Abstract
The global emergence of carbapenemase-producing bacteria capable of hydrolyzing the once effective carbapenem antibiotics is considered a contemporary public health concern. Carbapenemase enzymes, once constrained to isolates of Klebsiella pneumoniae, are now routinely reported in different bacteria within the Enterobacterales order of bacteria, creating the acronym CRE which now defines Carbapenem-Resistant Enterobacterales. CRE harboring different types of enzymes, including the most prevalent types KPC, VIM, IMP, NDM, and OXA-48, are now routinely reported and more importantly, are now frequently present in many infections world-wide. Defining and updating the contemporary epidemiology of both the US and global burden of carbapenem-resistant infections is now more important than ever. This review describes the global distribution and continued evolution of carbapenemases which continue to spread at alarming rates. Informed understanding of the current epidemiology of CRE, coupled with advances in antibiotic options, and the use rapid diagnostics offers the potential for rapid identification and management of carbapenem-resistant infections.
Keywords: Carbapenem-resistant enterobacterales (CRE), Enterobacterales, Epidemiology, Global
Plain Language Summary
Carbapenems are a subclass of antibiotic used to treat infections caused by Gram-negative bacteria, particularly in resistant and multidrug-resistant (MDR) infections where penicillin and cephalosporins are no longer effective. However, carbapenem-resistant Enterobacterales (CRE) have emerged due to acquisition of carbapenemase enzymes, most prevalent types are KPC, VIM, IMP, NDM, and OXA-48; infections caused by these bacteria have disseminated globally in both the healthcare and community setting. Resulting in a significant public health issue and clinical burden, these CRE infections are associated with increased morbidity and mortality, in part because carbapenems are the last therapeutic line of defense against resistant and MDR bacterial infections. The author wanted to investigate current US and global epidemiology of carbapenem-resistant infections, identify factors driving changes, as well as diagnostic technologies, and reporting or surveillance methods in place to track trends and inform therapeutic protocols and development. Overall, carbapenemase enzymes originally only reported in one country or region in 2006-2007, by 2013 and onwards have spread not only to surrounding countries but to other continents, which has impacted antibiotic resistance patterns and susceptibility. Increasing human travel and environmental factors, such as livestock care, food distribution, sewage, and recreational water, have contributed to global dissemination of CRE. Active surveillance programs are key to tracking resistance in real time, in order to update susceptibility breakpoints and epidemiology, which can inform antibiotic treatment choices, management guidelines, and the development of new therapeutics. Together, these factors will help to identify, control, and treat the spread of carbapenem resistance.
Key Summary Points
| Carbapenemase enzymes which confer resistance to carbapenem antibiotics are no longer only found in Klebsiella pneumoniae isolates, but are now routinely documented among different Enterobacterales bacteria, now labeled Carbapenem-Resistant Enterobacterales (CRE). |
| Carbapenem antibiotics are often used as empiric therapy in patients with serious infections caused by gram-negative bacteria, including resistant isolates, but this treatment is threatened by the global dissemination of CRE, which are now present in many infections worldwide. |
| The contemporary epidemiology of both the US and global burden of carbapenem-resistant infections needs defining and updating in order to identify, control, and treat the spread of carbapenem resistance. |
| Susceptibility breakpoints need to reflect the changes in bacterial susceptibility in order to inform and guide prescribing clinical practices, and for implementation in laboratory diagnostic technologies. |
| Global and national surveillance programs are needed to continuously track global resistance rates in real time, to facilitate effective antibiotic stewardship efforts and direct development of new therapeutic agents. |
Digital Features
This article is published with digital features, including a summary slide and plain language summary, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.13469820.
Introduction
Beta-lactam drugs are a cornerstone of antibiotic therapy, and widely used for Gram-negative infections. However, inactivation of the core structural beta-lactam ring, by beta-lactamase enzymes, has gradually eroded the efficacy of once effective and life-saving therapies. Carbapenems, a subclass of beta-lactams, have often maintained efficacy in the setting of cephalosporin and penicillin resistance because they contain a unique beta-lactam core that renders them resistant, and slow, to hydrolysis by many beta-lactamase enzymes. Thus, for decades, carbapenems have remained active against Gram-negative infections, and are frequently used as empiric therapy in patients with serious infections caused by Gram-negative bacteria.
Unfortunately, the security of carbapenem therapy is now challenged because of the dissemination of carbapenemase enzymes, which have been acquired by members of Enterobacterales family. Globally, these enzymes are becoming more frequent in healthcare and community settings. The dissemination of carbapenem-resistant Enterobacterales (CRE) now frequently challenge the efficacy of carbapenem therapies as the last line of defense against multi-drug resistant (MDR) organisms. As the molecular mechanisms of resistance continue to evolve, so too must our understanding of the epidemiology of carbapenem-resistant organisms and the therapeutic options needed to combat the threat of antimicrobial resistance. This review will focus on the evolving epidemiology CRE and overview of enzymatic carbapenemase-mediated CRE.
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Classification and Evolution of Carbapenem Resistance
In 1913, 25 years before the release of penicillin, Ehrlich correctly assessed the capacity of bacteria to develop resistance to antibiotics[1]. Since Ehrlich’s initial observations in 1913, the challenge of predicting and minimizing the scope of bacterial resistance has been a goal of modern medicine. Hearing Ehrlich’s advice, Alexander Fleming himself predicted not only how useful antibacterial drugs would be but also how dangerous a world without them could be. In an interview shortly after winning the Nobel Prize in 1945 for discovering penicillin, Fleming said: "The thoughtless person playing with penicillin treatment is morally responsible for the death of the man who succumbs to infection with the penicillin-resistant organism." In 1940, Abraham and Chain described the first beta-lactamase enzyme (penicillinase) isolated from Bacillus (Escherichia) coli, well before the clinical use of penicillin [2]. Ironically, the enzyme was initially thought to present few clinical concerns because penicillin, largely developed to treat staphylococcal disease, was not found to be associated with beta-lactamase enzymes [3]. The global dissemination of methicillin-resistant Staphylococcus aureus, at its height from 1995–2005, provides perspective for the relevance of contemporary concerns over CRE. Today, over 2000 unique protein sequences coding for beta-lactamases have been in both Gram-negative and Gram-positive organisms [4].
Nomenclature for beta-lactamases are initially based on amino acid sequences, defined as the Ambler classification, which divides beta-lactamases into 4 major classes A–D [5].
A second classification system, first proposed by Richmond and Sykes [6], classified beta-lactamases based on functional characteristics. Further functional classification of key beta-lactamases was proposed by Bush, Jacoby, and Medeiros, and relies on substrate and inhibitor activity profiles to organize enzymes in a 1–4 system that can be readily identified with their phenotype in clinical isolates [5]. Table 1 outlines the classification of carbapenemase enzymes.
Table 1.
