Biol. Cell 14, 2292–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matsuo Y et al. (2007) Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J. Biol. Chem. 282, 25270–25277 [DOI] [PubMed] [Google Scholar]
- 86.Achila D et al. (2012) Biochemical characterization of ribosome assembly GTPase RbgA in Bacillus subtilis. Journal of Biological Chemistry 287, 8417–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gulati M et al. (2013) Mutational analysis of the ribosome assembly GTPase RbgA provides insight into ribosome interaction and ribosomestimulated GTPase activation. Nucleic Acids Research 41, 3217–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jomaa A et al. (2014) Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Research 42, 3419–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seffouh A et al. (2019) Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate. Nucleic Acids Research 47, 10414–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu D et al. (2019) Novel role of mitochondrial GTPases 1 in pathological cardiac hypertrophy. Journal of Molecular and Cellular Cardiology 128, 105–116 [DOI] [PubMed] [Google Scholar]
- 91.De Silva D et al. (2015) Mitochondrial ribosome assembly in health and disease. Cell Cycle 14, 2226–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim H-J et al. (2017) Mitochondrial ribosomes in cancer. Semin. Cancer Biol. 47, 67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bennison DJ et al. (2019) The Impact of the Stringent Response on TRAFAC GTPases and Prokaryotic Ribosome Assembly. Cells 8, 1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen SM et al. (1990) Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J. Biol. Chem. 265, 2888–2895 [PubMed] [Google Scholar]
- 95.Meier TI et al. (1999) 16S rRNA is bound to era of Streptococcus pneumoniae. J. Bacteriol. 181, 5242–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meier TI et al. (2000) Era GTPase of Escherichia coli: binding to 16S rRNA and modulation of GTPase activity by RNA and carbohydrates. Microbiology 146, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 97.Reyes A et al. (2020) RCC1L (WBSCR16) isoforms coordinate mitochondrial ribosome assembly through their interaction with GTPases. PLoS Genet 16, e1008923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saitou N and Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 99.Zuckerkandl E and Pauling L (1965) Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins pp. 97–166, Academic Press [Google Scholar]
- 100.Kumar S et al. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stecher G et al. (2020) Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]