Classification of carbapenemase enzymes
| Ambler Structural Class | Bush-Jacoby group | Enzyme name | Resistance profile | Examples | Examples of active antimicrobial agents | Common bacterial species involved |
|---|---|---|---|---|---|---|
| A | 2f | Serine carbapenemase | Carbapenems, cephalosporins, aztreonam | KPC, GES, SME-1 |
Ceftazadime-avibactam mipenem-relebactam eropenem-vaborbactam cefidercol |
KPC: K. pneumoniae, E. coli, Enterobacter spp. |
| B | 3a | Metallo-beta lactamase | All beta-lactams except aztreonam | IMP, NDM, VIM, IND | Cefidercol^ |
NDM: K. pneumoniae, E. coli VIM: K. pneumoniae |
| D | 2df | Carbapenemase |
Penicillins (variable hydrolysis) |
OXA | Cefidercol^ |
IMP: K. pneumoniae OXA48: K. pneumoniae, E. coli, Enterobacter spp. |
Kazmierczak et al. [58] reported activity of cefidercol against KPC-postive (n = 75); MIC50/90 1/2 μg/ml. NDM positive (n = 12); 4/8 μg/ml. OXA-48 (n = 32); 0.5/2/μg/ml. VIM positive (n = 27); MIC50/90 1/4 μg/ml
KPC Klebsiella pneumoniae carbapenemase, MBL metallo-β-lactamase, NDM New Delhi metallo-β-lactamase, VIM Verona integrin-encoded metallo-β-lactamase, IMP, imipenemase, OXA oxacillinase, GES Guiana-extended-spectrum β-lactamase, SEM Serratia marcescens enzyme
Class A Beta-Lactamases
Until recent concerns about carbapenemase enzymes, Enterobacterales were generalizable for their susceptibility to common beta-lactamase inhibitors, clavulanic acid, tazobactam, and sulbactam. In 1965, a class A beta-lactamase reported from E. coli described the first plasmid-mediated beta-lactamase from a patient from Greece, which now bears the name “TEM” from the index patient in which it was reported [7]. Additional class A beta-lactamases in the form of SHV (sulfhydryl reagent variable) variants are now commonly found in beta-lactamase-resistant isolates of E. coli and Klebsiella pneumoniae associated with bloodstream infections, as well as hospital-acquired respiratory and urinary tract infections [5]. Within 2 years of the introduction of cefotaxime and ceftazidime, novel extended spectrum β-lactamases (ESBLs) were reported in clinical isolates [8]. Mutations in pre-existing blaTEM-1 and blaSHV-1 genes gave rise to the emergence of ESBLs capable of hydrolyzing extended-spectrum cephalosporins with an oxyimino side chain. These cephalosporins include cefotaxime, ceftriaxone, and ceftazidime, as well as the oxyimino-monobactam aztreonam. Cefotaximases, characterized by CTX-M genes arose by plasmid transfer from pre-existing chromosomal ESBL genes from the bacteria Kluyvera spp., [9] CTX-M ESBLs now represent a large proportion of global ESBL strains and are the dominant ESBLs in the US, particularly among urinary tract infections [10, 10].
Class A Serine Carbapenemases
The non-metallo beta-lactamases can hydrolyze carbapenems as well as cephalosporins, penicillin, and aztreonam [9], marking the evolution of beta-lactamase activity from primarily targeting penicillins and cephalosporins to enzymes that hydrolyze carbapenems. Representative of class A serine carbapenemases include the enzymes NMC-A, IMI, SME, and KPC. Table 1 outlines the classification of beta-lactamases capable of hydrolyzing carbapenem antibiotics.
KPC enzymes have emerged as the most well-known, and well-described of the class A serine carbapenemases, which, while initially described in K. pneumoniae, are now readily detected in many members of the Enterobacteriaceae family. They confer resistance to all beta-lactams (excluding, carbapenems, and monobactams). Gram-negative bacteria harboring blaKPC genes can be carried on mobile genetic elements such as transposons (e.g., Tn4401b) and multiple plasmid (IncFII, IncL/M, and IncN) types [9]. Organisms expressing KPC genes are often concurrently resistant to other classes of antibiotics, such as quinolones and aminoglycosides, thus creating highly MDR organisms [12].
Class B Beta-Lactamases
Metallo-beta-lactamases (MBLs) have been isolated from Acinetobacter spp. and Pseudomonas spp., and are increasingly associated with members of the Enterobacterales. such as K. pneumoniae, K. oxytoca, E. coli, and Enterobacter spp. MBLs are commonly associated with the Verona integron–encoded metallo-β-lactamase (VIM), Pseudomonas (IMP)-type and the New Delhi MBL (NDM) [13, 13] (52). MBLs are commonly expressed from mobile genetic elements such as integrons, plasmids and transposons, which have contributed to the spread of MBLs. MBLs effectively hydrolyze beta-lactamases and are not inhibited by beta-lactamase inhibitors (clavulanic acid, tazobactam, vaborbactam). They also differ from KPCs (serine carbapenemases) because they contain a bivalent metal ion, commonly zinc (Zn2) ions which coordinate histidine/cytosine/asparagine residues at the active site [15]. Genetic insertion of entire sequences upstream of the blaNDM-1 gene among bacteria in the Enterobacterales seems to suggest that NDM variants likely evolved from Acinetobacter baumannii [16]. Currently, > 20 NDM types have been identified [17], yet expansion of NDMs do not appear to be due to proliferation of a single dominant clone. NDMs have been found in several epidemic clones, including K. pneumoniae ST11 and ST147 and E. coli ST131 and ST101, which are known to harbor other β-lactamase genes and antibiotic resistance determinants [12]. IMP-type metallo-β-lactamases are among the most common families of acquired carbapenemases detected from Enterobacteriaceae and have been reported mainly in East Asia, including Japan [18]. Although IMP-type metallo-β-lactamases are class B beta-lactamases, their hydrolytic properties for carbapenems are weak, often resulting in elevated resistance to meropenem while retaining imipenem susceptibility [19, 19]. This feature has been implicated in the spread of blaIMP harboring plasmids in Japan, as determined after screening the antibiotic susceptibility of infectious bacteria to imipenem as a representative carbapenem [21].
Class C, Serine-Based Cephalosporinases
AmpC β-lactamase expressors are common to members of the family are commonly associated with Serratia spp., Pseudomonas aeruginosa, Providencia spp., indole-positive Proteus mirabilis/vulgaris, Citrobacter spp., and Enterobacter spp., which collectively are often referred to as “SPICE” organisms. AmpC β-lactamase are generally resistant to penicillins, β-lactamase inhibitors, and cephalosporins such as cefoxitin, cefotetan, ceftriaxone and cefotaxime. In many Enterobacterales, AmpC expression is low but inducible in response to β-lactam exposure, and resistance occurs through two primary mechanisms. Disruption of peptidoglycan (murein) synthesis by beta-lactam agents leads to an accumulation of N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid oligopeptides. Displacement of the UDP-N-acetylmuramic acid peptides signals a conformational change in AmpR, which activates the transcription of ampC regulatory genes. Additionally, many Enterobacterales contain a cytoplasmic N-acetyl-muramyl-l-alanine amidase enzyme (AmpD) which removes peptides from the 1,6-anhydro-N-acetylmuramic acid and N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid oligopeptide derivatives. Under wild-type expression, AmpD aids in reducing their concentrations of the aforementioned enzymes, preventing the overexpression of AmpC. The most common cause of AmpC overexpression in clinical isolates is a mutation in ampD leading to AmpC hyper-inducibility and/or constitutive hyperproduction [22]. AmpR mutations are less common but can also result in high-constitutive or hyper-inducible phenotypes. Infrequently, AmpC induction can also occur via mutations in AmpG, an inner membrane permease enzyme, involved in cell wall turn-over and AmpC regulation into the cytosol [23, 23]. AmpC expression can often combine with other mechanisms of resistance often related to permeability such as porin loss and/or efflux pumps, resulting in clinically significant resistance [25].
Class D Serine Oxacillinases
Class D beta-lactamases, initially characterized as oxacillinases because of their hydrolytic activity against β-lactams have recently become a clinical concern. Class D beta-lactamases, and OXA variants, possess variable but significant carbapenemase activity and are generally not inhibited by clavulanic acid, tazobactam, and sulbactam (with some exceptions; e.g., OXA-2 and OXA-32 are inhibited by tazobactam but not by sulbactam and clavulanate, and OXA-53 is inhibited by clavulanate) [9]. Nearly one-third of OXA variants hydrolyze carbapenems. OXA-carbapenemases are predominantly isolated in Acinetobacter spp.; however, the OXA-48 carbapenemase was described in a MDR isolate of K. pneumoniae in 2004 in a patient with a UTI [26]. The initial cases of an OXA-48 producing K. pneumoniae presented as a highly drug-resistant strain because it co-expressed other beta-lactamases (class A extended-spectrum β-lactamase SHV-2a and the narrow-spectrum β-lactamases TEM-1 and OXA-47) as well as defects in outer membrane proteins [27]. While OXA-48 is a class D β-lactamase with the highest known catalytic efficiency for imipenem [28, 28], organisms containing the plasmid-mediated blaOXA-4 gene effectively hydrolyze penicillins, but hydrolyze carbapenems at low levels and show weak activity to extended-spectrum cephalosporins, and poor hydrolytic activity against ceftazidime and cefepime. Recently, OXA-48-positive organisms have spread to over 30 different countries and within various bacteria of the Enterobacterales [27, 27, 27].
This review will focus on enzymatic forms of CRE, attributed to carbapenemase-producing bacteria. However, it should be noted that carbapenem-resistance is often regarded as a phenotypic designation and encompasses non-enzymatic forms of carbapenem resistance. cell wall differences, external decreased membrane permeability, efflux pumps, and the presence of various broad-spectrum β-lactamases (e.g., AmpC cephalosporinase), often manifest in carbapenem resistance when measured phenotypically in laboratories.
Epidemiology of Carbapenem Resistance and Cre in the United States
Over a decade ago, the acronym ESKAPE pathogens was used to describe the names of six bacterial pathogens (Enterococcus faecium, S. aureus, K. pneumoniae. A.baumannii, P. aeruginosa and Enterobacter spp.) associated with declining clinical responses and rising rates of antimicrobial resistance [31]. Since this initial description, and plea for funding recognition to support antibiotic development, global expansion of carbapenem resistance among Gram-negative bacteria has steadily increased, prompting the US Centers for Disease Control and Prevention (CDC) to issue concerns over antibiotic resistance as an urgent public health threat [32].
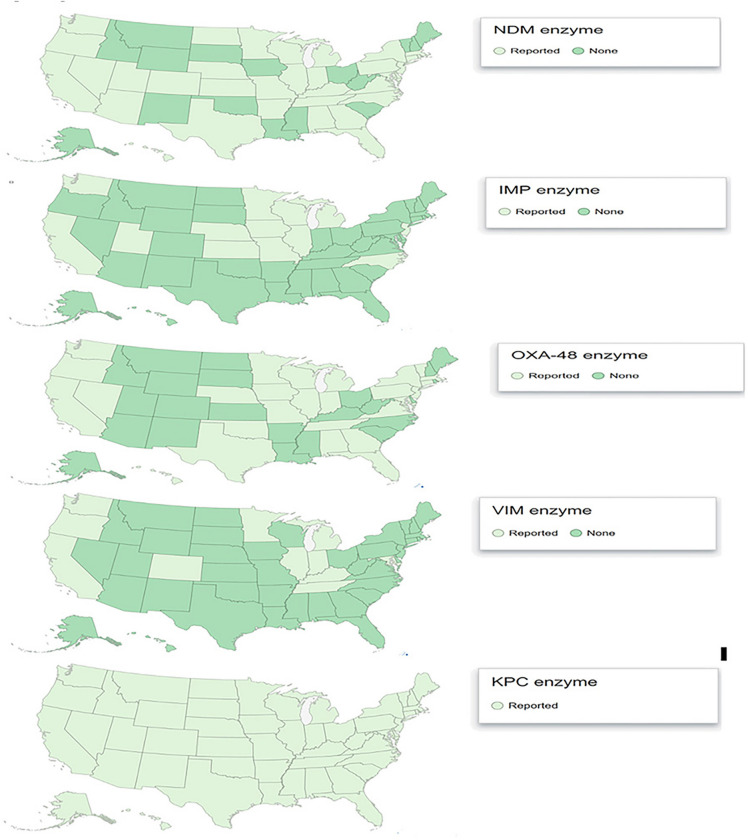
Much of the increase in CRE is likely related to the emergence and spread of the genes coding for carbapenemases. In the US, the most commonly identified carbapenemase is K. pneumoniae carbapenemase (KPC). This enzyme was first described in 2001 in an isolate obtained from a patient in North Carolina [33]. Cases involving KPC harboring organisms have been described from every US state (Fig. 1.), and other carbapenemases produced by Enterobacterales, including NDM, VIM, IMP, and OXA-48, have been reported in patients hospitalized in the US. CRE producing a carbapenemase other than KPC are generally uncommon in the US and often associated with healthcare exposures outside the US. However, recent reports suggest that patients may be colonized with (CR) organisms without traditional associated risk factors such as foreign travel [34] (personal communication, Minnesota Department of Health).
Fig. 1.
CRE reported cases, Centers for Disease Control as of 2017, reported by geographic US state reporting [55]. CRE defined by 2015 CDC case definition for CRE; resistant to imipenem, meropenem, doripenem, or ertapenem OR documentation that isolates possess a carbapenemase gene. Reference
Source CDC [56]
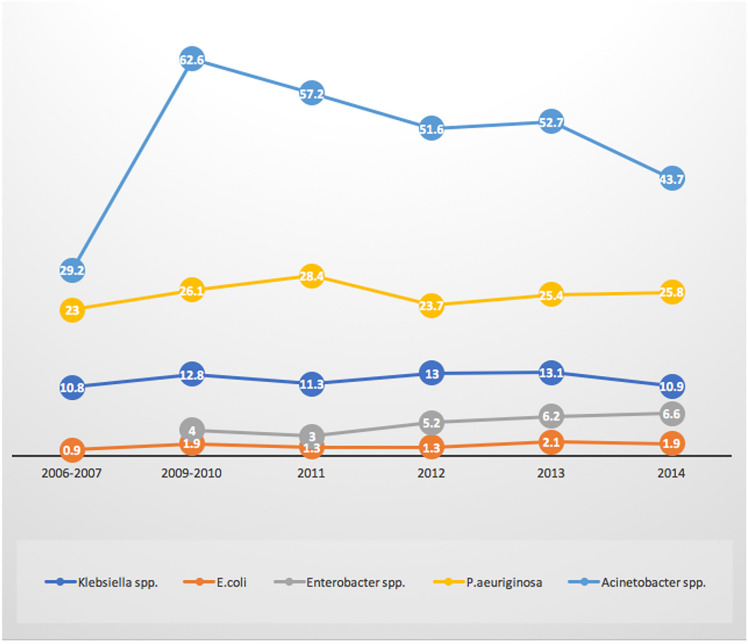
In response to concerns surrounding antibiotic resistance, the CDC launched the National Healthcare Safety Network (NHSN), a system which is used by the CDC, healthcare facilities, state health departments, and the Centers for Medicare and Medicaid Services, to provide surveillance of patient and healthcare safety, as well as a surveillance tool for tracking US antibiotic resistance trends. In 2006–2007, NHSN reported carbapenem resistance rates for E. coli and K. pneumoniae associated with central line-associated bloodstream infections (CLABSIs) at 0.9% and 10.8%, respectively [35]. Two years later, the 2009–2010 NHSN data showed CLABSIs CRE resistance rates in all tested isolates had doubled to 2% for E. coli, and K. pneumoniae increased to 12.8% [36] (Fig. 2.). Additionally, MDR among these E. coli isolates increased to 3.7% [36], mostly because of extended-spectrum cephalosporin resistance; this trend of MDR continued between 2011 and 2014 increasing from 11.1 to 14.1%, respectively [37].
Fig. 2.
Carbapenem resistance from Central Line–Associated Bloodstream Infections (CLABSIs). Figure
adapted from Sievert et al. [32]; values represent percent carbapenem resistant for respective bacteria isolated from postive blood cultures representative of CLABSI infection. Carbapenem resistance, as defined in this report, included all applicable pathogens with a result of I or R to imipenem, meropenem, or doripenem. 2006–2007 Klebsiella data inclusive of K. pneumoniae isolates only. Carbapenem resistance rates by pathogen differ depending on the site of infection [33][34]. Rates for P. aeruginosa and A. baumannii are lower for bloodstream infections that have been reported from other types of infections [e.g., ventilator-associated pneumonia (VAP)] [32][34] Thus, it is likely that studies reporting bloodstream and CLABSI infections may under-report true prevalence of carbapenem resistance rates
By 2014, carbapenem resistance in CLABSIs was readily detected among Klebsiella (pneumoniae/oxytoca.9%), E. coli (1.9%), Enterobacter spp. (6.6%), P. aeruginosa (25.8%) and Acinetobacter spp. 46.6%) [37]. Although carbapenem resistance rates are higher in P. aeruginosa and Acinetobacter spp., compared to members of the Enterobacteriaceae, the overall incidence of infections with P. aeruginosa and Acinetobacter spp. are less than that observed for the Enterobacteriaceae. Additionally, non-enzymatic forms of carbapenem resistance, such as efflux and porins, are common in P. aeruginosa.
National US-surveillance programs have focused on reports of carbapenem resistance from urine, CSF, and bloodstream infections, which is often a consequence of surveillance programs that focus on reporting from sterile sites. Among patients with VAP, rates of carbapenem resistance have risen steadily over the past decade The incidence of carbapenem resistance in K. pneumoniae and E. coli among VAP patients increased from 3.6 to 1.8% [35] to 11.2% to 10.1%, and 3.5% to 2.2% between 2013 and 2014 [36][36], respectively. Non-fermenting Gram-negative species including Acinetobacter spp., and P. aeruginosa associated with VAP continue to be problematic with reported carbapenem resistance rates of 55% and 28, respectively [37].
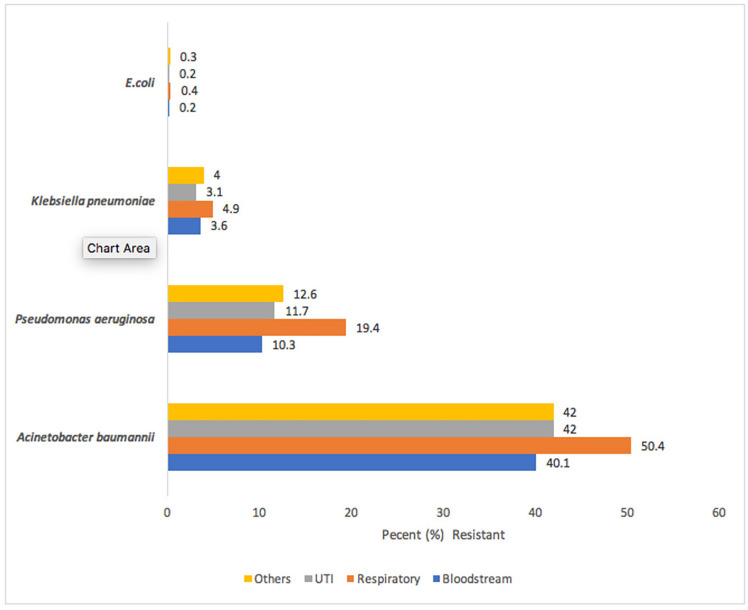
The incidence of CRE is increasing globally. Cai et al. evaluated carbapenem resistance rates for E. coli, K. pneumoniae, P. aeruginosa, and A. baumannii across 9 regions in the US, using Premiere Health Database data from 2009–2013, finding the overall carbapenem resistance rate among the 4 selected pathogens was 4.5% out of 292,742 isolates tested (Fig. 3) [38]. A. baumannii had the highest carbapenem resistance rates: blood 40.1%, respiratory 50.4%, urine 42.0%, and other sites 42%. Carbapenem resistance rates for P. aeruginosa ranked second highest, but varied more by infection site: 10.3% in blood, 19.4% in respiratory, 11.7% in urinary, and 12.6% in other. In the US, more than 80% of all carbapenem-resistant infections were caused by non-fermenters, A. baumannii or P. aeruginosa, which was a consistent finding across the 9 US regions sampled, except for the middle Atlantic region where only 67% were caused by these pathogens [38].
Fig. 3.
Prevalence of carbapenem-resistant Gram-negative infections in the United States.
Adapted from Cai et al. [38]. In the US, overall carbapenem resistance among the 4 selected pathogens (E. coli, K. pneumoniae, P. aeruginosa, A. baumannii) was 4.5% out of 292,742 isolates tested
Ceftazidime-avibactam-resistant strains of K. pneumoniae contain point mutations in KPC-2 and have now been described in the US. Nelson and colleagues, [39] phenotypically described a case in Los Angeles involving a resistant strain of K. pneumoniae isolated from blood cultures from a pancreatic cancer patient with an MIC of 32/4 μg/ml. While the authors failed to provide a genetic basis for ceftazidime-avibactam resistance, additional molecular testing for other known carbapenemase enzymes, and testing in the setting of an efflux pump inhibitor, phenyl-arginine β-naphthylamide, at 40 μg/ml, did not result in a decrease in the ceftazidime-avibactam MIC [40]. Follow-up subsequently revealed transposition of the Tn4401 transposon harboring blaKPC-3 into a second plasmid, pIncX3, which also harbored blaSHV-12, ultimately resulting in a higher copy number of blaKPC-3 in the resistant isolate [39]. KPC-3 gene mutations, D179Y, T243M, D179Y/T243M, and EL165-166, impacting the KPC Ω-loop, have also been described in the US as additional resistance mechanisms leading to ceftazidime-avibactam resistance in K. pneumoniae [41], further indicating that resistance mutations in KPC-genes can account for clinical resistance in some global strains [41].
Global Perspective on the Epidemiology of Carbapenem Resistance
Access to global travel has unquestionably led to the dissemination of antibiotic resistance throughout many countries [42]. The first carbapenemase was reported in Japan from an Aeromonas hydrophila isolate in the 1980s, which was subsequently followed by a report describing the Seoul imipenemase (SME-1) from Serratia marcescens, imipenemases (IMI-1) in California (1984) and NMC-A in France (1990), both from Enterobacter cloacae [43].
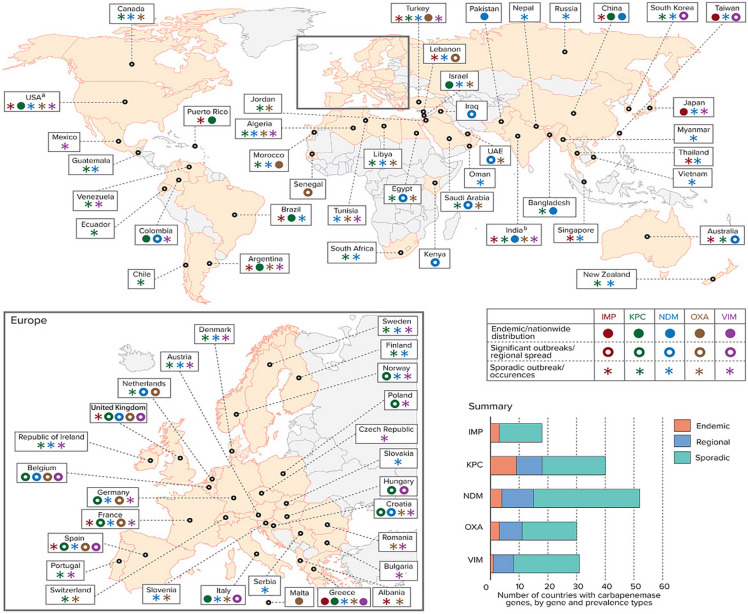
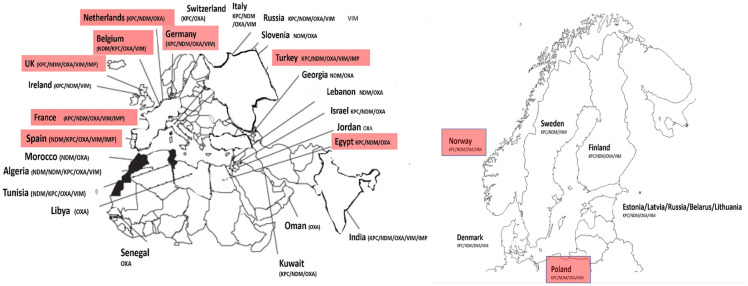
Globally, clinical characteristics of carbapenem-resistant carrying organisms vary with local conditions and across geographical regions (Fig. 4). Table 2 details the global distribution of reported enzymatic carbapenem resistance rates and published outbreak series by geographical region. Countries such as Israel, Greece, and Colombia have all reported significant outbreaks of CRE-associated infections [12]. No scenario better exemplifies the concerns surrounding the spread of carbapenem-resistant Gram-negative bacteria than the experience of Greece, which now has some of the highest carbapenem-resistance rates. Prior to 2001, the Greek System for the Surveillance of Antimicrobial Resistance reported a carbapenem-resistance prevalence of < 1%, and that CRE resistance was largely attributable to VIM-1–type MBL. In 2008, CRE in Greece had dramatically increased to 30% in hospital wards and 60% in ICUs [44]. By 2014, the European Centre for Disease Prevention and Control, EARS-Net, revealed that 62.3% of all K. pneumoniae isolates tested in Greece were now resistant to carbapenems [45, 45]. This spread in Greece is attributable to the introduction of the blaKPC-2 gene which spread through the country in 2007. Towards the end of the following year (2008), surveillance of 21 Greek hospitals revealed that 96% of carbapenem-resistant K. pneumoniae could be attributed to a single dominant sequence type (ST258 lineage) of K. pneumoniae harboring blaKPC-2 [44, 44]. Dissemination of carbapenemase genes and outbreak of CR infections in Europe are shown in Fig. 5. Following worldwide spread of K. pneumoniae ST258-type strain, including throughout the US, multi-loci sequence typing was used as a molecular tool to track the global epidemiology of CRE and the dissemination of KPC genes. K. pneumoniae ST258 continues to exist as a dominant global strain, which has been associated in outbreaks in [47] Poland, Columbia, and the United States, and is responsible for 90% of all K. pneumoniae infections in Israel [48].
Fig. 4.
Global distribution of Carbapenemase-producing bacteria: global distribution of carbapenemases in Enterobacteriaceae, by country and region. aKPCs are endemic in some US states; bOXA mainly refers to OXA-48, except in India, where it refers to OXA-181. IMP active on imipenem metallo-β-lactamase, KPC Klebsiella pneumoniae carbapenemase, NDM New Delhi metallo-β-lactamase, OXA oxacillinase-type carbapenem-hydrolyzing β-lactamase, VIM Verona integron-encoded metallo-β-lactamase. Reprinted with permission from: Logan, K., Weinstein, R. The epidemiology of carbapenem-resistant Enterobacteriacae: the impact and evolution of a global menace. J of Infect Disease. 2017;215(S1):S28–36. https://doi.org/10.1093/infdis/jiw282. (12)
Table 2.
Global distribution of reported enzymatic carbapenem resistance rates and published outbreak series by geographical region
| Region | Country | Carbapenemase genes described | Bacterial species implicated | Percent carbapenem resistance reported | Examples; Published outbreak | Reference |
|---|---|---|---|---|---|---|
| N. America | USA/Canada |
KPC, NDM, VIM, OXA, IMP |
Klebsiella pneumoniae | 10.3–58.5% | K. pneumoniae (KPC-2) | (57–60) (25) (30–34) |
| Escherichia coli | 0.2–4% | |||||
| Enterobacteralesa | 2–4% | |||||
| Acinetobacter baumannii | 11–50% | |||||
| Pseudomonas aeruginosa | 10–59% | |||||
| Asia -Pacific | Japan |
NDM, VIM, IMP, KPC, NDM, |
Klebsiella pneumoniae | 1.6–20% |
K. pneumoniae (KPC-16)) K. pneumoniae (KPC-15 K. pneumoniae (KPC-3) K. pneumoniae (OXA-48) |
(58, 61–65) |
| Thailand | ||||||
| Vietnam | Escherichia coli | 0–% | ||||
| Singapore | Enterobacteralesa | 0–32% | ||||
| Taiwan | Acinetobacter baumannii | 25%–91% | ||||
| S. Korea | Pseudomonas aeruginosa | 10–47% | ||||
| E. Asia | China |
KPC, NDM, VIM |
Klebsiella pneumoniae | 5–52% |
K. pneumoniae (NDM-5) K. pneumoniae (NDM-15 E. cloacae (NDM1,5) E. cloacae (KPC-2) E. cloacae (VIM-1) A. baumannii (OXA-23) P. aeruginosa (KPC-2) |
(66–69, 70–71) |
| Escherichia coli | ^ID (insufficient data) | |||||
| Enterobacteralesa | ^ID (insufficient data) | |||||
| Acinetobacter baumannii | 10–90% | |||||
| Pseudomonas aeruginosa | 10–100% | |||||
| S. Asia | India/Pakistan | Klebsiella pneumoniae | 0–52% |
K. pneumoniae (NDM-1) K. pneumoniae (NDM-2) |
(63, 72–76) | |
| Escherichia coli | 0–34% | |||||
| Enterobacteralesa | 3–21% | |||||
| Acinetobacter baumannii | 0–100% | |||||
| Pseudomonas aeruginosa | ^^ID (insufficient data) | |||||
| Europe | Klebsiella pneumoniae | 0.2–59% |
K. pneumoniae (NDM-1) K. pneumoniae (KPC-1–2-3–4-16) K. pneumoniae OXA-48 E. coli OXA-48 |
(16, 40, 42, 58, 77–79) | ||
| Escherichia coli | 0–7% | |||||
| Enterobacteralesa | ^ID (insufficient data) | |||||
| Acinetobacter baumannii | 3–100% | |||||
| Pseudomonas aeruginosa | 0–56% | |||||
| Latin America | Klebsiella pneumoniae | 1.3–20% |
K. pneumoniae (NDM-1) K. pneumoniae (KPC-2) P. aeruginosa (KPC) |
(21, 58, 61,62,80–83) | ||
| Escherichia coli | 0.5–9% | |||||
| Enterobacteralesa | 0.5–9% | |||||
| Acinetobacter baumannii | 21–90% | |||||
| Pseudomonas aeruginosa | 14–65% |
VIM Verona integron–encoded metallo-β-lactamase, NDM New Delhi MBL, KPC Klebsiella pneumoniae carbapenemase, IMP active-on-imipenem, OXA Oxacillinase
aSpecies not indicated
Fig. 5.
Countries in which carbapenemase genes have been described are shown. Countries shown in red boxes represents cases in which outbreaks involving carbapenemase genes have been described. Figure of Europe adapted from [12, 12, 12, 12]
KPC gene variants are the most commonly globally encountered carbapenemase, with the blaKPC-2 and blaKPC-3 gene variants being the most common. Countries with a high prevalence of KPC-producing K. pneumoniae include the US, Europe (Italy, Greece), the UK, and Asia (China), as well as some countries in the Middle East (Israel) and South America (Brazil, Colombia) [12].
The first identification of an OXA-48-producing K. pneumonia occurred as part of a reported outbreak in May 2006 to January 2007 [27]. In subsequent years, OXA-48-producing organisms remained largely restricted to healthcare systems in Turkey [27]. Since these initial reports, outbreaks of OXA-48-producing isolates have spread from Turkey to countries in the Middle East (Lebanon, Oman, Saudi Arabia, and Kuwait), as well as to countries in North Africa (Morocco, Tunisia, Egypt, and Libya). While OXA-48 harboring bacteria were initially geographically restricted to areas surrounding Turkey, OXA-48 producers have now been described in several European countries, such as France, Germany, Netherlands, Italy, Belgium, the UK, Ireland, Slovenia, Switzerland, and Spain. The first description of isolates with OXA-48-like (blaOXA-48-like) genes in the US was from a surveillance study in 2013, which incidentally reported 2 K. pneumonia isolates [49]. This description was followed shortly afterward by a report of 2 clinical K. pneumonia isolates with blaOXA-48-like genes in patients in a Virginia institution who had traveled to both India and Saudi Arabia [29, 29]. The rapid dissemination of OXA-48 producers among Enterobacterales species in addition to K. pneumoniae, including Citrobacter freundii, Enterobacter cloacae, Escherichia coli, K. oxytoca, Serratia marcescens, and Providencia rettgeri, suggests that the clinical significance of OXA-48 on antibiotic resistance may be more important that originally perceived. The OXA-48 enzyme hydrolyzes penicillins efficiently, carbapenems slowly, and extended-spectrum cephalosporins poorly, often resulting in variable antibiotic resistance patterns. Thus, recognizing the production of OXA-48 in isolates recovered from infections, and lack of routine screening processes for enterobacterial isolates with MIC values of ertapenem ≥ 0.5 mg/L, or imipenem or meropenem ≥ 1 mg/L, may result in an underestimation of the true prevalence and incidence of OXA-48-harboring organisms.
Environmental vectors of CR transmission also substantiate this suggestion. There is recent evidence in the US of wild bird (gull) feces harboring both plasmid-encoded blaKPC-2 or chromosomally-encoded blaOXA-48 genes [50] A recent report linking the association of CRE to livestock continues to inform the epidemiology of carbapenem-resistance. Kock et al. [51] describe the overall prevalence of CRE at < 1% among livestock and companion animals in Europe, but note that CRE can also be found in animals and livestock with a prevalence of 2–26% in Africa, and 1–15% in Asia. While wild birds (gulls) in Australia and Europe carried CRE in 16–19% [51], indicating that transmission of CRE between animals and humans may be an unknown reservoir and birds may become a vector for transmission of CRE, further research linking animal to human transmission is needed. Environmental sources of carbapenemases linked to recreational waters and sewage have also been reported as reservoirs for CRE [52]. Mahon and colleagues [52]identified NDM-producing E. coli and K. pneumoniae in recreational waters and sewers in Ireland. NDM-producing E. coli were indistinguishable between environment and human isolates, and K. pneumoniae isolated from sewage and waterways were closely related by pulsed-field gel [52].
Mani et al. [53] found KPC-producing Enterobacterales in Mytilus galloprovincialis (a filter-feeding Mediterranean mussel) bought at a retail market in Tunisia, the E. coli isolate contained blaKPC genes and was resistant to all β-lactams, ertapenem, meropenem, fluroquinolones, sulphonamides, and trimethoprim. The isolate is most likely of human origin as the sampled mussels were grown in an area receiving hospital effulents; this is also supported by the global epidemiology of blaKPC plasmids and KPC-producing E. coli clones, while carbapenems are not used in animals [53]. KPC-producing K. pneumonia have also been reported from wastewater in Austria and Brazil [54, 54]. These findings suggest that aquatic organisms may act as a secondary reservoir of KPC enzymes.
Future Perspectives
Carbapenemases continue to evolve, and the global dissemination of mobile genes encoding for carbapenem-resistance results in infections that have increased morbidity and mortality [56].
Does carbapenem resistance foreshadow a return to a pre-antibiotic era? Carbapenemase-encoding genes are already widespread in certain parts of the world, with carbapenem-resistance rates in some European and Asian countries already exceeding 50%. In the US, infections with KPC-positive bacteria are a major health concern, validated by the reality that every state in the US has now reported infections with KPC-expressing K. pneumoniae (Fig. 1). However, unlike the pre-antibiotic era, newer therapies, based on carbapenemase inhibitors, such as vaborbactam, avibactam, and most recently relebactam, provide a response to the threat of carbapenemase-based resistance. Coupling carbapenemase inhibitors to existing antimicrobial back backbone molecules, such as third-generation cephalosporins or carbapenems, offer therapeutic options and extend the clinical lifespan of existing antibiotics.
It is likely that effective control and management of carbapenem-resistant infections will require multifaceted efforts rooted in active stewardship, ongoing surveillance, optimizing dosing, and rapid diagnostics. Rapid molecular testing is required to supplement phenotypic approaches to traditional susceptibility testing. The ability to quickly identify carbapenemase genes within Gram-negative bacteria, or direct from clinical specimens, decreases the time to appropriate therapy, which has been shown to decrease mortality [56, 56]. Coupling molecular testing with phenotypic testing allows the laboratory diagnosis to differentiate carbapenemases across different types, which is important, as novel beta-lactamases and beta-lactamase inhibitor combinations demonstrate differential activity against different classes of carbapenemases. While both Ehrlich and Fleming predicted the scourge of antibiotic resistance, contemporary tools and data can help to slow the insidious nature of carbapenem resistance.
Acknowledgments
The author wishes to thank Glenn Tillotson and Nicolette Theriault at GST Micro for their assistance throughout the editing and publication of this manuscript.
Funding
No funding was received in the writing of this manuscript. The journal’s Rapid Service fee was funded by Melinta Therapeutics.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Glen T. Hansen has nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Ehrlich P. Address in pathology on chemotherapeutics: scientific principles, methods and results. Lancet. 1913;181:445–451. [Google Scholar]
- 2.Abraham E, Chain E. An enzyme from bacteria able to destroy penicillin. Nature. 1940;46:837. [PubMed] [Google Scholar]
- 3.Abraham EP, Chain E, Fletcher CM, et al. Further observations on penicillin. Eur J Clin Pharmacol. 1992;42:3–9. [PubMed] [Google Scholar]
- 4.Bush K, Fisher JF. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu Rev Microbiol. 2010;65:455–478. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- 5.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richmond MH, Sykes RB. The beta-lactamases of gram-negative bacteria and their possible physiological role. Adv Microb Physiol. 1973;9:31–88. doi: 10.1016/s0065-2911(08)60376-8. [DOI] [PubMed] [Google Scholar]
- 7.Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in enterobacteriaceae. Nature 1965:239–241. [DOI] [PubMed]
- 8.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 9.Drawz SM, Bonomo RA. Three decades of β-lactamase inhibitors. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surgers L, Boersma P, Girard P-M, et al. Molecular epidemiology of ESBL-producing E. coli and K. pneumoniae: establishing virulence clusters. Infect Drug Resist. 2018;309:13–18. doi: 10.2147/IDR.S179134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isgren CM, Edwards T, Pinchbeck GL, et al. Emergence of carriage of CTX-M-15 in faecal Escherichia coli in horses at an equine hospital in the UK; increasing prevalence over a decade (2008–2017) BMC Vet Res. 2019;15:268. doi: 10.1186/s12917-019-2011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier L, Dortet L, Jousset AB, et al. Molecular characterization of plasmid-encoded Tripoli MBL 1 (TMB-1) in Enterobacteriaceae. J Antimicrob Chemother. 2019;74:42–47. doi: 10.1093/jac/dky372. [DOI] [PubMed] [Google Scholar]
- 14.Tato M, Coque TM, Baquero F, Cantón R. Dispersal of carbapenemase blaVIM-1 gene associated with different Tn402 variants, mercury transposons, and conjugative plasmids in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:320–327. doi: 10.1128/AAC.00783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in gram-negative bacteria. Biomed Res Int. 2014;2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter R, D’Souza A, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Drug Resist Updat. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nordman P, L. P. he difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 2014; 20:821–830. [DOI] [PubMed]
- 19.Tada T, Nhung P, Miyoshi-Akiyama T, et al. IMP-51, a novel IMP-type metallo-β-lactamase with increased doripenem- and meropenem-hydrolyzing activities, in a carbapenem-resistant Pseudomonas aeruginosa clinical isolate. 2015; 59:7090–7093. [DOI] [PMC free article] [PubMed]
- 20.H. Y, A K, R O, H K, T K, M. I. Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother 2001; 45:1343–1348. [DOI] [PMC free article] [PubMed]
- 21.S K, T S, N K, T I-H, M T, Y Y. No Title. Antimicrob Agents Chemother 2017; 6:eoo576–17.
- 22.Schmidtke AJ, Hanson ND. Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob Agents Chemother. 2006;50:2030–2037. doi: 10.1128/AAC.01458-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guérin F, Isnard C, Cattoir V, Giard JC. Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob Agents Chemother. 2015;59:7753–7761. doi: 10.1128/AAC.01729-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacoby GA. AmpC -Lactamases. Clin Microbiol Rev. 2009;22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavez M, Neves P, Dropa M, et al. Emergence of carbapenem-resistant Escherichia coli producing CMY-2-type AmpC beta-lactamase in Brazil. J Med Microbiol. 2008;57:1590–1592. doi: 10.1099/jmm.0.2008/002774-0. [DOI] [PubMed] [Google Scholar]
- 26.Poirel L, Héritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 28.Docquier J-D, Calderone V, De Luca F, et al. Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases. Chem Biol. 2009;16:540–547. doi: 10.1016/j.chembiol.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Mathers AJ, Hazen KC, Carroll J, et al. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the ‘menace’ arrives in the new world. J Clin Microbiol. 2013;51:680–683. doi: 10.1128/JCM.02580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Al Laham N, Chavda KD, et al. First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza. Palestine Antimicrob Agents Chemother. 2015;59:4305–4307. doi: 10.1128/AAC.00565-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J Infect Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 32.CDC. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. 2013: 1–12.
- 33.Yigit H, Biddle JW, Alberti S, et al (2002) Novel carbapenem-hydrolyzing -lactamase, KPC-1, from a carbapenem-resistant strain of klebsiella pneumoniae. Antimicrob Agents Chemother 20:440–458. [DOI] [PMC free article] [PubMed]
- 34.Walters MS, Witwer M, Lee Y-K, et al. Notes from the field: carbapenemase-producing carbapenem-resistant enterobacteriaceae from less common enterobacteriaceae genera—United States, 2014–2017. MMWR Morb Mortal Wkly Rep. 2018;67:668–6695. doi: 10.15585/mmwr.mm6723a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hidron AI, Edwards JR, Patel J, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 36.Sievert DM, Ricks P, Edwards JR, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2009–2010. Infect Control Hosp Epidemiol 2013 [DOI] [PubMed]
- 37.Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37:1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai B, Echols R, Magee G, et al. Prevalence of carbapenem-resistant gram-negative infections in the United States predominated by acinetobacter baumannii and pseudomonas aeruginosa. Open Forum Infect Dis. 2017;4:176. doi: 10.1093/ofid/ofx176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson K, Hemarajata P, Sun D, et al. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother. 2017;61(10):e00989–e1017. doi: 10.1128/AAC.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphries RM, Yang S, Hemarajata P, et al. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2015;59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob Agents Chemother. 2017;61:e02534–e2616. doi: 10.1128/AAC.02534-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frost I, Van Boeckel TP, Pires J, Craig J, Laxminarayan R. Global geographic trends in antimicrobial resistance: the role of international travel. J Travel Med 2019; [DOI] [PubMed]
- 43.Codjoe F, Donkor E. Carbapenem resistance: a review. Med Sci. 2017;6(10):3390. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Souli M, Galani I, Antoniadou A, et al. An Outbreak of Infection due to β‐Lactamase Klebsiella pneumoniae Carbapenemase 2–Producing K. pneumoniae in a Greek University Hospital: Molecular Characterization, Epidemiology, and Outcomes. Clin Infect Dis 2009; 50:364–73. [DOI] [PubMed]
- 45.Pérez-Grajera I, Lara-Fuella N, Fernández-Romero S, et al. (2016) Rápido aumento de la resistencia a cefalosporinas de 3 a generación, imipenem y de la corresistencia en 7.140 aislados de Klebsiella pneumoniae en hemocultivos (2010–2014) según datos de EARS-Net en España. Enferm Infecc Microbiol Clin 35:480–486. [DOI] [PubMed]
- 46.CDC E. European Centre for Disease Prevention and Control (EARS-Net). Available at: https://ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/ears-net. Accessed 7 Feb 2019.
- 47.Baraniak A, Izdebski R, Zabicka D, et al. Multiregional dissemination of KPC-producing Klebsiella pneumoniae ST258/ST512 genotypes in Poland, 2010–14. J Antimicrob Chemother. 2017;72:1610–1616. doi: 10.1093/jac/dkx054. [DOI] [PubMed] [Google Scholar]
- 48.Rojas LJ, Weinstock GM, De La Cadena E, et al. An Analysis of the epidemic of klebsiella pneumoniae carbapenemase-producing K. pneumoniae: convergence of two evolutionary mechanisms creates the ‘perfect storm’. J Infect Dis. 2017;217:82–92. doi: 10.1093/infdis/jix524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JDD. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–136. doi: 10.1128/AAC.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahlstrom CA, Ramey AM, Woksepp H, Bonnedahl J. Repeated Detection of carbapenemase-producing Escherichia coli in gulls inhabiting alaska. Antimicrob Agents Chemother 2019; 63. [DOI] [PMC free article] [PubMed]
- 51.Köck R, Daniels-Haardt I, Becker K, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect. 2018;24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Mahon BM, Brehony C, McGrath E, et al. Indistinguishable NDM-producing Escherichia coli isolated from recreational waters, sewage, and a clinical specimen in Ireland, 2016 to 2017. Euro Surveill. 2017;22(10):2807. doi: 10.2807/1560-7917.ES.2017.22.15.30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mani Y, Mansour W, Mammeri H, et al. KPC-3-producing ST167 Escherichia coli from mussels bought at a retail market in Tunisia. J Antimicrob Chemother. 2017;72:2403–2404. doi: 10.1093/jac/dkx124. [DOI] [PubMed] [Google Scholar]
- 54.De Araujo CFM, Silva DM, Carneiro M, et al. Detection of carbapenemase genes in aquatic environments in Rio de Janeiro. Brazil Antimicrob Agents Chemother. 2016;60:4380–4383. doi: 10.1128/AAC.02753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galler H, Feierl G, Petternel C, et al. KPC-2 and OXA-48 carbapenemase-harbouring Enterobacteriaceae detected in an Austrian wastewater treatment plant. Clin Microbiol Infect 2014; 20. [DOI] [PubMed]
- 56.Martin A, Fahrbach K, Zhao Q, Lodise T. Association between carbapenem resistance and mortality among adult, hospitalized patients with serious infections due to enterobacteriaceae: Results of a systematic literature review and meta-analysis. Open Forum Infect Dis 2018; 5:ofy150. [DOI] [PMC free article] [PubMed]
- 57.Weir NJM, Pattison SH, Kearney P, et al. Criteria required for an acceptable point-of-care test for UTI detection: obtaining consensus using the Delphi technique. PLoS ONE 2018 [DOI] [PMC free article] [PubMed]
- 58.Kazmierczak KM, Tsuji M, Wise MG, et al. In vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-non-susceptible Gram-negative bacilli, including serine carbapenemase- and metallo-β-lactamase-producing isolates (SIDERO-WT-2014. Int J Antimicrob Agents. 2019;53:177–184. doi: 10.1016/j.ijantimicag.2018.10.007. [DOI] [PubMed] [Google Scholar]